- 1College of Medicine, Jinhua University of Vocational Technology, Jinhua, Zhejiang, China
- 2Department of Gastroenterology, Affiliated Jinhua Hospital, Zhejiang University School of Medicine, Jinhua, Zhejiang, China
- 3Department of Urology, Affiliated Jinhua Hospital, Zhejiang University School of Medicine, Jinhua, Zhejiang, China
Bladder cancer is one of the leading causes of mortality globally. The development of bladder cancer is closely associated with alternative splicing, which regulates human gene expression and enhances the diversity of functional proteins. Alternative splicing is a distinctive feature of bladder cancer, and as such, it may hold promise as a therapeutic target. This review aims to comprehensively discuss the current knowledge of alternative splicing in the context of bladder cancer. We review the process of alternative splicing and its regulation in bladder cancer. Moreover, we emphasize the significance of abnormal alternative splicing and splicing factor irregularities during bladder cancer progression. Finally, we explore the impact of alternative splicing on bladder cancer drug resistance and the potential of alternative splicing as a therapeutic target.
Introduction
Bladder cancer is a prevalent form of cancer worldwide, and it ranks among the top 10 most commonly observed types of cancer (1). Due to the high recurrence rate of bladder cancer, it has a strong tendency to metastasize and has limited treatment options. As such, patients with bladder cancer suffer from a short survival time and a poor quality of life. Moreover, bladder cancer is often diagnosed in the advanced stages because of its insidious onset. For these reasons, bladder cancer is a significant healthcare burden.
Chemotherapy remains the primary treatment option for recurrent and metastatic urothelial carcinoma; however, the efficacy of chemotherapy is limited. In recent years, the advent of immunotherapy, particularly programmed cell death protein 1 (PD-1) and programmed-death ligand 1 (PD-L1) inhibitors, has presented new therapeutic opportunities for patients with bladder cancer (2). Consequently, identifying biomarkers that can assist in the diagnosis and treatment of bladder cancer is of critical importance.
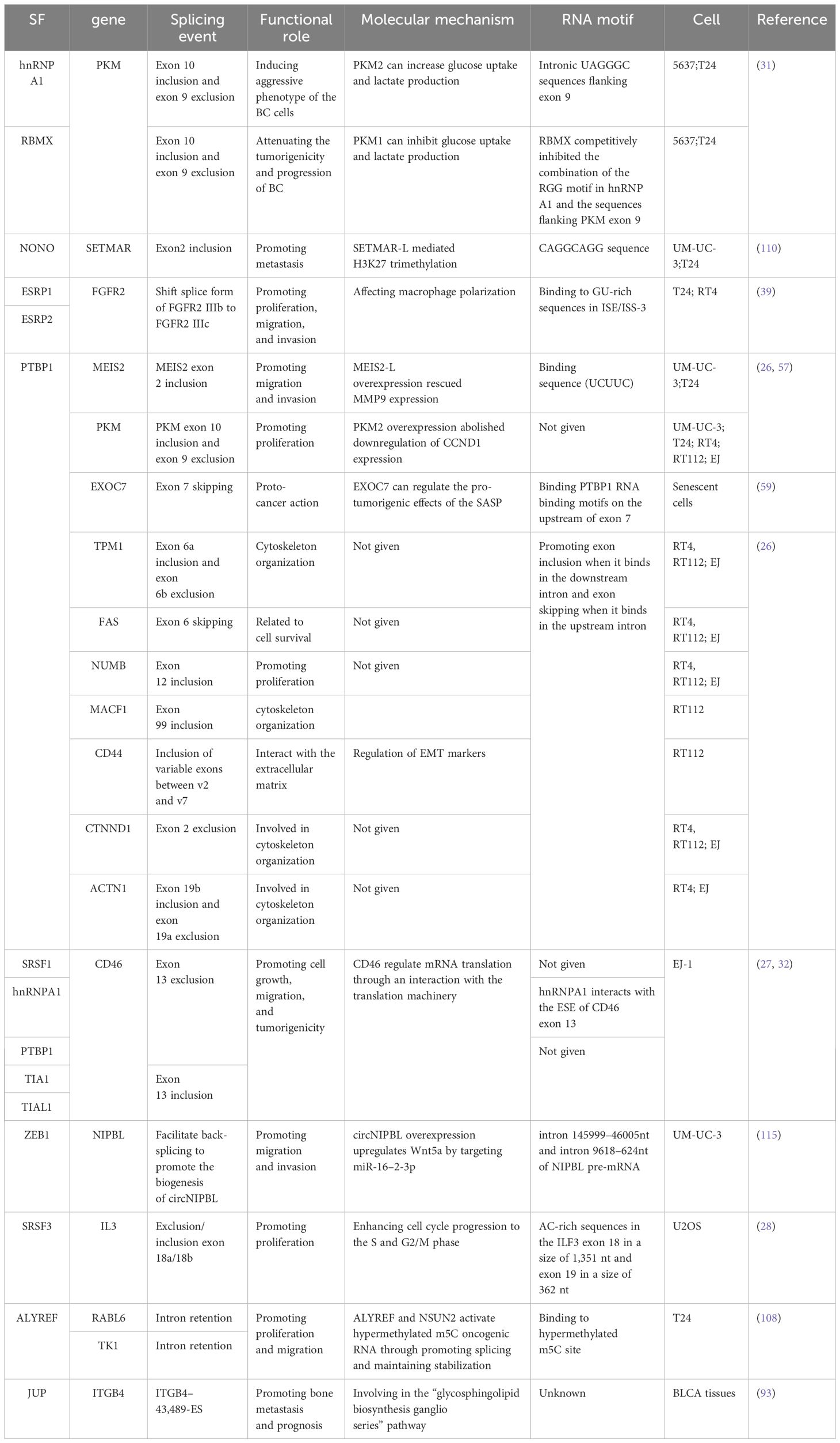
Alternative splicing involves the conversion of pre-mRNA into mature mRNA. Through different splicing methods, a single gene can produce multiple distinct mature mRNAs, resulting in the production of distinct proteins (3, 4). Alternative splicing significantly contributes to the genetic and proteomic diversity of eukaryotes (5, 6) and is mainly catalyzed by spliceosomes (7). After identifying splicing signals (8) within pre-mRNA, spliceosomes engage with more than 300 proteins, referred to as splicing factors. Splicing factors bind to specific motifs or elements in the pre-mRNA, named splicing regulatory elements (SREs). Depending on their roles in splicing and their locations within the pre-mRNA, SREs can be divided into four subtypes: exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs) (8–10) (Figure 1). These splicing complexes mediate the formation of multiple splice variants during different splicing events, such as exon skipping (SE), mutually exclusive exons (MXE), alternative 5′ splice sites (A5SS), alternative 3’ splice sites (A3SS), and intron retention (RI) (11) (Figure 2). Alternative splicing events occur in more than 90% of human genes and are precisely regulated within cells.
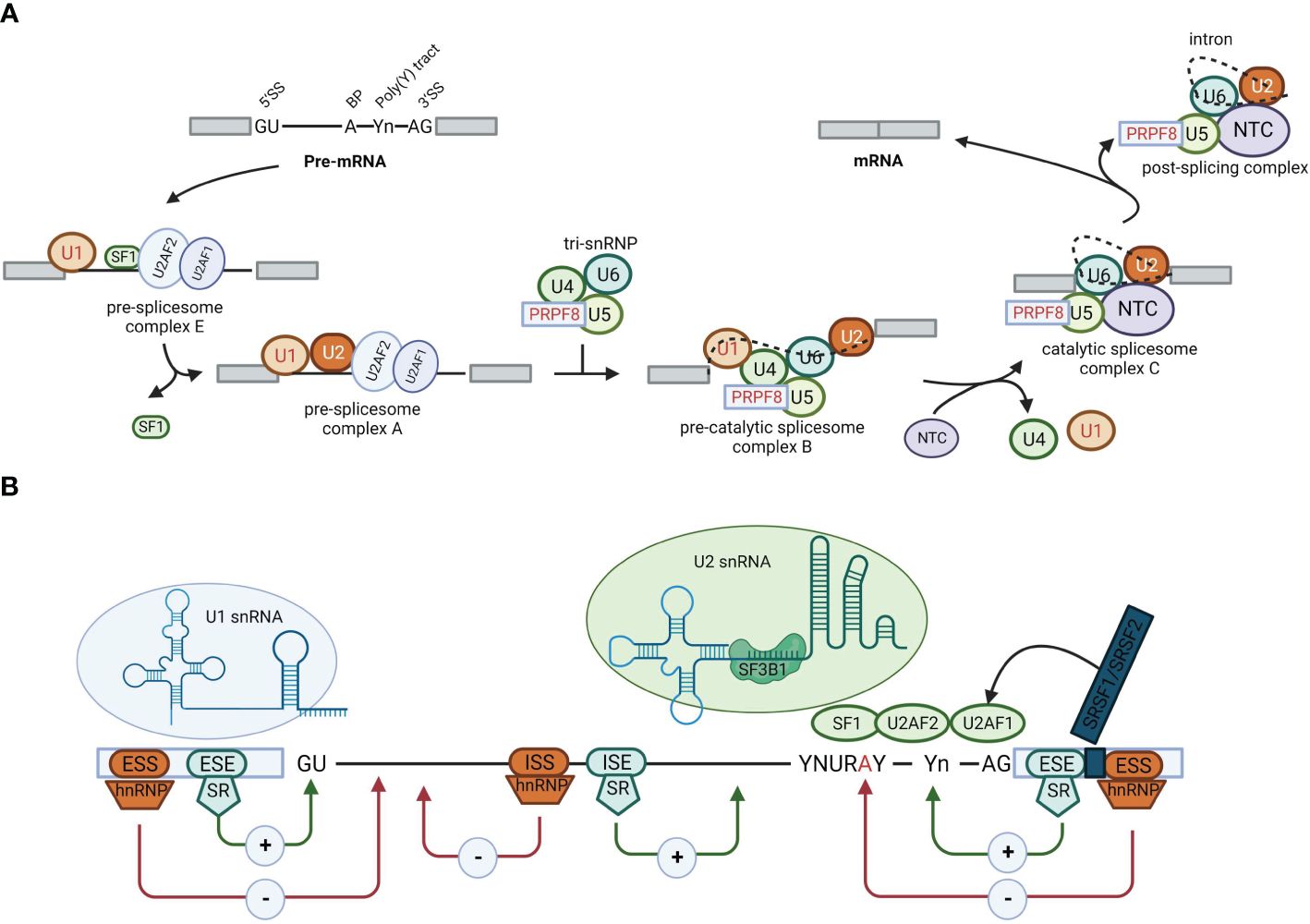
Figure 1 Alternative splicing. (A) Schematic representation of the stepwise assembly of the spliceosome from its small nuclear ribonucleoprotein (snRNP) components. U1 snRNP recognizes and binds to the 5′ splicing site, U2 small nuclear RNA auxiliary factor 2 (U2AF2) binds to the polypyrimidine poly (Y) tract, U2AF1 binds to the 3′ splicing site, and splicing factor 1 binds to the branch point, forming the pre-spliceosome complex E. Next, U2 snRNP replaces splicing factor 1 on the branch point to form pre-spliceosome complex (A) The tri-snRNP, consisting of U5, U4, and U6 snRNPs, joins complex A to form the pre-catalytic spliceosome complex (B) Thereafter, the U1 and U4 snRNPs leave, the U6 snRNP binds to the 5′ splicing site, and NTC is recruited so that the U6 snRNP and the U2 snRNP can pair, thereby generating the catalytic spliceosome complex C. Two subsequent transesterification reactions result in the creation of a post-splicing complex with intron exclusion and the formation of mature mRNA with interconnected exons. (B) Roles of cis-acting sequences and trans-acting factors in determining the splicing code for splice site selection. RNA-binding motif proteins, serine/arginine-rich family proteins (including SRSF2), and heterogeneous nuclear ribonucleoproteins bound to exonic or intronic regulatory elements can promote (+) or prevent (−) the recognition of the 5′ splicing site by the U1 snRNP or the 3′ splicing site by splicing factor 1, U2AF2, U2AF1, or U2 snRNP, thus affecting splice site choices and therefore alternative splicing decisions.
Figure 2 Summary of constitutive and alternative splicing events. The boxes represent exons, and the gray regions represent introns. The basic patterns of alternative splicing include exon skipping, alternative 5′ and 3′ splice sites, mutually exclusive exons, and intron retention.
Alternative splicing frequently occurs in various disease states, including tumors, and it is thought to be involved in tumor progression. In fact, recent studies have revealed that alternative splicing plays a pivotal role in tumorigenesis, progression, invasion, metastasis, angiogenesis, and drug resistance (12–15). Multiple computational methods have been used to predict the roles of alternative splicing in tumors (16). Notably, several tumor-associated alternative splicing events have been identified in bladder cancer (17–19), indicating that a correlation may exist between alternative splicing and bladder cancer deterioration.
This review discusses the expression and function of splicing factors and the alternative splicing events in bladder cancer at cell lines, human or animal model level, and recent advancements in the therapeutic approaches targeting alternative splicing.
Dysregulation of splicing-related proteins in bladder cancer
Alternative splicing is a remarkably intricate biological process that is meticulously regulated by the interplay between cis-elements (splicing signals and SREs) and trans-elements (spliceosomes and splicing factors) (Figure 1). These components give rise to a sophisticated regulatory network that operates in a coordinated and context-dependent manner.
Trans-elements are splicing-associated proteins that bind to specific RNA sequences or motifs. Splicing factors are a group of RNA binding factors that participate in the splicing of RNA precursors (20). Based on their functions, splicing factors are divided into two categories: small nuclear ribonucleoproteins (snRNPs) and non-snRNPs (21). After binding to pre-mRNAs, splicing factors either facilitate or hamper the interaction between spliceosomes and pre-mRNAs, suggesting their duality to either activate or inhibit alternative splicing. The evidence pertaining to protein changes associated with bladder cancer during RNA splicing is discussed herein.
Serine/arginine-rich family
The serine/arginine-rich (SR) proteins, which include 12 evolutionarily conserved members (SRSF1–SRSF12), play a crucial role in protein synthesis. These proteins are characterized by an N-terminal RNA recognition motif and a C-terminal SR domain. SR proteins are involved in mRNA trafficking, peptide synthesis, and translation initiation complex formation (22) as well as pre-mRNA splicing and alternative splicing (23).
A previous study showed that the SR proteins are among the most dysregulated splicing factors in pan-cancers (24). Another study demonstrated that SRSF9 is upregulated in several cancers, including bladder cancer (25). Interestingly, patients with higher SRSF9 expression tended to have better outcomes after immunotherapy for bladder cancer (25). Furthermore, it has been shown that SRSF7 and SRSF10 expressions were associated with G3 tumors in non-muscle-invasive bladder cancer (26). More recently, Zeng et al. reported that SRSF1 promoted bladder cancer cell growth and migration by inducing exon 13 skipping of CD46 (27). Another study reported that SRSF3 expression was significantly elevated in bladder cancer tissues. Moreover, SRSF3 knockdown or removal of the SRSF3-binding motifs suppressed tumor cell growth by triggering A3SS activation within exon 18 of ILF3 and promoting the skipping of exons 18 and 19, resulting in the production of interleukin enhancer-binding factor 3 (ILF3) isoforms 5 and 7 (28).
Taken together, this evidence demonstrates the potential of SR proteins as promoters in bladder cancer. However, the understanding of their role is still limited, and more in-depth research is needed.
Heterogeneous nuclear ribonucleoprotein family
The heterogeneous nuclear ribonucleoprotein (hnRNP) family comprises a minimum of 20 members. These proteins possess common domains that bind to splicing silencers, in turn affecting splicing events, including constitutive splicing and alternative splicing, across the entire human genome (29, 30). In bladder cancer, hnRNPs participate in proliferation, apoptosis, angiogenesis, and invasion, contributing to cancer development and progression.
Protein hnRNP-A1 functions as an RNA-binding protein in mRNA splicing to regulate intron removal, exon joining, and mature mRNA formation. The multifaceted function of hnRNP-A1 in regulating the alternative splicing of numerous gene variants underscores its significance in bladder cancer—for instance, hnRNP-A1 mediates the c-Myc-enhanced pyruvate kinase (PK)M2/PKM1 isoform switch by selectively including exon 10 and excluding exon 9 during the alternative splicing of PKM pre-mRNA (31). Meanwhile, hnRNP-A1 interacts with the ESE element of CD46 mRNA, leading to exon 13 exclusion during splicing (32).
hnRNP-F is an RNA-binding protein that participates in various biological processes, including alternative splicing (33) and post-transcriptional modification (33, 34). Elevated hnRNP-F has been observed in bladder cancer, suggesting that it may play a role in tumorigenesis—for instance, Li et al. found that hnRNP-F was required for tumor growth and induced metastasis in bladder cancer. Specifically, hnRNP-F was significantly upregulated in bladder cancer tissues and was correlated with a poor prognosis in 103 patients with bladder cancer. When hnRNP-F was silenced or enhanced, Snail1 expression changed at both the mRNA and protein levels, implying that Snail1 may be a downstream target of hnRNP-F (35). Recent studies have also indicated a connection between hnRNP-F and phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling. Inhibition of the PI3K/Akt pathway using the specific inhibitor LY294002 led to a significant decrease in hnRNP-F expression (36).
Epithelial splicing regulatory protein (ESRP)1 and ESRP2 are two RNA-binding proteins that belong to the hnRNP family. They have been shown to regulate the alternative splicing of fibroblast growth factor receptor 2 (FGFR2) pre-mRNA in epithelial tissues (37). The role of ESRP1 and ESRP2 in promoting splicing is reliant on the presence of ISE/ISS elements or the UGCAUG motif, which enhance FGFR2 transcripts from exon IIIc to exon IIIb splicing (38). Zhao et al. suggested that a decrease in ESRP1 or ESRP2 expression is related to lung metastasis in bladder cancer as it affects the splicing of FGFR2 and macrophage polarization (39). The hnRNP-F, hnRNP-H, and hnRNP-M proteins, which are closely related to ESRPs, also contribute to FGFR2 splicing regulation. Previous studies have shown that hnRNP-F, in association with hnRNP-M and hnRNP-H, suppresses FGFR2 exon IIIc, but it does not directly influence exon IIIb splicing (40, 41). However, no studies have addressed the FGFR2 splicing regulation of hnRNP-F, hnRNP-H, and hnRNP-M in bladder cancer.
According to previous studies, the complicated mechanisms underlying hnRNP-L alternative splicing regulation have been well studied in pan-cancers (42–45). Lv et al. (46) found that hnRNP-L expression was negatively correlated with the overall survival rate of 155 patients with bladder cancer. HNRNPL knockdown significantly inhibited cell proliferation and suppressed tumor development by promoting cell cycle arrest and apoptosis and inhibiting epithelial–mesenchymal transition. Moreover, it has been shown that hnRNP-L inhibits mitogen-activated protein kinase (MAPK) signaling. Therefore, the high expression of hnRNP-L in bladder cancer may lead to a poor prognosis and cancer development by suppressing intrinsic apoptotic signaling and promoting MAPK signaling (47).
In terms of cancer diagnosis and treatment, several studies have suggested that hnRNP-U may serve as a diagnostic marker for various cancers (48–51). An analysis of The Cancer Genome Atlas (TCGA) bladder cancer cohort also revealed a negative correlation between high hnRNP-U expression and patient survival, highlighting its detrimental impact—for instance, Shi et al. reported that hnRNP-U is highly expressed in bladder cancer, and inhibited hnRNP-U may improve chemotherapy sensitivity by enhancing cisplatin-induced apoptosis of bladder cancer cells (52). Similarly, hnRNP-K has been identified as a key factor associated with a poor prognosis in bladder cancer by influencing the proliferation and chemoresistance of bladder cancer cells (53).
Polypyrimidine tract-binding protein 1 (PTBP1) plays an essential role in splicing. It has demonstrated a positive association with tumor growth and is associated with a poor prognosis in various types of cancer (54–56). In bladder cancer, PTBP1 expression was found to be positively related to lymphatic metastasis, tumor stage, histological grade, and a poor patient prognosis in 104 patients with bladder cancer (57). Moreover, mechanistic studies have revealed that PTBP1 regulates massive splicing events in bladder cancer. Georgilis et al. found that PTBP1 regulates the development of inflammation-driven cancers by controlling exon 7 skipping of EXOC7 (58, 59). Xie reported that PTBP1 promotes bladder cancer proliferation and metastasis by modulating the alternative mRNA splicing of MEIS2-L and PKM2 (57). Moreover, PTBP1 regulates the corresponding splicing events of numerous genes that are involved in tumor cell proliferation, growth, and metastasis, such as TPM1, FAS, NUMB, MACF1, CD44, CTNND1, and ACTN1(60).
Spliceosome components
The spliceosome is a dynamic macromolecular complex that is responsible for pre-mRNA splicing, including internal intron removal and orderly exon connections (61–63). It is composed of five snRNPs (U1, U2, U4, U5, and U6) and a group of spliceosome-associated proteins. An snRNP comprises a small nuclear RNA (snRNA) molecule and a specific group of SNRP proteins (64). The SNRP proteins collectively form a structure that envelops the RNA molecule. The SNRP proteins share a conserved SNRP domain that regulates the step-by-step assembly of the snRNA (65, 66). Mutations in SNRP proteins can affect splicing patterns, potentially contributing to tumor development (67, 68).
The Sm proteins (B/B’, D1, D2, D3, E, F, and G) include seven members that form a ring-like structure that encloses the RNA molecule (64). Leili et al. (69) found that, of the 33 genes analyzed using the elastic net method, eight had an impact on the survival of 1,381 patients with bladder cancer. Among these, the expression of SNRPE was negatively correlated with survival time. However, to date, there are no new findings detailing the potential functional role of SNRP-E in bladder cancer.
Splicing factor 3A subunit 3 (SF3A3), which is encoded by SF3A3, is correlated with tumor stage and the prognosis of 49 patients with bladder cancer (70). Mechanistically, the transcription factor E2F6 interacts with KDM5C and binds to the promoter of SF3A3, leading to the demethylation of the GpC island associated with H3K4me2 and high SF3A3 expression.
Splicing factor 3B subunit 1 (SF3B1) is encoded by SF3B1 and is a component of the U2 complex. It binds to the branch point nucleotide along with SF3B3 and PHF5A in the pre-catalytic spliceosome (71). Somatic mutations in SF3B1 were initially identified by whole-exome sequencing in myelodysplastic syndrome (72), and they have since been identified in other hematological malignancies and solid tumors (73–79). Notably, Seiler et al. identified a specific SF3B1 mutation (p.E902K), which was associated with A3SS events in bladder cancer samples (80). However, no subsequent studies have analyzed the aberrant splicing events caused by SF3B1 mutations in bladder cancer.
RNA-binding motif proteins
RNA-binding motif (RBM) proteins are a diverse family of proteins that contain an RNA recognition motif (RRM) domain that is involved in RNA binding. Several RBM proteins have been studied in the context of bladder cancer, including RBM3, RBM10, RBM24, RBM5, and RBMX. Among them, RBM3 has been identified as a marker of a poor prognosis in urothelial carcinoma (81, 82). Moreover, Seiler et al. reported a loss-of-function mutation in RBM10, which led to the induction of exon inclusion events (80). Other studies have shown that high RBM24 expression is associated with a poor prognosis in 32 patients with bladder cancer via the Runx1t1/TCF4/miR-625–5p positive feedback loop (83, 84). RBM5 is downregulated in bladder cancer, and its overexpression promotes apoptosis and inhibits tumor growth. Mechanistically, RBM5 is thought to promote tumor growth through a feedback loop involving miR-432–5p and β-catenin (85). The most thoroughly researched RBM protein in bladder cancer is RBMX, which is a ubiquitously expressed nuclear RNA-binding protein. Truncation mutations in RBMX have been observed in lung cancer through genome sequencing, suggesting its potential as a tumor suppressor (86). A previous study showed that RBMX competitively inhibits the binding of hnRNP-A1 to sequences flanking exon 9 of PKM through its RGG motif, resulting in a reduction in PKM2 expression and an increase in PKM1 expression. The attenuation of PKM2 expression suppresses tumorigenicity and bladder cancer progression. Additionally, RBMX inhibits aerobic glycolysis via hnRNP-A1-dependent PKM alternative splicing, and it counteracts the aggressive phenotype induced by PKM2 overexpression in bladder cancer cells (31).
Others
Junction plakoglobin (JUP, γ-catenin) is a member of the Armadillo protein family (87). JUP forms complexes with adhesins and desmosomal adhesins, key components of the EXM (88). These desmosomal proteins mediate cell–cell interactions and signal transduction (89). JUP deletion leads to reduced cell contact, increased invasion, and bladder cancer cell metastasis (90). In line with this, the restoration of JUP expression could inhibit bladder cancer cell migration and tumor progression, and patients with bladder cancer with low JUP expression consistently exhibit poor survival rates (91, 92). JUP has also been reported as a key splicing factor that affects the metastasis and prognosis of other types of cancer (87). In bladder cancer, Huang et al. (93) showed that JUP is one of the splicing factors associated with overall survival-related alternative splicing events by the TCGA database.
Small nucleolar RNA (snoRNA) is a type of non-coding RNA that ranges in length from 60 to 300 nt. snoRNAs mainly originate from the introns of vertebrate host genes (94, 95). Initially considered to be transcriptional noise, snoRNAs are now known to regulate various biological processes (96–99). snoRNA dysregulation has been implicated in the development and progression of numerous diseases (100–104). Rui (105) investigated the correlation between five candidate snoRNAs (U49A, U3, SNORD19B, SNORD114–1, and SNORD113–9) and alternative splicing. Protein–protein interaction network analysis revealed that the related alternative splicing mRNAs clustered well. Moreover, Gene Ontology enrichment analysis revealed a significant enrichment of these snoRNAs related to EXM and focal adhesion. These findings suggest that snoRNAs may have an impact on alternative splicing events and potentially contribute to bladder cancer progression. In a case–control study involving 580 patients with bladder cancer and 1,101 control subjects, the single-nucleotide polymorphism rs978416 G>A in RBFOX3 was potentially associated with bladder cancer predisposition in the Chinese population (106), indicating its potential as a novel biomarker of bladder cancer risk.
ALYREF, also known as RNA methyltransferase Aly/REF export factor, is an important “reader” protein that is located in the nucleus and that is involved in RNA processing and transport in cancer biology. It specifically recognizes and binds to m5C (methylated cytosine) sites within RNA molecules, thereby facilitating efficient RNA export from the nucleus to the cytoplasm. Notably, ALYREF interacts with various splicing factors, including SRSF3, PRPF3, and DHX16, suggesting its involvement in the intricate regulation of mRNA splicing (107). The function of ALYREF has been reported in the context of bladder cancer, where it leads to the occurrence of intron retention events in RABL6 and TK1. Furthermore, an intriguing observation has been made regarding the m5C-dependent interplay between ALYREF and NOP2 Sun methyltransferase 2. Their cross-regulation promotes malignancy in urothelial bladder cancer, primarily by facilitating mRNA splicing and stabilization (108). These findings provide valuable insights into the underlying molecular mechanisms governing bladder cancer malignancy.
NONO, which is a member of the Drosophila behavior human splicing (DBHS) family, is an RNA-binding protein that is involved in diverse gene expression processes. Alongside its paralogs splicing factor proline and glutamine rich (SFPQ) and paraspeckle component 1 (PSPC1), NONO is predominantly localized to the nucleus and possesses two RRMs. These multifunctional proteins exert their regulatory roles by impacting various aspects of gene expression, such as transcriptional activation, inhibition, RNA splicing, stabilization, and export (109). NONO modulates the splicing events of SETMAR in bladder cancer by directly binding to its specific motifs, primarily through the RRM2 domain. Meanwhile, NONO interacts with the splicing factor SFPQ to further influence SETMAR alternative splicing. The activity of NONO is also associated with the suppression of bladder cancer cell metastasis (110). These findings illustrate the importance of NONO in bladder cancer progression and metastasis, shedding light on its potential as a therapeutic target in combating this disease.
Poly (U) binding specificity factor 60 (PUF60) participates in transcriptional regulation and splicing through direct binding to DNA or RNA (111). Although PUF60 is poorly studied in the context of cancer, its high expression has been detected in bladder cancer. Moreover, Long et al. identified a direct association between PUF60 and bladder cancer prognosis and aggressiveness by TCGA database (112). Mechanistically, this relationship was attributed to the transcriptional regulation of aurora kinase A (AURKA) by PUF60 on bladder cancer cell behavior (112). However, further research is needed to explore the precise mechanisms of PUF60 in bladder cancer and its potential as a therapeutic target.
Alternative splicing–splicing factor regulatory network analysis has identified several splicing factors, including pre-mRNA processing factor 39 (PRPF39), LUC7-like 3 pre-mRNA splicing factor (LUC7L), heat shock protein family A member 8 (HSPA8), and DEAD-box helicase 21 (DDX21), as potential biomarkers for bladder cancer (113). Moreover, eukaryotic translation initiation factor 3 subunit A (EIF3A), DDX21, SDE2, transportin 1 (TNPO1), and ring finger protein 40 (RNF40) are thought to mediate multiple alternative splicing events in bladder cancer, implying their essential roles in the development and progression of this disease (114). It is noteworthy that DDX21 has been associated with bladder cancer in several studies, but there are no mechanistic studies evaluating its involvement.
Aberrant alternative splicing events of cancer-related genes associated with bladder cancer
It has been shown that there is usually a dysregulation of alternative splicing events in tumor, that is, a gene due to the occurrence of abnormal splicing leads to the large production of its pro-cancerous variants, which affects all aspects of the tumor. This section summarizes the aberrant splicing events that affect the TIME, prognosis, and disease progression of bladder cancer and summarizes them in Table 1.
Alternative splicing events associated with tumor immune microenvironment
The tumor immune microenvironment (TIME) is composed of tumor cells, stromal cells, micro-vessels, interstitial fluid, and infiltrating cells, and it plays a crucial role in tumor occurrence, progression, and treatment (116–118). Evidence has suggested that the abnormal regulation of alternative splicing events has a significant impact on the composition and function of the TIME, ultimately influencing the prognosis of patients with bladder cancer. In bladder cancer, aberrant alternative splicing events have emerged as powerful biomarkers for evaluating the immune status, predicting prognosis, and identifying potential targeted therapies specifically in muscle-invasive bladder cancer (18, 85). A few notable studies have investigated the alternative splicing events of immune-related genes in bladder cancer—for example, the study of Li et al. (113) identified significant differences in the alternative splicing of PTGER3 between bladder cancer tissues and healthy tissues using gene differential expression analysis (Figure 3). Patients with higher prostaglandin E receptor 3 (PTGER3) expression exhibited shorter overall survival but had higher immune checkpoint-related gene expression, more significant enrichment of immune signature-related genes, and a greater number of infiltrating immune cells compared with the low-expression group. Furthermore, Yu et al. confirmed the relationship between TRMU expression and immune cells and checkpoint genes using the TIMER database (18). These studies have provided insights into the potential roles of alternative splicing events in immune-related targets and their associations with immune activation and prognosis in bladder cancer. However, further research is still needed to explore the full extent of alternative splicing events in immune-related genes and their functional consequences in bladder cancer.
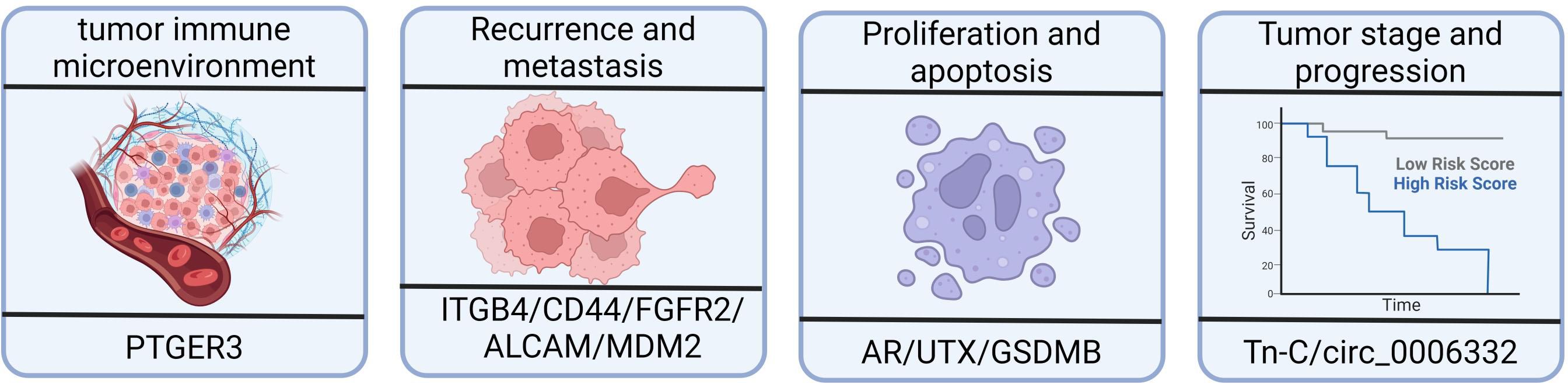
Figure 3 Effects of alternative splicing dysregulation on cancer progression. High prostaglandin E receptor 3 (PTGER3) expression may be associated with the immune response and the overall survival of patients with bladder cancer. Integrin subunit β4 (ITGB4) and CD44 are redistributed in bladder cancer tissues, impacting tumor migration and invasion. There is a higher proportion of shorter MDM2 alternative isoforms in bladder cancer patients with recurrence compared to patients without recurrence. ALCAM-Iso2 contributes to metastasis by increasing shedding and reducing cellular cohesion. The androgen receptor, UTX, and GSDMB isoforms play important roles in bladder cancer, including cell proliferation, viability, and cell death mechanisms. The expression of tenascin-C(L) variants is significantly increased in higher-stage and higher-grade tumors.
Alternative splicing events associated with bladder cancer proliferation and apoptosis
Splicing defects in many vital genes are associated with tumor cell proliferation (119–121). The androgen receptor, which is a member of the steroid receptor family of transcription factors, is crucial for mediating androgen signaling, specifically testosterone and dihydrotestosterone signaling (122, 123). The study of Kimberley et al. (124) found that the majority of bladder cancer cells expressed different levels of low-molecular-weight (LMW) androgen receptor isoforms, while a small number expressed the full-length androgen receptor. Knockdown of total androgen receptor isoforms using silencing RNA reduced cell survival and triggered apoptosis, along with increasing the expression of nuclear androgen receptors. This finding indicates that LMW androgen receptor isoforms are normally present in bladder cancer cells and contribute to cell viability.
The C-terminal JmjC domain of UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) is historically recognized for its histone demethylation function, contributing to its pathophysiological significance (125). In a previous study, multiple isoforms of UTX were identified in 5,637 cells. The three most abundant isoforms were lacking exon 14 (39% of UTX transcripts), exons 14 and 16 (27% of UTX transcripts), and the “long” isoform comprising all 30 exons (14% of UTX transcripts). Other identified isoforms included the isoforms lacking exon 16 (8%), exon 13 (6%), and exons 13 and 16 (4%), and there were several other isoforms with frequencies below 1%. The isoform lacking exon 14 exhibited weaker binding to chromatin, potentially due to its reduced nuclear abundance. The isoforms lacking exon 14 and exon 16 were unable to bind the polycomb repressive–deubiquitinase complex (PR-DUB) and mitotic deacetylase complex (MiDAC), which impaired the functional properties of the encoded protein (126).
Granzyme A, which is produced by killer lymphocytes, cleaves gasdermin B (GSDMB) and induces pyroptosis in tumor cells, leading to antitumor immune responses (Figure 3). Kong et al. discovered that different GSDMB isoforms have distinct functional properties. In their study, the N-terminal fragments of GSDMB isoforms 3 and 4, which are generated through cleavage, triggered pyroptosis. In contrast, isoforms 1, 2, and 5 failed to induce pyroptosis. The non-functional isoforms lacked a stable belt motif, which was either deleted or modified at exon 6. The belt motif facilitated the insertion of oligomeric GSDMB N-termini into the cell membrane. The expression of GSDMB3/4 isoforms, rather than GSDMB1/2, was commonly found to be upregulated in bladder cancers with better outcomes, indicating a protective role mediated by GSDMB3/4 in those tumors (127).
Alternative splicing events associated with bladder cancer recurrence and metastasis
Patients with non-muscle invasive bladder cancer exhibit a high recurrence rate (60%) (128), with approximately 20% progressing to muscle-invasive bladder cancer (129). Recurrence or metastasis occurs in about half of patients with muscle-invasive bladder cancer who undergo radical cystectomy, leading to a low 5-year mortality rate of 50% (130). Prognosis is particularly poor in patients with locally progressive or recurrent muscle-invasive bladder cancer (131), highlighting the importance of predicting bladder cancer recurrence for effective management and treatment. Abnormal alternative splicing has been extensively studied in relation to bladder cancer recurrence, and the findings of these studies are summarized below.
Huang et al. (93) identified significant associations between specific alternative splicing events (SMOX-58,619-AP, INO80C-45,170-AP, and ITGB4–43,489-ES) and bone metastasis, splicing factors, and survival (Figure 3). In normal epithelial cells, integrin subunit β4 (ITGB4) binds to hemidesmosomes and promotes epithelial cell anchoring to the basement membrane. However, in tumors, ITGB4 is located at the front of the cell, which is rich in lamellae and filopodia, thereby enhancing tumor migration and invasion (132, 133). Another study showed that ITGB4 redistribution facilitated tumor migration and invasion, making it a valuable prognostic marker for bladder cancer (134). Moreover, JUP has been found to downregulate ITGB4–43,489-ES through the glycosphingolipid biosynthetic pathway, which is also associated with prognosis of bladder cancer (93).
CD44, a cell surface glycoprotein, has been extensively studied as a marker of bladder cancer aggressiveness and stemness. Alternative splicing and glycosylation generate multiple CD44 isoforms with distinct functional roles. Transcriptomics analysis has revealed significant heterogeneity among CD44 isoforms in 75 patients with bladder cancer, which is thought to be associated with tumor invasion and poor prognosis (135). Moreover, another study showed that a reduction in ESRP1 or ESRP2 promoted bladder cancer cell growth and lung metastasis by altering FGFR2 splicing and macrophage polarization (39).
MDM2, which is involved in the regulation of tumor suppressor p53, exhibits differential alternative splicing in patients with bladder cancer with or without recurrence. The expression of specific MDM2 exons is higher in patients with recurrence, indicating a higher proportion of shorter MDM2 alternative isoforms (136) (Figure 3).
Spliced variants of periostin, an EXM protein, have been detected in bladder cancer tissues and cell lines. In one study, wild-type periostin was reduced by downregulation or alternative splicing, particularly in isoforms lacking exon 18, and was strongly associated with bladder cancer progression. However, the lack of exon 18 did not suppress cancer cell invasiveness and metastasis (137). This indicates that the pro-metastatic function is achieved by aberrant alternative splicing to downregulate wild-type periostin expression.
ALCAM-Iso2, a splice variant of ALCAM, has been identified as a driver of metastasis. In one study, loss of the membrane-proximal region of ALCAM (exon 13) mediated by a novel matrix metallopeptidase 14-dependent membrane distal cleavage site increased shedding and decreased cellular cohesion, contributing to bladder tumor metastasis (138) (Figure 3).
Most studies have shown that the splicing abnormalities of some EXM-related proteins are closely associated with bladder cancer recurrence, providing valuable insights for mechanistic studies on relapse and metastasis.
Alternative splicing events associated with tumor stage and progression
The primary transcript of the EXM protein, namely, tenascin-C (Tn-C), undergoes alternative splicing, resulting in the inclusion or omission of nine type III homology repeats. In bladder cancer, there is a significant increase in the expression of Tn-C(L) variants with a higher tumor stage and grade. Furthermore, the expression of Tn-C splicing variants differs depending on the tumor type, indicating differential regulation of Tn-C splicing in bladder cancer, which may contribute to its pathogenesis (139) (Figure 3).
Circ_0006332, which is derived from the splicing of MYBL2 exons 8 and 9, has been implicated in bladder cancer progression. In a previous study, circ_0006332 knockdown inhibited the proliferation, colony formation, and invasiveness of bladder cancer cells. Moreover, E-cadherin was upregulated, while vimentin, cyclin B1, and P21 were downregulated, indicating Circ_0006332’s role in promoting epithelial–mesenchymal transition and cell cycle progression in bladder cancer (140).
Targeted therapy of bladder cancer based on alternative splicing
Alternative splicing has emerged as a hallmark of cancer, and the development of therapeutic approaches targeting splicing holds great promise. Various tools have been developed to manipulate splicing, facilitating the identification of significant differential alternative splicing events associated with bladder cancer. These differential alternative splicing events, along with splicing factors, are thought to impact patients’ overall survival and chemotherapy drug sensitivity (141). Additionally, the prognostic signature based on alternative splicing is correlated with the response to common chemotherapeutic agents, indicating its potential as an indicator for treatment selection (140). Inhibitors of FGFR rearrangement, such as erdafitinib, infigratinib, and pemigatinib, have shown promise as targeted therapies against bladder cancer. Clinical trials are currently underway to explore the efficacy and safety profiles of these compounds in patients with bladder cancer with FGFR3 alterations (142). Perhaps increasing the sensitivity of chemotherapy drugs through targeted splicing is a relatively good way to achieve this.
Some drugs that may act by targeting alternative splicing are available. Enzalutamide, a second-generation therapeutic drug targeting the androgen receptor ligand-binding domain, has demonstrated effectiveness in inhibiting bladder cancer cells expressing the full-length androgen receptor (143, 144). However, the response to enzalutamide treatment appears to depend on the presence and localization of the full-length androgen receptor in the nuclear compartment rather than the absolute level of the full-length androgen receptor (124).
Targeted immunotherapy, particularly immune checkpoint inhibitor therapy, such as PD-1, PD-L1, and cytotoxic T-lymphocyte-associated protein 4 inhibitors, has demonstrated positive therapeutic effects in advanced bladder cancer (145–147). However, the effectiveness of immunotherapy varies among patients due to the heterogeneity of bladder cancer (148). Alternative splicing events not only regulate organ development, tissue identity acquisition, and tissue homeostasis but also play a significant role in tumor occurrence and development (149, 150). Furthermore, mutations that are tumor-specific to splicing factors have been identified as risk factors for tumor progression and maintenance (151), further emphasizing the potential of alternative splicing in immunotherapy (152).
Further research is warranted to unravel the complex mechanisms underlying splicing dysregulation in bladder cancer as well as to explore its therapeutic implications and advance precision medicine for this disease.
Conclusions
RNA splicing is a crucial process that regulates gene expression and contributes to proteome diversity. Abnormal alternative splicing has been implicated in tumorigenesis in various cancers, including bladder cancer. This review summarizes splicing dysregulation in bladder cancer, particularly in terms of specific splicing variants and splicing factors, which have potential clinical value as diagnostic/prognostic biomarkers or therapeutic targets. Meanwhile, the impact of splicing dysregulation on TIME, disease progression, recurrence, and therapy resistance are emphasized. The review collates the findings of studies investigating splicing-related factors associated with bladder cancer, with a focus on splicing factors, particularly the hnRNP family of proteins. These factors are aberrantly expressed in bladder cancer and are associated with specific splicing events and clinical outcomes (Table 1). However, the underlying mechanisms remain largely unknown, necessitating further research to elucidate the oncogenic and tumor-suppressing functions of alternative splicing events and splicing factors in bladder cancer. Pre-clinical studies have illustrated the contribution of splicing events to treatment resistance and disease progression in bladder cancer, illustrating that these events could serve as therapeutic targets. Nonetheless, due to the lack of clinical implementation in patients with bladder cancer, the development of drugs targeting spliceosomes remains challenging. Future investigations should focus on unraveling the mechanisms underlying splicing abnormalities and the consequences of inhibiting these factors, thereby enabling the full potential of splicing-based therapies in bladder cancer to be recognized.
Author contributions
LL: Writing – original draft, Writing – review & editing. TJ: Writing – review & editing, Writing – original draft. LH: Writing – review & editing. JD: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Project of Social Development of Jinhua Science and Technology Bureau of Zhejiang Province, China (No. 2021–3-079) and the Key Project of Social Development of Jinhua Science and Technology Bureau of Zhejiang Province, China (No. 2023–3-165).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Li X, Feng J, Sun Y, Li X. An exploration of the tumor microenvironment identified a novel five-gene model for predicting outcomes in bladder cancer. Front Oncol. (2021) 11:642527. doi: 10.3389/fonc.2021.642527
2. Stenehjem DD, Tran D, Nkrumah MA, Gupta S. PD1/PDL1 inhibitors for the treatment of advanced urothelial bladder cancer. OncoTargets Ther. (2018) 11:5973–89. doi: 10.2147/OTT
3. Montes M, Sanford BL, Comiskey DF, Chandler DS. RNA splicing and disease: animal models to therapies. Trends Genet. (2019) 35:68–87. doi: 10.1016/j.tig.2018.10.002
4. Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. (2015) 84:291–323. doi: 10.1146/annurev-biochem-060614-034316
5. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. (2003) 72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720
6. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJ, Giovannetti E, et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resistance Updates. (2020) 53:100728. doi: 10.1016/j.drup.2020.100728
7. Patel AA, Steitz J. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. (2003) 4(12):960–70. doi: 10.1038/nrm1259
8. Fu X-D, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. (2014) 15(10):689–701. doi: 10.1038/nrg3778
9. Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. (2009) 10(11):741–54. doi: 10.1038/nrm2777
10. Dvinge H. Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett. (2018) 592(17):2987–3006. doi: 10.1002/1873-3468.13119
11. Park E, Pan Z, Zhang Z, Lin L, Xing Y. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet. (2018) 102(1):11–26. doi: 10.1016/j.ajhg.2017.11.002
12. Black A, Gamarra J, Giudice J. More than a messenger: Alternative splicing as a therapeutic target. Biochim Biophys Acta (BBA)-Gene Regul Mech. (2019) 1862:194395. doi: 10.1016/j.bbagrm.2019.06.006
13. Paik PK, Drilon A, Fan P-D, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discovery. (2015) 5:842–9. doi: 10.1158/2159-8290.CD-14-1467
14. Huang S, Luo K, Jiang L, Zhang X-D, Lv Y-H, Li R-F. PCBP1 regulates the transcription and alternative splicing of metastasis−related genes and pathways in hepatocellular carcinoma. Sci Rep. (2021) 11:23356. doi: 10.1038/s41598-021-02642-z
15. Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. (2016) 26:732–44. doi: 10.1101/gr.199935.115
16. Liu Z, Rabadan R. Computing the role of alternative splicing in cancer. Trends Cancer. (2021) 7(4):347–58. doi: 10.1016/j.trecan.2020.12.015
17. He R-q, Zhou X-g, Yi Q-y, Deng C-w, Gao J-m, Chen G, et al. Prognostic signature of alternative splicing events in bladder urothelial carcinoma based on spliceseq data from 317 cases. Cell Physiol Biochem. (2018) 48:1355–68. doi: 10.1159/000492094
18. Yu X, Luo B, Lin J, Zhu Y. Alternative splicing event associated with immunological features in bladder cancer. Front Oncol. (2023) 12:966088. doi: 10.3389/fonc.2022.966088
19. Ye F, Liang Y, Cheng Z, Liu Y, Hu J, Li W, et al. Immunological characteristics of alternative splicing profiles related to prognosis in bladder cancer. Front Immunol. (2022) 13:911902. doi: 10.3389/fimmu.2022.911902
20. Du J-X, Zhu G-Q, Cai J-L, Wang B, Luo Y-H, Chen C, et al. Splicing factors: Insights into their regulatory network in alternative splicing in cancer. Cancer Lett. (2021) 501:83–104. doi: 10.1016/j.canlet.2020.11.043
21. Gonçalves V, Pereira JF, Jordan P. Signaling pathways driving aberrant splicing in cancer cells. Genes. (2017) 9(1):9. doi: 10.3390/genes9010009
22. Huang H, Kapeli K, Jin W, Wong YP, Arumugam TV, Koh JH, et al. Tissue-selective restriction of RNA editing of CaV1. 3 by splicing factor SRSF9. Nucleic Acids Res. (2018) 46:7323–38. doi: 10.1093/nar/gky348
23. Tacke R, Manley JL. Determinants of SR protein specificity. Curr Opin Cell Biol. (1999) 11:358–62. doi: 10.1016/S0955-0674(99)80050-7
24. Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. (2016) 16(7):413–30. doi: 10.1038/nrc.2016.51
25. Liu J, Wang Y, Yin J, Yang Y, Geng R, Zhong Z, et al. Pan-cancer analysis revealed SRSF9 as a new biomarker for prognosis and immunotherapy. J Oncol. (2022) 2022:3477148. doi: 10.1155/2022/3477148
26. Bielli P, Panzeri V, Lattanzio R, Mutascio S, Pieraccioli M, Volpe E, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non–muscle-invasive bladder cancer. Clin Cancer Res. (2018) 24(21):5422–32. doi: 10.1158/1078-0432.CCR-17-3850
27. Zeng J, Xu H, Huang C, Sun Y, Xiao H, Yu G, et al. CD46 splice variant enhances translation of specific mRNAs linked to an aggressive tumor cell phenotype in bladder cancer. Mol Ther Nucleic Acids. (2021) 24:140–53. doi: 10.1016/j.omtn.2021.02.019
28. Jia R, Ajiro M, Yu L, McCoy P Jr, Zheng ZM. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA. (2019) 25:630–44. doi: 10.1261/rna.068619.118
29. Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Reviews: RNA. (2012) 3:1–12. doi: 10.1002/wrna.100
30. Silipo M, Gautrey H, Tyson-Capper A. Deregulation of splicing factors and breast cancer development. J Mol Cell Biol. (2015) 7:388–401. doi: 10.1093/jmcb/mjv027
31. Yan Q, Zeng P, Zhou X, Zhao X, Chen R, Qiao J, et al. RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene. (2021) 40:2635–50. doi: 10.1038/s41388-021-01666-z
32. Tang SJ, Luo S, Ho JXJ, Ly PT, Goh E, Roca X. Characterization of the regulation of CD46 RNA alternative splicing. J Biol Chem. (2016) 291(27):14311–23. doi: 10.1074/jbc.M115.710350
33. Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. (2016) 135:851–67. doi: 10.1007/s00439-016-1683-5
34. Du J, Wang Q, Ziegler SF, Zhou B. FOXP3 interacts with hnRNPF to modulate pre-mRNA alternative splicing. J Biol Chem. (2018) 293:10235–44. doi: 10.1074/jbc.RA117.001349
35. Li F, Hongfan Z, Su M, Xie W, Fang Y, Du Y, et al. HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to its 3′ UTR. EBioMedicine. (2019) 45:208–19. doi: 10.1016/j.ebiom.2019.06.017
36. Li F, Xie W, Fang Y, Xie K, Liu W, Hou L, et al. HnRNP-F promotes the proliferation of bladder cancer cells mediated by PI3K/AKT/FOXO1. J Cancer. (2021) 12:281–91. doi: 10.7150/jca.50490
37. Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol Elsevier. (2012) pp:417–27. doi: 10.1016/j.semcancer.2012.04.003
38. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. (2009) 33:591–601. doi: 10.1016/j.molcel.2009.01.025
39. Zhao Y, Li M, Wu W, Miao W, Liu H. Downregulated ESRP1/2 promotes lung metastasis of bladder carcinoma through altering FGFR2 splicing and macrophage polarization. Front Immunol. (2023) 14:1161273. doi: 10.3389/fimmu.2023.1161273
40. Hovhannisyan RH, Carstens RP. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem. (2007) 282:36265–74. doi: 10.1074/jbc.M704188200
41. Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. (2008) 28:5403–19. doi: 10.1128/MCB.00739-08
42. Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest. (2010) 120:3923–39. doi: 10.1172/JCI43552
43. Hope NR, Murray GI. The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum Pathol. (2011) 42:393–402. doi: 10.1016/j.humpath.2010.08.006
44. D'Agostino L, Caracciolo V, Giordano A. NSP 5a3a’s link to nuclear-cyto proteins B23 and hnRNP-L between normal and aberrant breast cell lines. Cell Cycle. (2010) 9(6):1131–42. doi: 10.4161/cc.9.6.10991
45. Yau W-Y, Shih H-C, Tsai M-H, Sheu J-C, Chen C-H, Chow L-P. Autoantibody recognition of an N-terminal epitope of hnRNP L marks the risk for developing HBV-related hepatocellular carcinoma. J Proteomics. (2013) 94:346–58. doi: 10.1016/j.jprot.2013.10.003
46. Daojun L, Haoyue W, Rongwei X, Fangpeng S, Bin L, Chengyong L, et al. HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget. (2017) 8(8):13586–99. doi: 10.18632/oncotarget.14600
47. Gaudreau M-C, Grapton D, Helness A, Vadnais C, Fraszczak J, Shooshtarizadeh P, et al. Heterogeneous Nuclear Ribonucleoprotein L is required for the survival and functional integrity of murine hematopoietic stem cells. Sci Rep. (2016) 6(1):0–0. doi: 10.1038/srep27379
48. Shen Q, Yu M, Jia J-K, Li W-X, Tian Y-W, Xue H-Z. Possible molecular markers for the diagnosis of pancreatic ductal adenocarcinoma. Med Sci Monitor: Int Med J Exp Clin Res. (2018) 24:2368. doi: 10.12659/MSM.906313
49. Liu Z, Goldstein AM, Hsu W-L, Yu KJ, Chien Y-C, Ko J-Y, et al. Evaluation of rare and common variants from suspected familial or sporadic nasopharyngeal carcinoma (NPC) susceptibility genes in sporadic NPC. Cancer Epidemiol Biomarkers Prev. (2019) 28:1682–6. doi: 10.1158/1055-9965.EPI-19-0007
50. Xing C, Gong J, Zhou J, Cai Z, Xu J, Guo F. Identification of potential biomarkers involved in gastric cancer through integrated analysis of Non-Coding RNA associated competing endogenous RNAs network. Clin Lab. (2018) 64(10/2018):0–0. doi: 10.7754/Clin.Lab.2018.180419
51. Chen T, Zheng W, Chen J, Lin S, Zou Z, Li X, et al. Systematic analysis of survival-associated alternative splicing signatures in clear cell renal cell carcinoma. J Cell Biochem. (2020) 121:4074–84. doi: 10.1002/jcb.29590
52. Zengliang S, Lin H, Xing H, Zhuo-Xun W, Kun P, Yun D, et al. Targeting HNRNPU to overcome cisplatin resistance in bladder cancer. Mol Cancer. (2022) 21(1):0–0. doi: 10.1186/s12943-022-01517-9
53. Chen X, Gu P, Xie R, Han J, Liu H, Wang B, et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. (2017) 21:1266–79. doi: 10.1111/jcmm.12999
54. Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. (2016) 35:2031–9. doi: 10.1038/onc.2015.270
55. He X, Arslan A, Ho T, Yuan C, Stampfer M, Beck W. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and Malignant properties. Oncogenesis. (2014) 3:e84–4. doi: 10.1038/oncsis.2013.47
56. Wang K, Pan S, Zhao P, Liu L, Chen Z, Bao H, et al. PTBP1 knockdown promotes neural differentiation of glioblastoma cells through UNC5B receptor. Theranostics. (2022) 12:3847. doi: 10.7150/thno.71100
57. Xie R, Chen X, Chen Z, H. M, D. W, G. P, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. (2019) 449:31–44. doi: 10.1016/j.canlet.2019.01.041
58. Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon Y-S, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. (2009) 36:996–1006. doi: 10.1016/j.molcel.2009.12.003
59. Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, et al. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. (2018) 34:85–102. doi: 10.1016/j.ccell.2018.06.007
60. Bielli P, Panzeri V, Lattanzio R, Mutascio S, Pieraccioli M, Volpe E, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non-muscle-invasive bladder cancer. Clin Cancer Res. (2018) 24(21):5422–32. doi: 10.1158/1078-0432.ccr-17-3850
61. Quidville V, Alsafadi S, Goubar A, Commo F, Scott V, Pioche-Durieu C, et al. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res. (2013) 73:2247–58. doi: 10.1158/0008-5472.CAN-12-2501
62. Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harbor Perspect Biol. (2011) 3:a003707. doi: 10.1101/cshperspect.a003707
63. Wan R, Bai R, Shi Y. Molecular choreography of pre-mRNA splicing by the spliceosome. Curr Opin Struct Biol. (2019) 59:124–33. doi: 10.1016/j.sbi.2019.07.010
64. Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. (1999) 18:3451–62. doi: 10.1093/emboj/18.12.3451
65. Dvinge H, Guenthoer J, Porter PL, Bradley RK. RNA components of the spliceosome regulate tissue-and cancer-specific alternative splicing. Genome Res. (2019) 29:1591–604. doi: 10.1101/gr.246678.118
66. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. (2016) 539:452–5. doi: 10.1038/nature20149
67. Lee SC-W, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. (2016) 22:976–86. doi: 10.1038/nm.4165
68. Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood J Am Soc Hematol. (2018) 132:1225–40. doi: 10.1182/blood-2018-04-843771
69. Tapak L, Saidijam M, Sadeghifar M, Poorolajal J, Mahjub H. Competing risks data analysis with high-dimensional covariates: an application in bladder cancer. Genomics Proteomics Bioinf. (2015) 13:169–76. doi: 10.1016/j.gpb.2015.04.001
70. Liu K-L, Yin Y-W, Lu B-S, Niu Y-L, Wang D-D, Shi B, et al. E2F6/KDM5C promotes SF3A3 expression and bladder cancer progression through a specific hypomethylated DNA promoter. Cancer Cell Int. (2022) 22:109. doi: 10.1186/s12935-022-02475-4
71. Yan C, Wan R, Bai R, Huang G, Shi YJS. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. (2016) 353(6302):904–11. doi: 10.1126/science.aag0291
72. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. (2011) 478(7367):64–9. doi: 10.1038/nature10496
73. Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood J Am Soc Hematol. (2012) 119:3203–10. doi: 10.1182/blood-2011-12-399774
74. Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood J Am Soc Hematol. (2013) 122:3616–27. doi: 10.1182/blood-2013-08-518886
75. Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. (2014) 28:241–7. doi: 10.1038/leu.2013.336
76. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood J Am Soc Hematol. (2015) 125:1367–76. doi: 10.1182/blood-2014-11-610543
77. Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. (2014) 28:108–17. doi: 10.1038/leu.2013.263
78. Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. (2015) 526:525–30. doi: 10.1038/nature15395
79. Patnaik MM, Lasho TL, Finke CM, Hanson CA, Hodnefield JM, Knudson RA, et al. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: prevalence, clinical correlates, and prognostic relevance. Am J Hematol. (2013) 88:201–6. doi: 10.1002/ajh.23373
80. Patnaik MM, Lasho TL, Finke CM, Hanson CA, Hodnefield JM, Knudson RA, et al. Spliceosome mutations involving SRSF2, SF3B1 and U2AF35 in world health organization defined chronic myelomonocytic leukemia; prevalence, clinical correlates and prognosis. Blood. (2012) 120(21):1711–1. doi: 10.1182/blood.v120.21.1711.1711
81. Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. (2018) 23(1):282–96.e4. doi: 10.1016/j.celrep.2018.01.088
82. Hafez AM, Seleem MM, Alattar AZ, Elshorbagy S, Elsayed WSH. RNA-binding proteins RBM-HuR, RBM3 and PODXL expression in urothelial carcinoma of the urinary bladder. Prognostic Clin implications. Wspolczesna Onkologia-Contemporary Oncol. (2021) 25(4):279–90. doi: 10.5114/wo.2021.112371
83. Boman K, Segersten U, Ahlgren G, Eberhard J, Uhlén M, Jirström K, et al. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urology. (2013) 13(1):0–0. doi: 10.1186/1471-2490-13-17
84. Yin Y, Liu K, Lu BS, Li W, Niu YL, Zhao CM, et al. RBM24 exacerbates bladder cancer progression by forming a Runx1t1/TCF4/miR-625-5p feedback loop. Exp Mol Med. (2021) 53(5):933–46. doi: 10.1038/s12276-021-00623-w
85. AZhaTi B, Wu G, Zhan H, Liang W, Song Z, Lu L, et al. Alternative splicing patterns reveal prognostic indicator in muscle-invasive bladder cancer. World J Surg Oncol. (2022) 20:1–16. doi: 10.1186/s12957-022-02685-0
86. Renieri A, Mencarelli MA, Cetta F, Baldassarri M, Mari F, Furini S, et al. Oligogenic germline mutations identified in early non-smokers lung adenocarcinoma patients. Lung Cancer. (2014) 85:168–74. doi: 10.1016/j.lungcan.2014.05.020
87. Fang J, Xiao L, Zhang Q, Peng Y, Wang Z, Liu Y. Junction plakoglobin, a potential prognostic marker of oral squamous cell carcinoma, promotes proliferation, migration and invasion. J Oral Pathol Med. (2020) 49:30–8. doi: 10.1111/jop.12952
88. Lewalle J-M, Bajou K, Desreux J, Mareel M, Dejana E, Noël A, et al. Alteration of interendothelial adherens junctions following tumor cell–endothelial cell interactionin vitro. Exp Cell Res. (1997) 237:347–56. doi: 10.1006/excr.1997.3799
89. Cowin P, Kapprell H-P, Franke WW, Tamkun J, Hynes RO. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. (1986) 46:1063–73. doi: 10.1016/0092-8674(86)90706-3
90. Holen I, Whitworth J, Nutter F, Evans A, Brown HK, Lefley DV, et al. Loss of plakoglobin promotes decreased cell-cell contact, increased invasion, and breast cancer cell dissemination in vivo. Breast Cancer Res. (2012) 14:1–14. doi: 10.1186/bcr3201
91. Syrigos K, Harrington K, Waxman J, Krausz T, Pignatelli M. Altered gamma-catenin expression correlates with poor survival in patients with bladder cancer. J Urol. (1998) 160:1889–93. doi: 10.1016/S0022-5347(01)62438-8
92. Rieger-Christ K, Ng L, Hanley R, Durrani O, Ma H, Yee A, et al. Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br J Cancer. (2005) 92:2153–9. doi: 10.1038/sj.bjc.6602651
93. Huang R, Zheng Z, Xian S, Zhang J, Jia J, Song D, et al. Identification of prognostic and bone metastatic alternative splicing signatures in bladder cancer. Bioengineered. (2021) 12:5289–304. doi: 10.1080/21655979.2021.1964252
94. Bachellerie J-P, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. (2002) 84:775–90. doi: 10.1016/S0300-9084(02)01402-5
95. Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, et al. An updated human snoRNAome. Nucleic Acids Res. (2016) 44:5068–82. doi: 10.1093/nar/gkw386
96. Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol. (2016) 13:734–52. doi: 10.1038/nrurol.2016.193
97. Flippot R, Beinse G, Boilève A, Vibert J, Malouf GG. Long non-coding RNAs in genitourinary Malignancies: a whole new world. Nat Rev Urol. (2019) 16:484–504. doi: 10.1038/s41585-019-0195-1
98. Cong L, Yang Q, Hu C, Yu Q, Hao S, Li D. Current status of functional studies on circular RNAs in bladder cancer and their potential role as diagnostic and prognostic biomarkers: a review. Med Sci Monitor: Int Med J Exp Clin Res. (2019) 25:3425. doi: 10.12659/MSM.916697
99. Guo B, Li D, Du L, Zhu X. piRNAs: biogenesis and their potential roles in cancer. Cancer Metastasis Rev. (2020) 39:567–75. doi: 10.1007/s10555-020-09863-0
100. J.m. Cavaille H, Paulsen M, Ferguson-Smith AC, Bachellerie J-P. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader–Willi/Angelman syndrome region. Hum Mol Genet. (2002) 11(13):1527–38. doi: 10.1093/hmg/11.13.1527
101. Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. (2011) 14:33–44. doi: 10.1016/j.cmet.2011.04.009
102. Chang L-S, Lin S-Y, Lieu A-S, Wu T-L. Differential expression of human 5S snoRNA genes. Biochem Biophys Res Commun. (2002) 299:196–200. doi: 10.1016/S0006-291X(02)02623-2
103. Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, et al. SNORA21–an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine. (2017) 22:68–77. doi: 10.1016/j.ebiom.2017.07.009
104. Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Köhn M, et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol. (2017) 19:844–55. doi: 10.1038/ncb3563
105. Cao R, Bo M, Yuan L, Wang G, Tian Y. Small nucleolar RNAs signature (SNORS) identified clinical outcome and prognosis of bladder cancer (BLCA). Cancer Cell Int. (2020) 20(1):0–0. doi: 10.1186/s12935-020-01393-7
106. Wang X, Guo Z, Zhu H, Xin J, Yuan L, Qin C, et al. Genetic variants in splicing factor genes and susceptibility to bladder cancer. Gene. (2022) 809:146022. doi: 10.1016/j.gene.2021.146022
107. Yang X, Yang Y, Sun B-F, C. YS, X. JW, L. WY, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res. (2017) 27:606–25. doi: 10.1038/cr.2017.55
108. Wang N, Chen R-x, Deng M-h, Wei W-s, Zhou Z-h, Ning K, et al. m5C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer Malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis. (2023) 14:139. doi: 10.1038/s41419-023-05661-y
109. Knott GJ, Bond CS, Fox AH. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. (2016) 44(9):3989–4004. doi: 10.1093/nar/gkw271
110. Xie R, Chen X, Cheng L, Huang M, Zhou Q, Zhang J, et al. NONO inhibits lymphatic metastasis of bladder cancer via alternative splicing of SETMAR. Mol Ther. (2021) 29:291–307. doi: 10.1016/j.ymthe.2020.08.018
111. Královičová J, Ševčíková I, Stejskalová E, Obuća M, Hiller M, Staněk D, et al. PUF60-activated exons uncover altered 3′ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. (2018) 46(12):6166–87. doi: 10.1093/nar/gky389
112. Long Q, An X, Chen M, Wang N, Sui S, Li Y, et al. PUF60/AURKA axis contributes to tumor progression and Malignant phenotypes in bladder cancer. Front Oncol. (2020) 10:568015. doi: 10.3389/fonc.2020.568015
113. Li X, Lin Y, Huang W, Jia B, Lai Y. Immunological significance of alternative splicing prognostic signatures for bladder cancer. Heliyon. (2022) 8:e08994. doi: 10.1016/j.heliyon.2022.e08994
114. Chen J, Liao Y, Li R, Luo M, Wu G, Tan R, et al. Immunotherapeutic significance of a prognostic alternative splicing signature in bladder cancer. Technol Cancer Res Treat. (2022) 21:15330338221090093. doi: 10.1177/15330338221090093
115. Yuanlong L, Yao K, Mingjie A, Yuming L, Hanhao Z, Yunfeng L, et al. ZEB1-mediated biogenesis of circNIPBL sustains the metastasis of bladder cancer via Wnt/β-catenin pathway. J Exp Clin Cancer Res. (2023) 42(1):191. doi: 10.1186/s13046-023-02757-3
116. Fidler IJ. The pathogenesis of cancer metastasis: the'seed and soil'hypothesis revisited. Nat Rev Cancer. (2003) 3:453–8. doi: 10.1038/nrc1098
117. Weber CE, Kuo PC. The tumor microenvironment. Surg Oncol. (2012) 21:172–7. doi: 10.1016/j.suronc.2011.09.001
118. van Dijk N, Funt SA, Blank CU, Powles T, Rosenberg JE, van der Heijden MS. The cancer immunogram as a framework for personalized immunotherapy in urothelial cancer. Eur Urol. (2019) 75:435–44. doi: 10.1016/j.eururo.2018.09.022
119. Brinkman BMN. Splice variants as cancer biomarkers. Clin Biochem. (2004) 37(7):584–94. doi: 10.1016/j.clinbiochem.2004.05.015
120. Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. (2004) 64(21):7647–54. doi: 10.1158/0008-5472.CAN-04-1910
121. Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. (2006) 119(13):2635–41. doi: 10.1242/jcs.03053
122. Lombard AP, Mudryj M. The emerging role of the androgen receptor in bladder cancer. Endocrine-related Cancer. (2015) 22:R265–77. doi: 10.1530/ERC-15-0209
123. Messner EA, Steele TM, Tsamouri MM, Hejazi N, Gao AC, Mudryj M, et al. The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines. (2020) 8:422. doi: 10.3390/biomedicines8100422
124. Katleba KAL, Alan P, Tsamouri M-M, Baek HB, Nishida KS, Libertini SJ, et al. Depletion of androgen receptor low molecular weight isoform reduces bladder tumor cell viability and induces apoptosis. Cancer Lett. (2021) 504:49–57. doi: 10.1016/j.canlet.2021.01.029
125. Schulz WA, Lang A, Koch J, Greife A. The histone demethylase UTX/KDM6A in cancer: Progress and puzzles. Int J Cancer. (2019) 145:614–20. doi: 10.1002/ijc.32116
126. Fotouhi O, Nizamuddin S, Falk S, Schilling O, Knüchel-Clarke R, Biniossek ML, et al. Alternative mRNA splicing controls the functions of the histone H3K27 demethylase UTX/KDM6A. Cancers. (2023) 15:3117. doi: 10.3390/cancers15123117
127. Kong Q, Xia S, Pan X, Ye K, Li Z, Li H, et al. Alternative splicing of GSDMB modulates killer lymphocyte–triggered pyroptosis. Sci Immunol. (2023) 8:eadg3196. doi: 10.1126/sciimmunol.adg3196
128. Van Rhijn BW, Burger M, Lotan Y, Solsona E, Stief CG, Sylvester RJ, et al. Recurrence and progression of disease in non–muscle-invasive bladder cancer: from epidemiology to treatment strategy. Eur Urol. (2009) 56(3):430–42. doi: 10.1016/j.eururo.2009.06.028
129. Li F, Zeng J, Gao Y, Guan Z, Ma Z, Shi Q, et al. G9a inhibition induces autophagic cell death via AMPK/mTOR pathway in bladder transitional cell carcinoma. PLOS ONE. (2015) 10(9):e0138390. doi: 10.1371/journal.pone.0138390
130. Yoshida K, Tsuda M, Matsumoto R, Semba S, Wang L, Sugino H, et al. Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer Sci. (2019) 110(7):2119–32. doi: 10.1111/cas.14080
131. van der Vos K, Vis D, Nevedomskaya E, Kim Y, Choi W, McConkey D, et al. Epigenetic profiling demarcates molecular subtypes of muscle-invasive bladder cancer. Sci Rep. (2020) 10(1):10952. doi: 10.1038/s41598-020-67850-5
132. Kashyap T, Germain E, Roche M, Lyle S, Rabinovitz I. Role of β4 integrin phosphorylation in human invasive squamous cell carcinoma: regulation of hemidesmosome stability modulates cell migration. Lab Invest. (2011) 91:1414–26. doi: 10.1038/labinvest.2011.104
133. Gagnoux-Palacios L, Dans M, Van't Hof W, Mariotti A, Pepe A, Meneguzzi G, et al. Compartmentalization of integrin α6β4 signaling in lipid rafts. J Cell Biol. (2003) 162:1189–96. doi: 10.1083/jcb.200305006
134. Liu S, Ge D, Chen L, Zhao J, Su L, Zhang S, et al. A small molecule induces integrin β4 nuclear translocation and apoptosis selectively in cancer cells with high expression of integrin β4. Oncotarget. (2016) 7:16282. doi: 10.18632/oncotarget.v7i13
135. Gaiteiro C, Soares J, Relvas-Santos M, Peixoto A, Ferreira D, Paulo P, et al. Glycoproteogenomics characterizes the CD44 splicing code associated with bladder cancer invasion. Theranostics. (2022) 12:3150. doi: 10.7150/thno.67409
136. Dueñas M, Pérez-Figueroa A, Oliveira C, Suárez-Cabrera C, Sousa A, Oliveira P, et al. Gene expression analyses in non muscle invasive bladder cancer reveals a role for alternative splicing and Tp53 status. Sci Rep. (2019) 9:10362. doi: 10.1038/s41598-019-46652-4
137. Kim CJ, Isono T, Tambe Y, Chano T, Okabe H, Okada Y, et al. Role of alternative splicing of periostin in human bladder carcinogenesis. Int J Oncol. (2008) 32:161–9. doi: 10.3892/ijo
138. Hebron KE, Li EY, Arnold Egloff SA, Von Lersner AK, Taylor C, Houkes J, et al. Alternative splicing of ALCAM enables tunable regulation of cell-cell adhesion through differential proteolysis. Sci Rep. (2018) 8:3208. doi: 10.1038/s41598-018-21467-x
139. Berndt A, Anger K, Richter P, Borsi L, Brack S, Silacci M, et al. Differential expression of tenascin-C splicing domains in urothelial carcinomas of the urinary bladder. J Cancer Res Clin Oncol. (2006) 132:537–46. doi: 10.1007/s00432-006-0106-8
140. Li M, Liu Y, Liu J, Li W, Li N, Xue D, et al. Circ_0006332 promotes growth and progression of bladder cancer by modulating MYBL2 expression via miR-143. Aging (Albany NY). (2019) 11:10626. doi: 10.18632/aging.v11i22
141. Fan Z, Zhang Z, Piao C, Liu Z, Wang Z, Kong C. Genome-wide analyses of prognostic and therapeutic alternative splicing signatures in bladder urothelial carcinoma. Front Oncol. (2021) 11:626858. doi: 10.3389/fonc.2021.626858
142. Pederzoli F, Bandini M, Marandino L, Ali SM, Madison R, Chung J, et al. Targetable gene fusions and aberrations in genitourinary oncology. Nat Rev Urol. (2020) 17:613–25. doi: 10.1038/s41585-020-00379-4
143. Kawahara T, Ide H, Kashiwagi E, El-Shishtawy KA, Li Y, Reis LO, et al. Enzalutamide inhibits androgen receptor–positive bladder cancer cell growth. Urol Oncol: Semin Original Investigations Elsevier. (2016) 34:432.e15–e23. doi: 10.1016/j.urolonc.2016.05.016
144. Tyagi A, Chandrasekaran B, Kolluru V, Rai S, Jordan AC, Houda A, et al. Combination of androgen receptor inhibitor and cisplatin, an effective treatment strategy for urothelial carcinoma of the bladder. Urol Oncol. (2019) pp:492–502. doi: 10.1016/j.urolonc.2019.03.008
145. Petrylak DP, Powles T, Bellmunt J, Braiteh F, Loriot Y, Morales-Barrera R, et al. Atezolizumab (MPDL3280A) monotherapy for patients with metastatic urothelial cancer: long-term outcomes from a phase 1 study. JAMA Oncol. (2018) 4:537–44. doi: 10.1001/jamaoncol.2017.5440
146. Sharma P, Callahan MK, Bono P, Kim J, Spiliopoulou P, Calvo E, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. (2016) 17:1590–8. doi: 10.1016/S1470-2045(16)30496-X
147. Bellmunt J, De Wit R, Vaughn DJ, Fradet Y, Lee J-L, Fong L, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. New Engl J Med. (2017) 376:1015–26. doi: 10.1056/NEJMoa1613683
148. Eckstein M, Wirtz RM, Pfannstil C, Wach S, Stoehr R, Breyer J, et al. A multicenter round robin test of PD-L1 expression assessment in urothelial bladder cancer by immunohistochemistry and RT-qPCR with emphasis on prognosis prediction after radical cystectomy. Oncotarget. (2018) 9:15001. doi: 10.18632/oncotarget.v9i19
149. Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. (2017) 18:437–51. doi: 10.1038/nrm.2017.27
150. Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. (2017) 16:1–19. doi: 10.1186/s12943-016-0579-2
151. El Marabti E, Younis I. The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci. (2018) 5:80. doi: 10.3389/fmolb.2018.00080
Keywords: alternative splicing, splicing factor irregularity, aberrant splicing events, bladder cancer, cancer therapy
Citation: Li L, Jin T, Hu L and Ding J (2024) Alternative splicing regulation and its therapeutic potential in bladder cancer. Front. Oncol. 14:1402350. doi: 10.3389/fonc.2024.1402350
Received: 17 March 2024; Accepted: 05 July 2024;
Published: 26 July 2024.
Edited by:
Qinong Ye, Beijing Institute of Biotechnology, ChinaReviewed by:
Sabahattin Cömertpay, Kahramanmaras Sütçü Imam University, TürkiyePhilipp Erben, University of Heidelberg, Germany
Copyright © 2024 Li, Jin, Hu and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Ding, jhDingJin@zju.edu.cn; Lina Li, 20221054@jhc.edu.cn
†These authors have contributed equally to this work
 Lina Li
Lina Li Ting Jin2†
Ting Jin2† Jin Ding
Jin Ding