
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 25 June 2024
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1382369
This article is part of the Research Topic Molecular Markers for Pancreatic Cancers: New Technologies and Applications in the Clinical Practice View all 7 articles
Background: The diagnostic and prognostic clinical value of circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA) in pancreatic malignancies are unclear. Herein, we aimed to perform a meta-analysis to evaluate ctDNA and cfDNA as potential diagnostic and prognostic biomarkers.
Methods: PRISMA reporting guidelines were followed closely for conducting the current meta-analysis. The PubMed/Medline, Scopus, and Web of Science (WoS) databases were scanned in detail to identify eligible papers for the study. A quality assessment was performed in accordance with the REMARK criteria. The risk ratios (RRs) of the diagnostic accuracy of ctDNA compared to that of carbohydrate antigen 19.9 (CA 19.9) in all disease stages and the hazard ratios (HRs) of the prognostic role of ctDNA in overall survival (OS) were calculated with 95% confidence intervals (CIs).
Results: A total of 18 papers were evaluated to assess the diagnostic accuracy and prognostic value of biomarkers related to pancreatic malignancies. The pooled analysis indicated that CA19.9 provides greater diagnostic accuracy across all disease stages than ctDNA or cfDNA (RR = 0.64, 95% CI: 0.50–0.82, p < 0.001). Additionally, in a secondary analysis focusing on prognosis, patients who were ctDNA-positive were found to have significantly worse OS (HR = 2.00, 95% CI: 1.51–2.66, p < 0.001).
Conclusion: The findings of this meta-analysis demonstrated that CA19-9 still has greater diagnostic accuracy across all disease stages than KRAS mutations in ctDNA or cfDNA. Nonetheless, the presence of detectable levels of ctDNA was associated with worse patient outcomes regarding OS. There is a growing need for further research on this topic.
Systematic review registration: https://doi.org/10.37766/inplasy2023.12.0092, identifier INPLASY2023120092.
It is well known that tumors arising in the pancreas constitute one of the most prevalent malignancies affecting the upper gastrointestinal (GI) tract, following esophageal and gastric cancers. Moreover, it is one of the most difficult malignancies to manage due to its rapid progression and resistance to conventional treatments (1–3). The early stages of the disease typically show minimal or no symptoms (1, 2). Despite the introduction of new therapeutic approaches, the overall prognosis is generally unfavorable due to the aggressive nature of the tumor and remarkable recurrence rates. This emphasizes the importance of the crucial need for developing more effective diagnostic methods (3–6).
In clinical diagnostics for upper gastrointestinal malignancies, traditional serum protein markers such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19.9 (CA19.9) are increasingly being recognized. However, these markers have important limitations, especially in terms of sensitivity and specificity (4, 7). Therefore, circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA) can be used to detect the dynamic and genomic alterations associated with pancreatic malignancies (5, 6).
Liquid biopsy techniques such as ctDNA and cfDNA are promising methods for improving diagnostic accuracy and treatment strategies (8). A growing evidence base emphasizes the superiority of liquid biopsies compared to traditional tissue biopsies. Their noninvasive nature, increased safety, and improved ability to manage the complexities of intratumor heterogeneity are well documented (5, 6, 9). Additionally, it has been suggested that ctDNA provides faster and more accurate predictions than radiological imaging (6, 9). Furthermore, these studies also highlight the ability of ctDNA to detect real-time tumor dynamics with advances in technology.
Previous reports of primary research on the diagnostic accuracy of ctDNA and CA 19.9 have presented various contradictory findings. Several studies have proposed that CA 19.9 has notably greater diagnostic accuracy than ctDNA (10, 11). These studies also suggest that CA 19.9 may be more reliable. It has been well validated in many patient groups and is commonly used in clinical practice. Conversely, other studies propose no or negligible diagnostic difference between them (12, 13). Notably, numerous factors may have contributed to these inconsistent results, including the methodologies of the studies, the specific patient populations in the studies, and the technological platforms. Therefore, it is important and necessary to conduct more robust large-scale comparative studies, especially meta-analyses, to assess the current landscape associated with these biomarkers.
Herein, we performed a comprehensive meta-analysis to evaluate ctDNA and cfDNA as potential diagnostic and prognostic biomarkers for pancreatic malignancies.
Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) reporting guidelines were followed closely for documenting the current meta-analysis (14, 15). To validate adherence to established guidelines, the PRISMA checklist has been incorporated into Supplementary Table S1, serving as a key tool to ascertain compliance. The protocol for the meta-analysis was registered at the International Platform of Registered Systematic Review and Meta-analysis Protocols (INPLASY) and assigned the registration number INPLASY2023120092. The PubMed/Medline, Scopus, and Web of Science (WoS) databases were scanned in detail to identify eligible papers for the meta-analysis. The relevant databases were searched until December 2023, and there was an update in January 2024. Only articles published in English and with full-text access were evaluated, and papers published in other languages were not included in the initial search. The following main keyword combinations were used to develop the search strategy: ((Pancreatic[Title/Abstract]) AND (((((Carcinoma[Title/Abstract]) OR (cancer[Title/Abstract])) OR (neoplasm[Title/Abstract])) OR (tumor[Title/Abstract])) OR (tumour[Title/Abstract]))) AND ((circulating cfDNA[Title/Abstract]) OR (circulating ctDNA[Title/Abstract])). Medical Subject Headings (MeSH) and text terms were used to associate keywords, which were then combined using Boolean operators (AND/OR). The search method was developed in PubMed/Medline and then modified for use with additional databases (WoS and Scopus). The search procedures and related keyword combinations are documented in Supplementary Table S2.
The PICOS framework was as follows:
Population: This study involved patients who were diagnosed with pancreatic malignancies.
Intervention: The intervention under consideration was the presence of ctDNA and cfDNA.
Comparison: The comparison included comparing the presence of ctDNA or cfDNA with CA 19.9 for diagnostic accuracy and assessing the presence of ctDNA for overall survival.
Outcomes: OS in the presence of ctDNA and the diagnostic accuracy of ctDNA and cfDNA.
Study design: Cohort, case−control.
Original papers that provided reports related to ctDNA-cfDNA in pancreatic cancer were included in this meta-analysis. Two researchers independently assessed the titles and abstracts of the studies included in the initial review. Papers that were considered irrelevant based on their title and abstract were quickly omitted from the analysis. Disagreements were settled by discussion until consensus was achieved. Possible duplications were isolated after downloading potential studies that could be included in the study. The first two authors summarized the findings provided in the initial investigations after separating potential duplicate papers. Then, the studies were processed into a preprepared Microsoft Excel spreadsheet.
Studies had to meet the following selection criteria: i) the study population should include patients with pancreatic malignancies; ii) at least one of these patient groups must have had ctDNA or cfDNA analysis; and iii) the study population should contain information about the diagnostic, prognostic or predictive values of ctDNA and cfDNA. Studies that did not meet the related criteria were omitted from the meta-analysis. Moreover, conference reports, unpublished preprints, reviews, or case reports were not included in this meta-analysis.
All the data were processed independently by the first two authors (MEA and AI) according to a preprepared protocol. The following features of the studies were reviewed: first author name, year of publication, number of cases and controls, type and time of sampling, marker(s) or gene(s) of interest, cfDNA isolation, and sequencing method. In addition, the hazard ratios (HRs) for overall survival (OS) with 95% confidence intervals (CIs) were also calculated. All data obtained in the study were cross-checked by two authors via a standard spreadsheet to reach a consensus.
Quality assessment was performed in accordance with the REporting recommendations for Tumor MARKer Prognostic Studies (REMARK) criteria and is presented in Supplementary Table S3 (16). Each study can be given one point out of seven criteria. In the case of uncertainty, it was scored by giving half a point. Studies scoring ≥ 5.5 were included in the meta-analysis for further analysis.
Previous studies evaluating ctDNA, cfDNA, and CA 19.9 levels, as well as the prognostic role of ctDNA in OS, were considered in primary and secondary meta-analyses. The diagnostic accuracy of ctDNA or cfDNA compared to CA 19.9 in all disease stages was included in the primary meta-analysis. The prognostic role of ctDNA in OS was also included in the secondary meta-analysis. The risk ratios (RRs) were calculated for the diagnostic accuracy of ctDNA/cfDNA compared to CA 19.9 In the evaluation of the prognostic role of ctDNA, data regarding the relationship between the presence of ctDNA and OS were pooled and reported as hazard ratios (HRs). The outcomes of the meta-analysis are graphically illustrated via forest plots. Cochran’s Q test and I2 statistics were calculated to measure the statistical heterogeneity between studies in performing all the meta-analyses. The presence of significant heterogeneity was evaluated as follows: an I2 greater than 50% and a p value < 0.05 according to Cochran’s Q test (17). When it was determined that the outputs were heterogeneous, the analyses were carried out using the random effects model. Otherwise, the fixed-effects model was used to conduct the meta-analyses.
To demonstrate the robustness of the outcomes in this meta-analysis, sensitivity analyses were performed. The approach involved reassessing the effect size (ES)—a measure used to quantify the strength of the relationship or association—by sequentially omitting each study from the pooled meta-analysis. By removing one study at a time and recalculating the pooled effect size, the analysis helps identify whether any particular study disproportionately impacts the results, ensuring that the conclusions drawn are not unduly influenced by any single study and that the results are indeed consistent and reliable.
The possibility of publication bias in the meta-analysis was investigated using Egger’s linear regression test, Begg and Mazumdar’s rank correlation test, and funnel plots. The statistical significance level was defined as p < 0.05 for all outcomes. The Review Manager (v.5.4, Copenhagen, Denmark) (18) and ProMeta-3® (19) meta-analysis software packages were used to perform all analyses in our study.
A total of 751 papers were identified through structured literature searches of the PubMed/Medline (n = 132), Scopus (n = 357), and WoS (n = 262) databases. After eliminating irrelevant and duplicate records, the full texts of the remaining 79 studies were meticulously examined in detail. A total of 20 studies were evaluated for further analysis. After detailed quality assessment, one study was excluded from the meta-analysis because of an insufficient quality score (REMARK score < 5.5) (20). After all the reviews were completed, a total of 18 articles (10–12, 21–35) that met the criteria for the current meta-analysis were included in the study. The PRISMA flowchart, which maps out the number of records identified, included, and excluded, as well as the reasons for exclusions, is presented visually in Figure 1.
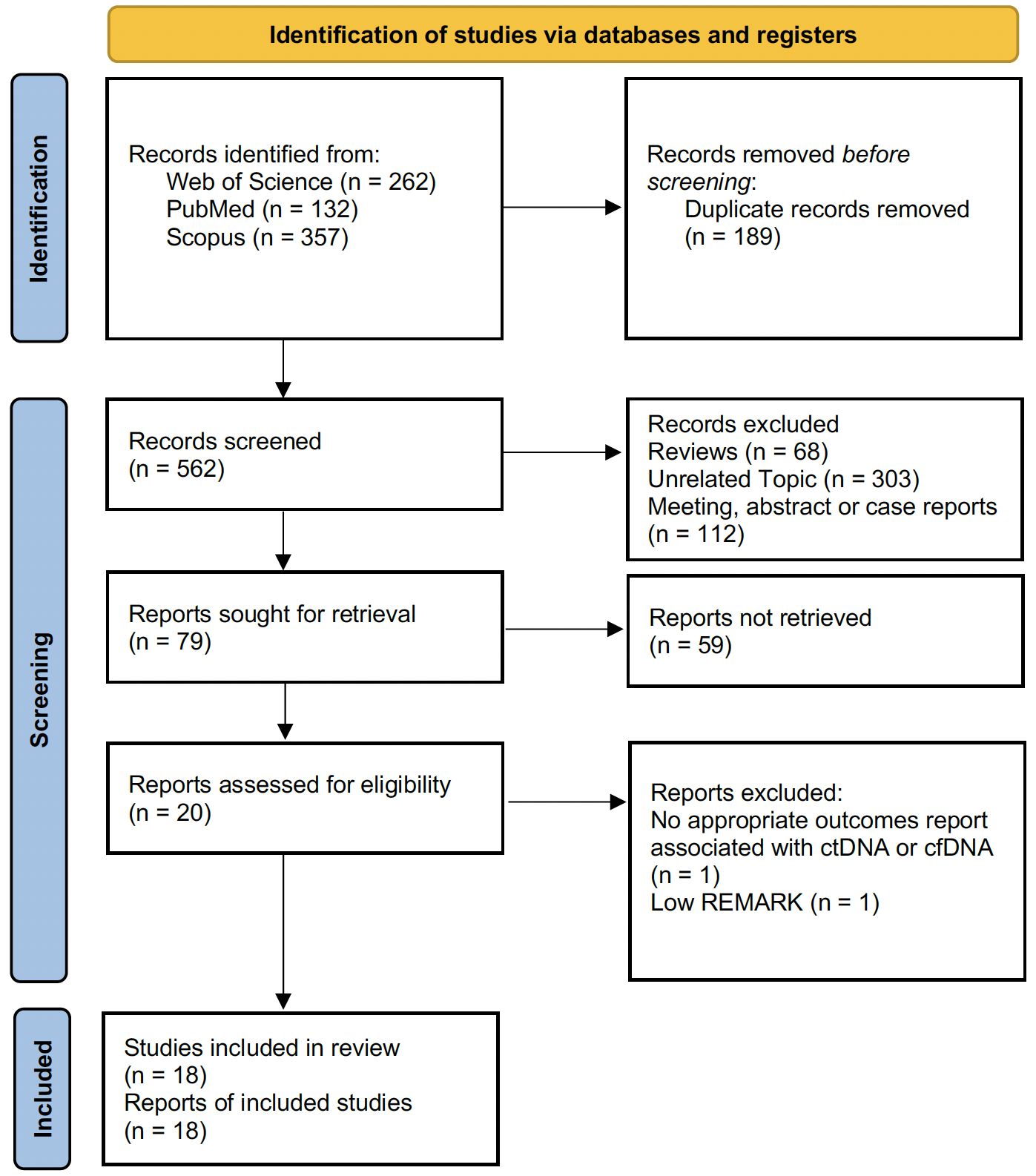
Figure 1 PRISMA flow diagram showing the number of records identified, included, and excluded, as well as the reasons for exclusions.
A total of 2,310 patients from 18 published papers (10–12, 21–35) were included in this meta-analysis. The sample sizes in the relevant studies ranged from 14 to 437. The main characteristics of the included studies are available in Table 1. The prognostic role of ctDNA in OS information was included in seven (12, 22, 25–27, 31, 33) of eighteen studies. The publication years of the studies included in the meta-analysis changed between 2002 and 2022. KRAS mutations were detected in almost all of the studies.
In 18 included studies, the REMARK criteria (16) were used to assess the study quality. Overall, the studies included in the meta-analysis were of high quality (≥5.5). Table 2 also illustrates the methodological quality evaluation of the included studies in detail.
A total of 15 eligible studies (16–20, 22, 24–30, 32, 33) containing data on CA 19.9 and the diagnostic accuracy of ctDNA/cfDNA were included in the primary meta-analysis. As shown in Figure 2, the results of the pooled meta-analysis confirmed that CA19–9 provides greater diagnostic accuracy than KRAS ctDNA at all disease stages (RR = 0.58, 95% CI = 0.42–0.81, p = 0.001). Significant heterogeneity was found between studies (I2 = 95%, p < 0.001). Therefore, the analysis was carried out using the random-effects model. Similarly, a pooled analysis of three primary studies indicated that CA19–9 provided significantly greater diagnostic accuracy than cfDNA at all disease stages (RR = 0.86, 95% CI = 0.74–1.00, p = 0.04). No significant publication bias was detected between studies (Egger’s test: p > 0.05; Begg’s test: p > 0.05) (Supplementary Figure S1).
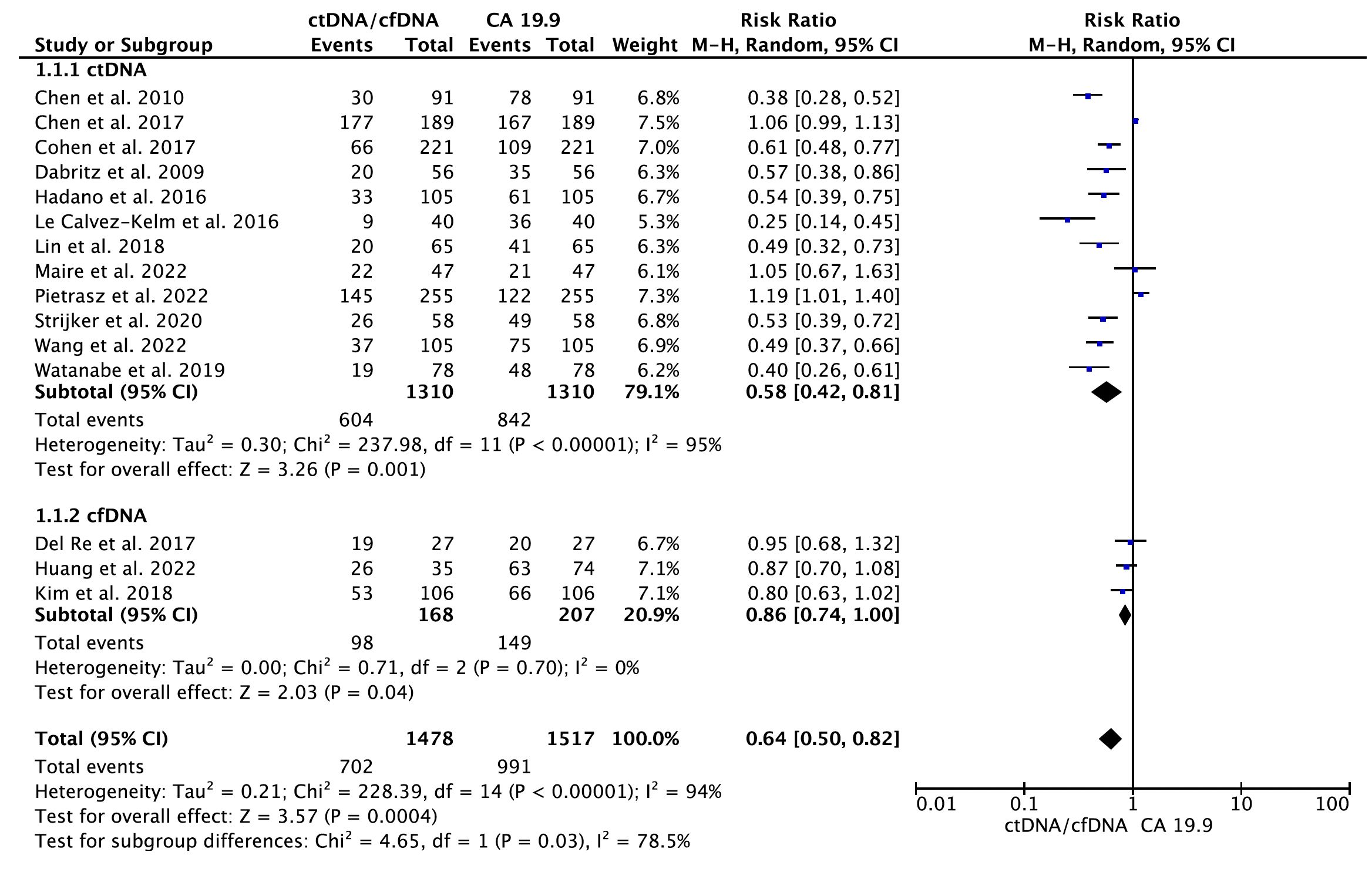
Figure 2 Meta-analysis of the diagnostic accuracy of circulating tumor DNA (ctDNA)/cell-free DNA (cfDNA) compared to CA 19-9 in all disease stages of pancreatic cancer.
In the secondary meta-analysis, data from a total of seven eligible papers (12, 22, 25–27, 31, 33) on the prognostic role of ctDNA in OS were evaluated. In the pooled analysis, ctDNA-positive patients had a worse OS (HR = 2.00, 95% CI = 1.51–2.66, p < 0.001) (Figure 3). Significant heterogeneity was observed among the included studies. Hence, the meta-analysis was carried out using the random-effects model (I2 = 74%, p < 0.001) (Supplementary Figure S2).
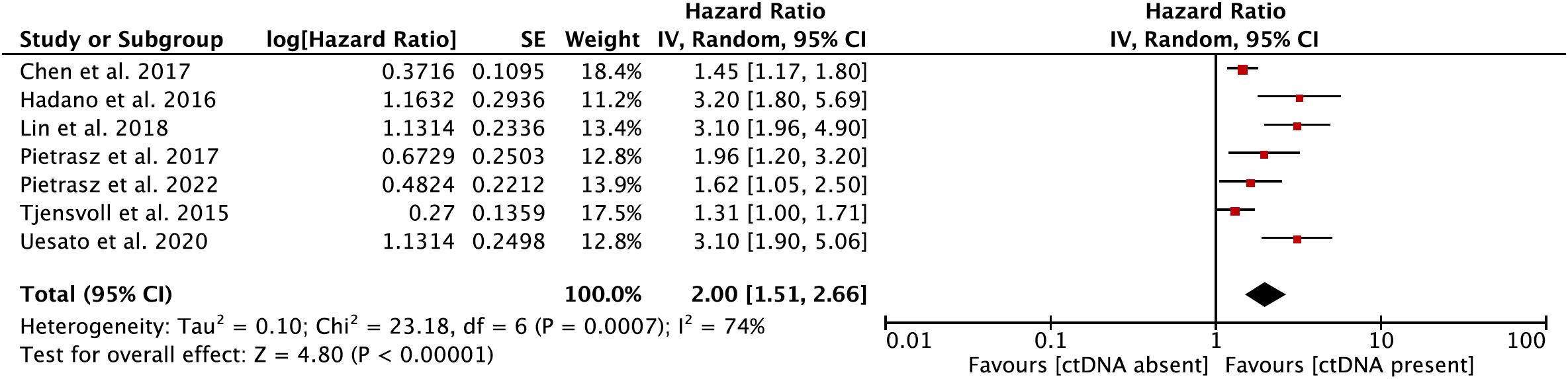
Figure 3 Forest plot showing the effect size of a meta-analysis evaluating the prognostic role of circulating tumor DNA (ctDNA) on overall survival.
To evaluate the robustness of our meta-analysis findings, sensitivity analyses were performed by systematically removing each study from the overall pool and observing the impact on the effect size (ES). The consistent ES across these recalculations underscores the reliability of the overall conclusions, as demonstrated in Supplementary Figures S3 and S4.
This current meta-analysis provides a comprehensive evaluation of ctDNA and cfDNA as potential diagnostic and prognostic biomarkers for pancreatic malignancies. Hence, this study provides a comprehensive perspective on research suggesting that liquid biopsies may be a potential instrument for the diagnosis and prognosis of pancreatic malignancies.
Reports of previous studies evaluating ctDNA and CA 19.9 for diagnostic accuracy have provided conflicting reports on the topic. In a study conducted by Chen et al., it was reported that the diagnostic accuracy of CA 19.9 was greater than that of ctDNA (21). Similarly, other studies conducted with various sample sizes have also reported that CA 19.9 has significantly greater diagnostic accuracy than ctDNA (10, 11, 32). However, several studies have proposed that the diagnostic accuracy of ctDNA is greater than that of CA 19.9 or that there is no significant difference between them (12, 13, 26). The observed discrepancy may be attributed to varying ctDNA detection techniques and patient cohorts. In the present meta-analysis, 12 primary studies provided information on the diagnostic accuracy of ctDNA against CA 19-9 (10–12, 21–27, 31–35). The detailed analysis revealed that CA19–9 offers greater diagnostic accuracy than ctDNA across all disease stages. Although the diagnostic accuracy of ctDNA is lower than that of the CA19–9 protein biomarker, the development of high-sensitivity and multigene NGS techniques is important for diagnosing several cancers, including pancreatic malignancies. It is critical to highlight that CA19–9 analysis has now become complementary to ctDNA analysis. The integration of both biomarkers in diagnostic and prognostic assessments offers a more comprehensive understanding of pancreatic cancer progression. While ctDNA provides real-time insights into tumor dynamics and genetic alterations, CA19–9 serves as an established marker that can enhance diagnostic accuracy when used alongside ctDNA. Taken together, these biomarkers should be considered for monitoring treatment response, detecting early relapses, and developing personalized treatment strategies, especially when surgical resection is not an option. In terms of cfDNA, reports from a total of three studies (28–30) were obtained in our study. The diagnostic accuracy of cfDNA was significantly lower than that of CA19.9 in the pooled analysis. However, the number of studies included in the meta-analysis was relatively small, and there were few statistical analyses. Therefore, it is essential to conduct more studies with larger sample sizes focusing on cfDNA to better understand its potential role in diagnosing pancreatic cancer.
Various studies have investigated the prognostic value of ctDNA in patients with early-stage or advanced-stage pancreatic cancer. In a study focusing on patients with resectable and detectable ctDNA, patients who were ctDNA-positive had a significantly worse prognosis (OS: 13.6 months) than those who were ctDNA-negative (OS: 27.6 months) (22). Similarly, in another study conducted by Chen et al., the K-ras mutation found in plasma DNA served as a predictive biomarker for poor prognosis in patients with unresectable pancreatic cancer (21). These findings suggest that it could serve as a valuable tool for determining the prognosis and developing personalized treatment strategies in patients with pancreatic cancer who cannot undergo surgical removal. In contrast to prior studies, another study indicated that the presence of ctDNA was not significantly associated with survival outcomes in a cohort of pancreatic cancer patients at all disease stages (36). This dissimilarity underlines the variability and complexity in the prognostic value of ctDNA across different patient populations and disease stages. The findings also suggest that the role of ctDNA in prognosis may be influenced by various factors, such as cancer stage, treatment methods, and individual patient characteristics. In this study, patients who were ctDNA positive were found to have poorer OS. Our meta-analysis highlights the prognostic clinical value of ctDNA in pancreatic malignancies, increasing the evidence level of existing prior studies. Therefore, these findings could have significant implications for patient management, influence treatment strategies, potentially guide treatment decisions, and enable more personalized therapeutic approaches for ctDNA-positive patients.
The potential role of ctDNA in screening and diagnosing malignant diseases remains unclear. Notably, the current situation in ctDNA parallels concerns regarding the use of CA19–9, another biomarker, which is a screening test for cancer according to international guidelines (37). There are several challenges to the widespread adoption of ctDNA. These challenges are related to the specificity, sensitivity, and variability of ctDNA levels across different cancers and cancer stages. Additionally, cost-effectiveness, ethical considerations, and the need for a comprehensive understanding of its implications across various types of malignancies contribute to the ongoing discussion. However, CA19–9 has limited application due to similar issues. Therefore, the future role of ctDNA in early cancer detection and screening has yet to be determined, pending further research and consensus in the scientific community.
Our meta-analysis had several limitations that should be discussed and highlighted. In this meta-analysis, a substantial amount of heterogeneity was observed, which can be attributed to large variations in the study populations and the methods used for detecting ctDNA. This variability may affect the consistency and comparability of the results, making it difficult to draw definitive conclusions from the pooled data. We utilized a random effects model in our statistical analyses to accommodate the variations among different patient groups. However, this approach does not eliminate the potential bias introduced by the diverse study populations. In particular, the limited number of studies examining the association between ctDNA and OS limits the generalizability of the results. This limited number of studies also restricted our capacity to conduct subgroup analyses, which could have provided deeper insights into the effects of ctDNA across different patient subpopulations or stages of the disease. Future studies with larger sample sizes and more homogeneous patient groups are needed to verify the findings of this meta-analysis and refine the understanding of the prognostic value of ctDNA in pancreatic cancer patients.
Moreover, the detection rate of ctDNA is affected by several uncertain factors, such as the tumor’s ability to release ctDNA into the bloodstream. This capability varies with the tumor’s size, type, stage, metabolic activity, and surrounding tissue environment. Another crucial factor is the rate at which ctDNA is cleared from the blood. This process is influenced by physiological factors such as degradation by nucleases and removal by organs such as the liver and kidneys. These variables contribute to the observed differences in ctDNA levels among patients and can significantly impact biomarker sensitivity and specificity in cancer detection and monitoring. Hence, the need for further understanding of these biological mechanisms in the context of this topic should be highlighted.
Despite these limitations, pooling data from primary studies has significantly increased the level of evidence. Therefore, this paper provides a strong perspective for future studies.
Taken together, the findings of this meta-analysis indicate that CA19.9 still provides greater diagnostic accuracy across all disease stages than KRAS mutations in ctDNA or cfDNA. However, the presence of detectable levels of ctDNA was associated with worse patient outcomes regarding OS in pancreatic malignancies. Recognizing the prognostic significance of ctDNA could significantly influence treatment decisions, enabling healthcare providers to tailor more personalized and effective therapeutic approaches. As the field of oncology continues to evolve, the role of these biomarkers in improving diagnosis, prognosis, and treatment monitoring may become more important. It is highly recommended that future research continue to investigate the potential role of ctDNA and cfDNA as biomarkers in larger samples and multicenter studies.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
MA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Aİ: Data curation, Investigation, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. YB: Conceptualization, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. NO: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1382369/full#supplementary-material
1. Zhao Z, Liu W. Pancreatic cancer: A review of risk factors, diagnosis, and treatment. Technol Cancer Res Treat. (2020) 19:1533033820962117. doi: 10.1177/1533033820962117
2. Bazeed AY, Day CM, Garg S. Pancreatic cancer: Challenges and opportunities in locoregional therapies. Cancers (Basel). (2022) 14:4257. doi: 10.3390/cancers14174257
3. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. (2020) 395:2008–20. doi: 10.1016/s0140–6736(20)30974–0
4. Ermiah E, Eddfair M, Abdulrahman O, Elfagieh M, Jebriel A, Al-Sharif M, et al. Prognostic value of serum CEA and CA19–9 levels in pancreatic ductal adenocarcinoma. Mol Clin Oncol. (2022) 17:126. doi: 10.3892/mco.2022.2559
5. Jang MA. Next-generation sequencing-based molecular profiling using cell-free DNA: A valuable tool for the diagnostic and prognostic evaluation of patients with gastric cancer. Ann Lab Med. (2024) 44:119–21. doi: 10.3343/alm.2023.0391
6. Malkawi W, Lutfi A, Afghan MK, Shah LM, Costandy L, Ramirez AB, et al. Circulating tumour cell enumeration, biomarker analyses, and kinetics in patients with colorectal cancer and other GI Malignancies. Front Oncol. (2023) 13:1305181. doi: 10.3389/fonc.2023.1305181
7. Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19–9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. (2007) 33:266–70. doi: 10.1016/j.ejso.2006.10.004
8. Goksel T, Özgür S, Vardarlı AT, Koç A, Karakuş HS, Özdemir TR, et al. Prognostic and predictive role of liquid biopsy in lung cancer patients. Front Oncol. (2024) 13:1275525. doi: 10.3389/fonc.2023.1275525
9. Xu C, Jun E, Okugawa Y, Toiyama Y, Borazanci E, Bolton J, et al. A circulating panel of circRNA biomarkers for the noninvasive and early detection of pancreatic ductal adenocarcinoma. Gastroenterology. (2023) 166(1):178–190.e16. doi: 10.1053/j.gastro.2023.09.050
10. Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA. (2017) 114:10202–7. doi: 10.1073/pnas.1704961114
11. Däbritz J, Preston R, Hänfler J, Oettle H. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19–9. Pancreas. (2009) 38:534–41. doi: 10.1097/MPA.0b013e31819f6376
12. Chen I, Raymond VM, Geis JA, Collisson EA, Jensen BV, Hermann KL, et al. Ultrasensitive plasma ctDNA KRAS assay for detection, prognosis, and assessment of therapeutic response in patients with unresectable pancreatic ductal adenocarcinoma. Oncotarget. (2017) 8:97769–86. doi: 10.18632/oncotarget.22080
13. Creemers A, Krausz S, Strijker M, van der Wel MJ, Soer EC, Reinten RJ, et al. Clinical value of ctDNA in upper-GI cancers: A systematic review and meta-analysis. Biochim Biophys Acta Rev Cancer. (2017) 1868:394–403. doi: 10.1016/j.bbcan.2017.08.00
14. Liberati A, Altman DG, Tetzla J, Mulrow C, Gotzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PloS Med. (2009) 6:e1000100. doi: 10.1016/j.jclinepi.2009.06.006
15. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PloS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
16. McShane LM, Altman DG, Sauerbrei W, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. (2005) 93:387–91. doi: 10.1038/sj.bjc.6602678
17. Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
18. The Cochrane Collaboration. Review Manager (RevMan) (2020). Available online at: www.revman.cochrane.org.
19. ProMeta-3 professional statistical software for conducting meta-analysis (2015). Available online at: https://idostatistics.com/prometa3/.
20. Theodor L, Melzer E, Sologov M, Idelman G, Friedman E, Bar-Meir S. Detection of pancreatic carcinoma: diagnostic value of K-ras mutations in circulating DNA from serum. Dig Dis Sci. (1999) 44:2014–9. doi: 10.1023/a:1026618317716
21. Chen H, Tu H, Meng ZQ, Chen Z, Wang P, Liu LM. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol. (2010) 36:657–62. doi: 10.1016/j.ejso.2010.05.014
22. Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. (2016) 115:59–65. doi: 10.1038/bjc.2016.175
23. Maire F, Micard S, Hammel P, Voitot H, Lévy P, Cugnenc PH, et al. Differential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNA. Br J Cancer. (2002) 87:551–4. doi: 10.1038/sj.bjc.6600475
24. Le Calvez-Kelm F, Foll M, Wozniak MB, Delhomme TM, Durand G, Chopard P, et al. KRAS mutations in blood circulating cell-free DNA: a pancreatic cancer case−-control. Oncotarget. (2016) 7:78827–40. doi: 10.18632/oncotarget.12386
25. Pietrasz D, Pécuchet N, Garlan F, Didelot A, Dubreuil O, Doat S, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res. (2017) 23:116–23. doi: 10.1158/1078–0432.CCR-16–0806
26. Pietrasz D, Wang-Renault S, Taieb J, Dahan L, Postel M, Durand-Labrunie J, et al. Prognostic value of circulating tumour DNA in metastatic pancreatic cancer patients: post-hoc analyses of two clinical trials. Br J Cancer. (2022) 126:440–8. doi: 10.1038/s41416–021-01624–2
27. Tjensvoll K, Lapin M, Buhl T, Oltedal S, Steen-Ottosen Berry K, Gilje B, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. (2016) 10:635–43. doi: 10.1016/j.molonc.2015.11.012
28. Del Re M, Vivaldi C, Rofi E, Vasile E, Miccoli M, Caparello C, et al. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci Rep. (2017) 7:7931. doi: 10.1038/s41598-017-08297-z
29. Huang CJ, Huang WY, Chen CY, Chao YJ, Chiang NJ, Shan YS. Cancer-cell-derived cell-free DNA can predict distant metastasis earlier in pancreatic cancer: a prospective cohort study. Ther Adv Med Oncol. (2022) 14:17588359221106558. doi: 10.1177/17588359221106558
30. Kim MK, Woo SM, Park B, Yoon KA, Kim YH, Joo J, et al. Prognostic implications of multiplex detection of KRAS mutations in cell-free DNA from patients with pancreatic ductal adenocarcinoma. Clin Chem. (2018) 64:726–34. doi: 10.1373/clinchem.2017.283721
31. Lin M, Alnaggar M, Liang S, Chen J, Xu K, Dong S, et al. Circulating tumor DNA as a sensitive marker in patients undergoing irreversible electroporation for pancreatic cancer. Cell Physiol Biochem. (2018) 47:1556–64. doi: 10.1159/000490874
32. Strijker M, Soer EC, de Pastena M, Creemers A, Balduzzi A, Beagan JJ, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int J Cancer. (2020) 146:1445–56. doi: 10.1002/ijc.32586
33. Uesato Y, Sasahira N, Ozaka M, Sasaki T, Takatsuki M, Zembutsu H. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PloS One. (2020) 15:e0235623. doi: 10.1371/journal.pone.0235623
34. Wang R, Zhao Y, Wang Y, Zhao Z, Chen Q, Duan Y, et al. Diagnostic and prognostic values of KRAS mutations on EUS-FNA specimens and circulating tumor DNA in patients with pancreatic cancer. Clin Transl Gastroenterol. (2022) 13:e00487. doi: 10.14309/ctg.0000000000000487
35. Watanabe F, Suzuki K, Tamaki S, Abe I, Endo Y, Takayama Y, et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PloS One. (2019) 14:e0227366. doi: 10.1371/journal.pone.0227366
36. Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest. (2015) 33:78–85. doi: 10.3109/07357907.2014.1001894
Keywords: CtDNA, cfDNA, pancreatic malignancy, meta-analysis, clinical value
Citation: Arayici ME, İnal A, Basbinar Y and Olgun N (2024) Evaluation of the diagnostic and prognostic clinical values of circulating tumor DNA and cell-free DNA in pancreatic malignancies: a comprehensive meta-analysis. Front. Oncol. 14:1382369. doi: 10.3389/fonc.2024.1382369
Received: 05 February 2024; Accepted: 10 June 2024;
Published: 25 June 2024.
Edited by:
Terence Moyana, The Ottawa Hospital, CanadaCopyright © 2024 Arayici, İnal, Basbinar and Olgun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehmet Emin Arayici, bWVobWV0LmUuYXJheWljaUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.