- 1Fourth Department of Orthopedic Surgery, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, China
- 2Shenyang 242 Hospital, Shenyang, Liaoning, China
In the contemporary epoch, cancer stands as the predominant cause of premature global mortality, necessitating a focused exploration of molecular markers and advanced therapeutic strategies. N6-methyladenosine (m6A), the most prevalent mRNA modification, undergoes dynamic regulation by enzymes referred to as methyltransferases (writers), demethylases (erasers), and effective proteins (readers). Despite lacking methylation activity, RNA-binding motif protein 15 (RBM15), a member of the m6A writer family, assumes a crucial role in recruiting the methyltransferase complex (MTC) and binding to mRNA. Although the impact of m6A modifications on cancer has garnered widespread attention, RBM15 has been relatively overlooked. This review briefly outlines the structure and operational mechanism, and delineates the unique role of RBM15 in various cancers, shedding light on its molecular basis and providing a groundwork for potential tumor-targeted therapies.
1 Introduction
Cancer is the primary cause of premature mortality worldwide in the 21st century, posing a substantial barrier to improving human life expectancy (1). Calculations and data analysis from the Global Cancer Observatory (GCO) database underscore the substantial burden cancer will impose on both low- and middle-income nations and the global population over the next five decades (2). Therefore, the prioritization of efficient molecular markers to elucidate tumorigenic and progression mechanisms, along with the exploration of advanced early diagnostic and therapeutic approaches, is of utmost importance (3). Recently, m6A modification has emerged as a key player in cancer development, propelling ongoing epigenetic investigations into its association with cancer (4–6).
More than 150 chemically distinct RNA modifications have been recognized (7). m6A, first identified in the 1970s in eukaryotic messenger RNAs (mRNAs) and viral nuclear RNAs, is one of the most widespread and evolutionarily conserved internal cotranscriptional modifications in eukaryotic RNAs (8–11). Operating at the post-transcriptional level, this modification is dynamically regulated by enzymes referred to as writers, erasers, and readers (8, 12–14). The MTC consists of proteins known as “writers”, including methyltransferase-like 3 (METTL3), METTL14, Wilms’ tumor 1-associating protein (WTAP), RBM15, vir-like m6A methyltransferase associated (VIRMA; also known as KIAA1429), METTL16, zinc finger CCCH-type containing protein 13 (Zc3h13), and Hakai (15–21). Particularly noteworthy is the ability of METTL3 and METTL14 to form a stable heterodimer in a 1:1 ratio, underscoring their pivotal roles within the MTC (22). WTAP facilitates the recruitment of the METTL3-METTL14 heterodimer to specific target mRNAs (22–24). Recent studies have extensively reported that MTCs, including RBM15, play a crucial role in the development of various cancers (25–27).
RBM15, also recognized as OTT or OTT1 is a member of the split-end (SPEN) family of proteins involved in cell fate determination (28). Initially identified as an ectopic gene in pediatric acute megakaryocytic leukemia (29, 30), RBM15, despite lacking methylation activity, plays a critical role in recruiting MTCs and facilitating their binding to target mRNAs as a member of the m6A writers (31). Beyond its implications in hematopoiesis and cardio-splenic development in mice (32, 33), RBM15 is implicated in various biological functions, including alternative splicing, nuclear export, and X chromosome inactivation (19, 34, 35). Recent studies have revealed the involvement of RBM15 in cellular biological behaviors such as proliferation, invasion, migration, and apoptosis in various cancers, like acute megakaryocytic leukemia (36), colorectal cancer (37), ovarian cancer (38), laryngeal squamous cell carcinoma (25) and osteosarcoma (39). High expression levels of RBM15 are typically correlated with a poor prognosis in patients with malignancies (40). In this review, we will focus on the individual function of RBM15 and its related mechanisms, highlighting the current status of RBM15 regulatory mechanisms in tumors and providing researchers with new ideas for tumor therapy.
2 Structural features of the RBM15
RBM15, an RNA-binding protein, is situated within the 1p13.2 region of the human chromosome, characterized by 19 exons and 18 introns (41). RBM15 exhibits a typical SPEN family protein-like structure, comprising three highly conserved N-terminal RNA recognition motifs (RRM) and a spen orthologue and paralogue C-terminal (SPOC) domain (42, 43) (Figure 1A). The RRM folding structure includes four antiparallel β-strands and two α-helices (44) (Figure 1B). The RRM domain is highly prevalent in eukaryotes and plays an essential role in post-transcriptional splicing, translation, nuclear export, and mRNA stabilization (45, 46). The SPOC domain, characterized by seven β-strands and four α-helices (Figure 1C), has been demonstrated to influence several facets of mammalian gene expression, encompassing transcription, RNA modification, RNA export, and X-chromosome inactivation (47, 48). Recent discovery revealed that the SPOC domain serves as a phosphoserine binding module, with conserved motifs on its surface specifically recognizing the C-terminal domain (CTD) phosphorylation tag of RNA polymerase II (RNA Pol II) (42). RBM15 SPOC domain was shown to predominantly regulate m6A modification and enhance mRNA stability by binding to the m6A reader (48). Furthermore, the SPOC domain engages with a range of factors, including histone lysine methyltransferase SETD1B, nuclear RNA export factor 1 (NXF1), and DEAD-box protein 5 (DBP5), thereby participating in RNA transcription, export, and RNA-related metabolism (35, 49, 50). In conclusion, the RRM domain and the SPOC domain of RBM15 can mediate a variety of protein interactions and play a key role as a bridge in the regulation of gene expression.
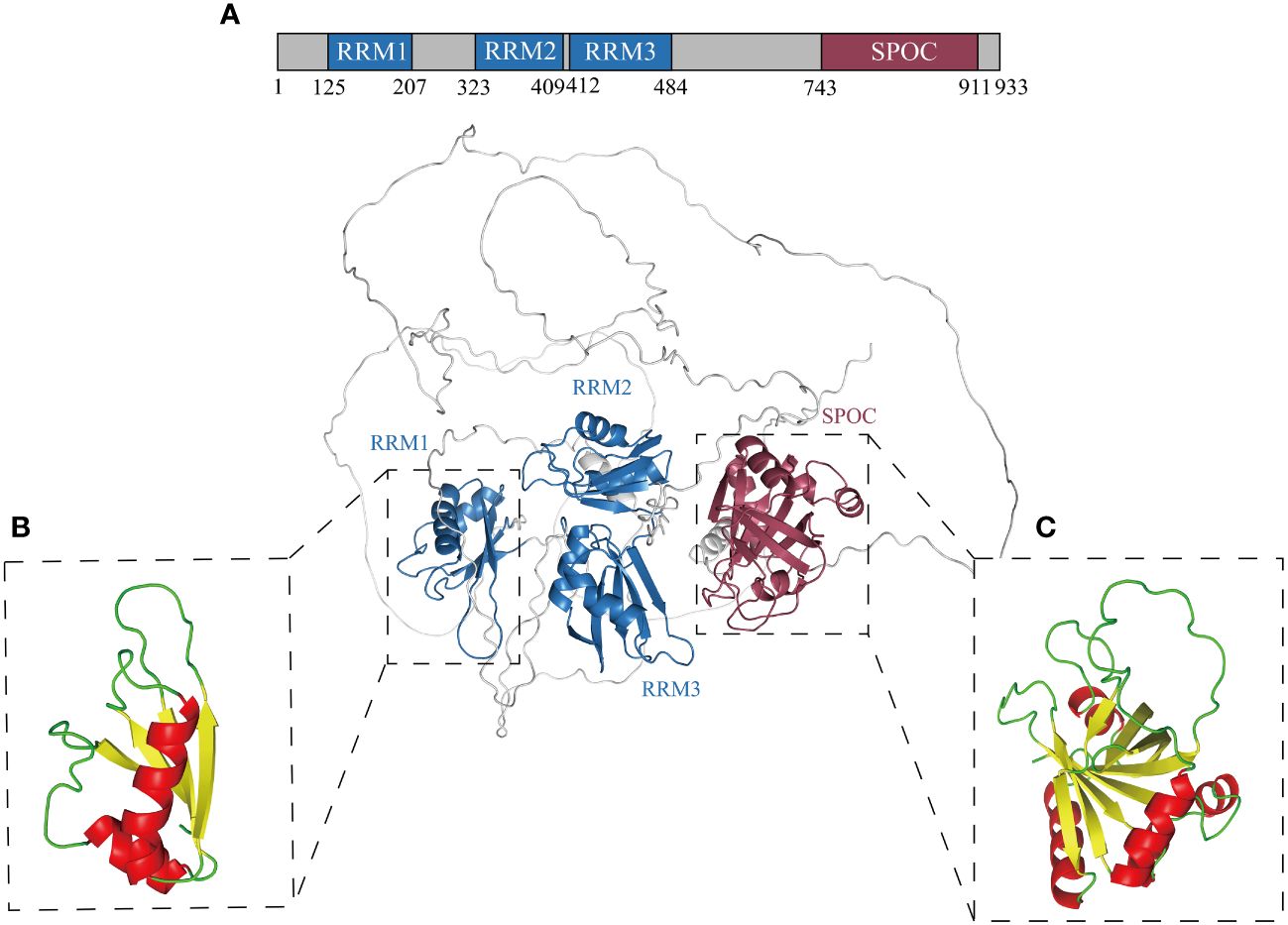
Figure 1 Structural model of RBM15 comprising three RRMs and one SPOC domain. (A) Schematic representation of RBM15 protein domains: RRM1 (aa 125 ~ 207), RRM2 (aa 323 ~ 409), RRM3 (aa 412 ~ 484), Spen orthologue and paralogue C-terminal (SPOC, aa 743 ~ 911). (B) Protein structural model of RBM15 RRM1: composed of 2 α-helices (red) and 4 β-strands (yellow). (C) Protein structure model of RBM15 SPOC: composed of 4 α-helices (red) and 7 β-strands (yellow). All structures were generated and colored using PyMOL version 2.5.4 (www.pymol.org).
3 Mechanisms underlying the function of RBM15
3.1 RBM15 serves as a writer in m6A
In the context previously discussed, RBM15 assumes a pivotal role in the constitution of the MTC, guiding the METTL3/METTL14 complex to specific mRNA target sites (51) (Figure 2). Furthermore, RBM15 exhibits a selective binding affinity to U-rich sequences on mRNAs, directing them to distinct localization sites and thereby promoting m6A adenosine ribonucleotide methylation consensus motifs (19, 51).
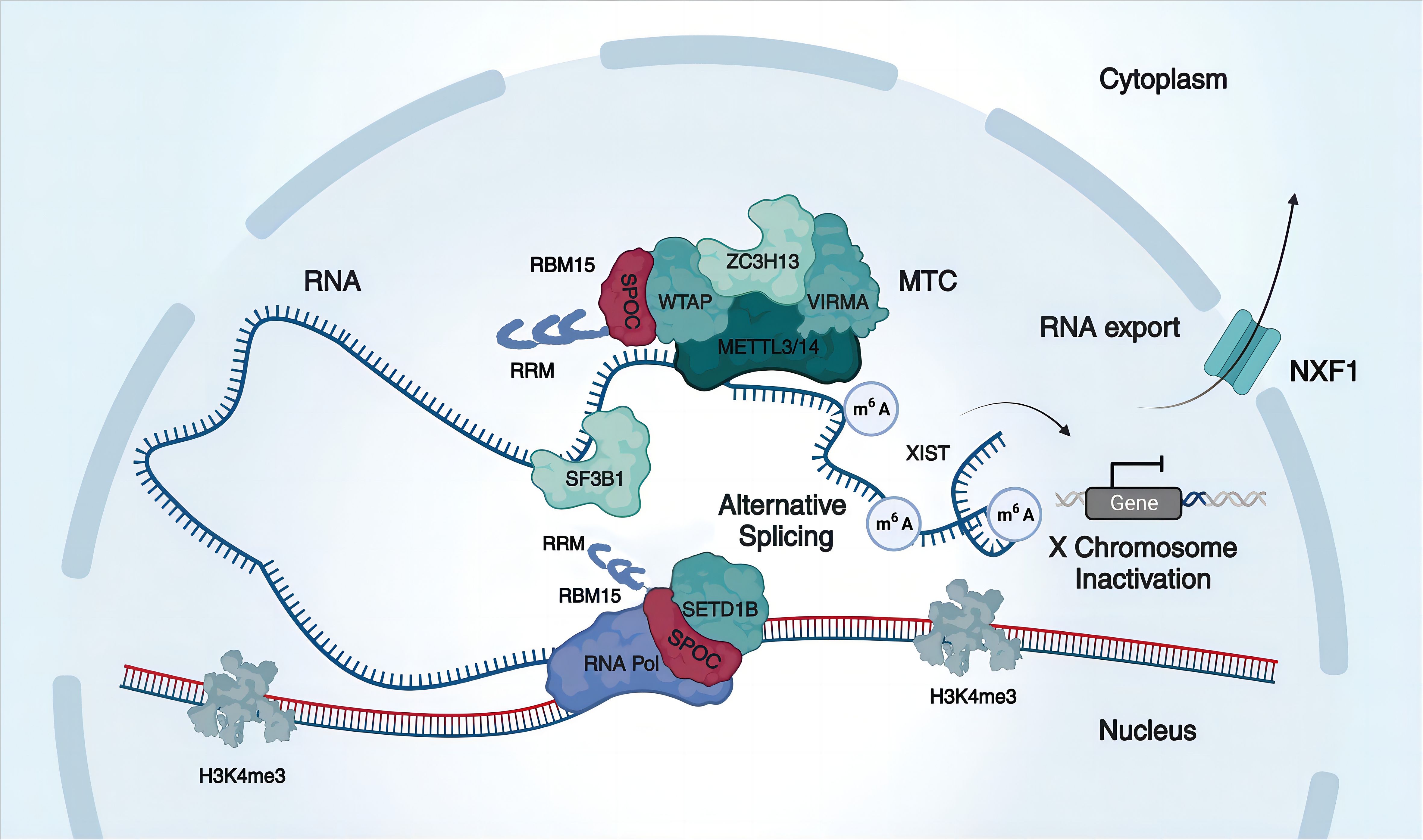
Figure 2 Mechanisms of RBM15 in m6A modification, alternative splicing, and RNA export. RBM15 comprises three RRMs and a SPOC domain. It collaborates with other m6A writers, including METTL3, METTL14, WTAP, VIRMA, and ZC3H13, to form a methyltransferase complex (MTC) that promotes methylation. For example, RBM15 promotes X-chromosome inactivation by enhancing XIST methylation levels. Additionally, RBM15 recruits the splicing factor SF3B1 to participate in alternative splicing, and RBM15 can target the histone H3K4me3 methyltransferase SETD1B to RNA via the SPOC domain, thereby regulating selective splicing via histone modifying enzymes. Furthermore, RBM15 promotes the nuclear export of mRNA by binding to the nuclear export factor NXF1.
Moreover, WTAP emerges as a robust candidate for interaction with RBM15, serving as an essential link in MTC recruitment by RBM15 (52). The interaction dynamics between the METTL3-METTL14 complex and RBM15 are intricately dependent on WTAP levels (19). It is noteworthy that the depletion of WTAP significantly diminishes or interrupts the binding interaction between RBM15 and the METTL3-METTL14 complex (19, 53–55). Subsequent experiments have elucidated that WTAP harbors a phosphorylated LSETD motif, emphasizing the potential dependence of the critical link between the RBM15 SPOC domain and WTAP on this phosphorylated motif (42).
RBM15, acting as a crucial mediator of m6A modifications, plays a multifaceted role in various biological processes. It collaborates with its analog RBM15b to recruit WTAP and METTL3-METTL14 to the long non-coding RNA X-inactive specific transcript (XIST) m6A regions, elevating XIST methylation levels and facilitating XIST-mediated gene silencing (19, 47, 56–58) (Figure 2). Interestingly, in cases of reduced RBM15 expression, RBM15b demonstrates functional compensation due to substantial sequence and structural domain similarity (19). Noteworthy XIST methylation reduction is observed only when both RBM15 and RBM15b are concurrently knocked down (19). Moreover, RBM15 orchestrates the m6A-mediated regulation of BAF155, contributing to the normal development of the mammalian cerebral cortex (54). RBM15 also regulates the expression of CLDN4 in mice, exerting influence on insulin sensitivity and promoting insulin resistance in gestational diabetic mice (59).
The intricate interplay between RBM15 and m6A readers plays a crucial role in orchestrating the intricate m6A regulatory mechanism. For instance, RBM15 can interact with IGF2BP1 to facilitate the post-transcriptional activation of YES proto-oncogene 1 (YES1), thus contributing to the regulation of hepatocellular carcinoma progression (60). Additionally, RBM15 may interact with Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) and collaborate in the m6A modification of TMBIM6, consequently promoting the malignant progression of laryngeal squamous cell carcinoma (25). In conclusion, RBM15 is involved in the composition of MTCs in a WTAP-dependent manner and recruits MTCs to specific sites to promote m6A methylation, which plays a key regulatory role in a variety of biological functions.
3.2 RBM15 controls alternative splicing
Alternative splicing, facilitated by spliceosome complexes binding to RNA Pol II transcripts, constitutes a pivotal factor contributing to the intricate complexity of the transcriptome in multicellular eukaryotes (61, 62). Within nuclear speckles, recognized as crucial depots for numerous splicing factors, both RBM15 and RBM15b are situated (52). RBM15 exhibits its influence by binding to specific intronic sites in pre-mRNA and modulating alternative splicing through the recruitment of splicing factors (34) (Figure 2). A notable instance involves RBM15 orchestrating alternative splicing by enlisting the splicing factor SF3B1 to a distinct splice site in the c-Mpl intron, leading to the upregulation of the c-Mpl truncation isoform upon RBM15 knockdown (34). Additionally, RBM15 may govern alternative splicing via chromatin modifications. Its interactions with Hdac3 and the histone methyltransferase SETD1B exemplify this (63), influencing c-Mpl RNA and chromatin interactions, thereby regulating H4 acetylation and H3K4me3 marks (64) (Figure 2). Noteworthy is the observation that inhibiting histone deacetylase or histone methyltransferase levels significantly heightens the abundance of truncated isoforms of c-Mpl (64, 65). Upstream in the regulatory cascade, protein arginine methyltransferase 1 (PRMT1) methylates RBM15, instigating its ubiquitination and subsequent degradation by subunit 4 of the CCR4-NOT transcriptional complex (CNOT4) (34). This intricate process potentially serves as a pathogenetic mechanism in hematopoietic malignancies.
Alternative splicing orchestrated by RBM15 is essential for megakaryocyte differentiation. For instance, the depletion of RBM15 disrupts the selective splicing of GATA1, leading to the generation of truncated GATA1 isoforms (34). These truncated isoforms hinder the differentiation of progenitors into mature megakaryocytes, a crucial process in the pathogenesis of leukemia (34). Additionally, the transcription factor TAL1 holds a critical role in the differentiation of megakaryocyte-erythroid progenitors. The strong interaction between the SF3B1K700E mutant and RBM15 can result in the dysregulation of alternative RNA splicing of TAL1, ultimately impeding erythropoiesis (66). In summary, RBM15 regulates alternative splicing by recruiting splicing factors and interacting with histone-modifying enzymes, thereby governing the splicing process and participating in diverse functions (34, 64).
3.3 RBM15 promotes the nuclear export of mRNA
RBM15, a member of the SPEN family, possesses a distinctive SPOC domain responsible for governing nuclear export through interactions with diverse proteins (28) (Figure 2). For example, EB2 interacts with RBM15/b and the SPEN SPOC domain, thus facilitating the nuclear export of viral mRNA (28). The RNA transport element (RTE) enhances RNA binding to the mRNA export receptor NXF1, with this process mediated by the interaction with RBM15 (35). Acting as a bridge, RBM15 connects RTE-containing RNA to NXF1, consequently amplifying the nuclear export of RNA (35). Moreover, the DBP5 plays a crucial role in providing the basic direction of nuclear export by specifically recognizing NXF1 through RBM15, allowing NXF1 to traverse the nuclear pore complex and enter the cytoplasm (35, 49). In summary, RBM15 actively engages in and facilitates cellular nuclear export. Nevertheless, when RBM15/b is subjected to knockdown, the nuclear export of mRNA appears to persist, raising questions about whether RBM15 solely aids export factors in enhancing the stability of their interactions (35, 49).
4 Role of RBM15 in cancer
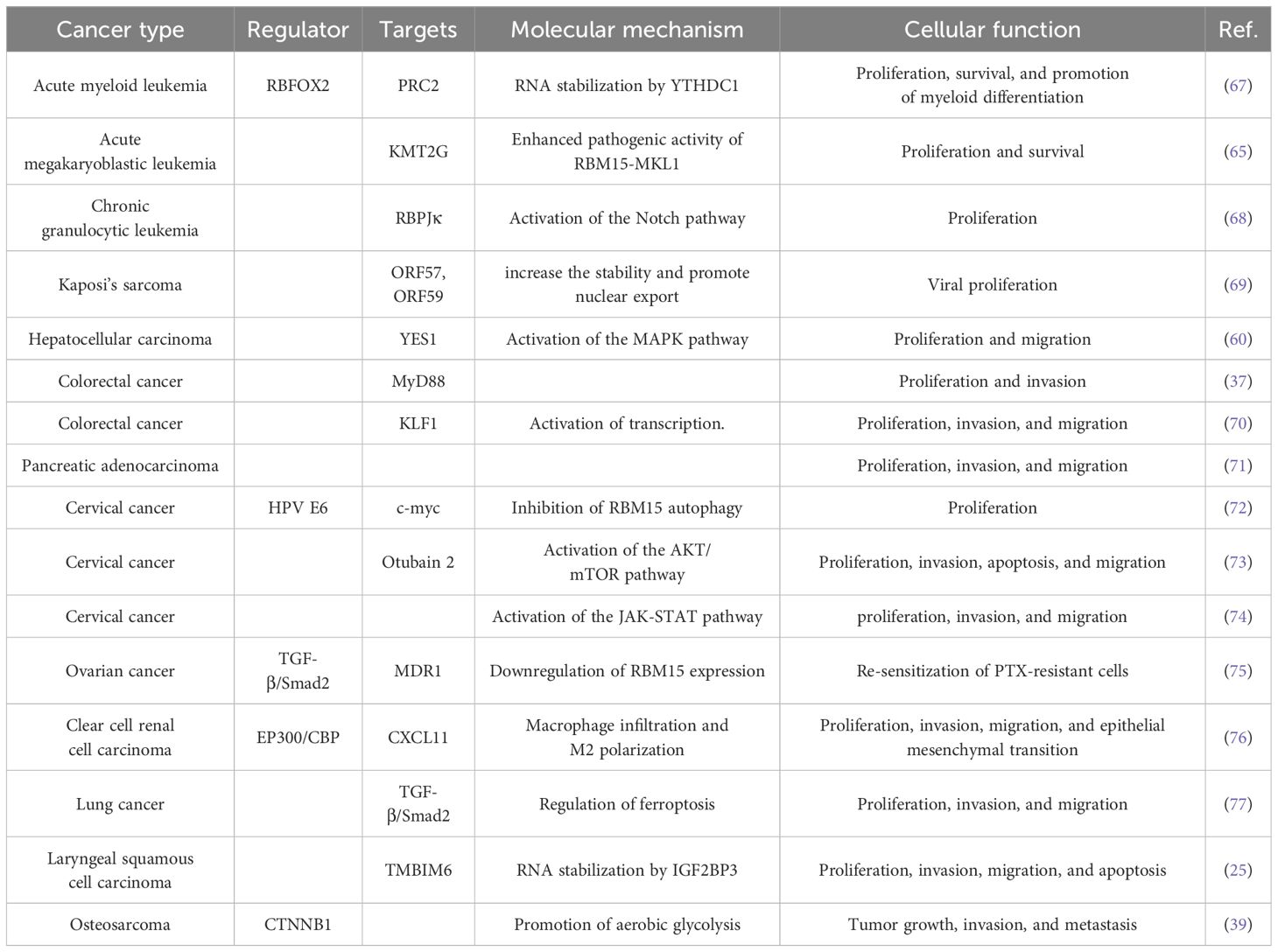
Recent evidence underscores the pivotal role of RBM15 in cancer, where it predominantly functions as a methyltransferase, enhancing the stability of target mRNAs through m6A modification, thereby contributing to the initiation and progression of diverse cancers. In addition, RBM15 also regulates cancer through signaling pathways or other modifications. In this review, we present a succinct summary of recent discoveries elucidating the expression of RBM15 in different tumors and its corresponding molecular regulatory mechanisms (Table 1 and Figure 3).
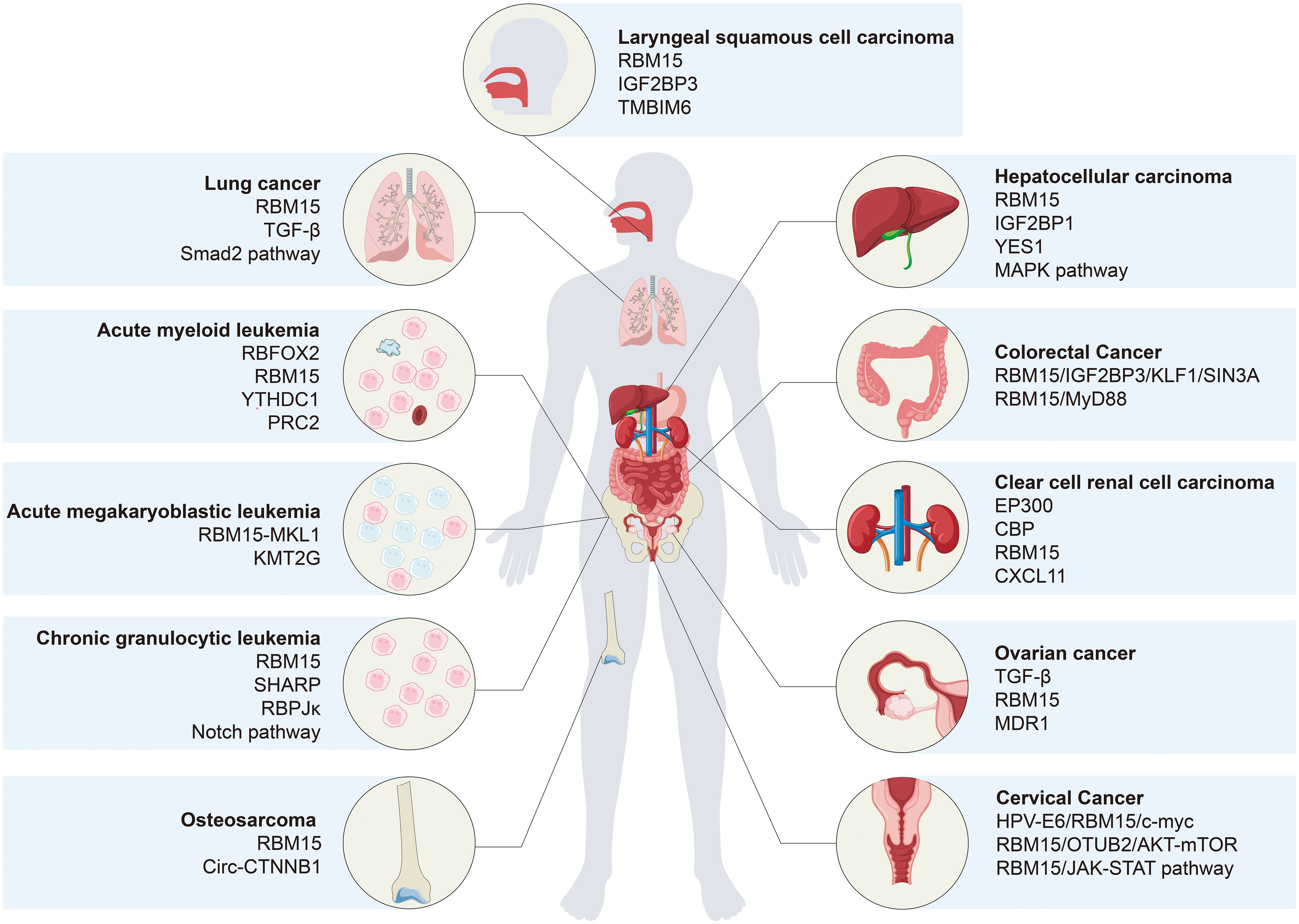
Figure 3 Molecular regulatory mechanisms of RBM15 in various types of cancer. In addition to conventional targeted regulation, RBM15 collaborates with m6A readers, such as IGF2BP1, IGF2BP3, and YTHDC1, to govern the malignant progression of cancer. Furthermore, RBM15 directs cancer progression by modulating various pathways, including TGF-β/Smad2, Notch, MAPK, and AKT/mTOR pathways.
4.1 Acute myeloid leukemia
Acute myeloid leukemia (AML) stands out as the preeminent malignancy affecting hematopoietic stem cells, marked by a notably unfavorable prognosis (78). Recent research has highlighted a substantial correlation between RBM15 expression levels and survival in AML patients, associating elevated RBM15 expression with shorter survival (67). Notably, a critical aspect of RBM15 function was uncovered in that RBM15 was recruited by RBFOX2 and established an interaction with the m6A reader YTH domain-containing proteins 1 (YTHDC1) (67). This interaction facilitated the recruitment of polycomb repressive complex 2 (PRC2) to the binding site of RBFOX2, leading to chromatin silencing and transcriptional repression (67). Of note, the expression levels of RBM15 and RBFOX2 were positively correlated in cancer patients. Furthermore, down-regulation of RBFOX2 significantly impedes the survival and proliferation of AML cells and induces myeloid differentiation (67).
4.2 Acute megakaryoblastic leukemia
Acute megakaryocytic leukemia (AMKL) is a subtype of acute myeloid leukemia primarily characterized by the presence of platelet-producing megakaryocytes within the bone marrow (79). This disease is prevalent in children and is associated with an unfavorable prognosis (79–81). The gene fusion product involving RBM15 and megakaryocytic leukemia 1 (MKL1), termed the RBM15-MKL1 fusion protein (also referred to as OTT-MAL), was initially identified in a pediatric patient with acute megakaryoblastic leukemia harboring the t(1;22)(p13;q13) translocation (82). The etiology of this malady is notably intricate, and investigations have revealed that the RBM15-MKL1 fusion protein interacts with Setd1b histone H3-Lys4 methyltransferase (also recognized as KMT2G). This interaction is contingent on the intact RBM15 SPOC domain and enhances its leukemic activity in megakaryocytes (65).
Furthermore, RBM15 plays multiple roles in the hematopoietic system and may also contribute to disease pathogenesis (83, 84). As an illustration, RBM15 interacts with SMRT/HDAC1-related inhibitory protein (SHARP) and is associated with the recombinant signal-binding protein RBP-Jκ. This interaction activates Notch-regulated gene expression, thereby inhibiting myeloid differentiation in hematopoietic cells (83). Additionally, RBM15 plays a role in hematopoietic stem cells (HSC) and contributes to megakaryocyte development by regulating the downstream target c-myc (85, 86). Furthermore, It has been shown that antisense RBM15 (AS-RBM15) finely modulates megakaryocyte differentiation by elevating the translation level of the RBM15 protein (87). Up-regulation of AS-RBM15 expression promotes terminal differentiation of megakaryocytes, while down-regulation has the opposite effect. Nevertheless, the connection of these findings to the pathogenesis of AMKL requires further investigation.
4.3 Chronic granulocytic leukemia
Chronic granulocytic leukemia (CML) is one of the most malignant diseases in the hematopoietic system, presenting a grave threat to patients (88). Related researchers revealed that the average expression level of RBM15 was notably higher in acute-phase CML cells when compared to those in the chronic and accelerated phases of the disease (68). Diminishing RBM15 levels demonstrated the capacity to impede the growth and proliferation of CML cells, impede the cell cycle, and induce apoptosis (68). Furthermore, evidence suggested that RBM15 might, in part or entirely, facilitate the malignant progression of CML through the Notch signaling pathway mediated by RBPJκ. This pathway is postulated to wield a crucial influence in the etiology of hematopoietic malignancies (68, 83).
4.4 Kaposi’s sarcoma
Kaposi’s sarcoma (KS) is a multicentric tumor arising from the endothelial cells of lymphatic vessels (89). The virulence genes ORF57 and ORF59 are vital contributors to the growth and proliferation of KS (90), and their expression levels are closely associated with RBM15 (69). Research has revealed that RBM15 plays a key role in enhancing the production of ORF57 nuclear transcripts. Inhibition of RBM15 hampers the production of ORF57 mRNA, resulting in a decrease in the overall RNA level of ORF57 (91). In addition, RBM15 and ORF57 interacted with the 5’ MRE of ORF59 to increase the stability of ORF59 mRNA and promote its nuclear export (90), preventing the overaccumulation of ORF59 mRNA in the nucleus and maintaining the balance between the nuclear and cytoplasmic levels of ORF59 mRNA (69). However, further experiments are needed to confirm that RBM15 regulates the expression of ORF57 and ORF59, thereby promoting the growth and proliferation of KS.
4.5 Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the predominant primary liver malignancy, distinguished by elevated malignancy, morbidity, and mortality rates (92). RBM15 exhibits high expression levels in HCC, indicative of an unfavorable prognosis (60, 93). The depletion of RBM15 significantly impedes the growth of HCC cells (60). Notably, it uncovered a crucial aspect of RBM15 function, demonstrating its pivotal role in the post-transcriptional activation of the YES1 through interaction with the m6A reader IGF2BP1 (60). This intricate interplay subsequently activates the mitogen-activated protein kinase (MAPK) pathway, thereby fostering the progression of HCC (60).
4.6 Colorectal cancer
Colorectal cancer (CRC) is the third most common malignancy and the second most lethal cancer in the world, and its incidence is increasing at an alarming rate (94). Research indicates that the expression of RBM15 in CRC tissues is significantly higher than that in nearby non-tumor tissues, and elevated expression of RBM15 is closely associated with poor prognosis, while inhibition of RBM15 expression significantly suppresses the proliferation and invasion of CRC cells (70). Recently, more and more studies have focused on the mechanistic investigation of RBM15 in CRC. For example, RBM15 increased the methylation level of MyD88 through m6A modification, which promoted the proliferation and invasion of CRC cells (37). In addition, RBM15 regulates the expression and enhances the stability of KLF1 mRNA by interacting with the m6A reader IGF2BP3, which activates the transcription of the downstream target SIN3A and ultimately promotes the proliferation, invasion, and migration of CRC cells (70).
4.7 Pancreatic cancer
Pancreatic adenocarcinoma (PAAD) is a prominent and exceptionally malignant neoplasm of the digestive system, characterized by a 5-year survival rate of approximately 10% following diagnosis (95). RBM15 exhibited high expression across various pancreatic cancer cell lines, frequently coexisting with a propensity for T lymphocyte aggregation (41). Studies have shown that elevated RBM15 expression emerged as a significant contributor to unfavorable prognosis, and conversely, the suppression of the RBM15 demonstrates the potential to inhibit cancer cell proliferation, invasion, and metastasis to varying extents (41, 71). Intriguingly, elevated blood glucose levels may enhance RBM15 expression in pancreatic cancer. It is yet to be determined whether this phenomenon is associated with the increased energy demands of pancreatic cancer malignancy (41).
4.8 Cervical cancer
Cervical cancer is the fourth most prevalent gynecological cancer, and more than 99% of cases are attributed to human papillomavirus (HPV) (96). HPV-E6, one of the eight protein-coding genes associated with cervical carcinogenesis (97), has been shown to maintain high expression of RBM15 in cervical cancer cells by preventing its autophagic degradation (72). Recent studies have focused on the specific regulatory mechanisms of RBM15 in cervical cancer. For example, RBM15 interacts with the downstream target c-myc, enhancing its m6A modification, a process crucial in cervical cancer development (72). Intriguingly, the knockdown of HPV-E6 resulted in a reduction of c-myc mRNA expression and m6A modification levels in cervical cancer cells, which were subsequently reversed by the overexpression of RBM15 (72). In addition, RBM15-mediated m6A modification facilitated the expression of the oncogene Otubain 2 (OTUB2) in cervical cancer cells, which further activated AKT/mTOR signaling, thereby promoting the proliferation, migration, and invasion of cervical cancer cells (73). Furthermore, it has been hypothesized that RBM15 might play a role in promoting the proliferation, invasion, and migration of cervical cancer cells through its interaction with the JAK-STAT pathway. Nevertheless, additional validation is required (74).
4.9 Ovarian cancer
Ovarian cancer (OC) is the third most common gynecologic malignancy worldwide but accounts for the highest mortality rate among these cancers (98). Studies have shown that the expression of RBM15 is higher in OC tissues than in normal tissues, and that high expression of RBM15 is closely associated with the propensity to metastasize in OC (38, 75). Interestingly, RBM15 is overexpressed in paclitaxel (PTX)-resistant cells and depletion of RBM15 also restores the sensitivity of PTX-resistant cells (75). Activation of the TGF-β/Smad pathway was shown to interact with the RBM15 promoter and directly inhibit RBM15 expression in PTX-resistant ovarian cancer cells (75). Subsequent low expression of RBM15 resulted in a reduction in the m6A level of MDR1, a recognized major target for overcoming OC resistance (75, 99). In summary, RBM15, serving as a tissue biomarker for OC and PTX resistance, may open new therapeutic avenues for the treatment of PTX-resistant OC in the future.
Most evidence has unveiled the oncogenic role of RBM15 in OC, however, a different voice also indicated a contradictory view: ubiquitin-like modification activating enzyme 6 antisense RNA 1 (UBA6-AS1) can m6A-regulate UBA6 mRNA by enlisting the aid of RBM15, followed by the m6A reader IGF2BP1, which bolsters the stability of UBA6 mRNA (100). UBA6-AS1 curbed UBA6 self-degradation through m6A modification mediated by RBM15, thereby effectively impeding ovarian cancer proliferation, invasion, and metastasis (100). This suggests to us that RBM15 may be associated with a favorable prognosis in ovarian cancer. However, further experiments are warranted to validate this association. While controversy surrounds RBM15’s role in ovarian cancer, these findings suggest the potential for a future role wherein RBM15 acts as a tumor suppressor or a tumor suppressor cofactor.
4.10 Clear cell renal cell carcinoma
Clear cell renal cell carcinoma (ccRCC) is a prevalent adenocarcinoma originating from renal tubular epithelial cells, frequently associated with an unfavorable prognosis (68). Most investigations have revealed a heightened expression of RBM15 in both ccRCC cells and tissues, correlating with augmented proliferation, invasion, and metastasis of ccRCC cells (76, 101). Creb-binding protein (CBP) and EP300 are crucial transcriptional co-regulators implicated in cancer progression (102). It has been shown that they can facilitate the enrichment of the RBM15 promoter, inducing histone 3 acetylation modification and the subsequent upregulation of RBM15 expression. Furthermore, RBM15 plays a role in enhancing the stability of CXCL11 mRNA through m6A modification, consequently promoting macrophage recruitment and M2 polarization (76). Noteworthy is the observation that the knockdown of RBM15 led to a significant reduction in the m6A level of CXCL11 mRNA, thereby restraining the malignant behavior of ccRCC cells (76).
4.11 Lung cancer
Lung cancer stands out as the most prevalent global malignancy and the leading cause of cancer-related mortality among men (103), Among its subtypes, lung adenocarcinoma (LUAD) constitutes approximately half of the total incidence (104). LUAD cells exhibit significantly heightened levels of RBM15 expression, and this upregulation strongly correlates with diminished overall survival rates (105, 106). Notably, RBM15’s potential to promote the malignant behavior of lung cancer is linked to its antagonistic relationship with SETD2, a recognized favorable prognostic indicator for LUAD (107). A recent study has further proposed that the knockdown of RBM15 effectively reduces the levels of TGF-β and Smad2, and promotes ferroptosis by regulating genes related to the iron concentration process, thus inhibiting proliferation, migration, invasion, and tumor growth (77). The proposition of targeting RBM15 opens new avenues for future directions in lung cancer treatment.
4.12 Laryngeal squamous cell carcinoma
Laryngeal squamous cell carcinoma (LSCC), a highly malignant tumor of the respiratory tract, holds the unenviable position of being the second most common head and neck cancer with a particularly dismal prognosis (108). LSCC tissues conspicuously exhibit elevated levels of RBM15 expression, significantly associated with a poor prognosis (20). Interestingly, inhibition of RBM15 by knockdown significantly impedes the invasion and migration capabilities of LSCC cells (25). Recently, a remarkable discovery demonstrated that RBM15 plays a critical role in promoting the methylation process of TMBIM6, consequently facilitating the malignant progression of LSCC (25). In addition, the m6A reader IGF2BP3 recognizes the m6A tag and exerts its influence by fortifying the stability of TMBIM6 mRNA. Notably, when the expression of RBM15 and IGF2BP3 was knocked down, a significant decrease in the expression level of TMBIM6 mRNA was observed (25).
4.13 Osteosarcoma
Osteosarcoma is an exceedingly uncommon primary malignancy of the skeletal system, primarily afflicting adolescents between the ages of 10 and 25 years (109). In osteosarcoma cells, RBM15 exhibits elevated expression levels, a phenomenon notably linked to an unfavorable prognosis (110). Mechanistic studies on how RBM15 regulates osteosarcoma are scarce, but a recent study suggested that RBM15 directly interacts with Circ-CTNNB1, thereby increasing the level of m6A modification of genes associated with aerobic glycolysis, and ultimately facilitating the glycolytic process (39). This augmentation in aerobic glycolysis unequivocally provides a survival advantage to osteosarcoma cells (39). Yet, the precise regulatory mechanism underpinning this phenomenon remains to be comprehensively elucidated.
5 Conclusions and perspectives
Cancer is characterized by numerous hallmark behaviors such as uncontrolled proliferation, evasion of cell death, angiogenesis, invasion, metastasis, metabolic dysregulation, and immune evasion (111). The most prevalent RNA modification in eukaryotes is m6A, which can determine the fate of the modified RNA (112). Methyltransferases have garnered substantial research interest due to their ability to catalyze RNA modifications, their involvement in tumor initiation and progression, and their potential as therapeutic targets in cancer (12). For instance, METTL3 may govern colorectal cancer metastasis by modulating the METTL3/miR-1246/SPRED2 axis (113), Additionally, METTL14 regulates USP48, enhancing SIRT6 stability via m6A modification, thereby restraining the malignancy of HCC (114).
RBM15, belonging to the SPEN family and distinguished by its specific SPOC domain, assumes a key role in recruiting MTC to specific sites, thereby facilitating m6A methylation (42, 51). Additionally, RBM15 extends its impact beyond alternative mRNA splicing and nuclear transport, encompassing a spectrum of biological functions mediated by m6A methylation. These functions encompass Xist-mediated chromosome inactivation and the mediation of the degradation of the chromatin remodeling factor BAF155 (19, 34, 35, 54). In addition, RBM15 is an oncogene in most cancers, and down-regulation of RBM15 can effectively inhibit cancer progression (37, 39, 81). Based on emerging evidence of their role in cancer and molecular mechanisms, m6A regulators have attracted increasing attention from researchers as therapeutic targets.
Certainly, RBM15 also assumes a significant role in non-neoplastic diseases. For instance, RBM15 can accelerate the progression of diabetic nephropathy by modulating cell proliferation, inflammation, and oxidative stress through activation of the AGE-RAGE pathway (82). In addition, RBM15 triggers abnormal immune responses and lymphopenia, thereby exacerbating inflammatory reactions in COVID-19 through the regulation of multiple downstream target genes (83). Consequently, RBM15 holds promise as a prospective target for the treatment of malignancies and a wide range of diseases. Manipulation of RBM15 levels, whether through direct or indirect means, is expected to improve patient prognosis in the future.
In this article, we provide a comprehensive review of RBM15 expression in various cancer types, exploring its impact on prognosis and the underlying molecular mechanisms. However, RBM15-targeted therapies are still in their infancy and there are still significant gaps in the understanding of its upstream regulation and downstream targeting. It remains unclear whether it occurs through the m6A pathway in various tumor models, which is crucial for clinical translational applications as well as the development of disease therapies. It should be emphasized that although RBM15 has a specific SPOC domain, the relevance of its structure to disease and the specific upstream and downstream regulatory mechanisms are still rarely mentioned. In the future, we can pay more attention to how the SPOC domains are removed or lose their functions, understand how the SPOC domain regulates the synthesis and fate of mRNAs at the molecular level, and potentially discover potential targets for new therapeutic approaches. In addition, RBM15b is an analog of RBM15, which together with RBM15 plays a key recruiting role in the process of m6A methylation. However, the synergistic effects of RBM15b on RBM15 in disease regulation, as well as their potential to jointly induce cancer and promote tumor cell growth, have rarely been analyzed in detail. In the future, we may need additional transgenic mouse models to test the specific roles of these two analogs and their synergistic effects in vivo.
In conclusion, RBM15 is involved in a wide range of biological processes and its importance in cancer regulation is increasing. Therefore, it is imperative to elucidate the complex roles of RBM15 in cancer and to exploit its potential in targeted tumor therapy to bridge the gap between research findings and clinical translation, ultimately improving the prognosis of cancer patients.
Author contributions
YC: Conceptualization, Investigation, Software, Visualization, Writing – original draft, Writing – review & editing. GQ: Supervision, Validation, Writing – review & editing. YD: Supervision, Validation, Writing – review & editing. WZ: Supervision, Validation, Writing – review & editing. YW: Conceptualization, Investigation, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81972522), and SMC Students’ scientific research projects (grant no. Y20210515 and Y20211009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Soerjomataram I, Bray F. Planning for tomorrow: global cancer incidence and the role of prevention 2020–2070. Nat Rev Clin Oncol. (2021) 18:663–72. doi: 10.1038/s41571-021-00514-z
3. de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. (2023) 41:374–403. doi: 10.1016/j.ccell.2023.02.016
4. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. (2019) 12:121. doi: 10.1186/s13045-019-0805-7
5. Lin H, Wang Y, Wang P, Long F, Wang T. Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: impacts on therapeutic resistance. Mol Cancer. (2022) 21:148. doi: 10.1186/s12943-022-01620-x
6. Chen Y, Miao L, Lin H, Zhuo Z, He J. The role of m6A modification in pediatric cancer. Biochim Biophys Acta Rev Cancer. (2022) 1877:188691. doi: 10.1016/j.bbcan.2022.188691
7. Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. (2022) 50:D231–d235. doi: 10.1093/nar/gkab1083
8. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. (2019) 18:176. doi: 10.1186/s12943-019-1109-9
9. Adams JM, Cory S. Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature. (1975) 255:28–33. doi: 10.1038/255028a0
10. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U.S.A. (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
11. Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. (1977) 113:165–79. doi: 10.1016/0022-2836(77)90047-X
12. An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. (2022) 21:14. doi: 10.1186/s12943-022-01500-4
13. Bi Z, Liu Y, Zhao Y, Yao Y, Wu R, Liu Q. A dynamic reversible RNA N(6) -methyladenosine modification: current status and perspectives. J Cell Physiol. (2019) 234:7948–56. doi: 10.1002/jcp.28014
14. You Y, Fu Y, Huang M, Shen D, Zhao B, Liu H. Recent advances of m6A demethylases inhibitors and their biological functions in human diseases. Int J Mol Sci. (2022) 23:1–2. doi: 10.3390/ijms23105815
15. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
16. Ianniello Z, Paiardini A, Fatica A. N(6)-methyladenosine (m(6)A): A promising new molecular target in acute myeloid leukemia. Front Oncol. (2019) 9:251. doi: 10.3389/fonc.2019.00251
17. Yue Y, Liu J, Cui X, Cao J, Luo GZ, Zhang ZZ. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discovery. (2018) 4:10. doi: 10.1038/s41421-018-0019-0
18. Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. (2020) 37:270–88. doi: 10.1016/j.ccell.2020.02.004
19. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
20. Mendel M, Chen KM, Homolka D, Gos P. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol Cell. (2018) 71:986–1000.e11. doi: 10.1016/j.molcel.2018.08.004
21. Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. (2017) 215:157–72. doi: 10.1111/nph.14586
22. Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna. (2018) 24:499–512. doi: 10.1261/rna.064063.117
23. Wu B, Li L, Huang Y, Ma J, Min J. Readers, writers and erasers of N(6)-methylated adenosine modification. Curr Opin Struct Biol. (2017) 47:67–76. doi: 10.1016/j.sbi.2017.05.011
24. Huang Q, Mo J, Liao Z, Chen X, Zhang B. The RNA m(6)A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis. (2022) 13:852. doi: 10.1038/s41419-022-05268-9
25. Wang X, Tian L, Li Y, Wang J, Yan B, Yang L. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. (2021) 40:80. doi: 10.1186/s13046-021-01871-4
26. Melstrom L, Chen J. RNA N(6)-methyladenosine modification in solid tumors: new therapeutic frontiers. Cancer Gene Ther. (2020) 27:625–33. doi: 10.1038/s41417-020-0160-4
27. Zhang N, Zuo Y, Peng Y, Zuo L. Function of N6-methyladenosine modification in tumors. J Oncol. (2021) 2021:6461552. doi: 10.1155/2021/6461552
28. Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem. (2005) 280:36935–45. doi: 10.1074/jbc.M501725200
29. Xiao N, Jani K, Morgan K, Okabe R, Cullen D. Hematopoietic stem cells lacking Ott1 display aspects associated with aging and are unable to maintain quiescence during proliferative stress. Blood. (2012) 119:4898–907. doi: 10.1182/blood-2012-01-403089
30. Ayllón V, González M, Pozas F, Reinés J, Montes R, Cacho L. New hPSC-based human models to study pediatric Acute Megakaryoblastic Leukemia harboring the fusion oncogene RBM15-MKL1. Stem Cell Res. (2017) 19:1–5. doi: 10.1016/j.scr.2016.12.019
31. Jiang X, Liu B, Nie Z, Duan L, Xiong Q. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. (2021) 6:74. doi: 10.1038/s41392-020-00450-x
32. Raffel GD, Chu GC, Jesneck J, Cullen D, Bronson R. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol Cell Biol. (2009) 29:333–41. doi: 10.1128/MCB.00370-08
33. Raffel GD, Mercher T, Shigematsu H, Williams I, Cullen E, Akashi K. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci U.S.A. (2007) 104:6001–6. doi: 10.1073/pnas.0609041104
34. Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang H. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. (2015) 4:2–3. doi: 10.7554/eLife.07938
35. Lindtner S, Zolotukhin AS, Uranishi H, Bear J, Kulkarni V, Smulevitch S. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J Biol Chem. (2006) 281:36915–28. doi: 10.1074/jbc.M608745200
36. Schuschel K, Helwig M, Hüttelmaier S, Heckl D, Klusman JH, Hoell JI. RNA-binding proteins in acute leukemias. Int J Mol Sci. (2020) 21:11–2. doi: 10.3390/ijms21103409
37. Zhang Z, Mei Y, Hou M. Knockdown RBM15 inhibits colorectal cancer cell proliferation and metastasis via N6-methyladenosine (m6A) modification of myD88 mRNA. Cancer Biother Radiopharm. (2022) 37:976–86. doi: 10.1089/cbr.2021.0226
38. Wang Q, Zhang Q, Li Q, Zhang J, Zhang J. Clinicopathological and immunological characterization of RNA m(6) A methylation regulators in ovarian cancer. Mol Genet Genomic Med. (2021) 9:e1547. doi: 10.1002/mgg3.1547
39. Yang F, Liu Y, Xiao J, Li B, Chen Y, Hu A. Circ-CTNNB1 drives aerobic glycolysis and osteosarcoma progression via m6A modification through interacting with RBM15. Cell Prolif. (2023) 56:e13344. doi: 10.1111/cpr.13344
40. Jiang A, Zhang S, Wang X, Li D. RBM15 condensates modulate m(6)A modification of STYK1 to promote tumorigenesis. Comput Struct Biotechnol J. (2022) 20:4825–36. doi: 10.1016/j.csbj.2022.08.068
41. Dong H, Zhang H, Mao X, Liu S, Xu W, Zhang Y. RBM15 promates the proliferation, migration and invasion of pancreatic cancer cell lines. Cancers (Basel). (2023) 15:2–3. doi: 10.3390/cancers15041084
42. Appel LM, Franke V, Benedum J, Grishkovskaya I, Strobl X, Polyansky A. The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators. Nat Commun. (2023) 14:166. doi: 10.1038/s41467-023-35853-1
43. Sánchez-Pulido L, Rojas AM, van Wely KH, Martinez AC, Valencia A. SPOC: a widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinf. (2004) 5:91. doi: 10.1186/1471-2105-5-91
44. Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. (2005) 272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x
45. Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics. (2000) 154:695–712. doi: 10.1093/genetics/154.2.695
46. Wiellette EL, Harding KW, Mace KA, Ronshaugen MR, Wang FY, McGinnis W. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development. (1999) 126:5373–85. doi: 10.1242/dev.126.23.5373
47. Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. (2020) 578:455–60. doi: 10.1038/s41586-020-1974-9
48. Appel LM, Benedum J, Engl M, Platzer S, Schleiffer A, Strobl X. SPOC domain proteins in health and disease. Genes Dev. (2023) 37:140–70. doi: 10.1101/gad.350314.122
49. Zolotukhin AS, Uranishi H, Lindtner S, Bear J, Pavlakis GN, Felber BK. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. (2009) 37:7151–62. doi: 10.1093/nar/gkp782
50. Uranishi H, Zolotukhin AS, Lindtner S, Warming S, Zhang GM, Bear J. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J Biol Chem. (2009) 284:26106–16. doi: 10.1074/jbc.M109.040113
51. Liu X, Wang H, Liu B, Qi Z, Li J, Xu B. The latest research progress of m(6)A modification and its writers, erasers, readers in infertility: A review. Front Cell Dev Biol. (2021) 9:681238. doi: 10.3389/fcell.2021.681238
52. Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. (2013) 288:33292–302. doi: 10.1074/jbc.M113.500397
53. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. (2018) 32:415–29. doi: 10.1101/gad.309146.117
54. Xie Y, Castro-Hernández R, Sokpor G, Pham L, Narayanan R, Rosenbusch J. RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Mol Neurobiol. (2019) 56:7305–20. doi: 10.1007/s12035-019-1595-1
55. Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PloS Genet. (2012) 8:e1002732. doi: 10.1371/journal.pgen.1002732
56. Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G. A pooled shRNA screen identifies Rbm15, spen, and wtap as factors required for xist RNA-mediated silencing. Cell Rep. (2015) 12:562–72. doi: 10.1016/j.celrep.2015.06.053
57. Coker H, Wei G, Moindrot B, Mohammed S, Nesterova T, Brockdorff N. The role of the Xist 5’ m6A region and RBM15 in X chromosome inactivation. Wellcome Open Res. (2020) 5:31. doi: 10.12688/wellcomeopenres
58. Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun. (2019) 10:3129. doi: 10.1038/s41467-019-11171-3
59. Fang J, Wu X, He J, Zhang H, Chen X, Zhang H. RBM15 suppresses hepatic insulin sensitivity of offspring of gestational diabetes mellitus mice via m6A-mediated regulation of CLDN4. Mol Med. (2023) 29:23. doi: 10.1186/s10020-023-00615-8
60. Cai X, Chen Y, Man D, Yang B, Feng X, Zhang D. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discovery. (2021) 7:315. doi: 10.1038/s41420-021-00703-w
61. Marasco LE, Kornblihtt AR. The physiology of alternative splicing. Nat Rev Mol Cell Biol. (2023) 24:242–54. doi: 10.1038/s41580-022-00545-z
62. Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. (2013) 14:153–65. doi: 10.1038/nrm3525
63. Sawada T, Nishiyama C, Kishi T, Sasazuki T, Komazawa-Sakon S, Xue X. Fusion of OTT to BSAC results in aberrant up-regulation of transcriptional activity. J Biol Chem. (2008) 283:26820–8. doi: 10.1074/jbc.M802315200
64. Xiao N, Laha S, Das SP, Morlock K, Jesneck JL, Raffel GD. Ott1 (Rbm15) regulates thrombopoietin response in hematopoietic stem cells through alternative splicing of c-Mpl. Blood. (2015) 125:941–8. doi: 10.1182/blood-2014-08-593392
65. Lee JH, Skalnik DG. Rbm15-Mkl1 interacts with the Setd1b histone H3-Lys4 methyltransferase via a SPOC domain that is required for cytokine-independent proliferation. PloS One. (2012) 7:e42965. doi: 10.1371/journal.pone.0042965
66. Jin S, Su H, Tran NT, Song J, Lu SS, Li Y. Splicing factor SF3B1K700E mutant dysregulates erythroid differentiation via aberrant alternative splicing of transcription factor TAL1. PloS One. (2017) 12:e0175523. doi: 10.1371/journal.pone.0175523
67. Dou X, Xiao Y, Shen C, Wang K, Wu T, Liu C. RBFOX2 recognizes N(6)-methyladenosine to suppress transcription and block myeloid leukaemia differentiation. Nat Cell Biol. (2023) 25:1359–68. doi: 10.1038/s41556-023-01213-w
68. Yang Y, Wang S, Zhang Y, Zhu X. Biological effects of decreasing RBM15 on chronic myelogenous leukemia cells. Leuk Lymphoma. (2012) 53:2237–44. doi: 10.3109/10428194.2012.684350
69. Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J. Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J Virol. (2011) 85:1528–40. doi: 10.1128/JVI.01709-10
70. Chen J. Regulatory mechanism of RNA binding motif protein 15-mediated N(6) methyladenosine modification in proliferation, invasion, and migration of colorectal cancer cells. Environ Toxicol. (2023) 38:2545–59. doi: 10.1002/tox.23883
71. Zhao Z, Ju Q, Ji J, Li Y, Zhao Y. N6-methyladenosine methylation regulator RBM15 is a potential prognostic biomarker and promotes cell proliferation in pancreatic adenocarcinoma. Front Mol Biosci. (2022) 9:842833. doi: 10.3389/fmolb.2022.842833
72. Nie G, Ju Q, Ji J, Li Y, Zhao Y. HPV E6 promotes cell proliferation of cervical cancer cell by accelerating accumulation of RBM15 dependently of autophagy inhibition. Cell Biol Int. (2023) 47:1327–43. doi: 10.1002/cbin.12020
73. Song Y, Wu Q. RBM15 m(6) A modification-mediated OTUB2 upregulation promotes cervical cancer progression via the AKT/mTOR signaling. Environ Toxicol. (2023) 38:2155–64. doi: 10.1002/tox.23852
74. Zhang C, et al. Knockdown of RBM15 inhibits tumor progression and the JAK-STAT signaling pathway in cervical cancer. BMC Cancer. (2023) 23:684. doi: 10.1186/s12885-023-11163-z
75. Yuan J, Guan W, Li X, Wang F, Liu H, Xu G. RBM15−mediating MDR1 mRNA m(6)A methylation regulated by the TGF−β signaling pathway in paclitaxel−resistant ovarian cancer. Int J Oncol. (2023) 63:3–7. doi: 10.3892/ijo
76. Zeng X, Chen K, Li L, Tian J, Ruan W, Hu Z. Epigenetic activation of RBM15 promotes clear cell renal cell carcinoma growth, metastasis and macrophage infiltration by regulating the m6A modification of CXCL11. Free Radic Biol Med. (2022) 184:135–47. doi: 10.1016/j.freeradbiomed.2022.03.031
77. Feng J, Li Y, He F, Zhang F. RBM15 silencing promotes ferroptosis by regulating the TGF-β/Smad2 pathway in lung cancer. Environ Toxicol. (2023) 38:950–61. doi: 10.1002/tox.23741
78. Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet. (2018) 392:593–606. doi: 10.1016/S0140-6736(18)31041-9
79. McNulty M, Crispino JD. Acute megakaryocytic leukemia. Cold Spring Harb Perspect Med. (2020) 10:1–2. doi: 10.1101/cshperspect.a034884
80. Shimony S, Stahl M, Stone RM. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol. (2023) 98:502–26. doi: 10.1002/ajh.26822
81. de Rooij JD, Masetti R, van den Heuvel-Eibrink MM, Cayuela JM, Trka J, Reinhardt D. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood. (2016) 127:3424–30. doi: 10.1182/blood-2016-01-695551
82. Mercher T, Coniat MB, Monni R, Mauchauffe M, Nguyen Khac F, Gressin L. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U.S.A. (2001) 98:5776–9. doi: 10.1073/pnas.101001498
83. Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. (2007) 27:3056–64. doi: 10.1128/MCB.01339-06
84. Hu M, Yang Y, Ji Z, Luo J. RBM15 functions in blood diseases. Curr Cancer Drug Targets. (2016) 16:579–85. doi: 10.2174/1568009616666160112105706
85. Niu C, Zhang J, Breslin P, Onciu M, Ma Z, Morris SW. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood. (2009) 114:2087–96. doi: 10.1182/blood-2009-01-197921
86. Moon H, Park H, Ro SW. c-Myc-driven hepatocarcinogenesis. Anticancer Res. (2021) 41:4937–46. doi: 10.21873/anticanres.15307
87. Tran NT, Su H, Khodadadi-Jamayran A, Lin S, Zhang L, Zhou D. The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep. (2016) 17:887–900. doi: 10.15252/embr.201541970
88. Pan YL, Zeng SX, Hao RR, Liang MH, Shen ZR, Huang WH. The progress of small-molecules and degraders against BCR-ABL for the treatment of CML. Eur J Med Chem. (2022) 238:114442. doi: 10.1016/j.ejmech.2022.114442
89. Lebbe C, Garbe C, Stratigos AJ, Harwood C, Peris K, Marmol VD. Diagnosis and treatment of Kaposi’s sarcoma: European consensus-based interdisciplinary guideline (EDF/EADO/EORTC). Eur J Cancer. (2019) 114:117–27. doi: 10.1016/j.ejca.2018.12.036
90. Massimelli MJ, Majerciak V, Kang JG, Liewehr DJ, Steinberg SM, Zheng ZM. Multiple regions of Kaposi’s sarcoma-associated herpesvirus ORF59 RNA are required for its expression mediated by viral ORF57 and cellular RBM15. Viruses. (2015) 7:496–510. doi: 10.3390/v7020496
91. Majerciak V, Deng M, Zheng ZM. Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology. (2010) 407:206–12. doi: 10.1016/j.virol.2010.08.014
92. Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. (2021) 18:525–43. doi: 10.1038/s41575-021-00438-0
93. Jiang H, Ning G, Wang Y, Lv W. Identification of an m6A-related signature as biomarker for hepatocellular carcinoma prognosis and correlates with sorafenib and anti-PD-1 immunotherapy treatment response. Dis Markers. (2021) p:5576683. doi: 10.1155/2021/5576683
94. Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for colorectal cancer screening. Gastroenterology. (2020) 158:418–32. doi: 10.1053/j.gastro.2019.06.043
95. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. (2021) 18:493–502. doi: 10.1038/s41575-021-00457-x
96. Yuan Y, Cai X, Shen F, Ma F. HPV post-infection microenvironment and cervical cancer. Cancer Lett. (2021) 497:243–54. doi: 10.1016/j.canlet.2020.10.034
97. Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. (2018) 26:158–68. doi: 10.1016/j.tim.2017.07.007
98. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs. (2019) 35:151–6. doi: 10.1016/j.soncn.2019.02.001
99. Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. (2016) 115:431–41. doi: 10.1038/bjc.2016.203
100. Wang Y, Chen Z. Long noncoding RNA UBA6-AS1 inhibits the Malignancy of ovarian cancer cells via suppressing the decay of UBA6 mRNA. Bioengineered. (2022) 13:178–89. doi: 10.1080/21655979.2021.2011640
101. Chen J, Yu K, Zhong G, Shen W. Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. (2020) 20:157. doi: 10.1186/s12935-020-01238-3
102. Wilson JE, Patel G, Patel C, Brucelle F, Huhn A, Gardberg AS. Discovery of CPI-1612: A potent, selective, and orally bioavailable EP300/CBP histone acetyltransferase inhibitor. ACS Med Chem Lett. (2020) 11:1324–9. doi: 10.1021/acsmedchemlett.0c00155
103. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. (2020) 41:1–24. doi: 10.1016/j.ccm.2019.10.001
104. Behrend SJ, Giotopoulou GA, Spella M, Stathopoulos GT. A role for club cells in smoking-associated lung adenocarcinoma. Eur Respir Rev. (2021) 30:1–2. doi: 10.1183/16000617.0122-2021
105. Ma M, Wang W, Wang B, Yang Y, Huang Y, Zhao G. The prognostic value of N6-methyladenosine RBM15 regulators in lung adenocarcinoma. Cell Mol Biol (Noisy-le-grand). (2022) 68:130–9. doi: 10.14715/cmb/2022.68.1.17
106. Zhang D, Zhang D, Wang C, Yang X, Zhang R, Li Q. Gene and prognostic value of N6-methyladenosine (m6A) modification regulatory factors in lung adenocarcinoma. Eur J Cancer Prev. (2022) 31:354–62. doi: 10.1097/CEJ.0000000000000717
107. Zeng Z, Zhang J, Li J, Li Y, Huang Z, Han L. SETD2 regulates gene transcription patterns and is associated with radiosensitivity in lung adenocarcinoma. Front Genet. (2022) 13:935601. doi: 10.3389/fgene.2022.935601
108. Song L, Zhang S, Yu S, Ma F, Wang B, Zhang C. Cellular heterogeneity landscape in laryngeal squamous cell carcinoma. Int J Cancer. (2020) 147:2879–90. doi: 10.1002/ijc.33192
109. Eaton BR, Schwarz R, Vatner R, Yeh B, Claude L, Indelicato DJ. Osteosarcoma. Pediatr Blood Cancer. (2021) 68 Suppl 2:e28352. doi: 10.1002/pbc.28355
110. Jiang J, Qu H, Zhan X, Liu D, Liang T, Chen L. Identification of osteosarcoma m6A-related prognostic biomarkers using artificial intelligence: RBM15. Sci Rep. (2023) 13:5255. doi: 10.1038/s41598-023-28739-1
111. Abolhassani H, Wang Y, Hammarström L, Pan-Hammarström Q. Hallmarks of cancers: primary antibody deficiency versus other inborn errors of immunity. Front Immunol. (2021) 12:720025. doi: 10.3389/fimmu.2021.720025
112. Huang W, Chen TQ, Fang K, Zeng ZC, Ye H, Chen YQ. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J Hematol Oncol. (2021) 14:117. doi: 10.1186/s13045-021-01129-8
113. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. (2019) 38:393. doi: 10.1186/s13046-019-1408-4
Keywords: RBM15, methyltransferase, m 6 A, cancer, regulatory mechanisms
Citation: Cao Y, Qiu G, Dong Y, Zhao W and Wang Y (2024) Exploring the role of m 6 A writer RBM15 in cancer: a systematic review. Front. Oncol. 14:1375942. doi: 10.3389/fonc.2024.1375942
Received: 24 January 2024; Accepted: 17 May 2024;
Published: 10 June 2024.
Edited by:
Junchao Shi, University of California, Riverside, United StatesCopyright © 2024 Cao, Qiu, Dong, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhao, emhhb3dlaTMzMkAxMjYuY29t; Yong Wang, d3lfc21jQDE2My5jb20=
 Yuan Cao1
Yuan Cao1 Yong Wang
Yong Wang