
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 17 June 2024
Sec. Head and Neck Cancer
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1373821
 Marian Constantin1,2
Marian Constantin1,2 Mariana Carmen Chifiriuc2,3,4*
Mariana Carmen Chifiriuc2,3,4* Coralia Bleotu2,5
Coralia Bleotu2,5 Corneliu Ovidiu Vrancianu2,3,6
Corneliu Ovidiu Vrancianu2,3,6 Roxana-Elena Cristian2,6,7
Roxana-Elena Cristian2,6,7 Serban Vifor Bertesteanu8
Serban Vifor Bertesteanu8 Raluca Grigore8
Raluca Grigore8 Gloria Bertesteanu8
Gloria Bertesteanu8The substantial heterogeneity exhibited by head and neck cancer (HNC), encompassing diverse cellular origins, anatomical locations, and etiological contributors, combined with the prevalent late-stage diagnosis, poses significant challenges for clinical management. Genomic sequencing endeavors have revealed extensive alterations in key signaling pathways that regulate cellular proliferation and survival. Initiatives to engineer therapies targeting these dysregulated pathways are underway, with several candidate molecules progressing to clinical evaluation phases, including FDA approval for agents like the EGFR-targeting monoclonal antibody cetuximab for K-RAS wild-type, EGFR-mutant HNSCC treatment. Non-coding RNAs (ncRNAs), owing to their enhanced stability in biological fluids and their important roles in intracellular and intercellular signaling within HNC contexts, are now recognized as potent biomarkers for disease management, catalyzing further refined diagnostic and therapeutic strategies, edging closer to the personalized medicine desideratum. Enhanced comprehension of the genomic and immunological landscapes characteristic of HNC is anticipated to facilitate a more rigorous assessment of targeted therapies benefits and limitations, optimize their clinical deployment, and foster innovative advancements in treatment approaches. This review presents an update on the molecular mechanisms and mutational spectrum of HNC driving the oncogenesis of head and neck malignancies and explores their implications for advancing diagnostic methodologies and precision therapeutics.
From the very heterogeneous group of head and neck cancers (HNC) (1), the head and neck squamous cell carcinomas (HNSCC) produced by the transformation of the squamous cell epithelia lining the oral cavity, pharynx and larynx are the sixth most common type of cancer (2–4), with 700,000 - 900,000 new cases recorded annually (2, 4), responsible for 0.5% (in case of oropharynx cancers) to 1.9% (for lip and oral cavity cancers) of deaths from all cancers combined (2).
The most invoked etiological factors are smoking, alcohol consumption (5, 6) and HPV (human papillomavirus) infections (7a; 7b; 8–14). In addition, several factors contribute more or less to HNC development, including EBV (Epstein-Barr virus) infections (15, 16), laryngopharyngeal reflux (17), chewing betel quid (Areca nuts) (18, 19), poor oral hygiene (20), oral dysbiosis (21), pro-inflammatory diet (22, 23), and inhalation of airborne pollutants (24, 25). With the development of molecular diagnostic techniques, many genetic aberrations (4, 26–28) and epigenetic factors (29) have been revealed.
HNC presents significant challenges in terms of both diagnosis and treatment. Late diagnosis due to the lack of effective screening strategies often complicates treatment options and reduces overall survival rates (30). The heterogeneous nature of HNC further adds to the complexity, requiring tailored approaches for different subtypes (24, 31–34). Moreover, the aggressive nature of traditional treatments like chemoradiotherapy can lead to significant side effects and reduced quality of life for patients (35).
The choice of the appropriate therapeutic strategy must consider the anatomical site, tumor stage, and etiological factors and could include surgery, radiotherapy, chemotherapy, immunotherapy, and, more recently, targeted therapies. Despite the poor outcomes in the advanced stages (36), the latter is expected to appropriately address the high heterogeneity of HNC. Consequently, it will contribute to better patient outcomes and improved 5-year survival rates, which currently average is around 50% (2, 33, 37).
Various clinical trials have been undertaken around the world on patients with HNSCC. In the clinicaltrials.gov database, 1266 clinical studies with the condition/disease ‘HNSCC’ have been registered since 2022. Recently, Goel and collaborators summarized the results of a large number of clinical trials examining the efficacy of immunotherapy and molecular-targeted treatments. A total of 393 studies were completed out of 1266 trials registered, while 590 studies were still recruiting. Docetaxel, cisplatin, and 5-fluorouracil (5-FU) have been identified as the most often used medications in clinical trials worldwide (38), but monoclonal antibody-based medicines such as nivolumab, pembrolizumab, cetuximab, panitumumab, zalutumumab, and nimotuzumab also hold significant promise for future therapeutic applications (39–41). However, the poor prognosis for many HNC patients and high rates of locoregional recurrence and metastasis advocate for better screening methods, more effective and tolerable, personalized treatment strategies (42). This paper reviews the molecular mechanisms involved in HNC, summarizing the signaling pathways harboring genetic aberrations (RAS–RAF–MEK–ERK, PIK3–AKT–mTOR, WNT/beta-catenin, JAK-STAT, NOTCH, and HIF–VEGF), epigenetic mechanisms and the roles of ncRNAs and tumor microenvironment in neoplastic progression. We also tried to summarize the therapies targeting the abnormal functioning of these signaling pathways and their efficacy.
The development of HNC is a multistep process in which mutations are accumulating in the main cellular signaling pathways that regulate proliferation, cell death, angiogenesis and immune system functions (3) (Figure 1).
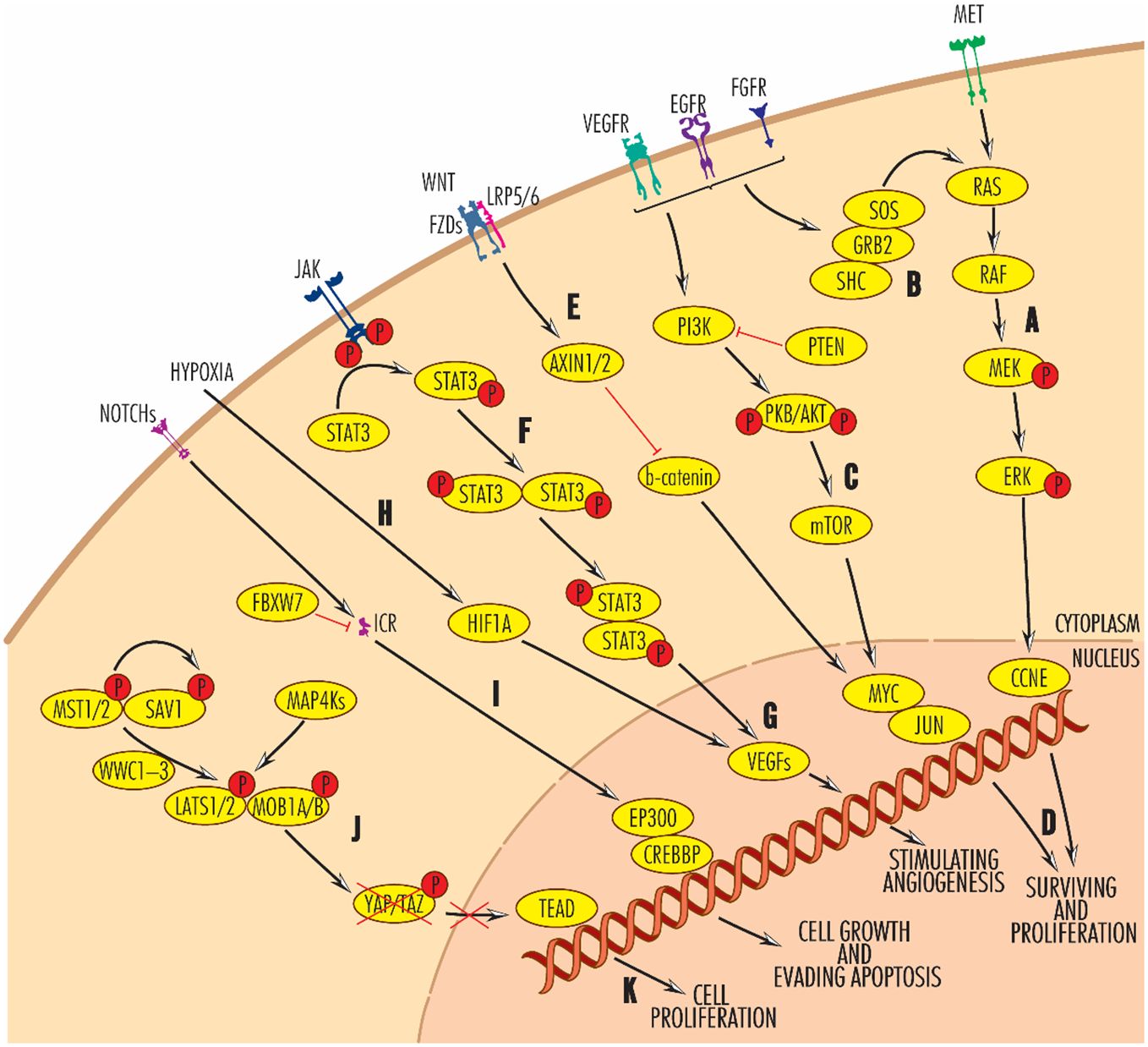
Figure 1 Main signaling pathways disregulated in HNC at the genomic or transcriptional level. Black arrow-ended lines indicate activation and red bar-ended lines indicate inhibition. RAS–RAF–MEK–ERK (A, B) and PIK3–AKT–mTOR (C) signaling pathways are involved in promoting cell survival and proliferation (D), antagonistically to the WNT pathway (E), in which AXN1/2 blocks beta-catenin activity. By enhancing JAK/STAT signaling (F), VEGF proteins (G), the main angiogenic molecules, are activated. Alternatively, VEGF expression is also modulated by hypoxia-induced factor 1A (H). Frequently reported in HNCs, angiogenesis builds the vasculature through which the tumors are supplied with oxygen and allows their progression and a worse prognosis. On the other hand, the NOTCH signalling pathway is involved in cell growth and evasion of apoptosis (I). Very important in the control of tumour processes in head and neck cancers is the Hippo pathway (J), which, when activated, phosphorylates YAP/TAZ, causing its intracytoplasmic sequestration and degradation, preventing its translocation to the nucleus and activation of TEAD. TEAD activation leads to cell proliferation. For further details, please see the text.
The most affected cell cycle regulating genes in HNC are TP53, RB, CCND1, CDKN2A/INK4, and CDK6, unblocking the G1/S transition. Thus, TP53 protein function is impaired in 50–80% of cases, the mutations in the TP53 gene in HNSCC being the seventh most frequent in cancer diagnosis worldwide (43). Several types of mutations (deletions, insertions, and frameshift mutations, and point mutations) in the TP53 protein are associated with increased risk of progression from mild dysplasia to invasive carcinoma and unfavorable tumor progression (33, 44–47). Specifically, TP53 missense mutations in the DNA binding region are significantly enriched in metastases and are associated with a common fragile site in chromosome 11, leading to amplification and overexpression of genes with established role in metastasis (45). In HPV-positive HNC, TP53 mutations are sporadic, as the function of this gene is selectively abrogated by viral proteins E6 and E7.
The RB1 gene function is altered by unilateral or bilateral deletions, methylation of the RB1 promoter, or point mutations (48).
The CCND1 gene is overexpressed in 30%-46% of cases, leading to G1 phase shortening and rapid entry into the S phase, bypassing the influence of growth factors and increasing the proliferation rate of gene-defective cells. These alterations are reported in cancers with frequent recurrence, lymph node metastasis, and reduced survival rate (49, 50).
After CCND1, the third most frequent alteration in HNC is the potent inhibitor CDKN2A, a regulator of cyclin activity and progression to S phase. CDKN2A alterations including deletions (more common in HPV-negative tumors), hypermethylation, and, less commonly, mutations in exon 2 (rarest in oropharyngeal tumors) are associated with metastatic cancers with poor prognosis (51). In laryngeal squamous cell carcinomas, mutations in the CDKN2A gene occur in approximately 14% of cases (52). CDK6 is overexpressed in some oral tumors and correlates with tumor stage advancement and progression (36).
The DNA repair pathway is a mechanism by which cells in the G1 or G2 phase are halted at checkpoints to identify and correct errors in the nucleotide sequence of nuclear genetic material. This process is initiated by the detection of DNA lesions and is activated by the ATM (ataxia-telangiectasia mutated) and ATR (ataxia-telangiectasia and Rad3 related) kinases, along with proteins such as BRCA1, BRCA2, (Mediator of DNA Damage Checkpoint 1) and T53BP1 (Tumor Protein P53 Binding Protein 1). Additionally, the synthesis of PALB2 (partner and locator of BRCA2), which interacts with BRCA1 and BRCA2, plays a crucial role in the DNA repair process.
In HNC, the most mutated genes in this signaling pathway are ATM (25%), BRCA2 (9.2%), BRCA1 (5.75%), and ATR (4.36%) (53–56). Recent studies have revealed that the disrupted expression of specific components within the DNA repair machinery, stemming from mutations in key pathway genes like ATM and BRCA1, can yield valuable prognostic insights for HNC patients (56, 57).
The alterations in critical cellular signaling pathways governing proliferation and cell survival have been exploited to create customized treatments for HNC individuals. The increasing interest in exploring the cell cycle (CDK4/6, CCND1, CDKN2A) and the DNA repair pathways (BRCA, ATM, ATR) is demonstrated by clinical trials (NCT03356223, NCT03065062, NCT03024489, NCT05878964, NCT04576091, NCT04491942, NCT02567422) actively investigating their potential in the broader context of HNC such as CDK4/6 inhibitors for HPV-negative tumors (42).
This pathway involves numerous proteins and receives biological signals from the extracellular space through various ligand-receptor pairs, including TGFα and EGF–EGFR/ERBB1/HER1, ERBB2/HER2, PDGF–PDGFRA and PDGFRB, IGF–IGF1R, FITL–KIT/c-KIT, FLT3L–FLT3, HGF–MET, and FGF–FGFR. These signals are transmitted into the nucleus, where they activate genes involved in cell proliferation and differentiation, inflammation, evasion of apoptosis, and support of angiogenesis (58–63). Interaction with cytokine ligands activates transmembrane receptors and recruits the growth factor receptor-bound protein 2 (GRB2) adaptor protein, which interacts with SOS1 protein (the human homolog of Drosophila son of sevenless 1). This is a RAS-specific guanine nucleotide exchange factor and reacts with RAS family members, the core proteins of this signaling pathway (64, 65). The RAS family of GTPases comprises three members, KRAS (Kirsten RAS oncogene homolog), HRAS (Harvey RAS oncogene homolog), and NRAS (Neuroblastoma RAS oncogene homolog), which transmit the signals downstream to RAFs (from the canonical RAS–RAF–MEK–ERK pathway), RALGDS (from the RALGDS–RAL–PLD1 signaling pathway), RASSF1 (from the RASSF1–MST1 signaling pathway) or PI3K (from the PI3K–AKT–mTOR signaling pathway) (66, 67). In the canonical RAS–RAF–MEK–ERK pathway, RAS proteins are the first members of a four-step cascade of cytoplasmic protein kinase kinases, which include: (1) RAF (rapidly accelerated fibrosarcoma kinase), RAF1/c-RAF, BRAF and ARAF family of kinases, designated MAPKKK or MAP3K; (2) MEK (mitogen-activated protein kinases); (3) ERK (extracellular signal-regulated kinase) (68, 69). In the RAS–RAF–MEK–ERK signaling pathway, overexpression of EGFR/ERBB1 occurs early in the progression of HNSCC, leading to poor prognosis (70). In tumor xenografts of HPV-positive HNSCC, it enhances the response to radiotherapy by decreasing the expression of the HPV protease E6 and affecting DNA repair mechanisms (71).
In most HNC, members of the RAS gene family are most frequently mutated, followed at a long distance by those of the RAF gene family (72). In a study of 51 patients with HNSCC (with higher prevalence in the larynx and trachea area), mutations of the KRAS gene (sometimes designated KRAS1 or KRAS2) were detected in 35% of cases, and mutations of the HRAS gene in 33% of cases, with the caveat that KRAS mutations, HRAS mutations, and HPV infection are mutually exclusive (68). The most frequent mutations (7%) occur in the HRAS gene (mainly in the oral cavity and salivary gland tumors and associated with advanced stages of tumors), with the other two family members, KRAS, at 2.89% (mostly in syn-nasal tumors and often associated with HPV infection), and NRAS, at 2.20% (predominantly in nasopharyngeal tumors) (69). Members of the RAF gene family undergo fewer mutations (in about 3% of cases) compared to RAS genes, with the most known mutations reported in the BRAF gene (V599/600E, G468/469A, and Q257R) (58). In HNC, 6% of cases have heterozygous mutations in exons 12 and 13. Because KRAS activates only the wild-type BRAF gene, mutations in KRAS and BRAF genes never occur together in HNC and are redundant (58).
Many inhibitors targeting this pathway have been developed, essentially modifying the therapeutic strategy of cancers. Starting from compound 12, first reported by the Schokat group in 2013 (73), a series of inhibitors based on the compound 12 structure are developed, such as ARS-853 and ARS-1620 (74, 75). The KRASG12C-specific drug, AMG510 (storasib), first went into clinical trial in 2019 and was subsequently proven by the FDA in 2021 (76, 77) (Table 1).
Another KRASG12C-specific covalent inhibitor, MRTX849 (adagrasib), developed by the Mirati group, also went into clinical trial in 2019 (92). Cetuximab is a chimeric mouse-human monoclonal IgG1 antibody against the extracellular domain of EGFR that can inhibit the functions of EGFR and induce cancer cell death via antibody-dependent NK cell-mediated cytotoxicity (93). In 2006, FDA approved the combination of cetuximab with radiotherapy for the treatment of locally advanced (LA) - HNSCC (94). In a study published in 2023, the conjugate of cetuximab with IRdye700DX, which is activated by illumination at 690 nm, has been used successfully on near infrared photoimmunotherapy to a patient with local recurrence of nasopharyngeal squamous cell carcinoma (95). Cetuximab, received full FDA approval for the treatment of patients with K-RAS wild-type, EGFR-mutant HNSCC following reports that its addition to radiation therapy results in significant improvements in disease control and overall survival (96, 97). Other inhibitors of EGFR/ERBB1 tested for HNC treatment are dacomitinib and vandetanib. When used with radiotherapy, dacomitinib reduces tumor volume in HNSCC, while vandetanib, and cisplatin radiosensitizes tumor cells. Combining vandetanib with radiotherapy is more efficient than other monotherapies or combination therapies (98, 99).
Several monoclonal antibodies of EGFR are still under intensive investigation, including panitumumab, nimotuzumab, and zalutumumab. According to the results from CONCERT-2 and HN.6 trials, panitumumab cannot replace cisplatin when combined with radiotherapy for LA-HNSCC (78, 79). In addition, combining panitumumab with the standard chemoradiotherapy strategy failed to provide any benefit (100). Adding nimotuzumab to radiotherapy with or without cisplatin provided long-term survival benefits for up to five years and improved the complete response rate in LA-HNSCC patients (101). In a phase 3 clinical trial involving 536 LA-HNSCC patients, nimotuzumab plus cisplatin and radiotherapy significantly improved the locoregional control rate without negatively impacting the quality of life (102). The promising results strongly supported the addition of nimotuzumab to LA-HNSCC patients who are treated with cisplatin and radiotherapy. Another monoclonal antibody, zalutumumab, extended the survival time from 8.4 to 9.9 weeks in recurrent or metastatic (R/M) HNSCC patients who had failed platinum-based chemotherapy (103). Meanwhile, moderate-to-severe skin rash during zalutumumab treatment was related to superior OS, independent of HPV infection and p16 status (104).
Some small molecular inhibitors of EGFR are also under investigation for the management of HNSCC, including selective inhibitors (e.g., gefitinib, erlotinib) (80, 81, 105, 106) and dual-target inhibitors (e.g., afatinib, lapatinib, and dacomitinib) (82, 107).
The PIK3–AKT–mTOR signaling pathway is very complex, being activated by extracellular signals represented by hormones, cytokines, and growth factors via receptors common with those of the RAS–RAF–MEK–ERK signaling pathway or via the enzyme PI3K (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase), directly or indirectly, after PI3K activation by the GRB2, SOS, and RAS. This pathway is involved in cell growth, differentiation, survival, migration and proliferation, apoptosis evasion, and glucose metabolism (108). The central actor of this signaling pathway is the class I PI3K enzyme, which is part of the PI3K family of lipid and protein kinases, classified into three classes (I, II, and III). The class I enzymes function as secondary messengers in the intracellular transduction of biological signals and are more commonly associated with cancer (109, 110). The PI3K signals are transmitted through a phosphorylation cascade, represented by PIP2 (phosphatidylinositol-4,5-bisphosphate), which becomes PIP3 (phosphatidylinositol-3,4,5-trisphosphate), AKT/PKB (Serine/Threonine Kinase), and mTOR–RICTOR (Mechanistic Target Of Rapamycin Kinase–Rapamycin Insensitive Companion of mTOR) complex. Phosphorylation of AKT/PKB is antagonized by PTEN (phosphatase and tensin homolog) activity, which dephosphorylates the latter (111) and functions as an essential tumor suppressor (112). Subsequently, the phosphorylated AKT/PKB activates multiple downstream targets, promoting cell survival, activating anti-apoptotic pathways, and blocking apoptotic ones. PI3K and AKT can induce chromosome instability MET-dependently, an event suppressed by PI3K/mTOR inhibition, AKT depletion or PTEN overexpression (113). Interacting with HGF (Hepatocyte Growth Factor), MET can activate the PI3K–AKT–mTOR signaling pathway in endosomes independently of EGFR (114) or in the presence of TP53 mutants, as is mainly the case in patients with HPV-positive tumors (115). The MET gene is overexpressed in over 75% of HNSCC and has an increased copy number of 13%, associated with tumor progression and tumor dissemination in the early stages (116). Mutations of the MET gene are reported less frequently, but they seem to be involved in lymph node metastasis (117, 118). HNC may also carry somatic mutations in PIK3R1 (~7%) and PTEN genes (51, 119), with loss of function in approximately 30% of HNC (more common than other cancers) due to mutations, loss of heterozygosity in the 10q region (which includes PTEN), detected in more than 70% of HNSCC, or hypermethylation, reported in ∼5% of these. In addition, TSC1/2 and LKB1 genes could be inactivated by loss of heterozygosity (TSC1/2), methylation (TSC2) and somatic mutations (LKB1) (119) (Figure 2).
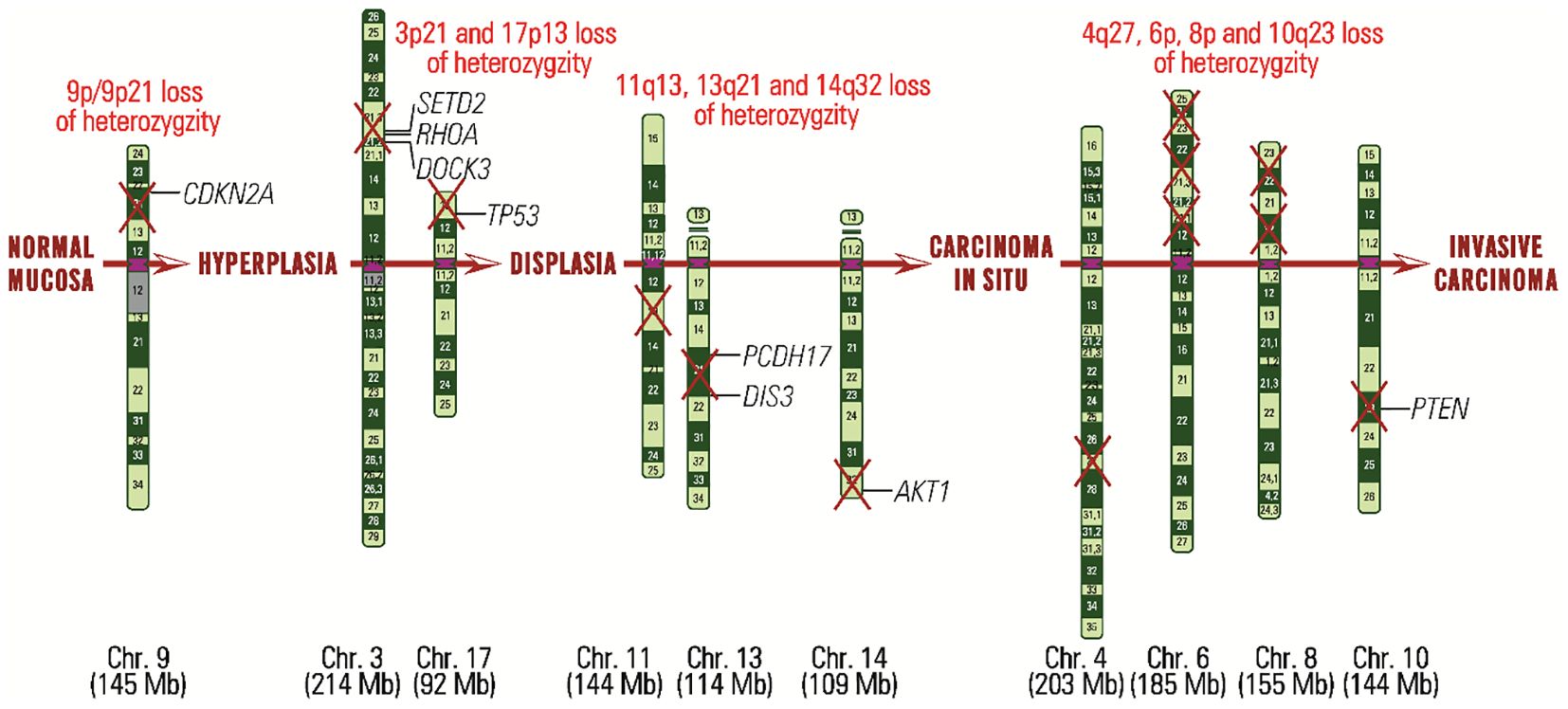
Figure 2 Carcinogenesis in HNC from normal mucinous stage to invasive carcinoma stage, highlighting each stage, chromosomal regions affected by loss of heterozygosity and genes that may be defective.
Although Akt is recognized as a key player in cell migration and metastasis, its role remains controversial. Akt1 promotes cell migration in fibroblasts but inhibits it in breast cancer, while Akt2 has the opposite effects. The deletion of Akt isoforms can impact cancer progression differently depending on whether it is systematic or cell-autonomous (120, 121). Akt regulates cell migration through various mechanisms, such as phosphorylating PAK1, Girdin/APE, and ACAP1, which are involved in cytoskeletal dynamics and integrin trafficking, crucial for cell motility (122, 123). Further research is needed to fully understand these phosphorylation events and their implications in cancer metastasis.
Currently, drugs that target PI3K for NOTCH1-mutant tumors (124) or mTOR are undergoing clinical trials. Everolimus (RAD001), an allosterically inhibitor of only mTORC1 but not of mTORC2, is clinically used to treat various cancers, including previously treated recurrent or metastatic HNSCC (38, 83). BKM120, an oral, highly specific pan-Class I PI3K inhibitor, has strong antiproliferative characteristics in tumor cell lines (125). Previous studies with the PI3Kα inhibitors taselisib, TAK-117, and alpelisib in patients with solid tumors have reported promising clinical results (84, 126). Taselisib treatment showed an overall rate response (ORR) of 36% in patients with PIK3CA mutations but 0% in patients without PIK3CA mutations (84). The phase I trial with alpelisib specifically enrolled patients with PIK3CA mutations and showed an ORR of 6% and a stable disease rate of 52% (127, 128). Two phase II clinical trials, including to date 47 patients, are currently evaluating the clinical efficacy of alpelisib as monotherapy in HPV-positive HNSCC (NCT03601507) and alpelisib in combination with the farnesyltransferase inhibitor tipifarnib in HRAS- and PIK3CA-mutant HNSCC (NCT04997902; 129). Buparlisib (BKM120), another pan-PI3K inhibitor, exhibited limited antitumor activity in patients with HNSCC, with a disease control rate of 49% and an ORR of only 3% (NCT01527877; NCT01737450) (85, 130). Another phase II trial, including patients with HNSCC, revealed a modestly longer median in patients receiving a combination of buparlisib and paclitaxel than in the paclitaxel and placebo group (NCT01852292) (131). The buparlisib/paclitaxel combination is currently in phase III trial in patients with HNSCC (NCT04338399). Understanding the intricate interplay between Akt, Rho GTPases, and their downstream effectors, such as PAK1, as well as the regulatory networks controlling RhoA expression, is essential for uncovering novel therapeutic targets and strategies for combating metastatic cancer.
The WNT(Wingless-Type)/beta-catenin signaling pathway is an alternative to the PIK3–AKT–mTOR pathway for promoting cell proliferation and avoiding apoptosis that includes three main pathways, the canonical WNT/b-catenin signaling pathway and the WNT/Ca2C and WNT/PCP non-canonical pathways (132–134). They also promote dysplastic transformation [WNT3, in oral leukoplakia (135), deterioration in histological grade, progression of clinical stage, and heightened metastatic potential in cervical lymph nodes [WNT3A, in laryngeal squamous cell carcinoma (136)], amplification of migration and invasiveness [WNT5A (137) and WNT5B (138), in OSCC], tumorigenesis and metastasis [WNT5A, in nasopharyngeal carcinoma (139) and laryngeal squamous cell carcinoma (137)], migration [WNT7A, in OSCC (140)], proliferation and invasiveness [WNT7B, in OSCC (141)], cell growth and survival and inhibition of apoptosis [WNT10B, in HNSCC (142)]. In some OSCC, WNT11 is involved in tumor suppression (143). Defects in the WNT11 gene are found in 5% of OSCC, consisting of duplications, deep deletions or punctiform mutations affecting its tumor suppressor function. Other WNT genes are affected in smaller proportions (144).
Porcupine (Porc) is a membrane-bound O-acyltransferase (MBOAT) by whose Wnt ligands palmitoylation takes place, a process allowing them to be further secreted and recognized (145, 146). Therefore, repressing Porc can be a promising solution against tumors with aberrant Wnt/β-catenin activation (147). WNT974 is a potent, selective, and orally bioavailable first-in-class inhibitor of Porcupine with preclinical activity in Wnt-dependent HNC (86). Recently, Rodon and collaborators conducted a phase 1 study (NCT01351103) to investigate the safety and efficacy of the WNT974 in patients with solid tumors. The results of this clinical trial revealed that this inhibitor could be tolerated and may influence immune cell recruitment to tumors and enhance checkpoint inhibitor activity (86). In a phase I clinical trial, OMP-18R5 (vantictumab), a monoclonal antibody targeting FZD receptors, inhibited tumor growth in HNC (148). In a phase Ib clinical trial, 54 patients with locally recurrent or metastatic HER2-negative breast cancer who were treated with weekly paclitaxel in combination with escalating doses of vantictumab were enrolled. The combination of vantictumab and paclitaxel was generally well tolerated and had promising efficacy. However, the incidence of fractures limits future clinical development of this particular WNT inhibitor in metastatic breast cancer (149). XAV939, a tankyrase inhibitor, inhibited β-catenin signaling attenuated cancer stem cells progression, consequently eliminating the chemical resistance in HNSCC (60, 150).
The JAK–STAT signaling pathway is involved in diverse physiological processes such as hematopoetic cell responses to cytokines (151), cell growth, proliferation and differentiation, survival, angiogenesis, and inflammatory or immune responses, but also in pathological conditions like tumor processes (152–154). The main proteins are JAKs (Janus kinases, named after the Roman god Janus, known to have two faces), with for members (JAK1, JAK2, JAK3 and TYK2, tyrosine kinase 2), and STAT (signal transducer and activator of transcription), with seven members (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6) (155). The JAK–STAT signaling pathway is activated in many cancers, including HNC, mainly through overexpression of STAT1, STAT3, and STAT5, with the first correlating with favorable outcomes and the latter two with unfavorable outcomes (33, 156). In OSCC, overexpression of STAT3 and its accumulation in the nucleus is associated with reduced survival or favorable prognosis (157), promoting tumor angiogenesis by stabilizing and modulating the activity of HIF1 (hypoxia-inducible factor 1), which promotes the synthesis of VEGFs (158), a key protein involved in tumor invasiveness and metastasis. However, in breast cancers and some colorectal, spine, head, and neck cancers, activation of the JAK–STAT signaling pathway appears to result in a more favorable prognosis (159).
Several studies in human tumors and HNC cell lines have identified the JAK/STAT signaling pathway as a potential therapeutic target (160, 161). The group of Kowshik demonstrated that astaxanthin could hinder tumor progression by attenuating JAK/STAT signaling and its target molecules, including VEGF, cyclin D1, and MMP in the HPV-induced tumor models (162). The JAK1/2-selective inhibitor ruxolitinib is FDA-approved for several diseases, including myelofibrosis and graft versus host disease (163, 164). Currently, two multicenter, phase 1b clinical trials are undergoing to investigate the safety and efficacy of ruxolitinib (NCT03153982) and pembrolizumab (NCT02646748) in patients with HNC.
The NOTCH signaling pathway is activated by two families of ligands, Jagged (JAG1 and JAG2) and Delta-like (DLL1, DLL3 and DLL4). Upon interaction with NOTCH receptors (NOTCH1, NOTCH2, NOTCH3, and NOTCH4) they influence cell self-renewal capacity, cell cycle exit, survival, proliferation, and angiogenesis in a cell- and biological context-dependent manner (165–167). NOTCH1 signaling suppresses tumor development by promoting terminal keratinocyte differentiation and is probably protective in advanced stages of HPV-induced carcinogenesis, by reducing transcription of viral E6 and E7 genes (168). Conversely, NOTCH signaling causes FGF1-mediated tumor invasiveness in OSCC and increases mortality (167), probably through activation of MDM2, which ubiquitinates TP53 and primes it for degradation (169, 170). NOTCH function is negatively regulated by EGFR-activated C-JUN and inhibited by TP63, the latter being overexpressed in numerous cases of HNSCC (171). Notch signaling pathway is altered in 66% cases of HNSCC (172), NOTCH1 gene presenting different percentages of mutations and genetics variants, some of which being nonsense and missense mutations (166, 173).
Several clinical trials were designed to target Notch signaling in patients with advanced solid tumors. An open-label phase 1a dose escalation clinical trial study (NCT01778439) of brontictuzumab, a monoclonal antibody, was designed to assess the safety, immunogenicity, pharmacokinetics, biomarkers, and efficacy of brontictuzumab in subjects with relapsed or refractory solid tumors. The group around Ferrarotto, who contributed to this clinical trial, reported significant clinical benefits in 6 of 36 patients, with four subjects having prolonged (≥ 6 months) disease stabilization. In addition, brontictuzumab was well tolerated at the maximum tolerated dose (174). More recently, two phase 1b studies with parallel dose-escalations (NCT02784795; NCT02836600) were designed to investigate the Notch inhibitor crenigacestat in patients with advanced or metastatic cancer from a variety of solid tumors. These clinical trials revealed that crenigacestat was poorly tolerated, leading to lowered dosing and limited clinical activity in patients with advanced or metastatic solid tumors (89, 175, 176).
The hypoxia and angiogenesis (HIF-VEGF) pathway is essential for oxygen supply supplementation in solid tumors. The intense metabolic activities of solid tumors require an increased oxygen supply, which cannot be provided by the physiologically existing capillary structure of tissues. Thus, small tumors a few millimeters in diameter can be supplied by diffusion, but larger tumors with increased oxygen requirements rapidly enter hypoxia (177). In HNSCC, hypoxia is a common condition associated with poor prognosis and 5-year survival approaching 0% (178). Recently, Matic and collaborators searched potential biomarkers in HNSCC by examining mRNA expression of five highly upregulated (CA9, CASP14, LOX, GLUT3, SERPINE1) and four highly downregulated (AREG, EREG, CCNB1, and KIF14) hypoxia-responsive genes in 32 HNSCC tumors and six adjacent normal oral tissue. The results showed a significantly higher mRNA expression of the hypoxia marker CA9 and SERPINE1 in all tumor biopsies compared to normal tissue. Regarding the hypoxia-downregulated genes, the authors observed higher KIF14 and AREG mRNA expression in HNSCC patients than in the the control group. In conclusion, the mRNA expression of KIF14 could be a potential diagnostic marker and might serve as a predictor of treatment response in HNSCC (Matic et al., 2024). Signaling via the hypoxia and angiogenesis pathway begins with HIF1–2 proteins (hypoxia-inducible factors 1–2), which heterodimerize, and the HIF1–HIF2 heterodimers are translocated to the nucleus, where HIF1 promotes transcription of some genes, including the angiogenesis inductors VEGFA-D (Vascular Endothelial Growth Factors A-D) (67). Activation of HIF1 and HIF2 and overexpression of VEGFA-D in HNSCC result in carcinogenesis progression, increased aggressiveness, and poor prognosis, with a 2-fold increase of 2-year mortality risk (161).
Bevacizumab, a monoclonal antibody, is an FDA-approved VEGF inhibitor used in treating numerous cancers, either as a single agent or combined with chemotherapy or radiotherapy (179). In a phase III trial including 403 patients with HNSCC, adding bevacizumab to platinum-based chemotherapy significantly improved the response rate and progression-free survival. However, it did not increase the median survival rate (91). Several clinical trials have recently investigated the benefits of bevacizumab in combination with immune and chemotherapy. A phase II multicenter study (NCT03818061) aims to assess the effects of atezolizumab, a PD-L1 inhibitor, and bevacizumab in recurrent or metastatic HNSCC, considering the ORR. Another phase II trial (NCT00409565) compared bevacizumab with cetuximab, which has an immune-mediated activity. The results published by Argiris and collaborators revealed a significant reduction in tumor vascularization, with an ORR of 16%, a disease control rate of 73%, and a generally well-tolerated response (90). Three phase II trials are currently investigating the combination of bevacizumab, cetuximab, and chemoradiation in HNSCC (NCT00968435; NCT00703976; NCT01588431).
Tyrosine kinase inhibitors are small molecules acting by inhibiting several targets within angiogenic signaling pathways, including VEGFRs, EGFR, FGFR, and PDGFRs. In two phase II clinical trials, sorafenib and sunitinib, a multi-kinase inhibitor, were investigated in HNSCC patients and revealed minimal response rates (180, 181). A phase II trial of axitinib demonstrated a low objective response rate but a favorable disease control rate of 77% and median overall survival (OS) of 10.9 months with an acceptable toxicity profile (182, 183). Aurora kinase inhibitors are tested for RB-deficient, HPV-positive HNSCCs (184).
Identified nearly three decades ago during tissue growth screening in Drosophila melanogaster, the Hippo signaling pathway is evolutionarily conserved in mammals (185, 186). Under physiological conditions, the Hippo signaling pathway restricts tissue growth in adult organisms by modulating cell proliferation, differentiation, and migration in growing organs (185). Its deregulation plays an important role in several diseases, including cancer and various organ-specific diseases (187, 188). In mammals, the pathway involves over 30 proteins, such as MST1/2, SAV1, MOB1A/B, and LATS1/2. These proteins phosphorylate YAP and TAZ, tagging them for cytoplasmic degradation and preventing their nuclear translocation, thus inhibiting transcription via TEADs and SMADs. Activation occurs through FAT1, KIBRA, AJUBA, NF2, RHO, AMPK, or by inactivation of STRIPAK complexes, which regulate MST1/2 and MAP4Ks (189, 190). In head and neck squamous cell carcinoma, common aberrations include mutations in FAT1, WWTR1/TAZ, YAP1, and MST2 (191, 192). Since 2016, several molecules targeting the Hippo pathway have been developed. In nasopharyngeal carcinoma, MGH-CP1, an inhibitor of TEAD2/4 auto-palmitoylation, is in preclinical trials, reducing TEAD4-mediated AKT signaling and inhibiting cell migration, invasion, and resistance to cisplatin (50, 189).
Acetylation is the transfer of an acetyl group from acetyl-CoA to the amino epsilon group of lysine residues by a histone acetyltransferase, neutralizing the positive charge of lysine residues and exposing DNA to transcriptional complexes (193). Other modifications consist of ubiquitination of lysine residues, phosphorylation of serine residues, SUMOylation of lysine residues (covalent interaction of a member of the SUMO, small ubiquitin-like modifier, protein family through an enzymatic cascade analogous but not similar to ubiquitination), and methylation of lysine and arginine residues (194, 195).
In HNSCC, global hypomethylation is more common in HPV-negative tumors. It is associated with genetic instability, including genome-wide loss-of-heterozygosity (LOH), single nucleotide polymorphisms (SNP), and alternative oncogenic pathways (196). Global hypomethylation was also linked with female gender and worse survival, predominantly for older patients with a stage I or II AJCC (American Joint Committee on Cancer) tongue squamous cell carcinoma without lymph node involvement and with postoperative radiotherapy (197). Assessment of the methylation status of CALML5, DNAJC5G, and LY6D genes identified in ctDNA from HNSCC patients demonstrated substantial predictive value in early cancer diagnosis (198). FAM135B (Family with Sequence Similarity 135 Member B) methylation appears to be associated with good prognosis, while APBA1/MINT1 (Amyloid Beta Precursor Protein Binding Family A Member 1), MINT31 (Methylated IN Tumors locus 31) DCC (Deleted In Colorectal Carcinoma Netrin 1 Receptor) methylation with poor prognosis, the latter one being also associated with bone invasiveness in the mandible (199). Binding KRAS and having a role in apoptosis induction, RASSF2 (Ras association domain-containing protein 1) is intensely methylated. Other intensely methylated genes are EDNRB (Endothelin Receptor Type B), methylated in 97% of primary HNC tissues, and RARB (Retinoic Acid Receptor Beta), involved in transcriptional control (199, 200), and the tumor suppressor genes PTEN, DAPK (death-associated protein kinase), MGMT (O6-methylguanine-DNA methyltransferase), involved in DNA repair, CDH1/ECAD (E-cadherin), involved in cell adhesion, and RASSF1 (Ras association domain-containing protein 1), involved in cell cycle control, apoptosis, and cell adhesion, inactivation of which is present in several cancers (201).
Normally, cells of HNSCC are hypoacetylated (e.g., H3K9ac in OSCC) (201) compared to normal mucosal cells, but histones in these cells can be acetylated by factors secreted by endothelial cells, in a paracrine manner. Consequently, acetylation induces the amplification of BMI1, a transcriptional repressor associated with poor survival and tumor aggressiveness, and vimentin, which marks the epithelial-mesenchymal transition (202). Deacetylation is mediated by histone deacetylases (HDACs), and HDACs inhibition in vitro results in fewer cancer stem cells (CSCs) in HNC. Thus, HDACs inhibition seems a promising strategy to disrupt the population of CSCs in HNC to create a homogeneous population of tumor cells characterized by well-defined biological traits and predictable behaviors (203). Several HDAC inhibitors are currently under evaluation in clinical trials for their efficiency in HNC treatment. When combined with chemoradiation therapy, vorinostat showed positive effects in HNC (NCT01064921). Additionally, abexinostat is under evaluation with pembrolizumab in an ongoing phase 1b dose-escalation trial for advanced solid tumors, including metastatic HNSCC (NCT03590054).
Non-coding RNAs do not encode proteins but have enzymatic, structural, or regulatory functions and can control gene expression. Depending on the number of nucleotides, they are short (microRNA, miRNA) or long (lncRNA, long ncRNA). miRNA are 21–23 nucleotides in size and bind partially complementary regions of the 3’ untranslated regions of several hundred mRNAs, potentially interfering with gene expression and cell differentiation, proliferation, and apoptosis. Although their aberrant expression can trigger the malignant process, miRNAs can be used as tumor suppressors due to their function in neoplastic development (194). In HNC, the expression of numerous miRNAs is associated with poor prognosis, decreased survival time, metastasis, and other tumor processes. Thus, miR34 and miR17–92 overexpression and miR137 underexpression are involved in apoptosis. miR210 overexpression and miR29 underexpression are associated with genetic instability, miR21 overexpression, and miR210 underexpression are involved in evasion of the immune response, miR26, and miR218 underexpression are associated with inflammation. miR26, and miR125b underexpression are involved in metabolism, overexpression of miR21 and miR155 and underexpression of miR29 and miR139 promote proliferation. Overexpression of miR26, miR125b, miR200b, miR96 and let-7d and underexpression of miR139, miR218, miR29 and miR200 are associated with metastasis. Overexpression of miR31, miR96, miR205 and miR96 and underexpression of miR210, miR125b and let-7d induce resistance to radiotherapy and chemotherapy (204). Polymorphisms in miR-146, miR-149, miR-196, and miR-499 may increase the risk of non-smokers infected with HPV, overexpression of miR-21, miR-181b, miR-184, and miR-345 is associated with malignant transformation, and overexpression of miR-21, miR-34c, 184 and miR-155 promotes proliferation and evasion of apoptosis (205), with miR-21 targeting the tumor suppressor genes PTEN (Phosphatase and Tensin Homolog) and PDCD4 (Programmed Cell Death 4) in some cancers (206).
Long ncRNAs are sequences of more than 200 nucleotides that carry methyl-guanosine ends, are often spliced or polyadenylated, and may be involved in chromatin remodeling. Being regulated by associated transcription factors, they control the transcription or can guide chromatin modification complexes to bind to specific loci, silencing or activating gene expression (194). For example, MIR31HG lncRNA appears to promote HIF1A and P21 expression, inducing proliferation and tumorigenesis, and LINC00460 lncRNA promotes the proliferation of HNSCC cells and epithelial-mesenchymal transition-mediated metastasis. On the other hand, SLC26A4-AS1 lncRNA interferes with cell invasiveness, migration, and metastasis, with a tumor suppressor role (207)
Besides their utility as biomarkers, ncRNAs are also very good therapeutic targets because they interact with numerous molecules when altering different cellular processes within the tumor microenvironment (208). The miRNA-based treatment relies on the premise that diseases disrupt the miRNA profiles which can be restored to normal (208).
Interestingly, the use of anti-miRNAs (miRNA sponges, miRNA masks, or miRNA antagonists) to deplete oncogenic miRNA as well as of miRNA mimetic molecules to simulate endogenous tumor suppressor miRNAs, showed promising results in limiting cancer cell growth in different HNC experimental models (208–211). Several ncRNA therapeutics have reached clinical trials in other malignancies. For instance, MRG-106 (cobomarsen), a miR-155 inhibitor, showed remarkable efficiency and tolerability in a phase I clinical trial (NCT02580552) involving 15 patients with cutaneous T-cell lymphoma (212). Therefore, although a nascent field of research, administering certain RNA-based formulations alone or in conjunction with systemic therapies seems to be a promising strategy for combating the burden of HNC.
The highly heterogeneous nature of HNC poses significant challenges in patient management due to cellular origin and anatomical site diversity, multiple etiological factors, and often late-stage diagnosis, which limits therapeutic options and affects survival and quality of life. Recent advancements in understanding the pathogenesis and drug resistance mechanisms have led to the development of various therapies.
Chemotherapy, chemoradiotherapy, targeted therapy, and immunotherapy show varied efficacy based on HNC stage, comorbidities, age, and previous treatments. New small molecule inhibitors, developed as monotherapies or in combination with other treatments, have shown promising results with moderate adverse effects by targeting specific gene expressions. These inhibitors have demonstrated fewer side effects compared to traditional therapies like chemotherapy and radiotherapy, enhancing patient tolerance. Notable drugs, such as the EGFR-directed monoclonal antibody cetuximab, pembrolizumab, and nivolumab, have achieved full FDA approval.
Despite these advancements, the complex interplay of multiple cell-signaling pathways limits therapeutic responses. Continuous clinical trials are necessary to confirm the effectiveness of these therapies in diverse patient groups and stages of HNC, and to identify suitable prognostic biomarkers for better therapeutic strategies. Comprehensive genomic sequencing studies have revealed numerous mutations in key signaling pathways, highlighting the potential of ncRNAs as biomarkers in HNC management.
Further improvements in treatment responses are needed, and the clinical translation of new inhibitors remains crucial. Combining these new agents with traditional treatments holds significant potential. Advances in molecular approaches are expected to enhance the success rate of targeted therapies, offering better evaluations of their efficacy and opening new research directions in personalized medicine for HNC diagnosis and treatment.
Ongoing studies aim to refine the use of novel compounds in therapeutic strategies, enabling precise identification of patients likely to benefit from these treatments, thus improving outcomes and innovating HNC treatment approaches.
MC: Writing – original draft, Writing – review & editing. MCC: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. CB: Writing – review & editing. COV: Writing – original draft, Writing – review & editing. REC: Writing – original draft, Writing – review & editing. SB: Writing – review & editing. RG: Writing – review & editing. GB: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge the financial support of C1.2.PFE-CDI.2021-587/ Contract no. 41PFE/30.12.2021; CNFIS-FDI-2024-F-0484; “The core program within the National Research Development and Innovation Plan, 2022–2027”, carried out with the support of the Ministry of Research, Innovation and Digitalization (MCID), project no. 23020101, Contract no. 7N from 3 January 2023; Project No. RO1567-IBB05/2023 from the Institute of Biology of the Romanian Academy; PN-III-P4-PCE-2021-0549 awarded by Romanian Executive Agency for Higher Education, Research, Development, and Innovation, and the support of the MRID, project PNRR-I8 no 842027778, contract no 760096. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Iftikhar A, Islam M, Shepherd S, Jones S, Ellis I. What is behind the lifestyle risk factors for head and neck cancer? Front Psychol. (2022) 13:960638. doi: 10.3389/fpsyg.2022.960638
2. Farah CS. Molecular landscape of head and neck cancer and implications for therapy. Ann Transl Med. (2021) 9:915. doi: 10.21037/atm-20–6264
3. Gormley M, Creaney G, Schache A, Ingarfield K, Conway DI. Reviewing the epidemiology of head and neck cancer: definitions, trends and risk factors. Br Dent J. (2022) 233:780–6. doi: 10.1038/s41415-022-5166-x
4. Miserocchi G, Spadazzi C, Calpona S, De Rosa F, Usai A, De Vita A, et al. Precision medicine in head and neck cancers: genomic and preclinical approaches. J Pers Med. (2022) 12:854. doi: 10.3390/jpm12060854
5. Moyses RA, López RV, Cury PM, Siqueira SA, Curioni OA, Gois Filho JF, et al. Significant differences in demographic, clinical, and pathological features in relation to smoking and alcohol consumption among 1,633 head and neck cancer patients. Clinics (Sao Paulo). (2013) 68:738–44. doi: 10.6061/clinics/2013(06)03
6. Chang CP, Siwakoti B, Sapkota A, Gautam DK, Lee YA, Monroe M, et al. Tobacco smoking, chewing habits, alcohol drinking and the risk of head and neck cancer in Nepal. Int J Cancer. (2020) 147:866–75. doi: 10.1002/ijc.32823
7. Hashibe M, Boffetta P, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Mates D, et al. Contribution of tobacco and alcohol to the high rates of squamous cell carcinoma of the supraglottis and glottis in Central Europe. Am J Epidemiol. (2007) 165:814–20. doi: 10.1093/aje/kwk066
8. Castellsagué X, Alemany L, Quer M, Halec G, Quirós B, Tous S, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Institute. (2016) 108:djv403. doi: 10.1093/jnci/djv403
9. Aboagye E, Agyemang-Yeboah F, Duduyemi BM, Obirikorang C. Human papillomavirus detection in head and neck squamous cell carcinomas at a tertiary hospital in sub-saharan africa. TheScientificWorldJournal. (2019) 2561530. doi: 10.1155/2019/2561530
10. de Sanjosé S, Serrano B, Tous S, Alejo M, Lloveras B, Quirós B, et al. Burden of human papillomavirus (HPV)-related cancers attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. (2019) 2:pky045. doi: 10.1093/jncics/pky045
11. Bulane A, Goedhals D, Seedat RY, Goedhals J, Burt F. Human papillomavirus DNA in head and neck squamous cell carcinomas in the Free State, South Africa. J Med Virol. (2020) 92:227–33. doi: 10.1002/jmv.25556
12. Sabatini ME, Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer. (2020) 122:306–14. doi: 10.1038/s41416–019-0602–7
13. Mahmutović L, Bilajac E, Hromić-Jahjefendić A. Meet the insidious players: review of viral infections in head and neck cancer etiology with an update on clinical trials. Microorganisms. (2021) 9:1001. doi: 10.3390/microorganisms9051001
14. Vani NV, Madhanagopal R, Swaminathan R, Ganesan TS. Dynamics of oral human papillomavirus infection in healthy population and head and neck cancer. Cancer Med. (2023) 12:11731–45. doi: 10.1002/cam4.5686
15. Prabhu SR, Wilson DF. Evidence of Epstein-Barr virus association with head and neck cancers: a review. J Can Dent Assoc. (2016) 82:g2.
16. Mulder FJ, Klufah F, Janssen FME, Farshadpour F, Willems SM, de Bree R, et al. Presence of human papillomavirus and epstein-barr virus, but absence of merkel cell polyomavirus, in head and neck cancer of non-smokers and non-drinkers. Front Oncol. (2021) 10:560434. doi: 10.3389/fonc.2020.560434
17. Eells AC, Mackintosh C, Marks L, Marino MJ. Gastroesophageal reflux disease and head and neck cancers: A systematic review and meta-analysis. Am J Otolaryngol. (2020) 41:102653. doi: 10.1016/j.amjoto.2020.102653
18. Sharan RN, Mehrotra R, Choudhury Y, Asotra K. Association of betel nut with carcinogenesis: revisit with a clinical perspective. PloS One. (2012) 7:e42759. doi: 10.1371/journal.pone.0042759
19. Yu MZ, Wu MM, Chien HT, Liao CT, Su MJ, Huang SF, et al. Risk prediction models for patients with head and neck cancer among the Taiwanese population. Cancers (Basel). (2022) 14:5338. doi: 10.3390/cancers14215338
20. Chang CC, Lee WT, Hsiao JR, Ou CY, Huang CC, Tsai ST, et al. Oral hygiene and the overall survival of head and neck cancer patients. Cancer Med. (2019) 8:1854–64. doi: 10.1002/cam4.2059
21. Pandey D, Szczesniak M, Maclean J, Yim HCH, Zhang F, Graham P, et al. Dysbiosis in head and neck cancer: determining optimal sampling site for oral microbiome collection. Pathogens. (2022) 11:1550. doi: 10.3390/pathogens11121550
22. Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. (2008) 371:1695–709. doi: 10.1016/S0140-6736(08)60728-X
23. Mazul AL, Shivappa N, Hébert JR, Steck SE, Rodriguez-Ormaza N, Weissler M, et al. Proinflammatory diet is associated with increased risk of squamous cell head and neck cancer. Int J Cancer. (2018) 143:1604–10. doi: 10.1002/ijc.31555
24. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. (2020) 6:92. doi: 10.1038/s41572–020-00224–3
25. Constantin M. Epidemiology, diagnosis, symptoms and TNM classification of head and neck cancers. Rom Biotechnol Lett. (2022) 27:3699–712. doi: 10.25083/rbl/27.5/3699.3712
26. Aung KL, Siu LL. Genomically personalized therapy in head and neck cancer. Cancers Head Neck. (2016) 1:2. doi: 10.1186/s41199-016-0004-y
27. Ali J, Sabiha B, Jan HU, Haider SA, Khan AA, Ali SS. Genetic etiology of oral cancer. Oral Oncol. (2017) 70:23–8. doi: 10.1016/j.oraloncology.2017.05.004
28. Huang Y, Zhao J, Sanghee MG, Lee G, Zhang J, Bi J, et al. Identification of novel genetic variants predisposing to familial oral squamous cell carcinomas. Cell Discovery. (2019) 5:57. doi: 10.1038/s41421–019-0126–6
29. Mali SB. Epigenetics: Promising journey so far but ways to go in head neck cancer. Oral Oncol. (2022) 135:106194. doi: 10.1016/j.oraloncology.2022.106194
30. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. (2018) 18:269–82. doi: 10.1038/nrc.2018.11
31. Economopoulou P, de Bree R, Kotsantis I, Psyrri A. Diagnostic tumor markers in head and neck squamous cell carcinoma (HNSCC) in the clinical setting. Front Oncol. (2019) 9:827. doi: 10.3389/fonc.2019.00827
32. Haider SP, Burtness B, Yarbrough WG, Payabvash S. Applications of radiomics in precision diagnosis, prognostication and treatment planning of head and neck squamous cell carcinomas. Cancers Head Neck. (2020) 5:6. doi: 10.1186/s41199–020-00053–7
33. Kordbacheh F, Farah CS. Molecular pathways and druggable targets in head and neck squamous cell carcinoma. Cancers (Basel). (2021) 13:3453. doi: 10.3390/cancers13143453
34. Bernacchi V, DeGuzman PB. Cancer-related distress: symptom clusters in rural head and neck cancer survivors. Clin J Oncol Nurs. (2023) 27:55–61. doi: 10.1188/23.CJON.55–61
35. Van den Bossche V, Zaryouh H, Vara-Messler M, Vignau J, Machiels JP, Wouters A, et al. Microenvironment-driven intratumoral heterogeneity in head and neck cancers: clinical challenges and opportunities for precision medicine. Drug Resist Updat. (2022) 60:100806. doi: 10.1016/j.drup.2022.100806
36. Poomsawat S, Sanguansin S, Punyasingh J, Vejchapipat P, Punyarit P. Expression of cdk6 in head and neck squamous cell carcinoma. Clin Oral Investig. (2016) 20:57–63. doi: 10.1007/s00784–015-1482–8
37. Angadi PV, Krishnapillai R. Cyclin D1 expression in oral squamous cell carcinoma and verrucous carcinoma: correlation with histological differentiation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2007) 103:e30–5. doi: 10.1016/j.tripleo.2006.09.011
38. Goel B, Tiwari AK, Pandey RK, Singh AP, Kumar S, Sinha A, et al. Therapeutic approaches for the treatment of head and neck squamous cell carcinoma-An update on clinical trials. Transl Oncol. (2022) 21:101426. doi: 10.1016/j.tranon.2022.101426
39. Samra B, Tam E, Baseri B, Shapira I. Checkpoint inhibitors in head and neck cancer: current knowledge and perspectives. J Invest. Med. (2018) 66:1023–30. doi: 10.1136/jim-2018–000743
40. Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G Jr, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. (2019) 394:1915–28. doi: 10.1016/S0140–6736(19)32591–7
41. De Felice F, Cattaneo CG, Franco P. Radiotherapy and systemic therapies: focus on head and neck cancer. Cancers (Basel). (2023) 15:4232. doi: 10.3390/cancers15174232
42. Li Q, Tie Y, Alu A, Ma X, Shi H. Targeted therapy for head and neck cancer: signaling pathways and clinical studies. Signal Transduct Target Ther. (2023) 8:31. doi: 10.1038/s41392–022-01297–0
43. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. (2013) 502:333–9. doi: 10.1038/nature12634
44. Lyu H, Li M, Jiang Z, Liu Z, Wang X. Correlate the TP53 mutation and the HRAS mutation with immune signatures in head and neck squamous cell cancer. Comput Struct Biotechnol J. (2019) 17:1020–30. doi: 10.1016/j.csbj.2019.07.009
45. Klinakis A, Rampias T. TP53 mutational landscape of metastatic head and neck cancer reveals patterns of mutation selection. EBioMedicine. (2020) 58:102905. doi: 10.1016/j.ebiom.2020.102905
46. Basyuni S, Nugent G, Ferro A, Barker E, Reddin I, Jones O, et al. Value of p53 sequencing in the prognostication of head and neck cancer: a systematic review and meta-analysis. Sci Rep. (2022) 12:20776. doi: 10.1038/s41598–022-25291–2
47. Nathan CA, Khandelwal AR, Wolf GT, Rodrigo JP, Mäkitie AA, Saba NF, et al. TP53 mutations in head and neck cancer. Mol Carcinog. (2022) 61:385–91. doi: 10.1002/mc.23385
48. Mandigo AC, Tomlins SA, Kelly WK, Knudsen KE. Relevance of pRB loss in human Malignancies. Clin Cancer Res. (2022) 28:255–64. doi: 10.1158/1078–0432.CCR-21–1565
49. Montalto FI, De Amicis F. Cyclin D1 in cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells. (2020) 9:2648. doi: 10.3390/cells9122648
50. Wang J, Su W, Zhang T, Zhang S, Lei H, Ma F, et al. Aberrant Cyclin D1 splicing in cancer: from molecular mechanism to therapeutic modulation. Cell Death Dis. (2023) 14:244. doi: 10.1038/s41419–023-05763–7
51. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. (2015) 517:576–82. doi: 10.1038/nature14129
52. Golabek K, Rączka G, Gaździcka J, Miśkiewicz-Orczyk K, Zięba N, Krakowczyk Ł, et al. Expression profiles of CDKN2A, MDM2, E2F2 and LTF genes in oral squamous cell carcinoma. Biomedicines. (2022) 10:3011. doi: 10.3390/biomedicines10123011
53. Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. (2006) 22:719–29. doi: 10.1016/j.molcel.2006.05.022
54. Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. (2015) 21:632–41. doi: 10.1158/1078–0432.CCR-13–3310
55. Feldman R, Gatalica Z, Knezetic J, Reddy S, Nathan CA, Javadi N, et al. Molecular profiling of head and neck squamous cell carcinoma. Head Neck. (2016) 38 Suppl 1:E1625–38. doi: 10.1002/hed.24290
56. Papalouka C, Adamaki M, Batsaki P, Zoumpourlis P, Tsintarakis A, Goulielmaki M, et al. DNA damage response mechanisms in head and neck cancer: significant implications for therapy and survival. Int J Mol Sci. (2023) 24:2760. doi: 10.3390/ijms24032760
57. Wang YC, Lee KW, Tsai YS, Lu HH, Chen SY, Hsieh HY, et al. Downregulation of ATM and BRCA1 predicts poor outcome in head and neck cancer: implications for ATM-targeted therapy. J Pers Med. (2021) 11:389. doi: 10.3390/jpm11050389
58. Weber A, Langhanki L, Sommerer F, Markwarth A, Wittekind C, Tannapfel A. Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene. (2003) 22:4757–9. doi: 10.1038/sj.onc.1206705
59. McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, et al. Roles of the Raf/MEK/ERK pathway in cell growth, Malignant transformation and drug resistance. Biochim Biophys Acta. (2007) 1773:1263–84. doi: 10.1016/j.bbamcr.2006.10.001
60. Roy M, Datta A. Cancer: genetics and important pathways. In: Cancer genetics and therapeutics. Springer, Singapore (2019). doi: 10.1007/978-981-13-9471-3
61. KEGG. Pathways in cancer - Reference pathway (2020). Available online at: https://www.genome.jp/kegg-bin/show_pathway?map05200 (Accessed 2023 Ian 09).
62. Rong C, Muller MF, Xiang F, Jensen A, Weichert W, Major G, et al. Adaptive ERK signaling activation in response to therapy and in silico prognostic evaluation of EGFR-MAPK in HNSCC. Br J Cancer. (2020) 123:288–97. doi: 10.1038/s41416–020-0892–9
63. Dillon M, Lopez A, Lin E, Sales D, Perets R, Jain P. Progress on ras/MAPK signaling research and targeting in blood and solid cancers. Cancers (Basel). (2021) 13:5059. doi: 10.3390/cancers13205059
64. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. (2018) 173:321–337.e10. doi: 10.1016/j.cell.2018.03.035
65. Kazemein Jasemi NS, Ahmadian MR. Allosteric regulation of GRB2 modulates RAS activation. Small GTPases. (2022) 13:282–6. doi: 10.1080/21541248.2022.2089001
66. Fernandez-Medarde A, De Las Rivas J, Santos E. 40 years of RAS-A historic overview. Genes (Basel). (2021) 12:681. doi: 10.3390/genes12050681
67. Antra P, Hungyo H, Jain A, Ahmad S, Tandon V. Unraveling molecular mechanisms of head and neck cancer. Crit Rev Oncol Hematol. (2022) 178:103778. doi: 10.1016/j.critrevonc.2022.103778
68. Sasaki E, Masago K, Fujita S, Hanai N, Yatabe Y. Frequent KRAS and HRAS mutations in squamous cell papillomas of the head and neck. J Pathol Clin Res. (2020) 6:154–9. doi: 10.1002/cjp2.157
69. Novoplansky O, Jagadeeshan S, Regev O, Menashe I, Elkabets M. Worldwide prevalence and clinical characteristics of RAS mutations in head and neck cancer: A systematic review and meta-analysis. Front Oncol. (2022) 12:838911. doi: 10.3389/fonc.2022.838911
70. Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL. ERK/MAPK signaling pathway and tumorigenesis. Exp Ther Med. (2020) 19:1997–2007. doi: 10.3892/etm.2020.8454
71. Alsahafi EN, Thavaraj S, Sarvestani N, Novoplansky O, Elkabets M, Ayaz B, et al. EGFR overexpression increases radiotherapy response in HPV-positive head and neck cancer through inhibition of DNA damage repair and HPV E6 downregulation. Cancer Lett. (2021) 498:80–97. doi: 10.1016/j.canlet.2020.10.035
72. Ngan HL, Law CH, Choi YCY, Chan JY, Lui VWY. Precision drugging of the MAPK pathway in head and neck cancer. NPJ Genom Med. (2022) 7:20. doi: 10.1038/s41525–022-00293–1
73. Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. (2013) 503:548–51. doi: 10.1038/nature12796
74. Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discovery. (2016) 6:316–29. doi: 10.1158/2159–8290.CD-15–1105
75. Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. (2018) 172:578–589.e17. doi: 10.1016/j.cell.2018.01.006
76. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. (2019) 575:217–23. doi: 10.1038/s41586–019-1694–1
77. Arbour KC, Khurana M, Dai T, Skoulidis F. Trial in progress: A phase 2 study of sotorasib as first-line treatment in patients with stage IV non–small cell lung cancer (NSCLC) whose tumors harbor a KRAS p.G12C mutation (CodeBreaK 201). JCO. (2022) 40:TPS9150–TPS9150. doi: 10.1200/JCO.2022.40.16_suppl.TPS9150
78. Giralt J, Trigo J, Nuyts S, Ozsahin M, Skladowski K, Hatoum G, et al. Panitumumab plus radiotherapy versus chemoradiotherapy in patients with unresected, locally advanced squamous-cell carcinoma of the head and neck (CONCERT-2): a randomised, controlled, open-label phase 2 trial. Lancet Oncol. (2015) 16:221–32. doi: 10.1016/S1470–2045(14)71200–8
79. Ringash J, Waldron JN, Siu LL, Martino R, Winquist E, Wright JR, et al. Quality of life and swallowing with standard chemoradiotherapy versus accelerated radiotherapy and panitumumab in locoregionally advanced carcinoma of the head and neck: A phase III randomised trial from the Canadian Cancer Trials Group (HN. 6). Eur J Cancer. (2017) 72:192–9. doi: 10.1016/j.ejca.2016.11.008
80. Irshad R, Haider G, Hashmi M, Hassan A. Efficacy of gefitinib and methorexate in patients with advanced stage and recurrent head and neck cancer. Cureus. (2021) 13:e15451. doi: 10.7759/cureus.15451
81. Baa AK, Sharma A, Bhaskar S, Biswas A, JeeBharti S, Thakar A, et al. A single-arm feasibility phase II study of EMF (erlotinib + methotrexate + 5-fluorouracil) regimen in platinum-refractory recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC). Ecancermedicalscience. (2022) 16:1451. doi: 10.3332/ecancer.2022.1451
82. Kao HF, Liao BC, Huang YL, Huang HC, Chen CN, Chen TC, et al. Afatinib and pembrolizumab for recurrent or metastatic head and neck squamous cell carcinoma (ALPHA study): A phase II study with biomarker analysis. Clin Cancer Res. (2022) 28:1560–71. doi: 10.1158/1078–0432.CCR-21–3025
83. Evrard D, Dumont C, Gatineau M, Delord JP, Fayette J, Dreyer C, et al. Targeting the Tumor Microenvironment through mTOR Inhibition and Chemotherapy as Induction Therapy for Locally Advanced Head and Neck Squamous Cell Carcinoma: The CAPRA Study. Cancers (Basel). (2022) 14:4509. doi: 10.3390/cancers14184509
84. Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez SM, et al. Phase I dose-escalation study of taselisib, an oral PI3K inhibitor, in patients with advanced solid tumors. Cancer Discovery. (2017) 7:704–15. doi: 10.1158/2159–8290.CD-16–1080
85. Kim HR, Kang HN, Yun MR, Ju KY, Choi JW, Jung DM, et al. Mouse-human co-clinical trials demonstrate superior anti-tumour effects of buparlisib (BKM120) and cetuximab combination in squamous cell carcinoma of head and neck. Br J Cancer. (2020) 123:1720–9. doi: 10.1038/s41416–020-01074–2
86. Rodon J, Argilés G, Connolly RM, Vaishampayan U, de Jonge M, Garralda E, et al. Phase 1 study of single-agent WNT974, a first-in-class Porcupine inhibitor, in patients with advanced solid tumours. Br J Cancer. (2021) 125:28–37. doi: 10.1038/s41416–021-01389–8
87. Qureshy Z, Li H, Zeng Y, Rivera J, Cheng N, Peterson CN, et al. STAT3 activation as a predictive biomarker for ruxolitinib response in head and neck cancer. Clin Cancer Res. (2022) 28:4737–46. doi: 10.1158/1078–0432.CCR-22–0744
88. Munster P, Iannotti N, Cho DC, Kirkwood JM, Villaruz LC, Gibney GT, et al. Combination of itacitinib or parsaclisib with pembrolizumab in patients with advanced solid tumors: A phase I study. Cancer Res Commun. (2023) 3:2572–84. doi: 10.1158/2767–9764.CRC-22–0461
89. Azaro A, Massard C, Tap WD, Cassier PA, Merchan J, Italiano A, et al. A phase 1b study of the Notch inhibitor crenigacestat (LY3039478) in combination with other anticancer target agents (taladegib, LY3023414, or abemaciclib) in patients with advanced or metastatic solid tumors. Invest New Drugs. (2021) 39:1089–98. doi: 10.1007/s10637–021-01094–6
90. Argiris A, Kotsakis AP, Hoang T, Worden FP, Savvides P, Gibson MK, et al. Cetuximab and bevacizumab: preclinical data and phase II trial in recurrent or metastatic squamous cell carcinoma of the head and neck. Ann Oncol. (2013) 24:220–5. doi: 10.1093/annonc/mds245
91. Argiris A, Li S, Savvides P, Ohr JP, Gilbert J, Levine MA, et al. Phase III randomized trial of chemotherapy with or without bevacizumab in patients with recurrent or metastatic head and neck cancer. J Clin Oncol. (2019) 37:3266–74. doi: 10.1200/JCO.19.00555
92. Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discovery. (2020) 10:54–71. doi: 10.1158/2159–8290.CD-19–1167
93. Fasano M, Della Corte CM, Viscardi G, Di Liello R, Paragliola F, Sparano F, et al. Head and neck cancer: the role of anti-EGFR agents in the era of immunotherapy. Ther Adv Med Oncol. (2021) 13:1758835920949418. doi: 10.1177/1758835920949418
94. Azoury SC, Gilmore RC, Shukla V. Molecularly targeted agents and immunotherapy for the treatment of head and neck squamous cell cancer (HNSCC). Discovery Med. (2016) 21:507–16.
95. Omura G, Honma Y, Matsumoto Y, Shinozaki T, Itoyama M, Eguchi K, et al. Transnasal photoimmunotherapy with cetuximab sarotalocan sodium: Outcomes on the local recurrence of nasopharyngeal squamous cell carcinoma. Auris Nasus Larynx. (2023) 50:641–5. doi: 10.1016/j.anl.2022.06.004
96. Taberna M, Oliva M, Mesía R. Cetuximab-containing combinations in locally advanced and recurrent or metastatic head and neck squamous cell carcinoma. Front Oncol. (2019) 9:383. doi: 10.3389/fonc.2019.00383
97. Chen TH, Pan YY, Lee TL, Wang LW, Tai SK, Chu PY, et al. Treatment outcomes of cetuximab-containing regimen in locoregional recurrent and distant metastatic head and neck squamous cell carcinoma. BMC Cancer. (2022) 22:1336. doi: 10.1186/s12885–022-10440–7
98. Chu PL, Shihabuddeen WA, Low KP, Poon DJJ, Ramaswamy B, Liang ZG, et al. Vandetanib sensitizes head and neck squamous cell carcinoma to photodynamic therapy through modulation of EGFR-dependent DNA repair and the tumour microenvironment. Photodiagnosis Photodyn Ther. (2019) 27:367–74. doi: 10.1016/j.pdpdt.2019.06.008
99. Hintelmann K, Kriegs M, Rothkamm K, Rieckmann T. Improving the efficacy of tumor radiosensitization through combined molecular targeting. Front Oncol. (2020) 10:1260. doi: 10.3389/fonc.2020.01260
100. Mesia R, Henke M, Fortin A, Minn H, Yunes Ancona AC, Cmelak A, et al. Chemoradiotherapy with or without panitumumab in patients with unresected, locally advanced squamous-cell carcinoma of the head and neck (CONCERT-1): a randomised, controlled, open-label phase 2 trial. Lancet Oncol. (2015) 16:208–20. doi: 10.1016/S1470–2045(14)71198–2
101. Reddy BK, Lokesh V, Vidyasagar MS, Shenoy K, Babu KG, Shenoy A, et al. Nimotuzumab provides survival benefit to patients with inoperable advanced squamous cell carcinoma of the head and neck: a randomized, open-label, phase IIb, 5-year study in Indian patients. Oral Oncol. (2014) 50:498–505. doi: 10.1016/j.oraloncology.2013.11.008
102. Menon N, Patil V, Noronha V, Joshi A, Bhattacharjee A, Satam BJ, et al. Quality of life in patients with locally advanced head and neck cancer treated with concurrent chemoradiation with cisplatin and nimotuzumab versus cisplatin alone - Additional data from a phase 3 trial. Oral Oncol. (2021) 122:105517. doi: 10.1016/j.oraloncology.2021.105517
103. Machiels JP, Subramanian S, Ruzsa A, Repassy G, Lifirenko I, Flygare A, et al. Zalutumumab plus best supportive care versus best supportive care alone in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck after failure of platinum-based chemotherapy: an open-label, randomised phase 3 trial. Lancet Oncol. (2011) 12:333–43. doi: 10.1016/S1470–2045(11)70034–1
104. Brondum L, Alsner J, Sørensen BS, Maare C, Johansen J, Primdahl H, et al. Associations between skin rash, treatment outcome, and single nucleotide polymorphisms in head and neck cancer patients receiving the EGFR-inhibitor zalutumumab: results from the DAHANCA 19 trial. Acta Oncol. (2018) 57:1159–64. doi: 10.1080/0284186X.2018.1464664
105. Tang X, He J, Li B, Zheng Y, Li K, Zou S, et al. Efficacy and safety of gefitinib in patients with advanced head and neck squamous cell carcinoma: A meta-analysis of randomized controlled trials. J Oncol. (2019) 2019:6273438. doi: 10.1155/2019/6273438
106. Jain AP, Radhakrishnan A, Pinto S, Patel K, Kumar M, Nanjappa V, et al. How to achieve therapeutic response in erlotinib-resistant head and neck squamous cell carcinoma? New insights from stable isotope labeling with amino acids in cell culture-based quantitative tyrosine phosphoproteomics. OMICS. (2021) 25:605–16. doi: 10.1089/omi.2021.0057
107. Harrington K, Berrier A, Robinson M, Remenar E, Housset M, de Mendoza FH, et al. Randomised Phase II study of oral lapatinib combined with chemoradiotherapy in patients with advanced squamous cell carcinoma of the head and neck: rationale for future randomised trials in human papilloma virus-negative disease. Eur J Cancer. (2013) 49:1609–18. doi: 10.1016/j.ejca.2012.11.023
108. Rascio F, Spadaccino F, Rocchetti MT, Castellano G, Stallone G, Netti GS, et al. The pathogenic role of PI3K/AKT pathway in cancer onset and drug resistance: an updated review. Cancers (Basel). (2021) 13:3949. doi: 10.3390/cancers13163949
109. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. (2019) 18:26. doi: 10.1186/s12943-019-0954-x
110. He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. (2021) 6:425. doi: 10.1038/s41392–021-00828–5
111. Khezri MR, Ghasemnejad-Berenji M, Moloodsouri D. Hesperetin and the PI3K/AKT pathway: Could their interaction play a role in the entry and replication of the SARS-CoV-2? J Food Biochem. (2022) 46:e14212. doi: 10.1111/jfbc.14212
112. Naderali E, Valipour B, Khaki AA, Soleymani Rad J, Alihemmati A, Rahmati M, et al. Positive effects of PI3K/akt signaling inhibition on PTEN and P53 in prevention of acute lymphoblastic leukemia tumor cells. Adv Pharm Bull. (2019) 9:470–80. doi: 10.15171/apb.2019.056
113. Nam HJ, Chae S, Jang SH, Cho H, Lee JH. The PI3K-Akt mediates oncogenic Met-induced centrosome amplification and chromosome instability. Carcinogenesis. (2010) 31:1531–40. doi: 10.1093/carcin/bgq133
114. Hervieu A, Kermorgant S. The role of PI3K in met driven cancer: A recap. Front Mol Biosci. (2018) 5:86. doi: 10.3389/fmolb.2018.00086
115. Gougis P, Moreau Bachelard C, Kamal M, Gan HK, Borcoman E, Torossian N, et al. Clinical development of molecular targeted therapy in head and neck squamous cell carcinoma. JNCI Cancer Spectr. (2019) 3:pkz055. doi: 10.1093/jncics/pkz055
116. Raj S, Kesari KK, Kumar A, Rathi B, Sharma A, Gupta PK, et al. Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in head and neck cancer. Mol Cancer. (2022) 21:31. doi: 10.1186/s12943–022-01503–1
117. Fujino T, Suda K, Koga T, Hamada A, Ohara S, Chiba M, et al. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J Hematol Oncol. (2022) 15:79. doi: 10.1186/s13045-022-01299-z
118. Khedkar HN, Chen LC, Kuo YC, Wu ATH, Huang HS. Multi-omics identification of genetic alterations in head and neck squamous cell carcinoma and therapeutic efficacy of HNC018 as a novel multi-target agent for c-MET/STAT3/AKT signaling axis. Int J Mol Sci. (2023) 24:10247. doi: 10.3390/ijms241210247
119. Hudeckova M, Koucký V, Rottenberg J, Gál B. Gene mutations in circulating tumour DNA as a diagnostic and prognostic marker in head and neck cancer-A systematic review. Biomedicines. (2021) 9:1548. doi: 10.3390/biomedicines9111548
120. Tsai PJ, Lai YH, Manne RK, Tsai YS, Sarbassov D, Lin HK, et al. Akt: a key transducer in cancer. J BioMed Sci. (2022) 29:76. doi: 10.1186/s12929–022-00860–9
121. Islam M, Jones S, Ellis I. Role of akt/protein kinase B in cancer metastasis. Biomedicines. (2023) 11:3001. doi: 10.3390/biomedicines11113001
122. Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, et al. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. (2005) 9:389–402. doi: 10.1016/j.devcel.2005.08.001
123. Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. (2010) 12:457–67. doi: 10.1038/ncb2047
124. Wise-Draper TM, Bahig H, Tonneau M, Karivedu V, Burtness B. Current therapy for metastatic head and neck cancer: evidence, opportunities, and challenges. Am Soc Clin Oncol Educ Book. (2022) 42:1–14. doi: 10.1200/EDBK_350442
125. Liu WL, Gao M, Tzen KY, Tsai CL, Hsu FM, Cheng AL, et al. Targeting Phosphatidylinositide3-Kinase/Akt pathway by BKM120 for radiosensitization in hepatocellular carcinoma. Oncotarget. (2014) 5:3662–72. doi: 10.18632/oncotarget.1978
126. Jhaveri K, Chang MT, Juric D, Saura C, Gambardella V, Melnyk A, et al. Phase I basket study of taselisib, an isoform-selective PI3K inhibitor, in patients with PIK3CA-mutant cancers. Clin Cancer Res. (2021) 27:447–59. doi: 10.1158/1078–0432.CCR-20–2657
127. Juric D, Rodon J, Tabernero J, Janku F, Burris HA, Schellens JHM, et al. Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: results from the first-in-human study. J Clin Oncol. (2018) 36:1291–9. doi: 10.1200/JCO.2017.72.7107
128. Batalini F, Xiong N, Tayob N, Polak M, Eismann J, Cantley LC, et al. Phase 1b clinical trial with alpelisib plus olaparib for patients with advanced triple-negative breast cancer. Clin Cancer Res. (2022) 28:1493–9. doi: 10.1158/1078–0432.CCR-21–3045
129. Ghosh S, Shah PA, Johnson FM. Novel systemic treatment modalities including immunotherapy and molecular targeted therapy for recurrent and metastatic head and neck squamous cell carcinoma. Int J Mol Sci. (2022) 23:7889. doi: 10.3390/ijms23147889
130. Fayette J, Digue L C, Ségura-Ferlay C, Treilleux I, Wang Q, Lefebvre G, et al. Buparlisib (BKM120) in refractory head and neck squamous cell carcinoma harbouring or not a PI3KCA mutation: A phase II multicenter trial. Ann Oncol. (2019) 30:v455. doi: 10.1093/annonc/mdz252.012
131. Soulieres D, Licitra L, Mesía R, Remenár É, Li SH, Karpenko A, et al. Molecular alterations and buparlisib efficacy in patients with squamous cell carcinoma of the head and neck: biomarker analysis from BERIL-1. Clin Cancer Res. (2018) 24:2505–16. doi: 10.1158/1078–0432.CCR-17–2644
132. Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. (2022) 7:3. doi: 10.1038/s41392–021-00762–6
133. Pascual-Carreras E, Marín-Barba M, Castillo-Lara S, Coronel-Córdoba P, Magri MS, Wheeler GN, et al. Wnt/β-catenin signalling is required for pole-specific chromatin remodeling during planarian regeneration. Nat Commun. (2023) 14:298. doi: 10.1038/s41467-023-35937-y
134. Werner J, Boonekamp KE, Zhan T, Boutros M. The roles of secreted wnt ligands in cancer. Int J Mol Sci. (2023) 24:5349. doi: 10.3390/ijms24065349
135. Chowdhury P, Nagamalini BR, Singh J, Ashwini BK, Sharada, Swaminathan U. Expression of β-catenin in oral leukoplakia and oral submucous fibrosis: An immunohistochemical study. J Oral Maxillofac Pathol. (2021) 25:124–30. doi: 10.4103/jomfp.JOMFP_41_20
136. Xie J, Huang L, Lu YG, Zheng DL. Roles of the wnt signaling pathway in head and neck squamous cell carcinoma. Front Mol Biosci. (2021) 7:590912. doi: 10.3389/fmolb.2020.590912
137. Prgomet Z, Andersson T, Lindberg P. Higher expression of WNT5A protein in oral squamous cell carcinoma compared with dysplasia and oral mucosa with a normal appearance. Eur J Oral Sci. (2017) 125:237–46. doi: 10.1111/eos.12352
138. Zhang W, Yan Y, Gu M, Wang X, Zhu H, Zhang S, et al. High expression levels of Wnt5a and Ror2 in laryngeal squamous cell carcinoma are associated with poor prognosis. Oncol Lett. (2017) 14:2232–8. doi: 10.3892/ol.2017.6386
139. Qin L, Yin YT, Zheng FJ, Peng LX, Yang CF, Bao YN, et al. WNT5A promotes stemness characteristics in nasopharyngeal carcinoma cells leading to metastasis and tumorigenesis. Oncotarget. (2015) 6:10239–52. doi: 10.18632/oncotarget.3518
140. Sakamoto T, Kawano S, Matsubara R, Goto Y, Jinno T, Maruse Y, et al. Critical roles of Wnt5a-Ror2 signaling in aggressiveness of tongue squamous cell carcinoma and production of matrix metalloproteinase-2 via ΔNp63β-mediated epithelial-mesenchymal transition. Oral Oncol. (2017) 69:15–25. doi: 10.1016/j.oraloncology.2017.03.019
141. Xie H, Ma Y, Li J, Chen H, Xie Y, Chen M, et al. WNT7A promotes EGF-induced migration of oral squamous cell carcinoma cells by activating β-catenin/MMP9-mediated signaling. Front Pharmacol. (2020) 11:98. doi: 10.3389/fphar.2020.00098
142. Shiah SG, Hsiao JR, Chang WM, Chen YW, Jin YT, Wong TY, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. (2014) 74:7560–72. doi: 10.1158/0008–5472.CAN-14–0978
143. Purwaningsih NMS, Khor GH, Nik Mohd Rosdy NMM, Abdul Rahman EO. Wnt pathway in oral cancer: A review update. Saudi Dent J. (2021) 33:813–8. doi: 10.1016/j.sdentj.2021.08.002
144. Aditya J, Smiline Girija AS, Paramasivam A, Vijayashree Priyadharsini J. Genetic alterations in Wnt family of genes and their putative association with head and neck squamous cell carcinoma. Genomics Inform. (2021) 19:e5. doi: 10.5808/gi.20065
145. Gao X, Hannoush RN. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat Chem Biol. (2014) 10:61–8. doi: 10.1038/nchembio.1392
146. Galli LM, Anderson MO, Gabriel Fraley J, Sanchez L, Bueno R, Hernandez DN, et al. Determination of the membrane topology of PORCN, an O-acyl transferase that modifies Wnt signalling proteins. Open Biol. (2021) 11:200400. doi: 10.1098/rsob.200400
147. Flanagan DJ, Woodcock SA, Phillips C, Eagle C, Sansom OJ. Targeting ligand-dependent wnt pathway dysregulation in gastrointestinal cancers through porcupine inhibition. Pharmacol Ther. (2022) 238:108179. doi: 10.1016/j.pharmthera.2022.108179
148. Le PN, McDermott JD, Jimeno A. Targeting the Wnt pathway in human cancers: therapeutic targeting with a focus on OMP-54F28. Pharmacol Ther. (2015) 146:1–11. doi: 10.1016/j.pharmthera.2014.08.005
149. Diamond JR, Becerra C, Richards D, Mita A, Osborne C, O’Shaughnessy J, et al. Phase Ib clinical trial of the anti-frizzled antibody vantictumab (OMP-18R5) plus paclitaxel in patients with locally advanced or metastatic HER2-negative breast cancer. Breast Cancer Res Treat. (2020) 184:53–62. doi: 10.1007/s10549-020-05817-w
150. Pećina-Šlaus N, Aničić S, Bukovac A, Kafka A. Wnt signaling inhibitors and their promising role in tumor treatment. Int J Mol Sci. (2023) 24:6733. doi: 10.3390/ijms24076733
151. Lai SY, Johnson FM. Defining the role of the JAK–STAT pathway in head and neck and thoracic Malignancies: implications for future therapeutic approaches. Drug Resist Updat. (2010) 13:67–78. doi: 10.1016/j.drup.2010.04.001
152. Sen M, Pollock NI, Black J, DeGrave KA, Wheeler S, Freilino ML, et al. JAK kinase inhibition abrogates STAT3 activation and head and neck squamous cell carcinoma tumor growth. Neoplasia. (2015) 17:256–64. doi: 10.1016/j.neo.2015.01.003
153. Cedars E, Johnson DE, Grandis JR. Jak/STAT signaling in head and neck cancer. Curr Cancer Res. (2018), 155–84. doi: 10.1007/978–3-319–78762-6_6
154. Gutiérrez-Hoya A, Soto-Cruz I. Role of the JAK/STAT pathway in cervical cancer: its relationship with HPV E6/E7 oncoproteins. Cells (2020) 9:2297. doi: 10.3390/cells9102297
155. Majoros A, Platanitis E, Kernbauer-Hölzl E, Rosebrock F, Müller M, Decker T. Canonical and non-canonical aspects of JAK–STAT signaling: lessons from interferons for cytokine responses. Front Immunol. (2017) 8:29. doi: 10.3389/fimmu.2017.00029
156. Anderson K, Ryan N, Nedungadi D, Lamenza F, Swingler M, Siddiqui A, et al. STAT1 is regulated by TRIM24 and promotes immunosuppression in head and neck squamous carcinoma cells, but enhances T cell antitumour immunity in the tumour microenvironment. Br J Cancer. (2022) 127:624–36. doi: 10.1038/s41416-022-01853-z
157. Nakagawa T, Oda G, Kawachi H, Ishikawa T, Okamoto K, Uetake H. Nuclear expression of p-STAT3 is associated with poor prognosis in ER(-) breast cancer. Clin Pract. (2022) 12:157–67. doi: 10.3390/clinpract12020020
158. Manuelli V, Pecorari C, Filomeni G, Zito E. Regulation of redox signaling in HIF-1-dependent tumor angiogenesis. FEBS J. (2022) 289:5413–25. doi: 10.1111/febs.16110
159. Park H, Lee S, Lee J, Moon H, Ro SW. Exploring the JAK/STAT signaling pathway in hepatocellular carcinoma: unraveling signaling complexity and therapeutic implications. Int J Mol Sci. (2023) 24:13764. doi: 10.3390/ijms241813764
160. Rah B, Rather RA, Bhat GR, Baba AB, Mushtaq I, Farooq M, et al. JAK/STAT signaling: molecular targets, therapeutic opportunities, and limitations of targeted inhibitions in solid Malignancies. Front Pharmacol. (2022) 13:821344. doi: 10.3389/fphar.2022.821344
161. Hu Q, Bian Q, Rong D, Wang L, Song J, Huang HS, et al. JAK/STAT pathway: Extracellular signals, diseases, immunity, and therapeutic regimens. Front Bioeng Biotechnol. (2023) 11:1110765. doi: 10.3389/fbioe.2023.1110765
162. Kowshik J, Baba AB, Giri H, Deepak Reddy G, Dixit M, Nagini S. Astaxanthin inhibits JAK/STAT-3 signaling to abrogate cell proliferation, invasion and angiogenesis in a hamster model of oral cancer. PloS One. (2014) 9:e109114. doi: 10.1371/journal.pone.0109114
163. Stover DG, Gil Del Alcazar CR, Brock J, Guo H, Overmoyer B, Balko J, et al. Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer. (2018) 4:10. doi: 10.1038/s41523-018-0060-z
164. von Bubnoff N, Ihorst G, Grishina O, Röthling N, Bertz H, Duyster J, et al. Ruxolitinib in GvHD (RIG) study: a multicenter, randomized phase 2 trial to determine the response rate of Ruxolitinib and best available treatment (BAT) versus BAT in steroid-refractory acute graft-versus-host disease (aGvHD) (NCT02396628). BMC Cancer. (2018) 18:1132. doi: 10.1186/s12885–018-5045–7
165. Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. (2011) 138:3593–612. doi: 10.1242/dev.063610
166. Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. (2014) 74:1091–104. doi: 10.1158/0008–5472.CAN-13–1259
167. Weaver AN, Burch MB, Cooper TS, Della Manna DL, Wei S, Ojesina AI, et al. Notch signaling activation is associated with patient mortality and increased FGF1-mediated invasion in squamous cell carcinoma of the oral cavity. Mol Cancer Res. (2016) 14:883–91. doi: 10.1158/1541–7786.MCR-16–0114
168. Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of Malignant transformation. Genes Dev. (2002) 16:2252–63. doi: 10.1101/gad.988902
169. Luo Z, Mu L, Zheng Y, Shen W, Li J, Xu L, et al. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J Mol Cell Biol. (2020) 12:345–58. doi: 10.1093/jmcb/mjz088
170. Henningsen KM, Manzini V, Magerhans A, Gerber S, Dobbelstein M. MDM2-driven ubiquitination rapidly removes p53 from its cognate promoters. Biomolecules. (2021) 12:22. doi: 10.3390/biom12010022
171. Porcheri C, Meisel CT, Mitsiadis T. Multifactorial contribution of notch signaling in head and neck squamous cell carcinoma. Int J Mol Sci. (2019) 20:1520. doi: 10.3390/ijms20061520
172. Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discovery. (2013) 3:770–81. doi: 10.1158/2159–8290.CD-12–0537
173. Shah PA, Huang C, Li Q, Kazi SA, Byers LA, Wang J, et al. NOTCH1 signaling in head and neck squamous cell carcinoma. Cells. (2020) 9:2677. doi: 10.3390/cells9122677
174. Ferrarotto R, Mishra V, Herz E, Yaacov A, Solomon O, Rauch R, et al. AL101, a gamma-secretase inhibitor, has potent antitumor activity against adenoid cystic carcinoma with activated NOTCH signaling. Cell Death Dis. (2022) 13:678. doi: 10.1038/s41419–022-05133–9
175. Doi T, Tajimi M, Mori J, Asou H, Inoue K, Benhadji KA, et al. A phase 1 study of crenigacestat (LY3039478), the Notch inhibitor, in Japanese patients with advanced solid tumors. Invest New Drugs. (2021) 39:469–76. doi: 10.1007/s10637–020-01001–5
176. Massard C, Cassier PA, Azaro A, Anderson B, Yuen E, Yu D, et al. A phase 1b study of crenigacestat (LY3039478) in combination with gemcitabine and cisplatin or gemcitabine and carboplatin in patients with advanced or metastatic solid tumors. Cancer Chemother Pharmacol. (2022) 90:335–44. doi: 10.1007/s00280-022-04461-z
177. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. (2020) 77:1745–70. doi: 10.1007/s00018–019-03351–7
178. Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol. (2005) 77:18–24. doi: 10.1016/j.radonc.2005.06.038
179. De Aguiar RB, de Moraes JZ. Exploring the immunological mechanisms underlying the anti-vascular endothelial growth factor activity in tumors. Front Immunol. (2019) 10:1023. doi: 10.3389/fimmu.2019.01023
180. Fountzilas G, Fragkoulidi A, Kalogera-Fountzila A, Nikolaidou M, Bobos M, Calderaro J, et al. A phase II study of sunitinib in patients with recurrent and/or metastatic non-nasopharyngeal head and neck cancer. Cancer Chemother Pharmacol. (2010) 65:649–60. doi: 10.1007/s00280–009-1070–1
181. Williamson SK, Moon J, Huang CH, Guaglianone PP, LeBlanc M, Wolf GT, et al. Phase II evaluation of sorafenib in advanced and metastatic squamous cell carcinoma of the head and neck: Southwest Oncology Group Study S0420. J Clin Oncol. (2010) 28:3330–5. doi: 10.1200/JCO.2009.25.6834
182. Swiecicki PL, Zhao L, Belile E, Sacco AG, Chepeha DB, Dobrosotskaya I, et al. A phase II study evaluating axitinib in patients with unresectable, recurrent or metastatic head and neck cancer. Invest New Drugs. (2015) 33:1248–56. doi: 10.1007/s10637–015-0293–8
183. Swiecicki PL, Bellile EL, Brummel CV, Brenner JC, Worden FP. Efficacy of axitinib in metastatic head and neck cancer with novel radiographic response criteria. Cancer. (2021) 127:219–28. doi: 10.1002/cncr.33226
184. Ghosh S, Mazumdar T, Xu W, Powell RT, Stephan C, Shen L, et al. Combined TRIP13 and aurora kinase inhibition induces apoptosis in human papillomavirus-driven cancers. Clin Cancer Res. (2022) 28:4479–93. doi: 10.1158/1078–0432.CCR-22–1627
185. Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. (2016) 30:1–17. doi: 10.1101/gad.274027.115
186. Chen Y, Han H, Seo G, Vargas RE, Yang B, Chuc K, et al. Systematic analysis of the Hippo pathway organization and oncogenic alteration in evolution. Sci Rep. (2020) 10:3173. doi: 10.1038/s41598–020-60120–4
187. Zheng Y, Pan D. The hippo signaling pathway in development and disease. Dev Cell. (2019) 50:264–82. doi: 10.1016/j.devcel.2019.06.003
188. Fu M, Hu Y, Lan T, Guan KL, Luo T, Luo M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther. (2022) 7:376. doi: 10.1038/s41392–022-01191–9
189. Lv L, Zhou X. Targeting Hippo signaling in cancer: novel perspectives and therapeutic potential. MedComm. (2020) 2023) 4:e375. doi: 10.1002/mco2.375
190. Peng Z, Gong Y, Liang X. Role of FAT1 in health and disease. Oncol Lett. (2021) 21:398. doi: 10.3892/ol.2021.12659
191. Santos-de-Frutos K, Segrelles C, Lorz C. Hippo pathway and YAP signaling alterations in squamous cancer of the head and neck. J Clin Med. (2019) 8:2131. doi: 10.3390/jcm8122131
192. Faraji F, Ramirez SI, Anguiano Quiroz PY, Mendez-Molina AN, Gutkind JS. Genomic hippo pathway alterations and persistent YAP/TAZ activation: new hallmarks in head and neck cancer. Cells. (2022) 11:1370. doi: 10.3390/cells11081370
193. Liu R, Wu J, Guo H, Yao W, Li S, Lu Y, et al. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm. (2023) 2020):e292. doi: 10.1002/mco2.292
194. Osorio JC, Castillo A. Epigenetic mechanisms in head and neck cancer. In: Bulgin D, editor. New aspects in molecular and cellular mechanisms of human carcinogenesis. InTech, Rijeka (2016). p. 67–95.
195. Bais MV. Impact of epigenetic regulation on head and neck squamous cell carcinoma. J Dent Res. (2019) 98:268–76. doi: 10.1177/0022034518816947
196. Camuzi D, Buexm LA, Lourenço SQC, Esposti DD, Cuenin C, Lopes MSA, et al. HPV infection leaves a DNA methylation signature in oropharyngeal cancer affecting both coding genes and transposable elements. Cancers (Basel). (2021) 13:3621. doi: 10.3390/cancers13143621
197. Chen HC, Yang CM, Cheng JT, Tsai KW, Fu TY, Liou HH, et al. Global DNA hypomethylation is associated with the development and poor prognosis of tongue squamous cell carcinoma. J Oral Pathol Med. (2016) 45:409–17. doi: 10.1111/jop.12381
198. Misawa K, Mochizuki D, Imai A, Mima M, Misawa Y, Mineta H. Analysis of site-specific methylation of tumor-related genes in head and neck cancer: potential utility as biomarkers for prognosis. Cancers (Basel). (2018) 10:27. doi: 10.3390/cancers10010027
199. Liouta G, Adamaki M, Tsintarakis A, Zoumpourlis P, Liouta A, Agelaki S, et al. DNA methylation as a diagnostic, prognostic, and predictive biomarker in head and neck cancer. Int J Mol Sci. (2023) 24:2996. doi: 10.3390/ijms24032996
200. Zhou C, Ye M, Ni S, Li Q, Ye D, Li J, et al. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics. (2018) 13:398–409. doi: 10.1080/15592294.2018.1465790
201. Gazdzicka J, Gołąbek K, Strzelczyk JK, Ostrowska Z. Epigenetic modifications in head and neck cancer. Biochem Genet. (2020) 58:213–44. doi: 10.1007/s10528–019-09941–1
202. Le JM, Squarize CH, Castilho RM. Histone modifications: Targeting head and neck cancer stem cells. World J Stem Cells. (2014) 6:511–25. doi: 10.4252/wjsc.v6.i5.511
203. Giudice FS, Pinto DS Jr, Nör JE, Squarize CH, Castilho RM. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PloS One. (2013) 8:e58672. doi: 10.1371/journal.pone.0058672
204. Romanowska K, Sobecka A, Rawłuszko-Wieczorek AA, Suchorska WM, Golusiński W. Head and neck squamous cell carcinoma: epigenetic landscape. Diagnostics (Basel). (2020) 11:34. doi: 10.3390/diagnostics11010034
205. Gupta S, Kumar P, Maini J, Kaur H, Das BC. Epigenetic biomarkers in head and neck cancer. Epigenetic biomarkers in head and neck cancer. J Cancer Genet And Biomarkers. (2018) 1:41–50. doi: 10.14302/issn.2572–3030.jcgb-18–2428
206. Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P, et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer. (2019) 18:148. doi: 10.1186/s12943–019-1075–2
207. Gougousis S, Petanidis S, Poutoglidis A, Tsetsos N, Vrochidis P, Skoumpas I, et al. Epigenetic editing and tumor-dependent immunosuppressive signaling in head and neck Malignancies. Oncol Lett. (2022) 23:196. doi: 10.3892/ol.2022.13317
208. Bhattacharjee R, Prabhakar N, Kumar L, Bhattacharjee A, Kar S, Malik S, et al. Crosstalk between long noncoding RNA and microRNA in Cancer. Cell Oncol (Dordr). (2023) 46:885–908. doi: 10.1007/s13402–023-00806–9
209. Barrera LN, Ridley PM, Bermejo-Rodriguez C, Costello E., Perez-Mancera P.A. The role of microRNAs in the modulation of cancer-associated fibroblasts activity during pancreatic cancer pathogenesis. J Physiol Biochem. (2023) 79:193–204. doi: 10.1007/s13105–022-00899–0
210. Chakrabortty A, Patton DJ, Smith BF, Agarwal P. miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes (Basel). (2023) 14:1375. doi: 10.3390/genes14071375
211. Sadeghi MS, Lotfi M, Soltani N, Farmani E, Fernandez JHO, Akhlaghitehrani S, et al. Recent advances on high-efficiency of microRNAs in different types of lung cancer: a comprehensive review. Cancer Cell Int. (2023) 23:284. doi: 10.1186/s12935-023-03133-z
Keywords: head and neck cancer, molecular pathways, targeted therapy, monoclonal antibodies, genomic, epigenetic, ncRNA
Citation: Constantin M, Chifiriuc MC, Bleotu C, Vrancianu CO, Cristian R-E, Bertesteanu SV, Grigore R and Bertesteanu G (2024) Molecular pathways and targeted therapies in head and neck cancers pathogenesis. Front. Oncol. 14:1373821. doi: 10.3389/fonc.2024.1373821
Received: 20 January 2024; Accepted: 03 June 2024;
Published: 17 June 2024.
Edited by:
Natalia Isaeva, University of North Carolina at Chapel Hill, United StatesReviewed by:
John Morton, University of Colorado Anschutz Medical Campus, United StatesCopyright © 2024 Constantin, Chifiriuc, Bleotu, Vrancianu, Cristian, Bertesteanu, Grigore and Bertesteanu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Carmen Chifiriuc, Y2FybWVuLmNoaWZpcml1Y0BiaW8udW5pYnVjLnJv
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.