- 1Department of Neurosurgery, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Advanced Molecular Pathology Institute of Soochow University and SANO, Suzhou, China
- 3Sano Precision Medicine Ltd., Suzhou, China
- 4Department of Pathology, The First Affiliated Hospital of Soochow University, Suzhou, China
- 5Department of Biological Sciences, Xi An Jiaotong-Liverpool University, Suzhou, China
- 6The College of Informatics, Huazhong Agricultural University, Wuhan, China
- 7Department of Pathology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States
Giant cell glioblastoma often exhibits genome instability and is frequently associated with mutations in genes involved in DNA repair pathways including TP53 and DNA mismatch repair genes. Several germline mutations have been identified in giant cell glioblastoma, including mutations of MSH1 and MSH2, TP53, and POLE. We have documented a case of a germline mutation in CHEK2, another gene crucial to DNA repair, in a patient with giant cell glioblastoma. The CHEK2 mutation was inherited from the patient’s father, who had a history of gastric cancer and renal cell carcinoma. In addition to the germline CHEK2 mutation, the giant cell glioblastoma exhibited a genome-wide loss of heterozygosity, a characteristic observed in a subset of giant cell glioblastomas. Additional mutations detected in the tumor included TP53, PTEN, and a PTPRZ1-MET fusion. This represents the first reported case of a CHEK2 germline mutation in giant cell glioblastoma, further supporting the significance of impaired DNA repair mechanisms in the development of this disease.
Introduction
Giant cell glioblastoma is a rare subtype of glioblastoma characterized by the presence of numerous large and morphologically bizarre tumor cells. Clinically, giant cell glioblastoma tends to have a better prognosis compared to classic glioblastoma. The genomic profile of giant cell glioblastoma differs from that of classic glioblastoma, which likely contributes to its distinct clinical behavior. While classic glioblastoma commonly exhibits a combination of TERT mutation, EGFR amplification, CDKN2A/B loss, and +7/−10 chromosome copy-number alterations, giant cell glioblastoma often shows TP53 and PTEN mutations, with TP53 mutation being the most frequently observed alteration, present in 90% of cases. Recently, a subgroup of giant cell glioblastoma with a near-haploid genome has been identified (4).
Germline mutations in several DNA repair-related genes, including TP53, POLE, and DNA mismatch repair genes MSH1 and MLH2 (1–3), have been associated with giant cell glioblastoma, indicating the significance of DNA repair deficiency in the development of this tumor subtype. Here, we present a case involving a germline mutation in the CHEK2 gene, which is also involved in the DNA damage response pathway. CHEK2 is part of the ATM-CHEK2-p53 axis, which plays a crucial role in the cellular response to DNA damage. Upon detection of DNA damage by ATM, CHEK2 is activated and phosphorylates p53, leading to cell cycle arrest for DNA repair or apoptosis to prevent malignant transformation.
Germline mutations in TP53 are associated with Li-Fraumeni syndrome, characterized by an increased risk of various cancers, including brain tumors, breast cancer, leukemia, sarcoma, adrenocortical carcinoma, and colon cancer (5). Some individuals with Li-Fraumeni syndrome do not have TP53 mutations but harbor other mutations, including CHEK2 mutations. In our patient, who has giant cell glioblastoma, the CHEK2 mutation was inherited from their father, who had gastric cancer and renal cell carcinoma. The CHEK2 p.H371Y mutation, located in the kinase domain, results in reduced kinase activity in functional assays [PMID: 21618645] and is associated with breast cancer [PMID: 24390236]. Additionally, breast cancer patients carrying the CHEK2 p.H371Y mutation are more likely to respond to neoadjuvant chemotherapy compared to non-carriers [PMID: 25884806]. To the best of our knowledge, this is the first documented case of a germline CHEK2 mutation in giant cell glioblastoma, thereby providing additional evidence for the importance of compromised DNA repair mechanisms in the development of this disease.
Materials and methods
Immunohistochemical studies
Immunohistochemistry (IHC) was performed on 5-μm tissue sections using the following protocol: (1) the slides were baked at 60 °C for 1 hour (2), deparaffinized and rehydrated with 100% xylene, 100% ethanol, 70% ethanol, and running water (3), blocked in a solution of 10% normal serum and 1% bovine serum albumin (BSA) in Tris-buffered saline (4), incubated with primary antibodies for 2 hours (5), endogenous peroxidase was blocked with 0.3% hydrogen peroxide, and (6) slides were incubated with horseradish peroxidase-labeled polymer (DAKO) according to the manufacturer’s instructions. The tissue sections were developed using 3,3’-diaminobenzidine (DAKO) as the chromogen and counterstained with Mayer’s hematoxylin.
Antibodies and their working concentrations are listed in Supplementary Table 1.
Targeted DNA next-generation sequencing (NGS)
DNA was extracted from Formalin-Fixed Paraffin-Embedded (FFPE) tumor tissue and peripheral blood, fragmented with a Bioruptor Pico (Diagenode, Denville, NJ) to 200-300 bp, subjected to end-polishing, phosphorylation, and DNA extension by incubating with the end-repair mix, Klenow exo- and Taq polymerase (Enzymatics, Beverly, MA) for 15 min at 12°C, 15 min at 37°C, and 15 min at 72°C, and ligated to a UMl-containing adaptor. Four cycles of polymerase chain reaction (PCR) were performed with adaptor-specific primers and the PCR products were incubated with a pool of biotin-labeled bait oligos targeting 638 genes commonly involved in tumors for 16 h. Targeted regions were enriched by pull-down with streptavidin beads, amplified by PCR and sequenced in an illumina NovaSeq sequencer (San Diego, CA, USA). Sequencing results were analyzed with SeqNext software (JSI, Ettenheim, Germany).
Targeted RNA sequencing and nested RT-PCR
Total RNA was extracted from fresh tumor sample using the TRIZOL reagent (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions.
The reverse transcription, end repairing, dA-tailing, and adaptor ligation were performed following standard NGS protocols (NEB, Cat E7771 and E6111, Ipswich, MA, USA). A group of 60 genes commonly involved in solid tumors was targeted using PCR enrichment with primers specific to these genes. The PCR products were sequenced using an Illumina NovaSeq sequencer. The sequencing results were analyzed using SeqNext software and laboratory-developed pipelines (Sano Medical Laboratories, China).
For nested RT-PCR, cDNA synthesis from total RNA was performed using random priming and the SuperScript™ IV reverse transcriptase (ThermoFisher). PCR amplification was then carried out using specific primers for PTPRZ1::MET (forward primer PTPRZ1: 5’-CACTCTGAGAAGCAGAGGAGCC and reverse primer MET: 5’-GGTGTTTCCGCGGTGAAGTT; nested PCR forward primer PTPRZ1: 5’-CCGTCTGGAAATGCGAATCC and reverse primer MET: 5’-ATTGCTCCTCTGCACCAAGG). The PCR was performed with one cycle at 95°C for 3 minutes followed by 30 cycles at 95°C for 30 seconds, 60°C for 1 minute and 72°C for 1 minute. The initial PCR product (0.1 µl) served as the template DNA for the subsequent nested PCR. This nested PCR comprised an initial cycle at 95°C for 3 minutes followed by 30 cycles at 95°C for 30 seconds, 58°C for 1 minute, and 72°C for 1 minute. The PCR products were then subjected to Sanger sequencing.
MGMT methylation analysis
The CpG islands of the MGMT promoter region from tumor DNA was amplified by using FAM fluorescently labeled primers (forward primer 5’- TTTGTGTTTTGATGTTTGTAGGTTTTTGT, reverse primer 5’- TTTCGACGTTCGTAGGTTTTCGC). The PCR protocol consisted of one cycle at 95°C for 10 minutes followed by 40 cycles at 95°C for 45 seconds, 60°C for 45 seconds and 72°C for 1 minute. The FAM-labeled PCR products were analyzed for fragments in a capillary electrophoresis instrument. Control groups included gDNA specimens known to be methylated and unmethylated at the MGMT promoter.
This study was approved by the institutional review board at respective institutions.
Results
A 53-year-old male presented with symptoms of delirium, abnormal behavior, and unresponsiveness. Magnetic Resonance Imaging (MRI) revealed a 75 x 50 mm irregular abnormal signal in the left temporal lobe. T1-weighted images showed hypo-to-isointense signals, while T2-weighted images displayed heterogeneous hyperintense signals. Diffusion-weighted imaging (DWI) indicated diffuse restriction, with surrounding edema and ring enhancement (Figures 1A–D). These radiological findings were suggestive of glioblastoma. A surgical procedure successfully removed a tumor measuring 50 x 60 x 70 mm. Histopathological analysis of the FFPE tumor tissue section revealed the presence of pleomorphic cells with numerous bizarre multinucleated giant cells and frequent mitotic figures. Immunohistochemical staining showed positive results for GFAP, Olig-2, MGMT, MAP2, ATRX, P53, EGFR, S100, focal vimentin and negative for IDH-1, NeuN, H3K27M, Syn, and CD34. Ki-67 staining demonstrated a high proliferation index of 60% (Figures 1E–H; Supplementary Figure 1). Based on these findings, a diagnosis of giant cell glioblastoma was made.
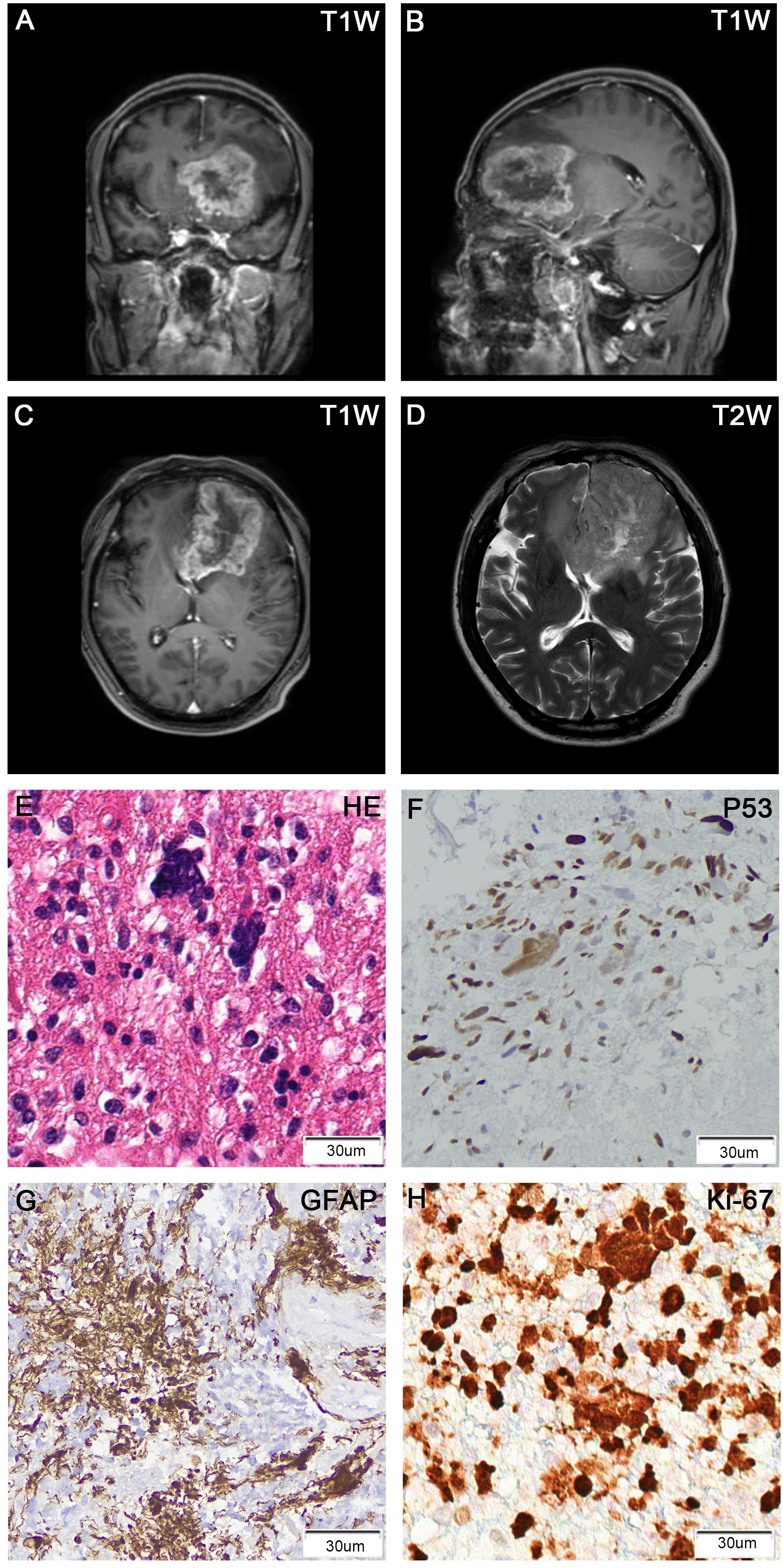
Figure 1. Brain MRI showed a ring-enhancing tumor in the left frontal lobe, with the largest cross-section of 75 x 50 mm. (A) T1-weighted contrast-enhanced (coronal view); (B) T1-weighted contrast-enhanced (sagittal view); (C) T1-weighted contrast-enhanced (axial view); (D) T2-weighted (axial view). H&E stain of FFPE tumor sections showed numerous large and multinucleated tumor cells (E). IHC of tumors cells was positive for p53 (F), GFAP (G), and Ki-67 (H).
Targeted DNA NGS of 638 cancer genes coupled with whole-genome single-nucleotide polymorphism (SNP) analysis was performed on tumor DNA. The results revealed a near-haploid genome with widespread loss of heterozygosity throughout the genome, except for chromosomes 7 and 18, which maintained heterozygosity. Notably, both copies of chromosome 13 were lost (Figure 2A). Additionally, three mutations were detected: TP53 p.T125K, PTEN p.D52M fs*2, and CHEK2 p.H371Y. While the TP53 and PTEN mutations were identified as somatic mutations, the CHEK2 mutation was also observed in the patient’s peripheral blood specimen, confirming a germline CHEK2 mutation.
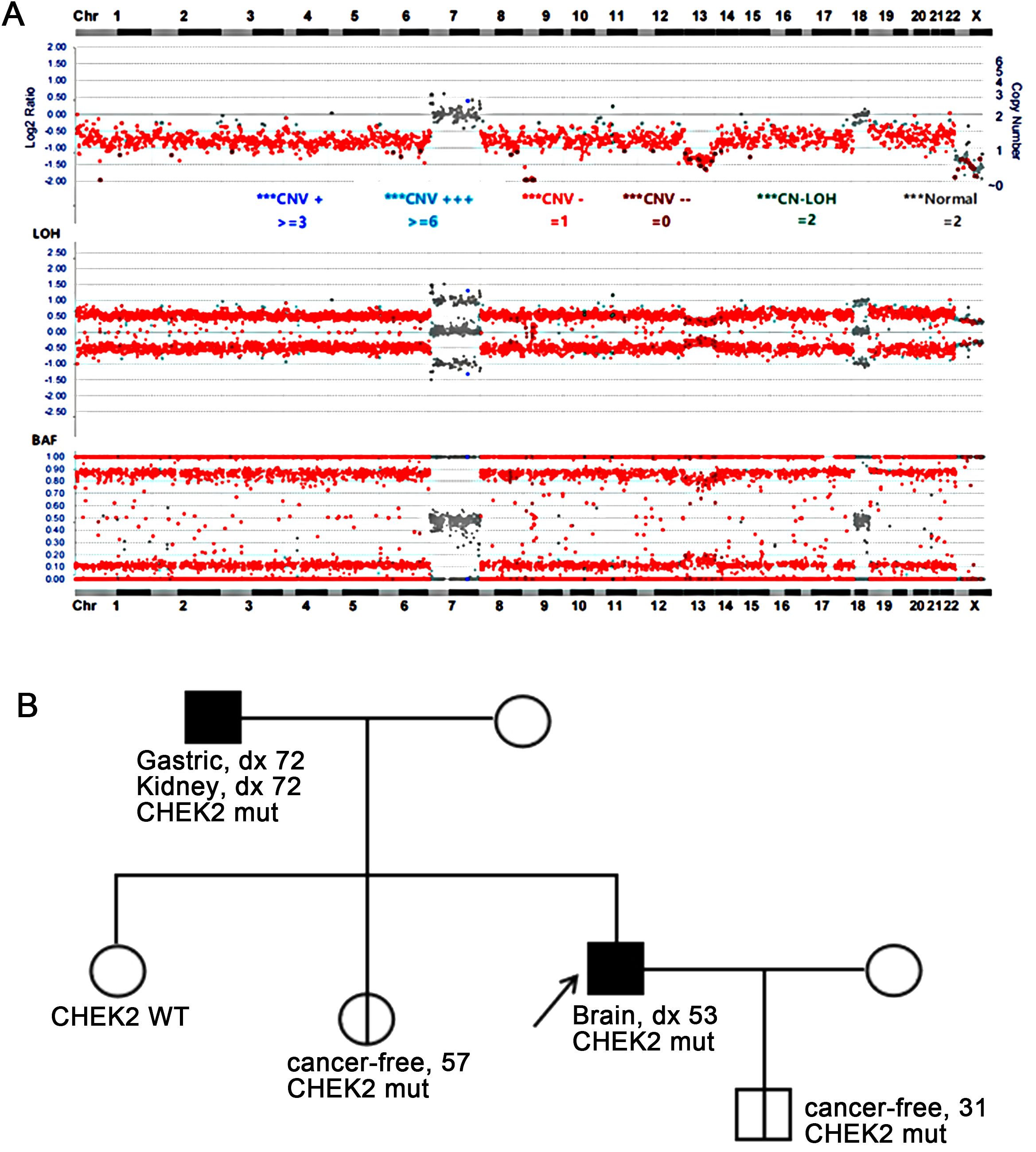
Figure 2. (A) Targeted DNA NGS revealed a near-haploid genome with genome-wide loss of heterozygosity, except for chromosomes 7 and 18. Additionally, a homozygous loss of chromosome 13 was observed. (B) The family pedigree showed the presence of the germline CHEK2 p.H371Y mutation in the patient (proband) as well as in his father, one of his sisters, and his son. The father was diagnosed with gastric cancer and renal cell carcinoma at the age of 72, the proband with giant cell glioblastoma at the age of 53, while no tumors were reported in the son at age 31 and the sister at age 57. WT, wild-type; mut, mutation.
The patient’s family history revealed that the patient’s father had a history of gastric cancer and renal cell carcinoma, both diagnosed at the age of 72. Subsequent CHEK2 mutation screening in the family confirmed that the patient’s father carried the same CHEK2 mutation. Additionally, the patient’s sister and son were found to carry the same CHEK2 mutation, although they have not reported any tumors at the ages of 57 and 31, respectively (Figure 2B).
Further analysis was conducted using targeted RNA NGS on a fresh tumor sample, which identified a PTPRZ1-MET rearrangement. Specifically, exon 2 of PTPRZ1 was fused with exon 2 of MET, resulting in a chimeric protein with an intact reading frame (Figure 3A). The chimeric protein retained a partial alpha-carbonic anhydrase domain from PTPRZ1 and all functional domains from MET, similar to previously described rearrangements (6). The PTPRZ1-MET fusion was confirmed an RT-PCR assay using primers specific to PTPRZ1 and MET followed by Sanger sequencing (Figures 3B, C). Methylation-specific PCR (MSP) and capillary electrophoresis were performed by evaluating MGMT promoter CpG methylation. The results of this analysis demonstrated methylated MGMT status (Supplementary Figure 2).
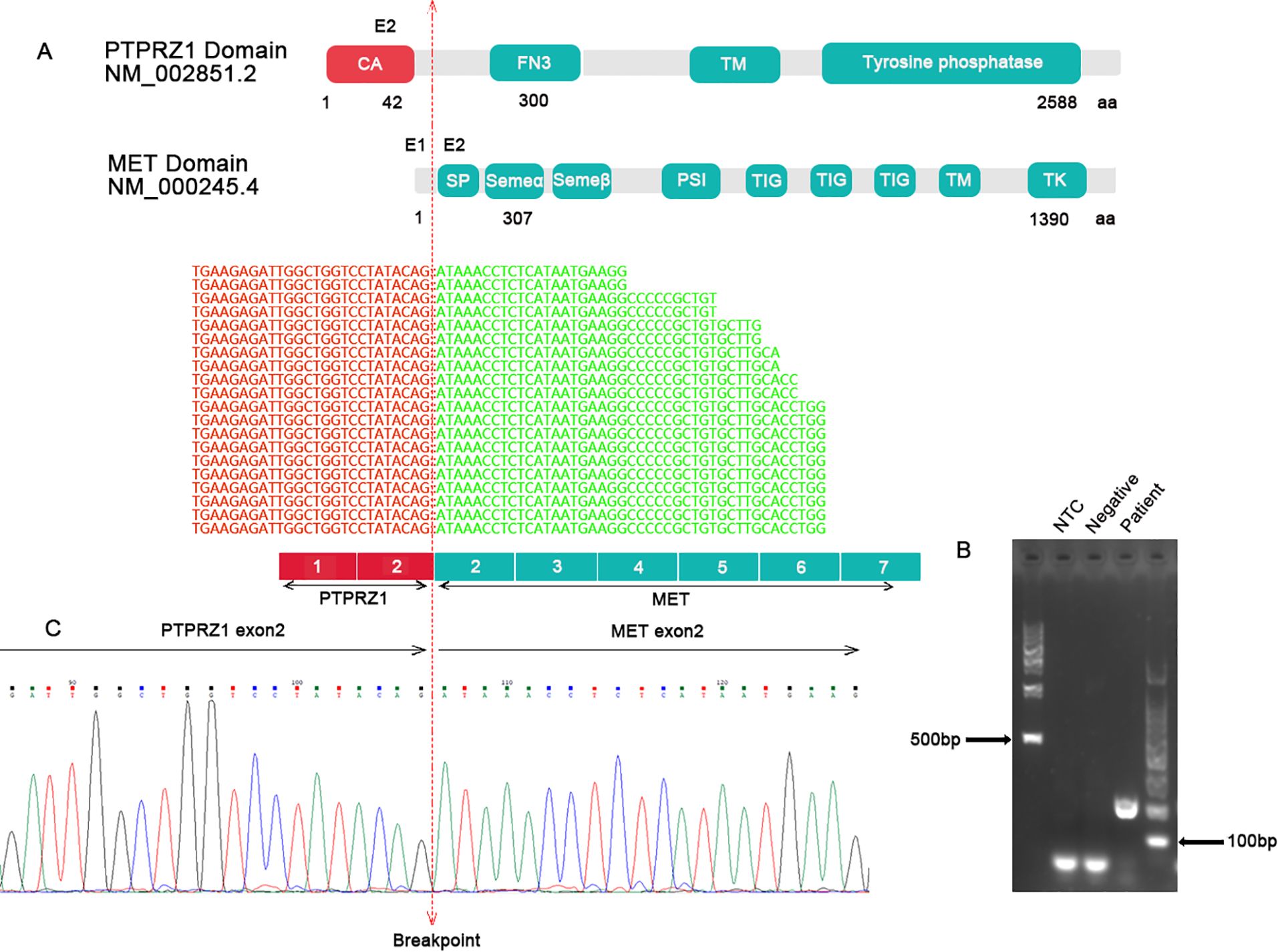
Figure 3. (A) Targeted RNA NGS showed an in-frame fusion between PTPRZ1 exon 2 and MET exon 2. The schematic of functional domains of PTPRZ1 and MET and their exon location were shown. The breakpoint was marked by a vertical dotted line. (B) RT-PCR with specific primers for PTPRZ1 and MET produced a band at expected size for the PTPRZ1::MET rearrangement. (C) Sanger sequencing of the PCR product confirmed the PTPRZ1::MET fusion. CA, alpha-carbonic anhydrase; FN3, fibronectin-like domain; TM, transmembrane domain; SP, signal peptide; SEMA, Semaphorin domain; PSI, Plexin semaphorin domain; TIG like plexins transcription factor; TM, transmembrane region; TK, tyrosine kinase.
Two cycles of temozolomide (TMZ) therapy and one cycle of external beam radiation therapy (EBRT) were administered. Unfortunately, the patient did not respond to these treatments and succumbed to the disease 10 months after the initial diagnosis.
Discussion
The presence of numerous giant cells is considered a classification-defining feature for giant cell glioblastoma. Giant cells can be seen in various physiological and pathological conditions. The multinucleated osteoblasts, macrophages, and muscle cells are formed by cell fusion. The hepatocytes and trophoblasts during placental development are cells of endoduplication, i.e., cells with repeated rounds of DNA replication without subsequent cell division. Giant cells can be observed in various types of tumors. Alongside giant cell glioblastoma, they are also commonly found in other tumors including bone tumors such as giant cell tumor of bone, chondroblastoma, and aneurysmal bone cyst (7–9). Additionally, giant cells can be present in soft tissue tumors such as dedifferentiated liposarcoma and undifferentiated pleomorphic sarcoma (10, 11). Furthermore, the Hodgkin and Reed-Sternberg cells are a characteristic feature of Hodgkin’s lymphoma (12). The mechanisms underlying giant cell formation in malignancies are likely disease-specific. In the case of giant cell glioblastoma, DNA repair deficiency and genome instability may play a role. DNA repair mechanisms are tightly linked to cell cycle checkpoints, which ensure proper DNA replication and repair prior to cell division. If these checkpoints fail due to DNA repair deficiencies, cells with damaged DNA can proceed through the cell cycle and divide, resulting in abnormal chromosome numbers, including polyploidy. Additionally, DNA repair defects can disrupt the normal process of cytokinesis, the division of replicated genetic material into two daughter cells. Failed cytokinesis can lead to polyploidy. DNA repair defects also promote the occurrence of breakage-fusion-bridge (BFB), a mechanism that accumulates chromosome aberrations through consecutive cell divisions, however, giant cell glioblastoma generally does not exhibit intrachromosomal gains or losses (13). Therefore, the BFB mechanism is not likely to be involved in the formation of giant polyploid cells in giant cell glioblastoma.
Giant cell glioblastoma commonly exhibits two genomic changes: TP53 mutation and a haploid genome. Interestingly, a subset of B-cell acute lymphoblastic leukemia (B-ALL) patients with a near-haploid genome also carries a germline TP53 mutation (14). The concurrent occurrence of TP53 mutation and haploid genome in both giant cell glioblastoma and B-ALL suggests a potential causative association. The haploid genome in cancer cells is thought to result from mitotic catastrophe, leading to the simultaneous loss of multiple suppressor genes, which can drive cancer cell evolution efficiently. However, such a dramatic genomic alteration would typically trigger apoptosis. P53 is a key regulator of cell death in response to genome instability, and its loss of function may help cancer cells evade cell death, even in the presence of mitotic catastrophe (15), thereby facilitating further malignant evolution.
The PTPRZ1-MET fusion has been identified in several types of cancer, mostly in glioblastoma and lung cancer (16, 17). The chimeric PTPRZ1-MET protein functions as a constitutively activated tyrosine kinase involved in processes such as cell proliferation, survival, and migration (18). MET-specific inhibitors, such as Tepotinib and Capmatinib, have been FDA-approved for the treatment of metastatic non-small cell lung cancer (NSCLC) with MET exon 14 skipping activation mutations. These inhibitors have shown efficacy in vitro against glioblastoma with PTPRZ1-MET rearrangement; however, their clinical efficacy in glioblastoma is currently unclear, with ongoing clinical trials investigating their potential.
In conclusion, we present a rare case of giant cell glioblastoma with an inherited germline CHEK2 mutation. Both CHEK2 and p53 are integral components of the ATM-CHEK2-p53 axis, and the simultaneous occurrence of a germline CHEK2 mutation and somatic TP53 mutation in this case strongly suggests the involvement of DNA repair deficiency in the development of this disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by The First Affiliated Hospital of Soochow University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
YB: Writing – review & editing. DW: Writing – original draft. SC: Writing – original draft. HC: Data curation, Methodology, Writing – original draft. LG: Investigation, Software, Writing – original draft. XH: Investigation, Software, Writing – original draft. RR: Conceptualization, Writing – original draft. JX: Data curation, Writing – review & editing. WG: Writing – review & editing. SX: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
Authors DW, SC, and HC were employed by Suzhou Sano Precision Medicine Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1361928/full#supplementary-material
References
1. Barresi V, Simbolo M, Mafficini A, Martini M, Calicchia M, Piredda ML, et al. IDH-wild type glioblastomas featuring at least 30% giant cells are characterized by frequent RB1 and NF1 alterations and hypermutation. Acta Neuropathol Commun. (2021) 9:200. doi: 10.1186/s40478-021-01304-5
2. Vande Perre P, Siegfried A, Corsini C, Bonnet D, Toulas C, Hamzaoui N, et al. Germline mutation p.N363K in POLE is associated with an increased risk of colorectal cancer and giant cell glioblastoma. Fam Cancer. (2019) 18:173–8. doi: 10.1007/s10689-018-0102-6
3. Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. (2006) 13:1017–26. doi: 10.1038/sj.cdd.4401913
4. Baker TG, Alden J, Dubuc AM, Welsh CT, Znoyko I, Cooley LD, et al. Near haploidization is a genomic hallmark which defines a molecular subgroup of giant cell glioblastoma. Neurooncol Adv. (2020) 2:vdaa155. doi: 10.1093/noajnl/vdaa155
5. Guha T, Malkin D. Inherited TP53 mutations and the li-fraumeni syndrome. Cold Spring Harb Perspect Med. (2017) 7(4):a026187. doi: 10.1101/cshperspect.a026187
6. Papadimitriou E, Kanellopoulou VK. Protein tyrosine phosphatase receptor zeta 1 as a potential target in cancer therapy and diagnosis. Int J Mol Sci. (2023) 24(9):8093. doi: 10.3390/ijms24098093
7. Sobti A, Agrawal P, Agarwala S, Agarwal M. Giant cell tumor of bone - an overview. Arch Bone Jt Surg. (2016) 4:2–9.
8. Ma JL, Wu Y, Wen JX, Zhong ZW, Yu BH, Liu C, et al. Images of giant cell tumor and chondroblastoma around the knee: retrospective analysis of 99 cases. Quant Imaging Med Surg. (2023) 13:787–800. doi: 10.21037/qims-22-616
9. Park HJ, Kwon SY, Cho SG, Kim J, Song HC, Kim SS, et al. Giant cell tumor with secondary aneurysmal bone cyst shows heterogeneous metabolic pattern on (18)F-FDG PET/CT: A case report. Nucl Med Mol Imaging. (2016) 50:348–52. doi: 10.1007/s13139-016-0423-z
10. Nimura F, Nakasone T, Matsumoto H, Maruyama T, Matayoshi A, Maruyama N, et al. Dedifferentiated liposarcoma of the oral floor: A case study and literature review of 50 cases of head and neck neoplasm. Oncol Lett. (2018) 15:7681–8. doi: 10.3892/ol.2018.8274
11. Lupo S, Berini C, Canepa C, Santini Araujo E, Biglione M. Undifferentiated pleomorphic sarcoma and the importance of considering the oncogenic and immune-suppressant role of the human T-cell lymphotropic virus type 1: A case report. Front Oncol. (2017) 7:91. doi: 10.3389/fonc.2017.00091
12. Gopas J, Stern E, Zurgil U, Ozer J, Ben-Ari A, Shubinsky G, et al. Reed-Sternberg cells in Hodgkin’s lymphoma present features of cellular senescence. Cell Death Dis. (2016) 7:e2457. doi: 10.1038/cddis.2016.185
13. Thomas R, Marks DH, Chin Y, Benezra R. Whole chromosome loss and associated breakage-fusion-bridge cycles transform mouse tetraploid cells. EMBO J. (2018) 37:201–18. doi: 10.15252/embj.201797630
14. Qian M, Cao X, Devidas M, Yang W, Cheng C, Dai Y, et al. TP53 germline variations influence the predisposition and prognosis of B-cell acute lymphoblastic leukemia in children. J Clin Oncol. (2018) 36:591–9. doi: 10.1200/JCO.2017.75.5215
15. Ozaki T, Nakagawara A. Role of p53 in cell death and human cancers. Cancers (Basel). (2011) 3:994–1013. doi: 10.3390/cancers3010994
16. Zeng AL, Yan W, Liu YW, Wang Z, Hu Q, Nie E, et al. Tumour exosomes from cells harbouring PTPRZ1-MET fusion contribute to a Malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene. (2017) 36:5369–81. doi: 10.1038/onc.2017.134
17. Chai RC, Liu X, Pang B, Liu YQ, Li JJ, Li YF, et al. Recurrent PTPRZ1-MET fusion and a high occurrence rate of MET exon 14 skipping in brain metastases. Cancer Sci. (2022) 113:796–801. doi: 10.1111/cas.v113.2
Keywords: CHEK2, glioblastoma, haploidy, MET, germline
Citation: Bi Y, Wan D, Chen S, Chen H, Guo L, He X, Rong R, Xiao J, Gao W and Xiao S (2024) Case report: Germline CHEK2 mutation is associated with a giant cell glioblastoma. Front. Oncol. 14:1361928. doi: 10.3389/fonc.2024.1361928
Received: 27 December 2023; Accepted: 02 September 2024;
Published: 01 October 2024.
Edited by:
Yusuke Suenaga, Chiba Cancer Center, JapanReviewed by:
Yohko Yamaguchi, Toho University, JapanCrismita Dmello, Northwestern University, United States
Copyright © 2024 Bi, Wan, Chen, Chen, Guo, He, Rong, Xiao, Gao and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Gao, Z2Fvd2VpQHN1ZGEuZWR1LmNu; Sheng Xiao, c3hpYW9AcGFydG5lcnMub3Jn
†These authors have contributed equally to this work
 Yongfeng Bi1†
Yongfeng Bi1† Dong Wan
Dong Wan Huafei Chen
Huafei Chen Rong Rong
Rong Rong Sheng Xiao
Sheng Xiao