- 1Liaoning Laboratory of Cancer Genomics and Department of Cell Biology, College of Basic Medical Sciences, Dalian Medical University, Dalian, China
- 2Translational Medicine Research Institute, First People’s Hospital of Foshan, Foshan, China
- 3Department of Breast Surgery, The First People’s Hospital of Foshan, Foshan, China
Breast cancer (BC) is the most prevalent malignancy among women worldwide. Traditional research models such as primary cancer cell and patient-derived tumor xenografts (PDTXs) have limitations. Cancer cells lack a tumor microenvironment (TME) and genetic diversity, whereas PDTXs are expensive and have a time-consuming preparation protocol. Therefore, alternative research models are warranted. Patient-derived organoids (PDOs) are a promising in vitro model. They mimic the TME, gene expression, and cell types of original cancer tissues. PDOs have been successfully developed from various cancers, including BC. In this review, we focused on the value and limitations of PDOs in BC research, including their characteristics and potential in drug development, personalized therapy, immunotherapy, and the application prospects of PDOs in drug testing and prognosis.
1 Introduction
Breast cancer (BC) is a prevalent malignancy among women globally. In 2020, BC surpassed lung cancer to become the most frequently diagnosed malignancy (1). Rapid advances in drug testing and high-throughput sequencing technology in the past few decades have resulted in considerable progress in the early-stage screening and clinical treatment of BC (2). Approximately 70%–80% of early-stage BCs can be cured with surgery and targeted therapy, with the mortality rate of patients with BC decreasing each year. However, approximately 685,000 patients still succumb to late-stage BC each year; therefore, it remains the leading reason for mortality among women worldwide (3).
At present, two-dimensional (2D) tumor cell lines and patient-derived tumor xenografts (PDTXs) remain the commonly used cancer research models (4). Although these experimental models have played a substantial role in basic BC research in the past, limitations remain.
BC emergence and progression involve processes other than tumor cell proliferation; they are intricately associated with the tumor microenvironment (TME) (5, 6), including tumor-associated fibroblasts (7), tumor-associated macrophages (8), adipocytes (9, 10), immune cells (11, 12) and the extracellular matrix (ECM) (13) (Figure 1). However, 2D-cultured tumor cells lack not only the TME but also the genetic heterogeneity of the original tumor owing to the substantial genetic changes that occur in multiple clones (14). Furthermore, although PDTXs can preserve the TME and the complex tumor structure, they also have many limitations, including higher experimental costs, longer experimental time, increased technical requirements, low success rate, and ethical issues (4). Therefore, alternative models for BC research are warranted.
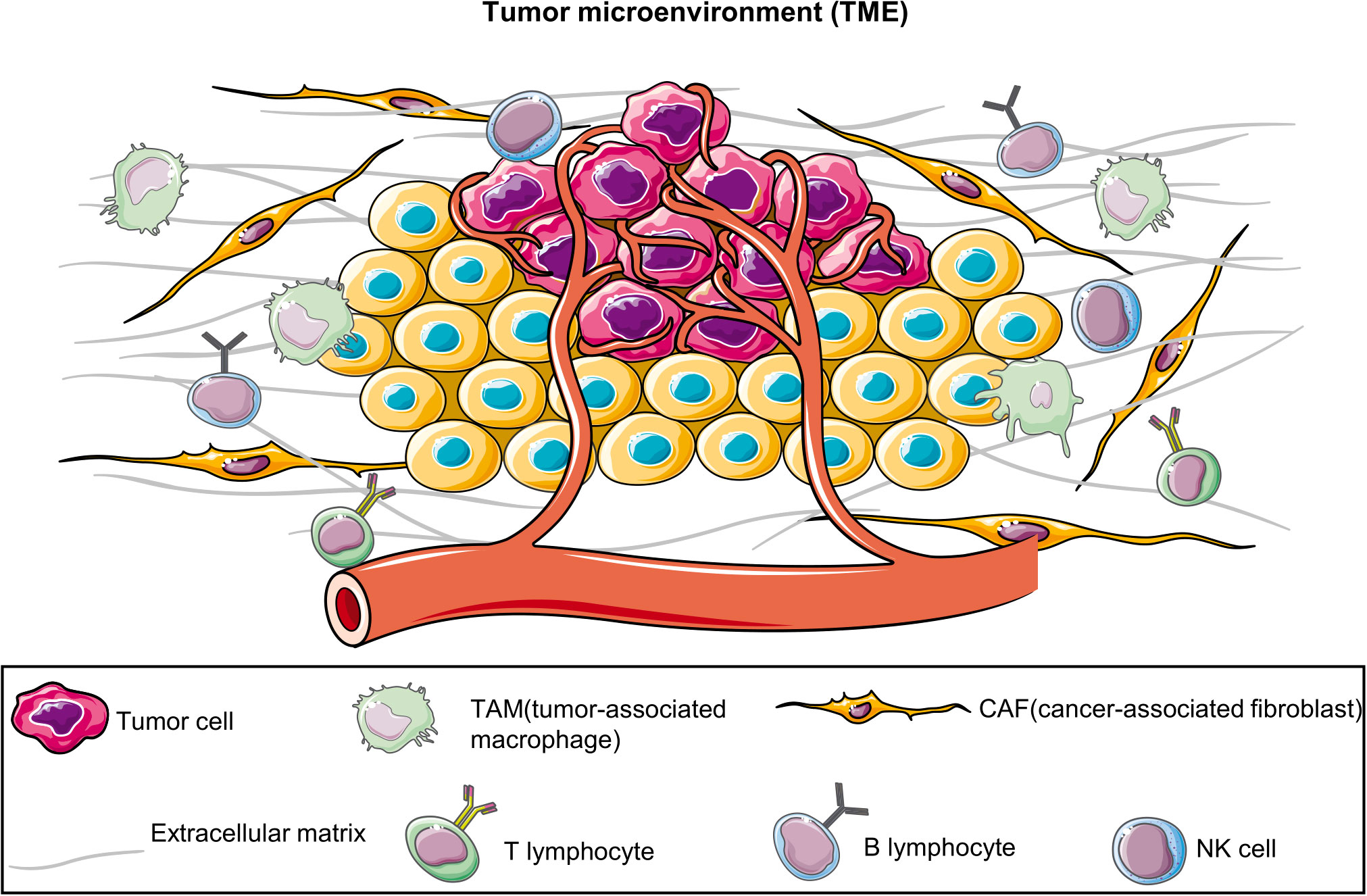
Figure 1 Tumor microenvironment (TME). TME comprises diverse cell types, including tumor cells, immune cells, vascular endothelial cells, fibroblasts, and the extracellular matrix. They collectively interact to affect tumor growth, metastasis, and treatment responses. The establishment of patient-derived organoids can effectively preserve these components.
In 2009, Sato et al. illustrated for the first time that a solitary leucine-rich repetitive G-protein-coupled receptor 5 (LGR5)-positive intestinal stem cell can form three-dimensional (3D) epithelial structures. These structures exhibit distinct polarized epithelial characteristics, featuring proliferative crevices and differentiated villous sections, and can maintain stable growth in vitro (15). Subsequently, this culture technique has facilitated the formation of other kinds of epithelial organoids, and patient-derived organoid (PDO) models have been developed from different organs and cancer types (4), including the liver (16), breast (17), colon (18), lungs (19), and stomach (20). Recently, 3D culture technology has emerged as a promising in vitro model for cancer. Different from monolayer cells and PDTXs, PDOs are 3D cultures that are formed in vitro from surgically resected or biopsy tissues, which are structurally and functionally similar to in vivo organs. They not only provide a good summary of the gene expression profiles, personalized characteristics, and cell types of native tissues but also preserve the TME of a patient’s tumor. In addition, organoids possess self-organization and self-renewal properties (14, 21, 22).
2 Overview of organoid development in BC
In 1979, Emerman et al. cultured normal mammary epithelial cells in collagen gel and observed that different from the 2D culture model, the collagen gel matrix allowed the entry of nutrients to the basolateral cell surface, cells to approach the medium surface and gas phase, the interaction of epithelial cells with matrix elements, and sublaminar flexibility, allowing changes in cell shape. This approach provided a unique microenvironment for breast epithelial cells in a 3D environment (23). In 1982, Bissell et al. studied BC organoids for the first time and noted that the presence of the ECM in 3D models can better simulate gene expression in BC cells (24). This study not only demonstrated the importance of the TME but also shifted the understanding of BC cells from 2D to 3D. Furthermore, in 1982, M Hiratsuka et al. isolated and cultured breast organoids with duct-like structures from breast biopsy tissues (25). In 1990, a study revealed the varying effects of distinct ECM compositions on murine mammary tissue-derived cells (26). In 1992, Bissell reported for the first time that normal primary mammary epithelial cells and biopsied cancer cells could be cultured to generalize the growth behavior and structural and functional differentiation of these cells in vivo (27). In 2007, Bissell et al. revealed that vital cellular signals disappeared when cells were cultured outside their natural environment on flat 2D plastic surfaces. However, a significant portion of these essential microenvironmental cues were reinstated using 3D laminin-rich extracellular matrix cultures (28). Since Sato et al. first cultured intestinal organoids from a single intestinal epithelial stem cell in 2009, BC stem cells have also gradually gained importance as the key to organoid formation (17). In 2012, breast Lgr5+ cells not only served as breast stem cells and survived for a long time in the passage process but also developed normal ductal structures similar to the breast via cytokine arrangement (29). Sachs et al. cultured 95 BC organoids from 155 surgical BC tissues and successfully established the pioneering BC organoid repository; they observed that the histopathology and hormone receptor status of BC organoids were similar to those of the original tumor; furthermore, they noted that BC organoids can be used for in vitro drug screening and respond similarly as in vivo xenografts and patients (19). Recently, with the continuous improvement and optimization of experimental techniques, the success rate of BC organoid culture has reached 87.5% (30). Therefore, BC organoid technology is widely employed in basic research, drug screening, and individualized therapy (Figure 2).
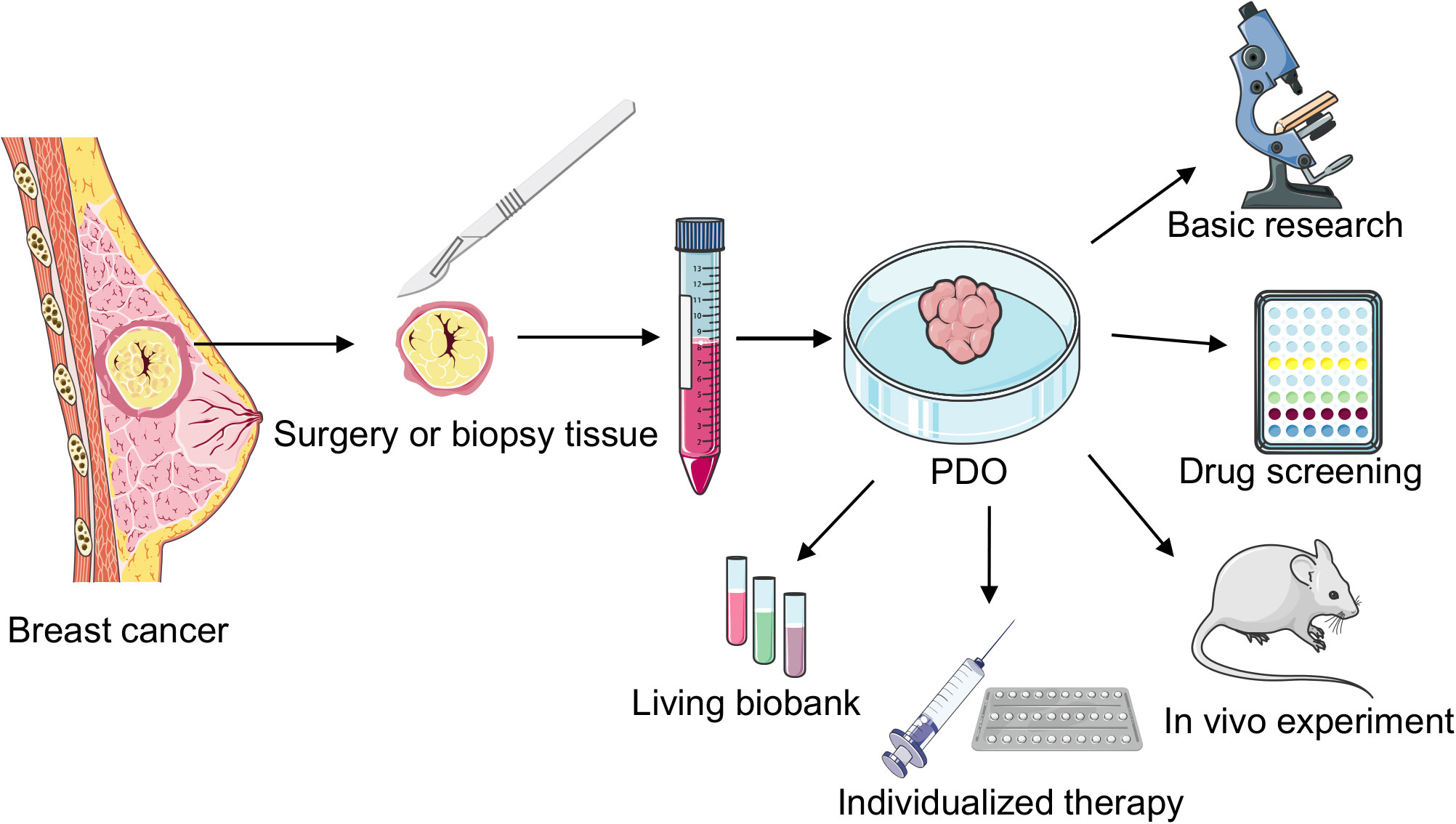
Figure 2 Cultivation and application of breast cancer (BC) patient-derived organoids (PDOs). PDOs derived from surgically resected or biopsied tumor tissues undergo enzymatic digestion to generate intricate organoid structures. These organoids are cultured after centrifugation, finding applications in diverse areas such as fundamental research, rapid drug screening, biobank development, in vivo transplantation studies, and individualized clinical treatments.
3 Advantages of PDO models
PDO models exhibit various captivating advantages, which transcend those offered by traditional cell and patient-derived xenograft (PDX) models (4, 14, 19).
3.1 Advantages of PDO over cell models
1. Simulated TME: Using the 3D model, the interactions between tumor cells and epithelial cells, immune cells, and fibroblasts, all of which are present in the TME but absent in 2D cell lines, can be studied. Therefore, PDOs more accurately replicate the in vivo TME, including cell–cell interactions, the ECM, growth factors, and cell signaling pathways. Furthermore, they more precisely reflect the biological characteristics of tumors, making it a research platform that resembles biological reality (31).
2. Preservation of heterogeneity: Heterogeneity is gradually lost during 2D cell culture; however, the TME remains highly heterogeneous. This suggests that the 2D culture microenvironment is less heterogeneous than the original TME. Compared with 2D models, PDOs exhibit higher heterogeneity (32).
3. Individualized treatment: Tumor-derived cell lines cannot fundamentally interact with the original tumor, which provides stromal support to cancer cells in the form of fibroblast elements, blood vessels, and immune mediators. For example, in a previous study, only seven prostate cancer cell lines were generated, with only four expressing androgen receptors (33). Furthermore, only a few cell lines have been generated for glioblastoma; these established glioma cell lines do not represent primary human gliomas (34). Therefore, cell lines cannot be a model to study personalized treatments. However, because PDOs originate from patient tissues, they provide robust support for individualized treatment, helping in formulating optimal treatment strategies based on the pathological characteristics and gene expression patterns of patients.
4. Biological complexity: PDOs can simulate more intricate biological processes, including cell–cell interactions and signal transduction, helping researchers gain deeper insights into the mechanisms underlying tumor development.
3.2 Advantages of PDO over PDX models
1. No need for animal experiments: Animal experiments are not needed to establish PDO; therefore, ethical and legal concerns related to animals and potential adaptational changes from interspecies transplantation are avoided.
2. Convenient sample acquisition: The collection of patient samples is relatively straightforward; in contrast, the establishment of PDX models requires intricate animal experiments and sample processing procedures.
3. Long-term cultivation: PDO can be maintained for an extended period in vitro, facilitating the long-term observation and evaluation of treatment effects.
4 BC PDOs recapitulate the heterogeneity of original tissues
Tumor heterogeneity refers to the concept that tumor cells can exhibit distinct genetic and phenotypic characteristics, including gene mutation, cell morphology, and gene, protein, and marker expression. Furthermore, because of genetic and phenotypic differences, tumor cells exhibit differences in proliferation, transfer ability, and drug resistance. Tumor heterogeneity can be categorized into two types: inter- and intratumor heterogeneity. Intertumor heterogeneity refers to the diversity observed across distinct tissues. For example, tumor occurrence and development are different in each patient; furthermore, the treatment process is different, which leads to tumor heterogeneity among different patients. On the other hand, intratumor heterogeneity primarily depends on the TME and intrinsic characteristics of tumor cells (35). Compared with monolayer cultures, PDOs can better summarize the heterogeneity of original tumor tissues. Diermeier et al. performed RNA sequencing (RNA-seq) to compare the tumor and organoid transcriptome profiles of luminal B (MMTV-PyMT) and HER2/nue-amplified (MMTV-Neu-NDL) mouse models. They observed that, in most instances, the organoids could effectively replicate the expression profile of the initial tumor tissue. Only a small fraction, approximately 1% of the gene transcriptome (142 of 13,854 expressed genes), exhibited statistically significant differences between the tumor and day 6 organoids. The affected genes were predominantly protein-coding genes (85%), affecting their connected signaling and metabolic pathways. Simultaneously, noncoding RNA levels were largely unaltered (36). Sachs et al. successfully created 95 organoid biobanks from patients’ tumors, representing all BC subtypes. Then, they compared the whole genome sequences of organoids and primary BC tissues via whole genome sequencing and RNA-seq and observed that the transcriptomic, histological, and genomic fingerprints of the original tumor tissue were largely unchanged in the organoids after 20 passages. Furthermore, they performed H&E staining to histopathologically analyze tumor tissues and organoid sections and observed that the phenotype of BC organoids was often similar to that of the original BC tissue. For example, solid organoids are produced for ductal carcinoma, whereas loose organoids are produced for lobular carcinoma. In addition, nuclear atypia is preserved in organoids, including enlarged and polymorphic nuclei and high mitotic activity (19). In the context of the BC model, preserving the key and common specific biomarkers, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2), is vital. These biomarkers serve as vital targets for drugs against BC (37). Chen et al. successfully established organoids from 132 patients with BC and evaluated the status of ER, PR, HER2, and Ki-67 on organoids and tumors via immunohistochemistry. They observed that the expression patterns of these BC markers were well preserved, regardless of whether the organoids had received systemic anticancer therapy (38). Bhatia et al. generated an organoid bank from 83 BC tissues. They not only elucidated that the organoids accurately replicated the traits of the patient’s tumor but also observed that when triple-negative breast cancer (TNBC) organoids were implanted into NOD/SCID mice, the resultant tumors were morphologically similar to the original tumor (39). In a recent study, Saeki et al. developed PDOs from 10 patients with BC and observed that these BC PDOs had different growth rates and patterns. Subsequently, they subjected these PDOs to transcriptome analysis at the single-cell level, innovating novel analytical techniques: (1) clustering cells within each PDO, (2) discerning genes specifically expressed in each cluster, and (3) quantifying their resemblance to subsequently categorize the clusters. These analyses revealed that each PDO comprises multiple subpopulations with different cell states and cycles, estrogenic responses, and epithelial–mesenchymal transition-like gene expression programs. This suggests that PDO-specific cell clusters reflect different PDO properties. Moreover, they subjected two other BC tissues or PDO pairs to RNA-seq and revealed that although a complete one-to-one correspondence was not achieved, PDOs retained some degree of tumor heterogeneity of the original tissue after successive passages (40).
5 Clinical application of organoids
5.1 PDOs as a preclinical BC model for drug discovery
PDOs can be used as a preclinical model for drug testing to elucidate the medicinal value of some compounds or small-molecule antagonists, thereby opening new avenues for developing novel pharmacological treatment modalities for cancer (41). Wu et al. identified that MS023, a type I protein arginine methyltransferase (PRMT) inhibitor that is presently in clinical development and not yet approved for use, exhibits antitumor growth activity in TNBC cell lines. However, they observed that TNBC PDOs exhibited different drug responses to MS023, with only 50% of TNBC PDOs being sensitive to this inhibitor and the remaining PDOs exhibiting resistance. Furthermore, they noted that the TNBC cells most sensitive to MS023 treatment exhibited PRMT1, which may be a therapeutic target for MS230. This study provides important insights into the pharmacological mechanisms underlying MS230 to develop targeted therapies for TNBC and other cancers and potential novel opportunities for cancer immunotherapy (42). Moreover, Sun et al. revealed an association between elevated SOST expression and bone metastasis in BC; this correlated with decreased survival rates among patients. Interestingly, SOST inhibition markedly decreased the bone metastatic capacity of SCP2 cells. Mechanically, SOST interacts with STAT3 to amplify the TGF-β/KRAS signaling pathway, thereby promoting BC growth and facilitating bone metastasis. To explore novel agents for patients with BC and bone metastasis, Sun et al. screened ~120,000 compounds from a small-molecule chemical library and observed that compound S6 significantly inhibited SCP2 cell proliferation in a dose- and time-dependent manner and effectively inhibited BC PDO growth. As a result, they identified S6 as a candidate compound for further development as a novel cancer therapeutic agent (43). Ma et al. performed a high-throughput screening of large tumor suppressor (LATS) kinase inhibitors using an in vitro LATS kinase assay and identified one effective LATS inhibitor, i.e., VT02956, from ~17,000 compounds. They observed that VT02956 inhibits ESR1 expression by targeting the Hippo pathway, resulting in the growth of ER+ BC cells and patient-derived tumor organoids, with little cytotoxicity in other cells. They concluded that LATS is an unexpected target for cancer therapy, particularly for endocrine-resistant breast cancer, and provided a novel idea for treating ER+ BC (44). JNK-IN-8 is a c-Jun N-terminal kinase (PRMTs) inhibitor that plays a tumor suppressor role in osteosarcoma, colorectal cancer, and pancreatic cancer; however, it has not been used in clinical trials. Soleimani et al. noted the significant inhibitor effect of JNK-IN-8 on TNBC cells, with translucent cytoplasmic vacuoles with lysosomal characteristics. Furthermore, JNK-IN-8 effectively inhibited the growth of TNBC PDOs, exhibiting rupture and disintegration phenotypes. Except for inhibiting JNK, JNK-IN-8 activates lysosomal biogenesis and autophagy by targeting TFEB/TFE3. Their results suggest that JNK-IN-8 is a novel and promising treatment for TNBC in the future (45). BC stem-like cells (BCSCs) are the potential reason for the recurrence, metastasis, and drug resistance of TNBC. In 2023, Liu et al. observed that CRM1 expression was significantly increased in the tumor tissue samples of patients with BC; this was closely associated with a poor prognosis. They developed LFS-1107, a highly effective small-molecule synthetic analog that targets CRM1 in cells. LFS-1107 can inhibit TNBC cells at low concentrations and selectively remove CD44+CD24− BCSCs. Furthermore, in a mouse xenotransplantation model, LFS-1107 strongly inhibited tumor growth and removed BCSCs in residual tumor tissues. LFS-1107 also prompted the nuclear retention of survivin, inhibiting the transactivation capabilities of STAT3 and subsequently decreasing the downstream expression of stemness regulators. In addition, Liu et al. cultured PDOs from the tumor tissues of patients with TNBC and observed that LFS-1107 inhibited TNBC PDOs more significantly compared with approved cancer drugs. Finally, they demonstrated that LFS-1107 can enhance the killing effect of chemotherapeutic agents and downregulate multidrug resistance-associated protein targets (46). Although PDO provides valuable tools for drug discovery, challenges in standardization and scalability may limit its widespread applicability.
5.2 PDOs can predict the response of patients with BC to drugs
To date, several organoids have been cultured from tumors directly sourced from patients and healthy breast tissues, facilitating the establishment of an organoid repository. In addition to capturing the diverse characteristics and histology of a patient’s tumor, these organoids can also predict the responsiveness of an individual patient to specific drugs. Subsequently, they offer insights into the expected drug reactions of patients (19, 47, 48) (Figure 3). Shu et al. collected 17 biopsy samples from suspected patients with BC; five organoids were successfully constructed from the tissue biopsies. They tested the sensitivity of the organoids to different neoadjuvant chemotherapy (NAC) drugs and observed that different BC PDOs responded differently to the NAC drugs. For example, BCb20, BCb27, and BCb30 belonged to HR-positive organoids; however, BCb20 was sensitive to epirubicin, and BCb27 and BCb30 were resistant to epirubicin. Furthermore, BCb22 and BCb21 belonged to HER2-positive organoids, whereas BCb22 was sensitive to carboplatin, but BCb21 was resistant to carboplatin. Finally, Su et al. compared the clinical responses of patients with BC with the results of in vitro experiments and observed that the responses of PDOs to each drug were consistent with the clinical responses of patients. Therefore, they concluded that PDOs can be utilized to predict a patient’s response to monotherapy and a patient’s efficacy to a combination of drugs or NAC, eventually leading to individualized treatment. To compare drug responses in both in vitro and in vivo contexts, Sachs et al. generated 12 BC organoids from the needle biopsies of 13 patients diagnosed with metastatic BC. They reported how these patients responded to standard-of-care treatments. For drug susceptibility tests, they selected two BC organoids sensitive and resistant to tamoxifen. The in vitro responses of BC organoids to tamoxifen were consistent with the responses of corresponding patients; this suggests that BC organoids can be utilized as predictive in vitro substitutes for BC in vivo (19). In addition, Guillen et al. observed that the drug responses of PDX-derived organoids (PDxO) are consistent with responses in vivo. They used 16 PDxO models to screen 45 compounds. Thereafter, they selected the drug that exhibited a unique pattern of response or resistance in the PDxO models and tested the drug in vivo on the PDX model. Most PDX models exhibited drug responses similar to the PDO models (49). Collectively, BC PDOs provide a more accurate and promising research model for predicting drug responses in patients with BC, leveraging its ability to simulate patient conditions. However, while the prospects are optimistic, further validation is warranted to ensure reliability. Nevertheless, PDO holds promise in advancing individualized cancer treatment strategies.
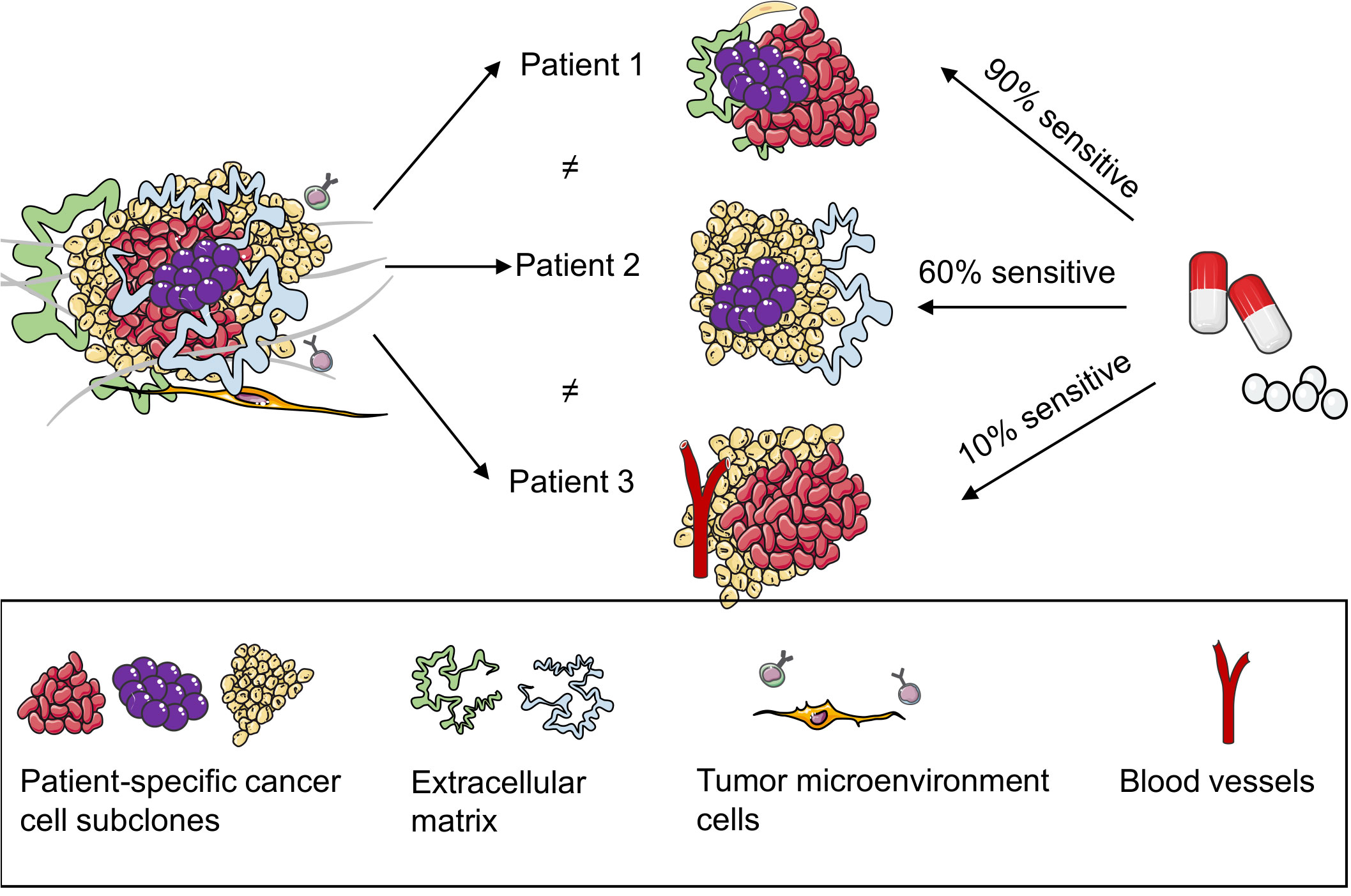
Figure 3 Patient-derived organoids (PDOs) can predict the responses of patients with breast cancer to drugs. PDOs present a model for elucidating the heterogeneity within patient tumors, adept at preserving multiple cell types and subpopulations inherent to the patient’s tumor. Subsequently, they proficiently retain the drug response characteristics of the patient’s tumor, thereby promoting the accuracy of predicting the drug efficacy for individual patients.
5.3 TME and immunotherapy
TME refers to a complex network of cells, stroma, and signaling molecules that surround tumor cells and play an essential role in tumor growth, invasion, metastasis, and therapy responses. The fundamental constituents within the TME are as follows (50–52):
1. Incorporating immune components: Immune cells such as T cells, B cells, natural killer cells, macrophages, and dendritic cells have a pivotal effect on the TME. Their interactions significantly shape tumor immune reactions and subsequently affect the efficacy of therapeutic interventions. Stromal cells such as fibroblasts, endothelial cells, and smooth muscle cells constitute the tumor stroma; they provide support for tumor growth and invasion.
2. ECM: The ECM comprises proteins, polysaccharides, and other substances, and it forms the tumor stroma. The ECM plays a vital role in tumor cell migration, invasion, and cellular signaling.
3. Blood vessels: Neovascularization formation plays a vital role in tumor growth and nourishment; this is closely associated with the malignancy and prognosis of tumors.
4. Cytokines and growth factors: Cytokines and growth factors such as tumor necrosis factor-α, interferons (IFNs), and vascular endothelial growth factor regulate immune responses, angiogenesis, and tumor growth.
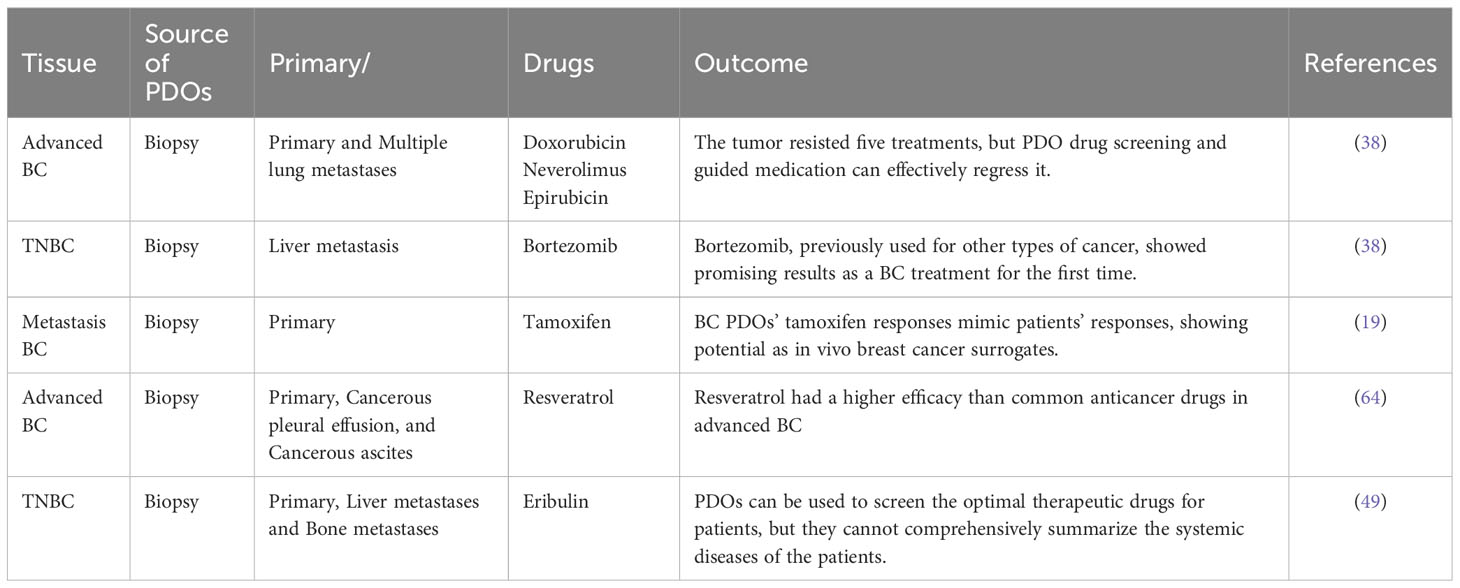
5. Interstitial fluid: The interstitial fluid is present in the interstitial spaces surrounding the tumor. It contains information on the tumor’s condition, including cellular fragments, DNA, and RNA.
6. Immune suppressive factors: Factors that inhibit immune responses are present in the TME, including immune checkpoint molecules, which help the tumor in evading immune attacks.
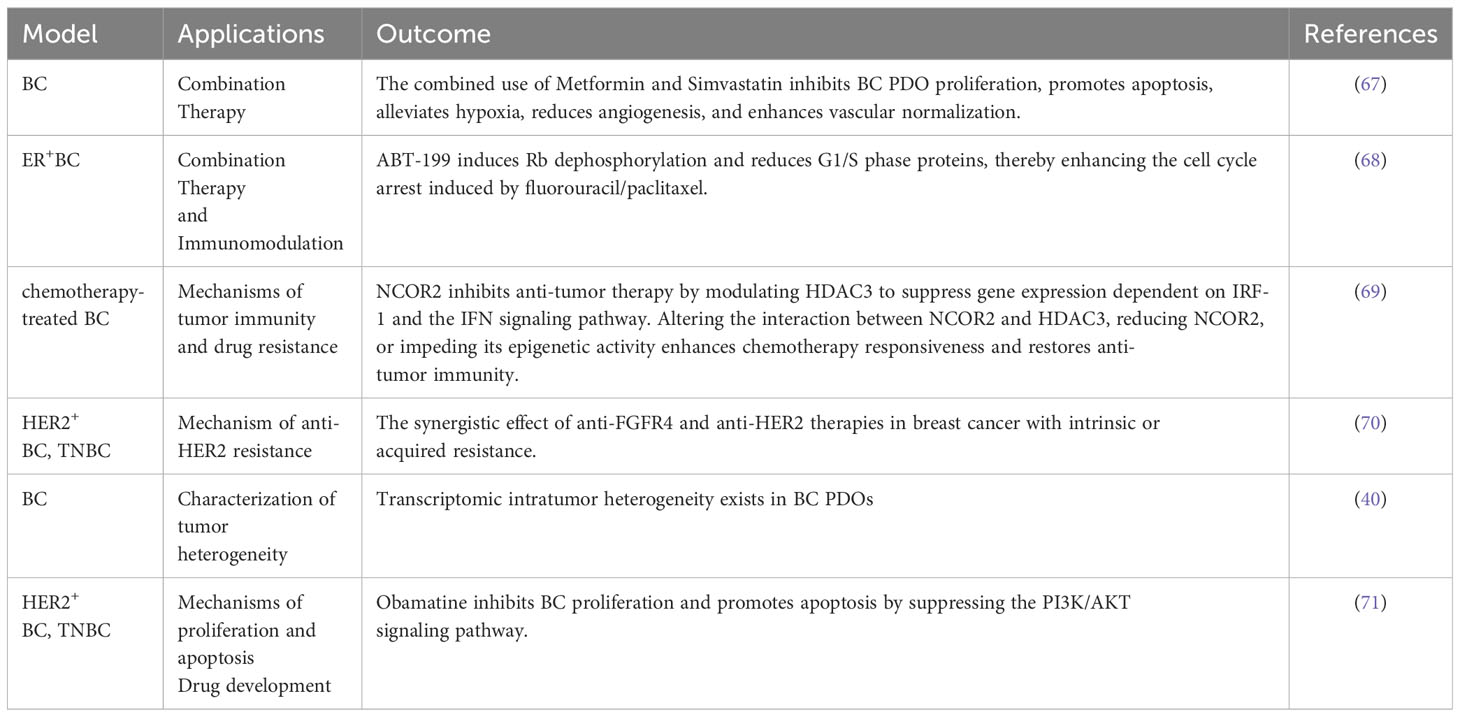
These components interact with each other, collectively forming the intricate TME network, which affects tumor behavior and treatment responses. Among them, immune cells and molecules can affect tumor growth, metastasis, invasion, and therapy responses, playing pivotal roles in controlling and inhibiting tumor development (53).
Recently, tumor immunotherapy has gained considerable attention as a prominent approach to cancer treatment. It activates or rallies the patient’s immune system, amplifying antitumor responses in the TME and regulating the elimination of tumor cells (54). However, owing to the complexity of the human immune system and its ineffective reconstruction, the efficacy of in vitro immunotherapy drug models does not match clinical responses in many cases (55). Therefore, there are considerable challenges associated with immune-tumor models, and in vitro models that can be used to verify the efficacy in individuals are urgently warranted in clinical settings.
Organoids provide a new opportunity for tumor immunotherapy. Owing to the precise recapitulation of immune cell and molecular interactions in the TME, PDOs offer a more authentic platform to elucidate the mechanisms underlying immunotherapy. Furthermore, leveraging PDO models derived from specific patients facilitates the prediction of individual responses to diverse immune-based therapeutic approaches. In addition, conducting drug screening within PDOs allows efficacy and toxicity assessment of various immunotherapy agents, thereby offering crucial insights into the development of novel immunotherapies (56, 57). This approach can expedite the drug development process for immunotherapies (Table 1).
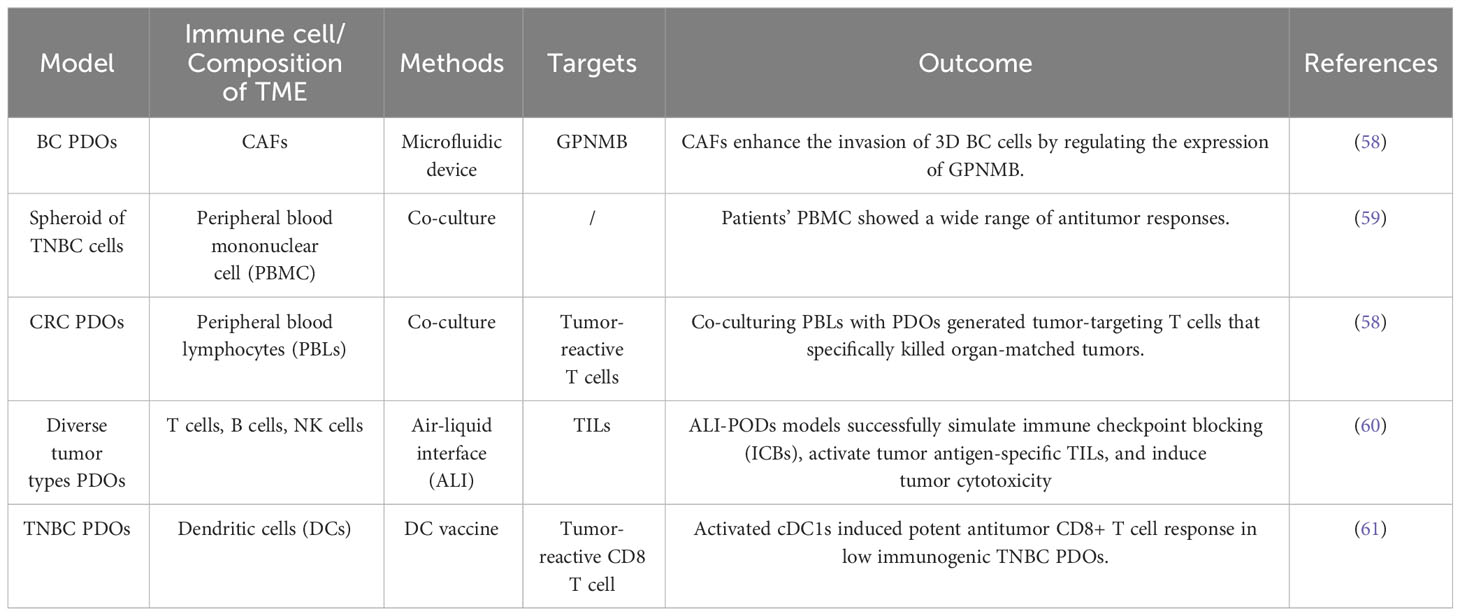
Table 1 Related studies on immunotherapy and the tumor microenvironment of patient-derived organoids (PDOs).
5.4 Drug screening and individualized therapy
In the field of oncology, the importance of individualized therapy is increasingly being emphasized, and PDOs have emerged as an innovative disease model, offering remarkable potential and opportunities for individualized treatment strategies. PDOs offer a distinct edge because they are developed from patient-derived tissue samples, facilitating the precise in vitro emulation of the distinct biological traits exhibited by individual tumors. This fidelity in replicating the unique characteristics and disease status of patients facilitates the generation of laboratory models that mimic the primary ailment of a patient. Therefore, they significantly help deepen our understanding of the fundamental nature of diseases and expedite the exploration of potential avenues for treatment. Furthermore, PDOs can be used to predict a patient’s response to specific treatment modalities, helping clinicians devise optimal therapeutic strategies. By evaluating diverse drugs or treatment protocols, we can identify the most effective therapeutic approach at an individual level. This process contributes to the increased treatment success rates while minimizing unnecessary drug exposure and potential side effects, thereby providing robust support for developing and refining individualized treatment strategies (38, 62, 63). With the continuous optimization of the 3D culture system of PDOs, many laboratories can culture organoids in large quantities. Therefore, PDO models are also employed for the high-throughput screening of different anticancer drugs to rapidly and accurately obtain individualized treatment regimens for patients (41). Chen et al. collected 132 BC tissues from 125 patients, including 77 who did not receive anticancer therapy and 55 who did. They developed 99 personalized cancer models (PDOs) from these tissues and established a drug library with 49 anticancer drugs. By screening the PDOs, they identified the most sensitive drugs for each patient, resulting in the successful development of patient-specific treatment strategies (38). More relevant studies on the applications of PDOs in drug screening and individualized therapy are presented in Table 2. BC PDOs represent a tailored treatment model to meet specific patient needs. However, there is potential variability in drug efficacy, and rigorous validation is warranted to ensure the reliability of individualized therapeutic strategies. Furthermore, the widespread application of PDOs remains limited. Despite these challenges, the prospect of improving treatment precision with PDOs remains a driving force in advancing individualized medicine.
5.5 Studies on the mechanisms underlying tumor progression
Compared with traditional cell lines, PDOs have the advantage of preserving tumor heterogeneity and maintaining a higher degree of TME. Therefore, by studying PDOs, researchers can achieve a more comprehensive understanding of the molecular and cellular mechanisms underlying BC development (4, 65). For example, Priscilla et al. established a BC PDO migration model. Using a 3D microfluidic system and computational model, they discovered that during the collective migration of heterogeneous tumor and tumor stromal cell clusters, the leading migrating cells identify and segregate a specific subset that is characterized by the collective migratory potential, notably the keratin-14 and calponin-3-positive subgroups. This process allows the migrating cells to control their protrusion dynamics by locally producing laminin; this emphasizes how these cells guide collective migration by interacting with the microenvironment (66). Moreover, a study revealed that metformin and simvastatin, two metabolism-related drugs, play crucial roles in modulating hypoxic TME and angiogenesis. Furthermore, the combined use of metformin and simvastatin inhibits BC PDO proliferation, promotes apoptosis, alleviates hypoxia, decreases angiogenesis, and enhances vascular normalization (67). Table 3 summarizes the studies on other PDOs.
6 Limitations and potential applications
In this review, we primarily discussed the research and application prospects of BC PDO models. PDOs can preserve a part of the TME of the original tissue, cell morphology, tumor heterogeneity, and the patient’s responses to drugs. Moreover, even after several passages, most of their features are preserved. However, the current PDO models remain immature and imperfect. One limitation of the PDO model is that it cannot generalize the entire TME of the patient and lacks a tumor matrix, blood vessels, and immune cells. In response, Neal et al. co-cultured PDOs with other cellular components to reconstruct the TME (60). Furthermore, Davaadelger et al. added fibroblasts to PDOs to investigate the effect of BRCA1 mutations on progesterone in breast cells (72). In addition, extracellular elements and microorganisms may be co-cultured with PDOs in the future (73, 74).
Organoids can be established from patient-derived tumor tissues and can be efficiently expanded and passaged for further drug screening. This makes PDO a valuable model for translating into practical applications and tailoring individualized cancer treatments. One of the most promising applications of PDOs is their ability to predict a patient’s drug response, possibly guiding individualized cancer treatment. For example, Chen et al. successfully guided the individualized treatment of six patients with BC via the drug sensitivity experiments of their PDOs; all patients achieved good efficacy (38). However, PDOs cannot predict the drug responses of the patient’s whole body.
In contrast, organoids include microfluidic systems with advanced 3D bioprinting technologies as the new technology. By employing various bioprinting techniques (such as extrusion, inkjet, photocuring, etc.), the use of bioinks, such as natural or synthetic polymers, cells, or biomolecules, creates complex and anatomically accurate structures and makes each organoid more uniform in volume, which can simulate the biophysical properties of tumors more realistically (75, 76). For example, Heinrich et al. used 3D printing technology to build mini-brain models of glioblastoma and macrophages to study cell interactions and tested the therapies targeting this interaction (77). In addition, the fluid of the microfluidic device can flow and connect to organoids of different tissue origins, further simulating the exchange of nutrients and substances and the formation of vascular networks between tumors. Therefore, the behavior of tumor metastasis and spread can be better simulated (78, 79). Skardal et al. established a “metastasis-on-a-chip” system and simulated the liver metastasis of colon cancer (80). In BC, Swaminathan et al. used bioprinting to directly print 3D spheres of breast epithelial cells and co-culture them with vascular endothelial cells, thereby successfully creating tissue models that could be used for drug efficacy studies (81). However, the accuracy of 3D printing, the cell survival rate, and how to accurately replicate the heterogeneity of the tumor in the model requires further research and validation.
Finally, organoids exhibit considerable potential in regenerative medicine. In addition to being derived from a patient’s tumor tissues, PDOs can also be developed from a patient’s stem cells. Dai et al. noted that breast organoids can be established from human-induced pluripotent stem cells and can differentiate into breast components in immunodeficient mice to further regenerate into breast-like structures. In the future, the issue of BC cell repair and regeneration may be solved after resection (82).
In general, as a new clinical model, PDOs are not only important to study BC mechanisms but also to screen drugs and guide treatment regimens. In the future, research directions can focus on updating breast organoid culture techniques and how to simulate the TME more comprehensively. In conclusion, PDO technology will continue to be an important tool in BC research and clinical drug screening.
Author contributions
YS: Writing – original draft. ZG: Writing – review & editing. GC: Writing – original draft. YN: Writing – original draft. CZ: Writing – review & editing. WL: Writing – review & editing. JL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by a grant from the project of “Dengfeng Plan” High-level Hospital Construction Project of First People’s Hospital of Foshan (No. 2021B001), the Natural Science Foundation of Guangdong Province, China (2022A1515140183).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Trapani D, Ginsburg O, Fadelu T, Lin NU, Hassett M, Ilbawi AM, et al. Global challenges and policy solutions in breast cancer control. Cancer Treat Rev (2022) 104:102339. doi: 10.1016/j.ctrv.2022.102339
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Duggan C, Trapani D, Ilbawi AM, Fidarova E, Laversanne M, Curigliano G, et al. National health system characteristics, breast cancer stage at diagnosis, and breast cancer mortality: a population-based analysis. Lancet Oncol (2021) 22(11):1632–42. doi: 10.1016/S1470-2045(21)00462-9
4. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer (2018) 18(7):407–18. doi: 10.1038/s41568-018-0007-6
5. Danenberg E, Bardwell H, Zanotelli VRT, Provenzano E, Chin SF, Rueda OM, et al. Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat Genet (2022) 54(5):660–9. doi: 10.1038/s41588-022-01041-y
6. Mittal S, Brown NJ, Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Rev Mol Diagn (2018) 18(3):227–43. doi: 10.1080/14737159.2018.1439382
7. Houthuijzen JM, Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev (2018) 37(4):577–97. doi: 10.1007/s10555-018-9768-3
8. Qiu SQ, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schröder CP. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat Rev (2018) 70:178–89. doi: 10.1016/j.ctrv.2018.08.010
9. Lyu X, Zhang Q, Fares HM, Wang Y, Han Y, Sun L. Contribution of adipocytes in the tumor microenvironment to breast cancer metabolism. Cancer Lett (2022) 534:215616. doi: 10.1016/j.canlet.2022.215616
10. Wu Q, Li B, Li Z, Li J, Sun S, Sun S. Cancer-associated adipocytes: key players in breast cancer progression. J Hematol Oncol (2019) 12(1):95. doi: 10.1186/s13045-019-0778-6
11. Wang S, Xiong Y, Zhang Q, Su D, Yu C, Cao Y, et al. Clinical significance and immunogenomic landscape analyses of the immune cell signature based prognostic model for patients with breast cancer. Brief Bioinform (2021) 22(4):bbaa311. doi: 10.1093/bib/bbaa311
12. Dieci MV, Miglietta F, Guarneri V. Immune infiltrates in breast cancer: recent updates and clinical implications. Cells (2021) 10(2):223. doi: 10.3390/cells10020223
13. Gonzalez-Molina J, Moyano-Galceran L, Single A, Gultekin O, Alsalhi S, Lehti K. Chemotherapy as a regulator of extracellular matrix-cell communication: Implications in therapy resistance. Semin Cancer Biol (2022) 86(Pt 3):224–36. doi: 10.1016/j.semcancer.2022.03.012
14. Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling physiological events in 2D vs. 3D Cell Culture Physiol (Bethesda) (2017) 32(4):266–77. doi: 10.1152/physiol.00036.2016
15. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature (2009) 459(7244):262–5. doi: 10.1038/nature07935
16. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell (2015) 160(1–2):299–312. doi: 10.1016/j.cell.2014.11.050
17. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell (2018) 172(1–2):373–386.e10. doi: 10.1016/j.cell.2017.11.010
18. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology (2011) 141(5):1762–72. doi: 10.1053/j.gastro.2011.07.050
19. Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA (2009) 106(31):12771–5. doi: 10.1073/pnas.0906850106
20. Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology (2015) 148(1):126–36. doi: 10.1053/j.gastro.2014.09.042
21. Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol (2016) 18(3):246–54. doi: 10.1038/ncb3312
22. Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov (2017) 7(5):462–77. doi: 10.1158/2159-8290.CD-16-1154
23. Emerman JT, Burwen SJ, Pitelka DR. Substrate properties influencing ultrastructural differentiation of mammary epithelial cells in culture. Tissue Cell (1979) 11(1):109–19. doi: 10.1016/0040-8166(79)90011-9
24. Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol (1982) 99(1):31–68. doi: 10.1016/0022-5193(82)90388-5
25. Hiratsuka M, Senoo T, Kimoto T, Namba M. An improved short-term culture method for human mammary epithelial cells. Gan (1982) 73(1):124–8.
26. Parry G, Cullen B, Kaetzel CS, Kramer R, Moss L. Regulation of differentiation and polarized secretion in mammary epithelial cells maintained in culture: extracellular matrix and membrane polarity influences. J Cell Biol (1987) 105(5):2043–51. doi: 10.1083/jcb.105.5.2043
27. Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and Malignant human breast epithelial cells. Proc Natl Acad Sci USA (1992) 89(19):9064–8. doi: 10.1073/pnas.89.19.9064
28. Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and Malignant breast epithelial cells. Nat Methods (2007) 4(4):359–65. doi: 10.1038/nmeth1015
29. de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, et al. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol (2012) 228(3):300–9. doi: 10.1002/path.4096
30. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science (2019) 364(6444):952–5. doi: 10.1126/science.aaw6985
31. Asghar W, El Assal R, Shafiee H, Pitteri S, Paulmurugan R, Demirci U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater Today (Kidlington) (2015) 18(10):539–53. doi: 10.1016/j.mattod.2015.05.002
32. Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature (2018) 560(7718):325–30. doi: 10.1038/s41586-018-0409-3
33. Caponigro G, Sellers WR. Advances in the preclinical testing of cancer therapeutic hypotheses. Nat Rev Drug Discovery (2011) 10(3):179–87. doi: 10.1038/nrd3385
34. Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res (2008) 6(1):21–30. doi: 10.1158/1541-7786.MCR-07-0280
35. Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med (2015) 21(8):846–53. doi: 10.1038/nm.3915
36. Diermeier SD, Chang KC, Freier SM, Song J, El Demerdash O, Krasnitz A, et al. Mammary tumor-associated RNAs impact tumor cell proliferation, invasion, and migration. Cell Rep (2016) 17(1):261–74. doi: 10.1016/j.celrep.2016.08.081
37. Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell (2016) 167(1):260–274.e22. doi: 10.1016/j.cell.2016.08.041
38. Chen P, Zhang X, Ding R, Yang L, Lyu X, Zeng J, et al. Patient-derived organoids can guide personalized-therapies for patients with advanced breast cancer. Adv Sci (Weinh) (2021) 8(22):e2101176. doi: 10.1002/advs.202101176
39. Bhatia S, Kramer M, Russo S, Naik P, Arun G, Brophy K, et al. Patient-derived triple-negative breast cancer organoids provide robust model systems that recapitulate tumor intrinsic characteristics. Cancer Res (2022) 82(7):1174–92. doi: 10.1158/0008-5472.CAN-21-2807
40. Saeki S, Kumegawa K, Takahashi Y, Yang L, Osako T, Yasen M, et al. Transcriptomic intratumor heterogeneity of breast cancer patient-derived organoids may reflect the unique biological features of the tumor of origin. Breast Cancer Res (2023) 25(1):21. doi: 10.1186/s13058-023-01617-4
41. Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol (2017) 24(9):1092–100. doi: 10.1016/j.chembiol.2017.06.012
42. Wu Q, Nie DY, Ba-Alawi W, Ji Y, Zhang Z, Cruickshank J, et al. PRMT inhibition induces a viral mimicry response in triple-negative breast cancer. Nat Chem Biol (2022) 18(8):821–30. doi: 10.1038/s41589-022-01024-4
43. Sun L, Zhang Y, Chen G, Ji Y, Ma Q, Qiao X, et al. Targeting SOST using a small-molecule compound retards breast cancer bone metastasis. Mol Cancer (2022) 21(1):228. doi: 10.1186/s12943-022-01697-4
44. Ma S, Tang T, Probst G, Konradi A, Jin C, Li F, et al. Transcriptional repression of estrogen receptor alpha by YAP reveals the Hippo pathway as therapeutic target for ER+ breast cancer. Nat Commun (2022) 13(1):1061. doi: 10.1038/s41467-022-28691-0
45. Soleimani M, Somma A, Kaoud T, Goyal R, Bustamante J, Wylie DC, et al. Covalent JNK inhibitor, JNK-IN-8, suppresses tumor growth in triple-negative breast cancer by activating TFEB- and TFE3-mediated lysosome biogenesis and autophagy. Mol Cancer Ther (2022) 21(10):1547–60. doi: 10.1158/1535-7163.MCT-21-1044
46. Liu C, Zhang Y, Gao J, Zhang Q, Sun L, Ma Q, et al. A highly potent small-molecule antagonist of exportin-1 selectively eliminates CD44+CD24- enriched breast cancer stem-like cells. Drug Resist Update (2023) 66:100903. doi: 10.1016/j.drup.2022.100903
47. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (2018) 359(6378):920–6. doi: 10.1126/science.aao2774
48. Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov (2018) 8(9):1112–29. doi: 10.1158/2159-8290.CD-18-0349
49. Guillen KP, Fujita M, Butterfield AJ, Scherer SD, Bailey MH, Chu Z, et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer (2022) 3(2):232–50. doi: 10.1038/s43018-022-00337-6
50. Shu D, Shen M, Li K, Han X, Li H, Tan Z, et al. Organoids from patient biopsy samples can predict the response of BC patients to neoadjuvant chemotherapy. Ann Med (2022) 54(1):2581–97. doi: 10.1080/07853890.2022.2122550
51. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature (2013) 501(7467):346–54. doi: 10.1038/nature12626
52. Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol (2015) 25(4):198–213. doi: 10.1016/j.tcb.2014.11.006
53. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol (2020) 17(8):807–21. doi: 10.1038/s41423-020-0488-6
54. Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science (2015) 348(6230):62–8. doi: 10.1126/science.aaa4967
55. Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer (2017) 16(1):41. doi: 10.1186/s12943-017-0600-4
56. Grönholm M, Feodoroff M, Antignani G, Martins B, Hamdan F, Cerullo V. Patient-derived organoids for precision cancer immunotherapy. Cancer Res (2021) 81(12):3149–55. doi: 10.1158/0008-5472.CAN-20-4026
57. Ye W, Luo C, Li C, Huang J, Liu F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett (2020) 477:31–40. doi: 10.1016/j.canlet.2020.02.027
58. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell (2018) 174(6):1586–1598.e12. doi: 10.1016/j.cell.2018.07.009
59. Saraiva DP, Matias AT, Braga S, Jacinto A, Cabral MG. Establishment of a 3D co-culture with MDA-MB-231 breast cancer cell line and patient-derived immune cells for application in the development of immunotherapies. Front Oncol (2020) 10:1543. doi: 10.3389/fonc.2020.01543
60. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell (2018) 175(7):1972–1988.e16. doi: 10.1016/j.cell.2018.11.021
61. Huang L, Rong Y, Tang X, Yi K, Qi P, Hou J, et al. Engineered exosomes as an in situ DC-primed vaccine to boost antitumor immunity in breast cancer. Mol Cancer (2022) 21(1):45. doi: 10.1186/s12943-022-01515-x
62. Veninga V, Voest EE. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell (2021) 39(9):1190–201. doi: 10.1016/j.ccell.2021.07.020
63. Bonaventura P, Alcazer V, Mutez V, Tonon L, Martin J, Chuvin N, et al. Identification of shared tumor epitopes from endogenous retroviruses inducing high-avidity cytotoxic T cells for cancer immunotherapy. Sci Adv (2022) 8(4):eabj3671. doi: 10.1126/sciadv.abj3671
64. Ye HS, Gao HF, Li H, Nie JH, Li TT, Lu MD, et al. Higher efficacy of resveratrol against advanced breast cancer organoids: A comparison with that of clinically relevant drugs. Phytother Res (2022) 36(8):3313–24. doi: 10.1002/ptr.7515
65. Clevers H. Modeling development and disease with organoids. Cell (2016) 165(7):1586–97. doi: 10.1016/j.cell.2016.05.082
66. Hwang PY, Mathur J, Cao Y, Almeida J, Ye J, Morikis V, et al. A Cdh3-β-catenin-laminin signaling axis in a subset of breast tumor leader cells control leader cell polarization and directional collective migration. Dev Cell (2023) 58(1):34–50.e9. doi: 10.1016/j.devcel.2022.12.005
67. Liu J, Wang H, Zhang M, Li Y, Wang R, Chen H, et al. Metformin and simvastatin synergistically suppress endothelin 1-induced hypoxia and angiogenesis in multiple cancer types. Cancer Sci (2023) 114(2):640–53. doi: 10.1111/cas.15602
68. Li B, Qi F, Zhu F, Lu Z, Wang M, Chu T, et al. Nanoparticle-based combination therapy enhances fulvestrant efficacy and overcomes tumor resistance in ER-positive breast cancer. Cancer Res (2023) 83(17):2924–37. doi: 10.1158/0008-5472.CAN-22-3559
69. Tsai KK, Huang SS, Northey JJ, Liao WY, Hsu CC, Cheng LH, et al. Screening of organoids derived from patients with breast cancer implicates the repressor NCOR2 in cytotoxic stress response and antitumor immunity. Nat Cancer (2022) 3(6):734–52. doi: 10.1038/s43018-022-00375-0
70. Zou Y, Zheng S, Xie X, Ye F, Hu X, Tian Z, et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat Commun (2022) 13(1):2672. doi: 10.1038/s41467-022-30217-7
71. Lin X, Chen D, Chu X, Luo L, Liu Z, Chen J. Oxypalmatine regulates proliferation and apoptosis of breast cancer cells by inhibiting PI3K/AKT signaling and its efficacy against breast cancer organoids. Phytomedicine (2023) 114:154752. doi: 10.1016/j.phymed.2023.154752
72. Davaadelger B, Choi MR, Singhal H, Clare SE, Khan SA, Kim JJ. BRCA1 mutation influences progesterone response in human benign mammary organoids. Breast Cancer Res (2019) 21(1):124. doi: 10.1186/s13058-019-1214-0
73. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell (2016) 18(1):25–38. doi: 10.1016/j.stem.2015.12.005
74. Dai X, Wang X, Yang C, Huang M, Zhou Z, Qu Y, et al. Human fibroblasts facilitate the generation of iPSCs-derived mammary-like organoids. Stem Cell Res Ther (2022) 13(1):377. doi: 10.1186/s13287-022-03023-7
75. Miserocchi G, Bocchini M, Cortesi M, Arienti C, De Vita A, Liverani C, et al. Combining preclinical tools and models to unravel tumor complexity: Jump into the next dimension. Front Immunol (2023) 14:1171141. doi: 10.3389/fimmu.2023.1171141
76. Monteiro MV, Zhang YS, Gaspar VM, Mano JF. 3D-bioprinted cancer-on-a-chip: level-up organotypic in vitro models. Trends Biotechnol (2022) 40(4):432–47. doi: 10.1016/j.tibtech.2021.08.007
77. Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J. 3D-bioprinted mini-brain: A glioblastoma model to study cellular interactions and therapeutics. Adv Mater (2019) 31(14):e1806590. doi: 10.1002/adma.201806590
78. Neufeld L, Yeini E, Pozzi S, Satchi-Fainaro R. 3D bioprinted cancer models: from basic biology to drug development. Nat Rev Cancer (2022) 22(12):679–92. doi: 10.1038/s41568-022-00514-w
79. Sharifi M, Bai Q, Babadaei MMN, Chowdhury F, Hassan M, Taghizadeh A, et al. 3D bioprinting of engineered breast cancer constructs for personalized and targeted cancer therapy. J Control Release (2021) 333:91–106. doi: 10.1016/j.jconrel.2021.03.026
80. Skardal A, Devarasetty M, Forsythe S, Atala A, Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng (2016) 113(9):2020–32. doi: 10.1002/bit.25950
81. Swaminathan S, Hamid Q, Sun W, Clyne AM. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication (2019) 11(2):025003. doi: 10.1088/1758-5090/aafc49
Keywords: breast cancer, patient-derived organoids, tumor microenvironment, drug screening, individualized therapy
Citation: Shi Y, Guan Z, Cai G, Nie Y, Zhang C, Luo W and Liu J (2024) Patient-derived organoids: a promising tool for breast cancer research. Front. Oncol. 14:1350935. doi: 10.3389/fonc.2024.1350935
Received: 06 December 2023; Accepted: 11 January 2024;
Published: 26 January 2024.
Edited by:
Preethi Korangath, Johns Hopkins University, United StatesReviewed by:
Geeta Upadhyay, Uniformed Services University of the Health Sciences, United StatesMichele Zanoni, Scientific Institute of Romagna for the Study and Treatment of Tumors (IRCCS), Italy
Copyright © 2024 Shi, Guan, Cai, Nie, Zhang, Luo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Luo, bHVvd2VpXzQyMUAxNjMuY29t; Jia Liu, bWNsaXVqaWFAc2N1dC5lZHUuY24=
†These authors have contributed equally to this work
 Yixin Shi
Yixin Shi Zhanwen Guan
Zhanwen Guan Gengxi Cai3
Gengxi Cai3 Yichu Nie
Yichu Nie Wei Luo
Wei Luo