- 1Institute for Brain Research and Rehabilitation, South China Normal University, Guangzhou, China
- 2Department of General Medicine, Shenzhen Longhua District Central Hospital, Shenzhen, China
- 3Xianning Medical College, Hubei University of Science and Technology, Xianning, China
- 4Department of Geriatric Medicine, Shenzhen Longhua District Central Hospital, Shenzhen, China
The core of tumor cell metabolism is the management of energy metabolism due to the extremely high energy requirements of tumor cells. The purine nucleotide synthesis pathway in cells uses the purinosomes as an essential spatial structural complex. In addition to serving a crucial regulatory role in the emergence and growth of tumors, it contributes to the synthesis and metabolism of purine nucleotides. The significance of purine metabolism in tumor cells is initially addressed in this current article. The role of purinosomes as prospective therapeutic targets is then reviewed, along with a list of the signaling pathways that play in the regulation of tumor metabolism. A thorough comprehension of the function of purinosomes in the control of tumor metabolism can generate fresh suggestions for the creation of innovative cancer treatment methods.
Introduction
Metabolic regulation of tumor cells
Tumor cells have different metabolic properties than normal cells, the most notable of which is a high reliance on nutrients, particularly glucose and glutamine. Increased glucose absorption is a typical feature in tumor cells. The Warburg effect exists in tumor cells and is shown by enhanced glycolysis and lactic acid production, which are hallmarks of tumor cell metabolism (1). Many tumor cells are glutamine-dependent, because glutamine provides cells with the carbon and nitrogen they require for biosynthesis (2). Glutamine is a significant mitochondrial substrate and glutamine catabolism gives cells a potent source of NADPH synthesis. In addition, tumor cells are able to utilize other metabolic pathways for energy, such as fatty acid oxidation and amino acid metabolism (3, 4).
Tumor cells require constant energy and biomolecule acquisition to support their aberrant proliferative activity during growth and reproduction. Purines are a type of biomolecule that is primarily utilized to construct nucleic acids and energy transfer molecules (such as ATP and GTP). Purine metabolism in tumor cells has a unique requirement for their rapid growth and proliferation. Tumor cells often oversynthesize purines to meet their rapid proliferation needs, and they also supply the precursor molecules such as glucose-6-phosphate and phosphoribosyl to produce purines through the glycolytic pathway. As a result of their high metabolic demands, tumor cells may modify purine metabolic pathways to support their growth, division, and proliferation. For example, tumor cells directly regulate several genes encoding key enzymes in the purine biosynthesis pathway, such as ADSL and IMPDH2, through the transcription factor MYC, and then up-regulate enzymes related to purine synthesis to maintain cell self-renewal and proliferation (5–7). Thus, purine metabolic pathways in tumor cells can be studied to learn more about how to design treatment approaches that specifically target cancer cells.
Tumor metabolic regulation not only affects the metabolic state of tumor cells themselves, but also affects the activation of immune cells, the generation of tumor-related blood vessels, as well as the invasion and metastasis of tumor cells by regulating the metabolic state of tumor microenvironment (8). Lactate acidification is a metabolic characteristic found in tumor tissues. Purine metabolism in tumor cells can influence lactate generation and acidification processes. Purine metabolism disorders may result in lower intracellular ATP levels, which activate the acidification pathway and allow tumor cells to adapt to the malignant environment by lactate acidification. Because of the accumulation of the glycolytic metabolite lactate, it can lower the pH value in the microenvironment and provide a favorable survival environment for tumor cells, which promotes tumor growth, invasion, and development (9, 10) as well as immune escape (11).
The management of tumor metabolism is critical for energy supply, biosynthesis, tumor cell growth and transformation, and tumor microenvironment regulation. An in-depth understanding of tumor metabolic regulatory mechanisms is extremely valuable for developing new cancer therapy techniques and improving treatment outcomes.
Beginning with an overview of the significance of purine metabolism in tumor cells, this paper goes on to discuss the identification of purinosomess and the current state of research into their composition and roles. Next, in an effort to generate fresh concepts for the creation of novel tumor treatment approaches, we enumerated the signaling pathways associated with purinosomes in the control of tumor metabolism, including the AMPK, mTOR, and HIF-1 signaling pathways. We also talked about the potential of purinosomes as therapeutic targets.
Discovery and function of purinosome
De novo purine synthesis is a ten-step process that produces inosine 5’ -monophosphate (IMP), adenosine 5’ -monophosphate (AMP), and guanosine 5’ -monophosphate (GMP). It is catalyzed by six different enzymes and can be split into two stages: the synthesis of IMP comes first, and then AMP and GMP are produced. Two phases come together to form the IMP. Three monofunctional enzymes phosphoribosyl pyrophosphate amidotransferase (PPAT), formylglycinamidine ribonucleotide synthase (FGAMS), and adenylosuccinate lyase (ADSL), two bifunctional enzymes phosphoribosyl aminoimidazole succinocarboxamide synthetase (PAICS) and IMP cyclohydrolase (ATIC), and a trifunctional enzyme GARS-GAR transformylase-aminoimidazole ribonucleotide synthetase (TrifGART) are among the enzymes involved in de novo purine synthesis. The de novo purine synthesis route involves several different enzymes, each of which is crucial to the process. Thus, some scientists started to question whether there was anything that organisms could do to ensure that this chain of events occurred frequently enough to guarantee the effective production of purines for cells.
Back in 2008, Songon An et al. published an article in Science on reversible compartmentalization of de novo purine biosynthesis complexes, which first proposed the concept of purinosomes. Purinosomes are punctate multienzyme complexes that are reversibly assembled by enzymes in the de novo purine synthesis pathway (12). Hong Zhao et al. found higher levels of IMP in cells rich in purinosomes when comparing cells treated with 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT), a casein kinase 2 (CK2) inhibitor that induces the development of purinosomes, to cells cultivated in purine removal media. Furthermore, when cells were cultivated under purine depletion conditions, de novo manufacture of IMP rose by 50%. Purinosomes, it is determined, are functional compounds that increase de novo metabolic flux when purine demand is high (13). Minjoung Kyoung et al. promptly inserted GFP-labeled purine remedy production pathway enzyme human hypoxanthine phosphoribosyltransferase 1 (HPRT1) into Hs578T cells, but under purine depletion circumstances, HPRT1-GFP did not create purinosomes in Hs578T cells. HPRT1-GFP co-transfection with FGAMS-OFP did not form purinosomes in purine-depleted Hs578T cells. Under purine depletion conditions, the human purine remedy production route is shown to be independent of purinosome assembly (14).
Purinosomes are now characterized using a range of biological approaches. Anthony M. Pedley and Stephen J. Benkovic developed a method to visualize purinosomes in living cells and a method to detect purinosome in fixed cells with the help of immunofluorescence (15). Meanwhile, In addition to the immunofluorescence technique, the study by Chung Yu Chan et al. also referred to the basic morphological characterization, that is, the average size and average quantity of purinosomes in a cell were used as the physical criteria to distinguish purinosomes from other cell bodies (16). Yijun Deng et al. probed protein-protein interactions within purinosomes with a novel application of the Tango PPI reporter system and found that three enzymes in the de novo purine biosynthesis pathway, PPAT, TrifGART, and FGAMS, form the core of purinosomes (17). Based on diffusion coefficient data obtained for six de novo purine biosynthesis enzymes in purine depleted medium, Minjoung Kyoung et al. discovered that PPAT, TrifGART, and FGAMS had similar diffusion coefficients, and PAICS and ADSL had similar diffusion coefficients. Three intermediates are proposed for enzymes involved in de novo purine biosynthesis in cells during purine deficiency (14).
Some investigations have discovered that in the presence of purine deficit, treating cells with nocodazole, a small molecule inhibitor of microtubule polymerization, causes microtubule depolymerization, which is not beneficial to the creation of purinosomes. Even if the medium is poor in purine, the amount of de novo purine biosynthesis by cells is reduced. Purinosomes were clearly integrated in microtubule networks, according to cellular imaging of the cytoskeleton structure in the presence of purinosomes. In conclusion, a microtubule network organizes the spatial distribution of purinosomes, and tubulin disintegration inhibits de novo purine biosynthesis (18).
De novo purine biosynthesis necessitates ATP consumption, and scientists are intrigued by the existence of correlations in the subcellular localization between purinosomes and mitochondria. Jarrod B. French et al. used 3D random optical reconstruction microscopy to show that purinosomes colocalize with mitochondria by imaging HeLa cells under conditions that encourage purinosomes production (19). Julie Williamson et al. found that under normal physiological conditions purine biosynthetic enzymes are present in all compartments of hippocampal neurons and that these enzymes are localized around mitochondria (20). Colleen A. Mangold et al. used HSV-1 virus infection as a natural cellular stressor to initiate high purine demand, discovering that de novo purine biosynthesis may occur at specific local sites within neurons and that this phenotype is conserved across species. Colocalization of FGAMS with mitochondria was also identified in neurons, implying that the functional relationship between mitochondria and de novo purine biosynthesis is conserved across cell types (21).
Vidhi Pareek et al. proved the existence of purinosomes directly in 2020 by using metabolomics and gas cluster ion beam secondary ion mass spectrometry to perform in situ three-via microchemical imaging of single cells to directly visualize multienzyme complexes of purinosomes that act synergistically to increase pathway flux by approximately 7-fold (22).
Characteristics of purinosome
Cell cycle dependence
Purinosomes self-assemble in response to the cell’s purine requirements. Purinosomes assemble and deconstruct themselves during the phase of cell division because it has the largest need for purines. This is known as cell cycle dependence. Studies demonstrated that the amount of purinosomes in Hela cells rises during the G1 phase, when the cells prepare the material for DNA replication (16). The availability of phosphoribosyl pyrophosphate (PRPP) and Pi affects purine synthesis that is reliant on the cell cycle. Since PRPP is the rate-limiting substrate in the first step of the de novo purine biosynthesis pathway and PRPP synthetase activity increases between G1 and S phases, purine synthesis increases when cells move from G1 to S phase (23).
Reversibility of assembly
Purinosome construction is reversible, allowing it to be carried out in response to the need for intracellular purine. A research shows that the proportion of purinosome-positive cells in Hela cells peaked in the G1 phase and gradually declined in the S and G2/M phases (16). Due to their purine-deficient development environment, the G1 phase cells have the highest purine demand, which stimulates the de novo purine biosynthesis pathway and produces a large amount of purinosomes. However, as cell growth continues, the need for purine gradually declines. Because the purine metabolites already present in the cell might activate the rescue purine synthesis pathway, this results in a reduction in the number of positive cells. After 12 hours, there were less positive cells because there were fewer cells in the G1 phase. This offers more proof that purinosome assembly is reversible and that it may be dynamically modified to meet the needs of the cell (16). It has been found that sequential addition of two CK2 inhibitors, DMAT and 4,5,6,7-tetrabromobenzotriazole (TBB), to HeLa cells can reverse purinosomes assembly. This finding directly demonstrated the reversibility of purinosomes (24, 25).
Mechanism of purinosome in the regulation of tumor metabolism
AMPK signaling pathway
AMP-activated protein kinase (AMPK) is an important cellular energy-sensing protein kinase and a key regulator of purine synthesis. It directly regulates nucleic acid synthesis in tumor cells (26). The role of AMPK as the cell’s primary metabolic hub, which is activated whenever the AMP/ATP ratio is high, has been found to suppress the growth of a number of cancer cells as well as the mammalian target of rapamycin (mTOR) signaling pathway. The metabolite 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) is an allosteric activator of AMPK and the substrate of the last enzyme in the de novo purine synthesis pathway. Increased de novo synthesis pathways of purine nucleotides often imply increased assembly of purinosomes, which will lead to the accumulation of AICAR, and increased endogenous AICAR activates AMPK and its downstream signaling pathways (27–29).
Based on the information provided earlier, purinosomes are made up of a number of enzymes involved in de novo purine synthesis, each of which is essential and very important. The research demonstrated that after treatment with an AMPK activator, FGAMS, one of the six pathway enzymes involved in de novo purine synthesis, exhibited single-enzyme self-assembly. Lower levels of purine metabolites such as IMP, AMP, GMP, and ATP in HeLa cells show a downregulation of de novo purine synthesis caused by AMPK’s encouragement of FGAMS self-assembly. The study revealed that the downregulation of purine production in human cells is driven by AMPK’s spatial isolation of FGAMS, and indirectly demonstrated that AMPK signaling pathway has a certain influence on purinosomes assembly (30). The actual pathway can be referred to Figure 1. Furthermore, it has been discovered that by raising the quantity of AMP within cells, ADSL activation might lead to AMPK activation (31). Additionally, it has been demonstrated that AICAR buildup brought on by ATIC enzyme inhibition might indirectly activate AMPK (32, 33). When considered collectively, these findings point to AMPK’s significant regulatory function in the de novo purine synthesis.
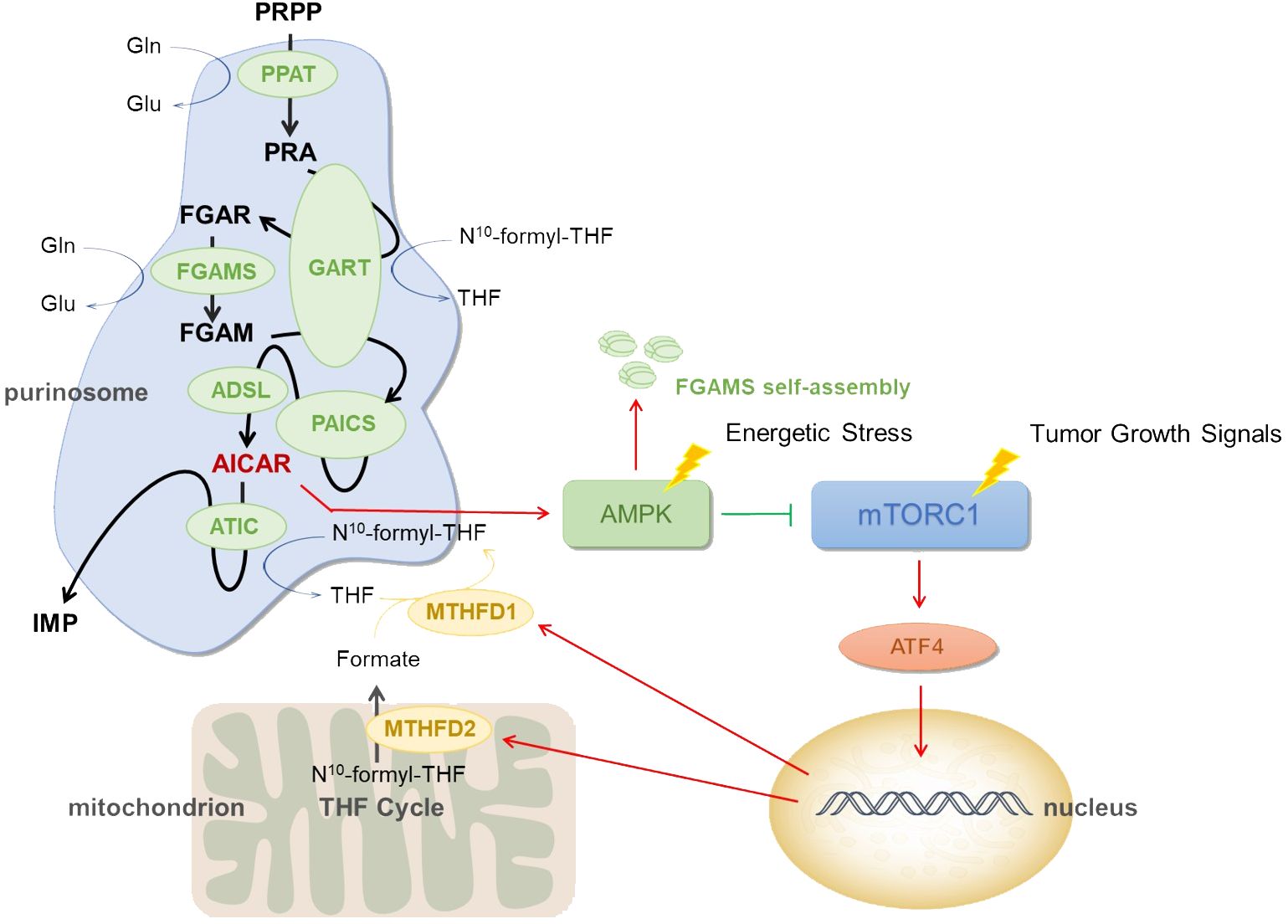
Figure 1 Mechanism of purinosome. Low blood sugar, hypoxia, ischemia, and other conditions are examples of energy stress. When cells are subjected to these energy stresses, they may cause purinosomes assembly and increase de novo purine synthesis. PRPP, phosphoribosyl pyrophosphate; PPAT, phosphoribosyl pyrophosphate amidotransferase; PRA, phosphoribosyl amine; GART, GARS-GAR transformylase-aminoimidazole ribonucleotide synthetase; FGAR, formylglycineamide ribotide; FGAMS, formylglycinamidine ribonucleotide synthase; FGAM, formylglycineamidine ribotide; PAICS, phosphoribosyl aminoimidazole succinocarboxamide synthetase; ADSL, adenylosuccinate lyase; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; ATIC, IMP cyclohydrolase; IMP, inosine 5’ -monophosphate; N10 -formyl-THF, N10 -formyl tetrahydrofolate; THF, tetrahydrofolate; Gln, glutamine; Glu, glutamate; MTHFD1, methylenetetrahydrofolate dehydrogenase 1; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; AMPK, AMP-activated protein kinase; mTORC1, mammalian target of rapamycin complex 1.
mTOR signaling pathway
Mammalian target of rapamycin is a key protein that controls cell proliferation and metabolism, and overactivation of this protein has been linked to the incidence and growth of numerous cancers. By controlling the mitochondrial tetrahydrofolate cycle, mTOR influences how purinosomes are produced and disassembled (34). mTORC1 signaling is connected to the expression of the mitochondrial enzyme methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) in both healthy and malignant cells. Activating transcription factor 4 (ATF4) stimulates the gene expression and serine biosynthesis necessary for the mitochondrial tetrahydrofolate (mTHF) pathway to produce formate (35). In order to increase the generation of formyl units needed for de novo purine biosynthesis and upregulate the de novo purine biosynthesis pathway, mTORC1 activates ATF4, boosts the translation of MTHFD2, and increases the expression of serine synthesis and other enzymes in the mTHF cycle (36). MTHFD2 expression regulates how much formates are released from the mitochondria into the cytoplasm. Tetrahydrofolate is finally incorporated by formate via methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) once they are in the cytosol, producing the 10-formyltetrahydrofolate cofactors necessary for GART and ATIC activity and subsequent IMP synthesis (37). See Figure 1.
Purinosomes are formed in cells as a result of mitochondrial dysregulation, and the reduction of MTHFD2 transcription following mitochondrial DNA depletion leads to a decrease in serine processed in one-carbon metabolism. The study found that FGAMS and ADSL in purinosome composition enzymes co-isolate from mitochondria in a rapamycin-dependent manner, implying that inhibiting mTOR disrupts purinosome-mitochondrial colocalization and inhibits purinosome formation stimulated by mitochondrial dysregulation (19).
mTOR has been demonstrated to stimulate the expression of genes involved in the pentose phosphate pathway, which results in the manufacture of PRPP, the substrate for the first step in the purine biosynthesis pathway (38–40). These findings imply that mTOR plays a critical regulatory role in the de novo purine biosynthesis pathway.
HIF-1 signaling pathway
Purinosome plays a role in tumor metabolism regulation by influencing the hypoxia inducible factor 1 (HIF-1) signaling pathway. As previously stated, tumor cells exhibit the Warburg effect, and glycolysis is active, with one of the causes being the activation of hypoxic-inducing molecules (41). Purinosome buildup in low oxygen environments can stabilize HIF-1α subunits and promote HIF-1 activation. HIF-1 can influence tumor cell metabolic adaptability and enhance glycolysis and lactic acid generation.
It has been discovered that cells have enhanced purinosome assembly when exposed to hypoxia. A class of proteins called hypoxia-inducible factors regulates how cells react to low oxygen levels and is essential for the construction of purinosome complexes under hypoxia. In cells lacking ATIC, hypoxia-driven purinosome assembly is suppressed. The glycolytic flux shifts into the pentose phosphate and serine biosynthesis pathways to provide enough PRPP and serine to maintain purine production, not the upregulation of transcription or translation of the enzymes within the de novo purine biosynthesis pathway, which would otherwise be the cause of the increase in purinosome formation under hypoxia (42).
Prospects
Only a small portion of the purine that the human body needs to synthesize on its own comes from diet. The primary purine synthesis mechanism in vivo is de novo purine biosynthesis. To keep the purine pool homeostasis in organisms, this process works in coordination with the rescue synthesis pathway of purine nucleotides. De novo purine synthetase builds purinosomes, which are structures that are focused on transferring and processing substrates quickly along the enzymatic pathway. Purinosomes are important because they boost intracellular metabolic flow effectiveness when there is a strong demand for purines. Purinosomes have been used as a therapeutic target as research has progressed, and signaling mechanisms connected to purinosome synthesis and breakdown, such as the mTOR signaling pathway, have been gradually identified. However, as current studies focus more on the formation conditions and components of purinosomes, the reference content of this paper is limited.
Due to its important role in tumor development, purinosome has become a potential target for cancer therapy. Anticancer medicines that target the de novo purine biosynthesis pathway are now classified into three types. For starters, purine analogues like 6-mercaptopurine are structurally identical to inoxanthine, which interferes with purine nucleotide synthesis and prevents cell development. The second type of analogue is amino acid analogues, such as aza-serine, which are structurally similar to glutamine and can disrupt the reaction in which glutamine participates in purine nucleotide synthesis. Third, folate analogues, such as methotrexate, down-regulate purine nucleotide synthesis by competitively inhibiting dihydrofolate reductase and lowering the level of tetrahydrofolate, the substrate for purine nucleotide synthesis. Anticancer drugs targeting enzymes related to purine de novo synthesis pathway collected from DrugBank include fluorouracil, mercaptopurine, etc. See Table 1 for details.
The argument made in this article is that future cancer treatments will increasingly focus on purine metabolism and target the construction and disassembly of purinosomes. Drugs made for purinosomes are still being developed, despite the fact that there are numerous therapeutic targets for the de novo purine biosynthesis pathway. Further research and discussion are needed to determine whether medications targeting the purinosome, a multi-enzyme complex, can be targeted by medications targeting the de novo purine biosynthesis pathway. There may also be distinctions between medications targeting the two. Some medications thought to target the de novo purine biosynthesis pathway may not be able to enter the purinosome structure and may only be able to act prior to the purinosome’s formation. However, there should also be medications that prevent the synthesis of purinosomes and subsequently purine metabolism. These medications may act as substrates or enzyme analogues in the construction of purinosomes, which would then directly participate in the competitive inhibition of purine synthesis by inhibiting the compartmentalization of purinosomes. It’s also feasible that the pathway for de novo purine synthesis is disrupted by the direct suppression of purinosome assembly. These hypotheses pave the path for further research on targeted purinosome medicines. Of course, the fact that the purinosomes mentioned above rely on microtubule networks makes them a possible target for anticancer drug development. At the same time, the AMPK, mTOR, and HIF-1 signaling pathways must be given more consideration when developing anti-cancer medications that target purinosomes. It is anticipated that these medications will have a very promising future.
Author contributions
CZ: Funding acquisition, Supervision, Writing – review & editing. JX: Investigation, Methodology, Resources, Software, Writing – original draft, Writing – review & editing. JL: Writing – review & editing. XC: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This present study was supported in part by the National Natural Science Foundation of China (grant numbers: 82270940). The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADSL, adenylosuccinate lyase; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; AMP, adenosine 5’ –monophosphate; AMPK, AMP-activated protein kinase; ATF4, activating transcription factor 4; ATIC, IMP cyclohydrolase; CK2, casein kinase 2; DMAT, 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole; FGAMS, formylglycinamidine ribonucleotide synthase; GART, GARS-GAR transformylase-aminoimidazole ribonucleotide synthetase; GMP, guanosine 5’ –monophosphate; HIF-1, hypoxia inducible factor 1; HPRT1, human hypoxanthine phosphoribosyltransferase 1; IMP, inosine 5’ –monophosphate; MTHFD1, methylenetetrahydrofolate dehydrogenase 1; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; mTHF, mitochondrial tetrahydrofolate; mTOR, mammalian target of rapamycin; PAICS, phosphoribosyl aminoimidazole succinocarboxamide synthetase; PPAT, phosphoribosyl pyrophosphate amidotransferase; PRPP, phosphoribosyl pyrophosphate; TBB, 4,5,6,7-tetrabromobenzotriazole.
References
1. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. (2016) 41:211–8. doi: 10.1016/j.tibs.2015.12.001
2. Yoo HC, Yu YC, Sung Y, Han JM. Glutamine reliance in cell metabolism. Exp Mol Med. (2020) 52:1496–516. doi: 10.1038/s12276-020-00504-8
3. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. (2013) 18:153–61. doi: 10.1016/j.cmet.2013.05.017
4. Engelen MP, Com G, Wolfe RR, Deutz NE. Dietary essential amino acids are highly anabolic in pediatric patients with cystic fibrosis. J Cyst Fibros. (2013) 12:445–53. doi: 10.1016/j.jcf.2012.12.011
5. Wang X, Yang K, Xie Q, Wu Q, Mack SC, Shi Y, et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat Neurosci. (2017) 20:661–73. doi: 10.1038/nn.4537
6. Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. (2009) 19:32–7. doi: 10.1016/j.gde.2009.01.002
7. Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. (2008) 7:2392–400. doi: 10.4161/cc.6390
8. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. (2012) 196:395–406. doi: 10.1083/jcb.201102147
9. Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, et al. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. (2009) 69:358–68. doi: 10.1158/0008-5472.CAN-08-2470
10. Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev. (2007) 26:299–310. doi: 10.1007/s10555-007-9064-0
11. Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. (2016) 24:657–71. doi: 10.1016/j.cmet.2016.08.011
12. An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. (2008) 320:103–6. doi: 10.1126/science.1152241
13. Zhao H, Chiaro CR, Zhang L, Smith PB, Chan CY, Pedley AM, et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J Biol Chem. (2015) 290:6705–13. doi: 10.1074/jbc.M114.628701
14. Kyoung M, Russell SJ, Kohnhorst CL, Esemoto NN, An S. Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry. (2015) 54:870–80. doi: 10.1021/bi501480d
15. Pedley AM, Benkovic SJ. Detecting purinosome metabolon formation with fluorescence microscopy. Methods Mol Biol. (2018) 1764:279–89. doi: 10.1007/978-1-4939-7759-8_17
16. Chan CY, Zhao H, Pugh RJ, Pedley AM, French J, Jones SA, et al. Purinosome formation as a function of the cell cycle. Proc Natl Acad Sci U S A. (2015) 112:1368–73. doi: 10.1073/pnas.1423009112
17. Deng Y, Gam J, French JB, Zhao H, An S, Benkovic SJ. Mapping protein-protein proximity in the purinosome. J Biol Chem. (2012) 287:36201–7. doi: 10.1074/jbc.M112.407056
18. An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci U S A. (2010) 107:12872–6. doi: 10.1073/pnas.1008451107
19. French JB, Jones SA, Deng H, Pedley AM, Kim D, Chan CY, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. (2016) 351:733–7. doi: 10.1126/science.aac6054
20. Williamson J, Petralia RS, Wang YX, Mattson MP, Yao PJ. Purine biosynthesis enzymes in hippocampal neurons. Neuromolecular Med. (2017) 19:518–24. doi: 10.1007/s12017-017-8466-6
21. Mangold CA, Yao PJ, Du M, Freeman WM, Benkovic SJ, Szpara ML. Expression of the purine biosynthetic enzyme phosphoribosyl formylglycinamidine synthase in neurons. J Neurochem. (2018) 144:723–35. doi: 10.1111/jnc.14304
22. Pareek V, Tian H, Winograd N, Benkovic SJ. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science. (2020) 368:283–90. doi: 10.1126/science.aaz6465
23. Fridman A, Saha A, Chan A, Casteel DE, Pilz RB, Boss GR. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem J. (2013) 454:91–9. doi: 10.1042/BJ20130153
24. Verrier F, An S, Ferrie AM, Sun H, Kyoung M, Deng H, et al. GPCRs regulate the assembly of a multienzyme complex for purine biosynthesis. Nat Chem Biol. (2011) 7:909–15. doi: 10.1038/nchembio.690
25. An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. (2010) 285:11093–9. doi: 10.1074/jbc.M110.101139
26. Qian X, Li X, Tan L, Lee JH, Xia Y, Cai Q, et al. Conversion of PRPS hexamer to monomer by AMPK-mediated phosphorylation inhibits nucleotide synthesis in response to energy stress. Cancer Discovery. (2018) 8:94–107. doi: 10.1158/2159-8290.CD-17-0712
27. Asby DJ, Cuda F, Beyaert M, Houghton FD, Cagampang FR, Tavassoli A. AMPK activation via modulation of de novo purine biosynthesis with an inhibitor of ATIC homodimerization. Chem Biol. (2015) 22:838–48. doi: 10.1016/j.chembiol.2015.06.008
28. Visnjic D, Lalic H, Dembitz V, Tomic B, Smoljo T. AICAr, a widely used AMPK activator with important AMPK-independent effects: A systematic review. Cells. (2021) 10. doi: 10.3390/cells10051095
29. Camici M, Allegrini S, Tozzi MG. Interplay between adenylate metabolizing enzymes and AMP-activated protein kinase. FEBS J. (2018) 285:3337–52. doi: 10.1111/febs.14508
30. Schmitt DL, Cheng YJ, Park J, An S. Sequestration-mediated downregulation of de novo purine biosynthesis by AMPK. ACS Chem Biol. (2016) 11:1917–24. doi: 10.1021/acschembio.6b00039
31. Sadasivan SK, Vasamsetti B, Singh J, Siddaraju N, Khan KM, Oommen AM, et al. Modulation of de novo purine biosynthesis leads to activation of AMPK and results in improved glucose handling and insulin sensitivity. J Diabetes Metab Disord. (2014) 13:51. doi: 10.1186/2251-6581-13-51
32. Racanelli AC, Rothbart SB, Heyer CL, Moran RG. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Res. (2009) 69:5467–74. doi: 10.1158/0008-5472.CAN-08-4979
33. Spurr IB, Birts CN, Cuda F, Benkovic SJ, Blaydes JP, Tavassoli A. Targeting tumour proliferation with a small-molecule inhibitor of AICAR transformylase homodimerization. Chembiochem. (2012) 13:1628–34. doi: 10.1002/cbic.201200279
34. Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. (2016) 351:728–33. doi: 10.1126/science.aad0489
35. Ma EH, Jones RG. CELL GROWTH. (TORC)ing up purine biosynthesis. Science. (2016) 351:670–1. doi: 10.1126/science.aaf1929
36. Yin J, Ren W, Huang X, Deng J, Li T, Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front Immunol. (2018) 9:1697. doi: 10.3389/fimmu.2018.01697p
37. Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem Sci. (2017) 42:141–54. doi: 10.1016/j.tibs.2016.09.009
38. Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. (2010) 39:171–83. doi: 10.1016/j.molcel.2010.06.022
39. Jing X, Wang XJ, Zhang T, Zhu W, Fang Y, Wu H, et al. Cell-cycle-dependent phosphorylation of PRPS1 fuels nucleotide synthesis and promotes tumorigenesis. Cancer Res. (2019) 79:4650–64. doi: 10.1158/0008-5472.CAN-18-2486
40. Pareek V, Pedley AM, Benkovic SJ. Human de novo purine biosynthesis. Crit Rev Biochem Mol Biol. (2021) 56:1–16. doi: 10.1080/10409238.2020.1832438
41. Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. (2008) 13:472–82. doi: 10.1016/j.ccr.2008.05.005
42. Doigneaux C, Pedley AM, Mistry IN, Papayova M, Benkovic SJ, Tavassoli A. Hypoxia drives the assembly of the multienzyme purinosome complex. J Biol Chem. (2020) 295:9551–66. doi: 10.1074/jbc.RA119.012175
43. Kunicka T, Prochazka P, Krus I, Bendova P, Protivova M, Susova S, et al. Molecular profile of 5-fluorouracil pathway genes in colorectal carcinoma. BMC Cancer. (2016) 16:795. doi: 10.1186/s12885-016-2826-8
44. Haddad FG, Kourie HR, Saleh K. Moving away from chemotherapy in newly diagnosed Philadelphia chromosome-positive B-cell acute lymphoblastic leukaemia. Br J Haematol. (2023) 202:1084–6. doi: 10.1111/bjh.19012
45. Jin S, Xu J, Zou Y, Li X, Yu B, Han J, et al. Microbiome changes involves in mercaptopurine mediated anti-inflammatory response in acute lymphoblastic leukemia mice. Int Immunopharmacol. (2023) 123:110782. doi: 10.1016/j.intimp.2023.110782
46. Sato Y, Matsuda S, Maruyama A, Nakayama J, Miyashita T, Udagawa H, et al. Metabolic characterization of antifolate responsiveness and non-responsiveness in Malignant pleural mesothelioma cells. Front Pharmacol. (2018) 9:2018.01129. doi: 10.3389/fphar.2018.01129
47. Tang C, Ke M, Yu X, Sun S, Luo X, Liu X, et al. GART functions as a novel methyltransferase in the RUVBL1/beta-catenin signaling pathway to promote tumor stemness in colorectal cancer. Adv Sci (Weinh). (2023) 10:e2301264. doi: 10.1002/advs.202301264
Keywords: purinosome, De novo purine synthesis, tumor metabolism, AMPK signaling pathway, mTOR signaling pathway, HIF-1 signaling pathway
Citation: Xie J, Liu J, Chen X and Zeng C (2024) Purinosomes involved in the regulation of tumor metabolism: current progress and potential application targets. Front. Oncol. 14:1333822. doi: 10.3389/fonc.2024.1333822
Received: 06 November 2023; Accepted: 16 April 2024;
Published: 26 April 2024.
Edited by:
Gary Piazza, Auburn University, United StatesReviewed by:
João Pessoa, University of Aveiro, PortugalMalvika Sharma, Memorial Sloan Kettering Cancer Center, United States
Copyright © 2024 Xie, Liu, Chen and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiehui Chen, eGhjaGVuNjZAMTI2LmNvbQ==; Changchun Zeng, emVuZ2NoY2hAZ2xtYy5lZHUuY24=
†ORCID: Jiaqi Xie, orcid.org/0000-0001-7567-1616
 Jiaqi Xie1,2†
Jiaqi Xie1,2† Xiehui Chen
Xiehui Chen Changchun Zeng
Changchun Zeng