
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 10 April 2024
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 14 - 2024 | https://doi.org/10.3389/fonc.2024.1332499
This article is part of the Research TopicDisparities in Early Onset Colorectal CancerView all 7 articles
Purpose: This study aimed to identify prognostic factors and develop a nomogram for predicting overall survival (OS) in stage III/IV early-onset colorectal cancer (EO-CRC).
Methods: Stage III/IV EO-CRC patients were identified from the Surveillance, Epidemiology, and End Results (SEER) database between 2010 and 2015. The datasets were randomly divided (2:1) into training and validation sets. A nomogram predicting OS was developed based on the prognostic factors identified by Cox regression analysis in the training cohort. Moreover, the predictive performance of the nomogram was assessed using the receiver operating characteristic (ROC) curves, calibration plots, and decision curve analysis (DCA). Subsequently, the internal validation was performed using the validation cohort. Finally, a risk stratification system was established based on the constructed nomogram.
Results: Of the 10,387 patients diagnosed with stage III/IV EO-CRC between 2010 and 2015 in the SEER database, 8,130 patients were included. In the training cohort (n=3,071), sex, marital status, race/ethnicity, primary site, histologic subtypes, grade, T stage, and N stage were identified as independent prognostic variables for OS. The 1-, 3-, and 5-year area under the curve (AUC) values of the nomogram were robust in both the training (0.751, 0.739, and 0.723) and validation cohorts (0.748, 0.733, and 0.720). ROC, calibration plots, and DCA indicated good predictive performance of the nomogram in both the training and validation sets. Furthermore, patients were categorized into low-, middle-, and high-risk groups based on the nomogram risk score. Kaplan-Meier curve showed significant survival differences between the three groups.
Conclusion: We developed a prognostic nomogram and risk stratification system for stage III/IV EO-CRC, which may facilitate clinical decision-making and individual prognosis prediction.
Colorectal cancer (CRC) is the second most deadly and third most common cancer worldwide (1). While CRC incidence has decreased in individuals ≥50 years of age, it has increased globally in individuals younger than 50 years in the past decades, which has been defined as early-onset CRC (EO-CRC) (2–7). EO-CRC has been in the spotlight recently, which would account for about 11% of colon cancers and 23% of rectal cancers in 2030 (8). Unfortunately, the reasons of this increase in EO-CRC remain unclear and probably multifactorial (9). Additionally, clinical features of EO-CRC, often diagnosed with advanced stage disease (6, 10–12), differ from those of later-onset disease (3, 4). Overall, EO-CRC contributes significantly to the global cancer burden. Hence, to facilitate clinical decision-making, it is important to predict the prognosis of EO-CRC patients.
Although widely used to examine the survival of CRC, the American Joint Committee on Cancer (AJCC) staging system is far from perfect. Several studies have identified prognostic risk factors for EO-CRC and developed the prognostic nomograms. However, some problems remain. First, flow chat was not available in two studies (13, 14). Second, three nomograms (15–17) were associated with too many prognosis factors (≥ 12), which reduced its practicability. Last, and most importantly, several nomograms were unreasonable in clinical context. For example, it was paradoxical that patients with grade I had a worse prognosis than those with grade II (15, 16, 18). Similar to the “grade paradox”, there were also “race/ethnicity paradox” (13, 14, 19, 20), “T stage paradox” (16, 18, 19), “histologic subtypes paradox” (21), and “primary site paradox” (15, 18, 21, 22). Thus, more high‐quality research is urgently needed.
To the best of our knowledge, to date, no nomogram has been constructed to predict the prognosis of patients with stage III/IV EO-CRC. Therefore, this study aimed to develop and validate a prognostic nomogram predicting overall survival (OS) for stage III/IV EO-CRC based on the Surveillance, Epidemiology, and End Results (SEER) database.
The clinicopathological data of patients with EO-CRC between 2010 and 2015 were extracted using SEER*Stat 8.4.1 software. Primary tumor sites were divided into anatomical subsites: cecum (C18.0), ascending colon (C18.2), hepatic flexure of colon (C18.3), transverse colon (C18.4), splenic flexure of colon (C18.5), descending colon (C18.6), sigmoid colon (C18.7), rectosigmoid junction (C19.9), and rectum (C20.9). Based on the third edition of the International Classification of Diseases Oncology Special Edition, the histologic subtypes included adenocarcinoma (8140/3, 8144/3, 8201/3,8210/3, 8211/3, 8213/3, 8220/3, 8221/3, 8255/3,8260/3, 8261/3, 8262/3, 8263/3, 8310/3, 8323/3), mucinous adenocarcinoma (MA) (8480/3, 8481/3), and signet ring cell carcinoma (SRCC) (8490/3).
The inclusion criteria were patients diagnosed with stage III/IV EO-CRC (pathologically confirmed) between 2010 and 2015. Exclusion criteria included patients younger than 18 years, and those lacking complete clinicopathological and survival information.
The collected variables included age, sex, marital status, race/ethnicity, primary site, histologic subtypes, grade, AJCC stage, T stage, N stage, survival time, and vital status record. Primary sites comprise right-sided colon (cecum, ascending colon, hepatic flexure, and transverse colon), left-sided colon (splenic flexure, descending colon, sigmoid colon, and rectosigmoid junction), and rectum (23). Race/ethnicity was divided into five categories: Hispanic, Non-Hispanic White (NHW), Non-Hispanic Black (NHB), Non-Hispanic Asian or Pacific Islander (NHAPI), and Non-Hispanic American Indian/Alaska Native (NHAIAN). The tumor-node-metastasis (TNM) stage was determined according to the AJCC seventh edition criteria. OS is defined as the time from diagnosis to death from any cause or the time of the last follow-up.
The datasets were randomly divided (2:1) into the training and validation sets. In the training cohort, univariate and multivariate Cox regression analyses were performed to identify the prognostic factors of patients with stage III/IV EO-CRC. The independent risk factors were used to construct the prognostic nomogram.
The discrimination ability of the prognostic nomogram was examined by the receiver operating characteristic (ROC) curves and the area under the curves (AUC). The calibration ability was evaluated by the calibration plot. Additionally, decision curve analysis (DCA) was performed to assess clinical utility by quantifying the net benefits at different threshold probabilities.
Based on the constructed nomogram, the total risk score was calculated for each patient. X-tile software (version 3.6.1, Yale University) was used to identify the optimal cutoff values for the total risk score. According to the cutoff values of risk score, patients were classified into low-, middle-, and high-risk groups. Kaplan–Meier curves and the log-rank test were used to compare the survival differences between different groups.
R software (version 4.2.1, https://www.r-project.org) and relevant packages were used to construct and validate the prognostic nomogram. A p-value < 0.05 was considered significant.
Overall, 10,387 patients with stage III/IV EO-CRC were identified between 2010 and 2015 from the SEER database. According to the inclusion and exclusion criteria, 8,130 patients with stage III/IV EO-CRC were included in the final analysis. The study screening flow chart is shown in Figure 1. Subsequently, the datasets were randomly divided (2:1) into a training set (n=3071) and a validation set (n=1535), with no statistical difference. The demographic and clinicopathological characteristics of stage III/IV EO-CRC patients are summarized in Table 1. The baseline characteristics of patients stratified by AJCC stage are shown in Supplementary Table S1.
The univariate and multivariate Cox regression analysis revealed that sex, marital status, race/ethnicity, primary site, histologic subtypes, grade, T stage, and N stage were identified as independent prognostic factors for OS in the training cohort (Table 2). Accordingly, these independent prognostic factors were utilized to construct the nomogram for predicting the 1-, 3-, and 5-year OS of stage III/IV EO-CRC patients (Figure 2).
The ROC curves demonstrated that the discriminating ability of the prediction model was robust both in the training and validation sets. The AUC values of the nomogram predicting 1-, 3-, and 5-year OS in the training cohort were 0.751, 0.739, and 0.723, respectively. Similarly, in the validation cohort, the corresponding values were 0.748, 0.733, and 0.720, respectively (Figure 3). Additionally, the calibration plots demonstrated the prognostic nomogram’s strong calibration capability (Figure 4). Furthermore, the DCA curves suggested good clinical utility (Figure 5).
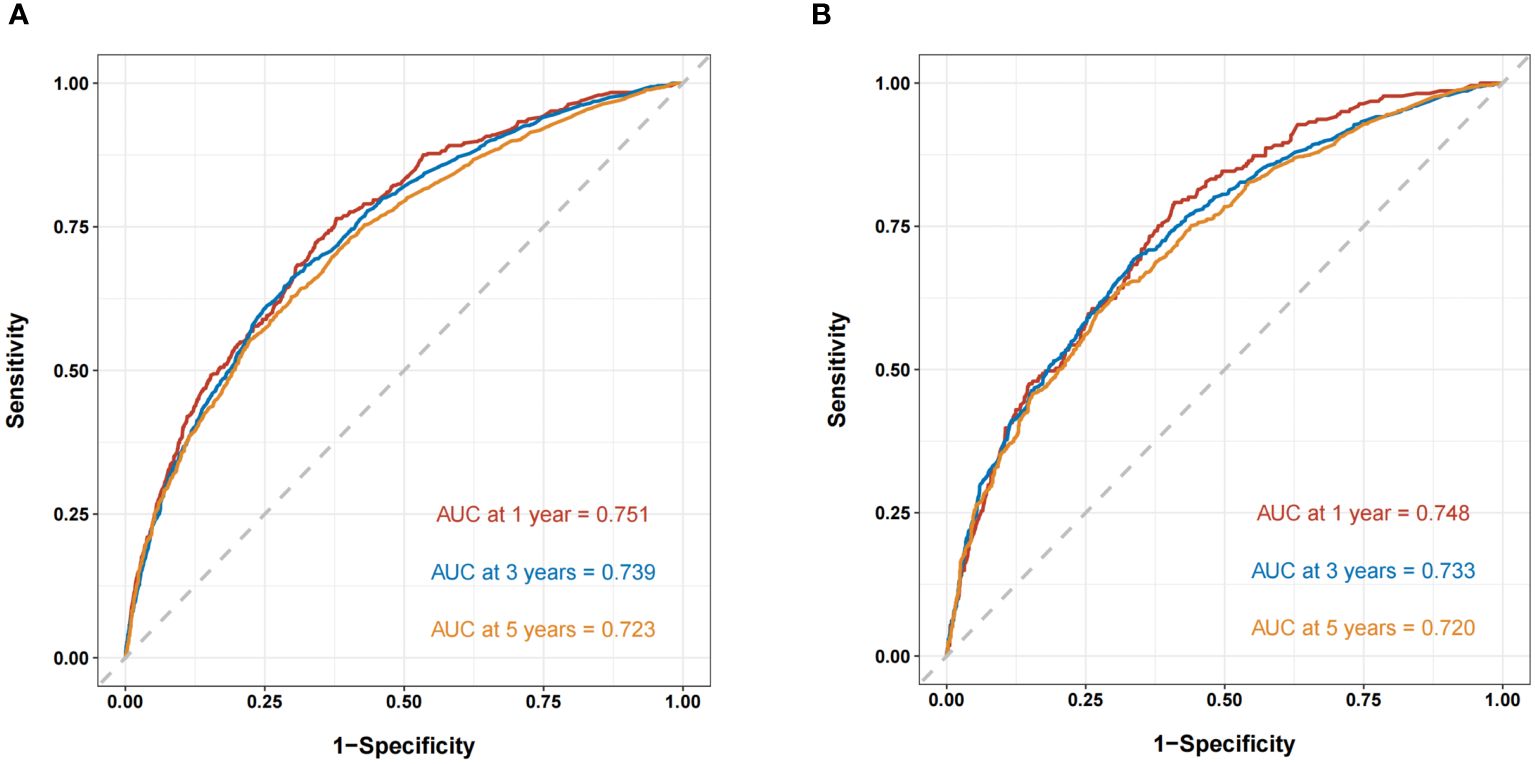
Figure 3 The ROC curves of the nomogram predicting OS in the training cohort (A) and validation cohort (B).
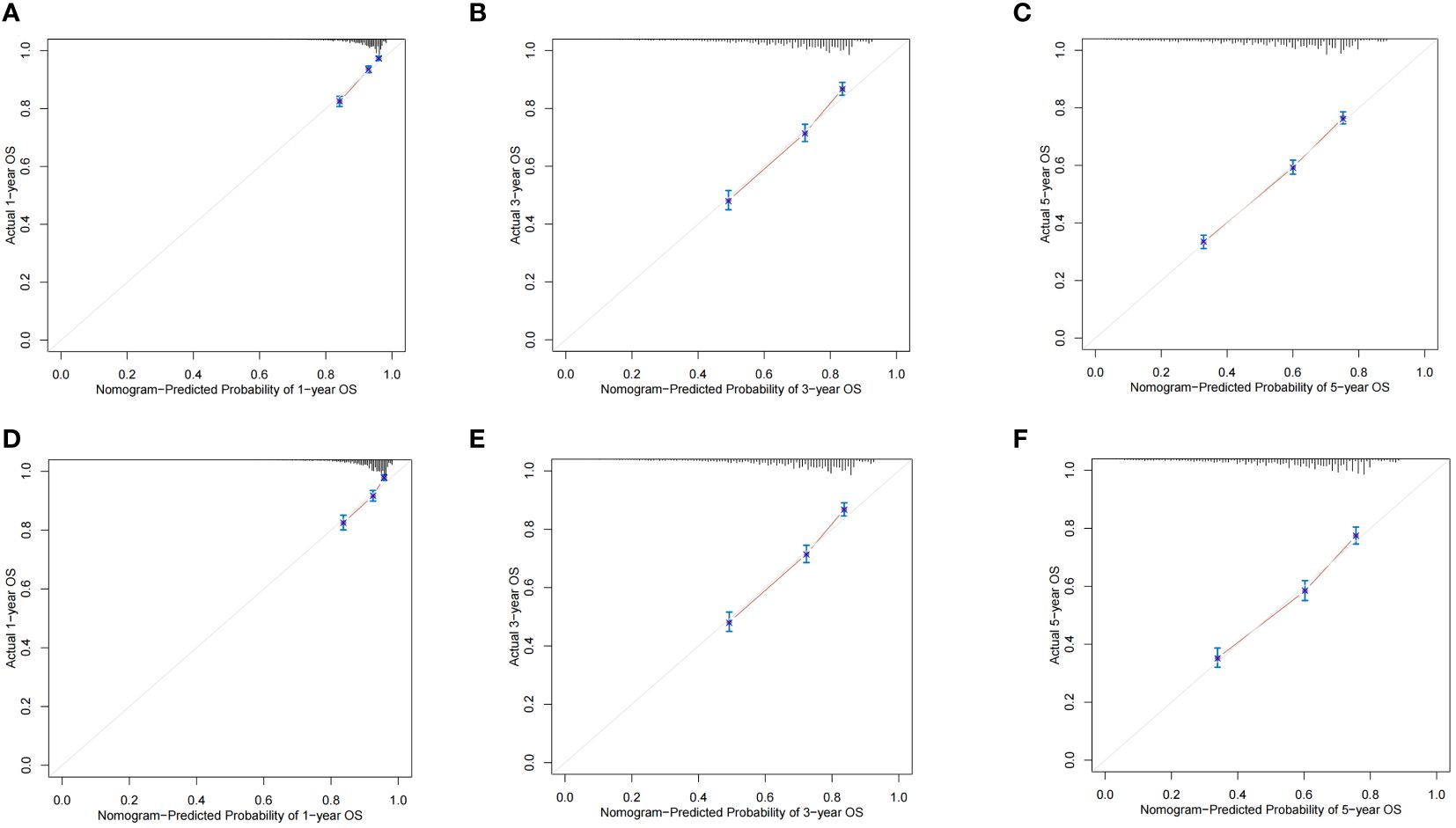
Figure 4 The calibration plots of the nomogram predicting 1-,3- and 5-year OS in the training cohort (A–C) and validation cohort (D–F).
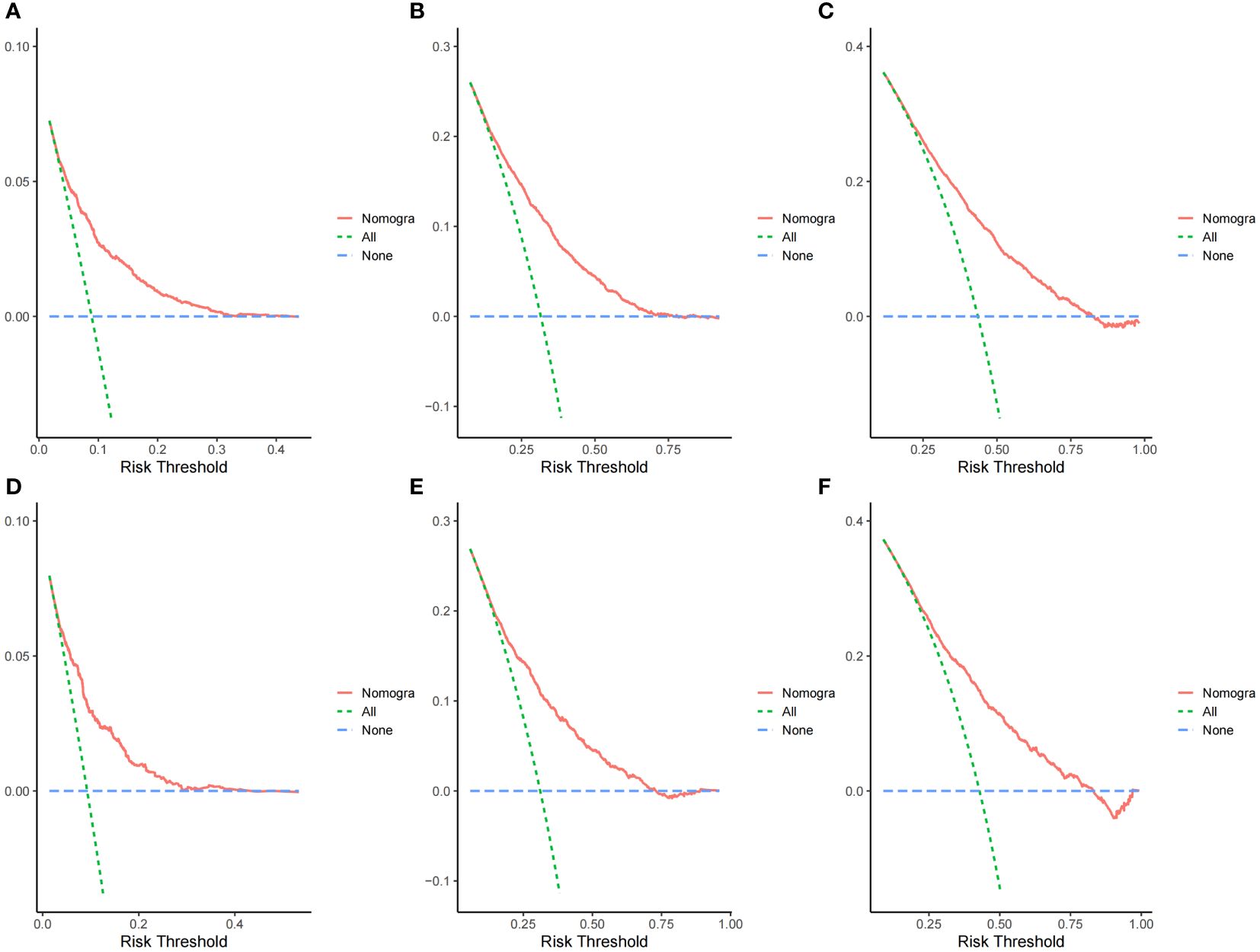
Figure 5 Decision curve analysis curves of the nomogram predicting 1-,3- and 5-year OS in the training cohort (A–C) and validation cohort (D–F).
Based on the optimal cutoff values for the total risk score, patients in the training cohort were grouped into the low-risk (≤95.5), middle-risk (95.5-162.5), and high-risk groups (>162.5) (Supplementary Figure S1). Similarly, patients in the validation cohort were categorized into low-risk (≤96), middle-risk (96-162.5), and high-risk groups (>162.5) (Supplementary Figure S2). Kaplan–Meier curves, using the log-rank test, revealed significant differences in OS among the three risk subgroups in both the training and validation cohorts (Figure 6).
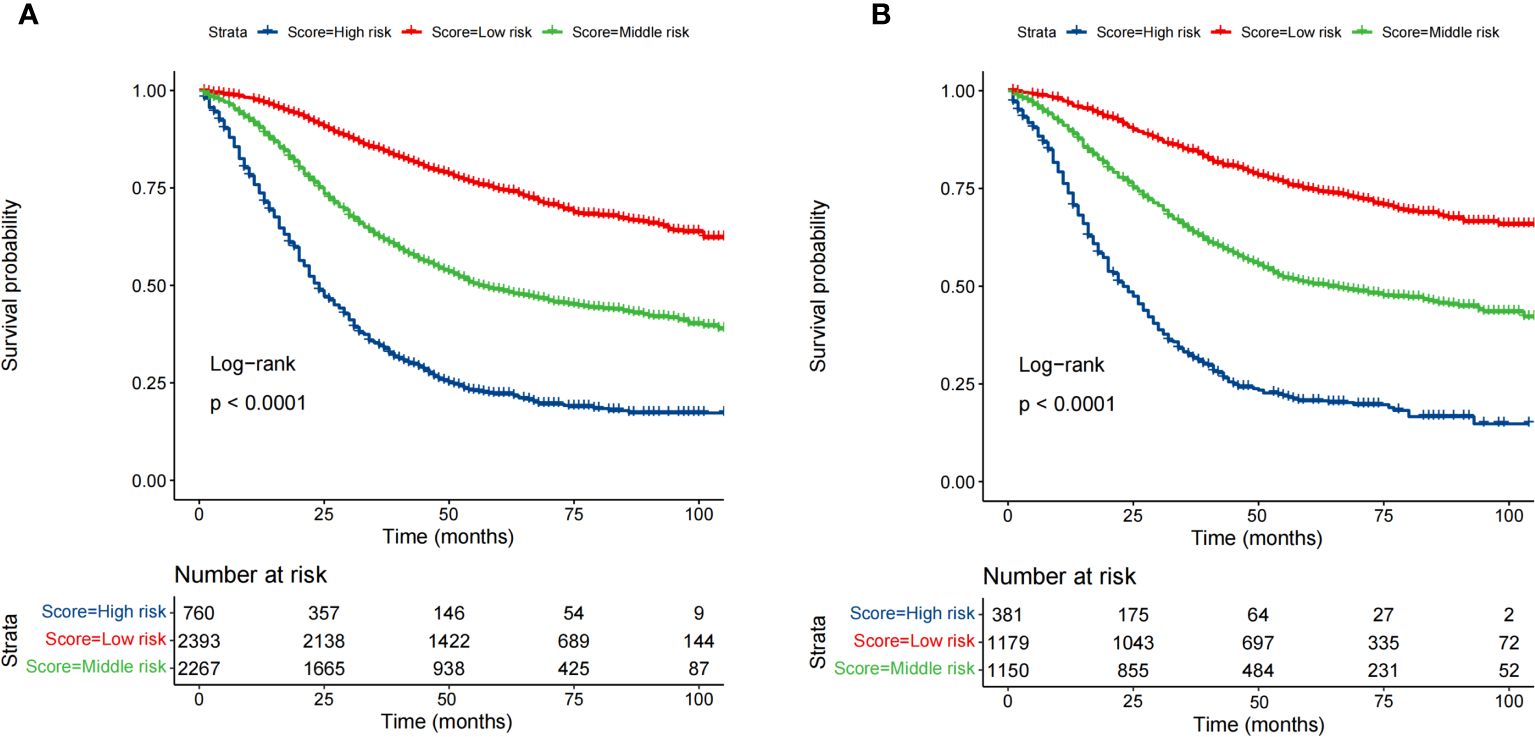
Figure 6 Survival analysis of the training cohort (A) and the validation cohort (B) by the risk score calculated by the nomogram.
Based on the SEER database, 8,130 stage III/IV EO-CRC patients were included. The present study identified that eight variables, including sex, marital status, race/ethnicity, primary site, histologic subtypes, grade, T stage, and N stage, were independent predictors for OS of patients with stage III/IV EO-CRC. Utilizing these variables, a nomogram with favorable performance predicting 1-, 3-, and 5-year OS was developed and validated. Furthermore, a risk stratification was successfully established based on the total risk score determined by the nomogram.
The initial nomogram demonstrated that AJCC stage had the most substantial impact on OS of the patients with stage III/IV EO-CRC. Based on our previous nomogram, risk score for stage III and IV is 0 and 100, respectively. This limits its clinical application. Therefore, AJCC stage was not included when constructing the new nomogram. Our new nomogram demonstrated that T stage contributed the most to prognosis, followed by N stage, grade, race, marital status, histologic subtypes, primary site, and sex. All eight prognostic variables are readily available and clinically reasonable. Our findings suggest that right colon cancer had a worse OS than rectum cancer, and rectum cancer had a worse OS than left colon cancer in stage III/IV EO-CRC. These results are in accord with published studies (16, 22).
Consistent with the literature (13, 15, 16, 18–20, 24), this study showed that age was not a significant prognostic factor for the survival of patients with stage III/IV EO-CRC. However, a few studies have presented contrasting views. A survival nomogram for stage I–III EO-CRC patients revealed that patients with older age had a worse survival than those with younger age (14). In contrast, another nomogram for combined lymphatic metastases in EO-CRC patients demonstrated that patients with younger age had a worse survival than those with older age (21). These divergent findings necessitate cautious interpretation.
Our study demonstrated that sex had a minor influence on prognosis. These results are in accord with three studies (20, 22, 25) indicating that male EO-CRC patients exhibited slightly poorer survival compared to their female counterparts. However, several studies (13, 15, 16, 18, 19, 24) indicated that sex was not a significant prognostic factor for the survival of EO-CRC patients. Collectively, the influence of sex on the survival of EO-CRC patients warrants further investigation.
In the present study, race/ethnicity was divided into five categories: Hispanic, NHW, NHB, NHAPI, and NHAIAN. Our nomogram showed that NHW patients had the best prognosis in stage III/IV EO-CRC, followed by NHAPI, Hispanic, NHB, and NHAIAN patients. A study using data from the SEER database demonstrated that the cancer-specific survival at five years for NHW, NHAPI, Hipanic, NHAIAN, and NHB patients with EO-CRC was 70.11, 68.70, 68.31, 65.67, and 65.58 months, respectively (26). These results were almost the same as ours. Additionally, many studies have investigated the racial/ethnic disparities in survival among EO-CRC patients. While the criteria for racial/ethnic classification may vary, the findings of these studies consistently indicate that NHB patients have the poorest prognosis, whereas NHW patients exhibit the most favorable prognosis (27–32). Our results also support this view.
Generally speaking, a higher T stage means a worse prognosis (13, 14, 21). Surprisingly, our study indicated that, compared to T1 patients, those with T2 (adjusted hazard ratio [aHR], 0.28; 95% confidence interval [CI], 0.21-0.36), T3 (aHR, 0.52; 95% CI, 0.45-0.61) and > T4 (aHR, 1.07; 95% CI, 0.91-1.26) had better OS independent of other variables. This seems a rather paradoxical finding. Nevertheless, we believe this to be an important clinical finding rather than a paradox. The most possible reason is that advanced tumors with light intestinal wall invasion may represent a biologically aggressive phenotype. More studies are needed to elucidate the underlying mechanism.
Similar to T stage, a higher N stage means a worse prognosis (14, 20, 24). In contrast, our study suggested another paradoxical finding. Compared to N0 patients, those with N1 (aHR, 0.36; 95% CI, 0.32-0.42), and N2 (aHR, 0.57; 95% CI, 0.50-0.65) had a better prognosis. The participants of our study were patients with stage III/IV EO-CRC. Therefore, the patients either present with lymph node metastasis or distant metastasis. Patients with N1 and N2 may not have distant metastasis, while patients with N0 must have distant metastasis. Thus, although paradoxical, this finding is logical in stage III/IV EO-CRC.
A higher tumor grade is commonly associated with a poorer prognosis, as confirmed in our study. However, a nomogram predicting OS for metastatic EO-CRC revealed counterintuitive trends: grade I patients had a worse prognosis than grade II, and grade III worse than grade IV (15). Clearly, these results are illogical. Based on their nomogram, the risk scores for grade I, II, III, and IV were 20, 0, 81, and 60, respectively. In the training cohort, the sample size for patients with grade I, II, III, and IV were 72, 1212, 378, and 67, respectively. We speculate that the small sample size for patients with grade I and IV (<100) may lead to the “grade paradox”. Notably, several other studies were also limited by “grade paradox” (16, 18, 21). We speculated that this survival paradox resulted from the small sample sizes.
There are several strengths to the present study. First, to the best of our knowledge, this is the first predictive nomogram focusing on patients with stage III/IV EO-CRC. Second, the AUC values of the nomogram predicting 1-, 3-, and 5-year OS were all greater than 0.71. The calibration plots showed a good calibration ability and the DCA curves indicated a good clinical utility. Moreover, the internal validation also demonstrated satisfactory results. Third, of the 10,387 patients with stage III/IV EO-CRC, only 2257 patients (lacking essential clinical and survival information) were excluded. Thus, 8,130 patients were included in the final analysis, suggesting a strong representativeness. Fourth, based on the nomogram, a risk stratification system was successfully established to identify high risk patients.
Of course, some limitations must be recognized in the present study. First, it was a retrospective study, limiting the generalizability of the results. In the future, prospective studies are warranted to verify the findings. Second, there was no detailed information on CEA, radiotherapy, chemotherapy, and surgical treatment in the SEER database. Third, the SEER database did not include potential prognostic factors for EO-CRC, such as systemic immune inflammation index (33), geriatric nutrition risk index (33), symptom duration of 3 months or more (34), carbohydrate antigen 19-9 (35), and cell cycle-related genes (36). Fourth, our data was exclusively from the SEER database, representing only a portion of the US population, potentially limiting the applicability of our results to other regions or countries. Last, it should be stated that although the internal validation demonstrated satisfactory results, external validation was not performed. This problem will be addressed in our future studies.
Patients with EO-CRC differ from those with later-onset CRC in underlying molecular mechanisms (37–40), clinical features, and treatment (41). This special population warrants further attention in the future.
Independent risk factors for OS in stage III/IV EO-CRC patients included sex, marital status, race/ethnicity, primary site, histologic subtypes, grade, T stage, and N stage. An effective nomogram and risk stratification system were established, potentially enhancing clinical decision-making and individual prognosis prediction.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The data sets analyzed in this study are available in the SEER database.
The requirement of ethical approval was waived by Tianjin Medical University General Hospital for the studies involving humans because de-identified data were used in this study. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/ institutional review board also waived the requirement of written informed consent for participation from the participants or the participants' legal guardians/next of kin because of the retrospective nature of the study.
WY: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. WP: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. TY: Methodology, Software, Visualization, Writing – review & editing. QZ: Methodology, Software, Visualization, Writing – review & editing. SZ: Data curation, Validation, Writing – review & editing. MZ: Data curation, Validation, Writing – review & editing. GL: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was sponsored by Tianjin Health Research Project (TJSJMYXYC-D2-027).
We are very grateful to the SEER program for approving the registration and the SEER database.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1332499/full#supplementary-material
Supplementary Figure 1 | Identification of the best cut-off point of total risk score in the training cohort through X-tile software.
Supplementary Figure 2 | Identification of the best cut-off point of total risk score in the validation cohort through X-tile software.
EO-CRC, early-onset colorectal cancer; AJCC, American joint committee on cancer; OS, overall survival; SEER, Surveillance, Epidemiology, and End Results; NHW, Non-Hispanic White; NHB, Non-Hispanic Black; NHAPI, Non-Hispanic Asian or Pacific Islander; NHAIAN, Non-Hispanic American Indian/Alaska Native; TNM, tumor-node-metastasis; ROC, receiver operating characteristic; AUC, area under the curves; DCA, decision curve analysis; SDW, separated, single, divorced, domestic partner or unmarried, widowed; MA, mucinous adenocarcinoma; SRCC, signet ring cell carcinoma; HR, Hazard ratio; CI, confidence interval.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Giannakis M, Ng K. A common cancer at an uncommon age. Science. (2023) 379:1088–90. doi: 10.1126/science.ade7114
3. Sinicrope FA. Increasing incidence of early-onset colorectal cancer. N Engl J Med. (2022) 386:1547–58. doi: 10.1056/NEJMra2200869
4. REACCT Collaborative, Zaborowski AM, Abdile A, Adamina M, Aigner F, d’Allens L, et al. Characteristics of early-onset vs late-onset colorectal cancer: A review. JAMA Surg. (2021) 156:865–74. doi: 10.1001/jamasurg.2021.2380
5. Probing early-onset colorectal cancer. Cancer Discovery. (2022) 12:OF2. doi: 10.1158/2159-8290.CD-NB2022-0011
6. Tom CM, Mankarious M, Jeganathan NA, Deutsch M, Koltun WA, Berg AS, et al. Characteristics and outcomes of right- versus left-sided early onset colorectal cancer. Dis Colon Rectum. (2022) 66:498–510. doi: 10.1097/DCR.0000000000002273
7. The Lancet Gastroenterology Hepatology. Addressing the rise of early-onset colorectal cancer. Lancet Gastroenterol Hepatol. (2022) 7:197. doi: 10.1016/S2468-1253(22)00003-6
8. Spaander MCW, Zauber AG, Syngal S, Blaser MJ, Sung JJ, You YN, et al. Young-onset colorectal cancer. Nat Rev Dis Primer. (2023) 9:21. doi: 10.1038/s41572-023-00432-7
9. Eng C, Jácome AA, Agarwal R, Hayat MH, Byndloss MX, Holowatyj AN, et al. A comprehensive framework for early-onset colorectal cancer research. Lancet Oncol. (2022) 23:e116–28. doi: 10.1016/S1470-2045(21)00588-X
10. Goh SS, Loo EX, Lee DJ. Trends and clinical outcomes in young-onset colorectal cancer patients. Ann Acad Med. (2020) 49:848–56. doi: 10.47102/annals-acadmedsg.20207
11. Gausman V, Dornblaser D, Anand S, Hayes RB, O’Connell K, Du M, et al. Risk factors associated with early-onset colorectal cancer. Clin Gastroenterol Hepatol. (2020) 18:2752–2759.e2. doi: 10.1016/j.cgh.2019.10.009
12. Cheng E, Blackburn HN, Ng K, Spiegelman D, Irwin ML, Ma X, et al. Analysis of survival among adults with early-onset colorectal cancer in the national cancer database. JAMA Netw Open. (2021) 4:e2112539. doi: 10.1001/jamanetworkopen.2021.12539
13. Li D. Establishment and validation of a prognostic nomogram for patients with early-onset stage I-II colon cancer. World J Surg Oncol. (2023) 21:103. doi: 10.1186/s12957-023-02988-w
14. Chen M, Chen T. Individualized conditional survival nomograms for stage I-III early onset colorectal cancer patients. Jpn J Clin Oncol. (2023) 53:115–21. doi: 10.1093/jjco/hyac169
15. Xu C, Zhang F, Cheng W, Zhu Y. Prediction models for overall and cancer-specific survival in patients with metastatic early-onset colorectal cancer: a population-based study. Int J Colorectal Dis. (2023) 38:99. doi: 10.1007/s00384-023-04369-x
16. Wu J, Lu L, Chen H, Lin Y, Zhang H, Chen E, et al. Prognostic nomogram to predict the overall survival of patients with early-onset colorectal cancer: a population-based analysis. Int J Colorectal Dis. (2021) 36:1981–93. doi: 10.1007/s00384-021-03992-w
17. Li Y, Liu D, Zhao L, Güngör C, Song X, Wang D, et al. Accurate nomograms with excellent clinical value for locally advanced rectal cancer. Ann Transl Med. (2021) 9:296. doi: 10.21037/atm-20-4144
18. Ding X, Yang X, Wu D, Huang Y, Dai Y, Li J, et al. Nomogram predicting the cancer-specific survival of early-onset colorectal cancer patients with synchronous liver metastasis: a population-based study. Int J Colorectal Dis. (2022) 37:1309–19. doi: 10.1007/s00384-022-04175-x
19. Dong B, Chen Y, Lyu G. Prognostic nomograms for predicting overall survival and cancer-specific survival of patients with very early-onset colorectal cancer: A population-based analysis. Bosn J Basic Med Sci. (2022) 22:803–17. doi: 10.17305/bjbms.2021.7035
20. Li Y, Liu W, Zhou Z, Ge H, Zhao L, Liu H, et al. Development and validation of prognostic nomograms for early-onset locally advanced colon cancer. Aging. (2020) 13:477–92. doi: 10.18632/aging.202157
21. Liu Y, Sun Z, Guo Y, Liu C, Tian S, Dong W. Construction and validation of a nomogram of risk factors and cancer-specific survival prognosis for combined lymphatic metastases in patients with early-onset colorectal cancer. Int J Colorectal Dis. (2023) 38:128. doi: 10.1007/s00384-023-04432-7
22. Cheng X, Li Y, Chen D, Xu X, Liu F, Zhao F. Nomogram predicting the survival of young-onset patients with colorectal cancer liver metastases. Diagnostics. (2022) 12:1395. doi: 10.3390/diagnostics12061395
23. Jacobs D, Zhu R, Luo J, Grisotti G, Heller DR, Kurbatov V, et al. Defining early-onset colon and rectal cancers. Front Oncol. (2018) 8:504. doi: 10.3389/fonc.2018.00504
24. Yu C, Zhang Y. Development and validation of a prognostic nomogram for early-onset colon cancer. Biosci Rep. (2019) 39:BSR20181781. doi: 10.1042/BSR20181781
25. Liu J, Liu Z, Li J, Tian S, Dong W. Personalizing prognostic prediction in early-onset Colorectal Cancer. J Cancer. (2020) 11:6727–36. doi: 10.7150/jca.46871
26. Aloysius MM, Goyal H, Shah NJ, Pallav K, John N, Gajendran M, et al. Impact of race and socioeconomics disparities on survival in young-onset colorectal adenocarcinoma—A SEER registry analysis. Cancers. (2021) 13:3262. doi: 10.3390/cancers13133262
27. Chen Y, He L, Lu X, Tang Y, Luo G, Chen Y, et al. Causes of death among early-onset colorectal cancer population in the United States: a large population-based study. Front Oncol. (2023) 13:1094493. doi: 10.3389/fonc.2023.1094493
28. Martinez ME, Gomez SL, Canchola AJ, Oh DL, Murphy JD, Mehtsun W, et al. Changes in cancer mortality by race and ethnicity following the implementation of the affordable care act in California. Front Oncol. (2022) 12:916167. doi: 10.3389/fonc.2022.916167
29. Zaki TA, Liang PS, May FP, Murphy CC. Racial and ethnic disparities in early-onset colorectal cancer survival. Clin Gastroenterol Hepatol. (2023) 21:497–506.e3. doi: 10.1016/j.cgh.2022.05.035
30. Arshad HMS, Kabir C, Tetangco E, Shah N, Raddawi H. Racial disparities in clinical presentation and survival times among young-onset colorectal adenocarcinoma. Dig Dis Sci. (2017) 62:2526–31. doi: 10.1007/s10620-017-4653-4
31. Murphy CC, Wallace K, Sandler RS, Baron JA. Racial disparities in incidence of young-onset colorectal cancer and patient survival. Gastroenterology. (2019) 156:958–65. doi: 10.1053/j.gastro.2018.11.060
32. Holowatyj AN, Ruterbusch JJ, Rozek LS, Cote ML, Stoffel EM. Racial/ethnic disparities in survival among patients with young-onset colorectal cancer. J Clin Oncol. (2016) 34:2148–56. doi: 10.1200/JCO.2015.65.0994
33. Xiang S, Yang Y-X, Pan W-J, Li Y, Zhang J-H, Gao Y, et al. Prognostic value of systemic immune inflammation index and geriatric nutrition risk index in early-onset colorectal cancer. Front Nutr. (2023) 10:1134300. doi: 10.3389/fnut.2023.1134300
34. Kim TJ, Kim ER, Hong SN, Chang DK, Kim Y-H. Long-term outcome and prognostic factors of sporadic colorectal cancer in young patients: A large institutional-based retrospective study. Med (Baltimore). (2016) 95:e3641. doi: 10.1097/MD.0000000000003641
35. Li Y-T, Wang X-Y, Zhang B, Tao B-R, Chen Z-M, Ma X-C, et al. The prognostic significance of clinicopathological characteristics in early-onset versus late-onset colorectal cancer liver metastases. Int J Colorectal Dis. (2023) 38:19. doi: 10.1007/s00384-023-04317-9
36. Xiang M, Gao Y, Zhou Y, Wang M, Yao X. A novel nomogram based on cell cycle-related genes for predicting overall survival in early-onset colorectal cancer. BMC Cancer. (2023) 23:595. doi: 10.1186/s12885-023-11075-y
37. Teng X, Dai Y, Li J. Methodological advances of bioanalysis and biochemical targeting of intracellular G-quadruplexes. Explor (Beijing). (2022) 2:20210214. doi: 10.1002/EXP.20210214
38. Lyu N, Pedersen B, Shklovskaya E, Rizos H, Molloy MP, Wang Y. SERS characterization of colorectal cancer cell surface markers upon anti-EGFR treatment. Explor (Beijing). (2022) 2:20210176. doi: 10.1002/EXP.20210176
39. Xu T, Zhang Y, Zhang J, Qi C, Liu D, Wang Z, et al. Germline profiling and molecular characterization of early onset metastatic colorectal cancer. Front Oncol. (2020) 10:568911. doi: 10.3389/fonc.2020.568911
40. Holowatyj AN, Perea J, Lieu CH. Gut instinct: a call to study the biology of early-onset colorectal cancer disparities. Nat Rev Cancer. (2021) 21:339–40. doi: 10.1038/s41568-021-00356-y
Keywords: early-onset, colorectal cancer, overall survivals, prognostic factors, nomogram
Citation: Yin W, Pei W, Yu T, Zhang Q, Zhang S, Zhang M and Liu G (2024) Construction and validation of a nomogram for predicting overall survival of patients with stage III/IV early−onset colorectal cancer. Front. Oncol. 14:1332499. doi: 10.3389/fonc.2024.1332499
Received: 03 November 2023; Accepted: 26 March 2024;
Published: 10 April 2024.
Edited by:
Caitlin Murphy, University of Texas Health Science Center at Houston, United StatesCopyright © 2024 Yin, Pei, Yu, Zhang, Zhang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Liu, bGcxMDU5QHRtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.