- 1School of Dentistry, University of California, Los Angeles, Los Angeles, CA, United States
- 2Stanford Cancer Institute, Stanford University, Stanford, CA, United States
- 3Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
In clinical oncology, cell-free DNA (cfDNA) has shown immense potential in its ability to noninvasively detect cancer at various stages and monitor the progression of therapy. Despite the rapid improvements in cfDNA liquid biopsy approaches, achieving the required sensitivity to detect rare tumor-derived cfDNA still remains a challenge. For next-generation sequencing, the perceived presentation of cfDNA is strongly linked to the extraction and library preparation protocols. Conventional double-stranded DNA library preparation (dsDNA-LP) focuses on assessing ~167bp double-stranded mononucleosomal (mncfDNA) and its other oligonucleosomal cell-free DNA counterparts in plasma. However, dsDNA-LP methods fail to include short, single-stranded, or nicked DNA in the final library preparation, biasing the representation of the actual cfDNA populations in plasma. The emergence of single-stranded library preparation (ssDNA-LP) strategies over the past decade has now allowed these other populations of cfDNA to be studied from plasma. With the use of ssDNA-LP, single-stranded, nicked, and ultrashort cfDNA can be comprehensively assessed for its molecular characteristics and clinical potential. In this review, we overview the current literature on applications of ssDNA-LP on plasma cfDNA from a potential cancer liquid biopsy perspective. To this end, we discuss the molecular principles of single-stranded DNA adapter ligation, how library preparation contributes to the understanding of native cfDNA characteristics, and the potential for ssDNA-LP to improve the sensitivity of circulating tumor DNA detection. Additionally, we review the current literature on the newly reported species of plasma ultrashort single-stranded cell-free DNA plasma, which appear biologically distinct from mncfDNA. We conclude with a discussion of future perspectives of ssDNA-LP for liquid biopsy endeavors.
1 Introduction
1.1 Cell-free DNA
Liquid biopsy, which harnesses biomolecules within biofluids to infer the characteristics and activity of a distant primary tumor of cancer within the body, has emerged from its infancy into a bustling field of study (1). Although tumor-derived cells, proteins, or metabolites spearheaded the initial liquid-biopsy interest, cell-free DNA (cfDNA) has now become the most highly focused analyte. Cell-free DNA is thought to be derived from degraded DNA from a variety of mechanisms (apoptosis, necrosis, or secretion (2)) and is detectable by many technologies, especially next-generation sequencing (NGS). There are various conformations of cfDNA present in blood plasma and serum, saliva, urine, and cerebral spinal fluid with unique characteristics. Therefore, cell-free DNA has become an important part of liquid biopsy workflow. In cancer diagnosis, one potential attribute of cfDNA is the ability to assay for tumor-derived cfDNA, referred to as circulating tumor DNA (ctDNA). Studying ctDNA involves the examination of cfDNA fragments that contain signature mutant signals such as single base pair mutations (3), amplifications (4, 5), fragment-size changes (6), methylation (7), or other discrimination topological features (8).
Despite its many virtues, due to its rarity, the detection of ctDNA is challenging and has been alluded to finding a needle in a haystack. Since non-cancer cells undergo constant replication and controlled death cycles, ctDNA is present at extremely low concentrations compared to cfDNA of non-tumor origin (9). For cfDNA, reported tumor DNA to wildtype sequences ratios range from >5-10% at later stages, which is feasible for detection, to increasingly rare ratios of <0.01 to 0.1% at early stages (or after surgical intervention) (10).
Adding to this complexity, the overall understanding of the biology and size distribution has not been established. The predominant type of cfDNA analyzed by current assays is the double-stranded 167-bp mononucleosomal cell-free DNA (mncfDNA) molecule and those derived from di- or tri- nucleosomes (11). The 167-bp cell-free DNA link to histones has been well established (12–14). The observed cfDNA structure and size can be dependent on the mechanism of release from cells. The appearance of cfDNA can be altered depending on if it is derived from cell necrosis, apoptosis, phagocytosis, and extracellular versicles release (15). During apoptosis, DNA processing creates the iconic pattern of fragments presenting in multiples of 180-200bp. DNA wrapped around histone octomers is 147bp in length, with a linker DNA ranging from 20-90bp. However, the cfDNA population also contains other conformations of DNA, including single-stranded, nicked, and jagged DNA of different sizes. These may not be comprehensively documented in all assays depending on the inherent nature of the analytical strategies.
2 cfDNA library preparations
2.1 Double-stranded DNA library preparation
NGS which provides basepair resolution of each incorpable DNA molecule in the sample, has been an effective method to assess the fragment size nature and associated sequence of cell-free DNA molecules. Traditionally, cfDNA analysis has been focused on double-stranded DNA (16–18). Double-stranded library preparation (dsDNA-LP) is accessible and affordable per sample and since its debut, it has been progressively optimized. During adapter ligation, for double-stranded library preparation, the overhangs of each dsDNA molecule must be polished, causing the dsDNA molecule to lose a portion of its original sequence (19, 20) (Figure 1A). Another aspect of the dsDNA-LP is that it is unable to incorporate short, degraded single-stranded DNA or those with single-strand breaks (nicks) (19, 22). Therefore, although it is established as a biomolecular tool, it is unable to assess all possible populations within each biological sample.
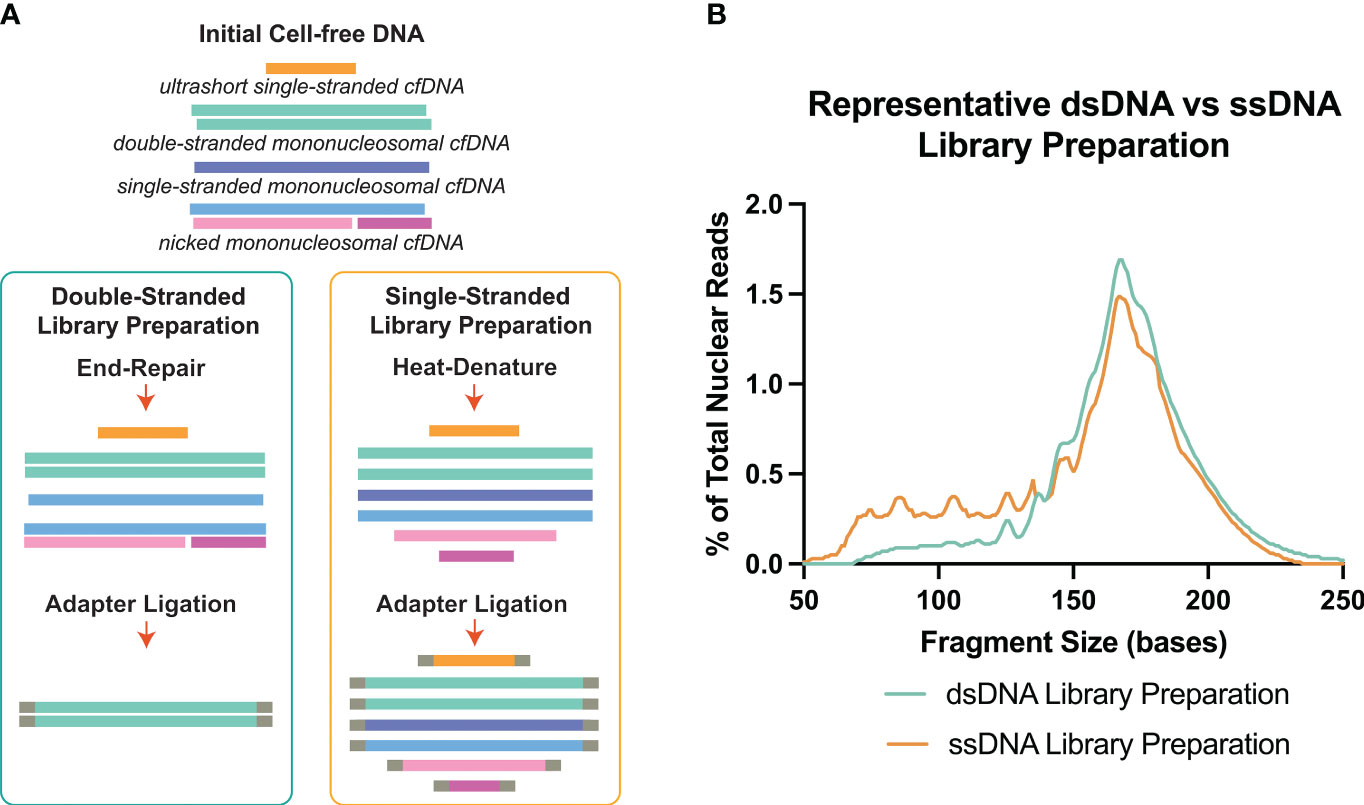
Figure 1 Double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) library preparation incorporates different DNA species from cell-free DNA. (A) Principal differences between the two library preparation methodologies. The initial heat-denature step in single-stranded library preparation allows the inclusion of multiple conformations of cell-free DNA. (B) Representative fragment profiles generated by double-stranded and single-stranded library preparations show that single-stranded library preparations are more sensitive for representing shorter cfDNA fragments below 80bp. Data has been derived from (11, 21).
2.2 Single-stranded DNA library preparation
The emergence of ssDNA-LP protocols, initially arose from the need for ancient DNA analysis, which dealt with greatly fragmented and degraded DNA samples within fossilized remains (22, 23). By utilizing a single-stranded library preparation, investigators were able to sequence the genome of a fossilized extracted DNA, which, through time, frequently become fragmented and single-stranded (22, 24). Svante Pääbo, who led these studies, eventually received the Nobel Prize for Physiology or Medicine in 2022 (25).
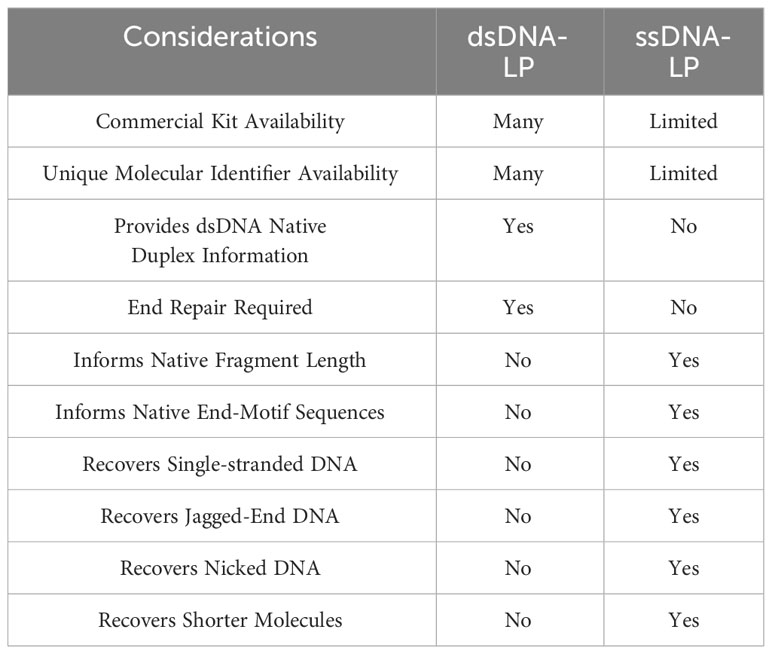
There are certain considerations when choosing dsDNA-LP vs ssDNA-LP workflows (Table 1). Single-stranded DNA library preparations require the heat denaturation of duplex template DNA, separating the molecules into two single-stranded templates prior to adapter ligation (Figure 1A). This denaturation allows for the incorporation of both blunt end and nicked dsDNA and ssDNA molecules. Therefore, by default, single-stranded libraries do not exclusively incorporate single-stranded molecules since they convert all DNA molecules to their single-stranded form. Additionally, since no end-repair is performed, the ends remain unaltered, making it possible to explore the native patterns of DNA fragmentation (26). In cell-free DNA, the use of single-stranded libraries has demonstrated an elevation in cfDNA molecules shorter than 100bp (11, 27–29) (Figure 1B).
3 Single-stranded DNA ligation strategies
A common protocol of all ssDNA-LP protocols is a heat-denature step (Figure 1A). Subsequently, in order to prepare the library for downstream sequencing, a method is required to attach the sequencing primer sequences to one end of the ssDNA molecule. The ligation strategy is essentially what unlocks the ability to fabricate complete NGS libraries. Many research groups have developed sophisticated methods for ssDNA library preparation, improving on limitations and caveats (Figure 2). The following are the current strategies for adapter ligation for single-stranded library preparation:
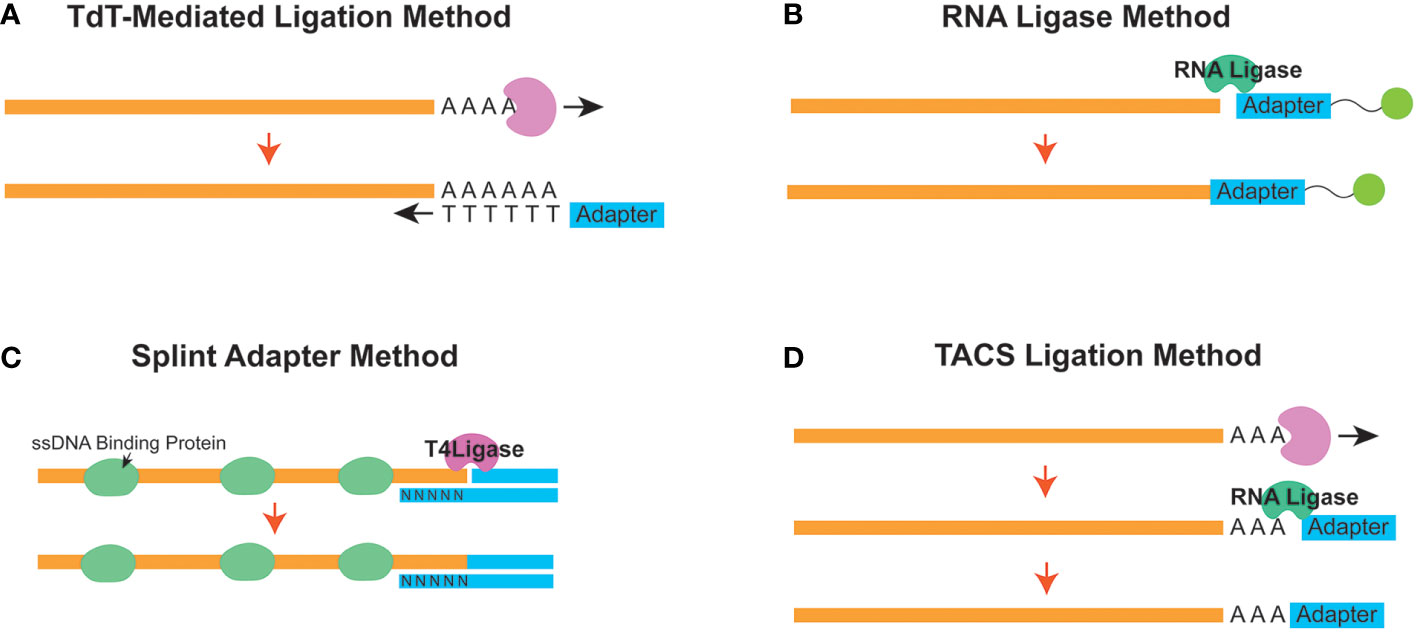
Figure 2 Different strategies to append the initial adapter to a single-stranded molecule in a single-stranded DNA library preparation workflow. Schematic diagrams for the (A) Terminal Deoxynucleotidyl Transferase (TDT) - Mediated Ligation, (B) RNA Ligase, (C) Splint Adapter, and (D) TDT-assisted adenylate connector-mediated single-stranded (ss) DNA (TACS) methods are shown.
3.1 Terminal deoxynucleotidyl transferase-mediated tailing
Terminal deoxynucleotidyl transferase (TdT)-mediated tailing is a strategy (Figure 2A) where the TdT enzyme is used to append a homopolymeric tail of adenosine or thymine nucleotides to the 3’-end of a ssDNA molecule (30). This resulting homopolymeric nucleotide tail can be used as a hybridization priming site for a complementary primer (31). Once the tail hybridizes with the primer, the cfDNA can be converted from a ssDNA molecule into one that is double-stranded. Subsequently, once converted, a sequencing second adapter on the 5’-end can be ligated using T4 DNA ligase (32). However, the homopolymeric tails can cause confusion downstream during downstream analysis since the investigator will need to be able to differentiate between the native and synthetically introduced nucleotides.
3.2 RNA ligase-based ligation
Another adapter ligation strategy uses the ability of the RNA ligase enzyme to conjoin a 5’-phosphorylated adapter to the 3’ end of the ssDNA molecule. This strategy was first introduced from ancient DNA workflows and has been implemented to assess cfDNA (11, 27) (Figure 2B). CircLigase II is one known enzyme that can attach the ssDNA to another ssDNA before using a primer to convert the molecule into dsDNA. Next, a second adapter ligation is performed using T4 DNA ligase (22). This strategy, although effective, can be expensive and time-consuming (33) since the efficiency of ssDNA ligation to ssDNA is reportedly low (34).
3.3 Splinted adapter method
As a follow-up to the ssDNA-LP method for ancient DNA, Gansuage et al. introduced the “ssDNA2.0” (34), which reduced some of the caveats of the earlier RNA Ligase-based ligation (22) by replacing the single-stranded ligation step with a splinted adapter (Figure 2C). Here, one side of the double-stranded adapter anneals with the target ssDNA strand through random hybridization with the single-stranded random nucleotide splint (35). This creates a nicked DNA scenario, allowing the use of the T4 DNA ligase instead of relying on the expensive and inefficient CircLigase-based reaction (34).
This method has been adapted to cell-free DNA (19). Similarly, the cell-free DNA application also uses a splinted double-stranded adapter where the bottom strand has a degenerate (or randomer) sequence to hybridize with the single-stranded target on both ends. Single-stranded binding proteins are added to stabilize the ssDNA conformation to facilitate better ligation. Once stably attached, a nick repair ligase (usually T4 DNA ligase) can be used to seal the nick and ligate the adapter. A heat denature step can then be used to remove the bottom strand for downstream reactions. Additionally, the initial adapter ligation can be designed so that it is performed all in one reaction, which reduces the need for multiple clean-up steps.
3.4 TDT-assisted adenylate connector-mediated single-stranded DNA ligation
Miura et al. published an improved method for adapter tagging technique using a TdT-assisted adenylate connector to mediate single-stranded ssDNA (TACS) ligation (Figure 2D). Similar to TDT-assisted approaches, this technique begins with the initial ribotailing of adenosines (up to 3 bases) at the 3’ end of the ssDNA. However, instead of using the polyA tail for complimentary hybridization, it creatively uses a particular RNA ligase (TS2126 RNA Ligase) (36) to append the desired adapter (37). This strategy is based on the observation that T4 RNA ligase has no preference between using DNA or RNA molecules as the donor molecule during a ligation reaction. In contrast, when considering the acceptor nucleic acid fragment, T4 RNA ligases prefer ligating nucleic acids onto RNA versus DNA (38). Therefore, modifying the ssDNA to become more “RNA-like” appeared to improve efficiency for ligation (39).
Although the TACS method improved upon the concept from the ancient DNA ligase strategy (24), they observed that this method was prone to forming adapter dimers. In situations with low DNA input, these adapter dimers would affect the proportion of useful information acquired from sequencing experiments. Additionally, they realized there were opportunities to improve the efficacy of second adapter ligation. To this end, they elected to forgo T4 DNA ligase and instead used vaccinia virus topoisomerase I (VTopoI) as a ligase enzyme for the second adapter ligation (40). Titling this method TACS-TOPO, they showed that, unlike T4 DNA ligase, VTopoI does not connect the 5’ phosphorylated end of the ssDNA molecule to the 3’ hydroxyl terminal. Instead, it ligates the 3’ phosphorylated end to a 5’ hydroxy end of a target DNA. Therefore, preventing the ligation of an available DNA oligo to a free 5’ phosphorylated end could reduce the occurrence of adapter-dimer formation.
3.5 CLAMP-Seq
An alternative atypical method for assessing single-stranded cell-free DNA has been developed, titled circular ligation amplification and sequencing (CLAMP-Seq). In this strategy, the cell-free DNA molecules are first separated by heat denaturation and then circularized (41). Next, using gene-specific primers pre-attached with sequencing adapters, they selectively replicated sequences from genes of interest. The investigators showed that constantly replicating from the original circularized strand reduced the propagation of potential PCR mistakes. This method appears ideal for enriching signals from targeted gene regions if effective primers can be designed to extract signals from all fragment permutations of the genes prior to sequencing.
All in all, the development of various ligation strategies is crucial for the initial step of ssDNA-LP workflow. As techniques and approaches evolve, the efficiency of the ligation step will improve. With these two different library preparation workflows available, researchers can consider which protocol would be suitable for their scientific questions (Table 1).
4 Observations in cell-free DNA using ssDNA library kit
Several studies have attempted to evaluate differences in cfDNA when processed by ssDNA-LP approach compared to a dsDNA-LP approach for the same DNA extracts (11, 27–29). These initial forays demonstrated that the ssDNA-LP is more inclusive to cfDNA for a broad range of types and lengths (Figure 1B). These reports suggested that a considerable fraction of cfDNA is non-nucleosomal in fragment size and could be the result of nuclease degradation (29). Both library preparation techniques showed a similar peak in mncfDNA at a dominant peak at 167bp. A 10.4 bp periodicity was also observed in the ssDNA kit but was offset by a 3bp rightward shift. This was attributed to the non-end repair nature of the ssDNA ligation, which may better showcase the nature of the ends of the original fragments.
4.1 The presence of ultrashort single-stranded cell-free DNA in plasma
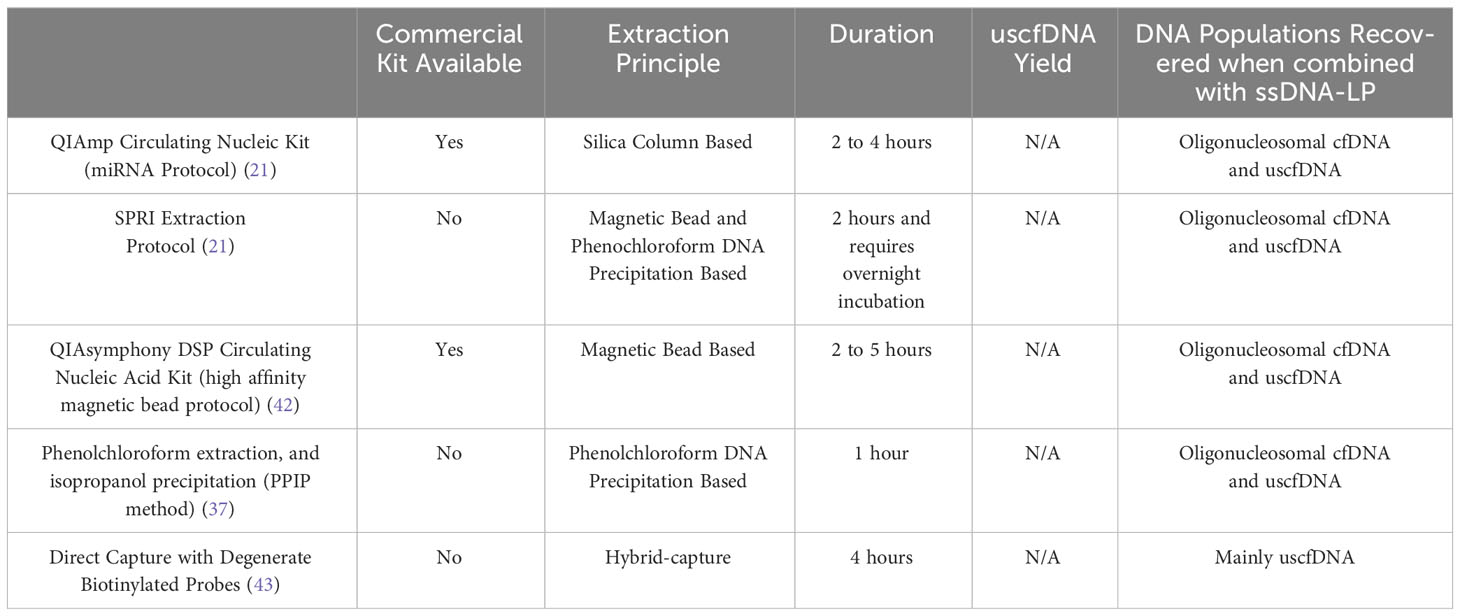
In addition to the major impact that ssDNA-LP had on the presentation of cfDNA, another important aspect is the effect of DNA extraction. Recently, multiple research groups reported the presence of ~50nt ultrashort single-strand DNA (uscfDNA) fragments in both the plasma from non-cancer and cancer patients (21, 37, 42, 43) (Figure 3). This novel population of cell-free DNA was revealed by pairing a low-molecular-weight optimized DNA extraction method (Table 2) with an ssDNA-LP (Figure 3).
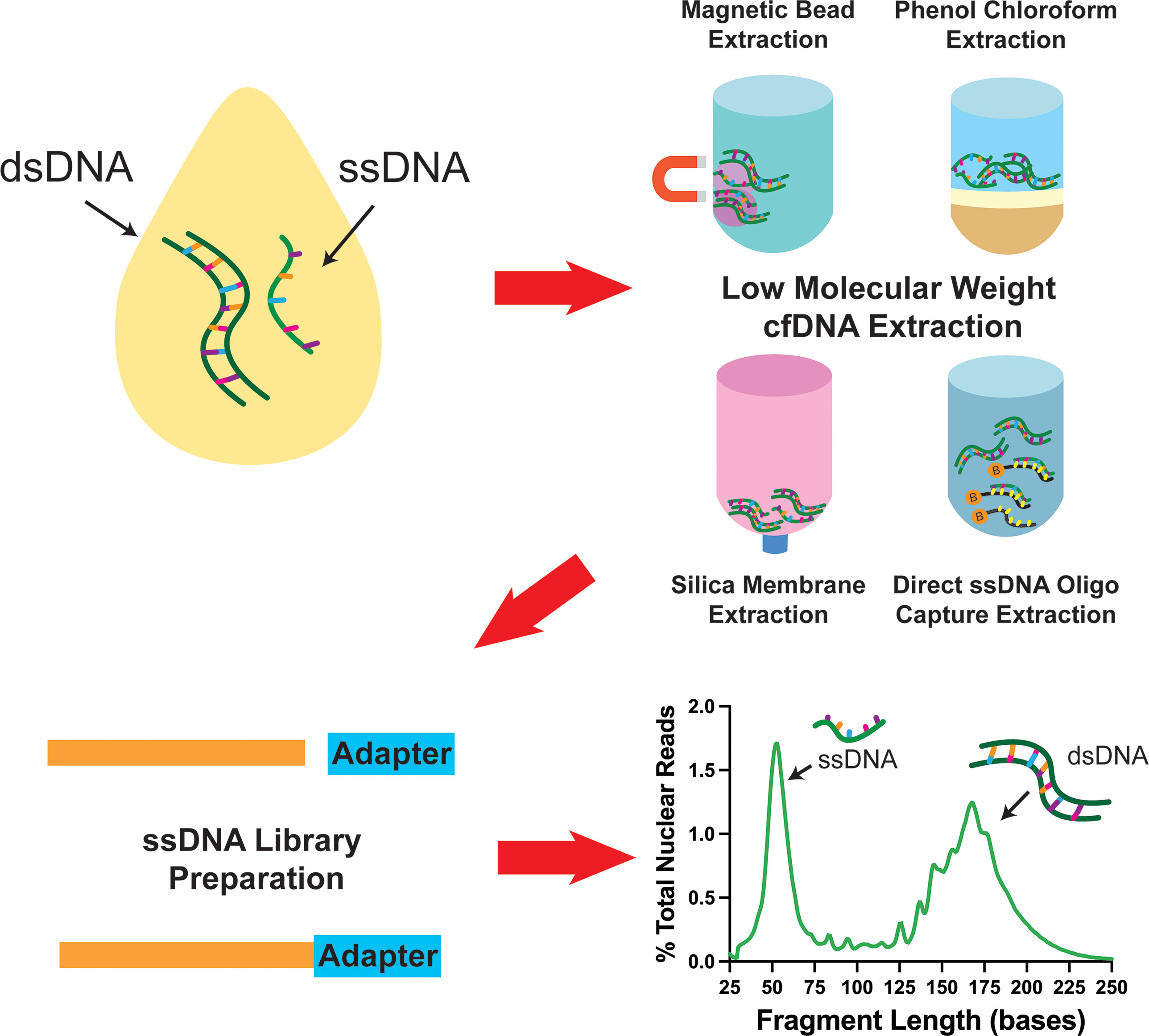
Figure 3 Schematic diagram showing that pairing various low molecular weight enriched extraction methods with ssDNA library preparation reveals the presence of ultrashort single-stranded cell-free DNA in plasma. Data has been derived from (11, 21).
4.2 Characteristics of uscfDNA
Our group demonstrated that using either the microRNA protocol (referred to as QiaM) of the commonly used Qiagen Circulating Nucleic Acid Kit (44), which uses additional isopropanol and buffer volume, is able to greatly enrich short and single-stranded nucleic acids (characteristics of miRNA) (21). Additionally, we showed that using solid phase reversible immobilization beads (SPRI) with high volumes of isopropanol and crowding agent (polyethylene glycol (PEG)) and salt with phenol-chloroform also promotes the retention of short single-stranded molecules during extraction. Similarly, other investigators showed various methods such as conventional phenol-chloroform-based extraction method (37) or magnetic beads with a commercial nucleic acid extraction kit (42) could also retain these species of cfDNA (Table 2). Another unique method reported used 10nt biotinylated capture probes with randomized nucleotide bases to directly capture random single-stranded cell-free DNA in plasma (43).
4.3 uscfDNA to mncfDNA ratio quantification challenges
If the size-distribution ratio of uscfDNA and mncfDNA are considered, the phenol-chloroform method (37) apparently recovers uscfDNA at similar efficiencies observed in the QiaM and SPRI extraction methods (21). In contrast, due to this bias toward short single-stranded molecules, the direct hybrid capture method resulted in a very high uscfDNA: mncfDNA ratio (43). This could be explained by the nature of the method, which has a lower affinity for double-stranded mncfDNA. The magnetic bead protocol (42) demonstrated a ratio where the peak of uscfDNA was slightly lower than the mncfDNA. These similar but varied results indicate that although key principals are required to visualize uscfDNA, their representation is still contingent on the method of extraction and library preparation. Therefore, evaluating the efficacy of the extraction between the five methods would be valuable. Currently, there are no methods to quantify uscfDNA from the heterogeneous pool of purified DNA specifically. Commonly used fluorescent-based DNA quantification methods measure total DNA (45), which would not provide any ratio relationships between uscfDNA and mncfDNA. NGS can inform on the ratio between these two species but requires careful spike-in experiments to clarify the recovered concentration compared to the spiked-in amount. In one study, spike-in with oligos of various sizes as a reference suggested that the uscfDNA are present at a concentration of 2.0 ng/ml (43). Total cell-free DNA has been reported to range from 0 to 2000ng/ml (46). Therefore, it is unclear if the concentration of 2.0ng/ml for uscfDNA should be viewed as a minor or major contributor. Hence, at this time, it is difficult to assess the actual uscfDNA concentration without developing new strategies.
4.4 Strandedness
Interestingly, through an assortment of deductive experiments, multiple groups inferred that the ~50nt uscfDNA is single-stranded in nature (Figure 4A). This was determined by performing strand-specific nuclease digestions on the extracted cfDNA (37) and prior to library preparation (21, 43), revealing that uscfDNA was digestible by ssDNA-specific nucleases (S1 Nuclease and Exo 1 nuclease) but remains intact with dsDNA-specific enzymes (dsDNase) (47). When extracted DNA was processed with the dsDNA-LP, the uscfDNA was not observable, whereas excluding the heat-denature retained the uscfDNA but not double-stranded mncfDNA (21, 37, 42, 43). These experiments provided strong evidence that uscfDNA exists as a single-stranded DNA molecule in circulation.
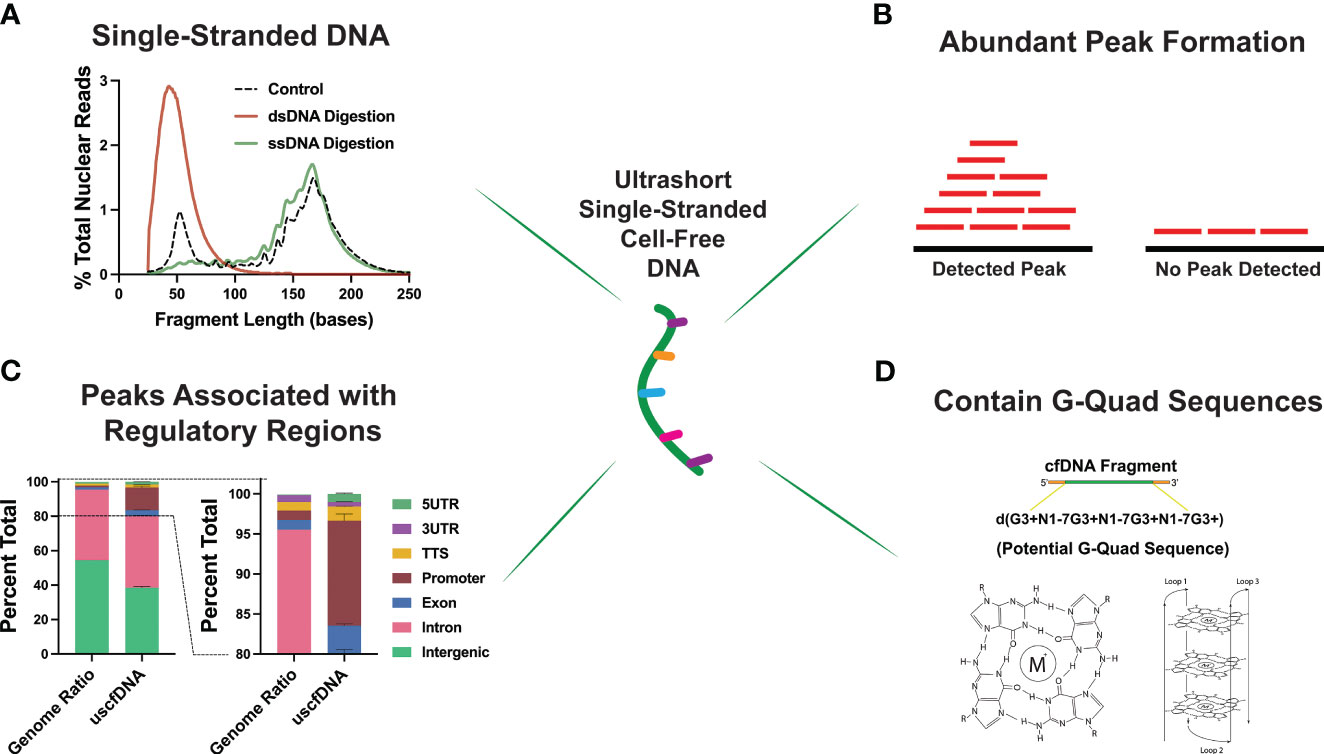
Figure 4 Unique properties of plasma ultrashort single-stranded cell-free DNA (uscfDNA). (A) Digestion assays suggest ultrashort cell-free DNA is single-stranded. (B) Peak detection bioinformatic tools indicate that uscfDNA maps as abundant peaks along the genome, and these peaks are enriched in (C) regulatory regions. (D) Sequences of uscfDNA contain potential G-Quad secondary structures. Data has been derived from (11, 21).
4.5 Genomic characteristics of uscfDNA differ from mncfDNA
As uscfDNA is present in non-cancer individuals, it is physiological but demonstrates distinct characteristics from mncfDNA. Karyograms of the normalized coverage of uscfDNA and mncfDNA populations showed significantly different coverage patterns with uscfDNA mapping to more hotspots within the body of chromosomes and telomeres than the mncfDNA (21, 43, 48). Once aligned, fragments of uscfDNA appear to congregate as peaks in open chromatin regions of the genome, most notably in regions with close proximity to the transcription start sites (TSS), intron, and exonic regions (37, 42, 48) (Figures 4B,C). Additionally, compared to mncfDNA, uscfDNA fragments are more colocalized with transcription binding factor sites and histone modification sites (37, 49).
These regulatory regions are also enriched in sequences with a high potential to form secondary structures such as GQuadruplexes (Figure 4D). G-Quadruplex structures are observable secondary structures within the chromatin regions of the genome and are correlated to expression levels of oncogenes in the tumor tissue (50, 51). Interestingly, uscfDNA contain a greater abundance of these sequences compared to mncfDNA. Lastly, annotation of the fragment end-motif profiles of uscfDNA reflects the non-random process of nuclease activity (52) and analysis shows that the end-motif profiles are dissimilar between uscfDNA and mncfDNA. Therefore, the properties of uscfDNA (in peak formation in regulatory regions and association with secondary structures) are different from mncfDNA and thus should be considered a separate sub-species of cfDNA. Further exploration could be performed by assessing the animal models of different nuclease knock-down models to observe how they impact the uscfDNA (53, 54).
5 Cancer-related differences of ssDNA-LP vs dsDNA-LP
5.1 Global fragment size changes in cancer
Early investigations suggested that the fragment profile of tumor-derived cell-free DNA (ctDNA) differs in length compared to those originating from wild-type cells (16, 55). A study showed that cell-free DNA fragments from 90-150bp are enriched in mutation-containing sequences, and by examining these binned sizes in isolation, they can improve ctDNA detection compared to looking at fragments of all lengths (18). The global cfDNA fragment profile can appear aberrated, and analyzing these global changes can be an effective metric for cancer detection (56).
Several studies have performed whole genome sequencing using ssDNA-LP to study the fragment profile of plasma cancer samples (29, 49, 57, 58). They hypothesized that the ssDNA-LP protocol would enrich the diversity of cfDNA molecules and potentially enhance the global fragment differences. To this end, one study compared the dsDNA-LP and ssDNA-LP approach to plasma from lung, breast, liver, and colorectal cancer individuals, showing that the apparent fragment patterns were different than the dsDNA library (although they did not examine paired individuals (58). Here, they established that the ssDNA-LP enriched and revealed a cfDNA fragment population from 30-80bp that was not previously detectable by the double-stranded library. In a follow-up study, they looked at metastatic colorectal cancer to examine if the pattern was able to see differences (29). Different cancer specimens with decreasing amounts of mutant allele fractions (MAF) (68.6%, 54.7%, 47.3%, 23.3%, 14.4%, 3.2%, 0.9%, and healthy) were evaluated, and they observed that both library preparation strategies showed clear differences between non-cancer and cancer subjects. However, their data suggested that ssDNA-LP could show a more pronounced difference. The samples processed with ssDNA-LP had a 10-fold greater number of reads in the small fragment region <100 bases. For example, the highest MAF specimen had a much larger proportion of reads between 30-143bp bases versus the dsDNA-LP, which also showed the trend in fragmentomics but as stated earlier, was apparently less pronounced.
In another study, samples processed with a ssDNA-LP workflow were deeply sequenced at (30-fold of the genome) and also showed a similar fragmentomics difference between colorectal cancer samples compared to non-cancer individuals (49). Therefore, the ability of ssDNA-LP to enrich smaller or single-stranded DNA may provide a better ability for fragmentomic analysis.
5.2 Circulating tumor DNA hotspot mutation detection
A report by Liu et al. showed an early attempt to combine ssDNA-LP with hybrid capture to enrich specific mutation-containing fragments in the plasma of 112 pancreatic cancer patients of varying stages (59). Using a custom panel built for 62 pancreatic cancer genes, they found cancer specific mutations in 88% of the samples, and KRAS-specific mutations in 70% of the samples, which was consistent with the tissue-based sequencing. Regarding fragment size, they showed that in pancreatic cancer samples, a substantial proportion of the mutated KRAS fragments were shorter than 100 bases. At the same time, the wild type version of those sequences retained their ~167bp modal size. Interestingly, they identified that the decreased footprints were more pronounced in the early stages of pancreatic such as those with intraductal papillary mucinous neoplasm cancer, compared to late stages.
Another paper using Clamp-Seq demonstrated excellent concordance of the detection of hotspot mutations between droplet digital PCR results of 97.4%. Similarly, an analysis of 134 NSCLC patients showed a 94.8% concordance with the tissue genotyping (41).
These studies showed that the ssDNA-LP methods could potentially provide equivalent ctDNA information to the dsDNA-LP methods.
However, one of the caveats of the ssDNA-LP protocol is that it requires the separation of double-stranded molecules prior to adapter ligation. Additionally, natively single-stranded DNA molecules may not have a clear duplex-mate. Therefore, with the current ssDNA-LP workflow, the native duplex information would be inaccessible. Duplex molecule information is often helpful for identifying and removing errors in sequenced reads (17, 60). If only one strand of the duplex reports a variant but not the other, it may be suggestive that the variant arose synthetically during the library preparation, potentially through oxidative DNA damage (61) or cytosine deamination (62). However, other forms of error suppression are still potentially eligible for future development of nonduplex reads. These strategies would likely utilize unique molecular identifier (UMI) correction or bioinformatic in silico error suppression models based on stereotyping experimental data (17).
5.3 Copy number variation inference for ctDNA burden
In another body of work, the investigators examined ten samples with high tumor DNA content from eight colorectal patients with high ctDNA % (63). They prepared three kinds of libraries: dsDNA-LP, ssDNA-LP, and pure ssDNA library (no heat denaturation), and the ssDNA-LP was constructed using a TdT-mediated ligation strategy. To evaluate the ctDNA tumor fraction, they developed an algorithm called the plasma genomic abnormality 841 score (PGA) (64). They observed that the ssDNA-LP, pure ssDNA library and ssDNA-LP had greater ctDNA content as per the PGA score. They suggested that the reason for the increased observed ctDNA signal (through PGA) was due to ssDNA-LP’s ability to ligate smaller DNA, pre-existing ssDNA, or nicked DNA. These conformations of DNA were abundant in the plasma of cancer samples, and inclusion could improve cancer signals from plasma.
In contrast, in a letter to the editor, Moser et al. questioned if, when compared to dsDNA-LP, ssDNA-LP could provide a greater ctDNA sensitivity (57). In their pilot study, they applied the ssDNA-LP through the use of RNA ligase-based strategy (27) and assessed the copy number variation signal of the cfDNA from five patients with various cancers (breast, colon, and prostate). However, their experiment failed to detect any significant difference or preferential enrichment in ctDNA.
In conclusion, the assessment of whether ssDNA-LP improves over dsDNA-LP for ctDNA detection is still dynamically ongoing. The preliminary papers are promising, but definitive studies have yet to be carried out. Since large-scale comparative studies have not yet been published, it is still unclear if the ssDNA-LP approach has greater sensitivity or specificity for cancer detection. However, there will likely be attempts to apply the creative approaches for ctDNA detection designed from dsDNA-LP to ssDNA-LP.
5.4 Clinical cancer detection potential of uscfDNA
A couple of studies have examined the utility of uscfDNA as a novel biomarker for cancer detection (42, 48). Hudecova et al. explored the properties of uscfDNA between plasma from 21 pan-cancer samples (breast, lung, thymoma, rectal colorectal, and ovarian) and 28 healthy individuals (42), whereas our group investigated alterations in the uscfDNA between 14 late-stage lung cancer and 18 healthy controls.
Regarding changes in the ratio of uscfDNA to mncfDNA changes, despite contrasting directionalities, there appears to be a change in uscfDNA abundance in cancer samples. In samples with higher ctDNA load [using copy number variation as an inference (18)] demonstrated the most observably decrease in uscfDNA abundance compared to other samples (42). In contrast, our group observed an increase in uscfDNA content in late-stage lung cancer samples compared to non-cancer individuals (48). Using copy number variation, Hudecova et al. observed that uscfDNA appeared to contain but was not enriched in the tumor-derived signals (42). Interestingly, both groups found that uscfDNA fragments that promoter regions were enriched in G-quadruplex secondary structure sequences and that this decreased in cancer patients. Additionally, changes in the composition of specific functional element peaks, end-motif profiles, and fragment-size distributions were observed in the uscfDNA population between lung cancer and non-cancer subjects (48). These early studies suggest that the accompanying uscfDNA with the conventional nucleosomal cell-free DNA appears to be a potentially new biomarker for cancer detection.
6 Other cfDNA applications of ssDNA-LP
6.1 Effect of ssDNA-LP on other biofluids
In the cell-free DNA of other biofluids, such as urine and saliva, single-stranded libraries have also been shown to alter the perceived fragment size characteristics. In cell-free saliva, with similarity to the observations made in plasma, compared to the dsDNA-LP, the ssDNA-LP demonstrated a 3bp rightward shift in the fragment periodicities. Additionally, there was a slightly greater retention of shorter fragments below 100 bases (65). For urine, the ssDNA-LP revealed that the cfDNA was short and fragmented, with a large proportion of fragments below 100 bases. However, in that report, the samples were not directly compared with dsDNA-LP (66).
6.2 Effect of ssDNA-LP on non-human species
Using the ssDNA-LP approach, Burnham et al. observed an increase in the proportion of bacteria and mitochondrial (cfmitDNA) content from samples (27). Other groups showed that the low molecular weight DNA extraction also helped enrich the cfmitDNA (21). The enhanced ability to track the profile of bacteria species using cell-free DNA has been effective in monitoring organ transplant outcomes (27, 67).
7 Conclusions and future directions
The introduction of robust ssDNA-LP technology has opened new avenues in the realm of liquid biopsy, illustrating the impact that different methodologies have on the perceived observations. The ability to assess a greater variety of cfDNA species in plasma as well as other biofluids has increased the pool of DNA species to be examined in plasma. Both biomolecular and bioinformatic techniques will need to be developed to harness these new populations of cell-free DNA. More studies will be needed to show if ssDNA-LP pushes the needle of sensitivity compared to dsDNA-LP. Additionally, ultrashort single-stranded cell-free DNA, which appears to have different properties and biological origins compared to mncfDNA, is now added to the toolbox as another potential biomarker for cancer detection. Resultingly, many opportunities are readily available for the development of novel strategies to examine the biological and clinical relevance of the diverse cell-free DNA populations uncovered by the ssDNA-LP approach.
Author contributions
JC: Conceptualization, Writing – original draft. NS: Conceptualization, Writing – review & editing. DW: Conceptualization, Writing – review & editing. DC: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NCI K00CA264398-03 to JC, NIH grants 2U01CA233370-06 to DW and 1R90DE031531 to NS.
Conflict of interest
DW is a consultant to Avellino/AIONCO, Colgate Palmolive, and has equity in Liquid Diagnostics LLC. JC, NS, and DW have filed the U.S. Provisional Patent Application No. 63/373,369 titled NEXT-GENERATION SEQUENCING PIPELINE TO DETECT ULTRASHORT SINGLE-STRANDED CELL-FREE DNA filed on 8/24/2022.
The remaining author declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic — implementation issues and future challenges. Nat Rev Clin Oncol. (2021) 18:297–312. doi: 10.1038/s41571-020-00457-x
2. Rostami A, Lambie M, Yu CW, Stambolic V, Waldron JN, Bratman SV. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. (2020) 31:107830. doi: 10.1016/j.celrep.2020.107830
3. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. (2013) 10:472–84. doi: 10.1038/nrclinonc.2013.110
4. Chan KCA, Jiang P, Chan CWM, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U.S.A. (2013) 110:18761–8. doi: 10.1073/pnas.1313995110
5. Mouliere F, Mair R, Chandrananda D, Marass F, Smith CG, Su J, et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med. (2018) 10:e9323. doi: 10.15252/emmm.201809323
6. Thierry AR. Circulating DNA fragmentomics and cancer screening. Cell Genom. (2023) 3:100242. doi: 10.1016/j.xgen.2022.100242
7. Luo H, Wei W, Ye Z, Zheng J, Xu R-H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol Med. (2021) 27:482–500. doi: 10.1016/j.molmed.2020.12.011
8. Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. (2021) 372:eaaw3616. doi: 10.1126/science.aaw3616
9. Mattox AK, Douville C, Wang Y, Popoli M, Ptak J, Silliman N, et al. The origin of highly elevated cell-free DNA in healthy individuals and patients with pancreatic, colorectal, lung, or ovarian cancer. Cancer Discovery. (2023) 13:2166–79. doi: 10.1158/2159-8290.CD-21-1252
10. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human Malignancies. Sci Transl Med. (2014) 6:224ra24. doi: 10.1126/scitranslmed.3007094
11. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. (2016) 164:57–68. doi: 10.1016/j.cell.2015.11.050
12. Holdenrieder S, Stieber P, Bodenmüller H, Busch M, Von Pawel J, Schalhorn A, et al. Circulating nucleosomes in serum. Ann N Y Acad Sci. (2001) 945:93–102. doi: 10.1111/j.1749-6632.2001.tb03869.x
13. Holdenrieder S, Mueller S, Stieber P. Stability of nucleosomal DNA fragments in serum. Clin Chem. (2005) 51:1026–9. doi: 10.1373/clinchem.2005.048454
14. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PloS Genet. (2016) 12:e1006162. doi: 10.1371/journal.pgen.1006162
15. Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. (2016) 35:347–76. doi: 10.1007/s10555-016-9629-x
16. Jiang P, Chan CWM, Chan KCA, Cheng SH, Wong J, Wong VW-S, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci. (2015) 112:E1317–25. doi: 10.1073/pnas.1500076112
17. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. (2016) 34:547–55. doi: 10.1038/nbt.3520
18. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. (2018) 10:eaat4921. doi: 10.1126/scitranslmed.aat4921
19. Troll CJ, Kapp J, Rao V, Harkins KM, Cole C, Naughton C, et al. A ligation-based single-stranded library preparation method to analyze cell-free DNA and synthetic oligos. BMC Genomics. (2019) 20:1023. doi: 10.1186/s12864-019-6355-0
20. Avgeris M, Marmarinos A, Gourgiotis D, Scorilas A. Jagged ends of cell-free DNA: rebranding fragmentomics in modern liquid biopsy diagnostics. Clin Chem. (2021) 67:576–8. doi: 10.1093/clinchem/hvab036
21. Cheng J, Morselli M, Huang W-L, Heo YJ, Pinheiro-Ferreira T, Li F, et al. Plasma contains ultrashort single-stranded DNA in addition to nucleosomal cell-free DNA. iScience. (2022) 25:104554. doi: 10.1016/j.isci.2022.104554
22. Gansauge M-T, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. (2013) 8:737–48. doi: 10.1038/nprot.2013.038
23. Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc R Soc B: Biol Sci. (2012) 279:4724–33. doi: 10.1098/rspb.2012.1745
24. Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. (2012) 338:222–6. doi: 10.1126/science.1224344
25. Klein RG. Profile of Svante Pääbo: 2022 Nobel laureate in physiology or medicine. Proc Natl Acad Sci U.S.A. (2023) 120:e2217025119. doi: 10.1073/pnas.2217025119
26. Jiang P, Sun K, Peng W, Cheng SH, Ni M, Yeung PC, et al. Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discovery. (2020) 10:664–73. doi: 10.1158/2159-8290.CD-19-0622
27. Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. (2016) 6:srep27859. doi: 10.1038/srep27859
28. Vong JSL, Tsang JCH, Jiang P, Lee W-S, Leung TY, Chan KCA, et al. Single-stranded DNA library preparation preferentially enriches short maternal DNA in maternal plasma. Clin Chem. (2017) 63:1031–7. doi: 10.1373/clinchem.2016.268656
29. Sanchez C, Roch B, Mazard T, Blache P, Dache ZAA, Pastor B, et al. Circulating nuclear DNA structural features, origins, and complete size profile revealed by fragmentomics. JCI Insight. (2021) 6:144561. doi: 10.1172/jci.insight.144561
30. Motea EA, Berdis AJ. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta. (2010) 1804:1151–66. doi: 10.1016/j.bbapap.2009.06.030
31. Turchinovich A, Surowy H, Serva A, Zapatka M, Lichter P, Burwinkel B. Capture and Amplification by Tailing and Switching (CATS). An ultrasensitive ligation-independent method for generation of DNA libraries for deep sequencing from picogram amounts of DNA and RNA. RNA Biol. (2014) 11:817–28. doi: 10.4161/rna.29304
32. Kuhn H, Frank-Kamenetskii MD. Template-independent ligation of single-stranded DNA by T4 DNA ligase. FEBS J. (2005) 272:5991–6000. doi: 10.1111/j.1742-4658.2005.04954.x
33. Kapp JD, Green RE, Shapiro B. A fast and efficient single-stranded genomic library preparation method optimized for ancient DNA. J Heredity. (2021) 112:241–9. doi: 10.1093/jhered/esab012
34. Gansauge M-T, Gerber T, Glocke I, Korlevic P, Lippik L, Nagel S, et al. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. (2017) 45:e79. doi: 10.1093/nar/gkx033
35. Kwok CK, Ding Y, Sherlock ME, Assmann SM, Bevilacqua PC. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Analytical Biochem. (2013) 435:181–6. doi: 10.1016/j.ab.2013.01.008
36. Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. (2005) 33:135–42. doi: 10.1093/nar/gki149
37. Hisano O, Ito T, Miura F. Short single-stranded DNAs with putative non-canonical structures comprise a new class of plasma cell-free DNA. BMC Biol. (2021) 19:225. doi: 10.1186/s12915-021-01160-8
38. Bullard DR, Bowater RP. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem J. (2006) 398:135–44. doi: 10.1042/BJ20060313
39. Miura F, Shibata Y, Miura M, Sangatsuda Y, Hisano O, Araki H, et al. Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. (2019) 47:e85. doi: 10.1093/nar/gkz435
40. Miura F, Kanzawa-Kiriyama H, Hisano O, Miura M, Shibata Y, Adachi N, et al. A highly efficient scheme for library preparation from single-stranded DNA. Sci Rep. (2023) 13:13913. doi: 10.1038/s41598-023-40890-3
41. Wang L, Hu X, Guo Q, Huang X, Lin C-H, Chen X, et al. CLAmp-seq: A novel amplicon-based NGS assay with concatemer error correction for improved detection of actionable mutations in plasma cfDNA from patients with NSCLC. Small Methods. (2020) 4:1900357. doi: 10.1002/smtd.201900357
42. Hudecova I, Smith CG, Hänsel-Hertsch R, Chilamakuri CS, Morris JA, Vijayaraghavan A, et al. Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res. (2022) 32(2):215–27. doi: 10.1101/gr.275691.121
43. Cheng LY, Dai P, Wu LR, Patel AA, Zhang DY. Direct capture and sequencing reveal ultra-short single-stranded DNA in biofluids. iScience. (2022) 25:105046. doi: 10.1016/j.isci.2022.105046
44. Lampignano R, Neumann MHD, Weber S, Kloten V, Herdean A, Voss T, et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin Chem. (2020) 66:149–60. doi: 10.1373/clinchem.2019.306837
45. Mardis E, McCombie WR. Library quantification: fluorometric quantitation of double-stranded or single-stranded DNA samples using the qubit system. Cold Spring Harb Protoc. (2017) 2017:pdb.prot094730. doi: 10.1101/pdb.prot094730
46. Bryzgunova OE, Konoshenko MY, Laktionov PP. Concentration of cell-free DNA in different tumor types. Expert Rev Mol Diagn. (2021) 21:63–75. doi: 10.1080/14737159.2020.1860021
47. Nilsen IW, Øverbø K, Jensen Havdalen L, Elde M, Gjellesvik DR, Lanes O. The Enzyme and the cDNA Sequence of a Thermolabile and Double-Strand Specific DNase from Northern Shrimps (Pandalus borealis). PloS One. (2010) 5:e10295. doi: 10.1371/journal.pone.0010295
48. Cheng J, Swarup N, Li F, Kordi M, Lin C-C, Yang S-C, et al. Distinct features of plasma ultrashort single-stranded cell-free DNA as biomarkers for lung cancer detection. Clin Chem. (2023) 69(11):1270–87. doi: 10.1093/clinchem/hvad131
49. Wang F, Li X, Li M, Liu W, Lu L, Li Y, et al. Ultra-short cell-free DNA fragments enhance cancer early detection in a multi-analyte blood test combining mutation, protein and fragmentomics. Clin Chem Lab Med. (2023) 62(1):168–77. doi: 10.1515/cclm-2023-0541
50. Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. (2016) 48:1267–72. doi: 10.1038/ng.3662
51. Hänsel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet. (2020) 52:878–83. doi: 10.1038/s41588-020-0672-8
52. Serpas L, Chan RWY, Jiang P, Ni M, Sun K, Rashidfarrokhi A, et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. PNAS. (2019) 116:641–9. doi: 10.1073/pnas.1815031116
53. Han DSC, Ni M, Chan RWY, Chan VWH, Lui KO, Chiu RWK, et al. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. (2020) 106:202–14. doi: 10.1016/j.ajhg.2020.01.008
54. Han DSC, Lo YMD. The nexus of cfDNA and nuclease biology. Trends Genet. (2021) 37:758–70. doi: 10.1016/j.tig.2021.04.005
55. Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumor-derived circulating DNA. PloS One. (2011) 6:e23418. doi: 10.1371/journal.pone.0023418
56. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. (2019) 1:385–89. doi: 10.1038/s41586-019-1272-6
57. Moser T, Ulz P, Zhou Q, Perakis S, Geigl JB, Speicher MR, et al. Single-stranded DNA library preparation does not preferentially enrich circulating tumor DNA. Clin Chem. (2017) 63:1656–9. doi: 10.1373/clinchem.2017.277988
58. Sanchez C, Snyder MW, Tanos R, Shendure J, Thierry AR. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. NPJ Genom Med. (2018) 3:31. doi: 10.1038/s41525-018-0069-0
59. Liu X, Liu L, Ji Y, Li C, Wei T, Yang X, et al. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine. (2019) 41:345–56. doi: 10.1016/j.ebiom.2019.02.010
60. Cohen JD, Douville C, Dudley JC, Mog BJ, Popoli M, Ptak J, et al. Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands. Nat Biotechnol. (2021) 39:1220–7. doi: 10.1038/s41587-021-00900-z
61. Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. (2013) 41:e67. doi: 10.1093/nar/gks1443
62. Chen G, Mosier S, Gocke CD, Lin M-T, Eshleman JR. Cytosine deamination is a major cause of baseline noise in next generation sequencing. Mol Diagn Ther. (2014) 18:587–93. doi: 10.1007/s40291-014-0115-2
63. Zhu J, Huang J, Zhang P, Li Q, Kohli M, Huang C-C, et al. Advantages of single-stranded DNA over double-stranded DNA library preparation for capturing cell-free tumor DNA in plasma. Mol Diagn Ther. (2020) 24:95–101. doi: 10.1007/s40291-019-00429-7
64. Xia S, Kohli M, Du M, Dittmar RL, Lee A, Nandy D, et al. Plasma genetic and genomic abnormalities predict treatment response and clinical outcome in advanced prostate cancer. Oncotarget. (2015) 6(18):16411–21. doi: 10.18632/oncotarget.3845
65. Swarup N, Cheng J, Choi I, Heo YJ, Kordi M, Aziz M, et al. Multi-faceted attributes of salivary cell-free DNA as liquid biopsy biomarkers for gastric cancer detection. biomark Res. (2023) 11:90. doi: 10.1186/s40364-023-00524-2
66. Burnham P, Dadhania D, Heyang M, Chen F, Westblade LF, Suthanthiran M, et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun. (2018) 9:2412. doi: 10.1038/s41467-018-04745-0
Keywords: cell-free DNA, liquid biopsy, single-stranded library preparation, fragment-size, ultrashort single-stranded cell-free DNA
Citation: Cheng JC, Swarup N, Wong DTW and Chia D (2024) A review on the impact of single-stranded library preparation on plasma cell-free diversity for cancer detection. Front. Oncol. 14:1332004. doi: 10.3389/fonc.2024.1332004
Received: 02 November 2023; Accepted: 07 February 2024;
Published: 06 March 2024.
Edited by:
David T. Miyamoto, Harvard Medical School, United StatesReviewed by:
Shervin Tabrizi, Massachusetts General Hospital and Harvard Medical School, United StatesMin Pan, Southeast University, China
Copyright © 2024 Cheng, Swarup, Wong and Chia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Chia, dchia@mednet.ucla.edu
 Jordan C. Cheng
Jordan C. Cheng Neeti Swarup1
Neeti Swarup1 David Chia
David Chia