- 1Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, Ganzhou, China
- 2The First School of Clinical Medicine, Gannan Medical University, Ganzhou, China
Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and extracellular vehicles (EVs) have received significant attention in recent times as emerging biomarkers and subjects of transformational studies. The three main branches of liquid biopsy have evolved from the three primary tumor liquid biopsy detection targets—CTC, ctDNA, and EVs—each with distinct benefits. CTCs are derived from circulating cancer cells from the original tumor or metastases and may display global features of the tumor. ctDNA has been extensively analyzed and has been used to aid in the diagnosis, treatment, and prognosis of neoplastic diseases. EVs contain tumor-derived material such as DNA, RNA, proteins, lipids, sugar structures, and metabolites. The three provide different detection contents but have strong complementarity to a certain extent. Even though they have already been employed in several clinical trials, the clinical utility of three biomarkers is still being studied, with promising initial findings. This review thoroughly overviews established and emerging technologies for the isolation, characterization, and content detection of CTC, ctDNA, and EVs. Also discussed were the most recent developments in the study of potential liquid biopsy biomarkers for cancer diagnosis, therapeutic monitoring, and prognosis prediction. These included CTC, ctDNA, and EVs. Finally, the potential and challenges of employing liquid biopsy based on CTC, ctDNA, and EVs for precision medicine were evaluated.
1 Introduction
Cancer is a highly lethal ailment that poses a significant threat to human existence. Key components of cancer prevention, detection, and therapy encompass the assessment of the effectiveness of cancer treatment, the surveillance of patient’s post-treatment, and the provision of timely alerts regarding the risk of tumor metastasis and recurrence. There is no accurate biomarker for early cancer diagnosis due to the complexity of tumor incidence and tumor heterogeneity, Despite the development of multiple technologies aimed at the early detection of tumor biomarkers from diverse clinical samples and the significant research conducted in this field (1). Technological developments have led to alternate methods of tumor liquid biopsy, which have seen significant success and promise (2). The current understanding in the scientific community is that a tumor can release many components, such as tumor cells proteins, extracellular vehicles (EVs), and nucleic acids, into the peripheral circulation (3). Since the original identification of tumor cells in patient peripheral blood in the 1860s, significant advancements have been made in separating circulating tumor cells (CTCs) from various blood cell types (4). Furthermore, in 1996, the identification of microsatellite alterations in the circulating tumor DNA of individuals with tumors demonstrated a precise correlation with the microsatellite changes noticed in the primary tumor (5).In 2011, the concept of liquid biopsy based on CTC detection was proposed (4), and after that, the technology of using ctDNA to obtain tumor biological information was included in the liquid biopsy’s scope (5).In 2015, the Massachusetts Institute of Technology Review named liquid biopsy one of the top ten breakthrough technologies of the year. Afterward, liquid biopsy entered a rapid development stage, and detection technologies such as extracellular vehicles (EVs) (6) and tumor-educated platelets (7) were also included in the scope of liquid biopsy. A method of in vitro diagnosis called liquid biopsy uses body fluids as the test material to obtain tumor biological information relative to tissue biopsy. Liquid biopsy, which obtains a sample of bodily fluids such as saliva, blood, and cerebrospinal fluid, has gained broad attention due to its little invasion and has increased prospects for cancer detection and continuous monitoring (6).
Each of the three techniques in liquid biopsy has its own strengths. CTCs detection monitors the trend of changes in the type and number of CTCs by capturing CTCs present in the peripheral blood in order to monitor tumor dynamics in real time, assess treatment efficacy, and enable real-time individual therapy. ctDNA is a fragment of DNA that is necrotic, apoptotic, or normally secreted by tumor cells into the bloodstream, and carries information about cancer-related genetic mutations, and can be used for early detection of cancer when the it can be used for early detection of cancer because it can be detected when mutations occur at the molecular level of the tumor. ctDNA is capable of fusion and remapping in different types of cancers (8). ctDNA has a number of clinical applications, for example, sequential ctDNA assays can be used to efficiently monitor patients and detect tiny residual lesions, which can help in the early detection of disease progression and the adjustment of the Adjuvant therapeutic regimens for ovarian cancer treatment (9). Plasma preoperative ctDNA testing in ovarian cancer patients is expected to serve as a biomarker for tumor staging and prognosis prediction (10). Early changes in circulating tumor DNA (ctDNA) predict treatment response in patients with metastatic KRAS-mutant colorectal cancer (mCRC) (11). ctDNA and radiated tumor volume identify patients with resected early-stage non-small-cell lung cancers who are at risk of recurrence (12). EVs have been linked to driving malignant cell behavior, including stimulating tumor cell growth, suppressing immune responses, inducing angiogenesis, facilitating tumor cell migration, and establishing metastasis, making them particularly attractive as cancer biomarkers (13). EVs contain cargoes of miRNAs,mRNAs, and proteins. It has been shown that microRNAs carried by EVs are associated with colorectal cancer (14), hepatocellular carcinoma (15), lung cancer (16), oral cancer (17), and ovarian cancer (18). The contents of EVs such as mRNA can promote epithelial-mesenchymal transition and chemoresistance in colorectal cancer (19). Therefore, not only CTCs can be used for tumor liquid biopsy, but also ctDNA,EVs can be used for cancer detection.
In this comprehensive analysis, we thoroughly examine the latest developments in the rapidly evolving domain of liquid biopsy. Our main objective is to explore the potential uses of liquid biopsy techniques in cancer clinical diagnosis, therapy monitoring, and prognosis prediction. These techniques encompass circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and extracellular vesicles (EVs). We also discussed the benefits and drawbacks of the most recent CTC, ctDNA, and EV detection, capture, and downstream analysis technologies. The application of comprehensive liquid biopsy combined with CTC, ctDNA, and EVs detection in cancer monitoring. Finally, we discussed liquid biopsy’s potential applications and difficulties in precision medicine. Figure 1 shows CTCs、ctDNA, and EVs detection techniques and their clinical applications.
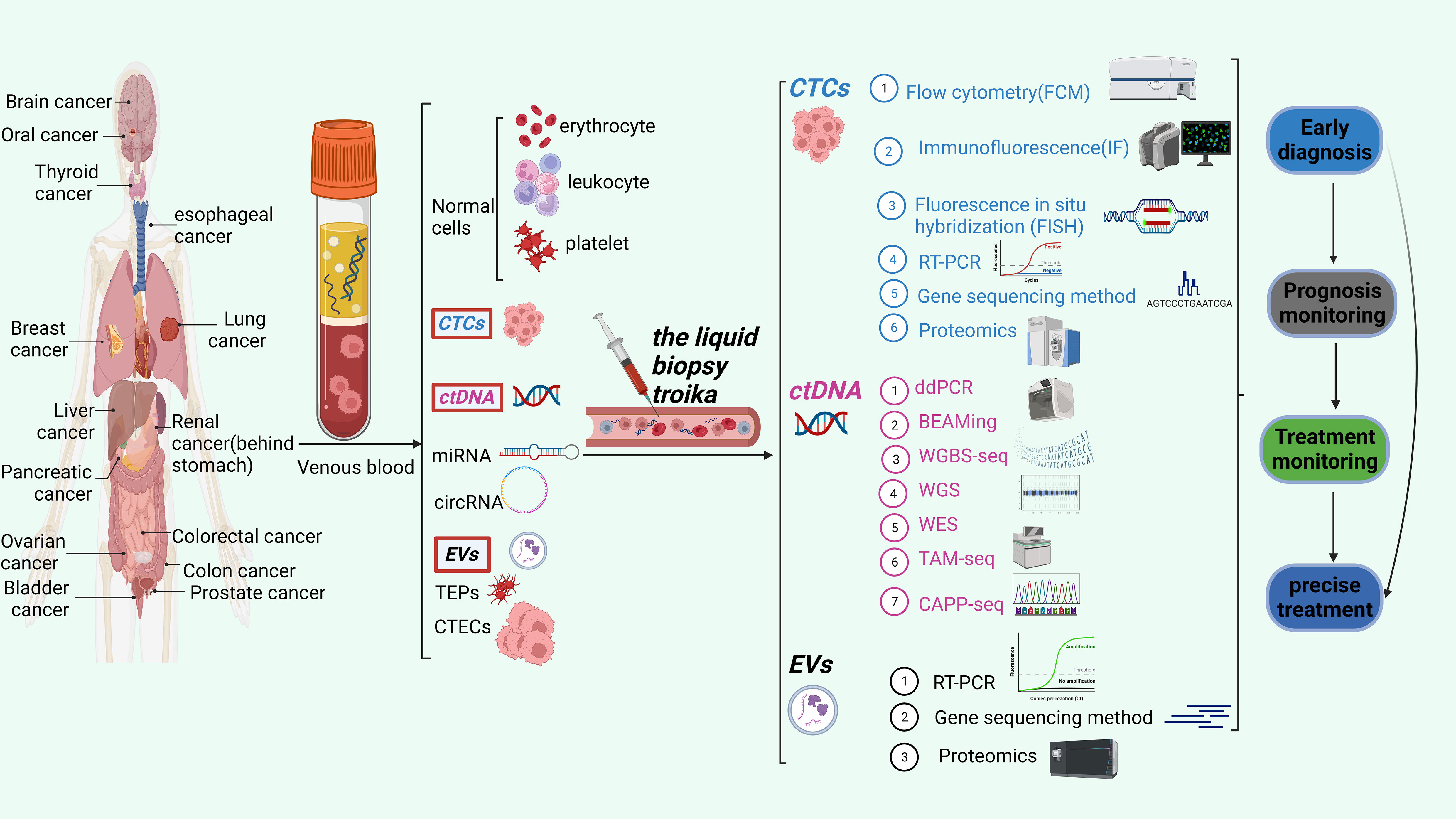
Figure 1 The liquid biopsy troika include CTCs, ctDNA and EVs. This figure shows the three detection techniques and their clinical applications.
2 Technologies for CTCs、ctDNA, and EV detection
2.1 CTCs
CTCs are tumor cells that, spontaneously or potentially resulting from surgical procedures, invade the peripheral blood circulation system after separating from the metastatic site or primary tumor. Circulating tumor cells (CTCs) have been detected in various malignancies, including breast, lung, ovarian, prostate, lung, and colon cancers. As a nearly non-invasive detection method, CTC can dynamically monitor the progression and changes of tumor conditions, has significant value for tumor diagnosis, treatment, and monitoring (20), and promises to advance precision medicine and help fundamental cancer research (21).
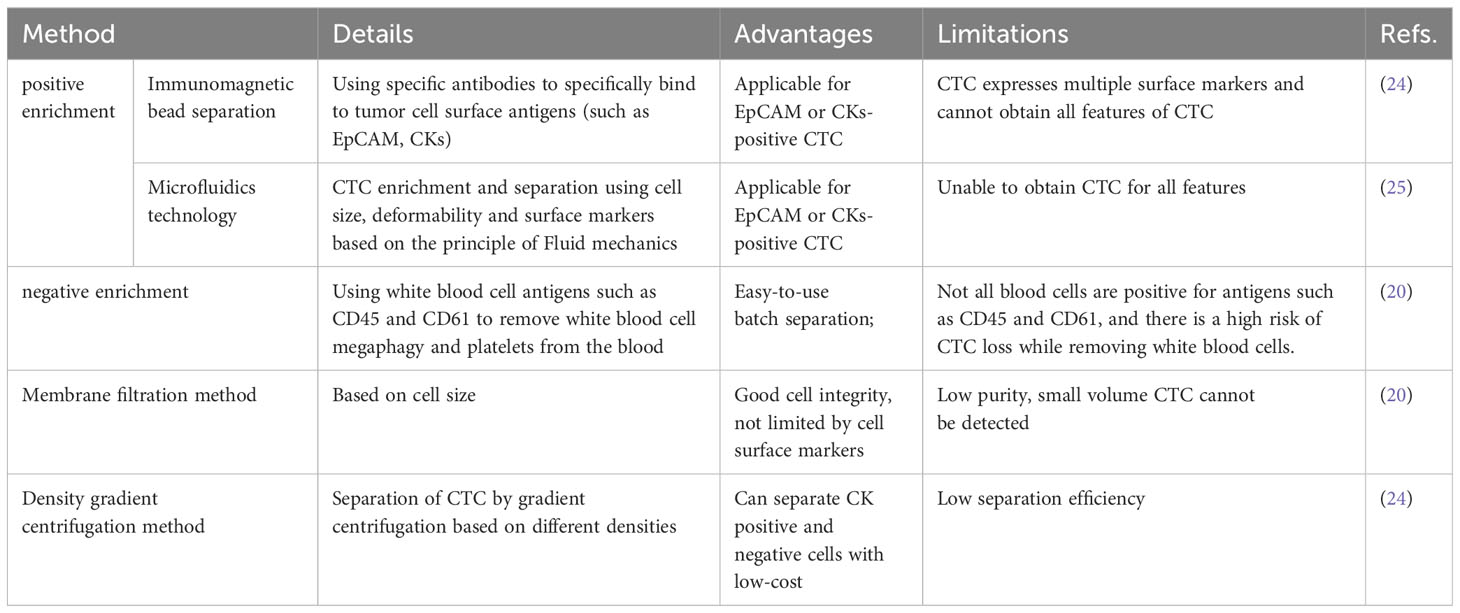
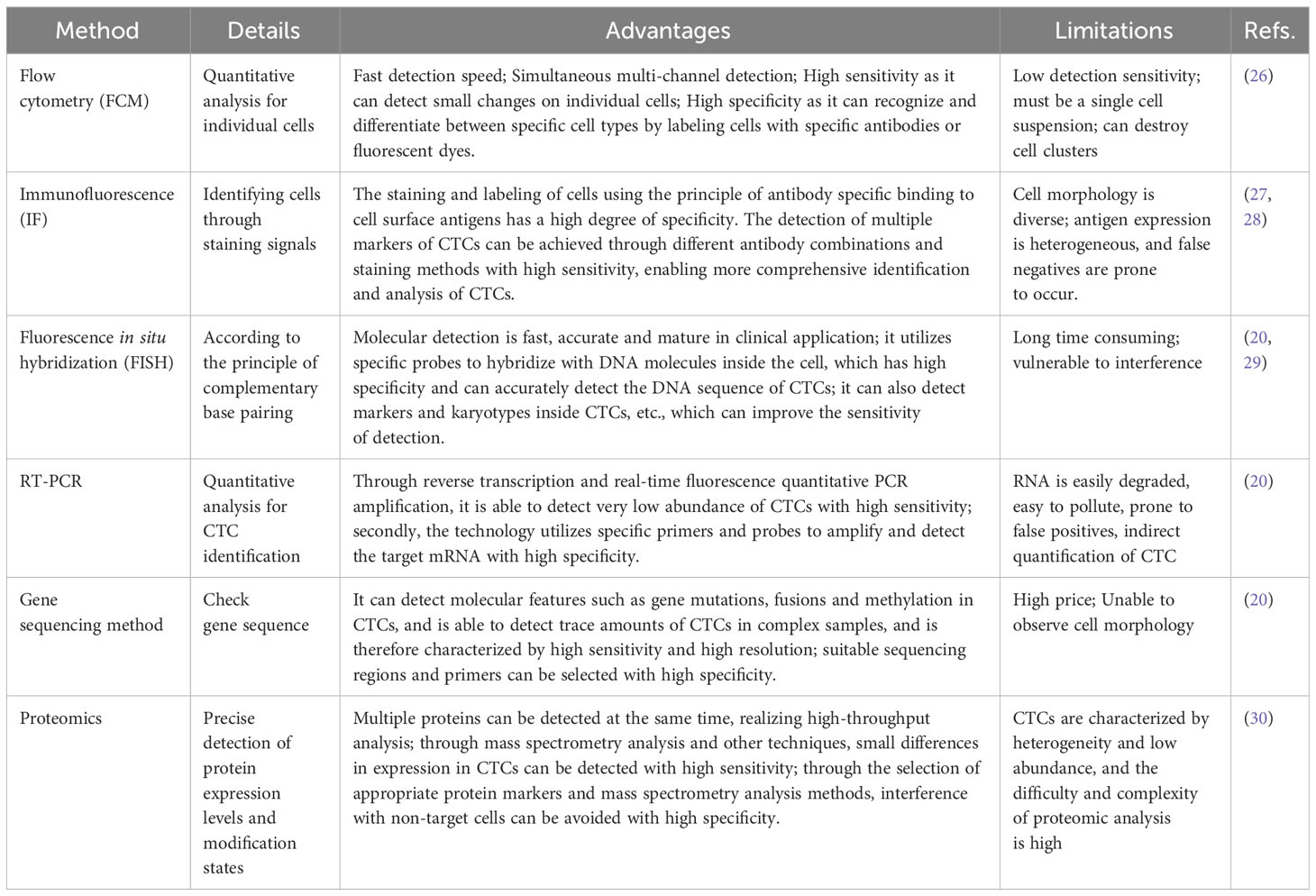
Cells are a more comprehensive biological entity and can provide dynamic information on RNA, proteins, DNA, and other biological molecules compared to ctDNA and EVs. This gives them an unmatched edge in assessments of transcriptomics, proteomics, and signal colocalization (22). Additionally, they can be cultivated in vitro to produce CTC lines for further study into therapy, the mechanisms underlying metastatic disease, and prevention (23). It has been found that CTC has differences in various aspects, such as cell size and morphology, molecular phenotype, activity level, metastasis potential, and proliferation potential. CTC detection has gradually moved from earlier cell counting to complete analysis of cell counting mixed with molecular typing due to the advancement of CTC research and increased clinical demand. CTC is extremely rare, with approximately one billion blood cells possibly being CTC. This poses significant technical challenges to isolate CTCs (23) effectively. Two counting processes comprise most CTC detection processes: CTC enrichment and separation and CTC identification. The term “CTC enrichment and separation technology” describes the process of concentrating and separating CTC from a large number of blood cells in peripheral blood based on one or more distinctive properties of CTC. CTCs were separated through biomarker-driven cell capture, employing various enrichment techniques rooted in biophysical and other characteristics, including immunogenicity, positive and negative enrichment, as well as enrichment strategies relying on size, density, dielectric properties, and deformability (14) (Table 1). CTC identification methods mainly include cell morphology identification, immunofluorescence staining, Fluorescence in situ hybridization, PCR etc (Table 2). CellSearch® is the first FDA-approved CTC separation technique that uses antibody-labeled magnetic nanoparticles to select cells that express EpCAM and fluorescence microscopy to identify keratin and DAPI positive CD45 negative cells (31). The isolation methods of CTCs are shown in Table 1.
After enriching and separating CTC in the blood, effective methods must be combined for analysis and identification. The detection of CTC has evolved from counting to analysis and type, single-cell sequencing, and cell function analysis due to the ongoing development of CTC enrichment, detection, and analysis methods. By directly examining the mechanisms of drug resistance, migration, and recurrence in each CTC, single-cell genomics and transcriptome analysis can aid in early cancer detection and provide better recommendations for early cancer diagnosis. It can also help to personalize treatment. These benefits include discovering the dynamic changes of tumors, analyzing tumor growth rules, and more (32). Table 2 lists CTC identification techniques.
2.2 ctDNA
Apoptosis and necrosis of human cells will release fragmented DNA into body fluid, called cfDNA, which includes the DNA of tumor cells and the DNA of normal cells; the latter is called ctDNA. Cancer patients’ cell-free DNA has been found to include ctDNA, allowing for the analysis of the genetic abnormalities that characterize each patient’s cancer (22). Over the past 10 years, research has shown considerable promise as a possible clinical tool to promote personalized therapy. Most ctDNA fragments are double-stranded and have lengths between 160 and 200 base pairs, or roughly the same as a nucleosome (33). Numerous aspects of the tumor, including its location, metastasis, size, tumor status, vascular infiltration and staging, impact the amount of ctDNA that enters the bloodstream. As a result, there is a wide variation in the proportion of ctDNA (32). Free DNA released by blood cells and another somatic cell may produce an interference phenomenon that must be effectively distinguished. Therefore, distinguishing ctDNA from normal cfDNA is essential to reflect the disease status (34, 35) accurately.
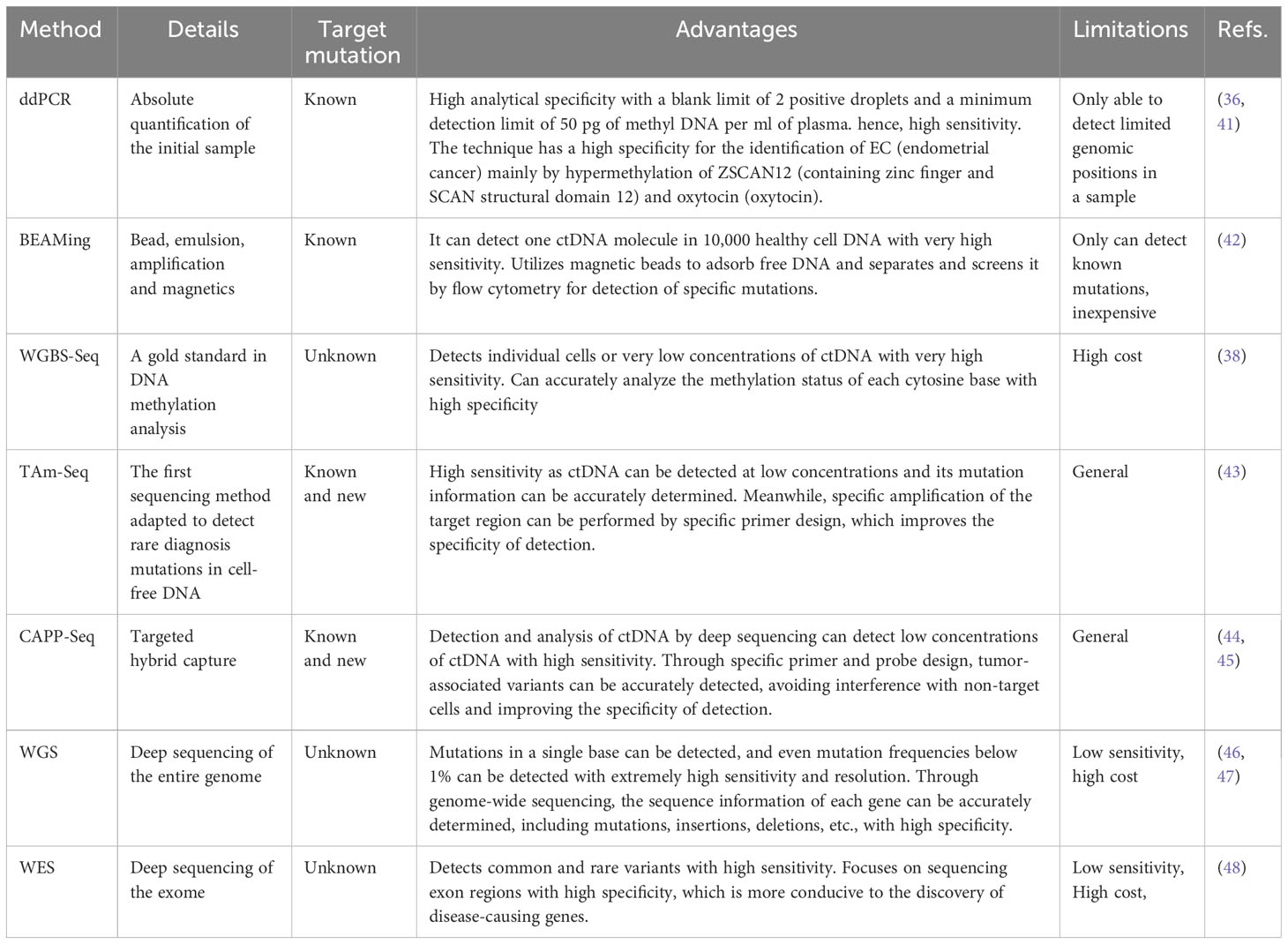
ctDNA can provide tumor-specific genetic variation (Point mutation, insertion and deletion, rearrangement, fusion, etc.) and epigenetics (methylation, etc.) and copy number variation information; its application value in the whole process management of tumor patients has been recognized and has been written into several clinical diagnosis and treatment guidelines. The main obstacles facing ctDNA detection technology are increasing the sensitivity of detection technology and suppressing background noise signals because ctDNA has been isolated and analyzed using a variety of techniques, including Tagged-amplicon deep sequencing (TAm-Seq), cancer personalized profiling by deep sequencing (CAPP-Seq), whole genome bisulfite sequencing (WGBS-Seq), and droplet digital polymerase chain reaction (ddPCR) sequencing technologies.
Tumor Circulating Among the new liquid biopsy technologies, DNA analysis is the most advanced, although meaningful comparisons between studies are made difficult by the range of ctDNA assays. In addition to being beneficial for calculating copy number variants and discovering possibly rare mutations, ddPCR is only useful for screening for known mutations (36). DdPCR (32) is one of the most useful and applicable methods today for analyzing genomic mutations. BEAMing, like ddPCR, can only test for known mutations. The BEAMing screening method is reasonably sensitive and affordable (36). In addition, studies have shown inconsistent clinical diagnostic testing between BEAMing and ddPCR, but to a low extent (37). The most accurate and precise DNA methylation analysis method is WGBS-Seq, which provides resolution for a single cytosine measurement. Increasing the scale of this method for large population studies, on the other hand, is prohibitively expensive and poses major bioinformatic challenges (38). Although the approach may detect partly methylated regions in cancer cells, it has several practical limitations. The detection approach may show a reduced sensitivity during detection, for example, if DNA is present but has undergone various degrees of degradation (27, 28). Its capacity for high sequencing throughput distinguishes tag-amplicon deep sequencing (TAm-Seq), decreased sequencing duration and expenses, and the capability to concurrently sequence millions of DNA molecules. All primary mutation types, involving single nucleotide variations, deletions, rearrangements, insertions, and copy number alterations can be found using cancer-personalized profiling by deep sequencing (CAPP-Seq). It’s crucial to bear in mind that CAPP-Seq has limitations regarding the types of mutations it can discover (16, 25). Whole genome sequencing (WGS) is a comprehensive approach that examines a tumor’s complete genomic content, which can greatly aid therapeutic decision-making and reveal novel therapy plans, unusual mutations, and invisible oncogenes. However, WGS is constrained by difficulties with quality control, ethics, cost, and timing (39). To identify frequent or rare disease-related abnormalities, whole exome sequencing (WES) characterizes and analyzes all currently reported tumor mutations. However, it might have a lower sensitivity than other methods (36, 40). Table 3 lists the benefits and drawbacks of the various detection strategies.
2.3 EVs
EVs are membrane vesicles containing lipid bilayer secreted by cells into the extracellular space (30–2,000 nm), which nearly all cells actively secrete (6). Extracellular vesicles (EVs) can be divided into three classes according to the International Society for Extracellular Vesicles’ (MISEV’s) recommendations, depending on their size: microvesicles, apoptotic bodies, and tiny extracellular vesicles. The classification of these EVs is determined by their respective dimensions (49). It is reported that EVs derived from cancer cells are important in changing the tumor microenvironment and promoting tumor progression (50). They also carry distinctive molecules from parental cells, including proteins, miRNAs, mRNAs, lncRNAs, DNA, and lipids, indicating the state of the disease at the time. Surface markers mostly include CD63, CD81, CD9, TSG101, and HSP27 (51). Multiple studies have shown that EVs may be reliably found in a variety of bodily fluids, including blood (44), saliva (45), urine (46), bronchoalveolar fluid (47), breast milk (48), and semen (49). As a result, EV cargo targeting enables us to evaluate essential molecular data concerning illness status (50).
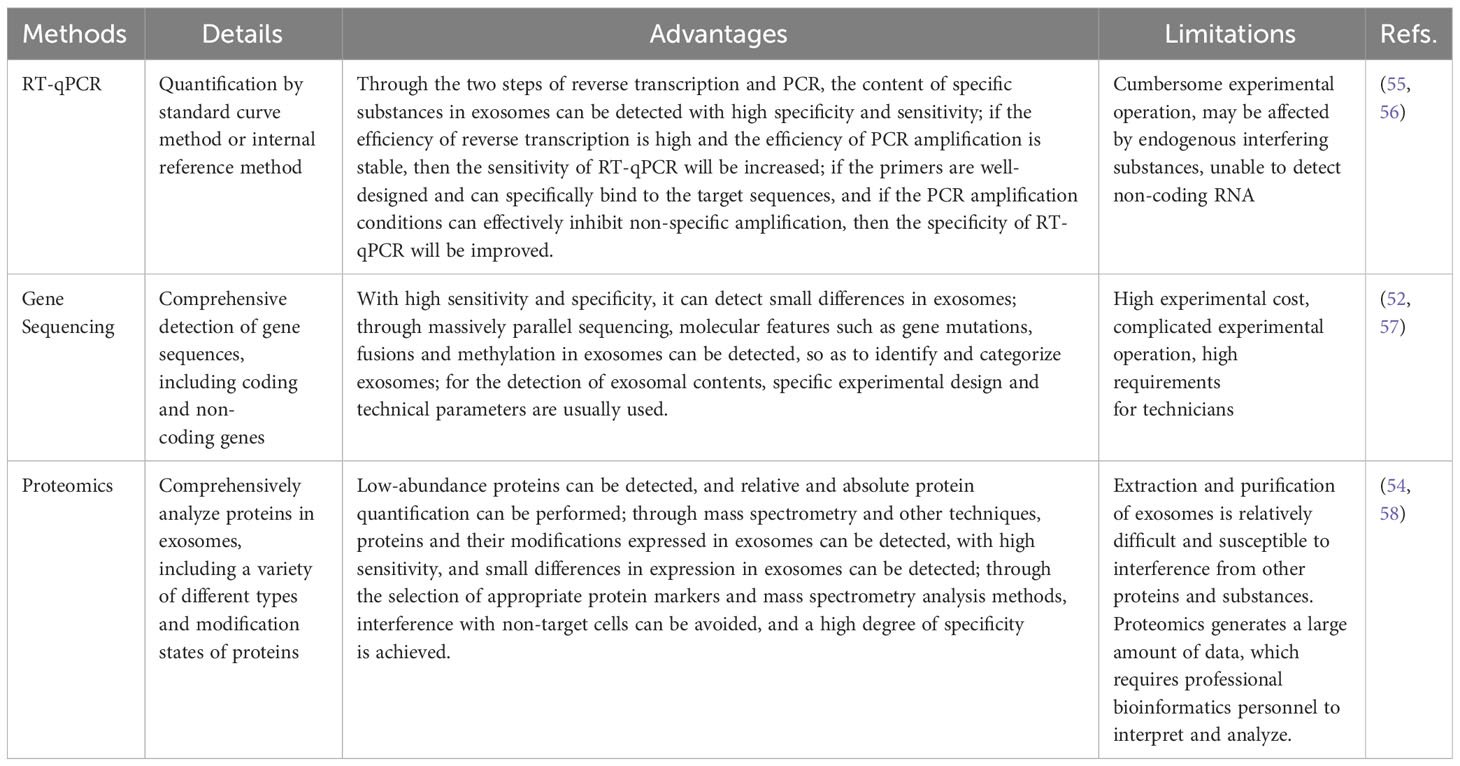
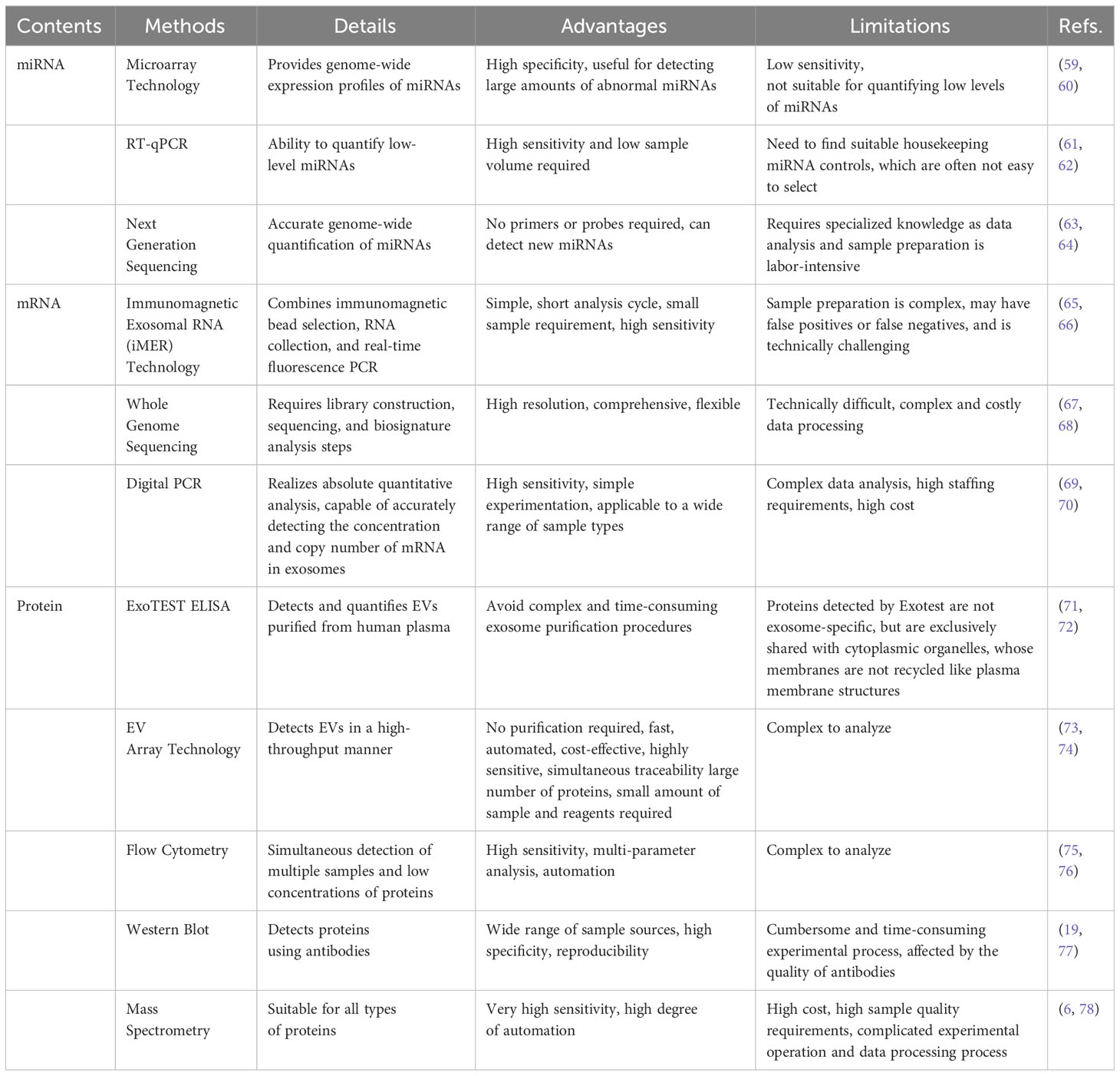
In recent years, there have been numerous methods for the detection of exosome contents, such as RT-PCR (52),genome sequencing (53),and proteomics (54). Table 4 lists the detections of exosomal contents. Table 5 lists the comparison of miRNA,mRNA and protein detection methods in EVs.
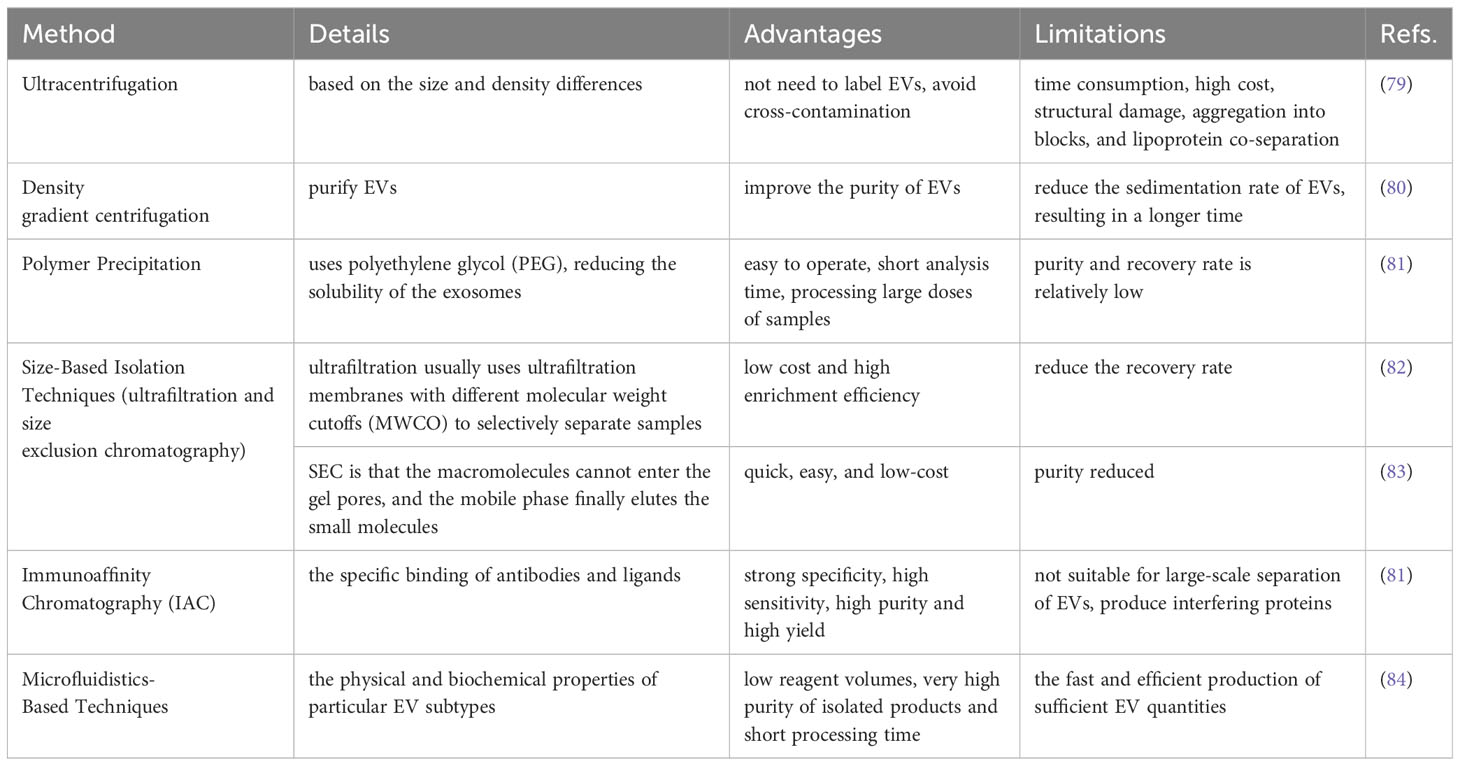
EVs are an important research direction in liquid biopsy; the value of its potential applications has been continually tapped. Repeatable EV separation and enrichment will enable the evaluation of their biological functions. However, the key obstacles in this sector are the separation and purification of EVs, which are heterogeneous in function, size, content, and source (6). An important step in experimental research is effective extraction. Size exclusion chromatography (SEC), polymerization, the process of precipitation immunoaffinity chromatography, and microfluidics-based methods are the primary methods used to isolate and refine extracellular vesicles (EVs) (51, 52). For different purposes and objectives, different separation methods are utilized. However, polymer precipitation, size-based isolation, immunoaffinity capture and ultracentrifugation are routinely used. Every approach and strategy is employed based on EVs’ size and origins. Each has advantages and disadvantages. The advantages and disadvantages of the various separation methods are listed in Table 6.
3 Importtant areas of clinical applications of CTCs、ctDNA and EVs
CTCs, ctDNA, and extracellular vesicles (EVs) are now used in clinical settings as substitute biomarkers for various solid tumors, and numerous research have been conducted, indicating enormous potential in therapeutic applications.
3.1 Clinical applications of CTCs
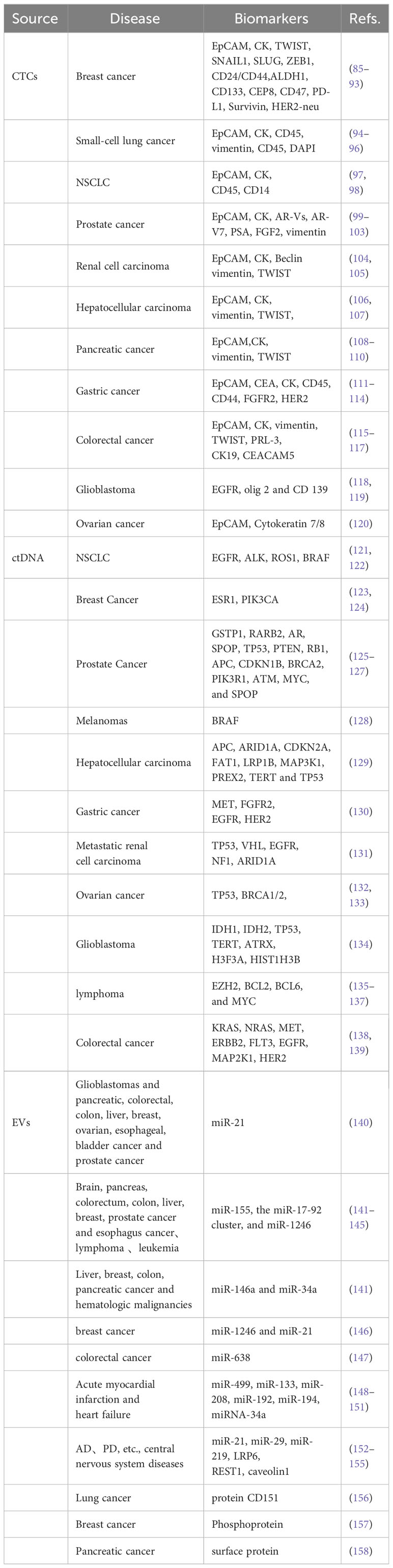
The increased demand for CTCs as precision oncology biomarkers. CTC detection is an appealing, non-invasive method for cancer diagnosis. CTC detection has important value in early diagnosis, efficacy determination, prognosis evaluation, and treatment monitoring of tumors. CTC analysis makes real-time monitoring, further analysis, and identification of protein, RNA, and DNA molecules possible by effectiveness, non-invasiveness, and high repeatability (58, 59). Many clinical studies have evaluated the potential of CTCs to use blood samples from known cancer diagnosis patients for cancer testing (Table 7).
3.1.1 Early diagnosis
Studies have shown that CTCs can diagnose or detect clinically related cancers early. CTC, for example, has been identified in individuals with early stages (stage I-IIIA) cancer of the breast (60, 61); more than one form of CTC was found in 20% of stage I disease patients, 26.8% of stage II disease patients, and 26.7% of stage III disease patients (62). CTC has been detected using cell search technology in patients with non-metastatic colorectal cancer (including stage I and II) and non-metastatic prostate cancer (159, 160). According to Barriere et al. (65), 41% of T1-stage breast cancer patients and 47% of axillary lymph node-negative patients had CTC. A study conducted by Thery et al. (66) found that the incidence of positive circulating tumor cells (CTCs) was 21% in lymph node-negative breast cancer and 24% in instances of lymph node-positive breast cancer cases. Using the EpCAM-based NanoVelcro CTC chip, CTC can be detected in 60% of patients with stage II diseases, and positive CTC distinguishes pancreatic ductal adenocarcinoma patients from non-adenocarcinoma pancreatic disease patients (161). A meta-analysis comprising 18 prospective studies has demonstrated that a positive CTC (circulating tumor cell) result serves as a valuable biomarker for forecasting adverse overall survival outcomes in individuals diagnosed with early-stage non-small cell lung cancer (NSCLC) (68). In another study, 98 suspected prostate cancer patients were predicted to undergo biopsy diagnosis before biopsy, and Clinically significant cancer was highly associated with CTC detection utilizing the Parsortix isolation technique (99).
Collectively, these findings demonstrate that CTCs can be identified ahead of primary tumors in imaging investigations, highlighting the fact that the greatest obstacle facing the use of CTCs in early cancer diagnosis is, in fact, their paucity and isolation. The PROLIPSY examination, as exemplified by the study identifier NCT04556916, along with analogous examinations related to breast cancer (NCT03511859), non-small cell lung cancer (NCT02380196), colorectal cancer (CRC; NCT05127096), and pancreatic cancer (PANCAID) (70), represent a subset of the numerous clinical trials exploring the application of CTCs (circulating tumor cells) in the early detection of various cancers.
3.1.2 Prognosis and treatment monitoring
CTCs have been examined extensively for their prognostic usefulness and are now recognized as an independent prognostic factor. CTCs have been found to have prognostic significance in various malignancies, including breast, prostate, colorectal, small cell, and non-small cell lung cancers (71–74), and their discovery in the bloodstream has been connected to various diseases. In addition to the only clinical CTC testing system approved by the FDA, CellSearch, there are other CTC testing systems, such as CanPatol and CTC chips (162).
The examination’s primary focus is the CTC count, with a positive threshold of ≥ 5, typically representing a poor prognosis. The persistence of CTC following therapy is associated with a poorer prognosis, and studies have demonstrated that changes in the quantity of CTC provide better predictive information than baseline CTC status (76, 77). The number of CTCs present before beginning neoadjuvant therapy hurts survival, according to a meta-analysis of 2239 breast cancer patients from 21 trials (246) (60). Furthermore, patient CTC analysis can aid in predicting low residual disease and late disease recurrence because CTCs can be seen 7-9 weeks before the disease’s clinical manifestation (163) and provide tools for early cancer detection (164, 165). A prognostic factor for a lower survival rate is an increased CTC count, or a failure to eliminate CTC during treatment (80, 81), and reduction or clearance of CTC count is associated with good treatment response (166). In addition, evaluating the abundance of CTC clusters can significantly improve the prognostic value of patients receiving treatment (167). CTC clusters indicate a bad prognosis and elevated baseline CTC levels are linked to decreased survival rates (168). Studies have shown that the molecular phenotype of CTC, including HER2 (159), CD47, and PD-L1, has significant prognostic value (85). CTC has been utilized as a helpful biomarker for assessing the effectiveness of cancer treatment in numerous clinical trials (99, 161), assisting clinical doctors in personalized treatment and drug resistance selection during tumor progression. CTCs can be used to track the progress of metastatic disease in patients with breast and colorectal cancer and prognosis, and a current NIH-sponsored clinical trial (NCT02973204) is looking into the utility of CTCs as clinical support tools in hepatocellular carcinoma (169).
Currently, many ongoing clinical trials also are studying the application of CTC in cancer prognosis and treatment monitoring, including breast cancer (NCT00382018、NCT02101385、NCT00601900、NCT01745757、NCT01701050、NCT00785291), prostate cancer (NCT01942837), pancreatic cancer (NCT01919151), colorectal cancer(NCT01442935、NCT01640405、NCT01640444) (162).
3.2 Clinical applications of ctDNA
With advantages such as being non-invasive, sensitive, comprehensive, and dynamic, ctDNA has shown great clinical value in tumor diagnosis and treatment. Although tissue biopsy is still the preferred method for diagnosis, non-invasive, real-time liquid biopsy has shown an increasingly important role and has been supported by guidelines in advanced lung cancer, breast cancer and other tumors. Currently, ctDNA testing has shown clinical value in initial molecular typing, prognostic staging of early tumors, predicting treatment responses, detecting MRD, guiding treatment, and discovering drug resistance mechanisms (170–173).
3.2.1 Early diagnosis
ctDNA detection has also shown great promise in the early identification of cancer, opening up new avenues for developing highly targeted therapeutic therapies for cancer patients (174). Future clinical trials may further explore predictive biomarkers found using ctDNA analysis (168). The ctDNA of cancer patients can be examined for genetic changes such as mutations, loss of heterozygosity, microsatellite instability, methylation, and copy number variations (CNVs) (85, 169). These molecular hallmarks of tumors serve as useful biomarkers for cancer diagnosis, staging, and treatment. Cancers of the breast, colon, pancreas, and esophagus were all found to have increased ctDNA levels in their advanced stages compared to their preliminary stages, and this was true regardless of tumor-specific molecular characteristics (175).
3.2.2 Prognosis and treatment monitoring
CtDNA collection from blood is non-invasive and repeatable over time compared to conventional tumor biopsy. It can monitor treatment response and predict real-time prognoses by assisting in early cancer diagnosis and recognizing small residual diseases or recurrence rates (176). Analyzing ctDNA from patients with metastatic colorectal cancer allows for monitoring of the disease’s temporal heterogeneity and individualized treatment, as more and more studies are showing (172, 174). Wyatt et al. found numerous genetic abnormalities, such as amplifications, mutations, and gene inactivation, in Prostate Cancer ctDNA by comparing it to matched tissue, which may be further examined in these patients. In order to prognostically and predictively stratify individuals based on their DNA, ctDNA assays may be used (175). In a prospective study of 69 patients with advanced NSCLC, those with high ctDNA levels fared significantly worse overall than those with low ctDNA levels and progression-free survival. Higher ctDNA than baseline indicates a poor prognosis (177). In various large patient cohorts, including prospective screening cohorts in high-risk groups for colorectal cancer, Luo and associates examined the methylation patterns on ctDNA. Methylation-based diagnostic scores were found and validated to distinguish colorectal cancer patients from healthy control groups in addition to prognosis ratings linked to patient survival (177).
Additionally, tumor cells from various sources may have different methylation profiles, enabling ctDNA analysis to reveal location information (129). Cell-free DNA (ctDNA) analysis has the potential to provide transcriptional information that could aid in the early diagnosis of cancers like prostate and colorectal by inferring transcription factor binding. Furthermore, ctDNA can predict tumor immune infiltration and response to systemic treatment following tumor recurrence after radical hepatectomy and help solve monitoring decisions and post-recurrence treatments (129), and tumor immune infiltration and responsiveness to systemic treatment after tumor recurrence after ctDNA can predict radical hepatectomy.
Currently, many ongoing clinical trials also are studying the application of ctDNA in cancer prognosis and treatment monitoring regarding colorectal cancer, such as TRACC (NCT04050345) and ADNCirc (NCT02813928), IMPROVE-IT2 (NCT04084249), NCI–sponsored randomized phase II/III COBRA study (NCT0406810), CIRCULATE trial (NCT04120701) and the DYNAMIC-II study (ACTRN12615000381583), the phase II/III DYNAMIC-III study (ACTRN12617001566325), and the phase II/III PROSPECT trial (NCT01515787) and in the OPRA trial (NCT02008656), CHRONOS (NCT03227926) and FIRE-4 (NCT02934529) (178). And Gastric Cancer clinical trials (NCT04947995、NCT04665687、NCT04511559、NCT05027347、NCT05029869、NCT04943406、NCT04510285、NCT03957564、NCT04817826、NCT04520295、NCT04576858) (179) and so on.
3.3 Clinical Applications of EVs
Exosome biology in disease is a relatively new area of research. EVs provide a rich source of biomarkers for the diagnosis and prognosis of disease. Tumor EVs are largely used in cancer because identifying cancer-predictive biomarkers in them can assist in increasing the early tumor diagnosis specificity and sensitivity. They have also made some headway in treating conditions affecting the heart, lungs, and central nervous system (53), and their application is increasingly being used to treat conditions affecting the liver (180) (180), kidney (181), and lung (181), among others. Due to their exceptional characteristics for transporting functional cargoes to diseased cells, exosomes can be therapeutic carriers at both the basic and clinical levels.
3.3.1 Diagnostic potential of EVs
EVs are desirable as minimally invasive liquid biopsies because they are present in all biological fluids, are substantially concentrated in biofluids, and are secreted by all cells. According to some research, there is a small amount of DNA in EVs; this can be used to detect mutations in serum EVs associated with cancer (180, 182). EVs may include particular miRNAs or miRNA sets with diagnostic or prognostic value for cancer identification. Since EVs include miRNAs produced differently in cancer cells than normal cells, they may have a high diagnostic value and aid in early detection. Bladder and prostate cancer have been associated with increased levels of exosomal miR-21 in the urine, whereas glioblastomas, colorectal, colon, liver, breast, ovarian, and esophageal cancer have all been connected to increased levels of circulating exocrine miR-21. Tumor-suppressing miRNAs like miR-1 4 6a and miR-3 4a are associated with hematological, liver, breast, colon, and pancreatic cancer (183, 184). Colorectal cancer can be diagnosed with miR-638 (185), while exosomes generated from breast cancer cells are greatly enriched in miR-1246 and miR-21 (186). Furthermore, those experiencing acute myocardial infarction and heart failure have shown increased secretory microRNAs linked to cardiovascular ailments. MicroRNAs such as miR-499, miR-133, miR-208, miR-192, miR-194, and miR-34a (86–88, 187) belong to this group. There is great potential for the clinical identification of disorders of the central nervous system based on the unique expression patterns of miR-21, miR-29, miR-219, LRP6, REST1, and caveolin1 in exosomes (89–92). Combining different microRNAs may increase EV miR characteristics’ diagnostic and prognostic value, which are constantly associated with cancer diagnosis and prognosis (93, 94). Breast cancer patients’ circulating exosomes have been studied for their surface proteins (96), and their phosphoproteins have been suggested to have diagnostic potential (95). Also, lung cancer patients have significant EV protein CD151 expression (97).
3.3.2 Therapeutic potential of EVs
Currently actively exploring EVs or as carriers for drug payloads as therapeutic agents (6). Compared to liposomes, injected exosomes can efficiently enter other cells and deliver functional cargo with minimal immune clearance when administered exogenously in mice (181, 188, 189). Tumor stroma-derived exosomes have also been associated with cancer chemotherapy resistance (183, 190), and thus targeting specific functions of exosomes can enhance response to therapy. EVs have the potential to serve as a targeted medication carrier and benefit from the advantages of natural drug delivery due to their capacity to cross biological barriers like the blood-brain barrier. Chinese herbal medicine Western medicine uses paclitaxel (PTX) (98), curcumin (100), berry anthocyanins (101), -element (102) triptolide (103), and compound Buyanghuanwu Decoction (104). Catalase (CAT) (106), doxorubicin (105), and others (53). Additionally, EVs can employ gene therapy techniques and transport gene therapy materials like DNA and RNA. For instance, oligonucleotides can mute particular genes to treat various human diseases, such as cancer or neurological disorders (107, 108). MicroRNAs (miRNAs) have been found to efficiently contribute to the degradation of target mRNA and the suppression of gene expression in receptor cells; as a result, EVs have been developed for CNS disorders and tumors to transport miRNA or small interfering RNA (siRNA) payloads. For example, preclinical tests using EVs to deliver miRNA or siRNA payloads focused on anti-cancer treatment and exploratory brain targeting in rodents with breast cancer (184), glioma (185)and pancreatic cancer (186, 187). Additionally, ligand enrichment on modified EVs can direct EVs to particular cell types or trigger or inhibit signal transduction events in receptor cells (113, 114).
4 Comprehensive liquid biopsy
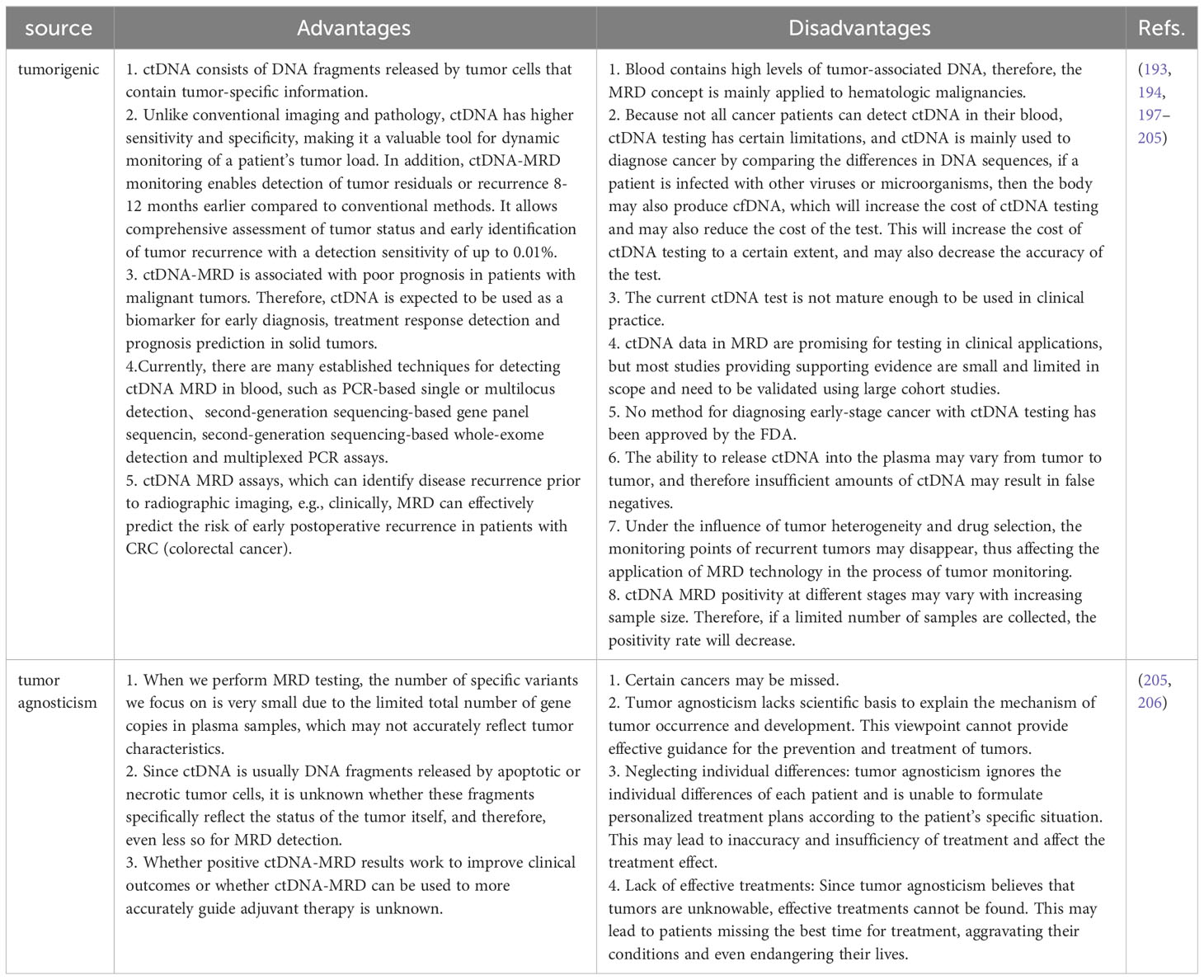
With the advent of precision medicine, researchers are looking into liquid biopsy to monitor tumor growth in real time and direct treatment accordingly. Molecular residual disease (MRD), also called a minimum residual disease or, in some circumstances, quantifiable residual disease, refers to solid tumors that, following therapy, cannot be detected by conventional imaging or other laboratory techniques. However, detecting tumor molecular abnormalities through liquid biopsy methods such as ctDNA and CTC indicates the persistence and clinical progression of the tumor (191). According to studies, liquid biopsies can detect metastatic diseases at least 4 years before they are clinically detected, indicating that thorough liquid biopsy evaluation of CTC combined with ctDNA monitoring MRD can provide very important information for treating and treating patients with breast cancer (192). Clinical studies have also shown the combination of CTC and ctDNA to improve the sensitivity and specificity of MRD detection in hepatic carcinoma (193). A new study examined the prognostic significance of combining CTC and ctDNA analyses. The results showed that CTC and total cfDNA levels were separately and together related to PFS and OS in MBC (194). In addition, combining ctDNA analysis based on Guardant360 NGS with CTC counting in MBC can help determine the site of metastasis (195). In a recent pilot study, 16 patients with metastatic urothelial cancer were compared using matched CTC and ctDNA samples, demonstrating that CTC and ctDNA offered complementing information (196). Therefore, CTC and ctDNA can be jointly tested for clinical tumor MRD monitoring services. There are two techniques in the field of ctDNA MRD, tumor native and tumor agnostic. Table 8 shows the advantages and disadvantages of these two techniques.
In recent years, it has been discovered that EVs miRNAs can act as vital information intermediaries for cancer cells and other cells, having a significant impact on the pathological development of tumors (205) and being potentially appropriate from a material aspect for different medication delivery and therapeutic uses (206). EVs can assess drug resistance, diagnose early cancers, and provide a prognosis. The key to understanding cancer may be found in the miRNAs found in EVs. A potential novel approach to treating lung cancer may involve preventing the production and release of EVs. EVs can be employed for additional study of ctDNA and miRNA discovered in The stability of nucleic acid molecules is successfully maintained by EVs, protecting ctDNA from degradation (17). ExoDX Lung (ALK), the first cancer diagnosis product ever released by EVs Diagnostics, was introduced in 2016. Real-time screening for EML4-ALK mutations in non-small cell lung cancer patients is made possible by the device’s EV detection technique, which can concurrently detect EVs RNA and ctDNA. According to the information supplied by EVs diagnostic (53), ExoDx lung (ALK), which can be utilized to assist physicians in determining if patients are amenable to targeted therapy with ALK inhibitors, has 88% sensitivity and 100% specificity in identifying non-small cell lung cancer. This is particularly true for those who cannot or do not want to perform a tissue biopsy.
Long-term high cancer mortality rates have motivated domestic and international research teams to create effective cancer therapy strategies. Liquid biopsy aims to collect biomarkers from biological tissues that are not solid in order to assess the disease status. It can sample and analyze different biogenic substances such as CTCs, nucleic acids, proteins or EVs in bodily fluids such as blood or urine (207). Currently, researchers are considering combining CTCs, ctDNA, RNA, miRNA, protein, and lipid EV cargos in diagnosing and prognosticating cancer and other diseases. A thorough understanding of the whole course of disease for each cancer patient might be provided if these technologies are combined to boost the positive and negative predictive value. Table 9 lists the applications of CTCs, ctDNA and EVs in the diagnosis, prognosis and therapeutic monitoring of different diseases.
5 Conclusions and perspectives
Precision therapy is individualized pharmacological treatment based on specific characteristics of tumors (17). Tumor heterogeneity, variations in gene expression, and polymorphisms may all affect a patient’s response to medication differently. Precision medicine and molecular diagnosis have developed quickly due to current diagnostic technology (208). Table 10 looks at the prospects of CTCs, ctDNA and EVs in tumor diagnostics in terms of cost, operability, simplicity and industrialization. Despite potential and challenges, liquid biopsies are at the forefront of this revolutionary approach, and the future of precision oncology is bright. It is also a relatively recent method to assist clinical decision-making for preventing, identifying, and treating human cancer.
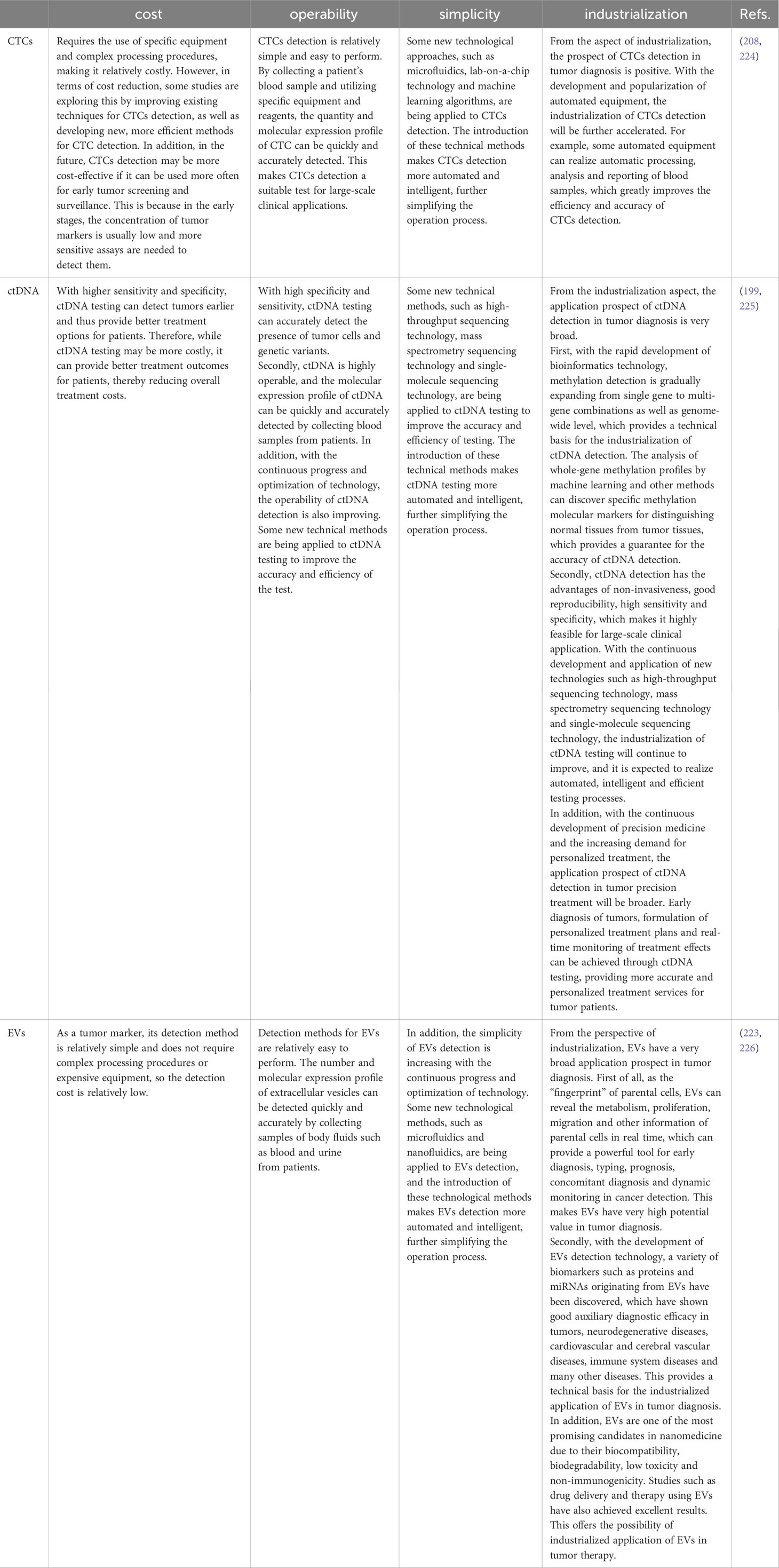
Table 10 Outlook of CTCs, ctDNA and EVs in tumor diagnostics in terms of cost, operability, simplicity and industrialization.
Although CTC analysis has several clinical limitations, it has the potential to deliver more thorough and accurate information on tumors. Individual differences and the extremely low CTC content in peripheral blood necessitate further research into effective CTC enrichment approaches to increase the reliability of CTC detection (23). Secondly, it is not possible to determine the appropriate CTC detection time. The morphological, genetic, and functional properties of CTCs differ due to various timeframes and treatment modalities, which impacts their therapeutic application (17). Therefore, the monitoring method for CTC is significantly challenged by its rarity and heterogeneity, which limits its clinical application to a certain extent. In addition, there are unanswered questions in CTC biology, unresolved concerns regarding the application of CTC biology in clinical settings (227) and so on. The advantage of ctDNA is that it is widely and uniformly distributed in body fluids, and the standardization of detection techniques is relatively easy. However, like CTCs, tumor patients’ ctDNA content is extremely low, making missed detection possible and necessitating highly sensitive detection technology (17). After DNA amplification, it is now possible to directly, qualitatively and quantitatively examine ctDNA using well-established detection techniques such as NGS and digital PCR owing to the rapid improvement of DNA sequencing technology. The therapeutic application value of ctDNA has to be further validated through many pertinent clinical trials and standardize the detection process (209). EVs have generated much attention in liquid biopsy of cancer as a possible biomarker for cancer detection. However, their clinical limits are also impacted by the number of EVs, separation tools, and purity (210). Even though researchers from both home and abroad have made some headway in exploring different methods for separation and purification, EV research and application are still constrained by cost and efficiency; as a result, developing a trustworthy EV extraction method will be advantageous for the clinical application of EVs (211).
In conclusion, a successful combination of these technologies may be useful for tumor research, notably for in-depth analysis and possible therapeutic applications. This is true despite the advantages and disadvantages of each technology. Blood is one of the windows for studying health and diseases. If the information in organs and blood is transformed into biological data and linked to the health trajectory of the human body, it will greatly benefit human health. Leroy E. Hood’s team found metastatic biomarkers of breast cancer, pancreatic cancer, lung cancer and hematopoietic system malignancies from the proteomic changes of plasma samples of health project participants, which advanced the diagnosis of metastatic cancer by more than 10 months (228). He has always been a proposer and practitioner of 4P medicine, distinguished by participative, personalized preventative, and predictive medicine. The core premise of 4P medicine is to identify early markers of disease transformation (229). Many of these biomarker techniques, including liquid biopsy as represented by CTCs (214), are now being researched、ctDNA (5) and EVs (65). Tumor formation and incidence are complex processes, and the underlying laws are not fully understood. One significant example is the early detection of cancers, which allows localized treatment choices to eradicate primary cancer and improves survival while lowering the likelihood of recurrence. Whether it is tissue biopsy or liquid biopsy represented by CTC, ctDNA, and EVs, it is a way to gain a deeper knowledge and understanding of tumors. Suppose these methods are integrated according to the concept of integrative medicine, and comprehensive detection and integrated analysis are carried out as far as possible when conditions permit the development of a more targeted treatment plan. In that case, it may help patients benefit from the survival time and quality of life.
Author contributions
XW: Writing – original draft. LW: Data curation, Writing – original draft. HL: Writing – review & editing. YZ: Writing – review & editing. DH: Data curation, Writing – review & editing. ML: Investigation, Writing – review & editing. XX: Formal analysis, Writing – review & editing. JH: Formal analysis, Writing – review & editing. WZ: Funding acquisition, Writing – review & editing. TZ: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Plan of Jiangxi Provincial Health Commission (202210863), Municipal level scientific research plan project of Ganzhou Municipal Health Commission (2022-2-62), Science and Technology Plan of Jiangxi Provincial Administration of Traditional Chinese Medicine (2022B495).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B (2015) 5(5):402–18. doi: 10.1016/j.apsb.2015.07.005
2. Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer discovery (2021) 11(4):858–73. doi: 10.1158/2159-8290.CD-20-1311
3. Junqueira-Neto S, Batista IA, Costa JL, Melo SA. Liquid biopsy beyond circulating tumor cells and cell-free DNA. Acta cytologica (2019) 63(6):479–88. doi: 10.1159/000493969
4. van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JM. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res (2011) 71(18):5955–60. doi: 10.1158/0008-5472.CAN-11-1254
5. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer (2011) 11(6):426–37. doi: 10.1038/nrc3066
6. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science (2020) 367(6478):1–15. doi: 10.1126/science.aau6977
7. Best MG, Sol N, In 't Veld S, Vancura A, Muller M, Niemeijer AN, et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell (2017) 32(2):238–52.e9. doi: 10.1016/j.ccell.2017.07.004
8. Kasi PM, Lee JK, Pasquina LW, Decker B, Vanden Borre P, Pavlick DC, et al. Circulating tumor DNA enables sensitive detection of actionable gene fusions and rearrangements across cancer types. Clin Cancer Res (2023) OF1–13. doi: 10.1158/1078-0432.CCR-23-2693
9. Heo J, Kim YN, Shin S, Lee K, Lee JH, Lee YJ, et al. Serial circulating-tumor DNA analysis with a tumor naïve next-generation sequencing panel detects minimal residual disease and predicts outcome in ovarian cancer. Cancer Res (2023). doi: 10.1158/0008-5472.CAN-23-1429
10. Dobilas A, Chen Y, Brueffer C, Leandersson P, Saal LH, Borgfeldt C. Preoperative ctDNA levels are associated with poor overall survival in patients with ovarian cancer. Cancer Genomics Proteomics (2023) 20(6suppl):763–70. doi: 10.21873/cgp.20423
11. Lavacchi D, Gelmini S, Calabri A, Rossi G, Simi L, Caliman E, et al. Early changes in circulating tumor DNA (ctDNA) predict treatment response in metastatic KRAS-mutated colorectal cancer (mCRC) patients. Heliyon (2023) 9(11):e21853. doi: 10.1016/j.heliyon.2023.e21853
12. Tran HT, Heeke S, Sujit S, Vokes N, Zhang J, Aminu M, et al. Circulating tumor DNA and radiological tumor volume identify patients at risk for relapse with resected, early-stage non-small cell lung cancer. Ann Oncol (2023) 1–7. doi: 10.1016/j.annonc.2023.11.008
13. Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol (2018) 15(10):617–38. doi: 10.1038/s41571-018-0036-9
14. Li F, Zhang M, Yin X, Zhang W, Li H, Gao C. Exosomes-derived miR-548am-5p promotes colorectal cancer progression. Cell Mol Biol (Noisy-le-grand) (2023) 69(12):104–10. doi: 10.14715/cmb/2023.69.12.17
15. Jouve M, Carpentier R, Kraiem S, Legrand N, Sobolewski C. MiRNAs in alcohol-related liver diseases and hepatocellular carcinoma: A step toward new therapeutic approaches? Cancers (Basel) (2023) 15(23):1–42. doi: 10.3390/cancers15235557
16. Letelier P, Saldías R, Loren P, Riquelme I, Guzmán N. MicroRNAs as potential biomarkers of environmental exposure to polycyclic aromatic hydrocarbons and their link with inflammation and lung cancer. Int J Mol Sci (2023) 24(23):1–26. doi: 10.3390/ijms242316984
17. Cheng YC, Chang KW, Pan JH, Chen CY, Chou CH, Tu HF, et al. Cold atmospheric plasma jet irradiation decreases the survival and the expression of oncogenic miRNAs of oral carcinoma cells. Int J Mol Sci (2023) 24(23):1–15. doi: 10.3390/ijms242316662
18. Zhang Y, Tedja R, Millman M, Wong T, Fox A, Chehade H, et al. Adipose-derived exosomal miR-421 targets CBX7 and promotes metastatic potential in ovarian cancer cells. J Ovarian Res (2023) 16(1):233. doi: 10.1186/s13048-023-01312-0
19. Hu JH, Tang HN, Wang YH. Cancer-associated fibroblast exosome LINC00355 promotes epithelial-mesenchymal transition and chemoresistance in colorectal cancer through the miR-34b-5p/CRKL axis. Cancer Gene Ther (2023). doi: 10.1038/s41417-023-00700-4
20. Hao SJ, Wan Y, Xia YQ, Zou X, Zheng SY. Size-based separation methods of circulating tumor cells. Advanced Drug delivery Rev (2018) 125:3–20. doi: 10.1016/j.addr.2018.01.002
21. Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol (2011) 192(3):373–82. doi: 10.1083/jcb.201010021
22. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol (2017) 14(9):531–48. doi: 10.1038/nrclinonc.2017.14
23. Liu J, Lian J, Chen Y, Zhao X, Du C, Xu Y, et al. Circulating tumor cells (CTCs): A unique model of cancer metastases and non-invasive biomarkers of therapeutic response. Front Genet (2021) 12:734595. doi: 10.3389/fgene.2021.734595
24. Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol (2019) 12(1):48. doi: 10.1186/s13045-019-0735-4
25. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol (2022) 15(1):131. doi: 10.1186/s13045-022-01351-y
26. Muchlińska A, Smentoch J, Żaczek AJ, Bednarz-Knoll N. Detection and characterization of circulating tumor cells using imaging flow cytometry-A perspective study. Cancers (2022) 14(17):1–10. doi: 10.3390/cancers14174178
27. Sabath DE, Perrone ME, Clein A, Tam M, Hardin M, Trimble S, et al. Clinical validation of a circulating tumor cell assay using density centrifugation and automated immunofluorescence microscopy. Am J Clin pathol (2022) 158(2):270–6. doi: 10.1093/ajcp/aqac040
28. Sulaiman R, De P, Aske JC, Lin X, Dale A, Vaselaar E, et al. A laboratory-friendly CTC identification: comparable double-immunocytochemistry with triple-immunofluorescence. Cancers (2022) 14(12):1–29. doi: 10.3390/cancers14122871
29. Sato K. Microdevice in cellular pathology: microfluidic platforms for fluorescence in situ hybridization and analysis of circulating tumor cells. Analytical Sci (2015) 31(9):867–73. doi: 10.2116/analsci.31.867
30. Zavarykina TM, Lomskova PK, Pronina IV, Khokhlova SV, Stenina MB, Sukhikh GT. Circulating tumor DNA is a variant of liquid biopsy with predictive and prognostic clinical value in breast cancer patients. Int J Mol Sci (2023) 24(23):1–34. doi: 10.3390/ijms242317073
31. Vasseur A, Kiavue N, Bidard FC, Pierga JY, Cabel L. Clinical utility of circulating tumor cells: an update. Mol Oncol (2021) 15(6):1647–66. doi: 10.1002/1878-0261.12869
32. Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol cancer (2022) 21(1):25. doi: 10.1186/s12943-021-01462-z
33. Fujisawa R, Iwaya T, Endo F, Idogawa M, Sasaki N, Hiraki H, et al. Early dynamics of circulating tumor DNA predict chemotherapy responses for patients with esophageal cancer. Carcinogenesis (2021) 42(10):1239–49. doi: 10.1093/carcin/bgab088
34. Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J (2018) 16:370–8. doi: 10.1016/j.csbj.2018.10.002
35. Lin C, Liu X, Zheng B, Ke R, Tzeng CM. Liquid Biopsy, ctDNA Diagnosis through NGS. Life (Basel Switzerland) (2021) 11(9):1–12. doi: 10.3390/life11090890
36. Li H, Jing C, Wu J, Ni J, Sha H, Xu X, et al. Circulating tumor DNA detection: A potential tool for colorectal cancer management. Oncol letters (2019) 17(2):1409–16. doi: 10.3892/ol.2018.9794
37. O'Leary B, Hrebien S, Beaney M, Fribbens C, Garcia-Murillas I, Jiang J, et al. Comparison of BEAMing and droplet digital PCR for circulating tumor DNA analysis. Clin Chem (2019) 65(11):1405–13. doi: 10.1373/clinchem.2019.305805
38. Wardenaar R, Liu H, Colot V, Colomé-Tatché M, Johannes F. Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in Arabidopsis. Methods Mol Biol (Clifton NJ) (2013) 1067:203–24. doi: 10.1007/978-1-62703-607-8_13
39. McGuire AL, Caulfield T, Cho MK. Research ethics and the challenge of whole-genome sequencing. Nat Rev Genet (2008) 9(2):152–6. doi: 10.1038/nrg2302
40. Imperial R, Nazer M, Ahmed Z, Kam AE, Pluard TJ, Bahaj W, et al. Matched whole-genome sequencing (WGS) and whole-exome sequencing (WES) of tumor tissue with circulating tumor DNA (ctDNA) analysis: complementary modalities in clinical practice. Cancers (2019) 11(9):1–16. doi: 10.3390/cancers11091399
41. Link-Lenczowska D, Pallisgaard N, Cordua S, Zawada M, Czekalska S, Krochmalczyk D, et al. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann hematol (2018) 97(12):2299–308. doi: 10.1007/s00277-018-3451-1
42. Nakamura K. [Circulating tumor DNA (ctDNA) detection using BEAMing and its clinical significance.]. Rinsho byori Japanese J Clin Pathol (2016) 64(4):400–6.
43. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Trans Med (2012) 4(136):136ra68. doi: 10.1126/scitranslmed.3003726
44. Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol diagnostics (2015) 15(6):715–9. doi: 10.1586/14737159.2015.1019476
45. Przybyl J, Chabon JJ, Spans L, Ganjoo KN, Vennam S, Newman AM, et al. Combination approach for detecting different types of alterations in circulating tumor DNA in leiomyosarcoma. Clin Cancer Res (2018) 24(11):2688–99. doi: 10.1158/1078-0432.CCR-17-3704
46. Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med (2013) 5(4):30. doi: 10.1186/gm434
47. Dolzhenko E, van Vugt J, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res (2017) 27(11):1895–903. doi: 10.1101/gr.225672.117
48. Hintzsche JD, Robinson WA, Tan AC. A survey of computational tools to analyze and interpret whole exome sequencing data. Int J Genomics (2016) 2016:7983236. doi: 10.1155/2016/7983236
49. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J extracellular vesicles (2018) 7(1):1535750. doi: 10.1080/20013078.2018.1535750
50. Marar C, Starich B, Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat Immunol (2021) 22(5):560–70. doi: 10.1038/s41590-021-00899-0
51. Labani-Motlagh A, Naseri S, Wenthe J, Eriksson E, Loskog A. Systemic immunity upon local oncolytic virotherapy armed with immunostimulatory genes may be supported by tumor-derived exosomes. Mol Ther oncolytics (2021) 20:508–18. doi: 10.1016/j.omto.2021.02.007
52. Mu Y, Huang X, Yang Y, Huang Z, Chen J, Li S, et al. Study of serum exosome miRNA as a biomarker for early onset adult ouclar myastthenia gravis. Gene (2023) 896:148034. doi: 10.1016/j.gene.2023.148034
53. Chen Y, Chen L, Zhu S, Yang H, Ye Z, Wang H, et al. Exosomal derived miR-1246 from hydroquinone-transformed cells drives S phase accumulation arrest by targeting cyclin G2 in TK6 cells. Chem Biol Interact (2023) 387:110809. doi: 10.1016/j.cbi.2023.110809
54. Zhao Y, Liu X, Zhang Y, Zhang J, Liu X, Yang G. [Tandem mass tag-based quantitative proteomics analysis of plasma and plasma exosomes in Parkinson's disease]. Se Pu. (2023) 41(12):1073–83. doi: 10.3724/SP.J.1123.2022.12022
55. Wang J, Gan L, Li F, Li Q, Wu T, Wu Z, et al. Tracheal epithelial cell-exosome-derived MiR-21-5p inhibits alveolar macrophage pyroptosis to resist pulmonary bacterial infection through PIK3CD-autophagy pathway. Life Sci (2023) 122340. doi: 10.2139/ssrn.4409494
56. Bashi A, Lekpor C, Hood JL, Thompson WE, Stiles JK, Driss A. Modulation of heme-induced inflammation using microRNA-loaded liposomes: implications for hemolytic disorders such as malaria and sickle cell disease. Int J Mol Sci (2023) 24(23):1–20. doi: 10.3390/ijms242316934
57. Lin J, Jia S, Jiao Z, Chen J, Li W, Cao F, et al. Global research trends in CRISPR-related technologies associated with extracellular vesicles from 2015 to 2022: a bibliometric, dynamic, and visualized study. Cell Mol Biol Lett (2023) 28(1):99. doi: 10.1186/s11658-023-00507-z
58. Ferreira GA, Thomé CH, Izumi C, Grassi ML, Lanfredi GP, Smolka M, et al. Proteomic analysis of exosomes secreted during the epithelial-mesenchymal transition and potential biomarkers of mesenchymal high-grade serous ovarian carcinoma. J Ovarian Res (2023) 16(1):232. doi: 10.1186/s13048-023-01304-0
59. Zhao JJ, Hua YJ, Sun DG, Meng XX, Xiao HS, Ma X. Genome-wide microRNA profiling in human fetal nervous tissues by oligonucleotide microarray. Childs Nerv Syst (2006) 22(11):1419–25. doi: 10.1007/s00381-006-0173-9
60. Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods (2014) 11(8):809–15. doi: 10.1038/nmeth.3014
61. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U.S.A. (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
62. Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, et al. Real-time PCR quantification of precursor and mature microRNA. Methods (2008) 44(1):31–8. doi: 10.1016/j.ymeth.2007.09.006
63. Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed (2013) 98(6):236–8. doi: 10.1136/archdischild-2013-304340
64. Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One (2009) 4(4):e5311. doi: 10.1371/journal.pone.0005311.65
65. Urabe F, Kosaka N, Ito K, Kimura T, Egawa S, Ochiya T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am J Physiol Cell Physiol (2020) 318(1):C29–c39. doi: 10.1152/ajpcell.00280.2019
66. Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun (2015) 6:6999. doi: 10.1038/ncomms7999
67. Li M, Li J, Kang L, Gong C, Luo M, Wang X, et al. Genome sequencing reveals molecular epidemiological characteristics and new recombinations of adenovirus in Beijing, China, 2014-2019. J Med Virol (2023) 95(12):e29284. doi: 10.1002/jmv.29284
68. Yu MC, Huang CK, Hung CS, Wang CH, Lin JC. Emerging insight of whole genome sequencing coupled with protein structure prediction into the pyrazinamide-resistance signature of Mycobacterium tuberculosis. Int J Antimicrob Agents (2023) 107053. doi: 10.1016/j.ijantimicag.2023.107053
69. Fan W, Zhao L, Yu L, Zhou Y. Chip-based digital PCR as a direct quantification method for residual DNA in mRNA drugs. J Pharm BioMed Anal (2024) 238:115837. doi: 10.1016/j.jpba.2023.115837
70. Bruno SM, Blaconà G, Lo Cicero S, Castelli G, Virgulti M, Testino G, et al. Quantitative evaluation of CFTR gene expression: A comparison between relative quantification by real-time PCR and absolute quantification by droplet digital PCR. Genes (Basel) (2023) 14(9):1–12. doi: 10.3390/genes14091781
71. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol (2002) 2(8):569–79. doi: 10.1038/nri855
72. Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One (2009) 4(4):e5219. doi: 10.1371/journal.pone.0005219
73. Jørgensen M, Bæk R, Pedersen S, Søndergaard EK, Kristensen SR, Varming K. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles (2013) 2:1–9. doi: 10.3402/jev.v2i0.20920
74. Jørgensen MM, Bæk R, Varming K. Potentials and capabilities of the extracellular vesicle (EV) array. J Extracell Vesicles (2015) 4:26048. doi: 10.3402/jev.v4.26048
75. Malvicini R, De Lazzari G, Tolomeo AM, Santa-Cruz D, Ullah M, Cirillo C, et al. Influence of the isolation method on characteristics and functional activity of mesenchymal stromal cell-derived extracellular vesicles. Cytotherapy (2023). doi: 10.1016/j.jcyt.2023.11.001
76. Chen M, Cao C, Ma J. Tumor-related exosomal circ_0001715 promotes lung adenocarcinoma cell proliferation and metastasis via enhancing M2 macrophage polarization by regulating triggering receptor expressed on myeloid cells-2. Thorac Cancer (2023). doi: 10.1111/1759-7714.15182
77. Zhang J, Pan Y, Jin L, Yang H, Cao P. Exosomal-miR-522-3p derived from cancer-associated fibroblasts accelerates tumor metastasis and angiogenesis via repression bone morphogenetic protein 5 in colorectal cancer. J Gastroenterol Hepatol (2023). doi: 10.1111/jgh.16345
78. Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol (2021) 32(4):466–77. doi: 10.1016/j.annonc.2021.01.074
79. Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J nanomed (2020) 15:6917–34. doi: 10.2147/IJN.S264498
80. Ford T, Graham J, Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Analytical Biochem (1994) 220(2):360–6. doi: 10.1006/abio.1994.1350
81. Rider MA, Hurwitz SN, Meckes DG Jr. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep (2016) 6:23978. doi: 10.1038/srep23978
82. Vergauwen G, Dhondt B, Van Deun J, De Smedt E, Berx G, Timmerman E, et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci Rep (2017) 7(1):2704. doi: 10.1038/s41598-017-02599-y
83. Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J extracellular vesicles (2014) 3:1–11. doi: 10.3402/jev.v3.23430
84. Tschuschke M, Kocherova I, Bryja A, Mozdziak P, Angelova Volponi A, Janowicz K, et al. Inclusion biogenesis, methods of isolation and clinical application of human cellular exosomes. J Clin Med (2020) 9(2):1–19. doi: 10.3390/jcm9020436
85. Papadaki MA, Koutsopoulos AV, Tsoulfas PG, Lagoudaki E, Aggouraki D, Monastirioti A, et al. Clinical relevance of immune checkpoints on circulating tumor cells in breast cancer. Cancers (2020) 12(2):1–20. doi: 10.3390/cancers12020376
86. Mego M, Karaba M, Minarik G, Benca J, Silvia J, Sedlackova T, et al. Circulating tumor cells with epithelial-to-mesenchymal transition phenotypes associated with inferior outcomes in primary breast cancer. Anticancer Res (2019) 39(4):1829–37. doi: 10.21873/anticanres.13290
87. Strati A, Nikolaou M, Georgoulias V, Lianidou ES. Prognostic significance of TWIST1, CD24, CD44, and ALDH1 transcript quantification in epCAM-positive circulating tumor cells from early stage breast cancer patients. Cells (2019) 8(7):1–16. doi: 10.3390/cells8070652
88. Zhang Y, Qi J, Li J, Jia S, Wang Y, Sun Q, et al. Utility of circulating tumor cells for detection of early-stage luminal A breast cancer. Am J Med Sci (2020) 360(5):543–51. doi: 10.1016/j.amjms.2020.01.020
89. Shliakhtunou YA. CTCs-oriented adjuvant personalized cytostatic therapy non-metastatic breast cancer patients: continuous non-randomized prospective study and prospective randomized controlled study. Breast Cancer Res Treat (2021) 186(2):439–51. doi: 10.1007/s10549-020-06036-z
90. Stefanovic S, Deutsch TM, Riethdorf S, Fischer C, Hartkopf A, Sinn P, et al. The lack of evidence for an association between cancer biomarker conversion patterns and CTC-status in patients with metastatic breast cancer. Int J Mol Sci (2020) 21(6):1–12. doi: 10.3390/ijms21062161
91. Magbanua MJM, Hendrix LH, Hyslop T, Barry WT, Winer EP, Hudis C, et al. Serial analysis of circulating tumor cells in metastatic breast cancer receiving first-line chemotherapy. J Natl Cancer Institute (2021) 113(4):443–52. doi: 10.1093/jnci/djaa113
92. Paoletti C, Regan MM, Niman SM, Dolce EM, Darga EP, Liu MC, et al. Circulating tumor cell number and endocrine therapy index in ER positive metastatic breast cancer patients. NPJ Breast cancer. (2021) 7(1):77. doi: 10.1038/s41523-021-00281-1
93. Wang C, Mu Z, Ye Z, Zhang Z, Abu-Khalaf MM, Silver DP, et al. Prognostic value of HER2 status on circulating tumor cells in advanced-stage breast cancer patients with HER2-negative tumors. Breast Cancer Res Treat (2020) 181(3):679–89. doi: 10.1007/s10549-020-05662-x
94. Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol (2012) 30(5):525–32. doi: 10.1200/JCO.2010.33.3716
95. Messaritakis I, Nikolaou M, Koinis F, Politaki E, Koutsopoulos A, Lagoudaki E, et al. Characterization of DLL3-positive circulating tumor cells (CTCs) in patients with small cell lung cancer (SCLC) and evaluation of their clinical relevance during front-line treatment. Lung Cancer (Amsterdam Netherlands) (2019) 135:33–9. doi: 10.1016/j.lungcan.2019.06.025
96. Wang PP, Liu SH, Chen CT, Lv L, Li D, Liu QY, et al. Circulating tumor cells as a new predictive and prognostic factor in patients with small cell lung cancer. J Cancer (2020) 11(8):2113–22. doi: 10.7150/jca.35308
97. Chemi F, Rothwell DG, McGranahan N, Gulati S, Abbosh C, Pearce SP, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat Med (2019) 25(10):1534–9. doi: 10.1038/s41591-019-0593-1
98. Li Z, Xu K, Tartarone A, Santarpia M, Zhu Y, Jiang G. Circulating tumor cells can predict the prognosis of patients with non-small cell lung cancer after resection: a retrospective study. Trans Lung Cancer Res (2021) 10(2):995–1006. doi: 10.21037/tlcr-21-149
99. Xu L, Mao X, Grey A, Scandura G, Guo T, Burke E, et al. Noninvasive detection of clinically significant prostate cancer using circulating tumor cells. J urology (2020) 203(1):73–82. doi: 10.1097/JU.0000000000000475
100. Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol (2009) 10(3):233–9. doi: 10.1016/S1470-2045(08)70340-1
101. Sieuwerts AM, Onstenk W, Kraan J, Beaufort CM, Van M, De Laere B, et al. AR splice variants in circulating tumor cells of patients with castration-resistant prostate cancer: relation with outcome to cabazitaxel. Mol Oncol (2019) 13(8):1795–807. doi: 10.1002/1878-0261.12529
102. Graf RP, Hullings M, Barnett ES, Carbone E, Dittamore R, Scher HI. Clinical utility of the nuclear-localized AR-V7 biomarker in circulating tumor cells in improving physician treatment choice in castration-resistant prostate cancer. Eur urology (2020) 77(2):170–7. doi: 10.1016/j.eururo.2019.08.020
103. Cieślikowski WA, Budna-Tukan J, Świerczewska M, Ida A, Hrab M, Jankowiak A, et al. Circulating tumor cells as a marker of disseminated disease in patients with newly diagnosed high-risk prostate cancer. Cancers (2020) 12(1):1–16. doi: 10.3390/cancers12010160
104. Zhang P, Wang Z, Yang X, Gao K, Li M, Chong T. The significance of detection of circulating tumor cells and beclin1 in peripheral blood of patients with renal cell carcinoma. Crit Rev eukaryotic Gene expression (2020) 30(6):483–92. doi: 10.1615/CritRevEukaryotGeneExpr.2020036246
105. Basso U, Facchinetti A, Rossi E, Maruzzo M, Conteduca V, Aieta M, et al. Prognostic role of circulating tumor cells in metastatic renal cell carcinoma: A large, multicenter, prospective trial. oncologist (2021) 26(9):740–50. doi: 10.1002/onco.13842
106. Cheng Y, Luo L, Zhang J, Zhou M, Tang Y, He G, et al. Diagnostic value of different phenotype circulating tumor cells in hepatocellular carcinoma. J gastrointestinal Surg (2019) 23(12):2354–61. doi: 10.1007/s11605-018-04067-y
107. Lei Y, Wang X, Sun H, Fu Y, Tian Y, Yang L, et al. Association of preoperative NANOG-positive circulating tumor cell levels with recurrence of hepatocellular carcinoma. Front Oncol (2021) 11:601668. doi: 10.3389/fonc.2021.601668
108. Wei T, Zhang X, Zhang Q, Yang J, Chen Q, Wang J, et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer letters (2019) 452:237–43. doi: 10.1016/j.canlet.2019.03.009
109. Zhao XH, Wang ZR, Chen CL, Di L, Bi ZF, Li ZH, et al. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J gastroenterol (2019) 25(1):138–50. doi: 10.3748/wjg.v25.i1.138
110. Hugenschmidt H, Labori KJ, Brunborg C, Verbeke CS, Seeberg LT, Schirmer CB, et al. Circulating tumor cells are an independent predictor of shorter survival in patients undergoing resection for pancreatic and periampullary adenocarcinoma. Ann surg (2020) 271(3):549–58. doi: 10.1097/SLA.0000000000003035
111. Szczepanik A, Sierzega M, Drabik G, Pituch-Noworolska A, Kołodziejczyk P, Zembala M. CD44(+) cytokeratin-positive tumor cells in blood and bone marrow are associated with poor prognosis of patients with gastric cancer. Gastric Cancer (2019) 22(2):264–72. doi: 10.1007/s10120-018-0858-2
112. Miki Y, Yashiro M, Kuroda K, Okuno T, Togano S, Masuda G, et al. Circulating CEA-positive and EpCAM-negative tumor cells might be a predictive biomarker for recurrence in patients with gastric cancer. Cancer Med (2021) 10(2):521–8. doi: 10.1002/cam4.3616
113. Kuroda K, Yashiro M, Miki Y, Sera T, Yamamoto Y, Sugimoto A, et al. Circulating tumor cells with FGFR2 expression might be useful to identify patients with existing FGFR2-overexpressing tumor. Cancer science (2020) 111(12):4500–9. doi: 10.1111/cas.14654
114. Matsushita D, Uenosono Y, Arigami T, Yanagita S, Okubo K, Kijima T, et al. Clinical significance of circulating tumor cells in the response to trastuzumab for HER2-negative metastatic gastric cancer. Cancer chemotherapy Pharmacol (2021) 87(6):789–97. doi: 10.1007/s00280-021-04251-z
115. Messaritakis I, Sfakianaki M, Vogiatzoglou K, Koulouridi A, Koutoulaki C, Mavroudis D, et al. Evaluation of the role of circulating tumor cells and microsatellite instability status in predicting outcome of advanced CRC patients. J personalized Med (2020) 10(4):1–13. doi: 10.3390/jpm10040235
116. Su P, Lai W, Liu L, Zeng Y, Xu H, Lan Q, et al. Mesenchymal and phosphatase of regenerating liver-3 status in circulating tumor cells may serve as a crucial prognostic marker for assessing relapse or metastasis in postoperative patients with colorectal cancer. Clin Trans gastroenterol (2020) 11(12):e00265. doi: 10.14309/ctg.0000000000000265
117. Pan RJ, Hong HJ, Sun J, Yu CR, Liu HS, Li PY, et al. Detection and clinical value of circulating tumor cells as an assisted prognostic marker in colorectal cancer patients. Cancer Manage Res (2021) 13:4567–78. doi: 10.2147/CMAR.S300554
118. Krol I, Castro-Giner F, Maurer M, Gkountela S, Szczerba BM, Scherrer R, et al. Detection of circulating tumour cell clusters in human glioblastoma. Br J cancer. (2018) 119(4):487–91. doi: 10.1038/s41416-018-0186-7
119. Soffietti R, Bettegowda C, Mellinghoff IK, Warren KE, Ahluwalia MS, De Groot JF, et al. Liquid biopsy in gliomas: A RANO review and proposals for clinical applications. Neuro-oncology (2022) 24(6):855–71. doi: 10.1093/neuonc/noac004
120. Ntouroupi TG, Ashraf SQ, McGregor SB, Turney BW, Seppo A, Kim Y, et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. Br J cancer. (2008) 99(5):789–95. doi: 10.1038/sj.bjc.6604545
121. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J Thorac Oncol (2018) 13(9):1248–68. doi: 10.1016/j.jtho.2018.05.030
122. Berger LA, Janning M, Velthaus JL, Ben-Batalla I, Schatz S, Falk M, et al. Identification of a high-level MET amplification in CTCs and cfTNA of an ALK-positive NSCLC patient developing evasive resistance to crizotinib. J Thorac Oncol (2018) 13(12):e243–e6. doi: 10.1016/j.jtho.2018.08.2025
123. Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Trans Med (2015) 7(313):313ra182. doi: 10.1126/scitranslmed.aac7551
124. O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun (2018) 9(1):896. doi: 10.1038/s41467-018-03215-x
125. Kwee S, Song MA, Cheng I, Loo L, Tiirikainen M. Measurement of circulating cell-free DNA in relation to 18F-fluorocholine PET/CT imaging in chemotherapy-treated advanced prostate cancer. Clin Trans science (2012) 5(1):65–70. doi: 10.1111/j.1752-8062.2011.00375.x
126. Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Institute (2017) 109(12):1–9. doi: 10.1093/jnci/djx118
127. Chen G, Jia G, Chao F, Xie F, Zhang Y, Hou C, et al. Urine- and blood-based molecular profiling of human prostate cancer. Front Oncol (2022) 12:759791. doi: 10.3389/fonc.2022.759791
128. Haselmann V, Gebhardt C, Brechtel I, Duda A, Czerwinski C, Sucker A, et al. Liquid profiling of circulating tumor DNA in plasma of melanoma patients for companion diagnostics and monitoring of BRAF inhibitor therapy. Clin Chem (2018) 64(5):830–42. doi: 10.1373/clinchem.2017.281543
129. Fu Y, Yang Z, Hu Z, Yang Z, Pan Y, Chen J, et al. Preoperative serum ctDNA predicts early hepatocellular carcinoma recurrence and response to systemic therapies. Hepatol Int (2022) 16(4):868–78. doi: 10.1007/s12072-022-10348-1
130. Maron SB, Chase LM, Lomnicki S, Kochanny S, Moore KL, Joshi SS, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res (2019) 25(23):7098–112. doi: 10.1158/1078-0432.CCR-19-1704
131. Bergerot PG, Hahn AW, Bergerot CD, Jones J, Pal SK. The role of circulating tumor DNA in renal cell carcinoma. Curr Treat options Oncol (2018) 19(2):10. doi: 10.1007/s11864-018-0530-4
132. Park YR, Kim YM, Lee SW, Lee HY, Lee GE, Lee JE, et al. Optimization to detect TP53 mutations in circulating cell-free tumor DNA from patients with serous epithelial ovarian cancer. Obstetrics gynecology science (2018) 61(3):328–36. doi: 10.5468/ogs.2018.61.3.328
133. Ratajska M, Koczkowska M, Żuk M, Gorczyński A, Kuźniacka A, Stukan M, et al. Detection of BRCA1/2 mutations in circulating tumor DNA from patients with ovarian cancer. Oncotarget (2017) 8(60):101325–32. doi: 10.18632/oncotarget.20722
134. Martínez-Ricarte F, Mayor R, Martínez-Sáez E, Rubio-Pérez C, Pineda E, Cordero E, et al. Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumor DNA from cerebrospinal fluid. Clin Cancer Res (2018) 24(12):2812–9. doi: 10.1158/1078-0432.CCR-17-3800
135. Nagy Á, Bátai B, Balogh A, Illés S, Mikala G, Nagy N, et al. Quantitative analysis and monitoring of EZH2 mutations using liquid biopsy in follicular lymphoma. Genes (2020) 11(7):1–11. doi: 10.3390/genes11070785
136. Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Trans Med (2016) 8(364):364ra155. doi: 10.1126/scitranslmed.aai8545
137. Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood (2018) 131(22):2413–25. doi: 10.1182/blood-2017-11-812073
138. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med (2015) 21(7):827. doi: 10.1038/nm0715-827b
139. Siravegna G, Lazzari L, Crisafulli G, Sartore-Bianchi A, Mussolin B, Cassingena A, et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell (2018) 34(1):148–62.e7. doi: 10.1016/j.ccell.2018.06.004
140. Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J extracellular vesicles (2016) 5:31292. doi: 10.3402/jev.v5.31292
141. Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J Cell Physiol (2018) 233(9):6370–80. doi: 10.1002/jcp.26481
142. Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res (2018) 78(7):1833–44. doi: 10.1158/0008-5472.CAN-17-2069
143. Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal microRNA miR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem (2017) 44(5):1741–8. doi: 10.1159/000485780
144. Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and Malignant mammary epithelial cells. PLoS One (2010) 5(10):e13515. doi: 10.1371/journal.pone.0013515
145. Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep (2016) 6:38750. doi: 10.1038/srep38750
146. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res BCR. (2016) 18(1):90. doi: 10.1186/s13058-016-0753-x
147. Yan S, Dang G, Zhang X, Jin C, Qin L, Wang Y, et al. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget (2017) 8(42):72220–6. doi: 10.18632/oncotarget.19689
148. Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J (2010) 31(6):659–66. doi: 10.1093/eurheartj/ehq013
149. Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res (2013) 113(3):322–6. doi: 10.1161/CIRCRESAHA.113.301209
150. Gidlöf O, Andersson P, van der Pals J, Götberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology (2011) 118(4):217–26. doi: 10.1159/000328869
151. Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet (2010) 3(6):499–506. doi: 10.1161/CIRCGENETICS.110.957415
152. Goetzl EJ, Boxer A, Schwartz JB, Abner EL, Petersen RC, Miller BL, et al. Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann Clin Trans neurol (2015) 2(7):769–73. doi: 10.1002/acn3.211
153. Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U States A (2013) 110(18):7312–7. doi: 10.1073/pnas.1220998110
154. Pusic AD, Kraig RP. Youth and environmental enrichment generate serum exosomes containing miR-219 that promote CNS myelination. Glia (2014) 62(2):284–99. doi: 10.1002/glia.22606
155. Vella LJ, Hill AF, Cheng L. Focus on extracellular vesicles: exosomes and their role in protein trafficking and biomarker potential in Alzheimer's and Parkinson's disease. Int J Mol Sci (2016) 17(2):173. doi: 10.3390/ijms17020173
156. Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol (2016) 11(10):1701–10. doi: 10.1016/j.jtho.2016.05.034
157. Chen IH, Xue L, Hsu CC, Paez JS, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A (2017) 114(12):3175–80. doi: 10.1073/pnas.1618088114
158. Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol (2018) 29(1):223–9. doi: 10.1093/annonc/mdx542
159. Abdalla TSA, Meiners J, Riethdorf S, König A, Melling N, Gorges T, et al. Prognostic value of preoperative circulating tumor cells counts in patients with UICC stage I-IV colorectal cancer. PLoS One (2021) 16(6):e0252897. doi: 10.1371/journal.pone.0252897
160. Loh J, Jovanovic L, Lehman M, Capp A, Pryor D, Harris M, et al. Circulating tumor cell detection in high-risk non-metastatic prostate cancer. J Cancer Res Clin Oncol (2014) 140(12):2157–62. doi: 10.1007/s00432-014-1775-3
161. Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J cancer. (2016) 114(12):1367–75. doi: 10.1038/bjc.2016.121
162. Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, et al. Circulating tumor cells: biology and clinical significance. Signal transduction targeted Ther (2021) 6(1):404. doi: 10.1038/s41392-021-00817-8
163. Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol (2009) 27(31):5153–9. doi: 10.1200/JCO.2008.20.6664
164. Liang DH, Hall C, Lucci A. Circulating tumor cells in breast cancer. Recent results Cancer Res Fortschr der Krebsforschung Progres dans les recherches sur le cancer. (2020) 215:127–45. doi: 10.1007/978-3-030-26439-0_7
165. Krol I, Schwab FD, Carbone R, Ritter M, Picocci S, De Marni ML, et al. Detection of clustered circulating tumour cells in early breast cancer. Br J cancer. (2021) 125(1):23–7. doi: 10.1038/s41416-021-01327-8
166. Li Y, Wu S, Bai F. Molecular characterization of circulating tumor cells-from bench to bedside. Semin Cell Dev Biol (2018) 75:88–97. doi: 10.1016/j.semcdb.2017.09.013
167. Larsson AM, Jansson S, Bendahl PO, Levin Tykjaer Jörgensen C, Loman N, Graffman C, et al. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res (2018) 20(1):48. doi: 10.1186/s13058-018-0976-0
168. Murlidhar V, Reddy RM, Fouladdel S, Zhao L, Ishikawa MK, Grabauskiene S, et al. Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. Cancer Res (2017) 77(18):5194–206. doi: 10.1158/0008-5472.CAN-16-2072
169. Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatol (Baltimore Md) (2021) 73(1):422–36. doi: 10.1002/hep.31165
170. Lipsyc-Sharf M, de Bruin EC, Santos K, McEwen R, Stetson D, Patel A, et al. Circulating tumor DNA and late recurrence in high-risk hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer. J Clin Oncol (2022) 40(22):2408–19. doi: 10.1200/JCO.22.00908
171. Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. New Engl J Med (2022) 386(24):2261–72. doi: 10.1056/NEJMoa2200075
172. Xu J, Liu Z, Bai H, Dong G, Zhong J, Wan R, et al. Evaluation of clinical outcomes of icotinib in patients with clinically diagnosed advanced lung cancer with EGFR-sensitizing variants assessed by circulating tumor DNA testing: A phase 2 nonrandomized clinical trial. JAMA Oncol (2022) 8(9):1328–32. doi: 10.1001/jamaoncol.2022.2719
173. Hua G, Zhang X, Zhang M, Wang Q, Chen X, Yu R, et al. Real-world circulating tumor DNA analysis depicts resistance mechanism and clonal evolution in ALK inhibitor-treated lung adenocarcinoma patients. ESMO Open (2022) 7(1):100337. doi: 10.1016/j.esmoop.2021.100337
174. Chen H, Ma Y, Liu Z, Li J, Li X, Yang F, et al. Circulating microbiome DNA: An emerging paradigm for cancer liquid biopsy. Cancer letters (2021) 521:82–7. doi: 10.1016/j.canlet.2021.08.036
175. Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget (2016) 7(30):48832–41. doi: 10.18632/oncotarget.9453
176. Alekseeva LA, Sen'kova AV, Zenkova MA, Mironova NL. Targeting circulating SINEs and LINEs with DNase I provides metastases inhibition in experimental tumor models. Mol Ther Nucleic Acids (2020) 20:50–61. doi: 10.1016/j.omtn.2020.01.035
177. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature (2017) 545(7655):446–51. doi: 10.1038/nature22364
178. Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB 3rd, Boland P, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol (2020) 17(12):757–70. doi: 10.1038/s41571-020-0392-0
179. Mencel J, Slater S, Cartwright E, Starling N. The role of ctDNA in gastric cancer. Cancers (2022) 14(20):1–20. doi: 10.3390/cancers14205105
180. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res (2014) 24(6):766–9. doi: 10.1038/cr.2014.44
181. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest (2016) 126(4):1208–15. doi: 10.1172/JCI81135
182. Kalluri R, LeBleu VS. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harbor Symp quantitative Biol (2016) 81:275–80. doi: 10.1101/sqb.2016.81.030932
183. Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One (2015) 10(5):e0125625. doi: 10.1371/journal.pone.0125625
184. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther (2013) 21(1):185–91. doi: 10.1038/mt.2012.180
185. Katakowski M, Buller B, Zheng X, Lu Y, Rogers T, Osobamiro O, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer letters (2013) 335(1):201–4. doi: 10.1016/j.canlet.2013.02.019
186. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature (2017) 546(7659):498–503. doi: 10.1038/nature22341
187. Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight (2018) 3(8):1–22. doi: 10.1172/jci.insight.99263
188. Fitts CA, Ji N, Li Y, Tan C. Exploiting exosomes in cancer liquid biopsies and drug delivery. Adv Healthc Mater (2019) 8(6):e1801268. doi: 10.1002/adhm.201801268
189. Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther (2017) 174:63–78. doi: 10.1016/j.pharmthera.2017.02.020
190. Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One (2014) 9(4):e95240. doi: 10.1371/journal.pone.0095240
191. Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol (2019) 16(7):409–24. doi: 10.1038/s41571-019-0187-3
192. Stergiopoulou D, Markou A, Strati A, Zavridou M, Tzanikou E, Mastoraki S, et al. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci Rep (2023) 13(1):1258. doi: 10.1038/s41598-022-25400-1
193. Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol (2019) 5(8):1124–31. doi: 10.1001/jamaoncol.2019.0528
194. Loupakis F, Sharma S, Derouazi M, Murgioni S, Biason P, Rizzato MD, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol (2021) 5. doi: 10.1200/PO.21.00101
195. Gerratana L, Davis AA, Polano M, Zhang Q, Shah AN, Lin C, et al. Understanding the organ tropism of metastatic breast cancer through the combination of liquid biopsy tools. Eur J Cancer (Oxford Engl 1990) (2021) 143:147–57. doi: 10.1016/j.ejca.2020.11.005
196. Chalfin HJ, Glavaris SA, Gorin MA, Kates MR, Fong MH, Dong L, et al. Circulating tumor cell and circulating tumor DNA assays reveal complementary information for patients with metastatic urothelial cancer. Eur Urol Oncol (2021) 4(2):310–4. doi: 10.1016/j.euo.2019.08.004
197. Dawood ZS, Alaimo L, Lima HA, Moazzam Z, Shaikh C, Ahmed AS, et al. Imaging, and carcinoembryonic antigen: comparison of surveillance strategies among patients who underwent resection of colorectal cancer-A systematic review and meta-analysis. Ann Surg Oncol (2023) 30(1):259–74. doi: 10.1245/s10434-022-12641-7
198. Wang Y, Li L, Cohen JD, Kinde I, Ptak J, Popoli M, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol (2019) 5(8):1118–23. doi: 10.1001/jamaoncol.2019.0512
199. Chen Y. [Research progress on circulating tumor DNA as a biomarker for minimal residual disease in solid tumors]. Zhongguo Dang Dai Er Ke Za Zhi (2023) 25(10):1072–7. doi: 10.7499/j.issn.1008-8830.2304040
200. Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol (2019) 5(12):1710–7. doi: 10.1001/jamaoncol.2019.3616
201. Chen G, Peng J, Xiao Q, Wu HX, Wu X, Wang F, et al. Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. J Hematol Oncol (2021) 14(1):80. doi: 10.1186/s13045-021-01089-z
202. Wang DS, Yang H, Liu XY, Chen ZG, Wang Y, Fong WP, et al. Dynamic monitoring of circulating tumor DNA to predict prognosis and efficacy of adjuvant chemotherapy after resection of colorectal liver metastases. Theranostics (2021) 11(14):7018–28. doi: 10.7150/thno.59644
203. Qi Z, Li Y, Wang Z, Tan X, Zhou Y, Li Z, et al. Monitoring of gastrointestinal carcinoma via molecular residual disease with circulating tumor DNA using a tumor-informed assay. Cancer Med (2023) 12(16):16687–96. doi: 10.1002/cam4.6286
204. Wen X, Coradduzza D, Shen J, Scanu AM, Muroni MR, Massidda M, et al. Harnessing minimal residual disease as a predictor for colorectal cancer: promising horizons amidst challenges. Medicina (Kaunas) (2023) 59(10):1–12. doi: 10.3390/medicina59101886
205. Zhu L, Xu R, Yang L, Shi W, Zhang Y, Liu J, et al. Minimal residual disease (MRD) detection in solid tumors using circulating tumor DNA: a systematic review. Front Genet (2023) 14:1172108. doi: 10.3389/fgene.2023.1172108
206. Bachet JB, Bouché O, Taieb J, Dubreuil O, Garcia ML, Meurisse A, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol (2018) 29(5):1211–9. doi: 10.1093/annonc/mdy061
207. Keup C, Kimmig R, Kasimir-Bauer S. The diversity of liquid biopsies and their potential in breast cancer management. Cancers (Basel) (2023) 15(22):1–73. doi: 10.3390/cancers15225463
208. Zhao H, Wang L, Fang C, Li C, Zhang L. Factors influencing the diagnostic and prognostic values of circulating tumor cells in breast cancer: a meta-analysis of 8,935 patients. Front Oncol (2023) 13:1272788. doi: 10.3389/fonc.2023.1272788
209. Hamilton G, Rath B, Stickler S. Significance of circulating tumor cells in lung cancer: a narrative review. Transl Lung Cancer Res (2023) 12(4):877–94. doi: 10.21037/tlcr-22-712
210. Pizzutilo EG, Pedrani M, Amatu A, Ruggieri L, Lauricella C, Veronese SM, et al. Liquid biopsy for small cell lung cancer either de novo or transformed: systematic review of different applications and meta-analysis. Cancers (Basel) (2021) 13(9):1–20. doi: 10.3390/cancers13092265
211. Crocetto F, Russo G, Di Zazzo E, Pisapia P, Mirto BF, Palmieri A, et al. Liquid biopsy in prostate cancer management-current challenges and future perspectives. Cancers (Basel) (2022) 14(13):1–18. doi: 10.3390/cancers14133272
212. Ying M, Mao J, Sheng L, Wu H, Bai G, Zhong Z, et al. Biomarkers for prostate cancer bone metastasis detection and prediction. J Pers Med (2023) 13(5):1–20. doi: 10.3390/jpm13050705
213. Shuai Y, Ma Z, Ju J, Wei T, Gao S, Kang Y, et al. Liquid-based biomarkers in breast cancer: looking beyond the blood. J Transl Med (2023) 21(1):809. doi: 10.1186/s12967-023-04660-z
214. Thompson JC, Scholes DG, Carpenter EL, Aggarwal C. Molecular response assessment using circulating tumor DNA (ctDNA) in advanced solid tumors. Br J Cancer (2023) 129(12):1893–902. doi: 10.1038/s41416-023-02445-1
215. Duffy MJ. Circulating tumor DNA (ctDNA) as a biomarker for lung cancer: Early detection, monitoring and therapy prediction. Tumour Biol (2023) 1–13. doi: 10.3233/TUB-220044
216. Alahdal M, Perera RA, Moschovas MC, Patel V, Perera RJ. Current advances of liquid biopsies in prostate cancer: Molecular biomarkers. Mol Ther Oncolytics (2023) 30:27–38. doi: 10.1016/j.omto.2023.07.004
217. Earland N, Chen K, Semenkovich NP, Chauhan PS, Zevallos JP, Chaudhuri AA. Emerging roles of circulating tumor DNA for increased precision and personalization in radiation oncology. Semin Radiat Oncol (2023) 33(3):262–78. doi: 10.1016/j.semradonc.2023.03.004
218. Zhang X, Wang C, Yu J, Bu J, Ai F, Wang Y, et al. Extracellular vesicles in the treatment and diagnosis of breast cancer: a status update. Front Endocrinol (Lausanne) (2023) 14:1202493. doi: 10.3389/fendo.2023.1202493
219. Luo T, Kang Y, Liu Y, Li J, Li J. Small extracellular vesicles in breast cancer brain metastasis and the prospect of clinical application. Front Bioeng Biotechnol (2023) 11:1162089. doi: 10.3389/fbioe.2023.1162089
220. Mullen S, Movia D. The role of extracellular vesicles in non-small-cell lung cancer, the unknowns, and how new approach methodologies can support new knowledge generation in the field. Eur J Pharm Sci (2023) 188:106516. doi: 10.1016/j.ejps.2023.106516
221. Liang H, Zhang L, Zhao X, Rong J. The therapeutic potential of exosomes in lung cancer. Cell Oncol (Dordr) (2023) 46(5):1181–212. doi: 10.1007/s13402-023-00815-8
222. Ishizuya Y, Uemura M, Narumi R, Tomiyama E, Koh Y, Matsushita M, et al. The role of actinin-4 (ACTN4) in exosomes as a potential novel therapeutic target in castration-resistant prostate cancer. Biochem Biophys Res Commun (2020) 523(3):588–94. doi: 10.1016/j.bbrc.2019.12.084
223. Chen H, Pang B, Zhou C, Han M, Gong J, Li Y, et al. Prostate cancer-derived small extracellular vesicle proteins: the hope in diagnosis, prognosis, and therapeutics. J Nanobiotechnol (2023) 21(1):480. doi: 10.1186/s12951-023-02219-0
224. Nguyen TNA, Huang PS, Chu PY, Hsieh CH, Wu MH. Recent progress in enhanced cancer diagnosis, prognosis, and monitoring using a combined analysis of the number of circulating tumor cells (CTCs) and other clinical parameters. Cancers (Basel) (2023) 15(22):1–33. doi: 10.3390/cancers15225372
225. Shegekar T, Vodithala S, Juganavar A. The emerging role of liquid biopsies in revolutionising cancer diagnosis and therapy. Cureus (2023) 15(8):e43650. doi: 10.7759/cureus.43650
226. Boussios S, Devo P, Goodall ICA, Sirlantzis K, Ghose A, Shinde SD, et al. Exosomes in the diagnosis and treatment of renal cell cancer. Int J Mol Sci (2023) 24(18):1–23. doi: 10.3390/ijms241814356
227. Ring A, Nguyen-Sträuli BD, Wicki A, Aceto N. Biology, vulnerabilities and clinical applications of circulating tumour cells. Nat Rev Cancer (2023) 23(2):95–111. doi: 10.1038/s41568-022-00536-4
228. Magis AT, Rappaport N, Conomos MP, Omenn GS, Lovejoy JC, Hood L, et al. Untargeted longitudinal analysis of a wellness cohort identifies markers of metastatic cancer years prior to diagnosis. Sci Rep (2020) 10(1):16275. doi: 10.1038/s41598-020-73451-z
Keywords: CTC, ctDNA, EVs, cancer, liquid biopsy, biomarker, precision
Citation: Wang X, Wang L, Lin H, Zhu Y, Huang D, Lai M, Xi X, Huang J, Zhang W and Zhong T (2024) Research progress of CTC, ctDNA, and EVs in cancer liquid biopsy. Front. Oncol. 14:1303335. doi: 10.3389/fonc.2024.1303335
Received: 27 September 2023; Accepted: 04 January 2024;
Published: 25 January 2024.
Edited by:
Chiara Nicolazzo, Sapienza University of Rome, ItalyCopyright © 2024 Wang, Wang, Lin, Zhu, Huang, Lai, Xi, Huang, Zhang and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianyu Zhong, zhongtianyu@gmail.com; Wenjuan Zhang, wjzhang_kyuu@gmu.edu.cn
†These authors have contributed equally to this work
 Xiaoling Wang
Xiaoling Wang Lijuan Wang
Lijuan Wang Haihong Lin1,2
Haihong Lin1,2 Yifan Zhu
Yifan Zhu Defa Huang
Defa Huang Junyun Huang
Junyun Huang Wenjuan Zhang
Wenjuan Zhang Tianyu Zhong
Tianyu Zhong