- 1Department of Medical, Oral and Biotechnological Sciences (DSMOB), Ageing Research Center and Translational Medicine-CeSI-MeT, “G. d’Annunzio” University Chieti-Pescara, Chieti, Italy
- 2Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India
- 3ER Stress and Intestinal Mucosal Biology Lab, School of Health Sciences, University of Tasmania, Launceston, TAS, Australia
- 4Department of Biotechnology, National Institute of Technology, Durgapur, India
- 5Institute of Environmental Protection and Sensors (IOS), Maribor, Slovenia
- 6Cardiac Metabolic Disease Laboratory, Department Of Biochemistry, School of Biological Sciences, Madurai Kamaraj University, Madurai, India
- 7Radiological Sciences Department, College of Applied Medical Sciences, King Khalid University, Abha, Saudi Arabia
- 8BioImaging Unit, Space Research Centre, University of Leicester, Leicester, United Kingdom
- 9Electrical Engineering Department, College of Engineering, King Khalid University, Abha, Saudi Arabia
SKP2 (S-phase kinase-associated protein 2) is a member of the F-box family of substrate-recognition subunits in the SCF ubiquitin-protein ligase complexes. It is associated with ubiquitin-mediated degradation in the mammalian cell cycle components and other target proteins involved in cell cycle progression, signal transduction, and transcription. Being an oncogene in solid tumors and hematological malignancies, it is frequently associated with drug resistance and poor disease outcomes. In the current review, we discussed the novel role of SKP2 in different hematological malignancies. Further, we performed a limited in-silico analysis to establish the involvement of SKP2 in a few publicly available cancer datasets. Interestingly, our study identified Skp2 expression to be altered in a cancer-specific manner. While it was found to be overexpressed in several cancer types, few cancer showed a down-regulation in SKP2. Our review provides evidence for developing novel SKP2 inhibitors in hematological malignancies. We also investigated the effect of SKP2 status on survival and disease progression. In addition, the role of miRNA and its associated families in regulating Skp2 expression was explored. Subsequently, we predicted common miRNAs against Skp2 genes by using miRNA-predication tools. Finally, we discussed current approaches and future prospective approaches to target the Skp2 gene by using different drugs and miRNA-based therapeutics applications in translational research.
Introduction
Poly ubiquitination is the binding of numerous ubiquitin molecules into the same target protein. Generally, the polyubiquitination of proteins is induced by different signaling molecules and co-operates for protein degradation by the proteasomes. This post-translational modification process (Polyubiquitination) regulates numerous cellular events, including cell growth, proliferation, differentiation and apoptosis in mammalian cells (1). Any deregulation in the ubiquitination machinery and its components could disarrange the cellular homeostasis and initiate the process of neoplastic transformation in various cancers. The step by step action of the ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes associated with the ubiquitin-proteasome system (UPS), mediate ubiquitination by which they degrade targeted substrate proteins (2).
The SKP1, CUL1, F-box protein (SCF) complex consists of three core components that remain constant: RING-box 1 (RBX1), a RING-finger protein responsible for recruiting the E2 ubiquitin-conjugating enzyme; Cullin 1 (CUL1), acting as the scaffolding protein; and S-phase kinase-associated protein 1 (SKP1), an unchanging adaptor that links the core SCF complex with a variable F-box protein and its corresponding target protein (3). The specificity of the SCF complex for particular targets is determined by F-box proteins, with each F-box protein recognizing and binding a specific set of substrates. In humans, there are a total of 69 F-box proteins, categorized into three families based on their substrate recognition domains: (1) FBXW with WD40 repeats; (2) FBXL with leucine-rich repeats (e.g., FBXL1, also known as the S-phase kinase-associated protein 2 [SKP2]); and (3) FBXO with other domains (4). To regulate the levels of specific protein targets, each F-box protein recruits one of its substrates, often phosphorylated, to the core SCF complex, facilitating polyubiquitination and subsequent degradation by the 26S proteasome (5). With a total of 69 distinct F-box genes, it suggests the existence of up to 69 unique SCF complexes, each responsible for regulating a diverse array of protein targets (4).
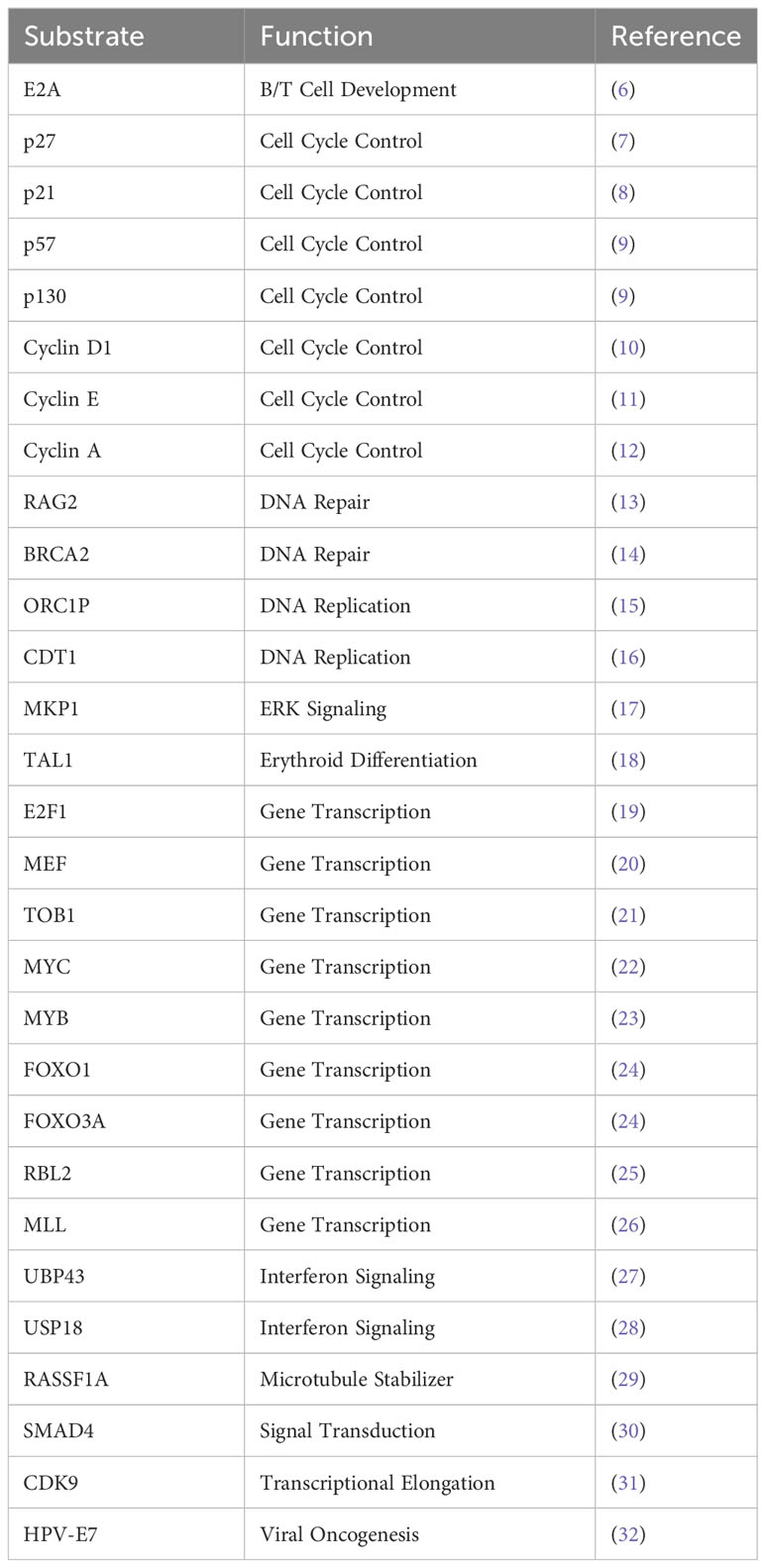
Few well-characterized F-box proteins regulate substrates which are involved in cell cycle regulation, signal transduction, and transcription (Table 1) (33). Among these, one of the E3 ligases called SKP2 (S-Phase Kinase Associated Protein 2 (~ 45kDa)), a member of the F box family (34), is recognized as a pro-oncogene. These F-box proteins are mostly composed of one of the four subunits of ubiquitin-protein ligases complex named SCFs but do not always recognize substrates in a phosphorylation-dependent manner. In this complex of SCF’s, the F-box is referred to as a subunit, which serves as the recognition site for protein substrates. The N-terminal F-Box domain of the F-box binds to SKP1 and thereby connects with the SCF complex. After that, C-terminal Leucine-rich repeat (LRR) and WD40 repeats support substrate binding. Association of SKP1-SKP2 is found in humans (35). SKP2 assembles to SCF-type E3 ubiquitin ligase complex along with Cullin-1, Skp1, and Rbx1 (36–39). In addition, the requirement of cell cycle regulator CDK subunit 1 [CKS1] is important for CF SKP2-mediated ubiquitinylation of p27 (7).
Skp2 gene plays a significant role in cell cycle progression and cell survival through ubiquitin-mediated degradation of many tumor suppressor proteins (p27, p21, p57, p130, FOXO1, BRCA2, RASSF1A, TOB1), cell cycle regulatory proteins (Cyclin D & E, E2F1, etc.) and oncogenes (c-MYC, MYB) (Figure 1) (40, 41). The target interruption of SKP2 leads to the accumulation of Cdk inhibitor p27, which leads to G1 phase cell cycle arrest. SKP2 mediates the degradation of p27 via ubiquitination through the 26S proteasome pathway (42, 43). Additionally, proteins like RING E3 ligases, are essential for the interaction of the E2-conjugating enzyme along with the SKP1adaptor protein (36). In addition, scaffold and ring finger proteins like Rbx1 are also required to target the substrate via its E3 ligase activity (36).
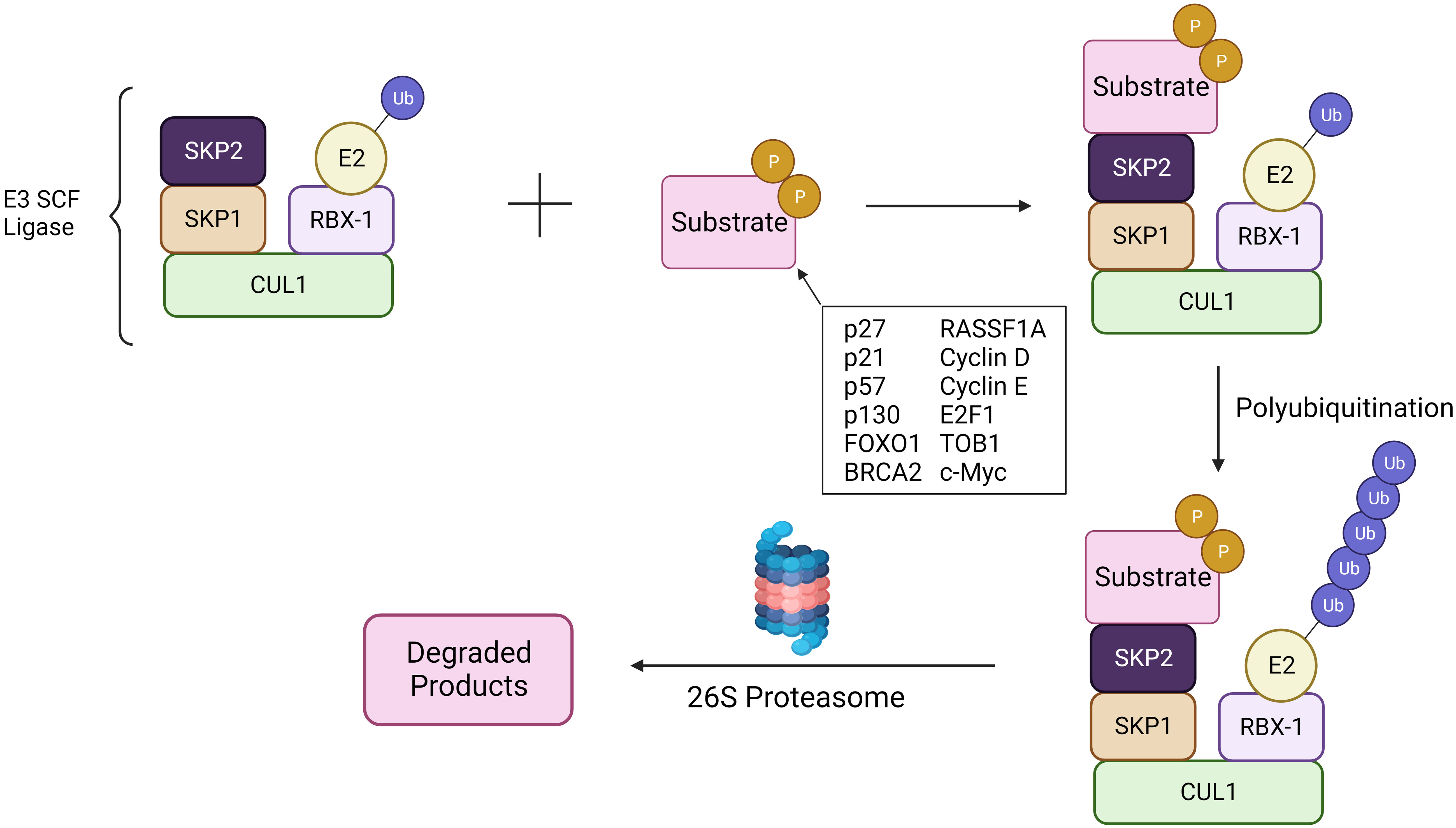
Figure 1 SCFSKP2 complex. The SCFSKP2 complex plays a pivotal role in regulating cell cycle progression and maintaining cellular homeostasis. SKP2, an F-box protein within the complex, acts as a substrate recognition component. It recognizes specific target substrates marking them for ubiquitination. Once ubiquitinated, the tagged proteins are targeted for degradation by the 26S proteasome. The SCF complex serves as an E3 ubiquitin ligase, facilitating the transfer of ubiquitin molecules to substrates. Ultimately, this polyubiquitination signals the proteasome to recognize and degrade the marked proteins, regulating key cellular processes and ensuring proper cell cycle dynamics.
SKP2 in cancers
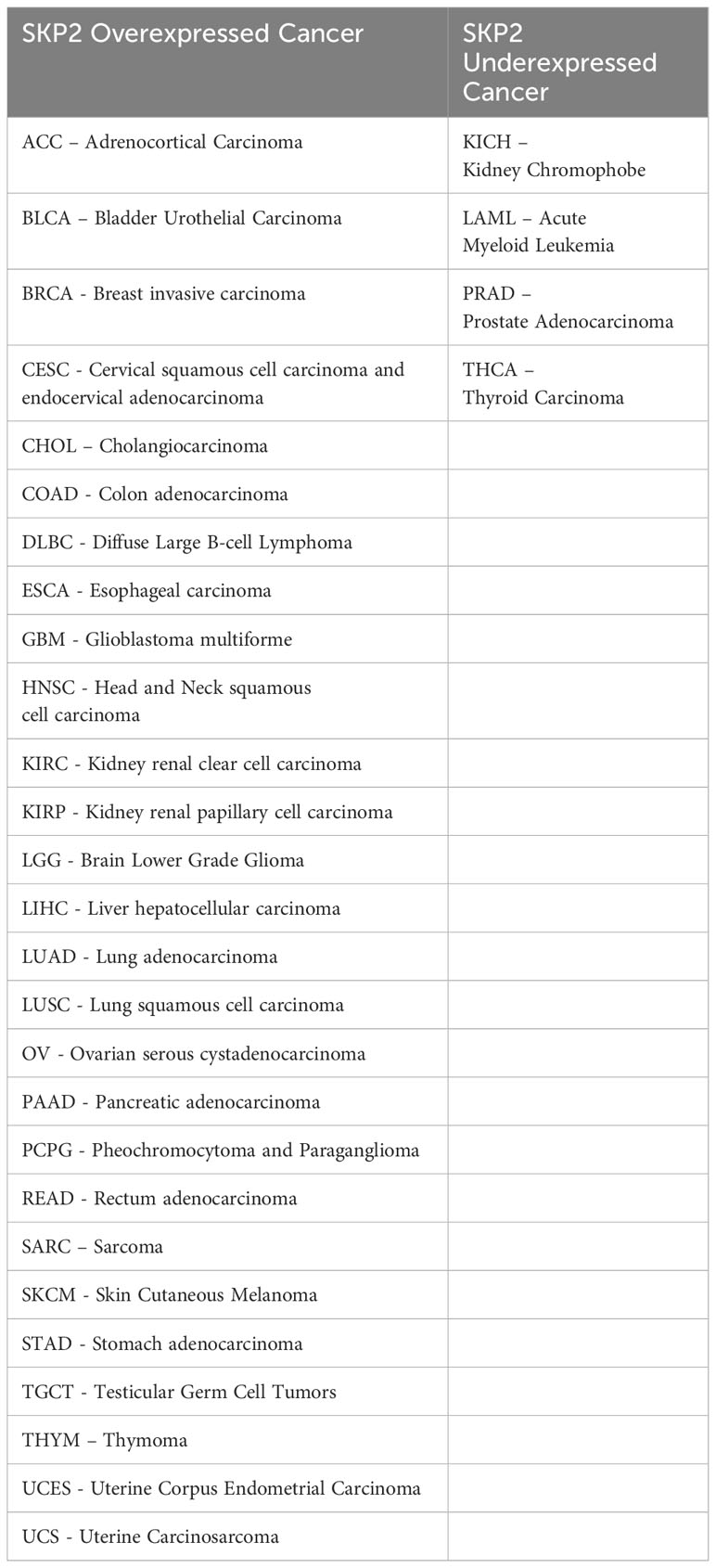
Higher expression of Skp2 is associated with tumor initiation and progression (Table 2) (44). Concurrently, the level of SKP2 oscillates during the cell cycle and is controlled by both transcriptional and post-transcriptional mechanisms. During cell cycle regulation, low expression of Skp2 is observed in both G0/G1 and late M/early G1, while a high level of SKP2 is found during G1/S transition, peaking at the S phase. Moreover, Cdk inhibitor p27 is usually stable in G0/G1 phase and unstable in the G1/S phase (8, 45). Cyclin E and E2F-1 proteolysis are essential for their rapid turnover during G1 to S phase progression, which directly increases the abundance of SKP2 during this time (46). Further, p300 acetylates SKP2 in the Nuclear Localization Signal (NLS) region, thereby mediates its localization in the cytoplasm, and enhances the stability of SKP2 (Figure 2) (43).
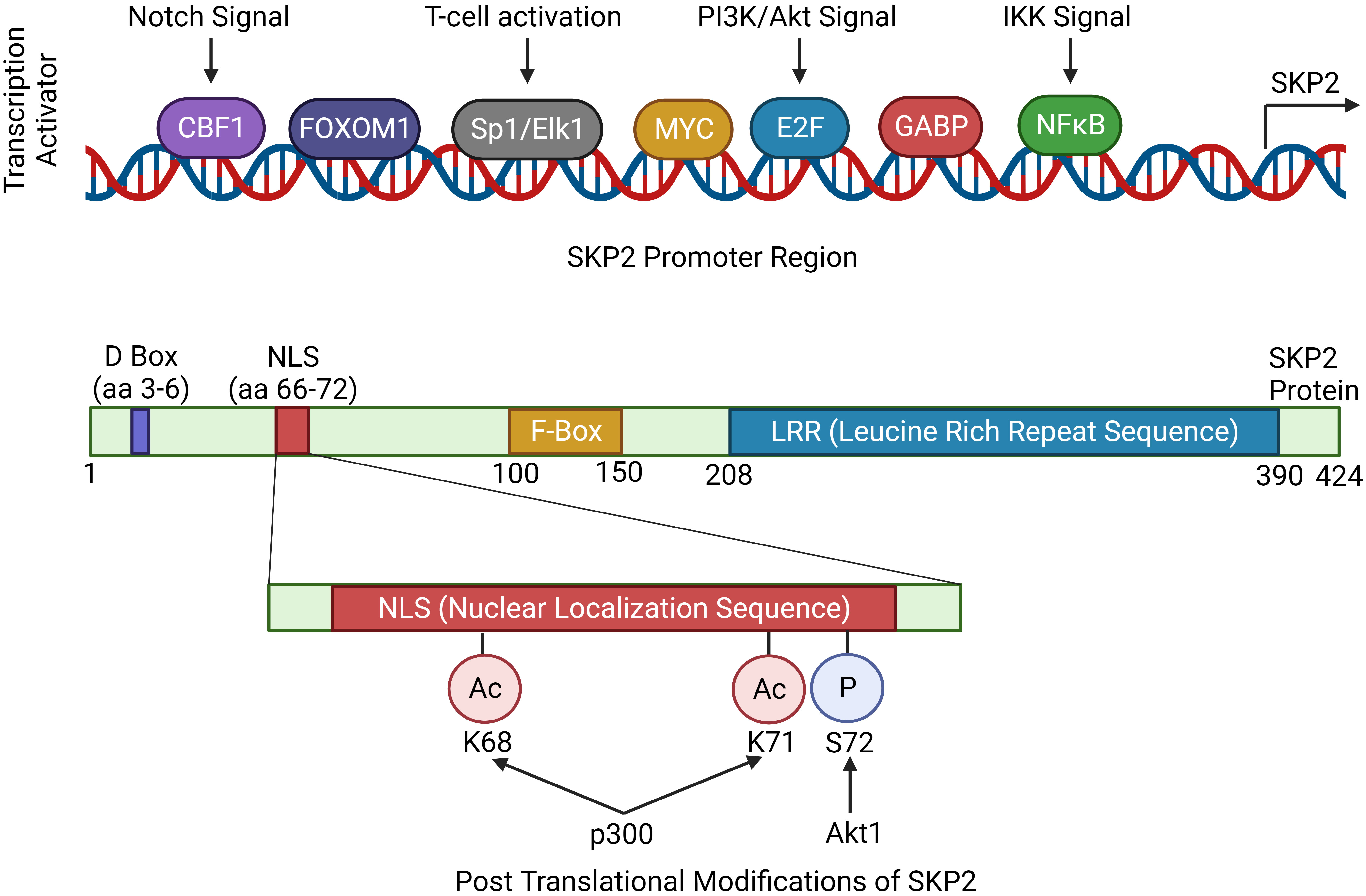
Figure 2 Regulation of Skp2 gene expression. The expression of Skp2 is intricately regulated by various signaling pathways, showcasing its significance in cellular homeostasis and proliferation. Key mitogenic signaling pathways like, Notch, PI3K/Akt and IKK, converges to SKP2 thereby modulating its expression. The coding region of Skp2 contains functional domains essential for its function. The D-box is crucial for recognition by the anaphase-promoting complex (APC/C), marking SKP2 for degradation during cell cycle progression. The NLS (nuclear localization signal) guides SKP2 into the nucleus. The F-box domain is characteristic of SKP2’s role in the SCF complex, facilitating substrate recognition. Finally, the LRR (leucine-rich repeat) domain contributes to protein-protein interactions, enabling SKP2 to engage with other components of the SCF complex and its target substrates, orchestrating precise control over cell cycle checkpoints and cellular processes.
A high level of Skp2 and a low level of p27 expressions are associated with poor prognosis in solid tumors. Similarly, an inverse correlation between Skp2 and p27 gene expression is also frequently found in hematological malignancies (47, 48). Thus an overexpression of the Skp2 gene concomitantly decreases the expression level of the p27 gene in diverse cancer types (Figure 3). However, the molecular mechanisms and the cause of p27 gene loss and elevated levels of Skp2 gene expression are not wholly investigated in all cancer types. To further support the significance of SKP2, an in-vivo xenograft mice model exhibiting high expression of the Skp2 gene was found to promote tumor growth (46). Surprisingly, following depletion of the SKP2, tumor development is dramatically reduced by inducing programmed cell death and cell senescence (49). Furthermore, another study on glioblastoma cells also demonstrated that depletion of SKP2 inhibits cancer progression via promoting cellular senescence (50). Similarly, transgenic mouse models overexpressing Skp2 have shown tumor growth in various tissues, but the cause of how SKP2 triggers neoplastic transformation is elusive (51).
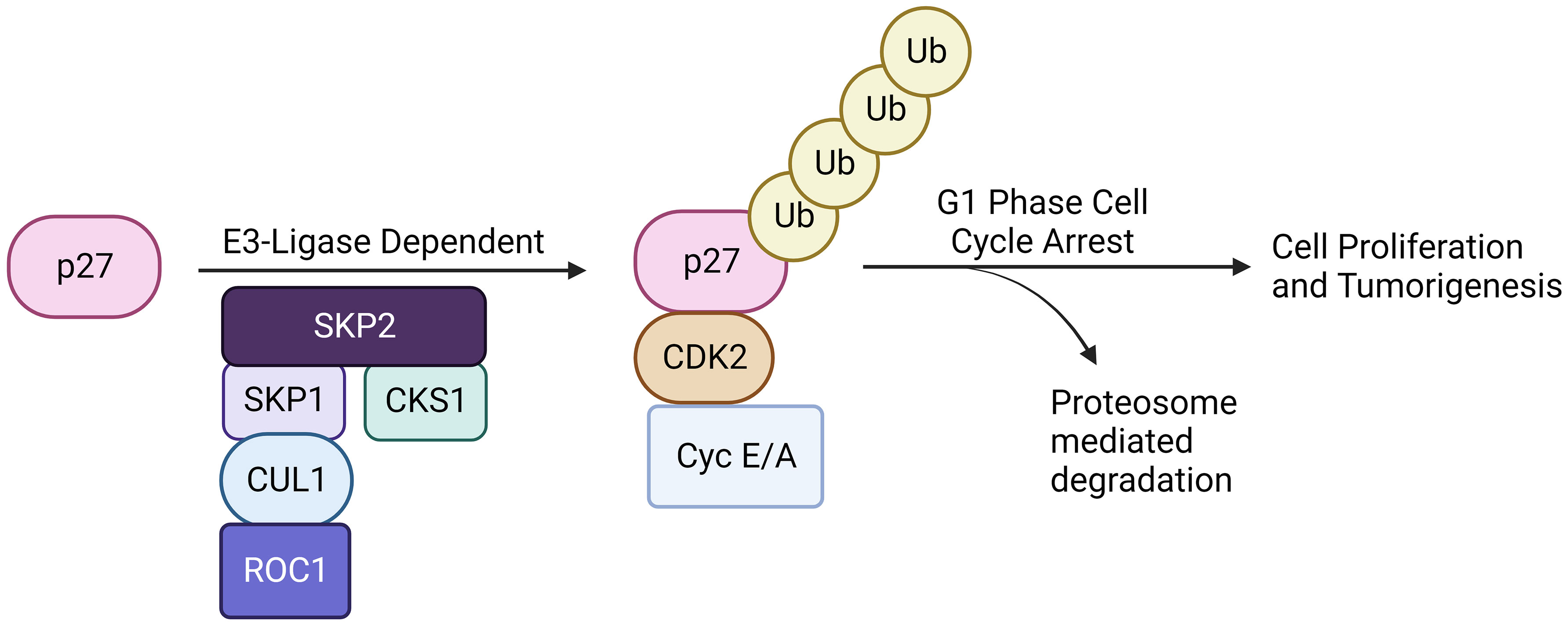
Figure 3 SKP2-p27 axis. The SKP2-p27 axis is a critical regulatory pathway governing cell cycle progression and proliferation. SKP2, a component of the SCF complex, targets the cyclin-dependent kinase inhibitor p27 for ubiquitination and subsequent proteasomal degradation. This ubiquitin-mediated destruction of p27 relieves its inhibitory effect on cell cycle progression, allowing cells to transition from G1 to S phase. Dysregulation of the SKP2-p27 axis is implicated in various cancers (solid and, including hematological malignancies), emphasizing its pivotal role in maintaining proper cell cycle control and highlighting its potential as a therapeutic targetin various malignancies.
While gene amplification may result in an enhanced Skp2 expression in cancers, oncogenic signals could also contribute to its elevated expression. Oncogenic alterations leading to higher expression of JAK2V617F mutation, BCR-ABL, and Her2/Neu, which further activates Jak/Stat, and PI3K/AKT signals thereby inducing Skp2 gene expression in malignant cells (33, 52). However, in the nucleus, BCR-ABL mediated transcription of Skp2 is associated with PI3K/AKT/SP1 pathway and mTORC2 via mTOR signaling pathways, implicating the modulation of p27 level expression. Mainly through PI3-kinase signaling, the mTORC2 pathway elevates the Skp2 expression, thereby reducing the p27 expression and initiating cancer progression (53, 54). Furthermore, p300 acetylates K68 and K71 residues of SKP2 during oncogenicity, sustaining their stability and enhancing retention in the cytoplasm (43). SKP2 promotes cellular invasion and migration by suppressing the tumor suppressor genes/protein expression and regulates its downstream targets, such as p21, p27Kip1, and FOXO1 (55). Aberrant regulation of the SKP2/p27 axis has also been noted in gastric cancer suppression, wherein MESP2 binds competitively to TCF4 (56). In addition, high SKP2 endorses cancer progression through the activation of various growth and survival-signaling pathways, for example, PTEN, ARF, pRB, FOXO1, and high Her2/Neu, etc. SKP2 acetylation and phosphorylation regulates its SCF E3 ligase activity in the cytoplasm during cancer progression, and AKT phosphorylates SKP2 at Ser72 during metastasis. The cytosolic SKP2 activates AKT and PTEN loss, implicating SKP2 translocation from the nucleus to the cytosol through Ser72 phosphorylation and induces tumor growth (57). Additionally, through neddylation, Cul-1 stabilizes the SKP2-SCF complex and negatively regulates the SKP2-SCF complex, Cul-1 dissociates from Cand1 by Cul1 neddylation and deneddylation of Cul1is mediated by Cop9-signalosome (CSN) protein complex (58). However, a complete SCF ligase activity is still largely unknown. Clinically, the elevated expression of Skp2 is recognized as a poor prognostic marker in many solid tumor cancers and hematological malignancies (1). Concerning hematological malignancies, SKP2 being a crucial regulator of the cell cycle, plays a multifaceted role. SKP2 aberrations have been implicated in malignancies like acute myeloid leukemia, chronic lymphocytic leukemia, T-cell acute lymphoblastic leukemia, chronic myelogenous leukemia, multiple myeloma, primary effusion lymphoma, Diffuse large B-cell lymphoma, extranodal natural killer (NK)/T-cell lymphoma, myeloproliferative diseases etc. (Figure 4), disrupting hematopoietic differentiation and fostering genomic instability. The following delineates the role of SKP2 in the above mentioned malignancies.
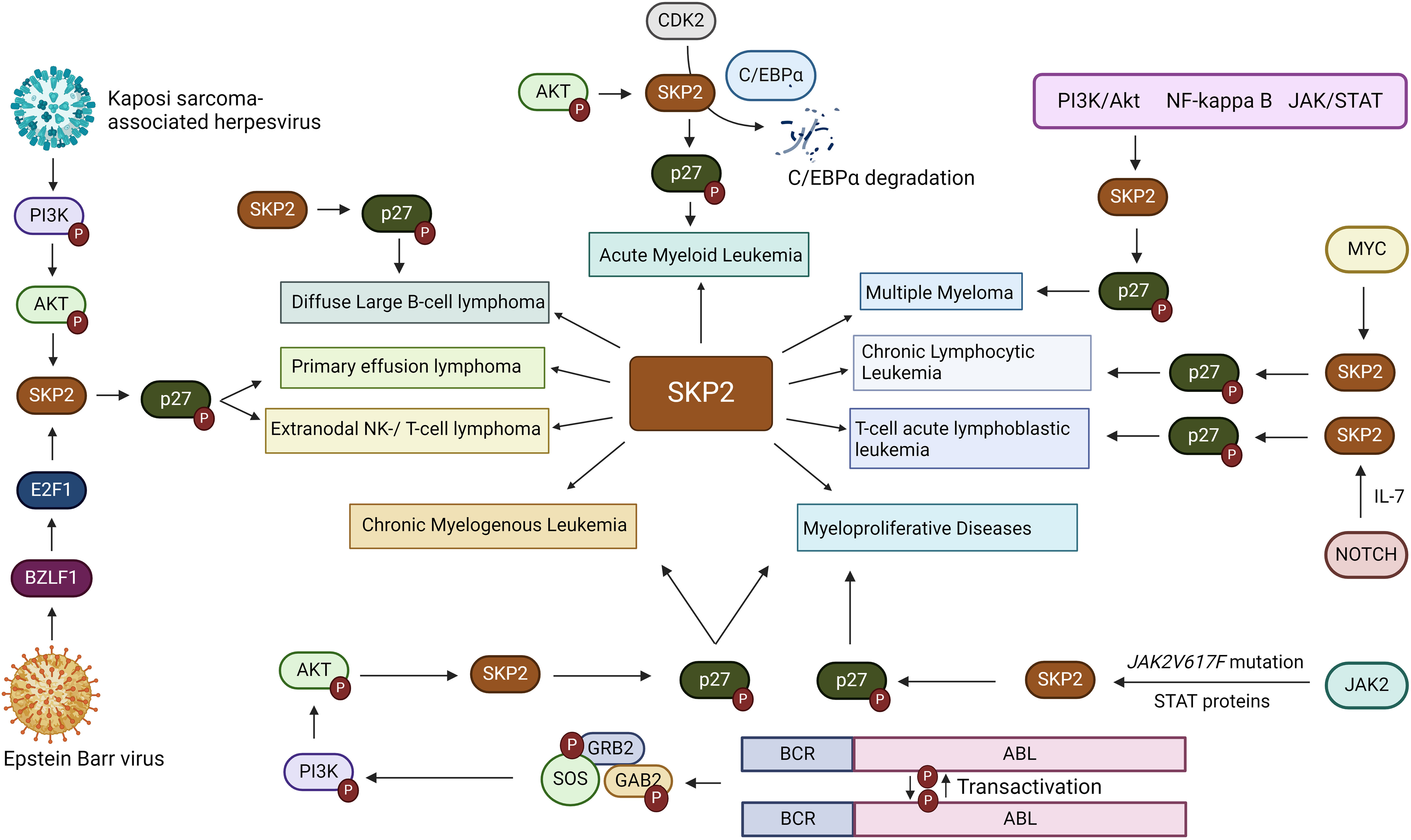
Figure 4 Involvement of SKP2 in hematological malignanices. SKP2, a critical player in hematological malignancies, features prominently in various cancers including acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), and T-cell acute lymphoblastic leukemia (T-ALL), chronic myelogenous leukemia (CML), multiple myeloma, primary effusion lymphoma (PEL), and diffuse large B-cell lymphoma (DLBCL), Extranodal natural killer (NK)/T-cell lymphoma and myeloproliferative diseases. Mediated through diverse pathways such as PI3K/Akt, NFkB, and MYC, overexpression of SKP2 is often correlated with aggressive disease and poor outcomes, highlighting its significance in cancer biology and emphasizing the need for targeted therapeutic interventions.
SKP2 in acute myeloid leukemia
Skp2 expression is recognized as an independent prognostic factor in AML. High expression of Skp2 is associated with shorter disease-free survival and overall survival. Interestingly, siRNA mediated knocking down of Skp2 in AML cell lines HL-60/A resulted in cell cycle arrest reversing the multidrug resistance by downregulating MRP gene expression (59). However, further studies are required to showcase the cause of Mrp gene modulation in AML. The RNAi-based disruption of anti-miR-196b activity or pharmacologic inhibition of the Cks1-Skp2-containing SCF E3-ubiquitin ligase complexes significantly elevated the level of p27Kip1, which induces monocytic differentiation (60), noticeable reduction of leukemogenic potential, induced apoptosis and suppressing human AML growth (48). SKP2 and p27Kip1 are localized in the cytoplasm (61), which hints that an aberrant regulatory pathway is conducted through SKP2-mediated p27Kip1 proteolysis in most AML cases (62). On the other hand, SKP2 is positively correlated with phosphorylated PTEN, suggesting that the pPTEN-SKP2 axis might be a promising therapeutic target in AML (63).
AML is a complex heterogeneous disease with diverse pathologies. There are conflicting reports on the Skp2 expression in AML. While results from TCGA shows a downregulation of SKP2 in AML (LAML), studies from other investigators reported an elevated SKP2 in AML (63). A possible reason for this apparent conflicting reports is due to the complex aetiology of AML. To improve the predictive value and therapeutic specificity of the Skp2 gene in solid and hematological malignancies, we analyzed the TCGA data (data not shown). We performed different statistical analyses on diverse populations. Our TCGA analysis relied on an online portal exploration, and the data was generated with an interactive web-portal (UALCAN tools (http://ualcan.path.uab.edu) through a TCGA-level setup. The three different RNA-sequence gene expression data and 31 different clinical cancer types’ data were used for analysis, such as 1). Relative expression of the gene(s) across tumor and normal samples, as well as in various tumor sub-groups based on individual cancer stages over and under-expressed genes in individual cancer types, tumor grade, race, body weight, or other clinical pathologic features 2) effect of gene expression level on patient survival. Finally, we used 3) in silico validation studies for target genes (derived from the GENT2 database). Results depict that leukemia showed a marginally increased trend in Skp2 expression as compared to lymphoma and Myeloma (Supplementary Table 1). Survival analysis also revealed a poor DFS (disease-free survival) with high Skp2 expression, as also seen in leukemia vs lymphoma.
Regulation of Skp2 gene in other hematological malignancies
SKP2 in Chronic Lymphocytic Leukemia (CLL)
Chronic Lymphocytic Leukemia (CLL) is the most commonly diagnosed leukemia in the Western world. CLL, also named B cell malignancy, is characterized by indolent lymph proliferative disorder, where immature B cells expressing CD5+, CD19, CD23, and CD20 B-cells progressively accumulate in the peripheral blood, bone marrow and lymph nodes (64). During the last decades, modern therapeutic approaches significantly improved to induce CLL apoptosis at various levels, but CLL remains incurable due to its drug resistance/relapse. Interestingly, the significantly higher expression [mean of 3 fold-protein] of a cell cycle inhibitor, p27, was detected in CLL tonsil and peripheral blood B lymphocyte samples as compared to healthy B cells. Besides, the expression of Myc is relatively low in CLL in comparison with normal healthy B cells. The inversely correlated MYC and p27 in CLL, and the larger set of CLL in cohort patient studies clearly demonstrated that the Skp2 gene is involved in p27 degradation. In a similar report, high Skp2 expression correlated with high Myc and low p27 expression in most of the CLL cases. On the other hand, low SKP2 samples showed high p27, and the mean MYC protein levels were significantly higher than high SKP2 levels in comparison with Tonsil and CLL. These findings demonstrated that through the MYC-SKP2-p27 axis pathway, MYC induces p27 degradation via upregulating the Skp2 gene in CLL (45).
SKP2 in T-cell acute lymphoblastic leukemia
T-cell acute lymphoblastic leukemia malignancy is a subtype of leukemia arising from thymocytes. In fact, T-ALL constitutes around 12-15% of newly diagnosed cases of ALL in pediatric patients, notable for its distinctive clinical and biological characteristics (65). Based on the current modern combination therapy, long-term therapies are needed to be improved, especially with aged group patients. The molecular mechanism of different gene functions in T-ALL is complex, including the chromosomal translocation of c-Myc, Hox 11, Tal1, and Lmo, with the T-cell receptor locus (66, 67). High prevalence activation of mutated Notch signaling pathway emerged as an important genetic component for T-ALL pathogenesis. Interestingly, in T-ALL Notch, signaling pathways are found to regulate the Skp2 expression and its protein target substrate p27. In T-ALL cells, the interaction of NOTCH 1 intracellular domain (ICD) with the Skp2 promoter triggers Skp2 expression levels and reduces p27Kip1 levels. The pharmacological agents blocking NOTCH signaling pathways reduce the expression of SKP2, and accumulate the p27Kip1, subsequently leading to G1 cell cycle arrest. Overall, NOTCH/SKP2/p27Kip1 axis might contribute to the pathogenesis of T-ALL (68).
SKP2 in Chronic Myelogenous Leukemia
Chronic Myelogenous Leukemia (CML) is the type of leukemia cancer subtype where dysregulation of myeloid cell growth in the bone marrow leads to the accumulation of undifferentiated white blood cells in the blood. CML is characterized by the translocation of BCR/ABL1 genes- chromosome t(9, 22)(q34;q11. 2). Almost 95% of CML patients have BCR/ABL translocation in the chromosomes (69). This translocation elevates the transcription level of SKP2 expression. SKP2-mediated p27Kip1 dysregulation has been observed in many types of cancers, and proteasome inhibitor BTZ reduces the expression of Skp2 in CML (70). On the other hand, the inverse relationship between SKP2 and p27Kip1 has been noticed after the gene silencing of Skp2 in CML (69, 71).
SKP2 in multiple myeloma
Multiple myeloma (MM) malignancy develops due to uncontrolled plasma cell proliferation and relapses in most patients, which remains a challenge for modern chemotherapeutic treatments. Interestingly, Myristoylated alanine-rich C-kinase substrate (MARCKS) overexpression plays an essential role in drug resistance in MM. Activated MARCKS (p-MACKS) modulates the SKP2/p27-signaling axis. SKP2 mediates E2F1-induced cell proliferation and cell cycle progression through the reduction of p27Kip1. MARCKS activation by siRNA/drug (enzastaurin) reduces the MM resistance cell growth and induces apoptosis. The current study demonstrated that targeting MARCKS-mediated SKP2 will be a more helpful therapy against MM resistance. Furthermore, cyclin-dependent kinases regulatory subunit 1 (CKS1, encoded in humans by the CKSB1 gene), cell cycle protein regulates p27Kip1, and p21CIP1 depends on Skp2 expression. Similar to the above study, SKP2/p27Kip1, and CKSB1 were also found to be inversely correlated in MM cell lines (46).
SKP2 in primary effusion lymphoma
Primary effusion lymphoma (PEL) is a rare, aggressive, immune-compromised type B cell lymphoma. It is associated with human herpesvirus type-8 infection, which commonly occurs in malignant effusions of the body cavities. In PEL, the LANA-2 gene (KSHV latent gene vIRF-3), binds to SKP2 and regulates c-MYC-dependent gene transcription by recruiting c-Myconin, its promoter regulatory region (72). Since c-MYC is a proto-oncogene, it regulates cell proliferation and survival in cancers. High expression of vIRF-3 induces the c-MYC ubiquitylation, plays a critical role in c-MYC mediated transcription, and stabilizes the c-MYC protein, leading to c-MYC-induced KSHV combined lymphomagenesis. Numerous studies have found that targeting SKP2 by proteasome inhibitor (MEG1320) or knocking down Skp2, stabilizes the p27Kip1, thereby triggering the mitochondrial-induced cell death by the caspase-dependent pathway (73). Interestingly, a plant compound Apigenin, also down-regulates the SKP2, stabilizes the p27Kip1 expression, and induces apoptosis in PEL cells (74).
Diffuse large B-cell lymphoma (DLBCL)
Diffuse large B-cell lymphoma (DLBCL) is a sub-type of B-cell cancer. In adults, 30-40% of Non-Hodgkin’s Lymphomas are DLBCL, thereby portraying DLBCL as the most common type of Non-Hodgkin Lymphona (75). One of the biggest challenges of DLBCL is that there is a relapse recorded in more than 50% of patients succeeding treatment with increased mortality (76). The cause of DLBCL resistance is still unclear. However, numerous studies demonstrate that dysregulation of oncogenic/tumor suppressor gene regulation and impairment of repair pathways contribute to developing DLBCL relapse. Interestingly, SKP2 is highly observed in DLBCL, which is significantly correlated with the worst clinical outcome compared to low SKP2-expressing patients. Further, high Skp2 in patients displayed a poor prognosis and less survival. High Skp2 correlated with Ki-67 but not with p27, demonstrating SKP2 as an independent prognostic marker of clinical outcome (77). Bortezomib (BTZ) treatment reduces SKP2 via escalation of p27Kip1protein, including XIAP, cIAP1, and survivin, implicating the SKP2/p27Kip1 signaling pathway in DLBCL pathogenesis (78). Unfortunately, in other studies, Rituximab via the CHOP-mediated pathway did not provide beneficial outcomes for DLBCL patients with high Skp2 and low p27 expression (79).
SKP2 in extranodal NK/T-cell lymphoma
Extranodal natural killer (NK)/T-cell lymphoma (ENKL) is a rare, aggressive type of malignancy in the lymph nodes besides GI tract, skin, and testis. ENKL shows poor survival among patients. The ENKT is often associated with Epstein–Barr virus (EBV) infection. The expression of Skp2 levels is significantly increased, and an inverse correlation between SKP2 and p27Kip1 was observed with patients infected with EBV and phenotype of SKP+/p27– in ENKL (80). Overall these studies suggest that SKP2 plays a major role in the pathogenesis of ENKL carcinogenesis mediated through EBV (80, 81).
Role of SKP2 in myeloproliferative diseases (MPD)
BCR-ABL induces MPD via impaired cell cycle regulation by destabilization of p27, which inhibits cyclin-dependent kinases (CDK). In contrast, BCR-ABL inhibition induces p27 and reduces Skp2, which leads to G1 arrest (33). A similar regulation pattern was also observed where leukemic cells were transformed by FLT3-ITD, JAK2V617F, and TEL-PDGFRβ, which suggests that SKP2/p27 passage may act as a common target for leukemogenic tyrosine kinases. The in vivo mice transplanted with BCR-ABL–infected SKP2_/_marrow resulted in myeloproliferative syndrome with an increased survival rate compared with recipients of BCR-ABL-expressing SKP2-/- marrow (33). At the same time, in the SKP2-/_model, the nuclear p27 expression is higher than SKP2-/-counterparts, demonstrating that leukemogenesis attenuation is regulated by high p27 levels in both MPD and CML (33, 82). The mutation of JAK2V617F commonly occurs in MPD, but in the case of its subset of polycythemia vera, homozygous JAK2V617F mutation is common. Therefore, mitotic recombination and duplication of the mutant allele are developed in MPD/CML. JAK2V617F mutation modulates the Skp2 expression through STAT3/STAT5 transcription factors on the SKP2 promoter regulatory region (52). Therefore, inhibiting SCF-SKP2 for p27 stabilization recognition may be more beneficial for a therapeutic approach in MPD/CML and other hematological malignancies.
Role of SKP2 in hematopoietic stem cells
The hematopoiesis process is a crucial step in producing diverse blood cells. This process undergoes long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs), compartments that are the primary sources of hematopoiesis. LT-HSCs self-renewal themselves in order to maintain HSC pool and differentiate into multipotent progenitors, and they can further differentiate into lymphoid progenitors and myeloid progenitors, which produce mature blood cells, whereas ST-HSCs have limited self-renewal ability to differentiate into multipotent progenitors (69, 82). However, the mechanism of HSCs quiescence is largely unknown, and re-entering the cell cycle by HSCs is very crucial. Interestingly, SKP2 is involved in regulating HSC quiescence, pool size, self-renewal capability, etc. (83). In HSCs, the SKP2 deletion stabilizes the CKIs p21Cip1, p27Kip2, P57Kip2, and p130, increasing proliferation and reducing the stem cell self-renewal capability (83). SKP2 targets SKI inhibitors that inhibit cell cycle progression from G1 to S phase (68, 84). Due to myelosuppression and post-transplantation occurrences, high expression of Skp2 is associated with neoplastic transformation, including HSC and its progenitors. High expression of Skp2 sufficiently provides hematopoietic stress.
On the other hand, depletion of SKP2 reduces HSC mitotic activity and enhances HSC quiescence, increasing pool size and maintenance (83). The depleted SKP2 results in HSC impairment during myeloablative stress because of their inability to enter the cell cycle, thereby protecting HSC regeneration. SKP2 negatively regulates cyclin D1, which might be responsible for SKP2 maintenance of HSC quiescence, pool size, and self-renewal capability (9, 85, 86). SKP2 acts as a critical regulator for HSC quiescence and self-renewal capability and gives a novel paradigm for HSCs. SKP2 maintains the HSC homing and residence in the endosteal niche. SKP2 deficiency reduces the expression of b-catenin and its target genes. Since SKP2 maintains homing of HSC succeeding the post-transplantation, SKP2 might be helpful as a predictive marker for monitoring transplantation efficiency (87). Depleted Skp2 expression enhances the sensitivity of HSCs and CMLs to chemotherapeutic drugs and triggers the long-term HSC reconstitution ability (9). Therefore, targeting SKP2 increases BM transplantation efficiency and sensitizes the cancer cell or CSC against chemotherapy. These model studies clearly demonstrate that future SKP2 targeting-based therapy will be an efficient approach against different cancer types, including HMs.
Relationship between microRNAs and Skp2 gene
MicroRNAs are small non-coding RNAs (10-24nts), and it regulate gene expression at the posttranscriptional level and play a critical role in cancer development (88). Mounting evidence displays the significant roles between miRNAs and Skp2 gene expression. MicroRNA-186 regulates Skp2 expression in pituitary tumors, induces p27Kip1-mediated cell cycle deregulation, and modulates cell proliferation. Similarly, human esophageal squamous carcinoma reduces cell proliferation and induces apoptosis (89, 90). In ovarian cancer, the expression of miR-30a-5p is low, but overexpression of miR-30a-5p reduces migration, invasion, and metastasis by posttranscriptional down-regulating SKP2 gene expression (91). Since the miR-34a is downregulated in prostate cancer, overexpressing miR-34a downregulates RhoA and suppresses the c-Myc-SKP2 -Miz1 transcriptional assembly complex c-Myc-pTEFB complex that elongates transcription of numerous genes and affects the cellular function (92). Nevertheless, the reason for Skp2 down-regulation through mir-34 has not been completely investigated yet in human renal carcinoma cells and prostate cancer (92). SKP2 mRNA is predicted to be a target of mir-7, but unfortunately, overexpressing miR-7 only reduces the SKP2 protein level but not at the transcriptional level. The SKP2-miR-7 mediated G1/S phase transition increases p27kip1 and reduces all G1 cell cycle indicators, such as Cks1, Cdk1/2, and CyclinD1/3, which suggests that overexpression of miR-7 arrests the CHO cell growth at G1 phase during cell undergoes stress (93, 94). miR-340 targets SKP2, inhibits non-small cell lung cancer tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 (95). Overexpressing miR-21-5p, miR-26-5p, and miR-30-5p in MCF-7 and tamoxifen-resistant MCF-7 cell lines showed marked reduction of SKP2 mRNA expression level (96). miR-203 targets SKP2 and regulates cell cycle and self-renewal in the hematopoietic stem cells and leukemia cells (97). Tumor suppressor miR-340 represses, the Skp2 expression, inhibits tumor cell proliferation, migration, and invasion, and induces apoptosis in hepatocellular carcinoma (95). miR-26, miR-182, miR-340, and miR-506 share the 3’UTR of both SKP2 and PCNX and suppress their expression in non-small cell lung cancer (NSCLC) (98). Ectopic expression of miR-21 down-regulates the SKP2 in ovarian cancer cells (60). Apart from miRNAs, the long noncoding RNA meg3 and miR-3163 also coordinately repress the Skp2 expression at the translation level and inhibit NSCLC cell growth, reducing NSCLC cell growth (99). miR-138 mimics or EZH2 inhibitor combined with a proteasome inhibitor, bortezomib-cavalcade, significantly reduces the MM tumors in a xenograft model by targeting RBPMS (100). To identify the SKP2 targeting miRNAs, we predicted through “TargetScan”, “MicroT-CDS” and “miRDB”: databases, where we found the seven most common miRNAs:` hsa-miR-21-5p, hsa-miR-590-5p, hsa-miR-26a-5p, hsa-miR-1297, hsa-miR-26b-5p, hsa-miR-30d-5p, hsa-miR-30a-5p (data not shown).
Possible role of SKP2 on drug resistance in hematological malignancies–HM
In HM, the patients undergoing chemotherapy don’t respond to drugs. The molecular mechanism behind cancer/tumor cell’s resistance to chemotherapy is elusive. Surprisingly, overexpression of Skp2 is associated with resistance and sensitization after pre-operative doxorubicin-based chemotherapeutically could aid in cancer cell death and successful chemotherapy in primary breast cancer patients (101, 102). SKP2 positively regulates the MAD2 via the p27-CDKs-E2F1 signaling pathway (103). Inhibition of SKP2 sensitizes paclitaxel-treated A549 and NCI-H1299 cells (103). SKP2 knockdown and/or inhibition sensitized the paclitaxel resistance prostate cancer cells, suggesting that SKP2 inhibitors might be the potential drugs against SKP2 upregulated cancers. Based on the SKP2 status in CML, USP10 inhibition significantly reduced the imatinib-sensitive and imatinib-resistant CML cell proliferation (71). Compound A (CpdA) interferes with SCF(SKP2) ligase by preventing the incorporation of SKP2 and induces G (1)/S cell-cycle arrest, SCF(SKP2)- and p27-dependent apoptosis, subsequently inducing p21 accumulation and other SCF(SKP2) substrates without affecting heat-shock protein response in MM (104). These studies indicate that SCF-SKP2 targeting agents may probably overcome the multidrug resistance mechanism and chemo-sensitize the MM cells (104). Furthermore, in breast cancer, SKP2 reactivates AKT-mediated resistance to PI3K inhibitors. Depletion of SKP2 reduces tumor growth in xenograft mice models (54). This study demonstrated that SKP2 plays a significant role in tumor progression and drug resistance. In lung cancer, small molecular inhibitors downregulated SKP2 and sensitized the lung cancer cells to paclitaxel. SKP2 has also been noted in stabilizing Mcl-1, conferring radioresistance in colorectal cancers (105).
In numerous malignancies, high SKP2 prevents apoptosis in a p53-dependent manner and promotes tumor progression and drug resistance (106). Combining SKP2 inhibitor C25 with bromocriptine sensitized the prolactinoma cells and induced apoptosis (107, 108). In multiple myeloma combinations of DT204, BTZ prevailed over drug resistance and induced apoptosis in proteasome inhibitors resistance cells. Both in vitro and in vivo model results strongly suggest that a combination of novel drug SCF-SKP2 inhibitor (DT204) and BTZ triggered synergistic anti-myeloma activity in the xenograft myeloma mouse model. Thus, targeting SCF-SKP2 by an SKP2 inhibitor combined with BTZ is a novel strategy to overcome drug resistance in MM. In addition, SKP2 inhibitor DT204 enhances the efficacy of BTZ-based therapies in multiple myeloma patients who are already BTZ-resistant (109). Proteosomal degradation of SKP2 also facilitates suppression of breast cancer growth by inducing autophagic cell death via F-box protein FBX041 (110). We previously showed that inhibition of pMARCKS potentiates BTZ-induced upregulation of p27 and p21 and downregulation of SKP2 (46). From a therapeutic perspective, it is noteworthy that targeting MARCKS can induce cell-cycle arrest and enhance apoptosis via E2F-1/SKP2/P27 axis in resistant MM cells (94). Therefore, identifying the molecular mechanism of drug resistance in HM is essential in the future. SCF-SKP2 inhibitors are the most widely used drugs to target the Ub++ proteasome system more precisely than PIs pharmacologically.
Role of epigenetic modifiers in drug resistance
SKP2 is a key regulatory protein involved in controlling cell cycle progression and the degradation of specific target proteins (111). It plays a crucial role in maintaining normal cell growth and proliferation (111). However, dysregulation of SKP2 has been implicated in various cancers, including leukemia, and is also associated with drug resistance (109). Overexpression of Skp2 in leukemia cells can contribute to drug resistance through several mechanisms (112). SKP2-mediated degradation of pro-apoptotic proteins may decrease the ability of cells to undergo apoptosis in response to chemotherapy (112). Enhanced cell cycle progression driven by SKP2 can lead to faster tumor cell growth, making it more challenging for drugs to keep pace with cell division (111, 113). SKP2 may influence DNA repair mechanisms, potentially reducing the effectiveness of DNA-damaging chemotherapeutic agents (114, 115). It is a critical player in regulating cell cycle progression and protein degradation, and its activity is intricately linked to epigenetic processes involving heritable changes in gene expression and chromatin structure without alterations in the DNA sequence (116). SKP2 can influence epigenetic regulation in multiple ways.
Regulation of epigenetic modifiers
SKP2 can target specific proteins for ubiquitin-mediated degradation. Some of these target proteins include epigenetic modifiers such as histone deacetylases (HDACs) and histone methyltransferases (84, 117). By controlling the levels of these epigenetic modifiers, SKP2 can indirectly impact the acetylation and methylation status of histones, leading to changes in chromatin structure and gene expression (118). SKP2-mediated degradation of certain epigenetic regulators can affect chromatin remodeling complexes. Alterations in chromatin structure can lead to changes in gene accessibility, potentially impacting gene expression patterns. SKP2 can interact with various transcription factors and co-factors involved in epigenetic regulation (22, 119). These interactions can modulate the activity of transcription factors, influencing their ability to bind to specific genomic regions and regulate gene expression (Figure 5).
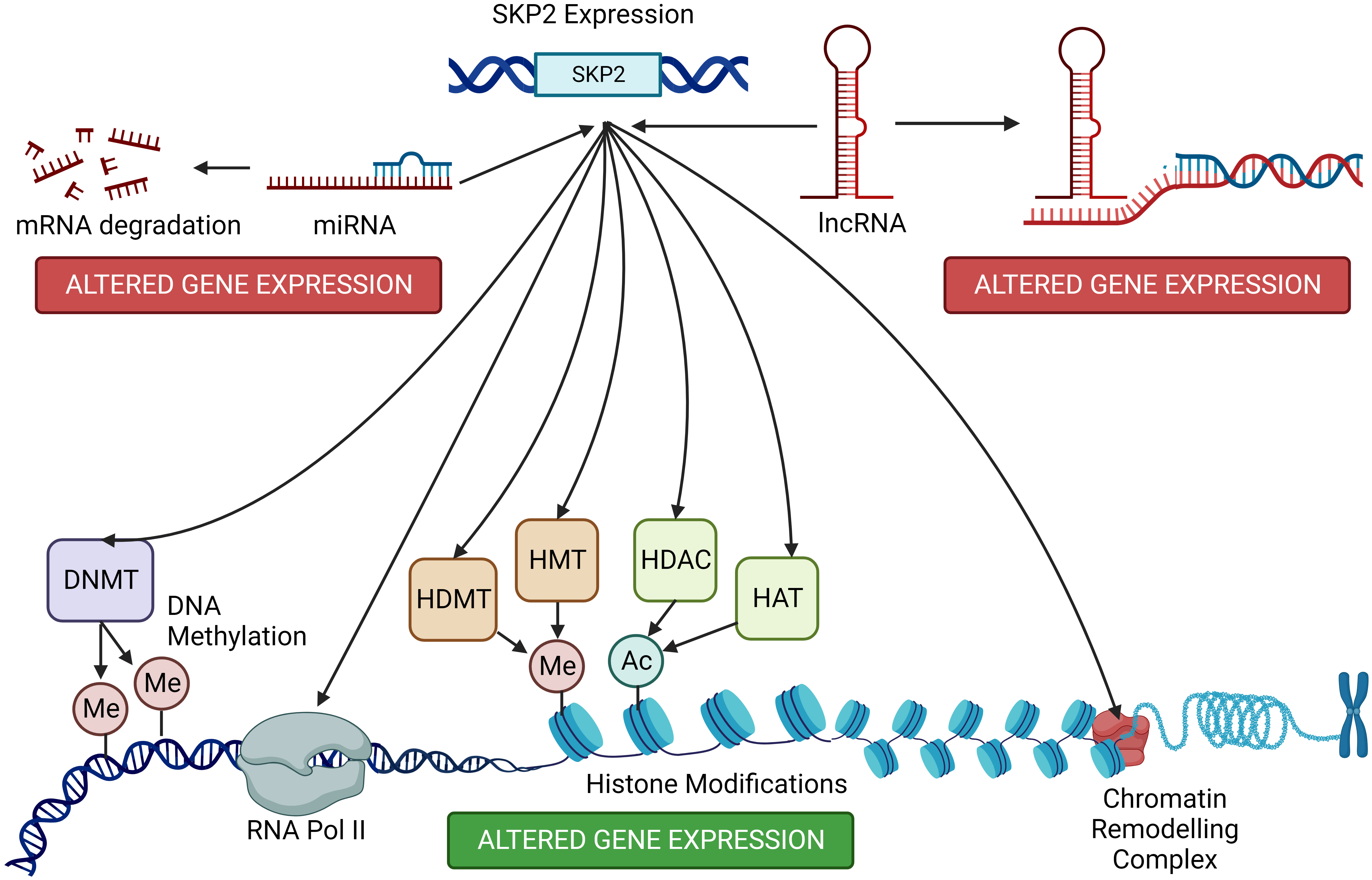
Figure 5 Host cell epigenetic modifications by SKP2. SKP2, beyond its canonical role in cell cycle regulation, influences host cell epigenetics through diverse mechanisms. Elevated SKP2 levels correlate with altered DNA methylation patterns and histone modifications, impacting gene expression. SKP2 promotes the degradation of key epigenetic regulators, disrupting the balance between chromatin modifications and transcriptional control. These modifications contribute to the development and progression of various diseases, including hematological malignancies. On the other hand, the expression of SKP2 is fine-tuned by a complex interplay of different miRNA and lnCNRA leading to its suppression or overexpression in different cancer types.
Epigenetic effects on Skp2 expression
Conversely, epigenetic modifications, such as DNA methylation and histone modifications, can also regulate the expression of Skp2. Aberrant epigenetic changes may result in dysregulated Skp2 expression, contributing to altered cell cycle control and tumorigenesis. Epigenetic modifications can directly impact the expression of genes that are targets of SKP2-mediated degradation. Altered epigenetic regulation of these genes may influence their susceptibility to SKP2-dependent degradation (22). Dysregulation of Skp2 and its interaction with epigenetic processes are associated with various diseases, including cancer (1). Aberrant Skp2 expression and epigenetic alterations can contribute to tumorigenesis, metastasis, and drug resistance (1, 120, 121). Understanding the interplay between SKP2 and epigenetics is critical for unraveling the complexities of cancer biology and other diseases. Targeting SKP2 and its associated epigenetic processes may hold promise for developing novel therapeutic strategies, especially in the context of cancers where Skp2 is dysregulated and contributes to disease progression. Additionally, research in this field continues to uncover the intricate mechanisms through which SKP2 and epigenetics intersect, providing insights into potential therapeutic targets and diagnostic markers. The regulation of SKP2 by epigenetic mechanisms plays a significant role in controlling its expression levels and activity. In the case of SKP2, several epigenetic mechanisms can in turn influence its expression.
The Skp2 gene promoter is reported to be hypermethylated in some cancer types, decreasing SKP2 expression (122). Reduced Skp2 expression due to DNA methylation can contribute to cell cycle dysregulation and impact cancer progression (122). There is also a cancer-grade specific methylation. Results from TCGA depict that leukemia showed a marginally increased trend in Skp2 expression compared to leukemia and myeloma. Survival analysis also revealed a poor DFS (disease-free survival) with high Skp2 expression, as also seen in leukemia vs lymphoma (63, 123).
Histone modifications, including acetylation and methylation of histone proteins, can influence chromatin structure and gene accessibility. While histone H3 lysine 4 (H3K4) methylation is linked to gene activation, H3K9 and H3K27 methylation are associated with gene repression. Epigenetic changes in histone modifications near the Skp2 gene are reported to modulate its transcriptional activity (124). In addition to histone modifications, specific miRNAs can target and degrade SKP2 mRNA or inhibit its translation, reducing SKP2 protein levels. Changes in miRNA expression profiles in cancer or other diseases can influence Skp2 expression through post-transcriptional regulation as discussed before (125, 126). lncRNAs have been identified as regulators of Skp2 expression, either by promoting its transcription or by destabilizing SKP2 mRNA (127).
Epigenetic changes can also influence the recruitment and activity of chromatin remodeling complexes that alter chromatin structure and gene accessibility. These complexes can either promote or inhibit the transcription of the Skp2 gene by modulating the chromatin landscape around its promoter region. Epigenetic regulation of Skp2 is particularly relevant in cancer, where dysregulated Skp2 expression can contribute to uncontrolled cell proliferation and tumorigenesis (112). Understanding the epigenetic modifications that affect SKP2 and their functional consequences is essential for developing targeted therapies that can restore normal SKP2 regulation in cancer cells. Additionally, research in this area continues to uncover the intricate details of SKP2 epigenetic regulation and its implications in various diseases. SKP2 can also promote immune evasion in cancer by regulating immune checkpoint molecules, immune response pathways, Treg function, antigen presentation, and the overall immune microenvironment (127). Understanding the role of SKP2 in immune evasion is crucial for developing strategies to enhance immune responses against cancer cells and improve the efficacy of immunotherapies (1). Targeting SKP2 or its downstream signaling pathways may represent a potential approach to mitigate immune evasion and enhance the immune system’s ability to recognize and eliminate cancer cells.
Future prospective of SKP2 inhibitors in HMs
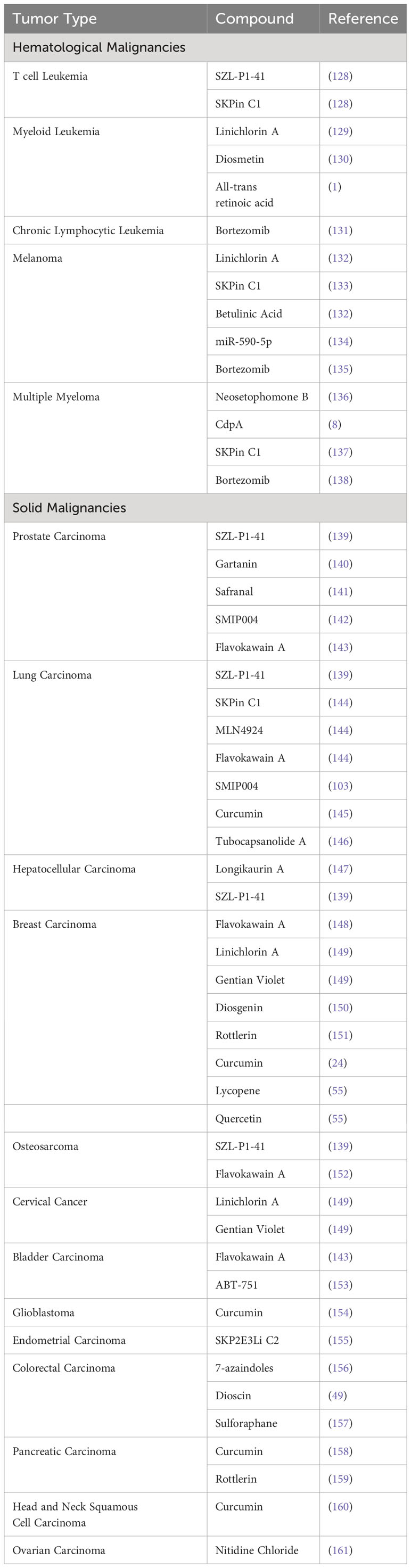
Based on several reports, downregulation of Skp2 induces the p27, promotes apoptosis, and sensitizes different types of cancers. However, further research is necessary to combat challenging tasks to identify the compound/inhibitors that are selectively employed for targeting the protein-protein interaction that holds the E3ligase together. Recently, the following inhibitors were developed against SKP2, named Bortezomib [FDA approved], Prodigiosin, Arsenic trioxide, Apigenin, curcumin, NSC689857, NSC681152, C1, C2, C16, C20, Compound A, Compound ZL25, and preclinical research compounds (Figure 6; Table 3) (8, 37, 74, 94, 109, 137, 162, 163).
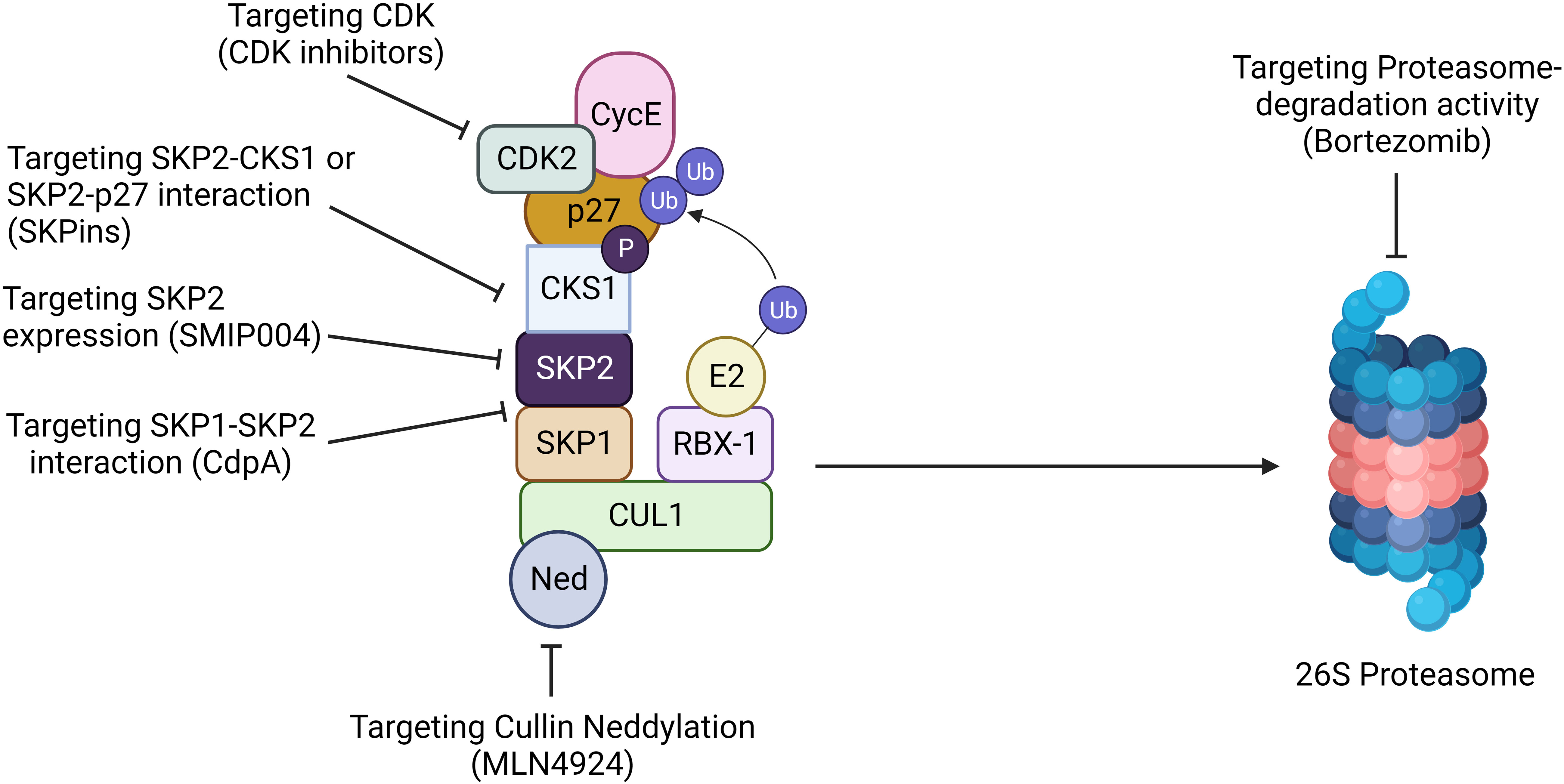
Figure 6 Targeting SKP2 for cancer therapy. Targeting SKP2 is emerging as a promising strategy for hematological malignancy. Inhibition of Cullin neddylation with MLN4924 disrupts SCF complex activity, leading to SKP2 degradation. CdpA hinders the SKP1-SKP2 interaction, while SMIP004 directly suppresses SKP2 expression. Compounds like SKPins disrupt SKP2-CKS1 or SKP2-p27 interactions, impeding cell cycle progression. Furthermore, targeting CDK inhibitors or using proteasome inhibitors like bortezomib prevents SKP2-mediated degradation of key proteins. These multifaceted approaches highlight the potential of SKP2 as a therapeutic target, offering diverse strategies to intervene in cancer progression and enhance treatment outcome.
BTZ, or pharmacological commercial name PS-341,/Valcade specifically, reversibly inhibits the 26S proteasome, an enzyme complex for regulating protein degradation under a controlled fashion. BTZ comprises of a peptide-like backbone and a boronate group, of which the latter exhibits a stronger binding affinity to the active site threonine, resulting in increased potency and selectivity toward the proteasome (164). In cancers, proteasomes’ inhibition leads to the building up of the protein substrates required for the cell cycle and apoptosis. Interestingly, BTZ suppresses the expression of Skp2 and increases the p27Kip1 expression in many types of cancers, and it has also been proven to significantly improve xenograft cancer cells in mice models. Furthermore, when BTZ combines with cisplatin, it suppresses cell proliferation and induces apoptosis by declining SKP2 and aiding in the accumulation of p27 expression. Recent studies denote combining a novel SKP2 inhibitor DT204 and BTZ synergistically induced anti-myeloma activity and sensitized drug resistance in MM. The antiproliferative effect of BTZ in CML implies that proteasomal inhibitors are highly potent, thus suggesting that it might be beneficial for a strategic intervention for CML (109).
Considering natural products, Apigenin (4′, 5, 7-trihydroxyflavone) is a natural plant product commonly found in dietary flavonoids found in various fruit and vegetables. Hussain et al. demonstrated that Apigenin triggers apoptosis in Primary effusion lymphoma (PEL) cells, suppressing the activation of AKT/PKB pathway via downregulating Skp2, hypo-phosphorylation of Rb, and accumulating p27Kip1 expression levels, which suggest that Apigenin may possibly have future therapeutic potential in PEL (74). Curcumin induces cell death by inhibiting PI3-Kinase/AKT Pathway in B-Precursor Acute Lymphoblastic Leukemia.
In addition, the pursuit of novel anti-HM therapeutic strategies does not just restrict to SKP2 inhibition, but also remains through selective inhibition of the ubiquitination–proteasome axis. To enhance specificity and selectivity in targeting, a promising avenue involves the focus on the E2–E3 complex (165, 166), given that the E2–E3 interaction imparts a high level of specificity and selectivity to the response by influencing specific ubiquitin bonds. Similarly, targeting the substrate binding domains of E3s offers a valuable opportunity. Inhibiting the interaction between a specific E3 and its target enhances specificity while minimizing off-target and side effects, potentially due to the limited impact on cellular events. As protein–protein interactions influence the specificity and selectivity of E3s, a deeper understanding of the three-dimensional structure of E3 enzymes through approaches like crystallography and cryo-electron microscopy will provide insights for developing novel inhibition strategies. Notably, Proteolysis Targeting Chimeras (PROTACs) (167) and molecular glues (168) represent effective approaches for promoting ubiquitination-mediated degradation of specific proteins, thereby contributing to increased substrate specificity.
Additionally, advancements in targeted drug delivery involve the use of cell membrane-coated nanoparticles (CNPs) and exosomes (169). CNPs, with their synthetic core containing anticancer drugs covered by a naturally derived cell membrane, offer a potential tool for precise delivery to disease sites. While promising, the translation of these strategies to clinical applications necessitates further technical improvements. Microenvironment-responsive drug-delivery systems based on nanoparticles and exosomes hold potential for guiding the release of SKP2/SCF component inhibitors in HM-specific microenvironments.
Moreover, the identification of synergistic combinations holds significant importance for advancing cancer treatment strategies. In the context of synthetic drug-target interaction, particularly those involving DNA damage-response genes and ubiquitination-mediated vulnerabilities, preclinical studies underscore the potential of combining these approaches with PARP inhibitors (170). CRISPR/Cas9- or RNAi- or shRNA-based whole genome screening, coupled with genomic and transcriptomic data analysis, contributes to predicting new synthetic lethal interactions for alternative anticancer therapeutic approaches. Recent discoveries, such as the identification of new substrates targeted by ubiquitination, including sugars alongside proteins, broaden the scope of this post-translational modification as a master regulator with potential implications for treating various pathologies, including HMs (171).
Discussion and concluding remarks
HMs account for a substantial number of newly diagnosed cases in most oncology settings. These malignancies are most common in the Western world. They occur due to several factors, such as abnormality of cytogenetic, epigenetic, gene mutations, and other environmental factors that influence the progression of HM (172, 173). In HM, blood cancer cells grow uncontrolled and function abnormally. Commonly, HM’s are categorized into three major subtypes: such as Lymphoma, Leukemia, and Myeloma (174). Among these, AML is developed among adults. More than 80% of cases re-occurred in patients over the age of 60, and 20-30% of cases are children. However, the average survival rate among all HM types is small, especially among elderly patients due to drug-resistant drug resistance or relapse after advanced chemotherapy and transplantation treatments. The molecular mechanism of drug resistance or relapse is not entirely understood in HMs. Compared to proteasomal inhibitors BTZ, E3 ligase drugs specifically block the entire protein degradation with less toxicity.
The components of E3 ligases such as MDM2, FBW7, RBX2/ROC2, RBX1/ROC1, Cullins, and many others are referred to as oncogenes or tumor suppressors; similarly, essential proteins such as p53 and Notch are associated during cancer development. The inverse correlation between SKP2 and p27 cell cycle regulators in HM and solid cancer demonstrated the shared mechanism of neoplastic transformation (175). However, Skp2 gene function has not been fully investigated in hematological malignancies. Hence, we analyzed in-silico RNA seq using available TCGA datasets. Our results (data not shown) demonstrated that the M7 AML within the French American classifications exhibited a high expression of Skp2, while there seems to be no significant difference in expression among different ethnic races. High Skp2 gene promoter methylation among African American populations illustrates the complexity of epigenetic aberration (data not shown), and increased expression of Skp2 across tumors demonstrates a common mechanism of SKP2 drug resistance. Poor survival rates among the African American population and FLT3 mutation suggested that common mutation patterns are linked with overexpression of the Skp2 gene (figure not shown). Our heatmap analysis demonstrated both positively and negatively correlated genes with SKP2; identifying the relationship with complex gene network functions that may support drug resistance mechanisms in various aspects (Supplementary Table 2). In addition, our STRING analysis (data not shown) demonstrated protein networks of SKP2 interaction, which will prompt the identification of the associated factors involved in the drug resistance mechanisms. Escalated expression of Skp2 in Basso Lymphoma results demonstrated strong evidence of a common mechanism involved in triggering the Skp2 gene in HM and a crucial player in Hematopoietic stem cells (data not shown).
Similarly, miRNAs are also found to be a crucial player in drug resistance affecting various genes. Hence, we highlighted the SKP2 gene function by projecting the essential role of miRNAs across various types of solid tumor malignancies. However in HM, only miR-203 was found to target SKP2 in Leukemia and hematopoietic stem cells.
Our miR target prediction and Venn diagram analysis demonstrated common miRNAs such as hsa-miR-21-5p, hsa-miR-590-5p, hsa-miR-26a-5p, hsa-miR-1297, hsa-miR-26b-5p, hsa-miR-30d-5p, hsa-miR-30a-5p Skp2 gene (data not shown). Overexpression of mir-21-5p induces apoptosis and cell cycle arrest by down-regulating SKP2 and overcoming Bortezomib resistance in Multiple Myeloma. Similarly, some reports demonstrated that targeting the EZH2/miR-138 axis might be a potential therapeutic target against MM (100). Nevertheless, more evidence is required to validate Skp2 gene regulation and its function in other HM, including AML (100). In the future, identifying the posttranscriptional and feedback regulatory loop mechanism of SKP2 will support the miRNA rational therapeutic approaches against relapse. Finally, we demonstrated the significance of SKP2 targeting drugs in HM, which strongly suggests a potential therapeutic strategy; however, miRNA mimics/miRNA inhibitors alongside natural products combination such as Apigenin, Dioscin, Arsenic trioxide, NSC689857, NSC681152, C1, C2 with, C16, C20, Compound A, and Compound ZL25 will open a new doorway for understanding the molecular mechanism of drug resistance or relapse in HM patients (49, 74, 160, 163, 176).
SKP2 remarkably promotes phosphorylation, ubiquitination, and degradation of PDCD4 (Programmed cell death protein 4), thereby facilitating cell proliferation and survival in breast cancer cells. SKP2 and PDCD4 displayed an inverse correlation in this cancer (177). Interestingly, its expression exhibits dynamic patterns, with some cases demonstrating overexpression in cancer samples compared to normal tissues, while others exhibit elevated levels in control samples relative to cancerous tissues. A high throughput screening identified SKP2 as a potentially novel cancer drug target (41), suggesting that pharmacologic SKP2 inactivation may limit tumor progression and overcome chemoresistance. However, in prostate cancer, SKP2 exhibits an opposite trend with high expression associated with a gain in mesenchymal and CSC-like phenotype compared with epithelial cells (178). This variability underscores the complexity of SKP2’s role in cancer progression and highlights the need for personalized therapeutic approaches. Given its diverse implications, personalized therapies targeting SKP2 may offer a tailored strategy to address the specific expression patterns observed in individual patients, potentially enhancing treatment efficacy and minimizing adverse effects. As researchers unravel the intricacies of SKP2’s involvement in hematological malignancies, the exploration of targeted interventions holds promise for advancing precision medicine in cancer therapy.
Thus, we can conclude that SKP2 is critical in regulating multiple cellular functions related to cell growth, differentiation, and cell cycle. These alterations could perturb the delicate balance and contribute to different pathological states like cancer. But the in-depth and detailed exploration of these aspects of SKP2 biology will be not only helpful in understanding cancer but also in discovering a therapeutic target. It has been observed that SKP2 dysregulation is one of the fundamental driver events for oncogenesis. These observations show that SKP2 is an oncogenic modulator; hence, its expression status is vital in cancer prognosis and determining treatment response. Small molecule activators or inhibitors for SKP2 hold tremendous promise against various cancers. It was reported that an expression of Skp2 confers drug resistance, and hence targeting SKP2 appears to be crucial for overcoming drug resistance in cancer chemotherapy. Therefore efforts have been made to develop novel inhibitors targeting SKP2 (43). However, more clinically relevant human tumor models, such as PDX and organoids and genetic mouse models should be applied to carefully evaluate the efficacy of Skp2 inhibitors. Further research is of utmost necessity to delineate the signaling pathway for SKP2 and identify its cellular target function to understand the molecular mechanism of drug resistance in different cancer types.
Author contributions
JW: Writing – original draft. RD: Data curation, Writing – review & editing. RG: Writing – review & editing. OS: Writing – review & editing. KP: Methodology, Writing – review & editing. AR: Methodology, Writing – review & editing. SR: Methodology, Writing – review & editing. MSA: Data curation, Funding acquisition, Writing – review & editing. MA: Data curation, Funding acquisition, Writing – review & editing. SK: Conceptualization, Project administration, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. SK thanks DBT (Department of Science and Technology), India for supporting the work. The authors thank the Deanship of Scientific Research at King Khalid University (KKU) for funding this research through the Research Group Program Under the Grant Number: R.G.P.2/591/44.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University (KKU) for funding this research through the Research Group Program Under the Grant Number:R.G.P.2/591/44.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1288501/full#supplementary-material
Supplementary Table 1 | SKP2 expression in Lymphoma, Leukemia and Myeloma.
Supplementary Table 2 | Positive and Negatively correlated genes with SKP2.
Abbreviations
SKP2, S-phase kinase-associated protein 2; CSC, Cancer Stem Cell; ALDH, Aldehyde dehydrogenase; AML, Acute myeloid leukemia; CDK, Cyclin dependent Kinase; SCF, Chemokine (C-X-C motif) ligand; DNMT, DNA methyltransferase; FAO, Fatty Acid Oxidation; GBM, Glioblastoma multiforme; HCC, Hepatocellular Carcinoma; PTEN, Phosphatase and tensin homolog; HIF, Hypoxia-inducible factor; IFN-γ, Interferon-gamma; TCGA, The Cancer Genome Atlas; BRCA1, Monoclonal antibody; MDSC, Myeloid-derived suppressor cells; BRCR-ABL, The breakpoint cluster region protein also known as renal carcinoma antigen NY-REN-26; mTORC, Mammalian Target of Rapamycin Complex; MAPK, Mitogen-activated protein Kinase; CUL1, Cullin1; JAK, Janus kinase; TAM, Tumor-associated macrophages; TAN, Tumor-associated neutrophils; TET, Ten-eleven translocation proteins; TGF β, Transforming growth factor β; TNBC, Triple-negative breast cancer; TNF α, Tumor necrosis factor α.
References
1. Cai Z, Moten A, Peng D, Hsu CC, Pan BS, Manne R, et al. The Skp2 pathway: A critical target for cancer therapy. Semin Cancer Biol. (2020) 67:16–33. doi: 10.1016/j.semcancer.2020.01.013
2. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. (1998) 67:425–79. doi: 10.1146/annurev.biochem.67.1.425
3. Chandra Dantu S, Nathubhai Kachariya N, Kumar A. Molecular dynamics simulations elucidate the mode of protein recognition by Skp1 and the F-box domain in the SCF complex. Proteins Struct Funct Bioinforma. (2016) 84:159–71. doi: 10.1002/prot.24963
4. Thompson LL, Rutherford KA, Lepage CC, McManus KJ. The SCF complex is essential to maintain genome and chromosome stability. Int J Mol Sci. (2021) 22(16):8544. doi: 10.3390/ijms22168544
5. Kulathu Y, Komander D. Atypical ubiquitylation-the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. (2012) 13:508–23. doi: 10.1038/nrm3394
6. Huang Z, Nie L, Xu M, Sun X-H. Notch-induced E2A degradation requires CHIP and Hsc70 as novel facilitators of ubiquitination. Mol Cell Biol. (2004) 24:8951–62. doi: 10.1128/mcb.24.20.8951-8962.2004
7. Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, et al. The cell-cycle regulatory protein Cks1 is required for SCF (Skp2) - mediated ubiquitinylation of p27. Nat Cell Biol. (2001) 3:321–4. doi: 10.1038/35060126
8. Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCFSkp2 results in p27and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. (2008) 111:4690–9. doi: 10.1182/blood-2007-09-112904
9. Tomiatti V, Istvánffy R, Pietschmann E, Kratzat S, Hoellein A, Quintanilla-Fend L, et al. Cks1 is a critical regulator of hematopoietic stem cell quiescence and cycling, operating upstream of Cdk inhibitors. Oncogene. (2015) 34:4347–57. doi: 10.1038/onc.2014.364
10. Yu ZK, Gervais JLM, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci U.S.A. (1998) 95:11324–9. doi: 10.1073/pnas.95.19.11324
11. Yeh KH, Kondo T, Zheng J, Tsvetkov LM, Blair J, Zhang H. The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem Biophys Res Commun. (2001) 281:884–90. doi: 10.1006/bbrc.2001.4442
12. Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27 (Kip1), polyploidy and centrosome overduplication. EMBO J. (2000) 19:2069–81. doi: 10.1093/emboj/19.9.2069
13. Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, et al. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. (2005) 18:699–709. doi: 10.1016/j.molcel.2005.05.011
14. Arbini AA, Greco M, Yao JL, Bourne P, Marra E, Hsieh JT, et al. Skp2 overexpression is associated with loss of BRCA2 protein in human prostate cancer. Am J Pathol. (2011) 178:2367–76. doi: 10.1016/j.ajpath.2011.01.050
15. Méndez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. (2002) 9:481–91. doi: 10.1016/S1097-2765(02)00467-7
16. Kondo T, Kobayashi M, Tanaka J, Yokoyama A, Suzuki S, Kato N, et al. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J Biol Chem. (2004) 279:27315–9. doi: 10.1074/jbc.M314023200
17. Lin YW, Yang JL. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. J Biol Chem. (2006) 281:915–26. doi: 10.1074/jbc.M508720200
18. Nie L, Wu H, Sun XH. Ubiquitination and degradation of Tal1/SCL are induced by Notch signaling and depend on Skp2 and CHIP. J Biol Chem. (2008) 283:684–92. doi: 10.1074/jbc.M704981200
19. Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. (1999) 1:14–9. doi: 10.1038/8984
20. Liu Y, Hedvat CV, Mao S, Zhu X-H, Yao J, Nguyen H, et al. The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF Skp2. Mol Cell Biol. (2006) 26:3114–23. doi: 10.1128/mcb.26.8.3114-3123.2006
21. Hiramatsu Y, Kitagawa K, Suzuki T, Uchida C, Hattori T, Kikuchi H, et al. Degradation of Tob1 mediated by SCFSkp2-dependent ubiquitination. Cancer Res. (2006) 66:8477–83. doi: 10.1158/0008-5472.CAN-06-1603
22. Von Der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. (2003) 11:1189–200. doi: 10.1016/S1097-2765(03)00193-X
23. Charrasse S, Carena I, Brondani V, Klempnauer KH, Ferrari S. Degradation of B-Myb by ubiquitin-mediated proteolysis: Involvement of the CdC34-SCF(p45Skp2) pathway. Oncogene. (2000) 19:2986–95. doi: 10.1038/sj.onc.1203618
24. Jia T, Zhang L, Duan Y, Zhang M, Wang G, Zhang J, et al. The differential susceptibilities of MCF-7 and MDA-MB-231 cells to the cytotoxic effects of curcumin are associated with the PI3K/Akt-SKP2- Cip/Kips pathway. Cancer Cell Int. (2014) 14:1–14. doi: 10.1186/s12935-014-0126-4
25. Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Genes Dev. (2002) 16:2946–57. doi: 10.1101/gad.1011202
26. Liu H, Cheng EHY, Hsieh JJD. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: A critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. (2007) 21:2385–98. doi: 10.1101/gad.1574507
27. Tokarz S, Berset C, La Rue J, Friedman K, Nakayama KI, Nakayama K, et al. The ISG15 isopeptidase UBP43 is regulated by proteolysis via the SCF Skp2 ubiquitin ligase. J Biol Chem. (2004) 279:46424–30. doi: 10.1074/jbc.M403189200
28. Vuillier F, Li Z, Commere PH, Dynesen LT, Pellegrini S. USP18 and ISG15 coordinately impact on SKP2 and cell cycle progression. Sci Rep. (2019) 9:1–11. doi: 10.1038/s41598-019-39343-7
29. Song MS, Song SJ, Kim SJ, Nakayama K, Nakayama KI, Lim DS. Skp2 regulates the antiproliferative function of the tumor suppressor RASSF1A via ubiquitin-mediated degradation at the G1-S transition. Oncogene. (2008) 27:3176–85. doi: 10.1038/sj.onc.1210971
30. Liang M, Liang Y, Wrighton K, Ungermannova D, Wang X, Brunicardi FC, et al. Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol. (2004) 24:7524–37. doi: 10.1128/MCB.24.17.7524
31. Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbé JC, Lorca T, et al. Interaction between cyclin T1 and SCF SKP2 targets CDK9 for ubiquitination and degradation by the proteasome. Mol Cell Biol. (2001) 21:7956–70. doi: 10.1128/mcb.21.23.7956-7970.2001
32. Oh K-J, Kalinina A, Wang J, Nakayama K, Nakayama KI, Bagchi S. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and cullin 1- and Skp2-containing E3 ligase. J Virol. (2004) 78:5338–46. doi: 10.1128/jvi.78.10.5338-5346.2004
33. Agarwal A, Bumm TGP, Corbin AS, O’Hare T, Loriaux M, Van Dyke J, et al. Absence of SKP2 expression attenuates BCR-ABL-induced myeloproliferative disease. Blood. (2008) 112:1960–70. doi: 10.1182/blood-2007-09-113860
34. Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. (2013) 328:198–206. doi: 10.1016/j.canlet.2012.09.017
35. Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ERE, Finnin MS, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. (2000) 408:381–6. doi: 10.1038/35042620
36. Spratt DE, Wu K, Kovacev J, Pan ZQ, Shaw GS. Selective recruitment of an E2∼ubiquitin complex by an E3 ubiquitin ligase. J Biol Chem. (2012) 287:17374–85. doi: 10.1074/jbc.M112.353748
37. Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of skp2-mediated p27 degradation. Chem Biol. (2012) 19:1515–24. doi: 10.1016/j.chembiol.2012.09.015
38. Xie J, Jin Y, Wang G. The role of SCF ubiquitin-ligase complex at the beginning of life. Reprod Biol Endocrinol. (2019) 17:1–9. doi: 10.1186/s12958-019-0547-y
39. Yao Z, Seeliger M, Itzhaki L, Robinson C. Investigation of protein interations between Cks1 and Skp2 using hydrogen exchange nano-electrospray mass spectrometry. Protein Sci. (2004) 13:86–6.
40. Huang Y, Zhao Q, Zhou CX, Gu ZM, Li D, Xu HZ, et al. Antileukemic roles of human phospholipid scramblase 1 gene, evidence from inducible PLSCR1-expressing leukemic cells. Oncogene. (2006) 25:6618–27. doi: 10.1038/sj.onc.1209677
41. Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: A novel potential therapeutic target for prostate cancer. Biochim Biophys Acta - Rev Cancer. (2012) 1825:11–7. doi: 10.1016/j.bbcan.2011.09.002
42. Kimura Y, Nagao A, Fujioka Y, Satou A, Taira T, Iguchi-Ariga SMM, et al. MM-1 facilitates degradation of c-Myc by recruiting proteasome and a novel ubiquitin E3 ligase. Int J Oncol. (2007) 31:829–36. doi: 10.3892/ijo.31.4.829
43. Lee Y, Lim H-S. Skp2 inhibitors: novel anticancer strategies. Curr Med Chem. (2016) 23:2363–79. doi: 10.2174/0929867323666160510122624
44. Yang G, Ayala G, De Marzo A, Tian W, Frolov A, Wheeler TM, et al. Elevated Skp2 protein expression in human prostate cancer: Association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin Cancer Res. (2002) 8:3419–26.
45. Caraballo JM, Acosta JC, Cortés MA, Albajar M, Teresa Gómez-Casares M, Batlle-López A, et al. High p27 protein levels in chronic lymphocytic leukemia are associated to low Myc and Skp2 expression, confer resistance to apoptosis and antagonize Myc effects on cell cycle. Oncotarget. (2014) 5:4694–708. doi: 10.18632/oncotarget.2100
46. Yang Y, Chen Y, Saha MN, Chen J, Evans K, Qiu L, et al. Targeting phospho-MARCKS overcomes drug-resistance and induces antitumor activity in preclinical models of multiple myeloma. Leukemia. (2015) 29:715–26. doi: 10.1038/leu.2014.255
47. Bretones G, Acosta JC, Caraballo JM, Ferrándiz N, Gómez-Casares MT, Albajar M, et al. SKP2 oncogene is a direct MYC target gene and MYC down-regulates p27 KIP1 through Skp2 in human leukemia cells. J Biol Chem. (2011) 286:9815–25. doi: 10.1074/jbc.M110.165977
48. Grey W, Ivey A, Milne TA, Haferlach T, Grimwade D, Uhlmann F, et al. The Cks1/Cks2 axis fine-tunes Mll1 expression and is crucial for MLL-rearranged leukaemia cell viability. Biochim Biophys Acta - Mol Cell Res. (2018) 1865:105–16. doi: 10.1016/j.bbamcr.2017.09.009
49. Zhou L, Yu X, Li M, Gong G, Liu W, Li T, et al. Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine. (2020) 51:1–11. doi: 10.1016/j.ebiom.2019.11.031
50. Seo SB, Lee JJ, Yun HH, Im CN, Kim YS, Ko JH, et al. 14-3-3β Depletion drives a senescence program in glioblastoma cells through the ERK/SKP2/p27 pathway. Mol Neurobiol. (2018) 55:1259–70. doi: 10.1007/s12035-017-0407-8
51. Hulit J, Lee RJ, Li Z, Wang C, Katiyar S, Yang J, et al. p27Kip1 repression of ErbB2-induced mammary tumor growth in transgenic mice involves Skp2 and Wnt/β-catenin signaling. Cancer Res. (2006) 66:8529–41. doi: 10.1158/0008-5472.CAN-06-0149
52. Furuhata A, Kimura A, Shide K, Shimoda K, Murakami M, Ito H, et al. p27 deregulation by Skp2 overexpression induced by the JAK2V617 mutation. Biochem Biophys Res Commun. (2009) 383:411–6. doi: 10.1016/j.bbrc.2009.04.015
53. Shapira M, Kakiashvili E, Rosenberg T, Hershko DD. The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res. (2006) 8:1–9. doi: 10.1186/bcr1533
54. Clement E, Inuzuka H, Nihira NT, Wei W, Toker A. Skp2-dependent reactivation of AKT drives resistance to PI3K inhibitors. Sci Signal. (2018) 11:521. doi: 10.1126/scisignal.aao3810
55. Huang H-C, Lin C-L, Lin J-K. 1,2,3,4,6-penta-O-galloyl-β-D-glucose, quercetin, curcumin and lycopene induce cell-cycle arrest in MDA-MB-231 and BT474 cells through downregulation of Skp2 protein. J Agric Food Chem. (2011) 59:6765–75. doi: 10.1021/jf201096v
56. Ge L, Zhao G, Lan C, Song H, Qi D, Huang P, et al. MESP2 binds competitively to TCF4 to suppress gastric cancer progression by regulating the SKP2/p27 axis. Cell Death Discovery. (2023) 9:1–13. doi: 10.1038/s41420-023-01367-4
57. Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APC-Cdh1-mediated Skp2 destruction. Nat Cell Biol. (2009) 11:397–408. doi: 10.1038/ncb1847
58. Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCF Skp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci U S A. (2006) 103:11515–20. doi: 10.1073/pnas.0603921103
59. Xiao J, Yin S, Li Y, Xie S, Nie D, Ma L, et al. SKP2 siRNA inhibits the degradation of P27kip1 and down-regulates the expression of MRP in HL-60/A cells. Acta Biochim Biophys Sin (Shanghai). (2009) 41:699–708. doi: 10.1093/abbs/gmp058
60. Meyer SE, Muench DE, Rogers AM, Newkold TJ, Orr E, O’Brien E, et al. miR-196b target screen reveals mechanisms maintaining leukemia stemness with therapeutic potential. J Exp Med. (2018) 215:2115–36. doi: 10.1084/jem.20171312
61. Sampson C, Wang Q, Otkur W, Zhao H, Lu Y, Liu X, et al. The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin Transl Med. (2023) 13(3):e1204. doi: 10.1002/ctm2.1204
62. Winteringham LN, Kobelke S, Williams JH, Ingley E, Klinken SP. Myeloid Leukemia Factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27Kip1 accumulation. Oncogene. (2004) 23:5105–9. doi: 10.1038/sj.onc.1207661
63. Min YH, Cheong JW, Lee MH, Kim JY, Lee ST, Hahn JS, et al. Elevated S-phase kinase-associated protein 2 protein expression in acute myelogenous leukemia: Its association with constitutive phosphorylation of phosphatase and tensin homologue protein and poor prognosis. Clin Cancer Res. (2004) 10:5123–30. doi: 10.1158/1078-0432.CCR-04-0136
64. Darwiche W, Gubler B, Marolleau JP, Ghamlouch H. Chronic lymphocytic leukemia B-cell normal cellular counterpart: Clues from a functional perspective. Front Immunol. (2018) 9:683. doi: 10.3389/fimmu.2018.00683
65. Raetz EA, Teachey DT. T-cell acute lymphoblastic leukemia. Pediatr Hematol Malig. (2016) 2016:580–8. doi: 10.1182/asheducation-2016.1.580
66. Curtis DJ, McCormack M. The molecular basis of Lmo2-induced T-cell acute lymphoblastic leukemia. Clin Cancer Res. (2010) 16:5618–23. doi: 10.1158/1078-0432.CCR-10-0440
67. Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. (2012) 122:3398–406. doi: 10.1172/JCI61269
68. Dohda T, Maljukova A, Liu L, Heyman M, Grandér D, Brodin D, et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res. (2007) 313:3141–52. doi: 10.1016/j.yexcr.2007.04.027
69. Chen JY, Wang MC, Hung WC. Bcr-Abl-induced tyrosine phosphorylation of Emi1 to stabilize Skp2 protein via inhibition of ubiquitination in chronic myeloid leukemia cells. J Cell Physiol. (2011) 226:407–13. doi: 10.1002/jcp.22346
70. Iskandarani A, Bhat AA, Siveen KS, Prabhu KS, Kuttikrishnan S, Khan MA, et al. Bortezomib-mediated downregulation of S-phase kinase protein-2 (SKP2) causes apoptotic cell death in chronic myelogenous leukemia cells. J Transl Med. (2016) 14:1–12. doi: 10.1186/s12967-016-0823-y
71. Andreu EJ, Lledó E, Poch E, Ivorra C, Albero MP, Martínez-Climent JA, et al. BCR-ABL induces the expression of Skp2 through the PI3K pathway to promote p27Kip1 degradation and proliferation of chronic myelogenous leukemia cells. Cancer Res. (2005) 65:3264–72. doi: 10.1158/0008-5472.CAN-04-1357
72. Baresova P, Pitha PM, Lubyova B. Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J Biol Chem. (2012) 287:16199–208. doi: 10.1074/jbc.M111.335216
73. Hussain AR, Ahmed M, Ahmed SO, Al-Thari S, Khan AS, Razack S, et al. Proteasome inhibitor MG-132 mediated expression of p27Kip1 via S-phase kinase protein 2 degradation induces cell cycle coupled apoptosis in primary effusion lymphoma cells. Leuk Lymphoma. (2009) 50:1204–13. doi: 10.1080/10428190902951799
74. Hussain AR, Khan AS, Ahmed SO, Ahmed M, Platanias LC, Al-Kuraya KS, et al. Apigenin induces apoptosis via downregulation of S-phase kinase-associated protein 2-mediated induction of p27Kip1 in primary effusion lymphoma cells. Cell Prolif. (2010) 43:170–83. doi: 10.1111/j.1365-2184.2009.00662.x
75. Susanibar-Adaniya S, Barta S. 2021 Update on Diffuse large B cell lymphoma: A review of current data and potential applications on risk stratification and management. Am J Hematol. (2021) 96:617–29. doi: 10.1002/ajh.26151
76. Chen B, Zou C, Wu J. Diffuse large B-cell non-Hodgkin lymphoma involving the unilateral carotid space in an elderly man: A case report. Mol Clin Oncol. (2017) 6:115–8. doi: 10.3892/mco.2016.1089
77. Seki R, Okamura T, Koga H, Yakushiji K, Hashiguchi M, Yoshimoto K, et al. Prognostic significance of the F-box protein Skp2 expression in diffuse large B-cell lymphoma. Am J Hematol. (2003) 73:230–5. doi: 10.1002/ajh.10379
78. Uddin S, Hussain A, Ahmed M, Belgaumi A, Al-Dayel F, Ajarim D, et al. S-phase kinase protein 2 is an attractive therapeutic target in a subset of diffuse large B-cell lymphoma. J Pathol. (2008) 216:483–94. doi: 10.1002/path.2433
79. Abdou AG, Asaad NY, Abd El-Wahed MM, Samaka RM, Gad Allah MS. The prognostic value of Skp2 expression in Egyptian diffuse large B-cell lymphoma. Appl Immunohistochem Mol Morphol. (2012) 20:47–55. doi: 10.1097/PAI.0b013e318219a19f
80. Xiang-Lan M, Zu-Lan S, Dan H, Bi-Hong S, Ya-Qin P, Han-Liang L. Skp2/p27 expression profile is correlated with Epstein-Barr virus status in extranodal nasal-type natural killer cell lymphoma. Transl Res. (2008) 151:303–8. doi: 10.1016/j.trsl.2008.04.004
81. Guo HQ, Pu XX, Guo CC, Rao HL, Li HR, Lin TY. The role of Skp2 in extranodal NK/T-cell lymphoma. Chin J Cancer. (2010) 29:567–71. doi: 10.5732/cjc.009.10645
82. Chen JY, Wang MC, Hung WC. Transcriptional activation of Skp2 by BCR-ABL in K562 chronic myeloid leukemia cells. Leuk Res. (2009) 33:1520–4. doi: 10.1016/j.leukres.2009.03.007
83. Rodriguez S, Wang L, Mumaw C, Srour EF, Lo Celso C, Nakayama KI, et al. The SKP2 E3 ligase regulates basal homeostasis and stress-induced regeneration of HSCs. Blood. (2011) 117:6509–19. doi: 10.1182/blood-2010-11-321521
84. Borriello A, Naviglio S, Bencivenga D, Caldarelli I, Tramontano A, Speranza MC, et al. Histone Deacetylase Inhibitors Increase p27Kip1 by Affecting Its Ubiquitin-Dependent Degradation through Skp2 Downregulation. Oxid Med Cell Longev. (2016) 2016:2481865. doi: 10.1155/2016/2481865
85. Wang J, Han F, Wu J, Lee SW, Chan CH, Wu CY, et al. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. (2011) 118:5429–38. doi: 10.1182/blood-2010-10-312785
86. Curiel-Lewandrowski C, Yamasaki H, Si CP, Jin X, Zhang Y, Richmond J, et al. Loss of nuclear pro-IL-16 facilitates cell cycle progression in human cutaneous T cell lymphoma. J Clin Invest. (2011) 121:4838–49. doi: 10.1172/JCI41769
87. Wang J, Han F, Lee S-W, Wu J, Chan C-H, Zhang X, et al. E3-ligase Skp2 regulates β-catenin expression and maintains hematopoietic stem cell homing. Biochem Biophys Res Commun. (2014) 445:566–671. doi: 10.1016/j.bbrc.2014.02.042
88. Mudduluru G, George-William JN, Muppala S, Asangani IA, Kumarswamy R, Nelson LD, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep. (2011) 31:185–97. doi: 10.1042/BSR20100065
89. He W, Feng J, Zhang Y, Wang Y, Zang W, Zhao G. MicroRNA-186 inhibits cell proliferation and induces apoptosis in human esophageal squamous cell carcinoma by targeting SKP2. Lab Investig. (2016) 96:317–24. doi: 10.1038/labinvest.2015.134
90. He Z, Chen L, Wang Q, Yin C, Hu J, Hu X, et al. MicroRNA-186 targets SKP2 to induce p27Kip1-mediated pituitary tumor cell cycle deregulation and modulate cell proliferation. Korean J Physiol Pharmacol. (2019) 23:171–9. doi: 10.4196/kjpp.2019.23.3.171
91. Qi F, He T, Jia L, Song N, Guo L, Ma X, et al. The miR-30 family inhibits pulmonary vascular hyperpermeability in the premetastatic phase by direct targeting of Skp2. Clin Cancer Res. (2015) 21:3071–80. doi: 10.1158/1078-0432.CCR-14-2785
92. Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Deng G, et al. Microrna-34a modulates c-Myc transcriptional complexes to suppress Malignancy in human prostate cancer cells. PLoS One. (2012) 7:1–12. doi: 10.1371/journal.pone.0029722
93. Sanchez N, Gallagher M, Lao N, Gallagher C, Clarke C, Doolan P, et al. MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including skp2 and Psme3. PLoS One. (2013) 8:1–10. doi: 10.1371/journal.pone.0065671
94. Feng S, Wang Y, Zhang R, Yang G, Liang Z, Wang Z, et al. Curcumin exerts its antitumor activity through regulation of miR-7/Skp2/p21 in nasopharyngeal carcinoma cells. Onco Targets Ther. (2017) 10:2377–88. doi: 10.2147/OTT.S130055
95. Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, et al. MiR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. (2015) 34:3240–50. doi: 10.1038/onc.2014.267
96. Lin YS, Lin YY, Yang YH, Lin CL, Kuan FC, Lu CN, et al. Antrodia cinnamomea extract inhibits the proliferation of tamoxifen-resistant breast cancer cells through apoptosis and skp2/microRNAs pathway. BMC Complement Altern Med. (2018) 18:1–11. doi: 10.1186/s12906-018-2204-y
97. Jackson SJ, Zhang Z, Feng D, Flagg M, O’Loughlin E, Wang D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Dev. (2013) 140:1882–91. doi: 10.1242/dev.089649
98. Li J, Tian H, Pan J, Jiang N, Yang J, Zhou C, et al. Pecanex functions as a competitive endogenous RNA of S-phase kinase associated protein 2 in lung cancer. Cancer Lett. (2017) 406:36–46. doi: 10.1016/j.canlet.2017.07.030
99. Su L, Han D, Wu J, Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumor Biol. (2016) 37:3925–31. doi: 10.1007/s13277-015-4151-2
100. Rastgoo N, Pourabdollah M, Abdi J, Reece D, Chang H. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia. (2018) 32:2471–82. doi: 10.1038/s41375-018-0140-y
101. Bar-On O, Shapira M, Hershko DD. Differential effects of doxorubicin treatment on cell cycle arrest and Skp2 expression in breast cancer cells. Anticancer Drugs. (2007) 18:1113–21. doi: 10.1097/CAD.0b013e3282ef4571
102. Davidovich S, Ben-Izhak O, Shapira M, Futerman B, Hershko DD. Over-expression of Skp2 is associated with resistance to preoperative doxorubicin-based chemotherapy in primary breast cancer. Breast Cancer Res. (2008) 10:1–10. doi: 10.1186/bcr2122
103. Huang T, Yang L, Wang G, Ding G, Peng B, Wen Y, et al. Inhibition of Skp2 sensitizes lung cancer cells to paclitaxel. Onco Targets Ther. (2017) 10:439–46. doi: 10.2147/OTT.S125789
104. Shi L, Wang S, Zangari M, Xu H, Cao TM, Xu C, et al. Over-expression of CKS1B activates both MEK/ERK and JAK/ STAT3 signaling pathways and promotes myeloma cell drugresistance. Oncotarget. (2010) 1:22–33. doi: 10.18632/oncotarget.105
105. Yu X, Zhou L, Liu W, Liu L, Gao F, Li W, et al. Skp2 stabilizes Mcl-1 and confers radioresistance in colorectal cancer. Cell Death Dis. (2022) 13:1–13. doi: 10.1038/s41419-022-04685-0
106. Kitagawa M, Lee SH, McCormick F. Skp2 Suppresses p53-Dependent Apoptosis by Inhibiting p300. Mol Cell. (2008) 29:217–31. doi: 10.1016/j.molcel.2007.11.036
107. Huang J, Zhang F, Jiang L, Hu G, Sun W, Zhang C, et al. Inhibition of SKP2 sensitizes bromocriptine-induced apoptosis in human prolactinoma cells. Cancer Res Treat. (2017) 49:358–73. doi: 10.4143/crt.2016.017
108. Lin Z, Fillmore GC, Um TH, Elenitoba-Johnson KSJ, Lim MS. Comparative microarray analysis of gene expression during activation of human peripheral blood T cells and leukemic Jurkat T cells. Lab Investig. (2003) 83:765–76. doi: 10.1097/01.LAB.0000073130.58435.E5
109. Malek E, Abdel-Malek MAY, Jagannathan S, Vad N, Karns R, Jegga AG, et al. Pharmacogenomics and chemical library screens reveal a novel SCF SKP2 inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia. (2017) 31:645–53. doi: 10.1038/leu.2016.258
110. Agrawal Y, Sharma T, Islam S, Nadkarni KS, Santra MK. F-box protein FBXO41 suppresses breast cancer growth by inducing autophagic cell death through facilitating proteasomal degradation of oncogene SKP2. Int J Biochem Cell Biol. (2022) 147:106228. doi: 10.1016/j.biocel.2022.106228
111. Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat Rev Cancer. (2008) 8:438–49. doi: 10.1038/nrc2396
112. Wu T, Gu X, Cui H. Emerging roles of skp2 in cancer drug resistance. Cells. (2021) 10:1–19. doi: 10.3390/cells10051147
113. Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. ScientificWorldJournal. (2010) 10:1001–15. doi: 10.1100/tsw.2010.89
114. Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, et al. Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell. (2012) 46:351–61. doi: 10.1016/j.molcel.2012.02.018
115. Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 ligase regulates akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. (2012) 149:1098–111. doi: 10.1016/j.cell.2012.02.065
116. Benanti JA. Coordination of cell growth and division by the ubiquitin-proteasome system. Semin Cell Dev Biol. (2012) 23:492–8. doi: 10.1016/j.semcdb.2012.04.005
117. Lu W, Liu S, Li B, Xie Y, Adhiambo C, Yang Q, et al. SKP2 inactivation suppresses prostate tumorigenesis by mediating JARID1B ubiquitination. Oncotarget. (2015) 6:771–88. doi: 10.18632/oncotarget.2718
118. Inuzuka H, Gao D, Finley LWS, Yang W, Wan L, Fukushima H, et al. Acetylation-dependent regulation of Skp2 function. Cell. (2012) 150:179–93. doi: 10.1016/j.cell.2012.05.038
119. Zhang L, Wang C. F-box protein Skp2: A novel transcriptional target of E2F. Oncogene. (2006) 25:2615–27. doi: 10.1038/sj.onc.1209286
120. Wang G, Chan CH, Gao Y, Lin HK. Novel roles of Skp2 E3 ligase in cellular senescence, cancer progression, and metastasis. Chin J Cancer. (2012) 31:169–77. doi: 10.5732/cjc.011.10319
121. Asmamaw MD, Liu Y, Zheng YC, Shi XJ, Liu HM. Skp2 in the ubiquitin-proteasome system: A comprehensive review. Med Res Rev. (2020) 40:1920–49. doi: 10.1002/med.21675
122. Roychowdhury A, Pal D, Basu M, Samadder S, Mondal R, Roy A, et al. Promoter methylation and enhanced SKP2 are associated with the downregulation of CDKN1C in cervical squamous cell carcinoma. Cell Signal. (2023) 109:110735. doi: 10.1016/j.cellsig.2023.110735
123. Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. (2008) 112:1415–24. doi: 10.1002/cncr.23317
124. Peng YT, Wu WR, Chen LR, Kuo KK, Tsai CH, Huang YT, et al. Upregulation of cyclin-dependent kinase inhibitors CDKN1B and CDKN1C in hepatocellular carcinoma-derived cells via goniothalamin-mediated protein stabilization and epigenetic modifications. Toxicol Rep. (2015) 2:322–32. doi: 10.1016/j.toxrep.2015.01.010
125. Ren H, Zhang Y, Zhu H. MiR-339 depresses cell proliferation via directly targeting S-phase kinase-associated protein 2 mRNA in lung cancer. Thorac Cancer. (2018) 9:408–14. doi: 10.1111/1759-7714.12597
126. Lin M, Xu Y, Gao Y, Pan C, Zhu X, Wang Zw. Regulation of F-box proteins by noncoding RNAs in human cancers. Cancer Lett. (2019) 466:61–70. doi: 10.1016/j.canlet.2019.09.008
127. Dan W, Zhong L, Yu L, Xiong L, Li J, Ye J, et al. Skp2 promotes APL progression through the stabilization of oncoprotein PML-RARα and the inhibition of JunB expression. Life Sci. (2022) 289:120231. doi: 10.1016/j.lfs.2021.120231
128. Rodriguez S, Abundis C, Boccalatte F, Mehrotra P, Chiang MY, Yui MA, et al. Therapeutic targeting of the E3 ubiquitin ligase SKP2 in T-ALL. Leukemia. (2020) 34:1241–52. doi: 10.1038/s41375-019-0653-z
129. Estévez-Sarmiento F, Saavedra E, Ruiz-Estévez M, León F, Quintana J, Brouard I, et al. Chlorinated guaiane-type sesquiterpene lactones as cytotoxic agents against human tumor cells. Int J Mol Sci. (2020) 21:1–14. doi: 10.3390/ijms21249767
130. Liu Y, Shao Z, Liao Y, Xia X, Huang C, He J, et al. Targeting SKP2/Bcr-Abl pathway with Diosmetin suppresses chronic myeloid leukemia proliferation. Eur J Pharmacol. (2020) 883:173366. doi: 10.1016/j.ejphar.2020.173366
131. Faderl S, Rai K, Gribben J, Byrd JC, Flinn IW, O’Brien S, et al. Phase II study of single-agent bortezomib for the treatment of patients with fludarabine-refractory B-cell chronic lymphocytic leukemia. Cancer. (2006) 107:916–24. doi: 10.1002/cncr.22097
132. Jing J, Rui L, Junyuan S, Jinfeng Y, Zhihao H, Weiguo L, et al. Small-molecule compounds inhibiting S-phase kinase-associated protein 2: A review. Front Pharmacol. (2023) 14:1122008. doi: 10.3389/fphar.2023.1122008
133. Zhao H, Pan H, Wang H, Chai P, Ge S, Jia R, et al. SKP2 targeted inhibition suppresses human uveal melanoma progression by blocking ubiquitylation of p27. Onco Targets Ther. (2019) 12:4297–308. doi: 10.2147/OTT.S203888
134. Tong Y, Jin L. MiR-590-5p targets Skp2 to inhibit the growth and invasion of Malignant melanoma cells. Dis Markers. (2022) 2022:8723725. doi: 10.1155/2022/8723725
135. Markovic SN, Geyer SM, Dawkins F, Sharfman W, Albertini M, Maples W, et al. A phase II study of bortezomib in the treatment of metastatic Malignant melanoma. Cancer. (2005) 103:2584–9. doi: 10.1002/cncr.21108
136. Kuttikrishnan S, Ahmad F, Mateo JM, Prabhu KS, El-Elimat T, Oberlies NH, et al. Neosetophomone B induces apoptosis in multiple myeloma cells via targeting of AKT/SKP2 signaling pathway. Cell Biol Int. (2023) 42(2):190–200. doi: 10.1002/cbin.12101
137. Yang Y, Yan W, Liu Z, Wei M. Skp2 inhibitor SKPin C1 decreased viability and proliferation of multiple myeloma cells and induced apoptosis. Braz J Med Biol Res. (2019) 52:1–10. doi: 10.1590/1414-431x20198412
138. San Miguel J, Bladé J, Boccadoro M, Cavenagh J, Glasmacher A, Jagannath S, et al. A practical update on the use of bortezomib in the management of multiple myeloma. Oncologist. (2006) 11:51–61. doi: 10.1634/theoncologist.11-1-51
139. Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. (2013) 154:556–68. doi: 10.1016/j.cell.2013.06.048
140. Pharm V, Rendon R, Le V, Tippin M, Fu D-J, Le TH, et al. Gartanin is a novel NEDDylation inhibitor for induction of Skp2 degradation, FBXW2 expression, and autophagy. Mol Carcinog. (2020) 59:193–201. doi: 10.1002/mc.23140
141. Jiang X, Li Y, Feng JL, Nik Nabil WN, Wu R, Lu Y, et al. Safrana l Prevents Prostate Cancer Recurrence by Blocking the Re-activation of Quiescent Cancer Cells via Downregulation of S-Phase Kinase-Associated Protein 2. Front Cell Dev Biol. (2020) 8:598620. doi: 10.3389/fcell.2020.598620
142. Rico-Bautista E, Yang CC, Lu L, Roth GP, Wolf DA. Chemical genetics approach to restoring p27Kip1 reveals novel compounds with antiproliferative activity in prostate cancer cells. BMC Biol. (2010) 8:1–18. doi: 10.1186/1741-7007-8-153
143. Tang Y, Simoneau AR, Xie J, Shahandeh B, Zi X. Effects of the kava chalcone flavokawain A differ in bladder cancer cells with wild-type versus mutant p53. Cancer Prev Res. (2008) 1:439–51. doi: 10.1158/1940-6207.CAPR-08-0165
144. Zhao H, Iqbal NJ, Sukrithan V, Nicholas C, Xue Y, Yu C, et al. Targeted inhibition of the E3 ligase SCFSkp2/Cks1 has antitumor activity in RB1-deficient human and mouse small-cell lung cancer. Cancer Res. (2020) 80:2355–67. doi: 10.1158/0008-5472.CAN-19-2400
145. Chiang IT, Wang WS, Liu HC, Yang ST, Tang NY, Chung JG. Curcumin alters gene expression-associated DNA damage, cell cycle, cell survival and cell migration and invasion in NCI-H460 human lung cancer cells in vitro. Oncol Rep. (2015) 34:1853–74. doi: 10.3892/or.2015.4159
146. Chang HC, Chang FR, Wang YC, Pan MR, Hung WC, Wu YC. A bioactive withanolide Tubocapsanolide A inhibits proliferation of human lung cancer cells via repressing Skp2 expression. Mol Cancer Ther. (2007) 6:1572–8. doi: 10.1158/1535-7163.MCT-06-0812
147. Liao YJ, Bai HY, Li ZH, Zou J, Chen JW, Zheng F, et al. a natural ent-kaurane, induces G2/M phase arrest via downregulation of Skp2 and apoptosis induction through ROS/JNK/c-Jun pathway in hepatocellular carcinoma cells. Cell Death Dis. (2014) 5:1–11. doi: 10.1038/cddis.2014.66
148. Jandial DD, Krill LS, Chen L, Wu C, Ke Y, Xie J, et al. Induction of G2M arrest by flavokawain a, a kava chalcone, increases the responsiveness of HER2-overexpressing breast cancer cells to herceptin. Molecules. (2017) 22(3):462. doi: 10.3390/molecules22030462
149. Ooi LC, Watanabe N, Futamura Y, Sulaiman SF, Darah I, Osada H. Identification of small molecule inhibitors of p27Kip1 ubiquitination by high-throughput screening. Cancer Sci. (2013) 104:1461–7. doi: 10.1111/cas.12246
150. Liu Y, Zhou Z, Yan J, Wu X, Xu G. Diosgenin exerts antitumor activity via downregulation of Skp2 in breast cancer cells. BioMed Res Int. (2020) 2020:1–10. doi: 10.1155/2020/8072639
151. Yin X, Zhang Y, Su J, Hou Y, Wang L, Ye X, et al. Rottlerin exerts its anti-tumor activity through inhibition of Skp2 in breast cancer cells. Oncotarget. (2016) 7:66512–24. doi: 10.18632/oncotarget.11614
152. Zhang Y, Zvi YS, Batko B, Zaphiros N, O’Donnell EF, Wang J, et al. Down-regulation of Skp2 expression inhibits invasion and lung metastasis in osteosarcoma. Sci Rep. (2018) 8:1–13. doi: 10.1038/s41598-018-32428-9
153. Dehghanian SZ, Pan CT, Lee JM, Shiue YL. ABT-751 induces multiple anticancer effects in urinary bladder urothelial carcinoma-derived cells: Highlighting the induction of cytostasis through the inhibition of SKP2 at both transcriptional and post-translational levels. Int J Mol Sci. (2021) 22:1–25. doi: 10.3390/ijms22020945
154. Wang L, Ye X, Cai X, Su J, Ma R, Yin X, et al. Curcumin suppresses cell growth and invasion and induces apoptosis by down-regulation of Skp2 pathway in glioma cells. Oncotarget. (2015) 6:18027–37. doi: 10.18632/oncotarget.4090
155. Pavlides SC, Huang KT, Reid DA, Wu L, Blank SV, Mittal K, et al. Inhibitors of SCF-Skp2/Cks1 E3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: Therapeutic potential for endometrial cancer. Endocrinology. (2013) 154:4030–45. doi: 10.1210/en.2013-1757
156. Altmann E, Erbel P, Renatus M, Schaefer M, Schlierf A, Druet A, et al. Azaindoles as zinc-binding small-molecule inhibitors of the JAMM protease CSN5. Angew Chemie - Int Ed. (2017) 56:1294–7. doi: 10.1002/anie.201608672
157. Chung YK, Chi-Hung Or R, Lu CH, Ouyang WT, Yang SY, Chang CC. Sulforaphane down-regulates SKP2 to stabilize p27KIP1 for inducing antiproliferation in human colon adenocarcinoma cells. J Biosci Bioeng. (2015) 119:35–42. doi: 10.1016/j.jbiosc.2014.06.009
158. Su J, Zhou X, Wang L, Yin X, Wang Z. Curcumin inhibits cell growth and invasion and induces apoptosis through down-regulation of Skp2 in pancreatic cancer cells. Am J Cancer Res. (2016) 6:1949–62.
159. Su J, Wang L, Yin X, Zhao Z, Hou Y, Ye X, et al. Rottlerin exhibits anti-cancer effect through inactivation of S phase kinase-associated protein 2 in pancreatic cancer cells. Am J Cancer Res. (2016) 6:2178–91.
160. Khan AQ, Siveen KS, Prabhu KS, Kuttikrishnan S, Akhtar S, Shaar A, et al. Curcumin-mediated degradation of S-phase kinase protein 2 induces cytotoxic effects in human papillomavirus-positive and negative squamous carcinoma cells. Front Oncol. (2018) 8:399. doi: 10.3389/fonc.2018.00399
161. Mou H, Guo P, Li X, Zhang C, Jiang J, Wang L, et al. Nitidine chloride inhibited the expression of S phase kinase-associated protein 2 in ovarian cancer cells. Cell Cycle. (2017) 16:1366–75. doi: 10.1080/15384101.2017.1327490
162. Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X. High-throughput screening alphascreen assay for identification of small-molecule inhibitors of ubiquitin E3 ligase SCFSkp2-Cks1. J Biomol Screen. (2013) 18:910–20. doi: 10.1177/1087057113485789
163. Gao J, Wang G, Wu J, Zuo Y, Zhang J, Jin X. Skp2 Expression Is Inhibited by Arsenic Trioxide through the Upregulation of miRNA-330-5p in Pancreatic Cancer Cells. Mol Ther - Oncolytics. (2019) 12:214–23. doi: 10.1016/j.omto.2019.01.006
164. Fricker LD. Proteasome inhibitor drugs. Annu Rev Pharmacol Toxicol. (2020) 60:457–76. doi: 10.1146/annurev-pharmtox-010919-023603
165. Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. (2020) 5(1):11. doi: 10.1038/s41392-020-0107-0
166. Celebi G, Kesim H, Ozer E, Kutlu O. The effect of dysfunctional ubiquitin enzymes in the pathogenesis of most common diseases. Int J Mol Sci. (2020) 21:1–24. doi: 10.3390/ijms21176335
167. Khan S, He Y, Zhang X, Yuan Y, Pu S, Kong Q, et al. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene. (2020) 39:4909–24. doi: 10.1038/s41388-020-1336-y
168. Sasso JM, Tenchov R, Wang DS, Johnson LS, Wang X, Zhou QA. Molecular glues: the adhesive connecting targeted protein degradation to the clinic. Biochemistry. (2023) 62:601–23. doi: 10.1021/acs.biochem.2c00245
169. Fang RH, Gao W, Zhang L. Targeting drugs to tumours using cell membrane-coated nanoparticles. Nat Rev Clin Oncol. (2023) 20:33–48. doi: 10.1038/s41571-022-00699-x
170. Spano D, Catara G. Targeting the ubiquitin–proteasome system and recent advances in cancer therapy. Cells. (2024) 13:29. doi: 10.3390/cells13010029
171. Kelsall IR, McCrory EH, Xu Y, Scudamore CL, Nanda SK, Mancebo-Gamella P, et al. HOIL-1 ubiquitin ligase activity targets unbranched glucosaccharides and is required to prevent polyglucosan accumulation. EMBO J. (2022) 41:1–17. doi: 10.15252/embj.2021109700
172. Mahlknecht U, Hoelzer D. Epigenetische Steuerelemente als neuartige therapeutische Zielstrukturen in der Hämato-/Onkologie. Dtsch Medizinische Wochenschrift. (2003) 128:2423–6. doi: 10.1055/s-2003-43593
173. Ross PM, Demirkan G, Birditt B, Bogatzi L, Carter W, Filanoski B, et al. An assay for simultaneous profiling of gene expression, phospho- and total protein abundance, and somatic DNA mutations for hematology-oncology research. Blood. (2017) 130:2481.
174. Jibu RM, Veeraraghavan VP, Gayathri R, Kavitha S. Awareness on hematological Malignancies among college students. J Adv Pharm Technol Res. (2022) 13:534–8. doi: 10.4103/japtr.japtr_175_22
175. Kullmann MK, Grubbauer C, Goetsch K, Jäkel H, Podmirseg SR, Trockenbacher A, et al. The p27-Skp2 axis mediates glucocorticoidinduced cell cycle arrest in T-lymphoma cells. Cell Cycle. (2013) 12:2625–35. doi: 10.4161/cc.25622
176. Morrow J, Lin H, Sun S, Zhang S. Targeting ubiquitination for cancer therapies. Future Med Chem. (2015) 7:2333–50. doi: 10.4155/fmc.15.148
177. Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D, et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J Exp Clin Cancer Res. (2019) 38:1–15. doi: 10.1186/s13046-019-1069-3
Keywords: AML, leukemia, hematological cancers, miRNA, Skp2, oncogene, in silico, cancer genomics
Citation: William JNG, Dhar R, Gundamaraju R, Sahoo OS, Pethusamy K, Raj AFPAM, Ramasamy S, Alqahtani MS, Abbas M and Karmakar S (2024) SKping cell cycle regulation: role of ubiquitin ligase SKP2 in hematological malignancies. Front. Oncol. 14:1288501. doi: 10.3389/fonc.2024.1288501
Received: 04 September 2023; Accepted: 15 February 2024;
Published: 15 March 2024.
Edited by:
Michael Liew, ARUP Laboratories, United StatesReviewed by:
Peter Wang, Zhejiang Zhongwei Medical Research Center, ChinaJavier Leon, University of Cantabria, Spain
Copyright © 2024 William, Dhar, Gundamaraju, Sahoo, Pethusamy, Raj, Ramasamy, Alqahtani, Abbas and Karmakar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subhradip Karmakar, subhradip.k@aiims.edu
†Present address: Jonahunnatha Nesson George William, Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy
‡These authors have contributed equally to this work
 Jonahunnatha Nesson George William
Jonahunnatha Nesson George William