- 1Department of Molecular Diagnosis, Northern Jiangsu People’s Hospital, Yangzhou University Clinical Medical College, Yangzhou, China
- 2Department of Emergency Medicine, Yizheng People’s Hospital, Yangzhou University Clinical Medical College, Yangzhou, China
Phosphoinositides, a small group of lipids found in all cellular membranes, have recently garnered heightened attention due to their crucial roles in diverse biological processes and different diseases. Among these, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), the most abundant bis-phosphorylated phosphoinositide within the signaling system, stands notably connected to breast cancer. Not only does it serve as a key activator of the frequently altered phosphatidylinositol 3-kinase (PI3K) pathway in breast cancer, but also its conversion to phosphatidylinositol-3,4,5-triphosphate (PI(3,4,5)P3) is an important direction for breast cancer research. The generation and degradation of phosphoinositides intricately involve phosphoinositide kinases. PI(4,5)P2 generation emanates from the phosphorylation of PI4P or PI5P by two lipid kinase families: Type I phosphatidylinositol-4-phosphate 5-kinases (PIP5Ks) and Type II phosphatidylinositol-5-phosphate 4-kinases (PIP4Ks). In this comprehensive review, we focus on these two lipid kinases and delineate their compositions and respective cellular localization. Moreover, we shed light on the expression patterns and functions of distinct isoforms of these kinases in breast cancer. For a deeper understanding of their functional dynamics, we expound upon various mechanisms governing the regulation of PIP5Ks and PIP4Ks activities. A summary of effective and specific small molecule inhibitors designed for PIP5Ks or PIP4Ks are also provided. These growing evidences support PIP5Ks and PIP4Ks as promising drug targets for breast cancer.
1 Introduction
Breast cancer, a disease of heterogeneity in the clinic, accounts for approximately 30% of all cancers and remains one of the most significant malignancies in women (1). Prior research has categorized breast cancers into four principal subtypes based on gene expression profiles and the expression of molecular biological markers, including luminal A (positive for estrogen or progesterone receptor ER+/PR+ with high Ki67 expression), luminal B (ER+/PR+ with low Ki67 or ER+/PR+/HER2+), HER2+(positive for human epidermal growth factor receptor 2) and triple negative (absence of estrogen, progesterone and HER2 receptors) (2, 3). Because of its frequent occurrence of tumor relapse and metastasis, comprehending the fundamental molecular mechanisms and identifying effective molecular biomarkers or anti-tumor targets for breast cancer is imperative.
Recent attention has increasingly focused on phosphoinositides, a small group of lipids found in all cellular membranes. They play crucial roles in diverse biological processes, including cell survival, proliferation, protein and membrane trafficking, cytoskeleton organization, as well as different diseases (4). Those lipid messengers are derived by the phosphorylation of the third, fourth, and fifth positions of the inositol headgroup of phosphatidylinositol (PI), resulting in the generation of seven different phosphoinositide species, like phosphatidylinositol-3-phosphate (PI3P), phosphatidylinositol-4-phosphate (PI4P), phosphatidylinositol-5-phosphate (PI5P), phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2), PI(4,5)P2, phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) and PI(3,4,5)P3 (5). Alterations in their metabolism underlie various human diseases including neurological disorders, channelopathies, diabetes and cancers (4). Notably, the most clinically advanced strategy targeting phosphoinositide signaling is the PI3K pathway in cancer. It is the most frequently altered pathway in breast cancer, with activation rates as high as 70%. It has been reported to mediate the occurrence of breast tumors, and is closely related to the degree of malignancy, metastasis and drug resistance of the breast tumors (6). In this pathway, PI3K phosphorylates PI(4,5)P2 to generate PI(3,4,5)P3 leading to the subsequent activation of protein kinase B (Akt) and its downstream signaling cascades, which lays the foundation for the application of phosphatidylinositol signaling in cancer research. Several molecules related to PI3K, PI(4,5)P2 and PI(3,4,5)P3 have been effective anti-tumor targets for breast cancer.
Crucially intertwined with phosphoinositide metabolism are phosphoinositide kinases. Multiple phosphoinositide kinases are also therapeutic targets in various human diseases, including cancer, viral infection, neurodegenerative diseases, developmental disorders, diabetes and inflammatory diseases (7). Thus, studies of these kinases can provide new insights into the mechanisms and the extent of their involvement in cancer. Currently, phosphoinositide kinases are broadly categorized into three general families: PI3K superfamily (encompassing all classes of PI3Ks), PIP kinase evolutionary family (including PIKfyve, PIP4Kα/β/γ and PIP5Kα/β/γ) and type II PI4Ks (PI4K2A/B) (7). All PI3Ks are considered therapeutic targets in human diseases, with certain PI3K inhibitors having advanced into clinical trials for breast cancer therapy (8). Beyond PI3Ks, PIP5Ks and PIP4Ks are emerging as an attractive target for therapeutics in inflammation and breast cancer, as they generate PI(4,5)P2 at the plasma membrane, critical for activating the PI3K/AKT signaling pathway. This review focuses on PIP4Ks and PIP5Ks within the PIP kinase evolutionary family, portraying them as intriguing potential targets for breast cancer.
2 The phosphoinositide signaling system and related kinases
In recent years, phosphoinositides have gained considerable attention due to their profound influence on a multitude of biochemical processes in eukaryotic cells. Each phosphoinositide localizes distinctly within the cell, performing specific functions. PI serves as the foundational component, primarily synthesized in the endoplasmic reticulum, and is subsequently transported to other membranes via vesicular or nonvesicular lipid transport mechanisms, then delivered by transfer proteins to specific membrane compartments for further phosphorylation by numerous lipid kinases (9, 10). PI3P, PI4P and PI5P are derived by the phosphorylation of PI at the 3rd, 4th and 5th positions, respectively. These phosphoinositides exhibit specific localizations and functions within the cell. For instance, PI3P localizes at early endosomes and acts as a ligand for numerous endosomal proteins. In addition to being catalyzed by PI3KII and PI3KIII, it is also generated from PI(3,4)P2 by 4-phosphatases (INPP4A and INPP4B) (11). Furthermore, PI3P is a substrate for generating PI(3,5)P2 by PIKfyve (12), which also produces the main pool of PI5P in the cell. PI4P, the most abundant monophosphorylated inositol phospholipid in mammalian cells, primarily undergoes phosphorylation in the Golgi complex and the plasma membrane. In the Golgi complex, it plays roles in the biogenesis of transport vesicles (13). However, at the plasma membrane, its primary function is as a substrate for further phosphorylation of PI(4,5)P2. Lastly, PI5P, the latest phosphoinositide discovery, is generated through multiple pathways and serves as a substrate for PI(4,5)P2 production, catalyzed by the enzyme PIP4K. However, the amount of PI(4,5)P2 synthesized from PI5P is small.
As an important bis-phosphorylated phosphoinositide in the phosphoinositide signaling system, PI(4,5)P2 exerts vital cellular functions, as elaborated in subsequent sections. It can be further phosphorylated by PI3K to PI(3,4,5)P3, another lipid with a major role in cell survival, proliferation and cell growth. The ensuing generation of PI(3,4,5)P3 activates Akt and its downstream signaling cascades, laying the foundation for phosphoinositide signaling research in cancer. Conversely, PI(3,4,5)P3 can be converted back to PI(4,5)P2 by the enzyme phosphate and tensin homolog deleted from chromosome 10 (PTEN), which is a tumor suppressor gene and thus becomes a common target of inactivation in breast cancer (14). Overall, PI(4,5)P2 and PI(3,4,5)P3 represent two critical phosphoinositides in cancer. While PI(3,4,5)P3 is typically absent in quiescent cells, it accumulates at the cell membrane upon stimulation with extracellular agonists (15).
Numerous studies have significantly advanced our understanding of the roles of these phosphoinositides. It is evident that their levels and functions are intricately regulated by the enzymatic activities of phosphatidylinositol kinases and phosphatases. These enzymes can regulate the synthesis and degradation of multiple phosphoinositides. This intricate interplay is depicted in Figure 1. Several kinases have emerged as promising targets for anticancer therapy, and in this context, our focus will be on PI(4,5)P2 and the regulation of its metabolic enzymes, PIP4Ks and PIP5Ks, in the context of breast cancer, as expounded in subsequent sections.
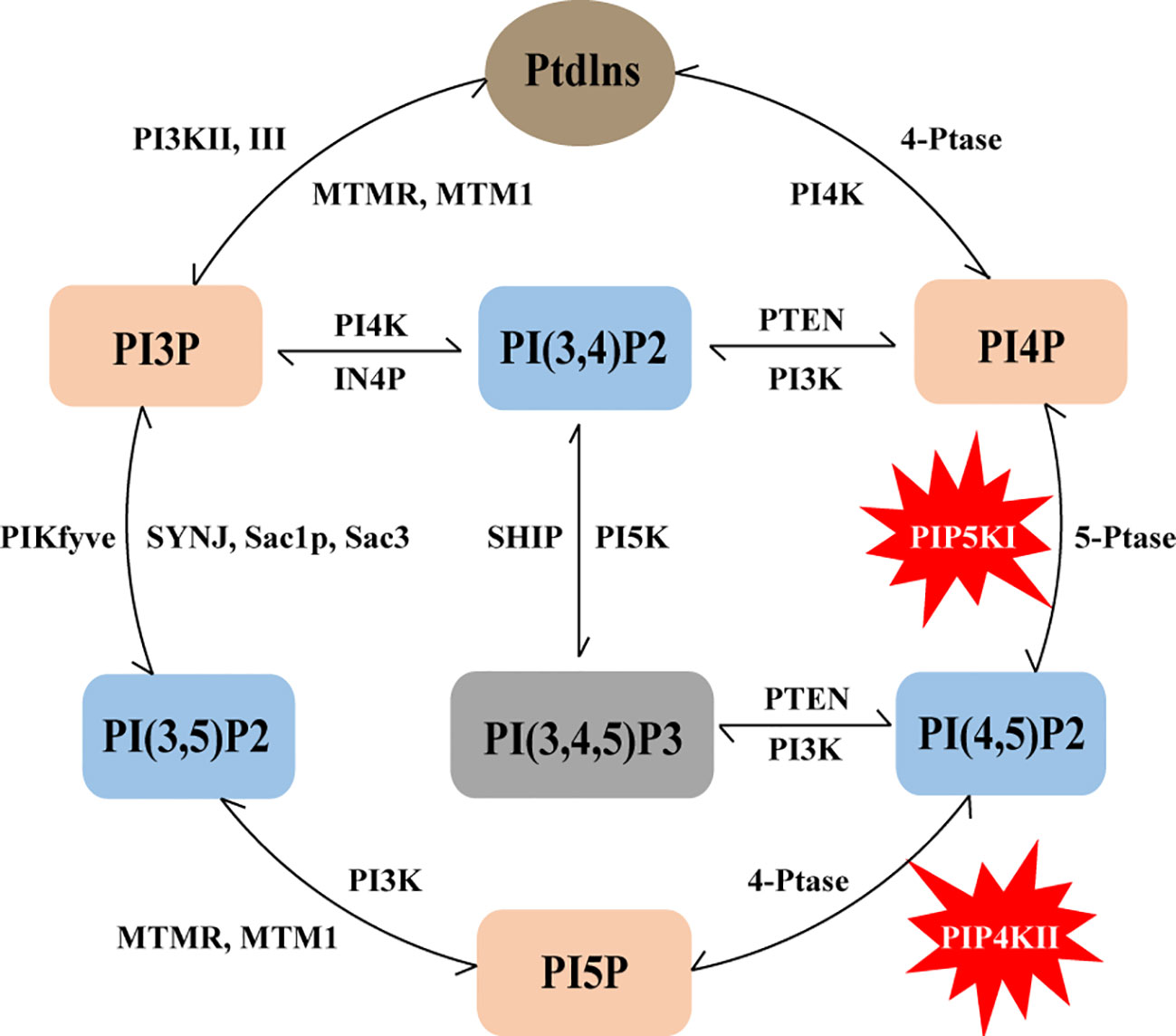
Figure 1 Schematic representation of the phosphoinositide metabolic cycle. PtdIns represents the core structure of phosphoinositides, it can lead to the generation of seven phosphoinositides, including three different mono-phosphorylated PIPs (PI3P, PI4P and PI5P), three bis-phosphorylated PIP2s (PI(3,4)P2, PI(3,5)P2 and PI(4,5)P2) and one tris-phosphorylated PIP3 PI(3,4,5)P3. The lipid kinases, phosphatases and lipases that can produce and convert these phosphoinositides are also indicated in the figure, and PIP5K and PIP4K are specifically labeled.
3 Insight into the PI(4,5)P2
As mentioned above, PI3K catalyzes PI(4,5)P2 into PI(3,4,5)P3, laying the foundation for the application of phosphatidylinositol signaling in cancer research. PI(4,5)P2 is the central component of the canonical phosphoinositide pathway, and its conversion to PI(3,4,5)P3 is a key event in activating the PI3K/Akt pathway. Conversely, PTEN facilitates the conversion of PI(3,4,5)P3 back to PI(4,5)P2. While PI(3,4,5)P3 is typically scarce in quiescent cells, PI(4,5)P2 abounds, and its relative levels remain relatively constant, barring minor fluctuations during agonist stimulation. The majority of PI(4,5)P2 resides in the plasma membrane (PM), with a substantial pool also observed in the nucleus (16). Within the nucleus, PI(4,5)P2 assumes a non-membranous structure, stabilizing nuclear PI(4,5)P2-binding proteins and participating in nuclear PI(4,5)P2 signaling (17).
PI(4,5)P2, recognized as the most abundant bis-phosphorylated phosphoinositide, serves as the substrate for PI3K, a vital precursor for second messenger inositol-1,4,5-triphosphate (PI(1,4,5)P3), diacylgycerol (DAG) and PI(3,4,5)P3 (7). Extensive studies have underscored the diverse cellular functions orchestrated by PI(4,5)P2, encompassing vesicular trafficking (18), membrane dynamics (15, 19), modulation of ion channel function (20–22), actin cytoskeleton assembly (23, 24), cell polarity (25) and phagocytosis (26). Notably, PI(4,5)P2 influences pre-mRNA splicing machinery and exhibits localization in nuclear speckles, potentially modulating transcriptional processes (27, 28). Furthermore, it has been implicated in cellular migration, a key feature of cancer progression (29, 30). Therefore, it is not surprising PI(4,5)P2 has recently been implicated in breast cancer metastasis. Reduction in plasma membrane PI(4,5)P2 levels, facilitated by specific phospholipases, can enhance cellular migration and metastatic capacity in breast cancer by increasing the abundance of active cytoplasmic cofilin (31). Accumulating evidences also indicate the involvement of PI(4,5)P2 production in the progression of various cancers such as leukemia, melanoma and glioblastoma (32–34).
PI(4,5)P2 generation occurs through the subsequent phosphorylation of mono phosphoinositides from PI, or via the dephosphorylation of PI(3,4,5)P3. It is noteworthy that PI(4,5)P2 can also be produced by the phosphorylation of PI4P or PI5P by two lipid kinase families: PIP5Ks and PIP4Ks. The Type I canonical pathway involves PIP5Ks phosphorylating the 5-position of PI4P to generate PI(4,5)P2, while the noncanonical pathway comprises PIP4Ks phosphorylating the 4-position of PI5P to yield PI(4,5)P2. In mammalian cells, the majority of PI(4,5)P2 is synthesized via the canonical pathway by PIP5Ks, with a smaller pool generated through the noncanonical pathway by PIP4Ks. Although the canonical pathway presumed to be the primary route for PI(4,5)P2 production, PIP4Ks may still generate significant amounts of PI(4,5)P2 at specific cellular locations. Despite generating the same lipid product, these two lipid kinases exhibit diverse biological and metabolic functions, each playing distinct roles, particularly in the context of breast cancer. A detailed discussion of their functions ensues in subsequent sections.
4 Insight into the PIP5K and PIP4K family
4.1 Location and functions of PIP5Ks and PIP4Ks
PIP5K and PIP4K are emerging as attractive targets for breast cancer due to their ability to produce PI(4,5)P2, which is required for activation of PI3K/AKT signaling pathway. Within this context, PIP5K stands out for its role in generating PI(4,5)P2 by phosphorylation of PI4P at the fifth position of the inositol ring. There are three type I PIP5K isoforms in mammals, including PIP5Kα, PIP5Kβ, and PIP5Kγ (35, 36). These isoforms are encoded by genes PIP5K1A, PIP5K1B and PIP5K1C, respectively. Each isoform exhibits several splice variants, although the specific functions of these variants remain inadequately understood. Remarkably, PIP5K isoforms have distinct cell localizations, translating into unique in vivo functions. For example, PIP5Kα is present in membrane ruffles and discrete regions in the nucleus (known as nuclear speckles), regulating the activity of Star-PAP a poly(A) polymerase that controlled the expression of select mRNAs (37). On the other hand, PIP5Kβ primarily resides at the plasma membrane, playing a critical role in receptor endocytosis and cell migration. It is also present on vesicles in the perinuclear region of the cell. PIP5Kγ localizes to focal adhesions and adherens junctions, where it regulates cell polarity and migration of adherent cells and leukocytes (38), Ca2+ flux (39), assembly of E-cadherin-based intercellular adhesions, epithelial polarization (40–42), and targets focal adhesions by associating with talin, thereby modulating nascent adhesion formation (43, 44). All three PIP5K isoforms play significant roles in actin remodeling events and various cellular functions, including protein trafficking, cell division and locomotion. Recent evidence has indicated the involvement of PIP5Ks in cancer progression, such as in prostate cancer (45, 46), ovarian cancer (47), and notably, breast cancer (48, 49). Given the critical role of PI(4,5)P2 in regulating cellular events and its contribution to tumor cell invasion and migration, the implication of PIP5Ks in breast cancer is a compelling avenue for investigation.
PIP4K, also comprising three lipid kinase members including PIP4Kα, PIP4Kβ and PIP4Kγ (encoded by PIP4K2A, PIP4K2B and PIP4K2C, respectively) (50, 51), exerts its enzymatic action by phosphorylating PI5P at the fourth position of the inositol ring, resulting in the generation of PI(4,5)P2. Predominantly located in intracellular membranes, the PIP4K isoforms exhibit distinctive cellular distributions and diverse functional roles. Specifically, PIP4Kα is involved in autophagosomes, lysosomes, and peroxisomes, PIP4Kβ primarily resides in the nucleus but can manifest in autophagosomes (52). PIP4Kγ is predominantly found in autophagosomes, Golgi apparatus, and membrane chambers (53). At a sequence level, the isoforms PIP4Kα and PIP4Kβ display a higher homology (83% protein homology), in contrast to PIP4Kγ, which shares approximately 60% homology (50). Due to their sequence differences, the PIP4K isoforms possess varying kinase activities, leading to diverse roles in insulin signaling, receptor recycling, gene regulation, cell stress response and cancers (54–56). PIP4Kα plays a critical role in intracellular cholesterol transport by modulating PI(4,5)P2 homeostasis on peroxisome membranes (57), and it is associated with mTORC2 regulation (58). PIP4Kβ is linked to determining insulin sensitivity and adiposity (59). Both PIP4Kα and PIP4Kβ are shown to be required for autophagosome-lysosome fusion during metabolic stress (60). Differently, PIP4Kγ positively regulates Notch1 signaling by facilitating receptor recycling (61). It contributes to increased inflammation characterized by decreased regulatory effectors cells (62), and plays crucial roles in T-cell signaling (63). Recent findings have established a link between PIP4Kγ and mTORC1, demonstrating their interaction in a self-regulated feedback loop to maintain low and tightly regulated mTORC1 activation during starvation (64). Besides the above functions that have already been discussed, these kinases have been implicated in several diseases, including acute leukemias, glioblastoma, soft tissue sarcomas, among others. Notably, there is a burgeoning interest in targeting PIP4Ks for therapeutic interventions in cancer (65). Among them, the first study to implicate PIP4Ks in cancer and for setting the stage for oncological research is in breast cancer (66).
4.2 The expression of PIP5Ks and PIP4Ks in breast cancer
To comprehensively understand the expression of PIP5K and PIP4K in multiple cancer (breast cancer included), we conducted an analysis comparing median expression levels between tumor samples and their paired normal tissue counterparts using the Gene Expression Profiling Interactive Analysis (GEPIA) dataset. As depicted in Figure 2A, PIP5K exhibits ubiquitous expression, with PIP5Kα demonstrating higher expression levels than the other two isoforms in breast cancer. PIP5Kα, a key upstream factor of PI3K/AKT pathway, is notably expressed at elevated levels in triple-negative breast cancers and associated with poor patient outcome (67). Its expression is imperative for invadopodia formation and promotes survival of breast cancer cells (68). Studies have also reported that the PIP5Kα-dependent pool of PI(4,5)P2 promotes breast cancer cell proliferation (49). Overexpression of PIP5K1B in MCF-7 breast cancer cells results in cell scattering and the stimulation of cell migration and invasion (69). Moreover, a significant correlation between PIP5Kγ expression and the progression of breast cancer is also observed (70). Notably, it is overexpressed in triple-negative breast cancer (71). Depletion of PIP5Kγ inhibits cell proliferation, MMP9 secretion, cell migration and invasion of breast cancer cells (48). Additionally, the expression of its 90 kD splice variant (PIP5Kγ90) also enhances the migration, invasion and proliferation of breast cancer cells (72).
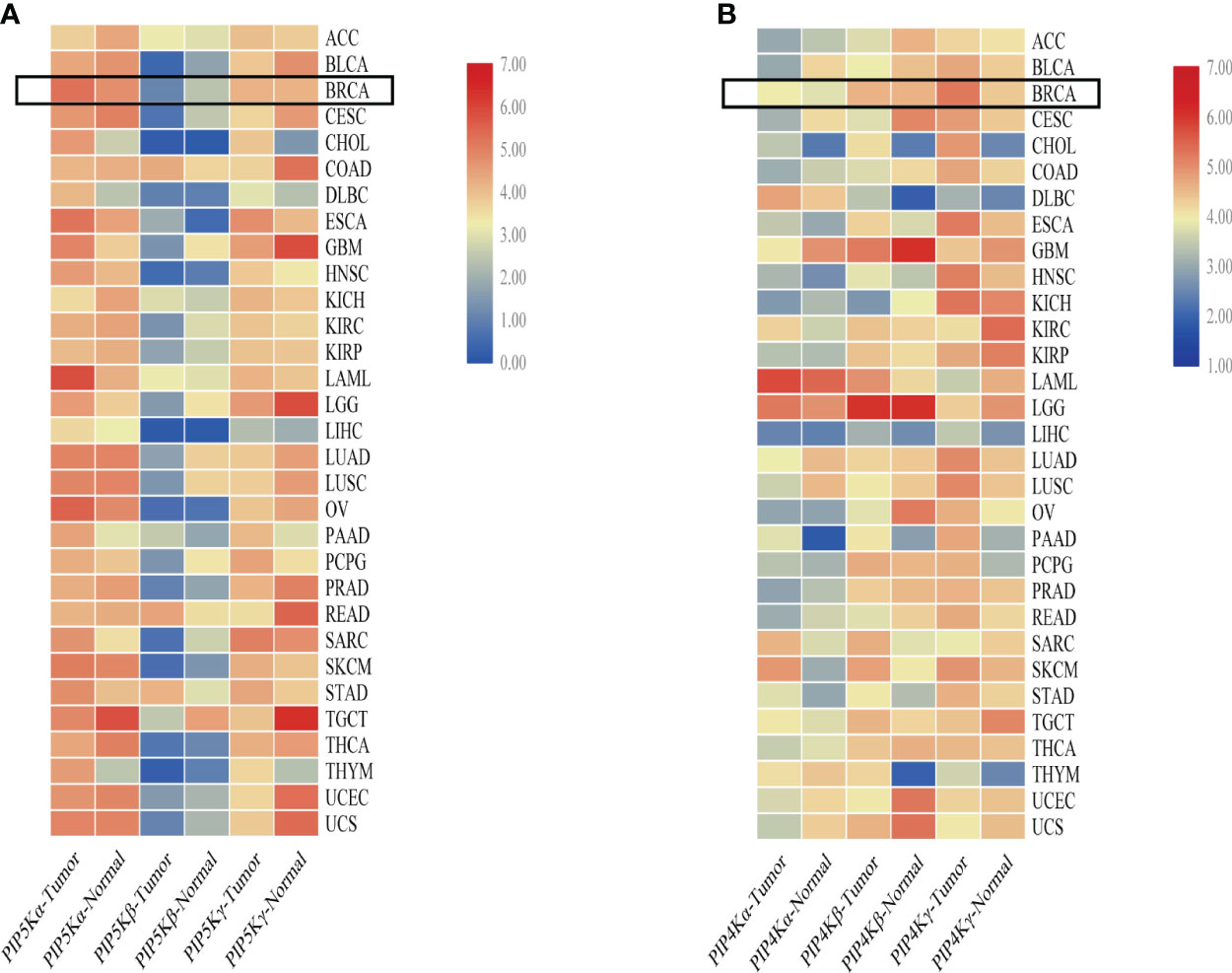
Figure 2 Comparative overview of PIP5K and PIP4K isoforms expression in multiple cancers. (A) shows the PIP5Ks expression in multiple cancers. (B) represents the expression of PIP4Ks. The heat map is organized based on the difference in median expression between the tumor samples and the paired normal tissue samples. The expressions of PIP5Ks and PIP4Ks in breast cancer are highlighted with a black box. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical and endocervical cancers; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma.
When studying the PIP4Ks, both PIP4Kα and PIP4Kβ exhibit elevated expression levels in breast cancer compared to normal tissues (66). Their downregulation significantly suppresses the proliferative capacity of triple negative breast cancer cell lines and dramatically diminishes the cellular viability. Intriguingly, deletion of these two isoforms in mice suppresses tumor formation in the presence of Trp53 deletion (66). For PIP4K2B, both excessively high and low levels are implicated in breast cancer and patient survival. Overexpression of PIP4K2B can increase breast cancer cell proliferation and anchorage-independent growth in different subsets of breast cancer cell lines (73). Additionally, a correlation between low PIP4K2B expression in human breast tumors and reduced patient survival is found (74). PIP4K2B is also reported to be co-amplified with the proto-oncogene ERBB2 gene. Notably, a striking co-occurrence between high PIP4K2A expression as well as PIP4K2B amplification with TP53 mutation/deletion is even showed. In contrast to PIP4Kα and PIP4Kβ, PIP4Kγ, while approximately 1% as active as the former isoforms, displays higher expression in breast cancer based on a comparative overview of PIP4K isoform expression in multiple cancers (Figure 2B). This underscores PIP4Kγ as a potential drug target for breast cancer treatment.
4.3 Regulation of PIP5Ks and PIP4Ks
Many studies have demonstrated various mechanisms of the regulation of PIP5K activity, including spatiotemporal regulation and other protein stability. When it comes to spatiotemporal regulation, PIP5K is intricately modulated by a cadre of upstream regulators, including Rho family of GTPases, binding partner proteins and post-translational modifications such as phosphorylation (75). For instance, elevated level of PIP5Kα increases expression of pSer-473 AKT and is in complexes with VEGFR2, acting as co-factor of ER-alpha to regulate activities of target genes, including cyclin D1 and CDK1, particularly in triple-negative and ER+ breast cancer cells (67). As a key player downstream of EGFR, a tight association between EGFR and PIP5Kγ in the progression of breast cancer is found. Phosphorylation modifications of PIP5Kγ by EGFR profoundly impacts tumor formation and metastasis. Cdk5-mediated PIPKIγ90 phosphorylation exerts spatiotemporal control over PI(4,5)P2 production and fibronectin secretion, intricately regulating cancer cell invasion (76). The interplay of PIP5Kγ and PI3K, orchestrated by Src-mediated regulation, spatially generates PI(4,5)P2 and PI(3,4,5)P3, culminating in subsequent Akt activation (71). Contrasting this perspective, a study proposed an alternative regulatory mechanism involving the neural precursor cell expressed, developmentally down-regulated gene 4 (NEDD4)-dependent control of PIP5K protein stability, affecting PIP5K-dependent PI(4,5)P2 generation. Specifically, they suggest that PIP5Kα increases the plasma membrane PI(4,5)P2 level and contributes to breast cancer cell proliferation through PI3K/Akt activation, while the whole process can be negatively controlled by the ubiquitin ligase NEDD4, which can interact with the C-terminal region and ubiquitinate PIP5Kα (49). In addition, PIP5Kγ is implicated in the transcriptional upregulation of the PD-L1 gene by activating the NF-κB pathway in the triple negative breast cancer (77).
Concerning PIP4Ks, they are often negatively regulated by phosphorylation. Protein kinase D (PKD)-mediated phosphorylation of PIP4Kα at Thr376 and its mutation to aspartate significantly reduces the enzymatic activity of PIP4Kα (78). Additionally, the protein kinase formerly casein kinase 2 (CK2) can phosphorylate the Ser304 site of PIP4Kα, intriguingly, mimicking phosphorylation by mutating the residue to aspartate induces a translocation from the cytoplasm to the plasma membrane instead of a change in the PIP kinase activity (79). PIP4Kβ, on the other hand, can be phosphorylated by the p38 stress-activated protein kinase at Ser326 directly, resulting in inhibition of PIP4Kβ activity (80). Furthermore, mTORC1 can phosphorylate PIP4Kγ at Ser324 and Ser328. In addition to the phosphorylation mentioned above, the activity of PIP4Ks is intricately modulated by interactions with other proteins, such as the proline isomerase Pin1 (81). Moreover, the regulation of PIP4Kβ is involved in ubiquitylation, where it acts as a key regulator of the p38 MAPK pathway, interacting with the ubiquitin ligase complex Cul3-SPOP (SPOP) and co-localizing at nuclear speckles (82).
4.4 Inhibitors of PIP5Ks and PIP4Ks
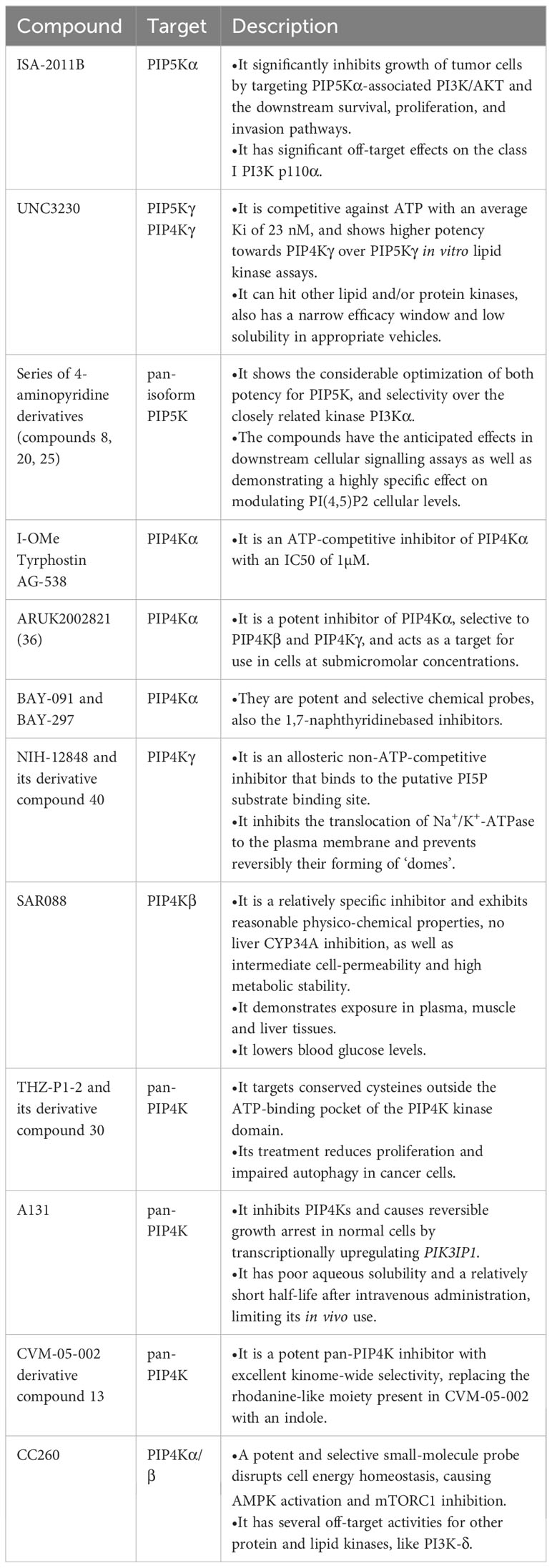
PIP5Ks has been hypothesized to provide a potential therapeutic target of interest in the treatment of cancers. To comprehensively comprehend and exploit the role of PIP5Ks in human cancers, some potent and specific PIP5K inhibitors have been reported, with some currently in early stages of preclinical development. Notably, ISA-2011B, a diketopiperazine fused C-1 indol-3-yl substituted tetra-hydro-isoquinoline, stands out as a well-described PIP5K inhibitor, specifically inhibiting the PIP5Kα-associated AKT pathways (45). However, further optimization is required due to its significant off-target effects. 5-(cyclohexanecarboxamido)-2-(phenylamino)thiazole-4-carboxamide, termed as UNC3230, has been identified as an inhibitor targeting PIP5Kγ (83). At the same time, it also targets PIP4K and shows higher potency towards PIP4Kγ over PIP5Kγ in in vitro lipid kinase assays. Moreover, several high-quality in vitro tool compounds have been identified (84). Despite the existence of some inhibitors, searching for more highly effective PIP5K inhibitors as tool compounds is still valuable.
Given the current roles of the PIP4Ks in cancers, they have also emerged as attractive drug targets in the context of cancers, with their inhibitors being actively explored in preclinical studies. At present, various PIP4K inhibitors, encompassing pan inhibitors and isoform specific inhibitors, have been reported. Compound CC260, a highly potent and selective noncovalent dual inhibitor for both PIP4Kα and PIP4Kβ, exhibits promising potential by disrupting cell energy homeostasis and activating AMPK while inhibiting mTORC1, selectively targeting p53 mutant breast cancer cells (85). However, it is important to note its off-target activity against PI3K-δ. THZ-P1-2, a pan-PIP4K inhibitor, targets cysteines in a disordered loop of isoforms PIP4Kα/β/γ, and it is shown to have anti-proliferative activity (86). In addition, PIP4K lipid kinases have been identified as the target of A131 and CVM-05-002 (87, 88). The former selectively regulates the cell cycle entry of Ras-activated cancer cells and induces reversible growth arrest in normal cells by transcriptionally upregulating PIK3IP1. Except for the inhibitors mentioned above, several isoform-specific PIP4K inhibitors have also been identified. For example, I-Ome Tyrphostin AG-538 is identified as the ATP-competitive inhibitor of PIP4Kα (89). BAY-091 and BAY-297 serve as the valuable inhibitors of the kinase PIP4K2A (90), ARUK2002821 is also identified as an selective PIP4Kα inhibitor and has broad selectivity against lipid and protein kinases (91). Another noteworthy inhibitor is SAR088, representing the first orally available and in vivo active PIP4K2B inhibitor (92). The quinazolin-4-amine compound NIH-12848 and its derivative compound 40 are putative PIP4Kγ inhibitors, and they are likely to interact with the PI5P-binding site of PIP4Kγ (93, 94). NCT-504 have also been disclosed (95). These diverse inhibitors collectively present a rich landscape for potential therapeutic interventions targeting PIP4Ks in cancer. These effective and specific small molecule inhibitors designed for PIP5Ks or PIP4Ks are summarized in Table 1.
5 Conclusion and perspective
In recent years, there has been a surge in research on phosphoinositide signaling, acknowledging its critical role in fundamental cellular processes and its close relationship with various diseases. However, for a comprehensive understanding and effective utilization of phosphoinositides, further cellular and physiological studies are imperative to unravel the intricacies of phosphoinositide signaling pathway and their associated enzymes. From the current perspective, lipid kinases PIP4Ks and PIP5Ks are emerging as promising therapeutic targets in breast cancer, but, clearly, a thorough exploration of their roles and mechanisms across diverse breast cancer subtypes necessitates extensive experimental investigation. Currently, distinct isoforms within the PIP5K/PIP4K family exhibit varying distribution and expression patterns in cells and tissues, implying their potentially divergent roles in different stages and subtypes of breast cancer. This underscores the importance of analyzing and approaching treatment strategies for triple-negative, ER+ or other breast cancer types differently, taking into account the unique characteristics of these isoforms. Among these isoforms, particularly, PIP4Kγ, despite its perceived lower activity, exhibits higher expression than the other PIP4K isoforms in breast cancer, warranting focused attention in future studies due to the limited research on this isoform in the context of breast cancer.
Further functional comprehension of PIP5Ks and PIP4Ks in breast cancer necessitates a deeper investigation into the regulation. While spatiotemporal regulation is a relatively well-studied mode of PIP4K/PIP5K regulation, in fact, attention should broaden to encompass the influence of other proteins and environmental factors on these kinases. At the same time, understanding their substrate PI5P and product PI(4,5)P2 is crucial, given their significance in the PI3K pathway. Especially the PI(4,5)P2, it is not only the key to activate the PI3K pathway, but also its conversion to PI(3,4,5)P3 hold substantial promise for breast cancer research. In the following studies, we can focus on the regulatory process between PIP5Ks/PIP4Ks and PI(4,5)P2.
Moreover, a key area of focus is the development of inhibitors targeting PIP5Ks and PIP4Ks. These inhibitors enable validation of the functions of these kinases and present promising avenues for future therapeutic interventions in breast cancer. Despite the existence of effective and specific small molecule inhibitors, enhancing specificity and overall pharmacokinetics while mitigating off-target activity is paramount. Considering targeting multiple PIPK families and isoforms, and designing inhibitors based on their unique properties, can address issues of off-target activity and drug resistance, thus enhancing the prospects of therapeutic efficacy. For example, PIP4Ks can bind to both ATP and GTP, but PIP4Kβ preferentially utilizes GTP rather than ATP for PI5P phosphorylation, which allows us to design ATP or GTP binding sites accordingly. In addition, the current common strategy for developing kinase inhibitors is to inhibit substrate phosphorylation by occupying the ATP-binding site of the drug molecule. However, ATP binding sites are highly conserved in kinases, which makes the development of competitive ATP inhibitors with high selectivity a huge challenge. Considering the highly differentiated substrates and the uniqueness of their binding sites in terms of geometric and electrostatic properties, targeting unique substrate binding sites and blocking substrate binding appears to be a superior strategy for inhibiting PIPK families. Meanwhile, we should also consider additive or synergistic effects between inhibitors with different working mechanisms.
In conclusion, the expression pattern and function of PIP5K and PIP4K in breast cancer underscore these two kinases as attractive targets for therapeutics in breast cancer. However, our current understanding of PIP5K and PIP4K is still limited, necessitating future studies aimed at unraveling the mechanisms specific to each isoform. This deeper understanding will pave the way for effective clinical applications of these kinases in breast cancer treatment.
Author contributions
JX: Writing – original draft. YJ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by YJ's Postdoctoral Excellence Program of Jiangsu Province (2022ZB895), China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistic. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
2. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U.S.A. (2001) 98(19):10869–74. doi: 10.1073/pnas.191367098
3. Moldovan L, Mitroi A, Petrescu CM, Aschie M. Classification of breast carcinomas according to gene expression profiles. J Med Life (2013) 6(1):14–7.
4. Hammond GRV, Burke JE. Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr Opin Cell Biol (2020) 63:57–67. doi: 10.1016/j.ceb.2019.12.007
5. Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev (2013) 93(3):1019–137. doi: 10.1152/physrev.00028.2012
6. Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev (2016) 35(4):515–24. doi: 10.1007/s10555-016-9637-x
7. Burke JE, Triscott J, Emerling BM, Hammond GRV. Beyond PI3Ks: targeting phosphoinositide kinases in disease. Nat Rev Drug Discovery (2023) 22(5):357–86. doi: 10.1038/s41573-022-00582-5
8. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med (2019) 380(20):1929–40. doi: 10.1056/NEJMoa1813904
9. Hammond GR, Balla T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim Biophys Acta (2015) 1851(6):746–58. doi: 10.1016/j.bbalip.2015.02.013
10. De Craene JO, Bertazzi DL, Bär S, Friant S. Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int J Mol Sci (2017) 18(3):634. doi: 10.3390/ijms18030634
11. Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev Mol Cell Biol (2019) 20(9):515–34. doi: 10.1038/s41580-019-0129-z
12. Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci U.S.A. (2012) 109(43):17472–7. doi: 10.1073/pnas.1203106109
13. Waugh MG. The Great Escape: how phosphatidylinositol 4-kinases and PI4P promote vesicle exit from the Golgi (and drive cancer). Biochem J (2019) 476(16):2321–46. doi: 10.1042/bcj20180622
14. Viennet T, Rodriguez Ospina S, Lu Y, Cui A, Arthanari H, Dempsey DR. Chemical and structural approaches to investigate PTEN function and regulation. Methods Enzymol (2023) 682:289–318. doi: 10.1016/bs.mie.2022.09.007
15. Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature (2006) 443(7112):651–7. doi: 10.1038/nature05185
16. Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4,5P2signaling specificity by association with effectors. Biochim Biophys Acta (2015) 1851(6):711–23. doi: 10.1016/j.bbalip.2015.01.009
17. Wen Y, Vogt VM, Feigenson GW. PI(4,5)P(2) clustering and its impact on biological functions. Annu Rev Biochem (2021) 90:681–707. doi: 10.1146/annurev-biochem-070920-094827
18. Li S, Ghosh C, Xing Y, Sun Y. Phosphatidylinositol 4,5-bisphosphate in the control of membrane trafficking. Int J Biol Sci (2020) 16(15):2761–74. doi: 10.7150/ijbs.49665
19. Mandal K. Review of PIP2 in cellular signaling, functions and diseases. Int J Mol Sci (2020) 21(21):8342. doi: 10.3390/ijms21218342
20. Hille B, Dickson EJ, Kruse M, Vivas O, Suh BC. Phosphoinositides regulate ion channels. Biochim Biophys Acta (2015) 1851(6):844–56. doi: 10.1016/j.bbalip.2014.09.010
21. Ye W, Han TW, Nassar LM, Zubia M, Jan YN, Jan LY. Phosphatidylinositol-(4, 5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F. Proc Natl Acad Sci U.S.A. (2018) 115(7):E1667–e1674. doi: 10.1073/pnas.1718728115
22. Ko W, Suh BC. Differential regulation of ca(2+)-activated cl(-) channel TMEM16A splice variants by membrane PI(4,5)P(2). Int J Mol Sci (2021) 22(8):4088. doi: 10.3390/ijms22084088
23. Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev (2010) 90(1):259–89. doi: 10.1152/physrev.00036.2009
24. Senju Y, Kalimeri M, Koskela EV, Somerharju P, Zhao H, Vattulainen I, et al. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc Natl Acad Sci U.S.A. (2017) 114(43):E8977–e8986. doi: 10.1073/pnas.1705032114
25. Hammond GR, Hong Y. Phosphoinositides and membrane targeting in cell polarity. Cold Spring Harb Perspect Biol (2018) 10(2):a027938. doi: 10.1101/cshperspect.a027938
26. Cheng S, Wang K, Zou W, Miao R, Huang Y, Wang H, et al. PtdIns(4,5)P2 and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J Cell Biol (2015) 210(3):485–502. doi: 10.1083/jcb.201501038
27. Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature (2008) 451(7181):1013–7. doi: 10.1038/nature06666
28. Harihar S. The role of phosphoinositide signaling in breast cancer metastasis suppressor 1-mediated metastasis suppression of human breast carcinoma cells. [dissertation]. Logan: Utah State University (2011).
29. Ling K, Schill NJ, Wagoner MP, Sun Y, Anderson RA. Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol (2006) 16(6):276–84. doi: 10.1016/j.tcb.2006.03.007
30. Al-Fahad D, Alyaseen F, Al-Amery A, Ibeas Bin C. Regulation of focal adhesion dynamics and cell migration by PLC/PI3K-mediated metabolism of ptdIns (4,5) P2 in a breast cancer cell line. Rep Biochem Mol Biol (2022) 11(2):270–81. doi: 10.52547/rbmb.11.2.270
31. Sengelaub CA, Navrazhina K, Ross JB, Halberg N, Tavazoie SF. PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J (2016) 35(1):62–76. doi: 10.15252/embj.201591973
32. Hamard-Peron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, et al. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol (2010) 84(1):503–15. doi: 10.1128/jvi.01134-09
33. Ramos AR, Elong Edimo W, Erneux C. Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved. Adv Biol Regul (2018) 67:40–8. doi: 10.1016/j.jbior.2017.09.001
34. Basappa J, Citir M, Zhang Q, Wang HY, Liu X, Melnikov O, et al. ACLY is the novel signaling target of PIP(2)/PIP(3) and Lyn in acute myeloid leukemia. Heliyon (2020) 6(5):e03910. doi: 10.1016/j.heliyon.2020.e03910
35. Ishihara H, Shibasaki Y, Kizuki N, Katagiri H, Yazaki Y, Asano T, et al. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem (1996) 271(39):23611–4. doi: 10.1074/jbc.271.39.23611
36. Loijens JC, Anderson RA. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem (1996) 271(51):32937–43. doi: 10.1074/jbc.271.51.32937
37. A PS, Laishram RS. Nuclear phosphatidylinositol-phosphate type I kinase α-coupled star-PAP polyadenylation regulates cell invasion. Mol Cell Biol (2018) 38(5):e00457–00417. doi: 10.1128/mcb.00457-17
38. Xu W, Wang P, Petri B, Zhang Y, Tang W, Sun L, et al. Integrin-induced PIP5K1C kinase polarization regulates neutrophil polarization, directionality, and in vivo infiltration. Immunity (2010) 33(3):340–50. doi: 10.1016/j.immuni.2010.08.015
39. Wang YJ, Li WH, Wang J, Xu K, Dong P, Luo X, et al. Critical role of PIP5KI{gamma}87 in InsP3-mediated Ca(2+) signaling. J Cell Biol (2004) 167(6):1005–10. doi: 10.1083/jcb.200408008
40. Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol (2007) 176(3):343–53. doi: 10.1083/jcb.200606023
41. Liu J, Zuo X, Yue P, Guo W. Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol Biol Cell (2007) 18(11):4483–92. doi: 10.1091/mbc.e07-05-0461
42. Xiong X, Xu Q, Huang Y, Singh RD, Anderson R, Leof E, et al. An association between type Iγ PI4P 5-kinase and Exo70 directs E-cadherin clustering and epithelial polarization. Mol Biol Cell (2012) 23(1):87–98. doi: 10.1091/mbc.E11-05-0449
43. Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature (2002) 420(6911):85–9. doi: 10.1038/nature01147
44. Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature (2002) 420(6911):89–93. doi: 10.1038/nature01082
45. Semenas J, Hedblom A, Miftakhova RR, Sarwar M, Larsson R, Shcherbina L, et al. The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc Natl Acad Sci U.S.A. (2014) 111(35):E3689–3698. doi: 10.1073/pnas.1405801111
46. Wang T, Sarwar M, Whitchurch JB, Collins HM, Green T, Semenas J, et al. PIP5K1α is required for promoting tumor progression in castration-resistant prostate cancer. Front Cell Dev Biol (2022) 10:798590. doi: 10.3389/fcell.2022.798590
47. Cao S, Chen C, Xue J, Huang Y, Yang X, Ling K. Silencing of type Iγ phosphatidylinositol phosphate kinase suppresses ovarian cancer cell proliferation, migration and invasion. Oncol Rep (2017) 38(1):253–62. doi: 10.3892/or.2017.5670
48. Chen C, Wang X, Xiong X, Liu Q, Huang Y, Xu Q, et al. Targeting type Iγ phosphatidylinositol phosphate kinase inhibits breast cancer metastasis. Oncogene (2015) 34(35):4635–46. doi: 10.1038/onc.2014.393
49. Tran MH, Seo E, Min S, Nguyen QT, Choi J, Lee UJ, et al. NEDD4-induced degradative ubiquitination of phosphatidylinositol 4-phosphate 5-kinase α and its implication in breast cancer cell proliferation. J Cell Mol Med (2018) 22(9):4117–29. doi: 10.1111/jcmm.13689
50. Clarke JH, Irvine RF. The activity, evolution and association of phosphatidylinositol 5-phosphate 4-kinases. Adv Biol Regul (2012) 52(1):40–5. doi: 10.1016/j.advenzreg.2011.09.002
51. Clarke JH, Irvine RF. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem J (2013) 454(1):49–57. doi: 10.1042/bj20130488
52. Bultsma Y, Keune WJ, Divecha N. PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J (2010) 430(2):223–35. doi: 10.1042/bj20100341
53. Clarke JH, Emson PC, Irvine RF. Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am J Physiol Renal Physiol (2008) 295(5):F1422–1430. doi: 10.1152/ajprenal.90310.2008
54. Fiume R, Stijf-Bultsma Y, Shah ZH, Keune WJ, Jones DR, Jude JG, et al. PIP4K and the role of nuclear phosphoinositides in tumour suppression. Biochim Biophys Acta (2015) 1851(6):898–910. doi: 10.1016/j.bbalip.2015.02.014
55. Jude JG, Spencer GJ, Huang X, Somerville TDD, Jones DR, Divecha N, et al. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene (2015) 34(10):1253–62. doi: 10.1038/onc.2014.77
56. Sharma S, Mathre S, Ramya V, Shinde D, Raghu P. Phosphatidylinositol 5 phosphate 4-kinase regulates plasma-membrane PIP(3) turnover and insulin signaling. Cell Rep (2019) 27(7):1979–1990.e1977. doi: 10.1016/j.celrep.2019.04.084
57. Hu A, Zhao XT, Tu H, Xiao T, Fu T, Wang Y, et al. PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P(2) homeostasis. J Lipid Res (2018) 59(3):507–14. doi: 10.1194/jlr.M082149
58. Bulley SJ, Droubi A, Clarke JH, Anderson KE, Stephens LR, Hawkins PT, et al. In B cells, phosphatidylinositol 5-phosphate 4-kinase-α synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc Natl Acad Sci U.S.A. (2016) 113(38):10571–6. doi: 10.1073/pnas.1522478113
59. Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB, Cantley LC. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta-/- mice. Mol Cell Biol (2004) 24(11):5080–7. doi: 10.1128/MCB.24.11.5080-5087.2004
60. Lundquist MR, Goncalves MD, Loughran RM, Possik E, Vijayaraghavan T, Yang A, et al. Phosphatidylinositol-5-phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol Cell (2018) 70(3):531–544.e539. doi: 10.1016/j.molcel.2018.03.037
61. Zheng L, Conner SD. PI5P4Kγ functions in DTX1-mediated Notch signaling. Proc Natl Acad Sci U.S.A. (2018) 115(9):E1983–1990. doi: 10.1073/pnas.1712142115
62. Shim H, Wu C, Ramsamooj S, Bosch KN, Chen Z, Emerling BM, et al. Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc Natl Acad Sci U.S.A. (2016) 113(27):7596–601. doi: 10.1073/pnas.1600934113
63. Poli A, Abdul-Hamid S, Zaurito AE, Campagnoli F, Bevilacqua V, Sheth B, et al. PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc Natl Acad Sci U.S.A. (2021) 118(31):e2010053118. doi: 10.1073/pnas.2010053118
64. Mackey AM, Sarkes DA, Bettencourt I, Asara JM, Rameh LE. PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci Signal (2014) 7(350):ra104. doi: 10.1126/scisignal.2005191
65. Arora GK, Palamiuc L, Emerling BM. Expanding role of PI5P4Ks in cancer: A promising druggable target. FEBS Lett (2022) 596(1):3–16. doi: 10.1002/1873-3468.14237
66. Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell (2013) 155(4):844–57. doi: 10.1016/j.cell.2013.09.057
67. Sarwar M, Syed Khaja AS, Aleskandarany M, Karlsson R, Althobiti M, Ødum N, et al. The role of PIP5K1α/pAKT and targeted inhibition of growth of subtypes of breast cancer using PIP5K1α inhibitor. Oncogene (2019) 38(3):375–89. doi: 10.1038/s41388-018-0438-2
68. Yamaguchi H, Yoshida S, Muroi E, Kawamura M, Kouchi Z, Nakamura Y, et al. Phosphatidylinositol 4,5-bisphosphate and PIP5-kinase Ialpha are required for invadopodia formation in human breast cancer cells. Cancer Sci (2010) 101(7):1632–8. doi: 10.1111/j.1349-7006.2010.01574.x
69. Azhunussova A, Shayakhmetov Y, Chao WT, Kunz J. PIPK1b regulates E- cadherin endocytosis in polarized breast cancer cells and promotes a switch to a migratory phenotype. Abstract retrieved from Nazarbayev University Repository (2014).
70. Sun Y, Turbin DA, Ling K, Thapa N, Leung S, Huntsman DG, et al. Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res (2010) 12(1):R6. doi: 10.1186/bcr2471
71. Thapa N, Choi S, Tan X, Wise T, Anderson RA. Phosphatidylinositol phosphate 5-kinase Iγ and phosphoinositide 3-kinase/akt signaling couple to promote oncogenic growth. J Biol Chem (2015) 290(30):18843–54. doi: 10.1074/jbc.M114.596742
72. Li X, Zhou Q, Sunkara M, Kutys ML, Wu Z, Rychahou P, et al. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I γ by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci (2013) 126(Pt 12):2617–28. doi: 10.1242/jcs.117044
73. Luoh SW, Venkatesan N, Tripathi R. Overexpression of the amplified Pip4k2beta gene from 17q11-12 in breast cancer cells confers proliferation advantage. Oncogene (2004) 23(7):1354–63. doi: 10.1038/sj.onc.1207251
74. Keune WJ, Sims AH, Jones DR, Bultsma Y, Lynch JT, Jirström K, et al. Low PIP4K2B expression in human breast tumors correlates with reduced patient survival: A role for PIP4K2B in the regulation of E-cadherin expression. Cancer Res (2013) 73(23):6913–25. doi: 10.1158/0008-5472.Can-13-0424
75. Weernink PA, Meletiadis K, Hommeltenberg S, Hinz M, Ishihara H, Schmidt M, et al. Activation of type I phosphatidylinositol 4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J Biol Chem (2004) 279(9):7840–9. doi: 10.1074/jbc.M312737200
76. Li L, Kołodziej T, Jafari N, Chen J, Zhu H, Rajfur Z, et al. Cdk5-mediated phosphorylation regulates phosphatidylinositol 4-phosphate 5-kinase type I γ 90 activity and cell invasion. FASEB J (2019) 33(1):631–42. doi: 10.1096/fj.201800296R
77. Xue J, Chen C, Qi M, Huang Y, Wang L, Gao Y, et al. Type Iγ phosphatidylinositol phosphate kinase regulates PD-L1 expression by activating NF-κB. Oncotarget (2017) 8(26):42414–27. doi: 10.18632/oncotarget.17123
78. Hinchliffe KA, Irvine RF. Regulation of type II PIP kinase by PKD phosphorylation. Cell Signal (2006) 18(11):1906–13. doi: 10.1016/j.cellsig.2006.02.005
79. Hinchliffe KA, Ciruela A, Letcher AJ, Divecha N, Irvine RF. Regulation of type IIalpha phosphatidylinositol phosphate kinase localisation by the protein kinase CK2. Curr Biol (1999) 9(17):983–6. doi: 10.1016/s0960-9822(99)80429-1
80. Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, et al. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell (2006) 23(5):685–95. doi: 10.1016/j.molcel.2006.07.014
81. Keune WJ, Jones DR, Bultsma Y, Sommer L, Zhou XZ, Lu KP, et al. Regulation of phosphatidylinositol-5-phosphate signaling by Pin1 determines sensitivity to oxidative stress. Sci Signal (2012) 5(252):ra86. doi: 10.1126/scisignal.2003223
82. Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem (2008) 283(13):8678–86. doi: 10.1074/jbc.M710222200
83. Wright BD, Simpson C, Stashko M, Kireev D, Hull-Ryde EA, Zylka MJ, et al. Development of a high-throughput screening assay to identify inhibitors of the lipid kinase PIP5K1C. J Biomol Screen (2015) 20(5):655–62. doi: 10.1177/1087057114564057
84. Andrews DM, Cartic S, Cosulich S, Divecha N, Faulder P, Flemington V, et al. Identification and optimization of a novel series of selective PIP5K inhibitors. Bioorg Med Chem (2022) 54:116557. doi: 10.1016/j.bmc.2021.116557
85. Chen S, Chandra Tjin C, Gao X, Xue Y, Jiao H, Zhang R, et al. Pharmacological inhibition of PI5P4Kα/β disrupts cell energy metabolism and selectively kills p53-null tumor cells. Proc Natl Acad Sci U.S.A. (2021) 118(21):e2002486118. doi: 10.1073/pnas.2002486118
86. Sivakumaren SC, Shim H, Zhang T, Ferguson FM, Lundquist MR, Browne CM, et al. Targeting the PI5P4K lipid kinase family in cancer using covalent inhibitors. Cell Chem Biol (2020) 27(5):525–537.e526. doi: 10.1016/j.chembiol.2020.02.003
87. Kitagawa M, Liao PJ, Lee KH, Wong J, Shang SC, Minami N, et al. Dual blockade of the lipid kinase PIP4Ks and mitotic pathways leads to cancer-selective lethality. Nat Commun (2017) 8(1):2200. doi: 10.1038/s41467-017-02287-5
88. Manz TD, Sivakumaren SC, Ferguson FM, Zhang T, Yasgar A, Seo HS, et al. Discovery and structure-activity relationship study of (Z)-5-methylenethiazolidin-4-one derivatives as potent and selective pan-phosphatidylinositol 5-phosphate 4-kinase inhibitors. J Med Chem (2020) 63(9):4880–95. doi: 10.1021/acs.jmedchem.0c00227
89. Davis MI, Sasaki AT, Shen M, Emerling BM, Thorne N, Michael S, et al. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PloS One (2013) 8(1):e54127. doi: 10.1371/journal.pone.0054127
90. Wortmann L, Bräuer N, Holton SJ, Irlbacher H, Weiske J, Lechner C, et al. Discovery and characterization of the potent and highly selective 1,7-naphthyridine-based inhibitors BAY-091 and BAY-297 of the kinase PIP4K2A. J Med Chem (2021) 64(21):15883–911. doi: 10.1021/acs.jmedchem.1c01245
91. Willems HMG, Edwards S, Boffey HK, Chawner SJ, Green C, Romero T, et al. Identification of ARUK2002821 as an isoform-selective PI5P4Kα inhibitor. RSC Med Chem (2023) 14(5):934–46. doi: 10.1039/d3md00039g
92. Voss MD, Czechtizky W, Li Z, Rudolph C, Petry S, Brummerhop H, et al. Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem Biophys Res Commun (2014) 449(3):327–31. doi: 10.1016/j.bbrc.2014.05.024
93. Clarke JH, Giudici ML, Burke JE, Williams RL, Maloney DJ, Marugan J, et al. The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem J (2015) 466(2):359–67. doi: 10.1042/bj20141333
94. Boffey HK, Rooney TPC, Willems HMG, Edwards S, Green C, Howard T, et al. Development of selective phosphatidylinositol 5-phosphate 4-kinase γ Inhibitors with a non-ATP-competitive, allosteric binding mode. J Med Chem (2022) 65(4):3359–70. doi: 10.1021/acs.jmedchem.1c01819
Keywords: breast cancer, phosphoinositide, PI(4,5)P2, phosphatidylinositol phosphate kinase, PIP5K, PIP4K
Citation: Jin Y and Xue J (2023) Lipid kinases PIP5Ks and PIP4Ks: potential drug targets for breast cancer. Front. Oncol. 13:1323897. doi: 10.3389/fonc.2023.1323897
Received: 18 October 2023; Accepted: 29 November 2023;
Published: 13 December 2023.
Edited by:
Bekir Cinar, Clark Atlanta University, United StatesReviewed by:
Irene Faenza, University of Bologna, ItalyPatrick Frost, University of California, Los Angeles, United States
Copyright © 2023 Jin and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Jin, eXl1ZV9raW5nQDE2My5jb20=
 Yue Jin
Yue Jin Jian Xue2
Jian Xue2