
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 01 December 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1322403
 Chengjun Hu1†
Chengjun Hu1† Qiuxin Dai1†
Qiuxin Dai1† Ruiyi Zhang2
Ruiyi Zhang2 Huanping Yang2
Huanping Yang2 Man Wang3
Man Wang3 Kaili Gu2
Kaili Gu2 Jiangang Yang1
Jiangang Yang1 Wenjun Meng1
Wenjun Meng1 Ping Chen2*
Ping Chen2* Maozhong Xu1*
Maozhong Xu1*Acute myeloid leukemia (AML) is a malignant disease of myeloid hematopoietic stem/progenitor cells characterized by the abnormal proliferation of primitive and naive random cells in the bone marrow and peripheral blood. Acute promyelocytic leukemia (APL) is a type (AML-M3) of AML. Most patients with APL have the characteristic chromosomal translocation t(15; 17)(q22; q12), forming PML::RARA fusion. The occurrence and progression of AML are often accompanied by the emergence of gene fusions such as PML::RARA, CBFβ::MYH11, and RUNX1::RUNX1T1, among others. Gene fusions are the main molecular biological abnormalities in acute leukemia, and all fusion genes act as crucial oncogenic factors in leukemia. Herein, we report the first case of LYN::LINC01900 fusion transcript in AML with a promyelocytic phenotype and TP53 mutation. Further studies should address whether new protein products may result from this fusion, as well as the biological function of these new products in disease occurrence and progression.
Acute myeloid leukemia (AML), the most common acute leukemia (AL) among adults, is a highly heterogeneous hematological malignant tumor characterized by the proliferation and abnormal differentiation of immature cloned myeloid cells (1). Acute promyelocytic leukemia (APL) is a type (AML-M3) of AML. Most patients have a specific chromosomal translocation t(15;17)(q22;q12), forming PML::RARA gene fusion, whose protein product leads to cell differentiation arrest and insufficient apoptosis, which is the main molecular mechanism of APL occurrence (2). Initial, prompt diagnosis of APL is made through morphological and flow cytometric analyses. APL typically displays a distinct immunophenotype that lacks HLA-DR, CD34, and CD11B expression (3, 4). However, similar immunophenotypic results have been observed in other AML types (5, 6). Thus, diagnosis of this APL-like AML has been a major challenge, and the lag between onset and diagnosis, as well as the resistance to the routine treatment of classical APL, results in an unfavorable prognosis (7, 8).
Many researchers have found that the onset of AML is usually accompanied by the appearance of different gene fusions such as CBFB::MYH11, PML::RARA and RUNX1::RUNX1T1 (9–11). Gene fusions are the main molecular markers for prognostic stratification, minimal residual disease (MRD) monitoring, and targeted therapy in patients with AML (12). Furthermore, interactions between gene fusions and mutated genes can play a crucial role in the prognosis and recurrence of AML (13). With an increasing number of new fusions identified, hematopoietic malignancies have been shown to have greater molecular diversity, which may have important implications for sophisticated subtyping with molecular markers.
Long non-coding RNA (lncRNA) is defined as a non-coding RNA molecule with a length exceeding 200 nucleotides that was once considered as transcriptional ‘junk’ DNA (14). However, with the development of molecular biology and sequencing technology, lncRNAs have been found to play an important role in tumor cell growth, apoptosis, invasion, and metastasis, as well as in the occurrence and progression of diseases (15, 16).
Herein, we identified the LYN::LINC01900 fusion transcript, which was predicted to express no fusion protein in an AML patient with TP53 mutation. Moreover, clinical evidence showed that the bone marrow cells of the patient retained a promyelocytic phenotype; but there was no PML::RARA fusion. This indicated the potential function of this fusion in a new AML subtype. Hence, we hope that this case report on LYN::LINC01900 transcript will provide a new perspective for understanding the occurrence and progression of AML.
A 76-year-old man was admitted to our hospital on October 12, 2022, with dizziness and fatigue for 6 months that had aggravated one month prior. The patient had a history of hypertension for over 20 years and type 2 diabetes mellitus and renal insufficiency for 8 and 2 years, respectively. After admission, a complete routine peripheral blood examination showed a white blood cell count of 1.26*10^9/L, red blood cell count of 1.44*10^12/L, monocyte cell count of 0.05*10^9/L, neutrophil cell count of 0.38*10^9/L, hemoglobin level of 51.0 g/L and platelet count of 32*10^9/L. The lymphocyte count was normal. The fibrinogen and D-dimer levels were 3.30 g/L (reference values [ref.] 2.00–4.00 g/L) and 0.55 mg/L (ref. 0.00–0.55 μg/mL), respectively. The prothrombin and activated partial thromboplastin time were 12.5 s (ref. 10.0–14.0 s) and 27.2 s (ref. 25–31.3 s), respectively.
To understand the cytological morphology, we observed the bone marrow smear of the patient and found that bone marrow cells proliferation and granulocyte proliferation were markedly high. The proportion of primitive granulocytes was 28%, and that of promyelocytes was as high as 56.5%. In addition, Wright Giemsa staining showed that erythrocyte and lymphocytic proliferation were inhibited (Figure 1A). We further performed peroxidase staining test and found that the results of peroxidase staining of the bone marrow cell smear were positive (Figure 1B). Immunophenotyping by flow cytometry showed that the granulocyte population accounted for 58.5%, and these cells were positive for CD117, CD33, and CD38; weakly expressed CD13, CD4, CD64, and CD45; and negative for CD7, CD34, HLA-DR, CD10, CD20, CD19, CD14, CD2, CD15, CD11B, CD56, CD8, and CD3.
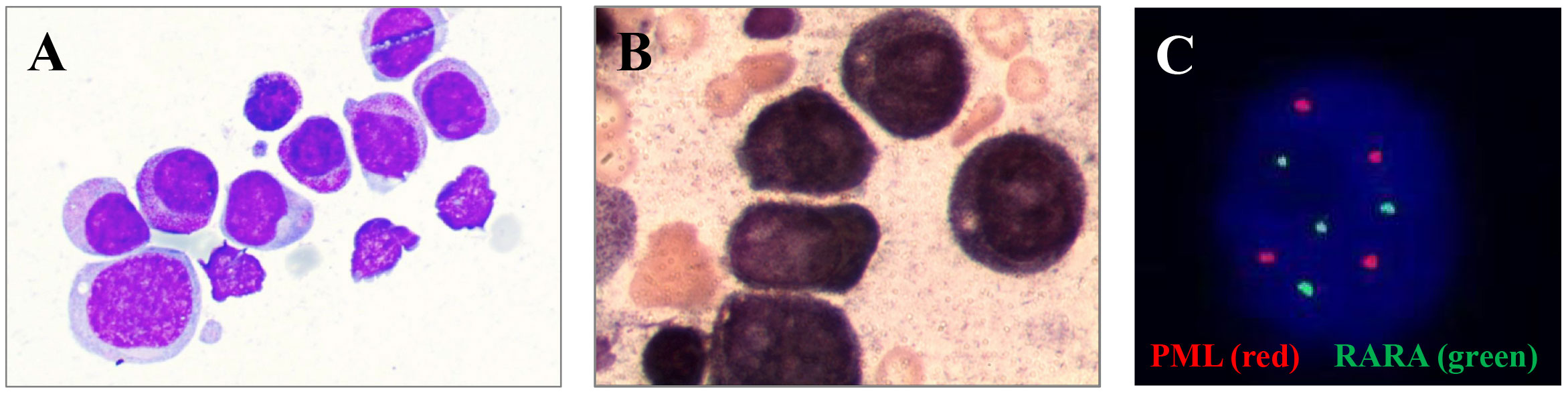
Figure 1 (A) Bone marrow smear of primitive granulocytes under a microscope. (B) Original granulocyte bone marrow smear after peroxidase staining. (C) FISH results of BM cells.
Subsequently, the karyotype of the patient was analyzed. The results revealed a complex karyotype of 73–80, XXY, +1, +2, –3, +4, +6,?der (7), +8, –9, –9, –11, –11, –12, +13, +14, +15, +15, +16, ?17, der (17)?i(17q), +19, +19, +20, +21, +22, +mar, inc[CP3]/45, X, –Y[5]/46, XY[2]. FISH showed no evidence of PML::RARA fusion invloving t (15, 17)(q24; q21) but showed a higher fluorescence signal (normally 2 red and 2 green in single cell, now 4 red and 4 green), indicating gene amplification or +15,+15,+17,+17, which may originate from aberrant cells (Figure 1C). To further identify whether there were molecular variations involving essential genes functioning in hematopoietic malignancies, we used a 45-gene panel to detect gene mutations and a 53-gene panel to conduct a more comprehensive fusion screening using targeted RNA sequencing. Gene mutation results showed that TP53 p.Arg273Cys was detected with a mutation allele frequency (MAF) of 81.4%. WT1 p.Thr277Ile was detected, and the WT1 mutation was suspected to be a germline mutation, based on the MAF of 67.10%. We used STAR software for bioinformatics analysis to predict fusion genes, with the reference genome being hg38. The LYN::LINC01900 transcript, which is a fusion of LYN (NM_001111097.3) exon 8 with LINC01900 (NC_000018.10) exon 2, was detected at a relative transcript expression level of 15.92% normalized to the reference gene ABL1 (Figures 2A, B). We then performed agarose gel electrophoresis on the amplified products (water as a blank control and healthy donor cDNA as a negative control). A band with a length of 230 bp was detected (100 bp of marker), while the reciprocal fusion LINC01900::LYN was not found, consistent with the RNA-targeted sequencing results (Figure 2C). Sanger sequencing of the targeted fusion product confirmed the presence of the LINC01900::LYN fusion transcript (Figure 2D).
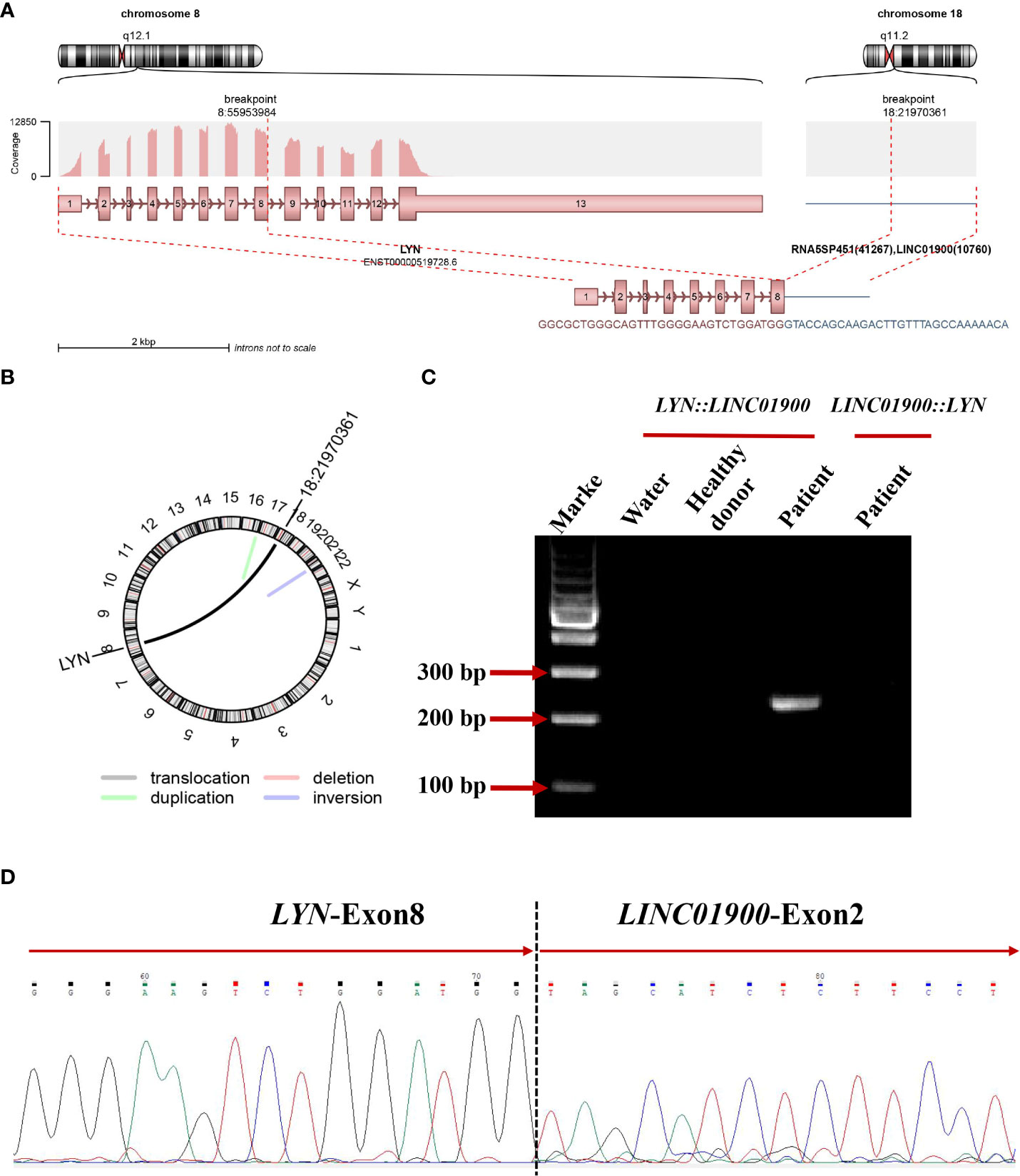
Figure 2 (A) Schematic diagram of the formation mechanism of LYN::LINC01900 transcript. (B) Circos plot indicating novel fusions between LYN and LINC01900. (C) Electrophoresis of RT-PCR products from patient showing the LYN::LINC01900 fusion transcript. (D) Partial nucleotide sequences surrounding the junctions of the LYN::LINC01900 fusion transcript.
The standard azacitidine + venetoclax therapy regimen, which consists of venetoclax (100 mg for day 1, 200 mg for day 2, and 400 mg from days 3 to 28), PO QD, and azacitidine (75 mg/m2 from days 1 to 7), subcutaneous injection (s.c.), was recommended according to the 2022 European LeukemiaNet (ELN) recommendations for the diagnosis and management of AML in adults (17). Considering the patient’s renal dysfunction, the treatment was finally adjusted to venetoclax (100 mg on day 1 and 200 mg from days 2 to 21), PO QD, and azacitidine (75 mg/m2 from days 1 to 7), s.c., supplemented with antiemetic treatment at the same time from November 02, 2022. The therapy regimen was adopted after communicating with the patient and their family members. The patient discharged due to economic reasons on November 09, 2022. After discharged, we conducted a follow-up finding that the patient continued the therapy of venetoclax (200 mg, PO QD) as we suggested until November 22, 2022. Unfortunately, no further clinical outcomes were monitored because the patient died of COVID-19 infection on December 2022 (Figure 3).
LYN, which belongs to the ABL/Src tyrosine kinase family, is a proto-oncogene expressed in many hematopoietic diseases, including AML (18). The functional domains of LYN include four major parts: Src Homology 2 (SH2), SH3, proline-rich hinge region (P), and tyrosine kinase domain (19). The tyrosine kinase encoded by LYN is mainly expressed in hematopoietic cells, nervous tissue, liver, and adipose tissue (20). It also plays an important role in B cell-related signaling, mast cell development, and erythrocyte differentiation (21). Currently, gene rearrangements involving LYN are rarely observed in hematological diseases. There are a total of 6 reported cases of rearrangement involving LYN in hematological diseases, among which ETV6::LYN has been reported in 3 cases across different disease types including AML, primary myelofibrosis (PMF), and myeloproliferative neoplasms (MPN) (22–24). The clinical outcomes of the patients after receiving allogeneic hematopoietic stem cell transplantation (allo-HSCT) and/or chemotherapy showed two deaths and one not available. In a study from the children’s oncology group, Shalini et al. reported the first GATAD2A::LYN transcript in B-cell acute lymphoblastic leukemia (B-ALL). However, no other relevant clinical data have been found in this paper (25). Yano et al. reported a NCOR1::LYN transcript accompanied with additional deletion of IKZF1, CDKN2A/2B, and BTG1 in B-ALL (26, 27). Also in B-ALL, Dai et al. found a NCOR1::LYN transcript accompanied with deletion of IKZF1 and CDKN2A. After receiving allo-HSCT and chemotherapy, the two patients received complete remission (CR) and complete molecular remission (CMR), respectively (28)(Table 1).
A new rearrangement of LYN was found in this patient. This molecule was an lncRNA named LINC01900, lncRNAs have been reported to play a role in tumor cell growth, apoptosis, invasion, and metastasis, as well as in the occurrence and development of diseases by acting as chromatin-modifying factors, X chromosome-inactivating factors, enhancers, transcription regulating factors, and post-transcription regulating factors (29–32). LYN::LINC01900 is formed through translocation, with LYN breaking at chr8:55953984:+ and LINC01900 breaking at chr18:22043217:+. This transcript was an out-of-frame fusion protein that did not produce chimeric proteins. However, it may produce a truncated or possibly non-produced LYN protein, indicating the pathogenic role of haploid LYN dysfunction in this patient. In addition, no evidence of the reciprocal fusion transcript LINC01900::LYN was found. This indicates a more complicated translocation process that may be coupled with further transcriptional or post-transcriptional regulation.
Most patients with APL have a specific chromosomal translocation, t (15, 17)(q22; q12), forming the PML::RARA fusion gene. In our case, the cells retained a promyelocytic phenotype based on blast morphology and flow cytometric analysis. However, the FISH results of PML::RARA fusion gene in bone marrow of this patient suggested that there was no sign of t (15, 17)(q24; q21), but there were higher fluorescence signals indicating gene amplification. Studies have reported that APL-like AML is often accompanied by mutations in NPM1 and/or FLT3 mutation (33, 34). However, no NPM1 mutation was detected. We found a TP53 (NM_000546.5) p.Arg273Cys mutation with a MAF of 81.4%. To the best of our knowledge, this is the first case of APL-like AML with TP53 mutation, which is the most important tumor suppressor factor and is crucial for maintaining cellular genomic integrity. These findings may be used to identify new AML subtypes. Studies have indicated that AML with TP53 mutations is often accompanied by complex karyotypes, which is consistent with the findings in our case (35, 36). Interactions between fusion and mutated genes can also play a crucial role in the prognosis and recurrence of AML (13). LYN::LINC01900 may affect AML progression by interacting with TP53. Interestingly, we found that the expression of LYN was significantly reduced in AML patients with TP53 mutation, compared to the AML patients without TP53 mutation. The transcript per million (TPM) values from RNA-sequencing of the present case appear to be higher than those of the AML patients with TP53 mutations, indicating that the LYN expression in this fusion may increase, which may cause the progression of diseases by regulating expression of other genes related to AML (Figure 4). Further clinical data and reports need to be analyzed to improve this study in the future.
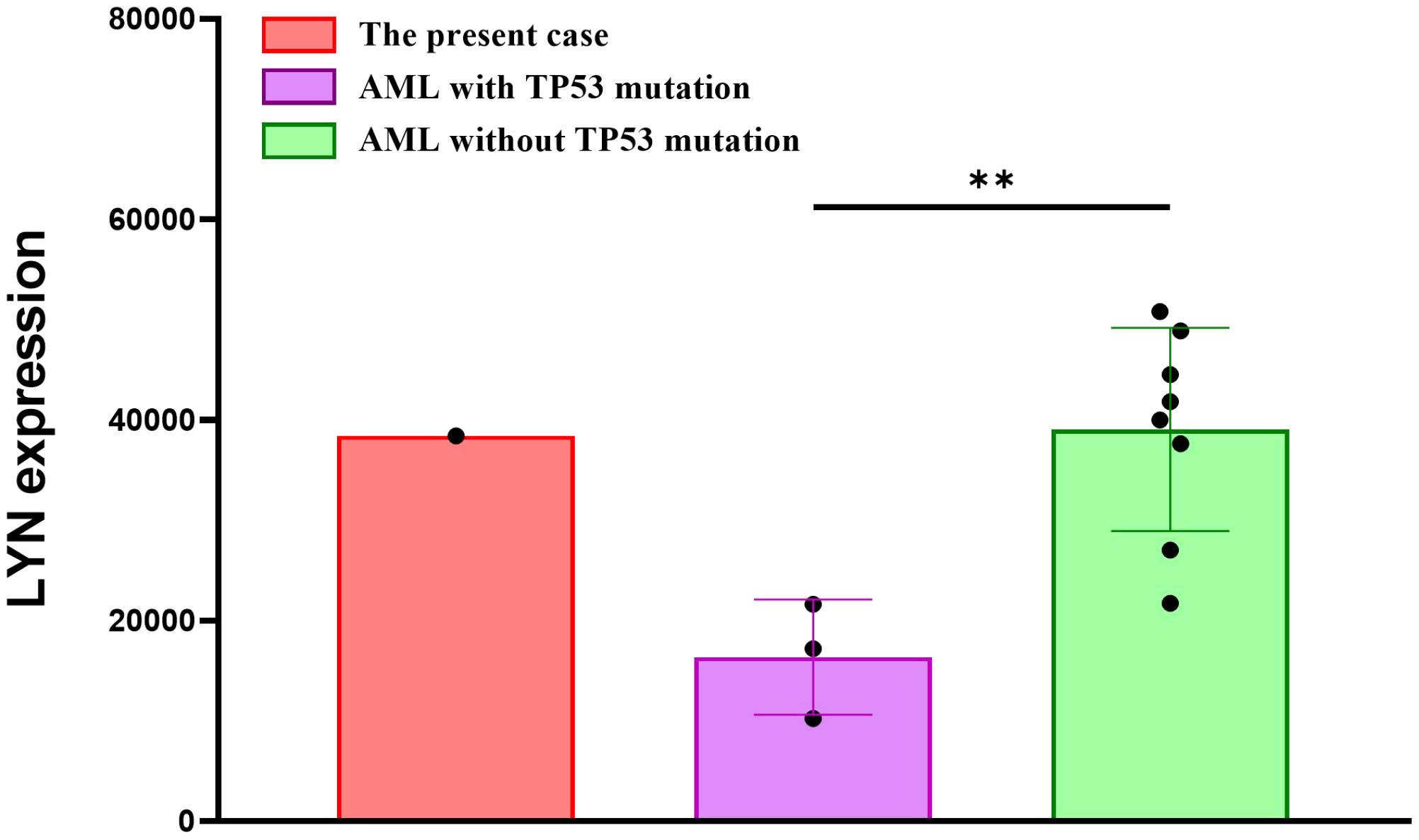
Figure 4 The mRNA expression of LYN in the AML patients with/without TP53 mutation, and the present case.
Although the new fusion transcript was confirmed using different methods, no significant differences in expression of the fusion partner genes were observed. Moreover, no fusion protein was predicted because there may have been no termination codons. Further studies should address whether new protein products may result from this fusion, as well as the biological function of these new products in disease occurrence and progression.
Here, we report the case of a 76-year-old man with AML and a significantly higher proportion of promyelocytes. Wright Giemsa compound staining and peroxidase staining showed that erythrocyte and lymphocytic proliferation were inhibited, and the results of peroxidase staining of the bone marrow cell smear were positive. Immunophenotyping showed that the granulocyte population was positive for CD117, CD33, and CD38; weakly expressing CD13, CD4, CD64, and CD45; and negative for CD7, CD34, HLA-DR, CD10, CD20, CD19, CD14, CD2, CD15, CD11B, CD56, CD8, and CD3. Although a complex chromosomal karyotype was identified, FISH, qPCR, and RNA-targeted sequencing confirmed no evidence of PML::RARA. Additionally, TP53 p.Arg273Cys was identified in the present case. Taken together, this patient was diagnosed with AML with TP53 mutation and a promyelocyte phenotype. A novel fusion LYN::LINC01900 was identified in this case. However, reciprocal fusion was not observed. Furthermore, the fusion was predicted to not express fusion proteins. Future studies should focus on the subgroups of fusions involving non-coding RNAs and their biological functions in hematopoietic malignancies. In conclusion, the LYN::LINC01900 fusion transcript from the patient is the first such fusion to be reported on a global scale, to our knowledge. Here, we share this case report a novel LYN::LINC01900 transcript with TP53 mutation in APL-like AML to provide a new perspective for understanding the molecular diversity of AML.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by Department of Hematology, The Affiliated Jiangyin Hospital of Southeast University Medical College, Jiangyin, Jiangsu, China. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
CH: Data curation, Writing – review & editing. QD: Data curation, Writing – review & editing. RZ: Data curation, Writing – original draft, Writing – review & editing. HY: Data curation, Writing – review & editing. MW: Data curation, Writing – review & editing. KG: Data curation, Writing – review & editing. JY: Data curation, Writing – review & editing. WM: Data curation, Writing – review & editing. PC: Data curation, Methodology, Writing – review & editing. MX: Data curation, Methodology, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
This work was co-finished by Department of Hematology, The Affiliated Jiangyin Hospital of Southeast University Medical College; Suzhou Jsuniwell Medical Laboratory; and Jiangsu Institute of Hematology, National Clinical Research Center for Hematologic Diseases, NHC Key Laboratory of Thrombosis and Hemostasis, The First Affiliated Hospital of Soochow University, Suzhou, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kantarjian H, Kadia T, Dinardo C, Daver N, Borthakur G, Jabbour E, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J (2021) 11(2):41. doi: 10.1038/s41408-021-00425-3
2. De Braekeleer E, Douet-Guilbert N, De Braekeleer M. RARA fusion genes in acute promyelocytic leukemia: a review. Expert Rev Hematol (2014) 7(3):347–57. doi: 10.1586/17474086.2014.903794
3. Orfao A, Chillon MC, Bortoluci AM, López-Berges MC, García-Sanz R, Gonzalez M, et al. The flow cytometric pattern Of CD34, CD15 and CD13 expression in acute myeloblastic leukemia is highly characteristic of the presence of PML-RARalpha gene rearrangements. Haematologica (1999) 84(5):405–12.
4. Paietta E, Andersen J, Gallagher R, Bennett J, Yunis J, Cassileth P, et al. The immunophenotype of acute promyelocytic leukemia (APL): an ECOG study. Leukemia (1994) 8(7):1108–12.
5. Scott AA, Head DR, Kopecky KJ, Appelbaum FR, Theil KS, Grever MR, et al. HLA-DR-, CD33+, CD56+, CD16- myeloid/natural killer cell acute leukemia: A previously unrecognized form of acute leukemia potentially misdiagnosed as french-american-british acute myeloid leukemia-M3. Blood (1994) 84:244–55. doi: 10.1182/blood.V84.1.244.244
6. Kaleem Z, Crawford E, Pathan MH, Jasper L, Covinsky MA, Johnson LR, et al. Flow Cytometric analysis of acute leukemias diagnostic utility and critical analysis of data. Arch Pathol Lab Med (2003) 127:42–8. doi: 10.5858/2003-127-42-FCAOA
7. Zhang X, Sun J, Yu W, Jin J. Current views on the genetic landscape and management of variant acute promyelocytic leukemia. biomark Res (2021) 9:33. doi: 10.1186/s40364-021-00284-x
8. Cicconi L, Testi AM, Montesinos P, Rego E, Zhu HH, Takahashi H, et al. Characteristics and outcome of acute myeloid leukemia with uncommon retinoic acid receptor-alpha (RARA) fusion variants. Blood Cancer J (2021) 11:167. doi: 10.1038/s41408-021-00561-w
9. Liu P, Tarlé SA, Hajra A, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science (1993) 261(5124):1041–4. doi: 10.1126/science.8351518
10. Kakizuka A, Miller WH Jr., Umesono K, Warrell RP Jr, Frankel SR, Murty VV, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell (1991) 66(4):663–74. doi: 10.1016/0092-8674(91)90112-c
11. Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J (1993) 12(7):2715–21. doi: 10.1002/j.1460-2075.1993.tb05933.x
12. Chen X, Wang F, Zhang Y, Ma X, Cao P, Yuan L, et al. Fusion gene map of acute leukemia revealed by transcriptome sequencing of a consecutive cohort of 1000 cases in a single center. Blood Cancer J (2021) 11(6):112. doi: 10.1038/s41408-021-00504-5
13. Guan W, Zhou L, Li Y, Yang E, Liu Y, Lv N, et al. Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns. Exp Hematol Oncol (2021) 10(1):27. doi: 10.1186/s40164-021-00207-4
14. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet (2015) 6:2. doi: 10.1101/gad.1800909
15. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
16. Carlevaro-fita J, Rahim A, Guigo R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA (2016) 22(6):867–82. doi: 10.1261/rna.053561.115
17. Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood (2022) 140(12):1345–77. doi: 10.1182/blood.2022016867
18. Sun Y, Yang Y, Zhao Y, Li X, Zhang Y, Liu Z. The role of the tyrosine kinase Lyn in allergy and cancer. Mol Immunol (2021) 131:121–6. doi: 10.1016/j.molimm.2020.12.028
19. Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity (2005) 22:9–18. doi: 10.1016/S1074-7613(04)00381-4
20. Dos Santos C, Demur C, Bardet V, Prade-Houdellier N, Payrastre B, Récher C, et al. A critical role for Lyn in acute myeloid leukemia. Blood (2008) 111(4):2269–79. doi: 10.1182/blood-2007-04-082099
21. Lamagna C, Hu Y, Defranco AL, Lowell CA. B cell-specific loss of Lyn kinase leads to autoimmunity. J Immunol (2014) 192(3):919–28. doi: 10.4049/jimmunol.1301979
22. Tanaka H, Takeuchi M, Takeda Y, Sakai S, Abe D, Ohwada C, et al. Identification of a novel TEL-Lyn fusion gene in primary myelofibrosis. Leukemia (2010) 24:197–200. doi: 10.1038/leu.2009.167
23. Telford N, Alexander S, McGinn OJ, Williams M, Wood KM, Bloor A, et al. Myeloproliferative neoplasm with eosinophilia and T-lymphoblastic lymphoma with ETV6-LYN gene fusion. Blood Cancer J (2016) 6:e412. doi: 10.1038/bcj.2016.11
24. Ma ESK, Wan TSK, Au CH, Ho DN, Ma SY, Ng MHL, et al. Next-generation sequencing and molecular cytogenetic characterization of ETV6-LYN fusion due to chromosomes 1, 8 and 12 rearrangement in acute myeloid leukemia. Cancer Genet (2017) 218-219:15–9. doi: 10.1016/j.cancergen.2017.09.001
25. Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, et al. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood (2017) 129:3352–61. doi: 10.1182/blood-2016-12-758979
26. Yano M, Imamura T, Asai D, Kiyokawa N, Nakabayashi K, Matsumoto K, et al. Identification of novel kinase fusion transcripts in paediatric B cell precursor acute lymphoblastic leukaemia with IKZF1 deletion. Br J Haematol (2015) 171:813–7. doi: 10.1111/bjh.13757
27. Imamura T, Kiyokawa N, Kato M, Imai C, Okamoto Y, Yano M, et al. Characterization of pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia with kinase fusions in Japan. Blood Cancer J (2016) 6(5):e419. doi: 10.1038/bcj.2016.28
28. Dai HP, Yin J, Li Z, Yang CX, Cao T, Chen P, et al. Rapid molecular response to dasatinib in a pediatric relapsed acute lymphoblastic leukemia with NCOR1-LYN fusion. Front Oncol (2020) 10:359. doi: 10.3389/fonc.2020.00359
29. Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF, Shan L, et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell (2020) 181(3):621–36.e22. doi: 10.1016/j.cell.2020.03.006
30. Mumbach MR, Granja JM, Flynn RA, Roake CM, Satpathy AT, Rubin AJ, et al. HiChIRP reveals RNA-associated chromosome conformation. Nat Methods (2019) 16(6):489–92. doi: 10.1038/s41592-019-0407-x
31. Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev (2016) 30(10):1224–39. doi: 10.1101/gad.276022.115
32. Oleary VB, Ovespian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep (2015) 11(3):474–85. doi: 10.1016/j.celrep.2015.03.043
33. Michael P, Brent T. Acute myeloid leukemia with NPM1 and FLT3 ITD mimicking acute promyelocytic leukemia. Blood (2020) 136(12):1467. doi: 10.1182/blood.2020007198
34. Arana Rosainz MJ, Nguyen N, Wahed A, Lelenwa LC, Aakash N, Schaefer K, et al. Acute myeloid leukemia with mutated NPM1 mimics acute promyelocytic leukemia presentation. Int J Lab Hematol (2021) 43(2):218–26. doi: 10.1111/ijlh.13357
35. Short NJ, Montalban-Bravo G, Hwang H, Ning J, Franquiz MJ, Kanagal-Shamanna R, et al. Prognostic and therapeutic impacts of mutant TP53 variant allelic frequency in newly diagnosed acute myeloid leukemia. Blood Adv (2020) 4(22):5681–9. doi: 10.1182/bloodadvances.2020003120
Keywords: acute myeloid leukemia, LYN::LINC01900, promyelocytic phenotype, lncRNA fusion, mutation
Citation: Hu C, Dai Q, Zhang R, Yang H, Wang M, Gu K, Yang J, Meng W, Chen P and Xu M (2023) Case Report: Identification of a novel LYN::LINC01900 transcript with promyelocytic phenotype and TP53 mutation in acute myeloid leukemia. Front. Oncol. 13:1322403. doi: 10.3389/fonc.2023.1322403
Received: 16 October 2023; Accepted: 15 November 2023;
Published: 01 December 2023.
Edited by:
Sebastien Malinge, University of Western Australia, AustraliaReviewed by:
Valeria Visconte, Cleveland Clinic, United StatesCopyright © 2023 Hu, Dai, Zhang, Yang, Wang, Gu, Yang, Meng, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maozhong Xu, eHVtYW96aG9uZ2p5QDE2My5jbQ==; Ping Chen, cGluZ19jaGVuQGZ1amlkYWJpby5jb20=
†These authors contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.