
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 18 December 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1273516
 Marian Constantin1,2
Marian Constantin1,2 Mariana Carmen Chifiriuc2,3,4*
Mariana Carmen Chifiriuc2,3,4* Grigore Mihaescu3
Grigore Mihaescu3 Corneliu Ovidiu Vrancianu2,3,5
Corneliu Ovidiu Vrancianu2,3,5 Elena-Georgiana Dobre2,6
Elena-Georgiana Dobre2,6 Roxana-Elena Cristian2,5,7
Roxana-Elena Cristian2,5,7 Coralia Bleotu2,8
Coralia Bleotu2,8 Serban Vifor Bertesteanu9
Serban Vifor Bertesteanu9 Raluca Grigore9
Raluca Grigore9 Bogdan Serban10
Bogdan Serban10 Catalin Cirstoiu10
Catalin Cirstoiu10Head and neck cancer (HNC) is the sixth most common type of cancer, with more than half a million new cases annually. This review focuses on the role of oral dysbiosis and HPV infection in HNCs, presenting the involved taxons, molecular effectors and pathways, as well as the HPV-associated particularities of genetic and epigenetic changes and of the tumor microenvironment occurred in different stages of tumor development. Oral dysbiosis is associated with the evolution of HNCs, through multiple mechanisms such as inflammation, genotoxins release, modulation of the innate and acquired immune response, carcinogens and anticarcinogens production, generation of oxidative stress, induction of mutations. Thus, novel microbiome-derived biomarkers and interventions could significantly contribute to achieving the desideratum of personalized management of oncologic patients, regarding both early diagnosis and treatment. The results reported by different studies are not always congruent regarding the variations in the abundance of different taxons in HNCs. However, there is a consistent reporting of a higher abundance of Gram-negative species such as Fusobacterium, Leptotrichia, Treponema, Porphyromonas gingivalis, Prevotella, Bacteroidetes, Haemophilus, Veillonella, Pseudomonas, Enterobacterales, which are probably responsible of chronic inflammation and modulation of tumor microenvironment. Candida albicans is the dominant fungi found in oral carcinoma being also associated with shorter survival rate. Specific microbial signatures (e.g., F. nucleatum, Bacteroidetes and Peptostreptococcus) have been associated with later stages and larger tumor, suggesting their potential to be used as biomarkers for tumor stratification and prognosis. On the other hand, increased abundance of Corynebacterium, Kingella, Abiotrophia is associated with a reduced risk of HNC. Microbiome could also provide biomarkers for differentiating between oropharyngeal and hypopharyngeal cancers as well as between HPV-positive and HPV-negative tumors. Ongoing clinical trials aim to validate non-invasive tests for microbiome-derived biomarkers detection in oral and throat cancers, especially within high-risk populations. Oro-pharyngeal dysbiosis could also impact the HNCs therapy and associated side-effects of radiotherapy, chemotherapy, and immunotherapy. HPV-positive tumors harbor fewer mutations, as well as different DNA methylation pattern and tumor microenvironment. Therefore, elucidation of the molecular mechanisms by which oral microbiota and HPV infection influence the HNC initiation and progression, screening for HPV infection and vaccination against HPV, adopting a good oral hygiene, and preventing oral dysbiosis are important tools for advancing in the battle with this public health global challenge.
Head and neck cancers (HNCs) encompass a group of malignancies that predominantly originate in the squamous cells (HNSCC) lining the upper aerodigestive mucosa (1) and rank as the sixth most common cancer globally, with an annual incidence of over half a million new cases, projected to exceed 900,000 new cases in 2020 (2).
HNCs have a multifactorial etiology, including genetic and epigenetic mechanisms (3), oral dysbiosis (4), infections with human papilloma virus (HPV), mostly oncogenic types 16 and 18 (5), and EBV (Epstein-Barr virus) (6), laryngopharyngeal reflux (7), prior exposure to radiotherapy (8) as well as various lifestyle features, such as heavy smoking and alcohol consumption (9, 10), chewing betel quid (Areca nuts) (11), marijuana use (12), poor oral hygiene (13, 14), pro-inflammatory diet (e.g., fried, smoked, or roasted meat) (15), oral dysbiosis (16), prolonged exposure to sunlight, inhalation of chemical pollutants (17–22) (Figure 1). These multifaceted risk factors highlight the need for comprehensive strategies in both prevention and management to combat HNCs effectively.
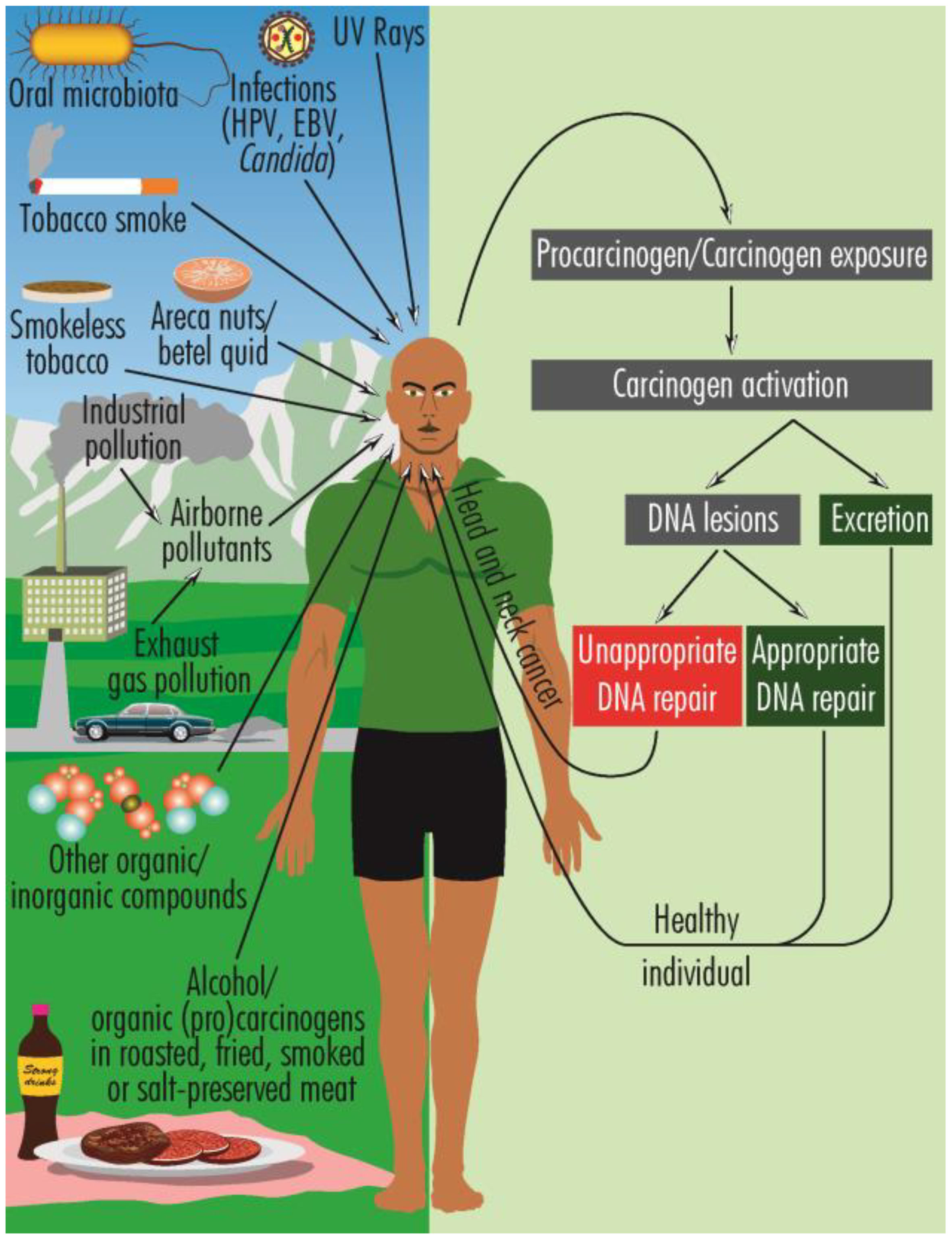
Figure 1 Schematic overview of the predisposing factors for HNCs and the general pattern of carcinogenesis. The activated carcinogens can produce DNA damage or are excreted. When the cellular repairing mechanisms are functioning properly, the DNA damage is repaired, but when these mechanisms are ineffective, genetic defects are perpetuated and can ultimately lead to HNCs.
Infectious agents, including bacteria, fungi, parasites and viruses are known to cause an important percentage of the total number of cancers. Among viruses, HPV is one of the main risk factors for HNCs, the HPV infection being among the several biomarkers that can be used for the early diagnosis of HNCs (the viral DNA being found in approximately 4.5% of all cancers and 25% of HNCs (23–27).
Recent evidence shows that human microbiome and dysbiosis are also associated with HNCs and could provide novel biomarkers for getting one step closer to the desideratum of personalized management of oncologic patients. The oral dysbiosis has been associated with chronic inflammation (28), genotoxins release (29), generation of carcinogens or inhibition of anticarcinogenic compounds synthesis (30), favoring the occurrence of a pro-tumor local microenvironment and causing tumor growth (29). All these effects could be involved in the genesis or progression of different malignancies such as upper aerodigestive tract, esophagus, stomach, pancreas, colorectum, liver, lung and breast cancer (31).
Several papers recently described the role of HPV infection and the composition of the host microbiome in HNCs carcinogenesis (4, 28, 32–35). However, there is still a lack of understanding of the association between human microbiota signatures and the risk of HNCs, designating new biomarkers in HNCs diagnosis, and the implication of dysbiosis in HNCs therapy. Therefore, in this review, we discussed first the most recent original paper and clinical trials aiming to investigate the human microbiota signatures associated with HNC and human microbiota signatures associated with a reduced risk of HNC. Secondly, we discussed the potential microbiome-derived biomarkers for HNCs diagnosis and implications of oro-pharyngeal dysbiosis in HNCs therapy, presenting the results from the very recent studies over the past two years (36–53). Finally, we discussed the contribution of HPV infection to HNCs initiation and progress and implications for prevention, early diagnosis, and treatment.
The oral cavity represents one of the most complex microbiomes in the human body (54), providing support and resources for an impressive number of microbial species (from 700-750 to several thousands) (28, 29), which include bacteria, archaea, fungi and protozoa (55), from at least 12 phyla (28), such as Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Spirochaetes, Synergistetes and Tenericutes (28). Oral microbes colonize both soft tissues (tongue, soft palate, oral mucosa, and tonsils) and hard tissues (teeth), developing specific biofilms (55). Under normal conditions, called eubiosis, the healthy microbiota shows relatively constant proportions of different taxons and stable diversity, which, in dysbiosis state, are disrupted in favor of commensal proinflammatory and pathogenic species (29).
At phylum level, Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, and Actinobacteria were the five most abundant phyla and accounted for > 90% of the bacterial community in aerodigestive tract cancers, including HNC (51). Other studies reveal a decrease of the phyla Actinobacteria and Cyanobacteria in HNCs (4, 29, 56).
At the genus level, Streptococcus, Abiotrophia, Prevotella and Leuconostoc were significantly reduced in the aerodigestive tract cancers group, Haemophilus increased, while Neisseria has been reported to have either high or low abundance (51, 57).
Other genera commonly associated with HNCs are Fusobacterium, Leptotrichia, Selenomonas, Treponema, Clostridium, and Pseudoalteromonas (40). In oral squamous cell carcinoma (OSCC), an increased abundance of Parvimonas, Fusobacterium (including F. nucleatum, which is reported to be the most abundant species in OSCCs samples, and F. periodonticum), Pseudomonas (Pseudomonas aeruginosa is reported to be the second most abundant species in OSCCs), Porphyromonas gingivalis (reported in gingival squamous cell carcinoma), Peptostreptococcus, Alloprevotella, Capnocytophaga, Prevotella, Bacteroidetes and Solobacterium, Actinomyces, Lactobacillus, Rothia, Haemophilus and Veillonella are reported (4, 29, 56–59).
Certain aerobic and facultative anaerobic bacteria, including Klebsiella, Citrobacter, Streptococcus, Enterobacter, and Serratia, have been found to affect the local tumor microenvironment in oral carcinoma (4).
Candida albicans is the dominant fungi found in oral carcinomas, a strong association between higher Candida carriage and a notably shorter overall survival (OS) being observed in patients with OSCC (60).
The abundance of different microbial species may change during tumor progression, and possible microbiota signatures can be associated with tumor stages and prognosis. High levels of serum class G antibodies against F. nucleatum were found in patients with gastrointestinal cancer and HNCs. However, in HNSCC developed by non-smokers, the abundance of F. nucleatum is associated with early tumor stages, but with reduced likelihood of recurrence, and increased survival duration (36).
During the progression of OSCC from stage I to IV, the abundance of F. periodonticum, Parvimonas micra, Streptococcus constellatus, Haemophilus influenzae, and Filifactor alocis gradually increases, while Actinobacteria phyla and Streptococcus mitis, Haemophilus parainfluenzae and Porphyromonas pasteri decrease (56, 61). Decrease of Parvimonas and increase of Fusobacterium (especially that of F. nucleatum), Rothia, Haemophilus, Veillonella, and Actinomyces is associated with early stages of tumor development (36, 56, 62, 63). A comprehensive systematic review concluded that F. nucleatum is present and in higher abundance in oral cancer samples when compared to non-cancer samples, suggesting that could contribute to oral cancer development (64). However, it is also possible that tumor colonization by F. nucleatum reflects its ability to exploit and replicate effectively in the hypoxic tumor microenvironment. To date, several F. nucleatum-carcinoma mechanisms have been discovered: promotion of the Wnt/β-catenin signaling pathway through FadA binding to E-cadherin (65), inhibition of the cytotoxicity of immune cells such as NK cells and T-cell activity (66), LPS binding to TLR4/MYD88 pathway and mediating downstream NF-κB expression (67) and the release of Fap2 that binding to Gal-GalNAc ligands (68). This evidence that F. nucleatum colonization begins early in the process of malignant transformation supports a potential role for microbiome changes in the pathogenesis of the disease (64). Consequently, Coppenhagen-Glazer and collaborators found that F. nucleatum is a key organism between early and late colonisers and its outer membrane adhesin Fap2 is partly responsible for facilitating multispecies biofilm formation (69). Also, it was revealed that the enrichment of F. nucleatum in OSSC is associated with host gene promoter methylation, including hypermethylation of tumor suppressor genes LXN and SMARCA2, a gene involved in ATP-dependent chromatin remodeling related to DNA repair and replication. This suggests that F. nucleatum enrichment may cause cell proliferation through epigenetic silencing (70).
Increasing abundance of Bacteroidetes and Peptostreptococcus has been associated with later stages and larger tumors (61). In terms of subsequent development, the presence of higher numbers of Stenophotromonas, Staphylococcus, Centipeda, Selenomonas, Alloscordovia, and Acinetobacter genera in the saliva of individuals with OSCC is associated with poor prognosis and poorer survival rate (63). Dou and collaborators observed that increased numbers of Schlegelella and Methyloversatilis in HNCs are associated with poor prognosis, while abundant Bacillus, Lactobacillus, and Sphingomonas are found in patients with favorable prognosis (37, 71).
Salivary bacteria, such as S. salivarius, Corynebacterium, and Stomatococcus, are associated with a strong oxidative stress that may contribute to oncogenesis and cancer development (4). The presence of P. gingivalis in the oral cavity tend to be associated with higher mortality rates (61, 72, 73). P. gingivalis is known to stimulate the production of myeloid-derived dendritic suppressor cells, which can inhibit the activity of cytotoxic T lymphocytes, a key component of the antitumoral immunity. Additionally, this bacterium can induce the overexpression of matrix metalloproteinase-9 and reduce the expression of the tumor suppressor gene TP53, thereby promoting cell proliferation and potentially contributing to cancer development (74). Some members of oral microbiota are metabolizing alcohol to acetaldehyde, a potent carcinogen and are reducing the synthesis of anticarcinogenic compounds, including siderophore group non-ribosomal peptides, 12-, 14- and 16-membered macrolides and monoterpenoids (30), The abundance of microorganisms in the phyla Actinobacteria is associated with mutations in TP53, while high numbers of Firmicutes with recurrent mutations in FAT1, FZR1, AXIN1 and WNT (29).
C. albicans was also discovered to play a role in accelerating the progression of OSCC in vitro. This acceleration was attributed to several mechanisms, including the increased synthesis of matrix metalloproteinases and oncometabolites, the promotion of pro-tumor signaling pathways, and the upregulation of genes associated with prognostic markers for metastatic events (44, 75). Given these insights, there is potential for interventions targeting C. albicans to serve as a therapeutic strategy for HNC, offering promising avenues for developing novel treatments.
The abundance of microorganisms of the genera Corynebacterium, Kingella (especially K. denitrificans), Neisseria, Abiotrophia, and Capnocytophaga is associated with a reduced risk of laryngeal cancers and the increased abundance of Actinomyces (A. oris), N. sicca and Veillonella denticariosi species with a reduced risk of pharyngeal and other HNCs (29, 74). Many commensal bacteria from the genera Corynebacterium and Kingella appear to have a preventive effect on developing HNCs (76), and the abundance of Veillonella is associated with better overall prognosis of OSCCs (63). In 2018, a nested case-control study including 129 HNC patients, revealed that Corynebacterium and Kingella can reduce the risk of developing HNCs by contributing to the breaking down and neutralization of harmful toxic substances, including compounds like toluene, styrene, and chlorobenzene (76).
Another recent nested case-control study aiming to investigate the relationship of oral microbiome with HNC demonstrated that the presence of oral fungi and relative abundance of multiple microbial species, including the red- and orange-complex periodontal pathogens (C. albicans, K. oralis, P. gingivalis), were associated with reduced risk of HNC (53).
The relationships between several types of HNCs and microorganisms is summarized in Table 1.
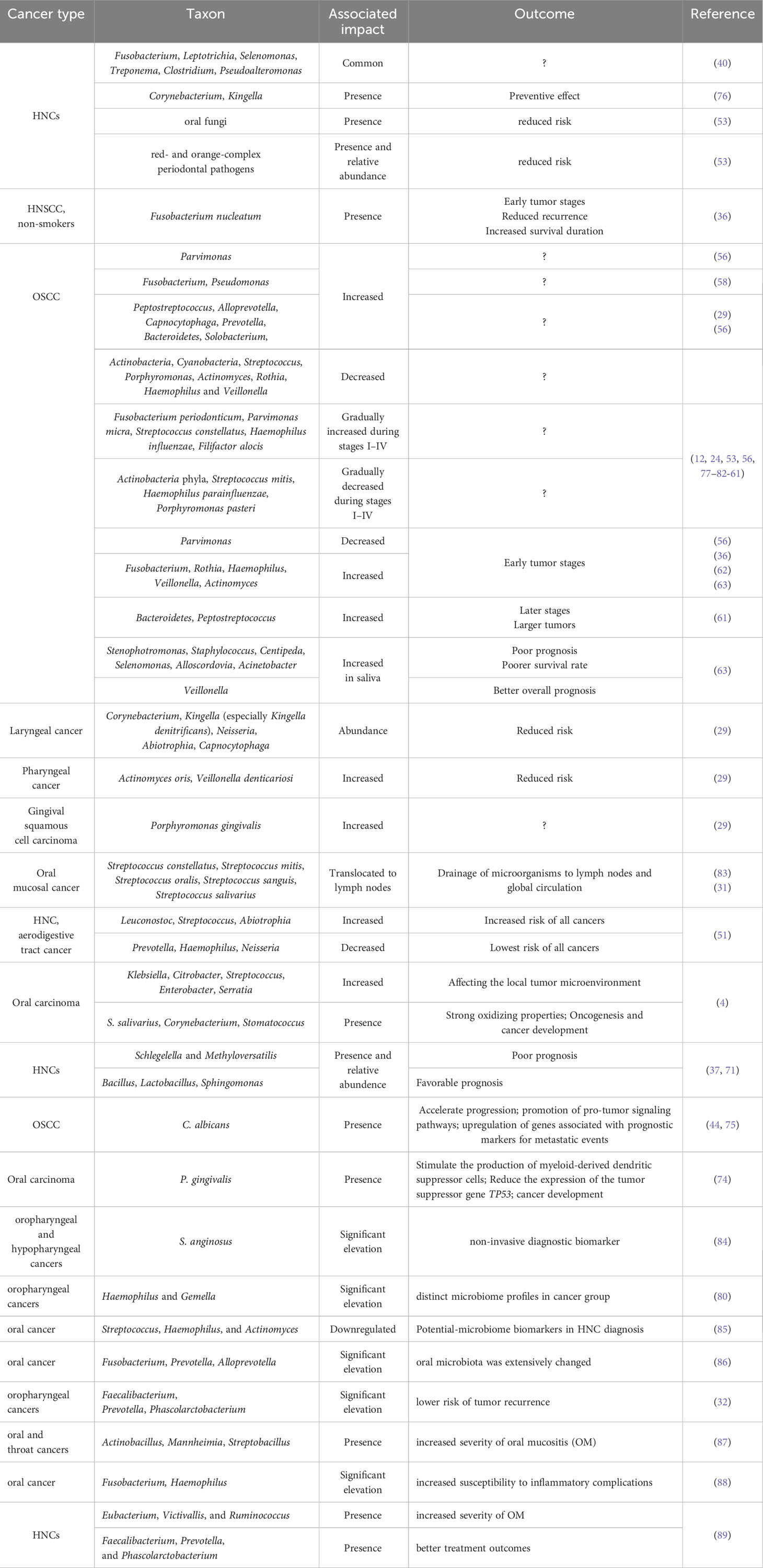
Table 1 Association of some types of HNCs with microorganisms present in the oral cavity and their outcomes.
In HNC, the current standard of screening and diagnosis relies on a physical exam and identification of lesions, followed by imaging, invasive biopsy, and histopathological evaluation (90). Current research aims to investigate and establish new microbiome signatures as potential microbiome-derived biomarkers for HNC diagnosis (45, 61, 84, 85, 91).
Differences in the microbiome profile between oropharyngeal and hypopharyngeal cancers were observed, with S. anginosus showing significant elevation in the saliva of oropharyngeal cancer patients (84). Mutational changes influence the abundance of bacterial groups like Firmicutes and Bacteroidetes, varying among different mutation profiles (61). In HPV-positive oropharyngeal cancers, a pilot study revealed distinct microbiome profiles compared to healthy controls, with a notable correlation between Haemophilus and Gemella genera in HPV-positive oropharyngeal cancer (80). Furthermore, certain bacterial species, including Actinomyces, Parvimonas, Selenomonas, and Prevotella, were more abundant in oral cavity cancers (91). At the species level, S. salivarius and S. vestibularis were identified as abundant in oral OSCC samples, while species from the vaginal microbiota, such as L. gasseri / johnsonii and L. vaginalis, were abundant in saliva (92). Banavar and collaborators conducted a study including 242 patients with oral cancer, aiming to develop and investigate machine-learning classifiers using metatranscriptomic data from saliva samples. The developed metatranscriptomic signatures incorporated both taxonomic and functional microbiome features, and revealed several taxa and functional pathways associated with oral cancers. The authors observed that several genera, such as Streptococcus, Haemophilus, and Actinomyces, are downregulated, while some other genera, like Fusobacterium, do not appear to be differentially expressed. At the genus level, the results revealed periodontal bacteria like Fusobacterium, Prevotella, and Porphyromonas in saliva samples from oral cancers. More recently, the same research group conducted a clinical trial aiming to develop and validate a non-invasive test for biomarkers detection in oral and throat cancers within a high-risk population. The authors collected saliva samples from 1175 patients and used machine learning methods to obtain a salivary microbial and human metatranscriptomic signature. This developed test, named CancerDetect for Oral and Throat Cancer (CDOT), has received the FDA’s breakthrough designation for accelerated review (45). These studies demonstrated the potential of a machine-learning tool for oral cancer diagnosing, opening a new era of non-invasive diagnostics, enabling early intervention, and improving patient outcomes.
In addition, Inchingolo and collaborators conducted a meta-analysis to evaluate the interplay between microbiota and oral cancer and the presence of biomarkers as risk predictors. The analysis of the results from 21 studies revealed the correlation between oral cancers and changes in the microbiota, which explains the paramount value of precision medicine in the diagnosis and treatment of HNCs (47). Ganly and collaborators observed that oral microbiota was extensively changed in oral cancer patients due to the increase of periodontal pathogens like Fusobacterium, Prevotella, and Alloprevotella and the reduction in commensal Streptococcus. Based on these marker genera, the oral microbiota was split into two types: periodontal-pathogen-low and periodontal-pathogen-high. This classification predicted oral cancer with 80% accuracy. In addition to the three periodontal pathogens discovered in the samples, the cumulative abundance of 14 periodontal pathogens increased gradually throughout the sequence of negative controls. These data consistently indicate that periodontal infections are an independent risk factor in patients who do not have substantial oral risk factors (86). Furthermore, in 5 patients with OSCC, saliva metaproteomics indicated a substantial rise in Prevotella and the adhesion and virulence factors linked to S. gordonii, as well as oral pathogens like Fusobacterium (93). In a study by Li and collaborators, the microbial composition in three distinct groups of samples from patients with oral cancer was investigated using metagenomic sequencing. The study found that while there was limited variation in the microbial diversity of the three groups, the oral microbiome of patients with precancerous lesions exhibited greater diversity than that of both oral cancer patients and healthy controls. Notably, a specific strain of Bacteroidetes within the phylum displayed differential enrichment in the oral cancer samples. Furthermore, at the genus level, the primary differentially enriched taxa included Prevotella, Peptostreptococcus, Carnobacterium, and Diastella. P. intermedia and Peptostreptococcus stomatis were identified as having distinct species-level enrichment patterns, suggesting that these profiles can be employed as diagnostic markers (67). A rise in potentially pathogenic bacteria, such as Capnocytophaga, and other LPS-producing bacteria, such as Neisseria, were seen in the oral microbiome of 56 HNC patients. The study concluded that HNC-related symptoms in conjunction with salivary microorganisms such as Capnocytophaga may be employed as a noninvasive technique for screening, identification, and treatment monitoring of HNC (94). In patients with OSCC, significant increase of Fusobacterium and a concomitant reduction in Firmicutes and Actinobacteria phyla have been found. Significant distinctions were also revealed in Actinobacteria, Firmicutes, Fusobacteriia, Fusobacteriales, Fusobacteriaceae, and Fusobacterium. These findings brought into light five unique oral microorganisms with high confidence and may be used to predict clinical diagnosis and prognosis (48).
The composition of a patient’s gut microbiota impacts the effectiveness and side effects of radiotherapy, chemotherapy, and immunotherapy, playing a significant role in HNC outcomes. A prospective pilot study including 20 HNC patients has shown that a pre-treatment microbiota enriched with Eubacterium, Victivallis, and Ruminococcus is associated with a higher risk of experiencing OM, a common side effect of cancer treatment that affects the mouth and throat. Conversely, when the gut microbiota has a higher relative abundance of immunomodulatory microbes such as Faecalibacterium, Prevotella, and Phascolarctobacterium, patients are at a lower risk of tumor recurrence (32). These microbes seem to play a role in modulating the response to immunotherapy by potentially enhancing the expansion and function of CD8+ T cells, which are crucial for mounting an effective antitumor immune response. However, it is essential to note that more extensive research is required to validate these associations and determine whether modifying the gut microbiota can predict and optimize treatment outcomes for HNC patients. Dysbiosis has been shown to promote the persistence of ulcers and delay the healing process (88). The presence of certain bacteria, such as Actinobacillus, Mannheimia, and Streptobacillus, has been associated with increased severity of OM (87). Fusobacterium and Haemophilus, when dominant in the oral microbiome before radiotherapy, are associated with an increased susceptibility to inflammatory complications. Specific bacteria, including Prevotella, Fusobacterium, Streptococcus, Megasphaera, and Cardiobacterium, have been considered as prognostic biomarkers for the onset of OM (88). Research by Jiang and collaborators demonstrated that patients who received probiotics during chemoradiotherapy experienced a lower incidence of oral mucositis compared to those who did not receive probiotics (95). Similarly, a study by Ma and collaborators found that patients who received probiotic therapy were more likely to complete radiotherapy without complications, in contrast to those without probiotics, where patients had to discontinue treatment due to complications (96). The research conducted by Al-Qadami and collaborators has revealed significant associations between specific bacterial genera and the severity of OM and treatment outcomes in cancer patients. Three bacterial genera, namely Eubacterium, Victivallis, and Ruminococcus, were found to be linked to more severe OM. On the other hand, the presence of bacterial genera Faecalibacterium, Prevotella, and Phascolarctobacterium was associated with better treatment outcomes (89). In conclusion, modifying the gut microbiota to align with more favorable treatment outcomes represents a promising avenue for future research and clinical practice.
According to Routy et al., patients who do not respond well to immunotherapy often have gut dysbiosis (97). Other studies show that addition of dietary supplementation with Bifidobacterium appears to have a comparable effect on tumor control compared to treatment with a specific antibody therapy targeting programmed death-ligand 1 (PD-L1). Furthermore, when combined, these therapies nearly eliminated the expansion of tumors, suggesting a synergistic or enhanced therapeutic effect (98). Similarly, Lactobacillus and Bacteroides species could trigger type I interferon production in dendritic cells, enhancing the cross-priming of antitumor CD8+ T cells having different impacts on the immunostimulatory effects (43). Additionally, those who have undergone antibiotic therapy, particularly immediately before or during cancer treatment, face a higher risk of rapid disease progression (97).
The severity of oral injuries in patients undergoing radiation therapy, such as changes in saliva quantity and composition, alterations in the oral microbiota, and tooth damage, is primarily linked to the radiation dose delivered to the oral cavity region (99). Radiation-induced acidification of the oral environment creates a favorable condition for the proliferation of acidogenic and cariogenic bacteria, such as S. mutans, Actinomyces, and Lactobacillus, while reducing the populations of Neisseria, Fusobacterium, and S. sanguinis. Untreated, these caries-associated bacteria contribute to developing radiation therapy-related dental caries (87). Furthermore, C. albicans can take advantage of these shifts in the oral microbiota, potentially leading to superinfections during and after therapy (99). A study by Huo et al. prospectively evaluated the dynamic changes in oral microbiota during radiation therapy and its association with the progression or aggravation of oropharyngeal mucositis in a cohort of nasopharyngeal carcinoma patients. The results showed that while the overall richness and evenness of mucosal bacterial diversity did not vary significantly during treatment, certain bacteria, such as Prevotella, Fusobacterium, Treponema, and Porphyromonas, exhibited noticeable synchronized shifts in their abundance throughout radiation therapy. These shifts often coincided with the onset of severe mucositis, suggesting that dysbiosis of the oral mucosal microbiota may play a role in exacerbating mucositis in nasopharyngeal carcinoma patients during radiation therapy (99). A clinical study by Mougeot and collaborators investigated the oral microbiome implications in developing post-radiotherapy caries in 31 HNC patients. The results suggested that baseline microbiome difference is an essential factor explaining dental caries outcomes in radiation-treated HNC patients. Also, the cariogenic role of P. melaninogenica and a potential protective role of specific bacterial species such as A. defectiva was reported (100).
In HNC patients undergoing chemoradiotherapy, treatment is often associated with challenging side effects, such as mucositis and dysphagia, to which oropharyngeal microbiota might contribute, but the precise causal relationship and clinical significance have remained unclear. To shed light on this matter, a prospective longitudinal observational study involving 47 HNC patients was conducted to determine if dysbiosis is present in HNC and to assess the impact of chemoradiotherapy on the dynamics of dysbiosis during and after treatment. The salivary microbiome in the HNC patients before initiating treatment exhibited notable differences in composition and decreased diversity compared to a control group of healthy individuals. During treatment, there was a significant decrease in α-diversity and a marked shift in β-diversity, suggesting a significant change in microbial composition compared to the pre-treatment state and healthy controls. The microbiome analysis showed no significant difference in α-diversity between HNC patients with severe mucositis and those with mild to moderate mucositis before treatment. However, marked differences in α-diversity emerged immediately after the completion of chemoradiotherapy (52). Omega-3 (ω-3) polyunsaturated fatty acids have recently gained a particular interest in dealing with oral diseases owing to their anti-inflammatory, antioxidant, and wound-healing properties (101). In a recent clinical study conducted by Morsy and collaborators, 34 HNC patients received radiotherapy and topical Omega-3 nanoemulgel. A significant reduction in Firmicutes/Bacteroidetes ratio was observed after six weeks in the test group, indicating less microbial dysbiosis. The results demonstrated that topical omega-3 nanoemulgel has a beneficial effect in preventing radiation-induced OM with the possibility of regulating oral microbial dysbiosis (50).
The success of immune checkpoint inhibitors in the context of palliative systemic therapy for HNSCC and the potential for combining these immunotherapies with radiotherapy have brought to the forefront the exploration of interactions between the tumor microenvironment and the immune landscape (39, 102). It has become increasingly clear that the regulation of the immune system by the microbiota is of paramount importance for both innate and adaptive immune surveillance against tumors and for the success of treatment with these immune checkpoint inhibitors (39, 103).
Recently, a meta-analysis conducted by Frey-Furtado and colleagues examined nine articles to evaluate the therapeutic effectiveness of probiotics in managing OM. Among these studies, four clinical trials reported a decrease in the severity of OM by using specific strains of bacteria, including Lactobacillus (L. casei and L. brevis CD2) and B. clausii UBBC07. Preclinical studies revealed the positive effects of L. lactis, L. reuteri, and S. salivarius K12 in reducing the severity of OM and the size of ulcers (46). Moreover, three distinct meta-analyses conducted by research teams from Taiwan, Italia and China, encompassing a total of 22 randomized clinical trials, investigated the potential of probiotics in preventing OM induced by cancer therapy and in managing the occurrence of chemotherapy-induced diarrhea and OM. These studies revealed the effectiveness of probiotics in preventing and alleviating cancer therapy-induced OM and addressing adverse reactions associated with chemotherapy (38, 41, 49).
Lactobacillus rhamnosus GG (LGG) is a naturally occurring gut commensal bacterium known for its anti-inflammatory properties and has been a pioneer in oncology research (104). LGG maintains the equilibrium of the intestinal mucosa by neutralizing harmful pathogens and toxins, effectively preventing breaches in the mucosal barrier through a high-affinity binding system (105). LGG is also recognized for enhancing the anticancer effects of geniposide, an anticancer molecule, and its potential as a beneficial adjuvant during cancer treatment (106). In the context of cancer treatment, Lactobacillus brevis CD2 lozenges have been found to reduce the occurrence of OM in patients undergoing high-dose chemotherapy (107). Additionally, L. brevis lozenges have shown benefits in reducing oral ulcers in individuals with recurrent aphthous stomatitis (108).
Xerostomia, a condition characterized by dry mouth, has a detrimental impact on the oral health of many patients undergoing radiotherapy for HNSCCs. In a pilot study, Vesty and collaborators explored the potential of using an oral probiotic to influence the oral bacterial community following radiotherapy positively. The authors conducted a four-week intervention involving oral probiotic lozenges containing Streptococcus salivarius M18 in seven patients and compared the changes in oral health and the composition of bacterial communities in plaque and saliva with a control group of six patients who received a placebo. Both groups improved periodontal screening and plaque index scores after the intervention. Surprisingly, the oral probiotic did not lead to significant alterations in the composition or diversity of bacterial communities in the oral cavity. Network analyses revealed potential negative interactions between administered probiotics and bacteria from genera known for their association with periodontal disease, such as Campylobacter, Fretibacterium, Selenomonas, and Treponema (109).
In a comprehensive meta-analysis, Lu and the research team investigated the impact of oral probiotics on the management of side effects induced by radiotherapy, chemotherapy, or chemoradiotherapy in cancer patients. Their study analyzed data from 16 randomized controlled trials involving 2,097 patients. The study’s findings revealed that when compared to placebo groups, the use of oral probiotics (Bifidobacterium longum,B. infantis, Lactobacillus acidophilus, Bacillus clausii, L. plantarum, L. rhamnosus, L. crispatus, Enterococcus faecium) yielded significant reductions in the occurrence of side effects associated with radiotherapy and chemotherapy across various cancer types, including HNSCCs. Additionally, the analysis indicated that the incidence of OM in HNSCCs patients significantly decreased following the oral administration of probiotics (42).
In a randomized clinical trial conducted by Doppalapudi and their research team, the primary objective was to evaluate the impact of probiotic bacteria on oral Candida counts in cancer patients undergoing head and neck radiotherapy at a tertiary care center. The study involved randomly allocating participants into three equal-sized groups: the probiotics group, the candid group, and the combination group. Participants in the probiotics group were administered probiotic sachets containing a minimum of 1.25 billion live cells consisting of a blend of four probiotic strains, namely L. acidophilus, L. rhamnosus, Bifidobacterium longum, and Saccharomyces boulardii. The study results unveiled a statistically significant reduction in the mean counts of Candida species (measured in colony-forming units per milliliter, CFU/ml) after the intervention. This notable reduction was primarily observed in both the probiotics group and the combination therapy group. Furthermore, besides a decrease in C. albicans, there was a significant reduction in C. glabrata and C. tropicalis following probiotic usage compared to the other groups (110). These findings strongly suggest that probiotic bacteria effectively reduce the presence of oral Candida species and could be recommended as a standalone approach or combined with traditional antifungal agents to effectively reduce oral Candida in patients undergoing head and neck radiotherapy.
From the >220 HPV viruses at least 12 are oncogenic (111, 112). Of all HNC cases caused by chronic, persistent HPV infection, approximately 85% are positive for the HPV16 or HPV18 types. The remaining approximately 15% are caused by HPV33, HPV35, HPV52, HPV45, HPV39, HPV58, HPV53, and HPV56 (26, 27). The HPV6, HPV11, HPV16, HPV18, HPV31, HPV33, HPV45, HPV52, and HPV58 strains are accounting for 90% of HNC cases (24, 26, 113). The percentage of HPV positivity varies with the type of HNCs and the different geographic regions (26, 27, 113–117), the highest incidence being reported for sub-Saharan African region (HPV has been identified in 50% of oropharyngeal cancers, 27% of laryngeal cancers, and 23% of oral cavity cancers, with the predominance of HPV16 (118).
Of the HPV viral components, the nonstructural E (early) proteins E5, E6 and E7 are associated with virus-mediated cellular transformation, the most active and expressed in HPV-positive tumor cells being E6 and E7 (27, 119). These viral proteins once accumulated intracellularly can initiate the carcinogenic process, affect the immune system, alter the activity of tumor suppressor proteins (e.g., E6 binds the TP53 protein via the cellular ubiquitin-protein ligase E6AP/E3A or UBE3A, mediating its proteasomes degradation) and circumvent cell-cycle checkpoints (5, 120) (Figure 2). TP53 activity is modulated by MDM2 (mouse double minute 2, also named retained in humans). According to an in silico study by Bouzid and collaborators, in HPV-positive HNSCC, MDM2 is overexpressed compared to HPV-negative tumors (33). The TP53 gene is only rarely mutated in HPV-positive, but very frequently in HPV-negative tumors, in which disruptive mutations are associated with reduced survival (27, 121). Actually, the number of mutations in HPV-positive tumors is twice as low as in HPV-negative tumors (122).
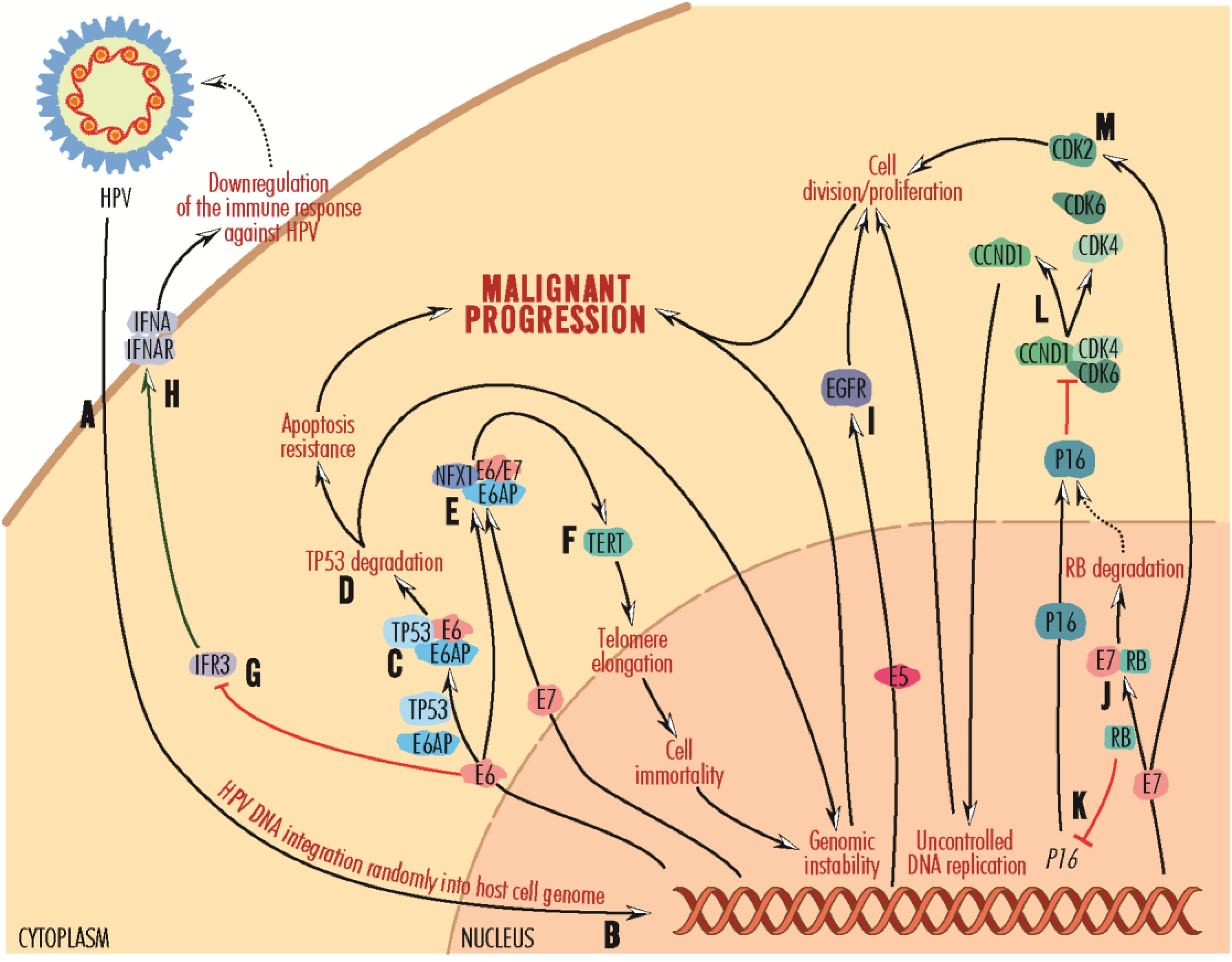
Figure 2 Mechanisms of carcinogenesis induced by persistent HPV infections. Viral particles infect epithelial cells in the oral or oropharyngeal mucosa (A), with HPV DNA randomly integrating into the host cell genome (B). It is replicated as the epithelial cells multiply, and the virus is activated when it reaches the surface. After integration into the host cell genome, viral DNA is copied into mRNA, and proteins are released into the nucleus and cytoplasm. The E6 protein recruits the cellular ubiquitin-protein ligase E6AP and targets the cellular protein TP53 (C), which is involved in maintaining the genetic health of cells. Complexed with E6 and E6AP, TP53 protein is degraded in proteasomes (D), an event that promotes resistance to apoptosis and malignant progression. Lacking the DNA integrity checkpoint mechanism, the cell can accumulate defects, leading to genomic instability and malignant progression. On the other hand, the E6 protein, and less E7, forms a complex with E6AP and NEX1 (neurogenic differentiation factor 6) (E), which activates TERT/hTERT (F). This telomerase reverse transcriptase promotes telomere elongation and cell immortalization. Further, cells with inactive TP53 due to proteasomal degradation may acquire genetic instability and be transformed toward malignant progression. The E6 protein inactivates IFR3 (G), normally promoting the IFNA-IFNAR complex (H) formation. By inhibiting the formation of this complex, E6 decreases immune recognition of HPV and helps the spreading of HPV infection. The E5 protein activates the EGFR-mediated signaling pathway (I), promoting cell division and proliferation toward malignant progression. The E7 protein forms a complex with RB, leading to proteasome degradation (J). In the absence of RB, P16 synthesis is activated (K), which binds and disrupts CCND1 complexes with CDK4 and CDK6 (L), CCND1 contributing to uncontrolled DNA replication and cell division, which can further lead to malignant progression. The E7 protein stimulates CDK2 activity (M), leading to cell proliferation and malignant progression.
HPV-positive tumors have TpC transversions and structural alterations of RNA and DNA, including insertion of the viral genes E6, E7, and E2F1 (the latter being amplified) and, in some cases, defects in the TRAF3 (TNF Receptor Associated Factor 3) gene. Defects in TP53 and CDKN2A genes are absent or rare, with frequent alterations in PIK3CA, PTEN (phosphatase and tensin homolog), FBXW7 (F-Box and WD Repeat Domain Containing 7), and KRAS genes.
HPV-positive tumors include two subtypes: HPV–KRT, with amplification of the 3q region, presence of mutations in PIK3CA, and overexpression of genes involved in keratinocyte differentiation (CDH3–Cadherin 3, and TP63– Transformation-Related Protein 63/Tumor Protein 63) and oxidation-reduction processes (CDH1 and KRT16), and HPV-IMU, characterized by deletion of the 16q region, differentiation of mesenchymal cells dictated by the BCL2 gene, activating mutations of the PIK3CA gene and a strong immune response based on activation of the NFKB (Nuclear Factor Kappa B), RELB (RELB Proto-Oncogene, NF-KB Subunit) and FOXP3 (Forkhead Box P3) genes (123).
Amplification of the 7p region containing the EGFR (Epidermal Growth Factor Receptor) gene, encoding a transmembrane receptor in the RAS–RAF–MEK–ERK and PIK3–AKT–mTOR signaling pathways, is also absent (124–128).
In HPV-positive premalignant tissue, the SYCP2 (synaptonemal complex protein 2), involved in the organization of chromatin is up-regulated (5), and in HPV-positive HNSCC recurrent mutations have been identified in the tumor suppressor genes PTEN and TRAF3, and in the PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha) gene, which promotes carcinogenesis (129). Also, Hinic and collaborators identified overexpression of PCNA (proliferating cell nuclear antigen) genes, associated with cell proliferation and transformation in cancer, TNFRSF14 (TNF receptor superfamily member 14), which promotes inflammatory and inhibitory T cell immune response, TRAF1 (TNF receptor-associated factor 1), TRAF2 (TNF receptor-associated factor 2), which mediate anti-apoptotic and pro-survival signals from TNF receptors, BIRC3 (baculoviral IAP repeat containing 3) and BCL2 (B-cell lymphoma 2), with anti-apoptotic functions (130). A set of genes associated with the extracellular matrix-receptor interaction pathway, which include ITGA5 (integrin alpha five subunits), ITGB1 (integrin beta 1 subunit), LAMB1 (laminin beta 1 subunit), and LAMC1 (laminin gamma 1 subunit), are overexpressed in HPV-positive HNCs (and cervical cancers). Genes associated with T lymphocyte function, CD3D (CD3 delta subunit of T-cell receptor complex), CD3E (CD3 epsilon subunit of T-cell receptor complex), CD8B (CD8 beta subunit), LCK (LCK proto-oncogene, SRC family tyrosine kinase), and ZAP70 (zeta chain of T-cell receptor-associated protein kinase 70kDa), are underexpressed in HPV-positive HNCs and cervical cancers. The observed dysregulation in the latter set of genes in both cancers indicates some expression specificity related to HPV infection, showing a significant prognostic impact on HPV-associated cancers (131).
The HPV-negative tumors frequently harbor CpG transversions, defects in TP53, CCND1, MYC, miR let-7c, TP63, and AJUBA genes, amplification of EGFR, ERBB2 (Erb-B2 Receptor Tyrosine Kinase 2), FGFR1 tyrosine kinase receptor genes, deletions in NSD1 (Nuclear Receptor Binding SET Domain Protein 1), CDKN2A, NOTCH1, SMAD4 (SMAD Family Member 4), FAT1 genes, NFE2L2, KEAP1 (Kelch Like ECH Associated Protein 1), CUL3 (Cullin 3), KMT2D/MLL2, HLA-A/CMH-IA, and co-amplifications of 11q13, with CCND1, CTTN (Cortactin) and FADD (Fas Associated via Death Domain) genes, and 11q22, with YAP1, BIRC2 (Baculoviral IAP Repeat Containing 2), and CASP8+/–HRAS genes, in some subsets of HPV-negative tumors (128). In a small number of HNCs, the MET (MET Proto-Oncogene, Receptor Tyrosine Kinase) gene transcript with missing exon 14, a form recognized as oncogenic in non-small cell lung cancer, and mRNAs of TP63 and KLK12 (Kallikrein Related Peptidase 12) genes as specific splicing variants have been reported (126). The harbor frequent inhibitory mutations in TP53 and the CDKN2A/B deletion associated or not with CCND1 amplification occur are leading to G1/S checkpoint abrogation (129). Activating mutations in the NOTCH1 gene and increased transcription of the FGF1 (Fibroblast Growth Factor 1) gene result in increased cell migration and invasiveness and increased mortality in patients with oral cancers (77). Depending on the presence or absence of gene copy number amplification, HPV-negative tumors are divided into two main subtypes: tumors without copy number amplification or class “M” (from mutations) tumors, which occur in the oral cavity and in which mutations are reported in the HRAS and CASP8 genes as well as in mismatch repair genes (in tumors developed by those who chew betel quid), but lack the TP53 gene, and tumors with copy number amplification. These are divided into three subtypes, basal, classical, and mesenchymal, found in frequent smokers. The basal subtype is characterized by deletions in the 9p arm, which includes the CDKN2A gene (9p21. 3), amplifications of genes in the 3q arm, 11q13/q22 co-amplification, coexistence of mutations in the HRAS and CASP8 (Caspase 8) genes, inactivation of NOTCH1, which has an oncogenic function but in HNCs appears to have a tumor suppressor function, and reduced activity of the SOX2 (SRY-Box Transcription Factor 2) gene (3q26.33). The classical subtype is present in high proportion in laryngeal squamous cell cancers, shares with the basal subtype the presence of deletions in the 9p arm, with loss of CDKN2A and apposition of genes in the 3q arm, and is characterized by mutations in the TP53 gene, changes in KEAP1, CUL3 and NFE2L2 genes, involved in oxidative stress management. In the mesenchymal subtype, epithelial-mesenchymal transitions, amplification of the 3q region, activation of the WNT-β-catenin pathway, mutations of the HLA-A/CMH-IA gene, and increased activity of the CD56/NCAM1 (Cluster of differentiation 56/Neural Cell Adhesion Molecule 1), VIM (Vimentin), DES (Desmin), TWIST1 (Twist Family BHLH Transcription Factor 1) and HGF (Hepatocyte Growth Factor) genes predominate. In both HNCs, amplification of the 3q26/28 region has been identified with TP63, SOX2, and PIK3CA genes, the product of the latter being part of the PIK3–AKT–mTOR signaling pathway with an important role in cell proliferation and evasion of apoptosis (128).
E6 also contributes to the downregulation of the immune response against HPV by suppressing IFR3 (Interferon Regulatory Factor 3), a transcription factor for interferons (IFNs) (132, 133), and by inhibiting IFNA (interferon alpha) interaction with its receptor (134). E7 acts synergically by inhibiting TLR9/CD289 (Toll-like receptor-9), present intracellularly in several immune cell types (135, 136).
Also, by stimulating CDK2 (cyclin-dependent kinase 2) activity and inactivating its inhibitors, P21CIP1 and P27KIP1, E7 supports cell division (135) and tumorigenesis. Viral protein E7 binds and degrades RB (retinoblastoma) tumor suppressor cell proteins (137). Further, RB inactivation induces expression of P16/P16INK4 (cyclin-dependent kinase inhibitor protein 16), which binds and disrupts the CCND (cyclin D) and CDK4 (cyclin-dependent kinase 4)/CDK6 (cyclin-dependent kinase 6) complexes, amplifying cyclin D1 and promoting uncontrolled DNA replication and the transition of cells from G1 to S phase (138).
In addition, E6 and E7 (to a lesser extent) are involved in stimulating cell division and cell immortalization by promoting TERT/hTERT (telomerase reverse transcriptase) expression, which is repressed in normal cells and elongates telomeres through replication and by preventing end-to-end-fusion-of-chromosomes-and-cell apoptosis.
P16 expression and HPV status influence the prognosis of oropharyngeal tumors. HPV+ and P16+ oropharyngeal tumors have a better prognosis than HPV+ and P16– or HPV– and P16+ tumors, while HPV– and P16– tumors have the poorest prognosis (139).
E5 mediates hyperactivation of the EGFR-mediated signaling pathway, stimulating cell proliferation (140).
Another mechanism by which viral proteins inhibit cell apoptosis is by blocking the FAS/FASL (Fas cell surface death receptor/Fas cell surface death receptor ligand) pathway and binding to TNFR1 (tumor necrosis factor receptor 1).
Depending on the affinity of viral proteins for inhibited cellular proteins, there are low-risk HPV strains and high-risk HPV strains that develop tumors. For example, the low-risk HPV11 is generally suppressed by innate immunity, and when this is overcome, it can cause benign lesions (5). On the contrary, high-risk strains, such as HPV16 and HPV18, are not suppressed by the innate immune system and are free to develop long-term infections and produce the majority of HNCs (27). However, HPV infections alone do not appear to be sufficient for the tumorigenic transformation of cells, this process requiring new mutations induced by other risk factors, including smoking and alcohol consumption, or infection with other oncoviruses, such as the polyomaviruses BKV and JCV (John Cunningham virus), the simian vacuolating virus 40 (SV40) and the most likely Epstein-Barr virus type B (141–145).
This could explain the different clinical manifestations of oropharyngeal cancer in HPV-positive versus HPV-negative patients (146, 147). Primary HPV-positive tumors are small in size but develop frequent, more extensive lymph node metastases (148), with frequent immune infiltrates rich in CD8+ cytotoxic T lymphocytes and PDL1 overexpression compared to HPV-negative tumors (82). Correlated with P16 expression, CD8+ T lymphocyte accumulation in the tumor microenvironment improves overall survival in OSCC (82, 149).
In addition to gene expression changes induced by nucleotide sequence alterations, in neoplastic development, an important role is also played by gene expression changes induced by epigenetic alterations, including DNA methylation, posttranslational covalent histone modifications, and non-coding RNA (150, 151). Methylation occurs through the DNMTs (DNA methyltransferases) activity in the presence of AdoMet (S-adenosylmethionine) as a cofactor. In cancer, DNMTs activities are altered, with tumor suppressor genes being silenced by hypermethylation while numerous other sequences spread throughout the genome are hypomethylated, leading to DNA double helix fragmentation and genomic instability (152). In HNCs, the methylation range of different genes is variable. It has been reported that there are significant differences in terms of DNA methylation between HPV-positive and HPV-negative HNCs, due to the fact that E6 and E7 viral proteins interfere with cellular DNA methylation complexes. For example, the inactivation of TP53 by E6 protein stops repression of DNMT1 (DNA (cytosine-5)-methyltransferase 1) promoter, altering the global cytosine methylation pattern (153, 154). Also, changes in cellular DNA-methylation machinery lead to altered gene expression. The affected gene classes are genes involved in cell cycle regulation and programmed cell death: CDKN2A (cyclin-dependent kinase inhibitor 2A, tumor suppressor which is hypomethylated in salivary samples of HPV-positive HNC patients) (155, 156), RASSF1 (Ras association domain family 1, tumor suppressor which is hypomethylated in NHCs) (156, 157), CCNA1 (cyclin A1, whose promoter is hypermethylated in HNCs) (155, 156); genes involved in cellular adhesion and communication: Cadherin Family Genes (involved in cell adhesion and playing important roles in cell signaling and communication, whose pşromoters are hypermethylated) (124, 156, 158), ITGA4 (integrin alpha 4, hypermethylated) (159, 160); genes involved in cellular migration and tumor progression: TIMP3 (tissue inhibitor of metalloproteinase and tumor suppressor, whose promoter methylation is reported in a few studies in HPV-driven HNCs) (78, 161), ELMO1 (engulfment and cell motility 1 protein, which is relatedto increased invasion and metastasis in several types of cancer and hypermethylated in HNCs) (81, 160); other genes: MEI1 (meiotic double-stranded break formation protein 1), whose promoter is hypomethylated in HNCs) (81, 124), and LINE1 (long interspersed nuclear element 1, an abundant retrotransposome found in human genome, which is hypermethylated in HNCs) (155, 162, 163). In HPV-negative tumors, LINE1 is hypomethylated (162). Also, the HPV integrated genomes become subjects of DNA methylation/hypermethylation (164). Even some studies have reported similar findings in matter of HPV-driven DNA-methylation signatures, making proposals for using them as biomarkers for HNCs, these patterns have not been comprehensively investigated. Their inconsistence requires further investigations for identifying the diagnostic methylation targets for HNCs (164).
Little progress has been made in using DNA demethylation as an approach in HVP-positive HNCs. Thus, Stich and collaborators reported that the demethylating agent 5-aza-2’-deoxycytidine can reduce E6 and E7 gene expression in HPV-infected HNCs and cervical cell lines effectively, with some restoration of TP53 and P21 function and increased tumor suppressor microRNA 375 levels, contributing to overall decrease in cancer cell growth and survival (165). These results lead to the conclusion that the ability to reverse DNA methylation makes it an attractive target for drug intervention in HNCs (independent of HPV status), unlike mutations and deletions, which are quite more difficult to correct (166, 167).
The tumor microenvironment is a complex entity that comprises diverse cellular components and extracellular matrix constituents and, through bidirectional interaction with tumor cells, can contribute to tumor progression (168). The cellular component of the tumor microenvironment is very diverse and comprises genetically transformed stromal cells, including endothelial cells, adipocytes, cancer-associated fibroblasts, peripheral nervous system-derived nerve fibers, blood or lymphatic cells, infiltrating immune cells (T lymphocytes, B lymphocytes, and NK cells), neuroendocrine cells, macrophages, neutrophils, antigen-presenting dendritic cells, and myeloid-derived suppressor cells. The immune component of the tumor microenvironment comprises cytotoxic T lymphocytes, regulatory T lymphocytes, B lymphocytes, NK cells, macrophages, neutrophils, antigen-presenting dendritic cells, and myeloid-derived suppressor cells infiltrating the tumor stroma. The NK cells target and induce apoptosis in transformed cells and virus-infected cells that escape the action of cytotoxic T lymphocytes. Macrophages maintain tumor progression by secretion of activating cytokines, angiogenesis, and metastasis. Myeloid-derived suppressor cells promote tumor growth, inhibition of T lymphocyte cytotoxicity, tumor angiogenesis, disintegration of extracellular matrix by secretion of MMPs, inhibition of NK cell activity, and activation of regulatory T lymphocytes with immunosuppressive function (169, 170). The tumor-associated B lymphocytes can trigger humoral antitumoral immunity through interactions with regulatory T lymphocytes and dendritic cells. Dendritic cells are among the most potent antigen-presenting cells, providing cytotoxic CD8+ T lymphocytes with recognition keys to tumor targets (171, 172).
In HNSCC, CD56dim NK cell infiltration is markedly higher in HPV-positive tumors and probably contributes to their more favorable prognosis. NK cells recognize unhealthy or foreign cells that expose inappropriate HLA/MHC class I molecules. Activation of NK cells requires that the proportion of activating signals exceeds that of inhibitory signals and occurs directly via membrane receptors in two pathways and indirectly via soluble factors. The first direct activation pathway requires binding FCGRIIIA/CD16, one of the most potent NK cell activating receptors, to the Fc region of immunoglobulins. The low affinity of the FCGRIIIA/CD16 receptor allows NK cells to recognize and release immunoregulatory cytokines against antibody-coated cellular targets (173). The second pathway of direct NK cell activation occurs via NKG2D and NCRs (Natural Cytotoxicity Receptors), such as NKP30 and NKP46, and is strongly induced under stress conditions in viral infections, including HPV infection and in tumor cells (174, 175). Indirect activation occurs via soluble cytokines, including the interleukins IL2, IL12, IL15, IL18, and IL21, TNFA (Tumor Necrosis Factor Alpha), and IFN (type I interferon) (170, 176).
Neutrophils are among the first immune cells recruited in infections (177) and inflammation in the microenvironment of HNSCC (178). Infiltration of neutrophils in the tumor stroma and an elevated neutrophil to lymphocyte ratio is associated with poor overall surviving for HNSCC patients (179). In HNSCC, the neutrophil increase is lower in HPV-positive than in HPV-negative tumors. However, in the former, an increased neutrophil count is associated with reduced survival duration (180).
Dendritic cells infiltrating HNSCC of the tonsil are of two types: plasmacytoid CD123+ dendritic cells, with characteristics of lymphocytes and classical dendritic cells, myeloid CD11c+ dendritic cells, with three subtypes, CD1c+ myeloid dendritic cells, CD141+ myeloid dendritic cells, and CD1c-CD141- myeloid dendritic cells (181). HNSCC significantly reduces the number of CD11c+ myeloid dendritic cells in the peripheral circulation, which increases after tumor resection. Due to antigen-presenting activity to T lymphocytes, CD1a+ myeloid dendritic cell clusters in the stroma of HNSCC are associated with favorable prognosis and increased survival duration, with some studies indicating them as a favorable prognostic marker for some HPV-positive but not HPV-negative tumors, but this is not always the case (182).
Cytotoxic CD8+T lymphocytes are the main cellular immune effectors against tumor cells. Activation of cytotoxic CD8+ T lymphocytes occurs through TCR (T cell receptor) recognition of HLA/MHC antigens presented by dendritic cells and interaction of co-stimulatory factors B7/CD80 on antigen-presenting cells and CD28 on T lymphocytes. CD28 activates CTLA4/CD152, expressed predominantly on cytotoxic CD8+ T lymphocytes and less on activated B lymphocytes, monocytes, dendritic cells, regulatory CD4+ T lymphocytes, and granulocytes, induces TGFB (Transforming Growth Factor Beta) synthesis with immunosuppressive effects (183). In tumor tissue, TGFB synthesis leads to overexpression of CTLA4/CD152, with depletion of T lymphocytes (184), which begin to release inhibitory molecules, including PD1, CTLA4, TIGIT (T Cell Immunoreceptor With Ig And ITIM Domains) and LAG3 (Lymphocyte Activating 3), which reduce their activity and production of cytokines and cytolytic molecules (185). PD1 is part of the CD28 family of receptors and has PDL1 and PDL2 ligands, and both are expressed on antigen-presenting cells, endothelial cells, and activated lymphocytes (186).
Increased expression of PD1 and PDL1 in the tumor microenvironment cells leads to the inactivation or depletion of cytotoxic CD8+ T lymphocytes, and even when these cells are present in large extent, it favors tumor survival (187). HPV infections are known to increase the number of specific CD 8+ T lymphocytes, whose proliferation is triggered mainly by L1 protein (188). In a similar manner, HPV-positive tumors attract an increased number of HPV-specific CD 8+ T lymphocytes, which account 0.1 to 10% among all the tumor-infiltrating CD8+ T lymphocytes, while their presence in peripheral blood is very low (0.02%), indicating a strong association with the tumor microenvironment. One subset of HPV-specific CD 8+ T lymphocytes is expressing the TCF7 and other genes associated with with PD1+ stem-like CD8 T lymphocytes, which are very important for maintaining T cell responses when HPV-antigen presence is prolonged. When stimulated with the HPV peptides, the PD1+TCF1+ stem-like subset of CD8+ T lymphocytes proliferates and differentiates into several subsets of effector cells, and the presence of functional and proliferative HPV-specific PD1+TCF1+CD45RO+ stem-like CD8 T proves that in HPV-positive NHC tumors there is active mechanisms to overcome the PD1 blockade (189), leading to chronic inflammatory reaction and poor prognosis in OSCC wif’th high expression of PD1 and its ligands (190, 191). On the other hand, 82 show that PDL1 expression on macrophages infiltrating HPV-positive tumors indicates a trend toward improved overall survival. Since HPV-positive HNCs are able to develop responsive mechanisms to PD1 blockade, the PD1–PDL1 pair could be an attractive target for antitumor therapies (192). In some cancer cases, PD1/PDL1 antibody therapies invigorate tumor-infiltrating CD8+ T lymphocytes, but their efficacy on heterogeneous CD8+ T cell populations is uneven (193). However, the tumor response to PD1 inhibitor therapy depends on tumor type. It is expected that, in the presence of CD8+ T lymphocytes, PD1+ tumors are more responsive compared to PD1-negative tumors (194–196), and anti-tumor therapeutic decisions may be guided by the results of immunohistochemical tests for PDL1 expression (197).
HNCs continue to be a global health challenge and a topic of significant contemporary importance. HNCs are aggressive tumors, with more than 90% of their origin in squamous cells from the mucosae of the upper aerodigestive tract, ranking sixth among the most common cancers. Recent advances in omics and bioinformatics technologies are vital for gaining insights into the biology and clinical behavior of HNCs, unveiling potential biomarkers and therapeutic targets with practical applications in this problematic disease (198–201).
The dysbiosis occurred in the complex oral microbiome is associated with the evolution of HNCs, through multiple mechanisms such as inflammation, genotoxins release, modulation of the innate and acquired immune response, of carcinogens and anticarcinogens productions, generation of oxidative stress, induction of mutations (34). Thus, novel microbiome-derived biomarkers and interventions could significantly contribute to achieving the desideratum of personalized management of oncologic patients, regarding both early diagnosis and treatment.
The most common microorganisms associated with HNCs are Porphyromonas gingivalis, Fusobacterium, Leptotrichia, Selenomonas, Treponema, Parvimonas, Pseudoalteromonas, Prevotella, Alloprevotella, Capnocytophaga, Bacteroidetes, Solobacterium, Clostridium and Peptostreptococcus. A higher abundance of Bacteroidetes and Peptostreptococcus are associated with later stages and larger tumors, while increased salivary levels of Stenophotromonas, Staphylococcus, Centipeda, Selenomonas, Alloscordovia, and Acinetobacter with poor prognosis and poorer survival in oral cancer (Figure 3).
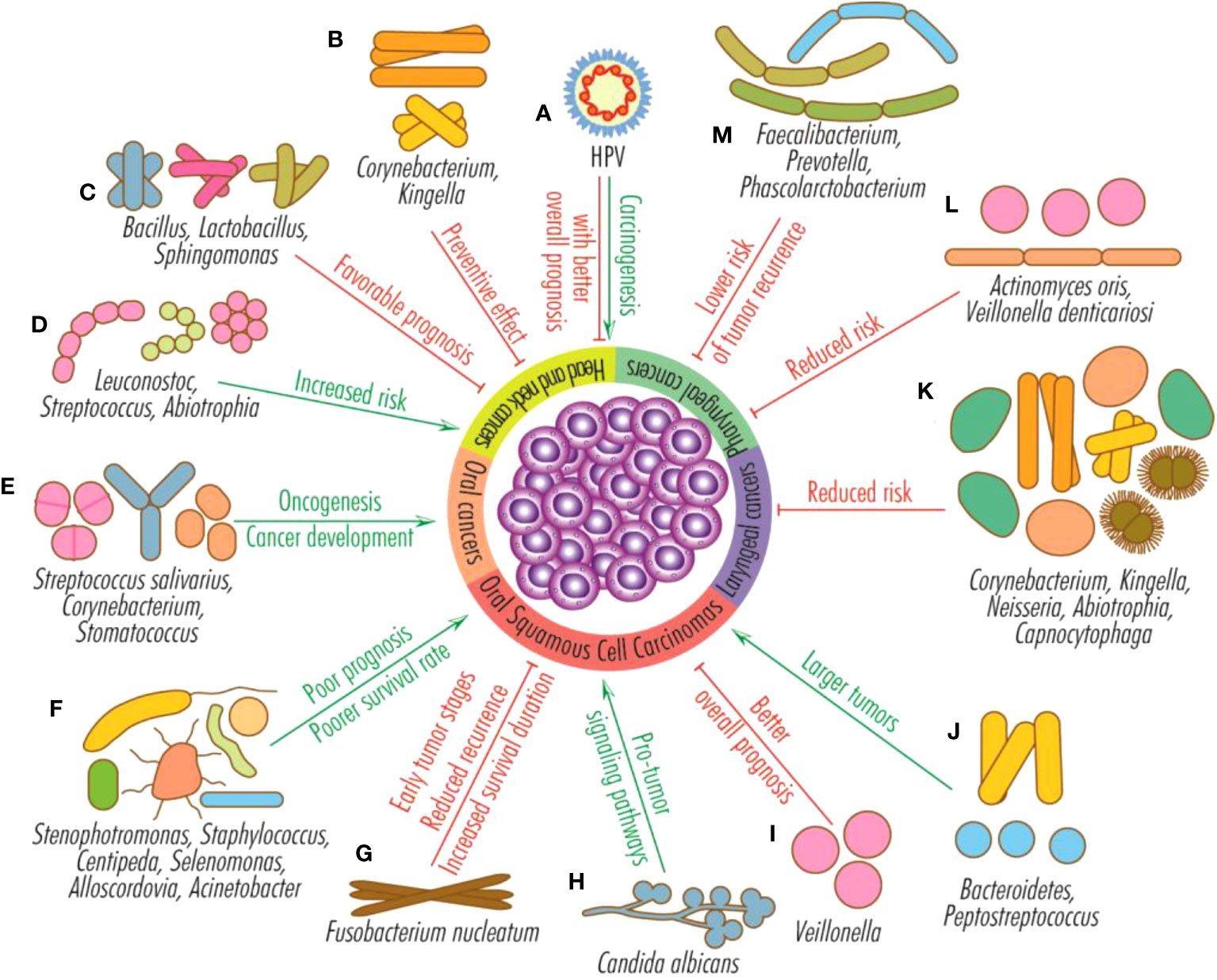
Figure 3 Effects of HPV infection and microbiota on head and neck tumors. HPV infection favors the development of head and neck cancers (mainly from the oral and oropharyngeal sphere), but their general prognosis is better than that of HPV-negative tumors (A); Corynebacterium and Kingella have a preventive effect on head and neck cancers (B); Bacillus, Lactobacillus and Sphingomonas sustain favorable prognosis (C); Leuconostoc, Streptococcus and Abiotrophia increase the risk of head and neck cancers (D); Streptococcus salivarius, Corynebacterium and Stomatococcus favor oncogenesis and the development of oral tumors (E); Stenophotromonas, Staphylococcus, Centipeda, Selenomonas, Alloscordovia and Acinetobacter predict poor prognosis and poorer survival rate for OSCCs (F); Fusobacterium nucleatum is generally associated with early oral squamous cell carcinomas stages, reduced recurrence and increased survival duration (G); the fungus Candida albicans stimulates pro-tumor signaling pathways (H); Veillonella is an indicator for better overall prognosis for OSCCs (I); Bacteroidetes and Peptostreptococcus are associated with large oral squamous cell carcinomas (J); the presence of the genera Corynebacterium, Kingella, Neisseria, Abiotrophia and Capnocytophaga reduce the risk of the appearance or progression of laryngeal tumors (K); the species Actinomyces oris and Veillonella denticariosi are associated with reduced risk of occurrence or progression of pharyngeal tumors (L); species from the genera Faecalibacterium, Prevotella and Phascolarctobacterium reduce the risk of pharyngeal cancer recurrence (M).
However, the results reported by different studies are not always congruent regarding the variations in the abundance of different taxons in HNCs. Thus, Actinobacteria phylum and Neisseria, Capnocytophaga, Veillonella genera are reported either with high or with low abundance in HNCs. The current studies are consistent in reporting a higher abundance of Gram-negative species such as Fusobacterium, Leptotrichia, Treponema, Porphyromonas gingivalis, Prevotella, Bacteroidetes, Haemophilus, Veillonella, Pseudomonas, Enterobacterales, which are probably responsible of chronic inflammation and modulation of tumor microenvironment. On the other side, a recent study shows that the presence of oral fungi and of red- and orange-complex periodontal pathogens was associated with reduced risk of HNCs. C. albicans is the dominant fungi found in oral carcinoma being also associated with shorter survival rate. The abundance of different microbial species such as F. nucleatum, Bacteroidetes and Peptostreptococcus has been associated with later stages and larger tumor, suggesting their potential to be used as biomarkers for tumor stratification and prognosis.
On the other side, some microbiota signatures, such as the abundance of microorganisms of the genera Corynebacterium, Kingella, Abiotrophia are associated with a reduced risk of HNCs.
Microbiome could also provide biomarkers for HNCs diagnosis, the profiles being different between oropharyngeal and hypopharyngeal cancers as well as between HPV-positive and HPV-negative tumors. Ongoing clinical trials aim to validate non-invasive tests for microbiome derived biomarkers detection in oral and throat cancers, especially within high-risk populations. These studies demonstrate the potential of machine-learning tools for oral cancer diagnosing, opening a new era of non-invasive diagnostics, enabling early intervention, and improving patient outcomes.
Oro-pharyngeal dysbiosis could also impact the HNCs therapy and associated side-effects of radiotherapy, chemotherapy, and immunotherapy, such as OM or tumor recurrence.
Elucidation of the molecular mechanisms by which oral microbiome and HPV infection influences the HNCs initiation and progression, screening for HPV infection and vaccination against HPV, adopting good oral hygiene, and preventing oral dysbiosis are important tools for advancing in the battle with this public health global challenge. Reducing pathogenic bacteria and promoting a healthy microbiome has been shown to enhance the effectiveness of both immunotherapy and chemotherapy. Furthermore, maintaining a balanced gut microbiome can help mitigate the side effects associated with these treatments, improving the overall experience for cancer patients (202, 203).
Current studies investigate the potential of probiotics in modulating the course of the disease and managing cancer therapy-related side effects in HNSCC and OSCC patients (38, 41, 42, 46, 49, 204). The current evidence underscores the potential of probiotics such as LGG and L. brevis in alleviating cancer associated oral mucosal problems and promoting overall health.
However, the current findings raise questions about the beneficial properties of particular oral probiotics, necessitating further investigation to better understand their effects and potential drawbacks. While the results regarding the use of microbiome-based interventions are promising, further research is recommended, specifically advocating for additional randomized, double-blind, multicenter trials conducted on a more extensive and diverse population. This approach will help provide a more robust understanding of the potential benefits of probiotics in managing cancer therapy-related side effects, such as the inflammation of the oral mucosa frequently encountered in individuals undergoing radiotherapy and chemotherapy (205).
As we gain a better understanding of the molecular traits distinguishing HPV-positive from HPV-negative HNSCCs, there is hope for developing novel early diagnosis markers (e.g., with methylated circulating tumor DNA) and targeted, personalized therapies. The specific genetic and epigenetic events occurred in each HNCs stage are influenced by the HPV infection status. HPV-positive tumors are characterized by fewer mutations, occurring predominantly in PIK3CA, PTEN, FBXW7, and KRAS genes, whereas HPV-negative tumors carry defects in a more extensive number of genes. In addition to inhibitory and activating mutations, in HNCs, gene expression is altered by several epigenetic changes, which, in the case of DNA methylation, are also influenced by the HPV status. The HPV status also influences the tumor microenvironment cellular components, particularly the NK, neutrophils, dendritic cells, and CD8 positive T cells. However, despite notable advancements in comprehending the molecular mechanisms by which HPV influences the HNCs evolution and response to treatment, the significant disparity in survival and clinical outcomes between HPV-positive and HPV-negative HNSCC patients following standard-of-care treatment remains enigmatic. Several research teams have proposed potential factors, including the elevated rates of cell proliferation and DNA damage in HPV-positive tumors compared to their HPV-negative counterparts (206). Future studies that address aberrant DNA methylation, histone post-translational modifications, non-coding RNAs, dysbiosis, and approaches to minimize immunosuppression within the tumor microenvironment will provide the science-based evidence for revolutionizing HNCs managment (207). Personalized treatments, encompassing targeted therapies, immunotherapies, cancer vaccines, and epigenetic inhibitors tailored to each individual’s molecular profile, hold great promise in overcoming the limitations of conventional therapies, offering patients more effective and precisely tailored care in clinical settings.
MC: Writing – original draft, Writing – review & editing. MCC: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition. GM: Writing – review & editing. COV: Writing – original draft, Writing – review & editing. E-GD: Writing – review & editing. R-EC: Writing – review & editing. CB: Writing – review & editing. SB: Writing – review & editing. RG: Writing – review & editing. BS: Writing – review & editing. CC: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge the financial support of C1.2.PFE-CDI.2021-587/Contract no. 41PFE/30.12.2021; FDI 0609/2023; EURO-MEDEX, Contract no. 33/PFE/2021; “The core program within the National Research Development and Innovation Plan, 2022–2027”, carried out with the support of the Ministry of Research, Innovation and Digitalization (MCID), project no. 23020101, Contract no. 7N from 3 January 2023; Project No. RO1567-IBB05/2023 from the Institute of Biology Bucharest of the Romanian Academy; PN-III-P4-PCE-2021-0549 awarded by Romanian Executive Agency for Higher Education, Research, Development, and Innovation, and the “Analysis of the potential for sustainable use of vegetation specific to the Danube-Danube Delta-Black Sea system” project, awarded by the European Regional Development Fund through the Competitiveness Operational Program 2014–2020, contract no. 108630. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gilyoma JM, Rambau PF, Masalu N, Kayange NM, Chalya PL. Head and neck cancers: a clinico-pathological profile and management challenges in a resource-limited setting. BMC Res Notes. (2015) 8:772. doi: 10.1186/s13104-015-1773-9
2. Gormley M, Creaney G, Schache A, Ingarfield K, Conway DI. Reviewing the epidemiology of head and neck cancer: definitions, trends and risk factors. Br Dent J (2022) 233:780–6. doi: 10.1038/s41415-022-5166-x
3. Suchanti S, Stephen BJ, Awasthi S, Awasthi SK, Singh G, Singh A, et al. Harnessing the role of epigenetic histone modification in targeting head and neck squamous cell carcinoma. Epigenomics (2022) 14(5):279–93. doi: 10.2217/epi-2020-0348
4. Dorobisz K, Dorobisz T, Zatoński T. The microbiome's influence on head and neck cancers. Curr Oncol Rep (2023) 25(3):163–71. doi: 10.1007/s11912-022-01352-7
5. Galati L, Chiocca S, Duca D, Tagliabue M, Simoens C, Gheit T, et al. HPV and head and neck cancers: Towards early diagnosis and prevention. Tumour Virus Res (2022) 14:200245. doi: 10.1016/j.tvr.2022.200245
6. Heawchaiyaphum C, Ekalaksananan T, Patarapadungkit N, Vatanasapt P, Pientong C. Association of human papillomavirus and epstein-barr virus infection with tonsil cancer in northeastern Thailand. Asian Pac J Cancer Prev (2022) 23(3):781–7. doi: 10.31557/APJCP.2022.23.3.781
7. Lechien JR, Saussez S, Cammaroto G, Hans S. Laryngopharyngeal reflux and head and neck cancers. Am J Otolaryngol (2021) 42(1):102815. doi: 10.1016/j.amjoto.2020.102815
8. Rocha PHP, Reali RM, Decnop M, Souza SA, Teixeira LAB, Júnior AL, et al. Adverse radiation therapy effects in the treatment of head and neck tumors. Radiographics (2022) 42(3):806–21. doi: 10.1148/rg.210150
9. Nam IC, Park JO, Kim CS, Park SJ, Lee DH, Kim HB, et al. Association of smoking status, duration and amount with the risk of head and neck cancer subtypes: a national population-based study. Am J Cancer Res (2022) 12(10):4815–24.
10. Katada C, Yokoyama T, Yano T, Suzuki H, Furue Y, Yamamoto K, et al. Alcohol consumption, multiple Lugol-voiding lesions, and field cancerization. DEN Open (2023) 4(1):e261. doi: 10.1002/deo2.261
11. Chuang HC, Tsai MH, Lin YT, Chou MH, Yang KL, Chien CY. Systemic and local effects among patients with betel quid-related oral cancer. Technol Cancer Res Treat (2022) 21:15330338221146870. doi: 10.1177/15330338221146870
12. Xie M, Gupta MK, Archibald SD, Stanley Jackson B, Young JEM, Zhang H. Marijuana and head and neck cancer: an epidemiological review. J Otolaryngol Head Neck Surg (2018) 47(1):73. doi: 10.1186/s40463-018-0319-2
13. Nishi H, Obayashi T, Ueda T, Ohta K, Shigeishi H, Munenaga S, et al. Head and neck cancer patients show poor oral health as compared to those with other types of cancer. BMC Oral Health (2023) 23(1):647. doi: 10.1186/s12903-023-03356-6
14. Tasoulas J, Farquhar DR, Sheth S, Hackman T, Yarbrough WG, Agala CB, et al. Poor oral health influences head and neck cancer patient survival: an International Head and Neck Cancer Epidemiology Consortium pooled analysis. J Natl Cancer Inst (2023) djad156. doi: 10.1093/jnci/djad156
15. Luo Z, Zhu X, Hu Y, Yan S, Chen L. Association between dietary inflammatory index and oral cancer risk: A systematic review and dose-response meta-analysis. Front Oncol (2022) 12:920452. doi: 10.3389/fonc.2022.920452
16. Pandey D, Szczesniak M, Maclean J, Yim HCH, Zhang F, Graham P, et al. Dysbiosis in head and neck cancer: determining optimal sampling site for oral microbiome collection. Pathogens (2022) 11(12):1550. doi: 10.3390/pathogens11121550
17. Leoncini E, Vukovic V, Cadoni G, Pastorino R, Arzani D, Bosetti C, et al. Clinical features and prognostic factors in patients with head and neck cancer: Results from a multicentric study. Cancer Epidemiol. (2015) 39(3):367–74. doi: 10.1016/j.canep.2015.02.004
18. Andersson BÅ, Löfgren S, Lewin F, Nilsson M, Laytragoon-Lewin N. Impact of cigarette smoking and head and neck squamous cell carcinoma on circulating inflammatory biomarkers. Oncology (2020) 98(1):42–7. doi: 10.1159/000502651
19. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers (2020) 6(1):92. doi: 10.1038/s41572-020-00224-3
20. Miranda-Galvis M, Loveless R, Kowalski LP, Teng Y. Impacts of environmental factors on head and neck cancer pathogenesis and progression. Cells (2021) 10(2):389. doi: 10.3390/cells10020389
21. Constantin M. Epidemiology, diagnosis, symptoms and TNM classification of head and neck cancers. Rom Biotechnol Lett (2022) 27(5):3699–712. doi: 10.25083/rbl/27.5/3699.3712
22. Jenwitheesuk K, Peansukwech U, Jenwitheesuk K. Predictive MERRA-2 aerosol diagnostic model for oral, oropharyngeal and laryngeal cancer caused by air pollution in Thai population. Toxicol Rep (2022) 9:970–6. doi: 10.1016/j.toxrep.2022.04.015
23. Economopoulou P, de Bree R, Kotsantis I, Psyrri A. Diagnostic tumor markers in head and neck squamous cell carcinoma (HNSCC) in the clinical setting. Front Oncol (2019) 9:827. doi: 10.3389/fonc.2019.00827
24. Wu J, Xiao F, Zheng Y, Lin Y, Wang HL. Worldwide trend in human papillomavirus-attributable cancer incidence rates between 1990 and 2012 and Bayesian projection to 2030. Cancer (2021) 127(17):3172–82. doi: 10.1002/cncr.33628
25. Goon P, Schürmann M, Oppel F, Shao S, Schleyer S, Pfeiffer CJ, et al. Viral and clinical oncology of head and neck cancers. Curr Oncol Rep (2022) 24(7):929–42. doi: 10.1007/s11912-022-01263-7
26. Liao CI, Francoeur AA, Kapp DS, Caesar MAP, Huh WK, Chan JK. Trends in human papillomavirus-associated cancers, demographic characteristics, and vaccinations in the US 2001-2017. JAMA Netw Open (2022) 5(3):e222530. doi: 10.1001/jamanetworkopen.2022.2530
27. Pinkiewicz M, Dorobisz K, Zatoński T. Human papillomavirus-associated head and neck cancers. Where are we now? A Systematic Review. Cancer Manage Res (2022) 14:3313–24. doi: 10.2147/CMAR.S379173
28. Benjamin WJ, Wang K, Zarins K, Bellile E, Blostein F, Argirion I, et al. Oral microbiome community composition in head and neck squamous cell carcinoma. Cancers (Basel) (2023) 15(9):2549. doi: 10.3390/cancers15092549
29. Metsaniitty M, Hasnat S, Salo T, Salem A. Oral microbiota-a new frontier in the pathogenesis and management of head and neck cancers. Cancers (Basel) (2021) 14(1):46. doi: 10.3390/cancers14010046
30. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT, et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis (2021) 42(1):127–35. doi: 10.1093/carcin/bgaa062
31. Irfan M, Delgado RZR, Frias-Lopez J. The oral microbiome and cancer. Front Immunol (2020) 11:591088. doi: 10.3389/fimmu.2020.591088
32. Al-Qadami G, Bowen J, Van Sebille Y, Secombe K, Dorraki M, Verjans J, et al. Baseline gut microbiota composition is associated with oral mucositis and tumour recurrence in patients with head and neck cancer: a pilot study. Support Care Cancer (2023) 31(1):98. doi: 10.1007/s00520-022-07559-5
33. Bouzid A, Al Ani M, de la Fuente D, Al Shareef ZM, Quadri A, Hamoudi R, et al. Identification of p53-target genes in human papillomavirus-associated head and neck cancer by integrative bioinformatics analysis. Front Oncol (2023) 13:1128753. doi: 10.3389/fonc.2023.1128753
34. Kozak M, Pawlik A. The Role of the oral microbiome in the development of diseases. Int J Mol Sci (2023) 24(6):5231. doi: 10.3390/ijms24065231
35. McKeon MG, Gallant JN, Kim YJ, Das SR. It takes two to tango: A review of oncogenic virus and host microbiome associated inflammation in head and neck cancer. Cancers (Basel) (2022) 14(13):3120. doi: 10.3390/cancers14133120
36. Chen Y, Huang Z, Tang Z, Huang Y, Huang M, Liu H, et al. More than just a periodontal pathogen -the research progress on Fusobacterium nucleatum. Front Cell Infect Microbiol (2022) 12:815318. doi: 10.3389/fcimb.2022.815318
37. Dou Y, Ma C, Wang K, Liu S, Sun J, Tan W, et al. Dysbiotic tumor microbiota associates with head and neck squamous cell carcinoma outcomes. Oral Oncol (2022) 124:105657. doi: 10.1016/j.oraloncology.2021.105657
38. Feng J, Gao M, Zhao C, Yang J, Gao H, Lu X, et al. Oral administration of probiotics reduces chemotherapy-induced diarrhea and oral mucositis: A systematic review and meta-analysis. Front Nutr (2022) 9:823288. doi: 10.3389/fnut.2022.823288
39. Reis Ferreira M, Pasto A, Ng T, Patel V, Guerrero Urbano T, Sears C, et al. The microbiota and radiotherapy for head and neck cancer: What should clinical oncologists know? Cancer Treat Rev (2022) 109:102442. doi: 10.1016/j.ctrv.2022.102442
40. Kim YK, Kwon EJ, Yu Y, Kim J, Woo SY, Choi HS, et al. Microbial and molecular differences according to the location of head and neck cancers. Cancer Cell Int (2022) 22(1):135. doi: 10.1186/s12935-022-02554-6
41. Liu G, Cao S, Liu X, Li Z, Tian Y, Zhang X, et al. Effect of perioperative probiotic supplements on postoperative short-term outcomes in gastric cancer patients receiving neoadjuvant chemotherapy: A double-blind, randomized controlled trial. Nutrition (2022) 96:111574. doi: 10.1016/j.nut.2021.111574
42. Lu Y, Luo X, Yang D, Li Y, Gong T, Li B, et al. Effects of probiotic supplementation on related side effects after chemoradiotherapy in cancer patients. Front Oncol (2022) 12:1032145. doi: 10.3389/fonc.2022.1032145
43. Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, et al. Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut (2022) 71(3):521–33. doi: 10.1136/gutjnl-2020-323426
44. Vadovics M, Ho J, Igaz N, Alföldi R, Rakk D, Veres É, et al. Candida albicans enhances the progression of oral squamous cell carcinoma in vitro and in vivo. mBio (2021) 13(1):e0314421. doi: 10.1128/mBio.03144-21
45. Banavar G, Ogundijo O, Julian C, Toma R, Camacho F, Torres PJ, et al. Detecting salivary host and microbiome RNA signature for aiding diagnosis of oral and throat cancer. Oral Oncol (2023) 145:106480. doi: 10.1016/j.oraloncology.2023.106480
46. Frey-Furtado L, Magalhães I, Azevedo MJ, Sampaio-Maia B. The role of biotics as a therapeutic strategy for oral mucositis - A systematic review. Probiotics Antimicrob Proteins (2023). doi: 10.1007/s12602-023-10116-z
47. Inchingolo AM, Malcangi G, Piras F, Palmieri G, Settanni V, Riccaldo L, et al. Precision medicine on the effects of microbiota on head-neck diseases and biomarkers diagnosis. J Pers Med (2023) 13(6):933. doi: 10.3390/jpm13060933
48. Li Z, Fu R, Wen X, Wang Q, Huang X, Zhang L. The significant clinical correlation of the intratumor oral microbiome in oral squamous cell carcinoma based on tissue-derived sequencing. Front Physiol (2023) 13:1089539. doi: 10.3389/fphys.2022.1089539
49. Minervini G, Franco R, Marrapodi MM, Fiorillo L, Badnjević A, Cervino G, et al. Probiotics in the treatment of radiotherapy-induced oral mucositis: systematic review with meta-analysis. Pharm (Basel) (2023) 16(5):654. doi: 10.3390/ph16050654
50. Morsy BM, El Domiaty S, Meheissen MAM, Heikal LA, Meheissen MA, Aly NM. Omega-3 nanoemulgel in prevention of radiation-induced oral mucositis and its associated effect on microbiome: a randomized clinical trial. BMC Oral Health (2023) 23(1):612. doi: 10.1186/s12903-023-03276-5
51. Nouri Z, Choi SW, Choi IJ, Ryu KW, Woo SM, Park SJ, et al. Exploring connections between oral microbiota, short-chain fatty acids, and specific cancer types: A study of oral cancer, head and neck cancer, pancreatic cancer, and gastric cancer. Cancers (Basel) (2023) 15(11):2898. doi: 10.3390/cancers15112898
52. Pandey D, Szcczesniak M, Maclean J, Zhang F, Yim H, El-Omar E, et al. Implications of oro-pharyngeal dysbiosis in head and neck cancer: oral microbiome and chemoradiation-related complications. Gut (2023) 72:A114–6. doi: 10.1136/gutjnl-2023-IDDF.101
53. Wu Z, Han Y, Wan Y, Hua X, Chill SS, Teshome K, et al. Oral microbiome and risk of incident head and neck cancer: A nested case-control study. Oral Oncol (2023) 137:106305. doi: 10.1016/j.oraloncology.2022.106305
54. Wade WG. The oral microbiome in health and disease. Pharmacol Res (2013) 69(1):137–43. doi: 10.1016/j.phrs.2012.11.006
55. Mosaddad SA, Tahmasebi E, Yazdanian A, Rezvani MB, Seifalian A, Yazdanian M, et al. Oral microbial biofilms: an update. Eur J Clin Microbiol Infect Dis (2019) 38(11):2005–19. doi: 10.1007/s10096-019-03641-9
56. Wang H, Funchain P, Bebek G, Altemus J, Zhang H, Niazi F, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med (2017) 9(1):14. doi: 10.1186/s13073-017-0405-5
57. Frank DN, Qiu Y, Cao Y, Zhang S, Lu L, Kofonow JM, et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene (2022) 41(9):1269–80. doi: 10.1038/s41388-021-02137-1
58. Al-Hebshi NN, Nasher AT, Maryoud MY, Homeida HE, Chen T, Idris AM, et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep (2017) 7(1):1834. doi: 10.1038/s41598-017-02079-3
59. Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol (2020) 9:476. doi: 10.3389/fcimb.2019.00476
60. Mohamed N, Litlekalsøy J, Ahmed IA, Martinsen EMH, Furriol J, Javier-Lopez R, et al. Analysis of salivary mycobiome in a cohort of oral squamous cell carcinoma patients from Sudan identifies higher salivary carriage of malassezia as an independent and favorable predictor of overall survival. Front Cell Infect Microbiol (2021) 11:673465. doi: 10.3389/fcimb.2021.673465
61. Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol (2018) 3:862(9). doi: 10.3389/fmicb.2018.00862
62. Takahashi Y, Park J, Hosomi K, Yamada T, Kobayashi A, Yamaguchi Y, et al. Analysis of oral microbiota in Japanese oral cancer patients using 16S rRNA sequencing. J Oral Biosci (2019) 61(2):120–8. doi: 10.1016/j.job.2019.03.003
63. Granato DC, Neves LX, Trino LD, Carnielli CM, Lopes AFB, Yokoo S, et al. Meta-omics analysis indicates the saliva microbiome and its proteins associated with the prognosis of oral cancer patients. Biochim Biophys Acta Proteins Proteom. (2021) 1869(8):140659. doi: 10.1016/j.bbapap.2021.140659
64. Bronzato JD, Bomfim RA, Edwards DH, Crouch D, Hector MP, Gomes BPFA. Detection of Fusobacterium in oral and head and neck cancer samples: A systematic review and meta-analysis. Arch Oral Biol (2020) 112:104669. doi: 10.1016/j.archoralbio.2020.104669
65. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
66. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
67. Li Z, Chen G, Wang P, Sun M, Zhao J, Li A, et al. Alterations of the oral microbiota profiles in chinese patient with oral cancer. Front Cell Infect Microbiol (2021) 11:780067. doi: 10.3389/fcimb.2021.780067
68. Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galNAc. Cell Host Microbe (2016) 20(2):215–25. doi: 10.1016/j.chom.2016.07.006
69. Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun (2015) 83(3):1104–13. doi: 10.1128/IAI.02838-14
70. Chen Z, Wong PY, Ng CWK, Lan L, Fung S, Li JW, et al. The intersection between oral microbiota, host gene methylation and patient outcomes in head and neck squamous cell carcinoma. Cancers (Basel) (2020) 12(11):3425. doi: 10.3390/cancers12113425
71. Orlandi E, Iacovelli NA, Tombolini V, Rancati T, Polimeni A, De Cecco L, et al. Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer. Oral Oncol (2019) 99:104453. doi: 10.1016/j.oraloncology.2019.104453
72. Mukherjee PK, Wang H, Retuerto M, Zhang H, Burkey B, Ghannoum MA, et al. Bacteriome and mycobiome associations in oral tongue cancer. Oncotarget (2017) 8(57):97273–89. doi: 10.18632/oncotarget.21921
73. Yost S, Stashenko P, Choi Y, Kukuruzinska M, Genco CA, Salama A, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int J Oral Sci (2018) 10(4):32. doi: 10.1038/s41368-018-0037-7
74. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat (2019) 18:1533033819867354. doi: 10.1177/1533033819867354
75. Zhang L, Chai D, Chen C, Li C, Qiu Z, Kuang T, et al. Mycobiota and C-type lectin receptors in cancers: Know thy neighbors. Front Microbiol (2022) 13:946995. 946995. doi: 10.3389/fmicb.2022.946995
76. Hayes RB, Ahn J, Fan X, Peters BA, Ma Y, Yang L, et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol (2018) 4(3):358–65. doi: 10.1001/jamaoncol.2017.4777
77. Weaver AN, Burch MB, Cooper TS, Della Manna DL, Wei S, Ojesina AI, et al. Notch signaling activation is associated with patient mortality and increased FGF1-mediated invasion in squamous cell carcinoma of the oral cavity. Mol Cancer Res (2016) 14(9):883–91. doi: 10.1158/1541-7786.MCR-16-0114
78. Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog. (2011) 50:680–8. doi: 10.1002/mc.20798
79. Wise-Draper TM, Bahig H, Tonneau M, Karivedu V, Burtness B. Current therapy for metastatic head and neck cancer: evidence, opportunities, and challenges. Am Soc Clin Oncol Educ Book. (2022) 42:1–14. doi: 10.1200/EDBK_350442
80. Wolf A, Moissl-Eichinger C, Perras A, Koskinen K, Tomazic PV, Thurnher D. The salivary microbiome as an indicator of carcinogenesis in patients with oropharyngeal squamous cell carcinoma: a pilot study. Sci Rep (2017) 7:5867. doi: 10.1038/s41598-017-06361-2
81. Worsham MJ, Chen KM, Datta I, Stephen JK, Chitale D, Gothard A, et al. The biological significance of methylome differences in human papilloma virus associated head and neck cancer. Oncol Lett (2016) 12:4949–56. doi: 10.3892/ol.2016.5303
82. Wuerdemann N, Gültekin SE, Pütz K, Wittekindt C, Huebbers CU, Sharma S, et al. PD-L1 expression and a high tumor infiltrate of CD8+ Lymphocytes predict outcome in patients with oropharyngeal squamous cells carcinoma. Int J Mol Sci (2020) 21(15):5228. doi: 10.3390/ijms21155228
83. Sakamoto H, Naito H, Ohta Y, Tanakna R, Maeda N, Sasaki J, et al. Isolation of bacteria from cervical lymph nodes in patients with oral cancer. Arch Oral Biol (1999) 44(10):789–93. doi: 10.1016/s0003-9969(99)00079-5
84. Panda M, Rai AK, Rahman T, Das A, Das R, Sarma A, et al. Alterations of salivary microbial community associated with oropharyngeal and hypopharyngeal squamous cell carcinoma patients. Arch Microbiol (2020) 202:785–805. doi: 10.1007/s00203-019-01790-1
85. Banavar G, Ogundijo O, Toma R, Rajagopal S, Lim YK, Tang K, et al. The salivary metatranscriptome as an accurate diagnostic indicator of oral cancer. NPJ Genom Med (2021) 6(1):105. doi: 10.1038/s41525-021-00257-x
86. Ganly I, Yang L, Giese RA, Hao Y, Nossa CW, Morris LGT, et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int J Cancer (2019) 145(3):775–84. doi: 10.1002/ijc.32152
87. Zhu X-X, Yang X-J, Chao Y-L, Zheng H-M, Sheng H-F, Liu H-Y, et al. The potential effect of oral microbiota in the prediction of mucositis during radiotherapy for nasopharyngeal carcinoma. EBioMedicine (2017) 18(101647039):23–31. doi: 10.1016/j.ebiom.2017.02.002
88. Reyes-Gibby CC, Wang J, Zhang L, Peterson CB, Do K-A, Jenq RR, et al. Oral microbiome and onset of oral mucositis in patients with squamous cell carcinoma of the head and neck. Cancer (2020) 126(23):5124–36. doi: 10.1002/cncr.33161
89. Al-Qadami G, Van Sebille Y, Le H, Bowen J. Gut microbiota: implications for radiotherapy response and radiotherapy-induced mucositis. Expert Rev Gastroenterol Hepatol (2019) 13(5):485–96. doi: 10.1080/17474124.2019.1595586
90. Owens D, Paleri V, Jones AV. Head and neck cancer explained: an overview of management pathways. Br Dent J (2022) 233(9):721–5. doi: 10.1038/s41415-022-5199-1
91. Lim Y, Fukuma N, Totsika M, Kenny L, Morrison M, Punyadeera C. The performance of an oral microbiome biomarker panel in predicting oral cavity and oropharyngeal cancers. Front Cell Infect Microbiol (2018) 8:267. doi: 10.3389/fcimb.2018.00267
92. Gougousis S, Mouchtaropoulou E, Besli I, Vrochidis P, Skoumpas I, Constantinidis I. HPV-related oropharyngeal cancer and biomarkers based on epigenetics and microbiome profile. Front Cell Dev Biol (2021) 8:625330. doi: 10.3389/fcell.2020.625330
93. Torralba MG, Aleti G, Li W, Moncera KJ, Lin YH, Yu Y, et al. Oral microbial species and virulence factors associated with oral squamous cell carcinoma. Microb Ecol (2021) 82(4):1030–46. doi: 10.1007/s00248-020-01596-5
94. Zuo HJ, Fu MR, Zhao HL, Du XW, Hu ZY, Zhao XY, et al. Study on the salivary microbial alteration of men with head and neck cancer and its relationship with symptoms in southwest China. Front Cell Infect Microbiol (2020) 10:514943. doi: 10.3389/fcimb.2020.514943
95. Jiang C, Wang H, Xia C, Dong Q, Chen E, Qiu Y, et al. A randomized, double-blind, placebo-controlled trial of probiotics to reduce the severity of oral mucositis induced by chemoradiotherapy for patients with nasopharyngeal carcinoma. Cancer (2019) 125(7):1081–90. doi: 10.1002/cncr.31907
96. Ma W, Mao Q, Xia W, Dong G, Yu C, Jiang F. Gut microbiota shapes the efficiency of cancer therapy. Front Microbiol (2019) 10:1050. doi: 10.3389/fmicb.2019.01050
97. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
98. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
99. Hou J, Zheng H, Li P, Liu H, Zhou H, Yang X. Distinct shifts in the oral microbiota are associated with the progression and aggravation of mucositis during radiotherapy. Radiother Oncol (2018) 129(1):44–51. doi: 10.1016/j.radonc.2018.04.023
100. Mougeot JC, Stevens CB, Almon KG, Paster BJ, Lalla RV, Brennan MT, et al. Caries-associated oral microbiome in head and neck cancer radiation patients: a longitudinal study. J Oral Microbiol (2019) 11(1):1586421. doi: 10.1080/20002297.2019.1586421
101. Shahidi F, Ambigaipalan P. Omega-3 polyunsaturated fatty acids and their health benefits. Annu Rev Food Sci Technol (2018) 9:345–81. doi: 10.1146/annurev-food-111317-095850
102. Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol (2015) 33(29):3293–304. doi: 10.1200/JCO.2015.61.1509
103. Zhou CB, Zhou YL, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. (2021) 7(7):647–60. doi: 10.1016/j.trecan.2021.01.010
104. Banna GL, Torino F, Marletta F, Santagati M, Salemi R, Cannarozzo E, et al. Lactobacillus rhamnosus GG: An overview to explore the rationale of its use in cancer. Front Pharmacol (2017) 8:603. doi: 10.3389/fphar.2017.00603
105. Okumura R, Takeda K. Maintenance of intestinal homeostasis by mucosal barriers. Inflammation Regener (2018) 38:5. doi: 10.1186/s41232-018-0063-z
106. Cheng Z, Xu H, Wang X, Liu Z. Lactobacillus raises in vitro anticancer effect of geniposide in HSC-3 human oral squamous cell carcinoma cells. Exp Ther Med (2017) 14(5):4586–94. doi: 10.3892/etm.2017.5105
107. Sharma A, Tilak T, Bakhshi S, Raina V, Kumar L, Chaudhary S, et al. S. Lactobacillus brevis CD2 lozenges prevent oral mucositis in patients undergoing high dose chemotherapy followed by haematopoietic stem cell transplantation. ESMO Open (2017) 1(6):e000138. doi: 10.1136/esmoopen-2016-000138
108. Abruzzo A, Vitali B, Lombardi F, Guerrini L, Cinque B, Parolin C, et al. Mucoadhesive buccal films for local delivery of Lactobacillus brevis. Pharmaceutics (2020) 12(3):241. doi: 10.3390/pharmaceutics12030241
109. Vesty A, Gear K, Boutell S, Taylor MW, Douglas RG, Biswas K. Randomised, double-blind, placebo-controlled trial of oral probiotic Streptococcus salivarius M18 on head and neck cancer patients post-radiotherapy: a pilot study. Sci Rep (2020) 10(1):13201. doi: 10.1038/s41598-020-70024-y
110. Doppalapudi R, Vundavalli S, Prabhat MP. Effect of probiotic bacteria on oral Candida in head- and neck-radiotherapy patients: A randomized clinical trial. J Cancer Res Ther (2020) 16(3):470–7. doi: 10.4103/jcrt.JCRT_334_18
111. Tempera I, Lieberman PM. Oncogenic viruses as entropic drivers of cancer evolution. Front Virol (2021) 1:753366. doi: 10.3389/fviro.2021.753366
112. Bruni L, Albero G, Rowley J, Alemany L, Arbyn M, Giuliano AR, et al. Global and regional estimates of genital human papillomavirus prevalence among men: a systematic review and meta-analysis. Lancet Global Health (2023) 11(9):e1345–62. doi: 10.1016/S2214-109X(23)00305-4
113. Castellsague X, Alemany L, Quer M, Halec G, Quirós B, Tous S, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Institute (2016) 108(6):djv403. doi: 10.1093/jnci/djv403
114. Syrjanen K, Syrjänen S, Lamberg M, Pyrhönen S, Nuutinen J. Morphological and immunohistochemical evidence suggesting human papillomavirus (HPV) involvement in oral squamous cell carcinogenesis. Int J Oral Surg (1983) 12(6):418–24. doi: 10.1016/s0300-9785(83)80033-7
115. de Sanjose S, Serrano B, Tous S, Alejo M, Lloveras B, Quirós B, et al. Burden of human papillomavirus (HPV)-related cancers attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr (2019) 2(4):pky045. doi: 10.1093/jncics/pky045
116. Bulane A, Goedhals D, Seedat RY, Goedhals J, Burt F. Human papillomavirus DNA in head and neck squamous cell carcinomas in the Free State, South Africa. J Med Virol (2020) 92(2):227–33. doi: 10.1002/jmv.25556
117. Vani NV, Madhanagopal R, Swaminathan R, Ganesan TS. Dynamics of oral human papillomavirus infection in healthy population and head and neck cancer. Cancer Med (2023) 12(10):11731–45. doi: 10.1002/cam4.5686
118. Aboagye E, Agyemang-Yeboah F, Duduyemi BM, Obirikorang C. Human papillomavirus detection in head and neck squamous cell carcinomas at a tertiary hospital in sub-Saharan Africa. TheScientificWorldJournal (2019), 2561530. doi: 10.1155/2019/2561530
119. Roman A, Munger K. The papillomavirus E7 proteins. Virology (2013) 445(1-2):138–68. doi: 10.1016/j.virol.2013.04.013
120. Li S, Hong X, Wei Z, Xie M, Li W, Liu G, et al. Ubiquitination of the HPV oncoprotein E6 is critical for E6/E6AP-mediated p53 degradation. Front Microbiol (2019) 10:2483. doi: 10.3389/fmicb.2019.02483
121. Huang Q, Li F, Ji M, Lin L, Hu C. Evaluating the prognostic significance of p53 and TP53 mutations in HPV-negative hypopharyngeal carcinoma patients: a 5-year follow-up retrospective study. BMC Cancer (2023) 23(1):324. doi: 10.1186/s12885-023-10775-9
122. Nwachuku K, Johnson DE, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma: opportunities for detection and monitoring via analysis of circulating tumor dna. In: El Assal R, Gaudilliere D, Connelly ST, editors. Early Detection and Treatment of Head & Neck Cancers. Switzerland AG: Springer, Cham (2021). doi: 10.1007/978-3-030-69852-2_5
123. Zhang Y, Koneva LA, Virani S, Arthur AE, Virani A, Hall PB, et al. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res (2016) 22(18):4735–45. doi: 10.1158/1078-0432.CCR-16-0323
124. Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med (2013) 5:15. doi: 10.1186/gm419
125. Feldman R, Gatalica Z, Knezetic J, Reddy S, Nathan CA, Javadi N, et al. Molecular profiling of head and neck squamous cell carcinoma. Head Neck (2016) 38,1(1):E1625–38. doi: 10.1002/hed.24290
126. Cho J, Johnson DE, Grandis JR. Therapeutic implications of the genetic landscape of head and neck cancer. Semin Radiat Oncol (2018) 28(1):2–11. doi: 10.1016/j.semradonc.2017.08.005
127. Johnson ME, Cantalupo PG, Pipas JM. Identification of head and neck cancer subtypes based on human papillomavirus presence and e2f-regulated gene expression. mSphere (2018) 3(1):e00580–17. doi: 10.1128/mSphere.00580-17
128. Farah CS. Molecular landscape of head and neck cancer and implications for therapy. Ann Transl Med (2021) 9(10):915. doi: 10.21037/atm-20-6264
129. Sastre-Garau X, Harlé A. Pathology of HPV-associated head and neck carcinomas: recent data and perspectives for the development of specific tumor markers. Front Oncol (2020) 10:528957. doi: 10.3389/fonc.2020.528957
130. Hinic S, Rich A, Anayannis NV, Cabarcas-Petroski S, Schramm L, Meneses PI. Gene expression and DNA methylation in human papillomavirus positive and negative head and neck squamous cell carcinomas. Int J Mol Sci (2022) 23(18):10967. doi: 10.3390/ijms231810967
131. Kwon E, Lee HR, Lee JH, Seo C, Ha M, Roh J, et al. Identification of differentially expressed genes and pathways for risk stratification in HPV-associated cancers governing different anatomical sites. Front bioscience (Landmark edition) (2022) 27(1):2. doi: 10.31083/j.fbl2701002
132. Shah M, Anwar MA, Park S, Jafri SS, Choi S. In silico mechanistic analysis of IRF3 inactivation and high-risk HPV E6 species-dependent drug response. Sci Rep (2015) 5:13446. doi: 10.1038/srep13446
133. Aarthy M, Singh SK. Interpretations on the interaction between protein tyrosine phosphatase and E7 oncoproteins of high and low-risk HPV: a computational perception. ACS omega (2021) 6(25):16472–87. doi: 10.1021/acsomega.1c01619
134. Castro-Munoz LJ, Rocha-Zavaleta L, Lizano M, Ramírez-Alcántara KM, Madrid-Marina V, Manzo-Merino J. Alteration of the IFN-pathway by human papillomavirus proteins: antiviral immune response evasion mechanism. Biomedicines (2022) 10(11):2965. doi: 10.3390/biomedicines10112965
135. Hasan UA, Zannetti C, Parroche P, Goutagny N, Malfroy M, Roblot G, et al. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the Toll-like receptor 9 promoter. J Exp Med (2013) 210(7):1369–87. doi: 10.1084/jem.20122394
136. Kang TH, Mao CP, Kim YS, Kim TW, Yang A, Lam B, et al. TLR9 acts as a sensor for tumor-released DNA to modulate anti-tumor immunity after chemotherapy. J immunotherapy Cancer (2019) 7(1):260. doi: 10.1186/s40425-019-0738-2
137. Tumban E. A current update on human papillomavirus-associated head and neck cancers. Viruses (2019) 11(10):922. doi: 10.3390/v11100922
138. Stephen JK, Divine G, Chen KM, Chitale D, Havard S, Worsham MJ. Significance of p16 in site-specific HPV positive and HPV negative head and neck squamous cell carcinoma. Cancer Clin Oncol (2013) 2(1):51–61. doi: 10.5539/cco.v2n1p51
139. Mehanna H, Taberna M, von Buchwald C, Tous S, Brooks J, Mena M, et al. Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol (2023) 24(3):239–51. doi: 10.1016/S1470-2045(23)00013-X
140. Gutierrez-Xicotencatl L, Pedroza-Saavedra A, Chihu-Amparan L, Salazar-Piña A, Maldonado-Gama M, Esquivel-Guadarrama F. Cellular functions of HPV16 E5 oncoprotein during oncogenic transformation. Mol Cancer Res MCR (2021) 19(2):167–79. doi: 10.1158/1541-7786.MCR-20-0491
141. Al-Thawadi H, Gupta I, Jabeen A, Skenderi F, Aboulkassim T, Yasmeen A, et al. Co-presence of human papillomaviruses and Epstein-Barr virus is linked with advanced tumor stage: a tissue microarray study in head and neck cancer patients. Cancer Cell Int (2020) 20:361. doi: 10.1186/s12935-020-01348-y
142. Gupta I, Ghabreau L, Al-Thawadi H, Yasmeen A, Vranic S, Al Moustafa AE, et al. Co-incidence of human papillomaviruses and epstein-barr virus is associated with high to intermediate tumor grade in human head and neck cancer in Syria. Front Oncol (2020) 10:1016. doi: 10.3389/fonc.2020.01016
143. Sabatini ME, Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer (2020) 122(3):306–14. doi: 10.1038/s41416-019-0602-7
144. Blanco R, Carrillo-Beltrán D, Corvalán AH, Aguayo F. High-risk human papillomavirus and epstein-barr virus coinfection: a potential role in head and neck carcinogenesis. Biology (2021) 10(12):1232. doi: 10.3390/biology10121232
145. Makvandi M, Jalilian S, Faghihloo E, Khanizadeh S, Ramezani A, Bagheri S, et al. Prevalence of human papillomavirus and co-infection with epstein-barr virus in oral and oropharyngeal squamous cell carcinomas. Asian Pacific J Cancer prevention: APJCP (2022) 23(11):3931–7. doi: 10.31557/APJCP.2022.23.11.3931
146. Carpen T, Sjöblom A, Lundberg M, Haglund C, Markkola A, Syrjänen S, et al. Presenting symptoms and clinical findings in HPV-positive and HPV-negative oropharyngeal cancer patients. Acta Otolaryngol (2018) 138(7):675. doi: 10.1080/00016489.2017.1405279
147. Lechner M, Liu J, Masterson L, Fenton TR. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol (2022) 19(5):306–27. doi: 10.1038/s41571-022-00603-7
148. Stenmark MH, Shumway D, Guo C, Vainshtein J, Mierzwa M, Jagsi R, et al. Influence of human papillomavirus on the clinical presentation of oropharyngeal carcinoma in the United States. Laryngoscope (2017) 127(10):2270–8. doi: 10.1002/lary.26566
149. Atipas K, Laokulrath N, Petsuksiri J, Ratanaprasert N, Pongsapich W. CD8+ T Cells and PD-L1 expression as prognostic indicators in a low prevalence of HPV-associated oropharyngeal squamous cell carcinoma. Curr Oncol (Toronto Ont.) (2023) 30(2):1450–60. doi: 10.3390/curroncol30020111
150. Osorio JC, Castillo A. Epigenetic mechanisms in head and neck cancer. In: Bulgin D, editor. New aspects in molecular and cellular mechanisms of human carcinogenesis. London, United Kinkdom: InTech, Rijeka (2016). p. 67–95. doi: 10.5772/61135
151. Liu J, Huang B, Ding F, Li Y. Environment factors, DNA methylation, and cancer. Environ Geochem Health (2023) 45(11):7543–68. doi: 10.1007/s10653-023-01749-8
152. Stenz L. The L1-dependant and Pol III transcribed Alu retrotransposon, from its discovery to innate immunity. Mol Biol Rep (2021) 48(3):2775–89. doi: 10.1007/s11033-021-06258-4
153. Peterson EJ, Bögler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res (2003) 63(20):6579–82.
154. Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res (2010) 70(14):5807–17. doi: 10.1158/0008-5472.CAN-09-4161
155. Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics (2011) 6:777–87. doi: 10.4161/epi.6.6.16216
156. Colacino JA, Dolinoy DC, Duffy SA, Sartor MA, Chepeha DB, Bradford CR, et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PloS One (2013) 8:e54742. doi: 10.1371/journal.pone.0054742
157. Dong SM, Sun DI, Benoit NE, Kuzmin I, Lerman MI, Sidransky D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin Cancer Res (2003) 9 Pt 1:3635–40.
158. Shen-Gunther J, Wang C-M, Poage GM, Lin C-L, Perez L, Banks NA, et al. Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology. Clin Epigenet. (2016) 8:96. doi: 10.1186/s13148-016-0263-9
159. Nakagawa T, Matsusaka K, Misawa K, Ota S, Takane K, Fukuyo M, et al. Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett (2017) 407:21–31. doi: 10.1016/j.canlet.2017.08.008
160. Ren S, Gaykalova D, Wang J, Guo T, Danilova L, Favorov A, et al. Discovery and development of differentially methylated regions in human papillomavirus-related oropharyngeal squamous cell carcinoma. Int J Cancer. (2018) 143:2425–36. doi: 10.1002/ijc.31778
161. Van Kempen PM, van Bockel L, Braunius WW, Moelans CB, van Olst M, de Jong R, et al. HPV-positive oropharyngeal squamous cell carcinoma is associated with TIMP3 and CADM1 promoter hypermethylation. Cancer Med (2014) 3:1185–96. doi: 10.1002/cam4.313
162. Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, et al. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PloS One (2009) 4:e4941. doi: 10.1371/journal.pone.0004941
163. Poage GM, Houseman EA, Christensen BC, Butler RA, Avissar-Whiting M, McClean MD, et al. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin Cancer Res (2011) 17:3579–89. doi: 10.1158/1078-0432.CCR-11-0044
164. Ekanayake Weeramange C, Tang KD, Vasani S, Langton-Lockton J, Kenny L, Punyadeera C. DNA methylation changes in human papillomavirus-driven head and neck cancers. Cells (2020) 9(6):1359. doi: 10.3390/cells9061359
165. Stich M, Ganss L, Puschhof J, Prigge E-S, Reuschenbach M, Guiterrez A, et al. 5-aza-2’-deoxycytidine (DAC) treatment downregulates the HPV E6 and E7 oncogene expression and blocks neoplastic growth of HPV-associated cancer cells. Oncotarget (2016) 8:52104–17. doi: 10.18632/oncotarget.10631
166. Burkitt K, Saloura V. Epigenetic modifiers as novel therapeutic targets and a systematic review of clinical studies investigating epigenetic inhibitors in head and neck cancer. Cancers (Basel) (2021) 13(20):5241. doi: 10.3390/cancers13205241
167. Yang SC, Wang WY, Zhou JJ, Wu L, Zhang MJ, Yang QC, et al. Inhibition of DNMT1 potentiates antitumor immunity in oral squamous cell carcinoma. Int Immunopharmacol. (2022) 111:109113. doi: 10.1016/j.intimp.2022.109113
168. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal (2020) 18(1):59. doi: 10.1186/s12964-020-0530-4
169. Parham P, Guethlein LA. Genetics of natural killer cells in human health, disease, and survival. Annu Rev Immunol (2018) 36:519–48. doi: 10.1146/annurev-immunol-042617-053149
170. Charap AJ, Enokida T, Brody R, Sfakianos J, Miles B, Bhardwaj N, et al. Landscape of natural killer cell activity in head and neck squamous cell carcinoma. J Immunother Cancer (2020) 8(2):e001523. doi: 10.1136/jitc-2020-001523
171. Jumaniyazova E, Lokhonina A, Dzhalilova D, Kosyreva A, Fatkhudinov T. Immune cells in head-and-neck tumor microenvironments. J Pers Med (2022) 12(9):1521. doi: 10.3390/jpm12091521
172. Venkatkumar S, Narayan M, Krishnan R. Recapitulating the tumor microenvironment in head-and-neck squamous cell carcinoma: A narrative review. Cancer Research Statistics Treat (2022) 5(3):499–506. doi: 10.4103/crst.crst_182_22
173. Mele D, Pessino G, Trisolini G, Luchena A, Benazzo M, Morbini P, et al. Impaired intratumoral natural killer cell function in head and neck carcinoma. Front Immunol (2022) 13:997806. doi: 10.3389/fimmu.2022.997806
174. Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol (2013) 31:413–41. doi: 10.1146/annurev-immunol-032712-095951
175. Chan IS, Ewald AJ. The changing role of natural killer cells in cancer metastasis. J Clin Invest. (2022) 132(6):e143762. doi: 10.1172/JCI143762
176. Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol (2018) 9:847. doi: 10.3389/fimmu.2018.00847
177. Decker AS, Pylaeva E, Brenzel A, Spyra I, Droege F, Hussain T, et al. Prognostic role of blood NETosis in the progression of head and neck cancer. Cells (2019) 8(9):946. doi: 10.3390/cells8090946
178. Ekstedt S, Piersiala K, Starkhammar M, Margolin G, Kumlien Georén S, Cardell LO. 998 Neutrophil subsets in head and neck cancer. J ImmunoTherapy Cancer (2022) 10. doi: 10.1136/jitc-2022-SITC2022.0998
179. Mascarella MA, Mannard E, Silva SD, Zeitouni A. Neutrophil-to-lymphocyte ratio in head and neck cancer prognosis: A systematic review and meta-analysis. Head Neck. (2018) 40(5):1091–100. doi: 10.1002/hed.25075
180. Ma SJ, Yu H, Khan M, Gill J, Santhosh S, Chatterjee U, et al. Evaluation of optimal threshold of neutrophil-lymphocyte ratio and its association with survival outcomes among patients with head and neck cancer. JAMA Netw Open (2022) 5(4):e227567. doi: 10.1001/jamanetworkopen.2022.7567
181. Abolhalaj M, Askmyr D, Sakellariou CA, Lundberg K, Greiff L, Lindstedt M. Profiling dendritic cell subsets in head and neck squamous cell tonsillar cancer and benign tonsils. Sci Rep (2018) 8(1):8030. doi: 10.1038/s41598-018-26193-y
182. Fialova A, Koucký V, Hajdušková M, Hladíková K, Špíšek R. Immunological network in head and neck squamous cell carcinoma-a prognostic tool beyond HPV status. Front Oncol (2020) 10:1701. doi: 10.3389/fonc.2020.01701
183. Mei Z, Huang J, Qiao B, Lam AK. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci (2020) 12(1):16. doi: 10.1038/s41368-020-0084-8
184. Ravi R, Noonan KA, Pham V, Bedi R, Zhavoronkov A, Ozerov IV, et al. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat Commun (2018) 9(1):741. doi: 10.1038/s41467-017-02696-6
185. Damasio MPS, Nascimento CS, Andrade LM, de Oliveira VL, Calzavara-Silva CE. The role of T-cells in head and neck squamous cell carcinoma: From immunity to immunotherapy. Front Oncol (2022) 12:1021609. doi: 10.3389/fonc.2022.1021609
186. Khan M, Arooj S, Wang H. Soluble B7-CD28 family inhibitory immune checkpoint proteins and anti-cancer immunotherapy. Front Immunol (2021) 12:651634. doi: 10.3389/fimmu.2021.651634
187. Banerjee S, Nahar U, Dahiya D, Mukherjee S, Dey P, Gupta R, et al. Role of cytotoxic T cells and PD-1 immune checkpoint pathway in papillary thyroid carcinoma. Front Endocrin. (2022) 13:931647. doi: 10.3389/fendo.2022.931647
188. Steele JC, Roberts S, Rookes SM, Gallimore PH. Detection of CD4(+)- and CD8(+)-T-cell responses to human papillomavirus type 1 antigens expressed at various stages of the virus life cycle by using an enzyme-linked immunospot assay of gamma interferon release. J Virol (2002) 76(12):6027–36. doi: 10.1128/jvi.76.12.6027-6036.2002
189. Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature (2021) 597:279–84. doi: 10.1038/s41586-021-03862-z
190. Economopoulou P, Perisanidis C, Giotakis EI, Psyrri A. The emerging role of immunotherapy in head and neck squamous cell carcinoma (HNSCC): anti-tumor immunity and clinical applications. Ann Transl Med (2016) 4(9):173. doi: 10.21037/atm.2016.03.34
191. de Vicente JC, Rodríguez-Santamarta T, Rodrigo JP, Blanco-Lorenzo V, Allonca E, García-Pedrero JM. PD-L1 expression in tumor cells is an independent unfavorable prognostic factor in oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev (2018) 28(3):546–54. doi: 10.1158/1055-9965.EPI-18-0779
192. Raskov H, Orhan A, Christensen JP, Gögenur I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br J Cancer (2021) 124(2):359–67. doi: 10.1038/s41416-020-01048-4
193. Wang X, Zha H, Wu W, Yuan T, Xie S, Jin Z, et al. CD200+ cytotoxic T lymphocytes in the tumor microenvironment are crucial for efficacious anti-PD-1/PD-L1 therapy. Sci Transl Med (2023) 15(679):eabn5029. doi: 10.1126/scitranslmed.abn5029
194. Fury M, Ou SI, Balmanoukian A, Hansen A, Massarelli E, Blake-Haskins A, et al. Clinical activity and safety of medi4736, an anti-pd-L1 antibody, in patients with head and neck cancer. Ann Oncol (2014) 25:4_suppl:iv341. doi: 10.1093/annonc/mdu340.3
195. Segal NH, Ou SHI, Balmanoukian AS, Fury. MG, Massarelli E, Brahmer J, et al. Safety and efficacy of MEDI4736, an anti-PD-L1 antibody, in patients from a squamous cell carcinoma of the head and neck (SCCHN) expansion cohort. J Clin Oncol (2015) 33:15_suppl:3011–1. doi: 10.1200/jco.2015.33.15_suppl.3011
196. Liu J, Chen Z, Li Y, Zhao W, Wu J, Zhang Z. PD-1/PD-L1 checkpoint inhibitors in tumor immunotherapy. Front Pharmacol (2021) 12:731798. doi: 10.3389/fphar.2021.731798
197. Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J immunotherapy Cancer (2019) 7(1):278. doi: 10.1186/s40425-019-0768-9
198. Dai X, Shen L. Advances and trends in omics technology development. Front Med (Lausanne). (2022) 9:911861. doi: 10.3389/fmed.2022.911861
199. Qi S, Wang J, Zhang Y, Naz M, Afzal MR, Du D, et al. Omics approaches in invasion biology: understanding mechanisms and impacts on ecological health. Plants (Basel). (2023) 12(9):1860. doi: 10.3390/plants12091860
200. Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, et al. Next-generation sequencing technology: current trends and advancements. Biology (Basel) (2023) 12(7):997. doi: 10.3390/biology12070997
201. Sokac M, Kjær A, Dyrskjøt L, Haibe-Kains B, Jwl Aerts H, Birkbak NJ. Spatial transformation of multi-omics data unlocks novel insights into cancer biology. Elife (2023) 12:RP87133. doi: 10.7554/eLife.87133
202. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol (2017) 28(6):1368–79. doi: 10.1093/annonc/mdx108
203. Oh B, Boyle F, Pavlakis N, Clarke S, Guminski A, Eade T, et al. Emerging evidence of the gut microbiome in chemotherapy: A clinical review. Front Oncol (2021) 11:706331. doi: 10.3389/fonc.2021.706331
204. Sankarapandian V, Venmathi Maran BA, Rajendran RL, Jogalekar MP, Gurunagarajan S, Krishnamoorthy R, et al. An update on the effectiveness of probiotics in the prevention and treatment of cancer. Life (Basel) (2022) 12(1):59. doi: 10.3390/life12010059
205. Pulito C, Cristaudo A, Porta C, Zapperi S, Blandino G, Morrone A, et al. Oral mucositis: the hidden side of cancer therapy. J Exp Clin Cancer Res (2020) 39(1):210. doi: 10.1186/s13046-020-01715-7
206. Powell SF, Vu L, Spanos WC, Pyeon D. The Key differences between human papillomavirus-positive and -negative head and neck cancers: biological and clinical implications. Cancers (Basel) (2021) 13(20):5206. doi: 10.3390/cancers13205206
Keywords: HNC, risk factors, signaling pathways, oral microbiota, HPV infection, tumor microenvironment
Citation: Constantin M, Chifiriuc MC, Mihaescu G, Vrancianu CO, Dobre E-G, Cristian R-E, Bleotu C, Bertesteanu SV, Grigore R, Serban B and Cirstoiu C (2023) Implications of oral dysbiosis and HPV infection in head and neck cancer: from molecular and cellular mechanisms to early diagnosis and therapy. Front. Oncol. 13:1273516. doi: 10.3389/fonc.2023.1273516
Received: 06 August 2023; Accepted: 30 November 2023;
Published: 18 December 2023.
Edited by:
Marcela Lizano, National Institute of Cancerology (INCAN), MexicoReviewed by:
Vui King Vincent-Chong, University at Buffalo, United StatesCopyright © 2023 Constantin, Chifiriuc, Mihaescu, Vrancianu, Dobre, Cristian, Bleotu, Bertesteanu, Grigore, Serban and Cirstoiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Carmen Chifiriuc, Y2FybWVuLmNoaWZpcml1Y0BiaW8udW5pYnVjLnJv
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.