
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 08 January 2024
Sec. Cancer Imaging and Image-directed Interventions
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1273414
 Shiming Zang1†
Shiming Zang1† Cuiping Jiang2†
Cuiping Jiang2† Lele Zhang1†
Lele Zhang1† Jingjing Fu1
Jingjing Fu1 Qingle Meng1
Qingle Meng1 Wenyu Wu1
Wenyu Wu1 Guoqiang Shao1
Guoqiang Shao1 Hongbin Sun3
Hongbin Sun3 Ruipeng Jia3*
Ruipeng Jia3* Feng Wang1*
Feng Wang1*Objectives: To explore the feasibility and importance of deep learning (DL) based on 68Ga-prostate-specific membrane antigen (PSMA)-11 PET/CT in predicting pathological upgrading from biopsy to radical prostatectomy (RP) in patients with prostate cancer (PCa).
Methods: In this retrospective study, all patients underwent 68Ga-PSMA-11 PET/CT, transrectal ultrasound (TRUS)-guided systematic biopsy, and RP for PCa sequentially between January 2017 and December 2022. Two DL models (three-dimensional [3D] ResNet-18 and 3D DenseNet-121) based on 68Ga-PSMA-11 PET and support vector machine (SVM) models integrating clinical data with DL signature were constructed. The model performance was evaluated using area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, and specificity.
Results: Of 109 patients, 87 (44 upgrading, 43 non-upgrading) were included in the training set and 22 (11 upgrading, 11 non-upgrading) in the test set. The combined SVM model, incorporating clinical features and signature of 3D ResNet-18 model, demonstrated satisfactory prediction in the test set with an AUC value of 0.628 (95% confidence interval [CI]: 0.365, 0.891) and accuracy of 0.727 (95% CI: 0.498, 0.893).
Conclusion: A DL method based on 68Ga-PSMA-11 PET may have a role in predicting pathological upgrading from biopsy to RP in patients with PCa.
Systematic biopsy guided by transrectal ultrasound (TRUS) has long been considered a standard diagnostic method for confirming prostate cancer (PCa) in patients with elevated prostate-specific antigen level and/or abnormal digital rectal examination (1). However, this method often misses clinically significant PCa, and some patients experience a pathological upgrade following radical prostatectomy (RP) (2–4). Considering the crucial influence accurate diagnosis has on treatment decisions and prognosis prediction (5, 6), there is a pressing need to develop reliable methods for predicting the pathological upgrading of PCa.
Prostate-specific membrane antigen (PSMA) is a highly specific prostatic epithelial cell transmembrane protein and is highly expressed in PCa cells (7, 8). The recent emergence of 68Ga-labeled PSMA inhibitors as promising agents for positron emission tomography/computed tomography (PET/CT) in patients with primary PCa has demonstrated diagnostic and staging capabilities that are superior to conventional imaging techniques (9–11). Moreover, these PET/CT images can be quantitatively analyzed using deep learning (DL), which involves automatically extracting complex and abstract information from medical images to achieve highly accurate detection or classification outcomes (12, 13). Previous studies have shown the potential feasibility of DL in PCa detection, risk assessment, and prognosis prediction (12–16). However, to the best of our knowledge, the predictive value of DL based on 68Ga-PSMA-11 PET/CT for assessing pathological upgrading in PCa has not been investigated.
Therefore, this study was aimed at exploring an efficient strategy with a PSMA PET/CT–based convolutional neural network (CNN) model for predicting pathological upgrading in patients with PCa.
We enrolled patients who had undergone 68Ga-PSMA-11 PET/CT prior to RP at our department between January 2017 and December 2022. The exclusion criteria were as follows: (1) lack of TRUS-guided systematic biopsy; (2) International Society of Urological Pathology grade group (ISUP GG) 5 on biopsy; (3) previous treatment for PCa before RP. The patients were randomly divided into a training cohort (n = 87) and a validation cohort (n = 22) with a ratio of 8:2 (Supplementary Figure 1).
All of the patients underwent PET/CT using a dedicated PET/CT system (United Imaging, uMI780, China) at 60 ± 5 min after intravenous injection of 2–2.3 MBq/kg 68Ga-PSMA-11 synthesized as previously described (17). A nonenhanced CT scan (120 kV, mA modulation, pitch 0.988, slice thickness 3.0 mm, increment 1.5 mm) was obtained, followed by a whole-body PET scan (3 min/bed, field of view 60 cm) in 3D mode (matrix 256 × 256) from the vertex to the proximal legs. The datasets were fully corrected for random coincidences, scatter radiation, and attenuation. For PET image reconstruction, the ordered-subsets expectation maximization method was used. Attenuation corrections of the PET images were performed using data from the CT scans. PET/CT fusion was performed using a workstation (uWS-MI, United Imaging).
The volumes of interest (VOIs) for the prostate gland were accurately delineated and segmented slice by slice using 3D Slicer software (version: 4.1.1.0; www.slicer.org) by a highly experienced nuclear medicine radiologist (FW) with 20 years of expertise in prostate PET/CT. The radiologist, blinded to the clinical information, performed this task by carefully analyzing the PET images and factoring in the corresponding CT scan for accurate localization and segmentation of the VOIs.
The full flow diagram for machine learning models is provided in Figure 1. DL models were developed based on a convolutional neural network (CNN), which is a typical and commonly used DL architecture used to have been extensively applied for image analysis. CNN achieves prediction tasks by iterating the low-level information retrieved from input data into more abstract high-level features. Two well-known CNNs—ResNet-18 and DenseNet-121—were selected in this study (18, 19). To extract context features comprehensively, three-dimensional (3D) ResNet-18 and 3D DenseNet-121 were used to learn and extract the relevant image features from the PET images.
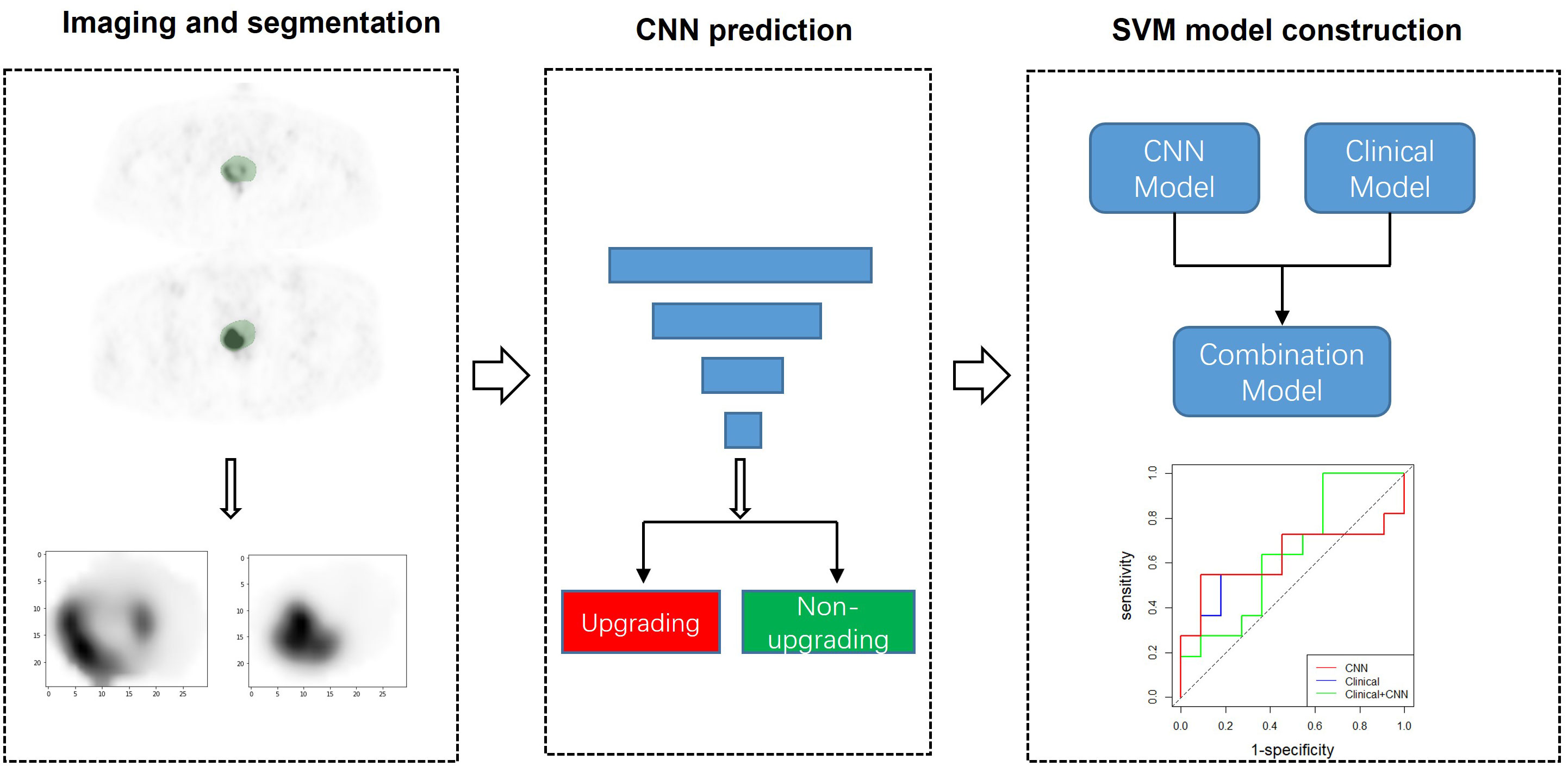
Figure 1 CNN modeling and analysis workflow. Using segmented 68Ga-PSMA-11 PET as input, two CNN models were trained to predict pathological upgrading. Additionally, the SVM models integrating clinical data with deep learning signature were constructed and evaluated. CNN, convolutional neural network; SVM, support vector machine.
For model training, each VOI image was normalized according to the z-score by subtracting the means and then dividing by the standard deviations. To reduce overfitting, data augmentation of random rotation, horizontal/vertical-flip, and affine transformation were employed. The Adam optimizer was used to upgrade the network parameters with a batch size of 4. The learning rate was set at 0.0001. An early stopping criterion was used to terminate training as there was no further improvement in loss. All training sessions were implemented using PyTorch (version 1.12.1; https://www.pytorch.org). The performance of the developed model was evaluated with cross-validation (five-fold) on the training cohort and then tested on the test cohort.
The support vector machine (SVM) algorithm was used to evaluate the performance of clinical features for predicting pathology upgrading. GridsearchCV was carried out to select optimal hyperparameters for the SVM model, including kernel function, C, and sigma. Moreover, the signature [predicted possibilities of upgrading) of the CNN achieving higher AUC and clinical data (age, prostate-specific antigen (PSA), percentage of positive cores (PPC), biopsy ISUP GG, prostate volume, prostate-specific antigen density (PSAD)]] were jointly employed by the SVM to perform the multimodality prediction.
All patients underwent 18-gauge needle biopsy under local anesthesia by experienced urologists. The biopsy was performed under the guidance of TRUS utilizing a 12-core extended scheme. The men subsequently underwent RP based on the joint decision between the treating surgeon and the patient, based on current clinical guidelines. Upgrading was defined as any increase of biopsy ISUP GG to RP ISUP GG.
The Mann–Whitney U test was used to compare continuous variables, while the chi-squared test was used for the comparison of categorical variables. The diagnostic performance of the models for predicting pathological upgrading was evaluated using receiver operating characteristic (ROC) curve analysis. The optimal cutoff values of the models were determined by maximizing the Youden index in the training set. The fixed model cutoff values from the training set were then applied to the test set. Statistical analysis was conducted using SPSS 22.0 (IBM, Armonk, NY, USA) and R software (version 4.1.3; http://www.Rproject.org). All statistical tests were two-sided, and a P value < 0.05 was considered statistically significant.
In total, 109 patients were finally included in the study and divided randomly into the training set (n = 87) and the test set (n = 22). The baseline characteristics of the patients in the training and test sets are summarized in Table 1. In the training set, 50.57% of the patients (44 of 87) were identified as upgrading and 49.43% (43 of 87) as non-upgrading. In the test set, 50.00% of the patients (11 of 22) were identified as upgrading and 50.00% of the patients (11 of 22) as non-upgrading. Based on those characteristics of the patients, no significant differences were observed between the training and test sets.
The AUC, accuracy, sensitivity, and specificity of the two CNNs measured in the cross-validation are illustrated in Supplementary Table 1 and Supplementary Figure 2. The overall performance of the 3D ResNet-18 model was slightly better than that of the 3D DenseNet-121 model (AUC: 0.621 vs. 0.559 [p = 0.443], accuracy: 0.644 vs. 0.609 [p = 0.638]). The 3D ResNet-18 model achieved sensitivity of 0.682 (95% confidence interval [CI]: 0.523, 0.809) and specificity of 0.605 (95% CI: 0.445, 0.706). Additionally, the signature of the 3D ResNet-18 model and clinical data were considered as input variables to build the combined model.
The performance of the CNN only, clinical data only, and combined model is summarized in Table 2 and Figure 2. The 3D ResNet-18 model yielded an AUC value of 0.612 (95% CI: 0.350, 0.873) and accuracy of 0.682 (95% CI: 0.546, 0.828), which was comparable to the clinical model (p = 0.863 and p = 1.000, respectively). The 3D ResNet-18 model performed comparable on specificity to the clinical model (0.818 vs. 0.364, p = 0.080), but worse on sensitivity (0.546 vs. 1.000, p = 0.035). The obtained CNN performance in the test set was comparable to cross-validation (p = 0.949).
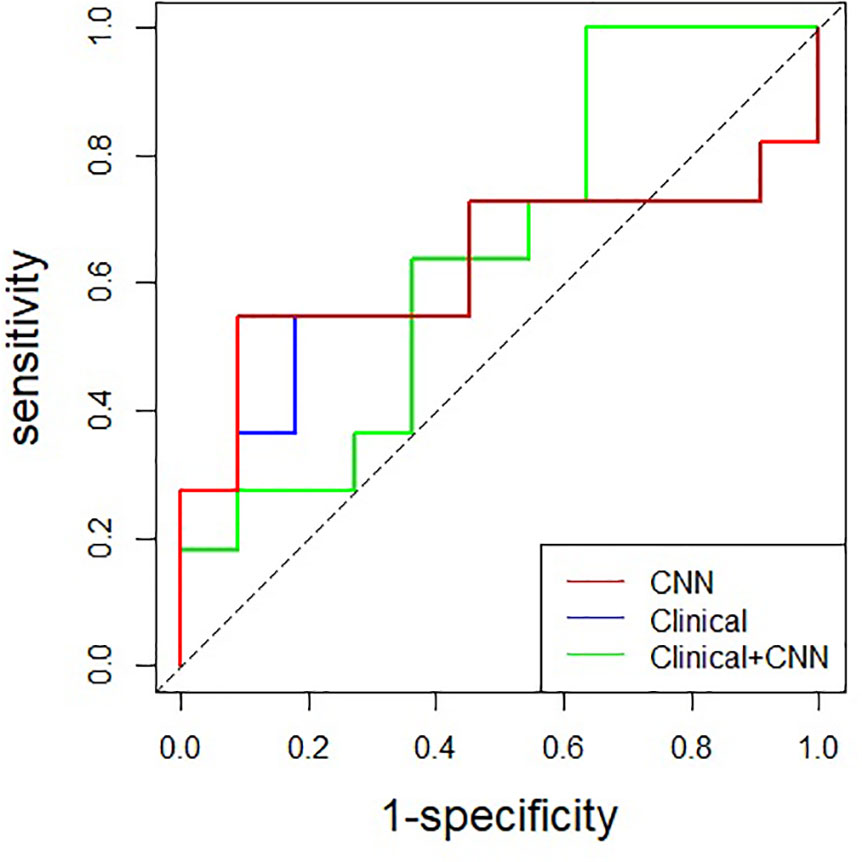
Figure 2 Comparison of receiver operating characteristic curves among CNN, clinical and combined model in the test set (AUC: 0.612, 0.645, and 0.628, respectively). CNN, convolutional neural network.
For multimodality analysis, the combined model achieved the highest accuracy (0.727 [95% CI: 0.498, 0.893]) among the models, although there were no significant differences in AUC (0.628 vs. 0.612, p = 0.406), accuracy (0.727 vs. 0.682, p = 0.741), sensitivity (0.546 vs. 0.546, p = 1.000), and specificity (0.909 vs. 0.818, p = 1.000) between the combined model and the 3D ResNet-18 model. No significant differences were found in AUC (0.628 vs. 0.645, p = 0.930) and accuracy (0.727 vs. 0.682, p = 0.741) when comparing the combined model with the clinical model. However, the combined model demonstrated lower sensitivity (0.546 vs. 1.000, p = 0.012) and higher specificity (0.909 vs. 0.364, p = 0.024) than the clinical model.
Histopathology results of biopsy samples and the biopsy Gleason score of the patients play a key role in clinical decision-making such as in choosing RP, extended lymphadenectomy, radiotherapy, or active surveillance (20–22). The present study demonstrated that the CNNs trained using PSMA PET/CT data performed well in predicting pathological upgrading from TRUS-guided systematic biopsy to final RP, which has never been investigated before. The 3D ResNet-18 model was successfully validated in the independent test set (AUC, 0.612; accuracy, 0.682; sensitivity, 0.546; specificity, 0.818).The combined model achieved significantly higher specificity but lower sensitivity than the clinical model.
Pathological upgrading from biopsy to RP is common in clinical practice. Serefoglu et al. (4) assessed 90 patients and reported an upgrading rate of 47.8% in those who underwent TRUS-guided systematic biopsy. Another study documented an upgrading rate of 31.5% in patients who had undergone the same procedure (23). On account of precise targeting, multiparametric magnetic resonance imaging targeted biopsy significantly improved the detection of clinically significant PCa and decreased the detection of insignificant PCa. In our study, we observed upgrading rates of 50.57% and 50.00% in the training and test sets, respectively. The variation in upgrading rates may be due to the exclusion of all patients with ISUP GG 5 at biopsy. Moreover, previous studies recorded different upgrading rates in patients with different risk groups or biopsy cores (24, 25). Considering that an adverse pathological outcome can potentially lead to erroneous decisions, the development of an efficient predictive model has great significance.
In recent years, there have been many studies assessing DL techniques based on PSMA PET/CT for detection and risk stratification of PCa. Zhao et al. (15) found that a CNN was able to accurately detect the bone and lymph node lesions on 68Ga-PSMA-11 PET/CT. Capobianco et al. (16) constructed a CNN model trained on 68Ga-PSMA PET/CT data to both classify PET/CT regions of interest as uptake suspicious or nonsuspicious for cancer and assign them an anatomical location classification. The evaluated algorithm showed satisfactory agreement with expert assessment for identification and anatomical location classification of suspicious uptake sites. In this study, we focused on predicting pathological upgrading and developed a CNN model based on 3D ResNet-18 net architecture for the prediction. The satisfactory performance of the CNN model suggests that DL has great potential for predicting pathological upgrading.
Previous studies have investigated the use of PSMA PET/CT in predicting pathological upgrading (26, 27). However, most of these studies focused on evaluating the standardized uptake value (SUV) of the prostate lesion, without considering the extensive spatial and morphological information available. Our study showcased the ability of 3D CNN models to extract comprehensive insights from the entire prostate to predict pathological upgrading. In order to eliminate the influence of surrounding organ tissues, the VOI of the prostate was segmented as the input volume for the CNN models. Moreover, we integrated clinical features with CNN to create a multimodality model so as to comprehensively predict pathological upgrading. Although the overall performance was satisfactory, no significant differences were found in AUC and accuracy when comparing the combined model with the CNN model, which could be due to the small size of our sample and consequent overfitting.
This study has some limitations. First, its single-center design and relatively small sample size may compromise the model’s generalization ability and affect its sensitivity and specificity. Therefore, in order to improve the robustness of the model, it is necessary to formulate a unified standard for multicenter studies, and identify and test multicenter data using DL methods. Second, further research using different PET/CT scanners is needed to validate the generalizability and robustness of the CNN model. Third, different observers who performed segmentation could have influenced the stability of the model. Automated and accurate tumor segmentation must be developed to facilitate the efficiency of the DL process.
The effective CNN model based on 68Ga-PSMA PET/CT may have a role in predicting pathological upgrading from biopsy to RP in patients with primary PCa.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the Ethic Committee of Nanjing First Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
SZ: Software, Writing – original draft, Conceptualization. CJ: Data curation, Methodology, Writing – original draft. LZ: Data curation, Methodology, Writing – original draft. JF: Formal Analysis, Validation, Writing – original draft. QM: Software, Writing – original draft. WW: Methodology, Writing – original draft. GS: Resources, Writing – original draft. HS: Resources, Writing – review & editing. RJ: Writing – review & editing. FW: Conceptualization, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1273414/full#supplementary-material
1. Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. Eau-eanm-estro-esur-siog guidelines on prostate cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol (2021) 79(2):243–62. doi: 10.1016/j.eururo.2020.09.042
2. Cohen MS, Hanley RS, Kurteva T, Ruthazer R, Silverman ML, Sorcini A, et al. Comparing the gleason prostate biopsy and gleason prostatectomy grading system: the lahey clinic medical center experience and an international meta-analysis. Eur Urol (2008) 54(2):371–81. doi: 10.1016/j.eururo.2008.03.049
3. Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric mri and trus biopsy in prostate cancer (Promis): A paired validating confirmatory study. Lancet (2017) 389(10071):815–22. doi: 10.1016/S0140-6736(16)32401-1
4. Serefoglu EC, Altinova S, Ugras NS, Akincioglu E, Asil E, Balbay MD. How reliable is 12-core prostate biopsy procedure in the detection of prostate cancer? Can Urol Assoc J (2013) 7(5-6):E293–8. doi: 10.5489/cuaj.11224
5. Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. Eau-eanm-estro-esur-siog guidelines on prostate cancer. Part ii-2020 update: treatment of relapsing and metastatic prostate cancer. Eur Urol (2021) 79(2):263–82. doi: 10.1016/j.eururo.2020.09.046
6. Brockman JA, Alanee S, Vickers AJ, Scardino PT, Wood DP, Kibel AS, et al. Nomogram predicting prostate cancer-specific mortality for men with biochemical recurrence after radical prostatectomy. Eur Urol (2015) 67(6):1160–7. doi: 10.1016/j.eururo.2014.09.019
7. Paschalis A, Sheehan B, Riisnaes R, Rodrigues DN, Gurel B, Bertan C, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol (2019) 76(4):469–78. doi: 10.1016/j.eururo.2019.06.030
8. Sheikhbahaei S, Afshar-Oromieh A, Eiber M, Solnes LB, Javadi MS, Ross AE, et al. Pearls and pitfalls in clinical interpretation of prostate-specific membrane antigen (Psma)-targeted pet imaging. Eur J Nucl Med Mol Imaging (2017) 44(12):2117–36. doi: 10.1007/s00259-017-3780-7
9. Fendler WP, Calais J, Eiber M, Flavell RR, Mishoe A, Feng FY, et al. Assessment of 68ga-psma-11 pet accuracy in localizing recurrent prostate cancer: A prospective single-arm clinical trial. JAMA Oncol (2019) 5(6):856–63. doi: 10.1001/jamaoncol.2019.0096
10. Donato P, Roberts MJ, Morton A, Kyle S, Coughlin G, Esler R, et al. Improved specificity with (68)Ga psma pet/ct to detect clinically significant lesions "Invisible" on multiparametric mri of the prostate: A single institution comparative analysis with radical prostatectomy histology. Eur J Nucl Med Mol Imaging (2019) 46(1):20–30. doi: 10.1007/s00259-018-4160-7
11. Donato P, Morton A, Yaxley J, Ranasinghe S, Teloken PE, Kyle S, et al. (68)Ga-psma pet/ct better characterises localised prostate cancer after mri and transperineal prostate biopsy: is (68)Ga-psma pet/ct guided biopsy the future? Eur J Nucl Med Mol Imaging (2020) 47(8):1843–51. doi: 10.1007/s00259-019-04620-0
12. Leung KH, Rowe SP, Leal JP, Ashrafinia S, Sadaghiani MS, Chung HW, et al. Deep learning and radiomics framework for psma-rads classification of prostate cancer on psma pet. EJNMMI Res (2022) 12(1):76. doi: 10.1186/s13550-022-00948-1
13. Tragardh E, Enqvist O, Ulen J, Hvittfeldt E, Garpered S, Belal SL, et al. Freely available artificial intelligence for pelvic lymph node metastases in psma pet-ct that performs on par with nuclear medicine physicians. Eur J Nucl Med Mol Imaging (2022) 49(10):3412–8. doi: 10.1007/s00259-022-05806-9
14. Johnsson K, Brynolfsson J, Sahlstedt H, Nickols NG, Rettig M, Probst S, et al. Analytical performance of apromise: automated anatomic contextualization, detection, and quantification of [(18)F]Dcfpyl (Psma) imaging for standardized reporting. Eur J Nucl Med Mol Imaging (2022) 49(3):1041–51. doi: 10.1007/s00259-021-05497-8
15. Zhao Y, Gafita A, Vollnberg B, Tetteh G, Haupt F, Afshar-Oromieh A, et al. Deep neural network for automatic characterization of lesions on (68)GA-PSMA-11 PET/CT. Eur J Nucl Med Mol Imaging (2020) 47(3):603–13. doi: 10.1007/s00259-019-04606-y
16. Capobianco N, Sibille L, Chantadisai M, Gafita A, Langbein T, Platsch G, et al. Whole-body uptake classification and prostate cancer staging in (68)GA-PSMA-11 PET/CT using dual-tracer learning. Eur J Nucl Med Mol Imaging (2022) 49(2):517–26. doi: 10.1007/s00259-021-05473-2
17. Bu T, Zhang L, Yu F, Yao X, Wu W, Zhang P, et al. (177)Lu-psma-I&T radioligand therapy for treating metastatic castration-resistant prostate cancer: A single-centre study in east asians. Front Oncol (2022) 12:835956. doi: 10.3389/fonc.2022.835956
18. He K, Zhang X, Ren S, Sun J. (2016). Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA: IEEE, 2016. pp. 770–8. doi: 10.1109/CVPR.2016.90
19. Huang G, Liu Z, van der Maaten L, Weinberger KQ. (2017). Densely connected convolutional networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA: IEEE, 2017. 2261–9. doi: 10.1109/CVPR.2017.243
20. Briganti A, Larcher A, Abdollah F, Capitanio U, Gallina A, Suardi N, et al. Updated nomogram predicting lymph node invasion in patients with prostate cancer undergoing extended pelvic lymph node dissection: the essential importance of percentage of positive cores. Eur Urol (2012) 61(3):480–7. doi: 10.1016/j.eururo.2011.10.044
21. Tosco L, Briganti A, D'Amico AV, Eastham J, Eisenberger M, Gleave M, et al. Systematic review of systemic therapies and therapeutic combinations with local treatments for high-risk localized prostate cancer. Eur Urol (2019) 75(1):44–60. doi: 10.1016/j.eururo.2018.07.027
22. Matoso A, Epstein JI. Defining clinically significant prostate cancer on the basis of pathological findings. Histopathology (2019) 74(1):135–45. doi: 10.1111/his.13712
23. Kayano PP, Carneiro A, Castilho TML, Sivaraman A, Claros OR, Baroni RH, et al. Comparison of gleason upgrading rates in transrectal ultrasound systematic random biopsies versus us-mri fusion biopsies for prostate cancer. Int Braz J Urol (2018) 44(6):1106–13. doi: 10.1590/S1677-5538.IBJU.2017.0552
24. Lendinez-Cano G, Alonso-Flores J, Beltran-Aguilar V, Cayuela A, Salazar-Otero S, Bachiller-Burgos J. Comparison of pathological data between prostate biopsy and radical prostatectomy specimen in patients with low to very low risk prostate cancer. Actas Urol Esp (2015) 39(8):482–7. doi: 10.1016/j.acuro.2015.02.005
25. D'Elia C, Cerruto MA, Cioffi A, Novella G, Cavalleri S, Artibani W. Upgrading and upstaging in prostate cancer: from prostate biopsy to radical prostatectomy. Mol Clin Oncol (2014) 2(6):1145–9. doi: 10.3892/mco.2014.370
26. Yin H, Chen M, Qiu X, Qiu L, Gao J, Li D, et al. Can (68)GA-PSMA-11 PET/CT predict pathological upgrading of prostate cancer from mri-targeted biopsy to radical prostatectomy? Eur J Nucl Med Mol Imaging (2021) 48(11):3693–701. doi: 10.1007/s00259-021-05217-2
Keywords: 68Ga-PSMA, pathological upgrading, prostate cancer, deep learning, PET/CT
Citation: Zang S, Jiang C, Zhang L, Fu J, Meng Q, Wu W, Shao G, Sun H, Jia R and Wang F (2024) Deep learning based on 68Ga-PSMA-11 PET/CT for predicting pathological upgrading in patients with prostate cancer. Front. Oncol. 13:1273414. doi: 10.3389/fonc.2023.1273414
Received: 06 August 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Sazan Rasul, Medical University of Vienna, AustriaCopyright © 2024 Zang, Jiang, Zhang, Fu, Meng, Wu, Shao, Sun, Jia and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wang, ZmVuZ3dhbmdjbjY4QDE2My5jb20=; Ruipeng Jia, cnJ1aXBlbmdqQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.