- 1Immune Health, Hunter Medical Research Institute, University of Newcastle, Newcastle, NSW, Australia
- 2University of Technology Sydney, Faculty of Science, School of Life Sciences, Sydney, NSW, Australia
- 3Centre for Inflammation, Centenary Institute and University of Technology Sydney, Faculty of Science, School of Life Sciences, Sydney, NSW, Australia
- 4Discipline of Pharmacy, Graduate School of Health, University of Technology Sydney, Sydney, NSW, Australia
- 5Department of Human Pathology in Adult and Developmental Age “Gaetano Barresi”, Section of Anatomic Pathology, University of Messina, Messina, Italy
- 6Pneumologia, Dipartimento di Scienze Biomediche, Odontoiatriche e delle Immagini Morfologiche e Funzionali (BIOMORF), Università degli Studi di Messina, Messina, Italy
- 7Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, University of Queensland, St. Lucia, QLD, Australia
- 8Department of Medicine, Boston University School of Medicine, Boston, MA, United States
- 9Department of Dermatology, The University of Sydney at Royal Prince Alfred Hospital, Sydney, Australia, and Centenary Institute, The University of Sydney, Sydney, NSW, Australia
Primary lung carcinoma or lung cancer (LC) is classified into small-cell or non-small-cell (NSCLC) lung carcinoma. Lung squamous cell carcinoma (LSCC) is the second most common subtype of NSCLC responsible for 30% of all LCs, and its survival remains low with only 24% of patients living for five years or longer post-diagnosis primarily due to the advanced stage of tumors at the time of diagnosis. The pathogenesis of LSCC is still poorly understood and has hampered the development of effective diagnostics and therapies. This review highlights the known risk factors, genetic and epigenetic alterations, miRNA biomarkers linked to the development and diagnosis of LSCC and the lack of therapeutic strategies to target specifically LSCC. We will also discuss existing animal models of LSCC including carcinogen induced, transgenic and xenograft mouse models, and their advantages and limitations along with the chemopreventive studies and molecular studies conducted using them. The importance of developing new and improved mouse models will also be discussed that will provide further insights into the initiation and progression of LSCC, and enable the identification of new biomarkers and therapeutic targets.
Introduction
Lung cancer (LC) is the leading cause of cancer death (11.6%), with an estimated 1.8 million deaths (18% of total cancer deaths) worldwide (1). Global age-standardised incidence rates of tracheal, bronchus, and lung cancer decreased by 7·4% in males but increased in females by 0·9%, 2010-19 (2). Although the survival rates of LC patients have improved in the last 10 years, over half of patients die within 1 year of diagnosis largely due to the majority being diagnosed with advanced stage disease, precluding treatment with curative intent (3). LC is classified as small-cell lung carcinoma, which accounts for <15% of LC cases, and non-small-cell lung carcinoma (NSCLC), which accounts for 85% of all reported LC (4). NSCLC is further divided into 3 main subtypes; lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LSCC), and large cell carcinoma. LUAD is the most common NSCLC and accounts for 40% of LC cases and occurs in the lung periphery whereas LSCC accounts for 30% of all the LC cases and mainly originates in the bronchial epithelium (5). The incidence of LC reported varies substantially worldwide, and in Asian countries, the incidence is affected by other secondary smoke exposure. Several findings may reflect the reason for the declining LSCC cases reported in Asian countries compared to western countries (6). LSCC was the most common histologic subtype amongst men prior to 1990s. In the developed and Industrialized nations, smoking rates peaked first in men, followed by women thus causing increased rates of LSCC than LUAD. LSCC incidence and mortality went up before declining following the initiation of comprehensive tobacco control program leading to declined LSCC rates in the US, Canada, many European countries, and Japan (7, 8). However, this was not the case in other countries such as Norway, Finland, Spain, and France where the LSCC cases are still stable (8). The incidence rates of LSCC on the Indian subcontinent and Taiwan has also declined gradually over recent years with an increased LUAD rates (9, 10). This decline in LSCC is attributed to high usage of smokeless tobacco. The rise of LUAD incidents compared to LSCC are greater in women and in overall population due to exposure to biofuels. Although they exhibit a lower incidence of LSCC but a higher mortality burden compared with developed countries due to unequal access to healthcare leading to delayed diagnosis and treatment, environmental contamination, and sociocultural barriers (6).
Cigarette smoke (CS) exposure is the major risk factor in the development of lung cancer, particularly LSCC as smoke injures the airway epithelium and can lead to pre-neoplastic changes that include hyperplasia (increased cell number), squamous metaplasia (expansion of squamous epithelium) and increasing grades of dysplasia (presence of abnormal cells) and ultimately carcinoma in situ (11). Whilst most pre-neoplastic lesions do not lead to the development of LSCC, a subset do progress on to invasive disease that is suggested to be in part attributed to breakdown in the to host immune surveillance (12, 13).
LSCC has lower 5-year survival rate largely due to detection at a late stage where therapeutic options are often ineffective (14). Despite the advancement of immunotherapies and regulatory approval of immune checkpoint inhibitors, such as PD-1 inhibitors, there remains an urgent need for additional and more effective treatment options for this important subset of LC patients. Here, we review current knowledge on LSCC pathogenesis, including risk factors, molecular alterations, current animal model, and highlighting the importance pre-clinical studies in the identification of biomarkers and novel therapies for improved diagnosis and treatment of patients.
LSCC risk factors in humans: CS & chronic obstructive pulmonary disease (COPD)
Extensive epidemiological data have clearly established CS exposure as a risk factor for LSCC, which is greater than for any other type of NSCLC (15–17). CS is comprised of >60 carcinogens, such as N-nitrosamines, polycyclic aromatic hydrocarbons, aldehydes, carbon monoxide, hydrogen cyanide, nitrogen oxides, benzene, toluene, phenols and other components (18–20). Some compounds require metabolic activation or spontaneous decomposition to have carcinogenic effects that lead to gene alterations and transformations (Figure 1) (18, 19). A pooled analysis of 13,169 cases and 16,010 controls revealed that LSCC was predominant in male heavy smokers compared to other groups, with an elevated odds ratio of 103.5 (95% Confidence Interval 74.8-143.2) (21). Current smoking men had higher risk of lung cancer than women irrespective of smoking quantity, duration or time since quitting (22). In addition to CS, other inspirable environmental pollutants/factors, such as bushfire/wildfire and solid fuel smoke, are also likely to be risk factors for LSCC development and warrant further research (23).
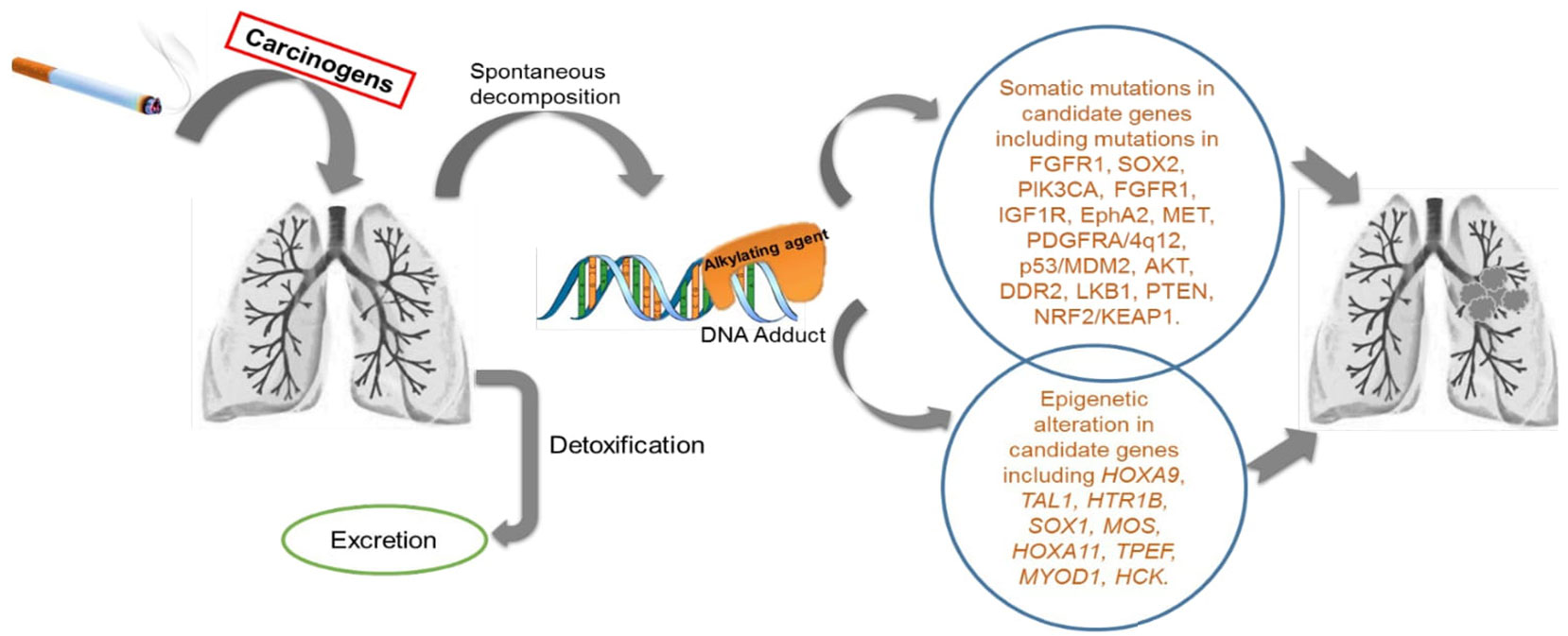
Figure 1 Schematic diagram of the activity of cigarette smoke exposure in human lungs: Cigarette smoke (CS) consists of more than 60 tumour initiating carcinogens tested in both laboratory animal models and in humans. There are 20 potentially known carcinogens found in a burning cigarette which are involved in lung carcinogenesis. These include polycyclic aromatic hydrocarbons (PAH), N- nitrosamines, 1,3-butadiene and ethylene oxide, cadmium, and the radioactive compound 210PO. Few of these compound needs to be metabolically activated or undergoes decomposition to exert the carcinogenic effect. There are detoxifying enzymes such as cytochrome P450, glutathione S-transferase and UDP-glucuronosyl transferases which helps in removal of metabolically activated carcinogens. DNA adducts are formed during this process. When these adducts are left unrepaired by the DNA repair enzymes in the body, they result in genetic changes in key genes for cellular function. These altered genes, eventually leads to carcinogenesis.
COPD is now the 3rd commonest cause of death globally (24). It is a chronic inflammatory lung disease of the lower airways and parenchyma often induced by chronic CS exposure and is characterized in advanced stages by severe breathing difficulties (25–27). Oxidative stress, increases in inflammatory cytokines, and an imbalance between protease and anti-protease levels, predispose individuals to developing COPD (28). The key pathological features of COPD include chronic bronchitis, remodelling and narrowing of the small airways, destruction of the lung parenchyma and emphysema. This results in increased airflow resistance, reduced lung elastic recoil, small airway closure and gas trapping ultimately leading to impaired lung function and gas exchange (14). Significantly, COPD patients have four-fold greater risk of developing LSCC than the general population (17). Thirty percent of patients diagnosed with moderate or even mild COPD subsequently develop LSCC, suggesting that COPD is an important contributing factor in the development and progression of this squamous subtype (17). Furthermore, chronic inflammation in COPD can increase the expression of growth factors, such as epithelial growth factor (EGFR), and activates transcription factors, such as NF-κB, that favor lung tumorigenesis (29). Studies of airway epithelial cells have also identified single nucleotide polymorphisms in specific genes in airway epithelial cells, including hedgehog-interacting protein, A disintegrin and metalloproteinase 19 (ADAM19), family with sequence similarity 13 member A (FAM13A), and cholinergic nicotinic receptor locus (CHRNA), that occur in both COPD and LSCC (30, 31). These findings highlight the similarities between genetic mechanisms underlying the development of both diseases. Epigenetic changes such as DNA methylation, histone modifications, non-coding RNA regulation are found to be similar in COPD and LSCC (32). Thus, in addition to CS exposure, there are genetic and epigenetic change that are shared factors for COPD and LSCC suggesting CS associated COPD is a major risk factor for LSCC.
Molecular drivers of human LSCC genomic alterations
Identifying the mutations responsible could lead to the development of effective targeted molecular therapies. A comprehensive characterization of genomic alterations in LSCC from a large cohort of 178 patients was conducted by The Cancer Genome Atlas (TCGA) that reported a mean of 360 exon mutations, 165 genomic rearrangements, and 323 segments of copy number alterations per tumor that defined the LSCC genomic landscape (33). TCGA reported recurrent mutations in 18 genes, including tumor protein p53 (TP53) mutations in 86% LSCCs. Mutations were also reported in oxidative stress genes including nuclear factor erythroid-2-related factor-2 (NFE2L2), cullin-3 (CUL3) and Kelch-like ECH-associated protein-1 (KEAP1) and genes associated with squamous differentiation including the amplification of tumor protein p63 (TP63) SRY (sex determining region Y)-box 2 (SOX2), and loss-of-function mutations in Notch homolog-1 and 2, (NOTCH1 and NOTCH2) and Achaete-Scute family BHLH transcription factor-4 (ASCL4), and focal deletions in Forkhead box protein P1 (FOXP1) were also reported in 44% of cases. A recent study was conducted on 39 lesions from 29 patients with pre-invasive lung carcinomas using whole genome sequencing and comparing with TCGA (34). A wide range of similar mutation burden in genes such as TP53, CDKN2A, SOX2, AKT2 and NOTCH1 was noted between pre-invasive lesions and LSCC data in TCGA.
Although TCGA set a precedent for mutational profiling of LSCC a number of additional studies have also identified somatic alterations in LSCC (35). The catalytic subunit (p110α) of PI3K, PI3KCA, which plays a significant role in the PI3K-AKT pathway, has copy number gains in LSCCs (36–39). Hot-spot mutations in phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit-α (PIK3CA) and B-raf proto-oncogene, serine/threonine kinase (BRAF)V600E promote LSCC development (40). Fibroblast growth factor receptor 1 (FGFR1) amplification is another key dysregulation in LSCCs and inhibition of FGFR1 in mouse models and cell lines inhibits tumor growth (35, 41). Activation of the insulin-like growth factor-1 receptor (IGF1R) is involved in triggering pathways such as RAS/RAF/MAPK, which are important in cell proliferation. Over-expression of IGF1R, located on chromosome 15q26, is often observed in both LUAD and LSCC (42, 43). Over-expression of EPHA2 is frequently observed in LSCCs but not other LCs (44). MET over-expression is associated with abnormal cell proliferation and invasion, which is common in LSCC compared to other NSCLCs (45, 46). Amplified PDGFRA is more frequent in LSCCs than ACs. An in vitro study with NCI-H1703, a LSCC cell line, showed that PDGFRA inhibition leads to anchorage-dependent cell growth, suggesting that PDGFRA is an oncogene in LSCC (47). p53 mutations are very common and occur in 65% of LSCC and ~50% of NSCLCs (48). Around 75% of p53 mutations are missense mutations and these often produce gain-of-function mutant p53 (35). p53 inactivation also results from over-expression of MDM2 that ubiquinates p53 and marks it for degradation (49), which is frequent in both LUAD and LSCC (50, 51). DDR2 kinase gene mutations are reported in 9 out of 277 (~3%) of LSCC cases (52). Altered PTEN and NRF2 expression is also observed in LSCC but not in LUAD (53, 54). p40 is a distinguishing marker in LSCC which differentiates it from LUAD (55). A recent comparative study concluded the overlapping and non-overlapping mutation burden between LAUD and LSCC. In this study, researchers identified 38 gene mutations in LAUD and 20 in LSCC using MutSig2CV. Out of these mutations, only 6 genes, TP53, RB1, ARID1A, CDKN2A, PIK3CA, and NF1, were observed to common in both tumor types although, the frequency of TP53, CDKN2A, and PIK3CA mutation was higher in LSCC (56).
Collectively, these studies show that different mutations may be common and/or unique to LSCC versus LUAD. These specific mutations may be novel targets for LSCC and represent key opportunities to develop LSCC-specific therapies in a landscape where many existing therapies for LUAD are ineffective for LSCC.
Epigenetic alterations
The induction of carcinogenic processes can also be driven by the accumulation of epigenetic changes, which results in the dysregulation of key oncogenes, tumor-suppressor genes, and DNA repair or housekeeping genes. Epigenetic alterations, demethylation, hypermethylation and hypomethylation are as frequent as genomic mutations in LC (57) (Table 1). Some epigenetic alterations can delineate between LSCC and AC, for example demethylation in the promoter region of Cancer/Testis Antigens (CTA) expression is highly associated with LSCC but not LUAD (57). Hypermethylation of gene promoters are also associated with transcriptional silencing of tumor suppressor genes and it is thought that this promotes the initiation and progression of carcinogenesis. Hypermethylation typically occurs in CpG rich regions/islands at or near gene promoters and promoter hypermethylation may be a marker for early diagnosis, disease stage and predictor of patient outcomes in LSCC. Notably, the methylation pattern of the CYTL1 promoter region changes between early and advanced stages of LSCC (57).
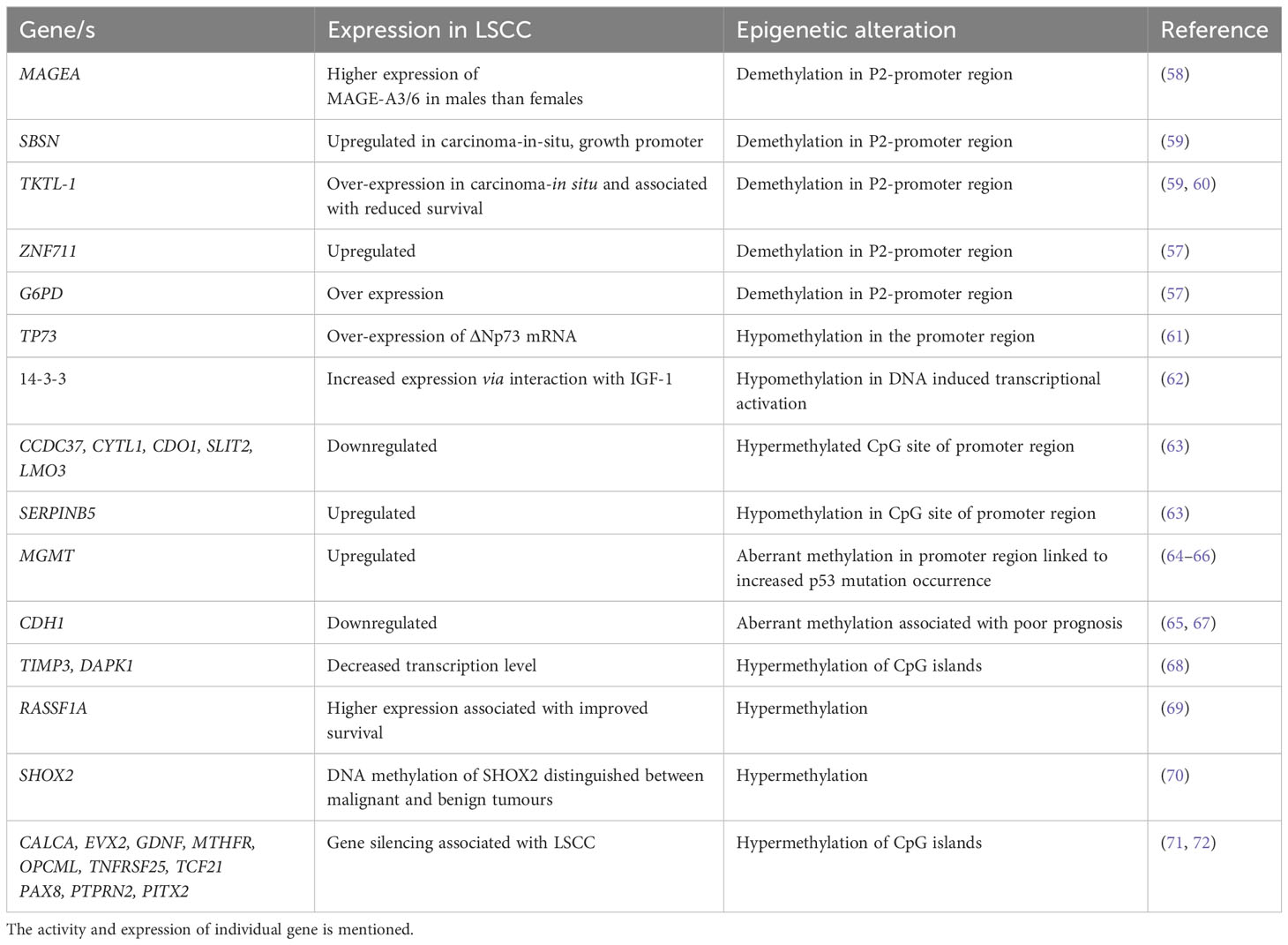
Table 1 Epigenetic alterations in squamous cell carcinoma: A detailed list of genes that are involved in epigenetic regulation in LSCC development.
Hypermethylation of the p16 and/or O6-methylguanine-DNA methyltransferase gene promoters was detected in sputum samples from LSCC patients up to 3 years preceding or at the time of diagnosis (64), and is a potential biomarker for early detection (73). Hypermethylated RASSF1A is associated with early recurrence of LSCC and is considered an informative diagnostic biomarker for remission (65, 71). It is important to assess methylation profiles in tumor versus non-tumor tissue. In one study, the methylation profile of 42 gene loci were analyzed in a collection of 45 LSCCs and compared with non-tumor lung tissues from the same patients (72). CALCA, EVX2, GDNF, MTHFR, OPCML, TNFRSF25, TCF21, PAX8, PTPRN2 and PITX2 were hypermethylated in LSCC compared to non-tumor tissues (71, 72). Whilst many epigenetically modified genes have been identified in LSCC that differentiate it from other LCs, the specific roles of these altered genes in the development, progression and recurrence of LSCC are yet to be elucidated.
Histone modifications and chromatin remodelling are critical in the pathogenesis of NSCLC and other CS-induced lung diseases including COPD. CS-induces post-translational histone modifications in H3 and H4 in lung cells. These are potential biomarkers as they play a key role in the epigenetic state in the pathogenesis of CS-induced LSCC and NSCLC (74). A study of 408 NSCLC tissues assessed the global modification status of histone H3 and H4 and their association to tumor recurrence with LSCC patients having lower levels of H3K4 dimethylation (73). Li et al. showed that in NSCLC higher expression of H3K4 histone demethylases (KDM1A, KDM5A, KDM5B and KDM5D) was associated with poor overall survival, while patients with low expression of H3K4 histone methyltransferases SMYD3 had worse prognosis (75). Another study by Leng et al. assessed KDM6A, a member of the mixed-lineage-leukemia (MLL2) H3K4 methyltransferase complex, and reported that expression of KDM6A protein was higher in NSCLC tissues than in corresponding para-cancer tissues and that higher expression was associated with poor prognosis (76). It should be noted that these studies were of NSCLC but none of them were on LSCC in particular.
Although studies have established the genetic and certain epigenetic alterations in advance stage LSCC, future studies assessing genetics and epigenetics in the developing early pre-malignant lesions would be important to understand the pathophysiology of LSCC development.
MicroRNAs (miRNAs) as potential biomarkers
LSCC antigen, neuron-specific enolase, and CYFRA 21-1 are reported biomarkers of NSCLC (74), however, they do not detect the early stages of these tumors and therefore early biomarkers are required. miRNAs are small non-coding RNAs that are emerging as reliable biomarkers for early diagnosis of respiratory diseases including LSCC (73, 75, 76). Thus, the identification of unique miRNAs or signatures would be a major advance in LSCC diagnosis. The miRNAs commonly identified in LSCC patients are summarised in Table 2. One study profiled miRNA expression in 61 LSCC compared to normal lung samples. A total of 15 differentially expressed miRNAs was identified, including increased expression of miR-17/92 clusters and paralogous miR-106a-363 and miR-93-106b, miR-182-183 clusters (84). When 474 human miRNAs were mapped onto the array comparative genomic hybridization database (a portal of databases with thorough data on human genome aberrations and variations), 77 were linked with DNA copy number changes in LSCC, which included 40 associated with amplified regions and 37 to regions deleted. In another study, mature miR-218 produced from two precursors hsa-mir-218-1; MI0000294 and hsa-mir-218-2; MI0000295, was downregulated in LSCC compared to normal lung tissue (77). An investigation of miRNA in Chinese LSCC patients revealed hsa-miR-31, which targets the tumor suppressor DICER1, as a potential prognostic biomarker as higher expression was associated with poor patient survival (78).
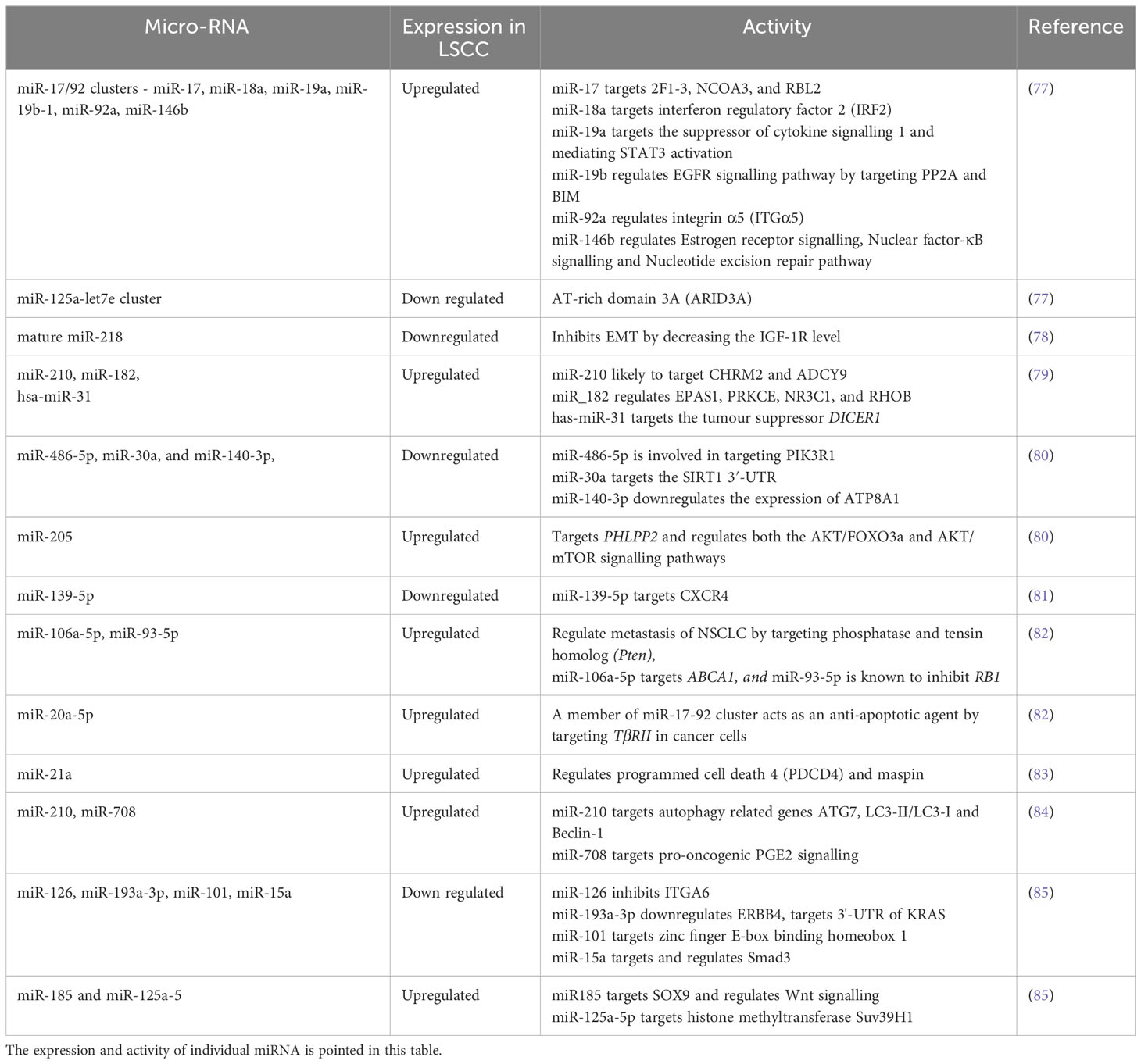
Table 2 Altered miRNAs in LSCC: A detailed list of all the miRNAs used as biomarkers are listed in Table 2.
In addition to diagnosis, miRNA profiling has also been used to differentiate LSCC from LUAD. miR-205 and miR-375 can distinguish the two types with 96% accuracy (79). Increased expression of miR-205 was noted in LSCC but was unaltered in LUAD, whereas miR-375 expression was high in LUAD and unchanged in LSCC compared to normal lung tissue (79). Furthermore, increased miR-139-3p and downregulated miR-139-5p in the sputum of LSCC patients may also be potential biomarkers (80). In another study, a panel of three miRNAs (miR-106a-5p, miR-20a-5p, miR-93-5p) were highly upregulated in LSCC patients compared to normal controls and may be potential diagnostic biomarkers (81). In other work miR-375 and miR-10b-5p expression was downregulated in the serum of LSCC patients compared to controls (82). A recent study screened 245 LSCC samples for genome-wide oncogenic miRNAs and compared them with the TCGA dataset (86). 231 of 1,001 miRNAs were associated with copy number alterations from which only 11 were increased in LSCC compared to adenocarcinoma and normal tissues. Three onco-miRNAs (miR-296-5p, miR-324-3p and miR-3928-3p) were specifically associated with poor prognosis (86). Another study showed that in particular, miRNA-21 expression was found in LSCC patients with short survival periods and was associated with poor prognosis (87). A biomarker panel of three miRNA from 15 sputum samples from LSCC patients was also created to detect early stage tumors (88). The study was validated in sputum from an independent set of 67 LSCC patients and 55 healthy controls. They identified over-expression of a three miRNA panel biomarker to detect early stage LSCC. Collectively, miRNAs common to all these studies show that miR-20a-5p, miR-93-5p, miR-106a-5p, miR-146b, miR-205, miR-210, miR-296-5p, miR-324-3p, miR-375, miR-708 and miR-3928-3p may be used as biomarkers and can differentiate LSCC over LUAD. Many of these miRNAs, such as miR-17/92 clusters, miR-20a-5p, miR-93-5p, miR-146b, miR-210 are involved in the regulation of the PTEN/PI3K/AKT/mTOR pathway, an essential regulatory pathway in NSCLC pathogenesis. However, some are also involved in the Wnt/β-catenin pathway such as miR-708, miR-324-3p, in the EMT pathway or are independently involved in LSCC. A detailed list of miRNAs along with their role in the LSCC are listed in Table 2.
Chemotherapeutic, targeted and immunotherapeutic approaches
Despite of understanding the genetic and epigenetic alterations identified in clinical samples of LSCC, there are many limitations in current therapeutic approaches. Chemotherapy, with or without radiotherapy, has remained the main treatment option for patients with LSCC (89–91), and many of these treatments were assessed in mouse models prior to clinical studies. A detailed list of treatment options are documented in Table 3. Increased anti-tumor activity of a combination of gemcitabine/cisplatin was established compared to the anti-tumor activity of only gemcitabine or cisplatin in preclinical studies using in vivo and in vitro approaches (101). LSCC patients undergoing cytotoxic therapy with gemcitabine/cisplatin had greater progression-free survival compared to pemetrexed/cisplatin treated patients (93). A phase III second-line trial revealed that patients treated with pemetrexed had a lower survival rate than those treated with docetaxel (98). Treatment with nab-paclitaxel/carboplatin improved the survival rate of LSCC patients compared to paclitaxel/carboplatin with fewer side effects of myalgia, neuropathy, and cytopenia (83). A different study showed that co-administration of necitumumab with gemcitabine/cisplatin improved overall median survival of LSCC patients (94). Bevacizumab (VEGF-A inhibitor) enhanced the anti-tumor activity of erlotinib in xenograft models (95). A study conducted on NSCLC with 25% LSCC patients showed that treatment with ramucirumab (VEGFR2 inhibitor)/docetaxel increased median progression-free survival in LSCC patients compared to placebo/docetaxel (96). Other targets such as FGFR and PI3K-AKT are under investigation.
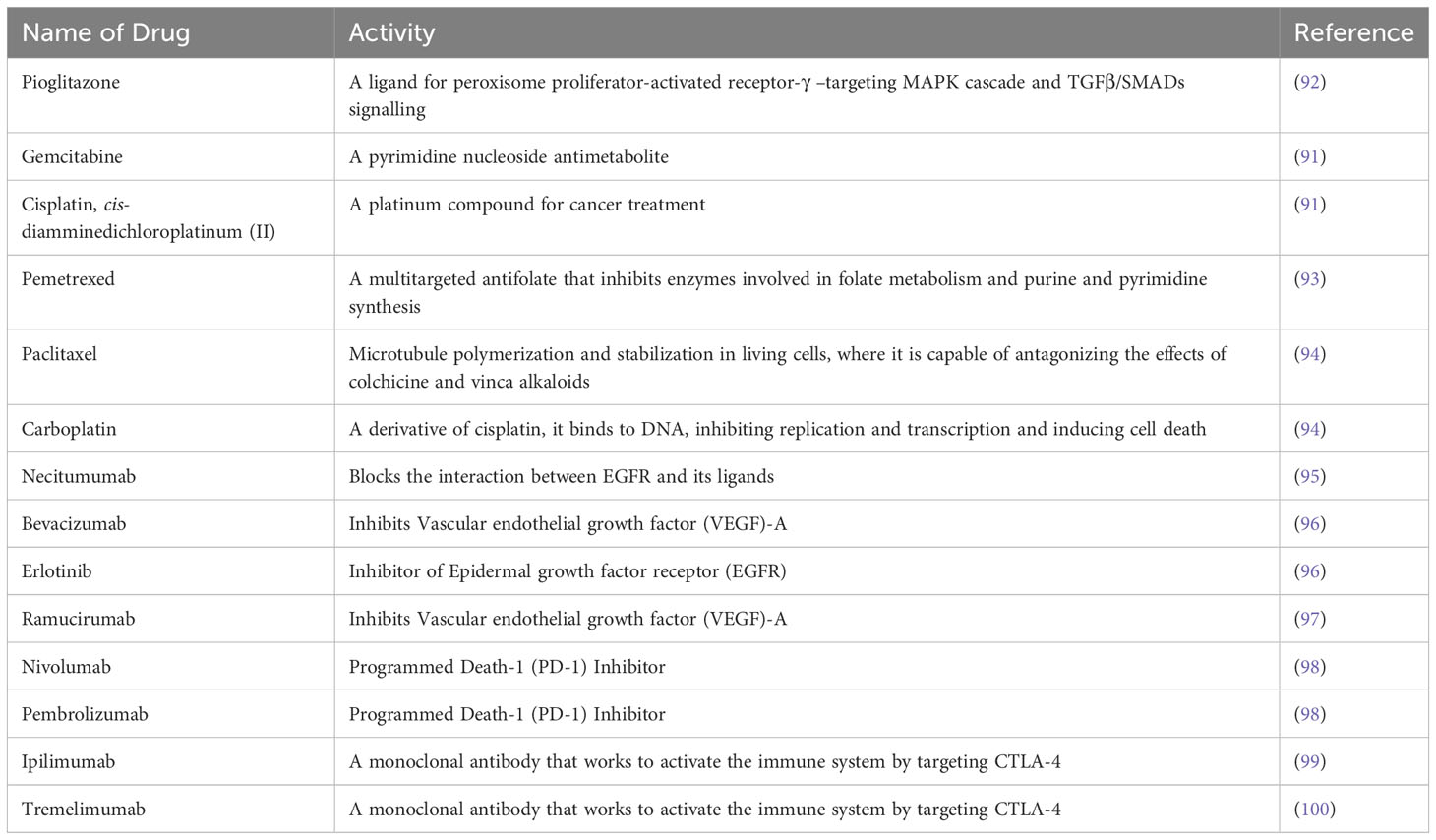
Table 3 Therapeutic drugs for NSCLC: A list of currently available therapeutic options for patients are noted in Table 3 with its activity in targeting pathway or immune-checkpoint in LSCC.
Immune checkpoint inhibitors (ICIs) such as targeting programmed death-1/programmed death ligand-1 (PD-1/PD-L1) and cytotoxic T-lymphocyte antigen-4 (CTLA-4) blockers have been integrated into standard-of-care regimens for patients with advanced LSCC (97).
LSCCs express PD-L1, enabling escape from immune responses by interacting with PD-1 on T-cells, which suppresses their anti-tumor effects (102). Yu H et. al., assessed the correlation of PD-L1 Expression with tumor mutation burden in early-stage LSCC (100). Nivolumab is a monoclonal antibody (mAb) which targets the PD-1 receptor on T-cells to activate the cells anti-tumor response. A phase II single-arm trial of nivolumab in LSCC patients showed an 11-month overall response rate of 15% (103). A recent phase III trial compared nivolumab to docetaxel as a second-line therapy in 272 advanced LSCC patients after having progressed on platinum-based chemotherapy. Nivolumab treatment increased overall survival by 41% over the docetaxel arm (103). Recent studies established that pembrolizumab (anti-PD-1) alone or combined with platinum-based chemotherapy, is valuable as a standard first-line treatment for LSCC and highly effective in clearing tumors in 20% of cases (104). However, once patients progress on such immune checkpoint inhibitors and chemotherapy, few options are available. A very recent Phase III LUX-Lung 8 study has established that the tyrosine kinase inhibitor (TKI) afatinib can significantly increase overall survival in LSCC patients and may be used as a second line treatment (104). A biomarker driven lung cancer master protocol (Lung-MAP; S1400) was recently performed to address the need of better therapies for LSCC (105). The study evaluated taselisib (targeting PIK3CA alterations), palbociclib (cell cycle gene alterations), AZD4547 (FGFR alterations), rilotumumab (hepatocyte growth factor/scatter factor:MET pathway inhibitor) plus erlotinib (TKI), talazoparib (homologous recombination repair deficiency inhibitor), telisotuzumab vedotin (targeting cMet) and evaluated durvalumab (blocks the interaction of PD-L-1 with the PD-1), and nivolumab (PD-1 Inhibitor) plus ipilimumab (anti-CTLA-4) for anti-PD-1 or anti-PD-L1-naive disease, and durvalumab (anti-PD-L1) plus tremelimumab (another anti-CTLA-4) for anti-PD-1 or anti-PD-L1 relapsed cancers. Seven % of patients responded to targeted therapy and 16·8% patients responded to anti-PD-1 or anti-PD-L1 therapy for immunotherapy-naive disease (105). A recent study has established that dual immunotherapy along with chemotherapy enhances clinical benefit such as longer overall as well as progression free survival. This study used nivolumab plus ipilimumab along with platinum doublet chemotherapy and chemotherapy alone. A significantly improved overall survival was noted compared to chemotherapy alone and also had a favorable risk–benefit profile (106). Another report showed that nivolumab plus ipilimumab with two cycles of chemotherapy has a long-term, durable efficacy up to 3-year minimum follow-up and survival benefit in LSCC patients with brain metastases (107).
Compared to LUAD, there has always been a lack of effective targeted treatments for LSCC and limited progress has been made in the systemic treatment of advanced disease. Despite of the several available options for the treatment of NSCLC, few of these treatments are specific to LSCC and therefore new therapies are urgently required. Importantly, most of these current therapies for NSCLC were developed in studies that used immunocompromised animals. The development of robust, short-term mouse models of LSCC in immunocompetent animals and that recapitulate human disease features by combining CS with carcinogens such as NTCU, is essential for increasing the understanding of disease pathogenesis as well as improving the efficacy of current therapies and informing novel therapeutic strategies for LSCC.
Murine models of LSCC
Pre-clinical animal models are crucial in cancer research and are valuable tools for understanding the underlying mechanisms involved in tumor initiation and progression. They also provide a platform to test the safety and efficacy of novel treatments. While an increasing number of naturally occurring cancers in animals, notably dogs, are being explored for these purposes (99, 108, 109), mouse models remain the mainstay of pre-clinical studies.
Chemical-induced mouse models
Chemical-induced mouse models can also be used to model human disease since they drive human cancers. In one of the earliest studies, BC3F1 (a cross between C57BL and C3H inbred strains) and DBA/2 mice were intratracheally administered 3-methylcholanthrene (MCA, 0.5mg) once a week for 6 and 4 weeks, respectively, and rested for up to 7 months (110). After 24 weeks, 86% of BC3F1 mice LSCC, however, only 3/50 DBA/2 mice developed tumors of squamous cell origin after 7 months (110). Intratracheal instillation of benzo[a]pyrene (BAP) with charcoal powder once a week for 8 weeks induced moderately to highly keratinized LSCC in C57BL/6 mice after 40 weeks (111). However, these models have not been used subsequently. In a different study, female Swiss mice were administered 40mM of N-nitroso compounds such as nitrosoalkylureas, nitrosoalkylcarbamates, and chlorinated nitrosotrialkylureas topically in the subscapular region, twice a week for 50 weeks. Eleven/20 mice administered nitroso-tris-(2-chloroethyl)urea (NTCU) developed LSCC after 110 weeks (112).
These NTCU model although exciting was challenging to reproduce until Wang et al., further developed the NTCU model treating eight inbred mouse strains (129/svJ, AKR/J, BALB/cJ, C57BL/6J, FVB/J, SWR/J, A/J,NIH Swiss) with NTCU using the same protocol and examined lung histology after 8 months to discover the NTCU was only able to indcuse LSCC in 5 of the 8 strains. The SWR/J, NIH Swiss, A/J, BALB/cJ, and FVB/J all developed tumors, where A/J, NIH Swiss and SWR/J mice were most susceptible to NTCU skin treatment, FVB/J and BALB/cJ mice had intermediate susceptibility, and AKR/J, 129/svJ and C57BL/6J mice were resistant (113). Most susceptible strains of mice progressively develop hyperplasia, metaplasia and invasive LSCC (75-100% incidence), whereas mice with intermediate susceptibility have ~45% incidence of invasive LSCC. Resistant strains had no evidence of invasive LSCC, although they had hyperplasia, metaplasia and carcinoma in situ. A more recent study though sought to limit some of the toxicity associated with NTCU has shown that A/J mice administered with NTCU (13 mM) for 2, 4, and 8 weeks, topically in the subscapular region and sacrificed 18 weeks had 25, 54 and 71% LSCC incidence, respectively, with LSCC originating in deltaNp63+CD44v+ club (Clara) cells (114). Whereas a study in FVB mice with topically administered NTCU (4, 8, or 40 mM) for 32 weeks, demonstrated that 4 mM and 8 mM NTCU although were well tolerated only induced flat atypia whereas 40mM lead to dysplasia and LSCC (115). Further, also in FVB mice, 20 mM NTCU twice a week for up to 32 weeks did not show any dysplastic changes until the 25th week and bronchial dysplasia and LSCC occurred after 32 weeks (116).
A recent study used transcriptome (RNA) sequencing (RNA-Seq) to profile bronchial airway gene expression in an NTCU-induced LSCC mouse model (117). Swiss mice were treated topically with 40 mM of NTCU (twice a week) for 24 weeks to induce early LSCC lesions. They found activation of oncogenic PI3K and Myc pathways in bronchial epithelial cells of mice with preneoplastic changes. The authors also showed that the expression of mouse microRNA (miRNA)-449c-5p (mmu-miR-449c-5p) was suppressed in cells with upregulated myc expression (117). Another recent study assessed and compared the effects of different topical doses of NTCU (20, 30 and 40 mM) in both female and male NIH Swiss, Black Swiss and FVB mice. NIH swiss mice had a higher LSCC incidence and lower mortality with 30mM of NTCU (118). One recent study has incorporated tobacco smoke with NTCU to enhance the efficacy of NTCU to develop pre-malignant lesions of LSCC. This study used a whole-body smoke system to expose A/J mice to CS for 3-6 weeks following 4-5 weeks of 20mM NTCU administration (twice a week) and concluded that NTCU combined with CS exposure leads to the development of LSCC in a shorter time period and would be more relevant to human disease (119). Although these models are more relevant to human disease compared to xenograft and transgenic models, they have their own limitations such as prolonged cytotoxic treatment leading to ethical challenges. Moreover, the recent combination of NTCU with CS exposure used whole-body smoke exposure systems which is more comparable to passive rather that active smoking. Although induce early neoplastic changes are induced, human LSCC occurs in active chronic smokers. Thus, nose-only/inhaled CS exposure combined with NTCU would give more relevant neoplastic changes in murine models.
Carcinogen-induced models used in chemoprevention studies
Carcinogen-induced murine SCC models have been employed in several chemoprevention studies. One of the earliest assessed the efficacy of anti-tumor B (ATB), which is also known as Zeng Sheng Ping (a Chinese herbal mixture), in treating SCCs in A/J mice that were induced by topical administration of NTCU (40 mM) twice a week for 22 weeks. Mice were treated with AIN76APurified Diet (normal control mouse chow) along with 250 g/kg ATB. ATB reduced the development of lung SCC (3.1-fold; p<0.05) but efficacy was not tested in human SCC patients (92). Another study assessed the anti-cancer activity of pioglitazone, a ligand for peroxisome proliferator-activated receptor-γ. The skin of female NIH Swiss mice was painted with of NTCU (30 mM/L) twice a week, with 3-day intervals for 32 weeks, and pioglitazone (15 mg/kg body weight) treatment was administered by oral gavage beginning 8 weeks after the first NTCU treatment. Treatment inhibited the progression of SCC by 35% (p<0.05) (120). The chemopreventive effect of Korea white ginseng (KWG) was assessed in NTCU-induced SCC in female Swiss mice. KWG was given orally with 30 mM/L NTCU administered topically twice a week with a 3.5-day interval for 24 weeks. KWG treatment blocked the progression of SCC (121). In another study, female A/J mice were given NTCU (0.5 mmol/L) and lipopolysaccharide (LPS, 4 µg) intranasally once a week for 26 weeks. Mice were fed a diet containing diindolylmethane (DIM, 10 µmol/g), which is one of the breakdown products of indole-3-carbinol, in AIN-93G/M powder (high protein and fat) throughout. LPS has pro-carcinogenic activity since mice treated with NTCU and LPS had 9-fold increases in bronchiolar SCCs, the chemopreventive capacity of DIM was demonstrated with a 2-fold reduction in SCC (122). Green tea provided as drinking water for A/J mice 2 weeks after the first NTCU treatment (40mM, twice a week for 32 weeks) reduced the development of SCC to 5/9 mice (56%) compared to 90% observed in NTCU only treated controls (123). In another study, the efficacy of pomegranate fruit extract (0.2% in drinking water throughout) as a chemoprevention agent in female A/J mice administered BAP or NTCU (140 and 240 days, respectively) was assessed. Treatment reduced tumor incidence by 53.9% and 65.9%, respectively, which was associated with down-regulated MAPKs, NF-κB, PI3K, and mTOR signalling networks, compared to untreated controls (124). The chemopreventive effect of dietary vitamin D was assessed in NTCU-treated SWR/J mice. Dysplastic changes were increased in NTCU mice on a low vitamin D diet compared to those on a normal/high vitamin D diet (125).
Recently C57BL/6 mice were used to examine the role of Rad52, which is involved in DNA repair mechanisms. In this study, NTCU (30 mM/L) was administered topically twice weekly with a 3.5-day interval for 38 weeks to Rad52-deficient (-/-) mice. Deletion of Rad52 increased tumor cell death and reduced growth compared to wild-type (WT) mice that appeared to be due to enhanced Rad52-/- NK and CD8+ T-cell effector functions (126).
Transgenic models
Mutations in serine/threonine kinase-11, also called Lkb1, are implicated in epithelial cancers. A Kras-mutant mouse model was used to analyse the role of Lkb1. Simultaneous activation of KrasG12D (Kras) and inactivation of Lkb1 led to LSCC and also other types of LC (127). However, inactivated Lkb1 did not lead to LSCC if KrasG12D was not activated. Nevertheless, Kras mutations are generally not observed in human LSCC (128). Inactivation of Lkb1 alone is insufficient for LSCC development but an incidence of 100% LSCC was induced in mice that lost Lkb1 and Pten after 40-50-weeks of latency (129). LSCC markers such as Sox2, were over-expressed with lentiviral delivery of Sox2 to the lungs of Lkb1-/- mice, which promotes LSCC by activating STAT and mTOR pathways. These mice express orthologues of human LSCC biomarkers, such as cytokeratin-5, 14, and p63 (130). A recent study defined the significance of Ikkα in LSCC development in mice. Kinase-dead IKKα knock-in (IkkαK44A/K44A, IkkαKA/KA) mice on an FVB background were generated to study the levels of IkkαKA/KA expression at different stages and its relationship to LSCC development. These mice started developing spontaneous lung tumors after 3 months of age. IKKα levels were high in newborn WT compared to IkkαKA/KA mice. IKKα levels were lowest in 4 month old IkkαKA/KA mice. The reduction in IKKα levels could contribute to LSCC (131). Recent studies have established that SOX2 overexpression in tracheobronchial basal cells combined with CDKN2AB and PTEN loss results in LSCC (132). This study also confirmed that although overexpression of FGFR1 transforms PTEN- and CDKN2AB-deficient tracheobronchial basal cells at a higher frequency, the tumors formed were heterogeneous with occasional squamous differentiation. It is also established that NKX2–1 suppresses SOX2-driven squamous tumorigenesis (133).
Xenograft models
Patient-derived xenografts (PDXs) have the potential to replicate the histopathological features, heterogeneity and gene expression pattern of the original tumor (134, 135). LSCCs have increased engraftment compared to other NSCLCs (135). Clinically relevant models of LSCC have been attempted by implanting human LSCCs from surgical resections and biopsy specimens subcutaneously into the flanks of immune-deficient NOD.SCIDprkdcIl2rγ−/− (NSG) mice, which resulted in 52% and 33% tumor take up, respectively (136). Engrafted LSCCs maintained the patient’s tumor phenotype for at least four passages. This model was used to show that FGFR1 mRNA detection is a better predictor of response to FGFR inhibitors than FGFR1 gene amplification. Sixty-two PDX LSCC models were developed that showed similar gene expression profiles, pY-proteome landscapes (profile to compare similarities between PDX models) and DNA methylation patterns as human tumors (137). Another study was only able to establish 35 PDX models out of 100 human tumors because of contamination by lymphomagenesis (138). In a separate study, 21 PDX models were established from 36 patients, with these xenograft tissues analyzed and determined to retain LSCC characteristics, including CK5/6, p63 and p40 expression (139). In the most recent study, 18 PDXs were established from 37 surgical specimens with 16/18 passaged to 2nd-3rd generations and were seen to develop LSCC. Histological markers, such as p53, p63, cytokeratin5/6, and E-cadherin, as well as response to cisplatin, were retained in the mouse tumors. Altered expression of Ki-67, long non-coding RNA and mRNA were observed in 3rd-generation xenografts (140).
Collectively, these studies further the understanding of human LSCC development and provide information for non-immune based therapies. However, there are limitations including the high mortality rate due to long-term cytotoxic treatment, low success rate of engraftment and unsuitability for studying anti-tumor immune responses, as the mice are immunodeficient. Thus, it is crucial to develop LSCC models in immunocompetent mice with reduced mortality rate and cytotoxic effects. Short term cytotoxic insults along with tumorigenic driving factors such as CS and CS-induced COPD in immunocompetent mice could provide a more robust murine model to study the pathogenesis and establish the diagnostic biomarkers of the disease.
Syngeneic mouse model
The lack of animal models that reflect human disease to assess the safety and efficacy of drugs and to explore the underlying molecular mechanisms is one of the major impediments in LSCC research. Syngeneic models are allografts immortalized from mouse cancer cell lines, which are then engrafted back into the same inbred immunocompetent mouse strain. Valencia et al., 2022 generated and characterized two syngeneic lung SCC cell lines i.e., UN-SCC679 and UN-SCC680 derived from NTCU carcinogen treated A/J mice. They observed similar genetic and transcriptomic patterns that may correspond to the classic LUSC human subtype. In addition, they compared the immune landscape generated by both tumor cells lines in vivo and assessed their response to immune checkpoint inhibition. The differences between the two cell lines are a good model of the broad heterogeneity of human SCC. Studies of the metastatic potential of these models revealed that both cell lines represent the organotropism of LUSC in humans, i.e. affinity for the brain, bones, liver and adrenal glands (141).
Azpilikueta et al., 2016 used the transplantable mouse lung SCC UN-SCC680AJ cell line to test the efficacy of immunotherapy (anti–PD-1/PD-L1 and anti-CD137 mAbs). In syngeneic mice, the tumors derived from UN-SCC680AJ cells were amenable to curative treatment with anti–PD-1, anti–PD-L1, or anti-CD137 immunostimulatory mAbs. Single-agent therapies lost curative efficacy when treatment was started beyond day +17, whereas a combination of anti–PD-1 plus anti-CD137 achieved complete rejection of the tumors. Tumor cells expressed weak baseline PD-L1 on the plasma membrane, but this could be readily induced by interferon-γ. Combined treatment efficacy required CD8 T cells and induced leukocyte infiltration in which T lymphocytes co-expressing CD137 and PD-1 were prominent. These promising results advocate the use of combined anti–PD-1/PD-L1 plus anti-CD137 mAb immunotherapy for the treatment of squamous non–small cell lung cancer in the clinical setting (142). Thus, these cell lines recapitulate the complexity of the human disease, and could be promising tools for lung SCC research.
Organoid models
These are the models that recapitulate the complex genetic profile of patients and could be another promising tool in lung SCC research. Hai et al., 2020 used CRISPR genome editing to delete multiple tumor suppressors in lung organoids derived from Cre-dependent SOX2 knock-in mice. They investigated both the therapeutic efficacy and immunological effects accompanying combination PD-1 blockade and WEE1 inhibition in mouse models and LSCC patient-derived cell lines. They show that multiplex gene editing of mouse lung organoids using the CRISPR-Cas9 system is an efficient and rapid means to generate LSCCs that closely mimic the human disease at the genomic and phenotypic level. Using this genetically defined mouse model and three-dimensional tumoroid culture system, they show that WEE1 inhibition induces DNA damage that primes for endogenous type I interferon responses and antigen presentation system in primary LSCC tumor cells. These events promote cytotoxic T cell-mediated clearance of tumor cells and reduce the accumulation of tumor-infiltrating neutrophils. Beneficial immunological features of WEE1 inhibition are further enhanced by the addition of anti-PD-1 therapy. They developed a mouse model to study a combination approach for immune checkpoint blockade with DNA damage-inducing therapies in the treatment of LSCC (143).
Genetic alterations involved in LSCC identified in various in murine models
Tumor genomic organisation differs between the subtypes of NSCLC. Profiling the genomic structure for each subtype of LC is essential to advance therapeutic treatments. An early study examined the effects of a resected human tumor xenograft (T1 lesion contained within the lung without spread to main bronchi with a moderately differentiated LSCC phenotype) in NSG mice (144). RNA was extracted from both CD133−/EpCAM+ and CD133+/EpCAM+ subpopulations of LSCC samples and whole transcriptome libraries were sequenced on the SOLiD platform. Both CD133− and CD133+ subpopulations involved gene copy number alterations. The expression of 992 genes was assessed. 124 moderately expressed isoforms (80 genes) were selected of which 24 genes were down-regulated and 19 were over-expressed in CD133+ versus CD133- cells. A signature of 22 genes encoding cell surface proteins was highly expressed in CD133+ cells. Results were compared to the Cancer Genome Atlas Program (TCGA) transcriptome data of 221 LSCCs. Most of the 22 cell surface genes were robustly expressed in all TCGA samples. Since the majority of these alterations are common between humans and mice, they can be readily assessed for potential targeted therapy.
Recently, LSCCs from NIH Swiss mice 32-weeks after NTCU challenge were subjected to whole exome and single cell (sc)RNA-seq. Sixteen resected tumors from mice and eight normal lungs were sequenced to a read depth of 129X (98-165X) and 137X (112-179X), respectively. A total of 5,664 somatic coding mutations were identified, including missense, nonsense, silent mutations, and small insertions and deletions. Fifty-nine genes were recurrently mutated the most frequently mutated were: Muc4, Prg4, Igf2r, Ctsll3, Dlgap1, Hspa9, Armcx3, Cdk1, Pcdhb15, Fus, Gga1, Il2rb, Kmt2d (Mll2), Mapk6, Myh1, Ncoa3 (Src3), Obscn, Runx2, Zmynd8, Ido1, Nkain2, Pyy, Stil, Tcl1b4, Tfeb and Trpv1. Murine LSCC mutations were compared to 191 human LSCC documented in the catalogue of somatic mutations in cancer (COSMIC) database (145), and of the 47 genes shared between mouse and human LSCC the most common were: KMT2D (MLL2), MYH1, OBSCN, ZEB2, BRAF, IGF2R, FLT1, HIVEP3, PRG4, ABCA1, ATR, DACH2, ABCB4, DST and MUC4. However, approximately 20% of recurrent mutations were not shared between human and mouse LSCC. scRNA-Seq of two mouse tumors revealed different sets of mutations (146). In one tumor a clonal mutation (R45P) in Igfbp7 was identified along with a mutation (R2457S) in Igf2r and others in Ahctf1, Notch4, Ncoa3, and Nfe2l2. The other tumor had a Trp53 somatic missense mutation along with mutations in the driver genes Myh9, Kmt2d and Keap1. Although the genomic alterations were documented, the controls were from different mice making it a limitation in this study. Taken together, these studies define a set of altered genes (such as Zeb2, Braf, Igf2r, Flt1, Atr, Muc4, Ncoa3 Src3) that occur in mouse models and are also present in the TCGA of LSCC and may be involved in pathogenesis. However, a wide range of alterations are not shared between the murine models and with TCGA. Moreover, these studies do not cover a wide range of other ‘omics (including proteomics, metabolomics and metagenomics) and epigenetic analysis. These limitations indicate the need for more accurate mouse models that better recapitulate the hallmark features of human LSCC. Table 4 determines a list of exclusive mutations found in murine models and certain common mutated genes between murine and human LSCC, indicating the dire need of a better murine model to study genetics of LSCC and also would include proteomics, metabolomics and metagenomics and epigenetic analysis.
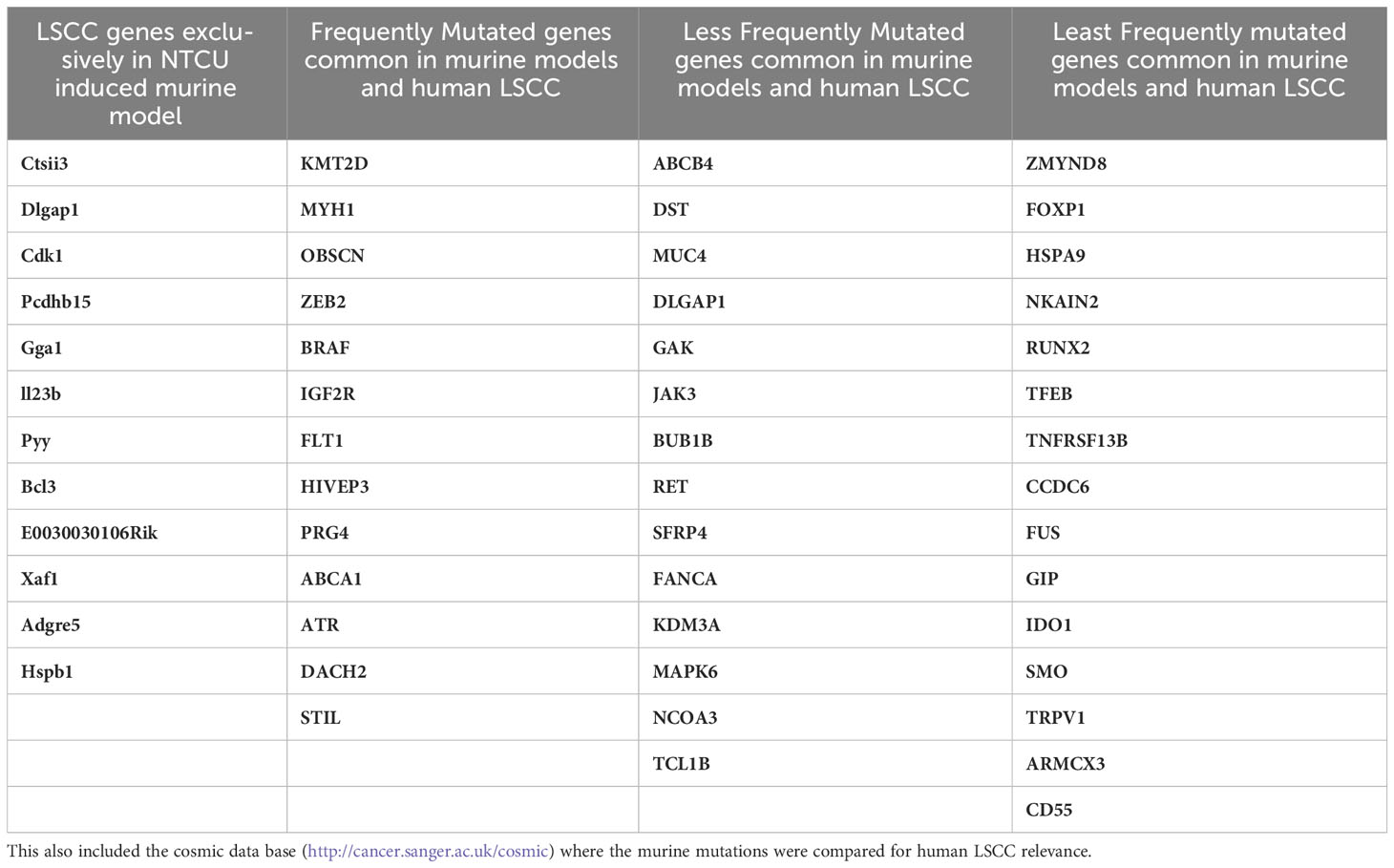
Table 4 Profiles of recurrently mutated LSCC genes in NTCU induced mouse: A list of 59 genes that were identified in NTCU induced mouse model are documented in Table 4 (146).
Conclusions
The global burden of LSCC is substantial and continues to increase, primarily due to the advanced stage of the disease at the time of diagnosis. CS exposure is the primary risk factor and creates a wide field of injury in the epithelium of the lower airways. Although several biomarkers are established for NSCLC, differentiating LSCC from the other NSCLCs remains a major challenge. There are only poorly effective therapies for LSCC, which are typically applied late due to late diagnosis that further reduces efficacy. Although many studies have been performed on resected human samples, there are multiple limitations. Longitudinal analysis of the developmental and prognostic stages of LSCC is not possible to study in humans. Thus, the use of murine models is important to define the early induction mechanism of LSCC and identify early biomarkers and therapeutic targets and develop and test new therapies that can be progressed into improved treatments.
Current established murine models of LSCC use chemical induction using NTCU, human xenografts and genetically-modified mice. The use of these models has contributed to elucidating the molecular alterations. However, they do have challenges such as high mortality from long-term cytotoxic treatment. They also have limitations where xenograft models do not recapitulate the hallmark features of human LSCC since immunocompromised mice are required, which reduces their applicability in elucidating the pathogenesis of neoplastic changes. Most current therapies were developed in immunocompromised animals. Although chemically-induced mice have challenges, they seem to better recapitulate the features of human LSCC. These models can be improved by combining with low-doses of carcinogens with CS, which is the primary cause of LSCCs in humans, in wild-type mice. This would provide an improved method of inducing LSCC in wild-type/non-genetically-modified mice that better replicate the changes in human LSCC. Using more resistant strains such as C57BL/6 mice to induce LSCC would lead to improved models to study the development and progression of LSCC. Although it is established that C57BL/6 mice are resistant to NTCU, which is widely used to induce LSCC in mice, they are susceptible to CS-induced COPD. Thus, C57BL/6 mice when challenged with both NTCU and CS, may develop LSCC.
The expression of some miRNAs, such as miR-106a-5p, miR-20a-5p and miR-93-5p, is increased in LSCC patients and are often useful diagnostic biomarkers for LSCC. These biomarkers are identified only in advanced stage of the disease. Murine models with low ethical challenges (in terms of time taken for approval, multiple rounds of amendments, strict regulations involved in use of carcinogen etc) that accurately replicates the hallmark features of LSCC would also enable the identification of early biomarkers of the disease.
Despite new therapies being tested for NSCLC, this has not yet improved the survival of LSCC patients. This can only be brought about by increasing the understanding of the disease-causing mechanisms and identifying new therapeutic targets particularly those that are altered early in tumorigenesis and the development and testing of new treatments. This can be achieved by developing murine models of LSCC that address and/or eliminate current limitations to identify and enable rigorous interrogation of underlying genetic and epigenetic changes, including miRNAs.
Author contributions
PS: Conceptualization, Writing – original draft, Writing – review & editing. CD: Conceptualization, Supervision, Validation, Writing – review & editing. KP: Visualization, Writing – review & editing. SP: Visualization, Writing – review & editing. VC: Visualization, Writing – review & editing. RK: Supervision, Visualization, Writing – review & editing. JH: Supervision, Visualization, Writing – review & editing. KD: Visualization, Writing – review & editing. AI: Visualization, Writing – review & editing. FN: Visualization, Writing – review & editing. HB-O: Investigation, Visualization, Writing – review & editing. SM: Visualization, Writing – review & editing. GC: Visualization, Writing – review & editing. JL: Visualization, Writing – review & editing. PH: Conceptualization, Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing.
Funding
PS is funded by University of Newcastle Postgraduate Research Scholarship (UNIPRS) and University of Newcastle Research Scholarship (UNRSE)/Lung Foundation Australia. CD is funded by a Fellowship and grant from the National Health and Medical Research Council (NHMRC) of Australia (1120152, 1138402). PMH is funded by a Fellowship and grants from the (NHMRC) of Australia (1079187, 1175134), The University of Technology Sydney, Lung Foundation of Australia and Cancer Council of NSW (1099119, 1157073), the International Association for the Study of Lung Cancer and the Maitland Cancer Appeal. KRP is funded by a fellowship from the International Association for the Study of Lung Cancer (IASLC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Global, regional, and national burden of respiratory tract cancers and associated risk factors from 1990 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Respir Med (2021) 9(9):1030–49.
3. Walters S, Maringe C, Coleman MP, Peake MD, Butler J, Young N, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax (2013) 68(6):551–64. doi: 10.1136/thoraxjnl-2012-202297
4. Malyla V, Paudel KR, Shukla SD, Donovan C, Wadhwa R, Pickles S, et al. Recent advances in experimental animal models of lung cancer. Future Med Chem (2020). doi: 10.4155/fmc-2019-0338
5. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol (2015) 10(9):1243–60. doi: 10.1097/JTO.0000000000000630
6. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Global Health (2019). doi: 10.5334/aogh.2419
7. Toyoda Y, Nakayama T, Ioka A, Tsukuma H. Trends in lung cancer incidence by histological type in osaka, Japan. Japanese J Clin Oncol (2008) 38(8):534–9. doi: 10.1093/jjco/hyn072
8. Lortet-Tieulent J, Soerjomataram I, Ferlay J, Rutherford M, Weiderpass E, Bray F. International trends in lung cancer incidence by histological subtype: Adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer (2014) 84(1):13–22. doi: 10.1016/j.lungcan.2014.01.009
9. Liu H-I, Chiang C-J, Su S-Y, Jhuang J-R, Tsai D-R, Yang Y-W, et al. Incidence trends and spatial distributions of lung adenocarcinoma and squamous cell carcinoma in Taiwan. Sci Rep (2023) 13(1):1655. doi: 10.1038/s41598-023-28253-4
10. Singh N, Agrawal S, Jiwnani S, Khosla D, Malik PS, Mohan A, et al. Lung cancer in India. J Thorac Oncol (2021) 16(8):1250–66. doi: 10.1016/j.jtho.2021.02.004
11. Auerbach O, Stout AP, Hammond EC, Garfinkel L. Changes in bronchial epithelium in relation to cigarette smoking and in relation to lung cancer. N Engl J Med (1961) 265:253–67. doi: 10.1056/NEJM196108102650601
12. Merrick DT, Gao D, Miller YE, Keith RL, Baron AE, Feser W, et al. Persistence of bronchial dysplasia is associated with development of invasive squamous cell carcinoma. Cancer Prev Res (Phila) (2016) 9(1):96–104. doi: 10.1158/1940-6207.CAPR-15-0305
13. Beane JE, Mazzilli SA, Campbell JD, Duclos G, Krysan K, Moy C, et al. Molecular subtyping reveals immune alterations associated with progression of bronchial preMalignant lesions. Nat Commun (2019) 10(1):1856. doi: 10.1038/s41467-019-09834-2
14. Bozinovski S, Vlahos R, Anthony D, McQualter J, Anderson G, Irving L, et al. COPD and squamous cell lung cancer: aberrant inflammation and immunity is the common link. Br J Pharmacol (2016) 173(4):635–48. doi: 10.1111/bph.13198
15. Hecht SS. Tobacco smoke carcinogens and lung cancer. JNCI: J Natl Cancer Institute (1999) 91(14):1194–210. doi: 10.1093/jnci/91.14.1194
16. Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer (2001) 31(2):139–48. doi: 10.1016/S0169-5002(00)00181-1
17. Papi A, Casoni G, Caramori G, Guzzinati I, Boschetto P, Ravenna F, et al. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax (2004) 59(8):679–81. doi: 10.1136/thx.2003.018291
18. Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer (2012) 131(12):2724–32. doi: 10.1002/ijc.27816
19. Takahashi H, Ogata H, Nishigaki R, Broide DH, Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKβ and JNK1 dependent inflammation. Cancer Cell (2010) 17(1):89.
20. Furrukh M. Tobacco smoking and lung cancer. Sultan Qaboos Univ Med J (2013) 13(3):345–58. doi: 10.12816/0003255
21. Pesch B, Kendzia B, Gustavsson P, Jöckel K-H, Johnen G, Pohlabeln H, et al. Cigarette smoking and lung cancer–relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int J Cancer (2012) 131(5):1210–9. doi: 10.1002/ijc.27339
22. Yu Y, Liu H, Zheng S, Ding Z, Chen Z, Jin W, et al. Gender susceptibility for cigarette smoking-attributable lung cancer: a systematic review and meta-analysis. Lung Cancer (2014) 85(3):351–60. doi: 10.1016/j.lungcan.2014.07.004
23. Kurmi OP, Arya PH, Lam KB, Sorahan T, Ayres JG. Lung cancer risk and solid fuel smoke exposure: a systematic review and meta-analysis. Eur Respir J (2012) 40(5):1228–37. doi: 10.1183/09031936.00099511
24. Prevalence and attributable health burden of chronic respiratory diseases, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med (2020) 8(6):585–96.
25. Fricker M, Deane A, Hansbro PM. Animal models of chronic obstructive pulmonary disease. Expert Opin Drug Discovery (2014) 9(6):629–45. doi: 10.1517/17460441.2014.909805
26. Jones B, Donovan C, Liu G, Gomez HM, Chimankar V, Harrison CL, et al. Animal models of COPD: What do they tell us? Respirology (2017) 22(1):21–32. doi: 10.1111/resp.12908
27. Ng Kee Kwong F, Nicholson AG, Harrison CL, Hansbro PM, Adcock IM, Chung KF. Is mitochondrial dysfunction a driving mechanism linking COPD to nonsmall cell lung carcinoma? Eur Respir Rev (2017) 26(146). doi: 10.1183/16000617.0040-2017
28. Brusselle GG, Joos GF, Bracke KR. New insights into the immunology of chronic obstructive pulmonary disease. Lancet (2011) 378(9795):1015–26. doi: 10.1016/S0140-6736(11)60988-4
29. Barnes PJ, Adcock IM. Chronic obstructive pulmonary disease and lung cancer: A lethal association. Am J Respir Crit Care Med (2011) 184(8):866–7. doi: 10.1164/rccm.201108-1436ED
30. Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer (2013) 13(4):233–45. doi: 10.1038/nrc3477
31. Gullón JA, Suárez I, Medina A, Rubinos G, Fernández R, González I. Role of emphysema and airway obstruction in prognosis of lung cancer. Lung Cancer (2011) 71(2):182–5. doi: 10.1016/j.lungcan.2010.05.018
32. Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer (2015) 90(2):121–7. doi: 10.1016/j.lungcan.2015.08.017
33. The Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature (2012) 489:519. doi: 10.1038/nature11404
34. Teixeira VH, Pipinikas CP, Pennycuick A, Lee-Six H, Chandrasekharan D, Beane J, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med (2019) 25(3):517–25. doi: 10.1038/s41591-018-0323-0
35. Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: A review. J Thorac Oncol (2012) 7(5):924–33. doi: 10.1097/JTO.0b013e31824cc334
36. Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res (2008) 68(17):6913–21. doi: 10.1158/0008-5472.CAN-07-5084
37. Okudela K, Suzuki M, Kageyama S, Bunai T, Nagura K, Igarashi H, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int (2007) 57(10):664–71. doi: 10.1111/j.1440-1827.2007.02155.x
38. Kawano O, Sasaki H, Okuda K, Yukiue H, Yokoyama T, Yano M, et al. PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer (2007) 58(1):159–60. doi: 10.1016/j.lungcan.2007.06.020
39. Hsu AC, Starkey MR, Hanish I, Parsons K, Haw TJ, Howland LJ, et al. Targeting PI3K-p110α Suppresses influenza virus infection in chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2015) 191(9):1012–23. doi: 10.1164/rccm.201501-0188OC
40. Nisa L, Häfliger P, Poliaková M, Giger R, Francica P, Aebersold DM, et al. PIK3CA hotspot mutations differentially impact responses to MET targeting in MET-driven and non-driven preclinical cancer models. Mol Cancer (2017) 16(1):93–3. doi: 10.1186/s12943-017-0660-5
41. Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med (2010) 2(62):62ra93. doi: 10.1126/scitranslmed.3001451
42. Dziadziuszko R, Merrick DT, Witta SE, Mendoza AD, Szostakiewicz B, Szymanowska A, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol (2010) 28(13):2174–80. doi: 10.1200/JCO.2009.24.6611
43. Cappuzzo F, Tallini G, Finocchiaro G, Wilson RS, Ligorio C, Giordano L, et al. Insulin-like growth factor receptor 1 (IGF1R) expression and survival in surgically resected non-small-cell lung cancer (NSCLC) patients. Ann Oncol (2010) 21(3):562–7. doi: 10.1093/annonc/mdp357
44. Faoro L, Singleton PA, Cervantes GM, Lennon FE, Choong NW, Kanteti R, et al. EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mamMalian target of rapamycin activation. J Biol Chem (2010) 285(24):18575–85. doi: 10.1074/jbc.M109.075085
45. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol (2003) 4(12):915–25. doi: 10.1038/nrm1261
46. Go H, Jeon YK, Park HJ, Sung SW, Seo JW, Chung DH. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol (2010) 5(3):305–13. doi: 10.1097/JTO.0b013e3181ce3d1d
47. Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther (2009) 8(21):2042–50. doi: 10.4161/cbt.8.21.9764
48. Kishimoto Y, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Aberrations of the p53 tumor suppressor gene in human non-small cell carcinomas of the lung. Cancer Res (1992) 52(17):4799–804.
49. Mandinova A, Lee SW. The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci Transl Med (2011) 3(64):64rv1. doi: 10.1126/scitranslmed.3001366
50. Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res (1998) 26(15):3453–9. doi: 10.1093/nar/26.15.3453
51. Higashiyama M, Doi O, Kodama K, Yokouchi H, Kasugai T, Ishiguro S, et al. MDM2 gene amplification and expression in non-small-cell lung cancer: immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br J Cancer (1997) 75(9):1302–8. doi: 10.1038/bjc.1997.221
52. Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discovery (2011) 1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005
53. Jin G, Kim MJ, Jeon HS, Choi JE, Kim DS, Lee EB, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer (2010) 69(3):279–83. doi: 10.1016/j.lungcan.2009.11.012
54. Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 abnorMalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res (2010) 16(14):3743–53. doi: 10.1158/1078-0432.CCR-09-3352
55. Affandi KA, Tizen NMS, Mustangin M, Zin RRMRM. p40 immunohistochemistry is an excellent marker in primary lung squamous cell carcinoma. J Pathol Trans Med (2018) 52(5):283–9. doi: 10.4132/jptm.2018.08.14
56. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet (2016) 48(6):607–16. doi: 10.1038/ng.3564
57. Brzezianska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep (2013) 40(1):309–25. doi: 10.1007/s11033-012-2063-4
58. Kim SH, Lee S, Lee CH, Lee MK, Kim YD, Shin DH, et al. Expression of cancer-testis antigens MAGE-A3/6 and NY-ESO-1 in non-Small-Cell lung carcinomas and their relationship with immune cell infiltration. Lung (2009) 187(6):401. doi: 10.1007/s00408-009-9181-3
59. Lonergan KM, Chari R, Coe BP, Wilson IM, Tsao M-S, Ng RT, et al. Transcriptome profiles of carcinoma-in-Situ and invasive non-small cell lung cancer as revealed by SAGE. PloS One (2010) 5(2):e9162. doi: 10.1371/journal.pone.0009162
60. Kayser G, Sienel W, Kubitz B, Mattern D, Stickeler E, Passlick B, et al. Poor outcome in primary non-small cell lung cancers is predicted by transketolase TKTL1 expression. Pathology (2011) 43(7):719–24. doi: 10.1097/PAT.0b013e32834c352b
61. Daskalos A, Logotheti S, Markopoulou S, Xinarianos G, Gosney JR, Kastania AN, et al. Global DNA hypomethylation-induced DeltaNp73 transcriptional activation in non-small cell lung cancer. Cancer Lett (2011) 300(1):79–86. doi: 10.1016/j.canlet.2010.09.009
62. Radhakrishnan VM, Jensen TJ, Cui H, Futscher BW, Martinez JD. Hypomethylation of the 14-3-3σ promoter leads to increased expression in non-small cell lung cancer. Genes Chromosomes cancer. (2011) 50(10):830–6. doi: 10.1002/gcc.20904
63. Kwon Y-J, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol (2012) 7(1):20–33. doi: 10.1097/JTO.0b013e3182307f62
64. Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res (2000) 60(21):5954–8.
65. Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, Carvalho AL, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res (2011) 17(13):4494–503. doi: 10.1158/1078-0432.CCR-10-3436
66. Lai JC, Cheng YW, Goan YG, Chang JT, Wu TC, Chen CY, et al. Promoter methylation of O(6)-methylguanine-DNA-methyltransferase in lung cancer is regulated by p53. DNA repair. (2008) 7(8):1352–63. doi: 10.1016/j.dnarep.2008.04.016
67. Nakata S, Sugio K, Uramoto H, Oyama T, Hanagiri T, Morita M, et al. The methylation status and protein expression of CDH1, p16INK4A, and fragile histidine triad in nonsmall cell lung carcinoma. Cancer (2006) 106(10):2190–9. doi: 10.1002/cncr.21870
68. Mashkova TD, Oparina NY, Zinov’eva OL, Kropotova ES, Dubovaya VI, Poltaraus AB, et al. Transcription of TIMP3, DAPK1, and AKR1B10 in squamous-cell lung cancer. Mol Biol (2006) 40(6):945–51. doi: 10.1134/S0026893306060148
69. Niklinska W, Naumnik W, Sulewska A, Kozlowski M, Pankiewicz W, Milewski R. Prognostic significance of DAPK and RASSF1A promoter hypermethylation in non-small cell lung cancer (NSCLC). Folia Histochemica Cytobiologica (2009) 47(2):275–80. doi: 10.2478/v10042-009-0091-2
70. Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer (2010) 10:600–. doi: 10.1186/1471-2407-10-600
71. Ji M, Guan H, Gao C, Shi B, Hou P. Highly frequent promoter methylation and PIK3CA amplification in non-small cell lung cancer (NSCLC). BMC Cancer (2011) 11:147–7. doi: 10.1186/1471-2407-11-147
72. Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, Campan M, et al. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer (2008) 7:62–2. doi: 10.1186/1476-4598-7-62
73. Foster PS, Plank M, Collison A, Tay HL, Kaiko GE, Li J, et al. The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. Immunol Rev (2013) 253(1):198–215. doi: 10.1111/imr.12058
74. Kulpa J, Wójcik E, Reinfuss M, Kołodziejski L. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21-1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem (2002) 48(11):1931–7. doi: 10.1093/clinchem/48.11.1931
75. Dua K, Hansbro NG, Foster PS, Hansbro PM. MicroRNAs as therapeutics for future drug delivery systems in treatment of lung diseases. Drug Delivery Transl Res (2017) 7(1):168–78. doi: 10.1007/s13346-016-0343-6
76. Awasthi R, Rathbone MJ, Hansbro PM, Bebawy M, Dua K. Therapeutic prospects of microRNAs in cancer treatment through nanotechnology. Drug Delivery Trans Res (2018) 8(1):97–110. doi: 10.1007/s13346-017-0440-1
77. Davidson MR, Larsen JE, Yang IA, Hayward NK, Clarke BE, Duhig EE, et al. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PloS One (2010) 5(9):e12560. doi: 10.1371/journal.pone.0012560
78. Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res (2011) 17(21):6802–11. doi: 10.1158/1078-0432.CCR-11-0419
79. Patnaik S, Mallick R, Kannisto E, Sharma R, Bshara W, Yendamuri S, et al. MiR-205 and miR-375 microRNA assays to distinguish squamous cell carcinoma from adenocarcinoma in lung cancer biopsies. J Thorac Oncol (2015) 10(3):446–53. doi: 10.1097/JTO.0000000000000423
80. Edmonds MD, Eischen CM. Differences in miRNA expression in early stage lung adenocarcinomas that did and did not relapse. PLoS One (2014) 9(7):e101802. doi: 10.1371/journal.pone.0101802
81. Zhang L, Shan X, Wang J, Zhu J, Huang Z, Zhang H, et al. A three-microRNA signature for lung squamous cell carcinoma diagnosis in Chinese male patients. Oncotarget (2017) 8(49):86897–907. doi: 10.18632/oncotarget.19666
82. Kumar S, Sharawat SK, Ali A, Gaur V, Malik PS, Kumar S, et al. Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients. Curr Probl Cancer (2020) 44(4):100540. doi: 10.1016/j.currproblcancer.2020.100540
83. Socinski MA, Bondarenko I, Karaseva NA, Makhson AM, Vynnychenko I, Okamoto I, et al. Weekly nab-paclitaxel in combination with carboplatin versus solvent-based paclitaxel plus carboplatin as first-line therapy in patients with advanced non-small-cell lung cancer: final results of a phase III trial. J Clin Oncol (2012) 30(17):2055–62. doi: 10.1200/JCO.2011.39.5848
84. Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res (2009) 69(14):5776–83. doi: 10.1158/0008-5472.CAN-09-0587
85. Yang Y, Li X, Yang Q, Wang X, Zhou Y, Jiang T, et al. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenetics (2010) 200(2):127–33. doi: 10.1016/j.cancergencyto.2010.03.014
86. Xia E, Kanematsu S, Suenaga Y, Elzawahry A, Kondo H, Otsuka N, et al. MicroRNA induction by copy number gain is associated with poor outcome in squamous cell carcinoma of the lung. Sci Rep (2018) 8(1):15363. doi: 10.1038/s41598-018-33696-1
87. Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol (2011) 137(4):557–66. doi: 10.1007/s00432-010-0918-4
88. Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Modern Pathol (2010) 23:1157. doi: 10.1038/modpathol.2010.111
89. Postmus PE, Kerr KM, Oudkerk M, Senan S, Waller DA, Vansteenkiste J, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol (2017) 28:iv1–iv21. doi: 10.1093/annonc/mdx222
90. Park K, Vansteenkiste J, Lee KH, Pentheroudakis G, Zhou C, Prabhash K, et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the management of patients with locally-advanced unresectable non-small-cell lung cancer: a KSMO-ESMO initiative endorsed by CSCO, ISMPO, JSMO, MOS, SSO and TOS. Ann Oncol (2020) 31(2):191–201. doi: 10.1016/j.annonc.2019.10.026
91. Passiglia F, Bertolaccini L, Del Re M, Facchinetti F, Ferrara R, Franchina T, et al. Diagnosis and treatment of early and locally advanced non-small-cell lung cancer: The 2019 AIOM (Italian Association of Medical Oncology) clinical practice guidelines. Crit Rev Oncology/Hematology (2020) 148:102862. doi: 10.1016/j.critrevonc.2019.102862
92. Wang Y, Zhang Z, Garbow JR, Rowland DJ, Lubet RA, Sit D, et al. Chemoprevention of lung squamous cell carcinoma in mice by a mixture of Chinese herbs. Cancer Prev Res (Philadelphia Pa.) (2009) 2(7):634–40. doi: 10.1158/1940-6207.CAPR-09-0052
93. Scagliotti GV, Parikh P, Pawel Jv Biesma B, Vansteenkiste J, Manegold C, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non–small-cell lung cancer. J Clin Oncol (2008) 26(21):3543–51. doi: 10.1200/JCO.2007.15.0375
94. Thatcher N, Hirsch FR, Szczesna A, Ciuleanu T-E, Szafranski W, Dediu M, et al. A randomized, multicenter, open-label, phase III study of gemcitabine-cisplatin (GC) chemotherapy plus necitumumab (IMC-11F8/LY3012211) versus GC alone in the first-line treatment of patients (pts) with stage IV squamous non-small cell lung cancer (sq-NSCLC). J Clin Oncol (2014) 32(15_suppl):8008–8. doi: 10.1200/jco.2014.32.15_suppl.8008
95. Li H, Takayama K, Wang S, Shiraishi Y, Gotanda K, Harada T, et al. Addition of bevacizumab enhances antitumor activity of erlotinib against non-small cell lung cancer xenografts depending on VEGF expression. Cancer chemotherapy Pharmacol (2014) 74(6):1297–305. doi: 10.1007/s00280-014-2610-x
96. Garon EB, Ciuleanu TE, Arrieta O, Prabhash K, Syrigos KN, Goksel T, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): a multicentre, double-blind, randomised phase 3 trial. Lancet (2014) 384(9944):665–73. doi: 10.1016/S0140-6736(14)60845-X
97. Yuan H, Liu J, Zhang J. The current landscape of immune checkpoint blockade in metastatic lung squamous cell carcinoma. Molecules (2021) 26(5). doi: 10.3390/molecules26051392
98. Ciuleanu T, Brodowicz T, Zielinski C, Kim JH, Krzakowski M, Laack E, et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small-cell lung cancer: a randomised, double-blind, phase 3 study. Lancet (2009) 374(9699):1432–40. doi: 10.1016/S0140-6736(09)61497-5
99. Atherton MJ, Morris JS, McDermott MR, Lichty BD. Cancer immunology and canine Malignant melanoma: A comparative review. Vet Immunol Immunopathol (2016) 169:15–26. doi: 10.1016/j.vetimm.2015.11.003
100. Yu H, Chen Z, Ballman KV, Watson MA, Govindan R, Lanc I, et al. Correlation of PD-L1 expression with tumor mutation burden and gene signatures for prognosis in early-stage squamous cell lung carcinoma. J Thorac Oncol (2019) 14(1):25–36. doi: 10.1016/j.jtho.2018.09.006
101. Teng JP, Yang ZY, Zhu YM, Ni D, Zhu ZJ, Li XQ. Gemcitabine and cisplatin for treatment of lung cancer in vitro and vivo. Eur Rev Med Pharmacol Sci (2018) 22(12):3819–25.
102. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer (2014) 14(8):535–46. doi: 10.1038/nrc3775
103. Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med (2015) 373(2):123–35. doi: 10.1056/NEJMoa1504627
104. Santos ES, Hart L. Advanced squamous cell carcinoma of the lung: current treatment approaches and the role of Afatinib. Onco Targets Ther (2020) 13:9305–21. doi: 10.2147/OTT.S250446
105. Redman MW, Papadimitrakopoulou VA, Minichiello K, Hirsch FR, Mack PC, Schwartz LH, et al. Biomarker-driven therapies for previously treated squamous non-small-cell lung cancer (Lung-MAP SWOG S1400): a biomarker-driven master protocol. Lancet Oncol (2020) 21(12):1589–601. doi: 10.1016/S1470-2045(20)30475-7
106. Paz-Ares L, Ciuleanu T-E, Cobo M, Schenker M, Zurawski B, Menezes J, et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol (2021) 22(2):198–211. doi: 10.1016/S1470-2045(20)30641-0
107. Paz-Ares LG, Ciuleanu T-E, Cobo M, Schenker M, Zurawski B, Menezes J, et al. First-line nivolumab plus ipilimumab with chemotherapy versus chemotherapy alone for metastatic NSCLC in checkMate 9LA: 3-year clinical update and outcomes in patients with brain metastases or select somatic mutations. J Thorac Oncol (2023) 18(2):204–22. doi: 10.1016/j.jtho.2022.10.014
108. Raposo TP, Arias-Pulido H, Chaher N, Fiering SN, Argyle DJ, Prada J, et al. Comparative aspects of canine and human inflammatory breast cancer. Semin Oncol (2017) 44(4):288–300. doi: 10.1053/j.seminoncol.2017.10.012
109. Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta veterinaria Scandinavica (2017) 59(1):71–1. doi: 10.1186/s13028-017-0341-9
110. Nettesheim P, Hammons AS. Induction of squamous cell carcinoma in the respiratory tract of mice. J Natl Cancer Inst (1971) 47(3):697–701.
111. Yoshimoto T, Hirao F, Sakatani M, Nishikawa H, Ogura T. Induction of squamous cell carcinoma in the lung of C57BL/6 mice by intratracheal instillation of benzo[a]pyrene with charcoal powder. Gan (1977) 68(3):343–52.
112. Lijinsky W, Reuber MD. Neoplasms of the skin and other organs observed in Swiss mice treated with nitrosoalkylureas. J Cancer Res Clin Oncol (1988) 114(3):245–9. doi: 10.1007/BF00405829
113. Wang Y, Zhang Z, Yan Y, Lemon WJ, LaRegina M, Morrison C, et al. A chemically induced model for squamous cell carcinoma of the lung in mice. Cancer Res (2004) 64(5):1647–54. doi: 10.1158/0008-5472.CAN-03-3273
114. Yamano S, Gi M, Tago Y, Doi K, Okada S, Hirayama Y, et al. Role of deltaNp63posCD44vpos cells in the development of N-nitroso-tris-chloroethylurea-induced peripheral-type mouse lung squamous cell carcinomas. Cancer Sci (2016) 107(2):123–32. doi: 10.1111/cas.12855
115. Hudish TM, Opincariu LI, Mozer AB, Johnson MS, Cleaver TG, Malkoski SP, et al. N-nitroso-tris-chloroethylurea induces pre-Malignant squamous dysplasia in mice. Cancer Prev Res (Philadelphia Pa.) (2012) 5(2):283–9. doi: 10.1158/1940-6207.CAPR-11-0257
116. Ghosh M, Dwyer-Nield LD, Kwon JB, Barthel L, Janssen WJ, Merrick DT, et al. Tracheal dysplasia precedes bronchial dysplasia in mouse model of N-nitroso trischloroethylurea induced squamous cell lung cancer. PloS One (2015) 10(4):e0122823. doi: 10.1371/journal.pone.0122823
117. Xiong D, Pan J, Zhang Q, Szabo E, Miller MS, Lubet RA, et al. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget (2017) 8(12):18885–900. doi: 10.18632/oncotarget.13806
118. Riolobos L, Gad EA, Treuting PM, Timms AE, Hershberg EA, Corulli LR, et al. The effect of mouse strain, sex and carcinogen dose on toxicity and the development of lung dysplasia and squamous cell carcinomas in mice. Cancer Prev Res (2019), canprevres.0442.2018. doi: 10.1158/1940-6207.CAPR-18-0442
119. Dwyer-Nield LD, McArthur DG, Tennis MA, Merrick DT, Keith RL. An Improved Murine Pre-Malignant Squamous Cell Model: Tobacco smoke exposure augments NTCU-induced murine airway dysplasia. Cancer Prev Res (Phila) (2020). doi: 10.1158/1940-6207.CAPR-20-0332
120. Wang Y, James M, Wen W, Lu Y, Szabo E, Lubet RA, et al. Chemopreventive effects of pioglitazone on chemically induced lung carcinogenesis in mice. Mol Cancer Ther (2010) 9(11):3074–82. doi: 10.1158/1535-7163.MCT-10-0510
121. Pan J, Zhang Q, Li K, Liu Q, Wang Y, You M. Chemoprevention of lung squamous cell carcinoma by ginseng. Cancer Prev Res (Phila) (2013) 6(6):530–9. doi: 10.1158/1940-6207.CAPR-12-0366
122. Song JM, Qian X, Teferi F, Pan J, Wang Y, Kassie F. Dietary diindolylmethane suppresses inflammation-driven lung squamous cell carcinoma in mice. Cancer Prev Res (Philadelphia Pa.) (2015) 8(1):77–85. doi: 10.1158/1940-6207.CAPR-14-0245
123. You MS, Rouggly LC, You M, Wang Y. Mouse models of lung squamous cell carcinomas. Cancer Metastasis Rev (2013) 32(1):77–82. doi: 10.1007/s10555-012-9406-4
124. Khan N, Afaq F, Kweon MH, Kim K, Mukhtar H. Oral consumption of pomegranate fruit extract inhibits growth and progression of primary lung tumors in mice. Cancer Res (2007) 67(7):3475–82. doi: 10.1158/0008-5472.CAN-06-3941
125. Mazzilli SA, Hershberger PA, Reid ME, Bogner PN, Atwood K, Trump DL, et al. Vitamin D repletion reduces the progression of preMalignant squamous lesions in the NTCU lung squamous cell carcinoma mouse model. Cancer Prev Res (Philadelphia Pa.) (2015) 8(10):895–904. doi: 10.1158/1940-6207.CAPR-14-0403
126. Lieberman R, Pan J, Zhang Q, You M. Rad52 deficiency decreases development of lung squamous cell carcinomas by enhancing immuno-surveillance. Oncotarget (2017) 8(21):34032–44. doi: 10.18632/oncotarget.16371
127. Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature (2007) 448:807. doi: 10.1038/nature06030
128. Farago AF, Snyder EL, Jacks T. SnapShot: Lung cancer models. Cell (2012) 149(1):246–.e1. doi: 10.1016/j.cell.2012.03.015
129. Xu C, Fillmore Christine M, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of lkb1 and pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell (2014) 25(5):590–604. doi: 10.1016/j.ccr.2014.03.033
130. Mukhopadhyay A, Berrett Kristofer C, Kc U, Clair Phillip M, Pop Stelian M, Carr Shamus R, et al. Sox2 cooperates with lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep (2014) 8(1):40–9. doi: 10.1016/j.celrep.2014.05.036
131. Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, et al. The pivotal role of IKKα in the development of spontaneous lung squamous cell carcinomas. Cancer Cell (2013) 23(4):527–40. doi: 10.1016/j.ccr.2013.03.009
132. Ferone G, Song J-Y, Sutherland Kate D, Bhaskaran R, Monkhorst K, Lambooij J-P, et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell (2016) 30(4):519–32. doi: 10.1016/j.ccell.2016.09.001
133. Mollaoglu G, Jones A, Wait SJ, Mukhopadhyay A, Jeong S, Arya R, et al. The lineage-defining transcription factors SOX2 and NKX2-1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity (2018) 49(4):764–79.e9. doi: 10.1016/j.immuni.2018.09.020
134. Hao C, Wang L, Peng S, Cao M, Li H, Hu J, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett (2015) 357(1):179–85. doi: 10.1016/j.canlet.2014.11.024
135. Russo MV, Faversani A, Gatti S, Ricca D, Del Gobbo A, Ferrero S, et al. A new mouse avatar model of non-small cell lung cancer. Front Oncol (2015) 5(52). doi: 10.3389/fonc.2015.00052
136. Weeden CE, Holik AZ, Young RJ, Ma SB, Garnier JM, Fox SB, et al. Cisplatin increases sensitivity to FGFR inhibition in patient-derived Xenograft models of lung squamous cell carcinoma. Mol Cancer Ther (2017) 16(8):1610–22. doi: 10.1158/1535-7163.MCT-17-0174
137. Wang D, Pham NA, Tong J, Sakashita S, Allo G, Kim L, et al. Molecular heterogeneity of non-small cell lung carcinoma patient-derived xenografts closely reflect their primary tumors. Int J Cancer (2017) 140(3):662–73. doi: 10.1002/ijc.30472
138. Ilie M, Nunes M, Blot L, Hofman V, Long-Mira E, Butori C, et al. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the preanalytical steps. Cancer Med (2015) 4(2):201–11. doi: 10.1002/cam4.357
139. Jiang Y, Zhao J, Zhang Y, Li K, Li T, Chen X, et al. Establishment of lung cancer patient-derived xenograft models and primary cell lines for lung cancer study. J Trans Med (2018) 16(1):138. doi: 10.1186/s12967-018-1516-5
140. Lu D, Luo P, Zhang J, Ye Y, Wang Q, Li M, et al. Patient-derived tumor xenografts of lung squamous cell carcinoma alter long non-coding RNA profile but not responsiveness to cisplatin. Oncol Lett (2018) 15(6):8589–603. doi: 10.3892/ol.2018.8401
141. Valencia K, Sainz C, Bertolo C, de Biurrun G, Agorreta J, Azpilikueta A, et al. Two cell line models to study multiorganic metastasis and immunotherapy in lung squamous cell carcinoma. Dis Model Mech (2022) 15(1). doi: 10.1242/dmm.049137
142. Azpilikueta A, Agorreta J, Labiano S, Perez-Gracia JL, Sanchez-Paulete AR, Aznar MA, et al. Successful immunotherapy against a transplantable mouse squamous lung carcinoma with anti-PD-1 and anti-CD137 monoclonal antibodies. J Thorac Oncol (2016) 11(4):524–36. doi: 10.1016/j.jtho.2016.01.013
143. Hai J, Zhang H, Zhou J, Wu Z, Chen T, Papadopoulos E, et al. Generation of genetically engineered mouse lung organoid models for squamous cell lung cancers allows for the study of combinatorial immunotherapy. Clin Cancer Res (2020) 26(13):3431–42. doi: 10.1158/1078-0432.CCR-19-1627
144. Barrett CL, Schwab RB, Jung H, Crain B, Goff DJ, Jamieson CHM, et al. Transcriptome sequencing of tumor subpopulations reveals a spectrum of therapeutic options for squamous cell lung cancer. PLoS One (2013) 8(3):e58714. doi: 10.1371/journal.pone.0058714
145. COSMIC, the Catalogue of Somatic Mutations in Cancer (2019). Available at: http://cancer.sanger.ac.uk/cosmic.
Keywords: lung cancer, squamous cell carcinoma, animal models, risk factors, genetic and epigenetic alterations
Citation: Sahu P, Donovan C, Paudel KR, Pickles S, Chimankar V, Kim RY, Horvart JC, Dua K, Ieni A, Nucera F, Bielefeldt-Ohmann H, Mazilli S, Caramori G, Lyons JG and Hansbro PM (2023) Pre-clinical lung squamous cell carcinoma mouse models to identify novel biomarkers and therapeutic interventions. Front. Oncol. 13:1260411. doi: 10.3389/fonc.2023.1260411
Received: 17 July 2023; Accepted: 29 August 2023;
Published: 25 September 2023.
Edited by:
James CM Ho, The University of Hong Kong, ChinaReviewed by:
Selena Sheng Yan, The University of Hong Kong, ChinaRoland Leung, Queen Mary Hospital, China
Copyright © 2023 Sahu, Donovan, Paudel, Pickles, Chimankar, Kim, Horvart, Dua, Ieni, Nucera, Bielefeldt-Ohmann, Mazilli, Caramori, Lyons and Hansbro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip M. Hansbro, cGhpbGlwLmhhbnNicm9AdXRzLmVkdS5hdQ==
 Priyanka Sahu
Priyanka Sahu Chantal Donovan
Chantal Donovan Keshav Raj Paudel
Keshav Raj Paudel Sophie Pickles3
Sophie Pickles3 Jay C. Horvart
Jay C. Horvart Kamal Dua
Kamal Dua Antonio Ieni
Antonio Ieni Francesco Nucera
Francesco Nucera Helle Bielefeldt-Ohmann
Helle Bielefeldt-Ohmann Sarah Mazilli
Sarah Mazilli Gaetano Caramori
Gaetano Caramori J. Guy Lyons
J. Guy Lyons Philip M. Hansbro
Philip M. Hansbro