
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 07 November 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1256054
This article is part of the Research TopicAcute Leukemias: Molecular Characterization, Leukemia-Initiating Cells, and Influence of the Microenvironment Vol. IIView all 9 articles
 Danna Lin1
Danna Lin1 Keyan Yang2
Keyan Yang2 Lihua Yu1
Lihua Yu1 Lulu Huang1
Lulu Huang1 Xiaorong Lai1
Xiaorong Lai1 Li Wu1
Li Wu1 Xiayu Xia2
Xiayu Xia2 Jingwen Zhang3
Jingwen Zhang3 Qinlong Zheng2*
Qinlong Zheng2* Lihua Yang1*
Lihua Yang1*Background: Overexpression of the cytokine receptor-like factor 2 (CRLF2) gene is the most common feature in the Philadelphia chromosome (Ph)-like subtype of B-cell acute lymphoblastic leukemia (B-ALL). However, the predictive value of CRLF2 overexpression for the prognosis of pediatric B-ALL patients remain controversial. The molecular mechanisms that upregulate CRLF2 expression level in patients has not been fully elucidated.
Methods: In this study, the prognostic impact of CRLF2 expression level on molecular types of B-ALL in pediatric patients from Zhujiang Hospital (n = 111) was retrospectively analyzed. Youden index analysis was used to categorize CRLF2 expression into 3 groups, and these categories more precisely described the differences in the prognosis of patients with varying expression levels of CRLF2 in both the Zhujiang Hospital cohort and the TARGET cohort.
Results: We used the Zhujiang Hospital cohort as a discovery cohort to determine the cutoff value of CRLF2 expression. CRLF2-high patients accounted for approximately 6%. In addition, the percentage of bone marrow blast cells and initial white blood cell count in CRLF2-high patients were higher than those in CRLF2-low patients, and MRD turned negative slower. The results were validated in the TARGET cohort and indicated that CRLF2 overexpression could be subdivided by CRLF2 expression levels into 2 categories: CRLF2-high with a poor survival and CRLF2-medium with a good OS and EFS. Such heterogeneity was attributed to the different molecular mechanisms leading to CLRF2 upregulation, where the CRLF2 overexpression level was high in Ph-like B-ALL and medium in high hyperdiploid B-ALL.
Conclusion: This study highlights the importance of the molecular mechanisms of the upregulation of CRLF2 expression in predicting the prognosis of pediatric B-ALL patients.
The cytokine receptor-like factor 2 (CRLF2) gene, located on chromosome Xp22.3 or Yp11.3, encodes cytokine receptor-like factors (CRLFs), which form heterodimers with IL-7Ra that receive and transmit signals from thymic stromal lymphopoietin and upregulated CRLF2 expression activates the Janus kinase/signal transducer and activator of transcription (JAK/STAT) or phosphatidylinositol 3-kinase/mammalian target of rapamycin (PI3K/mTOR) pathway to promote the occurrence and progression of leukemia (1–4). CRLF2 overexpression has been reported as a major feature in the Philadelphia chromosome (Ph)-like subtype of B-cell acute lymphoblastic leukemia(B-ALL), and approximately 50-60% of patients with Ph-like B-ALL have CRLF2 overexpression (1–4). Nonetheless, whether CRLF2 overexpression can be employed to predict the prognosis of B-ALL patients is controversial. The majority of previous studies have demonstrated that CRLF2 overexpression is associated with the poor prognosis of B-ALL patients (1–4). However, some studies have revealed that CRLF2 overexpression is not an independent prognostic factor for B-ALL (4, 5). The definition of CRLF2 overexpression varies in these studies, and the proportion of patients with CRLF2 overexpression among unscreened patients ranges from 4.7 to 25% (2, 4–7). Different criteria for CRLF2 overexpression may cover a variety of molecular mechanisms leading to the upregulation of CRLF2 expression; therefore, it is difficult to precisely characterize the factors driving disease progression (7–10).
In this study of Chinese pediatric patients, we identified varying levels of CRLF2 expression among different molecular subtypes. New cutoff value for CRLF2 overexpression associated with molecular types were proposed. Moreover, the prognoses of patients with different CRLF2 expression levels were validated using the B-ALL patient cohort from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) public database. Finally, the corresponding cutoffs for CRLF2-high in real-time quantitative polymerase chain reaction (RQ-PCR) were also determined, thereby providing a basis for the rapid identification of B-ALL patients with a potentially poor prognosis via CRLF2 expression level.
One hundred and eleven patients with B-ALL treated at the Department of Pediatrics, Southern Medical University, Zhujiang Hospital, between May 2020 and February 2022 were enrolled in this study; the final follow-up date was August 25, 2023. Children diagnosed before 1 year of age (n=5) were treated with the Inerfant-06 protocol (11). Among children diagnosed after 1 year of age, 1 was initially treated intermittently at another hospital, 1 was treated using the Guangdong Children’s Leukemia Group-ALL-2008 (GD-ALL-2008) protocol (12), and the others were treated using the South China Children’s Leukemia Group-ALL-2016 (SCCLG-ALL-2016) protocol (12) (n=104). The detailed clinical characteristics are shown in Table 1. Bone marrow (BM) samples were collected from patients at initial diagnosis. This study was approved by the central ethics committee of Zhujiang Hospital, Southern Medical University, in accordance with the Declaration of Helsinki (Approval number: 2022-KY-195-01).
Data for the TARGET cohort were initially generated for the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative (TARGET Phase II, phs000464). The data used for this analysis are available at (https://portal.gdc.cancer.gov/projects). Clinical and survival data were downloaded from cBioportal (http://www.cbioportal.org).
The TARGET cohort included 144 patients diagnosed with B-cell lymphoblastic leukemia and with diagnostic tumor RNA samples. FPKM values were calculated using the gene expression counts matrix from the RNA sequencing. The clinical characteristics are provided in Supplementary Table 2.
Genomic DNA was obtained from BM at diagnosis and subjected to hybridization-based capture by using a KAPA HyperPlus Kits (Roche Diagnostics, Basel, Switzerland) and then captured using a NimbleGen SeqCap EZ Library SR (Roche Diagnostics) kit probing the exon regions of 339 genes relevant to hematologic malignancies, followed by high throughput sequencing on an Illumina NextSeq 550 platform to produce PE 150 bp. Somatic variant calling was performed with the Broad Institute’s Genome Analysis Toolkit (GATK) v4.0.4.0 and Vardict v1.12 and annotated by Annovar v2018-04-16 (13–18). Selected pathogenic variants were manually inspected using the Integrative Genomics Viewer (IGV) software v2.8.0 (19).
Total RNA was extracted from the BM samples using a Cowin RNApure Blood kit (Cowin Bio., Jiangsu, China) and quantified using a Nanodrop one (Thermo Fisher). Stranded mRNA sequencing libraries were constructed using a VAHTS Stranded mRNA-seq Library Prep Kit for Illumina (Vazyme, Nanjing, China). The polyadenylated mRNAs were then purified and fragmented, followed by the generation of double-strand cDNA. The double stranded cDNAs were ligated to the VAHTS RNA Adapters (N803/N804, Vazyme, Nanjing, China) following end repair and A tailing. The purified ligation products (size 200-500 bps) were selected and quantified, followed by sequencing on an Illumina Novaseq 6000 to produce ≥30M 150 base pair paired-end reads. After performing quality control on the sample’s raw FASTQ data, reads were trimmed using fastp v0.19.4 to remove sequencing adaptors and low-quality bases or reads (13). The preserved clean reads were then mapped to the hg19 human reference genome using Hisat2 v2.2.1 and STAR v2.7.8a (20, 21). Gene fusions were detected using Arriba v2.3.0 and CICERO v1.7.0, with suspected pathogenic fusions confirmed by PCR and Sanger sequencing (22, 23). The read numbers for each gene were counted using HTSeq tool v0.13.5 (24). The gene expression counts for Zhujiang Hospital cohort samples and the TARGET cohort samples were input into ALLSorts (28 Jul 2021) (25). ALLSorts B-ALL subtype classifier predicted 18 subtypes and 5 meta-subtypes.
The MLPA assay was performed for all samples using 2 kits: Salsa MLPA Probemix P335 ALL-IKZF1 (MRC-Holland, Amsterdam, The Netherlands) to characterize the copy number alterations of the IKZF1, PAX5, ETV6, RB1, BTG1, EBF1, CDKN2A, CDKN2B genes and PAR1 region (SHOX area, CRLF2, CSF2RA, IL3RA and P2RY8 genes) and Salsa MLPA Probemix P327 iAMP21-ERG (MRC-Holland) to identify the copy number alterations of the RUNX1 and ERG genes.
Based on the MLPA results, patients were classified into 3 groups: No IKZF1 deletion, IKZF1 deletion (IKZF1del only) and IKZF1plus. IKZF1plus was defined as IKZF1del plus PAX5, CDKN2A, CDKN2B and/or CRLF2 with ERG not deleted (26).
RQ-PCR analysis of CRLF2 expression was performed following the reverse transcription of RNA samples using a RevertAid First Strand cDNA Synthesis Kit (K1622) (Thermo Fisher). RQ-PCR was performed using an ABI 7500 Real-Time PCR System (Thermo Fisher). The housekeeping gene GUS was used as an internal control. The primers and probes used for RQ-PCR analysis are listed in Table 2. PCR was performed in 20 μL reactions consisting of 1× TaqMan™ Fast Advanced Master Mix (Thermo Fisher), 300 nM primer, 200 nM probe and 5 μg of complementary DNA. The thermal cycling conditions were 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 3 s and 60°C for 30 s. The ratio of CRLF2 was calculates using CLRF2 copies normalized to GUS copies.
MFC for assessing MRD was performed on whole BM specimens obtained at the specified time intervals using a standard stain-lyse-wash procedure. A total of 1×106 cells were stained per analysis tube, acquired on a Navios flow cytometer using Navios software (Beckman Coulter) and analyzed using Kaluza (Beckman Coulter) or FCS Express (De Novo Software).
A 2-tube 7-colour panel was run. Tube A contained CD58-FITC (cLone: AICD58, Beckman Coulter), CD34-PE (cLone: 581, Beckman Coulter), CD10-PE-Cy7 (cLone: HI10a, BD), CD19-APC (cLone: SJ25C1, BD), CD38-eFluor 450 (cLone: HB7, eBioscience), CD45-BV510 (cLone: H130, BD) and 7AAD Viability Dye (Beckman Coulter). Tube B contained CD66c-FITC (cLone: B6.2/CD66, BD), CD13-PE (cLone: L138, BD), CD33-PE (cLone: D3HL60.251, Beckman Coulter), CD34-PerCP-Cy5.5 (cLone: 581, Biolegend), CD10-PE-Cy7 (cLone: HI10a, BD), CD19-APC (cLone: J25C1, BD), and CD45-BV510 (cLone: H130, BD).
MRD was identified using both the leukemia-associated immunophenotype (LAIP) and different-from-normal (DFN) method. To distinguish residual tumor B cells from normal B-progenitor cells, we used B-lymphocyte lineage and stage differentiation associated antigens such as CDl9, CDl0, CD34 and CD45, as backbone antibodies. Tumor B cells exhibit strong expression of CDl0, CDl9 and CD34, low expression of CD45, cross-line expression of myeloid antigens such as CD66c, CDl3 and CD33, overexpression of CD58 and low expression of CD38.
The short-term efficacy assessment indicators included the results for the response to prednisone and the results for the 15th and 33rd days of BM flow cytometry minimal residual disease (FCM-MRD). Response to prednisone is an independent prognostic indicator of acute lymphocyte leukemia. Prednisone good response (PGR) is considered the absolute blast cell count of peripheral blood on the 8th day ≤ 1000/μL (27). This study uses the scrag-al-2016 risk-layered system and the results of the response to prednisone, with the 15th- and 33rd-day BM FCM-MRD less than 1% and the 33rd-day FCM-MRD less than 0.01% (28). Complete remission (CR, Blasts cells in bone marrow <5% and no measurable extramedullary leukemia) was defined according to the U.S. National Comprehensive Cancer Network (NCCN) guidelines on day 33 (29).
Statistical analyses were conducted using R version 3.6.2 and IBM SPSS statistics 25. Data are presented as the median and interquartile range (IQR). The Wilcoxon rank-sum test and Kruskal-Wallis test were applied to assess differences between 2 groups and more than 2 groups. Receiver operating characteristic curves (ROCs) and the Youden index were used to identify optimal cutoff values for gene expression (30). Fisher’s exact test was used to analyze the clinical baseline of patients with different CRLF2 expression levels. Multivariate Cox regression, Kaplan-Meier curves and the log-rank test were used to analyze OS and EFS via the ‘survival’ and ‘survminer’ packages in R. P<0.05 was considered significant for two-sided tests.
The data from Zhujiang Hospital cohort used in this study can be found in Supplementary Table 1. The datasets an analyzed in the current study are publicly available from the ALL dataset of the TARGET database (TARGET Phase II, phs000464) [https://portal.gdc.cancer.gov/projects], and the clinical data were downloaded from cBioPortal [https://www.cbioportal.org/study/clinicalData?id=all_phase2_target_2018_pub]. The data for the Zhujiang Hospital cohort are available upon reasonable request.
A total of 111 newly diagnosed pediatric B-ALL patients admitted to and treated in the Department of Pediatric Hematology, Zhujiang Hospital, Southern Medical University, China, from May 2020 to February 2022 were enrolled in the Zhujiang Hospital cohort. The last follow-up was conducted on August 25, 2023, with a median follow-up time of 28.6 months (95%CI: 27.6-32.1 months). There were 67 (60%) males and 44 (40%) females, with a median age of 4 years (range: 0-14 years), including 81 patients (73%) aged ≤6 years and 30 patients (27%) aged >6 years. On the basis of the South China Children’s Leukemia Group-ALL-2016 (SCCLG-ALL-2016) risk stratification (28), 13 patients (12%) were standard risk, 36 (32%) were intermediate risk, 61 (55%) were high risk, and 1 (1%) had an uncompleted risk assessment due to the abandonment of treatment. At the initial visit, for all 111 patients, the median white blood cell count was 13.33×109/L [interquartile range (IQR): 5.23-43.5×109/L], and 24 patients had a white blood cell count ≥50×109/L. Moreover, the median proportion of bone marrow blast cells was 86% (IQR: 76-93.25%) at the initial visit among the 109 patients with records (Table 1). Next, molecular typing of B-ALL was implemented using ALLSorts, with the following results: DUX (4, 4%), ETV6-RUNX1 (12, 10%), ETV6-RUNX1-like (2, 2%), high hyperdiploid (38, 33%), KMT2A group (9, 8%), low hypodiploid (1, 1%), MEF2D (4, 4%), NUTM1 (2, 2%), PAX5 P80R (1, 1%), PAX alt (5, 5%), Philadelphia chromosome positive (Ph+, 4, 4%), Ph-like (5, 5%), TCF3-PBX1 (11, 10%), ZNF384 (1, 1%), and unclassified (13, 12%). Six patients had the Ph-like subtype, carrying somatic mutations of KRAS (n=1), FLT3 (n=1), PAX5 (n=1), and PHF6 (n=1) (Supplementary Figure 1A) as well as CRLF2-associated fusion (n=2) and EBF1-PDGFRB fusion (n=1).
All the 111 treatment-naïve B-ALL patients in the Zhujiang Hospital cohort underwent RNA-seq using pre-treatment bone marrow samples, and fragments per kilobase of transcript per million (FPKM) were calculated through bioinformatics analysis. The expression level of the CRLF2 gene was mainly within the interval of FPKM<1, i.e., generally low in the bone marrow samples of the treatment-naïve B-ALL patients. Some patients also manifested very high CRLF2 expression, with a maximum FPKM of 75.1 (Figure 1A, left). Patients with high CRLF2 expression mainly had the following B-ALL subtypes: KMT2A group, Ph+, Ph-like and high hyperdiploid (Figure 1A, right).
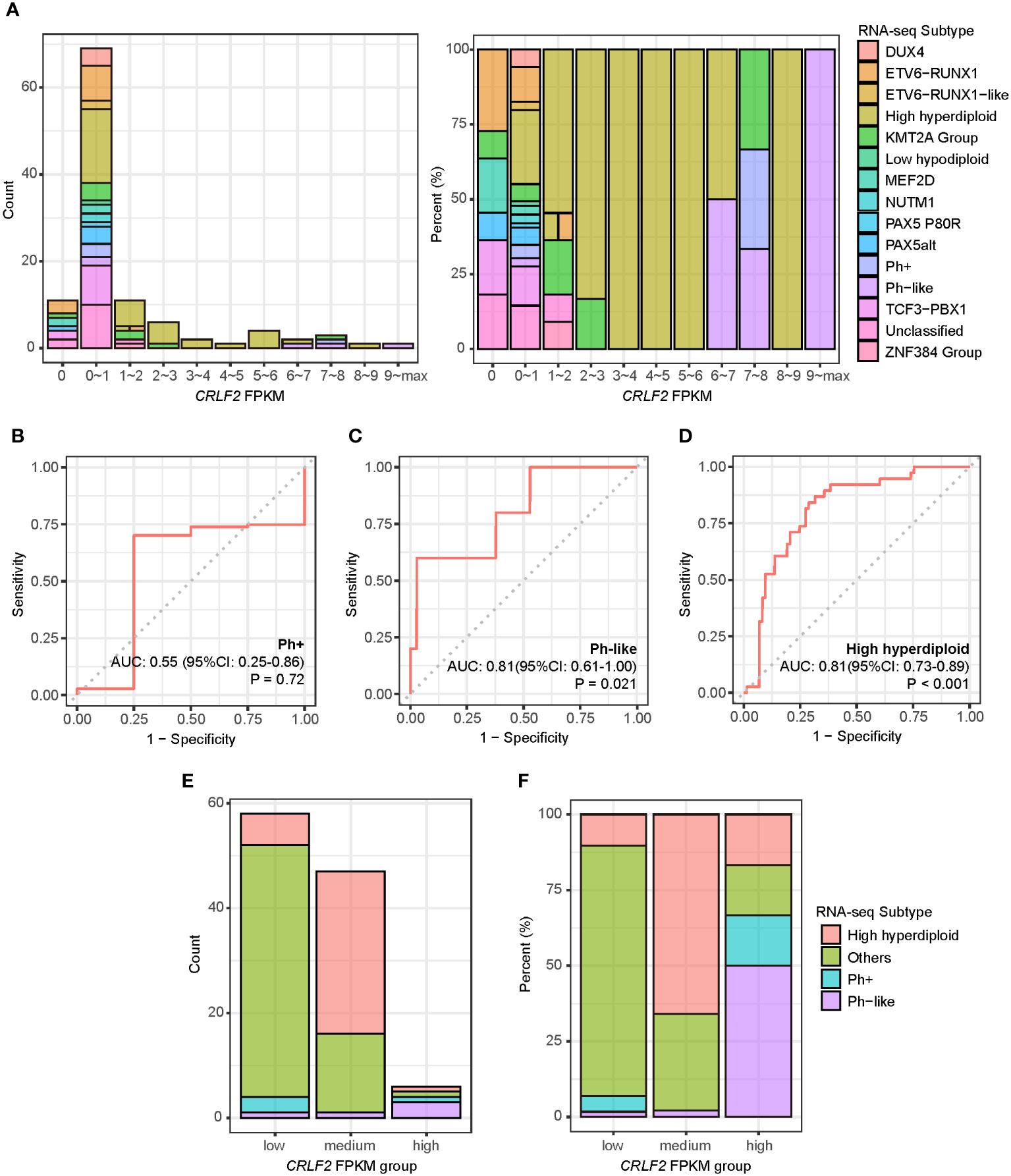
Figure 1 CRLF2 expression and B-ALL subtypes of pediatric B-ALL patients in the Zhujiang Hospital cohort. (A) CRLF2 expression in 111 B-ALL patients with RNA-seq B-ALL subtypes. (B–D) ROC curves for the correlation of CRLF2 expression with the Ph+, Ph-like and high hyperdiploid subtypes. (E, F) RNA-seq subtypes in CRLF2 FPKM groups. The Wilcoxon rank-sum test was applied to assess differences between 2 groups. The cutoff point between high/medium levels and medium/low levels were determined using the Youden index for CRLF2 expression with the Ph-like and high hyperdiploid subtypes.
Subsequently, the CRLF2 expression level was used to analyze the differences among the Ph+, Ph-like and high hyperdiploid subtypes. The expression level of the CRLF2 gene was not the same between the Ph+ and Ph-like subtypes despite similar expression profiles. In addition, the expression level of the CRLF2 gene in patients with the Ph-like and high hyperdiploid subtypes was significantly higher than that in other patients [Figures 1C, D, Ph-like: area under curve (AUC)=0.81, 95% confidence interval (CI): 0.611-1.000, P=0.021, high hyperdiploid: AUC=0.81, 95% CI: 0.728-0.894, p<0.001]. No correlation was found between the Ph+ subtype and CRLF2 expression level (Figure 1B, p=0.72). The Youden indexes of CRLF2 expression in predicting the Ph-like and high hyperdiploid subtypes were subsequently calculated (Figure 1E) (30), and the corresponding FPKM quantiles of CRLF2 expression were set as the cutoffs (Ph-like cutoff point=94%, high hyperdiploid cutoff point=52%). CRLF2 was assigned as a three-classification variable, namely CRLF2-high, CRLF2-medium and CRLF2-low (Figure 1E). Patients with CRLF2-high, CRLF2-medium and CRLF2-low accounted for 6% (Ph-like, n=3, high hyperdiploid, n=1, Ph+, n=1, KMT2A group, n=1), 42% (high hyperdiploid, n=31, Ph-like, n=1, other subtypes, n=15) and 52% (other subtypes, n=48, high hyperdiploid, n=6, Ph+, n=3, Ph-like, n=1), respectively, of all the patients (Figure 1F).
The clinical characteristics of the 3 groups of patients were analyzed. There were no significant differences in the incidence rates of concomitant testicular infiltration, central nervous system leukemia, intracranial hemorrhage, hepatosplenomegaly, and bone pain at the diagnosis of the disease in patients with different CRLF2 expression levels (Supplementary Table 1).
The proportion of bone marrow blast cells was not significantly different among the 3 groups of patients at diagnosis but was slightly higher in patients with CRLF2-high than in those with CRLF2-low (CRLF2-high vs. CRLF2-low, p=0.036, Kruskal-Wallis, p=0.06, Figure 2A). Similarly, the white blood cell count at initial visit was significantly higher in patients with CRLF2-high than in patients with CRLF2-low and CRLF2-medium (CRLF2-high vs. CRLF2-low, p=0.016, CRLF2-high vs. CRLF2-medium, p=0.016, Kruskal-Wallis, p= 0.042, Figure 2B).

Figure 2 Association of CRLF2 expression with blasts in BM, WBC, and flow cytometry MRD. (A) Proportion of bone marrow blast cells for patients with different CRLF2 expression levels. (B) Initial WBC of patients with different CRLF2 expression levels. (C) Flow cytometry MRD for different CRLF2 expression levels. The Wilcoxon rank-sum test and Kruskal-Wallis test were applied to assess differences between 2 groups and more than 2 groups.
The CR rate on 33rd day was significantly lower in patients with CRLF2-high (33%) compared to those with CRLF2-medium (80%, p = 0.03) and CRLF2-low (76%, p = 0.046, global p = 0.062, Figure S2). Furthermore, the MRD status of the patients at different treatment time points was dynamically monitored by means of flow cytometry. The time points for monitoring were set as the 15th day and 33rd day after the start of induction therapies as well as before the start of consolidation therapy. The proportion of tumor cells in flow cytometry MRD decreased rapidly during induction therapy in patients with CRLF2-medium, and MRD remained negative by the time of assessment before consolidation therapy (a total of 43 patients available for assessment via flow cytometry MRD<0.1%). However, the patients with CRLF2-high and CRLF2-low had varying degrees of residual MRD, and the proportion of positive patients before consolidation therapy was 33.3% (n=2) and 5.5% (n=3), respectively (Figure 2C).
To further validate the association between the CRLF2 expression level and poor prognosis, the publicly available TARGET cohort data were used as an externally independent validation cohort. The stratified cutoffs (quantiles) for the CRLF2 expression level obtained from the Zhujiang Hospital cohort were employed to stratify the FPKM data of CRLF2 in the TARGET cohort, followed by a survival analysis.
Because only pediatric patients aged ≤15 years were recruited for the Zhujiang Hospital cohort and the age range of patients in the TARGET cohort was 2-30 years, age was included as a stratification factor in the multivariate Cox regression analysis (Figures 3A, B). Both age and CRLF2 FPKM expression stratification served as independent prognostic factors for both event-free survival (EFS) and overall survival (OS) (EFS: C-index =0.61, p<0.001, OS: C-index =0.64, p=0.03). Age>15 years was a risk factor for both EFS and OS [EFS: hazard ratio (HR) =1.79, 95% CI: 1.03-3.10, p=0.037, OS: HR=2.74, 95% CI: 1.18-6.4, p=0.019]. However, CRLF2 expression stratification of CRLF2-medium was not a risk factor for EFS (HR=0.52, 95% CI: 0.35-0.78, p=0.002), and CRLF2-high was a distinct risk factor for OS (HR=4.33, 95% CI: 1.20-15.6, p=0.025).
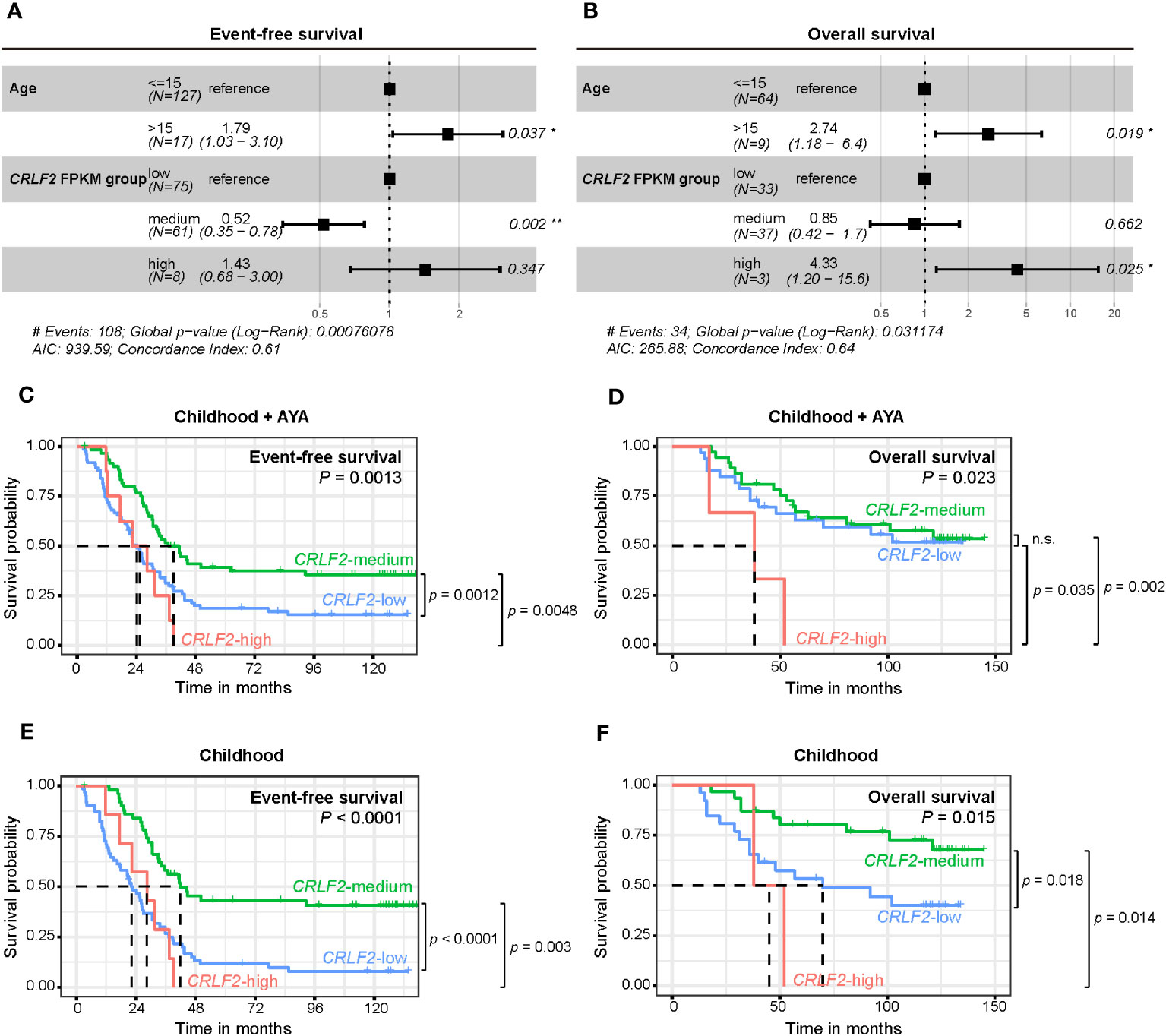
Figure 3 Survival analysis of pediatric B-ALL patients with CRLF expression in the TARGET cohort. (A, B) Effect of CRLF2 expression levels on event-free survival (EFS, A) and overall survival (OS, B) in the TARGET cohort. Estimates are based on multivariate Cox regression. (C, D) Kaplan-Meier plots of OS and EFS for patients with different CRLF2 expression levels in the TARGET cohort. (E, F) Kaplan-Meier plots of OS and EFS for pediatric patients with different CRLF2 expression levels in the TARGET cohort.
The effects of the CRLF2 expression level on EFS and OS were further validated when plotting K-M curves. Without considering the influence of age on prognosis, patients with CRLF2-high and CRLF2-low had notably shorter EFS than those with CRLF2-medium (CRLF2-high vs. CRLF2-medium, p=0.0048, CRLF2-low vs. CRLF2-medium, p=0.0012, global p=0.0013, Figure 3C). The patients with CRLF2-high manifested an overtly shorter OS than did those with CRLF2-low and CRLF2-medium (CRLF2-high vs. CRLF2-low, p=0.035; CRLF2-high vs. CRLF2-medium, p=0.002, global p=0.023, Figure 3D); there was no significant difference in OS between the patients with CRLF2-medium and CRLF2-low.
Then, patients in the TARGET B-ALL cohort were selected as a childhood group (age ≤ 15 years old). The age range for the children in the childhood group from the TARGET B-ALL cohort was consistent with that for the children in the Zhujiang Hospital cohort. In the childhood group, both EFS and OS were markedly superior for patients with CRLF2-medium, compared with those for patients with CRLF2-low and CRLF2-high (EFS: CRLF2-medium vs. CRLF2-high, p=0.003, CRLF2-medium vs. CRLF2-low, p<0.0001, global p<0.0001, Figure 3E, OS: CRLF2-medium vs. CRLF2-high, p=0.014, CRLF2-medium vs. CRLF2-low, p=0.018, global p=0.015, Figure 3F). The results are consistent with the MRD status observed in the Zhujiang Hospital cohort.
In our cohort, fusion genes were also detected in 58 out of the 111 patients (53.3%). The fusion genes detected in patients with varying CRLF2 gene expression levels are shown in Figure 4A. Although patients carried a wide range of fusion genes, CRLF2-related fusions were more prevalent in patients with CRLF2-high (2/6, 33.3%). In comparison, P2RY8-CRLF2 fusion was the predominant CRLF2-related fusion (6/7, 85.7%). Among the patients carrying the P2RY8-CRLF2 fusion, RQ-PCR was performed for 5 patients; the results are shown in Figure 4B. The FPKM of CRLF2 was consistent with the RQ-PCR results (CRLF2 ratio). The patient with the highest CRLF2 expression level, namely, P067, had the Ph-like subtype, had a flow cytometry MRD of 0.1% as of consolidation therapy, and failed to achieve negative flow cytometry MRD. Patients P009, P081 and P082 with CRLF2-medium all had the high hyperdiploid subtype, and flow cytometry MRD was negative on the 33rd day of induction therapy except for 1 patient who had an MRD of 0.1%; no residual tumor cells were detected in flow cytometry MRD in all 3 patients before consolidation therapy. Patient P085, with the lowest CRLF2 expression level, had the ETV6-RUNX1-like subtype and had negative flow cytometry MRD results on the 33rd day of induction therapy and thereafter. These results suggest that not all patients carrying the P2RY8-CRLF2 fusion were CRLF2-high. Additionally, patients with the high hyperdiploid subtype and P2RY8-CRLF2 fusion showed a better prognosis, as determined through flow cytometry MRD.

Figure 4 Association of CRLF2 expression and P2RY8-CRLF2 fusion. (A) Circos plot shows fusions in each CRLF2 expression level. (B) RNA-seq (FPKM) and RQ-PCR (ratio) of CRLF2 expression and dynamic changes in flow cytometry MRD for each P2RY8-CRLF2 positive patient (n = 5).
A total of 49 patients in the Zhujiang Hospital cohort underwent CRLF2 expression testing through RNA-seq and RQ-PCR, and relatively consistent RQ-PCR results for CRLF2 (CRLF2 ratio) were obtained (R=0.79, p<0.0001, Figure 5A). Moreover, the cutoff value for CRLF2-high in RQ-PCR was determined, and the point value corresponding to the Youden index was used as a cutoff point as well (AUC=0.988, 95% CI: 0.968-1.000, p<0.001, Youden index cutoff point =37.75%, sensitivity =100%, and specificity =97%, Figure 5B). Then, the CRLF2 ratios were arranged in descending order and classified by virtue of the RQ-PCR cutoff point, and 7 patients were classified as CRLF2-high (RQ-PCR data). There were 2 patients with the Ph-like subtype, 1 with the Ph+ subtype, 3 with the high hyperdiploid subtype, and 1 with the KMT2A subtype (Figure 5C). The 3 patients with the high hyperdiploid subtype had poor prognostic factors. Specifically, 1 patient had prednisone-poor response (PPR) on the 8th day of prednisone treatment, 1 had flow cytometry MRD results that were positive by the 15th day, and 1 carried mutations in genes (NRAS p.G13D, PTPN11 p.S502A) in the RTK-RAS pathway.
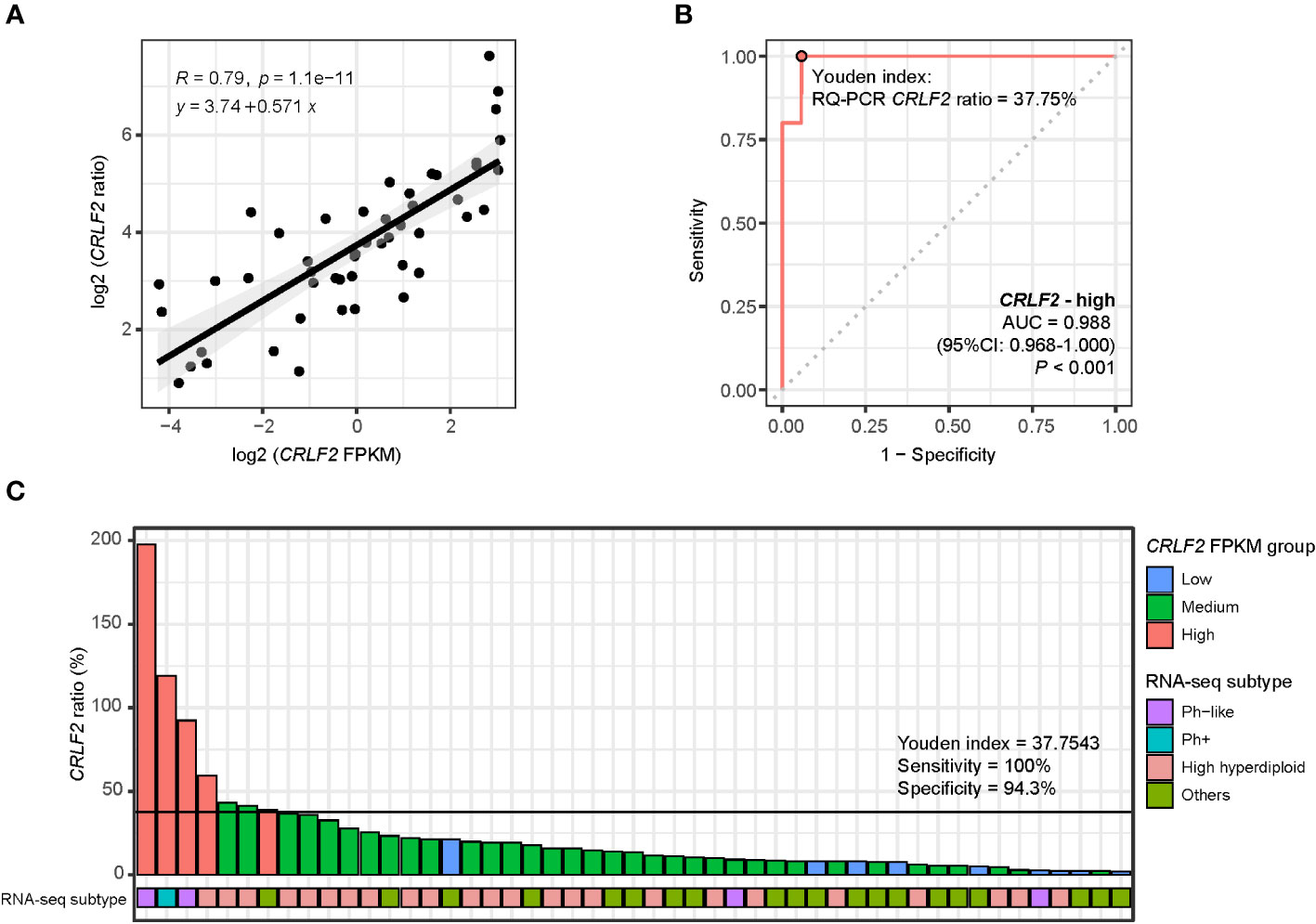
Figure 5 RQ-PCR for detection of CRLF2 expression. (A) Scatter diagram and regression line for CRLF2 expression measured by RQ-PCR (CRLF2_ratio) and RNA-seq (FPKM) (both in log 2 transformation). (B) ROC curves for the correlation of the RQ-PCR CRLF2 ratio with CRLF2-high (in RNA-seq). (C) RQ-PCR CRLF2 ratio and RNA-seq subtype for each patient (with RQ-PCR data, n = 49). The Wilcoxon rank-sum test and Kruskal-Wallis test were applied to assess differences between 2 groups and more than 2 groups. The cutoff points between CRLF2-high/-medium and CRLF2-medium/-low were determined using the Youden index of the RQ-PCR CRLF2 ratio with CRLF2-high and CRLF2-low (CRLF2 FPKM groups).
CRLF2 overexpression is involved in the pathogenesis and progression of B-ALL through the JAK/STAT signaling pathway (1–4). However, it is still unknown whether the prognosis of B-ALL patients can be predicted based on CRLF2 expression levels (5, 31, 32). The proportion of patients with CRLF2 overexpression varies substantially in previously published studies, and the proportion of patients with CRLF2 overexpression among unscreened patients ranges from a minimum of 4.7% to a maximum of 25% (2, 4–7), implying that CRLF2 overexpression needs to be defined by a more suitable cutoff value.
B-ALL can be categorized into different subtypes on the basis of genomic features and driver gene mutations, and such classification is of vital significance in risk stratification, treatment guidance and prognosis prediction. In this study, ALLSorts, defined by Gu et al., and RNA-seq analysis were used to identify 18 molecular subtypes (5, 31–33).
The exploration and analysis of the CRLF2 expression level and molecular types of B-ALL revealed that the molecular types were heterogeneous in patients with different CRLF2 expression levels. Patients with moderate upregulation of CRLF2 mostly had the high hyperdiploid subtype, while those with CRLF2 overexpression mainly had the Ph-like subtype. A close correlation between the Ph-like subtype and CRLF2 expression has been reported. The studies by Martínez-Anaya et al. and Harvey et al. reported that more than 50% of patients with CRLF2 overexpression have the Ph-like subtype (34, 35), a result that is consistent with the findings of this study. The high hyperdiploidy-associated upregulation of CRLF2 expression primarily manifested as CRLF2-medium in our cohort, where more than half of this subgroup had the high hyperdiploid subtype. Such upregulation may be associated with the amplification of the X/Y chromosome where the CRLF2 gene is located (Supplementary Figures 1B, C) (31, 36).
The majority of relevant studies have argued that CRLF2 overexpression is correlated with the poor prognosis of B-ALL patients, especially those with high-risk B-ALL (2, 3, 31, 34, 37). However, some studies have argued that CRLF2 overexpression has no impact on patient prognosis (5, 36). According to the baseline analysis of this cohort, the patients with CRLF2-high had a higher proportion of bone marrow blast cells and initial white blood cell counts. A high initial white blood cell count in childhood ALL is a recognized high-risk factor (38). Additionally, flow cytometry MRD decreased slowly or persistently positive during induction therapy in patients with CRLF2-high, indicating poor treatment response for such patients. The survival outcome for patients in the Zhujiang hospital cohort could not be analyzed due to the limited follow-up time. The publicly available childhood B-ALL data from the TARGET database were used to further validate the relationship between the CRLF2 expression level and prognosis. The CRLF2 expression level was defined in the TARGET cohort using the same methodology as that in the present cohort. Among B-ALL patients (pediatric + AYA), the patients with CRLF2-high had worse EFS and OS. Furthermore, pediatric patients with CRLF2-medium manifested a better prognosis, which is consistent with the better prognosis of pediatric patients with high hyperdiploid and X/Y chromosome amplification particularly (39–41). These results confirm the hypothesized relationship between CRLF2 expression level and prognosis in this study: There is heterogeneity in patients with elevated CRLF2 expression, that is, patients with CRLF2-high have a poor prognosis, and medium upregulation does not increase the prognostic risk.
CRLF2 rearrangement is the most studied class of genetic alteration leading to CRLF2 overexpression, mainly consisting of P2RY8-CRLF2 and IGH-CRLF2. The former alteration results from local deletion on pseudoautosomes, an event that is related to Down’s syndrome-associated pediatric ALL (DS-ALL) (42). It was previously reported that P2RY8-CRLF2 is mostly associated with a poor prognosis (2, 5, 37). Hence, the patients with P2RY8-CRLF2 fusion in our cohort were also analyzed. In our cohort, P2RY8-CRLF2 did not definitely lead to CRLF2-high. One patient with P2RY8-CRLF2 fusion and CRLF2-high had the Ph-like subtype, and the flow cytometry MRD decreased slowly and persistently, suggesting a possible poor prognosis. However, most of the P2RY8-CRLF2 patients with CRLF2-medium and CRLF2-low had the high hyperdiploid subtype and a better prognosis (3/4). It was speculated that P2RY8-CRLF2 may be a minor subclone in such patients (43). IGH-CRLF2 is another common type of CRLF2 rearrangement. Russell et al. identified an immunoglobulin heavy chain gene (IGH) translocated to CRLF2 at Xp22.3 or Yp11.3, forming the IGH-CRLF2 fusion, in approximately 5% of BCP-ALL patients (44). In our cohort, IGH-CRLF2 was found in only 1 (0.9%) patient. It has been reported that translocation patients (IGH-CRLF2) are older (median age: 16 years old), whereas deletion patients (P2RY8-CRLF2) are younger (median age: 4 years old) (2, 5, 37). In this study, the overall young age of our cohort (median age: 4 years old) may have led to fewer IGH-CRLF2 patients.
In the exploration of cutoffs for the 3 categories of CRLF2 expression, RNA-seq was adopted to detect the CRLF2 expression level (FPKM) and molecular types obtained from transcriptional profiling analysis. However, RNA-seq, as a research-based assay, is not practical to carry out in the clinic. In contrast, RQ-PCR is a more accessible assay in clinical practice. For detecting CRLF2 expression levels, RQ-PCR is faster, less expensive and more standardized than RNA-seq. The RNA-seq and RQ-PCR results were compared, and the detected CRLF2 expression levels were highly consistent. Furthermore, defining CRLF2-high using RQ-PCR CRLF2 ratio ≥37.75% as the cutoff yielded classification results that were consistent with those obtained using RNA-seq, thus laying a foundation for subsequent prospective studies on CRLF2 overexpression for stratified treatment.
By analyzing the data from 111 Chinese pediatric B-ALL patients in our center, the classification of the CRLF2 expression level into 3 categories more accurately described the prognosis and features of patients with various CRLF2 expression levels. In the external validation cohort (TARGET cohort), patients with CRLF2-high had a significantly poor prognosis. The three-classification system for the CRLF2 expression level detected by RQ-PCR in this study facilitates the rapid identification of patients with CRLF2-high in the clinic. For such patients, studies need to be conducted to further confirm and discover how to improve patient prognosis by intensifying chemotherapy or combining classic treatments with novel therapies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by the central ethics committee of Zhujiang Hospital, Southern Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
DL: Data curation, Methodology, Resources, Writing – original draft. KY: Formal Analysis, Software, Visualization, Writing – original draft. LYu: Data curation, Resources, Writing – review & editing. LH: Data curation, Resources, Writing – review & editing. XL: Data curation, Resources, Writing – review & editing. LW: Data curation, Resources, Writing – review & editing. XX: Formal Analysis, Software, Writing – original draft. JZ: Investigation, Writing – original draft. QZ: Conceptualization, Methodology, Supervision, Writing – review & editing. LYa: Conceptualization, Methodology, Supervision, Writing – review & editing.
The authors would like to thank all the participating patients and their families.
Author JZ is employed by Guangzhou KingMed Center for Clinical Lab. Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1256054/full#supplementary-material
Supplementary Figure 1 | Genomic alterations in pediatric B-ALL patients in the Zhujiang Hospital cohort. (A). Gene mutation profiles for individual patients. (B, C). CRLF2-related structural variants (B) and X chromosome structural variation (C) with different CRLF2 expression levels.
Supplementary Figure 2 | Survival and efficacy analysis in pediatric B-ALL patients in the Zhujiang Hospital cohort. CR rate in 33rd with different CRLF2 expression levels.
1. Cui L, Gao C, Wang CJ, Zhao XX, Li WJ, Li ZG, et al. Combined analysis of IKZF1 deletions and CRLF2 expression on prognostic impact in pediatric B-cell precursor acute lymphoblastic leukemia. Leuk Lymphoma (2021) 62(2):410–8. doi: 10.1080/10428194.2020.1832668
2. Dou H, Chen X, Huang Y, Su Y, Lu L, Yu J, et al. Prognostic significance of P2RY8-CRLF2 and CRLF2 overexpression may vary across risk subgroups of childhood B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer (2017) 56(2):135–46. doi: 10.1002/gcc.22421
3. Yano M, Imamura T, Asai D, Moriya-Saito A, Suenobu S, Hasegawa D, et al. An overall characterization of pediatric acute lymphoblastic leukemia with CRLF2 overexpression. Genes Chromosomes Cancer (2014) 53(10):815–23. doi: 10.1002/gcc.22190
4. Ensor HM, Schwab C, Russell LJ, Richards SM, Morrison H, Masic D, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood (2011) 117(7):2129–36. doi: 10.1182/blood-2010-07-297135
5. Palmi C, Vendramini E, Silvestri D, Longinotti G, Frison D, Cario G, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia (2012) 26(10):2245–53. doi: 10.1038/leu.2012.101
6. Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA (2010) 107(1):252–7. doi: 10.1073/pnas.0911726107
7. Maciel ALT, Barbosa TDC, Blunck CB, Wolch K, MaChado AAL, da Costa ES, et al. IKZF1 deletions associate with CRLF2 overexpression leading to a poor prognosis in B-cell precursor acute lymphoblastic leukaemia. Transl Oncol (2022) 15(1):101291. doi: 10.1016/j.tranon.2021.101291
8. Gu M, Jia Y, Xu M, Feng J, Tian Z, Ma X, et al. Effects of different aberrations in the CRLF2 gene on the biological characteristics and drug sensitivities of Nalm6 cells. Int J Lab Hematol (2021) 43(3):441–9. doi: 10.1111/ijlh.13419
9. Ge Z, Gu Y, Zhao G, Li J, Chen B, Han Q, et al. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget (2016) 7(31):49722–32. doi: 10.18632/oncotarget.10437
10. Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med (2011) 208(5):901–8. doi: 10.1084/jem.20110580
11. Pieters R, De Lorenzo P, Ancliffe P, Aversa LA, Brethon B, Biondi A, et al. Outcome of infants younger than 1 year with acute lymphoblastic leukemia treated with the interfant-06 protocol: results from an international phase III randomized study. J Clin Oncol (2019) 37(25):2246–56. doi: 10.1200/JCO.19.00261
12. Li XY, Li JQ, Luo XQ, Wu XD, Sun X, Xu HG, et al. Reduced intensity of early intensification does not increase the risk of relapse in children with standard risk acute lymphoblastic leukemia - a multi-centric clinical study of GD-2008-ALL protocol. BMC Cancer (2021) 21(1):59. doi: 10.1186/s12885-020-07752-x
13. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (2018) 34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
14. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
15. Picard. Available at: http://broadinstitute.github.io/picard.
16. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res (2010) 20(9):1297–303. doi: 10.1101/gr.107524.110
17. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res (2016) 44(11):e108. doi: 10.1093/nar/gkw227
18. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res (2010) 38(16):e164. doi: 10.1093/nar/gkq603
19. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform (2013) 14(2):178–92. doi: 10.1093/bib/bbs017
20. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol (2019) 37(8):907–15. doi: 10.1038/s41587-019-0201-4
21. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (2013) 29(1):15–21. doi: 10.1093/bioinformatics/bts635
22. Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res (2021) 31(3):448–60. doi: 10.1101/gr.257246.119
23. Tian L, Li Y, Edmonson MN, Zhou X, Newman S, McLeod C, et al. CICERO: a versatile method for detecting complex and diverse driver fusions using cancer RNA sequencing data. Genome Biol (2020) 21(1):126. doi: 10.1186/s13059-020-02043-x
24. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (2015) 31(2):166–9. doi: 10.1093/bioinformatics/btu638
25. Schmidt B, Brown LM, Ryland GL, Lonsdale A, Kosasih HJ, Ludlow LE, et al. ALLSorts: an RNA-Seq subtype classifier for B-cell acute lymphoblastic leukemia. Blood Adv (2022) 6(14):4093–7. doi: 10.1182/bloodadvances.2021005894
26. Stanulla M, Dagdan E, Zaliova M, Moricke A, Palmi C, Cazzaniga G, et al. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol (2018) 36(12):1240–9. doi: 10.1200/JCO.2017.74.3617
27. Gao J, Liu WJ. Prognostic value of the response to prednisone for children with acute lymphoblastic leukemia: a meta-analysis. Eur Rev Med Pharmacol Sci (2018) 22(22):7858–66. doi: 10.26355/eurrev_201811_16411
28. Xu LH, Geng X, Liao N, Yang LH, Mai HR, Wan WQ, et al. Prognostic significance of CNSL at diagnosis of childhood B-cell acute lymphoblastic leukemia: A report from the South China Children's Leukemia Group. Front Oncol (2022) 12:943761. doi: 10.3389/fonc.2022.943761
29. Brown PA, Shah B, Advani A, Aoun P, Boyer MW, Burke PW, et al. Acute lymphoblastic leukemia, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw (2021) 19(9):1079–109. doi: 10.6004/jnccn.2021.0042
30. Youden WJ. Index for rating diagnostic tests. Cancer (1950) 3(1):32–5. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3
31. Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood (2012) 119(15):3512–22. doi: 10.1182/blood-2011-11-394221
32. Hassan NM, Abdellateif MS, Radwan EM, Hameed SA, Desouky EDE, Kamel MM, et al. Prognostic significance of CRLF2 overexpression and JAK2 mutation in Egyptian pediatric patients with B-precursor acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk (2022) 22(6):e376–e85. doi: 10.1016/j.clml.2021.12.006
33. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet (2019) 51(2):296–307. doi: 10.1038/s41588-018-0315-5
34. Martinez-Anaya D, Moreno-Lorenzana D, Reyes-Leon A, Juarez-Figueroa U, Dean M, Aguilar-Hernandez MM, et al. Characterization of philadelphia-like pre-B acute lymphoblastic leukemia: experiences in mexican pediatric patients. Int J Mol Sci (2022) 23(17):9587. doi: 10.3390/ijms23179587
35. Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood (2010) 115(26):5312–21. doi: 10.1182/blood-2009-09-245944
36. Rasekh EO, Atef AM, Khalil M, Ebeid E, Madney Y, Hamdy N. Characterization of CRLF2 expression in pediatric B-cell precursor acute lymphoblastic leukemia. Clin Lab (2021) 67(1). doi: 10.7754/Clin.Lab.2020.200414
37. Chiaretti S, Brugnoletti F, Messina M, Paoloni F, Fedullo AL, Piciocchi A, et al. CRLF2 overexpression identifies an unfavourable subgroup of adult B-cell precursor acute lymphoblastic leukemia lacking recurrent genetic abnormalities. Leukemia Res (2016) 41:36–42. doi: 10.1016/j.leukres.2015.11.018
38. Schultz KR, Pullen DJ, Sather HN, Shuster JJ, Devidas M, Borowitz MJ, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG). Blood (2007) 109(3):926–35. doi: 10.1182/blood-2006-01-024729
39. Lee SHR, Antillon-Klussmann F, Pei D, Yang W, Roberts KG, Li Z, et al. Association of genetic ancestry with the molecular subtypes and prognosis of childhood acute lymphoblastic leukemia. JAMA Oncol (2022) 8(3):354–363. doi: 10.1001/jamaoncol.2021.6826
40. Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci USA (2018) 115(50):E11711–E20. doi: 10.1073/pnas.1814397115
41. Paulsson K, Lilljebjorn H, Biloglav A, Olsson L, Rissler M, Castor A, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet (2015) 47(6):672–6. doi: 10.1038/ng.3301
42. Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet (2009) 41(11):1243–6. doi: 10.1038/ng.469
43. Morak M, Attarbaschi A, Fischer S, Nassimbeni C, Grausenburger R, Bastelberger S, et al. Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2-harboring clones as main relapse-driving force in childhood ALL. Blood (2012) 120(26):5134–42. doi: 10.1182/blood-2012-07-443218
Keywords: B-ALL, CRLF2, Ph-like, pediatric, leukemia
Citation: Lin D, Yang K, Yu L, Huang L, Lai X, Wu L, Xia X, Zhang J, Zheng Q and Yang L (2023) Poor outcome of pediatric B-cell acute lymphoblastic leukemia associated with high level of CRLF2 gene expression in distinct molecular subtypes. Front. Oncol. 13:1256054. doi: 10.3389/fonc.2023.1256054
Received: 10 July 2023; Accepted: 23 October 2023;
Published: 07 November 2023.
Edited by:
Garrett Dancik, Eastern Connecticut State University, United StatesReviewed by:
Alexandra Kovach, Children’s Hospital of Los Angeles, United StatesCopyright © 2023 Lin, Yang, Yu, Huang, Lai, Wu, Xia, Zhang, Zheng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihua Yang, ZHJ5YW5nbGlodWFAMTYzLmNvbQ==; Qinlong Zheng, emhlbmdxbEBnb2Jyb2FkaGVhbHRoY2FyZS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.