
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 31 July 2023
Sec. Pharmacology of Anti-Cancer Drugs
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1226289
Evading apoptosis has been linked to tumor development and chemoresistance. One mechanism for this evasion is the overexpression of prosurvival B-cell lymphoma-2 (BCL-2) family proteins, which gives cancer cells a survival advantage. Mcl-1, a member of the BCL-2 family, is among the most frequently amplified genes in cancer. Targeting myeloid cell leukemia-1 (MCL-1) protein is a successful strategy to induce apoptosis and overcome tumor resistance to chemotherapy and targeted therapy. Various strategies to inhibit the antiapoptotic activity of MCL-1 protein, including transcription, translation, and the degradation of MCL-1 protein, have been tested. Neutralizing MCL-1’s function by targeting its interactions with other proteins via BCL-2 interacting mediator (BIM)S2A has been shown to be an equally effective approach. Encouraged by the design of venetoclax and its efficacy in chronic lymphocytic leukemia, scientists have developed other BCL-2 homology (BH3) mimetics—particularly MCL-1 inhibitors (MCL-1i)—that are currently in clinical trials for various cancers. While extensive reviews of MCL-1i are available, critical analyses focusing on the challenges of MCL-1i and their optimization are lacking. In this review, we discuss the current knowledge regarding clinically relevant MCL-1i and focus on predictive biomarkers of response, mechanisms of resistance, major issues associated with use of MCL-1i, and the future use of and maximization of the benefits from these agents.
The process of programmed cell death, apoptosis, is essential for cellular homeostasis, and its dysregulation has been incriminated in tumorigenesis and chemoresistance. Classically, apoptosis is triggered either intrinsically through the mitochondrial pathway or extrinsically through the death ligand-receptor pathway. The former is controlled by the BCL-2 family proteins that include antiapoptotic and proapoptotic molecules (1, 2). Myeloid cell leukemia-1 (MCL-1) is an anti-apoptotic protein that heterodimerizes with the proapoptotic members to prevent apoptotic cell death. MCL-1 is overexpressed in several cancers and mediates resistance to apoptosis triggered by chemotherapy and targeted therapy (3, 4). The development of MCL-1i inhibitors (MCL-1i) has been challenging owing to the shallow BCL-2 homology (BH3) binding groove of the MCL-1 protein (5). Using structure-based drug design, potent MCL-1i with in vivo activity have been developed and are now in clinical trials. Efforts are being made to identify biological markers of response or resistance to optimize and maximize the effect of the MCL-1i.
Apoptosis is a genetically programmed cell death that is important for development, tissue homeostasis, and immunity. Its dysregulation has been linked to various ailments in which uncontrolled apoptosis can contribute to neurodegenerative diseases, and decreased apoptosis can promote cancer and autoimmune diseases. Apoptosis can be triggered by 2 distinct routes: the mitochondrial and death receptor pathways. Both pathways eventually activate the effector caspases (caspases 3, 7, and 6) to initiate apoptosis (1, 2, 6).
The interactions between different BCL-2 family protein subgroups set the apoptotic threshold for the cells (7, 8). The increase in the BH3-only proapoptotic proteins (initiators) initiates apoptosis by binding to and inactivating the prosurvival proteins (guardians), thereby enabling activation of the proapoptotic effector proteins (BAK, BAX). BAX and BAK homo-oligomerize to disrupt the outer mitochondrial membrane, leading to the release of cytochrome c and second mitochondria-derived activator of caspase (SMAC) from the mitochondria. Cytochrome c activates caspase 9 on the scaffold protein apoptotic protease-activating factor 1 (APAF1), whereas SMAC blocks the caspase inhibitor X-linked inhibitor of apoptosis protein (XIAP) (9, 10). This eventually leads to caspase-mediated cleavage of cellular proteins and the initiation of cell death.
The BCL-2 family proteins share 1 or more of the 4 characteristic BH domains (designated as BH1, BH2, BH3, and BH4) (11). A common general structure in the BCL-2 family proteins is a central hydrophobic α-helix surrounded by amphipathic α-helices (12). These α-helices form a tertiary structure with a hydrophobic BH3 domain–binding groove that can bind to the BH3 domains of other family members. C- terminus transmembrane domains help some members to localize to the mitochondria. BCL-2 proteins can be classified into antiapoptotic proteins (BCL-2, MCL-1, BCL-extra-large [BCL-xL], BFL-1/BCL-2–related protein A1 [BCL-2A1], BCL-W, BCL-B), multidomain proapoptotic executioner proteins (BAX, BAK, and BOK) and BH3-only proapoptotic proteins (BIM, BAD, PUMA, Noxa, HRK, and BMF) (13). BH3-only proteins show specificity towards targeting their prosurvival partners. While BIM, PUMA, and BID act as nonselective general binders to prosurvival proteins, BAD specifically binds BCL-2, BCL-xL, and BCL-W. On the other hand, Noxa selectively binds to MCL-1 and BCL-2A1 (14–16).
In 1993, Kozopas et al. (17) identified Mcl-1 while exploring the genes that are induced by phorbol 12-myristate 13-acetate. The Mcl-1 gene is homologous to the BCL-2 gene. It is located at 1q21.2 with a promotor region that has several transcription factors binding sites; these include, but are not limited to, STAT, cAMP response elements, and nuclear factor kappa B binding sites. Mcl-1 is among the most amplified genes in cancer. A subset of multiple myeloma patients (40%) had amplification or gain of Mcl-1 gene (1q21) (18), with significantly shorter progression-free survival and lower overall survival (19). When 3000 samples from 26 different cancers were profiled, the Mcl-1 and BCL-2A1 genes were among the most amplified genes (3). Amplification was relatively higher in breast and non-small cell lung cancer (NSCLC) than in the other types of cancer and was correlated with higher amounts of Mcl-1 messenger RNA (mRNA) and unfavorable overall survival in patients (20).
The Mcl-1 gene encodes the full-length MCL-1 protein (MCL-1L, isoform 1) with antiapoptotic function. The alternatively spliced, shorter gene products (MCL-1S, isoform 2 and isoform 3) have a proapoptotic function, as they bind and inactivate MCL-1L (21–23). Switching from antiapoptotic to proapoptotic MCL-1 protein by controlling MCL-1 splicing appears to be a promising strategy in cancer therapy (24). Accordingly, splicing factor 3B subunit 1 (SF3B1) inhibitors have been recently explored as sensitizers that can switch on alternative splicing to generate proapoptotic MCL-1S isoforms (25). The MCL-1 transcript has adenylate-uridylate–rich elements that control the rapid turnover and short half-life of Mcl-1 mRNA (26).
MCL-1 protein is a unique member of the BCL-2 family of proteins. It is larger than its other prosurvival relatives (350 amino acid residues [37 kd] versus 239 residues for BCL-2 and 233 residues for BCL-xL). The C-terminus of MCL-1 protein (170-350 aa) has a transmembrane domain that is important for the mitochondrial localization of MCL-1 protein (27) (Figure 1). It also harbors 4 BH domains that regulate MCL-1’s interaction with other proteins (28). The BH3 domain of MCL-1 has a binding groove that interacts with other BH3-domain proteins (Noxa, BIM, BAK). This binding groove has four pockets (P1-P4) that binds to hydrophobic side chains (H1-H4) of the proapoptotic proteins (5, 29). In addition, the BH3 domain harbors the QRN motif that controls MCL-1 ubiquitination and degradation. The BH1 and BH3 domains also interact with KU70/KU80 dimers to repair DNA. The N-terminus of MCL-1 (1-170 residues) is long and unstructured, and it is absent in other BCL-2–related survival proteins. The N-terminus is rich in multiple proline (P), glutamate (E), serine (S), and threonine (T) residues (PEST regions) that regulate MCL-1’s stability and function (17) and result in rapid turnover and are characteristic of short-half-life proteins like MCL-1. These sequences also contain mitochondrial targeting signals.
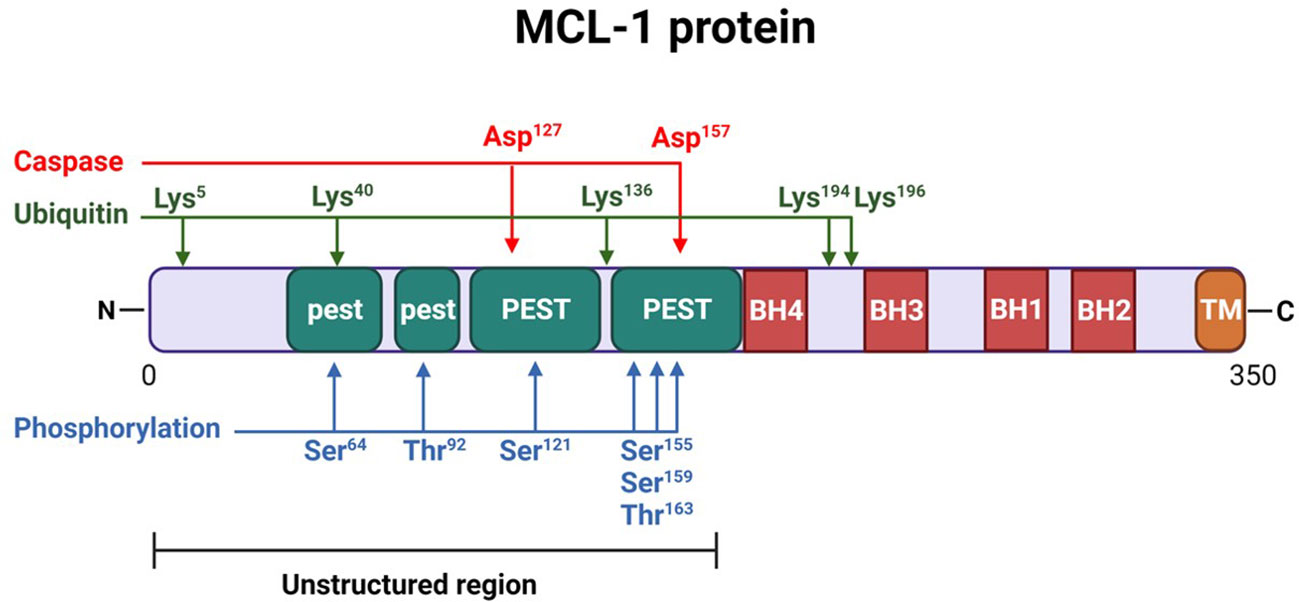
Figure 1 Structure of myeloid cell leukemia-1 (MCL-1) protein. MCL-1 protein consists of 350 amino acids, highlighting several important post-translational modification sites. Two caspase cleavage sites (aspartic acid [Asp] 127 and 157), 5 lysine [Lys] residues (5, 40, 136, 194, and 196) for ubiquitination, and 6 phosphorylation sites (serine [Ser] 64, threonine [Thr] 92, Ser 121, 155, and 159, and Thr 163) are indicated. The N-terminus of the protein is largely unstructured and includes 4 proline, glutamate, serine, and threonine (PEST) regions (2 major and 2 minor, labeled as “PEST” and “pest,” respectively). Four BCL-2 homology (BH) domains (BH1-BH4) are located near the C-terminus, which also contains the transmembrane (TM) domain necessary for mitochondrial localization.
The regulation of MCL-1 has been studied extensively (30–33), and its expression is controlled at the transcriptional, post-transcriptional, translational, and post-translational levels. Several modulators regulate MCL-1 protein levels in response to both internal and external stimuli such as cytokines and growth factors; these modulators induce signal transduction and the activation of transcription factors (34), endoplasmic reticulum stress (35), hypoxia (36), and microRNAs (37). Several post-translational modifications—including phosphorylation mediated by c-Jun N-terminal kinase, glycogen synthase kinase 3β (GSK-3β), and extracellular signal-regulated kinase (ERK)-1; ubiquitination (Mule, SCFβ-TrCP, SCFFbw7, APC/CCdc20, and Trim17 E3 ubiquitin ligases) and deubiquitination (USP9x); and caspase-mediated cleavage (38)—regulate the stability and functional activity of MCL-1. In addition, the interaction of MCL-1 protein with other BH3-only proteins can affect the stability and proteasomal degradation of MCL-1 protein. While BIM and PUMA binding leads to the stabilization of MCL-1 protein, Noxa binding enhances the degradation of MCL-1 protein (39, 40). MCL-1 is widely expressed in human tissue, including the bone marrow, lymph nodes, spleen, gall bladder, appendix, and heart (21, 22, 41, 42). In summary, MCL-1 is an early-response gene, and the presence of adenylate-uridylate–rich elements in transcripts and PEST domains in proteins result in fast transcript and protein turnover.
MCL-1 protein has antiapoptotic function that is important for cell viability. It inhibits apoptosis by binding and sequestering multidomain BH effector proteins (e.g., BAK, BAX), thus preventing their activation and mitochondrial outer membrane permeabilization. As mentioned before, MCL-1 protein binds preferentially to BAK and Noxa, while BIM and PUMA bind to all prosurvival proteins (43). MCL-1 is located in the nucleus, where it controls cell cycle progression and helps with DNA repair, and in the mitochondria, where it regulates other functions. Besides sequestering BAK, MCL-1 helps mitochondrial fragmentation by recruiting Drp1 and mitochondrial fusion via the stabilization of OPA1 in the inner mitochondrial membrane (44). MCL-1 also supports mitochondrial metabolic activities, it directly engages very-long-chain acyl-CoA dehydrogenase, preventing its stress induced excessive activity (45). It also promotes oxidative phosphorylation through uncharacterized mechanisms (46). MCL-1 inhibits beclin-1, and thus it can regulate autophagy. Its effect on autophagy depends on the cellular context. It also inhibits mitophagy through parkin/pink1, independently of beclin-1 (47). The role of MCL-1 in mitophagy and mitochondrial function may allow MCL-1 to be used therapeutically to target neurodegenerative diseases, including Alzheimer’s disease (48, 49). MCL-1 also has a role in cell senescence. In 2015, Demelash et al. (50) identified a loop domain responsible for inhibiting cellular senescence induced by chemotherapy, thus providing resistance to cancer therapy. MCL-1 also has an established role in the repair of DNA double-strand breaks (51). Collectively, these findings suggest a multidimensional role for MCL-1 protein.
As mentioned earlier, Mcl-1 gene amplification was highest among many tumors. Similarly, MCL-1 protein has been implicated in both tumorigenesis and chemotherapeutic resistance (4, 52). Consistent with this statement, it has been shown that, in xenograft models, knockdown of MCL-1 decreased the proliferation rate of cancer cells to a greater degree than that seen in controls (3). Conversely, increased incidence of B-cell lymphoma was noticed in transgenic mice overexpressing MCL-1 (53).
MCL-1 overexpression has been observed in several hematological and solid tumors (54–59). In clinical studies, multiple myeloma (MM) patients with high MCL-1 level had shorter event free survival (60). Similarly, MM patients overexpressing USP9x, which leads to increased MCL-1 stability, were shown to have a poor prognosis and can promote tumor survival (61). It has been shown that high levels of MCL-1 expression are required for B-lymphoma cell survival (62) and correlate with high-grade lymphoma (63), suggesting an association between high levels of MCL-1 expression and progressive disease (64). Interestingly, in glioblastoma, somatic mutations in the PEST region of MCL-1 (D155G, D155H, and L174S) were shown to stabilize MCL-1; these mutations may participate in gliomagenesis. Indeed, overexpressing these mutant plasmids in glioma cells accelerated the growth of glioma cells compared to wild type (WT) MCL-1 cells (65).
MCL-1 overexpression has also been implicated in resistance to both targeted therapy (such as venetoclax and navitoclax) (66–68) and conventional chemotherapy, including taxol, cisplatin, erlotinib, and cytarabine (69–71). In 2022, Zhang et al. (72) showed that there was selection for RAS-mutant clones in patients with acute myelogenous leukemia (AML) treated with venetoclax. These clones mediated resistance to venetoclax through MCL-1 upregulation, and cells were resensitized to the drug through the inhibition of MCL-1. Besides, concomitant use of MCL-1 inhibitors and venetoclax was better than other combination regimens in venetoclax resistant models (73). The overexpression of MCL-1 is related to cisplatin resistance (74). Depleting MCL-1 has reversed resistance to cisplatin and doxorubicin in osteosarcoma cell lines in vitro and xenograft tumors in vivo (75). MCL-1 amplification has been found in subsets of wild-type fibroblast growth factor receptor urothelial cancer, and its degradation by erdafitinib synergized BCL-xL/BCL-2 inhibitors (76).
Also, it has been shown that the knockdown of USP9X and expression of FBW7 in FBW7-deficient cells leads to increased MCL-1 protein turnover and sensitizes cells to ABT-737, suggesting the role of MCL-1 in the resistance to BCL-2/BCL-xl inhibitors (61, 77). MCL-1 inhibits chemotherapy induced senescence, thus mediating resistance to cancer therapy (50). MCL-1 is upregulated in senescent tumor cells and in cells that express low levels of BCL-2. While the BCL-2 inhibitor navitoclax successfully reduced metastases in mice with tumors, the MCL-1 inhibitor S63845 completely eradicated both senescent tumor cells and metastases (78). The direct inhibition of MCL-1 protein by AZD5991 or indirectly by CDK9 inhibitor (AZD4573), can overcome venetoclax resistance in AML cell lines and PDXs models (79). Besides, downregulating MCL-1 using PIK-75, a dual PI3K and CDK9 inhibitor, overcome venetoclax resistance in MCL cell lines (80).
MCL-1 protein can be targeted indirectly through modulating Mcl-1 gene transcription and translation or directly through inhibiting the functional interaction of MCL-1 with BAK (Figure 2). The cyclin-dependent kinases (CDKs) 7 and 9 play pivotal roles in the transcription of genes. MCL-1 protein expression has been modulated by inhibiting transcription using CDK 7/9 inhibitors such as flavopiridol, roscovitine, dinaciclib, and SNS-032 (81, 82).
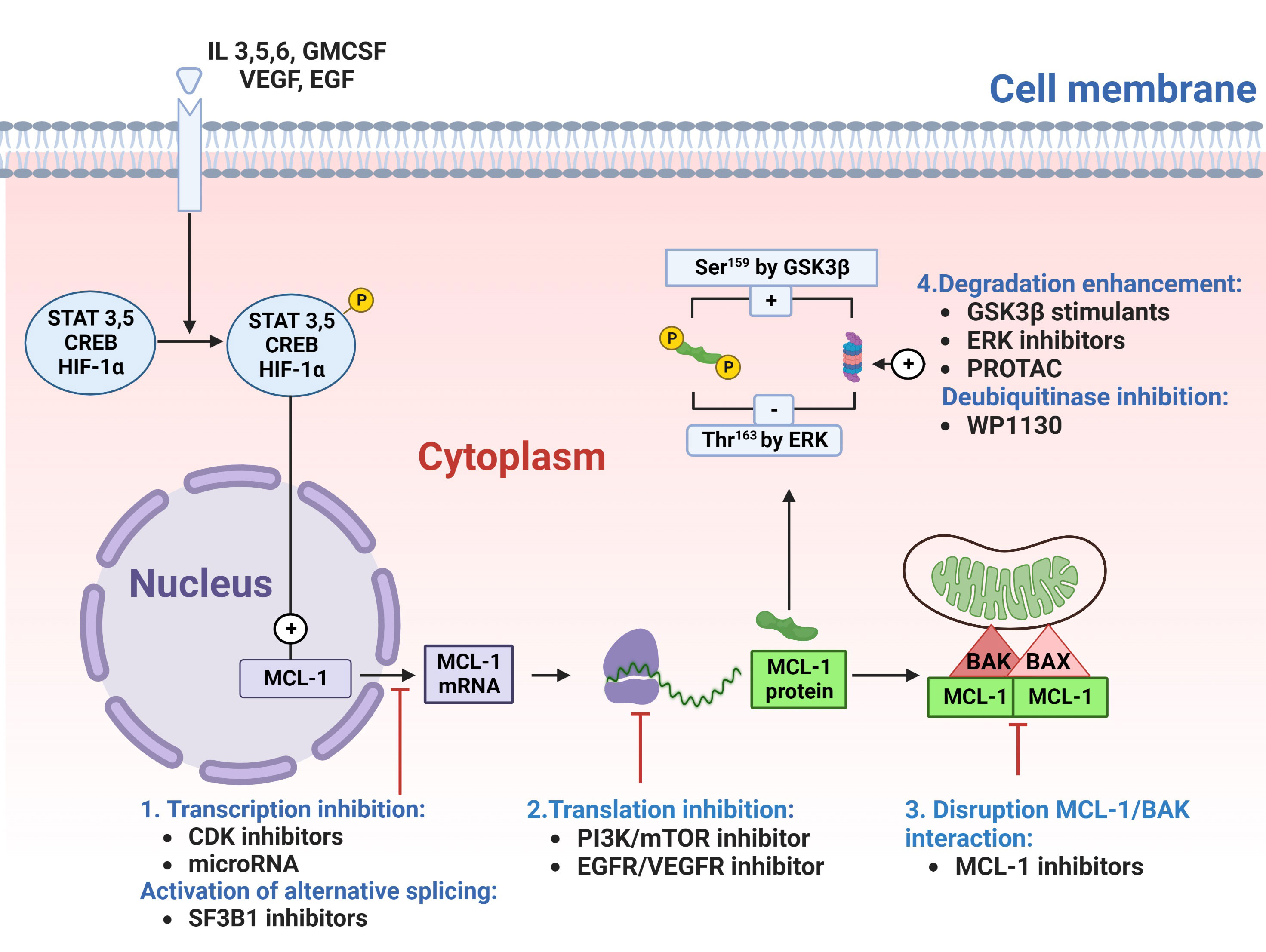
Figure 2 Targeting myeloid cell leukemia-1 (MCL-1) protein. MCL-1 protein can be targeted either directly, by disrupting MCL-1 interaction with proapoptotic proteins (#3), or indirectly by targeting MCL-1 protein at stages of transcription(#1), translation(#2) or degradation(#4). Cyclin-dependent kinase (CDK)7/9 inhibitors or microRNA can block the transcription of MCL-1 messenger RNA (mRNA). SF3B1 inhibitors can generate a proapoptotic MCL-1S isoform by switching on alternative splicing. Once transcribed, MCL-1 mRNA moves to the cytoplasm for translation into MCL-1 protein (depicted in green). This translation process can be prevented with phosphatidylinositol-3 kinase (PI3K)/mammalian target of rapamycin (mTOR) inhibitors or epidermal growth factor receptor (EGFR)/vascular endothelial growth factor receptor (VEGFR) inhibitors. After translation, MCL-1 protein binds and deactivates the BAK/BAX complex (depicted in red/pink) in the outer mitochondrial membrane. This binding can be disrupted with direct MCL-1 inhibitors), leading to BAX/BAK dissociation and apoptosis. Additionally, the proteasomal degradation of MCL-1 can be enhanced using glycogen synthase kinase 3β (GSK3β) stimulants, extracellular signal-regulated kinase (ERK) inhibitors, the proteolysis-targeting chimera (PROTAC), or deubiquitinating enzymes (DUB) inhibitors (e.g., WP1130), thereby increasing MCL-1 polyubiquitination and subsequent degradation. CREB, cAMP response element-binding protein; GMCSF, Granulocyte-macrophage colony-stimulating factor; HIF-1α, Hypoxia-inducible factor 1-alpha; IL, interleukin; Ser, serine; STAT, signal transducer and activator of transcription; Thr, threonine.
Similar to MCL-1 transcription inhibition, MCL-1 protein synthesis is blocked by inhibitors of protein translation such as omacetaxine. Omacetaxine binds to A-site cleft of ribosomes and is currently approved for the treatment of refractory chronic myelogenous leukemia. Its clinical success in producing cytogenetic and hematologic responses is based on its ability to decrease the oncoproteins Bcr-Abl, MCL-1, and c-Myc (83).
Other inhibitors of protein translation include the PI3K/mTOR inhibitors (BEZ235 and AZD8055), EGFR/VEGFR inhibitors (BAY43-9006), elF4F inhibitors (Silvestrol) or dephosphorylation of eukaryotic initiation factor 4G (benzyl isothiocyanate) (84–87). Also, MCL-1 protein can be targeted for proteasomal degradation by stimulating the GSK3β enzyme (with arsenic trioxide or bufalin) (88, 89), inhibiting the MEK/ERK pathway (with trametinib) (90), treating with deubiquitinase inhibitor WP1130 (91), or inducing selective intracellular proteolysis of MCL-1 protein via the proteolysis-targeting chimera (PROTAC) strategy (92). MCL-1 can also be targeted for degradation by upregulating Noxa. Alternatively, MCL-1 interaction can be targeted by the allosteric inhibition of MCL-1 protein (93) and direct inhibition of the MCL-1 protein-BH3 domain interaction (Figure 2).
Lee et al. (94) were the first researchers to target MCL-1 protein-protein interaction and to show that MCL-1 neutralization is an effective approach to MCL-1 targeting. This was an important observation for the development of small-molecule drugs directly targeting MCL-1.
Since then, researchers have focused on developing specific and potent MCL-1i. This work required overcoming 3 major challenges, that delayed the development of direct inhibitors of MCL-1. First, MCL-1’s shallow and relatively inflexible binding groove has delayed the development of high affinity MCL-1i. Besides, the structure of the BH3-binding grooves is without topological features and is very similar to various proteins of the prosurvival BCL-2 family, making it challenging to achieve selectivity within the family (95–97). Second, there is a lack of specificity to MCL-1 binding; because of this, early MCL-1i (gossypol (98), apogossypolone (99), antimycin A (100), obatoclax (101), and TW-37 (102)) were neither selective nor potent (103). Third, poor pharmacokinetic profiles and limited cell membrane permeability (104) were primary factors excluding MCL-1i from clinical use.
To overcome these challenges, researchers have tried to identify hot spots in MCL-1 protein that are critical for the stable binding of BH3 mimetics to the MCL-1 binding groove. Using nuclear magnetic resonance, X-ray crystallography, and alanine mutagenesis studies, researchers identified 4 hydrophobic pockets (P1-P4) and arginine 263 (Arg263) residue in the MCL-1 BH3 groove as hot spots required for peptide binding (105). Further studies revealed that, compared to BCL-2 and BCL-xL, the P2 and P3 pockets of MCL-1 have the most potential to bind MCL-1–binding peptides (106, 107). The P2 hydrophobic groove is relatively larger than the P3 hydrophobic groove and thus can accommodate ligands with larger structure moiety. Occupying the P3 and P4 pockets could further improve the binding affinity to MCL-1 (108). Additionally, the Arg263 residue was found to be an important hot spot that forms a hydrogen bond (salt bridge) with MCL-1i. Cocrystal structure analysis proved that this salt bridge formation was essential for MCL-1i’s efficacy; thus indole moiety may provide structural privilege for MCL-1i (109–111).
The direct, specific inhibitors of MCL-1’s protein-protein interactions have a similar structure to that of the BH3-only protein motifs and bind to MCL-1’s BH3 hydrophobic groove (112). Based on their structures, these inhibitors can be categorized into different types, including peptide inhibitors, marinopyrrole derivatives, gossypol derivatives, quinoline derivatives, S1 derivatives, and indole derivatives (113). The use of these inhibitors in MCL-1–dependent cell lines disrupted MCL-1:BAK interaction, thus paving the way for proapoptotic BH3 proteins (i.e. BIM) to bind BAK and initiate the intrinsic cell death pathway (5, 114, 115).
Since 2016, as many as 6 MCL-1i, including AMG-176 (114), AMG-397 (116), S64315 (5), AZD5991 (115), ABBV-467, and PRT1419, have been tested in the clinic. S63845 was the first MCL-1i reported to have potent in vivo activity and to enter clinical trials (NCT02992483) (5). Several clinical trials were initiated with AZD5991, AMG-176, AMG-397, and S64315 (Table 1), mostly in hematological malignancies owing to their preferential dependence on MCL-1 for growth. MCL-1i were tested in phase 1 trials as single agents or in combination with venetoclax or carfilzomib.
A-1210477 (in the indole-2-carboxylic acid group), developed by AbbVie, is the first selective high affinity inhibitor of MCL-1. It binds to MCL-1 with a Ki value of 0.454 nM, and has been shown to disrupt the MCL-1/BIM interactions that induce apoptosis only in cell lines that show dependency on MCL-1 protein (H929, H2110, and H23) (117). Moreover, it synergized with navitoclax in various cancer cell lines (118) and overcame resistance to navitoclax (119). Also, A-1210477 has been shown to be synergistic when combined with an inhibitor of Hedgehog signaling (120) (i.e., bromodomain extra-terminal protein inhibitor [ABBV-075] in CD34+ AML cells (121) or ABT-263 [navitoclax]) (122). MCL-1 has been shown to bind to and inhibit the transcriptional function of the tumor suppressor p73 through its BH3 domain. Consequently, A-1210477 has been used to induce p73 and thereby activate DNA double-strand break repair target gene expression, promoting cell cycle arrest and apoptosis (123). Because of its low potency, A1210477 remains a tool compound.
VU661013 is a potent MCL-1i that binds to MCL-1 with high affinity and was developed using a fragment-based screening and structure-based design to optimize a previously reported MCL-1i. VU661013 inhibited MCL-1 (Ki 97 ± 30 pmol/L) without significant inhibition of BCL-xL or BCL-2. Accordingly, VU661013 inhibited growth in AML cell lines, except those dependent on BCL-2 (124).
Venetoclax enhanced the cellular cytotoxicity of VU661013, even in AML cell lines and AML blasts with innate and/or acquired resistance to VU661013, when the cell lines or blasts were treated with venetoclax before or venetoclax and low dose cytarabine after VU661013 treatment. Further, an MV-4-11 cell line–based xenograft mouse model for AML showed a dose-dependent decrease in CD45+ MV-4-11 cells in the blood, bone marrow, and spleen; no evidence of toxicity in non-target tissues, and increased survival after VU661013 treatment (66). In addition to being effective in liquid tumors, the drug was effective in estrogen receptor–positive breast cancer cells in vitro and in a MCF7 xenograft model (125).
AZD5991 is a macrocyclic high affinity MCL-1i (Ki = 0.2 nM) developed by AstraZeneca from indole-2-carboxylic acids (115). It disrupts BAK/MCL-1 interaction to induce apoptosis, with preferential activities in hematological cancer cell lines and some NSCLC and breast cancer cell lines as well as primary myeloma cells. Besides, AZD5991 showed potent in vitro activity against primary leukemia cells and in vivo antitumor activity in various AML, non-Hodgkin lymphoma, and MM mouse xenograft models. The activity of AZD5991 against MM and AML in murine models was substantial and was further enhanced with bortezomib or venetoclax, respectively (115).
AZD5991 was also effective in solid tumor cells and demonstrated promise in colorectal carcinoma cell lines when combined with the multi kinase inhibitor, regorafenib (126). In 2019, Koch et al. (127) reported AZD5991’s activity in T-cell lymphoma cell lines and patient-derived xenograft models in vivo. In the models dependent on MCL-1, AZD5991 improved survival alone and in combination with cyclophosphamide, doxorubicin hydrochloride, vincristine sulfate, and prednisone (CHOP) chemotherapy. When used in acquired BRAFi + MEKi resistance model, AZD5991 enhanced the efficacy of ERK1/2 inhibitor (128). Interestingly, AZD5991 synergized with CB-839, a glutaminase inhibitor, to induce apoptosis and inhibit proliferation of CLL cell lines (129). The safety and clinical activities of AZD5991 are being investigated in phase 1 clinical trial (NCT03218683).
Caenpeel et al. (114) reported the discovery of AMG-176, a nonindole acid MCL-1i, which was identified using a “structure-based design and conformational restriction”. AMG-176 has a high affinity for MCL-1 protein (Ki = 0.06 nM) and a minimal binding affinity to BCL-2 and BCL-xL. In vitro studies with AM-8621 (a structural analogue of AMG-176) showed that it disrupted MCL-1/BAK interactions, induced BAX/BAK dependent apoptosis, and increased MCL-1 stability in various hematological malignancy cell lines. In vivo sensitivity was also observed in a subcutaneous xenograft model of MM and an orthotopic model of AML (114). AMG-176 treatment decreased peripheral blood and bone marrow cells including B cells, monocytes, neutrophils, eosinophils, basophils, and reticulocytes, which can be used as a pharmacodynamic endpoints to assess treatments (114).
In various models, AMG-176 has shown synergism in combination with different drugs (114). Similar to AZD5991, AMG-176 was also synergistic with carfilzomib (115). Moreover, the combination of venetoclax and AMG-176 was synergistic in AML orthotopic model and in ex vivo primary AML patient samples (114) and CLL primary lymphocytes (130). While AMG-176 showed robust antitumor activity in liquid neoplasms, it only showed modest antitumor activity in a few solid tumor cell lines (114).
The safety and clinical activity of AMG-176 is being evaluated in patients with relapsed or refractory MM or AML (NCT02675452). In addition, intervention studies on AMG-176, azacytidine, and itraconazole (NCT02675452) in patients with relapsed/refractory myeloma or AML are ongoing (131). A clinical trial is also evaluating AMG-397 (NCT03465540), the first MCL-1i given orally in the clinic. The use of AMG-397 for the treatment of various blood malignancies has been placed on clinical hold recently following incidences of cardiac toxicity (132).
S63845 is the first selective MCL-1i reported to have in vivo activity (5); it induced apoptosis in various hematological cell lines (MM, lymphoma, chronic myelogenous leukemia, AML, and T-cell acute lymphoblastic leukemia [T-ALL]) with half-maximal inhibitory concentration (IC50) values less than 100 nM (5, 133, 134). S63845 also showed potent antitumor activity in MM, lymphoma, and AML in vivo models (5). In CLL patient samples, S64315 induced apoptosis in a dose-dependent manner (135).
Venetoclax enhanced the S63845-induced apoptosis in AML in in vivo and in vitro models and in primary AML patient samples. This effect was even better than that for the combination of S63845 and standard-of-care AML chemotherapy, such as decitabine, idarubicin, and cytarabine (136). Similar findings were seen in T-ALL cell lines and in a zebrafish model of T-ALL (133). On the other hand, most solid tumors were resistant to S63845; only a few NSCLC, breast cancer, and melanoma cell lines had a modest response to the drug (5). In triple negative breast cancer, S63845 increased antitumor activity when combined with either chemotherapy or human epidermal growth factor receptor 2 (HER2)-targeted therapies (lapatinib, trastuzumab) (137).
S63845 had limited activity when tested in mice in initial studies. This was attributed to the lower affinity of S63845 to mouse MCL-1 (5). However, in human MCL-1 knock-in models, S63845 was tolerable and effective (138). S63845 was not selected for clinical evaluation, but second-generation S64315 (MIK665) was selected for clinical use in various hematological malignancies (NCT02979366 and NCT02992483) (Table 1).
PRT1419 is a new MCL-1i developed by Prelude Therapeutics. When given as a monotherapy in preclinical mouse models of MM, AML, and diffuse large B-cell lymphoma (DLBCL), and when given in combination with venetoclax in an AML model, it caused significant tumor regression (139). This agent is currently in clinical trials for patients with relapsed/refractory (R/R) hematologic malignancies, including lymphoid and myeloid disorders (NCT04543305), or with advanced solid tumors (NCT04837677), including melanoma, sarcoma, breast cancer, and lung cancer. It is also being tested in combination with either azacytidine or venetoclax in R/R myeloid and B-cell malignancies (NCT05107856). Additionally, AbbVie developed ABBV-467, which has been tested as monotherapy for MM patients (NCT04178902) in a phase 1 clinical trial. However, the trial has been terminated.
Interestingly, the MCL-1i upregulate MCL-1 protein in cell lines and in primary patient samples. This finding has not been seen with other BH3 mimetics that target BCL-2 or BCL-xL. Fluorescence resonance energy transfer studies have confirmed that this upregulation correlates with engagement of the MCL-1i into the MCL-1 protein. Thus, MCL-1 upregulation can be considered as a biomarker for MCL-1i target engagement (115).
This upregulation was related to the increased stability of MCL-1 rather than to increased transcription. Recently, Tantawy et al. (140) showed that the MCL-1i-induced stability of MCL-1 protein is mainly due to defective ubiquitination of MCL-1 (Figure 3). They showed that MCL-1i directly induced a state of MCL-1 that does not favor ubiquitination; rather, it favors deubiquitination by USP9x. The researchers also observed downregulation of Noxa and a transient decrease in the E3 ligase Mule following treatment with MCL-1i. The use of the deubiquitinase inhibitor WP1130 completely abrogated MCL-1 induction, reaffirming a critical role for deubiquitinases in stabilizing MCL-1 in response to MCL-1i (140). Despite the disruption of the BH3 proapoptotic members (e.g., BAK and Noxa) following MCL-1i treatment, the BH3 E3 ligase Mule was not disrupted. This finding was also seen with S64315 (141), AMG-176, and AZD5991 (140). Thus, MCL-1i can also be considered as anti-Noxa. Despite this upregulation of MCL-1, it did not confer any resistance to the MCL-1i. Besides decreased ubiquitination of MCL-1, another post-translation modification was noticed; MCL-1i induced ERK-mediated phosphorylation of Thr163 MCL-1 (140, 142). The effect of this upregulation and post-translation modification on the non-antiapoptotic function of MCL-1 needs to be elucidated.
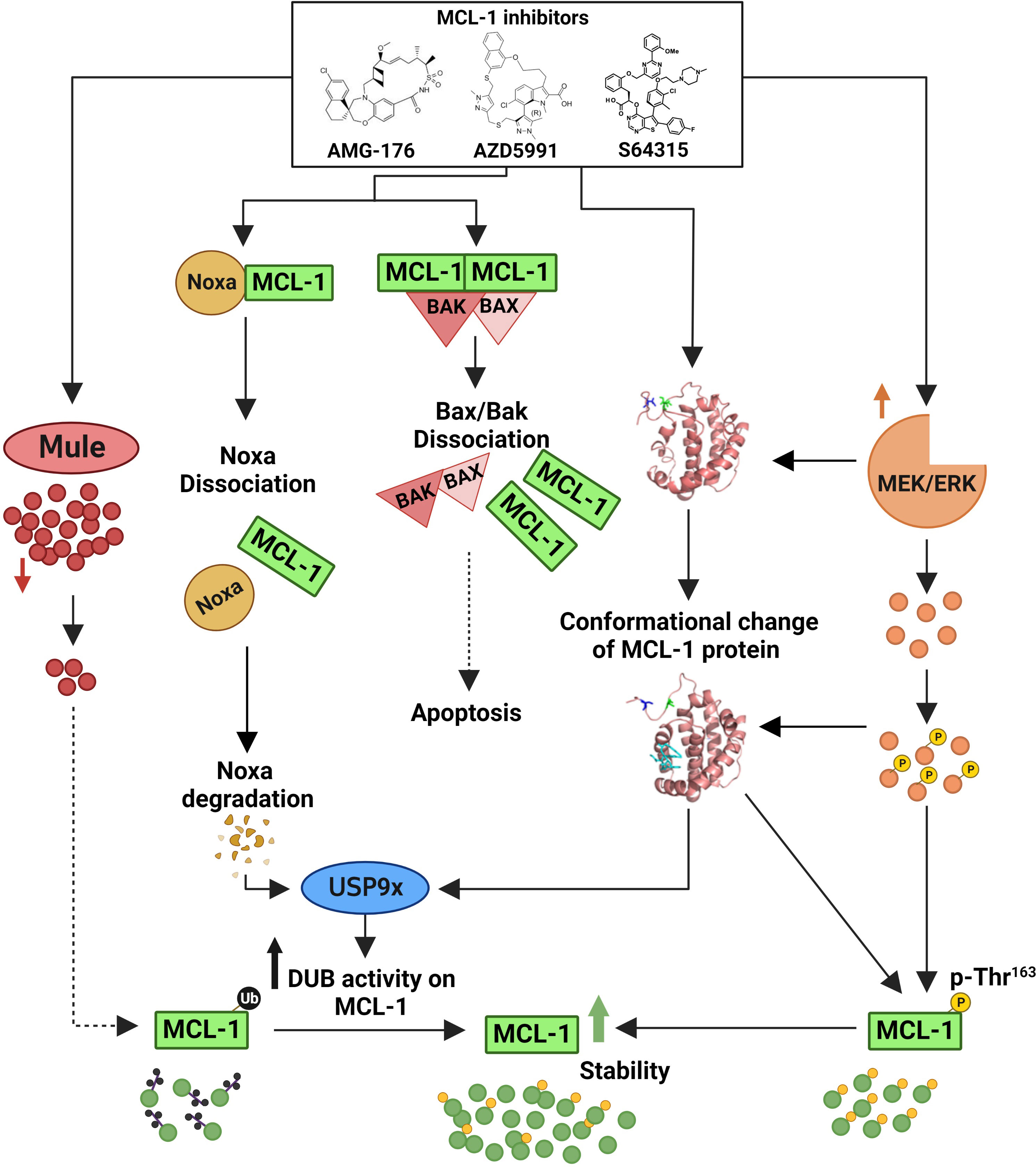
Figure 3 Mechanisms of myeloid cell leukemia-1 (MCL-1) protein upregulation by MCL-1 inhibitors. MCL-1i bind to the BCL-2 homology 3 (BH3) domain of MCL-1 and directly induce a conformation change/a state of MCL-1 that favors deubiquitination by deubiquitinating enzymes (DUBs [e.g., USP9x] rather than ubiquitination. This deubiquitination is further enhanced by Noxa disruption and rapid degradation, leading to enhanced DUBs activity on the MCL-1 protein. Additionally, the binding of MCL-1i transiently decreases the expression of the E3 ligase Mule and increases MEK/ERK-mediated threonine (Thr)163 phosphorylation of MCL-1, thus further contributing to the observed protein stability. Despite this upregulation of MCL-1 protein, MCL-1i disrupted the MCL-1–BAK/BAX interaction to induce apoptosis. The exact mechanism for MCL-1 upregulation was not studied using S63415, however, it is predicted to be the same owing to similar structure and function. Figure is adapted from Tantawy, et al, Clinical Cancer Research).
Most of the small molecule inhibitors of MCL-1 were discovered using structure-based design followed by fragment-based screening (143). This long and challenging pathway of optimizing MCL-1i has led S63845, AMG-176 and AZD5991 to be clinical candidates. Unlike AZD5991 and S63845, the AMG-176 compound lacks the salt bridge formation with MCL-1 Arg263 residue, and the chlorine atom in AMG-176 is buried in the hydrophobic pocket and does not interact with Ala227 (144). Understanding this difference is critical in the development of MCL-1i, as a recent study in CLL revealed that AZD5991 was more potent than AMG-176 in inducing apoptosis in primary CLL patient samples and the Mino cell line (140). Also, despite MCL-1 stabilization in cell lines following treatment with MCL-1i, cell-free in vitro ubiquitination studies revealed that, unlike AZD5991, AMG-176 induced in vitro ubiquitination of MCL-1 protein even in the absence of Mule (140). This observation raised 2 important questions. First, can the difference between AMG-176 and AZD5991 in binding MCL-1 protein explain their differences in potency in CLL, given that the salt bridge formation with Arg263 has been shown to be critical for the efficacy for some MCL-1i? Second, can the behavior of MCL-1i in cell-free in vitro ubiquitination assays (whether favoring or not favoring MCL-1 ubiquitination) predict the MCL-1i’s efficacy or potency, assuming that potent MCL-1i will behave similarly to AZD5991 in not favoring in vitro ubiquitination?
New approaches to MCL-1 inhibition-based therapy have focused on enhancing the activities of MCL-1i by designing molecules that target different moieties of MCL-1 or neutralize MCL-1 along with other prosurvival proteins of the BCL-2 family. Zhang et al. (39) demonstrated that it is possible to have a dual target for MCL-1; this is achieved by inhibiting MCL-1’s interaction with its proapoptotic BH3 counterparts and enhancing its ubiquitination and degradation. Zhang et al. discovered a hidden dynamic region—the Q221R222N223 (QRN) motif—in the BH3 domain of MCL-1 that controls MCL-1 ubiquitination and degradation. They reported that compound 5 binds to the H224 of MCL-1; thus, besides disrupting the MCL-1 interaction, it switches the BH3 domain towards a helical conformation, which facilitates MCL-1 ubiquitination and degradation. Similar observations have been made for maritoclax and UMI-77, which also are known to ubiquitinate and degrade MCL-1. In contrast, A-1210477 did not enhance in vitro ubiquitination of MCL-1 (39). Researchers have shown that lysine residue in the BH3 domain of MCL-1 can covalently bind an MCL-1i. NA1-115-7 has been shown to inhibit MCL-1 through a “covalent interaction between its 2 aldehyde functional groups and a lysine residue of the MCL-1 protein.” It triggered apoptosis in an MCL-1–dependent manner with no observed toxicity to peripheral blood cells or cardiomyocytes (145).
Dual MCL-1 and BCL-2 inhibitors have been designed to neutralize BCL-2 protein and partially inhibit MCL-1 to improve their safety and avoid MCL-1 upregulation (93). Both BCL-2 and MCL-1 share P2 as a common hot spot region and could be targeted by 2 novel compounds, IS20 and IS21, which induce apoptosis in melanoma and NSCLC cell lines. IS21 also decreased tumor growth in leukemic and melanoma mice models, comparable to ABT199 and ABT-263 (146). Additionally, Drennen et al. [148] developed an indazole-3-acylsulfonamide from a carboxylic acid core. The new compound inhibited both BCL-2 and MCL-1 and had a much lower affinity for BCL-xL. However, this compound was not tested in ex vivo or in vivo (147). Benzimidazole chalcone and flavonoid scaffold–derived bicyclic compounds were optimized to target both BCL-2 and MCL-1. These compounds exhibited significant cytotoxic activity against the AW13516 cell line in a caspase-dependent manner and displaced the BH3 binding partners of MCL-1 and BCL-2 (148). Dual inhibition of MCL-1 and BCL-xL has been proposed. However, dual-targeting inhibitors do not appear to be in clinical evaluation because of the increased hepatotoxicity seen when both MCL-1 and BCL-xL proteins are co-targeted systemically (149). Furthermore, the thrombocytopenia associated with BCL-xL inhibition may limit the use of MCL-1/BCL-xL dual inhibition (150, 151).
Although none of the direct inhibitors of MCL-1 have been successful in the clinic so far, targeting MCL-1 has remained attractive strategy. New MCL-1i patents for innovative agents derived from new chemicals or featuring new scaffolds are being filed and granted (152).
Cells can hinder the response to MCL-1i by becoming dependent on multiple antiapoptotic proteins; in this way, the cells can compensate for one another (5, 114, 153). In such instances, administering combination therapy is the best option to achieve optimum antitumor activity (Figure 4). It has been shown that CLL is dependent on BCL-2 and has a robust response to venetoclax (154), while AML exhibits variable dependency on MCL-1 and BCL-2 (136). The variable dependency of AML on both BCL-2 and MCL-1 and the recent approval of venetoclax and azacytidine as a combination therapy for AML (these drugs were shown to be synergistic, as azacytidine was able to downregulate MCL-1) had led the use of combination therapies with BCL-2 inhibitors and MCL-1i for the treatment of AML. This approach is now being tested in clinical trials (5, 114). The combination of S63845 or AZD5991 with venetoclax has been found to be synergistic in AML and to overcome cytarabine resistance in the disease (155). The loss of TP53 impairs BAX/BAK activation, resulting in prolonged and sublethal targeting of either BCL-2 or MCL-1. Thus, intact TP53 function was found to be necessary to the action of BH3 mimetics, and the combination of MCL-1i and BCL-2 inhibitors may enhance long-term outcomes in aberrant-TP53 AML (156). CLL patient samples were found to be more dependent on BCL-2 than MCL-1, and low-dose venetoclax combined with AMG-176 has been shown to be additive or synergistic (130).
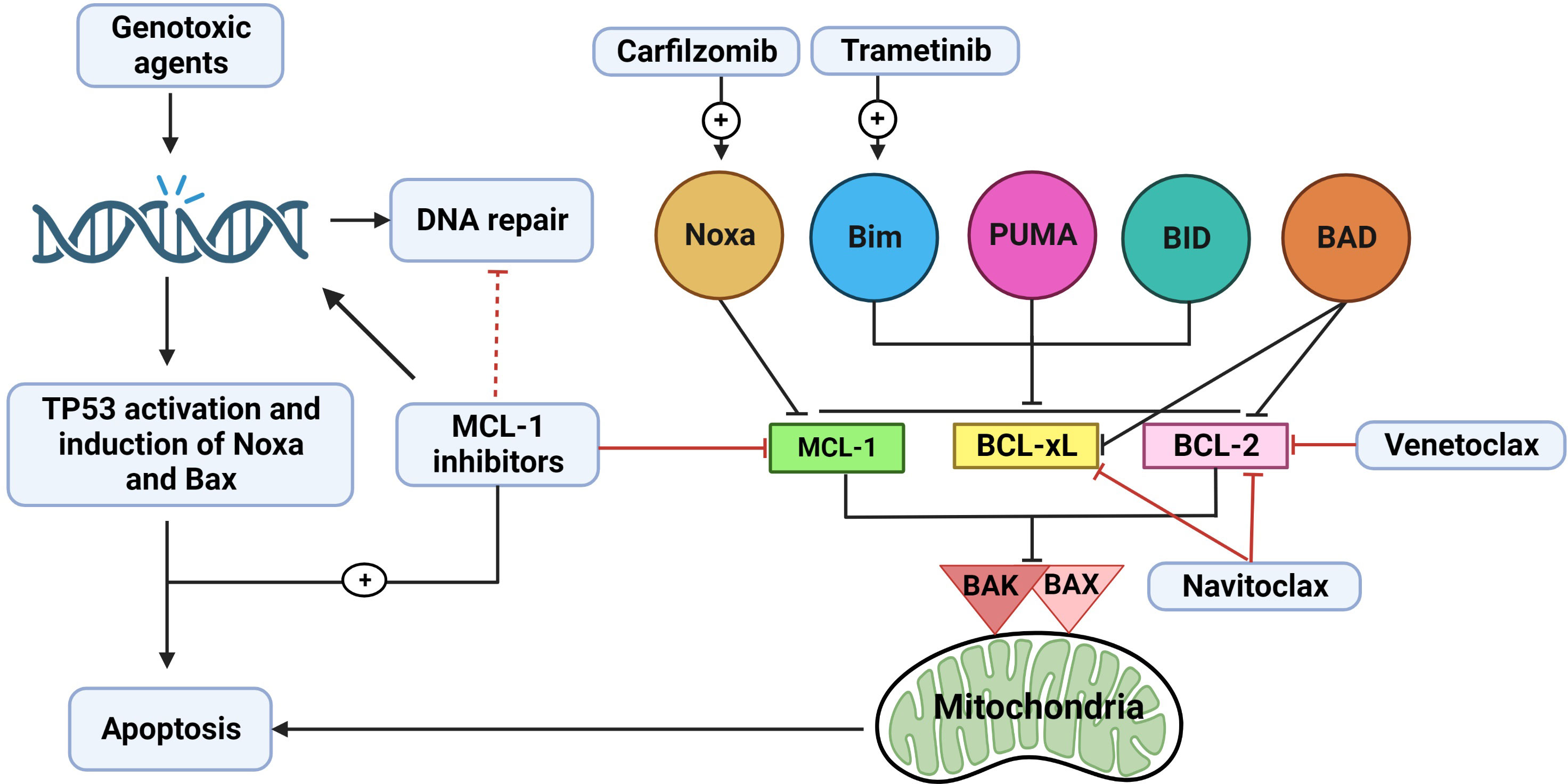
Figure 4 The rationale for combination therapy using MCL-1 inhibitors. Myeloid cell leukemia-1 (MCL-1), B-cell lymphoma (BCL)-2 extra large (BCL-xL), and BCL-2 prosurvival proteins cooperate to bind and inactivate BAX, BAK complex, and other proapoptotic BCL-2 homology (BH)3-only proteins (Noxa, BIM, PUMA, BID, and BAD). When one of these prosurvival proteins loses its function (e.g., from MCL-1 inhibition by MCL-1i), the other proteins (BCL-2 and BCL-xL) may at least partially compensate for this loss. Thus, combination therapy using MCL-1i with a BCL-2 inhibitor (e.g., venetoclax) or BCL-2/BCL-xL inhibitor (e.g., navitoclax) appears promising and has been shown to be effective. Another way to enhance the activity of MCL-1i is to increase the expression of BH3-only proteins. Trametinib can upregulate BIM, which can then bind to MCL-1, thus increasing dependency on MCL-1 protein and subsequently increasing sensitivity to MCL-1i. BIM can also bind to BCL-2 or BCL-xL, indirectly inactivating them. Carfilzomib can upregulate Noxa, which is a BH3-only sensitizer protein that binds to MCL-1, leading to more BH3-only activator proteins (BIM, PUMA, and BID) becoming available to bind to BAX/BAK and activate downstream apoptosis. Chemotherapy induces DNA damage with subsequent activation of TP53 and induction of BAX and Noxa. In addition, various chemotherapeutic agents can downregulate MCL-1. MCL-1i can synergize with chemotherapy by inducing more DNA damage, possibly affecting DNA repair, and neutralizing MCL-1. Effect of the clinically relevant MCL-1i (S63845, AMG-176 and AZD5991) on DNA repair mechanisms needs to be further explored.
Another approach to enhancing the response to MCL-1i in cell lines that show less addiction to MCL-1 is to elicit an increased dependency on MCL-1 proteins in these cell lines by increasing the association between MCL-1 and proapoptotic proteins. This strategy was shown by Nangia et al. (157), who studied KRAS-mutant NSCLC cell lines that show some sensitivity to trametinib. The use of trametinib increased BIM expression (by stabilizing BIM) in NSCLC cells that are buffered by either BCL-xL or MCL-1; thus, the cells became dependent on both BCL-xL and MCL-1 protein. The use of navitoclax or AM-8621 synergized MEK inhibition in these cell lines. Prior BCL-xL inhibition increased MCL-1 dependence and enhanced sensitivity to MCL-1i, but not vice versa.
In those cell lines that show sensitivity to MCL-1i, the combination of MCL-1i with standard-of-care chemotherapy or targeted therapy agents has been shown to be synergetic. MCL-1i were shown to be effective only in tumors dependent on MCL-1 protein. The growth of MM cells was found to rely on the presence of the MCL-1 protein, and they exhibited a more favorable response to MCL-1i and even superior to venetoclax (5, 114, 115). The MCL-1i were synergistic when combined with carfilzomib and dexamethasone, even in cell lines that were resistant to single-agent MCL-1i (114, 115). Interestingly, proteasome inhibitors upregulated Noxa protein, resulting in indirect inhibition of MCL-1 (158); this finding may explain the synergistic effect of combination therapy with MCL-1i and carfilzomib plus dexamethasone (159). A similar finding was reported in a murine double-hit lymphoma model, in which inhibiting MCL-1 protein synthesis by homoharringtonine and concomitant Noxa induction by bortezomib reduced tumor growth and increased survival significantly (160). Of note, the combination of AMG-176 with dexamethasone and carfilzomib in R/R MM is being studied in a clinical trial (NCT02675452).
Genotoxic agents induce DNA breaks with subsequent TP53, Noxa, and Bax upregulation and the induction of apoptosis (161). AZD5991 has been shown to induce DNA damage that is further increased when the drug is combined with cytarabine (162). MCL-1 depletion has been shown to induce genomic instability and impair DNA double-strand break repair (51). In addition, MCL-1 controls the switch between homologous recombination and nonhomologous end-joining DNA repair by binding to the Ku70/Ku80 dimers through its BH1 and BH3 domains; MCL-1 depletion reduces homologous recombination and favors nonhomologous end-joining repair. MI-223, which blocks the BH1 domain of MCL-1, impaired MCL-1–mediated homologous recombination DNA repair and sensitized cells to DNA-replication stress inducers (hydroxyurea and olaparib) (163). Studies are needed to explore the effects of the clinically relevant MCL-1i (S63845, AMG-176, and AZD5991) on DNA repair mechanisms. The potential effect of MCL-1 inhibition on DNA damage and DNA repair provides a rationale for combining MCL-1i with chemotherapy that upregulates TP53-mediated Noxa and Bax and depletes MCL-1, leading to apoptosis. It has also been shown that doxorubicin synergized with S63845 to induce apoptosis in BCP-ALL cell lines (164).
The synergism of ERK1/2 inhibition with MCL-1i has also been reported in melanoma, in which AZD5991 synergized with ERK inhibition and delayed the development of BRAFi/MEKi resistance, thereby improving “the efficacy of an ERK1/2 inhibitor in a model of acquired BRAFi + MEKi resistance” (128). Similarly, the combination of MCL-1i and trametinib was also synergistic in AML cell lines and in 6 of 12 primary patient samples. Interestingly, it has been reported that the cell lines that had higher levels of MCL-1 protein were more susceptible to 50-nM S63845 than were cell lines with lower levels of MCL-1 protein. suggesting that MCL-1 and MEK1/2-protein expression levels may predict responses to S63845 and trametinib, respectively (165). Ulixertinib, another ERK1/2 inhibitor, is highly synergistic in rhabdomyosarcoma in vitro and in vivo through upregulation of the proapoptotic proteins BIM and BMF (166).
High BAK expression and low BCL-xL expression predicted sensitivity to AM-8621 and ANJ810, while high BCL-xL expression predicted resistance to AM-8621 (114, 167). Response to S63845 inversely correlated with the expression level of BCL2L1 gene (5). MCL-1 protein level and mRNA expression correlated poorly with response to MCL-1i (5, 66, 114). Recently, some studies have revealed that the MCL-1/BCL-xL ratio predicted synergistic response to either AZD5991 or navitoclax in combination with an ERK1/2 inhibitor (128, 168). Current methods to predict the sensitivity to MCL-1i and dependency on MCL-1 protein include dynamic BH3 profiling or ex-vivo incubation by MCL-1i and observing the response (66, 127, 169). Clinical successes, genomics and multiomics data from MCL-1i clinical trials may provide clues to biomarkers of response.
It is well known that stromal microenvironment can provide resistance to the action of MCL-1i (79, 170, 171). Although the mechanism is not fully understood, the overexpression of MCL-1 and BCL-xL, in mantle cell lymphoma and CLL, is thought to mediate this resistance. Sensitivity can be restored upon the use of a BCL-xL inhibitor (173) or by AT-101, a pan-BCL-2 prosurvival protein inhibitor (172).
Cell lines resistant to MCL-1i has been used to identify mechanisms of resistance. Wang et al. (174) generated Mino and SUDHL cell lines resistant to MCL-1i (10× IC50). Compared to the parental cell lines, the resistant cell lines showed a higher level of BCL-2 protein and a slight increase in BCL-xL. Knocking down BCL-2 restored sensitivity to the MCL-1i S63845, confirming a role for BCL-2 in mediating resistance to MCL-1. Further kinome and transcriptome analysis revealed higher activity for the MEK, ERK, and BCR pathways, possibly impacting BCL-2. The resistant cell lines were more sensitive to MEK/ERK inhibition, further confirming a role for MEK and ERK pathways in the modulation of the BCL-2 family proteins.
In DLBCL cell lines, the loss of TP53 or BAX conferred resistance to AZD5991. The knockout of TP53 decreased BAX and PUMA expression, and BAX overexpression in TP53-deficient cell lines restored sensitivity to AZD5991, suggesting that a functional TP53 mediates sensitivity to AZD5991, at least partially, through BAX (175). Also, lower levels of BCL-2 and BCL-xL correlated with a higher sensitivity to S63845 in the HH cell line. Strikingly higher levels of BCL-W were found in the S63845-resistant MyLa and SeAx cell lines (176).
Bolomsky et al. (177) generated 2 myeloma cell lines (OPM2-R and KMS12BM-R) resistant to the MCL-1i S63845 to study MCL-1i resistance mechanisms. The resistant cell lines showed resistance to other MCL-1i (AZD5991 and AMG-176) and heterogenous modulation of cell type specific BCL-2 family proteins. BAK, BAX, and BIM were downregulated in the OPM2-R cells, MCL-1 and BCL-2 were upregulated in the KMS12BM-R cells, and there were no detectable alterations in the protein levels of the other BCL-2 family members (BAD, BID, Noxa, and PUMA). In addition,“high-throughput drug screening (n = 528 compounds) indicated alternative BH3 mimetics as best combination partners for MCL-1i in sensitive and resistant cells, particularly with BCL-xL inhibitors”.
Although BCL-2 mutations are known to drive resistance to venetoclax in CLL by inhibiting the binding of venetoclax to its target (178), very limited data are available regarding the potential mutations in MCL-1 that may hinder the activity of MCL-1i. Chen et al. (179) examined the frequency of mutations in BCL-2 family proteins in 982 MM patients (NCT0145429) and found that BCL-2 family protein mutations were generally rare. Interestingly, 10 patient samples were found to harbor MCL-1 mutations at baseline. These mutations were missense mutations in the N-terminal region (G32R; n = 1), in the PEST domain (n = 4; V140I, P142S, E149Q, and E173K), in an uncharacterized region between the PEST and BH1 domains (L186F), in BH1 (V249L and L267V), and within the BH3 (N223S and R214Q) domains. To functionally characterize the impact of these mutations, WT MCL-1 and mutant MCL-1 plasmids were overexpressed in an ALL murine cell line. The overexpressed human MCL-1 can replace the murine MCL-1 in this cell line. In cells with the V249L, N223S, and R214Q mutations, S63845 upregulated MCL-1, disrupted the MCL-1–BIM interaction, and showed a similar sensitivity to that in the WT MCL-1 cells. In contrast, the L267V mutation was resistant to both S63845 and AZD5991; in these cells, the MCL-1– BIM interaction was not disrupted, despite MCL-1 upregulation. This finding suggests that the L267V mutation did not hinder the binding of MCL-1i to MCL-1 protein, but that it blocked its ability to disrupt MCL-1’s interaction with BIM (179). Additional investigations are needed to assess clinical impact of these mutations. Also, MM cell lines with acquired carfilzomib resistance showed cross resistance to AZD5991, S63845, and A-1210477, but not to AMG-176. Immunoblot analysis revealed the upregulation of MDR1 protein, suggesting drug efflux as a mechanism of resistance to MCL-1i. The use of MDR1 inhibitors (tariquidar or verapamil) restored the sensitivity of these resistant cell lines to the action of MCL-1i (177). While some molecules are being identified for further studies of resistance, additional studies are needed.
In AML, c-Myc levels negatively correlated with the half-maximal effective concentration of AZD5991 in cell lines and primary patient samples. In the MOLM-13 and MV4-11 cell lines, which are resistant to AZD5991, the c-Myc and MCL-1 protein levels were upregulated. Targeting c-Myc by 10058-F partially overcome resistance to AZD5991 in resistant cell lines (180). A summary of predictive biomarkers and mechanisms of resistance is provided in Table 2.
There are 2 major issues to be overcome regarding the clinical application of current MCL-1i. First, reports of MCL-1i cardiotoxicity are alarming and raise safety concerns. Second, researchers must determine how to fit MCL-1i into treatment algorithms to identify the patients likely to benefit from the drugs.
Cardiotoxicity appears to be a class effect of MCL-1i. The FDA put both AMG-397 and AZD5991 on hold because of concerns regarding cardiovascular adverse effects. The incidence of cardiotoxicity was low, the number of patients enrolled in the study was small, and the exact nature of the cardiotoxicity was not known. The work on AMG-397 is now stopped, while a related compound, AMG-176, is being tested in a phase 1 clinical trial. (https://ashpublications.org/ashclinicalnews/news/4765/FDA-Places-Trials-of-MCL-1-Inhibitor-on-Clinical?searchresult=1). Similarly, AZD5991 was put on clinical hold after reporting an asymptomatic elevation of laboratory cardiac parameters in one patient with multiple comorbidities that occurred in the AZD5991 and venetoclax combination arm.
The mechanism of MCL-1i–mediated incident cardiotoxicity is not very clear. MCL-1 has high expression in the myocardium and is essential for maintaining cardiac homeostasis and inducing autophagy in the heart. It has been shown that “cardiac-specific deletion of MCL-1 in mice” led to mitochondrial dysfunction, impaired autophagy, hypertrophy, and cardiomyopathy with distorted ultrastructure of disorganized sarcomeres and swollen mitochondria (182–184). Interestingly, concomitant BAX/BAK knockout in these mice models largely rescued the lethality and impaired cardiac function but not the ultrastructure changes of the mitochondria caused by the MCL-1 deletion (183). Also, double knockout of MCL-1 and cyclophilin D, which controls the mitochondrial permeability transition pore, extended survival and delayed the progression to heart failure (184). These data may indicate that the cell death associated with MCL-1 deletion contributes to the observed cardiac dysfunction, independently of mitochondrial dysfunction. Although the authors did not specifically study the effect of this mitochondrial dysfunction on the MCL-1 deletion induced cardiomyopathy, they predicted that the observed mitochondrial dysfunction may also contribute to the observed cardiotoxicity, owing to the importance of mitochondria to the cardiac function (185). Similarly, Perciavalle et al. (46), showed that MCL-1 deletion in murine embryonic fibroblast and hepatocyte distorted mitochondrial morphology with abnormal cristae and defective electron transport system. However, the direct effects of MCL-1i are different because MCL-1i upregulate, rather than deplete, MCL-1 (5, 114, 115). It is not known whether the cardioprotective function of MCL-1 depends on MCL-1’s BH3 domain or not. S63845 has been shown to disrupt cytoskeleton formation, mitochondrial morphology and dynamics in “cardiomyocytes derived from human-induced pluripotent stem cells”, leading to overall poor cardiomyocyte performance (186). These effects have been seen with high doses of the drug and/or prolonged periods of treatment. Besides, S63845 decreased cardiomyocyte viability in a dose dependent manner, supporting the dependency of the cardiomyocyte on MCL-1 for survival. Treating the hiPSC-CMs with 100 nM S63845 for two weeks impaired cardiomyocyte beating with mitochondrial dysfunction and impaired calcium influx, despite live cells, indicating that the impaired cardiomyocyte beating might be independent of cell death. Again, the mitochondrial dysfunction and decreased calcium influx was more prominent with treatment with S63845 and AZD5991 as compared with AMG-176. The use of the necrosis inhibitor IM-54, but not the caspase inhibitor QVD, rescued the cell death induced by S63845. Similarly, in Mec1 and HG3 CLL cell lines, AZD5991 was shown to decrease oxygen consumption rate (OCR), decrease ATP production, and increase mitochondrial oxygen reactive species, suggesting mitochondrial dysfunction (129). Organized mitochondrion is required for adequate cardiac function (185) through regulating metabolic bioenergetics, calcium flux and modulating cardiac contraction (187, 188). Its dysfunction has been linked to different cardiovascular diseases including cardiomyopathy and arrhythmias (189–191). To circumvent these side effects, researchers are working to identify new MCL-1i with low potential toxicity. ANJ810, for example, is a new potent MCL-1i with a short half-life and rapid systemic clearance; thus, it provides short-term inhibition of MCL-1 and has a low potential for cardiotoxicity (167).
Noxa downregulation provides an alternative mechanism of MCL-1i-mediated cardiotoxicity. The MCL-1i S63845 (141), AZD5991, and AMG-176 (140) downregulate Noxa protein upon disruption of their interaction with MCL-1 protein. This raises the question of whether the cardiac toxicity of MCL-1i is related to the targeting of MCL-1’s BH3 domain or to the downregulation of Noxa. Noxa downregulation has been shown to mediate phenylephrine-induced cardiac hypertrophy. Also, rapamycin has been shown to inhibit cardiomyopathy by promoting autophagy through beclin-1 and Noxa (192). Autophagy is important for the heart, and its dysregulation mediates several drug-induced cardiotoxicities, such as those resulting from daunorubicin treatment (193). Like MCL-1, Noxa can regulate autophagy (194, 195); therefore, it is important to explore the impact of MCL-1i-induced Noxa downregulation. An understanding of this downregulation may open the door for new combination strategies that maintain Noxa levels and could potentially enhance the efficacy and safety of MCL-1i.
In CLL cell lines, MCL-1i have been shown to induce a transient decrease in Mule expression that is associated with a compensatory increase of Mule at later time points (140). However, it is not known if similar changes occur in cardiomyocytes. Mule also has been shown to have a cardioprotective role against oxidative stress, and its deletion leads to cardiomyopathy (196).
MCL-1i induced cardiotoxicity may be related to the high affinity of MCL-1i for MCL-1 protein, at least in part. A study of mouse lymphoma models knocked in with human MCL-1 showed that S63845 was efficacious at tolerable doses. “However, the maximum tolerated dose was lower in huMcl-1 mice than in the control group (138, 197), suggesting that S63845 toxicity depends on its immediate target, MCL-1, although the precise mechanism is unknown”. As discussed earlier, MCL-1 protein is engaged in various non-antiapoptotic functions, particularly autophagy, oxidative phosphorylation, and mitochondrial bioenergetics. It is not known whether these functions depend on the BH3 domain of MCL-1 protein or not. Thus, further studies to assess how MCL-1i can affect the non-antiapoptotic function of MCL-1 are needed to better understand the cardiotoxicity associated with MCL-1i. In summary, data regarding MCL-1i–induced cardiotoxicity are limited. Multiple factors may contribute to the observed cardiotoxicity, including the very high affinity of MCL-1i for MCL-1, high doses of and prolonged exposure to MCL-1i, mitochondrial dysfunction, impaired autophagy, and the induction of cell death. Accumulating evidence suggests that cardiotoxicity be related to the cardiomyocyte cell death and or/mitochondrial dysfunction and impaired calcium influx. Mitochondrial dysfunction is predicted to contribute to the observed cardiotoxicity either through the impaired bioenergetic metabolism, ATP production, impaired calcium influx, increased ROS production and/or through potentiating cell death. Current efforts to circumvent these unwanted events include designing inhibitors that provide partial inhibition of MCL-1 (dual MCL-1 and BCL-2 inhibitors) and MCL-1i that provide short-term inhibition of MCL-1 (e.g., ANJ810). Future studies are needed to explore the role of Noxa and Mule downregulation associated with the use of MCL-1i and observed cardiotoxicity and to determine the effect of MCL-1i on other non-antiapoptotic functions of MCL-1. The effect of MCL-1 deletion or MCL-1 inhibition on the heart is summarized in Figure 5.
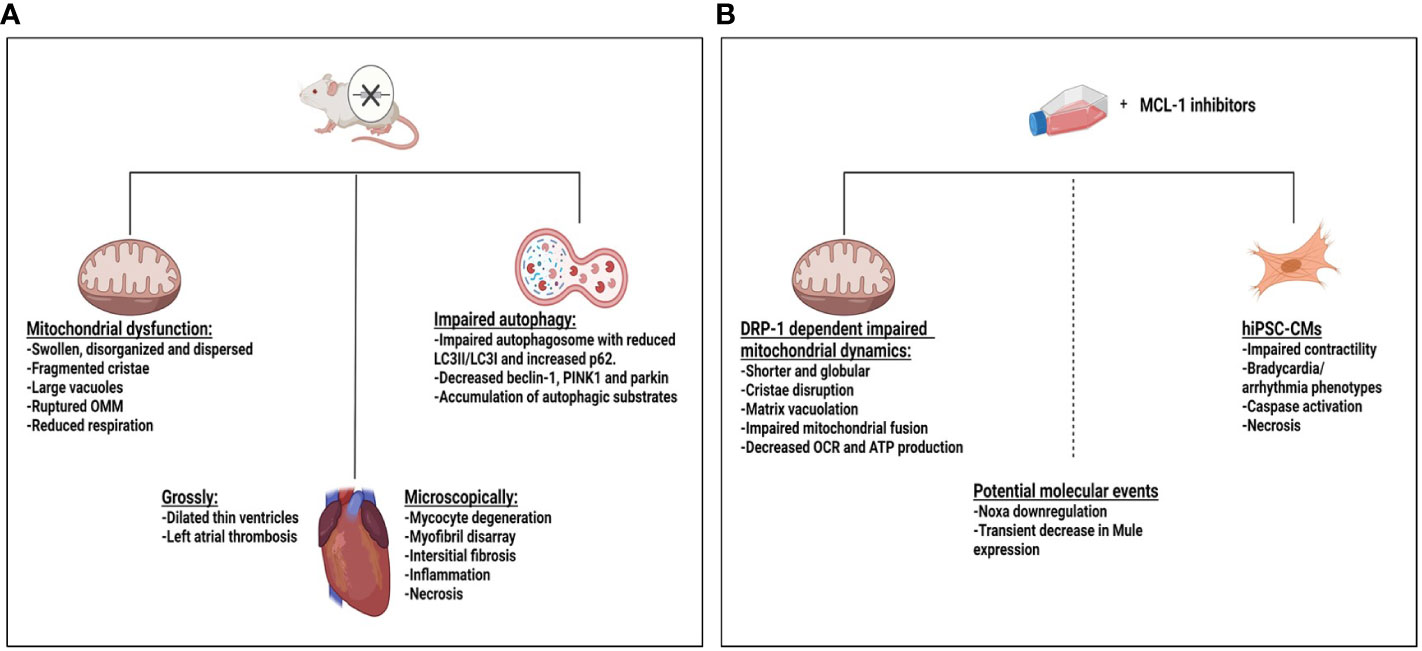
Figure 5 Effect of myeloid cell leukemia-1 (MCL-1) deletion or MCL-1 inhibitors on the heart. (A) Mcl-1 knockout in a mouse model led to dilated cardiomyopathy with thin ventricle walls and atrial thrombus, features of impaired cardiac hemodynamics. Ultrastructure changes in the mitochondria, myocyte degeneration, fibrosis, inflammation, and necrosis were observed. Mitochondrial dysfunction and impaired autophagy may have contributed to the observed toxicity. (B) Treating hiPSC-CMs with S68345 induced dynamin-related protein 1 (DRP-1–dependent mitochondrial dysfunction, cytoskeleton disruption, and impaired/irregular beating of the cardiomyocytes. The potential role of Noxa and Mule downregulation on cardiotoxicity needs to be further explored. ATP, adenosine triphosphate; OCR, oxygen consumption rate; OMM, outer mitochondrial membrane.
It is important to determine how to fit MCL-1i into treatment algorithms for optimal benefits to patients. It appears that, compared to solid tumors, hematologic malignancies show preferential sensitivity to MCL-1i (114, 115). Preclinical data indicate that MCL-1i are efficacious in cell lines and tumors that are dependent on MCL-1 for survival. The classic example of this is MM, in which MCL-1 appears to act as a gatekeeper against apoptosis. Accordingly, there is a good chance for MCL-1i to be successful in patients with MM (114). AML, on the other hand, shows dual or heterogenous dependency on BCL-2/MCL-1. In addition, MCL-1 appears to be a major driver of resistance to venetoclax (136, 198), underscoring the value of the combination of MCL-1i and venetoclax. In CLL, it is clear that the disease is mainly dependent on BCL-2 and that patients benefit the most from the use of venetoclax alone (199). However, an approach to testing combination therapies with venetoclax and MCL-1i is needed. In fact, declines in MCL-1 have been achieved via ibrutinib treatment followed by venetoclax treatment; this combination therapy appears to be successful in CLL (200, 201) and MCL. Future studies are needed to determine the optimum combination therapy approaches involving MCL-1i. It is feasible that the transient use of MCL-1i in mechanism-based combinations may benefit patients without untoward cardiotoxicity.
Laboratory cell line data, murine model systems, and clinical observations clearly underscore MCL-1 as a therapeutic target for many cancers. Although 6 MCL-1i that directly neutralize MCL-1’s function are being tested in phase 1 clinical trials, our knowledge about the use of MCL-1i in the clinic is currently limited and depends upon how successful the clinical trials are. Optimizing and designing new potent and specific MCL-1i is urgently needed in light of the emerging role of MCL-1 in tumorigenesis and therapeutic resistance. Identifying biomarkers of response and resistance will guide us to better uses of MCL-1i in the clinic. In addition, gaining a deeper understanding of the effects of MCL-1i on the non-antiapoptotic function of MCL-1 may help improve the safety profiles of MCL-1i. Efforts from chemists and pharmaceutical interests, the enthusiasm of scientists to carve out optimal MCL-1i usage and combination strategies, and clinical endeavors and observations are at their peak levels and will help make MCL-1 a clinical target.
ST researched the literature and outlined and wrote the review. NT reviewed the manuscript, created the figures, and wrote the figure legends. AS and VG supervised the project, researched the literature, reviewed, wrote, and edited the manuscript, and outlined the review. All authors contributed to the article and approved the submitted version.
This work was supported by a CLL Global Research Foundation Alliance and The University of Texas MD Anderson Cancer Center’s Moon Shot Program.
We thank Laura L. Russell, scientific editor, Research Medical Library, for editing this article.
Author VG has received research grants from Pharmacyclics, Acerta, Gilead, Sunesis, ClearCreek Bio, Infinity, and Loxo Oncology.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer (2002) 2(9):647–56. doi: 10.1038/nrc883
2. Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wideranging implications in tissue kinetics. Br J Cancer (1972) 26(4):239–57. doi: 10.1038/bjc.1972.33
3. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature (2010) 463(7283):899–905. doi: 10.1038/nature08822
4. Campbell KJ, Dhayade S, Ferrari N, Sims AH, Johnson E, Mason SM, et al. MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis (2018) 9(2):1–14. doi: 10.1038/s41419-017-0035-2
5. Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature (2016) 538(7626):477–82. doi: 10.1038/nature19830
6. Golstein P. Cell death in us and others. Am Assoc Adv Sci (1998). doi: 10.1126/science.281.5381.1283
7. Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for bcl-2 regulation of apoptosis. Science (1997) 275(5303):1132–6. doi: 10.1126/science.275.5303.1132
8. Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to c. elegans CED-4, participates in cytochrome c–dependent activation of caspase-3. Cell (1997) 90(3):405–13. doi: 10.1016/S0092-8674(00)80501-2
9. Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. cell (2000) 102(1):43–53. doi: 10.1016/S0092-8674(00)00009-X
10. Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c–dependent caspase activation by eliminating IAP inhibition. Cell (2000) 102(1):33–42. doi: 10.1016/S0092-8674(00)00008-8
11. Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol (2005) 5(3):189–200. doi: 10.1038/nri1568
12. Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu Rev Immunol (1998) 16(1):395–419. doi: 10.1146/annurev.immunol.16.1.395
13. Akgul C, Moulding DA, White M, Edwards SW. In vivo localisation and stability of human mcl-1 using green fluorescent protein (GFP) fusion proteins. FEBS Lett (2000) 478(1-2):72–6. doi: 10.1016/S0014-5793(00)01809-3
14. Carrington EM, Zhan Y, Brady JL, Zhang J-G, Sutherland RM, Anstee NS, et al. Anti-apoptotic proteins BCL-2, MCL-1 and A1 summate collectively to maintain survival of immune cell populations both in vitro and in vivo. Cell Death Differ (2017) 24(5):878–88. doi: 10.1038/cdd.2017.30
15. Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer (2015) 15(10):577–92. doi: 10.1038/nrc4000
16. Long GV, Stroyakovskiy D, Gogas H, Levchenko E, De Braud F, Larkin J, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet (2015) 386(9992):444–51. doi: 10.1016/S0140-6736(15)60898-4
17. Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci (1993) 90(8):3516–20. doi: 10.1073/pnas.90.8.3516
18. Slomp A, Peperzak V. Role and regulation of pro-survival BCL-2 proteins in multiple myeloma. Front Oncol (2018) 8:533. doi: 10.3389/fonc.2018.00533
19. Shah V, Sherborne AL, Walker BA, Johnson DC, Boyle EM, Ellis S, et al. Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia (2018) 32(1):102–10. doi: 10.1038/leu.2017.179
20. Yin J, Li Y, Zhao H, Qin Q, Li X, Huang J, et al. Copy-number variation of MCL1 predicts overall survival of non-small-cell lung cancer in a southern Chinese population. Cancer Med (2016) 5(9):2171–9. doi: 10.1002/cam4.774
21. Kim J-H, Sim S-H, Ha H-J, Ko J-J, Lee K, Bae J. MCL-1ES, a novel variant of MCL-1, associates with MCL-1L and induces mitochondrial cell death. FEBS Lett (2009) 583(17):2758–64. doi: 10.1016/j.febslet.2009.08.006
22. Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem (2000) 275(33):25255–61. doi: 10.1074/jbc.M909826199
23. Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK. Exon skipping in mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem (2000) 275(29):22136–46. doi: 10.1074/jbc.M909572199
24. Aird D, Teng T, Huang C-L, Pazolli E, Banka D, Cheung-Ong K, et al. Sensitivity to splicing modulation of BCL2 family genes defines cancer therapeutic strategies for splicing modulators. Nat Commun (2019) 10(1):1–15. doi: 10.1038/s41467-018-08150-5
25. Laetsch T, Liu X, Vu A, Sliozberg M, Vido M, Elci O, et al. Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis (2014) 5(2):e1072–e. doi: 10.1038/cddis.2014.40
26. Yang T, Buchan HL, Townsend KJ, Craig RW. MCL-1, a member of the BCL-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J Cell Physiol (1996) 166(3):523–36. doi: 10.1002/(SICI)1097-4652(199603)166:3<523::AID-JCP7>3.0.CO;2-R
27. Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol (2005) 37(2):267–71. doi: 10.1016/j.biocel.2004.04.007
28. Lutz R. Role of the BH3 (Bcl-2 homology 3) domain in the regulation of apoptosis and bcl-2-related proteins. Portland Press Ltd (2000) 28(2):51–6.
29. Yap JL, Chen L, Lanning ME, Fletcher S. Expanding the cancer arsenal with targeted therapies: disarmament of the antiapoptotic bcl-2 proteins by small molecules: miniperspective. J Med Chem (2017) 60(3):821–38. doi: 10.1021/acs.jmedchem.5b01888
30. Craig R. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia (2002) 16(4):444–54. doi: 10.1038/sj.leu.2402416
31. Thomas LW, Lam C, Edwards SW. Mcl-1; the molecular regulation of protein function. FEBS Lett (2010) 584(14):2981–9. doi: 10.1016/j.febslet.2010.05.061
32. Gores GJ, Kaufmann SH. Selectively targeting mcl-1 for the treatment of acute myelogenous leukemia and solid tumors. Genes Dev (2012) 26(4):305–11. doi: 10.1101/gad.186189.111
33. Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: unique regulation of an essential survival protein. Cells (2014) 3(2):418–37. doi: 10.3390/cells3020418
34. Jourdan M, De Vos J, Mechti N, Klein B. Regulation of bcl-2-family proteins in myeloma cells by three myeloma survival factors: interleukin-6, interferon-alpha and insulin-like growth factor 1. Cell Death Differ (2000) 7(12):1244–52. doi: 10.1038/sj.cdd.4400758
35. Jiang CC, Lucas K, Avery-Kiejda KA, Wade M, deBock CE, Thorne RF, et al. Up-regulation of mcl-1 is critical for survival of human melanoma cells upon endoplasmic reticulum stress. Cancer Res (2008) 68(16):6708–17. doi: 10.1158/0008-5472.CAN-08-0349
36. Piret J-P, Minet E, Cosse J-P, Ninane N, Debacq C, Raes M, et al. Hypoxia-inducible factor-1-dependent overexpression of myeloid cell factor-1 protects hypoxic cells against tert-butyl hydroperoxide-induced apoptosis. J Biol Chem (2005) 280(10):9336–44. doi: 10.1074/jbc.M411858200
37. Mott JL, Kobayashi S, Bronk SF, Gores GJ. Mir-29 regulates mcl-1 protein expression and apoptosis. Oncogene (2007) 26(42):6133–40. doi: 10.1038/sj.onc.1210436
38. Herrant M, Jacquel A, Marchetti S, Belhacene N, Colosetti P, Luciano F, et al. Cleavage of mcl-1 by caspases impaired its ability to counteract bim-induced apoptosis. Oncogene (2004) 23(47):7863–73. doi: 10.1038/sj.onc.1208069
39. Song T, Wang Z, Ji F, Feng Y, Fan Y, Chai G, et al. Deactivation of mcl-1 by dual-function small-molecule inhibitors targeting the bcl-2 homology 3 domain and facilitating mcl-1 ubiquitination. Angewandte Chemie Int Edition (2016) 55(46):14250–6. doi: 10.1002/anie.201606543
40. Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DCS, et al. Structural insights into the degradation of mcl-1 induced by BH3 domains. Proc Natl Acad Sci (2007) 104(15):6217–22. doi: 10.1073/pnas.0701297104
41. Ding Q, He X, Hsu J-M, Xia W, Chen C-T, Li L-Y, et al. Degradation of mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol (2007) 27(11):4006–17. doi: 10.1128/MCB.00620-06
42. Yang T, Kozopas KM, Craig RW. The intracellular distribution and pattern of expression of mcl-1 overlap with, but are not identical to, those of bcl-2. J Cell Biol (1995) 128(6):1173–84. doi: 10.1083/jcb.128.6.1173
43. Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell (2005) 17(3):393–403. doi: 10.1016/j.molcel.2004.12.030
44. Benard G, Karbowski M. Mitochondrial fusion and division: regulation and role in cell viability. In: Seminars in cell & developmental biology. (2009) 20(3):365–374. Academic Press
45. Escudero S, Zaganjor E, Lee S, Mill CP, Morgan AM, Crawford EB, et al. Dynamic regulation of long-chain fatty acid oxidation by a noncanonical interaction between the MCL-1 BH3 helix and VLCAD. Mol Cell (2018) 69(5):729–43.e7. doi: 10.1016/j.molcel.2018.02.005
46. Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M, et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol (2012) 14(6):575–83. doi: 10.1038/ncb2488
47. Hollville E, Carroll RG, Cullen SP, Martin SJ. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell (2014) 55(3):451–66. doi: 10.1016/j.molcel.2014.06.001
48. Cen X, Xu X, Xia H. Targeting MCL1 to induce mitophagy is a potential therapeutic strategy for Alzheimer disease. Autophagy (2021) 17(3):818–9. doi: 10.1080/15548627.2020.1860542
49. Cen X, Chen Y, Xu X, Wu R, He F, Zhao Q, et al. Pharmacological targeting of MCL-1 promotes mitophagy and improves disease pathologies in an alzheimer’s disease mouse model. Nat Commun (2020) 11(1):5731. doi: 10.1038/s41467-020-19547-6
50. Demelash A, Pfannenstiel LW, Tannenbaum CS, Li X, Kalady MF, DeVecchio J, et al. Structure-function analysis of the mcl-1 protein identifies a novel senescence-regulating domain. J Biol Chem (2015) 290(36):21962–75. doi: 10.1074/jbc.M115.663898
51. Mattoo AR, Pandita RK, Chakraborty S, Charaka V, Mujoo K, Hunt CR, et al. MCL-1 depletion impairs DNA double-strand break repair and reinitiation of stalled DNA replication forks. Mol Cell Biol (2017) 37(3):e00535–16. doi: 10.1128/MCB.00535-16
52. Perciavalle RM, Opferman JT. Delving deeper: MCL-1's contributions to normal and cancer biology. Trends Cell Biol (2013) 23(1):22–9. doi: 10.1016/j.tcb.2012.08.011
53. Zhou P, Levy NB, Xie H, Qian L, Lee C-YG, Gascoyne RD, et al. MCL1 transgenic mice exhibit a high incidence of b-cell lymphoma manifested as a spectrum of histologic subtypes. Blood J Am Soc Hematol (2001) 97(12):3902–9. doi: 10.1182/blood.V97.12.3902
54. Aichberger KJ, Mayerhofer M, Krauth M-T, Skvara H, Florian S, Sonneck K, et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood (2005) 105(8):3303–11. doi: 10.1182/blood-2004-02-0749
55. Breitenbuecher F, Markova B, Kasper S, Carius B, Stauder T, Böhmer FD, et al. A novel molecular mechanism of primary resistance to FLT3-kinase inhibitors in AML. Blood J Am Soc Hematol (2009) 113(17):4063–73. doi: 10.1182/blood-2007-11-126664
56. Fleischer B, Schulze-Bergkamen H, Schuchmann M, Weber A, Biesterfeld S, Müller M, et al. Mcl-1 is an anti-apoptotic factor for human hepatocellular carcinoma. Int J Oncol (2006) 28(1):25–32. doi: 10.3892/ijo.28.1.25
57. Henson ES, Hu X, Gibson SB. Herceptin sensitizes ErbB2–overexpressing cells to apoptosis by reducing antiapoptotic mcl-1 expression. Clin Cancer Res (2006) 12(3):845–53. doi: 10.1158/1078-0432.CCR-05-0754
58. Le Gouill S, Podar K, Amiot M, Hideshima T, Chauhan D, Ishitsuka K, et al. VEGF induces mcl-1 up-regulation and protects multiple myeloma cells against apoptosis. Blood (2004) 104(9):2886–92. doi: 10.1182/blood-2004-05-1760
59. Song L, Coppola D, Livingston S, Cress WD, Haura EB. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol Ther (2005) 4(3):267–76. doi: 10.4161/cbt.4.3.1496
60. Wuilleme-Toumi S, Robillard N, Gomez P, Moreau P, Le Gouill S, Avet-Loiseau H, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia (2005) 19(7):1248–52. doi: 10.1038/sj.leu.2403784
61. Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature (2010) 463(7277):103–7. doi: 10.1038/nature08646
62. Michels J, O'Neill JW, Dallman CL, Mouzakiti A, Habens F, Brimmell M, et al. Mcl-1 is required for Akata6 b-lymphoma cell survival and is converted to a cell death molecule by efficient caspase-mediated cleavage. Oncogene (2004) 23(28):4818–27. doi: 10.1038/sj.onc.1207648
63. Cho-Vega JH, Rassidakis GZ, Admirand JH, Oyarzo M, Ramalingam P, Paraguya A, et al. MCL-1 expression in b-cell non-hodgkin’s lymphomas. Hum Pathol (2004) 35(9):1095–100. doi: 10.1016/j.humpath.2004.04.018
64. Kuramoto K, Sakai A, Shigemasa K, Takimoto Y, Asaoku H, Tsujimoto T, et al. High expression of MCL1 gene related to vascular endothelial growth factor is associated with poor outcome in non-hodgkin's lymphoma. Br J Haematol (2002) 116(1):158–61. doi: 10.1046/j.1365-2141.2002.03253.x
65. Lv X, Tan G, Yao Y, Lv L, Deng X, Dong L, et al. Somatic mutations in myeloid cell leukemia−1 contribute to the pathogenesis of glioma by prolonging its half−life. Mol Med Rep (2015) 12(1):1265–71. doi: 10.3892/mmr.2015.3493
66. Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, et al. A novel MCL1 inhibitor combined with venetoclax rescues venetoclax-resistant acute myelogenous leukemia. Cancer Discov (2018) 8(12):1566–81. doi: 10.1158/2159-8290.CD-18-0140
67. Van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, et al. The BH3 mimetic ABT-737 targets selective bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if mcl-1 is neutralized. Cancer Cell (2006) 10(5):389–99. doi: 10.1016/j.ccr.2006.08.027
68. Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell (2018) 34(6):879–91. doi: 10.1016/j.ccell.2018.11.004
69. Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov (2014) 4(2):232–45. doi: 10.1158/2159-8290.CD-13-0286
70. Wei S-H, Dong K, Lin F, Wang X, Li B, Shen J-j, et al. Inducing apoptosis and enhancing chemosensitivity to gemcitabine via RNA interference targeting mcl-1 gene in pancreatic carcinoma cell. Cancer Chemother Pharmacol (2008) 62(6):1055–64. doi: 10.1007/s00280-008-0697-7
71. Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature (2011) 471(7336):110–4. doi: 10.1038/nature09779
72. Zhang Q, Riley-Gillis B, Han L, Jia Y, Lodi A, Zhang H, et al. Activation of RAS/MAPK pathway confers MCL-1 mediated acquired resistance to BCL-2 inhibitor venetoclax in acute myeloid leukemia. Signal Transduct Targeted Ther (2022) 7(1):51.
73. Bhatt S, Pioso MS, Olesinski EA, Yilma B, Ryan JA, Mashaka T, et al. Reduced mitochondrial apoptotic priming drives resistance to BH3 mimetics in acute myeloid leukemia. Cancer Cell (2020) 38(6):872–90.e6. doi: 10.1016/j.ccell.2020.10.010
74. Michels J, Obrist F, Vitale I, Lissa D, Garcia P, Behnam-Motlagh P, et al. MCL-1 dependency of cisplatin-resistant cancer cells. Biochem Pharmacol (2014) 92(1):55–61. doi: 10.1016/j.bcp.2014.07.029
75. Osaki S, Tazawa H, Hasei J, Yamakawa Y, Omori T, Sugiu K, et al. Ablation of MCL1 expression by virally induced microRNA-29 reverses chemoresistance in human osteosarcomas. Sci Rep (2016) 6:28953. doi: 10.1038/srep28953
76. Ohtsu A, Arai S, Sawada T, Kato M, Maeno Y, Miyazawa Y, et al. Fibroblast growth factor receptor inhibitor erdafitinib promotes mcl-1 degradation and synergistically induces apoptosis with bcl-xL/Bcl-2 inhibitor in urothelial cancer cells. Biochem Biophys Res Commun (2022) 628:76–83. doi: 10.1016/j.bbrc.2022.08.083
77. Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF FBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature (2011) 471(7336):104–9. doi: 10.1038/nature09732
78. Troiani M, Colucci M, D’Ambrosio M, Guccini I, Pasquini E, Varesi A, et al. Single-cell transcriptomics identifies mcl-1 as a target for senolytic therapy in cancer. Nat Commun (2022) 13(1):2177. doi: 10.1038/s41467-022-29824-1
79. Carter BZ, Mak PY, Tao W, Warmoes M, Lorenzi PL, Mak D, et al. Targeting MCL-1 dysregulates cell metabolism and leukemia-stroma interactions and re-sensitizes acute myeloid leukemia to BCL-2 inhibition. Haematologica (2022) 107(1):58. doi: 10.3324/haematol.2020.260331
80. Huang S, Liu Y, Chen Z, Wang M, Jiang VC. PIK-75 overcomes venetoclax resistance via blocking PI3K-AKT signaling and MCL-1 expression in mantle cell lymphoma. Am J Cancer Res (2022) 12(3):1102–1115.
81. Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ, et al. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood J Am Soc Hematol (2009) 113(19):4637–45. doi: 10.1182/blood-2008-12-190256
82. Kitada S, Zapata JM, Andreeff M, Reed JC. Protein kinase inhibitors flavopiridol and 7-hydroxy-staurosporine down-regulate antiapoptosis proteins in b-cell chronic lymphocytic leukemia. Blood J Am Soc Hematol (2000) 96(2):393–7. doi: 10.1182/blood.V96.2.393
83. Gandhi V, Plunkett W, Cortes JE. Omacetaxine: a protein translation inhibitor for treatment of chronic myelogenous LeukemiaOmacetaxine: a protein translation inhibitor in CML. Clin Cancer Res (2014) 20(7):1735–40. doi: 10.1158/1078-0432.CCR-13-1283
84. Liu T, Nair SJ, Lescarbeau A, Belani J, Sp P, Conley J, et al. Synthetic silvestrol analogues as potent and selective protein synthesis inhibitors. J Med Chem (2012) 55(20):8859–78. doi: 10.1021/jm3011542
85. Abulwerdi F, Liao C, Liu M, Azmi AS, Aboukameel A, Mady AS, et al. A novel small-molecule inhibitor of mcl-1 blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther (2014) 13(3):565–75. doi: 10.1158/1535-7163.MCT-12-0767
86. Lee JS, Tang SS, Ortiz V, Vo T-T, Fruman DA. MCL-1-independent mechanisms of synergy between dual PI3K/mTOR and BCL-2 inhibition in diffuse large b cell lymphoma. Oncotarget (2015) 6(34):35202. doi: 10.18632/oncotarget.6051
87. Rahmani M, Davis EM, Bauer C, Dent P, Grant S. Apoptosis induced by the kinase inhibitor BAY 43-9006 in human leukemia cells involves down-regulation of mcl-1 through inhibition of translation. J Biol Chem (2005) 280(42):35217–27. doi: 10.1074/jbc.M506551200
88. Kang X-h, Zhang J-h, Zhang Q-q, Cui Y-h, Wang Y, Kou W-z, et al. Degradation of mcl-1 through GSK-3β activation regulates apoptosis induced by bufalin in non-small cell lung cancer H1975 cells. Cell Physiol Biochem (2017) 41(5):2067–76. doi: 10.1159/000475438
89. Wang R, Xia L, Gabrilove J, Waxman S, Jing Y. Downregulation of mcl-1 through GSK-3β activation contributes to arsenic trioxide-induced apoptosis in acute myeloid leukemia cells. Leukemia (2013) 27(2):315–24. doi: 10.1038/leu.2012.180
90. Nifoussi SK, Vrana JA, Domina AM, De Biasio A, Gui J, Gregory MA, et al. Thr 163 phosphorylation causes mcl-1 stabilization when degradation is independent of the adjacent GSK3-targeted phosphodegron, promoting drug resistance in cancer. PloS One (2012) 7(10). doi: 10.1371/journal.pone.0047060
91. Thrane S, Pedersen A, Thomsen M, Kirkegaard T, Rasmussen B, Duun-Henriksen A, et al. A kinase inhibitor screen identifies mcl-1 and aurora kinase a as novel treatment targets in antiestrogen-resistant breast cancer cells. Oncogene (2015) 34(32):4199–210. doi: 10.1038/onc.2014.351
92. Papatzimas JW, Gorobets E, Maity R, Muniyat MI, MacCallum JL, Neri P, et al. From inhibition to degradation: targeting the antiapoptotic protein myeloid cell leukemia 1 (MCL1). J Med Chem (2019) 62(11):5522–40. doi: 10.1021/acs.jmedchem.9b00455
93. Lee S, Wales TE, Escudero S, Cohen DT, Luccarelli J, Gallagher CG, et al. Allosteric inhibition of antiapoptotic MCL-1. Nat Struct Mol Biol (2016) 23(6):600–7. doi: 10.1038/nsmb.3223
94. Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN, et al. A novel BH3 ligand that selectively targets mcl-1 reveals that apoptosis can proceed without mcl-1 degradation. J Cell Biol (2008) 180(2):341–55. doi: 10.1083/jcb.200708096
95. Harvey EP, Seo H-S, Guerra RM, Bird GH, Dhe-Paganon S, Walensky LD. Crystal structures of anti-apoptotic BFL-1 and its complex with a covalent stapled peptide inhibitor. Structure (2018) 26(1):153–60.e4. doi: 10.1016/j.str.2017.11.016
96. Winter A, Higueruelo AP, Marsh M, Sigurdardottir A, Pitt WR, Blundell TL. Biophysical and computational fragment-based approaches to targeting protein–protein interactions: applications in structure-guided drug discovery. Q Rev Biophys (2012) 45(4):383–426. doi: 10.1017/S0033583512000108
97. Rehman AU, Khurshid B, Ali Y, Rasheed S, Wadood A, Ng H-L, et al. Computational approaches for the design of modulators targeting protein-protein interactions. Expert Opin Drug Discovery (2023). doi: 10.1080/17460441.2023.2171396
98. Kitada S, Leone M, Sareth S, Zhai D, Reed JC, Pellecchia M. Discovery, characterization, and structure– activity relationships studies of proapoptotic polyphenols targeting b-cell lymphocyte/leukemia-2 proteins. J Med Chem (2003) 46(20):4259–64. doi: 10.1021/jm030190z
99. Arnold AA, Aboukameel A, Chen J, Yang D, Wang S, Al-Katib A, et al. Preclinical studies of apogossypolone: a new nonpeptidic pan small-molecule inhibitor of bcl-2, bcl-X l and mcl-1 proteins in follicular small cleaved cell lymphoma model. Mol Cancer (2008) 7(1):20. doi: 10.1186/1476-4598-7-20
100. Tzung S-P, Kim KM, Basañez G, Giedt CD, Simon J, Zimmerberg J, et al. Antimycin a mimics a cell-death-inducing bcl-2 homology domain 3. Nat Cell Biol (2001) 3(2):183–91. doi: 10.1038/35055095
101. Acoca S, Cui Q, Shore GC, Purisima EO. Molecular dynamics study of small molecule inhibitors of the bcl-2 family. Proteins: Struct Function Bioinf (2011) 79(9):2624–36. doi: 10.1002/prot.23083
102. Mohammad RM, Goustin AS, Aboukameel A, Chen B, Banerjee S, Wang G, et al. Preclinical studies of TW-37, a new nonpeptidic small-molecule inhibitor of bcl-2, in diffuse large cell lymphoma xenograft model reveal drug action on both bcl-2 and mcl-1. Clin Cancer Res (2007) 13(7):2226–35. doi: 10.1158/1078-0432.CCR-06-1574
103. Villalobos-Ortiz M, Ryan J, Mashaka TN, Opferman JT, Letai A. BH3 profiling discriminates on-target small molecule BH3 mimetics from putative mimetics. Cell Death Differ (2020) 27(3):999–1007. doi: 10.1038/s41418-019-0391-9
104. Wan Y, Dai N, Tang Z, Fang H. Small-molecule mcl-1 inhibitors: emerging anti-tumor agents. Eur J Med Chem (2018) 146:471–82. doi: 10.1016/j.ejmech.2018.01.076
105. Belmar J, Fesik SW. Small molecule mcl-1 inhibitors for the treatment of cancer. Pharmacol Ther (2015) 145:76–84. doi: 10.1016/j.pharmthera.2014.08.003
106. Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. Solution structure of prosurvival mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J Biol Chem (2005) 280(6):4738–44. doi: 10.1074/jbc.M411434200
107. Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol (2014) 21(9):1102–14. doi: 10.1016/j.chembiol.2014.09.001
108. Marimuthu P, Razzokov J, Eshonqulov G. Disruption of conserved polar interactions causes a sequential release of bim mutants from the canonical binding groove of Mcl1. Int J Biol Macromol (2020) 158:364–74. doi: 10.1016/j.ijbiomac.2020.04.243
109. Shaw S, Bian Z, Zhao B, Tarr JC, Veerasamy N, Jeon KO, et al. Optimization of potent and selective tricyclic indole diazepinone myeloid cell leukemia-1 inhibitors using structure-based design. J Med Chem (2018) 61(6):2410–21. doi: 10.1021/acs.jmedchem.7b01155
110. Dutta S, Gullá S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for mcl-1 versus bcl-xL. J Mol Biol (2010) 398(5):747–62. doi: 10.1016/j.jmb.2010.03.058
111. Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol (2010) 6(8):595. doi: 10.1038/nchembio.391
112. Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol (2014) 15(1):49–63. doi: 10.1038/nrm3722
113. Negi A, Murphy PV. Development of mcl-1 inhibitors for cancer therapy. Eur J Med Chem (2021) 210:113038. doi: 10.1016/j.ejmech.2020.113038
114. Caenepeel S, Brown SP, Belmontes B, Moody G, Keegan KS, Chui D, et al. AMG 176, a selective MCL1 inhibitor, is effective in hematologic cancer models alone and in combination with established therapies. Cancer Discov (2018) 8(12):1582–97. doi: 10.1158/2159-8290.CD-18-0387
115. Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, et al. Discovery of mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun (2018) 9(1):1–14. doi: 10.1038/s41467-018-07551-w
116. Caenepeel S, Karen R, Belmontes B, Verlinsky A, Tan H, Yang Y, et al. Discovery and preclinical evaluation of AMG 397, a potent, selective and orally bioavailable MCL1 inhibitor. Cancer Res (2020) 80(16_Supplement):6218–. doi: 10.1158/1538-7445.AM2020-6218
117. Leverson J, Zhang H, Chen J, Tahir S, Phillips D, Xue J, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis (2015) 6(1):e1590–e. doi: 10.1038/cddis.2014.561
118. Xiao Y, Nimmer P, Sheppard GS, Bruncko M, Hessler P, Lu X, et al. MCL-1 is a key determinant of breast cancer cell survival: validation of MCL-1 dependency utilizing a highly selective small molecule inhibitor. Mol Cancer Ther (2015) 14(8):1837–47. doi: 10.1158/1535-7163.MCT-14-0928
119. Wang Q, Hao S. A−1210477, a selective MCL−1 inhibitor, overcomes ABT−737 resistance in AML. Oncol Lett (2019) 18(5):5481–9. doi: 10.3892/ol.2019.10891
120. Meister MT, Boedicker C, Linder B, Kögel D, Klingebiel T, Fulda S. Concomitant targeting of hedgehog signaling and MCL-1 synergistically induces cell death in hedgehog-driven cancer cells. Cancer Lett (2019) 465:1–11. doi: 10.1016/j.canlet.2019.08.012
121. Fiskus W, Cai T, DiNardo CD, Kornblau SM, Borthakur G, Kadia TM, et al. Superior efficacy of cotreatment with BET protein inhibitor and BCL2 or MCL1 inhibitor against AML blast progenitor cells. Blood Cancer J (2019) 9(2):1–13. doi: 10.1038/s41408-018-0165-5
122. Ow TJ, Fulcher CD, Thomas C, Broin PÓ, López A, Reyna DE, et al. Optimal targeting of BCL-family proteins in head and neck squamous cell carcinoma requires inhibition of both BCL-xL and MCL-1. Oncotarget (2019) 10(4):494. doi: 10.18632/oncotarget.26563
123. Widden H, Kaczmarczyk A, Subedi A, Whitaker RH, Placzek WJ. MCL1 binds and negatively regulates the transcriptional function of tumor suppressor p73. Cell Death Dis (2020) 11(11):946. doi: 10.1038/s41419-020-03068-7
124. Pelz NF, Bian Z, Zhao B, Shaw S, Tarr JC, Belmar J, et al. Discovery of 2-indole-acylsulfonamide myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods. J Med Chem (2016) 59(5):2054–66. doi: 10.1021/acs.jmedchem.5b01660
125. Williams MM, Elion DL, Rahman B, Hicks DJ, Sanchez V, Cook RS. Therapeutic inhibition of mcl-1 blocks cell survival in estrogen receptor-positive breast cancers. Oncotarget (2019) 10(52):5389. doi: 10.18632/oncotarget.27070
126. Song X, Shen L, Tong J, Kuang C, Zeng S, Schoen RE, et al. Mcl-1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics (2020) 10(18):8098. doi: 10.7150/thno.45363
127. Koch R, Christie AL, Crombie JL, Palmer AC, Plana D, Shigemori K, et al. Biomarker-driven strategy for MCL1 inhibition in T-cell lymphomas. Blood J Am Soc Hematol (2019) 133(6):566–75. doi: 10.1182/blood-2018-07-865527
128. Sale MJ, Minihane E, Monks NR, Gilley R, Richards FM, Schifferli KP, et al. Targeting melanoma’s MCL1 bias unleashes the apoptotic potential of BRAF and ERK1/2 pathway inhibitors. Nat Commun (2019) 10(1):1–19. doi: 10.1038/s41467-019-12409-w
129. Timofeeva N, Ayres ML, Baran N, Santiago-O’Farrill JM, Bildik G, Lu Z, et al. Preclinical investigations of the efficacy of the glutaminase inhibitor CB-839 alone and in combinations in chronic lymphocytic leukemia. Front Oncol (2023) 13:1161254. doi: 10.3389/fonc.2023.1161254
130. Yi X, Sarkar A, Kismali G, Aslan B, Ayres M, Iles LR, et al. AMG-176, an mcl-1 antagonist, shows preclinical efficacy in chronic lymphocytic leukemia. Clin Cancer Res (2020) 26(14):3856–67. doi: 10.1158/1078-0432.CCR-19-1397
131. Sulkshane P, Teni T. Myeloid cell leukemia-1: a formidable barrier to anticancer therapeutics and the quest of targeting it. Explor Targeted Anti-tumor Ther (2022) 3(3):278. doi: 10.37349/etat.2022.00083
132. Roberts AW, Wei AH, Huang DC. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood J Am Soc Hematol (2021) 138(13):1120–36. doi: 10.1182/blood.2020006785
133. Li Z, He S, Look AT. The MCL1-specific inhibitor S63845 acts synergistically with venetoclax/ABT-199 to induce apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia (2019) 33(1):262–6. doi: 10.1038/s41375-018-0201-2
134. Manzano M, Patil A, Waldrop A, Dave SS, Behdad A, Gottwein E. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat Commun (2018) 9(1):1–14. doi: 10.1038/s41467-018-05506-9
135. Chen LS, Keating MJ, Wierda WG, Gandhi V. Induction of apoptosis by MCL-1 inhibitors in chronic lymphocytic leukemia cells. Leukemia Lymphoma (2019). doi: 10.1080/10428194.2019.1622098
136. Moujalled DM, Pomilio G, Ghiurau C, Ivey A, Salmon J, Rijal S, et al. Combining BH3-mimetics to target both BCL-2 and MCL1 has potent activity in pre-clinical models of acute myeloid leukemia. Leukemia (2019) 33(4):905–17. doi: 10.1038/s41375-018-0261-3
137. Merino D, Whittle JR, Vaillant F, Serrano A, Gong J-N, Giner G, et al. Synergistic action of the MCL-1 inhibitor S63845 with current therapies in preclinical models of triple-negative and HER2-amplified breast cancer. Sci Trans Med (2017) 9(401):eaam7049. doi: 10.1126/scitranslmed.aam7049
138. Brennan MS, Chang C, Tai L, Lessene G, Strasser A, Dewson G, et al. Humanized mcl-1 mice enable accurate preclinical evaluation of MCL-1 inhibitors destined for clinical use. Blood J Am Soc Hematol (2018) 132(15):1573–83. doi: 10.1182/blood-2018-06-859405
139. Bhagwat N, Grego A, Gowen-MacDonald W, Wang M, Cowart M, Wu X, et al. Preclinical characterization of PRT1419, a potent, selective and orally available inhibitor of MCL1. Cancer Res (2021) 81(13_Supplement):983–. doi: 10.1158/1538-7445.AM2021-983
140. Tantawy SI, Sarkar A, Hubner S, Tan Z, Wierda WG, Eldeib A, et al. Mechanisms of MCL-1 protein stability induced by MCL-1 antagonists in b-cell malignancies. Clin Cancer Res (2023) 29(2):446–57. doi: 10.1158/1078-0432.CCR-22-2088
141. Haschka MD, Karbon G, Soratroi C, O’Neill KL, Luo X, Villunger A. MARCH5-dependent degradation of MCL1/NOXA complexes defines susceptibility to antimitotic drug treatment. Cell Death Differ (2020) 27(8):2297–312. doi: 10.1038/s41418-020-0503-6
142. Tantawy SI, Timofeeva N, Hernandez A, Sarkar A, Gandhi V. Decoding the mechanism behind MCL-1 inhibitors: a pathway to understanding MCL-1 protein stability. Oncotarget (2023) 14:653. doi: 10.18632/oncotarget.28440
143. Friberg A, Vigil D, Zhao B, Daniels RN, Burke JP, Garcia-Barrantes PM, et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J Med Chem (2013) 56(1):15–30. doi: 10.1021/jm301448p
144. Hird AW, Tron AE. Recent advances in the development of mcl-1 inhibitors for cancer therapy. Pharmacol Ther (2019) 198:59–67. doi: 10.1016/j.pharmthera.2019.02.007
145. Daressy F, Séguy L, Favre L, Corvaisier S, Apel C, Groo A-C, et al. NA1–115—7, from zygogynum pancheri, is a new selective MCL-1 inhibitor inducing the apoptosis of hematological cancer cells but non-toxic to normal blood cells or cardiomyocytes. Biomed Pharmacother (2022) 154:113546. doi: 10.1016/j.biopha.2022.113546
146. Valentini E, D'Aguanno S, Di Martile M, Montesano C, Ferraresi V, Patsilinakos A, et al. Targeting the anti-apoptotic bcl-2 family proteins: machine learning virtual screening and biological evaluation of new small molecules. Theranostics (2022) 12(5):2427. doi: 10.7150/thno.64233
147. Drennen B, Goodis CC, Bowen N, Yu W, Vickers G, Wilder PT, et al. Scaffold hopping from indoles to indazoles yields dual MCL-1/BCL-2 inhibitors from MCL-1 selective leads. RSC Med Chem (2022) 13(8):963–9. doi: 10.1039/D2MD00095D
148. Uthale A, Anantram A, Sulkshane P, Degani M, Teni T. Identification of bicyclic compounds that act as dual inhibitors of bcl-2 and mcl-1. Mol Diversity (2022), 1–16. doi: 10.1007/s11030-022-10494-6
149. Hikita H, Takehara T, Shimizu S, Kodama T, Li W, Miyagi T, et al. Mcl-1 and bcl-xL cooperatively maintain integrity of hepatocytes in developing and adult murine liver. Hepatology (2009) 50(4):1217–26. doi: 10.1002/hep.23126
150. Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol (2012) 30(5):488. doi: 10.1200/JCO.2011.34.7898
151. Kipps TJ, Eradat H, Grosicki S, Catalano J, Cosolo W, Dyagil IS, et al. A phase 2 study of the BH3 mimetic BCL2 inhibitor navitoclax (ABT-263) with or without rituximab, in previously untreated b-cell chronic lymphocytic leukemia. Leukemia Lymphoma (2015) 56(10):2826–33. doi: 10.3109/10428194.2015.1030638
152. Fletcher S. MCL-1 inhibitors–where are we now (2019)? Expert Opin Ther Patents (2019) 29(11):909–19. doi: 10.1080/13543776.2019.1672661
153. Wei G, Margolin AA, Haery L, Brown E, Cucolo L, Julian B, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell (2012) 21(4):547–62. doi: 10.1016/j.ccr.2012.02.028
154. Letai A, Certo M, Brown JR, Moore V. The molecular basis for BCL-2 oncogene addiction in CLL. Am Soc Hematol (2005). doi: 10.1182/blood.V106.11.5008.5008
155. Liu F, Zhao Q, Su Y, Lv J, Gai Y, Liu S, et al. Cotargeting of bcl-2 and mcl-1 shows promising antileukemic activity against AML cells including those with acquired cytarabine resistance. Exp Hematol (2022) 105:39–49. doi: 10.1016/j.exphem.2021.10.006
156. Thijssen R, Diepstraten ST, Moujalled D, Chew E, Flensburg C, Shi MX, et al. Intact TP-53 function is essential for sustaining durable responses to BH3-mimetic drugs in leukemias. Blood J Am Soc Hematol (2021) 137(20):2721–35. doi: 10.1182/blood.2020010167
157. Nangia V, Siddiqui FM, Caenepeel S, Timonina D, Bilton SJ, Phan N, et al. Exploiting MCL1 dependency with combination MEK+ MCL1 inhibitors leads to induction of apoptosis and tumor regression in KRAS-mutant non–small cell lung cancer. Cancer Discov (2018) 8(12):1598–613. doi: 10.1158/2159-8290.CD-18-0277
158. Touzeau C, Maciag P, Amiot M, Moreau P. Targeting bcl-2 for the treatment of multiple myeloma. Leukemia (2018) 32(9):1899–907. doi: 10.1038/s41375-018-0223-9
159. Ponder KG, Matulis SM, Hitosugi S, Gupta VA, Sharp C, Burrows F, et al. Dual inhibition of mcl-1 by the combination of carfilzomib and TG02 in multiple myeloma. Cancer Biol Ther (2016) 17(7):769–77. doi: 10.1080/15384047.2016.1192086
160. Nguyen T, Parker R, Zhang Y, Hawkins E, Kmieciak M, Craun W, et al. Homoharringtonine interacts synergistically with bortezomib in NHL cells through MCL-1 and NOXA-dependent mechanisms. BMC Cancer (2018) 18(1):1–11. doi: 10.1186/s12885-018-5018-x
161. Happo L, Cragg MS, Phipson B, Haga JM, Jansen ES, Herold MJ, et al. Maximal killing of lymphoma cells by DNA damage–inducing therapy requires not only the p53 targets puma and noxa, but also bim. Blood J Am Soc Hematol (2010) 116(24):5256–67. doi: 10.1182/blood-2010-04-280818
162. Wang Y, Wang D, Wang Y, Yang H, Wang G, Wu S. Synergistic activity and mechanism of cytarabine and MCL-1 inhibitor AZD5991 against acute myeloid leukemia. Neoplasma (2023), 221217N1185–N1185. doi: 10.4149/neo_2023_221217N1185
163. Chen G, Magis AT, Xu K, Park D, David SY, Owonikoko TK, et al. Targeting mcl-1 enhances DNA replication stress sensitivity to cancer therapy. J Clin Invest (2018) 128(1):500–16. doi: 10.1158/1538-7445.AM2018-328
164. Ebrahimi E, Shabestari RM, Bashash D, Safa M. Synergistic apoptotic effect of mcl-1 inhibition and doxorubicin on b-cell precursor acute lymphoblastic leukemia cells. Mol Biol Rep (2022) 49(3):2025–36. doi: 10.1007/s11033-021-07021-5
165. Seipel K, Schmitter K, Bacher U, Pabst T. Rationale for a combination therapy consisting of MCL1-and MEK-inhibitors in acute myeloid leukemia. Cancers (2019) 11(11):1779. doi: 10.3390/cancers11111779
166. Winkler M, Friedrich J, Boedicker C, Dolgikh N. Co-Targeting MCL-1 and ERK1/2 kinase induces mitochondrial apoptosis in rhabdomyosarcoma cells. Trans Oncol (2022) 16:101313. doi: 10.1016/j.tranon.2021.101313
167. Rauh U, Wei G, Serrano-Wu M, Kosmidis G, Kaulfuss S, Siegel F, et al. ANJ810 is a highly selective novel MCL1 inhibitor with optimized in vivo clearance showing robust efficacy in preclinical solid and hematological tumor models. Cancer Res (2023) 83(7_Supplement):506–. doi: 10.1158/1538-7445.AM2023-506
168. Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell (2013) 23(1):121–8. doi: 10.1016/j.ccr.2012.11.007
169. Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell (2015) 160(5):977–89. doi: 10.1016/j.cell.2015.01.042
170. Gupta VA, Matulis SM, Conage-Pough JE, Nooka AK, Kaufman JL, Lonial S, et al. Bone marrow microenvironment–derived signals induce mcl-1 dependence in multiple myeloma. Blood J Am Soc Hematol (2017) 129(14):1969–79. doi: 10.1182/blood-2016-10-745059
171. Pei X-Y, Dai Y, Felthousen J, Chen S, Takabatake Y, Zhou L, et al. Circumvention of mcl-1-dependent drug resistance by simultaneous Chk1 and MEK1/2 inhibition in human multiple myeloma cells. PloS One (2014) 9(3):e89064. doi: 10.1371/journal.pone.0089064
172. Balakrishnan K, Burger JA, Wierda WG, Gandhi V. AT-101 induces apoptosis in CLL b cells and overcomes stromal cell–mediated mcl-1 induction and drug resistance. Blood J Am Soc Hematol (2009) 113(1):149–53. doi: 10.1182/blood-2008-02-138560
173. Dengler MA, Teh CE, Thijssen R, Gangoda L, Lan P, Herold MJ, et al. Potent efficacy of MCL-1 inhibitor-based therapies in preclinical models of mantle cell lymphoma. Oncogene (2020) 39(9):2009–23. doi: 10.1038/s41388-019-1122-x
174. Wang MY, Li T, Ren Y, Shah BD, Lwin T, Gao J, et al. MCL-1 dependency as a novel vulnerability for aggressive b cell lymphomas. Blood Cancer J (2021) 11(1):14. doi: 10.1038/s41408-020-00402-2
175. Liu T, Lam V, Thieme E, Sun D, Wang X, Xu F, et al. Pharmacologic targeting of mcl-1 induces mitochondrial dysfunction and apoptosis in b-cell lymphoma cells in a TP53-and BAX-dependent MannerMcl-1 inhibition in lymphoma. Clin Cancer Res (2021) 27(17):4910–22. doi: 10.1158/1078-0432.CCR-21-0464
176. Sumarni U, Zhu J, Sinnberg T, Eberle J. Sensitivity of cutaneous T-cell lymphoma cells to the mcl-1 inhibitor S63845 correlates with the lack of bcl-w expression. Int J Mol Sci (2022) 23(20):12471. doi: 10.3390/ijms232012471
177. Bolomsky A, Miettinen JJ, Malyutina A, Besse A, Huber J, Fellinger S, et al. Heterogeneous modulation of bcl-2 family members and drug efflux mediate MCL-1 inhibitor resistance in multiple myeloma. Blood Adv (2021) 5(20):4125–39. doi: 10.1182/bloodadvances.2020003826
178. Tausch E, Close W, Dolnik A, Bloehdorn J, Chyla B, Bullinger L, et al. Venetoclax resistance and acquired BCL2 mutations in chronic lymphocytic leukemia. Haematologica (2019) 104(9):e434. doi: 10.3324/haematol.2019.222588
179. Chen B, Pamela W, Auclair D, Keats JJ, Secrist P, Cidado J, et al. Myeloma patient-derived MCL1 point mutations can influence MCL1-inhibitor function. Blood (2018) 132:951. doi: 10.1182/blood-2018-99-113444
180. Liu S, Qiao X, Wu S, Gai Y, Su Y, Edwards H, et al. C-myc plays a critical role in the antileukemic activity of the mcl-1-selective inhibitor AZD5991 in acute myeloid leukemia. Apoptosis (2022), 1–16. doi: 10.1007/s10495-022-01756-7
181. Gomez-Bougie P, Maiga S, Tessoulin B, Bourcier J, Bonnet A, Rodriguez MS, et al. BH3-mimetic toolkit guides the respective use of BCL2 and MCL1 BH3-mimetics in myeloma treatment. Blood J Am Soc Hematol (2018) 132(25):2656–69. doi: 10.1182/blood-2018-03-836718
182. Moyzis AG, Leon LJ, Najor RH, Gustafsson AB. MCL-1 regulates mitochondrial fission and mitophagy. Circulation (2017) 136(suppl_1):A14178–A. doi: 10.1161/res.119.suppl_1.375
183. Wang X, Bathina M, Lynch J, Koss B, Calabrese C, Frase S, et al. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev (2013) 27(12):1351–64. doi: 10.1101/gad.215855.113
184. Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, et al. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev (2013) 27(12):1365–77. doi: 10.1101/gad.215871.113
185. Kuznetsov AV, Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci (2009) 10(4):1911–29. doi: 10.3390/ijms10041911
186. Rasmussen ML, Taneja N, Neininger AC, Wang L, Robertson GL, Riffle SN, et al. MCL-1 inhibition by selective BH3 mimetics disrupts mitochondrial dynamics causing loss of viability and functionality of human cardiomyocytes. Iscience (2020) 23(4), 101015. doi: 10.1016/j.isci.2020.101015
187. Walsh C, Barrow S, Voronina S, Chvanov M, Petersen OH, Tepikin A. Modulation of calcium signalling by mitochondria. Biochim Biophys Acta (BBA)-Bioenergetics (2009) 1787(11):1374–82. doi: 10.1016/j.bbabio.2009.01.007
188. Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic (2007) 8(12):1668–75. doi: 10.1111/j.1600-0854.2007.00644.x
189. Cadenas S, Aragones J, Landazuri MO. Mitochondrial reprogramming through cardiac oxygen sensors in ischaemic heart disease. Cardiovasc Res (2010) 88(2):219–28. doi: 10.1093/cvr/cvq256
190. Rosca MG, Hoppel CL. Mitochondria in heart failure. Cardiovasc Res (2010) 88(1):40–50. doi: 10.1093/cvr/cvq240
191. Verdejo HE, Del Campo A, Troncoso R, Gutierrez T, Toro B, Quiroga C, et al. Mitochondria, myocardial remodeling, and cardiovascular disease. Curr Hypertension Rep (2012) 14:532–9. doi: 10.1007/s11906-012-0305-4
192. Gu J, Hu W, Song Z-P, Chen Y-G, Zhang D-D, Wang C-Q. Rapamycin inhibits cardiac hypertrophy by promoting autophagy via the MEK/ERK/Beclin-1 pathway. Front Physiol (2016) 7:104. doi: 10.3389/fphys.2016.00104
193. Xiao B, Hong L, Cai X, Mei S, Zhang P, Shao L. The true colors of autophagy in doxorubicin−induced cardiotoxicity. Oncol Lett (2019) 18(3):2165–72. doi: 10.3892/ol.2019.10576
194. Wang J, Cui D, Gu S, Chen X, Bi Y, Xiong X, et al. Autophagy regulates apoptosis by targeting NOXA for degradation. Biochim Biophys Acta (BBA)-Mol Cell Res (2018) 1865(8):1105–13. doi: 10.1016/j.bbamcr.2018.05.007
195. Liu YL, Lai F, Wilmott JS, Yan XG, Liu XY, Luan Q, et al. Noxa upregulation by oncogenic activation of MEK/ERK through CREB promotes autophagy in human melanoma cells. Oncotarget (2014) 5(22):11237. doi: 10.18632/oncotarget.2616
196. Dadson K, Hauck L, Hao Z, Grothe D, Rao V, Mak TW, et al. The E3 ligase mule protects the heart against oxidative stress and mitochondrial dysfunction through myc-dependent inactivation of pgc-1α and Pink1. Sci Rep (2017) 7(1):1–14. doi: 10.1038/srep41490
197. Senichkin VV, Streletskaia AY, Zhivotovsky B, Kopeina GS. Molecular comprehension of mcl-1: from gene structure to cancer therapy. Trends Cell Biol (2019) 29(7):549–62. doi: 10.1016/j.tcb.2019.03.004
198. Guièze R, Liu VM, Rosebrock D, Jourdain AA, Hernández-Sánchez M, Zurita AM, et al. Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell (2019) 36(4):369–84.e13. doi: 10.1016/j.ccell.2019.08.005
199. Moore VDG, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest (2007) 117(1):112–21. doi: 10.1172/JCI28281
200. Cervantes-Gomez F, Lamothe B, Woyach JA, Wierda WG, Keating MJ, Balakrishnan K, et al. Pharmacological and protein profiling suggests venetoclax (ABT-199) as optimal partner with ibrutinib in chronic lymphocytic LeukemiaCombination of ABT-199 and ibrutinib for CLL. Clin Cancer Res (2015) 21(16):3705–15. doi: 10.1158/1078-0432.CCR-14-2809
201. Jain N, Keating M, Thompson P, Ferrajoli A, Burger J, Borthakur G, et al. Ibrutinib and venetoclax for first-line treatment of CLL. New Engl J Med (2019) 380(22):2095–103. doi: 10.1056/NEJMoa1900574
Keywords: MCL-1 protein, BCL-2 family proteins, MCL-1 inhibitors, apoptosis, BCL-2 family, cancer therapy
Citation: Tantawy SI, Timofeeva N, Sarkar A and Gandhi V (2023) Targeting MCL-1 protein to treat cancer: opportunities and challenges. Front. Oncol. 13:1226289. doi: 10.3389/fonc.2023.1226289
Received: 24 May 2023; Accepted: 03 July 2023;
Published: 31 July 2023.
Edited by:
Chris Pepper, Brighton and Sussex Medical School, United KingdomReviewed by:
Shruti Bhatt, National University of Singapore, SingaporeCopyright © 2023 Tantawy, Timofeeva, Sarkar and Gandhi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Varsha Gandhi, dmdhbmRoaUBtZGFuZGVyc29uLm9yZw==; Aloke Sarkar, YXNhcmthckBtZGFuZGVyc29uLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.