
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 11 October 2023
Sec. Breast Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1226118
This article is part of the Research Topic Epigenetic and Metabolic Regulators of Breast Carcinogenesis View all 13 articles
Claudin-low breast cancer (CLBC) is a subgroup of breast cancer discovered at the molecular level in 2007. Claudin is one of the primary proteins that make up tight junctions, and it plays crucial roles in anti-inflammatory and antitumor responses as well as the maintenance of water and electrolyte balance. Decreased expression of claudin results in the disruption of tight junction structures and the activation of downstream signaling pathways, which can lead to tumor formation. The origin of Claudin-low breast cancer is still in dispute. Claudin-low breast cancer is characterized by low expression of Claudin3, 4, 7, E-cadherin, and HER2 and high expression of Vimentin, Snai 1/2, Twist 1/2, Zeb 1/2, and ALDH1, as well as stem cell characteristics. The clinical onset of claudin-low breast cancer is at menopause age, and its histological grade is higher. This subtype of breast cancer is more likely to spread to lymph nodes than other subtypes. Claudin-low breast cancer is frequently accompanied by increased invasiveness and a poor prognosis. According to a clinical retrospective analysis, claudin-low breast cancer can achieve low pathological complete remission. At present, although several therapeutic targets of claudin-low breast cancer have been identified, the effective treatment remains in basic research stages, and no animal studies or clinical trials have been designed. The origin, molecular biological characteristics, pathological characteristics, treatment, and prognosis of CLBC are extensively discussed in this article. This will contribute to a comprehensive understanding of CLBC and serve as the foundation for the individualization of breast cancer treatment.
Breast cancer is one of the three most common cancers in the world. Hierarchical cluster analysis of the genes that vary more between tumors than between repeated samples of the same tumor has revealed the existence of four major breast cancer intrinsic subtypes (luminal A, luminal B, HER2-enriched, and basal-like), as well as a normal breast-like group (1, 2). In 2007, Herschkowitz et al. (3) found a breast cancer subtype called the claudin-low subtype upon the examination of murine and human breast tumors. This newly discovered subtype of breast cancer has features of low expression of tight junction proteins and adhesion proteins (Claudin3, 4, 7, and E-cadherin) and luminal markers but high expression of basal-related genes and lymphocyte- and endothelial cell-related markers. Recently, scientists have paid more attention to this intrinsic molecular subtype of breast cancer, which is known as claudin-low breast cancer (CLBC). A comprehensive analysis of CLBC will help us to better understand this intrinsic subtype of breast cancer.
Furuse and Tsukita first discovered claudin in 1998. Claudin derives from the Latin word ‘claudere’, which means to close (4, 5). Claudins (CLDN) are cell–cell adhesion proteins that are expressed at tight junctions (TJs), which are the most prevalent apical cell–cell adhesions (6). Tight junctions, together with adherens junctions and desmosomes, form the apical junctional complex in epithelial and endothelial cellular sheets. Adherens junctions and desmosomes are responsible for the mechanical adhesion between adjacent cells, whereas tight junctions are essential for the tight sealing of the cellular sheets, thus controlling paracellular ion flux and therefore maintaining tissue homeostasis (4, 7, 8). The tight junction proteins are diverse and include occludins, claudins, tricellulin, cingulin, and junctional adhesion molecules (JAMs). These proteins interact within themselves and with the cellular cytoskeleton to form a complex architecture. Among these TJ proteins, claudins are key proteins that act as both pores and barriers, aiding the paracellular pathway between epithelial cells (9). In mammals, it is composed of 27 members, and specific CLDN combinations are expressed in specific cell and tissue types. Claudins can be divided into classical claudins and nonclassical claudins according to the homology of the claudin sequence and its function (10). Claudin has four transmembrane proteins; in mammals, 27 members are between 20 and 34 kDa in size (11). Claudin has four transmembrane helices, and its amino- and carboxy-termini penetrate deep into the cytoplasm. The N-terminal domain of Claudins is relatively short, contain 7 amino acid sequences followed by a large extracellular loop (ECL1) of approximately 50 amino acid sequences. A short inner loop is separated by a small extracellular loop (ECL2) of approximately 25 amino acid residues. The main role of ECL1 is paracellular transport with selective ion permeability. The role of ECL2 is to participate in the interaction between Claudins. ECL2 of Claudin3 and Claudin4 contains binding receptors for Clostridium perfringens enterotoxin (CPE) (5, 10, 12–14). Studies have found that CPE can be used as a target for the development of claudin-targeted drugs (15–17). The localization and function of claudin are also regulated by the phosphorylation of the C-terminus, a target of serine, threonine, and tyrosine kinases. Claudin regulates functional changes in TJ proteins through posttranslational modifications, such as phosphorylation, ubiquitination, palmitoylation, and glycosylation, to regulate claudin conformation, stability, transport, and function (10). The C-terminus of claudins contains a PDZ-binding domain, for signal transduction. Claudin binds to Zonula occludens (ZO1, ZO2, and ZO3 (18)), Pals1-associated tight junction protein (PATJ) (19) and Multi-PDZ domain protein 1 (MUPP1) (20) through a PDZ-interacting domain, which plays an important role in cell function. ZO-1, ZO-2, and ZO-3 are TJ-related proteins (21–23). ZO-1 and ZO-2 are key proteins for cell junction assembly and permeability, respectively (24, 25). When ZO-1 and ZO-2 are missing, cells cannot assemble TJs (26, 27). The structure diagram of claudin is shown in Figure 1. The PDZ domain-binding motif located at the C-terminal end of claudin cytoplasm can directly interact with ZO family proteins, which plays important roles in many cellular processes.
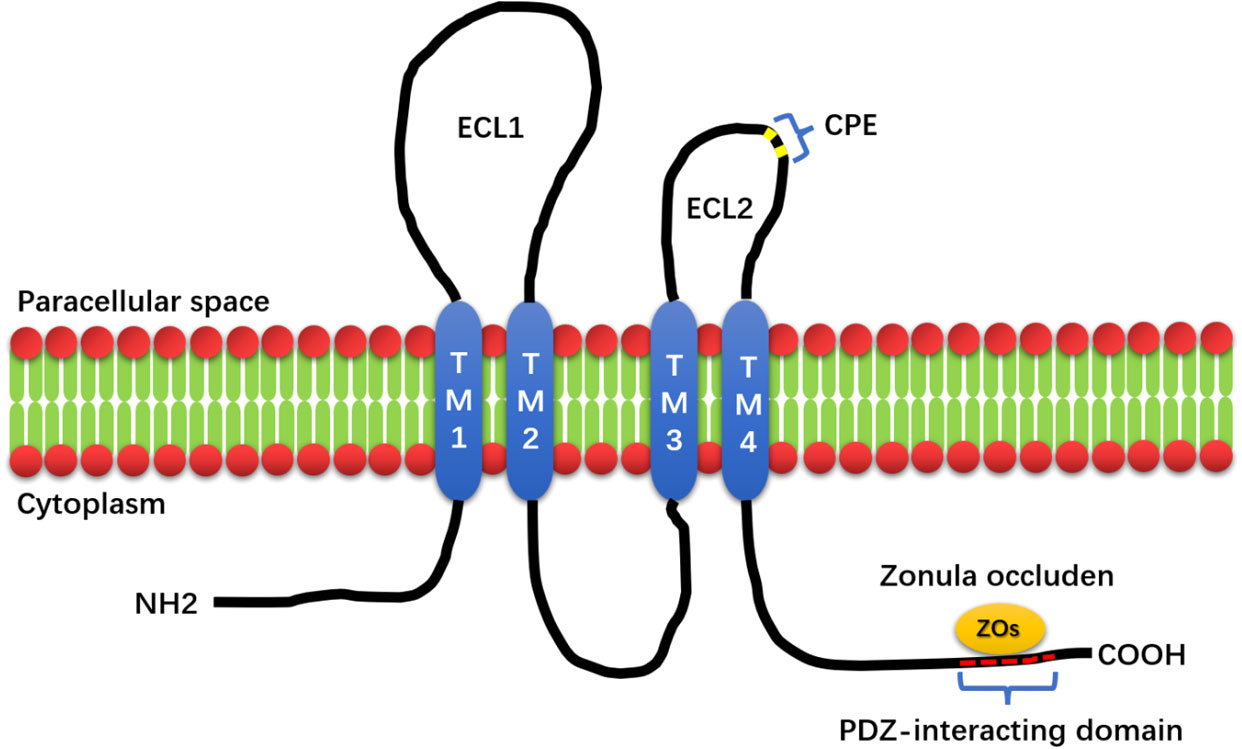
Figure 1 The structure of Claudin. The structure of Claudin mainly consists of four transmembrane structures (TM1, 2, 3, and 4), two extracellular loops, and one intracellular loop (ECL1, ECL2). The small extracellular loop contains the Clostridium perfringens enterotoxin-binding receptor (CPE), and the PDZ-interacting domain at the C-terminus of Claudin is capable of binding to Zonula occluden (Zos) to maintain cell homeostasis.
Whether different claudin proteins copolymerize to form tight junction chains as heteropolymers, and whether claudin interacts in a homogeneous manner, between two molecules of the same claudin member, or between two different claudin members remains unclear. According to a previous study, different claudin members can interact within and between tight junction strands, but these combinations were restricted to specific combinations of isoforms (28).
Claudin is widely expressed in many different tissues. The majority of claudin expression occurs in barrier-forming epithelial cells and endothelial cells. Not all claudins are expressed in all tissues at the same time, and these differences in how claudins are expressed control the functions of cells, especially paracellular barrier functions. Claudins play a key role in regulating the paracellular permeability selectivity. The overexpression of Claudin affects the resistance and permeability of epithelial cells to different ions, and ECL1 plays an important role in the selectivity of charge (29, 30). Various mouse models have shown that Claudin plays an important role in cellular barriers. For example, claudin-1-deficient mice are dehydrated due to increased cell permeability, leading to rapid death (31). Claudin shows abnormal expression in several cancers, which is related to the occurrence and progression of cancer. Claudin-1 is not expressed in breast cancer and colon cancer, which is related to the decomposition of TJs in tumor development (32, 33). Claudin is a double-edged sword. The loss or low expression of Claudin in some tumors is related to the progression, invasion and metastasis of cancer, such as gastric cancer (34), esophageal squamous cell carcinoma (35) and colorectal cancer (36). Claudin has the opposite effect in some cancers. Claudin can directly or indirectly activate various signal pathways or proteases, and promote the occurrence of tumors. Therefore, it has been observed that Claudin is highly expressed in some tumors. Claudin-3 and -4 have been found to be expressed in many types of tumors, and some studies have shown that the overexpression of Claudin in tumors is related to tumor growth and invasion (16, 37), such as ovarian cancer (38, 39), glioma (40) and pancreatic cancer (41, 42). The role of Claudin in cancer stem cell biology through the WNT pathway has attracted increasing attention. Claudin-1 and -2 transcription is regulated by WNT signaling, which is known to regulate the β-Catenin-T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) signaling pathway and stemness (43, 44).
CLBC was originally defined by a gene expression signature, which represents the low expression of claudin and cell-cell adhesion-related genes, including claudin 3, 4, and 7, and the high expression of EMT-related genes and breast cancer stem cell-related genes (45). Defining CLBC by genome is the foundation for most researchers to study CLBC. In addition, some researchers defined CLBC by immunohistochemistry. Most scholars define the low expression of claudins 3, 4, and 7 in the tumor cell membrane as CLBC. However, unlike HER2 in immunohistochemical staining, there is no unified standard to define the low expression and high expression of claudin in the interpretation of Claudin immunohistochemical (IHC) staining. Some scholars have multiplied the staining intensity and staining degree of claudin in tumor cells and delineated the corresponding boundary to define the high and low expression of claudin (46–50). Other researchers defined CLBC when less than 50% of tumor cells express claudins 3 and 4 and less than 5% of tumor cells express claudin 7 (51). However, some scholars have recently put forward a new view that Claudin-low is a biological characteristic that describes breast cancer and is not equivalent to the inherent molecular classification of breast cancer (52). Therefore, we believe that the definition of Claudin-low breast cancer should be a type of breast tumor characterized by low expression or absence of epithelial adhesion proteins (Claudin3, Claudin4, Claudin7 and E-cadherin, etc.). Compared with the intrinsic molecular classification of breast cancer, CLBC has its own unique characteristics in clinical pathology, molecular biology and other aspects.
The origin of CLBC has been disputed. At present, there are three main theories about the origin of CLBC. Some scholars have found that CLBC has features of tumor-initiating cells or stem cells. They speculated that the intrinsic subtype of breast cancer may reflect different stages of epithelial development. CLBC represents the most primitive tumors that are similar to mammary stem cells and may originate from mammary stem cells (MaSCs) (53–55). Previous studies have found that the active form of RAS and various cytokines, including transforming growth factor-β (TGF-β), promote epithelial-mesenchymal transformation (EMT), which transforms breast epithelial cells into malignant cells, which have all the characteristics of CLBC (56–59). Many factors regulate the transformation of luminal epithelium into CLBC through EMT. For example, oncogenic RAS signaling drives the occurrence and progression of triple-negative breast cancer from the luminal epithelium (60). Absence of the Notch signaling regulators Lunatic Fringe (Lfng) and p53 leads to CLBC (61). TbL1 interacts with ZEB1, which inhibits the activation of the E-cadherin (CDH-1) gene, activates the ZEB1 gene promoter, and promotes the transformation of mammary epithelial cells to CLBC (62). There is evidence that, under the influence of genetic mutations or environmental conditions, reactivation or dedifferentiation of luminal epithelial cells induces and accelerates the formation of more aggressive mammary tumors. Furthermore, the deletion of p53 can lead to clonal proliferation of the luminal epithelium, which promotes the development of breast tumors and the acquisition of MaSC characteristics, resulting in the formation of CLBC (63–65). In recent years, some scholars have deleted the Pten gene in the mouse mammary epithelium and induced the p53-R270H mutation, leading to CLBC (66). Other scholars have found that the synergistic effect of MET and p53 deletion can induce CLBC (67). However, according to the study by Pommier et al. (68), CLBC shows remarkable diversity. Based on the analysis of genetics, gene methylation, and gene expression, they found that CLBC arises from three subgroups, two of which are related to luminal subtype breast cancer and basal-like subtype breast cancer, which are transformed by activating the EMT process during tumor development. The third subgroup is closely related to normal human breast stem cells (MaSCs), showing genome distortion and a low frequency of the TP53 mutation. In short, the origin of CLBC is a complicated process.
According to previous studies, claudin-low breast cancer has always been considered to fall into the category of triple-negative breast cancer. According to Pommier et al. (68), claudin-low breast cancer may originate from various stages of breast cancer development, and this is likely the best explanation for the high expression of breast cancer stem cell markers and EMT-related markers in claudin-low breast cancer as well as its overlap in gene and immune expression with luminal A/B, HER2 overexpression, and triple-negative breast cancer. The Shanghai Cancer Center of Fudan University (FUSCC) used androgen receptor (AR), CD8, FOXC1, and DCLK1 as immunohistochemical markers and classified TNBCs into five subtypes based on the staining results: (a) IHC‐based luminal androgen receptor (IHC‐LAR; AR+), (b) IHC‐based immunomodulatory (IHC‐IM; AR−, CD8+), (c) IHC‐based basal‐like immune‐suppressed (IHC‐BLIS; AR−, CD8−, FOXC1+), (d) IHC‐based mesenchymal (IHC‐MES; AR−, CD8−, FOXC1−, DCLK1+), and (e) IHC‐based unclassifiable (AR−, CD8−, FOXC1−, DCLK1−) (69). Currently, there is no relevant research showing the relationship between claudin-low and FUSCC triple-negative breast cancer staging. Since claudin breast cancer is positive for EMT and stem cell-related immune markers, we believe it is more related to MES.
Claudin is highly expressed in normal tissues. Although the expression of Claudin varies across tumors, it is decreased in many tumors, indicating that Claudin can inhibit the function of tumors. The increased expression of Claudin significantly inhibits the proliferation of tumor cells and promotes EMT, migration and invasion of tumors. Low expression of claudins is associated with advanced disease, metastasis, and a poor prognosis (70). At present, the mechanism by which Claudin inhibits tumor occurrence is not clear, but some scholars have proposed the following three hypotheses: (A) Claudin is able to prevent microorganisms, toxins, and growth factors from passing through the paracellular channel, which are common oncogenic factors. In the absence of Claudin, these oncogenic factors enter the body and are likely to induce tumors (71–73). (B) Claudin directly binds or utilizes other scaffold proteins [mainly zonula occludens (ZOs)] to indirectly bind some key factors of signaling pathways, such as yes-associated protein/transcriptional coactivator (YAP/TAZ), pyruvate dehydrogenase kinase 1 (PDK-1), and β-catenin, which are all well-known carcinogenic factors. Claudin keeps them on the cell membrane and blocks carcinogenic signaling pathways, such as the PDK-AKT and YAP/TAZ-TEAD pathways (74–77). Reduced expression of ZO-1 in breast cancer destroys the structural integrity of the tight junction, leading to a loss of cell-cell adhesion. MUPP-1 expression is reduced in patients with poor prognosis and increased tumor grade. MUPP-1, like ZO-1, can be used as a crosslinker between claudin and tight-linked chains and tight-linked JAM oligomers, and other integral membrane proteins can be recruited into tight-linked claudin via MUPP-1. Studies have found that the transcription levels of ZO-1 and MUPP-1 in metastatic breast cancer are significantly reduced. The higher the expression of ZO-1 in breast cancer, the worse the prognosis of breast cancer patients (78). Normal tissues at the tight junction of mammary epithelial cells showed strong ZO-1 staining, and approximately 70% of breast cancers were found to have reduced or lost ZO-1. ZO-1 staining was positively correlated with tumor differentiation, and the reduction in ZO-1 staining was closely related to the reduction in E-cadherin staining. Therefore, ZO-1 may be directly involved in the malignant progression of breast cancer (79). A decrease in ZO-2 expression has also been observed in most breast cancers (80). (C) The basement membrane Claudin was found to colocalize with integrin β1 and form a protein complex to maintain epithelial cell attachment and inhibit cell proliferation. If claudin is depleted or expressed at low levels, epithelial cells become destabilized and proliferative (81, 82).
Apart from causing cancer in the above three aspects, Claudin also inhibits EMT. EMT is associated with tumorigenesis, progression and fibrosis (83). Transforming growth factor beta 1 (TGF-β1) has been shown to induce EMT during various stages of embryogenesis and progressive disease. Medici et al. (84) found that claudin protein expression was missing when TGF-β was used to induce EMT. SMAD3 and SMAD4 interact with SNAIL1, a transcriptional repressor and EMT promoter, and form a complex (SNAIL1-SMAD3/4) that targets the gene promoter of CAR, a tight junction protein, and E-cadherin. SNAIL1 and SMAD3/4 act as corepressors of the CAR, occludin, claudin-3, and E-cadherin promoters. In contrast, the combined silencing of SNAIL1 and SMAD4 with siRNA promoted the transcription of CAR and occludin during EMT (85). These studies indicate that Claudin is inhibited during EMT. Studies have found that the deletion of Claudin3 or Claudin4 leads to activation of the PI3K pathway, which is manifested as increased Akt phosphorylation, increased PIP3 content and PI3K activity, as well as up regulation of mRNA and protein levels of the transcription factor Twist. Claudin3 and Claudin4 maintain the epithelial phenotype, and their deletion promotes EMT (86).
Tumor metastasis is an important step in the process of tumor progression. The interaction between cancer cells and endothelial cells is the key for the distant metastasis of tumor cells. In the process of tumor metastasis, tightly connected Claudin participates in the defense of tumor cells, and tightly connected Claudin is the barrier of paracellular channels for epithelial and endothelial cell material exchange (87). Researchers believe that the loss of adhesion of tumor epithelial cells is a necessary condition for tumor invasion and metastasis. The expression of claudin in human cancer may decrease or increase in a tissue-specific manner. Claudin is responsible for the structural and functional integrity of the tight junction of the epithelial cell layer. The decrease or loss of claudin expression is accompanied by cell-cell adhesion and polarity damage. Tumorigenesis is accompanied by the destruction of tight junctions, a process that plays an important role in the loss of adhesion and the enhancement of invasiveness in tumor cells. The loss of Claudins and other tight junction proteins in cancer is considered a mechanism of cell adhesion loss and an important step in metastasis (88). Claudin constructs a complete biological system based on the function of the cell-side barrier. Generally, these claudin functional deficits are associated with water imbalance, inflammation, cancer, and brain disease, depending on the tissues and organs involved (31, 74, 89–92). Many studies have found that Claudin is expressed at low levels in many breast cancer tissues (93–95).
The feedback pathway of ZEB1/miR-200a promotes the invasion and metastasis of breast cancer cells (96, 97). It was found that miR-200a can regulate YAP1 in the HIPPO pathway and promote the survival and metastasis of CLBC (98). The ectopic expression of miR-200a can weaken the migration and invasion of CLBC by reducing the expression level of ELK3 mRNA (99). Compared with other subtypes of breast cancer, CLBC has the characteristics of a vascular system and endothelium and has higher vascular permeability, which promotes the metastasis of CLBC (100).
It has been proved that changes of protein glycosylation is involved in the regulation of EMT process of cancer cells (101). Claudin and low expression of Claudin play a role in carcinogenesis through multiple signaling pathways. Dedicator of cytokinesis 1 (DOCK1) down-regulates Claudin through ribosomal RNA processing 1B (RRP1B)/DNA methyltransferase (DNMT) to promote the growth and migration of breast cancer cells (102); Claudin through down-regulates the transforming growth factor-β(TGF-β)/Smad2/DNMT to promote EMT, metastasis and invasion of breast cancer cells (103). Low expression of Claudin promotes EMT by activating PI3K/AKt/mTOR, and regulates ERK/Sp1/CyclinD1 and ERK/IL-8 to promote the proliferation, migration and invasion of breast cancer cells (104). Claudin dysregulation promotes tumorigenesis by upregulating gp130/IL6/Stat3 signaling and activating the Wnt/β-catenin signaling pathway (74). Epithelial cell adhesion molecule (EpCAM) is a homophilic type I transmembrane glycoprotein belonging to the small GA733 protein family, EpCAM functions not only in physiological processes but also participates in the development and progression of cancer (105). The glycosylation of EpCAM is inhibited, which increases the expression of EpCAM in breast cancer. At the same time, the combination of Claudin and EpCAM increases, promotes the phosphorylation of PI3K/Akt and p38, promotes the activation of MAPK and PI3K/Akt pathways, promotes the process of EMT, reduces Apoptotic ability of breast cancer cells, promoting cell proliferation (106). In addition, Claudin and Integrin β1 inhibit the proliferation of tumor cells. If Claudin is missing, the combination of Claudin and Integrin β1 will be reduced, which will promote the tumor (81). The mechanism of claudin and claudin low-expression carcinogenic signal pathway is shown in Figure 2.
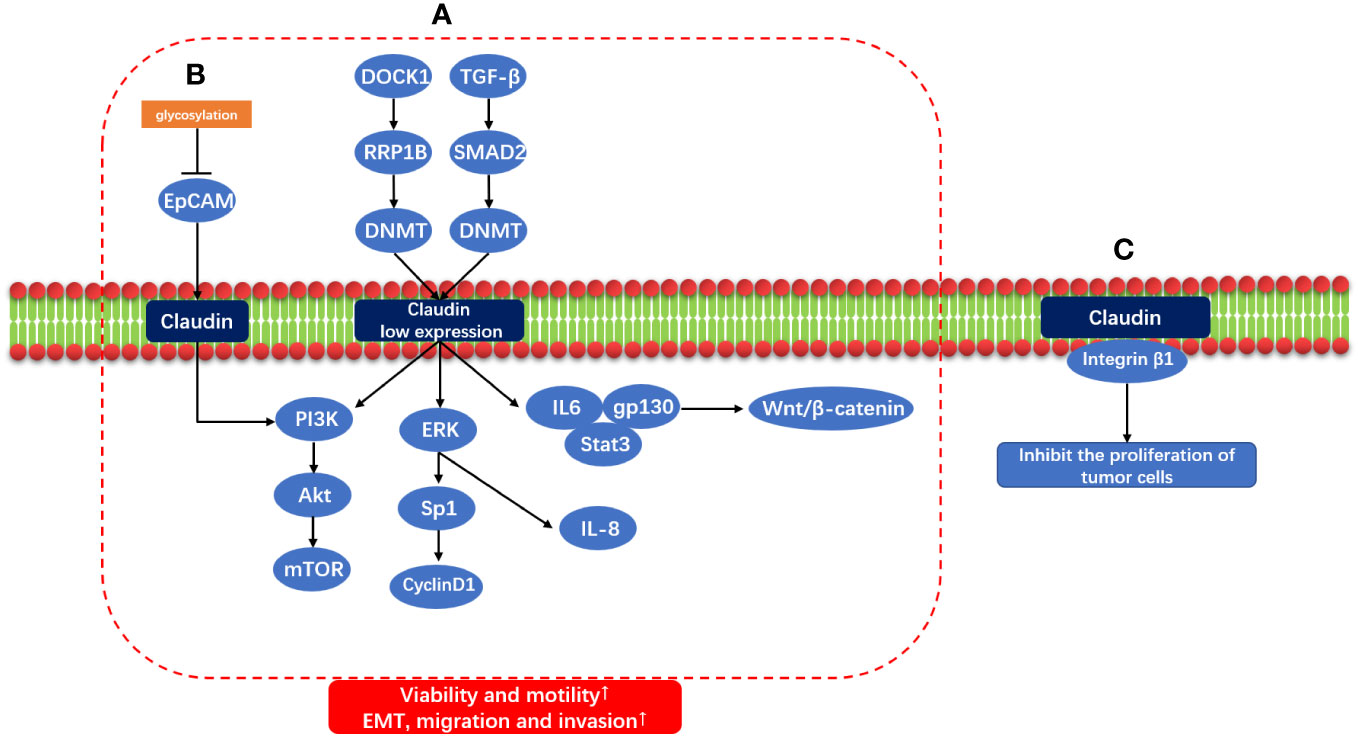
Figure 2 Claudin and Claudin low-expression carcinogenic signal pathway. (A) DOCK1/RRP1B/DNMT and TGF-β/SMAD2/DNMT induce Claudin low expression through methylation, and Claudin low expression through IL6/gp130/Stat3, Wnt/β-Catenin signal pathway; ERK/Sp1/CyclinD1 and ERK/IL8; PI3K/AKt/mTOR promotes EMT, migration and invasion of tumor. (B) Glycosylation of EPCAM was inhibited, and PI3K/AKt/mTOR was activated by combining with Claudin to promote EMT. (C) Claudin and Integrin β1 also inhibit the proliferation of tumor cells.
Currently, the following cell lines are used to study claudin-low breast cancer: MDA-MB157, MDA-MB436, BT549, MDA-MB231, HBL100, SUM159PT, Hs578T, MDA-MB435, and SUM1315. Some studies have extracted two cell lines, RM11A and RJ348, from breast tumors developed in MTB-IGFIR transgenic mice, which have the histological characteristics and gene expression pattern of claudin-low breast cancer and have been reinjected into the mouse breast fat pad to obtain high tumorigenicity (107). Fougner et al. (108) found that claudin-low-like MPA/DMBA-induced mouse mammary tumors represent a transcriptionally accurate model for human claudin-low breast cancer.
CLBC has the features of EMT and stem cells (109). Taube et al. (110) showed that tumor cells gain a certain degree of invasiveness and metastasize through EMT; at the same time, they also gain the potential for self-renewal, proliferating to form a macroscopically metastatic large cell population. Studies have shown that TGF-β induces EMT through the TGF-β/SMAD/LEF/PDGF pathway and the MAPK pathway (111). In addition, the homeobox proteins Goosecoid and the zinc finger proteins SNAIL and TWIST can also induce EMT. Another study found that miR-200 expression decreases during EMT (112). Surprisingly, Jones et al. (113) re-expressed miR-200c in murine claudin-low breast tumor cells to inhibit tumor cell proliferation and growth. Therefore, they believe that the expression of miR-200c can inhibit the growth of claudin-low breast tumor cells.
In addition, CLBC has a high degree of immune cell infiltration and high expression of T and B lymphoid cell markers (such as CD14 and CD79a) (45, 114, 115). Some scholars have compared CLBC with non-claudin-low breast cancer and found that CLBC has few gene mutations, low genetic instability, a low frequency of TP53 and PIK3CA mutations, and minimal MYC and MDM4 gain (52). Through Gene Ontology (GO) analysis, 165 genes were found to be differentially expressed in CLBC, among which 69 were upregulated and 64 downregulated. They analyzed 193 pathways and found that the inflammatory IL-13 signaling pathway was significantly enriched. Among them, five upregulated genes (IL6, CXCL8, VEGF-C, NRF1, and EREG) were mapped as hubs and may play an important role in CLBC (116). The comparison of Claudin-low breast cancer and other subtypes of breast cancer in immunohistochemistry and gene phenotype is shown in Table 1.
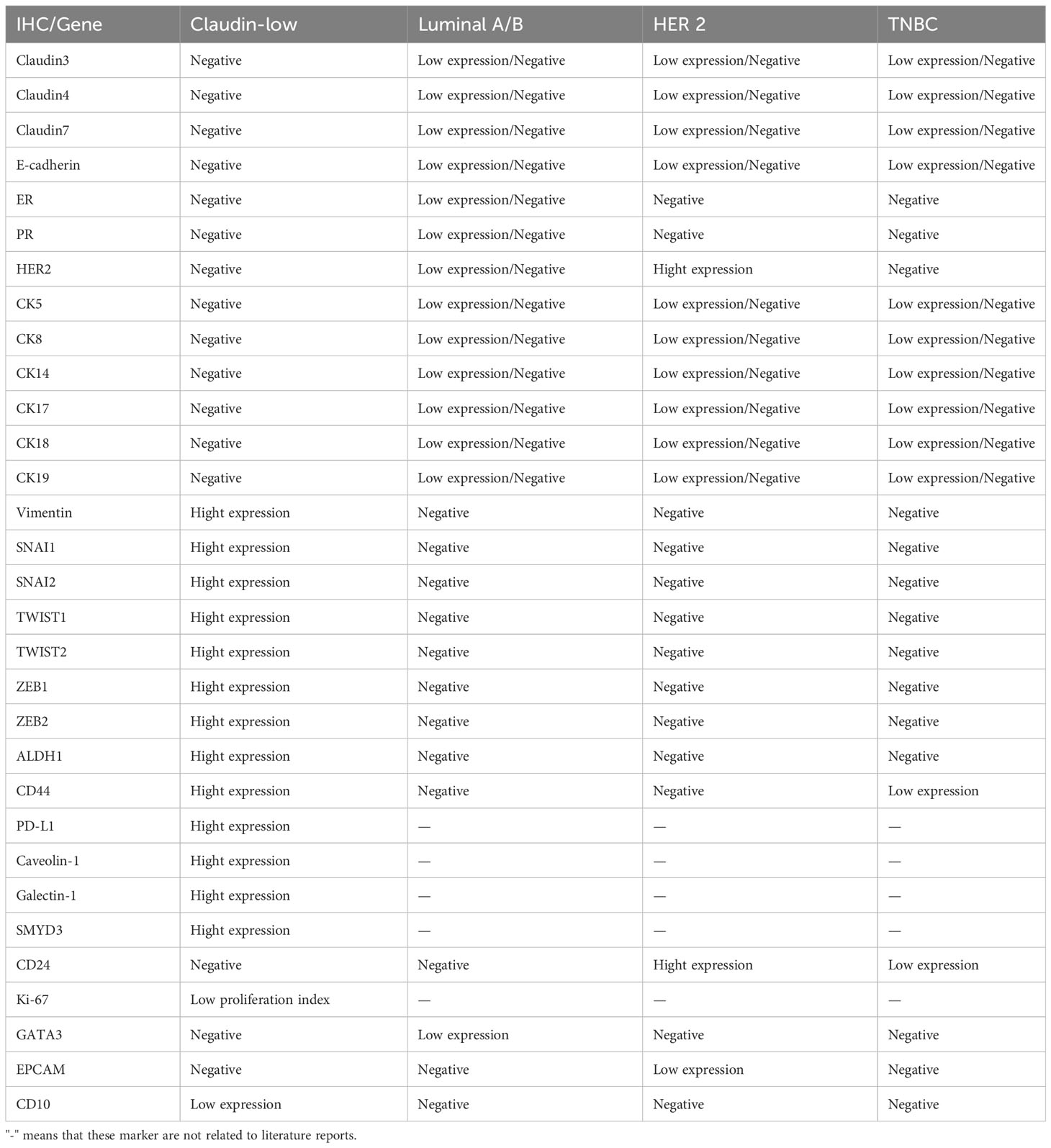
Table 1 Immunohistochemical and gene comparison of Claudin-low breast cancer and other subtypes of breast cancer.
Several studies have shown that RAS/MAPK kinases are highly activated in breast cancer (60, 68, 108). The dataset from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and TCGA cohorts was studied, and RAS signals were found to be highly expressed in CLBC. The GDSC database was also used to observe the IC50 of three MEK inhibitors (trametinib, selumetinib, and refametinib) in various subtypes of breast cancer, and CLBC was sensitive to three MEK inhibitors compared with other subtypes. This finding indicates that CLBC is driven by the MAPK pathway. Unfortunately, these findings were only performed on the genetic claudin-low breast cancer cell line and the corresponding mouse model, and clinical trial studies are lacking.
At present, there is still controversy about the age of CLBC. Some scholars have found that patients with CLBC are younger than patients with other subtypes (114, 115, 117, 118). However, Xu et al. (119) found that the age of CLBC patients is not significantly different from other subtypes, which may be related to their small sample size. Fougner et al. (52) found that the onset age of patients with negative ER, PR and HER in CLCB is younger than that of patients without negative ER, PR and HER. However, when they compared the CLBC and non-CLBC groups, the ages of the two groups of patients were similar.
The research results of Sabatier et al. (114) and Xu et al. (119) were consistent, in that CLBC was mainly larger than 2 cm (62%, 61.9%). In the tumor group larger than 2 cm, the proportion of CLBC was lower than that of the HER2-enriched subtype and the basal-like subtype and higher than that of the luminal subtype and the normal-like subtype. They also found that the proportion of histological grade 3 was higher than that of other subtypes (56%, 71.4%), except for the base-like subtype (85%, 56.6%). The rate of lymph node metastasis (46%, 38.1%) was higher than that of the base-like subtype (35%, 25%) and luminal A subtype (54%, 26.8%). This indicates that CLBC has a larger tumor diameter, a higher histological grade, and a higher rate of axillary lymph node metastasis. This may be related to the low expression of adhesion-related proteins in CLBC, which makes it easier to transfer.
Prat et al. (45) found that approximately 50% (8/16) of CLBCs are metaplastic carcinomas,
and Sabatier et al. (114) found that CLBCs are mainly nonspecialized breast cancer (77.7%), metaplastic carcinoma (71.4%), and medullary carcinoma (24%). Some scholars have also found that the histological types of CLBC are mainly invasive lobular carcinoma, metaplastic carcinoma, and medullary carcinoma (120). In short, the histological types of CLBC are mainly poorly adherent carcinomas, metaplastic carcinomas, and medullary carcinomas. Both CLBC and lobular carcinoma have down-regulation of E-cadherin, but CLBC does not always show the histological characteristics of lobular carcinoma. This may be along with the down-regulation of E-cadherin and other molecular changes, CLBC is more prone to ductal carcinoma. Characteristically, in contrast to lobular carcinoma, downregulation of E-cadherin in CLBC is associated with epigenetic or posttranscriptional dysregulation (121).
CLBC has low expression of tight junction proteins and adhesion proteins (Claudin3, 4, 7 and E-cadherin), and it also has low expression of cytokeratin (CK5, 8, 14, 17, 18, 19, and 24), HER-2 and luminal-related markers (45, 115). These features are similar to the basal-like subtype of breast cancer. CLBC has low expression of proliferation genes and Ki-67, which is inconsistent with the basal-like subtype of breast cancer (45, 115). Compared with other subtypes of breast cancer, CLBC has high expression of EMT-related markers (such as Vimentin, SNAI1 and SNAI2, TWIST1 and TWIST2, and ZEB1 and ZEB2) (114) and breast cancer stem cells/tumors with the initial cell markers ALDH1 and CD44hi/CD24-/low (115, 117) and high expression of PD-L1 (122). Recently, caveolin-1, galectin-1, and SMYD3 were found to be highly expressed in CLBC (123–125).
Some studies have found that CLBC is heterogeneous, as is its immunohistochemical expression. Fougner et al. (52) believed that they listed CLBC for analysis without considering the six intrinsic subtypes. This may have masked the intrinsic characteristics of CLBC. Pommier et al. (68) divided CLBC into three subgroups (CL-1, 2, and 3) according to FGA levels, which were derived from mammary stem cells (MaSC), mature luminal cells (mature luminal, mL), and luminal precursor cells (luminal progenitor, PL), respectively, which have the characteristics of expressing stem cell-related protein markers, luminal-related markers, and basal-like markers. This is in line with the findings of Fougner et al. (52) CLBC penetrates every subtype of breast cancer. Therefore, the immunohistochemical expression of CLBC is a complicated process, and different markers are expressed depending on the origin of the tumor.
Prat et al. (114) analyzed UNC337 and two independent gene expression databases (NKI1295 and MDACC133) with nine cell lines (claudin-low and PAM50 subtype predictors) and found that the incidence of CLBC was 7%~14%. In terms of prognosis, the Kaplan-Meier analysis of the two databases showed that the prognosis of CLBC (HR 2.83, RFS and OS 5.66) was significantly lower than that of luminal A subtype breast cancer (HR 4.71, RFS and OS 17.98). A study by Dias et al. (115) in 2016 showed that the overall survival (OR) of CLBC at 3, 5, and 10 years was higher than that of the basal-like subtype and lower than that of other subtypes, and the local recurrence rate was higher than that of other subtypes. Disease-free survival (DFS) is lower than that of luminal A breast cancer but higher than that of other subtypes. Sabatier et al. (114) found that the five-year local recurrence survival rate (67%) of CLBC is similar to that of luminal B (64%) and basal-like (60%) but higher than that of the HER2-enriched type (55%) and lower than that of luminal A (79%). Lu et al. (50) found that claudin-low has worse disease-free survival than non-CLBC, which is consistent with the conclusion found by Xu et al. (119). Overall, the prognosis for CLBC is poor.
Fougner et al. (52) conducted a study on the METABRIC database and found that, when CLBC was studied as a single group, the results showed that CLBC was associated with a poor prognosis, which was consistent with the findings mentioned above. However, when they analyzed CLBC according to the intrinsic molecular classification of breast cancer, the results showed that the basal-like Claudin-low had the worst prognosis, which was consistent with the prognostic results of the intrinsic molecular classification. In addition, the analysis of the claudin-low and non-claudin-low subtypes of the same intrinsic subtype of breast cancer showed that the difference was not statistically significant, indicating that there is no evidence that claudin-low can affect the prognosis of breast cancer.
Several studies have found that many immunohistochemical markers are helpful in evaluating the prognosis of CLBC. TBL1 is necessary for the mesenchymal phenotype of transformed breast epithelial cell lines and claudin-low subtype breast cancer cell lines. The high expression of the TBL1 gene is related to poor prognosis and an increased metastasis rate in breast cancer patients, indicating that the expression level of TBL1 can be used as a prognostic marker (62). According to the clinical data analysis of Fenizia C et al. (125), a higher level of SMYD3 is related to the poor prognosis of CLBC and the reduction in metastasis-free survival of breast cancer patients.
At present, research on CLBC treatment is still in the basic research stage, and most of the studies have not yet entered the animal experiment and clinical trial stages. At present, studies on the pathological complete remission of claudin-low breast cancer are based on the follow-up of the original cases. Prat et al. (45) followed up 133 patients with the MADCC Breast Cancer Database and found that the complete pathological remission rate (pCR rate, 38.9%) of CLBC was significantly lower than that of the basal subtype (pCR rate, 73.3%) but higher than that of luminal A and B. Sabatier et al. (110) studied 5,447 patients, and 1,294 patients received neoadjuvant chemotherapy with a pCR rate of 23%. The pCR rate of 228 CLBC patients was 32%, which was close to that of the basal sample (pCR rate, 33%), lower than that of the HER2-rich type (pCR rates, 37%), and much higher than that of the luminal A and luminal B subtypes (pCR rates, 7% and 18%). Both results showed that the pCR rate of CLBC is higher than that of other subtypes except basal-like breast cancer, indicating that CLBC is relatively sensitive to chemotherapeutic drugs.
Immunotherapy of CLBC. Fougner et al. (52) pointed out that CLBC showed high lymphocyte infiltration and high expression of PD-L1 and T lymphocyte reaction, which may provide opportunities for immunotherapy. At present, the means of immunotherapy mainly include immune checkpoint inhibitors and anti-PD1/PD-L1 therapy, etc. CLBC is rich in lymphocyte infiltration and drinks high expression of PD-L1 and IL6. These immunotherapy can provide new treatment schemes for CLBC patients (126). There is a lack of identification of CLBC from intrinsic molecular classification of breast cancer in clinical practice, so there is little research on immunotherapy of CLBC. In addition, some researchers have found that CLBC is rich in Tregs. Depletion of Tregs and suppression of immune checkpoints can weaken tumor growth and prolong survival, but they cannot lead to tumor regression (127).
Several novel targets have been proposed for the treatment of CLBC. Chang et al. (128) found that PIK3CG is a potential target for the treatment of CLBC. Inhibiting the activation of PIK3CG can enhance the therapeutic effect of PTX on CLBC. Stalker et al. (129) discovered that PDGFR inhibitors (sunitinib, regorafenib, and masitinib) can be used to inhibit the migration and metastasis of CLBC cells. Some scholars found that the combination of pyrazole derivatives and doxorubicin can increase the death of MDA-MB-231 cells (130). Panobinostat combined with gefitinib can synergistically inhibit the proliferation of CLBC cells and promote apoptosis (131, 132). CLBC is rich in EMT features, which may provide a new therapeutic target for chemotherapy. ZEB1, AP-1, and TEAD/YAP, the effector of the HIPPO pathway, form a transactivation complex to activate oncogenes, disrupting their molecular interaction may provide a promising treatment for CLBC (133). Recently, some new therapeutic targets have been found. AgNPs can selectively induce lipid peroxidation and cause irreversible proteotoxicity in CLBCs (134). SLC20A1 siRNA knockdown leads to suppression the activity of CLBC, and high expression of SLC20A1 indicates a poor prognosis (135). Studies have identified PVT-1 as a long noncoding RNA can regulate the expression of Claudin-4 in CLBC, indicating that it may be an important target for treating CLBC (136). Although some therapeutic targets for CLBC have been discovered, there is still a long way to go before clinical application.
In summary, CLBC is derived from breast cancer stem cells, luminal precursor cells, and luminal mature cells. The process of transforming to CLBC is related to EMT. We summarized the clinicopathological features of CLBC (Table 2), which is highly expressed in the immune response, breast cancer stem cells/tumor initiating cells, and EMT-related gene markers and is related to the MAPK pathway. CLBC is more aggressive because of its low expression of adhesion-related proteins, which show larger tumor size, a higher histological grade, and higher lymph node metastasis. The histological type is mainly invasive ductal carcinoma, and metaplastic carcinoma is associated with medullary carcinoma. CLBC is sensitive to chemotherapy, but its overall prognosis is poor. Currently, our knowledge of claudin-low breast cancer is inadequate due to the lack of uniformity in the classification of claudin-low breast cancer by previous investigators and the limited sample size of the study, thus leading to some bias in the results of the investigators. In the future, the origin of claudin-low breast cancer and the classification criteria by immunohistochemistry still need investigation. The potential therapeutic targets ans strategies for claudin-low breast cancer should be studied.
CP and AX wrote and edited the paper, they have made the same contribution to this work and share the identity of the first author. XM, YZ and YY generated the tables and figure. CC and CW revised the paper. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (grant number 82160461) and the Science, Technology Innovation Team of Pathological Diagnosis of Triple Negative Breast Cancer in Kunming Medical University (grant number CXTD202208) and Yunnan Provincial Health Commission Medical Discipline Leader Training Plan (grant number D2018058).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490(7418):61–70. doi: 10.1038/nature11412
2. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature (2000) 406(6797):747–52. doi: 10.1038/35021093
3. Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol (2007) 8(5):R76. doi: 10.1186/gb-2007-8-5-r76
4. Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol (1963) 17(2):375–412. doi: 10.1083/jcb.17.2.375
5. Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol (1998) 141(7):1539–50. doi: 10.1083/jcb.141.7.1539
6. Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol (2000) 149(1):13–6. doi: 10.1083/jcb.149.1.13
7. Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol (2004) 286(6):C1213–28. doi: 10.1152/ajpcell.00558.2003
8. Tsukita MF S, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol (2001) 2(4):285–93. doi: 10.1038/35067088
9. Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol (2001) 2(4):285–93. doi: 10.1038/35067088
10. Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta (2008) 1778(3):631–45. doi: 10.1016/j.bbamem.2007.10.018
11. Itallie C, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol (2006) 68(1):403. doi: 10.1146/annurev.physiol.68.040104.131404
12. Günzel D, Fromm M. Claudins and other tight junction proteins. Compr Physiol (2012) 2(3):1819–52. doi: 10.1002/cphy.c110045
13. Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol (1998) 143(2):391–401. doi: 10.1083/jcb.143.2.391
15. Kominsky SL, Tyler B, Sosnowski J, Brady K, Doucet M, Nell D, et al. Clostridium perfringens enterotoxin as a novel-targeted therapeutic for brain metastasis. Cancer Res (2007) 67(17):7977–82. doi: 10.1158/0008-5472.CAN-07-1314
16. Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res (2005) 65(21):9603–6. doi: 10.1158/0008-5472.CAN-05-2782
17. Kominsky SL. Claudins: emerging targets for cancer therapy. Expert Rev Mol Med (2006) 8(18):1–11. doi: 10.1017/S1462399406000056
18. Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol (1999) 147(6):1351–63. doi: 10.1083/jcb.147.6.1351
19. Roh MH, Liu CJ, Laurinec S, Margolis B. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem (2002) 277(30):27501–9. doi: 10.1074/jbc.M201177200
20. Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem (2002) 277(1):455–61. doi: 10.1074/jbc.M109005200
21. Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol (2003) 160(3):423–32. doi: 10.1083/jcb.200210020
22. Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci (1991) 88(8):3460–4. doi: 10.1073/PNAS.88.8.3460
23. Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol (1998) 141(1):199–208. doi: 10.1083/jcb.141.1.199
24. McNeil E, Capaldo CT, Macara IG. Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell (2006) 17(4):1922–32. doi: 10.1091/mbc.e05-07-0650
25. Umeda K, Matsui T, Nakayama M, Furuse K, Sasaki H, Furuse M, et al. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J Biol Chem (2004) 279(43):44785–94. doi: 10.1074/jbc.M406563200
26. Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell (2006) 126(4):741–54. doi: 10.1016/j.cell.2006.06.043
27. Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol (2016) 17(9):564–80. doi: 10.1038/nrm.2016.80
28. Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol (1999) 147(4):891–903. doi: 10.1083/jcb.147.4.891
29. Van Itallie JMA. Claudins CM. and epithelial paracellular transport. Annu Rev Physiol (2006) 68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404
30. Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol (2002) 283(1):C142–7. doi: 10.1152/ajpcell.00038.2002
31. Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol (2002) 156(6):1099–111. doi: 10.1083/jcb.200110122
32. Krämer F, White K, Kubbies M, Swisshelm K, Weber BH. Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Hum Genet (2000) 107(3):249–56. doi: 10.1007/s004390000375
33. Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol (2005) 18(4):511–8. doi: 10.1038/modpathol.3800301
34. Dottermusch M, Krüger S, Behrens HM, Halske C, Röcken C. Expression of the potential therapeutic target claudin-18.2 is frequently decreased in gastric cancer: results from a large Caucasian cohort study. Virchows Arch (2019) 475(5):563–71. doi: 10.1007/s00428-019-02624-7
35. Sung CO, Han SY, Kim SH. Low expression of claudin-4 is associated with poor prognosis in esophageal squamous cell carcinoma. Ann Surg Oncol (2011) 18(1):273–81. doi: 10.1245/s10434-010-1289-4
36. Shibutani M, Noda E, Maeda K, Nagahara H, Ohtani H, Hirakawa K. Low expression of claudin-1 and presence of poorly-differentiated tumor clusters correlate with poor prognosis in colorectal cancer. Anticancer Res (2013) 33(8):3301–6.
37. Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res (2005) 65(16):7378–85. doi: 10.1158/0008-5472.CAN-05-1036
38. Romani C, Cocco E, Bignotti E, Moratto D, Bugatti A, Todeschini P, et al. Evaluation of a novel human IgG1 anti-claudin3 antibody that specifically recognizes its aberrantly localized antigen in ovarian cancer cells and that is suitable for selective drug delivery. Oncotarget (2015) 6(33):34617–28. doi: 10.18632/oncotarget.5315
39. Uthayanan L, El-Bahrawy M. Potential roles of claudin-3 and claudin-4 in ovarian cancer management. J Egypt Natl Canc Inst (2022) 34(1):24. doi: 10.1186/s43046-022-00125-4
40. Yang F, Xu W, Tang X, Li Q, Hou X, Hui X. Claudin 4 enhances the Malignancy of glioma cells via NNAT/Wnt signaling. Am J Cancer Res (2023) 13(6):2530–9.
41. Wang C, Wu N, Pei B, Ma X, Yang W. Claudin and pancreatic cancer. Front Oncol (2023) 13:1136227. doi: 10.3389/fonc.2023.1136227
42. Kyuno D, Kojima T, Yamaguchi H, Ito T, Kimura Y, Imamura M, et al. Protein kinase Cα inhibitor protects against downregulation of claudin-1 during epithelial-mesenchymal transition of pancreatic cancer. Carcinogenesis (2013) 34(6):1232–43. doi: 10.1093/carcin/bgt057
43. Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res (2001) 12(11-12):469–76. doi: 10.3727/096504001108747477
44. Gowrikumar S, Ahmad R, Uppada SB, Washington MK, Shi C, Singh AB, et al. Upregulated claudin-1 expression promotes colitis-associated cancer by promoting β-catenin phosphorylation and activation in Notch/p-AKT-dependent manner. Oncogene (2019) 38(26):5321–37. doi: 10.1038/s41388-019-0795-5
45. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res (2010) 12(5):R68. doi: 10.1186/bcr2635
46. Gerhard R, Ricardo S, Albergaria A, Gomes M, Silva AR, Logullo ÂF, et al. Immunohistochemical features of claudin-low intrinsic subtype in metaplastic breast carcinomas. Breast (2012) 21(3):354–60. doi: 10.1016/j.breast.2012.03.001
47. Duarte GM, Almeida NR, Tocchet F, Espinola J, Barreto C, Pinto GA, et al. Claudin-4 expression is associated with disease-free survival in breast carcinoma-in-situ: mean follow-up of 8.2 years. Clin Breast Cancer (2018) 18(5):e1111–1111e1116. doi: 10.1016/j.clbc.2018.06.005
48. Logullo AF, Pasini FS, Nonogaki S, Rocha RM, Soares FA, Brentani MM. Immunoexpression of claudins 4 and 7 among invasive breast carcinoma subtypes: A large diagnostic study using tissue microarray. Mol Clin Oncol (2018) 9(4):377–88. doi: 10.3892/mco.2018.1685
49. Katayama A, Handa T, Komatsu K, Togo M, Horiguchi J, Nishiyama M, et al. Expression patterns of claudins in patients with triple-negative breast cancer are associated with nodal metastasis and worse outcome. Pathol Int (2017) 67(8):404–13. doi: 10.1111/pin.12560
50. Lu S, Singh K, Mangray S, Tavares R, Noble L, Resnick MB, et al. Claudin expression in high-grade invasive ductal carcinoma of the breast: correlation with the molecular subtype. Mod Pathol (2013) 26(4):485–95. doi: 10.1038/modpathol.2012.187
51. Jääskeläinen A, Soini Y, Jukkola-Vuorinen A, Auvinen P, Haapasaari KM, Karihtala P. High-level cytoplasmic claudin 3 expression is an independent predictor of poor survival in triple-negative breast cancer. BMC Cancer (2018) 18(1):223. doi: 10.1186/s12885-018-4141-z
52. Fougner C, Bergholtz H, Norum JH, Sørlie T. Re-definition of claudin-low as a breast cancer phenotype. Nat Commun (2020) 11(1):1787. doi: 10.1038/s41467-020-15574-5
53. Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U.S.A. (2009) 106(33):13820–5. doi: 10.1073/pnas.0905718106
54. Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist (2010) 15 Suppl 5:39–48. doi: 10.1634/theoncologist.2010-S5-39
55. Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol (2011) 5(1):5–23. doi: 10.1016/j.molonc.2010.11.003
56. Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PloS One (2008) 3(8):e2888. doi: 10.1371/journal.pone.0002888
57. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell (2008) 133(4):704–15. doi: 10.1016/j.cell.2008.03.027
58. Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze Malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PloS Genet (2012) 8(5):e1002723. doi: 10.1371/journal.pgen.1002723
59. Asiedu MK, Ingle JN, Behrens MD, Radisky DC, Knutson KL. TGFbeta/TNF(alpha)-mediated epithelial-mesenchymal transition generates breast cancer stem cells with a claudin-low phenotype. Cancer Res (2011) 71(13):4707–19. doi: 10.1158/0008-5472.CAN-10-4554
60. Rädler PD, Wehde BL, Triplett AA, Shrestha H, Shepherd JH, Pfefferle AD, et al. Highly metastatic claudin-low mammary cancers can originate from luminal epithelial cells. Nat Commun (2021) 12(1):3742. doi: 10.1038/s41467-021-23957-5
61. Chung WC, Zhang S, Challagundla L, Zhou Y, Xu K. Lunatic fringe and p53 cooperatively suppress mesenchymal stem-like breast cancer. Neoplasia (2017) 19(11):885–95. doi: 10.1016/j.neo.2017.08.006
62. Rivero S, Gómez-Marín E, Guerrero-Martínez JA, García-Martínez J, Reyes JC. TBL1 is required for the mesenchymal phenotype of transformed breast cancer cells. Cell Death Dis (2019) 10(2):95. doi: 10.1038/s41419-019-1310-1
63. Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature (2015) 525(7567):119–23. doi: 10.1038/nature14665
64. Suárez-Arriaga MC, Méndez-Tenorio A, Pérez-Koldenkova V, Fuentes-Pananá EM. Claudin-low breast cancer inflammatory signatures support polarization of M1-like macrophages with protumoral activity. Cancers (Basel) (2021) 13(9):2248. doi: 10.3390/cancers13092248
65. Tao L, Xiang D, Xie Y, Bronson RT, Li Z. Induced p53 loss in mouse luminal cells causes clonal expansion and development of mammary tumours. Nat Commun (2017) 8:14431. doi: 10.1038/ncomms14431
66. Wang S, Liu JC, Kim D, Datti A, Zacksenhaus E. Targeted Pten deletion plus p53-R270H mutation in mouse mammary epithelium induces aggressive claudin-low and basal-like breast cancer. Breast Cancer Res (2016) 18(1):9. doi: 10.1186/s13058-015-0668-y
67. Knight JF, Lesurf R, Zhao H, Pinnaduwage D, Davis RR, Saleh SM, et al. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc Natl Acad Sci U.S.A. (2013) 110(14):E1301–10. doi: 10.1073/pnas.1210353110
68. Pommier RM, Sanlaville A, Tonon L, Kielbassa J, Thomas E, Ferrari A, et al. Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution. Nat Commun (2020) 11(1):3431. doi: 10.1038/s41467-020-17249-7
69. Zhao S, Ma D, Xiao Y, Li XM, Ma JL, Zhang H, et al. Molecular subtyping of triple-negative breast cancers by immunohistochemistry: molecular basis and clinical relevance. Oncologist (2020) 25(10):e1481–1481e1491. doi: 10.1634/theoncologist.2019-0982
70. Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human Malignancy: a clinical perspective. Cancer Manag Res (2013) 5:367–75. doi: 10.2147/CMAR.S38294
71. Coussens LM, Werb Z. Inflammation and cancer. Nature (2002) 420(6917):860–7. doi: 10.1038/nature01322
72. Li J, Li YX, Chen MH, Li J, Du J, Shen B, et al. Changes in the phosphorylation of claudins during the course of experimental colitis. Int J Clin Exp Pathol (2015) 8(10):12225–33.
73. Kim BG, Lee PH, Lee SH, Park CS, Jang AS. Impact of ozone on claudins and tight junctions in the lungs. Environ Toxicol (2018) 33(7):798–806. doi: 10.1002/tox.22566
74. Ahmad R, Kumar B, Chen Z, Chen X, Müller D, Lele SM, et al. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer Malignancy by hyperactivating Wnt/β-catenin signaling. Oncogene (2017) 36(47):6592–604. doi: 10.1038/onc.2017.259
75. Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu FX, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest (2018) 128(3):970–84. doi: 10.1172/JCI90429
76. Shimobaba S, Taga S, Akizuki R, Hichino A, Endo S, Matsunaga T, et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and Akt in human lung adenocarcinoma A549 cells. Biochim Biophys Acta (2016) 1863(6 Pt A):1170–8. doi: 10.1016/j.bbamcr.2016.02.015
77. Akizuki R, Shimobaba S, Matsunaga T, Endo S, Ikari A. Claudin-5, -7, and -18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma. Biochim Biophys Acta Mol Cell Res (2017) 1864(2):293–302. doi: 10.1016/j.bbamcr.2016.11.018
78. Martin TA, Watkins G, Mansel RE, Jiang WG. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur J Cancer (2004) 40(18):2717–25. doi: 10.1016/j.ejca.2004.08.008
79. Hoover KB, Liao SY, Bryant PJ. Loss of the tight junction MAGUK ZO-1 in breast cancer: relationship to glandular differentiation and loss of heterozygosity. Am J Pathol (1998) 153(6):1767–73. doi: 10.1016/S0002-9440(10)65691-X
80. Chlenski A, Ketels KV, Korovaitseva GI, Talamonti MS, Oyasu R, Scarpelli DG. Organization and expression of the human zo-2 gene (tjp-2) in normal and neoplastic tissues. Biochim Biophys Acta (2000) 1493(3):319–24. doi: 10.1016/s0167-4781(00)00185-8
81. Lu Z, Kim DH, Fan J, Lu Q, Verbanac K, Ding L, et al. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer (2015) 14:120. doi: 10.1186/s12943-015-0387-0
82. Singh AB, Uppada SB, Dhawan P. Claudin proteins, outside-in signaling, and carcinogenesis. Pflugers Arch (2017) 469(1):69–75. doi: 10.1007/s00424-016-1919-1
83. Rastaldi MP. Epithelial-mesenchymal transition and its implications for the development of renal tubulointerstitial fibrosis. J Nephrol (2006) 19(4):407–12.
84. Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell (2006) 17(4):1871–9. doi: 10.1091/mbc.e05-08-0767
85. Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol (2009) 11(8):943–50. doi: 10.1038/ncb1905
86. Lin X, Shang X, Manorek G, Howell SB. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PloS One (2013) 8(6):e67496. doi: 10.1371/journal.pone.0067496
87. Tobioka H, Sawada N, Zhong Y, Mori M. Enhanced paracellular barrier function of rat mesothelial cells partially protects against cancer cell penetration. Br J Cancer (1996) 74(3):439–45. doi: 10.1038/bjc.1996.378
88. Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta (2009) 1788(4):872–91. doi: 10.1016/j.bbamem.2008.11.005
89. Tokumasu R, Yamaga K, Yamazaki Y, Murota H, Suzuki K, Tamura A, et al. Dose-dependent role of claudin-1 in vivo in orchestrating features of atopic dermatitis. Proc Natl Acad Sci U.S.A. (2016) 113(28):E4061–8. doi: 10.1073/pnas.1525474113
90. Menard C, Pfau ML, Hodes GE, Kana V, Wang VX, Bouchard S, et al. Social stress induces neurovascular pathology promoting depression. Nat Neurosci (2017) 20(12):1752–60. doi: 10.1038/s41593-017-0010-3
91. Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, et al. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology (2012) 142(2):305–15. doi: 10.1053/j.gastro.2011.10.025
92. Hagen SJ, Ang LH, Zheng Y, Karahan SN, Wu J, Wang YE, et al. Loss of tight junction protein claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology (2018) 155(6):1852–67. doi: 10.1053/j.gastro.2018.08.041
93. Hoevel T, Macek R, Mundigl O, Swisshelm K, Kubbies M. Expression and targeting of the tight junction protein CLDN1 in CLDN1-negative human breast tumor cells. J Cell Physiol (2002) 191(1):60–8. doi: 10.1002/jcp.10076
94. Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene (2003) 22(13):2021–33. doi: 10.1038/sj.onc.1206199
95. Swisshelm K, Machl A, Planitzer S, Robertson R, Kubbies M, Hosier S. SEMP1, a senescence-associated cDNA isolated from human mammary epithelial cells, is a member of an epithelial membrane protein superfamily. Gene (1999) 226(2):285–95. doi: 10.1016/s0378-1119(98)00553-8
96. Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep (2008) 9(6):582–9. doi: 10.1038/embor.2008.74
97. Zou Q, Zhou E, Xu F, Zhang D, Yi W, Yao J. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem (2018) 119(2):2189–99. doi: 10.1002/jcb.26380
98. Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res (2013) 19(6):1389–99. doi: 10.1158/1078-0432.CCR-12-1959
99. Kim HK, Park JD, Choi SH, Shin DJ, Hwang S, Jung HY, et al. Functional link between miR-200a and ELK3 regulates the metastatic nature of breast cancer. Cancers (Basel) (2020) 12(5):1225. doi: 10.3390/cancers12051225
100. Harrell JC, Pfefferle AD, Zalles N, Prat A, Fan C, Khramtsov A, et al. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clin Exp Metastasis (2014) 31(1):33–45. doi: 10.1007/s10585-013-9607-4
101. Greville G, Llop E, Howard J, Madden SF, Perry AS, Peracaula R, et al. 5-AZA-dC induces epigenetic changes associated with modified glycosylation of secreted glycoproteins and increased EMT and migration in chemo-sensitive cancer cells. Clin Epigenet (2021) 13(1):34. doi: 10.1186/s13148-021-01015-7
102. Chiang SK, Chang WC, Chen SE, Chang LC. DOCK1 regulates growth and motility through the RRP1B-claudin-1 pathway in claudin-low breast cancer cells. Cancers (Basel) (2019) 11(11):1762. doi: 10.3390/cancers11111762
103. Lu Y, Wang L, Li H, Li Y, Ruan Y, Lin D, et al. SMAD2 inactivation inhibits CLDN6 methylation to suppress migration and invasion of breast cancer cells. Int J Mol Sci (2017) 18(9):1863. doi: 10.3390/ijms18091863
104. Song P, Li Y, Dong Y, Liang Y, Qu H, Qi D, et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J Exp Clin Cancer Res (2019) 38(1):354. doi: 10.1186/s13046-019-1359-9
105. Liu Y, Wang Y, Sun S, Chen Z, Xiang S, Ding Z, et al. Understanding the versatile roles and applications of EpCAM in cancers: from bench to bedside. Exp Hematol Oncol (2022) 11(1):97. doi: 10.1186/s40164-022-00352-4
106. Wen R, Lin H, Li X, Lai X, Yang F. The regulatory mechanism of epCAM N-glycosylation-mediated MAPK and PI3K/akt pathways on epithelial-mesenchymal transition in breast cancer cells. Cell Mol Biol (Noisy-le-grand) (2022) 68(5):192–201. doi: 10.14715/cmb/2022.68.5.26
107. Campbell CI, Thompson DE, Siwicky MD, Moorehead RA. Murine mammary tumor cells with a claudin-low genotype. Cancer Cell Int (2011) 11:28. doi: 10.1186/1475-2867-11-28
108. Fougner C, Bergholtz H, Kuiper R, Norum JH, Sørlie T. Claudin-low-like mouse mammary tumors show distinct transcriptomic patterns uncoupled from genomic drivers. Breast Cancer Res (2019) 21(1):85. doi: 10.1186/s13058-019-1170-8
109. Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res (2009) 69(10):4116–24. doi: 10.1158/0008-5472.CAN-08-3441
110. Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA (2010) 107(35):15449–54. doi: 10.1073/pnas.1004900107
111. Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell (2001) 12(1):27–36. doi: 10.1091/mbc.12.1.27
112. Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle (2008) 7(20):3112–8. doi: 10.4161/cc.7.20.6851
113. Jones R, Watson K, Bruce A, Nersesian S, Kitz J, Moorehead R. Re-expression of miR-200c suppresses proliferation, colony formation and in vivo tumor growth of murine claudin-low mammary tumor cells. Oncotarget (2017) 8(14):23727–49. doi: 10.18632/oncotarget.15829
114. Sabatier R, Finetti P, Guille A, Adelaide J, Chaffanet M, Viens P, et al. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Mol Cancer (2014) 13:228. doi: 10.1186/1476-4598-13-228
115. Dias K, Dvorkin-Gheva A, Hallett RM, Wu Y, Hassell J, Pond GR, et al. Claudin-low breast cancer; clinical & Pathological characteristics. PloS One (2017) 12(1):e0168669. doi: 10.1371/journal.pone.0168669
116. Zayed H. The identification of highly upregulated genes in claudin-low breast cancer through an integrative bioinformatics approach. Comput Biol Med (2020) 127:103806. doi: 10.1016/j.compbiomed.2020.103806
117. de Beça FF, Caetano P, Gerhard R, Alvarenga CA, Gomes M, Paredes J, et al. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J Clin Pathol (2013) 66(3):187–91. doi: 10.1136/jclinpath-2012-201169
118. Okano M, Oshi M, Mukhopadhyay S, Qi Q, Yan L, Endo I, et al. Octogenarians’ Breast cancer is associated with an unfavorable tumor immune microenvironment and worse disease-free survival. Cancers (Basel) (2021) 13(12):2933. doi: 10.3390/cancers13122933
119. Jing Xu, Ketao L, Tianhui Su, Zhenfeng Li, Yue W, Qinqin Gu. Clinicopathologic and prognostic features of Claudin-low type breast cancer. Chin J Pathol (2017) 46(9):634–9. doi: 10.3760/cma.j.issn.0529-5807.2017.09.009
120. Kim S, Moon BI, Lim W, Park S, Cho MS, Sung SH. Feasibility of classification of triple negative breast cancer by immunohistochemical surrogate markers. Clin Breast Cancer (2018) 18(5):e1123–1123e1132. doi: 10.1016/j.clbc.2018.03.012
121. Voutsadakis IA. Comparison of clinical subtypes of breast cancer within the claudin-low molecular cluster reveals distinct phenotypes. Cancers (Basel) (2023) 15(10):2689. doi: 10.3390/cancers15102689
122. Alsuliman A, Colak D, Al-Harazi O, Fitwi H, Tulbah A, Al-Tweigeri T, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol Cancer (2015) 14:149. doi: 10.1186/s12943-015-0421-2
123. Thompson DE, Siwicky MD, Moorehead RA. Caveolin-1 expression is elevated in claudin-low mammary tumor cells. Cancer Cell Int (2012) 12:6. doi: 10.1186/1475-2867-12-6
124. Balestrieri K, Kew K, McDaniel M, Ramez M, Pittman HK, Murray G, et al. Proteomic identification of tumor- and metastasis-associated galectin-1 in claudin-low breast cancer. Biochim Biophys Acta Gen Subj (2021) 1865(2):129784. doi: 10.1016/j.bbagen.2020.129784
125. Fenizia C, Bottino C, Corbetta S, Fittipaldi R, Floris P, Gaudenzi G, et al. SMYD3 promotes the epithelial-mesenchymal transition in breast cancer. Nucleic Acids Res (2019) 47(3):1278–93. doi: 10.1093/nar/gky1221
126. Liu Y, Hu Y, Xue J, Li J, Yi J, Bu J, et al. Advances in immunotherapy for triple-negative breast cancer. Mol Cancer (2023) 22(1):145. doi: 10.1186/s12943-023-01850-7
127. Taylor NA, Vick SC, Iglesia MD, Brickey WJ, Midkiff BR, McKinnon KP, et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Invest (2017) 127(9):3472–83. doi: 10.1172/JCI90499
128. Chang J, Hong L, Liu Y, Pan Y, Yang H, Ye W, et al. Targeting PIK3CG in combination with paclitaxel as a potential therapeutic regimen in claudin-low breast cancer. Cancer Manag Res (2020) 12:2641–51. doi: 10.2147/CMAR.S250171
129. Stalker L, Pemberton J, Moorehead RA. Inhibition of proliferation and migration of luminal and claudin-low breast cancer cells by PDGFR inhibitors. Cancer Cell Int (2014) 14(1):89. doi: 10.1186/s12935-014-0089-5
130. Saueressig S, Tessmann J, Mastelari R, da Silva LP, Buss J, Segatto NV, et al. Synergistic effect of pyrazoles derivatives and doxorubicin in claudin-low breast cancer subtype. BioMed Pharmacother (2018) 98:390–8. doi: 10.1016/j.biopha.2017.12.062
131. Lyu H, Hou D, Liu H, Ruan S, Tan C, Wu J, et al. HER3 targeting augments the efficacy of panobinostat in claudin-low triple-negative breast cancer cells. NPJ Precis Oncol (2023) 7(1):72. doi: 10.1038/s41698-023-00422-8
132. Matossian MD, Burks HE, Elliott S, Hoang VT, Bowles AC, Sabol RA, et al. Panobinostat suppresses the mesenchymal phenotype in a novel claudin-low triple negative patient-derived breast cancer model. Oncoscience (2018) 5(3-4):99–108. doi: 10.18632/oncoscience.412
133. Feldker N, Ferrazzi F, Schuhwerk H, Widholz SA, Guenther K, Frisch I, et al. Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J (2020) 39(17):e103209. doi: 10.15252/embj.2019103209
134. Snyder CM, Rohde MM, Fahrenholtz CD, Swanner J, Sloop J, Donati GL, et al. Low doses of silver nanoparticles selectively induce lipid peroxidation and proteotoxic stress in mesenchymal subtypes of triple-negative breast cancer. Cancers (Basel) (2021) 13(16):4217. doi: 10.3390/cancers13164217
135. Onaga C, Tamori S, Motomura H, Ozaki A, Matsuda C, Matsuoka I, et al. High SLC20A1 expression is associated with poor prognoses in claudin-low and basal-like breast cancers. Anticancer Res (2021) 41(1):43–54. doi: 10.21873/anticanres.14750
Keywords: breast cancer, Claudin-low, immunohistochemistry, mammary stem cells, epithelial-mesenchymal transformation
Citation: Pan C, Xu A, Ma X, Yao Y, Zhao Y, Wang C and Chen C (2023) Research progress of Claudin-low breast cancer. Front. Oncol. 13:1226118. doi: 10.3389/fonc.2023.1226118
Received: 20 May 2023; Accepted: 26 September 2023;
Published: 11 October 2023.
Edited by:
Noha Mousaad Elemam, University of Sharjah, United Arab EmiratesCopyright © 2023 Pan, Xu, Ma, Yao, Zhao, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ceshi Chen, Y2hlbmNAa21tdS5lZHUuY24=; Chunyan Wang, NzQwNzI5Y2h1bnlhbkBzaW5hLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.