
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 12 September 2023
Sec. Thoracic Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1212788
Background: We investigated the biological predisposition to site of metastasis in patients with NSCLC based on their molecular profiling and program death ligand PD-L1 status. We sought to identify any association between metastatic site and molecular profile in NSCLC patients.
Methods: This was a retrospective analysis of patients with stage IV NSCLC who were newly diagnosed from January 2014 to June 2022. Clinical characteristics, pathology, molecular reports, and imaging were retrieved and analyzed.
Results: A total of 143 patients were included in the study. Median age was 65 years, with an equal number of men (n=71) and women (n=72). The most common histology was adenocarcinoma (81.8%). At least one genetic mutation was discovered in 100 patients. Mutations with a targetable drug were found in 86 patients. The most common mutations were TP53 (25.2%), EGFR (24.5%), KRAS/NRAS (20.3%), and CDKN2A/2B (7.7%). Patients with any mutation were significantly more likely to have metastatic disease to the brain (57% vs. 37%, p=0.03), but there was no difference in metastatic disease to bone (34% vs. 26%, p=0.32). Patients without a discoverable mutation were significantly more likely to have metastatic disease to other sites (e.g., adrenal gland 91% vs. liver 66%, p=0.002). There was no difference in progression-free survival (PFS) or overall survival (OS) between those with versus without mutations. Median PFS and OS were significantly longer in patients with an EGFR mutation than those with KRAS/NRAS or TP53 mutations. Patients with PD-L1 >1% or TP53 were significantly more likely to have metastatic disease to organs other than bone or brain (p=0.047 and p=0.023, respectively). We identified four prognostic groups in metastatic NSCLC. Patients with PD-L1 <1% and no actionable mutations have the poorest prognosis, with median survival of around 20 months.
Conclusion: Patients with mutations discoverable on NGS are more likely to have metastatic disease to the brain. KRAS/NRAS in particular has a predilection to metastasize to the brain and bone. PD-L1 expression and a TP53 mutation, on the other hand, tend to lead to metastasis of NSCLC to organs other than brain or bone. These results need to be corroborated in larger prospective studies.
Non-small cell lung cancer (NSCLC) comprises about 85% of lung cancer. NSCLC is a term that includes a variety of different lung cancers, most notably adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma. NSCLC also includes other subsets of lung cancer such as adeno-squamous carcinoma, sarcomatoid carcinoma, and non–small cell neuroendocrine tumors. Adenocarcinoma is the most common type of NSCLC (1). Since the introduction of next-generation sequencing (NGS), molecular genotyping has become essential in metastatic NSCLC, and the development of mutation-directed therapy has revolutionized the treatment of NSCLC. The overall prognosis for lung cancer continues to improve due to the evolving availability of targeted therapy and immunotherapy.
NSCLC can metastasize to adjacent tissues or organs by direct invasion or follow typical oncogenic metastasis by hematogenous, lymphatic or lympho-hematogenous pathway. NSCLC tends to metastasize to liver, brain, bone and adrenal glands. The process of cancer migration and metastasis is a sequential process, where cancer cells will either directly invade into surrounding tissues/organs or invade into vascular and lymphatic system and disseminate to other organs (2). Due to the complexity of lymphatic and vascular system, the metastatic pathway is always unpredictable. Hellman and Weichselbaum have once proposed that the probability and the sites of metastasis may reflect the inner state of tumor development in the process of acquiring necessity properties for dissemination (3). The discovery of molecular alterations especially in the metastasis/aggressive setting, may further revise the concept of oncogenic metastasis, highlighting the potential role of molecular alterations in directing the site of metastasis or dissemination. Cancer with mutations involving the epidermal growth factor receptor may have a distinct metastasis pattern highlighting the potential role of tumor genetics in the metastasis behavior of the tumor.
There is limited data to date to predict or assess the biological predisposition of site of metastasis in patients with NSCLC based on the molecular profiling. Kuijpers et al. reported in a retrospective analysis the potential association of molecular profiling of non-squamous NSCLC with the site of metastasis. They reported that about 54% of stage IV EGFR-positive tumors had bone metastasis at the time of diagnosis (4). Hsu et al. further highlighted a higher incidence of lung and brain metastasis in NSCLC patients with an EGFR mutation (5). Moreover, Hsu et al. also reported that the incidence of liver metastasis is significantly different between subtypes with the EGFR exon 19 deletion versus the exon 21 mutation, highlighting the potential association of molecular profiling in the spread of NSCLC tumors (5).
Based on these previous results, we propose that the biological predisposition to a metastatic site may differ between the molecular subgroups of NSCLC. Furthermore, differences in metastatic disposition could have a differential effect on morbidity, mortality, and the natural history of the disease. Hence, there is a paramount need to understand the association between the molecular landscape and metastatic site.
This was a retrospective analysis conducted at UTHealth Houston/Memorial Hermann Cancer Center. All patients ≥18 years old with a newly diagnosis of stage IV primary NSCLC diagnosed between January 2014 and June 2022 were identified and retrieved. Additional inclusion criteria were pathology-proved NSCLC as per AJCC staging, 8th edition, available mutation analysis information, and sufficient medical information for analysis. Exclusion criteria included recent history of non-NSCLC cancer (i.e., any malignancy within 5 years before NSCLC diagnosis, except for skin tumors other than melanoma), non-invasive tumors, progressive NSCLC and tumors with molecular alterations identified on pathology material obtained ≥3 months after diagnosis. The database that we retrieved included patients’ clinical characteristics, medical history including medications, pathology report, molecular profile, imaging, and clinical laboratory data.
The study protocol was approved by the Institutional Review Board of both Memorial Hermann Hospital and The University of Texas Health Science Center at Houston. All procedures were conducted in compliance with the ethical standards of the Memorial Hermann Hospital, The University of Texas Health Science Center at Houston, and the Declaration of Helsinki for research on human beings. A waiver of HIPAA privacy authorization was been obtained through the Ethical Review Board.
Biopsy results were obtained from either the primary lung tissue or metastatic site. Biopsies were reviewed and reports were authorized by a UTHealth pathologist. The molecular profile (NGS) results were obtained using commercially available assays (Foundation One or validated assays by UT Molecular Pathology). The NGS results were obtained from the initial biopsy tissue or metastatic site depending on original tissue sample availability. The choice of biopsy specimen (primary vs metastatic) chosen for NGS testing depended on 1) accessibility of tissue specimen for biopsy as well as 2) amount of tissue available to run NGS. For patients who did not provide a tissue sample for NGS testing, molecular profiles were obtained from peripheral blood. As for the PD-L1, the PD-L1 expression was determined by the Tumor Proportion Score and classified into TPS <1%, TPS 1 to 49% and TPS ≥50%.
The evaluation of the metastatic lesions was conducted based on imaging either via PET/CT scan, CT chest abdomen pelvis with contrast, MRI brain with/without contrast and or biopsy of the site of metastasis. There are about 59% of the patients in our study have the biopsy obtained from the distant metastatic sites. The assessment of presence of metastatic disease was first done by the treating physician and later by our research team giving a second layer of confidence in existence of metastatic disease
We analyzed the patients’ demographic characteristics, as well as clinical and biochemical data. Descriptive data were represented by percentages, mean ± standard deviation, medians, and numbers. Continuous-variable analysis was performed with Kruskal–Wallis one-way analysis of variance and t-test for non-normal and normal distribution, respectively. As for categorical variables, the χ2 or Fisher’s exact test was used. Progression-free survival (PFS) was calculated from the time of initial diagnosis until the time of disease progression. Overall survival (OS) was calculated from the time of initial diagnosis until censoring (death/events or date of last follow-up). PFS and OS were analyzed using the Kaplan–Meier test.
We did all data analysis using the statistical software GraphPad Prism version 14.0.2. Statistical significance was achieved if the null hypothesis could be rejected at P < 0.05 with a 95% confidence interval (CI). We also compared differences in clinical parameters, mutation profile, PFS, OS, and other parameters in patients who with OS <12 months versus OS ≥12 months.
After excluding patients who did not have complete clinical data, a total of 143 patients were included, and their NGS panels were analyzed (Figure 1). Fifty-seven (40%) NGS reports were obtained from primary lung tissue, 84 (59%) from metastatic sites, and 2 (1%) from peripheral blood (liquid biopsy). Median age was 65 years, with an equal number of men (n=71) and women (n=72). The most common histology was adenocarcinoma (81.8%), followed by squamous cell cancer (11.9%) and large cell carcinoma (3.5%). At least one genetic mutation was discovered in 100 patients (70%); 43 (30%) had no discoverable genetic mutation. Mutations with a targetable drug were found in 86 patients (60%), and many patients had >1 genetic mutation. The most common mutations were TP53 (25.2%), EGFR (24.5%, 63% with the classic targetable EGFR mutation exon 19 and L858R and 37% with atypical EGGR mutations such as exon 18, exon 20, L861Q, or T790M), KRAS/NRAS (20.3%), and CDKN2A/2B (7.7%). The most common metastatic sites were brain (51%), bone (31.5%), contralateral lung (23.1%), pleura (21%), and adrenal gland (14%). Program death ligand PD-L1 status was known in 117 (81.8%) patients. Of these, 45 (38%) had no PD-L1 expression, 41(35%) had 1%–49%, and 31 (21.7%) had >50% PD-L1 expression.
The median PFS and OS for entire cohort were 24 months and 41.2 months, respectively. Detailed clinical characteristics and demographic analysis are presented in Table 1.
Patients with any mutation were significantly more likely to have metastatic disease to the brain (57% vs. 37%, p=0.03), but there was no difference in rates of metastatic disease to the bone (34% vs. 26%, p=0.32). Patients without a discoverable mutation were significantly more likely to have metastatic disease to other sites (e.g., adrenal gland or liver; 91% vs. 66% for patients with discoverable mutations, p=0.002). In our study NGS testing was done on primary site (40%), metastatic (59%), and peripheral blood (1%). Branched evolution of tumor during the progression of disease has been reported to result in intratumor heterogeneity, nonetheless, single site NGS can accurately estimates genetic landscape of the tumor. To verify the validity of our results, we have performed a stratified analysis of mutations and sites of metastatic disease in biopsy obtained from primary and metastatic lesions. Based on this stratified analysis, there was no difference in sites of metastatic disease predicted based on presence of absence of discoverable mutation (Supplementary Table).
When stratifying based on the mutational profile, patients with TP53 mutations were significantly more likely to have metastatic disease to organs other than bone or brain (hazard ratio [HR] 0.29, p=0.023). With regard to single-organ metastasis, tumors with KRAS/NRAS mutations had a predilection to metastasize to brain and bone, (45% of patients, HR 3.47, p=0.04). A detailed analysis is given in Table 2. Tumors with EGFR and KRAS/NRAS mutations had a predilection for brain metastasis, which was observed in 60% and 59% of patients, respectively.
When comparing the survival outcomes (PFS and OS) in patients with or without mutations, there was no difference observed in PFS (24 vs. 22.9 months, respectively, log rank p=0.58) or OS (41.2 vs. 36.7 months, respectively, log rank p=0.71) between these two groups via Kaplan–Meier analysis (Figure 2). It might be expected that the survival outcome is better in the subgroup of patients with mutations in light of targeted therapies; however, approximately 45% of patients had either TP53 or KRAS mutations, which have been shown in multiple studies to indicate poorer outcomes in NSCLC.
Median PFS was significantly longer in patients with an EGFR mutation than those with KRAS/NRAS or TP53 mutations (36 vs. 16.2 vs. 11.9 months, p=0.03). Median OS for patients with an EGFR mutation was not reached and was significantly longer than in patients with TP53 (28.7 months) or KRAS/NRAS (26 months) mutations (p=0.003) (Figure 3).
With regard to PD-L1 status, patients with PD-L1 >1% were significantly more likely to have metastatic disease to organs other than bone or brain (HR 0.4, p=0.047). A detailed analysis is shown in Table 3.
When we compared the PFS and OS with regard to PD-L1 status and the presence of a discoverable mutation, 4 prognostic groups were identified: P0M0 (PD-L1 <1% and no actionable mutation), P0M1 (PD-L1 <1% with an actionable mutation), P1M0 (PD-L1 ≥1% and no actionable mutation), and P1M1 (PD-L1 ≥1% with an actionable mutation) (Table 4). The P0M0 subgroup (n=24) had the worst median PFS and OS (8.5 and 20.6 months, respectively). The P1M1 subgroup (n=41) had a median PFS of 13.1 months and the longest OS of 44.5 months. Patients in the P0M1 group had a longer PFS than those in the P1M1 group (25.2 vs. 13.1 months), likely due to the presence of more patients with an EGFR mutation (57% vs. 32%) However, this did not translate into improved OS (P0M1: 33.5 months, P1M1: 44.5 months, log-rank p=0.98), perhaps because of the durable long-term response to immunotherapy in patients with PD-L1 ≥1% or due to limited second-line options in P0M1 patients at the time of progression. The results were not significant, likely because of the small sample size in each category. Detailed survival analysis depicted in Figure 4 and Table 4.
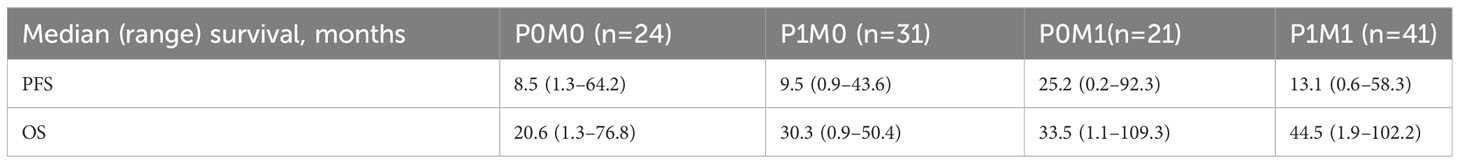
Table 4 Comparison of site of survival outcome between patients PD-L1 <1% vs PD-L1 >=1%, stratified by presence of targeted mutations.
When comparing patients with an OS >12 months to those with OS ≥12 months, younger age, adenocarcinoma subtype, the presence of an EGFR mutation, and contralateral lung metastasis were associated with longer OS (p-value 0.014, <0.01, 0.02, and 0.019, respectively).
By contrast, Black patients, those with squamous cell carcinoma, the presence of an HER2 mutation, and brain or bone metastasis were associated with poorer OS (p-value <0.01, 0.023, 0.028, 0.034, and 0.039, respectively). A detailed analysis is shown in Table 5.
This study has demonstrated varying results with respect to the common sites of metastasis for NSCLC patients with common mutations. EGFR mutation can be seen in up to 10% of white patients with NSCLC and up to 50% in NSCLC patients of Asian origin (6). In our study, EGFR was the most common actionable mutation seen, accounting for about 24% of all mutations. KRAS gene mutations accounted for about 25% of driver mutations, and other targetable activating gene mutations found included EML4-ALK, HER-2, RET, MET, and ROS1, among others (7). Classic EGFR mutations (exon 19 deletion and L858R) have been shown to be commonly associated with bone and pleural metastasis (4). Another single-institution study showed that EGFR exon 19 deletions were commonly associated with bone and intrapulmonary metastasis. That study also showed that patients with a T790M mutation had higher incidences of brain, bone, and liver metastasis compared with patients who had other EGFR gene mutations (8). In contrast, another retrospective study analyzing data from 105 patients did not find any statistically significant difference in the site of metastasis for EGFR mutant vs wild-type tumors (9). We also found that patients with a mutation were significantly more likely to have metastatic disease to the brain. Tumors with EGFR and KRAS/NRAS mutations specifically had a predilection for brain metastasis, which was observed in 60% and 59% of patients, respectively.
Our data showed that patients with an EGFR gene mutation had a longer OS than patients without this mutation or patients with a KRAS or TP53 mutation. This finding is consistent with existing literature describing a longer OS for patients with an activating EGFR mutation or those who received an EGFR-TKI (10). The original IPASS study was able to show improved 12-month PFS in patients who received an EGFR-TKI compared to patients who received chemotherapy (24.9% vs. 6.7%) (11). In the FLAURA trial, a newer generation EGFR-TKI, osimertinib, showed improved PFS compared to previous generations (HR for progression or death 0.46) and, for the first time, PFS also translated to an increase in OS (HR 0.8, 95% CI 0.64–1.00) (12). We also found a significantly decreased 12-month OS in patients with a HER-2 mutation, which is consistent with previous studies associating HER-2 mutation with a poor prognosis (13). Furthermore, although some studies have found shorter PFS and OS in KRAS-mutated tumor compared to non–KRAS-mutated tumors, we did not find a difference in 12-month OS by KRAS mutation status (14, 15).
TP53 is known to be the most frequent mutation found in NSCLC, and this was also demonstrated in our study (16). An analysis of 1441 patients with NSCLC showed that TP53 mutations are not only the most common mutation found but also that a TP53 gene mutation is an adverse prognostic factor (16). In our study, patients with a TP53 mutation had a significantly poorer OS and PFS than patients with EGFR mutations; however, this result was not statically significant, likely due to our small sample size. Recent data also suggest that activated TP53 can be associated with EGFR mutations and can be utilized in the stratification of tumor types (16). Studies have shown that these co-mutations are associated with a poorer prognosis (16, 17). Recent data also suggest that TP53 mutations sensitize chemo-resistant tumors with EGFR-activating mutations (16). In our retrospective analysis, only 8 patients had both EGFR and TP53 mutations, although their PFS was shorter than that of patients with an EGFR mutation alone (10.4 vs. 15 months, respectively), and there was no difference in OS (29.4 vs. 29.1 months, respectively). The data was not statistically significant due to our small sample size.
PD-L1 plays an important role in the immune checkpoint responsible for allowing tumor cells to evade the immune system. Currently, several monoclonal therapies are available that target tumors displaying the appropriate biomarkers. Thus, PD-L1 remains a clinically significant biomarker for the treatment of NSCLC. However, associations between the presence of PD-L1 markers and clinicopathologic features of NSCLC have not been well studied. We found no significant rate of metastatic disease to the brain or bone in patients with PD-L1 >1% compared to those with PD-L1 <1%, but we did find a significant difference in rates of metastatic disease to other organs. This is aligned with the results of a study by Zhang et al., which found an association between PD-L1 expression and abdominal organ metastasis but not brain metastasis (18). However, another study by Lin et al. found no significant correlation between PD-L1 expression and metastasis to the brain or other organs (19). Furthermore, there are mixed results on whether PD-L1 expression has any association with lymph node metastasis: some studies have shown that it does, while others have found the opposite results (20, 21). Overall, the role of PD-L1 expression and its correlation to metastatic sites is unclear due to the limited number of studies available, and further research is needed to determine its use as a prognostic factor.
In a systemic literature review by Brody et al., out of 10 studies, 4 reported a significantly shorter survival with higher PD-L1 expression, and 2 reported shorter survival with higher PD-L1 expression that was either not significant or not analyzed (22). In the same review, 3 studies found no significant association between PD-L1 levels and survival, and 1 study reported longer survival with higher levels of PD-L1 expression (22). In our study, we found a nonsignificant increase in OS associated with PD-L1 between the P0M0 group and the P1M0 group (20.6 vs. 30.3 months) and between the P0M1 group and the P1M1 group (33.5 vs. 44.5 months). The longer OS in the P1M1 groups was likely due to more targeted therapy options.
Based on our data, we identified 4 prognostic groups in metastatic NSCLC based on the current treatment paradigm. Patients with no PD-L1 expression (<1%) and no discoverable mutation had the worst outcomes, whereas patients with the presence of a targetable mutation and PD-L1 >1% had the best outcomes. These results need to be validated in larger studies to see whether they hold true.
There are a few strengths of this study. First, we not only analyzed molecular driver genes but also included PD-L1 expression levels in identifying the potential site of metastasis and its role in survival outcomes in patients with stage IV NSCLC. We also gathered data on non-targetable molecular mutations, including HER2, ALK, ROS, BRAF, NTRK, RET, CDKN2A, and others. Furthermore, up to 60% of patients had PET/CT scan either at the time of diagnosis or at progression, which allowed better detection of metastasis compared with CT scan alone.
Our study has a few limitations. First, this is single-center retrospective study, which may limit the generalizability of the results; however, our findings are consistent with those of other large population studies. Second, we have a small patient sample for each molecular driver cohort, which makes the analysis and interpretation difficult; thus, no definitive comparison or conclusions can be drawn for each of the cohorts. For example, we had only 4 patients with HER-2 mutations and only 8 patients with both TP53 and EGFR mutations. Third, there may be a bias due to the site of metastasis, given that we included the molecular profile obtained from primary and metastasis samples. There were also two liquid biopsies included in the analysis. Lastly, our study only includes patients with de novo Stage IV NSCLC and trying to identify the pattern of mutation in respect to site of metastasis. In our study, we reported the incidence and predilection of site of metastasis with the molecular profile, we were not able to use the molecular profile to predict the site of progression which require different research methodology.
This retrospective analysis provides greater insight into the correlation between site of metastasis and prognosis of patients with stage IV NSCLC and their molecular profiles and PD-L1 status. Patients with discoverable mutations on NGS are more likely to have metastatic disease to the brain. Tumors with KRAS/NRAS mutations in particular showed a predilection to metastasis to the brain and bone and were associated with poorer prognosis. Patients with EGFR mutations, despite these tumors having a great propensity to brain metastasis, have significantly better PFS and OS than patients with KRAS/NRAS and TP53 mutations, likely due to targeted therapy options. PD-L1 expression and TP53 mutation, on the other hand, tend to lead to disease metastatic to organs other than brain or bone. Our data suggested that four prognostic groups can be identified in metastatic NSCLC based on current treatment paradigm. Patients with PD-L1 <1% and no actionable mutations have the poorest prognosis, with a median survival of around 20 months. This is an important example of using real-world data to predict survival outcomes of patients with stage IV NSCLC, and these results need to be corroborated in larger studies.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The study protocol was approved by the Institutional Review Board of both Memorial Hermann Hospital and The University of Texas Health Science Center at Houston. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because This is a retrospective cohort analysis.
KC contributed to the conception ad design, acquisition, analysis, and interpretation of data, as well as participated in drafting and revision of the manuscript. AS and JL actively participated in collecting data, analysis of data, and drafting and revision of manuscript. SJ contributed to idea design, data analysis, and critically revision of the manuscript and as well as approving the final version of manuscript. All authors contributed to the article and approved the submitted version.
We would to express our sincere gratitude and appreciation to Ms. Virginia Mohlere, ELS, who helped revise the grammar of our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1212788/full#supplementary-material
1. Clark SB, Alsubait S. Non-small cell lung cancer. In: StatPearls. Treasure Island (FL: StatPearls Publishing (2021).
2. Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, et al. Metastatic patterns in adenocarcinoma. Cancer (2006) 106(7):1624–33. doi: 10.1002/cncr.21778
3. Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncol (1995) 13(1):8–10. doi: 10.1200/JCO.1995.13.1.8
4. Kuijpers CCHJ, Hendriks LEL, Derks JL, Dingemans AC, van Lindert ASR, van den Heuvel MM, et al. Association of molecular status and metastatic organs at diagnosis in patients with stage IV non-squamous non-small cell lung cancer. Lung Cancer (2018) 121:76–81. doi: 10.1016/j.lungcan.2018.05.006
5. Hsu F, De Caluwe A, Anderson D, Nichol A, Toriumi T, Ho C. Patterns of spread and prognostic implications of lung cancer metastasis in an era of driver mutations. Curr Oncol (2017) 24(4):228–33. doi: 10.3747/co.24.3496
6. Hirsch FR, Bunn PA Jr. EGFR Testing in lung cancer is ready for prime time. Lancet Oncol (2009) 10(5):432–3. doi: 10.1016/S1470-2045(09)70110-X
7. Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: characteristics, detection methods, and targeted therapies. Oncotarget (2017) 8(34):57680–92. doi: 10.18632/oncotarget.17016
8. Chen Y, Deng J, Liu Y, Wang H, Zhao S, He Y, et al. Analysis of metastases in non-small cell lung cancer patients with epidermal growth factor receptor mutation. Ann Transl Med (2021) 9(3):206. doi: 10.21037/atm-20-2925
9. Beypinar I, Demir H, Araz M, Uysal M. The relationship between EGFR mutation and metastasis pattern in lung adenocarcinoma. J Oncol Sci (2019) 5(2):65–9. doi: 10.1016/j.jons.2019.08.002
10. Zhao D, Chen X, Qin N, Su D, Zhou L, Zhang Q, et al. The prognostic role of EGFR-TKIs for patients with advanced non-small cell lung cancer. Sci Rep (2017) 7:40374. doi: 10.1038/srep40374
11. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med (2009) 361(10):947–57. doi: 10.1056/NEJMoa0810699
12. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med (2020) 382(1):41–50. doi: 10.1056/NEJMoa1913662
13. Mazières J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol (2013) 31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095
14. Marabese M, Ganzinelli M, Garassino MC, Shepherd FA, Piva S, Caiola E, et al. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy. Oncotarget (2015) 6(32):34014–22. doi: 10.18632/oncotarget.5607
15. Goulding RE, Chenoweth M, Carter GC, Boye ME, Sheffield KM, John WJ, et al. KRAS mutation as a prognostic factor and predictive factor in advanced/metastatic non-small cell lung cancer: a systematic literature review and meta-analysis. Cancer Treat Res Commun (2020) 24:100200. doi: 10.1016/j.ctarc.2020.100200
16. Canale M, Andrikou K, Priano I, Cravero P, Pasini L, Urbini M, et al. The role of TP53 mutations in EGFR-mutated non-small-cell lung cancer: clinical significance and implications for therapy. Cancers (Basel) (2022) 14(5):1143. doi: 10.3390/cancers14051143
17. Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 mutation on survival in patients with advanced EGFR-mutant non-small-cell lung cancer. JCO Precis Oncol (2018) 2018. PO.18.00107. doi: 10.1200/PO.18.00107
18. Zhang J, Gao J, Li Y, Nie J, Dai L, Hu W, et al. Circulating PD-L1 in NSCLC patients and the correlation between the level of PD-L1 expression and the clinical characteristics. Thorac Cancer (2015) 6(4):534–8. doi: 10.1111/1759-7714.12247
19. Lin C, Chen X, Li M, Liu J, Qi X, Yang W, et al. Programmed death-ligand 1 expression predicts tyrosine kinase inhibitor response and better prognosis in a cohort of patients with epidermal growth factor receptor mutation-positive lung adenocarcinoma. Clin Lung Cancer (2015) 16(5):e25–35. doi: 10.1016/j.cllc.2015.02.002
20. Sasaki H, Suzuki A, Shitara M, Hikosaka Y, Okuda K, Moriyama S, et al. PD-L1 gene expression in Japanese lung cancer patients. BioMed Rep (2013) 1(1):93–6. doi: 10.3892/br.2012.10
21. Mao Y, Li W, Chen K, Xie Y, Liu Q, Yao M, et al. B7-H1 and B7-H3 are independent predictors of poor prognosis in patients with non-small cell lung cancer. Oncotarget (2015) 6(5):3452–61. doi: 10.18632/oncotarget.3097
Keywords: non-small cell lung cancer, NSCLC, metastasis, molecular profile, mutations, survival
Citation: Chan KH, Sridhar A, Lin JZ and Jafri SHR (2023) Genomic profiling and sites of metastasis in non-small cell lung cancer. Front. Oncol. 13:1212788. doi: 10.3389/fonc.2023.1212788
Received: 26 April 2023; Accepted: 29 August 2023;
Published: 12 September 2023.
Edited by:
Alessandro Russo, A.O. Papardo, ItalyReviewed by:
Pasquale Pisapia, University of Naples Federico II, ItalyCopyright © 2023 Chan, Sridhar, Lin and Jafri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Syed Hassan Raza Jafri, c3llZC5oLmphZnJpQHV0aC50bWMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.