
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol., 06 July 2023
Sec. Thoracic Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1207892
This article is part of the Research TopicKRAS in Stage IV Non-Small Cell Lung CancerView all 10 articles
Background: The aim of this study was to investigate the diagnostic accuracy of KRAS mutation detection using plasma sample of patients with non-small cell lung cancer (NSCLC).
Methods: Databases of Pubmed, Embase, Cochrane Library, and Web of Science were searched for studies detecting KRAS mutation in paired tissue and plasma samples of patients with NSCLC. Data were extracted from each eligible study and analyzed using MetaDiSc and STATA.
Results: After database searching and screening of the studies with pre-defined criteria, 43 eligible studies were identified and relevant data were extracted. After pooling the accuracy data from 3341 patients, the pooled sensitivity, specificity and diagnostic odds ratio were 71%, 94%, and 59.28, respectively. Area under curve of summary receiver operating characteristic curve was 0.8883. Subgroup analysis revealed that next-generation sequencing outperformed PCR-based techniques in detecting KRAS mutation using plasma sample of patients with NSCLC, with sensitivity, specificity, and diagnostic odds ratio of 73%, 94%, and 82.60, respectively.
Conclusion: Compared to paired tumor tissue sample, plasma sample showed overall good performance in detecting KRAS mutation in patients with NSCLC, which could serve as good surrogate when tissue samples are not available.
Lung cancer is a leading cause of cancer-related death worldwide (1). As its most prevalent subtype, non-small cell lung cancer (NSCLC) represents approximately 85% of lung cancer cases (2). Treatments of NSCLC include surgery, radiotherapy, chemotherapy, immunotherapy, and targeted therapy in tumors harboring certain oncogenetic variations, e.g., anti-epidermal growth factor receptor (EGFR) therapy (2).
Kirsten rat sarcoma viral oncogene homologue (KRAS) is the most frequently mutated oncogene in many types of cancer (3), with an overall prevalence of 27.5% in NSCLC (4). Mutation of KRAS gene is associated with resistance to anti-EGFR therapies (5–7). In addition, although KRAS was thought to be an “undruggable” target, it has become “druggable” after the successful approval of KRAS (G12C) inhibitor (Sotorasib) for the treatment of KRAS G12C-mutated metastatic NSCLC (8). Due to these important roles of KRAS mutation in targeted therapies, accurate detection of KRAS gene mutations, especially G12C, is crucial for the success of anti-EGFR therapies and KRAS inhibitors.
The detection of KRAS mutations in tumors is usually performed using tumor tissue samples, e.g., formalin-fixed paraffin-embedded (FFPE) tumor tissue samples. However, tissue samples are sometimes not available, or may not reflect the real-time mutation status of tumor due to the existence of cancer evolution (9). Research efforts were therefore made to find possible surrogates for tumor tissue samples, which are mainly cell-free DNA (cfDNA)-containing samples, such as plasma, urine, saliva, feces, exhaled breath condensate, and etc (10, 11). Before their clinical application, however, those surrogate sample types needs to be validated for their accuracy performance in detecting KRAS mutations. Many such studies have been conducted. A recently-published systemic review and meta-analysis by Palmieri (12) summarized the results of 40 relevant studies and reported an overall adequate accuracy of cfDNA-containing samples. This meta-analysis by Palmieri focused on cfDNA, and involved studies using plasma, urine, or sputum samples. However, cfDNA levels in the three sample types are quite different, which could potentially influence accuracy performance. In addition, compared to urine or sputum samples which could be highly concentrated or diluted, cfDNA levels in plasma samples are considered to be more stable and therefore had potentially better stability in accuracy performance. Considering these advantages, we chose to focus on plasma, and aimed to better understand the accuracy performance of plasma sample in KRAS mutation detection in NSCLC, including potential impact of patient characteristics.
Literature search was performed by BY and JZ in June 2022. Online literature databases (Pubmed, Embase, Cochrane Library, and Web of Science) were searched using keywords: “KRAS”, “plasma”, and “NSCLC”. Alternative spelling or abbreviations were also included in the literature search, e.g., non-small-cell lung cancer, non-small-cell lung carcinoma, NSCLCs, NSCLC’s, plasmas, and plasma’s (please see detailed searching strategy in Supplementary Material). Searching results were exported from each database. Duplicated literatures were then identified by matching titles, names of first author, or identification numbers (e.g., Pubmed ID) of literatures from different databases. After removing the duplicated literatures, the abstracts of the searching results were firstly screened to exclude irrelevant literatures. The full texts of the rest literatures were then downloaded and screened for eligible studies. The criteria used for the two screening steps were as follows. Inclusion criteria: all original studies testing KRAS mutation in paired plasma and tumor tissue samples of NSCLC. Exclusion criteria: 1) not a human study; 2) missing plasma or tumor tissue samples; 3) plasma and tumor tissue samples were not paired; 4) not testing KRAS mutation in either plasma or tissue samples; 5) lacking KRAS wild-type or KRAS mutated samples; 6) not an original study; 7) un-interpretable data; 8) not NSCLC samples. Accuracy data were then extracted from the KRAS mutation testing results of paired plasma and tumor tissue samples in the eligible studies, including numbers of true positive, false positive, false negative, and true negative. In addition, characteristics of patients or techniques were also extracted, including region and population of studies, tumor stage, and techniques used to test KRAS mutation in plasma and in tissue samples. All the eligible studies were evaluated by quality assessment of diagnostic accuracy studies 2 (QUADAS-2) (13). Any disagreement between the two investigators (BY and JZ) were solved by a third investigator (PC). PRISMA 2009 Checklist is included in Supplementary Material.
Statistical analysis was performed using Meta-DiSc 1.4 (14) and STATA 12.0 (STATA Corp.). Sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under curve (AUC) of summary receiver operating characteristic (SROC) curve were pooled from the accuracy data extracted from the eligible studies. During the pooling, random effects model was used when significant heterogeneity was observed (I2 ≥ 50% and P < 0.05), and fixed effects model was used when no significant heterogeneity was observed (14). In case of significant heterogeneity, threshold analysis and meta-regression were performed to find its possible sources. Deek’s funnel plot asymmetry test was performed to find potential publication bias in the eligible studies. P < 0.05 was considered statistically significant.
As shown in Figure 1, a total of 622 publications were identified after the literature search (Pubmed: 114; Embase: 333; Cochrane Library: 29; Web of Science: 146). After removing 216 duplicated literatures, titles and abstracts of the rest 406 publications were screened, and 305 irrelevant studies were excluded. Full text of the rest 101 publications were downloaded and carefully evaluated for their eligibility, and another 58 publications were further excluded. From the 43 eligible studies, accuracy data and other relevant information were extracted.
Twenty-nine of the 43 eligible studies (Table 1) used next-generation sequencing (NGS) to detect KRAS mutation in plasma samples. In the rest 14 studies, 12 studies used PCR-based techniques, 1 study used pyrosequencing, and 1 study used MassARRAY.
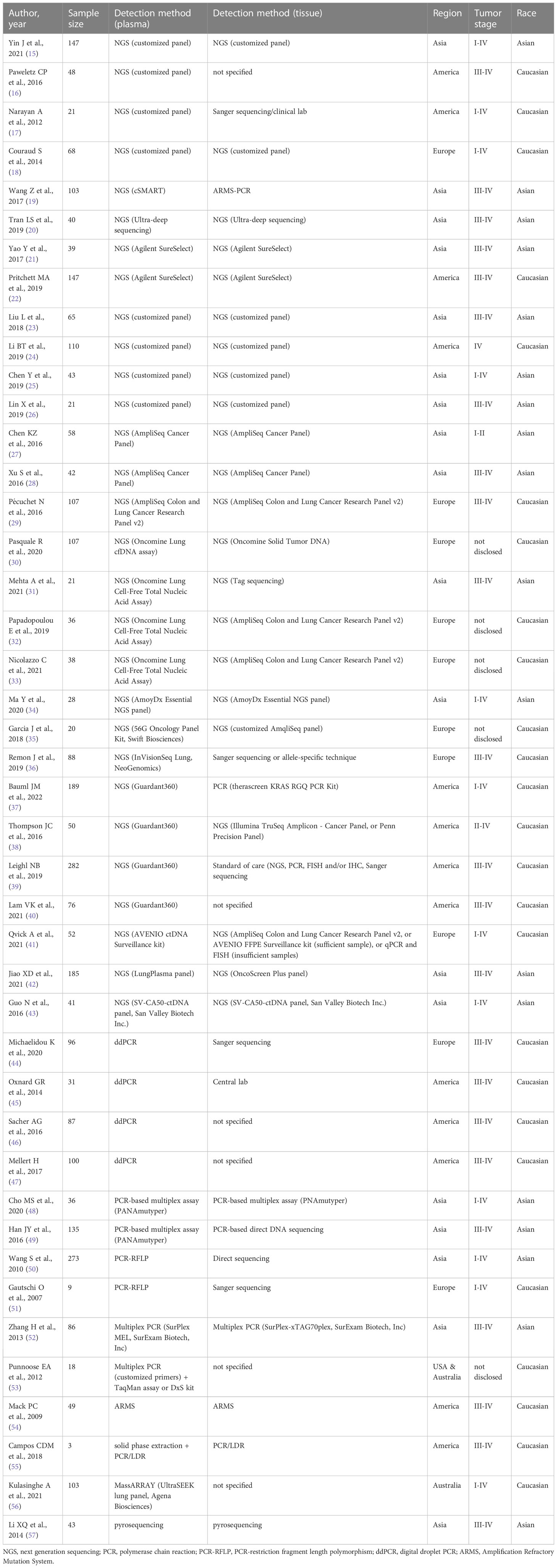
Table 1 Summary of studies detecting KRAS mutation in paired plasma and tissue samples from NSCLC patients.
In the eligible studies using NGS, sensitivities ranged from 25% to 100%, and specificities and concordance rates were relatively higher, ranging from 64% to 100% and from 52.63% to 100%, respectively.
Twelve studies used customized NGS panels, in which 5 studies used amplicon-based targeted sequencing (15–19). In the study by Yin (15), KRAS mutation detected in tumor tissue samples were all detected in paired plasma samples, resulting in 100% sensitivity. The specificity and concordance rate were 99.24% and 99.32%, respectively. Similarly, study by Narayan (17) showed perfect matching (100% concordance rate) of KRAS mutation results between plasma and tissue samples. However, study by Paweletz (16) and by Couraud (18) showed much lower sensitivity (54.55% and 75%, respectively), although high specificity (100%) was observed. In the study by Wang Z (19), circulating single-molecule amplification and resequencing technology (cSMART) showed sensitivity of 58.82%, specificity of 100%, and concordance rate of 93.20%. The large variations in the sensitivity of KRAS mutation detection in plasma samples may be due to the small number of patients included in these studies.
The rest 7 studies used hybridization-based targeted sequencing (20–26). A customized panel from xGen (Integrated DNA Technologies) showed perfect match between plasma and tumor tissue results (100% concordance rate) (20). Studies by Yao (21) and Pritchett (22) used a hybridization-based target enrichment method from Agilent Technologies (SureSelect). The two studies showed similar concordance rates (91.16% and 97.44%). Studies by Liu (23), Li BT (24), Chen Y (25), and Lin (26) also used hybridization-based capture methods to enrich customized gene panels for NGS sequencing of plasma samples. The concordance rates of those studies were all high, ranging from 93.02% to 96.92%.
Besides customized NGS panels, several commercial NGS panels were also used, such as AmpliSeq panels, Oncomine panels, AmoyDx Essential NGS panel, 56G Oncology Panel, InVisionSeq Lung, Guardant360, AVENIO ctDNA Surveillance kit, LungPlasma panel, and SV-CA50-ctDNA panel. AmpliSeq Cancer Panel (Thermo Fisher Scientific) was used in two studies (27, 28). However, the results varied greatly between them. Sensitivity, specificity, and concordance rate were 60%, 96.23%, and 93.10% in Chen KZ’s study (27), and 100%, 83.33%, and 85.71% in Xu’s study (28). AmpliSeq Colon and Lung Cancer Research Panel v2 showed sensitivity of 62.96%, specificity of 100%, and concordance rate of 90.65% (29). Oncomine Lung cfDNA Assay (Thermo Fisher Scientific) showed sensitivity, specificity, and concordance rate of 61.54%, 93.83%, and 85.98%, respectively (30). Oncomine Lung Cell-Free Total Nucleic Acid Assay (Thermo Fisher Scientific) was used in three studies, and accuracy results varied greatly: sensitivity from 30.77% to 81.82%, specificity from 64% and 100%, and concordance rate from 52.63% to 94.44% (31–33). AmoyDx Essential NGS panel (Amoy Diagnostics) was used in a 28-patient cohort, and the sensitivity, specificity, and concordance rate were 66.67%, 96%, and 92.86%, respectively (34). Studies by Garcia (35) and Remon (36) also used amplicon-based targeted sequencing techniques, including 56G Oncology Panel (Swift Biosciences), InVisionSeq Lung (NeoGenomics), respectively. Results showed sensitivity of 64.29% and 88%, specificity of 83.33% and 88.89%, and concordance rate of 70% and 88.64%.
Four studies validated the accuracy of Guardant360 in detecting KRAS mutation in plasma samples (37–40). Sensitivity ranged from 66.67% to 87.50%. Specificity ranged from of 74.81% to 100%, and concordance rate ranged from 75.89% to 98%. AVENIO ctDNA Surveillance kit (Roche) is also a commercial panel using hybridization-based target enrichment. A study using AVENIO ctDNA Surveillance kit showed sensitivity of 72.73%, specificity of 100%, and concordance rate of 94.23% (41).
In the rest two studies using commercial NGS panels, detailed target enrichment method was not disclosed. Studies by Jiao (42) used LungPlasma NGS panel (Burning Rock Biotech), and sensitivity, specificity, and concordance rate were 68.97%, 99.36%, and 94.59%. Guo (43) used SV-CA50-ctDNA panel (San Valley Biotech), and results showed 50% sensitivity, 97.44% specificity, and 95.12% concordance rate.
A total of 4 studies used digital droplet PCR (ddPCR) to detect KRAS mutation in plasma samples (44–47). Although ddPCR is a sensitive technique which could detect genetic mutations as low as 0.01%, the results of these studies did not show high accuracy of ddPCR in plasma-based KRAS mutation detection. Sensitivity ranged from 51.43% to 87.88%, and specificity ranged from 88.52% to 100%, resulting in concordance rates from 75% to 96%.
Other than ddPCR, several PCR-based techniques were also used to detect KRAS mutation in plasma samples, such as PANAmutyper, PCR-restriction fragment length polymorphism (PCR-RFLP), multiplex PCR, Amplification Refractory Mutation System (ARMS), and PCR/ligase detection reaction (LDR) technique. Overall, those PCR-based techniques were mostly used in early studies, which showed sensitivity ranging from 33.33% to 100%, specificity from 50% to 100%, and concordance rate from 55.56% to 100%.
PANAmutyper is a multiplex PCR method which increases sensitivity through suppressing amplification of wild-type DNA using specific peptide nucleic acids (PNA) (48). In the two studies using PANAmutyper, the sensitivity was 33.33% and 50%, and specificity was 100% and 89.43%, resulting in concordance rates of 88.89% and 85.93%, respectively (48, 49).
In the two studies using PCR-RFLP, accuracy results varied greatly. In Wang S’s study (50), the sensitivity, specificity, and concordance rate were 76.67%, 95.06%, and 93.04%, respectively. In the study of Gautschi (51), these numbers were 50%, 66.67%, and 55.56%, respectively.
Multiplex PCR was used in two studies. Study by Zhang (52) used SurExam MEL (SurExam Biotech), a typical commercial multiplex PCR, to detect KRAS mutation in plasma samples, and sensitivity, specificity, and concordance rate were 33.33%, 98.80%, and 96.51%. In the study by Punnoose (53), the KRAS mutation results of plasma samples matched perfectly with tissue samples (100% concordance rate).
An early study by Mack (54) used KRAS Scorpion-ARMS test kit (DxS Ltd), and results showed 50% sensitivity, 100% specificity, and 97.96% concordance rate.
Campos (55) and colleagues developed a microfluidic solid-phase extraction device to extract cfDNA, which were then analyzed using PCR/LDR technique. Only 3 NSCLC samples were tested in the study, and the results showed 100% sensitivity, 50% specificity, and 66.67% concordance rate.
UltraSEEK lung panel (Agena Biosciences), a commercial MassARRAY panel, was used in a 103-patient cohort, and sensitivity, specificity, and concordance rate were 62.96%, 92.11%, and 84.47%, respectively (56). Pyrosequencing was used in an early study (57), and sensitivity and specificity were 75% and 100%, respectively, resulting in a concordance rate of 97.67%.
In all, the 43 eligible studies compared KRAS mutation status in paired plasma and tissue samples from 3341 NSCLC patients. Thirty-nine of the 43 eligible studies (39/43) showed high specificity (≥ 80%), and 37 studies showed high concordance rate (≥ 80%). However, high sensitivity (≥ 80%) was only observed in 14 out of 43 studies.
Quality assessment of eligible studies was performed using QUADAS-2. As shown in Table 2, the 43 eligible studies showed overall good quality, with high risk observed in only 2 studies (both in flow and timing). In the assessment of risk of bias, percentage of low risk ranged from 46.51% (n = 20, Index test) to 69.77% (n = 30, both patient selection and reference standard). In the application concerns, no high risk was observed, and percentage of low risk ranged from 83.72% (n = 36, reference standard) to 86.05% (n = 37, both patient selection and index test).
From the 43 eligible studies, we pooled the KRAS mutation detection results from paired plasma and tissue samples of 3341 patients with NSCLC. The overall sensitivity and specificity were 0.71 [95% confidence interval (CI): 0.68-0.75] and 0.94 (95%CI: 0.93-0.95), respectively. The pooled DOR was 59.28 (95%CI: 34.37-102.25), and AUC of SROC curve was 0.8883. Please see Table 3 and Figure 2 for details.
Since significant heterogeneity (I2 ≥ 50% and P < 0.05) was observed, we further analyzed its possible sources. Analysis of diagnostic threshold showed no significant threshold effect (spearman correlation coefficient = 0.058, P = 0.714). Meta-regression revealed that inter-study heterogeneity was associated with techniques used for plasma sample (P = 0.0388), but not with techniques used for tissue sample (P = 0.1280), region of study (P = 0.3299), tumor stage (P = 0.3049), or race of patients (P = 0.7798).
Subgroup analysis was then performed on different techniques used for plasma sample. The 43 eligible studies were grouped into three subgroups: NGS, PCR-based techniques, and other techniques. Meta-analysis was performed in each subgroup except other techniques due to limited number (only two) of studies in that subgroup. As shown in Table 3, compared to PCR-based techniques, NGS showed overall better accuracy: sensitivity of 0.73 (95%CI: 0.69-0.77), specificity of 0.94 (95%CI: 0.93-0.95), DOR of 82.60 (95%CI: 40.62-167.96), and AUC of SROC curve of 0.9162. After further dividing the group of PCR-based techniques into two subgroups (ddPCR and other PCR-based techniques), ddPCR showed higher sensitivity [0.68 (95%CI: 0.59-0.77)], specificity [0.97 (95%CI: 0.93-0.99)], and DOR [85.60 (95%CI: 6.80-1978.05)], but much lower AUC of SROC curve (0.2741).
Subgroup analysis was also performed on the region of studies, including Asia, America, Australia, and Europe. Australia was excluded from the subgroup analysis due to limited number of studies in the subgroup. In the other three subgroups, studies performed in America showed overall best accuracy, with pooled sensitivity of 0.76 (95%CI: 0.71-0.81), specificity of 0.92 (95%CI: 0.90-0.94), DOR of 111.35 (95%CI: 56.05-221.20), and AUC of SROC curve of 0.9272.
Twenty-four of the 43 eligible studies used late-stage (stage III and IV) NSCLC samples, and 13 studies used NSCLC samples of any stage (stage I to IV). As shown in Table 3, pooled accuracy results of the two subgroups (stage III-IV versus stage I-IV) did not differ much from each other. However, this result should be treated carefully because although early-stage NSCLC samples were involved, majority of the samples were still late-stage in stage I-IV subgroup. The rest 6 studies were not involved in the subgroup analysis, including 1 study using early-stage (I and II) NSCLS samples only, and 5 studies which did not disclose the tumor stage of samples.
Majority of the 43 eligible studies were conducted using samples from Caucasian patients, and the rest studies used samples of Asian patients. Between the two subgroups, pooled accuracy data were similar (see Table 3).
Publication bias was evaluated using Deek’s funnel plot (Figure 3). The results indicated no significant publication bias (P = 0.097).
Before anti-EGFR therapies are given to NSCLC patients, it is important to determine whether the tumor carries KRAS mutation since it may lead to resistance to anti-EGFR therapies. Moreover, determination of KRAS mutation status is also required before the usage of KRAS (G12C) inhibitor, e.g., Sotorasib. Tumor tissue samples are the “gold standard” in the determination of KRAS mutation. However, tumor tissue samples are sometimes not available, and cfDNA-containing samples (e.g., plasma, urine, saliva, etc.) have been intensively investigated as surrogates for tissue samples. A recently-published systemic review and meta-analysis by Palmieri summarized the performance of cfDNA-containing samples in detecting KRAS mutation in NSCLC (12). Due to the higher and more stable levels of cfDNA in plasma compared to other cfDNA-containing sample types, we focused solely on plasma in this systemic review and meta-analysis, and investigated its accuracy in determining tumor KRAS mutation status in NSCLC.
In order to investigate the accuracy of KRAS mutation detection using plasma samples, several previous studies compared KRAS mutation results in paired plasma and tissue samples from patients with NSCLC. After database searching and screening, we identified 43 eligible studies. After pooling the KRAS mutation status from 3341 patients with NSCLC, the results showed overall moderate sensitivity (0.71) and high specificity (0.94). Other important indicators of diagnostic accuracy, DOR and AUC of SROC curve, were also high (59.28 and 0.8883, respectively). Although with moderate sensitivity, these results indicated overall high accuracy of plasma samples in detecting KRAS mutation. In the systemic review and meta-analysis by Palmieri (12), the pooled sensitivity and specificity were 0.71 and 0.93, respectively, and DOR was 35.24, which were similar to the findings of our study.
Since significant inter-study heterogeneity was observed during the pooling (I2 ≥ 50% and P < 0.05), we investigated its possible sources. Analysis of diagnostic threshold did not indicate significant threshold effect. Meta-regression revealed significant association between inter-study heterogeneity and techniques used for plasma sample. This is different from Palmieri’s study, in which detection method did not contribute to heterogeneity (12). No significant association was shown between heterogeneity and other covariates (techniques used for tissue sample, region of study, tumor stage, and race of patients).
Different from Palmieri’s study, we further conducted subgroup analysis. Subgroup analysis on technique used for plasma sample was firstly performed. After pooling the accuracy results, we found that NGS outperformed PCR-based techniques in many accuracy parameters, including sensitivity (0.73), DOR (82.60), and AUC of SROC curve (0.9162). We further divided the group of PCR-based techniques into two groups: ddPCR and other PCR-based techniques. Compared to NGS, ddPCR showed similar sensitivity (0.68), specificity (0.97), and DOR (85.60), except for surprisingly low AUC of SROC curve (0.2741) which was possibly due to the limited number of studies in this subgroup (Table 3).
We also performed subgroup analysis on region of study. Studies performed in Asia showed the highest AUC of SROC curve (0.9381). Studies performed in America showed the highest sensitivity (0.76) and DOR (111.35), and similar AUC of SROC curve with Asia (0.9272), indicating overall the highest accuracy of the studies from America.
Late-stage tumors was reported to be associated with significantly higher fraction of circulating tumor DNA (ctDNA) in cfDNA (58), which may indicate potentially better performance of genetic testing using these samples. In the 43 eligible studies, involvement of early-stage samples did not significantly influence the accuracy results. However, this result should be treated with care because numbers of early-stage samples were much smaller than late-stage samples in a large proportion of these studies. Race of patients also did not show significant impact on the accuracy results. The performance of KRAS mutation testing using plasma was similar between Asian and Caucasian patients. Significant publication bias was not observed using Deek’s funnel plot asymmetry test.
In summary, results of this systemic review and meta-analysis indicated overall high accuracy of plasma samples in predicting KRAS mutation results of paired NSCLC tumor tissue samples. Plasma could serve as surrogates when tissue samples are not available, although it may miss a small proportion of patients carrying KRAS mutation considering its moderate sensitivity. Among different techniques, NGS showed the best accuracy. Although majority of accuracy results were comparable to NGS, ddPCR suffered from its low AUC of SROC curve. Therefore, NGS is recommended in the detection of KRAS mutations in plasma samples of patients with NSCLC, especially when multiple genetic variations are tested considering the high-throughput of the technology. Limitation of this study may be the small number of studies in the ddPCR subgroup and limited numbers of early-stage tumor samples used in some studies, which must be treated carefully. In addition, although different techniques are generally thought to have similar performance in tumor samples considering the high abundance of DNA, it may still cause potential bias. Large prospective studies are required to further validate the results of this study.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
PC, BY, and DY contributed to conception and design of the study. BY and JZ organized the database. PY performed the statistical analysis. PC wrote the first draft of the manuscript. BY, JZ, and PY wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
A Project Supported by Center for Early Childhood Education Research, Sichuan (grant number CECER-2022-B01) and Chengdu Municipal Health Commission, 2022 Chengdu Medical Research Project (grant number 2022582).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1207892/full#supplementary-material
1. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol (2016) 893:1–19. doi: 10.1007/978-3-319-24223-1_1
2. Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, et al. Non-small-cell lung cancer. Nat Rev Dis Primers (2015) 1:15009. doi: 10.1038/nrdp.2015.9
3. Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal transduct targeted Ther (2021) 6(1):386. doi: 10.1038/s41392-021-00780-4
4. Judd J, Abdel Karim N, Khan H, Naqash AR, Baca Y, Xiu J, et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer. Mol Cancer Ther (2021) 20(12):2577–84. doi: 10.1158/1535-7163.MCT-21-0201
5. Del Re M, Tiseo M, Bordi P, D'Incecco A, Camerini A, Petrini I, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget (2017) 8(8):13611–9. doi: 10.18632/oncotarget.6957
6. Ye P, Wang Y, Li R, Chen W, Wan L, Cai P. The HER family as therapeutic targets in colorectal cancer. Crit Rev oncology/hematol (2022) 174:103681. doi: 10.1016/j.critrevonc.2022.103681
7. Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res (2007) 13(10):2890–6. doi: 10.1158/1078-0432.CCR-06-3043
8. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. New Engl J Med (2021) 384(25):2371–81. doi: 10.1056/NEJMoa2103695
9. Zhu X, Li S, Xu B, Luo H. Cancer evolution: a means by which tumors evade treatment. Biomed pharmacother = Biomed pharmacother (2021) 133:111016. doi: 10.1016/j.biopha.2020.111016
10. Freitas AJA, Causin RL, Varuzza MB, Calfa S, Hidalgo Filho CMT, Komoto TT, et al. Liquid biopsy as a tool for the diagnosis, treatment, and monitoring of breast cancer. Int J Mol Sci (2022) 23(17):9952. doi: 10.3390/ijms23179952
11. Ryan DJ, Toomey S, Smyth R, Madden SF, Workman J, Cummins R, et al. Exhaled breath condensate (EBC) analysis of circulating tumour DNA (ctDNA) using a lung cancer specific UltraSEEK oncogene panel. Lung Cancer (2022) 168:67–73. doi: 10.1016/j.lungcan.2022.04.013
12. Palmieri M, Zulato E, Wahl SGF, Guibert N, Frullanti E. Diagnostic accuracy of circulating free DNA testing for the detection of KRAS mutations in non-small cell lung cancer: a systematic review and meta-analysis. Front Genet (2022) 13:1015161. doi: 10.3389/fgene.2022.1015161
13. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Internal Med (2011) 155(8):529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
14. Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Method (2006) 6:31. doi: 10.1186/1471-2288-6-31
15. Yin JX, Hu WW, Gu H, Fang JM. Combined assay of circulating tumor DNA and protein biomarkers for early noninvasive detection and prognosis of non-small cell lung cancer. J Cancer (2021) 12(4):1258–69. doi: 10.7150/jca.49647
16. Paweletz CP, Sacher AG, Raymond CK, Alden RS, O'Connell A, Mach SL, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res (2016) 22(4):915–22. doi: 10.1158/1078-0432.CCR-15-1627-T
17. Narayan A, Carriero NJ, Gettinger SN, Kluytenaar J, Kozak KR, Yock TI, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res (2012) 72(14):3492–8. doi: 10.1158/0008-5472.CAN-11-4037
18. Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanche H, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res (2014) 20(17):4613–24. doi: 10.1158/1078-0432.CCR-13-3063
19. Wang Z, Cheng G, Han X, Mu X, Zhang Y, Cui D, et al. Application of single-molecule amplification and resequencing technology for broad surveillance of plasma mutations in patients with advanced lung adenocarcinoma. J Mol diagn JMD (2017) 19(1):169–81. doi: 10.1016/j.jmoldx.2016.09.008
20. Tran LS, Pham HT, Tran VU, Tran TT, Dang AH, Le DT, et al. Ultra-deep massively parallel sequencing with unique molecular identifier tagging achieves comparable performance to droplet digital PCR for detection and quantification of circulating tumor DNA from lung cancer patients. PloS One (2019) 14(12):e0226193. doi: 10.1371/journal.pone.0226193
21. Yao Y, Liu J, Li L, Yuan Y, Nan K, Wu X, et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget (2017) 8(2):2130–40. doi: 10.18632/oncotarget.12883
22. Pritchett MA, Camidge DR, Patel M, Khatri J, Boniol S, Friedman EK, et al. Prospective clinical validation of the InVisionFirst-lung circulating tumor DNA assay for molecular profiling of patients with advanced nonsquamous non-small-cell lung cancer. JCO Precis Oncol (2019) 3:PO.18.00299. doi: 10.1200/PO.18.00299
23. Liu L, Liu H, Shao D, Liu Z, Wang J, Deng Q, et al. Development and clinical validation of a circulating tumor DNA test for the identification of clinically actionable mutations in nonsmall cell lung cancer. Genes Chromosomes Cancer (2018) 57(4):211–20. doi: 10.1002/gcc.22522
24. Li BT, Janku F, Jung B, Hou C, Madwani K, Alden R, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the actionable genome consortium. Ann Oncol (2019) 30(4):597–603. doi: 10.1093/annonc/mdz046
25. Chen Y, Han T, Zhou Y, Mao B, Zhuang W. Comparing the efficacy of targeted next-generation sequencing in the identification of somatic mutations in circulating tumor DNA from different stages of lung cancer. Neoplasma (2019) 66(4):652–60. doi: 10.4149/neo_2018_181130N910
26. Lin X, Dong W, Lai X, Feng W, Yu X, Gu Q, et al. The clinical value of circulating tumor DNA detection in advanced non-small cell lung cancer. Trans Cancer Res (2019) 8(1):170–9. doi: 10.21037/tcr.2019.01.20
27. Chen KZ, Lou F, Yang F, Zhang JB, Ye H, Chen W, et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep (2016) 6:31985. doi: 10.1038/srep31985
28. Xu S, Lou F, Wu Y, Sun DQ, Zhang JB, Chen W, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett (2016) 370(2):324–31. doi: 10.1016/j.canlet.2015.11.005
29. Pecuchet N, Zonta E, Didelot A, Combe P, Thibault C, Gibault L, et al. Base-position error rate analysis of next-generation sequencing applied to circulating tumor DNA in non-small cell lung cancer: a prospective study. PloS Med (2016) 13(12):e1002199. doi: 10.1371/journal.pmed.1002199
30. Pasquale R, Forgione L, Roma C, Fenizia F, Bergantino F, Rachiglio AM, et al. Targeted sequencing analysis of cell-free DNA from metastatic non-small-cell lung cancer patients: clinical and biological implications. Trans Lung Cancer Res (2020) 9(1):61–70. doi: 10.21037/tlcr.2020.01.01
31. Mehta A, Kumar Sharma S, Kumar D, Vasudevan S. Plasma biopsy by tag-sequencing: an acceptable alternative to tumor tissue profiling in non-small-cell lung cancer. Polish J Pathol (2021) 72(2):117–25. doi: 10.5114/pjp.2021.109514
32. Papadopoulou E, Tsoulos N, Tsantikidi K, Metaxa-Mariatou V, Stamou PE, Kladi-Skandali A, et al. Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PloS One (2019) 14(12):e0226853. doi: 10.1371/journal.pone.0226853
33. Nicolazzo C, Gelibter A, Bottillo I, Belardinilli F, Pisegna S, De Renzi G, et al. Comparison of two blood-based genotyping tests to investigate the KRAS G12C mutation in patients with non-Small-Cell lung cancer at failure of first-line treatments. Diagn (Basel) (2021) 11(12):2196. doi: 10.3390/diagnostics11122196
34. Ma Y, Li Q, Du Y, Chen W, Zhao G, Liu X, et al. Oncogenic genetic alterations in non-Small-Cell lung cancer (NSCLC) in southwestern China. Cancer Manage Res (2020) 12:10861–74. doi: 10.2147/CMAR.S266069
35. Garcia J, Forestier J, Dusserre E, Wozny AS, Geiguer F, Merle P, et al. Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget (2018) 9(30):21122–31. doi: 10.18632/oncotarget.24950
36. Remon J, Lacroix L, Jovelet C, Caramella C, Howarth K, Plagnol V, et al. Real-world utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non-Small-Cell lung cancer. JCO Precis Oncol (2019) 3:PO.18.00211. doi: 10.1200/PO.18.00211
37. Bauml JM, Li BT, Velcheti V, Govindan R, Curioni-Fontecedro A, Dooms C, et al. Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib. Lung Cancer (2022) 166:270–8. doi: 10.1016/j.lungcan.2021.10.007
38. Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, Balli D, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res (2016) 22(23):5772–82. doi: 10.1158/1078-0432.CCR-16-1231
39. Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res (2019) 25(15):4691–700. doi: 10.1158/1078-0432.CCR-19-0624
40. Lam VK, Zhang J, Wu CC, Tran HT, Li L, Diao L, et al. Genotype-specific differences in circulating tumor DNA levels in advanced NSCLC. J Thorac Oncol (2021) 16(4):601–9. doi: 10.1016/j.jtho.2020.12.011
41. Qvick A, Stenmark B, Carlsson J, Isaksson J, Karlsson C, Helenius G. Liquid biopsy as an option for predictive testing and prognosis in patients with lung cancer. Mol Med (2021) 27(1):68. doi: 10.1186/s10020-021-00331-1
42. Jiao XD, Ding LR, Zhang CT, Qin BD, Liu K, Jiang LP, et al. Serum tumor markers for the prediction of concordance between genomic profiles from liquid and tissue biopsy in patients with advanced lung adenocarcinoma. Trans Lung Cancer Res (2021) 10(7):3236–50. doi: 10.21037/tlcr-21-543
43. Guo N, Lou F, Ma Y, Li J, Yang B, Chen W, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep (2016) 6:33519. doi: 10.1038/srep33519
44. Michaelidou K, Koutoulaki C, Mavridis K, Vorrias E, Papadaki MA, Koutsopoulos AV, et al. Detection of KRAS G12/G13 mutations in cell free-DNA by droplet digital PCR, offers prognostic information for patients with advanced non-small cell lung cancer. Cells (2020) 9(11):2514. doi: 10.3390/cells9112514
45. Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O'Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res (2014) 20(6):1698–705. doi: 10.1158/1078-0432.CCR-13-2482
46. Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O'Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol (2016) 2(8):1014–22. doi: 10.1001/jamaoncol.2016.0173
47. Mellert H, Foreman T, Jackson L, Maar D, Thurston S, Koch K, et al. Development and clinical utility of a blood-based test service for the rapid identification of actionable mutations in non-small cell lung carcinoma. J Mol diagn JMD (2017) 19(3):404–16. doi: 10.1016/j.jmoldx.2016.11.004
48. Cho MS, Park CH, Lee S, Park HS. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PloS One (2020) 15(3):e0230622. doi: 10.1371/journal.pone.0230622
49. Han JY, Choi JJ, Kim JY, Han YL, Lee GK. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer (2016) 16:627. doi: 10.1186/s12885-016-2678-2
50. Wang S, An T, Wang J, Zhao J, Wang Z, Zhuo M, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res (2010) 16(4):1324–30. doi: 10.1158/1078-0432.CCR-09-2672
51. Gautschi O, Huegli B, Ziegler A, Gugger M, Heighway J, Ratschiller D, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett (2007) 254(2):265–73. doi: 10.1016/j.canlet.2007.03.008
52. Zhang H, Liu D, Li S, Zheng Y, Yang X, Li X, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol diagn JMD (2013) 15(6):819–26. doi: 10.1016/j.jmoldx.2013.06.006
53. Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res (2012) 18(8):2391–401. doi: 10.1158/1078-0432.CCR-11-3148
54. Mack PC, Holland WS, Burich RA, Sangha R, Solis LJ, Li Y, et al. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. J Thorac Oncol (2009) 4(12):1466–72. doi: 10.1097/JTO.0b013e3181bbf239
55. Campos CDM, Gamage SST, Jackson JM, Witek MA, Park DS, Murphy MC, et al. Microfluidic-based solid phase extraction of cell free DNA. Lab chip (2018) 18(22):3459–70. doi: 10.1039/c8lc00716k
56. Kulasinghe A, O'Leary C, Monkman J, Bharti V, Irwin D, Dutta S, et al. The identification of circulating tumour DNA using MassARRAY technology in non-small-cell lung cancer (NSCLC). Lung Cancer (2021) 160:73–7. doi: 10.1016/j.lungcan.2021.08.005
57. Li X, Zhang L, Xu Y, Peng L, Sun J, Chen Z. A modified method for detecting the common mutations of K-ras gene from both plasma and cancer tissue samples of NSCLC patients. Med J Chin People's Liberation Army (2014) 39(3):202–5. doi: 10.11855/j.issn.0577-7402.2014.03.07
Keywords: KRAS, plasma, non-small cell lung cancer, diagnostic accuracy, meta-analysis
Citation: Cai P, Yang B, Zhao J, Ye P and Yang D (2023) Detection of KRAS mutation using plasma samples in non-small-cell lung cancer: a systematic review and meta-analysis. Front. Oncol. 13:1207892. doi: 10.3389/fonc.2023.1207892
Received: 18 April 2023; Accepted: 20 June 2023;
Published: 06 July 2023.
Edited by:
Wouter H. Van Geffen, Medisch Centrum Leeuwarden, NetherlandsReviewed by:
Oke Dimas Asmara, University of Groningen, NetherlandsCopyright © 2023 Cai, Yang, Zhao, Ye and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongmei Yang, ZG9uZ21laXkyMDIwQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.