- Department of Internal Medicine, Section of Hematology, Oncology & Rheumatology, University Hospital Heidelberg, Heidelberg, Germany
Inherited hematologic malignancies are linked to a heterogenous group of genes, knowledge of which is rapidly expanding using panel-based next-generation sequencing (NGS) or whole-exome/whole-genome sequencing. Importantly, the penetrance for these syndromes is incomplete, and disease development, progression or transformation has critical clinical implications. With the earlier detection of healthy carriers and sequential monitoring of these patients, clonal hematopoiesis and somatic driver variants become significant factors in determining disease transformation/progression and timing of (preemptive) hematopoietic stem cell transplant in these patients. In this review, we shed light on the detection of probable germline predisposition alleles based on diagnostic/prognostic ‘somatic’ NGS panels. A multi-tier approach including variant allele frequency, bi-allelic inactivation, persistence of a variant upon clinical remission and mutational burden can indicate variants with high pre-test probability. We also discuss the shared underlying biology and frequency of germline and somatic variants affecting the same gene, specifically focusing on variants in DDX41, ETV6, GATA2 and RUNX1. Germline variants in these genes are associated with a (specific) pattern or over-/underrepresentation of somatic molecular or cytogenetic alterations that may help identify the underlying germline syndrome and predict the course of disease in these individuals. This review is based on the current knowledge about somatic drivers in these four syndromes by integrating data from all published patients, thereby providing clinicians with valuable and concise information.
Introduction
In 1999, Song et al. characterized the first inherited leukemia syndrome by recognizing that germline RUNX1 variants lead to lifelong thrombocytopenia and an increased risk of myelodysplastic syndrome (MDS) and acute leukemia (AL) (1). Subsequently, many more genes have been associated with a germline predisposition to develop hematologic malignancies, which has led to the inclusion of the entity ‘myeloid neoplasms with germline predisposition’ into the revised 2016 World Health Classification (WHO) (2). In 2022, the WHO update incorporated additional newly associated genes and extended the phenotype of some of the syndromes to also include a predisposition to lymphoid malignancies (3). Generally, these syndromes are distinguished into three different categories (1): myeloid neoplasms without a preexisting disorder or organ dysfunction (e.g. CEBPA, DDX41), (2) myeloid neoplasms with a preexisting platelet disorder (ANKRD26, ETV6, RUNX1) and (3) myeloid neoplasms with potential other organ dysfunctions. The latter category covers a diverse spectrum of syndromes like bone marrow failure syndromes, telomere biology disorders and predisposition syndromes related to pathogenic variants in GATA2, SAMD9 and SAMD9L. Likewise, the International Consensus Classification of myeloid neoplasms and acute leukemias in 2022 also encompassed hematologic neoplasms with germline predisposition in a similar format (4). Germline predisposition to hematologic malignancies has also been integrated in clinical guidelines such as the European LeukemiaNet and the National Comprehensive Cancer Network (5, 6). The most frequent germline syndromes are caused by pathogenic variants in transcription factors like CEBPA, ETV6, GATA2, and RUNX1, in the RNA helicase DDX41, and a variety of genes associated with telomere biology disorders and inherited bone marrow failure (7).
With the advent of diagnostic/prognostic next-generation sequencing (NGS) panels designed for somatic variants, germline variants are invariably detected as well, and recognition of these germline syndromes has increased. Since sequencing is performed on DNA from bone marrow/peripheral blood representing the affected tissue, the presence of a variant at a germline variant allele frequency (VAF) alone is not sufficient to presume germline origin. A multi-tier approach including confirmation in true germline material is usually required and specific criteria may indicate a higher likelihood for the presence of a germline variant.
Additional cytogenetic and molecular alterations are requisite for the transformation of a clone with a pathogenic germline variant to MDS or AL. These alterations may include (1): Additional well-known driver variants such as loss-of-function (LOF) variants in tumor suppressors, gain-of-function variants in (proto-) oncogenes and variants in genes involved in DNA repair, chromatin modification, transcriptional activation, DNA methylation and numerous others. Patients with congenital neutropenia represent an example for a strong association of somatic variants truncating the cytoplasmic domain of CSF3R with an impending leukemic transformation in variant carriers (8); (2) Contributory cytogenetic alterations that are well-known to promote disease. For instance, the association of germline SAMD9/SAMD9L variants with monosomy 7 is significant and essential for leukemogenesis in most patients (9, 10); (3) Bi-allelic inactivation with a second somatic hit on the other allele has been frequently observed in DDX41, CEBPA, RUNX1, TP53, and to a much lesser extent, in GATA2 and is consistent with initiation or progression of disease (11–15). Additional factors such as an increase of the clonal VAF or an increase in somatic mutational burden also affect transformation or progression of disease (16–19).
Identifying the factors pointing to disease progression or transformation is critical for the follow-up of patients with germline syndromes and may affect the timing of treatment initiation, type of treatment and (preemptive) hematopoietic stem cell transplantation (HSCT). Current recommendations for patients with germline syndromes consist of a baseline bone marrow biopsy and additional bone marrow biopsies upon significant and persistent changes in blood counts (20). The timing and intervals of bone marrow biopsies and cytogenetic/molecular (re-) analyses may be tailored based on the individual risk of clonal alterations in these patients.
In this review, we discuss how germline variants can be identified using NGS-based panels primarily designed for somatic variants. We also assess somatic drivers in patients with germline variants in DDX41, ETV6, GATA2 and RUNX1 by reviewing the published evidence and analyzing the type and pattern of both germline and somatic variants in these genes and their role in malignant transformation.
Germline variants detected upon diagnostic/prognostic sequencing of tumor tissue
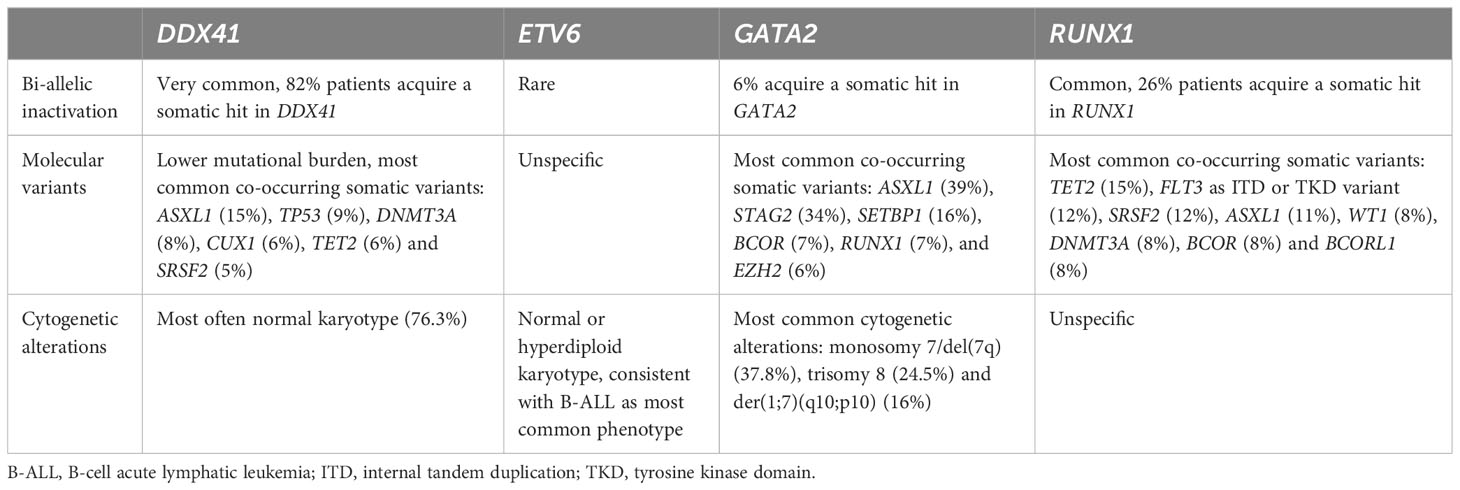
The current standard of diagnosis and risk-stratification for myeloid and lymphoid malignancies are NGS panels based on a variety of genes known to be frequently mutated in the malignant clone. Risk-stratification elicited from molecular genetics are implemented in numerous risk scores, among others: the European LeukemiaNet risk classification for acute myeloid leukemia (AML) (5) and the Molecular International Prognostic Scoring System (IPSS-M) for MDS (16). These bear significant weight in estimating the patient’s individual risk and prognosis. Although we generally use the term ‘somatic NGS-panel’, these panels can also detect germline variants. Several studies have reported that NGS-based prognostic panels performed at the time of diagnosis frequently detect germline variants (14, 15, 21–32). The spectrum of germline mutated genes depends on the genes covered by the NGS panel and whether the panel has been designed to include copy number variants (CNVs), which also represent common predisposition alleles (33, 34). The following criteria indicate a higher likelihood of an underlying germline syndrome: (1) VAF — a heterozygous variant would be expected at/near heterozygosity (VAF 40-60%) and a recessive variant at/near homozygosity (VAF 90-100%) or in the compound heterozygous state. Of note, the VAF threshold may differ depending on the panel used so that the afore mentioned thresholds do not exclude the presence of a germline variant outside of this range (Table 1). (2) Presence of bi-allelic inactivation — specifically, tumor suppressors are often inactivated by a second hit on the trans allele, a phenomenon that has been well-described for CEBPA, DDX41, RUNX1, and TP53 among others (Table 1) (11, 12, 14, 15, 37). (3) The persistence of a pathogenic/likely pathogenic variant at a germline VAF over the course of disease when remission is achieved is another clue and can be utilized when longitudinal sequencing data are present (Table 1). However, clonal hematopoiesis of unknown potential (CHIP) can also explain the persistence of a variant in remission/after therapy, when these variants were not part of the malignant clone. (4) Some variants are only reported in germline, for example LOF variants in DDX41 are usually considered of germline origin (Table 1). The association of the mutational burden, defined as the total number of somatic variants in a patient, as independent prognostic variable with worsening outcomes has been well-established in MDS (16–18). The recently implicated IPSS-M concludes that the additional number of somatic variants in so called residual genes, that were not individually weighted, does lead to an additive increase in risk from 0 to 2, followed by saturation of worsening outcomes when 3 or more variants are present (16). A higher than usual mutational burden has been linked to a germline syndrome affecting the mismatch-specific DNA N-glycosylase MBD4. MBD4 acts as safeguard against damage from 5mC deamination and its deficiency results in increased risk to develop CHIP/AML, colorectal polyposis and uveal melanoma with significantly increased mutational burden compared to sporadic cases (35, 36). In AML specifically, MBD4-deficiency displays a 33-fold higher mutational burden with a unique mutational signature, where 95% of variants are CG>TG substitutions (Table 1) (35). Only few patients with autosomal recessive MBD4-deficiency and evidence of CHIP/AML have been described to date. In contrast to this rather distinct germline syndrome, most other germline syndromes are associated with a mutational burden similar to patients with sporadic disease affecting the same gene. Therefore higher, or lower mutational burden is not per se indicative of a germline syndrome.
Importantly, performing panel-based NGS designed for somatic variants cannot be used to rule out an underlying germline syndrome. Non-coding regions such as the 5’UTR in ANKRD26, containing all known germline predisposition alleles for this gene or the deep intronic enhancer in GATA2 are not covered by a panel directed towards somatic variants. The same is true for CNVs, accounting for 10-15% of all predisposition alleles as mentioned earlier (33, 34), that are usually not detected either. Somatic and germline variant interpretation is based on a combination of different criteria (38, 39) and mechanisms of disease, location of variants and functional assessment may vary resulting in different evaluations of pathogenicity. Table 1 summarizes the criteria suggesting a germline variant related to sequencing data and the genes with higher pre-test probability of variants confirmed in germline.
Leukemogenesis in DDX41 germline-mutated patients relies on bi-allelic inactivation of the trans DDX41 allele
DDX41 is a multifunctional DEAD box helicase that operates as a DNA sensor, initiating an innate immune response, as a tumor suppressor through regulation of pre-mRNA splicing and RNA processing and as a modulator of gene expression of numerous oncogenes, tumor suppressor genes and genes involved in immune response and antigen presentation (40–42). DDX41 germline variants cause the most frequent hematopoietic germline syndrome yet known (12, 43). Based on published studies of DDX41 carriers, approximately 1.5% to 3.8% of unselected MDS/AML patients carry a germline DDX41 variant (12, 43, 44), indicating that variants in this gene carry a high pre-test probability for a germline origin. Presumably somatic DDX41 variants occur in approximately 2.4% of all MDS patients (16) and less than 1% of AML patients (45–47), strengthening the assumption that the majority of identified DDX41 variants may indeed be germline.
Based on a compilation of all published DDX41 germline variants (12, 21, 22, 25, 33, 43, 44, 48–67), LOF variants, including nonsense, frameshift and canonical splice variants in DDX41 amass to the majority of germline LOF variants (65.4%, n=641, Figure 1A). Almost all LOF variants (98,1%, n=629) occur early in the gene and are predicted or confirmed to undergo nonsense-mediated decay (NMD) (Figure 1A) with no functional protein product. Based on population data (https://gnomad.broadinstitute.org/) two founder variants, p.? (also known as p.M1I or p.M1?, NM_016222.4) and p.D140fs are common in the European (Non-Finnish) population at a VAF of 0.000156 and 0.000185, respectively, and p.A500fs is a founder variant most often detected in the East Asian subpopulation with a slightly lower VAF of 0.000109. Together, these three founder variants account for 39.9% (n=391) of all described germline patients so far (Figure 1A). Other variants like nonsynonymous substitutions (n=280, 28.6%) and indels (n=51, 5.2%) are also well-known predisposition alleles (Figure 1A). Few patients have been reported with non-canonical splice variants and one CNV has been described, encompassing exons 12 to 17 (67).
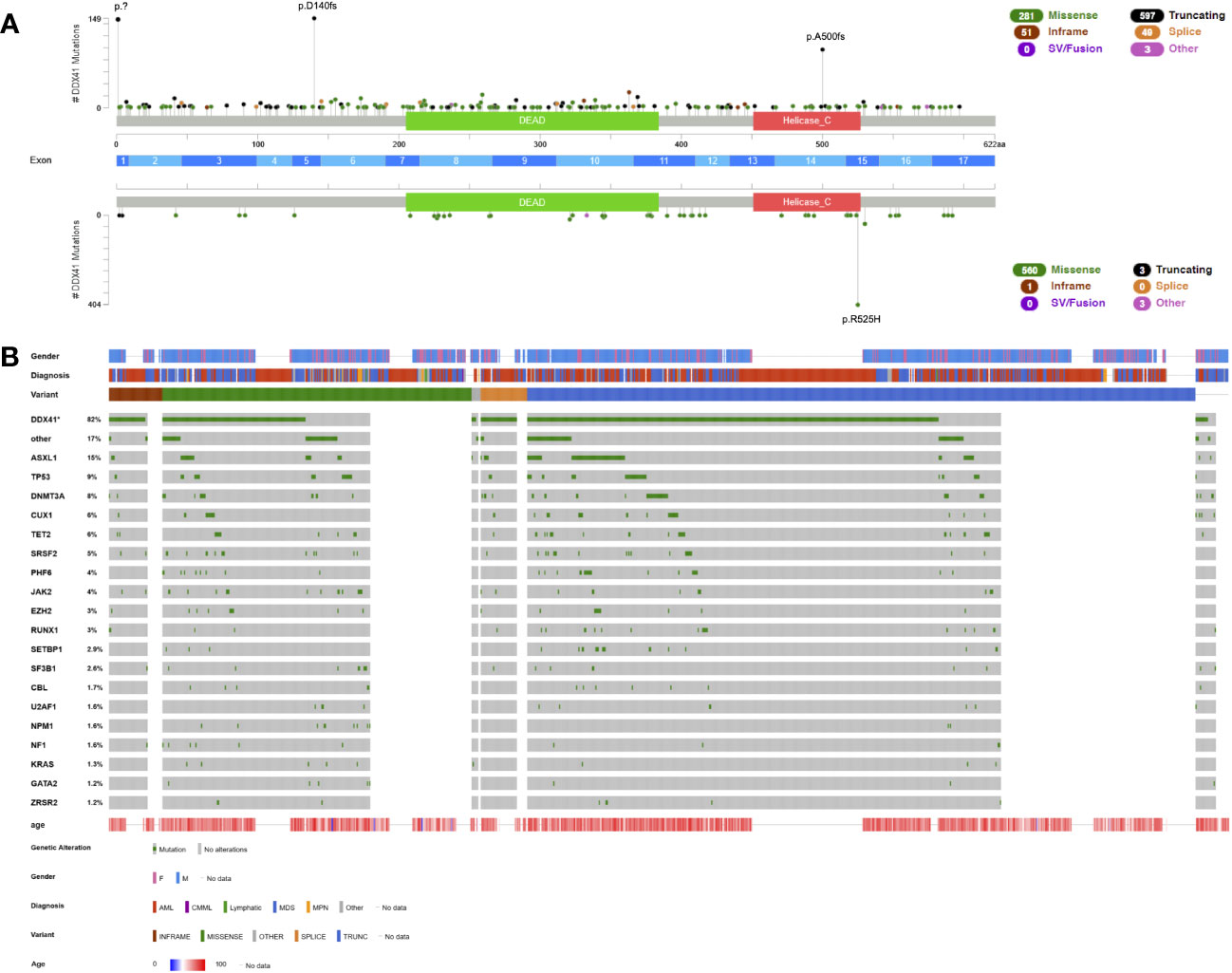
Figure 1 Lollipop plot and Oncoplot of all known DDX41 germline variants. (A) Schematic of the DDX41 transcript NM_016222.4 and its protein domains and exon distribution with the location of all reported variants. Germline variants are shown above the schematic and somatic variants below the schematic. The variant count is displayed on the x axis and the variant type is represented by different colors of the lollipops as outlined in the legend. (B) Oncoplot depicting DDX41 variant carriers and their associated gender, diagnosis, age at diagnosis, variant type and most common co-occurring somatic variants including percentages. AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm. This schematic was created using cBioPortal (https://www.cbioportal.org/) (68, 69).
Germline DDX41 variants predispose to myeloid malignancies, most often MDS and AML and to a lesser extent chronic myelomonocytic leukemia (CMML) or myeloproliferative neoplasms (MPNs) (Figure 1B). Lymphoid malignancies such as chronic lymphatic leukemia (65), large granular lymphocytic leukemia (62), lymphoplasmacytic lymphoma (65), plasma cell disorders (62, 65) and others have been reported in few patients with DDX41 germline variants (Figure 1B). However, a causative association has not been clearly established. The median age of onset — unlike most other germline syndromes — is similar to the expected disease onset in sporadic disease: 67 years (n=328) in DDX41 germline mutated AML patients and 68 years (n=326) in MDS patients (Figure 1B), which is congruent with the average age of onset for AML (68 years, https://seer.cancer.gov/statfacts/html/amyl.html) and MDS (71 years) patients (70). It is rarely diagnosed in patients before the age of 40 (1.6%, Figure 1B). The penetrance is reduced and currently estimated at roughly 50% (12). This germline syndrome often goes unnoticed as a result of this reduced penetrance, older age at diagnosis, and consequently fewer families with a positive family history.
Hematopoietic malignancies with a DDX41 variant — irrespective whether this is a germline or somatic variant — define a distinct subgroup with a lower allelic burden, frequent association with a normal karyotype (Table 2) and overall more favorable prognosis (12, 44, 64, 65, 71). The favorable prognosis is retained in patients with otherwise poor prognostic markers such as multi-hit TP53 variants when compared to age-matched controls (12, 43, 64). The disease shows a clear male predominance with a penetrance of hematologic malignancies that appears three-times higher in males than females (Figure 1B) (12, 25, 43).
Heterozygous DDX41 LOF, however, is not sufficient to initiate leukemogenesis, and the majority of AML and MDS patients with germline variants carry additional somatic nonsynonymous substitutions primarily in the helicase catalytic center of the other allele. Approximately 82% of all patients with germline DDX41 variants acquire a somatic hit with a mean VAF of 11%, 68.8% of those variants are located within the helicase domain and 19.7% within the DEAD box domain (Figure 1A, Table 2). The most common variant is p.R525H found in 71.7% of patients with a somatic hit and located within the helicase domain (Figure 1A). This hotspot variant perturbs ATPase activity and interferes with cell growth in a dominant-negative manner (72). Other common somatic variants include nonsynonymous substitutions p.T227M, p.P321L, p.E345D and p.G530D/S (Figure 1A). Curiously, while almost all LOF variants were confirmed germline with a somatic nonsynonymous substitution, three cases of an identical germline substitution within the DEAD box, p.R369G, with a confirmed LOF somatic DDX41 variant (p.E2* and p.S4*) have been described (12, 25, 44).
The most common co-occurring somatic variants were found in ASXL1 (15%), TP53 (9%), DNMT3A (8%), CUX1 (6%), TET2 (6%) and SRSF2 (5%) (Figure 1B, Table 2). Recurrent molecular markers such as variants in NMP1, CEBPA or internal tandem duplications (ITDs) of FLT3 have only been described in few patients (12, 65). A normal karyotype was described in most patients for whom cytogenetic data were available (76.3%, n=242, Table 2). A range of different cytogenetic alterations e.g. del(5q), -7/del(7q), +8, del (20q), loss of chromosome Y and others, all frequently reported in MDS and AML with myelodysplasia-related changes, has been detected in the remaining patients. A complex karyotype was reported in several patients, most often including a del(5q), presumably derived from clonal evolution of a pre-existing del(5q) clone (12, 44, 65). Recurrent, subgroup-defining cytogenetic aberrations have only been described in four cases, two patients with inversion inv(16) (43, 65) and another two with a translocation t(8;21) (43, 65), again highlighting the near-absence of recurrent molecular or cytogenetic aberrations in DDX41 germline-mutated patients. Little is known about clonal evolution in healthy carriers since most cases reported in the literature are already diseased. With the high percentage of bi-allelic inactivation of DDX41, low mutational burden and frequent absence of cytogenetic alterations, it stands to argue that bi-allelic inactivation is the main driver of disease. Prodromal features such as cytopenia with or without clonal markers and/or some level of dysplasia not meeting MDS criteria (yet) are infrequent (Figure 1B) and usually lead to an MDS/AML diagnosis down the line.
ETV6-deficiency is associated with predominantly lymphoid malignancies concomitant with a normal or hyperdiploid karyotype
ETV6, located on chromosome 12p13, is part of the large ETS transcription factor family, comprising 28 genes primarily controlling tumor initiation and development. Its functional domains consist of a highly conserved N-terminal PNT domain involved in protein-protein interactions with itself and other proteins including FLI1, another member of the ETS family implicated in megakaryocyte lineage commitment (73). A central regulatory domain mediates repressive complex recruitment (including SMRT, Sin3A and NCOR) and autoinhibitory activity (74), and the C-terminal ETS domain conveys DNA binding (75). In cancer, structural variants of this gene are common and occur in a wide variety of different hematologic and solid tumors — with more than 30 translocation partners known to date (76). The well-known recurrent translocation t(12, 21)(p13;q22), resulting in a RUNX1-ETV6 fusion, is identified in 20-25% of pediatric acute lymphatic leukemia (ALL) (77). Somatic variants of this gene are less frequent than structural alterations and have been reported in up to 5% of patients with T-ALL (78, 79), 2.7% of patients with MDS (16) and 1.1% in AML (39, 45, 46). Heterozygous germline variants of this gene were first reported in 2015 and go along with lifelong thrombocytopenia and a predisposition to both lymphoid and myeloid malignancies (80). The manner of inheritance is autosomal dominant with a near-complete penetrance exceeding 90% for thrombocytopenia but incomplete penetrance for hematologic malignancies, estimated in the range of 30% (75, 81). Based on the published studies to date (58, 75, 82–93), B-ALL is the most common malignant phenotype (n=26, 20%), followed by MDS/AML (n=8, 6.2%). At least two cases of mixed phenotype leukemia have been described (80, 82). There is no clear causal association between the few cases of patients with germline ETV6 variants diagnosed with diffuse large B-cell lymphoma and polycythemia vera (83, 85, 94). The median age at onset of a hematologic malignancy is 11 years. Thrombocytopenia is present in most individuals from birth — albeit at times only recognized later in life — and when present is of moderate severity (median of 85+/-28 G/l) accompanied with a mild to moderate bleeding propensity. Nonsynonymous substitutions and LOF variants, predicted to undergo NMD, depict most of the variants reported to date (n=85, 65.9% and n=31, 24%, respectively, Figure S1). The nonsynonymous substitution, p.P214L (NM_001987.5), represents the most common recurrent variant, located within the central region, while other substitutions are mostly scattered across the C-terminal ETS domain (Figure S1). Intragenic CNVs have been described in two families, spanning exons 2 and 5 (84, 89). A constitutional balanced translocation t (12,14)(p13.2;q23.1) was identified in one family with familial B-ALL without thrombocytopenia and breakpoints were located in intron 1 of ETV6 and RTN1, respectively (86). Bi-allelic inactivation of ETV6 in the leukemic clone is rare, be it somatic variants or cytogenetic deletions of the trans allele and have only been described in few cases (Table 2) (82, 95). Karyotypic abnormalities often include a normal or hyperdiploid karyotype, consistent with common alterations detected in (childhood) B-ALL (Table 2). The pattern of somatic co-occurring variants in the leukemic clone resembles a typical pattern seen in ALL or AML/MDS without any obvious association of specific molecular or cytogenetic alterations with germline ETV6 deficiency (Table 2). A longitudinal study of four ETV6 germline mutated carriers did not reveal any evidence of clonal hematopoiesis however, the number of patients was too small to draw conclusions at this time (88).
Somatic ASXL1-, STAG2- and SETBP1 variants, monosomy 7/del(7q) and trisomy 8 drive myeloid malignancies in GATA2-deficiency
GATA2 is a key zinc-finger transcription factor encompassing two zinc finger domains that regulate hematopoietic stem and progenitor cell self-renewal, survival, and differentiation (96). Somatic GATA2 variants occur in 1.9% of all MDS patients (16) and 3.3% of all AML patients (39, 45, 46), mainly affect the first zinc finger domain and often co-occur with CEBPA variants. In contrast, GATA2 germline variants are frequently found within the second zinc finger domain. These variants are common in childhood MDS, accounting for 15% of advanced and 7% of all primary pediatric MDS cases (97). GATA2-deficiency is strongly associated with monosomy 7 and is observed in more than half of all childhood MDS cases with monosomy 7 (97). Roughly 4 to 5% of MDS patients diagnosed as young adults carry GATA2 germline variants (34, 98), however, the likelihood of a germline GATA2-deficiency decreases significantly with the age at diagnosis. More than half of all variants arise de novo, resulting in the lack of a positive family history and evidence of segregation among affected family members (11, 99). The phenotype is highly variable and includes hematopoietic and non-hematopoietic features, encompassing immunodeficiency predominantly through reduced or absent monocytes, B cells, natural killer cells, neutrophils, and/or dendritic cells, MDS/AML, pulmonary disease, vascular/lymphatic dysfunction, and hearing loss (11, 100). The penetrance for any phenotypic features is near-complete, whereas penetrance for myeloid malignancies is variable and incomplete, and most likely lies within a range of 30 to 75% (10, 97, 101, 102).
Based on published GATA2 germline variants (11, 58, 89, 103–112), most variants are truncating LOF variants (46%, n=296) or nonsynonymous variants (44.1%, n=284, Figure 2A). CNVs account for 3.2% (n=20), with most of them being whole-gene deletions (21, 97, 104, 113–119). As opposed to somatic nonsynonymous variants located mainly within the first zinc finger domain, germline nonsynonymous substitutions are — with few exceptions — located within the second zinc finger domain (Figure 2A). Variants in the first versus second zinc fingers seem to harbour different functional consequences and co-occur with specific leukemic lesions (96). Hotspot amino acids within the second zinc finger are C349, C352, T354, T357, T358, L359, W360, R361, N371, A372, C373, L375, P385, M388, R396, and R398 (Figure 2A). A unique mechanism of disease are variants within a deep-intronic +9.5 44bp intronic enhancer element, consisting of an e-box, GATA and Ets/FLI1 motif and two spacers. Currently, causative variants have only been described in the e-box and ets/FLI1 motifs and make up roughly 6% of GATA2 germline variants (97, 120), abrogating normal steady-state hematopoiesis and embryonic development (121).
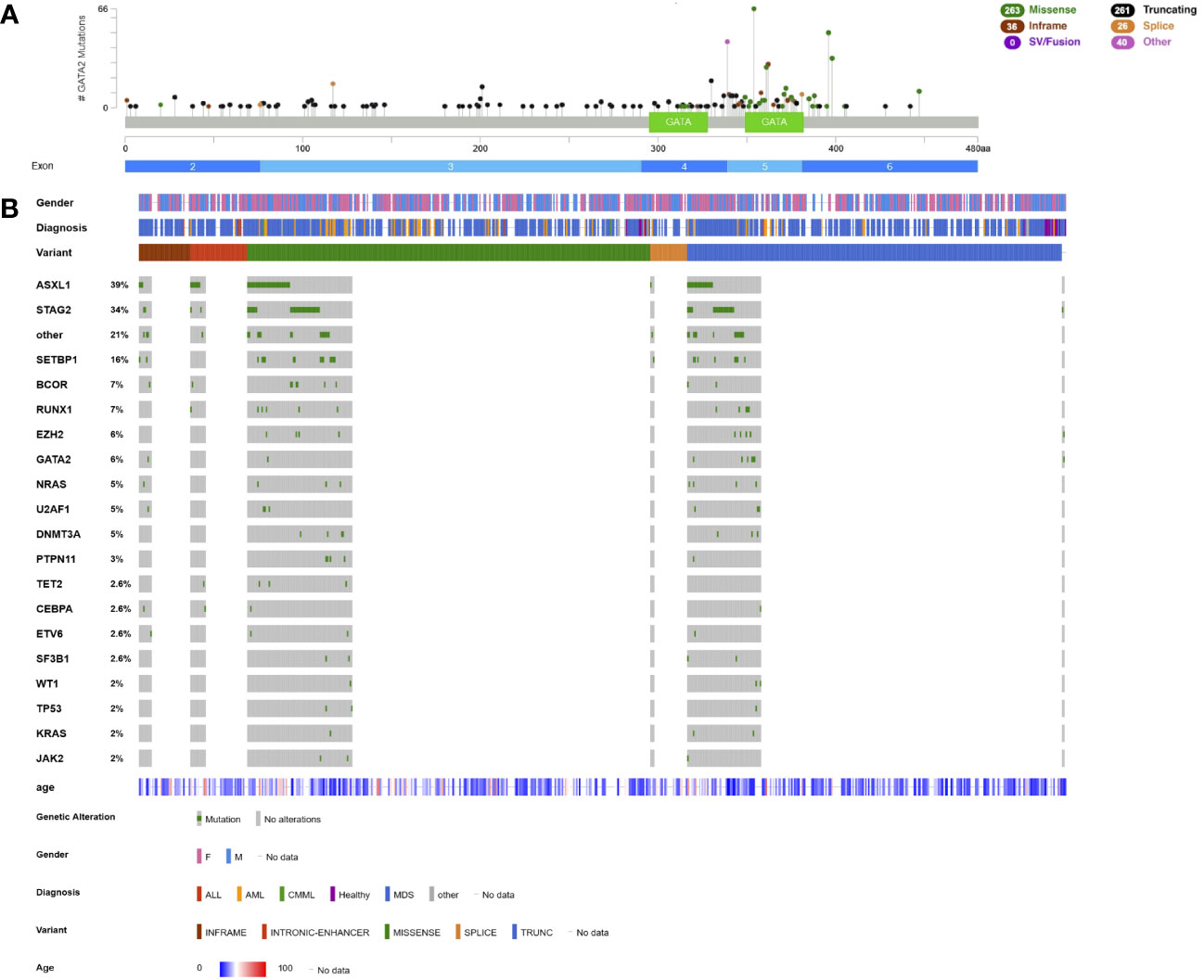
Figure 2 Lollipop plot and Oncoplot of all known GATA2 germline variants. (A) Schematic of the GATA2 transcript NM_032638.5 and its protein domains and exon distribution with the location of all reported germline variants. The variant count is displayed on the x axis and the variant type is represented by different colors of the lollipops as outlined in the legend. (B) Oncoplot depicting GATA2 variant carriers and their associated gender, diagnosis, age at diagnosis, variant type and most common co-occurring somatic variants including percentages. ALL, acute lymphatic leukemia; AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; MDS, myelodysplastic syndrome. This schematic was created using cBioPortal (https://www.cbioportal.org/) (68, 69).
Most patients with GATA2-deficiency have been diagnosed with MDS, CMML/juvenile myelomonocytic leukemia, MDS/MPN overlap, AML and bone marrow failure (78.3%, n=508, Figure 2B). The median age of onset for GATA2-related myeloid disease is 17 years (n=426, Figure 2B). Few cases of B- or T-ALL (105, 116, 122–124) and primary myelofibrosis (112) have been described (Figure 2B).
The most common co-occurring somatic variants are found in ASXL1 (39%), STAG2 (34%), SETBP1 (16%), BCOR (7%), RUNX1 (7%), and EZH2 (6%, Figure 2B, Table 2). A second hit on the other GATA2 allele was reported in 6% of patients (Figure 2B, Table 2). Compared to sporadic AML/MDS cases, variants in ASXL1, SETBP1 and STAG2 are statistically overrepresented in GATA2-deficiency, while variants in DNA methylation modifiers DNMT3A and TET2 seem to be less common (Figure 2B). Monosomy 7/del(7q) is the most common cytogenetic alteration and is found in 37.8% of patients (n=165), followed by trisomy 8 in 24.5% (n=107) of cases (Table 2). A derivative chromosome der(1;7)(q10;p10), resulting in an unbalanced chromosomal translocation with trisomy 1q and deletion 7q, is also commonly observed in GATA2-deficiency (16%, n=35, Table 2). In contrast to monosomy 7/del(7q) and trisomy 8, the der(1;7) is a rare chromosomal alteration and significantly enriched in patients with GATA2-deficiency (109). ASXL1- and STAG2 variants as well as monosomy 7 and trisomy 8 have been observed in several patients with no overt hematologic malignancy, however, these are also common driver variants in the patients with hematologic malignancies.
Germline RUNX1 variants frequently display bi-allelic inactivation in the malignant clone
The master transcription factor RUNX1 is located on chromosome 21q22 and acts as transcriptional regulator of normal hematopoiesis. Functional domains comprise a highly conserved Runt-homology domain (RHD) spanning 128 amino acids, a C-terminal transactivation and inhibitory domain and a VWRPY motif binding transcriptional repressors (125). Three main isoforms A, B, and C are expressed by the use of two promotors and alternative splicing and display isoform-specific functions controlling stem cell expansion and hematopoietic differentiation (126–128).
RUNX1 is frequently mutated in myeloid malignancies in approximately 14.2% of patients with MDS (16) and 10.5% of patients with AML (45–47). RUNX1 variants are considered poor prognostic factors in both MDS and AML and HSCT is mandated whenever possible (5, 16). Structural variants involving RUNX1 are frequent, in fact RUNX1 is the most common target of chromosomal translocations found in AL. The translocation t(8;21)(q22;q22) resulting in a RUNX1-RUNX1T1 fusion is a recurrent genetic abnormality in AML and accompanied by a favorable prognosis (5). The above mentioned translocation t (12; 21), giving rise to a ETV6-RUNX1 fusion protein defines a subtype of pediatric ALL and various other RUNX1 translocation partners have been described as well (125).
Germline RUNX1 variants were first linked to inherited thrombocytopenia and predisposition to myeloid malignancies in 1999 by Song et al. (1). It is currently estimated that in approximately 16% of RUNX1-mutated AML patients the identified RUNX1 variant is indeed of germline origin — higher percentages were reported but germline pathogenicity was not established for all variants (15, 29). This leads to the occurrence of a germline RUNX1 variant in about 1 to 2% of an unselected AML population (29, 31). The penetrance is near-complete for mild to moderate thrombocytopenia with normal sized platelets, potentially in combination with an additional bleeding propensity caused by platelet alpha or dense granule secretion defects and/or impaired platelet aggregation (129). Several patients with intermittent or transient thrombocytopenia have been described so that serial assessment of the platelet count should be considered in patients with presumed normal platelet count. The penetrance for hematologic malignancies is incomplete and estimated at about 40-50% (130, 131).
Based on literature research including all published patients with a causative RUNX1 germline variant (15, 34, 115, 132–170), most germline variants are LOF variants, including nonsense, frameshift, and canonical splice variants (n=244, 53.6%) and roughly one third of these variants are located in the C-terminal transactivation domain predicted to not undergo NMD but instead promoting decreased transactivation capacity (Figure 3A). Strikingly, CNVs account for 15.4% (n=70) of patients, including whole-gene deletions and both in-frame and out-of-frame recurrent intragenic deletions and duplications. A recurrent deletion of exons 1 and 2 or exons 1 to 3 (NM_001754.5), removing the N-terminal 20 to 33 amino acids of isoform C together with its distal promotor, has been detected in multiple unrelated families with a classic phenotype (37, 149, 155, 168, 171–174). Intragenic duplications are rare but have been independently discovered in two families (135, 169). Nonsynonymous substitutions comprise 29.9% (n=132) of all variants and are mostly located within the RHD that is essential for DNA binding (Figure 3A). Causative nonsynonymous variants are primarily found within amino acids 89 to 204 of the RHD, which is where the β-sheet portion of the core binding factor β heterodimerization domain starts, noted as functionally important. Hotspots within the RHD consist of amino acids R107, K110, A134, R162, R166, S167, R169, G170, K194, T196, D198, R201, and R204 (Figure 3A) (129, 175). Outside of these amino acid hotspots, most other variants are private and only occur in one index patient/family (Figure 3B). Germline indels and noncanonical splice variants are infrequent. A germline translocation t(16;21)(p13;q22) has been described, translocating the distal promotor used for isoform C and the +23 enhancer to chromosome 16 (176). The breakpoint is localized in intron 1 of RUNX1 with a high content of simple tandem repeats, consistent with the major breakpoint pattern in the recurrent somatic t(12;21) in ALL (177). Another germline translocation t(11;21)(q13;q22) is also primarily affecting isoform C with the breakpoint located within the same chromosomal region of 21q22 (178).
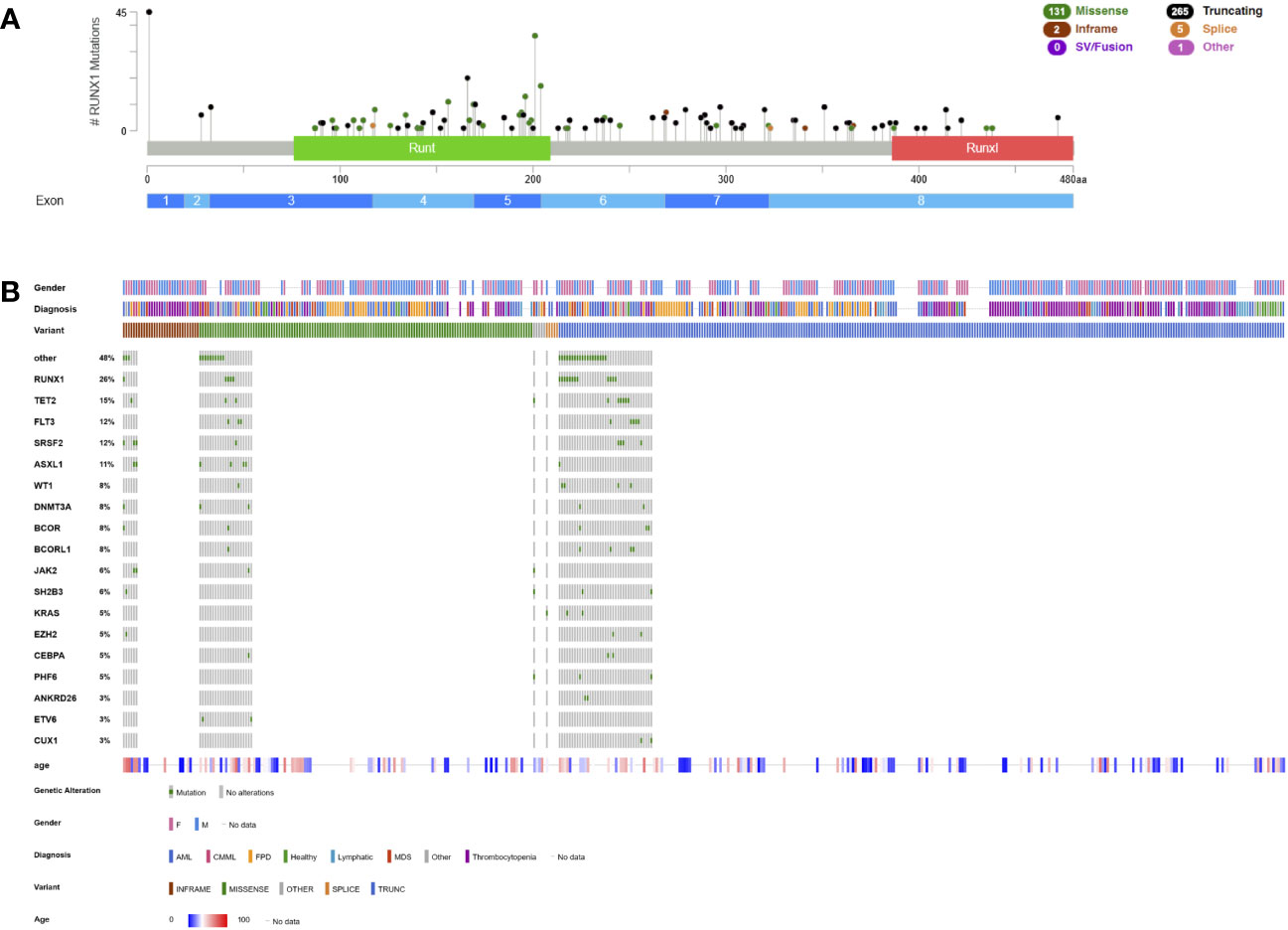
Figure 3 Lollipop plot and Oncoplot of all known RUNX1 germline variants. (A) Schematic of the RUNX1 transcript NM_032638.5 and its protein domains and exon distribution with the location of all reported germline variants. The variant count is displayed on the x axis and the variant type is represented by different colors of the lollipops as outlined in the legend. (B) Oncoplot depicting RUNX1 variant carriers and their associated gender, diagnosis, age at diagnosis, variant type and most common co-occurring somatic variants including percentages. AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; FPD, familial platelet disorder not otherwise specified; MDS, myelodysplastic syndrome; Runt, RUNX1 homology domain; RunxI, RUNX1 inhibition domain (aka transactivation domain). This schematic was created using cBioPortal (https://www.cbioportal.org/) (68, 69).
Most germline RUNX1-related malignancies are myeloid malignancies, in particular MDS, CMML, and AML that account for 37.3% (n=170) of patients (Figure 3B). Given its rarity in a sporadic population, T-ALL (4.4%, n=20) is overrepresented in these patients and has also been established as part of the phenotype (129, 179). Few cases diagnosed with B-ALL (137, 163, 167), non-Hodgkin lymphoma (37, 180), MPN (35), chronic lymphatic leukemia (181) or eosinophilic leukemia (155) have been identified but a causative relationship has not been established (Figure 3B). The median age at diagnosis is 42 years for MDS/AML and 22 years for T-ALL (Figure 3B).
Concomitant somatic variants frequently occur on the other RUNX1 allele (26% of patients) as bi-allelic inactivation and as discussed earlier, increase the pre-test probability of a germline allele (Figure 3B, Table 2). Other frequent co-occurring somatic variants were detected in TET2 (15%), FLT3 as ITD or tyrosine kinase domain variant (12%), SRSF2 (12%), ASXL1 (11%), WT1 (8%), DNMT3A (8%), BCOR (8%) and BCORL1 (8%) (Figure 3B, Table 2). Early-onset CHIP has been previously described in RUNX1 variant carriers and preleukemic individuals were found to carry variants in DNMT3A, TET2, KMT2A, KRAS and U2AF1, consistent with CHIP (37, 139, 153, 174, 182, 183). Recurrent CDC25C variants were reported in ~50% of RUNX1-mutated patients and hierarchical architecture analysis showed that these variants represent an early event during transformation, defining a pre-leukemic clone (140). Other studies have frequently detected variants in TET2, BCOR, PHF6, CDC25C, SRSF2, and GATA2 (13, 37, 184). In one study, BCOR variants were particularly common with up to four different variants per patients, however, presence, number or VAF did not correlate with clonal evolution or disease progression (185). Upon cytogenetic analyses, a normal karyotype was most frequently detected, followed by monosomy 7/del(7q), translocations with no apparent recurrent breakpoints/translocation partners and trisomy 8 (Table 2). Only few patients were investigated cytogenetically prior to the development of a hematologic malignancy.
Conclusion
Germline variants predisposing to inherited hematologic malignancies are detected more and more frequently due to increased recognition and detection using NGS-based diagnostic/prognostic panels. A multi-tier approach of several different criteria can be utilized to identify patients with a probable germline variant: (1) germline VAF, (2) bi-allelic inactivation, (3) longitudinal persistence of a pathogenic/likely pathogenic variant at germline VAF when remission is achieved, (4) variants that only occur in germline, and — in the case of MBD4 variants — (5) significantly higher mutational burden. The clone size in relationship to the blast count, the frequency of somatic variants in the gene in question, and whether the variant has been previously reported as somatic variant and/or is consistent with the mechanism of disease may also be considered (66).
Germline variants in DDX41, ETV6, GATA2, and RUNX1 have been well-described and may be uncovered upon initial diagnostic/prognostic work-up and subsequently confirmed in appropriate germline material. Importantly, the spectrum of germline predisposition alleles has been expanded over the past few years with the advent of whole-exome or whole-genome sequencing or NGS panel-based sequencing covering non-coding areas and functional testing of variants of unknown significance (VUS). CNVs are frequently identified as causative alleles in patients with RUNX1- and GATA2-deficiency at a frequency of 15.4% and 3.2% respectively, and few families with intragenic deletions of DDX41 and ETV6 have been reported as well (67, 84, 89). Germline translocations are infrequent but occur in genes that are otherwise known as common translocation partners in the leukemic clone, namely ETV6 (86) and RUNX1 (176, 178) and disrupt the promotor region. Other variants such as noncanonical splice variants and the recurrent variants in the deep-intronic enhancer element of GATA2 may be not detected or easily overlooked and require functional testing to establish pathogenicity. When suspicion is high based on family history and/or phenotypic criteria, upfront negative testing should be critically questioned, and further testing or re-analysis of the sequencing data considered.
Specific molecular or cytogenetic drivers have been identified and associated with distinct germline syndromes: The development of myeloid disease in DDX41 germline mutated patients most often requires bi-allelic inactivation of the DDX41 trans allele through nonsynonymous substitutions primarily within the helicase domain. Patients are more prone to have a normal karyotype and other recurrent, WHO-subgroup defining aberrations are exceedingly rare. Germline variants in ETV6, most often associated with B-ALL, have a normal or hyperdiploid karyotype that is consistent with this type of malignancy. Specific drivers of disease are not apparent but the number of patients with reported molecular/cytogenetic data is small. GATA2-deficiency displays a significant correlation with ASXL1-, SETBP1- and STAG2 variants, monosomy 7/del(7q), trisomy 8, and der(1;7)(q10;p10). The der(1;7)(q10;p10) is a specific aberration that only occurs in 0.4% of children and adolescents with MDS and wild-type GATA2 (109) and may be used as an indicator of a germline GATA2 variant. Lastly, a second somatic hit of the other allele is also common in patients with germline RUNX1 variants and somatic variants in BCOR, BCORL1 and CDC25C may be more prevalent in these patients. CHIP affecting genes such as DNMT3A, TET2, KMT2A, KRAS and U2AF1 and recurrent variants in CDC25C may represent early markers of clonal evolution and disease progression in carriers of a RUNX1 germline variant. Generally, early- versus late somatic alterations in the process of leukemic transformation are not well-studied and patients without obvious hematologic malignancy often do not receive a baseline bone marrow biopsy. Longitudinal pre-leukemic data are needed to identify early markers of clonal evolution and disease progression.
An emergent discussion is whether co-occurring somatic alterations can be used to predict the presence of a germline syndrome and help determining the pathogenicity of the variant when it is formally classified as a VUS. The above-mentioned markers may be applied in a multi-tier approach in addition to the VAF, bi-allelic inactivation, longitudinal persistence of variants and other factors to assess the likelihood of a germline variant in DDX41, ETV6, GATA2 and RUNX1. Most somatic alterations are not specific enough to be employed within the framework of American College of Medical Genetics and Genomics/Association for Molecular Pathology rules for germline variant interpretation (39). As an exception, bi-allelic DDX41 inactivations with nonsynonymous substitutions affecting hotspot amino acids or the presence of a der(1;7)(q10;p10) in patients with suspected GATA2-deficiency might confer enough significance, however, this would possibly only apply in combination with other phenotypic features.
In summary, this review provides new insights into the identification of germline syndromes by means of diagnostic/prognostic NGS data as well as specificity and pattern of somatic drivers in patients with germline DDX41, ETV6, GATA2, and RUNX1 variants based on large patient cohorts. These data will help to predict the clinical course of disease and thereby improve and individualize the clinical management for these patients.
Author contributions
SF designed and coordinated the study. JZ and DT provided conceptional input. JZ, DT and SF collected, analyzed and interpreted the data. JZ visualized the data. SF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
SF receives funding from the Olympia Morata Program of the Medical Faculty Heidelberg.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1205855/full#supplementary-material
References
1. Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet (1999) 23(2):166–75. doi: 10.1038/13793
2. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood (2016) 127(20):2391–405. doi: 10.1182/blood-2016-03-643544
3. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia Springer Nat (2022) 36:1703–19. doi: 10.1038/s41375-022-01613-1
4. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood (2022) 140(11):1200–28. doi: 10.1182/blood.2022015850
5. Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 ELN recommendations from an international expert panel. Blood (2022) 129(4):424–47. doi: 10.1182/blood.2022016867
6. Greenberg PL, Stone RM, Al-Kali A, Barta SK, Bejar R, Bennett JM, et al. Myelodysplastic syndromes, version 2.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw (2017) 15(1):60–87. doi: 10.6004/jnccn.2017.0007
7. Feurstein S, Drazer M, Godley LA. Germline predisposition to hematopoietic Malignancies. Hum Mol Genet (2021) 30(R2):R225–35. doi: 10.1093/hmg/ddab141
8. Xia J, Miller CA, Baty J, Ramesh A, Jotte MRM, Fulton RS, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia. Blood (2018) 131(4):408–16. doi: 10.1182/blood-2017-08-801985
9. Pastor VB, Sahoo SS, Boklan J, Schwabe GC, Saribeyoglu E, Strahm B, et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica (2018) 103(3):427–37. doi: 10.3324/haematol.2017.180778
10. Sahoo SS, Kozyra EJ, Wlodarski MW. Germline predisposition in myeloid neoplasms: Unique genetic and clinical features of GATA2 deficiency and SAMD9/SAMD9L syndromes. Best Pract Res Clin Haematol (2020) 33(3):101197. doi: 10.1016/j.beha.2020.101197
11. Homan CC, Venugopal P, Arts P, Shahrin NH, Feurstein S, Rawlings L, et al. GATA2 deficiency syndrome: A decade of discovery. Hum Mutat (2021) 42(11):1399–421. doi: 10.1002/humu.24271
12. Makishima H, Saiki R, Nannya Y, Korotev S, Gurnari C, Takeda J, et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasms. Blood (2023) 141(5):534–49. doi: 10.1182/blood.2022018221
13. Brown AL, Hahn CN, Scott HS. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA). Blood (2020) 136(1):24–35. doi: 10.1182/blood.2019000937
14. Tawana K, Wang J, Renneville A, Bodor C, Hills R, Loveday C, et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood (2015) 126(10):1214–23. doi: 10.1182/blood-2015-05-647172
15. Simon L, Spinella JF, Yao CY, Lavallée VP, Boivin I, Boucher G, et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood (2020) 135(21):1882–6. doi: 10.1182/blood.2019003357
16. Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Arango Ossa JE, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evidence (2022) 1(7):1–14. doi: 10.1056/EVIDoa2200008
17. Nazha A, Komrokji R, Meggendorfer M, Jia X, Radakovich N, Shreve J, et al. Personalized prediction model to risk stratify patients with myelodysplastic syndromes. J Clin Oncol (2021) 39(33):3737–46. doi: 10.1200/JCO.20.02810
18. Bersanelli M, Travaglino E, Meggendorfer M, Matteuzzi T, Sala C, Mosca E, et al. Classification and personalized prognostic assessment on the basis of clinical and genomic features in myelodysplastic syndromes. J Clin Oncol (2021) 39(11):1223–33. doi: 10.1200/JCO.20.01659
19. Jiang L, Ye L, Ma L, Ren Y, Zhou X, Mei C, et al. Predictive values of mutational variant allele frequency in overall survival and leukemic progression of myelodysplastic syndromes. J Cancer Res Clin Oncol (2022) 148(4):845–56. doi: 10.1007/s00432-021-03905-y
20. Churpek JE, Godley LA. How I diagnose and manage individuals at risk for inherited myeloid Malignancies. Blood (2016) 128(14):1800–13. doi: 10.1182/blood-2016-05-670240
21. Drazer MW, Kadri S, Sukhanova M, Patil SA, West AH, Feurstein S, et al. Prognostic tumor sequencing panels frequently identify germ line variants associated with hereditary hematopoietic Malignancies. Blood Adv (2018) 2(2):146–50. doi: 10.1182/bloodadvances.2017013037
22. DiNardo CD, Bannon SA, Routbort M, Franklin A, Mork M, Armanios M, et al. Evaluation of patients and families with concern for predispositions to hematologic Malignancies within the hereditary hematologic Malignancy clinic (HHMC). Clin Lymphoma Myeloma Leuk (2016) 16(7):417–28. doi: 10.1016/j.clml.2016.04.001
23. DiNardo CD, Routbort MJ, Bannon SA, Benton CB, Takahashi K, Kornblau SM, et al. Improving the detection of patients with inherited predispositions to hematologic Malignancies using next-generation sequencing-based leukemia prognostication panels. Cancer (2018) 124(13):2704–13. doi: 10.1002/cncr.31331
24. Yannakou CK, Jones K, Ryland GL, Thompson ER, Reid G, McBean M, et al. Incidental detection of germline variants of potential clinical significance by massively parallel sequencing in haematological Malignancies. J Clin Pathol (2018) 71(1):84–7. doi: 10.1136/jclinpath-2017-204481
25. Bannon SA, Routbort MJ, Montalban-Bravo G, Mehta RS, Jelloul FZ, Takahashi K, et al. Next-generation sequencing of DDX41 in myeloid neoplasms leads to increased detection of germline alterations. Front Oncol (2021) 10:582213/full(January). doi: 10.3389/fonc.2020.582213/full
26. Aguilera-Diaz A, Larrayoz MJ, Palomino-Echeverría S, Vazquez I, Ariceta B, Mañú A, et al. Strategy for identification of a potential inherited leukemia predisposition in a 299 patient’s cohort with tumor-only sequencing data. Leuk Res (2020) 95(April):106386. doi: 10.1016/j.leukres.2020.106386
27. Pabst T, Eyholzer M, Haefliger S, Schardt J, Mueller BU. Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol (2008) 26(31):5088–93. doi: 10.1200/JCO.2008.16.5563
28. Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CAJ, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood (2011) 117(8):2469–75. doi: 10.1182/blood-2010-09-307280
29. Feurstein S, Zhang L, DiNardo CD. Accurate germline RUNX1 variant interpretation and its clinical significance. Blood Adv (2020) 4(24):6199–203. doi: 10.1182/bloodadvances.2020003304
30. Fenwarth Laurène, Duployez N, Marceau-Renaut A, Chahla WA, Ducassou Stéphane, Gandemer V, et al. Germline pathogenic variants in transcription factors predisposing to pediatric acute myeloid leukemia: results from the French ELAM02 trial. Haematologica (2020) 106(3):908–12. doi: 10.3324/haematol.2020.248872
31. Ernst MPT, Kavelaars FG, Löwenberg B, Valk PJM, Raaijmakers MHGP. RUNX1 germline variants in RUNX1-mutant AML: how frequent? Blood (2021) 137(10):1428–31. doi: 10.1182/blood.2020008478
32. Kraft IL, Godley LA. Identifying potential germline variants from sequencing hematopoietic Malignancies. Hematology (2020) 2020(1):219–27. doi: 10.1182/hematology.2020006910
33. Feurstein S, Adegunsoye A, Mojsilovic D, Vij R, West DePersia AH, Rajagopal PS, et al. Telomere biology disorder prevalence and phenotypes in adults with familial hematologic and/or pulmonary presentations. Blood Adv (2020) 4(19):4873–86. doi: 10.1182/bloodadvances.2020001721
34. Feurstein S, Churpek JE, Walsh T, Keel S, Hakkarainen M, Schroeder T, et al. Germline variants drive myelodysplastic syndrome in young adults. Leukemia (2021) 35(8):2439–44. doi: 10.1038/s41375-021-01137-0
35. Sanders MA, Chew E, Flensburg C, Zeilemaker A, Miller SE, Al Hinai AS, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood (2018) 132(14):1526–34. doi: 10.1182/blood-2018-05-852566
36. Palles C, West HD, Chew E, Galavotti S, Flensburg C, Grolleman JE, et al. Germline MBD4 deficiency causes a multi-tumor predisposition syndrome. Am J Hum Genet (2022) 109(5):953–60. doi: 10.1016/j.ajhg.2022.03.018
37. Brown AL, Arts P, Carmichael CL, Babic M, Dobbins J, Chong CE, et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv (2020) 4(6):1131–44. doi: 10.1182/bloodadvances.2019000901
38. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagnostics (2017) 19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002
39. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17(5):405–23. doi: 10.1038/gim.2015.30
40. Jiang Y, Zhu Y, Liu ZJ, Ouyang S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell (2017) 8(2):83–9. doi: 10.1007/s13238-016-0303-4
41. Qin K, Jian D, Xue Y, Cheng Y, Zhang P, Wei Y, et al. DDX41 regulates the expression and alternative splicing of genes involved in tumorigenesis and immune response. Oncol Rep (2021) 45(3):1213–25. doi: 10.3892/or.2021.7951
42. Singh RS, Vidhyasagar V, Yang S, Arna AB, Yadav M, Aggarwal A, et al. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep (2022) 39(8):110856. doi: 10.1016/j.celrep.2022.110856
43. Sébert M, Passet M, Raimbault A, Rahmé R, Raffoux E, Sicre de Fontbrune F, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood (2019) 134(17):1441–4. doi: 10.1182/blood.2019000909
44. Quesada AE, Routbort MJ, DiNardo CD, Bueso-Ramos CE, Kanagal-Shamanna R, Khoury JD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol (2019) 94(7):ajh.25486. doi: 10.1002/ajh.25486
45. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell (2018) 173(2):321–37. doi: 10.1016/j.cell.2018.03.035
46. Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell (2018) 173(2):305–320.e10. doi: 10.1016/j.cell.2018.03.033
47. The Cancer Genome Atlas Research Network (TCGA). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Engl J Med (2013) 368(22):2059–74. doi: 10.1056/NEJMoa1301689
48. Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell (2015) 27(5):658–70. doi: 10.1016/j.ccell.2015.03.017
49. Lewinsohn M, Brown AL, Weinel LM, Phung C, Rafidi G, Lee MK, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid Malignancies. Blood (2016) 127(8):1017–23. doi: 10.1182/blood-2015-10-676098
50. Cardoso SR, Ryan G, Walne AJ, Ellison A, Lowe R, Tummala H, et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia (2016) 30(10):2083–6. doi: 10.1038/leu.2016.124
51. Vairo F, Ferrer A, Cathcart-Rake E, King RL, Howard MT, Viswanatha DS, et al. Novel germline missense DDX41 variant in a patient with an adult-onset myeloid neoplasm with excess blasts without dysplasia. Leuk Lymphoma (2019) 60(5):1337–9. doi: 10.1080/10428194.2018.1522443
52. Weinberg OK, Kuo F, Calvo KR. Germline predisposition to hematolymphoid neoplasia. Am J Clin Pathol (2019) 152(3):258–76. doi: 10.1093/ajcp/aqz067
53. Abou Dalle I, Kantarjian H, Bannon SA, Kanagal-Shamanna R, Routbort M, Patel KP, et al. Successful lenalidomide treatment in high risk myelodysplastic syndrome with germline DDX41 mutation. Am J Hematol (2020) 95(2):227–9. doi: 10.1002/ajh.25610
54. Polprasert C, Takeda J, Niparuck P, Rattanathammethee T, Pirunsarn A, Suksusut A, et al. Novel DDX41 variants in Thai patients with myeloid neoplasms. Int J Hematol (2020) 111(2):241–6. doi: 10.1007/s12185-019-02770-3
55. Qu S, Li B, Qin T, Xu Z, Pan L, Hu N, et al. Molecular and clinical features of myeloid neoplasms with somatic DDX41 mutations. Br J Haematol (2021) 192(6):1006–10. doi: 10.1111/bjh.16668
56. Yasuda T, Sanada M, Nishijima D, Kanamori T, Iijima Y, Hattori H, et al. Clinical utility of target capture-based panel sequencing in hematological Malignancies: A multicenter feasibility study. Cancer Sci (2020) 111(9):3367–78. doi: 10.1111/cas.14552
57. Kim B, Yun W, Lee ST, Choi JR, Yoo KH, Koo HH, et al. Prevalence and clinical implications of germline predisposition gene mutations in patients with acute myeloid leukemia. Sci Rep (2020) 10(1):14297. doi: 10.1038/s41598-020-71386-z
58. Andrés-Zayas C, Suárez-González J, Rodríguez-Macías G, Dorado N, Osorio S, Font P, et al. Clinical utility of targeted next-generation sequencing for the diagnosis of myeloid neoplasms with germline predisposition. Mol Oncol (2021) 15(9):2273–84. doi: 10.1002/1878-0261.12921
59. Choi EJ, Cho YU, Hur EH, Jang S, Kim N, Park HS, et al. Unique ethnic features of DDX41 mutations in patients with idiopathic cytopenia of undetermined significance, myelodysplastic syndrome, or acute myeloid leukemia. Haematologica (2021) 107(2):510–8. doi: 10.3324/haematol.2020.270553
60. Fazio F, Quintini M, Carmosino I, Matteucci C, Miulli E, Pellanera F, et al. New dead/H-box helicase gene (ddx41) mutation in an Italian family with recurrent leukemia. Leuk Lymphoma (2021) 62(9):2280–3. doi: 10.1080/10428194.2021.1910689
61. Singhal D, Hahn CN, Feurstein S, Wee LYA, Moma L, Kutyna MM, et al. Targeted gene panels identify a high frequency of pathogenic germline variants in patients diagnosed with a hematological Malignancy and at least one other independent cancer. Leukemia (2021) 35(11):3245–56. doi: 10.1038/s41375-021-01246-w
62. Goyal T, Tu ZJ, Wang Z, Cook JR. Clinical and pathologic spectrum of DDX41-mutated hematolymphoid neoplasms. Am J Clin Pathol (2021) 156(5):829–38. doi: 10.1093/ajcp/aqab027
63. Gibson CJ, Kim HT, Zhao L, Murdock HM, Hambley B, Ogata A, et al. Donor clonal hematopoiesis and recipient outcomes after transplantation. J Clin Oncol (2022) 40(2):189–201. doi: 10.1200/JCO.21.02286
64. Duployez N, Largeaud L, Duchmann M, Kim R, Rieunier J, Lambert J, et al. Prognostic impact of DDX41 germline mutations in intensively treated acute myeloid leukemia patients: an ALFA-FILO study. Blood (2022) 140(7):756–68. doi: 10.1182/blood.2021015328
65. Li P, Brown S, Williams M, White T, Xie W, Cui W, et al. The genetic landscape of germline DDX41 variants predisposing to myeloid neoplasms. Blood (2022) 140(7):716–55. doi: 10.1182/blood.2021015135
66. Feurstein S, Trottier AM, Estrada-Merly N, Pozsgai M, McNeely K, Drazer MW, et al. Germ line predisposition variants occur in myelodysplastic syndrome patients of all ages. Blood (2022) 140(24):2533–48. doi: 10.1182/blood.2022015790
67. Saygin C, Roloff G, Hahn CN, Chhetri R, Gill S, Elmariah H, et al. Allogeneic hematopoietic stem cell transplant outcomes in adults with inherited myeloid Malignancies. Blood Adv (2023) 7(4):549–54. doi: 10.1182/bloodadvances.2022008172
68. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
69. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal (2013) 6(269):pl1. doi: 10.1126/scisignal.2004088
70. Ma X, Does M, Raza A, Mayne ST. Myelodysplastic syndromes. Cancer (2007) 109(8):1536–42. doi: 10.1002/cncr.22570
71. Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol (2017) 106(2):163–74. doi: 10.1007/s12185-017-2260-y
72. Kadono M, Kanai A, Nagamachi A, Shinriki S, Kawata J, Iwato K, et al. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol (2016) 44(8):745–54. doi: 10.1016/j.exphem.2016.04.017
73. Tijssen MR, Ghevaert C. Transcription factors in late megakaryopoiesis and related platelet disorders. J Thromb Haemostasis (2013) 11(4):593–604. doi: 10.1111/jth.12131
74. Poggi M, Canault M, Favier M, Turro E, Saultier P, Ghalloussi D, et al. Germline variants in ETV6 underlie reduced platelet formation, platelet dysfunction and increased levels of circulating CD34+ progenitors. Haematologica (2016). 102(2):282–94. doi: 10.3324/haematol.2016.147694
75. Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological Malignancies. Int J Hematol (2017) 106(2):189–95. doi: 10.1007/s12185-017-2259-4
76. De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological Malignancies: A review. Leuk Res (2012) 36(8):945–61. doi: 10.1016/j.leukres.2012.04.010
77. Mullighan CG. The molecular genetic makeup of acute lymphoblastic leukemia. Hematol / Educ Program Am Soc Hematol Am Soc Hematol Educ Program (2012) 2012:389–96. doi: 10.1182/asheducation.V2012.1.389.3798360
78. Vicente C, Schwab C, Broux M, Geerdens E, Degryse S, Demeyer S, et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica (2015) 100(10):1301–10. doi: 10.3324/haematol.2015.130179
79. Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet (2017) 49(8):1211–8. doi: 10.1038/ng.3909
80. Zhang MY, Churpek JE, Keel SB, Walsh T, Lee MK, Loeb KR, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic Malignancy. Nat Genet 2015/01/13 (2015) 47(2):180–5. doi: 10.1038/ng.3177
81. Porter CC, Di Paola J, Pencheva B. ETV6 thrombocytopenia and predisposition to leukemia. In: Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al. editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle. (1993–2023).
82. Dirse V, Norvilas R, Gineikiene E, Matuzevičienė R, Griskevicius L, Preiksaitiene E. ETV6 and NOTCH1 germline variants in adult acute leukemia. Leuk Lymphoma (2018) 59(4):1022–4. doi: 10.1080/10428194.2017.1359742
83. Duployez N, Abou Chahla W, Lejeune S, Marceau-Renaut A, Letizia G, Boyer T, et al. Detection of a new heterozygous germline ETV6 mutation in a case with hyperdiploid acute lymphoblastic leukemia. Eur J Haematol (2018) 100(1):104–7. doi: 10.1111/ejh.12981
84. Paulsson K, Forestier E, Lilljebjörn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci (2010) 107(50):21719–24. doi: 10.1073/pnas.1006981107
85. Rampersaud E, Ziegler DS, Iacobucci I, Payne-Turner D, Churchman ML, Schrader KA, et al. Germline deletion of ETV6 in familial acute lymphoblastic leukemia. Blood Adv (2019) 3(7):1039–46. doi: 10.1182/bloodadvances.2018030635
86. Järviaho T, Bang B, Zachariadis V, Taylan F, Moilanen J, Möttönen M, et al. Predisposition to childhood acute lymphoblastic leukemia caused by a constitutional translocation disrupting ETV6. Blood Adv (2019) 3(18):2722–31. doi: 10.1182/bloodadvances.2018028795
87. Karastaneva A, Nebral K, Schlagenhauf A, Baschin M, Palankar R, Juch H, et al. Novel phenotypes observed in patients with ETV6-linked leukaemia/familial thrombocytopenia syndrome and a biallelic ARID5B risk allele as leukaemogenic cofactor. J Med Genet (2020) 57(6):427–33. doi: 10.1136/jmedgenet-2019-106339
88. Drazer MW, Homan CC, Cavalcante de Andrade Silva K, McNeely KE, Pozsgai MJ, et al. Clonal hematopoiesis in patients with ANKRD26 or ETV6 germline mutations. Blood Adv (2022) 6(15):4357–9. doi: 10.1182/bloodadvances.2022007211
89. Ovsyannikova G, Pavlova A, Deordieva E, Raykina E, Pshonkin A, Maschan A, et al. Single center experience with pediatric patients with GATA2 deficiency. Front Pediatr (2022) 10:801810/full. doi: 10.3389/fped.2022.801810/full
90. Bąk A, Skonieczka K, Jaśkowiec A, Junkiert-Czarnecka A, Heise M, Pilarska-Deltow M, et al. Germline mutations among Polish patients with acute myeloid leukemia. Hered Cancer Clin Pract (2021) 19(1):42. doi: 10.1186/s13053-021-00200-2
91. Kanamaru Y, Uchiyama T, Kaname T, Yanagi K, Ohara O, Kunishima S, et al. ETV6-related thrombocytopenia associated with a transient decrease in von Willebrand factor. Int J Hematol (2021) 114(2):297–300. doi: 10.1007/s12185-021-03136-4
92. de Smith AJ, Lavoie G, Walsh KM, Aujla S, Evans E, Hansen HM, et al. Predisposing germline mutations in high hyperdiploid acute lymphoblastic leukemia in children. Genes Chromosomes Cancer (2019) 58(10):723–30. doi: 10.1002/gcc.22765
93. Yoshino H, Nishiyama Y, Kamma H, Chiba T, Fujiwara M, Karaho T, et al. Functional characterization of a germline ETV6 variant associated with inherited thrombocytopenia, acute lymphoblastic leukemia, and salivary gland carcinoma in childhood. Int J Hematol (2020) 112(2):217–22. doi: 10.1007/s12185-020-02885-y
94. Melazzini F, Palombo F, Balduini A, De Rocco D, Marconi C, Noris P, et al. Clinical and pathogenic features of ETV6-related thrombocytopenia with predisposition to acute lymphoblastic leukemia. Haematologica (2016) 101(11):1333–42. doi: 10.3324/haematol.2016.147496
95. Topka S, Vijai J, Walsh MF, Jacobs L, Maria A, Villano D, et al. Germline ETV6 mutations confer susceptibility to acute lymphoblastic leukemia and thrombocytopenia. PloS Genet 2015/06/24 (2015) 11(6):e1005262. doi: 10.1371/journal.pgen.1005262
96. Leubolt G, Redondo Monte E, Greif PA. GATA2 mutations in myeloid Malignancies: Two zinc fingers in many pies. IUBMB Life (2020) 72(1):151–8. doi: 10.1002/iub.2204
97. Wlodarski MW, Hirabayashi S, Pastor V, Stary J, Hasle H, Masetti R, et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood (2016) 127(11):1387–97. doi: 10.1182/blood-2015-09-669937
98. Keel SB, Scott A, Sanchez-Bonilla M, Ho PA, Gulsuner S, Pritchard CC, et al. Genetic features of myelodysplastic syndrome and aplastic anemia in pediatric and young adult patients. Haematologica (2016) 101(11):1343–50. doi: 10.3324/haematol.2016.149476
99. Calvo KR, Hickstein DD. The spectrum of GATA2 deficiency syndrome. Blood (2023) 141(13):1524–32. doi: 10.1182/blood.2022017764
100. Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood (2014) 123(6):809–21. doi: 10.1182/blood-2013-07-515528
101. Oleaga-Quintas C, de Oliveira-Júnior EB, Rosain J, Rapaport F, Deswarte C, Guérin A, et al. Inherited GATA2 deficiency is dominant by haploinsufficiency and displays incomplete clinical penetrance. J Clin Immunol (2021) 41(3):639–57. doi: 10.1007/s10875-020-00930-3
102. Wlodarski MW, Collin M, Horwitz MS. GATA2 deficiency and related myeloid neoplasms. Semin Hematol (2017) 54(2):81–6. doi: 10.1053/j.seminhematol.2017.05.002
103. Nell E, Cornellissen H, Hodkinson K, Urban MF, Bassa FC, Fazel FB, et al. Infection and myelodysplasia: A case report of GATA2 deficiency in a South African patient. Clin Case Rep (2023) 11(3):e7075. doi: 10.1002/ccr3.7075
104. Largeaud L, Collin M, Monselet N, Vergez F, Fregona V, Larcher L, et al. Somatic genetic alterations predict haematological progression in GATA2 deficiency. Haematologica (2023). 108(6):1515–29. doi: 10.3324/haematol.2022.282250
105. Heropolitańska-Pliszka E, Piątosa B, Szmydki-Baran A, Kuczborska K, Miarka-Walczyk K, Pastorczak A, et al. Case report: Successful allogeneic stem cell transplantation in a child with novel GATA2 defect associated B-cell acute lymphoblastic leukemia. Front Immunol (2022) 13:928529. doi: 10.3389/fimmu.2022.928529
106. Basheer A, Padrao EMH, Huh K, Parker S, Shah T, Gerardi DA. Pulmonary Alveolar Proteinosis due to Familial Myelodysplastic Syndrome with resolution after stem cell transplant. Autopsy Case Rep (2022) 12:e2021382. doi: 10.4322/acr.2021.382
107. Jørgensen SF, Buechner J, Myhre AE, Galteland E, Spetalen S, Kulseth MA, et al. A nationwide study of GATA2 deficiency in Norway—the majority of patients have undergone allo-HSCT. J Clin Immunol (2022) 42(2):404–20. doi: 10.1007/s10875-021-01189-y
108. West RR, Calvo KR, Embree LJ, Wang W, Tuschong LM, Bauer TR, et al. ASXL1 and STAG2 are common mutations in GATA2 deficiency patients with bone marrow disease and myelodysplastic syndrome. Blood Adv (2022) 6(3):793–807. doi: 10.1182/bloodadvances.2021005065
109. Kozyra EJ, Göhring G, Hickstein DD, Calvo KR, DiNardo CD, Dworzak M, et al. Association of unbalanced translocation der(1;7) with germline GATA2 mutations. Blood (2021) 138(23):2441–5. doi: 10.1182/blood.2021012781
110. Mika T, Vangala D, Eckhardt M, La Rosée P, Lange C, Lehmberg K, et al. Case report: hemophagocytic lymphohistiocytosis and non-tuberculous mycobacteriosis caused by a novel GATA2 variant. Front Immunol (2021) 12:682934. doi: 10.3389/fimmu.2021.682934
111. Shen Y, Li Y, Li H, Liu Q, Dong H, Wang B, et al. Diagnosing monoMAC syndrome in GATA2 germline mutated myelodysplastic syndrome via next-generation sequencing in a patient with refractory and complex infection: case report and literature review. Infect Drug Resist (2021) Volume 14:1311–7. doi: 10.2147/IDR.S305825
112. Rütsche CV, Haralambieva E, Lysenko V, Balabanov S, Theocharides APA. A patient with a germline GATA2 mutation and primary myelofibrosis. Blood Adv (2021) 5(3):791–5. doi: 10.1182/bloodadvances.2020003401
113. Kazenwadel J, Secker GA, Liu YJ, Rosenfeld JA, Wildin RS, Cuellar-Rodriguez J, et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood (2012) 119(5):1283–91. doi: 10.1182/blood-2011-08-374363
114. Mir MA, Kochuparambil ST, Abraham RS, Rodriguez V, Howard M, Hsu AP, et al. Spectrum of myeloid neoplasms and immune deficiency associated with germline GATA2 mutations. Cancer Med (2015) 4(4):490–9. doi: 10.1002/cam4.384
115. Bluteau O, Sebert M, Leblanc T, Peffault de Latour R, Quentin S, Lainey E, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood (2018) 131(7):717–32. doi: 10.1182/blood-2017-09-806489
116. Donadieu J, Lamant M, Fieschi C, de Fontbrune FS, Caye A, Ouachee M, et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica (2018) 103(8):1278–87. doi: 10.3324/haematol.2017.181909
117. Abou Dalle I, Bannon SA, Patel KP, Routbort MJ, Cortes JE, Ferrajoli A, et al. Germline genetic predisposition to myeloid neoplasia from GATA2 gene mutations: lessons learned from two cases. JCO Precis Oncol (2019) 3):1–5. doi: 10.1200/PO.18.00301
118. Monif M, Huq A, Chee L, Kilpatrick T. MonoMac syndrome with associated neurological deficits and longitudinally extensive cord lesion. BMJ Case Rep (2018) 2018:bcr–2017-222872. doi: 10.1136/bcr-2017-222872
119. Bogaert DJ, Laureys G, Naesens L, Mazure D, De Bruyne M, Hsu AP, et al. GATA2 deficiency and haematopoietic stem cell transplantation: challenges for the clinical practitioner. Br J Haematol (2020) 188(5):768–73. doi: 10.1111/bjh.16247
120. Hsu AP, Johnson KD, Falcone EL, Sanalkumar R, Sanchez L, Hickstein DD, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood (2013) 121(19):3830–7. doi: 10.1182/blood-2012-08-452763
121. Soukup AA, Zheng Y, Mehta C, Wu J, Liu P, Cao M, et al. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J Clin Invest (2019) 129(3):1180–92. doi: 10.1172/JCI122694
122. Koegel AK, Hofmann I, Moffitt K, Degar B, Duncan C, Tubman VN. Acute lymphoblastic leukemia in a patient with MonoMAC syndrome/GATA2 haploinsufficiency. Pediatr Blood Cancer (2016) 63(10):1844–7. doi: 10.1002/pbc.26084
123. Esparza O, Xavier AC, Atkinson TP, Hill BC, Whelan K. A unique phenotype of T-cell acute lymphoblastic leukemia in a patient with GATA2 haploinsufficiency. Pediatr Blood Cancer (2019) 66(6):e27649. doi: 10.1002/pbc.27649
124. Hofmann I, Avagyan S, Stetson A, Guo D, Al-Sayegh H, London WB, et al. Comparison of outcomes of myeloablative allogeneic stem cell transplantation for pediatric patients with bone marrow failure, myelodysplastic syndrome and acute myeloid leukemia with and without germline GATA2 mutations. Biol Blood Marrow Transplantation (2020) 26(6):1124–30. doi: 10.1016/j.bbmt.2020.02.015
125. Lam K. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Bioscience (2012) 17(1):1120. doi: 10.2741/3977
126. Navarro-Montero O, Ayllon V, Lamolda M, López-Onieva L, Montes R, Bueno C, et al. RUNX1c regulates hematopoietic differentiation of human pluripotent stem cells possibly in cooperation with proinflammatory signaling. Stem Cells (2017) 35(11):2253–66. doi: 10.1002/stem.2700
127. Tsuzuki S, Seto M. Expansion of functionally defined mouse hematopoietic stem and progenitor cells by a short isoform of RUNX1/AML1. Blood (2012) 119(3):727–35. doi: 10.1182/blood-2011-06-362277
128. Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol (2010) 38(5):403–16. doi: 10.1016/j.exphem.2010.02.011
129. Luo X, Feurstein S, Mohan S, Porter CC, Jackson SA, Keel S, et al. ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv (2019) 3(20):2962–79. doi: 10.1182/bloodadvances.2019000644
130. Feurstein S, Drazer MW, Godley LA. Genetic predisposition to leukemia and other hematologic Malignancies. Semin Oncol (2016) 43(5):598–608. doi: 10.1053/j.seminoncol.2016.10.003
131. Homan CC, Scott HS, Brown AL. Hereditary platelet disorders associated with germ line variants in RUNX1, ETV6, and ANKRD26. Blood (2023) 141(13):1533–43. doi: 10.1182/blood.2022017735
132. Kellaway SG, Coleman DJL, Cockerill PN, Raghavan M, Bonifer C. Molecular basis of hematological disease caused by inherited or acquired RUNX1 mutations. Exp Hematol (2022) 111:1–12. doi: 10.1016/j.exphem.2022.03.009
133. Appelmann I, Linden T, Rudat A, Mueller-Tidow C, Berdel WE, Mesters RM. Hereditary thrombocytopenia and acute myeloid leukemia: a common link due to a germline mutation in the AML1 gene. Ann Hematol (2009) 88(10):1037–8. doi: 10.1007/s00277-009-0722-x
134. Ripperger T, Steinemann D, Göhring G, Finke J, Niemeyer CM, Strahm B, et al. A novel pedigree with heterozygous germline RUNX1 mutation causing familial MDS-related AML: can these families serve as a multistep model for leukemic transformation? Leukemia (2009) 23(7):1364–6. doi: 10.1038/leu.2009.87
135. Jongmans MCJ, Kuiper RP, Carmichael CL, Wilkins EJ, Dors N, Carmagnac A, et al. Novel RUNX1 mutations in familial platelet disorder with enhanced risk for acute myeloid leukemia: clues for improved identification of the FPD/AML syndrome. Leukemia (2010) 24(1):242–6. doi: 10.1038/leu.2009.210
136. Nishimoto N, Imai Y, Ueda K, Nakagawa M, Shinohara A, Ichikawa M, et al. T cell acute lymphoblastic leukemia arising from familial platelet disorder. Int J Hematol (2010) 92(1):194–7. doi: 10.1007/s12185-010-0612-y
137. Linden T, Schnittger S, Groll AH, Juergens H, Rossig C. Childhood B-cell precursor acute lymphoblastic leukaemia in a patient with familial thrombocytopenia and RUNX1 mutation. Br J Haematol (2010) 151(5):528–30. doi: 10.1111/j.1365-2141.2010.08370.x
138. Latger-Cannard V, Philippe C, Jonveaux P, Lecompte T, Favier R. Dysmegakaryopoiesis, a clue for an early diagnosis of familial platelet disorder with propensity to acute myeloid leukemia in case of unexplained inherited thrombocytopenia associated with normal-sized platelets. J Pediatr Hematol Oncol (2011) 33(7):e264–6. doi: 10.1097/MPH.0b013e31821754ac
139. Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood (2015) 126(22):2484–90. doi: 10.1182/blood-2015-04-641100
140. Yoshimi A, Toya T, Kawazu M, Ueno T, Tsukamoto A, Iizuka H, et al. Recurrent CDC25C mutations drive Malignant transformation in FPD/AML. Nat Commun (2014) 5(1):4770. doi: 10.1038/ncomms5770
141. Ok CY, Leventaki V, Wang SA, Dinardo C, Medeiros LJ, Konoplev S. Detection of an abnormal myeloid clone by flow cytometry in familial platelet disorder with propensity to myeloid Malignancy. Am J Clin Pathol (2016) 145(2):271–6. doi: 10.1093/ajcp/aqv080
142. Ouchi-Uchiyama M, Sasahara Y, Kikuchi A, Goi K, Nakane T, Ikeno M, et al. Analyses of genetic and clinical parameters for screening patients with inherited thrombocytopenia with small or normal-sized platelets. Pediatr Blood Cancer (2015) 62(12):2082–8. doi: 10.1002/pbc.25668
143. Schmit JM, Turner DJ, Hromas RA, Wingard JR, Brown RA, Li Y, et al. Two novel RUNX1 mutations in a patient with congenital thrombocytopenia that evolved into a high grade myelodysplastic syndrome. Leuk Res Rep (2015) 4(1):24–7. doi: 10.1016/j.lrr.2015.03.002
144. Fixter K, Rabbolini DJ, Valecha B, Morel-Kopp MC, Gabrielli S, Chen Q, et al. Mean platelet diameter measurements to classify inherited thrombocytopenias. Int J Lab Hematol (2018) 40(2):187–95. doi: 10.1111/ijlh.12763
145. Chisholm KM, Denton C, Keel S, Geddis AE, Xu M, Appel BE, et al. Bone marrow morphology associated with germline RUNX1 mutations in patients with familial platelet disorder with associated myeloid Malignancy. Pediatr Dev Pathol (2019) 22(4):315–28. doi: 10.1177/1093526618822108
146. Penkert J, Schmidt G, Hofmann W, Schubert S, Schieck M, Auber B, et al. Breast cancer patients suggestive of Li-Fraumeni syndrome: mutational spectrum, candidate genes, and unexplained heredity. Breast Cancer Res (2018) 20(1):87. doi: 10.1186/s13058-018-1011-1
147. Tawana K, Wang J, Király PA, Kállay K, Benyó G, Zombori M, et al. Recurrent somatic JAK-STAT pathway variants within a RUNX1-mutated pedigree. Eur J Hum Genet (2017) 25(8):1020–4. doi: 10.1038/ejhg.2017.80
148. Yoshimi A, Trippett TM, Zhang N, Chen X, Penson AV, Arcila ME, et al. Genetic basis for iMCD-TAFRO. Oncogene (2020) 39(15):3218–25. doi: 10.1038/s41388-020-1204-9
149. DiFilippo EC, Coltro G, Carr RM, Mangaonkar AA, Binder M, Khan SP, et al. Spectrum of abnormalities and clonal transformation in germline RUNX1 familial platelet disorder and a genomic comparative analysis with somatic RUNX1 mutations in MDS/MPN overlap neoplasms. Leukemia (2020) 34(9):2519–24. doi: 10.1038/s41375-020-0752-x
150. De Rocco D, Melazzini F, Marconi C, Pecci A, Bottega R, Gnan C, et al. Mutations of RUNX1 in families with inherited thrombocytopenia. Am J Hematol (2017) 92(6):E86–8. doi: 10.1002/ajh.24703
151. Ng IK, Lee J, Ng C, Kosmo B, Chiu L, Seah E, et al. Preleukemic and second-hit mutational events in an acute myeloid leukemia patient with a novel germline RUNX1 mutation. biomark Res (2018) 6(1):16. doi: 10.1186/s40364-018-0130-2
152. Staňo Kozubík K, Radová L, Pešová M, Réblová K, Trizuljak J, Plevová K, et al. C-terminal RUNX1 mutation in familial platelet disorder with predisposition to myeloid Malignancies. Int J Hematol (2018) 108(6):652–7. doi: 10.1007/s12185-018-2514-3
153. Duarte BKL, Yamaguti-Hayakawa GG, Medina SS, Siqueira LH, Snetsinger B, Costa FF, et al. Longitudinal sequencing of RUNX1 familial platelet disorder: new insights into genetic mechanisms of transformation to myeloid Malignancies. Br J Haematol (2019) 186(5):724–34. doi: 10.1111/bjh.15990
154. Vormittag-Nocito E, Ni H, Schmidt ML, Lindgren V. Thrombocytopenia and predisposition to acute myeloid leukemia due to mosaic ring 21 with loss of RUNX1: cytogenetic and molecular characterization. Mol Syndromol (2018) 9(6):306–11. doi: 10.1159/000494645
155. Tang C, Rabbolini DJ, Morel-Kopp M, Connor DE, Crispin P, Ward CM, et al. The clinical heterogeneity of RUNX1 associated familial platelet disorder with predisposition to myeloid Malignancy – A case series and review of the literature. Res Pract Thromb Haemost (2020) 4(1):106–10. doi: 10.1002/rth2.12282
156. Duployez N, Martin JE, Khalife-Hachem S, Benkhelil R, Saada V, Marzac C, et al. Germline RUNX1 intragenic deletion: implications for accurate diagnosis of FPD/AML. Hemasphere (2019) 3(3):e203. doi: 10.1097/HS9.0000000000000203
157. Abdelmoumen K, Fabre M, Ducastelle-Lepretre S, Favier R, Ballerini P, Bordet JC, et al. Eltrombopag for the treatment of severe inherited thrombocytopenia. Acta Haematol (2021) 144(3):308–13. doi: 10.1159/000509922
158. Bagla S, Regling KA, Wakeling EN, Gadgeel M, Buck S, Zaidi AU, et al. Distinctive phenotypes in two children with novel germline RUNX1 mutations - one with myeloid Malignancy and increased fetal hemoglobin. Pediatr Hematol Oncol (2021) 38(1):65–79. doi: 10.1080/08880018.2020.1814463
159. Gartstein E, Orr K, Eames G, Firan M, Howrey R, Ray A. A unique presentation of T-lymphoblastic lymphoma in a pediatric patient with a germline RUNX1 mutation. Pediatr Blood Cancer (2023) 70(5):e30184. doi: 10.1002/pbc.30184
160. Palma-Barqueros V, Bastida JM, López Andreo MJ, Zámora-Cánovas A, Zaninetti C, Ruiz-Pividal JF, et al. Platelet transcriptome analysis in patients with germline RUNX1 mutations. J Thromb Haemost (2023). 21(5):1352–65. doi: 10.1016/j.jtha.2023.01.023
161. Yamamori A, Hamada M, Muramatsu H, Wakamatsu M, Hama A, Narita A, et al. Germline and somatic RUNX1 variants in a pediatric bone marrow failure cohort. Am J Hematol (2023) 98(5):E102–5. doi: 10.1002/ajh.26874
162. Marconi C, Pecci A, Palombo F, Melazzini F, Bottega R, Nardi E, et al. Exome sequencing in 116 patients with inherited thrombocytopenia that remained of unknown origin after systematic phenotype-driven diagnostic workup. Haematologica (2022) 108(7):1909–19. doi: 10.3324/haematol.2022.280993
163. Fournier E, Debord C, Soenen V, Trillot N, Gonzales F, Tintiller V, et al. Baseline dysmegakaryopoiesis in inherited thrombocytopenia/platelet disorder with predisposition to haematological Malignancies. Br J Haematol (2020) 189(4):e119–22. doi: 10.1111/bjh.16543
164. Matsumura T, Nakamura-Ishizu A, Muddineni SSNA, Tan DQ, Wang CQ, Tokunaga K, et al. Hematopoietic stem cells acquire survival advantage by loss of RUNX1 methylation identified in familial leukemia. Blood (2020) 136(17):1919–32. doi: 10.1182/blood.2019004292
165. Lachowiez C, Bannon S, Loghavi S, Wang F, Kanagal-Shamanna R, Mehta R, et al. Clonal evolution and treatment outcomes in hematopoietic neoplasms arising in patients with germline RUNX1 mutations. Am J Hematol (2020) 95(11):E313–5. doi: 10.1002/ajh.25965
166. Almazni I, Chudakou P, Dawson-Meadows A, Downes K, Freson K, Mason J, et al. A novel RUNX1 exon 3 - 7 deletion causing a familial platelet disorder. Platelets (2022) 33(2):320–3. doi: 10.1080/09537104.2021.1887470
167. Six KA, Gerdemann U, Brown AL, Place AE, Cantor AB, Kutny MA, et al. B-cell acute lymphoblastic leukemia in patients with germline RUNX1 mutations. Blood Adv (2021) 5(16):3199–202. doi: 10.1182/bloodadvances.2021004653
168. St. Martin EC, Ferrer A, Wudhikarn K, Mangaonkar A, Hogan W, Tefferi A, et al. Clinical features and survival outcomes in patients with chronic myelomonocytic leukemia arising in the context of germline predisposition syndromes. Am J Hematol (2021) 96(9):1–716. doi: 10.1002/ajh.26250
169. Ovsyannikova GS, Fedorova DV, Tesakov IP, Martyanov AA, Ignatova AA, Ponomarenko EA, et al. Platelet functional abnormalities and clinical presentation in pediatric patients with germline RUNX1, ANKRD26, and ETV6 mutations. Haematologica (2022) 107(10):2511–6. doi: 10.3324/haematol.2022.281340
170. Brunet J, Badin M, Chong M, Iyer J, Tasneem S, Graf L, et al. Bleeding risks for uncharacterized platelet function disorders. Res Pract Thromb Haemost (2020) 4(5):799–806. doi: 10.1002/rth2.12374
171. Engvall M, Karlsson Y, Kuchinskaya E, Jörnegren Å, Mathot L, Pandzic T, et al. Familial platelet disorder due to germline exonic deletions in RUNX1: a diagnostic challenge with distinct alterations of the transcript isoform equilibrium. Leuk Lymphoma (2022) 63(10):2311–20. doi: 10.1080/10428194.2022.2067997
172. Cavalcante de Andrade Silva M, Krepischi ACV, Kulikowski LD, Zanardo EA, Nardinelli L, Leal AM, et al. Deletion of RUNX1 exons 1 and 2 associated with familial platelet disorder with propensity to acute myeloid leukemia. Cancer Genet (2018) 222–223:32–7. doi: 10.1016/j.cancergen.2018.01.002
173. Guidugli L, Johnson AK, Alkorta-Aranburu G, Nelakuditi V, Arndt K, Churpek JE, et al. Clinical utility of gene panel-based testing for hereditary myelodysplastic syndrome/acute leukemia predisposition syndromes. Leukemia (2017) 31(5):1226–9. doi: 10.1038/leu.2017.28
174. Sakurai M, Kasahara H, Yoshida K, Yoshimi A, Kunimoto H, Watanabe N, et al. Genetic basis of myeloid transformation in familial platelet disorder/acute myeloid leukemia patients with haploinsufficient RUNX1 allele. Blood Cancer J (2016) 6(2):e392–2. doi: 10.1038/bcj.2015.81
175. Feurstein S, Luo X, Shah M, Walker T, Mehta N, Wu D, et al. Revision of RUNX1 variant curation rules. Blood Adv (2022) 6(16):4726–30. doi: 10.1182/bloodadvances.2022008017
176. Buijs A, Poot M, van der Crabben S, van der Zwaag B, van Binsbergen E, van Roosmalen MJ, et al. Elucidation of a novel pathogenomic mechanism using genome-wide long mate-pair sequencing of a congenital t(16;21) in a series of three RUNX1-mutated FPD/AML pedigrees. Leukemia (2012) 26(9):2151–4. doi: 10.1038/leu.2012.79
177. Jin Y, Wang X, Hu S, Tang J, Li B, Chai Y. Determination of ETV6-RUNX1 genomic breakpoint by next-generation sequencing. Cancer Med (2016) 5(2):337–51. doi: 10.1002/cam4.579
178. Sakurai M, Nannya Y, Yamazaki R, Yamaguchi K, Koda Y, Abe R, et al. Germline RUNX1 translocation in familial platelet disorder with propensity to myeloid Malignancies. Ann Hematol (2021). 101(1):237–9. doi: 10.1007/s00277-021-04430-1
179. Li Y, Yang W, Devidas M, Winter SS, Kesserwan C, Yang W, et al. Germline RUNX1 variation and predisposition to childhood acute lymphoblastic leukemia. J Clin Invest (2021) 131(17):e147898. doi: 10.1172/JCI147898
180. Yoshimi A, Toya T, Nannya Y, Takaoka K, Kirito K, Ito E, et al. Spectrum of clinical and genetic features of patients with inherited platelet disorder with suspected predisposition to hematological Malignancies: a nationwide survey in Japan. Ann Oncol (2016) 27(5):887–95. doi: 10.1093/annonc/mdw066
181. Latger-Cannard V, Philippe C, Bouquet A, Baccini V, Alessi MC, Ankri A, et al. Haematological spectrum and genotype-phenotype correlations in nine unrelated families with RUNX1 mutations from the French network on inherited platelet disorders. Orphanet J Rare Dis (2016) 11(1):49. doi: 10.1186/s13023-016-0432-0
182. Manchev VT, Bouzid H, Antony-Debré I, Leite B, Meurice G, Droin N, et al. Acquired TET2 mutation in one patient with familial platelet disorder with predisposition to AML led to the development of pre-leukaemic clone resulting in T2-ALL and AML-M0. J Cell Mol Med (2017) 21(6):1237–42. doi: 10.1111/jcmm.13051
183. Antony-Debré I, Duployez N, Bucci M, Geffroy S, Micol JB, Renneville A, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia (2016) 30(4):999–1002. doi: 10.1038/leu.2015.236
184. Armes H, Rio-Machin A, Krizsán S, Bödör C, Kaya F, Bewicke-Copley F, et al. Acquired somatic variants in inherited myeloid Malignancies. Leukemia (2022) 36(5):1377–81. doi: 10.1038/s41375-022-01515-2
Keywords: inherited hematologic malignancies, next-generation sequencing somatic drivers, DDX41, ETV6, GATA2, RUNX1
Citation: Zoller J, Trajanova D and Feurstein S (2023) Germline and somatic drivers in inherited hematologic malignancies. Front. Oncol. 13:1205855. doi: 10.3389/fonc.2023.1205855
Received: 14 April 2023; Accepted: 15 September 2023;
Published: 13 October 2023.
Edited by:
Michael Drazer, The University of Chicago, United StatesReviewed by:
Courtney DiNardo, University of Texas MD Anderson Cancer Center, United StatesMarcin Wlodarski, University of Freiburg Medical Center, Germany
Copyright © 2023 Zoller, Trajanova and Feurstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simone Feurstein, c2ltb25la3Jpc3RpbmEuZmV1cnN0ZWluQG1lZC51bmktaGVpZGVsYmVyZy5kZQ==
 Julian Zoller
Julian Zoller Despina Trajanova
Despina Trajanova Simone Feurstein
Simone Feurstein