- 1Department of Urology, Affiliated Hospital of Qingdao University, Qingdao, China
- 2School of Basic Medicine, Qingdao University, Qingdao, China
Circular RNAs (circRNAs) are a class of single-stranded non-coding RNAs that form circular structures through irregular splicing or post-splicing events. CircRNAs are abnormally expressed in many cancers and regulate the occurrence and development of tumors. Circulating circRNAs are cell-free circRNAs present in peripheral blood, they are considered promising biomarkers due to their high stability. In recent years, more and more studies have revealed that circulating circRNAs participate in various cellular communication and regulate the occurrence and development of tumors, which involve many pathological processes such as tumorigenesis, tumor-related immunity, tumor angiogenesis, and tumor metastasis. Understanding the role of cell communication mediated by circulating circRNAs in tumor will further reveal the value and significance behind their use as biomarkers and potential therapeutic targets. In this review, we summarize the recent findings and provide an overview of the cell-cell communication mediated by circulating circRNAs, aiming to explore the role and application value of circulating circRNAs in tumors.
The structure, regulation and function of circRNA
Structure
Unlike linear RNAs terminated with 5’ caps and 3’ tails, circRNAs are characterized by covalently closed-loop structures with neither 5’ to 3’ polarity nor a polyadenylated tail. CircRNA is produced by pre-mRNA through back-splicing or alternative (back) splicing. No specific motifs are required for circularization except the splice sites (1, 2). A median exon length of 353 nucleotides is required for single-exon back-splicing, compared with only 112-130 nucleotides per exon for multiple-exon back-splicing in human cells (3). CircRNA can be divided into three types according to its composition: exon-derived circRNA (this type of circRNA is composed of single or multiple exons of a gene), intron-derived circRNA (consisting of a single intron, mainly derived from the lasso RNA and tRNA introns produced by pre-mRNA splicing) and exon−intron circRNA (including both exons and introns). Most endogenous human circRNAs contain several exons, usually two or three (3).
Regulatory mechanisms of circRNA expression
Back-splicing and alternative splicing mediate the formation of circRNA. According to reports, intronic complementary elements facilitate back-splicing to increase circRNA levels (2). Exon circularization efficiency can be regulated by competition between RNA pairings across flanking introns or within individual introns. Importantly, the alternative formation of inverted repeated Alu pairs and the competition between them can lead to alternative circularization, resulting in multiple circular RNA transcripts produced from a single gene. In addition, RNA binding proteins (RBPs) regulate the formation of circRNAs. The one hand, RNA binding proteins (RBPs) directly bridge distal splice sites to promote back-splicing (4). On the other hand, RBPs bind to intronic complementary elements to enhance (5) or suppress (6) back-splicing by enhancing or reducing the pairing capacity of intronic complementary elements. It has also been reported that circular RNA biogenesis can proceed through an exon-containing lariat precursor without relying on cis elements or trans factors (7). Abnormal circRNA expression may also be associated with changes in genomic DNA (8).
Functions of circRNA
The expression of circRNAs is cell-specific and developmental stage-specific (9, 10), and their biological functions are diverse. CircRNAs can negatively regulate pre-mRNA splicing, resulting in a reduction in linear mRNA levels and changing the composition of processed mRNA. Some circRNAs may be reverse transcribed to cDNA and incorporated into the genome. Stable circRNAs and EIcircRNAs are localized in the nucleus where they bind to the elongated RNA Pol II and promote transcription. They can act as sponges by competitively binding miRNAs to aid in miRNA transport or inhibit the binding of miRNAs to target genes (11). In addition, circRNAs can bind to RBPs and act as protein sponges (12). They can also bind two or more proteins to act as scaffolds and promote the interaction between enzymes and substrates (13). Besides, they can encode functional proteins (14).
Circulating circRNA and cell-cell communication
Circulating circulatory RNAs, as their name suggests, are cell-free circulatory RNAs present in peripheral blood. They mainly come from passive leakage of dead cells or active secretion of living cells (15, 16). They exist in two forms: free circRNAs that bind to proteins, and circRNAs in exosomes or microparticles. Compared to other circulating RNAs, circulating RNAs facilitate cell communication to a greater extent, reach a wider range of target cells, and have more opportunities for contact with other tissues and cells. These characteristics endow them with special advantages and important functions in cell-cell communication (Table 1).
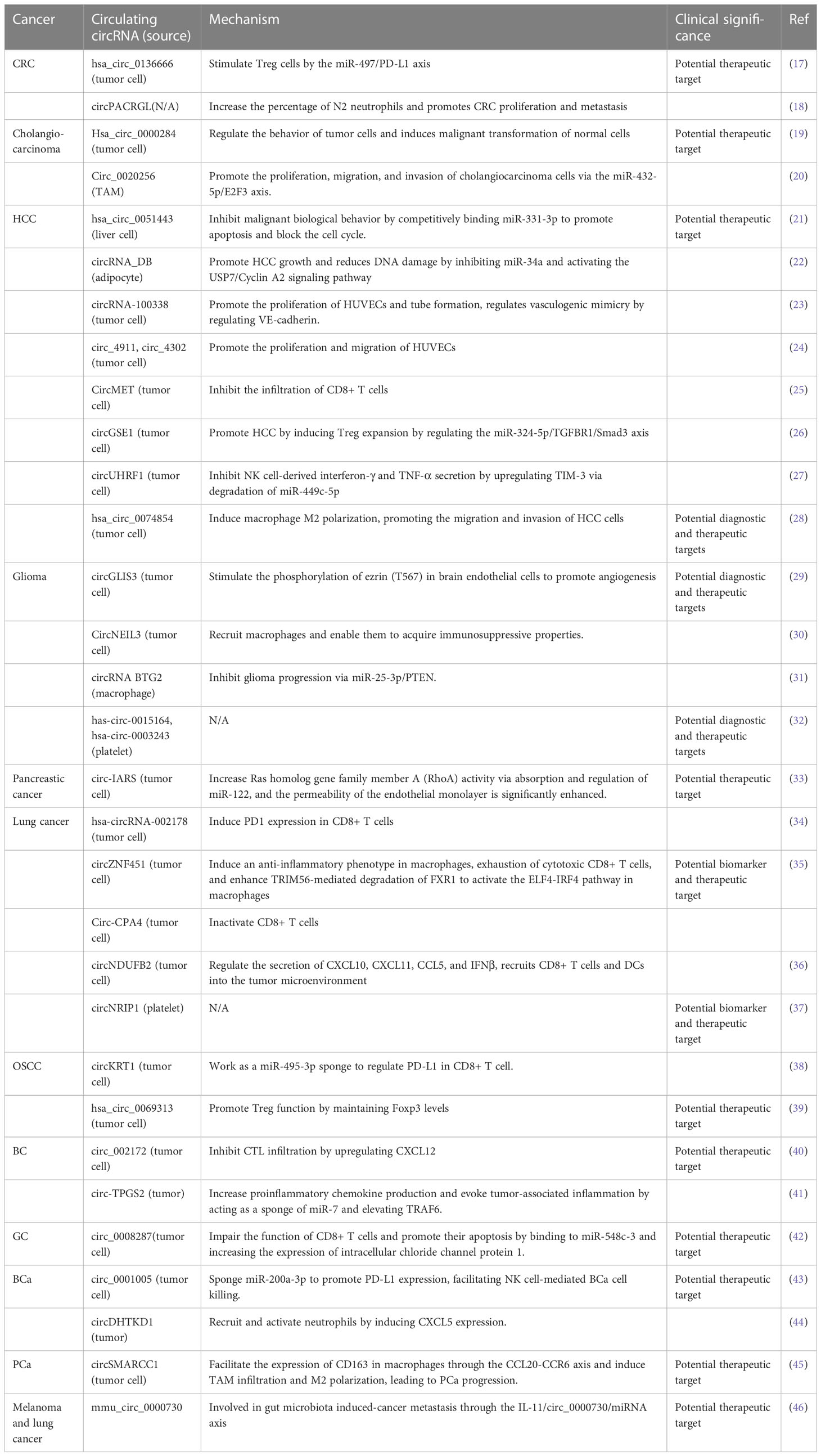
Table 1 The communication between tumor cells and nontumor cells is mediated by circulating circRNAs.
Communication between tumor cells
Cancer is a dynamic disease with high heterogeneity. Tumor heterogeneity may exist between individuals, tissues, sites, or cells, and contribute to the low efficacy or failure of therapies through the development of drug resistance. Tumor heterogeneity is derived from the genetic differences between cells and the differences in the microenvironment, and the information exchange between cells confers tumor plasticity and heterogeneity (47–49), as well as drug resistance. Cisplatin and 5-Fluorouracil (5-FU) are the most common anticancer drugs used for the treatment of a variety of solid tumors, such as ovarian cancer and lung cancer (LC), and adjuvant treatment of glioma (50–53). However, these drugs often result in the development of chemoresistance, leading to therapeutic failure. In esophageal cancer, circ_0000337-containing exosomes secreted by CDDP-resistant esophageal cancer cells could promote CDDP resistance in CDDP-sensitive esophageal cancer cells in vitro partly by regulating the miR-377-3p/JAK2 axis (54). Hon et al. studied the role of hsa-circ-0000338 in colorectal cancer (CRC) patients who were resistant to 5-FU and oxaliplatin (FOLFOX), they found exosomes transferred chemoresistance from FOLFOX-resistant HCT116-R cells into parental HCT116-P cells by selectively transferring hsa_circ_0000338 into recipient cells (55). Additionally, high expression of exosomal circ_0063526 in serum was associated with poor response to cisplatin treatment in gastric cancer (GC) patients. Exosomal circ_0063526 facilitated cisplatin resistance in GC by regulating the miR-449a/SHMT2 axis (56). Circulating circRNAs act as messengers between tumor cells, and targeting them would provide strategies for blocking intercellular communication and combating drug resistance.
In addition to drug resistance, the communication between tumor cells also involves symbiotic nutrient sharing, nutrient competition, and the transmission of oxidative stress (57). Reprogramming of energy metabolism is a hallmark of tumours caused by genomic instability. Recently, circRNAs have been reported to be associated with mutant glycolysis, lipogenesis and lipolysis, glutamate glycoside breakdown, and oxidative respiration in tumors (58). For example, silencing of circHIPK3, which is abundant in pancreatic islets, decreased Slc2a2 expression that encodes GLUT2 (59). CircHIPK3 could sponge miR-124, which represses the expression of several enzymes and transporters of glycolysis (60). However, it remains to be further investigated whether circRNAs are involved in metabolic regulation across different tumor cells.
Communication between tumor cells and normal cells
The malignant transformation of normal cells is an important factor to promote the rapid progression of tumors. In cholangiocarcinoma, hsa_circ_0000284 was evidently elevated in plasma exosomes compared to normal controls, it enhanced the migration and proliferation of cholangiocarcinoma cells in vitro. Exosomes from cholangiocarcinoma cells stimulated the migration and proliferation of surrounding normal cells by transferring circ-0000284 (19). At the same time, circRNAs from normal cell-derived exosomes can be delivered to tumor cells. In hepatocellular carcinoma (HCC), hsa_circ_0051443 was packaged primarily in exosomes and transmitted from normal cells to HCC cells via exosomes, and it inhibited malignant biological behavior by competitively binding miR-331-3p to promote apoptosis and block the cell cycle (21). Hsa_circ_0051443 expression was significantly lower in plasma exosomes and tissues from patients than in those from healthy controls (21). This is a strategy for tumor cell survival, but it also provides lessons for treating tumors from normal cells.
Communication between tumor cells and stromal cells
Stromal cells, together with extracellular matrix components, are critical components of the tumor microenvironment. Common stromal cells include fibroblasts, stellate cells, vascular endothelial cells, and adipocytes. The composition of stromal cells varies in different tumor tissues (61). The communication between stromal cells and tumor cells is the key to the dynamic shift of tumor microenvironment, which affects the occurrence and development of tumor.
Fibroblasts
Cancer-associated fibroblasts (CAFs) constitute a major portion of the tumor stroma and play crucial roles in tumor progression and metastasis. CAFs derived either from resident fibroblasts or tumor-infiltrating mesenchymal stem cells (MSCs). When recruited into the tumor stroma, bone-marrow-derived MSCs can promote cancer stem cell development by secreting a specific set of paracrine factors or transforming into pro-stem cell CAFs. CircRNAs are essential mediators of the intercommunication between the tumor and CAFs, supporting tumor growth, survival, invasion and metastasis. For instance, hypoxia could induce secretion of exosomal circEIF3K from CAFs, and exosomal circEIF3K promoted colorectal cancer (CRC) progression via miR-214/PD-L1 axis (62). Hsa_circ_0056686, derived from CAFs, promoted cell proliferation and suppressed apoptosis in uterine leiomyoma through inhibiting endoplasmic reticulum stress (63). Moreover, breast cancer cells exposed to CAF-derived exosomes carrying circHIF1A display significantly increased stemness (64). In addition, there is evidence that circRNA is involved in the communication between tumor cells and MSCs. For example, exosomal circ_0030167 derived from bone-marrow-derived MSCs inhibited the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/β-catenin axis (65). Tumor-associated exosomes play a significant role in the differentiation of fibroblasts into CAFs, as well as the differentiation of mesenchymal stem cells (66–68). Currently, there is evidence to show that circRNAs play a role in their differentiation (69–71), however, whether tumor-cell-derived circRNAs directly induce their differentiation still needs to be further revealed.
Stellate cell
Stellate cells are resting stem cells of mesenchymal origin located in the liver and pancreas. Hepatic stellate cells (HSCs) comprise a minor cell population in the liver but serve numerous critical functions in hepatic physiology and pathology (72). HSCs are primarily known for their activation upon liver injury and for producing the collagen- abundant extracellular matrix in liver fibrosis (72). During HCC progression, activated HSCs are thought to accelerate carcinogenesis by affecting proliferation (73), migration and invasion (74), and angiogenesis (75). Recently, several studies have revealed that circRNAs are involved in the communication between HCC cells and HSCs. For example, HSC exosome-derived circWDR25 promoted the progression of HCC via the miRNA-4474-3P-ALOX-15 and EMT axes (76). In addition, peritumoral circWDR25 secreted by HSCs affects the prognosis of HCC patients after radical hepatectomy, and their elevated expression in the adjacent tissues was closely related to a poor prognosis of the patients (77). In pancreatic cancer, upregulated circRNA chr7:154954255-154998784+ in cancer-associated pancreatic stellate cells promoted the growth and metastasis of pancreatic cancer by targeting the miR-4459/KIAA0513 axis (78). There are different subgroups of HSCs, and the subgroups have different roles in HCC (79). Whether circRNAs are involved in the activation of HSCs and the dynamical shift of HSC subpopulations still needs to be further revealed.
Endothelial cells
Endothelial cells are involved in angiogenesis and tumor metastasis. Huang et al. revealed that circRNA-100338 helped mediate the communication between HCC cells and human umbilical vein endothelial cells (HUVECs) via exosomes (23). On the one hand, exosomal circRNA-100338 promotes the proliferation of HUVECs and tube formation. On the other hand, exosomal circRNA-100338 regulates vasculogenic mimicry by modulating VE-cadherin. In glioma, circGLIS3 could be packed into exosomes and absorbed by human brain microvascular endothelial cells, stimulating the phosphorylation of ezrin (T567) to promote angiogenesis (29). During tumor metastasis, changes in endothelial permeability act as accelerators for tumor expansion. Circulating circRNAs act as the “key” that allows tumor cells to “open” the endothelial cell barrier. In pancreatic cancer, circ-IARS expression is upregulated in cancerous tissues and in plasma exosomes of patients with metastatic disease. Circ-IARS was found to enter HUVECs through exosomes, it significantly increased Ras homolog gene family member A activity via absorption and regulation of miR-122, and the permeability of the endothelial monolayer was significantly enhanced (33). It was reported that increased Ras homolog gene family member A activity in HUVECs promoted actin-cytoskeletal remodeling and cell contraction and reduced the expression of the tight junction ligand protein Zonula occludens-1, leading to endothelial barrier dysfunction (80, 81) and endothelial hyperpermeability (82, 83). Cell migration is driven by local membrane protrusion through directed polymerization of F-actin at the front (84). Circ-IARS also increased F-actin expression and focal adhesion in HUVECs (33).
Besides, pericytes are also one of the main cellular components, they are typically described as greatly elongated, slender, and branched cells, with projections that extend longitudinally and circumferentially around the vessel wall (85, 86). They support the formation and function of blood vessel, and can be recruited during tumor angiogenesis (87, 88). However, the mechanism by which they are recruited is unclear. Whether it involves communication between tumor cells and pericytes, endothelial cells and pericytes remains to be explored further.
Adipocyte
Adipocytes are specialized cells that regulate energy balance, store excess energy as fat, and play a regulatory role in tumors by secreting metabolites, enzymes, hormones, growth factors, and cytokines. During the development of cancer, tumor cells have a metabolic symbiosis with neighboring adipose tissue. For example, adipocytes provided adipokines and lipids to cancer cells and regulated therapeutic resistance (89). Zhang et al. reported that circRNAs secreted by adipocytes regulated the development of HCC. Exosomal circRNA_DB was upregulated in HCC patients with high body fat ratios. CircRNA_DB secreted by adipocytes promoted HCC growth and reduced DNA damage by inhibiting miR-34a and activating the USP7/Cyclin A2 signaling pathway (22). Increased adiposity contributes to carcinogenesis and tumor progression, but advanced stages of numerous cancers are associated with loss of white adipose tissue and wasting of the body, which complicates treatment and adversely affects patient survival. Exploring the communication between adipocytes and tumor cells may provide an answer to this question.
Communication between tumor cells and immune cells
T cells
Current immunotherapeutic methods mainly focus on T lymphocytes, especially restoring exhausted cytotoxic T cells (CTLs). An example of such an approach is immune-checkpoint blockade, in which monoclonal neutralizing antibodies block of receptors or ligands that inhibit the activation of CTLs, including programmed cell death protein 1 (PD1), PD1 ligand PD-L1 and lymphocyte-activation gene-3 (90). The use of PD1 antibodies decreases tumor progression and provides long-term clinical benefits in patients (91, 92). However, most patients inevitably acquire resistance after several cycles of treatment (93). More and more mechanisms have been revealed, including the role of circulating circulating RNA.
First, circulating circRNAs are important regulators of PD1 expression in CTLs. In lung adenocarcinoma (LUAD), hsa-circRNA-002178 was significantly upregulated in LUAD tissues and LUAD cancer cells. It can be detected in the exosome of plasma from LUAD patients and could serve as a biomarker for early diagnosis of LUAD. Hsa-circRNA-002178 could enhance PD-L1 expression by sponging miR-34 in cancer cells, and circRNA-002178 could be delivered into CD8+ T cells to induce PD1 expression via exosomes (34). Second, circulating circRNAs inactivate CTLs by up-regulating PD-L1 expression in tumor cells. By coculturing non-small cell lung cancer (NSCLC) cells with CD8+ T cells isolated from human peripheral blood mononuclear cells in a transwell coculturing system, Hong et al. found that Circ-CPA4 positively regulated exosomal PD-L1. NSCLC cells inactivated CD8+ T cells in a secreted PD-L1-dependent manner, and NSCLC cells with circ-CPA4 ablation reactivated CD8+ T cells in the coculturing system (94). Third, circulating circRNAs regulate T cell penetration. In HCC, high levels of circMET were significantly correlated with a low density of tumor-infiltrating CD8+ lymphocytes. CircMET promoted HCC development by inducing EMT and enhancing the immunosuppressive tumor microenvironment through the miR-30-5p/Snail/dipeptidyl peptidase 4 (DPP4)/CXCL10 axis (25). In NSCLC, circNDUFB2 regulated the secretion of CXCL10, CXCL11, CCL5, and IFNβ, it is recognized by RIG-I to activate RIG-I-MAVS signaling cascades and recruited CD8+ T cells and dendritic cells into the tumor microenvironment, and circNDUFB2 downregulation in NSCLC tissues was correlated with NSCLC malignant features (36). What’s more, circulating circRNAs regulate T cell apoptosis. Upregulation of circ_0008287 in GC impaired the function of CD8+ T cells and promoted their apoptosis by competitively binding to miR-548c-3 and increasing the expression of intracellular chloride channel protein 1 (42).
The adaptive immune system is modulated by an essential subset of CD4+ T lymphocytes called regulatory T cells (Tregs) that function in maintaining immune homeostasis by preventing excessive immune activation. In tumors, Tregs secrete immunosuppressive factors to inhibit the function of CD8+ T cells, causing tumor immune escape (26, 95). In OSCC, hsa_circ_0069313 was upregulated and predicts a poor7 prognosis. It is an exosomal circRNA, and the transfer of hsa_circ_0069313 to Treg cells promoted Treg function by maintaining Foxp3 levels (39). In conclusion, circulating circRNA affects the body’s immunity by regulating the function of T cells in various ways, and targeting circulating circRNA will provide a new strategy for tumor immunotherapy.
Natural killer cells
In recent years, the rapid and potent antitumor function of innate immunity, which occurs even at highly early stages of tumor progression, has attracted increasing attention. As a subset of innate lymphoid cells, natural killer (NK) cells, commonly considered type 1 innate-like cells, are currently defined as effector cells, exerting natural cytotoxicity against primary tumor cells and metastasis by inhibiting migration and colonization to distant tissues (96). They can distinguish abnormal cells from healthy cells, leading to more specific antitumor cytotoxicity and reduced off-target complications (97). In addition, they can produce cytokines, mainly interferon-γ, to modulate adaptive immune responses and participate in other related pathways (98). Recently, NK cell dysfunction has been demonstrated in various malignancies. Several studies have shed light on the role and mechanisms of circulating circRNAs in NK cell dysfunction. On the one hand, circulating circRNA levels are correlated with the number of NK cells and their penetration into tumor tissue. In plasma from HCC patients, circUHRF1 was predominantly secreted by HCC cells in an exosomal fashion. Elevated plasma exosome circulating UHRF1 levels were associated with a decreased NK cell fraction and decreased NK cell tumor infiltration. CircUHRF1 inhibited NK cell-derived interferon-γ and TNF-α secretion by upregulating TIM-3 via degradation of miR-449c-5p, thereby promoting immune evasion (27). On the other hand, circulating circRNA modulates the killing effect of NK cells on tumor cells. For example, in bladder cancer (BCa), androgen receptor (AR) influenced the initiation and progression of tumors. Both androgen therapy and AR knockdown effectively reduced PD-L1 expression, facilitating NK cell-mediated BCa cell killing. Mechanistically, androgen receptor upregulated circ_0001005 expression via the RNA-editing gene ADAR2. Circ_0001005 competitively sponged miR-200a-3p to promote PD-L1 expression (43). In addition, the abnormal expression of circRNAs also regulated the toxicity of NK cells to tumor cells in HCC (99), breast cancer (100), and renal cell carcinoma (101).
Macrophages
Macrophages include many cell types with complex and delicate regulatory networks. The type, density and location of macrophages have good prognostic value in various cancer types. Tumor-associated macrophages (TAMs), including both resident macrophages and circulating monocytes recruited to the tumor microenvironment, are a predominant cell type in tumors (102). Under the guidance of different microenvironmental signals, macrophages differentiate into two functional phenotypes, namely, classically activated macrophages (M1) and alternately activated macrophages (M2). In contrast to the anti-tumor effects of M1 macrophages, M2 macrophages have anti-inflammatory and tumorigenic properties. Consistent with macrophages, TAMs are also highly plastic and switch from one phenotype to another (103, 104).
CircRNAs mediate macrophage infiltration, facilitating carcinogenesis and cancer development. CircNEIL3 derived from NEIL3 increased with increasing glioma grade and was regulated by EWS RNA-binding protein 1 (EWSR1). Functionally, circNEIL3 promoted tumorigenesis and progression of gliomas in vitro and in vivo. Mechanically, circNEIL3 stabilized IGF2BP3, a known oncogen, by preventing HECTD4-mediated ubiquitination. CircNEIL3-overexpressing glioma cells drove macrophage penetration into the tumor microenvironment by activating YAP1 signaling and secreting CCL2 and LOX. Moreover, circNEIL3 could be packaged into exosomes by hnRNPA2B1 and transmitted to infiltrating TAMs, thereby enabling them to acquire immunosuppressive properties by stabilizing IGF2BP3, in turn promoting glioma progression (30). In prostate cancer (PCa), high expression of circSMARCC1 was positively associated with colonization of CD68+/CD163+/CD206+ TAMs in the tumor microenvironment. Overexpression of circSMARCC1 facilitated the expression of CD163 in macrophages through the CCL20-CCR6 axis and induces TAM infiltration and M2 polarization, thereby leading to PCa progression (45). CircRNAs are also involved in M2 macrophage polarization. CRITGB6, which was robustly upregulated in tumor tissue and sera from platinum-resistant OC patients, was associated with a poorer prognosis. CircITGB6 overexpression promoted M2 macrophage-dependent CDDP resistance both in vivo and in vitro. Mechanistic research determined that circITGB6 directly interacted with IGF2BP2 and FGF9 mRNA to form a circITGB6/IGF2BP2/FGF9 RNA−protein ternary complex in the cytoplasm, thereby stabilizing FGF9 mRNA and inducing polarization of TAMs toward the M2 phenotype. Importantly, blocking M2 macrophage polarization with an antisense oligonucleotide targeting circITGB6 markedly reversed the circITGB6-induced CDDP resistance of OC in vivo (105). In HCC, hsa_circ_0074854 can be transferred from HCC cells to macrophages via the exosomes. Exosomes with downregulated hsa_circ_0074854 suppressed macrophage M2 polarization, which in turn suppressed the migration and invasion of HCC cells both in vitro and in vivo (106). Interestingly, the exosomal circRNAs produced by TAMs also affect tumors. Circ_0020256 in TAM-secreted exosomes promoted the proliferation, migration, and invasion of cholangiocarcinoma cells via the miR-432-5p/E2F3 axis (20). In HCC, exosomal miR-628-5p from M1-polarized macrophages hindered N6-methyladenosine (m6A) modification of circFUT8 to suppress HCC progression (107). In summary, circulating circRNAs affect tumor development by modulating the interaction between macrophages and tumor cells. Targeting circRNA-mediated information exchange between macrophages and tumor cells may be one of the strategies to reshape the tumor microenvironment.
Neutrophils
The role and importance of neutrophils in cancer has received increasing attention over the past decade. Many patients with advanced cancer show elevated neutrophilia levels. Intratumoural neutrophils (also known as tumor-associated neutrophils (TANs)) are connected to a dismal prognosis, and the neutrophil-to-lymphocyte ratio has been introduced as a significant prognostic factor for survival in many types of cancer (108). TANs constitute an important portion of the infiltrating immune cells in the tumor microenvironment, and they are recruited to the tumor and can acquire either protumor or antitumor functions (109). In BCa, circDHTKD1 was positively associated with lymph node metastasis and significantly upregulated. CircDHTKD1 interacted directly with miR-149-5p and antagonized CXCL5 inhibition by miR-149-5p. CircDHTKD1-induced CXCL5 expression recruited and activated neutrophils, which participated in lymphangiogenesis by secreting VEGF-C (44). Like TAMs, TANs are also divided into two types, N1 and N2 (37). N1 TANs have cytotoxic and antitumor effects, and N2 TANs promote tumor progression (110). Transforming growth factor-beta (TGF-β) is a multifunctional cytokine implicated in tumor initiation, progression, and metastasis. TGF-β promotes the formation of TANs. In particularly, TGF-β inhibits N1 but promotes N2 neutrophil differentiation (111). It has been reported that circPACRGL served as a sponge for miR-142-3p/miR-506-3p to facilitate TGF-β1 expression in CRC. CRC-derived exosomal circPACRGL increased the percentage of N2 neutrophils and promotes CRC proliferation and metastasis (18).
Communication between tumor cells and platelets
Platelets participate in physiologic hemostasis and pathologic thrombus, and the role in tumors has also attracted considerable attention. They affect all aspects of cancer progression, most notably tumor cell metastasis. Platelets isolated from cancer patients frequently show different RNA and protein profiles without significant changes in hemostatic activity. This phenotype is unique to a population known as tumor-educated platelets. At present, the mechanism by which tumor cells educate platelets is not completely understood, but many studies have revealed the interaction between tumor cells and platelets. For example, platelets can interact with circulating tumor cells through receptors and ligands (112, 113), and platelets can be recruited to the tumor tissue microenvironment to play a regulatory role (114, 115). Of concern, platelets can directly take up and store RNAs (mRNAs, miRNAs, LncRNAs) from tumor cells, and tumor-educated platelets in liquid biopsies have emerged as valuable resources that can be used to assess cancer-related RNA profiles with tumor specificity, high sensitivity, and high accuracy (116–118). CircRNA levels in human platelets are 17- to 188-fold higher than those in nucleated tissues and 14- to 26-fold higher than those in samples digested with RNase R (119). Like other RNAs, platelet circRNAs have shown promising applications in human disease. For example, NRIP1 circRNA was identified to be pregnancy specific, with significant upregulation in maternal platelets in the first trimester compared to those from non-pregnant control participants. This finding allows NRIP1 circRNA to be used as a first-tier check (gold standard) in future efforts for diagnostic screening purposes using circRNAs as targets and maternal first-trimester platelets as a source (120). In NSCLC, 4732 circRNAs were detected in platelet samples from patients and controls, and 411 of these circRNAs were significantly differentially expressed; circNRIP1 is one representative of the differentially expressed circRNA. CircNRIP1, which was downregulated in the NSCLC group, could be employed as a potential biomarker for NSCLC (37). Chen et al. analyzed the platelet RNA profiles of 8 glioblastoma samples and 12 normal samples and constructed a ceRNA network (circRNA-mRNA−miRNA) based on 2 miRNAs (hsa-let-7a-5p, hsa-miR-1-3p), 2 mRNAs (CCR7 and FAM102A), and 2 circRNAs (has-circ-0015164, hsa-circ-0003243) to investigate the potential interactions (32). This study also sheds light on the potential relationship between platelet circRNAs and cancer, as well as the clinical value of platelet circRNAs. However, the cause of changes in platelet circRNA in patients with tumors remains unclear. These changes may be related to the uptake of circulating circRNA or may be the result of platelet-selective release (119). In addition, platelets are fragments from megakaryocytes, and tumor cells can also educate megakaryocytes (121). Whether these changes are the result of megakaryocytes being educated by tumor cells remains to be further revealed. What’s more, tumor-associated thrombosis is one of the common complications in tumor patients and is an important cause of death (122, 123), which is closely related to the abnormality of platelet quantity and function (124–126). Whether circulating circRNA is involved in platelet dysfunction remains to be further investigated.
Communication between tumor cells and erythrocytes
Erythrocytes account for ∼84% of the total blood cells count in the average adult (127). Recently, several studies show that abnormal red blood cell distribution width is associated with poor prognosis in cancers (128, 129). Erythrocytes can interact with tumor cells (130), and erythrocytes from cancer patients have a differential proteome profile compared with cancer-free controls (131). In fact, erythrocytes also have high circRNA content besides platelets (132–134). Recently, Nicolet et al. provided the first detailed analysis of circRNA expression during erythroid differentiation (135). They found that circRNA expression not only significantly increased upon enucleation but also had limited overlap between progenitor cells and mature cells, and only one out of 2531 (0.04%) circRNAs was associated with mRNA translation regulation. These results suggest that circRNA expression originates from regulatory processes rather than from mere accumulation, yet their contribution to regulatory cellular processes is still unknown. Interestingly, the levels of approximately 4% circRNAs in the erythrocytes did not overlap with the levels of mRNAs in differentiated erythrocytes. These circRNAs may be acquired from other cell types in the circulation or endogenous vascular-derived RNA (136). If this is the case, circRNAs taken up by erythrocytes could be novel biomarkers for tumors due to their high stability. In addition, tumor-associated anemia (137), is the key reason for leading to a decline in patients’ quality of life. And anemia in tumor patients is associated with a decrease in the number of red blood cells (138). Whether circulating circRNAs are involved in the reduction of red blood cells is also an urgent question to be explored.
Communication between tumor cells and the gut microbiota
The human gut harbors diverse microbes that play a fundamental role in the well-being of their host, including participating in energy collection and storage, fermenting and absorbing undigested carbohydrates (139), promoting the maturation of immune cells and the normal development of immune function (140). However, the influence of gut microbiota is not limited to the local part of the intestine but is instead systemic (141, 142). In recent years, the role of the gut microbiota in tumor-related immune regulation has attracted much attention. In particular, the gut microbiota regulates antitumor immunity through metabolites, which are small molecules that can be transported in the body and act on local and systemic antitumor immune responses to promote the efficacy of immune checkpoint inhibitor (ICI) therapy (143–146). Recently, Zhu et al. found that the levels of circulating miRNAs and circRNAs changed significantly with dysbiosis of the microbiota, and the gut microbiota regulated tumor metastasis via circRNA/miRNA networks. Specifically, the gut microbiota downregulated circulating mmu_circ_0000730 and upregulated circulating mmu-miR-466i-3p or mmu-miR-466 f-3p. SRY-box transcription factor 9 (SOX9) is a target gene of mmu-miR-466i-3p and mmu-miR-466 f-3p. Mmu_circ_0000730 upregulated SRY-box transcription factor 9 (SOX9) and activated epithelial-mesenchymal transition markers in cancer cells by targeting mmu-miR-466i-3p or mmu-miR-466 f-3p. Furthermore, intestinal flora reconstruction significantly decreased IL-11 expression. IL-11 treatment induced mmu_circ_0000730/SOX9 expression, while it downregulated mmu-miR-466i-3p and mmu-miR-466 f-3p. In cancer cells, IL-11 promoted SOX9 expression, induced cell invasion, and promoted the stemness of cancer stem cells (46). This study shows that there is a mechanistic link between the gut microbiota and cancer metastasis through the IL-11/circRNA/miRNA axis, which will help to pave the way to the clinical use of gut microbiota for cancer prevention or treatment in the future (147–149).
Clinical applications of circulating circRNAs involved in cell-cell communication
Biomarkers for tumors
Seeking specific molecules of tumor origin to improve the accuracy and sensitivity of early screening and prognosis assessment has consistently been an urgent issue. CircRNAs have attracted much attention in tumor biomarker studies due to their stability, tissue specificity of expression, and relatively high content (150). Circulating circRNAs are widely respected in the field of non-invasive liquid biopsy. Large numbers of clinical samples also reveal its huge application prospect (151–154). For example, AR is the key driver gene and a common target for the treatment of PCa. CircRNA AR3,derived from AR, is widely expressed in PC cells and prostate tumors. Plasma circRNA_AR3 level was positively associated with Gleason scores and lymph node metastasis of PCa, and it was undetectable in men after radical prostatectomy (151). It is considered a promising circulating RNA with tumor specificity. In fact, the circRNAs identified so far, tumor-specific circRNAs are very rare, and most of them are non-specific. However, these abnormally expressed circRNAs still have high diagnostic value (155–157). For example, in CRC, circRNA_PNN was found to have significant value for CRC diagnosis among 122 differentially expressed circRNAs. The area under the receiver operating characteristic curve of serum exosomal circ-PNN for early-stage CRC was 0.854 (sensitivity=91.7%, specificity=69.0%) (152). In addition, another study showed that exosomal HSA-circ-0004771 levels in serum were 14 times higher in patients with CRC than in healthy controls. Its level was correlated with stage and distant metastasis. The sensitivity and specificity values of exosomal hsa-circ-0004771 for differentiating CRC patients from HCs were 80.91% and 82.86%, respectively (153). To further improve diagnostic accuracy, several studies have analyzed multiple circulating circRNAs in combination, which performed well in tumor detection and showed a higher accuracy (158). For example, a plasma circRNA panel (CircPanel) (hsa_circ_0000976, hsa_circ_0007750 and hsa_circ_0139897) showed a higher accuracy than alpha-fetoprotein (AFP) in distinguishing individuals with HCC from controls. The circPanel also performed well in detecting small HCC (solitary, ≤3 cm), AFP-negative HCC and AFP-negative small HCC (158). These results indicate the high sensitivity and accuracy of circulating circRNAs in tumor diagnosis. The biological functions of these non-tumor specific circRNAs are not restricted to the tumor cells. Compared with tumor-specific circRNAs, those that promote malignant transformation of normal cells are also worthy of attention (19), they are the starting point of evil and the key point of evil progress. Moreover, angiogenesis is a prerequisite for rapid tumor development and a key to metastasis. Endothelial cell proliferation and angiogenesis based on the regulation of circulating circRNAs is another concern (24, 29), and the levels of these circulating circRNAs will provide a strong reference for evaluating tumor metastasis or prognosis.
Biomarkers for tumor immunity
Compared with normal people, numerous tumor patients have obvious abnormalities in peripheral blood immune cells, including changes in number and function. In addition to the self-regulation of immune cells, the role of circulating circRNA should not be ignored. First, circulating circRNAs reflect the functional state of T cells. The functional status of CD8+T cells is of primary concern in current immunotherapy. Abnormally expressed circulating circRNAs cause T cell dysfunction in several ways, such as affecting PD1/PDL1 expression (25, 34, 94) or reducing infiltration (34, 159) or promoting their exhaustion (25, 40). In addition, circulating circRNA enhances CD8+ T cytotoxicity (38). The detection of specific circulating circRNAs will provide reference and support for the analysis of T cell functional status. Second, circulating circRNAs reveal the function of NK cells. NK cells are known for their cytotoxic role, but they are dysfunctional in tumors. On the one hand, NK cell infiltration is regulated by circRNA. A study involving 240 patients with HCC reported that there was a negative correlation between plasma circUHRF1 levels and the proportion of NK cells. With higher expression of circUHRF1, NK cell penetration also decreased. CircUHRF1 inhibited NK cell function by reducing the expression of interferon-γ and TNF-α, which promoted immune evasion (27). On the other hand, the killing effect of NK cells is regulated by circRNAs. Circ_0001005 competed with miR-200a-3p in sponging to promote PD-L1 expression. and NK cell-mediated BCa cell killing could be facilitated by downregulating circ_0001005 (43). Third, cancer-associated systemic inflammation is frequently characterized by a high ratio of neutrophils to lymphocytes and is associated with a poor prognosis, it occurs in both the late stages of the tumor and in the early stages (stage I and stage II, separately) before treatment (160–162). Exosomal circRNAs are involved in recruitment and activation of neutrophils (18, 44), suggesting their potential value as biomarkers. Therefore, the screening and identification of circulating circRNAs involved in cell communication could be useful for the detection of tumor-related immunity and would also provide a reference for the early prevention and treatment of tumors.
Targets for therapy
In addition to acting as biomarkers, circulating circRNAs are promising therapeutic targets (163–166). For example, the expression of hsa_circ_0014235 was notably elevated in NSCLC serum derived exosomes. Hsa_circ_0014235 promoted proliferation and invasion of NSCLC cells. Hsa_circ_0014235 triggered the malignant development of NSCLC through the miR-520a-5p/CDK4 regulatory axis (163). In addition to the direct regulatory effect on tumor cells, the effect of circulating circRNA on other cells in the tumor background is also of interest. For example, in HCC, the exosomal circRNA-100338 enhanced communication between HCC cells and endothelial cells. It promoted the proliferation and tube formation of HUVECs and regulated vasculogenic mimicry (23). And circRNA_DB secreted by adipocytes promoted HCC growth and reduced DNA damage by inhibiting miR-34a and activating the USP7/Cyclin A2 signaling pathway (22). However, when exosomal circ_0051443 was transferred from hepatocytes to HCC cells, it inhibited HCC progression by promoting apoptosis and blocking the cell cycle (21). These studies further demonstrate that tumor development is a networked regulatory process based on multicellular communication, and that breaking cellular communication is key to blocking its development.
Signals of drug resistance
Medication is critical, especially for patients who miss the best time for surgery. However, drug resistance significantly reduces the effectiveness of treatment and increases mortality. Circulating circRNAs are deeply involved in resistance to antitumor drugs, from traditional chemotherapeutic drugs to targeted and immunotherapeutic drugs (167–170). Numerous experiments have demonstrated that drug resistance was never the sole event of individual cells (55, 171). For example, in CRC, exosomes from FOLFOX-resistant CRC cells selectively transferred hsa_circ_0000338 into recipient cells to confer chemoresistance (55). The transmissibility of chemotherapeutic drug resistance is due to the heterogeneity and communication among tumor cells. Communication between tumor cells and immune cells is closely related to tumor immunotherapy resistance. Immunotherapy, represented by anti-PD-1/PD-L1 antibodies, has shown great efficacy in the clinical treatment of cancers (172). Circulating circRNAs are involved in immunotherapeutic resistance through mediated dysregulation of PD-1/PD-L1 expression (34, 38). Additional exploration of the role of circulating circRNA on immune cells will shed further light on the mechanism of resistance.
Potential challenges in clinical application
Circulating circRNAs have a higher stability than other RNAs due to their circular structure and critical regulatory role in tumor development, making them a rising star in tumor detection. However, there are still some challenges and problems that need to be addressed in practical clinical application. First, the source of the samples. The samples collected in different studies varied widely, with some are exosomal circRNAs and others are cell-free circRNAs (including free circRNAs that bind with proteins and circRNAs in exosomes or microparticles). The difference in the samples may lead to difference in circRNA content and thus in the origin. In addition, some studies used serum and some used plasma. Serum and plasma have different sensitivities and specificities in tumor detection, and the RNAs they contain are also different (173, 174). The choice of sample will directly affect the type and content of circRNA. The second is the absence of standardized protocols for sample preparation, such as which anticoagulant should be chosen and at what temperature the sample should be stored. In addition, the accidental release of nucleic acid from peripheral blood cells should be fully considered (175, 176). Third, there is the problem of technical methods. The level of circulating circRNAs is extremely low compared to tissue circRNAs. Traditional experimental methods such as PCR, microarray technique and northern blot are no longer suitable for clinical use due to their low specificity and sensitivity. RNA sequencing (RNA-seq) has been widely used for the discovery and quantification of circRNAs. Despite the tremendous successes of short-read RNA-seq studies of circRNAs (177, 178), there are inherent limitations of this approach, especially the inability to capture the full length of circRNA completely and internal alternative splicing events within circRNAs, and the difficulty in accurately quantification of circRNA (179). Long-read RNA-seq (third-generation RNA-seq) technology is a newly developed transcriptome analysis technology (such as the Pacific biosciences (PacBio) and the Oxford nanopore technology company (permanent)), which can produce thousands to tens of thousands of bases in length (180), and can fundamentally eliminate the excessive fragmentation of short-read RNA-seq, bringing hope for full-length detection and accurate quantification of circRNAs. However, it has a higher error rate and lower throughput (181). Although the development of some technologies has made up for the loopholes of the third generation RNA-seq to some extent (181, 182), a technical means with high sensitivity, accuracy, repeatability and universality is still needed in the practical clinical application.
Conclusion and future prospect
Cancer is a rapidly progressive malignant disease, especially in its terminal stages, that can involve multiple systems and organs in the body. The exchange of information between tissues or cells underlies a range of processes and is key to the occurrence and rapid development of tumors. Due to their unique advantages, circulating circRNAs participate in multiple cell-cell communications and regulate the occurrence and development of tumors from many aspects, which gives them great value and significance in tumor detection and treatment. However, intercellular communication is diverse and complex, and many questions remain open, such as whether circulating circRNAs are involved in the communication between tumor cells and red blood cells, and whether tumor-associated anemia is related to circulating circRNAs. It is also unclear whether tumor-associated thrombosis is associated with platelet uptake of tumor-derived circRNAs (Figure 1). In addition, most of the MSCs and immune cells in the tumor microenvironment come from the circulatory system. The role and mechanism of circulating circRNAs or tumor-derived circulating RNAs in the recruitment of these cells remains to be investigated further. An in-depth understanding of cell-cell communication mediated by circulating circRNAs will establish a new mechanism of tumor development from point to surface, breaking the one-way relationship in previous studies and improving the accuracy and specificity of tumor diagnosis and enhancing the effectiveness of tumor-targeted therapies.
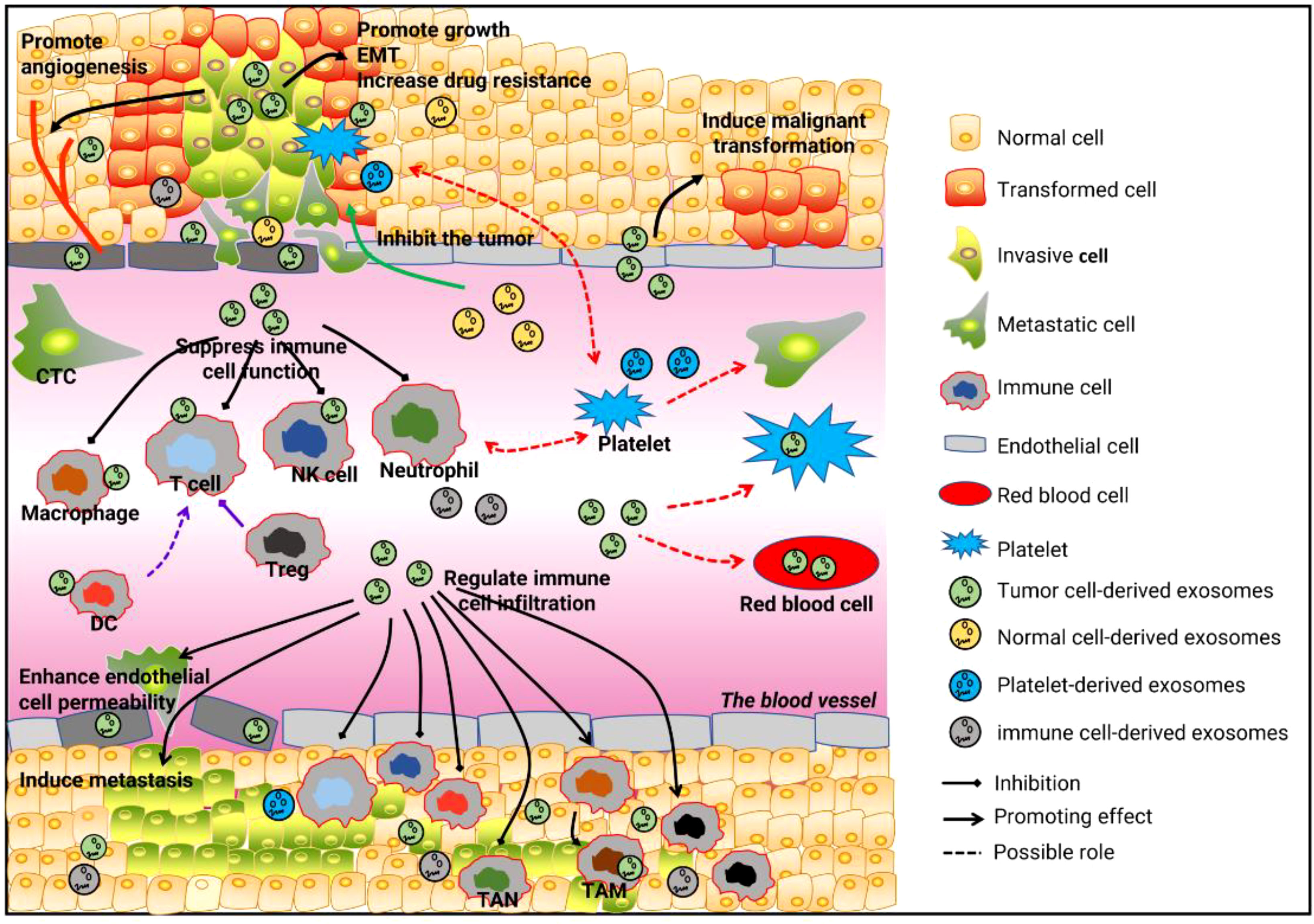
Figure 1 Cell-cell communication is mediated by circulating RNAs and tumor development. First, circulating cirRNAs affect tumor growth, metastasis, angiogenesis, and drug resistance by mediating communication between tumor cells and tumor cells, tumor cells and normal cells, and tumor cells and endothelial cells. Second, circulating cirRNAs mediate communication between tumor cells and immune cells, by which they suppress tumor-related immunity and reshape the tumor microenvironment. Third, the communication between tumor cells and platelets, and between tumor cells and red blood cells, may be involved in the occurrence and development of tumors.
Author contributions
SM drafted and wrote the manuscript. QZ design and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (Project No.:82100285) and Shandong Province Postdoctoral Innovation Research Project (Project No.:202003034).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CAFs, cancer-associated fibroblasts; AFP, alpha-fetoprotein; BCa, bladder cancer, circRNAs, circular RNAs; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; HSCs, hepatic stellate cells; HUVEC, human umbilical vein endothelial cells; LC, lung cancer; LUAD, lung adenocarcinoma; MSCs, mesenchymal stem cells; NK, natural killer; NSCLC, non-small-cell lung cancer; PCa, prostate cancer; PD1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; RBP, RNA binding protein; SOX9: SRY-box transcription factor 9, TANs: tumor-associated neutrophils; TAMs, tumor-associated macrophages; TGF-β, Transforming growth factor-beta; Tregs, regulatory T cells.
References
1. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
2. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
3. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase r resistant RNAs. Int J Mol Sci (2014) 15:9331–42. doi: 10.3390/ijms15069331
4. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
5. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell (2017) 67:214–227 e7. doi: 10.1016/j.molcel.2017.05.023
6. Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the alu invasion of the human genome. Nature (2017) 544:115–9. doi: 10.1038/nature21715
7. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife (2015) 4:e07540. doi: 10.7554/eLife.07540
8. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett (2017) 388:208–19. doi: 10.1016/j.canlet.2016.12.006
9. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell (2015) 58:870–85. doi: 10.1016/j.molcel.2015.03.027
10. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
11. Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res (2018) 78:2839–51. doi: 10.1158/0008-5472.CAN-17-2808
12. Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep (2016) 6:31313. doi: 10.1038/srep31313
13. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ (2017) 24:1609–20. doi: 10.1038/cdd.2017.86
14. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst (2018) 110. doi: 10.1093/jnci/djx166
15. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol (2011) 13:423–33. doi: 10.1038/ncb2210
16. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia (2006) 20:1487–95. doi: 10.1038/sj.leu.2404296
17. Xu YJ, Zhao JM, Gao C, Ni XF, Wang W, Hu WW, et al. Hsa_circ_0136666 activates treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell Signal (2021) 86:110095. doi: 10.1016/j.cellsig.2021.110095
18. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer (2020) 19:117. doi: 10.1186/s12943-020-01235-0
19. Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (Lond) (2019) 133:1935–53. doi: 10.1042/CS20190589
20. Chen S, Chen Z, Li Z, Li S, Wen Z, Cao L, et al. Tumor-associated macrophages promote cholangiocarcinoma progression via exosomal Circ_0020256. Cell Death Dis (2022) 13:94. doi: 10.1038/s41419-022-04534-0
21. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
22. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene (2019) 38:2844–59. doi: 10.1038/s41388-018-0619-z
23. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res (2020) 39:20. doi: 10.1186/s13046-020-1529-9
24. Yan K, Cheng W, Xu X, Cao G, Ji Z, Li Y. Circulating RNAs, circ_4911 and circ_4302, are novel regulators of endothelial cell function under a hepatocellular carcinoma microenvironment. Oncol Rep (2020) 44:1727–35. doi: 10.3892/or.2020.7702
25. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
26. Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory t cells. Cancer Sci (2022) 113:1968–83. doi: 10.1111/cas.15365
27. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer (2020) 19:110. doi: 10.1186/s12943-020-01222-5
28. Lu C, Shi W, Hu W, Zhao Y, Zhao X, Dong F, et al. Endoplasmic reticulum stress promotes breast cancer cells to release exosomes circ_0001142 and induces M2 polarization of macrophages to regulate tumor progression. Pharmacol Res (2022) 177:106098. doi: 10.1016/j.phrs.2022.106098
29. Li Y, Chen J, Chen Z, Xu X, Weng J, Zhang Y, et al. CircGLIS3 promotes high-grade glioma invasion via modulating ezrin phosphorylation. Front Cell Dev Biol (2021) 9:663207. doi: 10.3389/fcell.2021.663207
30. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer (2022) 21:16. doi: 10.1186/s12943-021-01485-6
31. Shi L, Cao Y, Yuan W, Guo J, Sun G. Exosomal circRNA BTG2 derived from RBP-j overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis (2022) 13:506. doi: 10.1038/s41419-022-04908-4
32. Chen X, Lin Q, Jiang Y, Wang C, Min F, Ou Y, et al. Identification of potential biomarkers of platelet RNA in glioblastoma by bioinformatics analysis. BioMed Res Int (2022) 2022:2488139. doi: 10.1155/2022/2488139
33. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res (2018) 37:177. doi: 10.1186/s13046-018-0822-3
34. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis (2020) 11:32. doi: 10.1038/s41419-020-2230-9
35. Gao J, Ao YQ, Zhang LX, Deng J, Wang S, Wang HK, et al. Exosomal circZNF451 restrains anti-PD1 treatment in lung adenocarcinoma via polarizing macrophages by complexing with TRIM56 and FXR1. J Exp Clin Cancer Res (2022) 41:295. doi: 10.1186/s13046-022-02505-z
36. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun (2021) 12:295. doi: 10.1038/s41467-020-20527-z
37. D'Ambrosi S, Visser A, Antunes-Ferreira M, Poutsma A, Giannoukakos S, Sol N, et al. The analysis of platelet-derived circRNA repertoire as potential diagnostic biomarker for non-small cell lung cancer. Cancers (Basel) (2021) 13. doi: 10.3390/cancers13184644
38. Yang Z, Chen W, Wang Y, Qin M, Ji Y. CircKRT1 drives tumor progression and immune evasion in oral squamous cell carcinoma by sponging miR-495-3p to regulate PDL1 expression. Cell Biol Int (2021) 45:1423–35. doi: 10.1002/cbin.11581
39. Chen Y, Li Z, Liang J, Liu J, Hao J, Wan Q, et al. CircRNA has_circ_0069313 induced OSCC immunity escape by miR-325-3p-Foxp3 axes in both OSCC cells and treg cells. Aging (Albany NY) (2022) 14:4376–89. doi: 10.18632/aging.204068
40. Li P, Ren X, Zheng Y, Sun J, Ye G. Tumor promoting effect of circ_002172 associates with induced immune escape in breast cancer via the miR-296-5p/CXCL12 axis. Int Immunopharmacol (2022) 106:108530. doi: 10.1016/j.intimp.2022.108530
41. Wang S, Feng X, Wang Y, Li Q, Li X. Dysregulation of tumour microenvironment driven by circ-TPGS2/miR-7/TRAF6/NF-kappaB axis facilitates breast cancer cell motility. Autoimmunity (2021) 54:284–93. doi: 10.1080/08916934.2021.1931843
42. Li B, Liang L, Chen Y, Liu J, Wang Z, Mao Y, et al. Circ_0008287 promotes immune escape of gastric cancer cells through impairing microRNA-548c-3p-dependent inhibition of CLIC1. Int Immunopharmacol (2022) 111:108918. doi: 10.1016/j.intimp.2022.108918
43. Liu Q, You B, Meng J, Huang CP, Dong G, Wang R, et al. Targeting the androgen receptor to enhance NK cell killing efficacy in bladder cancer by modulating ADAR2/circ_0001005/PD-L1 signaling. Cancer Gene Ther (2022) 29:1988–2000. doi: 10.1038/s41417-022-00506-w
44. Lu Q, Yin H, Deng Y, Chen W, Diao W, Ding M, et al. circDHTKD1 promotes lymphatic metastasis of bladder cancer by upregulating CXCL5. Cell Death Discovery (2022) 8:243. doi: 10.1038/s41420-022-01037-x
45. Xie T, Fu DJ, Li ZM, Lv DJ, Song XL, Yu YZ, et al. CircSMARCC1 facilitates tumor progression by disrupting the crosstalk between prostate cancer cells and tumor-associated macrophages via miR-1322/CCL20/CCR6 signaling. Mol Cancer (2022) 21:173. doi: 10.1186/s12943-022-01630-9
46. Zhu Z, Huang J, Li X, Xing J, Chen Q, Liu R, et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes (2020) 12:1788891. doi: 10.1080/19490976.2020.1788891
47. Pe'er D, Ogawa S, Elhanani O, Keren L, Oliver TG, Wedge D. Tumor heterogeneity. Cancer Cell (2021) 39:1015–7. doi: 10.1016/j.ccell.2021.07.009
48. Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol (2021) 18:79–92. doi: 10.1038/s41585-020-00400-w
49. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature (2013) 501:328–37. doi: 10.1038/nature12624
50. Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol (2014) 740:364–78. doi: 10.1016/j.ejphar.2014.07.025
51. Lugones Y, Loren P, Salazar LA. Cisplatin resistance: Genetic and epigenetic factors involved. Biomolecules (2022) 12. doi: 10.3390/biom12101365
52. Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, et al. Molecular mechanisms of cisplatin resistance. Oncogene (2012) 31:1869–83. doi: 10.1038/onc.2011.384
53. Wigmore PM, Mustafa S, El-Beltagy M, Lyons L, Umka J, Bennett G. Effects of 5-FU. Adv Exp Med Biol (2010) 678:157–64. doi: 10.1007/978-1-4419-6306-2_20
54. Zang R, Qiu X, Song Y, Wang Y. Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p. Front Cell Dev Biol (2021) 9:673237. doi: 10.3389/fcell.2021.673237
55. Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci Rep (2019) 9:16497. doi: 10.1038/s41598-019-53063-y
56. Yang G, Tan J, Guo J, Wu Z, Zhan Q. Exosome-mediated transfer of circ_0063526 enhances cisplatin resistance in gastric cancer cells via regulating miR-449a/SHMT2 axis. Anticancer Drugs (2022) 33:1047–57. doi: 10.1097/CAD.0000000000001386
57. Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol (2017) 27:863–75. doi: 10.1016/j.tcb.2017.06.003
58. Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol (2019) 12:90. doi: 10.1186/s13045-019-0776-8
59. Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab (2018) 9:69–83. doi: 10.1016/j.molmet.2018.01.010
60. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
61. Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H. Cancer-associated fibroblasts: Their characteristics and their roles in tumor growth. Cancers (Basel) (2015) 7:2443–58. doi: 10.3390/cancers7040902
62. Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer (2021) 21:933. doi: 10.1186/s12885-021-08669-9
63. Suo M, Lin Z, Guo D, Zhang A. Hsa_circ_0056686, derived from cancer-associated fibroblasts, promotes cell proliferation and suppresses apoptosis in uterine leiomyoma through inhibiting endoplasmic reticulum stress. PloS One (2022) 17:e0266374. doi: 10.1371/journal.pone.0266374
64. Zhan Y, Du J, Min Z, Ma L, Zhang W, Zhu W, et al. Carcinoma-associated fibroblasts derived exosomes modulate breast cancer cell stemness through exonic circHIF1A by miR-580-5p in hypoxic stress. Cell Death Discovery (2021) 7:141. doi: 10.1038/s41420-021-00506-z
65. Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, et al. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/beta-catenin axis. Cancer Lett (2021) 512:38–50. doi: 10.1016/j.canlet.2021.04.030
66. Maacha S, Bhat AA, Jimenez L, Raza A, Haris M, Uddin S, et al. Extracellular vesicles-mediated intercellular communication: roles in the tumor microenvironment and anti-cancer drug resistance. Mol Cancer (2019) 18:55. doi: 10.1186/s12943-019-0965-7
67. Baglio SR, Lagerweij T, Perez-Lanzon M, Ho XD, Leveille N, Melo SA, et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin Cancer Res (2017) 23:3721–33. doi: 10.1158/1078-0432.CCR-16-2726
68. Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D'Ippolito E, Cataldo A, et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis (2016) 7:e2312. doi: 10.1038/cddis.2016.224
69. Zheng S, Hu C, Lin H, Li G, Xia R, Zhang X, et al. circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-kappaB signaling pathway. J Exp Clin Cancer Res (2022) 41:71. doi: 10.1186/s13046-021-02237-6
70. Lin Z, Tang X, Wan J, Zhang X, Liu C, Liu T. Functions and mechanisms of circular RNAs in regulating stem cell differentiation. RNA Biol (2021) 18:2136–49. doi: 10.1080/15476286.2021.1913551
71. Zhang D, Ni N, Wang Y, Tang Z, Gao H, Ju Y, et al. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin alpha5 expression. Cell Death Differ (2021) 28:283–302. doi: 10.1038/s41418-020-0600-6
72. Kamm DR, McCommis KS. Hepatic stellate cells in physiology and pathology. J Physiol (2022) 600:1825–37. doi: 10.1113/JP281061
73. Xiao H, Zhang Y, Li Z, Liu B, Cui D, Liu F, et al. Periostin deficiency reduces diethylnitrosamine-induced liver cancer in mice by decreasing hepatic stellate cell activation and cancer cell proliferation. J Pathol (2021) 255:212–23. doi: 10.1002/path.5756
74. Liu C, Zhou X, Long Q, Zeng H, Sun Q, Chen Y, et al. Small extracellular vesicles containing miR-30a-3p attenuate the migration and invasion of hepatocellular carcinoma by targeting SNAP23 gene. Oncogene (2021) 40:233–45. doi: 10.1038/s41388-020-01521-7
75. Kang MJ, Lee S, Jung U, Mandal C, Park H, Stetler-Stevenson WG, et al. Inhibition of hepatic stellate cell activation suppresses tumorigenicity of hepatocellular carcinoma in mice. Am J Pathol (2021) 191:2219–30. doi: 10.1016/j.ajpath.2021.08.004
76. Liu L, Liao R, Wu Z, Du C, You Y, Que K, et al. Hepatic stellate cell exosome-derived circWDR25 promotes the progression of hepatocellular carcinoma via the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci Trends (2022) 16:267–81. doi: 10.5582/bst.2022.01281
77. Liu L, Du C, Wei X, Liao R. [Correlation of peritumoral circWDR25 expression with the prognosis of patients with hepatocellular carcinoma after curative resection]. Nan Fang Yi Ke Da Xue Xue Bao (2021) 41:1388–93. doi: 10.12122/j.issn.1673-4254.2021.09.14
78. Shao F, Cai M, Fan FF, Huang M, Tao Y, Wang C, et al. Overexpression of circRNA chr7:154954255-154998784+ in cancer-associated pancreatic stellate cells promotes the growth and metastasis of pancreatic cancer by targeting the miR-4459/KIAA0513 axis. Am J Transl Res (2020) 12:5048–63.
79. Filliol A, Saito Y, Nair A, Dapito DH, Yu LX, Ravichandra A, et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature (2022) 610:356–65. doi: 10.1038/s41586-022-05289-6
80. Schaeffer RC Jr., Gong F, Bitrick MS Jr., Smith TL. Thrombin and bradykinin initiate discrete endothelial solute permeability mechanisms. Am J Physiol (1993) 264:H1798–809. doi: 10.1152/ajpheart.1993.264.6.H1798
81. Lum H, Malik AB. Mechanisms of increased endothelial permeability. Can J Physiol Pharmacol (1996) 74:787–800. doi: 10.1139/y96-081
82. Breslin JW, Yuan SY. Involvement of RhoA and rho kinase in neutrophil-stimulated endothelial hyperpermeability. Am J Physiol Heart Circ Physiol (2004) 286:H1057–62. doi: 10.1152/ajpheart.00841.2003
83. van Nieuw Amerongen GP, van Hinsbergh VW. Endogenous RhoA inhibitor protects endothelial barrier. Circ Res (2007) 101:7–9. doi: 10.1161/CIRCRESAHA.107.156513
84. Bisaria A, Hayer A, Garbett D, Cohen D, Meyer T. Membrane-proximal f-actin restricts local membrane protrusions and directs cell migration. Science (2020) 368:1205–10. doi: 10.1126/science.aay7794
86. Ribeiro AL, Okamoto OK. Combined effects of pericytes in the tumor microenvironment. Stem Cells Int (2015) 2015:868475. doi: 10.1155/2015/868475
87. Raza A, Franklin MJ, Dudek AZ. Pericytes and vessel maturation during tumor angiogenesis and metastasis. Am J Hematol (2010) 85:593–8. doi: 10.1002/ajh.21745
88. Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol (2005) 7:452–64. doi: 10.1215/S1152851705000232
89. Lengyel E, Makowski L, DiGiovanni J, Kolonin MG. Cancer as a matter of fat: The crosstalk between adipose tissue and tumors. Trends Cancer (2018) 4:374–84. doi: 10.1016/j.trecan.2018.03.004
90. Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature (2014) 515:577–81. doi: 10.1038/nature13988
91. Mortezaee K, Majidpoor J, Najafi S, Tasa D. Bypassing anti-PD-(L)1 therapy: Mechanisms and management strategies. BioMed Pharmacother (2023) 158:114150. doi: 10.1016/j.biopha.2022.114150
92. Rouzbahani E, Majidpoor J, Najafi S, Mortezaee K. Cancer stem cells in immunoregulation and bypassing anti-checkpoint therapy. BioMed Pharmacother (2022) 156:113906. doi: 10.1016/j.biopha.2022.113906
93. Horn L, Spigel DR, Vokes EE, Holgado E, Ready N, Steins M, et al. Nivolumab versus docetaxel in previously treated patients with advanced non-Small-Cell lung cancer: Two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057). J Clin Oncol (2017) 35:3924–33. doi: 10.1200/JCO.2017.74.3062
94. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res (2020) 39:149. doi: 10.1186/s13046-020-01648-1
95. Takeuchi Y, Nishikawa H. Roles of regulatory t cells in cancer immunity. Int Immunol (2016) 28:401–9. doi: 10.1093/intimm/dxw025
96. Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol (2018) 18:671–88. doi: 10.1038/s41577-018-0061-z
97. Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol (2016) 17:1025–36. doi: 10.1038/ni.3518
98. Luetke-Eversloh M, Cicek BB, Siracusa F, Thom JT, Hamann A, Frischbutter S, et al. NK cells gain higher IFN-gamma competence during terminal differentiation. Eur J Immunol (2014) 44:2074–84. doi: 10.1002/eji.201344072
99. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis (2021) 12:94. doi: 10.1038/s41419-020-03334-8
100. Ke H, Zhang J, Wang F, Xiong Y. ZNF652-induced circRHOT1 promotes SMAD5 expression to modulate tumorigenic properties and nature killer cell-mediated toxicity in bladder cancer via targeting miR-3666. J Immunol Res (2021) 2021:7608178. doi: 10.1155/2021/7608178
101. Yang F, Chen Y, Luo L, Nong S, Li T. circFOXO3 induced by KLF16 modulates clear cell renal cell carcinoma growth and natural killer cell cytotoxic activity through sponging miR-29a-3p and miR-122-5p. Dis Markers (2022) 2022:6062236. doi: 10.1155/2022/6062236
102. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity (2014) 41:14–20. doi: 10.1016/j.immuni.2014.06.008
103. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest (2012) 122:787–95. doi: 10.1172/JCI59643
104. Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol (2015) 33:643–75. doi: 10.1146/annurev-immunol-032414-112220
105. Li H, Luo F, Jiang X, Zhang W, Xiang T, Pan Q, et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer (2022) 10. doi: 10.1136/jitc-2021-004029
106. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomedicine (2021) 16:2803–18. doi: 10.2147/IJN.S284560
107. Wang L, Yi X, Xiao X, Zheng Q, Ma L, Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett (2022) 27:106. doi: 10.1186/s11658-022-00406-9
108. Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol (2019) 16:601–20. doi: 10.1038/s41571-019-0222-4
109. Shaul ME, Fridlender ZG. Cancer-related circulating and tumor-associated neutrophils - subtypes, sources and function. FEBS J (2018) 285:4316–42. doi: 10.1111/febs.14524
110. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer (2020) 20:485–503. doi: 10.1038/s41568-020-0281-y
111. Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell (2009) 16:183–94. doi: 10.1016/j.ccr.2009.06.017
112. Schlesinger M. Role of platelets and platelet receptors in cancer metastasis. J Hematol Oncol (2018) 11:125. doi: 10.1186/s13045-018-0669-2
113. Mammadova-Bach E, Gil-Pulido J, Sarukhanyan E, Burkard P, Shityakov S, Schonhart C, et al. Platelet glycoprotein VI promotes metastasis through interaction with cancer cell-derived galectin-3. Blood (2020) 135:1146–60. doi: 10.1182/blood.2019002649
114. Miao Y, Xu Z, Feng W, Zheng M, Xu Z, Gao H, et al. Platelet infiltration predicts survival in postsurgical colorectal cancer patients. Int J Cancer (2022) 150:509–20. doi: 10.1002/ijc.33816
115. Miao S, Lu M, Liu Y, Shu D, Zhu Y, Song W, et al. Platelets are recruited to hepatocellular carcinoma tissues in a CX3CL1-CX3CR1 dependent manner and induce tumour cell apoptosis. Mol Oncol (2020) 14:2546–59. doi: 10.1002/1878-0261.12783
116. Miao S, Zhang Q, Chang W, Wang J. New insights into platelet-enriched microRNAs: production, functions, roles in tumors, and potential targets for tumor diagnosis and treatment. Mol Cancer Ther (2021). doi: 10.1158/1535-7163.MCT-21-0050
117. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
118. Wang H, Wei X, Wu B, Su J, Tan W, Yang K. Tumor-educated platelet miR-34c-3p and miR-18a-5p as potential liquid biopsy biomarkers for nasopharyngeal carcinoma diagnosis. Cancer Manag Res (2019) 11:3351–60. doi: 10.2147/CMAR.S195654
119. Preusser C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles (2018) 7:1424473. doi: 10.1080/20013078.2018.1424473
120. Oudejans C, Manders V, Visser A, Keijser R, Min N, Poutsma A, et al. Circular RNA sequencing of maternal platelets: A novel tool for the identification of pregnancy-specific biomarkers. Clin Chem (2021) 67:508–17. doi: 10.1093/clinchem/hvaa249
121. Zaslavsky A, Baek KH, Lynch RC, Short S, Grillo J, Folkman J, et al. Platelet-derived thrombospondin-1 is a critical negative regulator and potential biomarker of angiogenesis. Blood (2010) 115:4605–13. doi: 10.1182/blood-2009-09-242065
122. Poulose R, Derda AA, Omar M, von Falck C, Langer F, Tillmanns J, et al. [Uncommon tumor-associated deep vein thrombosis]. Internist (Berl) (2022) 63:545–50. doi: 10.1007/s00108-022-01272-5
123. Fang L, Xu Q, Qian J, Zhou JY. Aberrant factors of fibrinolysis and coagulation in pancreatic cancer. Onco Targets Ther (2021) 14:53–65. doi: 10.2147/OTT.S281251
124. Miao S, Shu D, Zhu Y, Lu M, Zhang Q, Pei Y, et al. Cancer cell-derived immunoglobulin g activates platelets by binding to platelet FcgammaRIIa. Cell Death Dis (2019) 10:87. doi: 10.1038/s41419-019-1367-x
125. Vismara M, Manfredi M, Zara M, Trivigno SMG, Galgano L, Barbieri SS, et al. Proteomic and functional profiling of platelet-derived extracellular vesicles released under physiological or tumor-associated conditions. Cell Death Discovery (2022) 8:467. doi: 10.1038/s41420-022-01263-3
126. Faria AVS, Andrade SS, Peppelenbosch MP, Ferreira-Halder CV, Fuhler GM. Platelets in aging and cancer-"double-edged sword". Cancer Metastasis Rev (2020) 39:1205–21. doi: 10.1007/s10555-020-09926-2
127. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
128. Seretis C, Seretis F, Lagoudianakis E, Gemenetzis G, Salemis NS. ) is red cell distribution width a novel biomarker of breast cancer activity? data from a pilot study. J Clin Med Res (2013) 5:121–6. doi: 10.4021/jocmr1214w
129. Takeuchi H, Abe M, Takumi Y, Hashimoto T, Miyawaki M, Okamoto T, et al. Elevated red cell distribution width to platelet count ratio predicts poor prognosis in patients with breast cancer. Sci Rep (2019) 9:3033. doi: 10.1038/s41598-019-40024-8
130. Helwa R, Heller A, Knappskog S, Bauer AS. Tumor cells interact with red blood cells via galectin-4 - a short report. Cell Oncol (Dordr) (2017) 40:401–9. doi: 10.1007/s13402-017-0317-9
131. Pereira-Veiga T, Bravo S, Gomez-Tato A, Yanez-Gomez C, Abuin C, Varela V, et al. Red blood cells protein profile is modified in breast cancer patients. Mol Cell Proteomics (2022) 21:100435. doi: 10.1016/j.mcpro.2022.100435
132. Alhasan AA, Izuogu OG, Al-Balool HH, Steyn JS, Evans A, Colzani M, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood (2016) 127:e1–e11. doi: 10.1182/blood-2015-06-649434
133. Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) (2017) 95:1179–89. doi: 10.1007/s00109-017-1582-9
134. Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res (2018) 46:8168–80. doi: 10.1093/nar/gky721
135. Nicolet BP, Jansen SBG, Heideveld E, Ouwehand WH, van den Akker E, von Lindern M, et al. Circular RNAs exhibit limited evidence for translation, or translation regulation of the mRNA counterpart in terminal hematopoiesis. RNA (2022) 28:194–209. doi: 10.1261/rna.078754.121
136. Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost (2017) 117:948–61. doi: 10.1160/TH16-11-0873
137. Humar R, Schaer DJ, Vallelian F. Erythrophagocytes in hemolytic anemia, wound healing, and cancer. Trends Mol Med (2022) 28:906–15. doi: 10.1016/j.molmed.2022.08.005
138. Lang E, Bissinger R, Qadri SM, Lang F. Suicidal death of erythrocytes in cancer and its chemotherapy: A potential target in the treatment of tumor-associated anemia. Int J Cancer (2017) 141:1522–8. doi: 10.1002/ijc.30800
139. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (2006) 312:1355–9. doi: 10.1126/science.1124234
140. Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol (2010) 107:243–74. doi: 10.1016/B978-0-12-381300-8.00008-3
141. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148:1258–70. doi: 10.1016/j.cell.2012.01.035
142. Pickard JM, Zeng MY, Caruso R, Nunez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev (2017) 279:70–89. doi: 10.1111/imr.12567
143. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342:967–70. doi: 10.1126/science.1240527
144. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol (2017) 14:356–65. doi: 10.1038/nrgastro.2017.20
145. Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med (2022) 28:690–703. doi: 10.1038/s41591-022-01779-2
146. Zhao H, Wang D, Zhang Z, Xian J, Bai X. Effect of gut microbiota-derived metabolites on immune checkpoint inhibitor therapy: Enemy or friend? Molecules (2022) 27. doi: 10.3390/molecules27154799
147. Zhu W, Miyata N, Winter MG, Arenales A, Hughes ER, Spiga L, et al. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J Exp Med (2019) 216:2378–93. doi: 10.1084/jem.20181939
148. Zitvogel L, Galluzzi L, Viaud S, Vetizou M, Daillere R, Merad M, et al. Cancer and the gut microbiota: an unexpected link. Sci Transl Med (2015) 7:271ps1. doi: 10.1126/scitranslmed.3010473
149. Zhu R, Lang T, Yan W, Zhu X, Huang X, Yin Q, et al. Gut microbiota: Influence on carcinogenesis and modulation strategies by drug delivery systems to improve cancer therapy. Adv Sci (Weinh) (2021) 8:2003542. doi: 10.1002/advs.202003542
150. Najafi S. Circular RNAs as emerging players in cervical cancer tumorigenesis; a review to roles and biomarker potentials. Int J Biol Macromol (2022) 206:939–53. doi: 10.1016/j.ijbiomac.2022.03.103
151. Luo J, Li Y, Zheng W, Xie N, Shi Y, Long Z, et al. Characterization of a prostate- and prostate cancer-specific circular RNA encoded by the androgen receptor gene. Mol Ther Nucleic Acids (2019) 18:916–26. doi: 10.1016/j.omtn.2019.10.015
152. Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-seq profiling of serum exosomal circular RNAs reveals circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol (2020) 10:982. doi: 10.3389/fonc.2020.00982
153. Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet (2019) 10:1096. doi: 10.3389/fgene.2019.01096
154. Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med (2018) 7:2783–91. doi: 10.1002/cam4.1514
155. Hu Y, Song Q, Zhao J, Ruan J, He F, Yang X, et al. Identification of plasma hsa_circ_0008673 expression as a potential biomarker and tumor regulator of breast cancer. J Clin Lab Anal (2020) 34:e23393. doi: 10.1002/jcla.23393
156. Li X, Ma F, Wu L, Zhang X, Tian J, Li J, et al. Identification of Hsa_circ_0104824 as a potential biomarkers for breast cancer. Technol Cancer Res Treat (2020) 19:1533033820960745. doi: 10.1177/1533033820960745
157. Li J, Li H, Lv X, Yang Z, Gao M, Bi Y, et al. Diagnostic performance of circular RNAs in human cancers: A systematic review and meta-analysis. Mol Genet Genomic Med (2019) 7:e00749. doi: 10.1002/mgg3.749
158. Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, et al. Plasma circular RNA panel to diagnose hepatitis b virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer (2020) 146:1754–63. doi: 10.1002/ijc.32647
159. Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ t cell exhaustion in bladder cancer microenvironments. Mol Ther (2022) 30:1054–70. doi: 10.1016/j.ymthe.2022.01.022
160. Li Z, Zhao R, Cui Y, Zhou Y, Wu X. The dynamic change of neutrophil to lymphocyte ratio can predict clinical outcome in stage i-III colon cancer. Sci Rep (2018) 8:9453. doi: 10.1038/s41598-018-27896-y
161. Orditura M, Galizia G, Diana A, Saccone C, Cobellis L, Ventriglia J, et al. Neutrophil to lymphocyte ratio (NLR) for prediction of distant metastasis-free survival (DMFS) in early breast cancer: a propensity score-matched analysis. ESMO Open (2016) 1:e000038. doi: 10.1136/esmoopen-2016-000038
162. Wu CN, Chuang HC, Lin YT, Fang FM, Li SH, Chien CY. Prognosis of neutrophil-to-lymphocyte ratio in clinical early-stage tongue (cT1/T2N0) cancer. Onco Targets Ther (2017) 10:3917–24. doi: 10.2147/OTT.S140800
163. Xu X, Tao R, Sun L, Ji X. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int (2020) 20:552. doi: 10.1186/s12935-020-01642-9
164. Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis (2019) 10:792. doi: 10.1038/s41419-019-2028-9
165. Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol (2020) 235:6929–41. doi: 10.1002/jcp.29589
166. Ghafouri-Fard S, Najafi S, Hussen BM, Basiri A, Hidayat HJ, Taheri M, et al. The role of circular RNAs in the carcinogenesis of bladder cancer. Front Oncol (2022) 12:801842. doi: 10.3389/fonc.2022.801842
167. Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med (2019) 11:55. doi: 10.1186/s13073-019-0663-5
168. Luo Y, Gui R. Circulating exosomal CircMYC is associated with recurrence and bortezomib resistance in patients with multiple myeloma. Turk J Haematol (2020) 37:248–62. doi: 10.4274/tjh.galenos.2020.2020.0243
169. Liu YT, Han XH, Xing PY, Hu XS, Hao XZ, Wang Y, et al. Circular RNA profiling identified as a biomarker for predicting the efficacy of gefitinib therapy for non-small cell lung cancer. J Thorac Dis (2019) 11:1779–87. doi: 10.21037/jtd.2019.05.22
170. Chen T, Luo J, Gu Y, Huang J, Luo Q, Yang Y. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer (2019) 10:930–41. doi: 10.1111/1759-7714.13032
171. Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol (2020) 31:e75. doi: 10.3802/jgo.2020.31.e75
172. Byun DJ, Wolchok JD, Rosenberg LM, Girotra M. Cancer immunotherapy - immune checkpoint blockade and associated endocrinopathies. Nat Rev Endocrinol (2017) 13:195–207. doi: 10.1038/nrendo.2016.205
173. Gyoba J, Shan S, Roa W, Bedard EL. Diagnosing lung cancers through examination of micro-RNA biomarkers in blood, plasma, serum and sputum: A review and summary of current literature. Int J Mol Sci (2016) 17:494. doi: 10.3390/ijms17040494
174. Max KEA, Bertram K, Akat KM, Bogardus KA, Li J, Morozov P, et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci USA (2018) 115:E5334–43. doi: 10.1073/pnas.1714397115
175. Parpart-Li S, Bartlett B, Popoli M, Adleff V, Tucker L, Steinberg R, et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res (2017) 23:2471–7. doi: 10.1158/1078-0432.CCR-16-1691
176. Ward Gahlawat A, Lenhardt J, Witte T, Keitel D, Kaufhold A, Maass KK, et al. Evaluation of storage tubes for combined analysis of circulating nucleic acids in liquid biopsies. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20030704
177. Ferrero G, Licheri N, Coscujuela Tarrero L, De Intinis C, Miano V, Calogero RA, et al. Docker4Circ: A framework for the reproducible characterization of circRNAs from RNA-seq data. Int J Mol Sci (2019) 21. doi: 10.3390/ijms21010293
178. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell (2019) 176:869–881 e13. doi: 10.1016/j.cell.2018.12.021
179. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet (2016) 17:679–92. doi: 10.1038/nrg.2016.114
180. Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol (2020) 21:30. doi: 10.1186/s13059-020-1935-5
181. Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol (2019) 37:1155–62. doi: 10.1038/s41587-019-0217-9
Keywords: circulating circRNAs, cancer, cell communication, diagnosis, therapy
Citation: Miao S and Zhang Q (2023) Circulating circRNA: a social butterfly in tumors. Front. Oncol. 13:1203696. doi: 10.3389/fonc.2023.1203696
Received: 11 April 2023; Accepted: 20 June 2023;
Published: 21 July 2023.
Edited by:
Brian D. Adams, Brain Institute of America, United StatesReviewed by:
Shahram Arsang-Jang, Zanjan University of Medical Sciences, IranZhaofeng Liang, Jiangsu University, China
Sajad Najafi, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2023 Miao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingsong Zhang, UWluZ3NvbmczMzMzQDE2My5jb20=
 Shuo Miao1,2
Shuo Miao1,2 Qingsong Zhang
Qingsong Zhang