
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 17 August 2023
Sec. Breast Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1198259
The mammary microbiome is a newly characterized bacterial niche that might offer biological insight into the development of breast cancer. Together with in-depth analysis of the gut microbiome in breast cancer, current evidence using next-generation sequencing and metabolic profiling suggests compositional and functional shifts in microbial consortia are associated with breast cancer. In this review, we discuss the fundamental studies that have progressed this important area of research, focusing on the roles of both the mammary tissue microbiome and the gut microbiome. From the literature, we identified the following major conclusions, (I) There are unique breast and gut microbial signatures (both compositional and functional) that are associated with breast cancer, (II) breast and gut microbiome compositional and breast functional dysbiosis represent potential early events of breast tumor development, (III) specific breast and gut microbes confer host immune responses that can combat breast tumor development and progression, and (IV) chemotherapies alter the microbiome and thus maintenance of a eubiotic microbiome may be key in breast cancer treatment. As the field expectantly advances, it is necessary for the role of the microbiome to continue to be elucidated using multi-omic approaches and translational animal models in order to improve predictive, preventive, and therapeutic strategies for breast cancer.
Breast cancer, apart from nonmelanoma skin cancer, is the most common cancer in women and affects approximately 12% of all women during their lifetimes (1, 2). Characterization of the mammary and gut microbiomes in breast cancer is well underway but much regarding the interaction of these microbiomes and their hosts begs to be clarified, with the breast tissue ecological niche only recently described (3, 4). Most recent research on the breast and gut microbiome suggests dysbiosis, an imbalance in the microflora, which may preclude the development of breast tumors (4–7). However, the mechanisms linking the microbiome to breast tumor development are still under investigation and the directionality of this relationship has yet to be resolved. Does the microbiome instigate breast cancer or is the changing tumor microenvironment responsible for alterations in microbial composition? How does the microbiome interact with its host to progress or prevent tumor development? In this review, we discuss the foundational and fundamental studies that have progressed these important areas of research, focusing on the roles of both the mammary tissue and gut microbiomes in breast cancer initiation, progression, and response to therapeutics.
Researchers recently characterized the microbiome within human breast tissue, identifying significant differences in bacterial composition in healthy tissue relative to tumor tissue (Table S1) (3, 8–10). Notably, the identification of microbes within the classical tumor environment is a replicated phenomenon (11, 12), with specific intracellular bacteria being implicated in a variety of unique tumor-type environments, including Fusobacterium nucleatum, a microbe associated with epigenetic changes in the cancer environment, in pancreatic and breast tumors, the pathogenic and clinically-relevant Enterobacter cloacae in pancreatic, breast and glioblastoma multiforme tumors, and Citrobacter freundii, known to cause urinary tract infections, diarrhea, pneumonia, and meningitis and intracranial abscesses, in pancreatic, lung, and breast tumors (13, 14). Even inter-tumor microbiome signatures can differ in microbiome composition. One group used whole genome and transcriptome amplification and pan-pathogen microarrays to compare the microbiome within tumor subtypes, i.e., groups of tumors that are different according to histological characteristics of their respective tissues, cancer cell features, or genetic alterations (15). The researchers used hierarchical clustering to group tissues based on their microbiome signatures derived from these multi-omics techniques, with common differential organisms including increased Actinomyces, Bartonella, Brevundimonas, Coxiella, Mobiluncus, Mycobacterium, Rickettsia, and Sphingomonas within tumor tissues as compared to non-cancerous control tissue (15).
More recent studies are also beginning to elucidate breast cancer-associated bacterial dysbiosis in relation to breast cancer subtypes and breast cancer stages between populations of different races/ethnicities. This is reflective of the difference in incidence and mortality within these populations as well, as White women are more likely than Black or Asian women to develop breast cancer, but Black women experience a higher rate of mortality and worse severity of disease (16). A study comparing non-Hispanic Black and White women emphasized the complexity of the tumor microbiota (17). 16S ribosomal RNA (rRNA) gene amplicon sequencing was conducted on normal tissue (n = 8), normal adjacent tissue (normal pairs, n = 11), and breast tumors (n = 64), with a total of 13 stage 1, 24 stage II, and 19 stage III and IV breast cancer tissues analyzed by 4 subtypes of luminal A, luminal B, human epidermal growth factor receptor 2 (HER2), and triple negative breast cancer (TNBC), from non-Hispanic Black and non-Hispanic White women (17). Results from this study showed that Proteobacteria was the most abundant phylum present within all analyzed tissues, while Firmicutes, Bacteroidetes, and Actinobacteria, were less abundant (17). Regarding specific stages, the family Ruminococcaceae and genus Hyphomicrobium were abundant in stage 1 breast tumors, while stage 2 breast tumors contained increased genus Sporosarcina and stage 3 and 4 breast tumors showed abundance of only genus Bosea (17). Tumor tissues from Black women also showed a higher abundance of the genus Ralstonia as compared to those from White women (17). One study of Mediterranean women identified Ralstonia as a key breast-cancer-associated bacterium (18). Species within Ralstonia have markedly been implicated in various diseases, including nosocomial bloodstream infection and bacteraemia (19, 20), highlighting this bacterial group as a potential breast cancer pathogen.
A key factor supporting the colonization of bacteria in the tumor microenvironment is that this microenvironment may offer essential nutrition and oxygen for bacterial survival (21). In this way, the tumor microenvironment can provide a niche for microbial populations, but these microbial populations may also negatively respond to the enhanced metabolic activity of host tumor cells, leading to bacterial dysbiosis in breast tissue following a breast cancer diagnosis (7). Moreover, the relationship between breast tissue bacteria and cancer cells may be bidirectional; dysbiosis in breast tissue appears to support tumor progression, and tumor progression also appears to negatively affect breast tissue bacteria, suggesting the need for further research to clarify the directionality of this relationship.
Characterization of the functional role of the local tumor microbiota in breast cancer is well underway. In the next sections we discuss host-breast tissue microbiome interactions in breast cancer. Furthering the concept of a bidirectional relationship between the host and mammary microbiome, we identified studies that highlight the microbiome’s influence on host cell proliferation, activity, and death, host DNA damage, and host immune function. We also identified studies that emphasize the metabolic response of the microbiome to breast tumor development.
Researchers have observed that tumor tissue expresses lower basal levels of antibacterial response gene expression as compared to healthy breast tissue (22). However, there are a number of studies in which breast tissue microbes associate and potentially instigate changes in host cell genetic programming and cell-cycle progression, either attenuating or promoting tumor development.
Hassan et al. investigated the potential of live, heat-killed cells and the cytoplasmic fractions of Enterococcus faecalis and Staphylococcus hominis as anti-breast cancer agents (23). They used human breast cancer cell lines with estrogen, progesterone and glucocorticoid receptors, and non-malignant epithelial cell lines and treated the cells with varying amounts of each of the live, heat-killed microbes, and the cytoplasmic fractions of the bacteria (23). They evaluated cytotoxicity using the MTT assay, a colorimetric assay for assessing cell metabolic activity, morphological features of the treated cells by fluorescence microscopy, and the stage of cell cycle arrest and apoptosis by flow cytometry (23). Notably, the authors report that all three forms of the bacteria caused a significant decrease in cancer cell proliferation in a concentration- and time-dependent manner, with morphological features of apoptosis (cell death, cell shrinkage and membrane blebbing) observed, and little to no effect on normal cells (23). This suggests that these bacteria can be used as an alternative nutraceutical for breast cancer because of their non-cytotoxic effects on normal cells.
Esfandiary et al. explored the role of Lactobacilli in breast cancer, specifically in relation to that of the human Hypoxia-Inducible Factor (HIF)-1 (24). HIF is a major player in the body’s response to low oxygen concentrations and regulates the expression of genes implicated in homeostasis, vascularization, anaerobic metabolism, and immunological responses; its increase is also associated with increased proliferation and more aggressive breast tumor development (24). The authors analyzed the expression of HIF-1α, SLC2A1, VHL, HSP90, XBP1, and SHARP1 genes from the HIF pathway in triple-negative breast cancer line cells before and after treatment with Lactobacillus crispatus and Lactobacillus rhamnosus culture supernatants by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (24). The Lactobacillus spp. were cytotoxic to the cancer cell line and down-regulated the expression levels of the measured genes from the HIF pathway (24). This analysis indicates an important interaction and communication between commensal microbiota and host pathways involved in the homeostatic regulation of human cells.
Conversely, other groups report that specific bacteria instigate tumor growth. In a mouse study, researchers report increasing Gal-GalNAc levels as human breast cancer progresses, and that occurrence of Fusobacterium nucleatum genomic DNA in breast cancer samples correlates with high Gal-GalNAc levels in breast cancer cells (25). Moreover, they demonstrate that inoculation with F. nucleatum in the breast suppresses the accumulation of tumor-infiltrating T cells and promotes tumor growth and metastatic progression, the latter two of which could be counteracted by antibiotic treatment in mice (25). In a large cohort study, researchers integrated 16S rRNA sequencing with host tumor expression profiles in 668 breast tumor tissues and 72 non-cancerous adjacent tissues (26). They observed an increase of Proteobacteria in tumor tissues, an increase of Actinobacteria in non-cancerous adjacent tissues, and, most interestingly, an association between gene set enrichment of Listeria spp. and expression profiles of host genes involved in with epithelial to mesenchymal transitions (26). The study also observed that H. influenza correlated with genes in the G2M checkpoint, E2F transcription, and mitotic spindle assembly pathways, while L. fleischmannii associated with genes involved in epithelial to mesenchymal transition in breast tumors (26). S. pyogenes also correlated with GUSBP4, GUSBP9, and GPA2 expression levels, implicating the microbe in the glucuronidation of estrogen and, indirectly, potential cell-cycle progression due to upregulation of estrogen (26).
This research suggests similar findings to the microbiome’s influence in other facets of health. There are certain bacteria that appear pathogenic in breast cancer, supporting breast tumor growth and progression, and there are those which appear to protect against breast tumor development.
Urbaniak et al. were among the first to report a distinct breast tissue microbiome and to distinguish the microbiome compositionally between healthy women and women with breast cancer (3). However, this study also reports the ability of mammary microbiome species, E. coli and Staphylococcus epidermidis, isolated from women with breast cancer, to induce DNA-double stranded breaks in Henrietta Lacks (HeLa) cells, alluding to another potential pathogenic mechanism (3). Microbial interference in host genetic material may lead to the later dysregulation of host cell cycles and, eventually, the development of breast cancer. Next we will explore the current research linking the mammary microbiome to host immune modulation.
Importantly, the largest study to date looking at the human mammary microbiome utilized 16S rRNA gene sequencing to characterize the microbiome of human breast tissue, studying a total of 221 patients with breast cancer, 18 individuals predisposed to breast cancer, referring to individuals with a genetic predisposition via a pathogenic gene carrier, first-degree relative with breast cancer, or past personal history of breast cancer, and 69 controls (8). This group identified decreased alpha diversity and altered microbiota composition in tumor and high risk tissues relative to controls and adjacent normal tissues (8). Expanding beyond compositional analysis, this study also developed microbiome-immune networks, correlating microbial profiles with host immune cell populations of different breast tissues, e.g., from healthy women, women at high risk of breast cancer, and women with active breast cancer (8). They observed a more disconnected immune-microbiome network structure in tumor tissues compared to benign tissue (healthy control and high risk tissues) (8). They also reported that tumor-depleted taxa, Streptococcus and Propionibacterium, were positively correlated with T-cell activation-related genes (8). This study suggests possible tumor-suppression via immune modulation and supports analysis of the breast tissue microbiome as a biomarker of breast cancer risk. In the next section, we explore further the bidirectional relationship between host and microbes, discussing the association of breast cancer with altered microbiome metabolic capacity.
Recent studies have investigated the metabolic function of the breast tissue microbiome using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt or PICRUSt2, most recent version) (27) These studies have identified a diverse array of metabolic contributions and responses of the microbiome in breast cancer. For example, one group conducted 16S rRNA sequencing on aseptically collected breast tissue from women with benign (n = 13) or malignant breast tumors (n = 15), identifying the breast tissue microbiome as distinguishable from skin or buccal cells and defining key bacterial genera, namely, Fusobacterium, Atopobium, Gluconacetobacter, Hydrogenophaga and Lactobacillus, in malignant cancerous tissue (28). This group also used PICRUSt to elucidate a decrease in inositol phosphate microbial metabolism in malignant cancer samples (28). Further, they identified a decrease in cysteine and methionine metabolism, glycosyltransferase, fatty acid biosynthesis, and C5-branched dibasic acid metabolism from the microbiome in these tissues (28). This is especially relevant to breast cancer as studies have identified cysteine and methionine as important and targetable players in cancer biology and glycosyltransferase gene profiles as prognostic biomarkers (29–31).
One group identified functional changes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, also inferred by PICRUSt from 16S rRNA gene amplicon sequencing, in the breast microbiome of 22 benign and 72 malignant breast cancer patients aseptically collected using a needle biopsy (32). Their analysis of the breast tissue functional bacteriome in relation to breast cancer showed upregulation of glycerophospholipid biosynthesis and ribosome biosynthesis processes (32). Conversely, flavonoid biosynthesis decreased as breast cancer grade worsened (32).
Notably, our research group identified a unique breast tissue bacterial compositional signature in pre-diagnostic tissue (n = 15), healthy tissue collected before the women were diagnosed with breast cancer, relative to healthy controls (n = 50), normal adjacent tissue (n = 49), and tumor tissue (n = 46) (7). This signature was enhanced in tumor and adjacent normal tissues (n = 49 and n = 46, respectively), suggesting bacterial dysbiosis as an early event in breast tumor development (7). We applied PICRUSt2 on the 16S rRNA amplicon sequences from these tissues and observed significant metabolic dysregulation in tissues from women diagnosed with breast cancer (pre-diagnostic, tumor, and adjacent normal), suggesting Warburg metabolism of the microbiome and a potential response of the breast tissue microbiome to a changing microenvironment during breast tumor development (7). Further, integral within the host-microbiota interactions discussed previously, we conducted preliminary correlation analyses between host transcriptome profiling and the microbial taxa and functionally-annotated genes in healthy and prediagnostic tissues (7). We identified altered associations between the host transcriptome and mammary bacterial taxa and functional bacterial KEGG orthologs in prediagnostic tissue compared with healthy tissue (7).
Characterization of the metabolic output of the bacteriome using tools such as PICRUSt and PICRUSt2 are resourceful methods to begin establishing breast tissue host-microbiome interactions in breast cancer (27). This may be expanded upon in future studies using shotgun metagenomic sequencing to evaluate the genes within the tissue as compared to those that are predicted to be present. Regardless, these studies implicate microbial functional dysbiosis in breast cancer, with most recent studies suggesting a bacterial response to the changing tumor microenvironment.
Altogether, the current literature suggests that breast tissue microbial compositional and functional dysbiosis are associated with and may be early events of breast cancer (4–7, 17, 32). Further, certain bacterial taxa act as pathogenic (e.g. Staphylococcus epidermidis, Ralstonia spp) (17, 18) or probiotic organisms (e.g. Lactobacillus) in breast tumor development (7, 17). The literature also suggests that these pathogenic or probiotic species can interact with the host in a variety of contexts; modulating the immune system to prevent tumor growth or enhancing cell-cycle progression, promoting tumor growth (8, 24–26). However, more research in these areas is necessary to corroborate these findings and identify potential bacterial species that could be used as microbial therapeutics or biomarkers for this disease. There is also much to be elucidated regarding host-microbiome interactions in the local breast tissue microenvironment that dictate breast tumor initiation. Future studies should also seek to elucidate the means by which bacteria identified in population-level studies alter or are altered by the formation of breast tumors in patients by coupling these clinical studies with translational animal models that can establish mechanisms. Additionally, it will be prudent to also consider the microbiomes of other body sites as they relate to breast cancer. The next section of this review focuses on the gut-breast cancer axis, highlighting studies key in linking the gut microbiome to breast cancer development.
Another facet of breast cancer microbiome research involves gut bacteria. The importance of this microbial consortium in human health has long since been established (33). In the next few sections, we outline the current perspectives surrounding the role of the gut microbiome in breast cancer, highlighting studies that characterize the gut-breast axis in both mice and humans, and the roles of gut-derived microbial metabolites and the estrobolome in breast cancer risk (Table S2). Though there is mounting evidence that gut bacteria can and do influence breast tumor development, which bacteria are the most influential and the mechanisms by which they promote or prevent tumor initiation are still under investigation.
Clinical studies applying next-generation sequencing and other genomics strategies are key in identifying microbial candidates for translational animal models and defining associations between common risk factors of breast cancer and gut microbial dysbiosis. Luu et al. applied 16S rRNA sequencing together with qPCR and characterized fecal samples from breast cancer patients in various stages of the disease (34). They observed differences in C. coccoides, F. prausnitzii, and Blautia, short-chain fatty acid (SCFA)-producing bacteria that have been associated with beneficial health outcomes (35, 36), according to the clinical stages of breast cancer and the histo-prognostic grades of corresponding donors (34). Zhu et al. reported on an analysis of the gut microbiomes of 18 premenopausal breast cancer patients, 25 premenopausal healthy controls, 44 postmenopausal breast cancer patients, and 46 postmenopausal healthy controls through shotgun metagenomics (37). They observed higher microbial diversity within breast cancer patients and 45 differential species between postmenopausal patients and controls, e.g., 38 species, including Escherichia coli, were enriched in breast cancer patients while 7, including Eubacterium and Lactobacillus spp., were depleted (37). They also noted changes in the functional genetic potential within breast cancer patients typified in the enrichment of lipopolysaccharide (LPS) biosynthesis, iron complex transport system, phosphotransferase system, secretion system, and beta-oxidation genes (37).
In contrast to this study, another group observed that the Shannon diversity index of the gut microbiome was lower in women with breast cancer as compared to healthy controls (38). This difference may be mediated by a relative enrichment in Firmicutes, as well as a depletion in Bacteroidetes in patients diagnosed with early breast cancer compared to that of healthy women (38). The malignant or benign nature of breast tumors may also be interrelated with the gut microbiome, as Yang et al. have initially begun to explore in a 16S rRNA-sequenced gut microbiome pilot study of 83 women with invasive ductal breast carcinoma and 19 women with benign breast tumors (39). They observed no differences in diversity metrics between the groups but did note significant dysregulation of metabolic pathways in the malignant tumor group, identified via PICRUSt (39). Further, a recent study of the gut microbiome, characterized by 16S rRNA sequencing, of 26 subjects with breast cancer, 20 with benign breast lesions, and 20 matched healthy controls by Ma et al. reported that breast cancer patients had significantly lower alpha diversity indices, with alterations in species, such as higher levels of Porphyromonas and Peptoniphilus spp., in breast cancer patients (40).
Observations of the naturally occurring human gut may elucidate its impact on or change following the development of breast cancer, but there are various perturbations within the gut which have also warranted investigation. For example, the type of breast tumor diagnosed can shape clinical prognosis, provide essential insight into how a patient will be impacted by breast cancer, and determine the mode of treatment (41). Depending on the stage of life patients experience breast cancer, the hormonal levels within the body and gut may also be variable and influence health (42) and microbiota statuses (43). Many patients also require surgical removal of tumors, which is typically accompanied by prophylactic antibiotics and/or chemotherapy, both of which can instigate and or exacerbate microbial compositional alterations (6, 44, 45); we explore these perturbations later in this review. In the next sections, we focus on animal model studies of the gut-breast cancer axis, wherein the mechanisms by which specific pathogenic or probiotic bacteria may influence or inhibit tumor development may be elucidated.
Similar to animal and cell-based studies of the local breast tissue microbiome, animal model studies of the gut microbiota in relation to breast cancer primarily focus on specific bacterial taxa and metabolites present within the gut and how these taxa correlate with either preventive or pathogenic effects (46–50). A number of these studies highlight the immune-modulatory effects of specific bacteria (47–50), characterizing these microbes as potential probiotics to combat breast cancer.
Lakritz et al. tested a mechanistic hypothesis of gut microbiota-modulation of distal breast cancer using Helicobacter hepaticus, a known pathogen (51), in a microbial infection animal model (46). Following orogastric infection of mice with Helicobacter hepaticus, researchers observed subsequent infiltration of myeloperoxidase-positive neutrophils in the mammary tissue and tumorigenesis (46). Tumorigenesis was inhibited by the depletion of these neutrophils, alluding to the important role of intestinal microbial balance in distal tumor development and the cancer promoting inflammatory response (46).
Conversely, certain probiotic bacteria, such as Lactobacillus spp., have been heavily studied in mouse models and the immunomodulatory capabilities of these species further elucidated in breast cancer. One study used FVB strain erbB2 (HER2) mutant mice with genetic prepositions to mammary tumors and found that when supplied with Lactobacillus reuteri to the gut, the mouse immune system triggered CD4+CD45RBloCD25+ lymphocyte (Treg cells) protective mechanisms to inhibit cancer progression (47).
Further, oral administration of L. acidophilus to Balb/C female mice was found to induce a decrease in breast tumor growth patterns and altered production of interferon (IFN)-γ, interleukin (IL)-4, transforming growth factor (TGF)-β, and lymphocyte proliferation, favoring antitumor immunity and reducing tumor growth (52). Dallal et al. studied the effects of oral administration of L. casei on natural killer cell cytotoxicity and production of cytokines in the spleen cell culture of BALB/c mice with concurrent invasive ductal carcinoma (n = 30 female in-bred BALB/c mice, divided into two groups of test and control each containing 15 mice) (48). Their results showed that oral administration of L. casei increased the production of IL-12 and IFN-γ, increased natural killer cell cytotoxicity in spleen cell culture, prolonged survival, and decreased the growth rate of tumors in the test mice (48). Building upon this, Yazdi et al. reported that Lactobacillus brevis treatment is more effective when the bacteria is combined with biogenic immunomodulating selenium nanoparticles (SeNPs) (n = 60 female inbred BALB/c mice, divided into 4 groups–either given oral PBS daily and injected by this buffer after tumor induction (control), given 100 μg/day of SeNPs as an oral supplement for 30 days, given no supplementation of SeNPs and injected with 4T1 cell crude antigens, or supplemented 100 μg/day SeNPs for 30 days and simultaneously injected with the crude antigens–and each n = 15 mice) (49). The authors identified increased levels of IFN-γ, IL-17, natural killer cytotoxicity, extended life span, and decrease in the tumor metastasis to the liver in SeNP-enriched L. brevis administered mice as compared to control mice or mice given L. brevis alone (49). This is most likely mediated by the SeNPs’ ability to directly interact with innate immune cells such as dendritic cells, macrophages, and natural killer cells and regulate innate immunity (53). Finally, one study focused on the antitumor effects of Lactobacillus by studying immune cells in mammary glands and the cytokine concentration in serum of mice fed with milk fermented by Lactobacillus helveticus (50). The mice were fed for 7 days prior to being injected or not with breast tumor cells, and then fed milk 4 days following injection (50). The researchers measured immunoglobulin (Ig) A, CD4, CD8, cytokines and Bcl-2 positive cells in mammary glands, and cytokines in serum (50). They found that mice fed with L. helveticus R389 fermented milk had a modulated immune response, including increased IgA and CD4 positive cells in mammary glands, increased anti-inflammatory IL-10, and decreased inflammatory IL-6 (associated with poor breast cancer prognostic outcomes), indicating that L. helveticus has a regulatory role in the breast tissue environment (50). The authors concluded that milk fermented by L. helveticus R389 could be used as an oral immune adjuvant to protect against mammary gland pathologies such as cancer (50). This study alludes to the use of dietary interventions, which may alter the gut microbiome composition and influence protection from or progression of breast tumors.
Murine models can also be used to incorporate a carefully controlled diet to assess its role in the gut-breast cancer axis. Ma et al. studied the effect of the combination of berberine, a chemical found in plants such as grapes, turmeric, and goldthread, and exercise as an anti-tumor treatment in mice (54). Together with a significantly slowed progression of breast cancer in 4T1 tumor-bearing mice, they observed significantly increased levels of SCFAs, microbial-derived metabolites, in mouse gastrointestinal tracts, which may influence general inflammation (54).
Lakritz et al. used two different mouse models to analyze the development of mammary cancer when eating a Westernized diet as compared to a genetic predilection to breast cancer, with the additional variable of Lactobacillus supplementation (47). Mammary carcinogenesis was inhibited by routine exposure to Lactobacillus reuteri ATCC-PTA-6475 in drinking water in the Westernized diet model (47). The second model (FVB strain erbB2 (HER2) mutant mice, genetically susceptible to mammary tumors fed regular chow) showed that oral supplementation with L. reuteri was also sufficient to inhibit features of mammary neoplasia (47). Specifically in relation to the development of a breast tumor itself, researchers determined the protective mechanism to be microbially-triggered CD4+CD25+ lymphocytes; the transplantation of these cells to other murine subjects generated similar results for the inhibition of mammary neoplasia and tumors (47). Finally, Zamberi et al. studied the effects of kefir, a cultured product containing probiotics, made from kefir grains cultured in Malaysia as an anti-inflammatory, immunomodulatory, and anticancer treatment (55). They first treated 4T1 cancer cells with kefir water in vitro to assess its effects; they then injected BALB/c mice with the cancer cells and treated them orally with kefir water for 28 days (55). Remarkably, the kefir water was cytotoxic toward the cancer cells at half-maximal inhibitory concentration of 12.5 and 8.33 mg/mL and a reduction in tumor size, reduction in weight, and a substantial increase in helper T cells and cytotoxic T cells were observed in the kefir water-treated group (55)
The animal studies described above highlight probiotic and diet-induced host immune responses, which are key in combating and preventing breast tumor growth and proliferation. These represent critical pathways to explore further in clinical trials of the microbiome and breast cancer.
A significant step forward in gut microbiome-breast cancer studies in humans is the establishment of clinical trials to assess whether the modulatory effects of certain taxa discussed in the previous sections can influence breast cancer development/progression. Recent studies include one by the Mayo Clinic in Jacksonville, Florida (recently completed in March 2023, results pending), evaluating whether engineering the gut microbiome via probiotics, and thereby facilitating a diverse microbiota population, might shape the immune system’s reaction to operable stage I-III breast or lung cancers (56). Another prospective study is still underway by Hackensack Meridian Health, Yale University, and Georgetown University (57). This is a study of newly diagnosed triple negative breast cancer patients undergoing neoadjuvant chemotherapy (57). Researchers are aiming to correlate gut and intratumoral microbiome composition and anti-tumor immune responses (57). Finally, clinical trials testing whether probiotics, containing bacteria such as Lactobacillus, will beneficially affect the immune system during the course of breast cancer have been carried out (58, 59), with pending results. It is exciting that research connecting current translational mouse model studies with human cohort studies is already underway. In the next section, we will discuss additional pathways within the gut-breast cancer axis, the relationship of gut microbial metabolites with breast tumor development and the estrobolome’s influence on breast cancer. Both pathways are relatively underexplored, highlighting additional research areas within this field that may yield key insights in breast cancer development and progression.
The gut microbiome is responsible for numerous metabolites utilized by our host cells as energy sources and immune-modulators. Literature suggests bacterial metabolites, such as bile acids, SCFAs, and cadaverine, as influential in inhibiting breast tumor development and progression.
Studies report the association of lithocholic acid (a bacterial derived bile acid) with Clostridiales spp. and its ability to reduce breast cancer proliferation and vascular endothelial growth factors (60–63). SCFA receptors are also reported to inhibit invasive phenotypes in breast cancer cells via the activation of cognate receptors (64). Sodium butyrate in particular facilitates histone deacetylase inhibition, modulation of glycolytic pathways, pyruvate kinase, lactate dehydrogenase, oxygen-consuming activity, and apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment (65, 66). Similarly, researchers report that SCFA-producing E. coli strains, namely KUB-36 which is a non-exotoxin producer, exhibit simultaneous cytotoxic and anti-inflammatory effects on cancer cells (67).
Indolepropionic acid (IPA) exhibits cytostatic properties and selectively targets breast cancer cells while having no effect on non-transformed, primary fibroblasts (68). IPA reduced the proportion, proliferation, and metastasis of cancer cells by inducing epithelial-to-mesenchymal transition and oxidative stress and facilitated anti-tumor immune activity (68). Akin to the mechanism of IPA, bacteria use diamines, in this case, cadaverine, to buffer the pH of their environment, and cadaverine has been seen to reverse endothelial-to-mesenchymal transition and inhibit cellular movement and invasion in breast cancer cell lines (63).
Altogether, these bacterial metabolites may play early and continuous regulatory roles in the progression of breast tumors. Yet, the origin of many of these metabolites have yet to be fully described or mechanistically interrogated due to the inherently complex interactions between members of the gut microbiota and between the microbiota and human body. Regardless, they represent a key mechanistic connection between the gut microbiome and breast cancer, which should be expanded on in further research.
One final area to be discussed in relation to the gut-breast cancer axis is the gut estrobolome and its potential influence on estrogen regulation as it relates to breast cancer. The functional gut estrobolome is defined as a collection of bacterial genes encoding enzymes essential in estrogen metabolism, which aids in modulating the enterohepatic circulation of estrogens (43). Endogenous estrogens are critically related to the risk of breast cancer in postmenopausal women (69, 70). The hormone receptor–positive (HR+)/HER2–negative subtype is also the most common subtype of breast cancer found in post-menopausal women (71, 72). The activation of estrogen receptors in breast tissues often causes an increase of cells entering the G0 and G1 phases and cell proliferation that is commonly seen in breast cancer patients (73). Thus, in connection with the alterations in bacterial populations seen within breast cancer, host hormonal metabolism changes seen during breast cancer may be due to the shifts in host gut estrobolome diversity and abundance, affecting overall levels of endogenous estrogen (74, 75).
The estrobolome is able to regulate endogenous estrogen through many pathways. One possible pathway is through the abundance modulation of bacterial species that produce β-glucuronidases (GUS) and β-glucuronides (43, 76). Ervin et al. conducted an in vitro study of gut microbial GUS enzymes and found that GUS can reactivate the inactivated estrogen form of estrone-3-glucuronide and estradiol-17-glucuronide to estrone and estradiol (77). The menopause-associated estrobolome dysbiosis in favor of bacterial species that produced GUS likely contributes to a higher estrogen load on the host and, thus, a greater risk for carcinogenesis (69, 70, 77). The estrobolome may also synthesize estrogen-like compounds through the breakdown of normally indigestible dietary fibers and polyphenols that exhibit varied estrogenic potency, contributing to the host’s estrogen load and breast cancer carcinogenesis to varying degrees (75). Little research relating the estrobolome to breast cancer has been conducted and this represents an area of research within the gut-breast cancer axis that begs further exploration. As risk factors such as age at menopause and obesity are closely linked to estrogenic shifts and the gut microbiome, our next section highlights the potential role of the microbiome as a mediator between these risk factors in breast cancer.
Breast cancer is a multifactorial disease, influenced by many environmental and genetic risk factors (78). Significant research has been conducted on the effects of therapeutic exposures, such as prophylactic antibiotics and chemotherapies, on the microbiome in relation to breast cancer. Since the microbiome represents a key component of the tumor microenvironment as well as a potential causal influence on breast tumor development and progression, assessment of this microbial consortia in cancer therapeutic strategies is critical to holistically treat patients with this disease.
Studies have shown that later age at menopause is significant in the development of breast cancer (79, 80). Intricately linked with this connection is age; it is an important factor in extreme variability and phylum proportions in the gut microbiome, potentially due to factors such as lowered nutrition availability to the gut (80–82). Goedert et al. observed decreased diversity at the community level of the gut microbiome using 16S rRNA sequencing data of microbial DNA present in the fecal samples of 48 postmenopausal breast cancer patients as compared to 48 control patients (83). This decrease in diversity was independent of total estrogens, which correlated with α-diversity in control patients (Spearman Rho = 0.37, P = 0.009) but not case patients (Spearman Rho = 0.04, P = 0.77) (83). For premenopausal women, one research group notes a reduction in gut microbiome α-diversity and alterations in β-diversity in breast cancer patients (80). The study reports 14 microbial markers identified in the different menopausal statuses of breast cancer, including Bacteroides fragilis in young women of premenopausal statuses and Klebsiella pneumoniae in older women of postmenopausal statuses (80). Zhu et al. studied 18 premenopausal breast cancer patients, 25 premenopausal healthy controls, 44 postmenopausal breast cancer patients, and 46 postmenopausal healthy controls: they found that the relative abundance of 45 species differed between postmenopausal patients and postmenopausal controls: 38 species were enriched in postmenopausal patients including a number of gram-negative species, and 7 species were less abundant in postmenopausal patients, including gram-positive Eubacterium eligens and Lactobacillus vaginalis (37).
Obesity, which involves low-grade inflammation related to gut dysbiosis, is another factor implicated within the development of breast cancer (84, 85). A systematic review of 12,496 studies up until August 2019 revealed that obese individuals have a greater Firmicutes/Bacteroidetes ratio, as well as less Verrucomicrobia (Akkermansia muciniphila), Faecalibacterium prausnitzii, Bacteroidetes, Methanobrevibacter smithii, and Lactobacillus plantarum and paracasei (86). They concluded that individuals with obesity showed gut microbiota profiles different from lean individuals (86). Houssain et al. conducted a metagenomic study of a syngeneic mouse model of triple-negative breast cancer (87). They found that obesity significantly lowers the alpha diversity of the gut microbiome in this mouse model and that tumor progression was significantly accelerated in obese mice compared to controls, suggesting a relationship between obesity-associated gut dysbiosis and breast cancer progression (87). However, more comprehensive studies are needed to establish a mechanistic relationship between breast cancer and obesity itself.
The risk factors of later age at menopause and obesity clearly affect the human microbiome, but the nuances of these effects need clarification through larger and more technologically extensive studies. Further, it is not yet clear the effects of these risk factors on the local breast tissue microbiome. The presence of certain microbiota alter health outcomes, but their utilization of available resources, metabolic output, and interaction with host cells and other bacterial organisms may provide insight into their role in breast cancer development. Thus, studies exploring the effect of co-morbidities upon differences in microbiota presence, their function, and metabolic output, in combination with breast cancer diagnoses and outcomes, will be essential in the progression and expansion of the field as a whole.
Antibiotics may positively or negatively alter the gut microbiome composition in breast cancer patients (44, 88, 89). Prophylactic antibiotics are often prescribed for breast-cancer associated surgeries (90). But population-based analyses of antibiotic usage in female breast cancer patients report modest positive associations between antibiotic usage and breast cancer development (90, 91). These antibiotics are orally administered and have the potential to directly affect the bacterial populations within the body, which then may affect bacterial modulation of immune and tumor cell development and subsequent cancer treatments (91, 92).
Antibiotic-treated mice injected with tumor cells are also reported to respond poorly to CpG-oligonucleotide treatment and chemotherapy: such mice lack a balanced commensal gut microbiome, have depleted myeloid cell counts, and are less responsive to oxaliplatin, a chemotherapeutic, for tumor regression (45). Notably, McKee et al. report that antibiotic-induced microbiota disturbances promote tumor growth in several breast cancer models, that increased tumor volume positively correlates with stromal mast cell density, and that supplementation with Faecalibacterium rodentium restores tumor growth to normal (93).
An interesting study by Rosean et al. tested how pre-cancer development of dysbiosis within the gut microbiome affects breast cancer (6). They found that the pre-established disruption of commensal microbiota homeostasis via antibiotic gavage (confirmed using 16S rRNA sequencing) resulted in enhanced circulating tumor cells, increased fibrosis and collagen deposition both systemically and locally within the tumor microenvironment, and significant myeloid infiltration into the mammary gland and breast tumor (6).
Interestingly, antibiotic exposure contributes to gut dysbiosis, which is reported to enhance tumor progression and worsen patient responses to cancer therapeutics. Further, per the study by Rosean et. al., antibiotic induced gut dysbiosis may be key in initiation of breast cancer (6). Though prophylactic antibiotics are often necessary prior to surgery, these studies emphasize the need to approach antibiotic exposure as it relates to breast cancer with caution. In the next section, we will discuss the microbiome in cancer therapies, highlighting a fragile relationship between our microbiome and our body’s response to cancer therapeutics.
The microbiome of cancer patients is also affected by chemotherapeutics. A recent study of shotgun metagenomic sequencing of 121 stool samples from 76 breast cancer patients (n = 45 for samples prior to and post-chemotherapy) by Terrisse et al. found that an overabundance of microbiota commensals may negatively influence the outcome and side effects of breast cancer treatments and validated these findings in murine models of breast cancer (94). Similarly, Aarnoutse et al. recently observed changes within the intestinal microbiota composition of 16S rRNA-sequenced stool samples from 44 patients undergoing chemotherapy, with the abundance of Proteobacteria, unclassified Enterobacterales, Lactobacillus, Ruminococcaceae NK4A214 group, Marvinbryantia, Christensenellaceae R7 group, and Ruminococcaceae UCG-005 changing significantly over time and decreased species richness when patients were affected with diarrhea (95). Moreover, in a 4T1 mammary carcinoma experimental mouse model and using 16S rRNA sequencing, researchers were able to investigate the effects of Vismodegib treatment on the gut microbiota (96). The researchers observed remodeling of the gut microbiota and an increase in proliferative CD8+ T cells in the colonic immune network (96).
In some cases, chemotherapies have been reported to instigate tumor-progressing microbiota, while in others, probiotic microbes can enhance tumor-targeting immune responses. Chiba et al. evaluated the effect of neoadjuvant chemotherapy on the human tumor microbiome (97). They used snap-frozen aseptically collected breast tumor tissue from women who underwent neoadjuvant chemotherapy (n = 15) or women with no prior therapy at the time of surgery (n = 18) and performed 16S rRNA sequencing to identify tumor bacterial populations (confirmed with staining of breast tumor microarrays) (97). They found that chemotherapy administration increased Pseudomonas spp. in breast tumors (97). Further, in patients that experience metastases, breast tumors showed increased Brevundimonas and Staphylococcus (97). Finally, they display the direct tumor modulating ability of bacteria via inoculation of tumor cells with Pseudomonas aeruginosa supplemented media and show how this bacterium can modulate doxorubicin (Dox) (chemotherapeutic)-mediated cell death, either enhancing or inhibiting the drug’s effect based upon the cell line under study (97). Metabolites that had consistent effects included LPS, which stimulated cancer cell proliferation, and pyocyanin (a P. aeruginosa-derived metabolite), which potentiated Dox effects and cancer cell death (97). Viaud et al. demonstrated that cyclophosphamide, a class of medications called alkylating agents that slows or stops the growth of cancer cells and suppresses the immune system, alters small intestine microbiota composition in mice, and induces the translocation of specific Gram-positive bacteria into secondary lymphoid organs (98). These bacteria then stimulate the generation of a specific subset of “pathogenic” T helper 17 (pTH17) cells and memory TH1 immune responses, thereby augmenting anti-tumor activity (98). Although antibiotics could disrupt the effectiveness of cyclophosphamide, its efficacy could be restored by Enterococcus hirae and Barnesiella intestinihominis (98).
Finally, Bawaneh et al. recently performed a similar analysis utilizing Dox, an anthracycline treatment that interrupts cancer cell growth by blocking topoisomerase 2, in female BALB/c mice (n = 115) injected with 4T1 cancer cells (99). The researchers treated these mice with Dox in combination with antibiotics, diet-derived fecal microbiota transplantation (FMT), and/or exogenous LPS (these groups were then stratified into Dox responders or Dox nonresponders) (99). One of the most notable results from this experiment was that Dox responders and groups treated with antibiotics in addition to Dox displayed reduced tumor weight and metastatic burden (99). Further, the researchers found that Dox was associated with increased Akkermansia muciniphila (99), a well-studied mucin-degrading bacterium that has been inversely associated with many adverse health outcomes (100). In contrast, when mice were treated with Dox in combination with a high-fat diet-derived FMT, tumor growth continued and Dox was not as effective (99). The researchers found that these reduced Dox effects were interrelated with microbial components of both the gut and plasma through the increase in LPS (99). When LPS was injected in its exogenous form, this was followed by intestinal inflammation, reduced Dox responsiveness, and increased lung metastasis in Dox nonresponding mice and those that were treated with Dox and an FMT (99).
These studies highlight, again, opposing effects of specific bacteria in breast tumor progression and the potential to target specific microbes as breast cancer therapeutics. Further, it is clear that microbiome interaction with host immune functions is critical to the host’s response to chemotherapeutics. In the future, it will be critical to conduct clinical trials that assess the effect of breast-cancer probiotic microbes before and after chemotherapy. These studies should prepare to analyze host immunological responses following probiotic treatment to further elucidate if the microbiome mediates tumor-targeting immunity when undergoing chemotherapy.
In this review, we outline studies analyzing the breast and gut microbiomes in breast cancer initiation and progression, and during chemotherapy. From the literature, we identified the following major conclusions, (I) There are unique breast and gut microbial signatures (both compositional and functional) that are associated with breast cancer, (II) breast and gut microbiome compositional and breast functional dysbiosis represent potential early events of breast tumor development, (III) specific breast and gut microbes confer host immune responses that can combat breast tumor development and progression, and (IV) chemotherapies alter the microbiome and thus maintenance of a eubiotic microbiome may be key in breast cancer treatment (Figure 1).
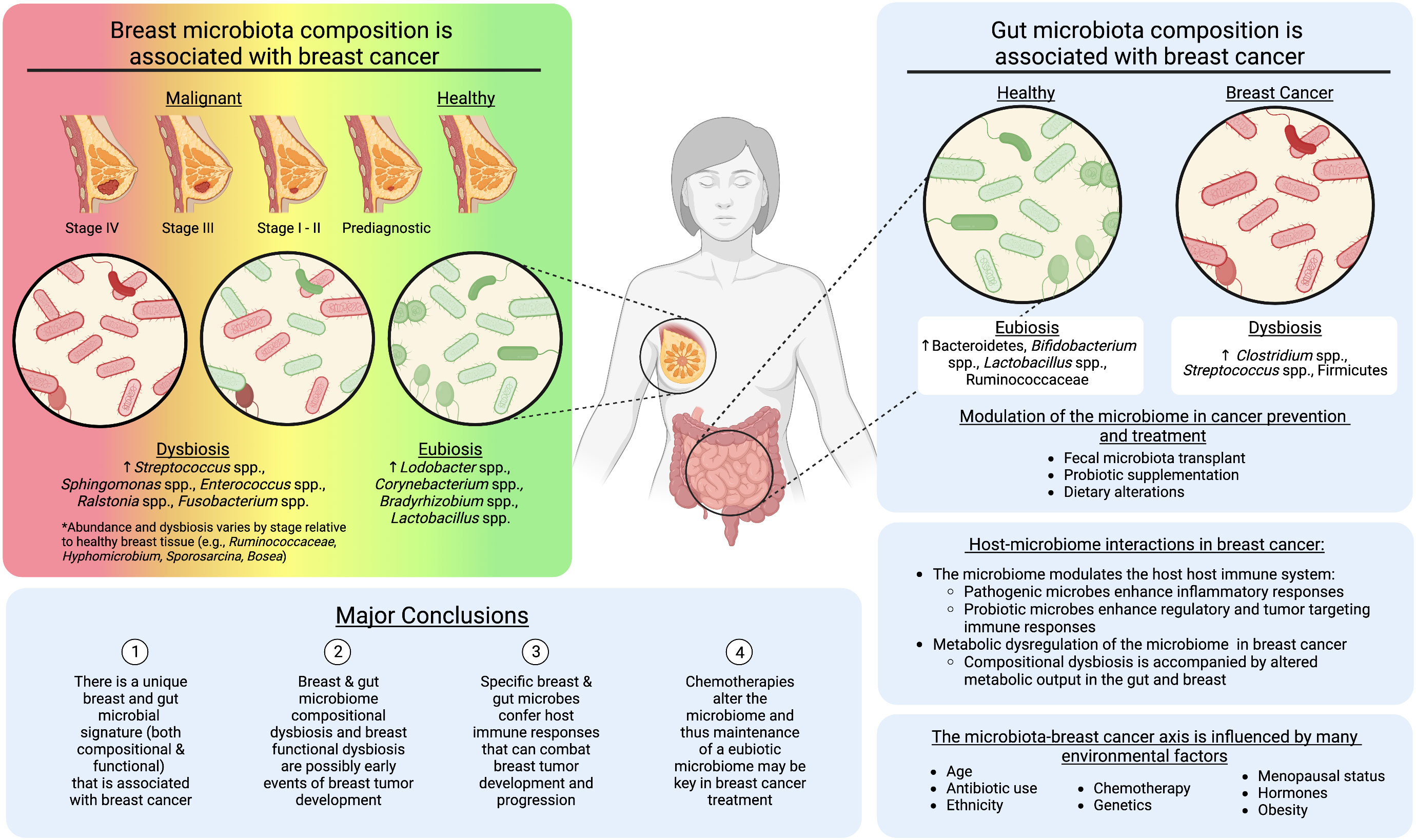
Figure 1 The role of the breast and gut microbiomes in breast cancer development. Created with BioRender.com.
The current research suggests an intricate, bidirectional relationship between the gut and breast microbiomes and breast cancer development. Based on our assessment of the current literature, there is evidence that the breast tissue and gut microbiome composition is altered following breast cancer diagnosis (4–7, 15, 17, 37, 46–50, 61). However, recent work suggests a progression toward enhanced dysbiosis as the breast tumor tissue progresses from early stages, prior to diagnosis, through later stages of breast cancer (6–8, 17) (Figure 1). These compositional shifts are also accompanied by changes in the metabolic output of the microbiome (7, 28, 32, 39). Metabolic dysregulation may be a by-product of compositional dysbiosis, due to the loss of specific bacterial taxa. However, it also suggests the microbiome is in functional disarray due to the changing microenvironment, which along with histological and transcriptomic abnormalities present in pre-diagnostic tissue (101), may be another early event of breast tumor initiation. For example, Marino et al. used microdissection and whole-transcriptome profiling of human breast epithelium prior to and post tumor diagnosis and revealed significant upregulation of genes involved in lipid metabolism, including fatty acid uptake/transport, lipolysis, and lipid peroxidation, which may create a more favorable environment for cancer cell transformation, proliferation, and survival (101). Our recent manuscript characterized the microbiome of pre-diagnostic tissue from some of the same women, highlighting a decreased abundance of microbial lipid metabolism genes, suggesting a response of the microbiome to the changing tumor microenvironment (7). A holistic understanding of the key events that establish and progress breast cancer will be important for the development of cancer therapeutics. However these works also highlight the potential to use bacteria and bacterial metabolites as biomarkers for this disease. Notably, the current work on the pre-diagnostic microbiome is specific to the local breast tissue bacteria (7, 8). It will be critical in future studies to include the breast and gut microbiomes in analyses of the breast tumor microenvironment before and during tumor initiation. This should involve multi-omic strategies within pre-diagnostic tissues, and translational animal models, wherein the microbiome is disrupted prior to breast tumor initiation.
We also determined through this literature review that specific bacterial taxa in the breast and gut are reported as pathogenic, enhancing tumor progression (e.g. Ralstonia spp., Streptococcus epidermidis), or probiotic, stalling tumor progression (e.g. Lactobacillus spp., Corynebacterium spp.) (3, 17, 18, 49, 50). Additionally, though the local and gut microbiomes interact with the host in a variety of ways, the current literature suggests that host immune-modulation is the most influential pathway by which microbes influence breast tumor progression or inhibition (8, 46–49). The majority of studies where immune factors have been assessed involve the gut microbiome (46–49), and thus there is a need to expand breast tissue microbiome studies to include host immune characteristics. Reported evidence that specific microbial taxa can be both pathogenic and probiotic in breast cancer is also highly related to the immune response elicited by the host (46–49). Specific taxa are pathogenic and instigate a response that can progress tumor development, while others act as probiotics, supporting regulatory or anti-inflammatory immune responses that slow or even stall tumor progression (46–49). These findings are mirrored in studies of chemotherapeutics that include analysis of the microbiome, suggesting that specific taxa are not only pathogenic, supporting cancer progression, but they also may stall treatment of breast cancer (98–100). Consequently, future studies analyzing host-microbiome interactions in breast cancer should focus on host immune markers among pre-diagnostic and post-diagnostic cases, which could be further divided into chemo-treated and untreated groups.
Aside from these avenues for further research in this field, we also note that there are highly underexplored research areas that will help bridge our understanding of host-microbiome interactions in breast cancer. Firstly, the role of the estrobolome in breast cancer has not been clearly elucidated. This will require multi-omics strategies and, potentially, a targeted or hypothesis driven approach and re-exploration of functional metagenomic data from the breast and/or gut microbiomes. Further, metabolomics is a natural addition to dietary intervention studies in breast cancer. Targeted and untargeted metabolomics in conjunction with microbiome and host-genome/transcriptome sequencing will be key in establishing additional mechanistic links between the local and gut microbiomes and breast tumor development. Lastly, analysis of breast cancer subtypes, stages, and effects on the microbiome stimulated by factors such as obesity, menopause, and age, should be further explored. This will require large cohort studies where appropriate statistical power can be maintained even after establishment of sub-groups.
Altogether, the current literature highlights the potential to identify probiotic bacterial taxa that could be used as breast cancer therapeutics or shifts in bacterial abundances that could be used as biomarkers of breast cancer. It is evident that certain bacteria within the gut and breast microbiomes do play roles in cancer pathogenicity, while others serve as probiotics, supporting host health and protecting against breast cancer. However, it is equally apparent, based on the current literature, that both of these bacterial consortia may be responding to breast cancer. The relationship between host and microbiome in breast cancer is bidirectional, but clarification of the bacterial taxa that fall into these two categories is necessary to develop microbe-based treatment strategies and clarify how response of the microbiome to cancer and cancer therapeutics might inhibit cancer treatment.
As we look toward future perspectives on the utilization of microbiome discoveries for the prevention and therapeutic treatment of breast cancer, the outlook for microbe-based therapeutic strategies to prevent or treat breast cancer is promising. Analysis of the breast tissue microbiome offers potential for predictive biomarkers, but more research is needed to determine how this microbial consortium is seeded and maintained before modulation of this microbiome can be applied as a breast cancer treatment. Alteration of the gut microbiota, however, shows promise for microbiota modulation in relation to breast cancer. Probiotic and prebiotic supplementation methods are attractive prospects, particularly as these supplementations are shown to induce immune modulations that protect against breast tumor development (49, 52). However, the persistent presence of these supplemented bacteria has not been demonstrated (102). Future clinical trials (discussed in this review) will be key in clarifying the effects of beneficial bacterial species in the prevention and treatment of breast cancer (56, 57). However, the wholesale transfer of a eubiotic microbiota via fecal microbiota transplant (FMT) from healthy individuals to those at high risk of developing, or those that have developed, cancer represents another avenue to harness the gut microbiome as a breast cancer therapeutic (103–105).
FMT has most successfully been applied in humans for the treatment of Clostridium difficile infections following the treatment of antibiotics (106). It has also been demonstrated as an effective cancer treatment in preliminary studies, particularly in colon cancer and potentially in melanoma skin cancers (103, 107, 108). Further, recent work presented at the American Society of Clinical Oncology highlights the use of FMTs from healthy individuals to patients receiving immune checkpoint inhibitors and who had subsequently developed colitis to improve clinical phenotypes (109). This highlights the close interplay of the immune system with the microbiota, an avenue we note in this review as key to explore in future studies. For breast cancer specifically, one mouse-model study suggests FMT from healthy individuals compared with mice humanized by FMT from breast cancer patients can enhance the anti-cancer effects of chemotherapy (94). Another group performed FMT between mice fed control or high fat diets demonstrating the transfer of protumorigenic effects from HFD-diet mice to control mice after HFD fecal transplant (108). This group also reports that FMT alters composition of both the mammary and gut microbiomes, suggesting FMT as a potential method to establish eubiotic breast and gut microbial communities, which may improve treatment of or even prevent breast cancer (108).
In conclusion, though the breast tissue microbiome is the most direct route of tumor-microbe interaction in breast cancer, modulation of the gut microbiome, whether it be with probiotic supplementations, dietary changes, or FMT, may support promotion of eubiotic states in both body niches (108) (Figure 1). Ultimately, the interplay of various microbial components, including bacteria, colonocytes, archaea, viruses, fungi, protists, and metabolites, of an FMT and their respective roles is still under study (110), but the immunostimulatory effects, competitive exclusion, and/or prevention of a self-potentiating dysbiotic inflammatory state supports the usage of FMTs cancer therapeutics. Further, transfer of a collective eubiotic microbial community, rather than specific probiotic species, which may be outcompeted by dysbiotic taxa, has the potential to synergistically support breast cancer chemotherapies through a wider array of immune-microbe interactions (108, 109). Altogether, current research supports the microbiome as a key physiological component that cannot be ignored in relation to gut and non-gut related cancers. It may be a critical missing piece in developing holistic breast cancer treatments and preventive strategies.
CH led the review outline and article collation and wrote the manuscript. RJ made the figure and tables and contributed to outlining, collating articles, and writing the manuscript. CH and RJ reviewed and edited the manuscript prior to submission. LS supervised the work and critically reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
RJ was supported by the Pepperdine’s Summer Undergraduate Research in Biology (SURB) program (DBI-1950350) and Pepperdine’s Faculty-Student Mentor Program. CH is supported by the University of British Columbia John Turner Fellowship in Microbiology and Immunology. LS was supported by a Breakthrough Fellowship Award from the Department of Defense Breast Cancer Research Program (W81XWH1810749).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1198259/full#supplementary-material
DOX, Doxorubicin; ER, Estrogen Receptor; FEC60, Fluorouracil, epirubicin, and cyclophosphamide; FMT, Fecal Microbiota Transplant; GUS, Gut microbial β-glucuronidase; HER2, Human epidermal growth factor receptor 2; HIF-1, Human hypoxia-Inducible Factor -1; HR+, Hormone receptor-positive; IPA, Indole propionic acid; KEGG, Kyoto Encyclopedia of Genes and Genomes; LeFSe, Linear Discriminant Analysis Effect Size; LCA, Litocholic acid; LPS, Exogenous lipopolysaccharide; PBS, Phosphate buffered saline; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; pTH17, “Pathogenic” T helper 17; qPCR, Quantitative PCR; qRT-PCR, Quantitative reverse transcription polymerase chain reaction; ROS, Reactive oxygen species; SCFA, Short chain fatty acids; SeNPs, Selenium nanoparticle.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
3. Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol (2014) 80(10):3007–14. doi: 10.1128/AEM.00242-14
4. Eslami-S Z, Majidzadeh-A K, Halvaei S, Babapirali F, Esmaeili R. Microbiome and breast cancer: new role for an ancient population. Front Oncol (2020) 10:120. doi: 10.3389/fonc.2020.00120
5. Wang N, Sun T, Xu J. Tumor-related microbiome in the breast microenvironment and breast cancer. J Cancer (2021) 12(16):4841–8. doi: 10.7150/jca.58986
6. Buchta Rosean C, Bostic RR, Ferey JCM, Feng TY, Azar FN, Tung KS, et al. Preexisting commensal dysbiosis is a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor–positive breast cancer. Cancer Res (2019) 79(14):3662–75. doi: 10.1158/0008-5472.CAN-18-3464
7. Hoskinson C, Zheng K, Gabel J, Kump A, German R, Podicheti R, et al. Composition and functional potential of the human mammary microbiota prior to and following breast tumor diagnosis. Gibbons SM editor mSystems (2022) 7(3):e01489–21. doi: 10.1128/msystems.01489-21
8. Tzeng A, Sangwan N, Jia M, Liu CC, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med (2021) 13(1):60. doi: 10.1186/s13073-021-00874-2
9. Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget (2017) 8(50):88122–38. doi: 10.18632/oncotarget.21490
10. Zhang J, Xia Y, Sun J. Breast and gut microbiome in health and cancer. Genes Dis (2021) 8(5):581–9. doi: 10.1016/j.gendis.2020.08.002
11. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature (2019) 574(7777):264–7. doi: 10.1038/s41586-019-1608-2
12. Apostolou P, Tsantsaridou A, Papasotiriou I, Toloudi M, Chatziioannou M, Giamouzis G. Bacterial and fungal microflora in surgically removed lung cancer samples. J Cardiothorac Surg (2011) 6(1):137. doi: 10.1186/1749-8090-6-137
13. Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell (2021) 12(5):426–35. doi: 10.1007/s13238-020-00813-8
14. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
15. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol (2018) 9:951. doi: 10.3389/fmicb.2018.00951
16. Hirko KA, Rocque G, Reasor E, Taye A, Daly A, Cutress RI, et al. The impact of race and ethnicity in breast cancer—disparities and implications for precision oncology. BMC Med (2022) 20(1):72. doi: 10.1186/s12916-022-02260-0
17. Smith A, Pierre JF, Makowski L, Tolley E, Lyn-Cook B, Lu L, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci Rep (2019) 9(1):11940. doi: 10.1038/s41598-019-48348-1
18. Costantini L, Magno S, Albanese D, Donati C, Molinari R, Filippone A, et al. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci Rep (2018) 8(1):16893. doi: 10.1038/s41598-018-35329-z
19. Fang Q, Feng Y, Feng P, Wang X, Zong Z. Nosocomial bloodstream infection and the emerging carbapenem-resistant pathogen Ralstonia insidiosa. BMC Infect Dis (2019) 19(1):334. doi: 10.1186/s12879-019-3985-4
20. Said M, Van Hougenhouck-Tulleken W, Naidoo R, Mbelle N, Ismail F. Outbreak of Ralstonia mannitolilytica bacteraemia in patients undergoing haemodialysis at a tertiary hospital in Pretoria, South Africa. Antimicrob Resist Infect Control (2020) 9(1):117. doi: 10.1186/s13756-020-00778-7
21. Goc J, Hepworth MR, Sonnenberg GF. Group 3 innate lymphoid cells: regulating host–commensal bacteria interactions in inflammation and cancer. Int Immunol (2016) 28(1):43–52. doi: 10.1093/intimm/dxv056
22. Xuan C, Shamonki JM, Chung A, DiNome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PloS One (2014) 9(1):e83744. doi: 10.1371/journal.pone.0083744
23. Hassan Z, Mustafa S, Rahim RA, Isa NM. Anti-breast cancer effects of live, heat-killed and cytoplasmic fractions of Enterococcus faecalis and Staphylococcus hominis isolated from human breast milk. Vitro Cell Dev Biol - Anim (2016) 52(3):337–48. doi: 10.1007/s11626-015-9978-8
24. Esfandiary A, TaherianEsfahani Z, AbedinDo A, Mirfakhraie R, Shirzad M, GhafouriFard S. Lactobacilli modulate hypoxia-inducible factor HIF)-1 regulatory pathway in triple negative breast cancer cell line. Cell J Yakhteh (2016) 18(2):237–44. doi: 10.22074/cellj.2016.4319
25. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun (2020) 11(1):3259. doi: 10.1038/s41467-020-16967-2
26. Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PloS One (2017) 12(11):e0188873. doi: 10.1371/journal.pone.0188873
27. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol (2020) 38(6):685–8. doi: 10.1038/s41587-020-0548-6
28. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and Malignant disease. Sci Rep (2016) 6(1):30751. doi: 10.1038/srep30751
29. Bonifácio VDB, Pereira SA, Serpa J, Vicente JB. Cysteine metabolic circuitries: druggable targets in cancer. Br J Cancer (2021) 124(5):862–79. doi: 10.1038/s41416-020-01156-1
30. Kaiser P. Methionine dependence of cancer. Biomolecules (2020) 10(4):568. doi: 10.3390/biom10040568
31. Ashkani J, Naidoo KJ. Glycosyltransferase gene expression profiles classify cancer types and propose prognostic subtypes. Sci Rep (2016) 6(1):26451. doi: 10.1038/srep26451
32. Meng S, Chen B, Yang J, Wang J, Zhu D, Meng Q, et al. Study of microbiomes in aseptically collected samples of human breast tissue using needle biopsy and the potential role of in situ tissue microbiomes for promoting Malignancy. Front Oncol (2018) 8:318. doi: 10.3389/fonc.2018.00318
33. Sadrekarimi H, Gardanova ZR, Bakhshesh M, Ebrahimzadeh F, Yaseri AF, Thangavelu L, et al. Emerging role of human microbiome in cancer development and response to therapy: special focus on intestinal microflora. J Transl Med (2022) 20(1):301. doi: 10.1186/s12967-022-03492-7
34. Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. Intestinal proportion of blautia sp. Is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr Cancer (2017) 69(2):267–75. doi: 10.1080/01635581.2017.1263750
35. Parsaei M, Sarafraz N, Moaddab SY, Ebrahimzadeh Leylabadlo H. The importance of Faecalibacterium prausnitzii in human health and diseases. New Microbes New Infect (2021) 43:100928. doi: 10.1016/j.nmni.2021.100928
36. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia —a new functional genus with potential probiotic properties? Gut Microbes (2021) 13(1):1875796. doi: 10.1080/19490976.2021.1875796
37. Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome (2018) 6(1):136. doi: 10.1186/s40168-018-0515-3
38. Bobin-Dubigeon C, Luu HT, Leuillet S, Lavergne SN, Carton T, Le Vacon F, et al. Faecal microbiota composition varies between patients with breast cancer and healthy women: A comparative case-control study. Nutrients (2021) 13(8):2705. doi: 10.3390/nu13082705
39. Yang P, Wang Z, Peng Q, Lian W, Chen D. Comparison of the gut microbiota in patients with benign and Malignant breast tumors: A pilot study. Evol Bioinforma (2021) 17:117693432110575. doi: 10.1177/11769343211057573
40. Ma Z, Qu M, Wang X. Analysis of gut microbiota in patients with breast cancer and benign breast lesions. Pol J Microbiol (2022) 71(2):217–26. doi: 10.33073/pjm-2022-019
41. Cao SS, Lu CT. Recent perspectives of breast cancer prognosis and predictive factors. Oncol Lett (2016) 12(5):3674–8. doi: 10.3892/ol.2016.5149
42. Endogenous Hormones and Breast Cancer Collaborative Group Sex hormones and risk of breast cancer in premenopausal women: a collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol (2013) 14(10):1009–19. doi: 10.1016/S1470-2045(13)70301-2
43. Plottel CS, Blaser MJ. Microbiome and Malignancy. Cell Host Microbe (2011) 10(4):324–35. doi: 10.1016/j.chom.2011.10.003
44. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing Long-term impacts on the human throat and gut microbiome. PloS One (2010) 5(3):e9836. doi: 10.1371/journal.pone.0009836
45. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
46. Lakritz JR, Poutahidis T, Mirabal S, Varian BJ, Levkovich T, Ibrahim YM, et al. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget (2015) 6(11):9387–96. doi: 10.18632/oncotarget.3328
47. Lakritz JR, Poutahidis T, Levkovich T, Varian BJ, Ibrahim YM, Chatzigiagkos A, et al. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int J Cancer (2014) 135(3):529–40. doi: 10.1002/ijc.28702
48. Soltan Dallal MM, Yazdi MH, Holakuyee M, Hassan ZM, Abolhassani M, Mahdavi M. Lactobacillus casei ssp.casei induced Th1 cytokine profile and natural killer cells activity in invasive ductal carcinoma bearing mice. Iran J Allergy Asthma Immunol (2012) 11(2):183–9.
49. Yazdi MH, Mahdavi M, Setayesh N, Esfandyar M, Shahverdi AR. Selenium nanoparticle-enriched Lactobacillus brevis causes more efficient immune responses in vivo and reduces the liver metastasis in metastatic form of mouse breast cancer. DARU J Pharm Sci (2013) 21(1):33. doi: 10.1186/2008-2231-21-33
50. de Moreno de LeBlanc A, Matar C, Thériault C, Perdigón G. Effects of milk fermented by Lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology (2005) 210(5):349–58. doi: 10.1016/j.imbio.2005.05.024
51. Falsafi T, Mahboubi M. Helicobacter hepaticus, a new pathogenic species of the Helicobacter genus: Similarities and differences with H. pylori. Iran J Microbiol (2013) 5(3):185–94.
52. Maroof H, Hassan ZM, Mobarez AM, Mohamadabadi MA. Lactobacillus acidophilus could modulate the immune response against breast cancer in murine model. J Clin Immunol (2012) 32(6):1353–9. doi: 10.1007/s10875-012-9708-x
53. Chen G, Yang F, Fan S, Jin H, Liao K, Li X, et al. Immunomodulatory roles of selenium nanoparticles: Novel arts for potential immunotherapy strategy development. Front Immunol (2022) 13:956181. doi: 10.3389/fimmu.2022.956181
54. Ma W, Zhang Y, Yu M, Wang B, Xu S, Zhang J, et al. In-vitro and in-vivo anti-breast cancer activity of synergistic effect of berberine and exercise through promoting the apoptosis and immunomodulatory effects. Int Immunopharmacol (2020) 87:106787. doi: 10.1016/j.intimp.2020.106787
55. Zamberi NR, Abu N, Mohamed NE, Nordin N, Keong YS, Beh BK, et al. The antimetastatic and antiangiogenesis effects of kefir water on murine breast cancer cells. Integr Cancer Ther (2016) 15(4):NP53–66. doi: 10.1177/1534735416642862
56. Chumsri S. Effects of probiotics on the gut microbiome and immune system in operable stage I-III breast or lung cancer. U.S. National Library of Medicine (2021). Available at: https://clinicaltrials.gov/ct2/show/NCT04857697?term=microbiome&cond=breast+cancer&draw=8&rank=24.
57. Pusztai L, Mainor C, Feinman R, Davidson M, Graham D, Starr G, et al. Gut and intratumoral microbiome effect on the neoadjuvant chemotherapy-induced immunosurveillance in triple negative breast cancer. U.S. National Library of Medicine (2017). Available at: https://clinicaltrials.gov/ct2/show/NCT03586297?term=microbiome&cond=breast+cancer&draw=2&rank=4.
58. Reid G, Brackstone M. Probiotics and breast health. U.S. National Library of Medicine (2019). Available at: https://clinicaltrials.gov/ct2/show/NCT03290651.
59. Chumsri S. Engineering gut microbiome to target breast cancer. U.S. National Library of Medicine (2017). Available at: https://clinicaltrials.gov/ct2/show/NCT03358511.
60. Mikó E, Vida A, Kovács T, Ujlaki G, Trencsényi G, Márton J, et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta BBA - Bioenerg (2018) 1859(9):958–74. doi: 10.1016/j.bbabio.2018.04.002
61. Luu TH, Bard JM, Carbonnelle D, Chaillou C, Huvelin JM, Bobin-Dubigeon C, et al. Lithocholic bile acid inhibits lipogenesis and induces apoptosis in breast cancer cells. Cell Oncol (2018) 41(1):13–24. doi: 10.1007/s13402-017-0353-5
62. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res (2006) 47(2):241–59. doi: 10.1194/jlr.R500013-JLR200
63. Kovács T, Mikó E, Vida A, Sebő É, Toth J, Csonka T, et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci Rep (2019) 9(1):1300. doi: 10.1038/s41598-018-37664-7
64. Thirunavukkarasan M, Wang C, Rao A, Hind T, Teo YR, Siddiquee AAM, et al. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PloS One (2017) 12(10):e0186334. doi: 10.1371/journal.pone.0186334
65. Rodrigues MF, Carvalho É, Pezzuto P, Rumjanek FD, Amoêdo ND. Reciprocal modulation of histone deacetylase inhibitors sodium butyrate and trichostatin a on the energy metabolism of breast cancer cells. J Cell Biochem (2015) 116(5):797–808. doi: 10.1002/jcb.25036
66. Salimi V, Shahsavari Z, Safizadeh B, Hosseini A, Khademian N, Tavakoli-Yaraki M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis (2017) 16(1):208. doi: 10.1186/s12944-017-0593-4
67. Nakkarach A, Foo HL, Song AAL, Mutalib NEA, Nitisinprasert S, Withayagiat U. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb Cell Factories (2021) 20(1):36. doi: 10.1186/s12934-020-01477-z
68. Sári Z, Mikó E, Kovács T, Jankó L, Csonka T, Lente G, et al. Indolepropionic acid, a metabolite of the microbiome, has cytostatic properties in breast cancer by activating ahr and pxr receptors and inducing oxidative stress. Cancers (2020) 12(9):2411. doi: 10.3390/cancers12092411
69. Woolcott CG, Shvetsov YB, Stanczyk FZ, Wilkens LR, White KK, Caberto C, et al. Plasma sex hormone concentrations and breast cancer risk in an ethnically diverse population of postmenopausal women: the Multiethnic Cohort Study. Endocr Relat Cancer (2010) 17(1):125–34. doi: 10.1677/ERC-09-0211
70. Samavat H, Kurzer MS. Estrogen metabolism and breast cancer. Cancer Lett (2015) 356(2):231–43. doi: 10.1016/j.canlet.2014.04.018
71. Kohler BA, Sherman RL, Howlader N, Jemal A, Ryerson AB, Henry KA, et al. Annual report to the nation on the status of cancer, 1975-2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. JNCI J Natl Cancer Inst (2015) 107(6). doi: 10.1093/jnci/djv048
72. Anderson WF, Katki HA, Rosenberg PS. Incidence of breast cancer in the United States: Current and future trends. JNCI J Natl Cancer Inst (2011) 103(18):1397–402. doi: 10.1093/jnci/djr257
73. Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer (2003) 10(2):179–86. doi: 10.1677/erc.0.0100179
74. Kwa M, Plottel CS, Blaser MJ, Adams S. The intestinal microbiome and estrogen receptor–positive female breast cancer. JNCI J Natl Cancer Inst (2016) 108(8). doi: 10.1093/jnci/djw029/2457487/The-Intestinal-Microbiome-and-Estrogen
75. Ruo SW, Alkayyali T, Win M, Tara A, Joseph C, Kannan A, et al. Role of gut microbiota dysbiosis in breast cancer and novel approaches in prevention, diagnosis, and treatment. Cureus (2021) 13(8):e17472. doi: 10.7759/cureus.17472
76. Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PloS One (2012) 7(6):e39745. doi: 10.1371/journal.pone.0039745
77. Ervin SM, Li H, Lim L, Roberts LR, Liang X, Mani S, et al. Gut microbial β-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens. J Biol Chem (2019) 294(49):18586–99. doi: 10.1074/jbc.RA119.010950
78. Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, et al. Risk factors and preventions of breast cancer. Int J Biol Sci (2017) 13(11):1387–97. doi: 10.7150/ijbs.21635
79. Dunneram Y, Greenwood DC, Cade JE. Diet, menopause and the risk of ovarian, endometrial and breast cancer. Proc Nutr Soc (2019) 78(3):438–48. doi: 10.1017/S0029665118002884
80. Hou MF, Ou-Yang F, Li CL, Chen FM, Chuang CH, Kan JY, et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp Mol Med (2021) 53(10):1636–46. doi: 10.1038/s12276-021-00686-9
81. Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci (2011) 108(supplement_1):4586–91. doi: 10.1073/pnas.1000097107
82. Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao Jz, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol (2016) 16(1):90. doi: 10.1186/s12866-016-0708-5
83. Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. JNCI J Natl Cancer Inst (2015) 107(8):djv147. doi: 10.1093/jnci/djv147
84. Muscogiuri G, Cantone E, Cassarano S, Tuccinardi D, Barrea L, Savastano S, et al. Gut microbiota: a new path to treat obesity. Int J Obes Suppl (2019) 9(1):10–9. doi: 10.1038/s41367-019-0011-7
85. Duan M, Wang Y, Zhang Q, Zou R, Guo M, Zheng H. Characteristics of gut microbiota in people with obesity. PloS One (2021) 16(8):e0255446. doi: 10.1371/journal.pone.0255446
86. Crovesy L, Masterson D, Rosado EL. Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr (2020) 74(9):1251–62. doi: 10.1038/s41430-020-0607-6
87. Hossain F, Majumder S, David J, Bunnell BA, Miele L. Obesity modulates the gut microbiome in triple-negative breast cancer. Nutrients (2021) 13(10):3656. doi: 10.3390/nu13103656
88. Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol (2016) 6. doi: 10.3389/fmicb.2015.01543
89. Konstantinidis T, Tsigalou C, Karvelas A, Stavropoulou E, Voidarou C, Bezirtzoglou E. Effects of antibiotics upon the gut microbiome: A review of the literature. Biomedicines (2020) 8(11):502. doi: 10.3390/biomedicines8110502
90. Gallagher M, Jones DJ, Bell-Syer SV. Prophylactic antibiotics to prevent surgical site infection after breast cancer surgery. Cochrane Database Syst Rev (2019) 2019(9). doi: 10.1002/14651858.CD005360.pub5
91. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
92. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
93. McKee AM, Kirkup BM, Madgwick M, Fowler WJ, Price CA, Dreger SA, et al. Antibiotic-induced disturbances of the gut microbiota result in accelerated breast tumor growth. iScience (2021) 24(9):103012. doi: 10.1016/j.isci.2021.103012
94. Terrisse S, Derosa L, Iebba V, Ghiringhelli F, Vaz-Luis I, Kroemer G, et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ (2021) 28(9):2778–96. doi: 10.1038/s41418-021-00784-1
95. Aarnoutse R, Ziemons J, Hillege LE, de Vos-Geelen J, de Boer M, Bisschop SMP, et al. Changes in intestinal microbiota in postmenopausal oestrogen receptor-positive breast cancer patients treated with (neo)adjuvant chemotherapy. NPJ Breast Cancer (2022) 8(1):89. doi: 10.1038/s41523-022-00455-5
96. Hinshaw DC, Swain CA, Chen D, Hanna A, Molina PA, Maynard CL, et al. Hedgehog blockade remodels the gut microbiota and the intestinal effector CD8+ T cells in a mouse model of mammary carcinoma. Lab Invest (2022) 102(11):1236–44. doi: 10.1038/s41374-022-00828-1
97. Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Mol Cancer Res (2020) 18(1):130–9. doi: 10.1158/1541-7786.MCR-19-0451
98. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (2013) 342(6161):971–6. doi: 10.1126/science.1240537
99. Bawaneh A, Wilson AS, Levi N, Howard-McNatt MM, Chiba A, Soto-Pantoja DR, et al. Intestinal microbiota influence doxorubicin responsiveness in triple-negative breast cancer. Cancers (2022) 14(19):4849. doi: 10.3390/cancers14194849
100. Zhou K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J Funct Foods (2017) 33:194–201. doi: 10.1016/j.jff.2017.03.045
101. Marino N, German R, Rao X, Simpson E, Liu S, Wan J, et al. Upregulation of lipid metabolism genes in the breast prior to cancer diagnosis. NPJ Breast Cancer (2020) 6(1):50. doi: 10.1038/s41523-020-00191-8
102. Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr (2019) 73(1):24–37. doi: 10.1038/s41430-018-0135-9
103. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J Cancer (2019) 145(8):2021–31. doi: 10.1002/ijc.32003
104. Xu H, Cao C, Ren Y, Weng S, Liu L, Guo C, et al. Antitumor effects of fecal microbiota transplantation: Implications for microbiome modulation in cancer treatment. Front Immunol (2022) 13:949490. doi: 10.3389/fimmu.2022.949490
105. Stoff R, Wolf Y, Boursi B. Fecal microbiota transplantation as a cancer therapeutic. Cancer J (2023) 29(2):102–8. doi: 10.1097/PPO.0000000000000651
106. Rohlke F, Stollman N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Ther Adv Gastroenterol (2012) 5(6):403–20. doi: 10.1177/1756283X12453637
107. Ma Y, Chen H. Faecal microbiota transplantation, a promising way to treat colorectal cancer. eBioMedicine (2019) 49:13–4. doi: 10.1016/j.ebiom.2019.10.015
108. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
109. Elkrief A, Nicholas W, Smith N, Dai A, Slingerland JB, Aleynick N, et al. Fecal microbiota transplantation for refractory immune-checkpoint-inhibitor colitis. J Clin Oncol (2023) 41(16_suppl):2657–7. doi: 10.1200/JCO.2023.41.16_suppl.2657
Keywords: mammary microbiome, gut microbiome, breast cancer, breast tumor, breast microbiome
Citation: Hoskinson C, Jiang RY and Stiemsma LT (2023) Elucidating the roles of the mammary and gut microbiomes in breast cancer development. Front. Oncol. 13:1198259. doi: 10.3389/fonc.2023.1198259
Received: 31 March 2023; Accepted: 26 July 2023;
Published: 17 August 2023.
Edited by:
Andrea Botticelli, Sapienza University of Rome, ItalyReviewed by:
Mohammed A. Al-Obaide, Texas Tech University Health Science Center Amarillo, United StatesCopyright © 2023 Hoskinson, Jiang and Stiemsma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leah T. Stiemsma, bGVhaC5zdGllbXNtYUBwZXBwZXJkaW5lLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.