
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 23 January 2024
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1192386
Hepatocellular carcinoma (HCC) is an extremely heterogeneous malignant tumor with a high morbidity and mortality. Circular RNAs (circRNAs) are noncoding RNAs with high stability, organ/tissue/cell-specific expression and are conserved across species. Accumulating evidence suggested that circRNAs play crucial roles as microRNA sponges, protein sponges, scaffolds, recruiters and could even polypeptide encoders. Many studies have since revealed that circRNAs were aberrantly expressed in HCC and acted as crucial modulators of HCC carcinogenesis and progression. Furthermore, circRNAs have also been identified as potential diagnostic and prognostic biomarkers for HCC. In this review, we thoroughly outline and evaluate the function of circRNAs in HCC development, with an emphasis on the specific molecular pathways by which they participated in the formation and progression of HCC, and we address their potential for serving as clinical biomarkers in HCC.
Hepatocellular carcinoma (HCC) is one of the most prevalent primary liver cancers with a poor prognosis and is the fourth leading cause of cancer-related death worldwide (1). Currently, the principal treatments for HCC are surgical resection along with radiotherapy and chemotherapy. However, postoperative recurrence, metastasis and acquired chemoresistance strongly reduce the therapeutic efficacy of the aforementioned treatments (2). Because the carcinogenesis and progression of HCC are exceedingly complex processes, the mechanisms underlying the development of HCC remain poorly understood. Although mounting evidence compelling demonstrated that numerous noncoding RNAs (ncRNAs) play prominent roles in a wide range of clinical and physiological processes, including the progression of HCC, the function of circRNAs remains to be clarified.
CircRNAs, a group of ncRNAs, are distinguished by covalent closed-loop structures without terminal 5’ and 3’ poly(A) tails, which are generated by a special noncanonical splicing mechanism known as back-splicing (3). CircRNAs are more stable than linear RNA because of their unusual architecture, which protects them from exonuclease degradation. In addition, circRNAs are characterized with organ/tissue/cell-specific expression and are conserved across species. There is growing evidence showed that aberrantly expressed circRNAs exert crucial effect on HCC cell proliferation, apoptosis, metastasis and chemoresistance. Moreover, circRNAs can be identified as non-invasive biomarkers because they can be transmitted into the blood by exosomes. Due to the lack of obvious symptoms at the early stage and lack of dependable and valid biomarkers, HCC patients are frequently diagnosed at the advanced stage and the overall survival rate is less than ideal. As a result, an increasing number of studies have focused on the potential function of circRNAs in HCC as noteworthy disease biomarkers. In this study, we extensively describe the circRNAs involved in the progression of HCC and recent clinical research findings associated to these circRNAs. Hopefully, this comprehensive review of HCC-associated circRNAs could help to improve understanding of complex mechanisms and provide valuable clues for future HCC therapy and diagnosis strategies.
CircRNAs, discovered in the 1990s, are a novel class of endogenously produced ncRNAs that were previously assumed to be the result of aberrant splicing. Unlike lncRNAs, which have 5′-end m7G caps and 3′-end poly(A) tails (4), circRNAs are covalent closed-loop structures without terminal 5’ and 3’ poly(A) tails that are produced from introns or exons via back-splicing (5). These characteristics are linked to numerous properties, including high abundance, stability, special conservation, and organic/tissue/cell-specific expression patterns, indicating that circRNAs extensively participate in a wide variety of physiological and pathophysiological processes (6).
CircRNA closed loop structures are formed via three main circularization mechanisms: 1) intron pairing, inverted complementary intronic sequences (such as Alu elements) flanking the exon junction could form a loop which bringing the 3’ extremity of a downstream exon (donor) proximity to the 5’ extremity of an upstream exon (acceptor), and thus facilitate the circRNAs looping structure (7). 2) RNA-binding proteins (RBPs)-mediated procedures, for example, the RBP Quaking (QKI) and FUS can recognize and interact with specific flanking intronic motifs, which driving back-splicing by direct interaction or dimerization (8). and 3) lariat intron-driven ways, during conventional splicing event, the skipped exonic and intronic sequences are integrated into a circular lariat by ‘head-to-tail’ junction, which can be processed again to form circRNAs (9). CircRNAs are classified into three types based on their origins and internal elements: exonic circRNAs (EcircRNAs), exon-intron circRNAs (EIciRNAs), and intronic circRNAs (ciRNAs). EcircRNAs are chiefly situated in the cytoplasm, whereas ciRNAs and EIciRNAs have higher content in the nucleus (10).
Accumulating studies indicated that circRNAs may play a vital role in various biological processes, including miRNA sponging, protein translation, RBP interactions, as well as other unknown processes.
CircRNAs harbor well-conserved nucleotides and canonical miRNA response elements (MREs), implying that some circRNAs can function as miRNA sequesters or sponges (11). CiRS-7 also referred to as cerebellar degeneration-related protein 1 transcript (CDR1as) and was the first EcircRNA acted as miRNA sponges. CiRS-7 contains over 70 miR-7 binding sites and can dramatically impair miR-7 activity (9). According to recent evidence, many functioning circRNAs have the latent capacity to operate as miRNA sponges and are related to HCC progression (Supplementary Table 1). These findings directly demonstrated that miRNA sponging is a common function for circRNAs. However, most circRNAs contain few binding sites for miRNAs, therefore it is unclear how circRNAs exert substantial effects.
Aside from binding miRNAs, circRNAs have been shown in a few studies to combine with several RBPs to form RNA-protein complexes that affect gene expression and ultimately influence the progression of various disorders. For instance, circZKSCAN1 was recently shown to be a suppressive cancer stem cells (CSCs) modulator via interacting with fragile X mental retardation protein (FMRP) rather than serving as a miRNA sponge (12). However, not all circRNAs bind to proteins and inhibit protein function. CircRHOT1, For example, has been discovered to accelerate HCC growth and metastasis by recruiting Tat-interacting protein 60 (TIP60) to upregulate nuclear receptor subfamily 2, group F, member 6 (NR2F6) expression (13). Interestingly, circRNA-SORE, a novel circRNA, played a momentous role in the maintenance and spread of sorafenib resistance in HCC by interacting with Y-box binding protein 1 (YBX1) to change its subcellular localization and sequentially preventing PRP19-mediated YBX1 ubiquitination and degradation (14). In the cases above, circRNAs acted as protein decoys, scaffolds, and recruiters in the progression of HCC. Nevertheless, bioinformatic analyses suggested that circRNAs had a lower abundance of RBP binding sites than linear RNAs, and aside from their mutual binding sites and/or sequences, the exact regulatory mechanisms connecting circRNA with RBPs and altering protein-protein interactions remain unclear and require further investigations.
Although circRNAs are classified as a type of ncRNA, there is growing evidence that some circRNAs are translatable. A recent study discovered that circβ-catenin was translatable and highly expressed in HCC, and translation of circβ-catenin generated a novel isoform named β-catenin-370aa that promoted HCC progression by protecting β-catenin from GSK3β-induced degradation and thus indirectly facilitating activation of the Wnt/β-catenin pathway (15). Another study revealed that circMRPS35 exerted its oncogenic function in HCC not only by sponging miR-148a to modulate the STX3-PTEN pathway, but also by being translated into circMRPS35-168aa, which resulted in cisplatin resistance, indicating that circMRPS35 might be a vital component in the progression and chemoresistance of HCC with different expression patterns under different conditions (16). The protein-coding function of circRNAs is a novel aspect in the development of diseases, and the function of protein-coding circRNAs may be more crucial than previously believed. Nevertheless, the regulatory frameworks of circRNA translation processes have received little attention, and whether circRNA derived proteins have discernible effects on the development of HCC remains controversial.
With the continuous improvement of detection technologies, a large number of circRNAs have been validated as considerable molecular regulators in the development of HCC. Supplementary Table 1 and Figure 1 list the expression of aberrant circRNAs in HCC along with their roles. These circRNAs are essential for several processes associated with HCC, such as cell proliferation, apoptosis, CSCs, metastasis, epithelial-mesenchymal transition (EMT), chemoresistance, angiogenesis, and have effects on the tumor microenvironment (TME), exosomes and other factors. Thus, we discussed these dysregulated circRNAs and their molecular and pathogenic mechanisms in HCC.
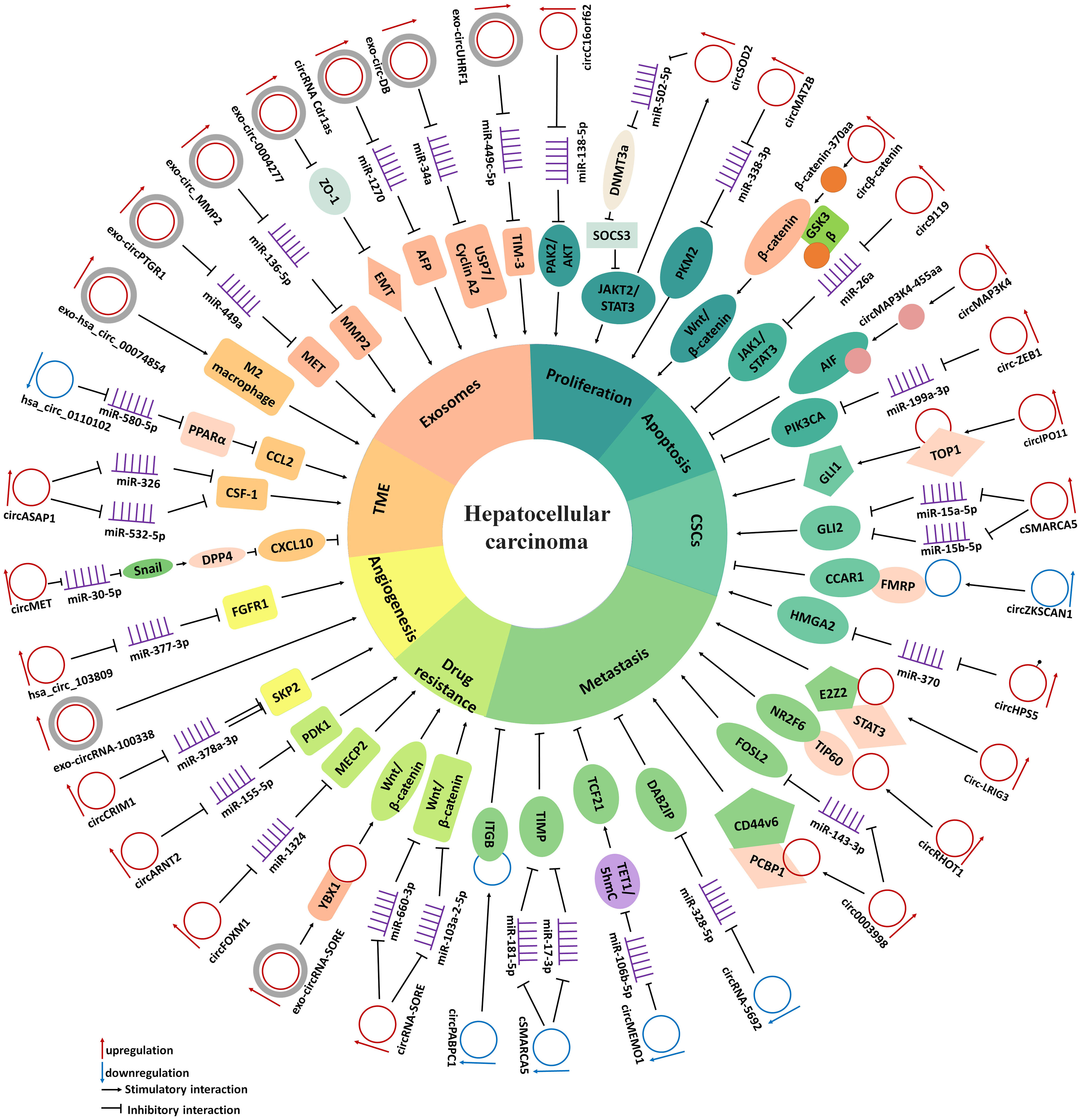
Figure 1 The diagram demonstrates the mechanism by which circRNAs in the regulate cell proliferation, apoptosis, cancer stem cells, metastasis, drug resistance, angiogenesis, tumor microenvironment and exosomes in HCC.
Tumor cells maintain proliferative conditions abnormally active by stimulating cell proliferation signaling pathways. The PI3K/AKT/mTOR axis is a crucial pathway in the advancement of HCC, affecting a wide range of malignant progression (17). The m6A-modified circMDK regulated the miR-346/miR-874-3p-ATG16L1 axis, activating the PI3K/AKT/mTOR signaling pathway and eventually accelerating the process of HCC carcinogenesis (18). Circ-CDYL was reported to influence the HDGF-NCL-PI3K-AKT and HIF1AN-NOTCH2 pathways as well as boost SURVIVIN and C-MYC expression, promoting stem-like characteristics and cell proliferation in HCC (19). Moreover, circC16orf62 sustained the proliferation of HCC cells through the circC16orf62/miR-138-5p/PTK2/AKT regulatory network (20). Because it regulated mTOR signaling by activating the miR-141-3p/RHEB axis, circRNA-100338 was strongly linked to the stimulation of HCC cell proliferation and metastasis (21). Unexpectedly, circSOD2 played a crucial role in accelerating HCC development via the circSOD2/SOCS3-JAK2/STAT3/circSOD2 innovative feedback pathway (22). Furthermore, circMTO1 competitively sponged miR-541-5p and decreased ZIC1 expression, inhibiting HCC progression by blocking the signaling pathway of Wnt/β-catenin (23). The Hippo pathway is a highly conserved pathway that takes a pivotal part in controlling stem cell self-renewal, cell proliferation and EMT by stimulating the downstream transcriptional factors YAP and TAZ. CircCPSF6 functioned as a novel m6A-modified circRNA accelerating tumorigenicity and cell metastasis of HCC by competitively binding to PCBP2, thus weakening the PCBP2-induced destabilization of YAP1 and triggering its expression, further activating its downstream cascade (24).
Glucose metabolism reprogramming, which is a generally recognized feature of many malignancies, happens to meet the greater rate of glycolysis required for macromolecular synthesis and to support rapid proliferation in hypoxia. CircMAT2B facilitated the progression of HCC by accelerating glycolysis through regulation of the miR-338-3p/PKM2 signaling pathway under hypoxia (12). Moreover, circRPN2 played a vital role in restraining the glycolytic reprogramming, progression and metastasis of HCC by directly interacting with ENO1 to facilitate its degradation and regulating the miR-183–5p/FOXO1 signaling pathway (25). Through its interaction with YTHDF1, circRHBDD1 accelerated the translation of PIK3R1 by modifying m6A, hence facilitating the glycolysis of HCC cells (26). Lipid metabolism is also essential for cancer cells and plays a vital role in adapting tumors to the local microenvironment. As the primary modulator of lipid metabolism, Peroxisome proliferator-activated receptor-α (PPARA) was deemed to be a promising HCC treatment. Hsa_circ_0098181 engaged in the miR-18a-3p/PPARA signaling pathway to exert anti-HCC effects (27). By physically attaching to CAPRIN1 and G3BP1, circVAMP3 exhibited tumor suppressor qualities in HCC by causing CAPRIN1 to phase separate and promoting the production of stress granules, which prevented the translation of c-Myc (28).
One factor that leads a normal cell to become malignant is dysregulation of the cell cycle machinery. The important cell cycle-related proteins known as Cyclin-dependent kinases 4 and 6 (CDK4/6) are essential for controlling the G1/S transition and the advancement of the G1 phase (29). As a miR-200a-3p sponge, circ-ZEB1.33 was a tumor promotor that stimulated CDK6 expression and HCC cell proliferation (30). Moreover, by controlling the cell cycle through the miR-1263/CDK6 signaling pathway, circERBIN accelerated the growth of HCC and subsequently accelerated the G1/S transition (31). In addition, by modifying the miR-486/CDK4 axis, hsa_circ_0016788 improved the cell cycle aberrations and migratory capabilities of HCC cells (32). CircSP3 promoted HCC growth by sponging miR-198 and upregulating CDK4 (33). Furthermore, it was showed that circCCNB1 silencing suppressed GPM6A expression to participate in the cell cycle regulation of HCC cells by upregulating DYNC1I1 expression and triggering the AKT/ERK signaling pathway (34). P21 takes a crucial part in growth arrest by inhibiting the activity of cyclin D-CDK2/4 complexes. For instance, circMTO1 functioned as a critical tumor suppressor by facilitating cell cycle arrest via the miR-9/p21 pathway (35).
In addition to stimulating cell proliferation, tumor cells frequently escape the growth-inhibitory effects of tumor suppressor pathways. PTEN functions as an important tumor suppressor by restraining cell growth and proliferation. CircHIAT1 restrained HCC cell proliferation by acting on the miR-3171/PTEN axis (36). MAPK14 could limit cell growth and cancer by engaging on cell cycle checkpoints. CircSETD3 had been identified as a tumor suppressor that suppressed the tumorigenesis of HCC by modulating the miR-421/MAPK14 pathway (37). In addition, cellular senescence is a stress-mediated, persistent cell cycle arrest of cells that were previously able to replicate (38). The p53/p21 pathway is an important pathway in senescence. CircLARP4 decreased cell proliferation and induces cell cycle arrest and senescence by suppressing miR-761, thereby increasing the RUNX3 expression and regulating activation of the downstream p53/p21 signaling pathway (39). In addition to miRNA modulation, an increasing number of studies have declared that some circRNAs promoted the proliferation of HCC cells and HCC tumorigenesis through efficient translation. For instance, circβ-catenin was translatable and highly expressed in HCC, and circβ-catenin translation generated a novel isoform named β-catenin-370aa, which subsequently promoted HCC progression by protecting β-catenin from GSK3β-induced degradation and thus facilitated the activation of the Wnt/β-catenin pathway (15).
Overall, the above examples clearly demonstrated that circRNAs had crucial regulatory functions in HCC tumorigenesis and progression, contributing to a deeper comprehension of HCC pathogenesis (Figure 2).
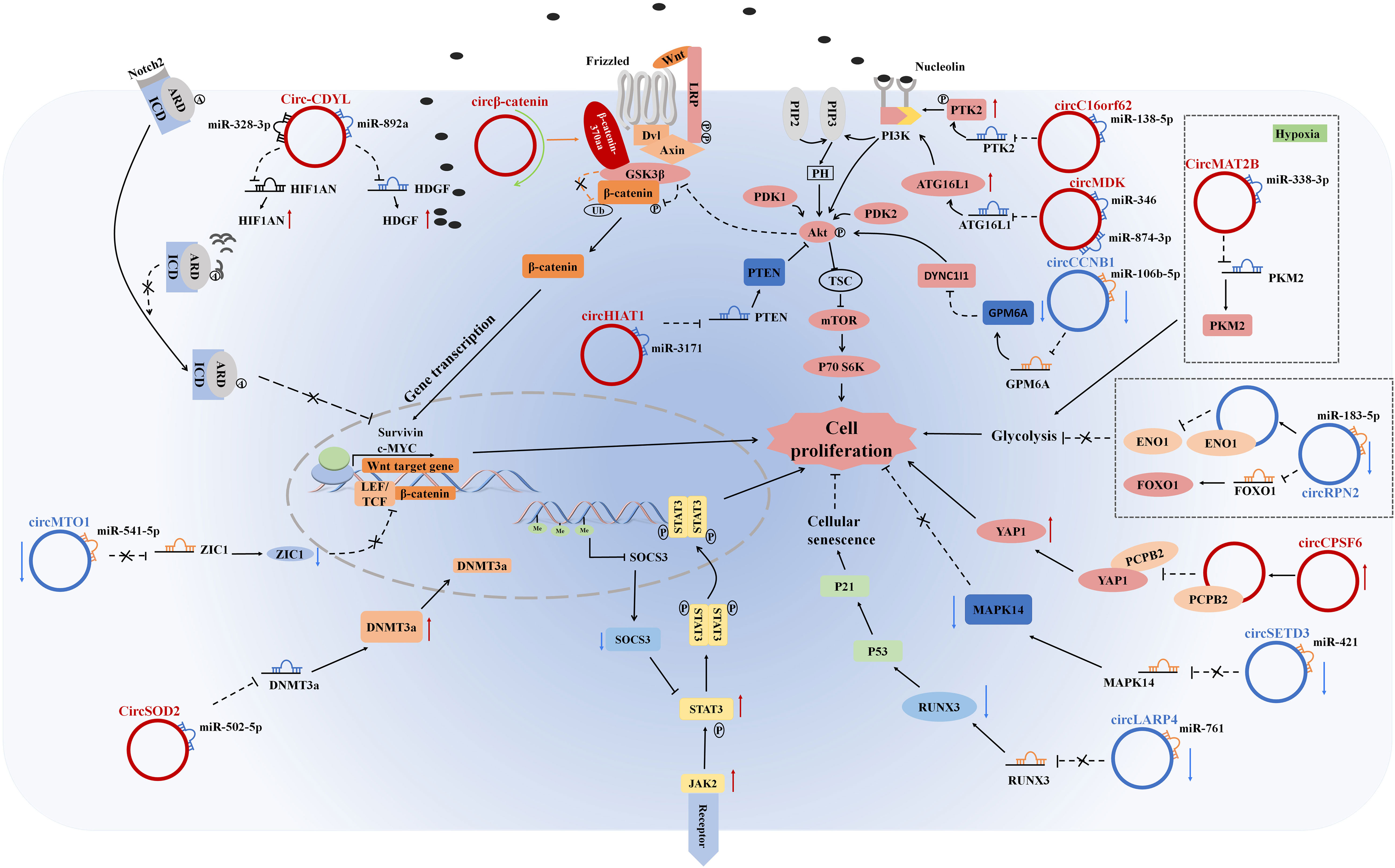
Figure 2 CircRNAs regulate cell proliferation, tumor growth suppression, glycolysis, and cellular senescence in HCC cells by stimulating/inhibiting cell proliferation signaling pathways.
Apoptosis, autophagy, and necrosis are three primary mechanisms contributing to programmed cell death that tumor cells are efficient at avoiding (40). Tumor cells can evade apoptosis, enabling cell immortality. STATs are latent transcription factors that participate in cell growth, development, apoptosis, and a series of other cellular events. Increasing evidence has demonstrated that STAT3 plays a critical role in apoptosis (41). As shown in previous studies, human HCC cells constantly stimulated the JAK1/STAT3 pathway to accelerate HCC progression. Circ9119 was a neoteric oncogene that increased cell propagation and inhibited apoptosis through the miR-26a/JAK1/STAT3 signaling pathway (42). Furthermore, m6A-modified circMAP3K4 can be translated into circMAP3K4-455, which decreased cisplatin-induced apoptosis and stimulated HCC progression by directly binding to apoptosis inducing factor mitochondria associated 1 (AIF), thus blocking its nuclear translocation (43). Clearly, circRNAs can regulate the apoptosis of HCC cells.
CSCs have been regarded as the origin of carcinogenesis, chemoresistance, tumor metastasis and recurrence. The Hedgehog signaling pathway plays a critical role in various biological processes including embryonic development, tissue homeostasis and regeneration. Irregular Hedgehog signaling may induce numerous human malignancies and the generation of CSCs. CircIPO11 interacted with TOP1 to trigger GLI1 transcription, thus initiating the self-renewal of liver CSCs and stimulating the development of HCC through mediating Hedgehog signals (44). In addition, CircZNF609 also promoted the proliferation, metastasis, and stemness of HCC cells by mediating the Hedgehog pathway by binding to miR-15a-5p/15b-5p and increasing the GLI2 expression (45). CircZKSCAN1 inhibited stemness by physically interacting with FMRP to prevent FMRP-CCAR1-induced signaling (12). Interestingly, m6A modification of circHPS5 facilitated its cytoplasmic output via an YTHDC1-dependent pattern and accelerated EMT and CSC phenotypes, facilitating tumorigenesis of HCC via the miR-370/HMGA2 axis (46).
Metastasis is characterized by a multistep process that allows tumor cells from the primary tumor to spread into circulation through lymphatic and blood vessels and colonize distant organs, ultimately leading to secondary tumors. MMP9 plays a crucial role in tumor progression and metastasis by degrading extracellular matrix (ECM). Recent studies have demonstrated that circUBAP2 accelerated the progression and metastasis of HCC by functioning as a sponge of miR-194-3p to upregulate MMP9 expression (47). MMP1 had also been demonstrated to play critical roles in migration and invasion in various types of cancers. CircDLC1 inhibited the proliferation and metastasis of HCC through the HuR-MMP1 axis (48). FBLIM1 is a crucial promotor of migration in a wide variety of cell types and is related to more aggressive HCC phenotypes. circFBLIM1 had been reported to promote HCC cell proliferation and invasion by acting as a ceRNA to upregulate FBLIM1 expression (49). SOX9 is a distinguished oncogenic transcription mediator in various human cancers that can activate diverse signaling pathways. Circ-FOXP1 facilitated HCC growth and metastasis via the SOX9/circ-FOXP1/miR-875-3p/miR-421 signaling pathway (50). Emerging evidence had shown that MAPK1 can function as an important tumor promoter in the progression of HCC. Consistent with these findings, circASAP1 accelerated HCC cell proliferation and invasion via miR-326/miR-532-5p MAPK1 signals (51). Focal adhesion kinase (FAK) is a nonreceptor tyrosine kinase which dysregulation had been noted in various types of tumors in relation to tumor metastasis. circRASGRF2 had been confirmed to be sharply increased in HCC and to accelerate HCC progression by sponging miR-1224 to upregulate FAK expression (52). The STAT3 signals is oncogenic and constantly hyperactivated in a variety of human cancers, including HCC. Circ-LRIG3 was notably overexpressed in HCC and functioned as a protein scaffold to promote EZH2-mediated STAT3 methylation and phosphorylation by physically interacting with EZH2 and STAT3. Furthermore, activated STAT3 immediately bound to the circ-LRIG3 promoter region to enhance the transcriptional activity of circ-LRIG3, thereby forming a positive feedback pathway to accelerate the metastasis of HCC (53). Moreover, circRHOT1 accelerated HCC growth and metastasis by recruiting TIP60 to upregulate NR2F6 expression (13).
EMT is an important developmental program for tumor metastasis. Twist1 is a critical EMT-related transcription factor that regulates the expression of EMT-related genes through promoter activation or suppression, inhibiting the transcription of epithelial phenotype-associated genes. For example, Twist1 enhanced vimentin expression and HCC tumorigenesis and metastasis via Twist/circ-10720/vimentin signaling pathway (54). PCBP1 participated in the EMT program through various cancer processes, particularly TGF-β signaling. Importantly, has_circ_0003998 was highly overexpressed in HCC and it participated in EMT via both miR-143-3p/FOSL2 signaling and the PCBP1/CD44v6 axis (55). Moreover, EIF4A3-induced circTOLLIP facilitated the progression of HCC by regulating the miR-516a-5p/PBX3/EMT signaling pathway (56). Hsa_circ_0003288 functioned as an oncogene to facilitate EMT and invasion of HCC by sponging miR-145 and increasing the expression of PD-L1 via the PI3K/AKT signaling pathway (57). The Wnt/β-catenin pathway is a crucial signaling cascade strongly related to tumor progression and its activation facilitates tumor invasion through the upregulation of factors modulating EMT. Circ_0067934, as an oncogene, facilitated HCC development by upregulating the miR-1324/FZD5/Wnt/β-catenin axis (58).
DAB2IP functions as a tumor suppressor for a variety of tumors. circRNA-5692 had been found to inhibit HCC development and EMT progression by accelerating demethylation of the DAB2IP gene and enhancing its expression and circRNA-5692 also acted as a tumor suppressor via the miR-328-5p/DAB2IP pathway (59). EZH2, a histone methyltransferase, is the enzymatically active core subunit of the polycomb repressive complex 2 (PRC2) and is a notable oncoprotein that plays an essential role in malignant cancer behaviors, especially metastasis (60). Circ-ADD3 facilitated the binding of CDK1 and EZH2 by directly interacting with them, leading to subsequent ubiquitination and degradation and ultimately restraining HCC metastasis (61). circTRIM33-12 had been found to be significantly reduced in HCC and functioned as an important tumor suppressor to restrain the proliferation, metastasis and immune evasion of HCC cells by sponging miR-191 and inducing TET1 expression (62). SMAD2, plays a critical role in accelerating EMT progression. However, circSMAD2, originating from SMAD2, was markedly decreased in HCC tissues and restrains the metastasis and EMT of HCC cells by directly interacting with miR-629 (63). Moreover, SMARCA5 acted as a tumor promoter and regulated the Wnt/β-catenin signaling pathway to stimulate the proliferation of HCC cells. However, cSMARCA5, generated by SMARCA5, a tumor suppressor, was notably reduced in HCC and may restrain the growth and metastasis of HCC through the DHX9-cSMARCA5-miR-17-3p/miR-181b-5p-TIMP pathway (64).
Taken together, circRNAs are critical molecular regulators of the EMT process and motility in HCC.
Chemoresistance is inevitable in HCC treatment. Sorafenib, a multi-target tyrosine kinase inhibitor (TKI), is a first-line treatment for advanced HCC. However, drug resistance consistently appears after long-term application. Numerous factors related to sorafenib resistance in HCC have been demonstrated, such as the Wnt/β-catenin pathway, EMT, the TME and epigenetic regulation (65). Increasing evidence have elucidated that circRNAs were participated in the sensitivity of HCC to chemotherapy. circRNA-SORE was overexpressed in sorafenib-resistant HCC cells and reduced the sensitivity of HCC cells to sorafenib by interacting with miR-103a-2-5p and miR-660-3p; in addition, circRNA-SORE competitively upregulated the Wnt/β-catenin pathway, thereby impairing chemotherapy effectiveness (66). YBX1 is tightly related to tumor progression, chemoresistance, and cancer prognosis. Interestingly, another study demonstrated that circRNA-SORE, which can be delivered extracellularly via exosomes, playing a vital role in prohibiting PRP19-induced YBX1 degradation and was thereby involved in the maintenance and spread of sorafenib resistance in HCC (14). CircFOXM1 was a crucial promotor of sorafenib resistance in HCC that can interact with miR-1324 to stimulate MECP2 expression and therefore strengthened sorafenib resistance in HCC (67). Lenvatinib, another multitarget TKI, had illustrated promising antitumor activity in HCC patients. However, the molecular mechanisms of primary and acquired resistance to lenvatinib remain to be revealed, creating a critical challenge for HCC targeted therapy. CircMED27 promoted lenvatinib resistance of HCC through sponging miR-655-3p and increasing the expression of USP28 (68). CircKCNN2, transcriptionally inhibited by NFYA, suppressed HCC recurrence and associated with lenvatinib resistance by reducing the FGFR4 expression through the miR-520c-3p/MBD2 axis (69).
In addition to sorafenib and lenvatinib, cisplatin is one of the most universally applied chemotherapy drugs to cure advanced HCC. However, acquired multidrug resistance (MDR) in response to cisplatin treatment remains the biggest obstacle to an ideal therapeutic effect in HCC patients (70). Indeed, circARNT2 was highly expressed in cisplatin-resistant HCC tissues and functions as a tumor promotor by inhibiting miR-155-5p, leading to PDK1 overactivation and eventually resistance to cisplatin in HCC cells (71). Another study revealed that circMRPS35 exerted its oncogenic function in HCC not only by sponging miR-148a to modulate the STX3-PTEN pathway, but also by being translated into circMRPS35-168aa, resulting in cisplatin resistance, which indicated that circMRPS35 could be a crucial factor in the progression and chemoresistance of HCC with different expression patterns under different conditions (16).
In summary, emerging reports have illustrated that circRNAs play a crucial role in the drug resistance of HCC. However, research on the role of circRNAs in chemoresistance is still at the early stage, and we must further elucidate the chemoresistance mechanism of countless circRNAs in HCC. Moreover, future translational studies and/or clinical trials are necessary to develop circRNA-targeted treatments, which may ultimately improve the prognosis of HCC patients by enabling patients to gradually overcome drug resistance.
Tumor angiogenesis, which is important for cancerous growth, continuously supplies malignant tissues with necessary oxygen and nutrients and removes metabolic wastes, playing an essential role in tumor progression and development. Without angiogenesis, tumors cannot grow beyond a size of 1-2 mm (72). FGF signaling pathways are some of the most potent stimulants of angiogenesis and can facilitate the proliferation, migration and differentiation of endothelial cells in culture. During hepatocarcinogenesis, FGFR1 disrupts DNA synthesis and cell proliferation at an early stage and further boosts neoangiogenesis at later stages, and these processes are mediated by vascular endothelial growth factor. According to Zhan et al. the hsa_circ_103809/miR-377-3p/FGFR1 axis may play an essential role in the development of HCC (73).
The TME is dependent on the crosstalk between various cell types, especially Cancer-associated fibroblasts (CAFs), ECM cells and infiltrating immune cells and plays a crucial role in the progression, metastasis and therapeutic treatment of cancer (74). A growing amount of evidence has revealed the intricate relationship between circRNAs and critical components in the TME.
Tumor cells develop and metastasize through a variety of mechanisms to evade recognition and attack by the immune system, a phenomenon known as tumor immune escape. Immunosuppression is one of the most vital mechanisms of tumor immune escape. Snail is a transcriptional mediator of DPP4 that promotes local immunosuppression by decreasing lymphocyte infiltrating via decreasing the expression of the chemokine CXCL10. A recent report claimed that circMET acted as a novel onco-circRNA that stimulated HCC progression and immune escape through the snail/DPP4/CXCL10 signaling pathway (75).
Natural killer (NK) cells are regarded as the major cell involved in host immune surveillance and play a substantial role in antitumor immunotherapy. Growing evidence illustrated that circRNAs secreted by tumor cells played an essential role in tumor immune surveillance by strengthening NK-cell activity and enhancing NK-cell-mediated immune surveillance. For instance, circARSP91 was found to enhance the ability of NK cells to recognize and attack target tumor cells by interacting with ULBP1 to increase the expression of ULBP1 (76). However, hsa_circ_0007456 may extremely impair HCC cells susceptibility to NK cells via the miR-6852-3p/ICAM-1 signaling pathway (77). Tumor-associated macrophages (TAMs), especially TAMs with the M2 phenotype are the most crucial immune cells in the TME that stimulate tumor progression and metastasis. CircASAP1 facilitated macrophage proliferation and chemotactic migration by sponging miR-326 and miR-532-5p and thus inducing overexpression of CSF-1, which was a crucial regulator of macrophage differentiation and function, resulting in macrophage recruitment to the tumor bed (51). CCL2, as a chemokine, bind to C-C motif chemokine receptor 2 (CCR2) and recruited monocytes and macrophages, and CCL2 was regulated by the activation of PPARα. Hsa_circ_0110102 regulated the miR-580-5p/PPARα/CCL2 signaling pathway in HCC. Interestingly, CCL2 released into the TME mediated the expression and delivery of COX-2 and PGE2 in macrophages to stimulate the proliferation of HCC cells (78). Moreover, hsa_circ_0003410 facilitated HCC progression by increasing the ratio of M2/M1 macrophages through stimulating the expression of CCL5 (79).
In addition, CAFs play an important role in tumor progression by modulating the inflammatory microenvironment. CXCL11 functioned as a critical regulator in regulating the interaction between CAFs and CAF-adjacent cancer cells and served as an extracellular remodeler to facilitate HCC cell migration and metastasis by upregulating the circUBAP2/miR-4756/IFIT1/3 pathway (80).
Growing evidence suggested that circRNAs could participate in HCC progression by affecting the immune system of HCC patients (Figure 3). However, research on circRNAs in the TME is still in its infancy, and the specific roles of circRNAs in the TME remain to be further explored.
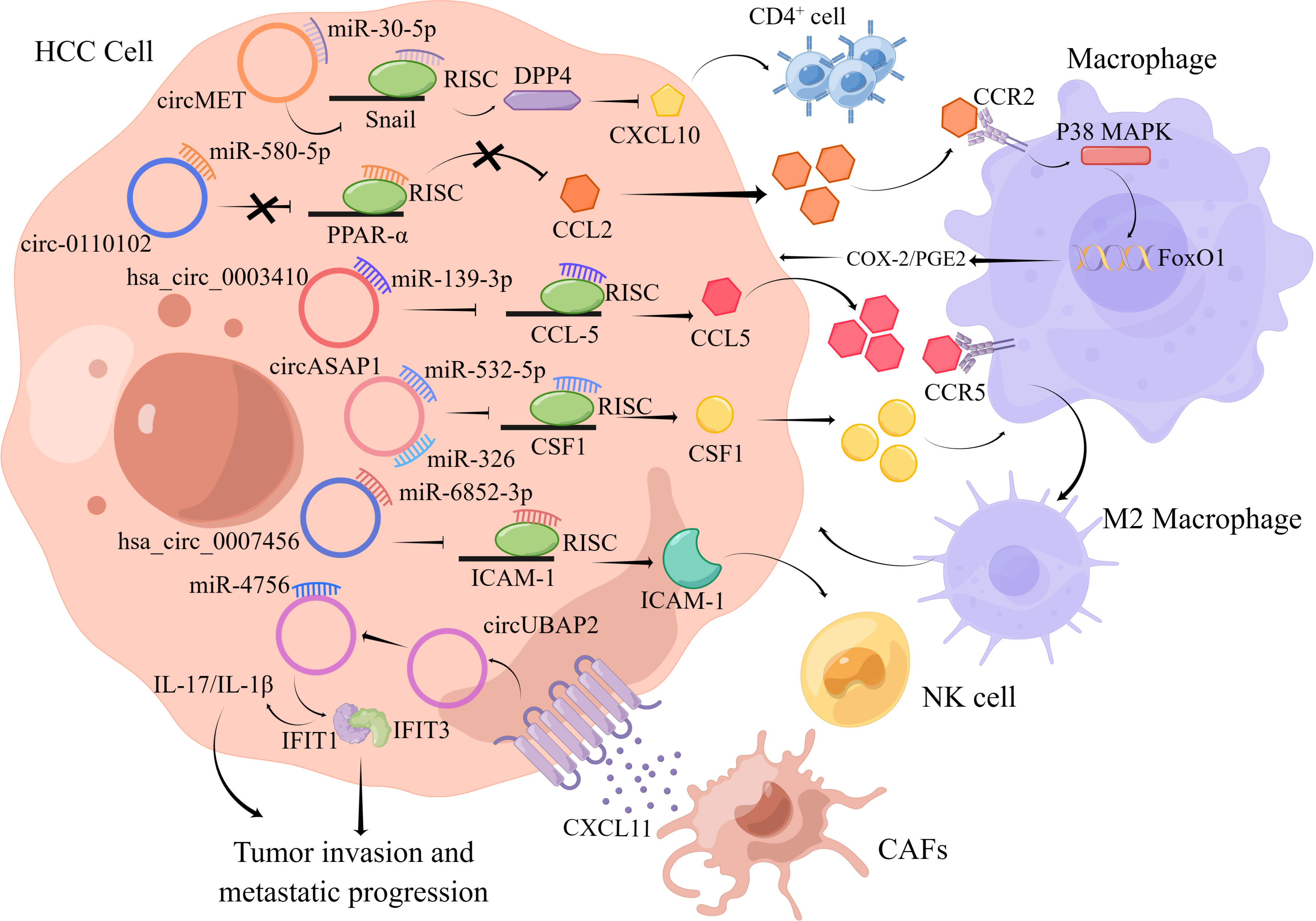
Figure 3 The figure illustrates the interaction between circRNAs and important components in the HCC tumor microenvironment.
Exosomes are small extracellular vesicular bodies with a diameter of 30-100 nm that are released from a variety of cells and contain mRNAs, miRNAs, circRNAs and proteins that may affect various biological functions of recipient cells (81). Emerging evidence have revealed that exosomes exert a crucial function in tumor development processes, including angiogenesis, metastasis, drug resistance and TME processes. Recently, exosome-derived circRNAs have been proven to be a potential factor related to tumorigenesis and progression. Evidence provided that exosomal circPTGR1 from highly metastatic cells could have a notable impact on cell metastatic potential by restraining the miR449a-MET interaction in recipient cells, resulting in lethal damage in TME homeostasis and accelerating HCC progression (82). In another study, circ_MMP2 was found to be transported by 97-Hderived exosomes stimulated HCC metastasis by sponging miR-136-5p to enhance the expression of its target gene, MMP2 (50). Tumor cell-derived exosomal circ-0072088 promoted the migration and invasion of HCC cells by sponging miR-375 and upregulating MMP-16 (83). Notably, circRNA-SORE was delivered by exosomes, which promoted transmission of sorafenib resistance among HCC cells (14).Moreover, exosomal circ-0004277 directly derived from HCC cells can be delivered to normal surrounding cells and may modulate their biological functions by suppressing the expression of ZO-1 and stimulating the development of EMT (84). In another study, circRNA Cdr1as functioned as a ceRNA to accelerate the development of HCC by inhibiting miR-1270 to increase AFP expression. In addition, the exosomal circRNA Cdr1as secreted by HCC cells, can be delivered to surrounding normal cells to ultimately promote the malignant processes of peripheral normal cells (85).Moreover, exosome circ-DB generated from adipocytes was increased in HCC patients with higher body fat values. Moreover, researchers have illustrated that exo-circ-DB increased HCC progression and reduced DNA damage by reducing the level of miR-34a and activating the deubiquitination-related USP7/cyclin A2 signaling pathway (86).
Interestingly, exosomes may generate an immunosuppressive environment favoring tumor cell survival by serving as messengers between tumor cells and peripheral recipient cells. HCC-secreted exosomal circUHRF1 increased the expression of the miR-449c-5p target gene TIM-3 in NK cells by absorbing miR-449c-5p, and thus stimulated immune escape and reduced sensitivity to anti-PD1 immunotherapy in HCC (87). HCC-derived exosomal circTMEM181 sponged miR-488-3p and elevated the expression of the CD39 in macrophages and the expression of the CD73 in HCC cells, which collaboratively stimulated the eATP-adenosine pathway and generated more adenosine, damaging CD8+ T cell function to indulge immunosuppression and acquire anti-PD1 resistance in HCC (88). Moreover, exosomal circGSE1 exerted tumor immunosuppressive functions by inducing the expansion of Tregs via activation of the miR-324-5p/TGFBR1/Smad3/FOXP3 signaling pathway (89). Interestingly, hsa_circ_00074854 may regulate the growth and metastasis of HCC cells by enhancing HuR protein stability and activating the ZEB1 axis. In addition, HCC cell-derived hsa_circ_00074854 may be transmitted to macrophages by exosomes and stimulated macrophage polarization toward the M2 type, which facilitated the metastasis and EMT of HCC cells (90). In addition, exosomal hsa_circ_0004658 secreted by RBPJ (recombination signal binding protein-Jκ) -overexpressing macrophages inhibited HCC progression through the miR-499b-5p/JAM3 pathway (91).
This evidence confirmed that exosomal circRNAs secreted by HCC cells, normal cells and immune cells exert critical functions in HCC progression, metastasis and drug resistance, suggesting that it is important to study the molecular mechanisms enabling exosomal circRNAs from different secreted sources to play an important role in HCC progression (Figure 4).
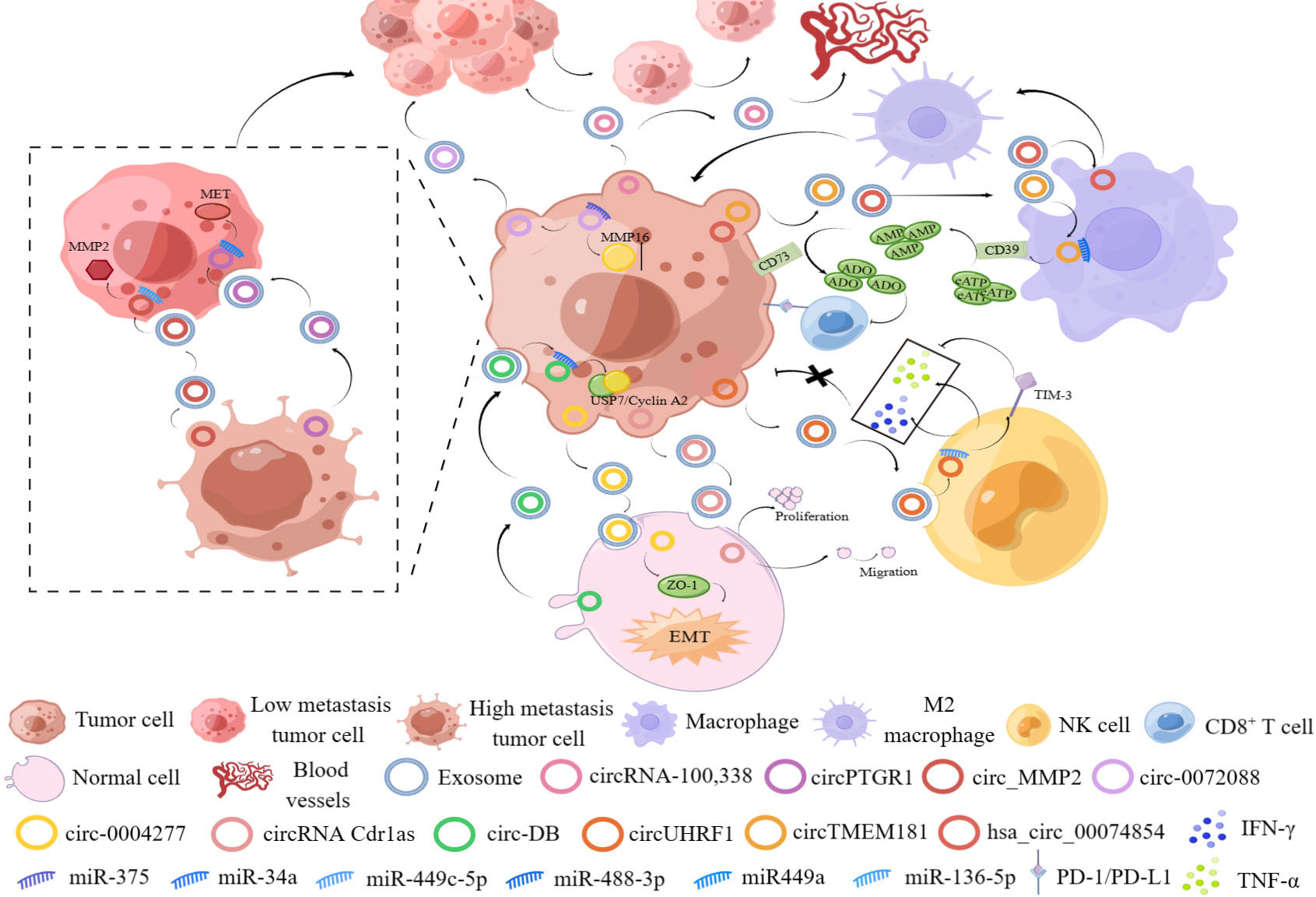
Figure 4 The figure shows the important role of exosomal circRNAs from different sources in the occurrence and development of HCC.
Due to the lack of obvious symptoms at the early stage and lack of dependable and valid biomarkers, HCC patients are frequently diagnosed at the advanced stage and the overall survival (OS) rate is less than ideal. Furthermore, current diagnostic biomarkers, including α-fetoprotein (AFP), α-fetoprotein-L3 (AFP-L3), and des-carboxy-prothrombin (DCP), show poor sensitivity in diagnosing HCC. CircRNAs are closely related to various biological processes in HCC, and circRNAs are different from their homologous linear RNAs, as they have a distinctive closed-loop construction structure that aids detection in tissues, saliva, plasma, serum, urine, exosomes and other biological samples. Furthermore, circRNAs have been proposed as novel early- stage diagnostic and prognostic biomarkers for HCC because of their abundance, stability, conservation nature and specificity in tissues and cells.
Emerging evidence utilizing clinical HCC samples have illustrated the aberrant expression, increased disease specificity and clinical significance of specific circRNAs, suggesting that they are attractive biomarker candidates for HCC diagnosis (Table 1). For example, circZNF566, which functioned as a tumor promoter, was increased significantly in HCC tissues. The AUC of circZNF566 was 0.834, with a sensitivity and specificity of 82.2% and 72.4%, respectively, showing that circZNF566 was a potential biomarker for the clinical diagnosis and evaluation of HCC (92). The AUC of circRNA_104075 for HCC diagnosis was 0.973, and the sensitivity and specificity were 96.0 and 98.3%, respectively, indicating remarkable diagnostic value (93). The AUC of hsa_circ_0091570 in diagnosing HCC was 0.736, suggesting that it could be used as a diagnostic biomarkers (94). CircCRIM1 was significantly elevated in HCC tissues and was related to large tumor size and advanced TNM stage and Edmondson grade. Thus, circCRIM1 was a potential prognostic biomarker for HCC (95). In addition to HCC tissue, serum exosomes can also be assessed to reveal expression changes of circRNAs as diagnostic biomarkers for HCC. Compared with nonmetastatic HCC samples, samples from HCC patients with pulmonary metastasis showed notable increases in exosome-derived circRNA-100338 in serum (96). Moreover, plasma hsa_circ_0005397 combined with serum AFP and AFP-L3 had more promising diagnostic performance (97).
CircRNAs can function as biomarkers to predict patient survival parameters as well, including overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS). To evaluate the prognostic function of circRNAs in HCC, we gathered data from reporter and analyzed the connections between circRNA expression and OS, DFS, and PFS (Table 2). Twenty increased circRNAs and eleven decreased circRNAs were illustrated to predict poor OS. Upregulated circMAP3K4, circRHBDD1 and circTOLLIP expressions were related to poor prognosis implying that these circRNAs were prospective prognostic markers for HCC (26, 43, 56). Furthermore, downregulated expression of circUBE2J2 and circKCNN2 can serve as independent predictors of poor OS in HCC (69, 98).
In summary, the abnormal expression of circRNAs can be monitored not only in tumor tissues but also in body fluids, such as blood, saliva and urine, as well as in exosomes in these fluids, suggesting that they are valuable noninvasive biomarker candidates. However, there are still many challenges ahead in exploring circRNAs as biomarkers for the clinical diagnosis and prognostication of HCC, especially those derived from blood and urine. In another way, a single circRNA class is unlikely to be able to predict HCC progression or therapy effects, as the expression may vary per patient and the heterogeneity of tumors in different patients.
In the emerging world of RNA drugs, circRNA, as a novel multifunctional therapeutic target that can transmit genetic information, is expected to be a potential alternative to mRNA due to its excellent stability. As mentioned above, circRNAs are more stable than mRNA and specific express in tissue or cell type which may exert potential therapeutic targets in HCC. Results of previous study illustrated that some potential treatments such as RNAi vectors, conditional circRNAs knockout or knockdown through the cre-lox system, CRISPR/Cas9 and CRISPR/Cas13 system (99). A troublesome in utilizing circRNAs as therapeutic targets is the conveyance of the circRNA-targeted-siRNA or circRNA overexpressed vector in vivo. In one way, exosomes are being explored as delivery platforms for circRNA-targeted agents. As previously mentioned, some exosome-circRNAs can facilitate the transformation of the tumor microenvironment and promote or suppress the invasion and drug resistance of HCC. However, low loading efficiencies and low exosome yields are capital challenge to overcome when using exosomal circRNAs for the treatment of HCC. Interestingly, in another way, the use of nanoparticles (NPs) as a delivery platform has dramatically enhanced the feasibility of circRNA-targeted treatment in vivo. The PAE-based si-circMDK nanoparticles effectively inhibited HCC proliferation and metastasis in vivo (18). PLGA-PEG (si-circROBO1) NPs exhibited excellent anti-HCC activity in vitro and in vivo (100). Lipid nanoparticles (LNPs) are clinically advanced carriers for delivering RNA to target organs. Since most LNPs have a propensity to accumulate in hepatocytes and limit their use to targets outside of the liver (101). This limitation may be of great aid in the delivery of circRNAs to the liver for the treatment of HCC. However, the treatment methods for circRNA are still in the preliminary stage, further clinical trials investigating the feasibility of them are required.
Initially, circRNAs were regarded as the results of random transcriptional RNA splicing errors in transcription. With the rapid advancement of high throughput sequencing technologies and bioinformatics tools, growing evidence has illustrated that numerous aberrantly expressed circRNAs have a strong correlation with the progression of HCC. Some differentially expressed circRNAs are correlated with the clinicopathological features in HCC patients. Moreover, aberrant circRNA expression exists, and increases in the expression of some circRNAs are related to the tumorigenesis, proliferation, metastasis, apoptosis and drug resistance of HCC. Accumulating studies have demonstrated the biological function of circRNAs, including miRNA sponges, RBP sponges, regulators of gene transcription and potential regulators of translation. However, the comprehensive biological functions and specific roles of circRNAs in HCC occurrence and development remain uncertain.
Novel insights into circRNAs have emerged rapidly. However, circRNAs in HCC are still relatively underexplored compared with miRNAs and lncRNAs in HCC and most studies have been cross-sectional association studies. Furthermore, the mechanisms of circRNA formation, cellular localization and degradation are still not clear. In addition, only little critical circRNAs in HCC have been distinguished and characterized. Innovative roles of circRNAs in addition to their role as miRNA sponges need to be revealed. Notably, recent studies have point to the importance of using tumor tissues, perhaps the exploration of circRNAs in tumor tissues is a more intuitive reflection of HCC progression than tumor cell lines. In the future, studies detecting the level of circRNAs in body fluids and analyzing the sensitivity and specificity of such circRNAs as biomarkers are needed. The ultimate purpose of various circRNA studies is to enable the application of circRNA in early clinical diagnosis and early treatment, and many experiments are still needed to accomplish this goal.
Z-DL and CZ wrote, revised and edited the manuscript. Z-DL, JL and SL prepared all tables and figures. Y-LL and L-HZ supervised the study. All authors participated in the design of the review and revised the manuscript. All authors have read and approved the final manuscript.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by National Natural Science Foundation of China [82273352], Zhejiang Provincial Natural Science Foundation of China [LTY21H160001, LY21H160017, 2021], Key Laboratory of Clinical Cancer Pharmacology and Toxicology Research of Zhejiang Province [2020E10021], Huadong Medicine Joint Funds of the Zhejiang Provincial Natural Science Foundation of China [HDMY22H160421, 2022], Public-service Technology Research Plan of Zhejiang Province [LGF21H310002, 2021], Zhejiang Provincial Medical and Health Technology Project [2021433724, 2021; 2020RC026, 2020].
We would like to thank Figdraw for its help in creating the figures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1192386/full#supplementary-material
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Raza A, Sood GK. Hepatocellular carcinoma review: current treatment, and evidence-based medicine. World J Gastroenterol (2014) 20(15):4115–27. doi: 10.3748/wjg.v20.i15.4115
3. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
4. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Bio (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
5. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J (2019) 38(16):e100836. doi: 10.15252/embj.2018100836
6. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene (2018) 37(5):555–65. doi: 10.1038/onc.2017.361
7. Zhang XO, Wang HB, Zhang Y, Lu XH, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
8. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
9. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol (2020) 21(8):475–90. doi: 10.1038/s41580-020-0243-y
10. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol (2022) 19(3):188–206. doi: 10.1038/s41571-021-00585-y
11. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
12. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics (2019) 9(12):3526–40. doi: 10.7150/thno.32796
13. Wang LY, Long HY, Zheng QH, Bo XT, Xiao XH, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer (2019) 18:119. doi: 10.1186/s12943-019-1046-7
14. Xu JJ, Ji L, Liang YL, Wan Z, Zheng W, Song XM, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Tar Therapy (2020) 5(1):298. doi: 10.1038/s41392-020-00375-5
15. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol (2019) 20(1):84. doi: 10.1186/s13059-019-1685-4
16. Li P, Song R, Yin F, Liu M, Liu H, Ma S, et al. CircMRPS35 promotes Malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther (2022) 30(1):431–47. doi: 10.1016/j.ymthe.2021.08.027
17. Wu Y, Zhang Y, Qin X, Geng H, Zuo D, Zhao Q. PI3K/AKT/mTOR pathway-related long non-coding RNAs: roles and mechanisms in hepatocellular carcinoma. Pharmacol Res (2020) 160:105195. doi: 10.1016/j.phrs.2020.105195
18. Du A, Li S, Zhou Y, Disoma C, Liao Y, Zhang Y, et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer (2022) 21(1):109. doi: 10.1186/s12943-022-01575-z
19. Wei YP, Chen X, Liang C, Ling Y, Yang XW, Ye XF, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology (2020) 71(1):130–47. doi: 10.1002/hep.30795
20. Zhang S, Lu Y, Jiang HY, Cheng ZM, Wei ZJ, Wei YH, et al. CircC16orf62 promotes hepatocellular carcinoma progression through the miR-138-5p/PTK2/AKT axis. Cell Death Dis (2021) 12(6):597. doi: 10.1038/s41419-021-03866-7
21. Huang XY, Huang ZL, Zhang PB, Huang XY, Huang J, Wang HC, et al. CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Front Oncol (2019) 9:392. doi: 10.3389/fonc.2019.00392
22. Zhao ZW, Song JJ, Tang BF, Fang SJ, Zhang DK, Zheng LY, et al. CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J Exp Clin Canc Res (2020) 39(1):259. doi: 10.1186/s13046-020-01769-7
23. Li D, Zhang J, Yang J, Wang J, Zhang R, Li J, et al. CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating Wnt/beta-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis (2021) 13(1):12. doi: 10.1038/s41419-021-04464-3
24. Chen Y, Ling Z, Cai X, Xu Y, Lv Z, Man D, et al. Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives Malignancy in hepatocellular carcinoma. Cancer Res (2022) 82(4):599–614. doi: 10.1158/0008-5472.CAN-21-1628
25. Li J, Hu ZQ, Yu SY, Mao L, Zhou ZJ, Wang PC, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res (2022) 82(6):1055–69. doi: 10.1158/0008-5472.CAN-21-1259
26. Cai J, Chen Z, Zhang Y, Wang J, Zhang Z, Wu J, et al. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol Ther Oncolytics (2022) 24:755–71. doi: 10.1016/j.omto.2022.02.021
27. Luo YY, Tao KG, Lu YT, Li BB, Wu KM, Ding CH, et al. Hsa_Circ_0098181 suppresses hepatocellular carcinoma by sponging miR-18a-3p and targeting PPARA. Front Pharmacol (2022) 13:819735. doi: 10.3389/fphar.2022.819735
28. Chen S, Cao X, Zhang J, Wu W, Zhang B, Zhao F. circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-myc translation. Adv Sci (Weinh) (2022) 9(8):e2103817. doi: 10.1002/advs.202103817
29. Dai M, Boudreault J, Wang N, Poulet S, Daliah G, Yan G, et al. Differential regulation of cancer progression by CDK4/6 plays a central role in DNA replication and repair pathways. Cancer Res (2021) 81(5):1332–46. doi: 10.1158/0008-5472.CAN-20-2121
30. Gong YH, Mao JZ, Wu D, Wang XM, Li L, Zhu L, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int (2018) 18:116. doi: 10.1186/s12935-018-0602-3
31. Yang S, Yu F, Ji Y, Shen Y, Lu H, Gao Y, et al. Circular RNA ERBIN Promotes Proliferation of Hepatocellular Carcinoma via the miR-1263/CDK6 Axis. Front Oncol (2022) 12:878513. doi: 10.3389/fonc.2022.878513
32. Guan Z, Tan J, Gao W, Li X, Yang YD, Li XG, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol (2019) 234(1):500–8. doi: 10.1002/jcp.26612
33. Li M, Chen H, Xia L, Huang P. Circular RNA circSP3 promotes hepatocellular carcinoma growth by sponging microRNA-198 and upregulating cyclin-dependent kinase 4. Aging (Albany NY) (2021) 13(14):18586–605. doi: 10.18632/aging.203303
34. Liu YM, Cao Y, Zhao PS, Wu LY, Lu YM, Wang YL, et al. CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway. Int J Biol Sci (2022) 18(2):637–51. doi: 10.7150/ijbs.66915
35. Han D, Li JX, Wang HM, Su XP, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (2017) 66(4):1151–64. doi: 10.1002/hep.29270
36. Wang ZM, Zhao YX, Wang Y, Jin CX. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. BioMed Pharmacother (2019) 116:108932. doi: 10.1016/j.biopha.2019.108932
37. Xu LL, Feng XF, Hao XY, Wang P, Zhang YF, Zheng XB, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Canc Res (2019) 38(1):98. doi: 10.1186/s13046-019-1041-2
38. Frey N, Venturelli S, Zender L, Bitzer M. Cellular senescence in gastrointestinal diseases: from pathogenesis to therapeutics. Nat Rev Gastroenterol Hepatol (2018) 15(2):81–95. doi: 10.1038/nrgastro.2017.146
39. Chen ZQ, Zuo XL, Pu LY, Zhang Y, Han GY, Zhang L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci (2019) 110(2):568–81. doi: 10.1111/cas.13901
40. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer (2015) 14:48. doi: 10.1186/s12943-015-0321-5
41. Chai EZ, Shanmugam MK, Arfuso F, Dharmarajan A, Wang C, Kumar AP, et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol Ther (2016) 162:86–97. doi: 10.1016/j.pharmthera.2015.10.004
42. Yang LX, Xue H, Sun YF, Zhang L, Xue F, Ge RL. CircularRNA-9119 protects hepatocellular carcinoma cells from apoptosis by intercepting miR-26a/JAK1/STAT3 signaling. Cell Death Dis (2020) 11(7):605. doi: 10.1038/s41419-020-02807-0
43. Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC, Liang H, et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer (2022) 21(1):93. doi: 10.1186/s12943-022-01537-5
44. Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N, et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer (2021) 20(1):132. doi: 10.1186/s12943-021-01435-2
45. He YK, Huang H, Jin L, Zhang F, Zeng M, Wei L, et al. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis (2020) 11(5):358. doi: 10.1038/s41419-020-2441-0
46. Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids (2021) 26:637–48. doi: 10.1016/j.omtn.2021.09.001
47. Liu B, Tian Y, Chen M, Shen H, Xia J, Nan J, et al. CircUBAP2 Promotes MMP9-Mediated Oncogenic Effect via Sponging miR-194-3p in Hepatocellular Carcinoma. Front Cell Dev Biol (2021) 9:675043. doi: 10.3389/fcell.2021.675043
48. Liu H, Lan T, Li H, Xu L, Chen X, Liao H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics (2021) 11(3):1396–411. doi: 10.7150/thno.53227
49. Bai N, Peng EM, Qiu XS, Lyu N, Zhang ZJ, Tao YM, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Canc Res (2018) 37(1):172. doi: 10.1186/s13046-018-0838-8
50. Wang W, Li Y, Li X, Liu B, Han S, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. BioMed Pharmacother (2020) 121:109517. doi: 10.1016/j.biopha.2019.109517
51. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies circASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology (2020) 72(3):906–22. doi: 10.1002/hep.31068
52. Wu D, Xia AL, Fan TL, Li GQ. circRASGRF2 functions as an oncogenic gene in hepatocellular carcinoma by acting as a miR-1224 sponge. Mol Ther-Nucl Acids (2021) 23:13–26. doi: 10.1016/j.omtn.2020.10.035
53. Sun SF, Gao J, Zhou S, Li Y, Wang Y, Jin L, et al. A novel circular RNA circ-LRIG3 facilitates the Malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Canc Res (2020) 39(1):252. doi: 10.1186/s13046-020-01779-5
54. Meng J, Chen S, Han JX, Qian BX, Wang XR, Zhong WL, et al. Twist1 regulates vimentin through cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res (2018) 78(15):4150–62. doi: 10.1158/0008-5472.CAN-17-3009
55. Song LN, Qiao GL, Yu J, Yang CM, Chen Y, Deng ZF, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Canc Res (2020) 39(1):114. doi: 10.1186/s13046-020-01576-0
56. Liu Y, Song J, Zhang H, Liao Z, Liu F, Su C, et al. EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway. J Exp Clin Cancer Res (2022) 41(1):164. doi: 10.1186/s13046-022-02378-2
57. Xu G, Zhang P, Liang H, Xu Y, Shen J, Wang W, et al. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int (2021) 21(1):212. doi: 10.1186/s12935-021-01902-2
58. Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun (2018) 497(2):626–32. doi: 10.1016/j.bbrc.2018.02.119
59. Liu ZG, Yu YQ, Huang ZB, Kong Y, Hu XW, Xiao W, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis (2019) 10(12):900. doi: 10.1038/s41419-019-2089-9
60. Laugesen A, Hojfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell (2019) 74(1):8–18. doi: 10.1016/j.molcel.2019.03.011
61. Sun SF, Wang W, Luo XY, Li Y, Liu BW, Li XF, et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am J Cancer Res (2019) 9(8):1695.
62. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer (2019) 18(1):105. doi: 10.1186/s12943-019-1031-1
63. Zhang XW, Luo P, Jing W, Zhou H, Liang CZ, Tu JC. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Oncotargets Ther (2018) 11:2853–63. doi: 10.2147/OTT.S158008
64. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol (2018) 68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012
65. Xia S, Pan Y, Liang Y, Xu J, Cai X. The microenvironmental and metabolic aspects of sorafenib resistance in hepatocellular carcinoma. Ebiomedicine (2020) 51:102610. doi: 10.1016/j.ebiom.2019.102610
66. Xu JJ, Wan Z, Tang MY, Lin ZJ, Jiang S, Ji L, et al. N-6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating beta-catenin signaling. Mol Cancer (2020) 19(1):163. doi: 10.1186/s12943-020-01281-8
67. Weng HQ, Zeng LX, Cao LH, Chen T, Li YS, Xu YXX, et al. circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Mol Ther-Nucl Acids (2021) 23:811–20. doi: 10.1016/j.omtn.2020.12.019
68. Zhang P, Sun H, Wen P, Wang Y, Cui Y, Wu J. circRNA circMED27 acts as a prognostic factor and mediator to promote lenvatinib resistance of hepatocellular carcinoma. Mol Ther Nucleic Acids (2022) 27:293–303. doi: 10.1016/j.omtn.2021.12.001
69. Liu D, Liu W, Chen X, Yin J, Ma L, Liu M, et al. circKCNN2 suppresses the recurrence of hepatocellular carcinoma at least partially via regulating miR-520c-3p/methyl-DNA-binding domain protein 2 axis. Clin Transl Med (2022) 12(1):e662. doi: 10.1002/ctm2.662
70. Kalyan A, Nimeiri H, Kulik L. Systemic therapy of hepatocellular carcinoma: current and promising. Clin Liver Dis (2015) 19(2):421–32. doi: 10.1016/j.cld.2015.01.009
71. Li YY, Zhang YJ, Zhang S, Huang DY, Li BS, Liang GC, et al. circRNA circARNT2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the miR-155-5p/PDK1 Axis. Mol Ther-Nucl Acids (2021) 23:244–54. doi: 10.1016/j.omtn.2020.08.037
72. Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis (2017) 20(4):409–26. doi: 10.1007/s10456-017-9562-9
73. Zhan W, Liao X, Chen ZS, Li LH, Tian T, Yu L, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol (2020) 235(2):1733–45. doi: 10.1016/j.omtn.2020.08.037
74. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer (2020) 19(1):14. doi: 10.1186/s12943-019-1125-9
75. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer (2020) 19(1):92. doi: 10.1186/s12943-020-01213-6
76. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett (2019) 17(1):388–97. doi: 10.3892/ol.2018.9606
77. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis (2021) 12(1):94. doi: 10.1038/s41419-020-03334-8
78. Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARalpha/CCL2 pathway. Aging (Albany NY) (2021) 13(8):11969–87. doi: 10.18632/aging.202900
79. Cao P, Ma B, Sun D, Zhang W, Qiu J, Qin L, et al. hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci (2021) 13(2):634–47. doi: 10.1111/cas.15238
80. Liu G, Sun J, Yang ZF, Zhou C, Zhou PY, Guan RY, et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis (2021) 12(3):260. doi: 10.1038/s41419-021-03545-7
81. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol (2002) 2(8):569–79. doi: 10.1038/nri855
82. Wang GY, Liu W, Zou Y, Wang GS, Deng YN, Luo JY, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. Ebiomedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
83. Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor cell-derived exosomal circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front Cell Dev Biol (2021) 9:726323. doi: 10.3389/fcell.2021.726323
84. Zhu CR, Su Y, Liu L, Wang SC, Liu YT, Wu JS. Circular RNA hsa_circ_0004277 stimulates Malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front Cell Dev Biol (2021) 8:585565. doi: 10.3389/fcell.2020.585565
85. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) (2019) 11(19):8183–203. doi: 10.18632/aging.102312
86. Zhang HY, Deng T, Ge SH, Liu Y, Bai M, Zhu KG, et al. : Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene (2019) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
87. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
88. Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol (2021) 14(1):200. doi: 10.1186/s13045-021-01207-x
89. Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci (2022) 113(6):1968–83. doi: 10.1111/cas.15365
90. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J Nanomedicine (2021) 16:2803–18. doi: 10.2147/IJN.S284560
91. Zhang L, Zhang J, Li P, Li T, Zhou Z, Wu H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis (2022) 13(1):32. doi: 10.1038/s41419-021-04345-9
92. Li SB, Weng JY, Song FB, Li L, Xiao C, Yang WQ, et al. Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis (2020) 11(6):452. doi: 10.1038/s41419-020-2616-8
93. Zhang X, Xu YF, Qian ZJ, Zheng WS, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis (2018) 9(11):1091. doi: 10.1038/s41419-018-1132-6
94. Wang YG, Wang T, Ding M, Xiang SH, Shi M, Zhai B. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett (2019) 460:128–38. doi: 10.1016/j.canlet.2019.06.007
95. Ji Y, Yang S, Yan X, Zhu L, Yang W, Yang X, et al. CircCRIM1 promotes hepatocellular carcinoma proliferation and angiogenesis by sponging miR-378a-3p and regulating SKP2 expression. Front Cell Dev Biol (2021) 9:796686. doi: 10.3389/fcell.2021.796686
96. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Canc Res (2020) 39(1):20. doi: 10.1186/s13046-020-1529-9
97. Liu R, Li Y, Wu A, Kong M, Ding W, Hu Z, et al. Identification of plasma hsa_circ_0005397 and combined with serum AFP, AFP-L3 as potential biomarkers for hepatocellular carcinoma. Front Pharmacol (2021) 12:639963. doi: 10.3389/fphar.2021.639963
98. Zhang L, Liu Y, Tao H, Zhu H, Pan Y, Li P, et al. Circular RNA circUBE2J2 acts as the sponge of microRNA-370-5P to suppress hepatocellular carcinoma progression. Cell Death Dis (2021) 12(11):985. doi: 10.1038/s41419-021-04269-4
99. He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduction Targeted Ther (2021) 6(1):185. doi: 10.1038/s41392-021-00569-5
100. Meng H, Li R, Xie Y, Mo Z, Zhai H, Zhang G, et al. Nanoparticles Mediated circROBO1 Silencing to Inhibit Hepatocellular Carcinoma Progression by Modulating miR-130a-5p/CCNT2 Axis. Int J Nanomedicine (2023) 18:1677–93. doi: 10.2147/IJN.S399318
Keywords: CircRNAs, hepatocellular carcinoma, carcinogenesis, progression, biomarker
Citation: Li Z-d, Li Y-l, Lu J, Liang S, Zhang C and Zeng L-h (2024) Recent research progress of circular RNAs in hepatocellular carcinoma. Front. Oncol. 13:1192386. doi: 10.3389/fonc.2023.1192386
Received: 23 March 2023; Accepted: 18 December 2023;
Published: 23 January 2024.
Edited by:
Mantang Qiu, Peking University People’s Hospital, ChinaReviewed by:
Rana A. Youness, German International University in Cairo (GIU), EgyptCopyright © 2024 Li, Li, Lu, Liang, Zhang and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling-hui Zeng, emVuZ2xoQGh6Y3UuZWR1LmNu; Chong Zhang, emhhbmdjaG9uZ0BoemN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.