- 1Department of Gastroenterology, Southern University of Science and Technology Hospital, Shenzhen, Guangdong, China
- 2Department of Gastroenterology, The First Affiliated Hospital of Naval Medical University, Shanghai, China
- 3Department of Gastroenterology, Hainan Cancer Hospital, Haikou, Hainan, China
- 4Department of Gastrointestinal Surgery, Hainan Cancer Hospital, Haikou, Hainan, China
Background: Colorectal cancer (CRC) is the third most common malignant tumor in the world. The morbidity and mortality rates in Western countries have decreased, but they are still on the rise in China. C10orf90 is associated with a variety of cancers, but the correlation between C10orf90 and CRC is not yet known.
Methods: A total of 1,339 subjects were randomly enrolled in our study. After extracting their DNA, three single-nucleotide polymorphisms (SNPs) of C10orf90 were genotyped to analyze the potential relationship between these variants and CRC risk. PLINK software packages (version 1.07) were used to evaluate multiple genetic models by calculating the odds ratio (OR) and 95% confidence interval (95% CI). The best SNP–SNP interaction model was defined by the multifactor dimensionality reduction (MDR) analysis.
Results: C10orf90 rs12412320 was significantly associated with CRC risk (p = 0.006) and might be associated with the lower CRC risk (OR: 0.78; 95% CI: 0.65–0.93). The relationship of rs12412320 with lower CRC risk was found in people aged >60 years and ≤60 years, women, non-smokers, or non-drinkers. Rs11245008 in people aged ≤60 years and rs11245007 among men had a higher CRC susceptibility. Rs12412320 was related to the lower risk of advanced stages (III/IV stage), while rs11245007 might be associated with the higher risk of advanced stages (III/IV stage). Moreover, rs12412320 had the most significant relationship with the susceptibility to rectal cancer.
Conclusion: This study is the first to report between C10orf90 gene polymorphisms and CRC risk in Chinese people, which suggests that C10orf90 rs12412320 might play a crucial role in preventing CRC occurrence.
Introduction
Colorectal cancer (CRC) is the third (3.11%) most common malignant tumor in the world and the second (3.5%) leading cause of cancer death (1). Globally, there are approximately 1 million new CRC patients every year, and more than 915,880 patients die each year (1). The CRC incidence and mortality rates in China, Europe, and North America account for more than half of the world's CRC incidence and mortality, respectively (2). Most recently, the incidence rate of CRC has been increasing and has become the second most common malignant tumor in China, which seriously threatens the life and health of residents (3). In China, the survival rate of CRC in the recent 5 years is significantly lower than that of many developed countries (4, 5). Despite the high incidence and low survival rate of CRC in China, the pathogenesis of CRC remains unclear. Genetics and environment are the major factors in the development of CRC (6, 7). Previously, hyperlipidemia, obesity, alcohol consumption, and smoking were suggested to be risk factors, and other potential risk factors included hypertension, metabolic syndrome, dietary factors, sedentary behavior, and occupational exposure (8). Furthermore, genetic predisposition is one of the key risk factors in the development of CRC (9).
C10orf90 (Chromosome 10 Open Reading Frame 90) is a protein coding gene and is known as the fragile-site associated tumor suppressor (FATS), which is also a regulator of the p53-p21 pathway (10). Studies have shown that in conjunctival melanoma, the deletion of the tumor suppressor gene C10orf90 is related to the significantly reduced metastasis-free survival of tumor patients (11). In addition, C10orf90 is a target gene of p53, and its overexpression can inhibit tumorigenicity in vivo, which is related to anti-tumor activity (12). FATS is an E2- and E3-independent ubiquitin ligase for promoting p53 stability and activation in response to DNA damage (13). The expression of C10orf90 gene is downregulated or silenced in many cancers, and it is related to non-small cell lung cancer, breast cancer, and others (14, 15). Furthermore, C10orf90 variants have been reported to be associated with the risk of various cancers, including breast cancer (16) and conjunctival melanomas (11). However, whether the genetic variants in C10orf90 may modulate CRC susceptibility remain unknown.
Single-nucleotide polymorphism (SNP) in the coding regions of genes may affect protein function. Here, three polymorphisms in the exon region of C10orf90 were genotyped to explore the relationship with CRC susceptibility in the Chinese Han population and to correlate these with demographic characteristics and clinical features.
Methods
Subjects
In this study, a total of 666 CRC patients at Hainan Province Cancer Hospital from August 2020 to December 2022 were randomly enrolled in the case group. A total of 673 healthy adults form the control group; they were from the same hospital during the same period without a history of cancer and chronic or severe diseases. The selection criteria of patients complied with the “Guidelines for diagnosis and treatment of colorectal cancer (2021 CSCO)” (17), and all patients were independent of each other. Patients suffering from inflammation, renal dysfunction, digestive system disease, and other chronic or endocrine disease, and who have been receiving any anti-cancer drugs or treatments were excluded. Demographic and clinical information of all subjects were gathered through standardized questionnaires and medical records, which include age, sex, smoking status, drinking status, body mass index (BMI), cancer stage, lymph node metastasis status, cancer style, carcinoembryonic antigen (CEA), alpha-fetoprotein (AFP), and cancer antigen-199 (CA199). The study was approved by the ethical committee of Hainan Province Cancer Hospital, and informed consent forms were signed by all subjects before the study, according to the Helsinki Declaration.
DNA extraction and SNP genotyping
Three SNPs (rs12412320, rs11245007, and rs11245008) in C10orf90 were selected for the study of their potential role in the risk of CRC based on a minor allele frequency (MAF) > 0.05 through the 1000 Genome Project. The potential biological functions of these loci were predicted through bioinformatics databases, including dbSNP, RegulomeDB, VannoPortal, and HaploReg v4.2.
Genomic DNA was extracted from peripheral blood samples (5 mL) of each subject using the Whole Blood Genomic DNA Isolation Kit (Xi’an Gold Mag Biotechnology, Xi'an, China). DNA was stored together with EDTA in a tube at −80°C. DNA concentrations were measured using NanoDrop 2000 (Ultra-fine ultraviolet spectrophotometer, Thermo, Waltham, MA, USA). SNP genotyping with a standard protocol was carried out using Agena MassARRAY RS1000. Agena Typer Software version 4.0 was used for data management.
Data analysis
Independent samples t-test and Chi-square test were used to assess the differences in demographic characteristics of the study participants. We used Fisher’s test to evaluate the Hardy–Weinberg equilibrium (HWE) of each SNP in the subjects. Odds ratio (OR) and 95% confidence interval (95% CI) were assessed to estimate the correlations of SNPs and CRC risk using logistic regression analysis. PLINK software packages (version 1.07) were used to evaluate multiple genetic models (allele model, genotype model, dominant model, recessive model, and additive model). Statistical analysis was performed using Microsoft Excel and SPSS 17.0 statistical packages (SPSS, Chicago, IL). A two-tailed p < 0.05 was considered statistically significant, and a Bonferroni-corrected p < 0.05/3 was considered significant. In addition, we used the multifactor dimensionality reduction (MDR) analysis to identify the best SNP–SNP interaction model.
Results
Characteristics of subjects
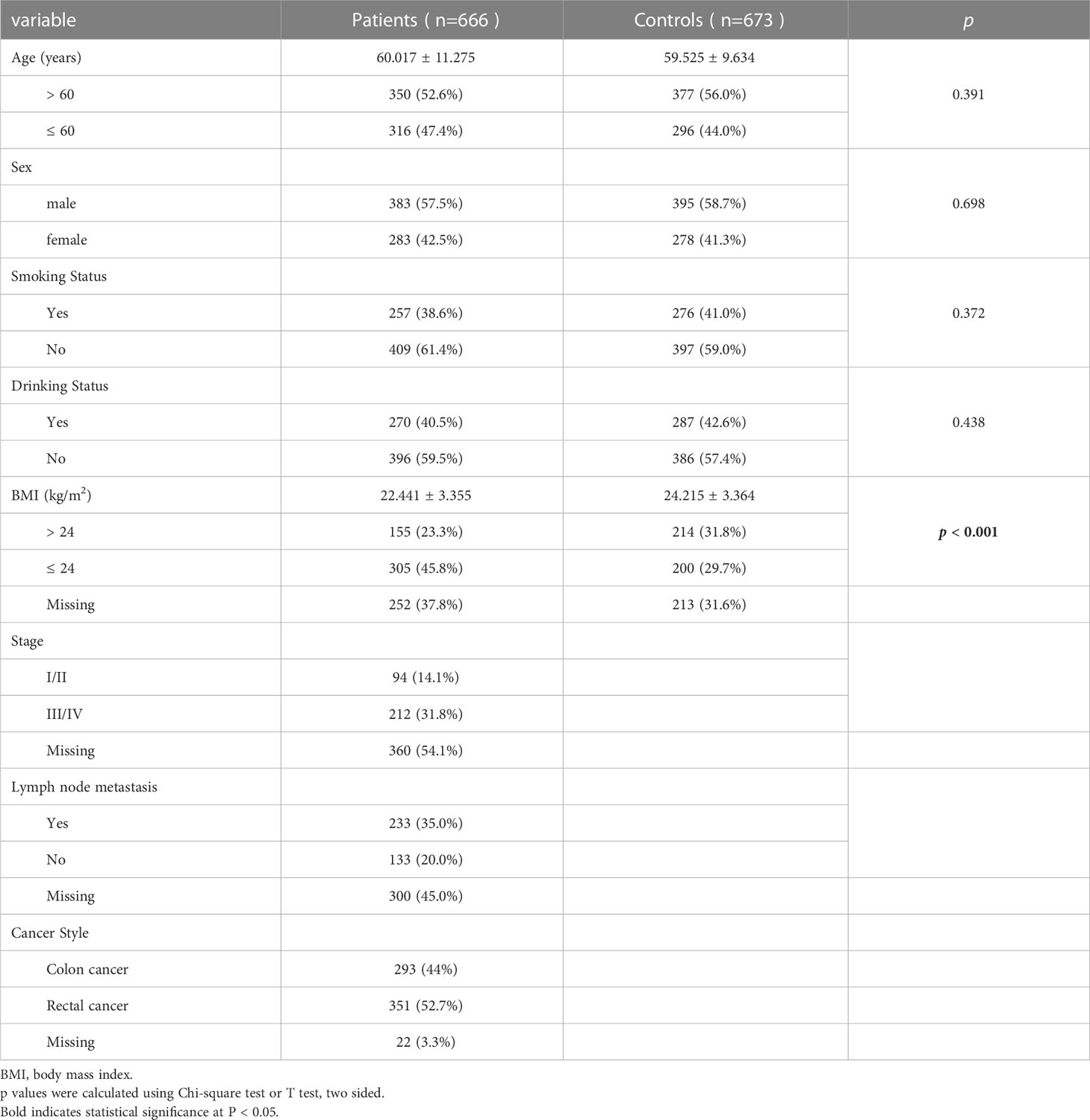
There were 1,339 subjects in this study, namely, 666 CRC patients (age: 60.02 ± 11.28 years) and 673 healthy controls (age: 59.53 ± 9.63 years). Table 1 shows the relevant characteristics of all subjects including the case group and the control group. It can be seen that there are no statistical differences between CRC patients and healthy controls in these indexes such as age (p = 0.391), sex (p = 0.698), smoking (p = 0.372), and drinking (p = 0.438). There was a significant difference in BMI between CRC patients and healthy controls (p < 0.001).
Relationship between C10orf90 SNPs and CRC risk
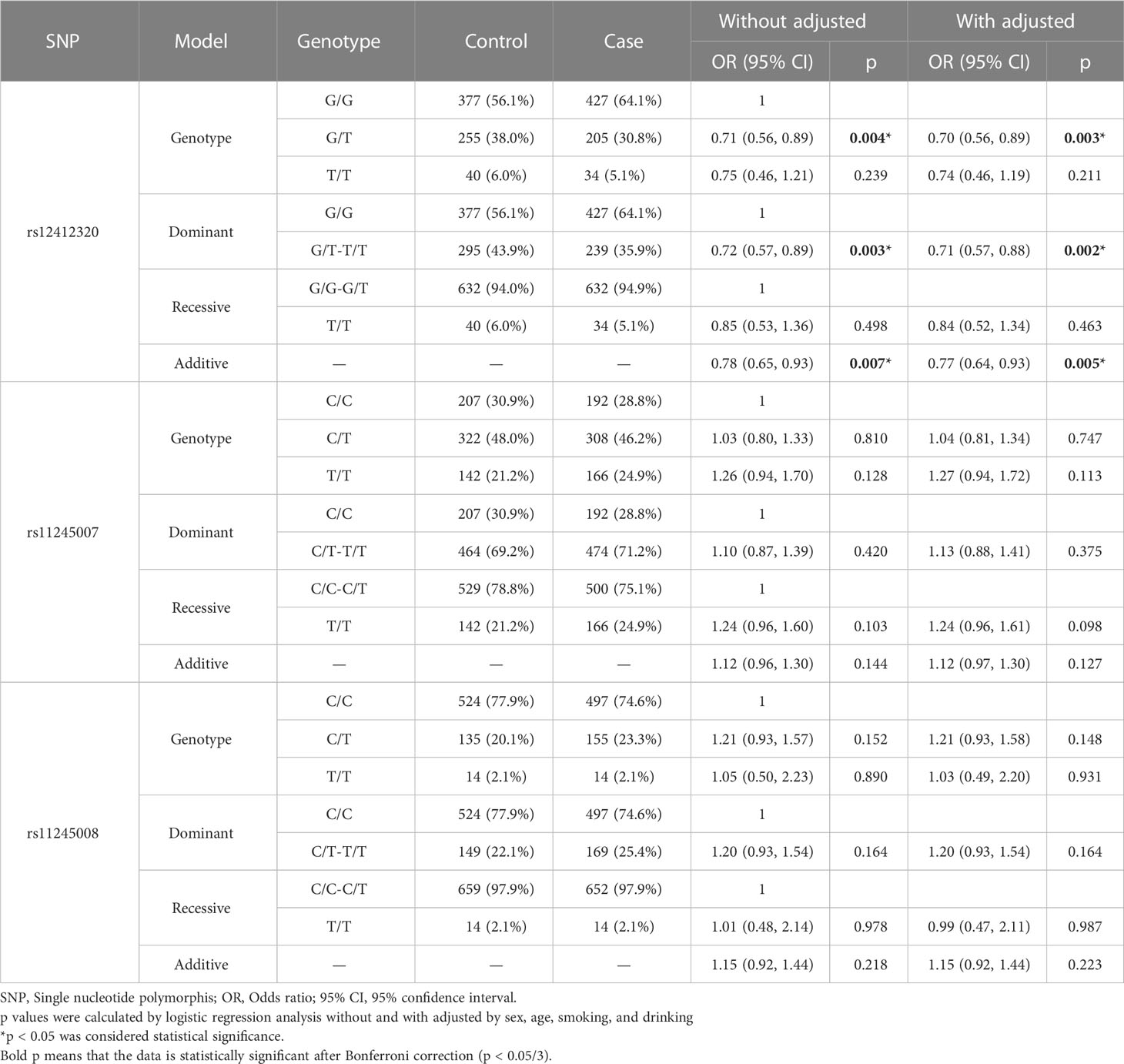
The relationship between SNPs of C10orf90 and CRC risk is listed in Table 2. All SNPs were missense variants. All SNPs of C10orf90 complied with the Hardy–Weinberg equilibrium (p > 0.05). The MAF of each SNP was above 5% in the Chinese Han population. C10orf90 rs12412320 was significantly associated with CRC risk (p = 0.006) and might be associated with the lower CRC risk (OR: 0.78; 95% CI: 0.65–0.93). Bioinformatics analysis found that these SNPs may be involved in promoter/enhancer histone marks, and protein-bound motifs changed the binding of transcription factors (TFs) and the action of DNase. Figure 1 shows the most significant Hi–C interactions between the variant locus and the target regions.
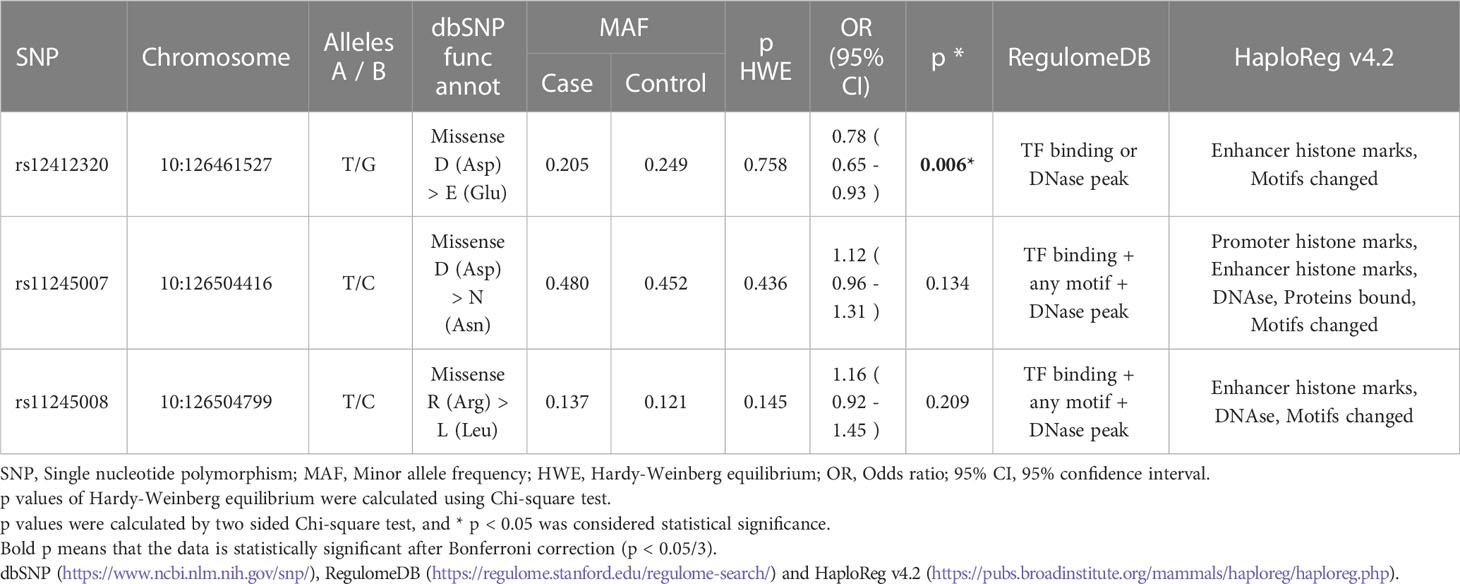
Table 2 The based information of selected SNPs in C10orf90 and the association with the risk of colorectal cancer in the allele model.
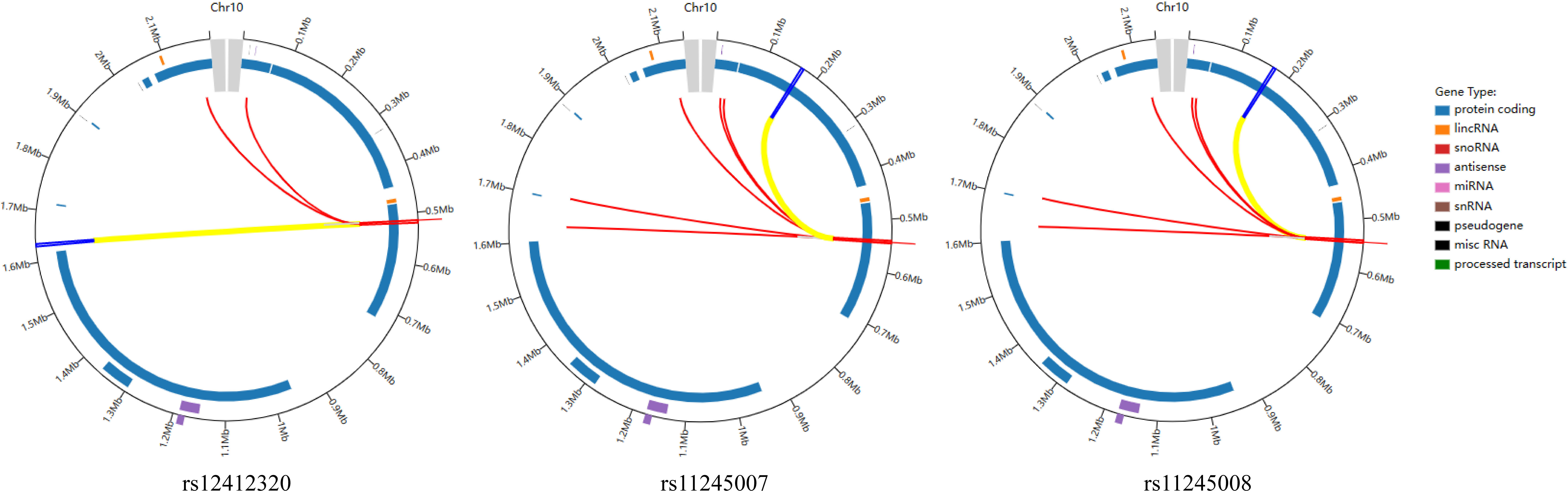
Figure 1 Virtual 4C circular plot for the most significant Hi–C interactions between the variant locus and the target regions.
Table 3 shows the relationship between CRC risk and the different genetic models of C10orf90 polymorphisms in the overall analysis. Logistic regression analysis showed that, whether corrected or not, there were significant differences in the correlation between SNPs of C10orf90 rs12412320 and the risk of CRC. Among them, three allele models of rs12412320 (Heterozygous: p = 0.003, OR: 0.70, 95% CI: 0.56, 0.89; Dominant: p = 0.002, OR:0.71, 95% CI: 0.57, 0.88; Additive: p = 0.005, OR: 0.77, 95% CI: 0.64, 0.93, adjusted) were significantly correlated with the risk of CRC. The protective significance of rs12412320 for CRC occurrence still existed after Bonferroni multiple correction (p < 0.05/3).
C10orf90 SNPs associated with CRC risk in the stratified analysis
To explore the relationship of three SNPs with CRC, we performed the subgroup stratification analysis by demographic characteristics (age, sex, smoking, drinking, and BMI), as shown in Supplementary Table S1 and Figure 2. After Bonferroni multiple correction, the relationship of rs12412320 in people aged >60 years (p = 0.008, OR: 0.65) and ≤60 years (p = 0.013, OR: 0.35, and p = 0.013, OR: 0.70) and that of rs11245008 in people aged ≤60 years (p = 0.011, OR: 1.57) were also remarkable. In the sex-stratified analysis, rs12412320 (p = 0.001, OR: 0.56; and p = 0.005, OR: 0.61) had a lower CRC risk in women, whereas rs11245007 (p = 0.011, OR: 1.30; p = 0.008, OR: 1.52; and p = 0.011, OR: 1.29) had a higher CRC susceptibility among men after Bonferroni multiple correction. After Bonferroni multiple correction, C10orf90 rs12412320 was also significantly associated with CRC in non-smokers (p = 0.001, OR: 0.58; p = 0.001, OR: 0.60; and p = 0.004, OR: 0.71) and non-drinkers (p = 0.003, OR: 0.70; p < 0.001, OR: 0.57; p < 0.001, OR: 0.58; and p = 0.002, OR: 0.69).
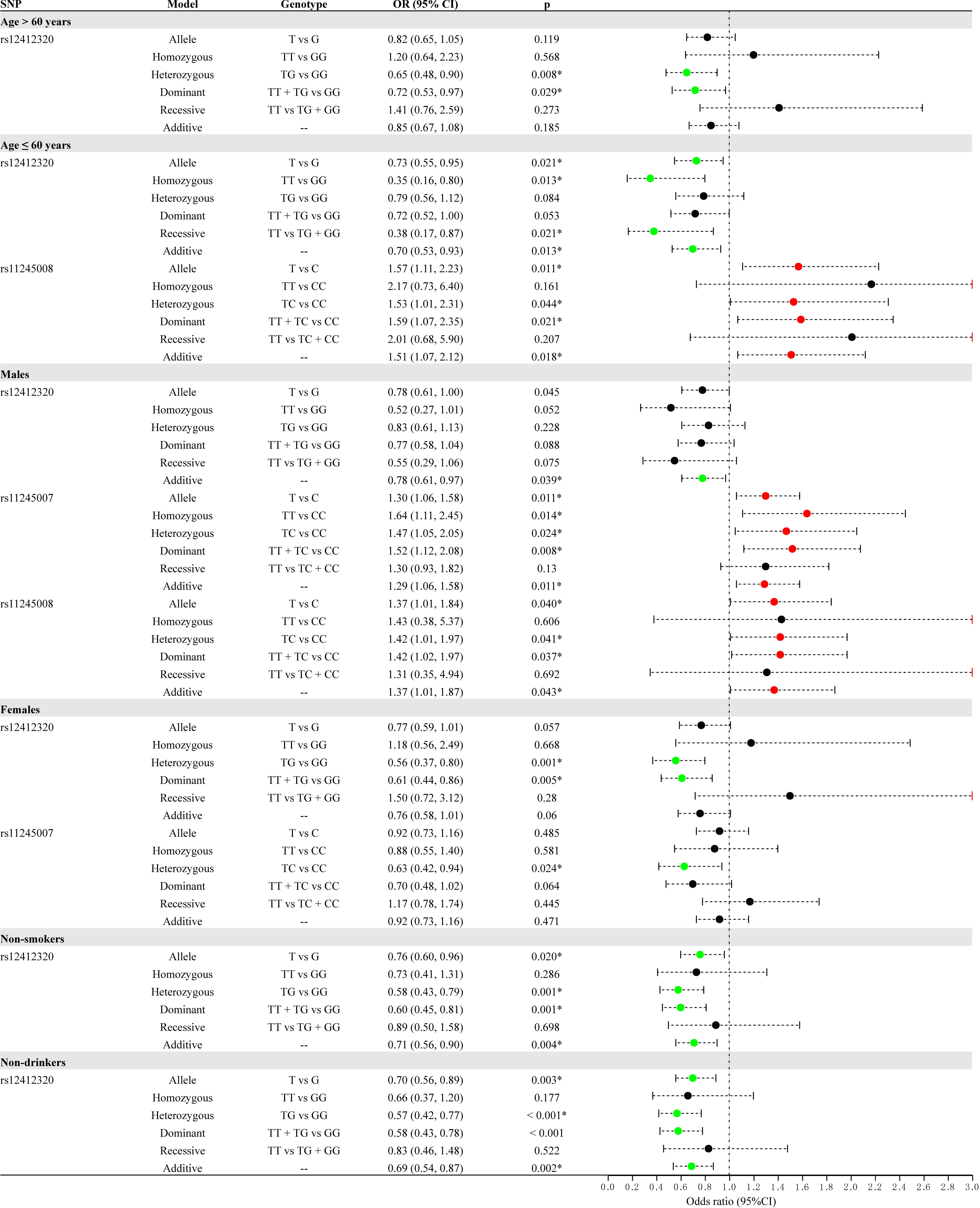
Figure 2 Forest map for the stratification analysis by demographic characteristics (age, sex, smoking, and drinking) for the association between C10orf90 variants and colorectal cancer (CRC) risk.
Stratified analysis by clinical features (stage, lymph node metastasis, and cancer style) for the association between C10orf90 variants and the risk of CRC is displayed in Supplementary Table S2 and Figure 3. After Bonferroni multiple correction, rs12412320 (p = 0.002, OR: 0.52; p = 0.008, OR: 0.23; p = 0.010, OR: 0.51; and p = 0.003, OR: 0.53) was related to the lower risk of advanced stages (III/IV stage), while rs11245007 (p = 0.001, OR: 1.80; p = 0.002, OR: 3.06; p = 0.003, OR: 2.53; and p = 0.002, OR: 1.71) might be associated with the higher risk of advanced stages (III/IV stage). Rs12412320 (p = 0.009, OR: 0.69; and p = 0.016, OR: 0.72) had the most significant relationship with the susceptibility of rectal cancer after Bonferroni multiple correction. Moreover, rs12412320 was associated with the risk of colon cancer, but no significance was found after Bonferroni multiple correction.
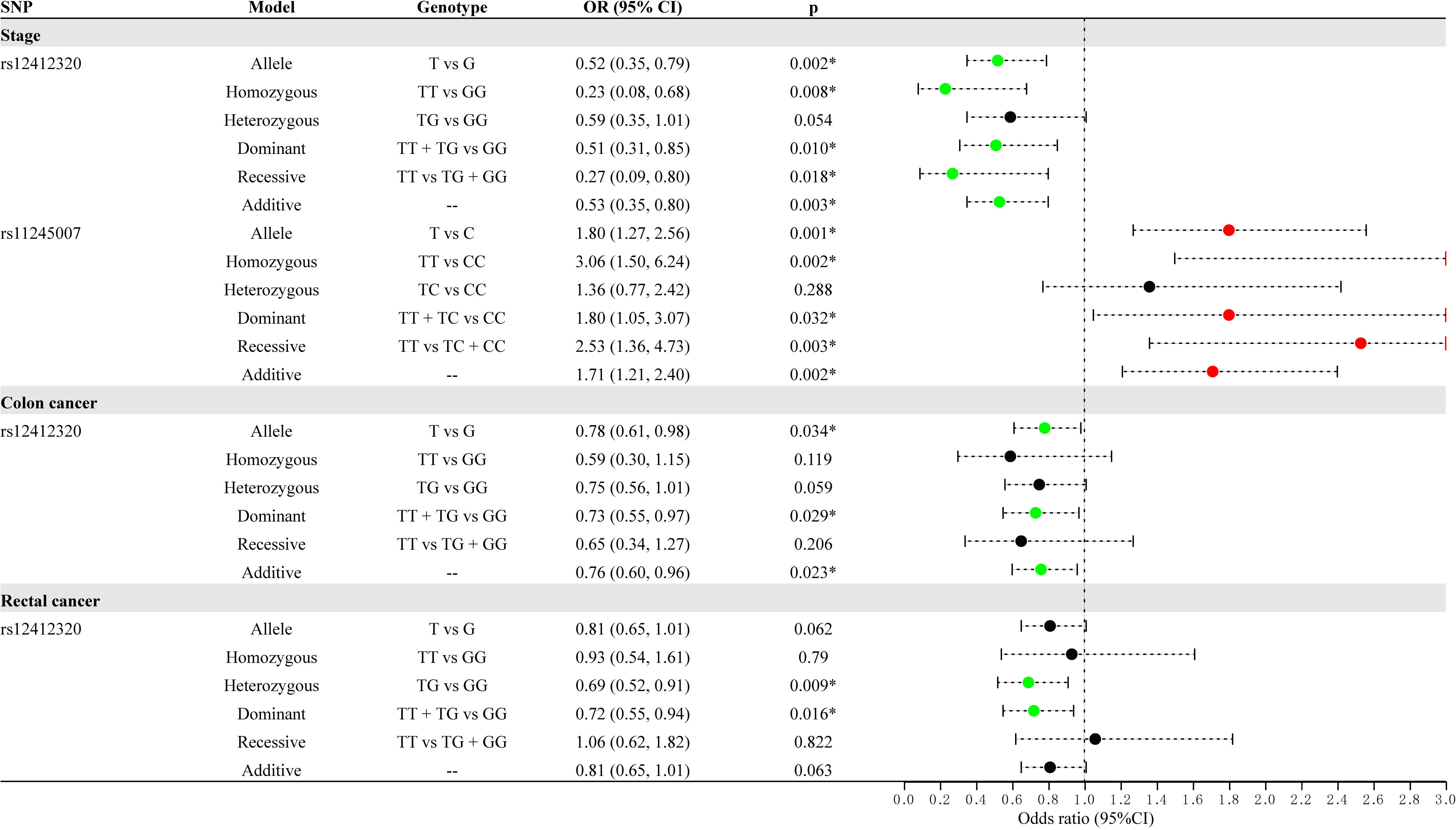
Figure 3 Forest map for the stratification analysis by clinical features (cancer type and stage) for the association between C10orf90 variants and colorectal cancer (CRC) risk.
MDR analysis for C10orf90 variants
Then, the relationship between the interaction of C10orf90 SNPs and CRC risk was analyzed by the MDR method. The results of the MDR model analysis of the SNP–SNP interactions are demonstrated in Table 4 and Figure 4. The dendrogram (Figure 4A) shows that loci with strong interactions were located very close to each other on the branches, while loci with weak interactions were far apart from each other. The most significant single-locus model was rs12412320 [testing accuracy: 0.5338, p = 0.0077, cross-validation consistency (CVC): 10/10] with an information gain of 0.50% (Figure 4B); the best two-locus models were rs12412320 and rs11245008 (testing accuracy: 0.5308, p = 0.0041, CVC: 6/10); and the best three-locus models were rs12412320, rs11245007, and rs11245008 (testing accuracy: 0.5300, p = 0.0007, CVC: 10/10), which is the best SNP–SNP interaction model. Therefore, the impact of the three candidate SNPs on the risk of CRC may be interdependent.
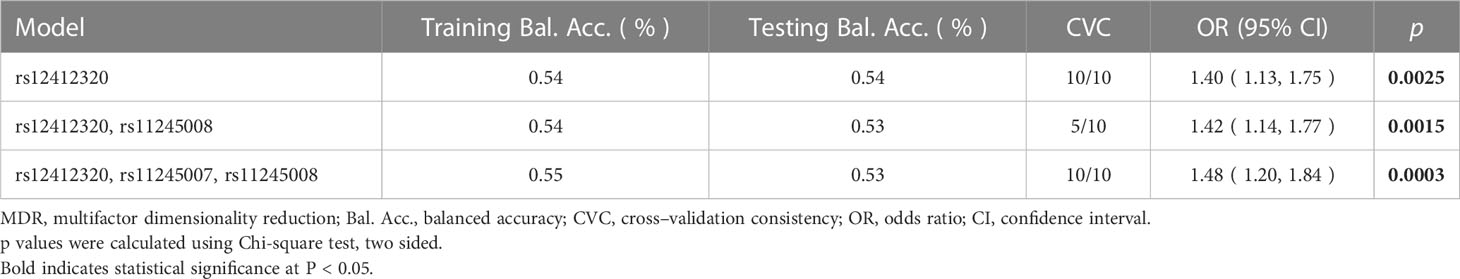
Table 4 Summary of SNP – SNP interactions on the risk of colorectal cancer analyzed through MDR method.
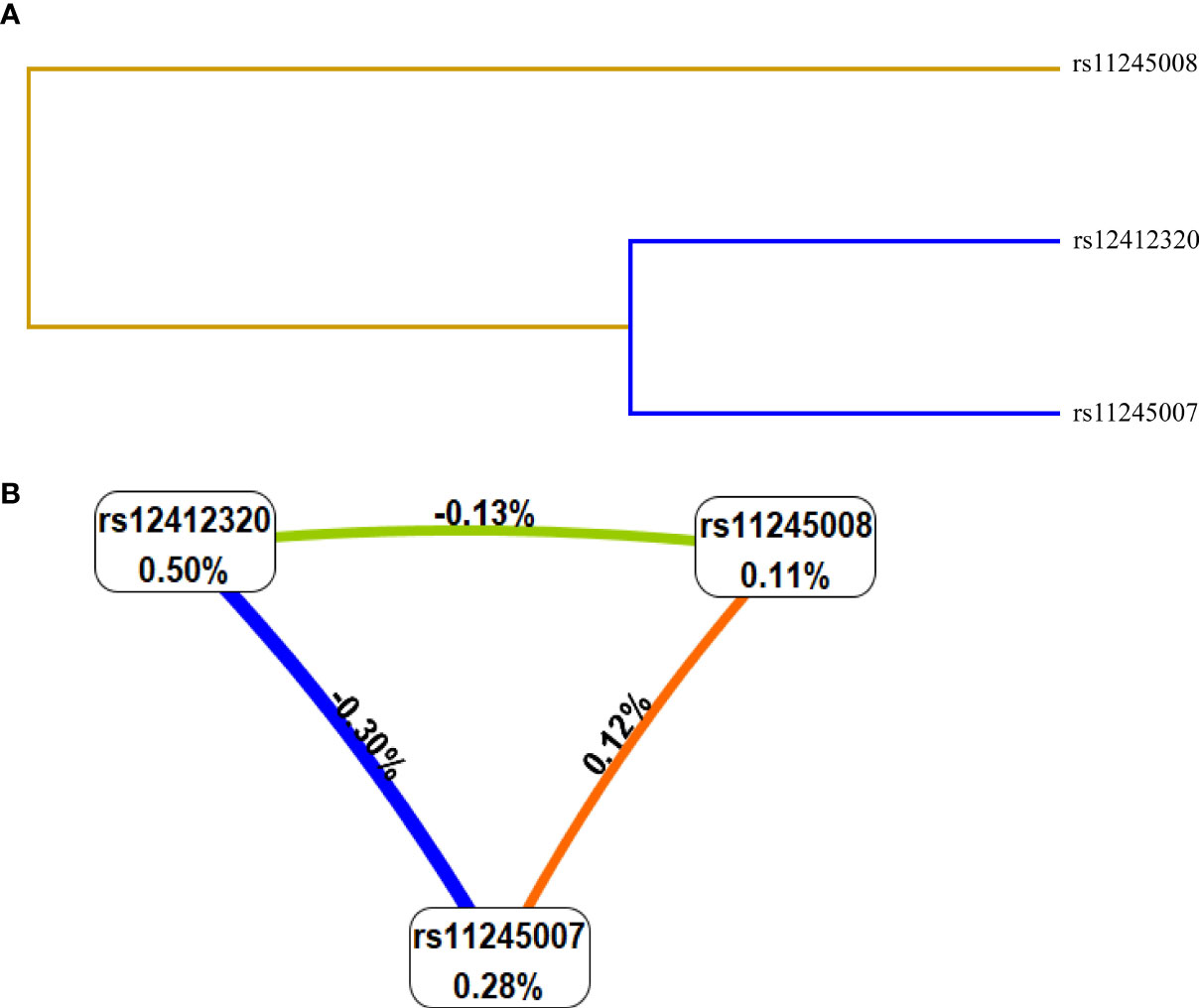
Figure 4 Interaction map among single-nucleotide polymorphisms (SNPs) of genes on the risk of colorectal cancer (CRC). Values in nodes represent the information gain (IGs) of individual attribute (main effects). Values between nodes are IGs of each pair of attributes (interaction effects). SNP-SNP interaction dendrogram (A) and Fruchterman-Reingold (B).
Discussion
The multi-disciplinary approach that combines genetics, immunology, and chemotherapy has the potential to revolutionize the treatment of CRC and other types of cancer as well. One of the major challenges in cancer treatment is the heterogeneity of tumors, which can make it difficult to develop effective therapies. However, by understanding the molecular mechanisms underlying cancer and the role of the immune system in cancer development and progression, researchers can develop personalized treatment approaches that target the specific characteristics of each patient’s tumor (18). Genetic factors are important influencing factors of CRC. Research shows that approximately 5% of CRC is caused by chromosomal variation, which is hereditary (19). Previous studies have reported many loci associated with the risk of CRC (20–22). However, the specific molecular mechanism of CRC has not been fully understood. There are still a large number of loci that may affect the risk of CRC that have not been reported. Therefore, further exploring the relationship between gene SNPs and CRC risk is much more significant and useful for the specific diagnosis on CRC. As a tumor suppressor associated with fragile sites, C10orf90 is involved in DNA damage-induced carcinogenesis. C10orf90 overexpression significantly enhances the sensitivity of non-small cell lung cancer (NSCLC) cells to cisplatin, and is related to the overall survival rate (23). In this study, we analyzed the association between genetic polymorphisms of C10orf90 and the risk of CRC in 1,339 Chinese people. The results displayed that the genetic polymorphisms of C10orf90 were significantly associated with the risk of CRC, especially SNP rs12412320. Here, we had reported for the first time that C10orf90 rs12412320 was associated with a reduced risk of CRC in the Chinese Han population. There are currently few reports on this locus. Bioinformatics analysis revealed that these SNPs may be related to promoter/enhancer histone marks, and protein-bound motifs changed the binding of TFs and the action of DNase. This indicates that C10orf90 rs12412320 may affect the risk of CRC by affecting the expression of the gene.
At present, it is universally recognized that the occurrence of CRC is related to immutable risk factors, including age, sex, genetic factors, environment, and lifestyle (6, 7, 24). CRC usually appears after 50 years of age, and the incidence rate of CRC in women is low, usually accounting for one-third of the total incidence rate (25). The combination of tobacco and alcohol increases the risk of cancer. Smoking increases the susceptibility to CRC in a dose-dependent manner with intensity and duration (26). Alcohol consumption and obesity are considered modifiable risk factors for CRC (27, 28). The stratification analysis was explored for the effect of demographic characteristics (age, sex, smoking, drinking, and BMI) on the relationship of three SNPs with CRC. After Bonferroni multiple correction, the relationship of rs12412320 with lower CRC risk was found in people aged >60 years and ≤60 years, women, non-smokers, or non-drinkers. Some studies have suggested that smoking, which created a hypoxic microenvironment that was quite common in solid tumors, might cooperate with genetic polymorphism to produce a superimposed effect on the progression of CRC (29). Previously, possible interactions between GWAS-identified CRC susceptibility SNPs and alcohol consumption were investigated, and genetic polymorphisms were associated with increased risk of CRC among ever drinkers and higher-level alcohol drinkers, suggesting that alcohol consumption could be a possible effect modifier (30). Our study found that C10orf90 rs12412320 was associated with a reduced risk of CRC overall. The stratification analysis showed that C10orf90 rs12412320 was also significantly associated with CRC in non-smokers and non-drinkers, but not smokers and drinkers. These results suggested that not smoking or not drinking was found to reduce the likelihood of CRC risk among the population who carried C10orf90 rs12412320-T allele. Moreover, rs11245008 in people aged ≤60 years and rs11245007 among men had a higher CRC susceptibility after Bonferroni multiple correction. Functional analysis demonstrated that rs11245007, a functional variant of C10orf90, can modulate p53 activation, resulting from the more pronounced polyubiquitination of p53 by rs11245007-T (mutant allele) (16). Song et al. showed that rs11245007 played a vital role in preventing the occurrence of breast cancer (16). Here, rs11245007 can increase the risk of CRC in men, which is opposite to its role in breast cancer. It may be caused by the different pathogenesis of various diseases, tumor heterogeneity, and so on. Our study provides evidence to clarify that the pathogenic effect on CRC may be partially attributed to the interaction between C10orf90 variants and age, sex, smoking, and alcohol consumption.
The clinical characteristics of CRC patients are related to prognosis, and the complex interaction between staging, metastasis, and genetic factors plays a role in guiding prognosis, risk stratification, and adjuvant treatment of CRC (31, 32). In the study, stratified analysis by clinical features (stage, lymph node metastasis, and cancer style) for the association between C10orf90 variants and the risk of CRC was investigated. After Bonferroni multiple correction, rs12412320 was related to the lower risk of advanced stages (III/IV stage), while rs11245007 might be associated with the higher risk of advanced stages (III/IV stage). Moreover, rs12412320 had the most significant relationship with the susceptibility of rectal cancer after Bonferroni multiple correction.
Unavoidably, this study has several limitations. Firstly, the study was conducted in a single hospital in Hainan Province, China, which limits the generalizability of the results to other populations. Secondly, because a proportion of the samples lack information on environmental factors (such as diet, physical activity, and environmental factors) and because of the relatively small sample size, our study did not explain the role of the interaction between C10orf90 variants and environmental factors on CRC risk. In the future, we would like to increase the sample size and complete the environmental factors to evaluate the relationship and to verify our findings. Thirdly, only three SNPs of C10orf90 were studied in this study, and other genetic variants that may play a role in CRC susceptibility were not investigated. Experimental design will continue to explore the correlation of other loci on this gene with CRC risk in the future. Fourthly, the mechanism of these SNPs is only predicted through bioinformatics analysis; therefore, functional experiments are needed to further explore the function of C10orf90 loci in CRC etiology. Fifthly, CRC patients’ tissues and normal tissues had not been explored in protein expression studies. In subsequent research, we plan to collect enough CRC patients’ tissues and normal tissues to examine them via immunohistochemistry (IHC) using protein expression studies and to conduct functional research of these SNPs in CRC.
Conclusion
In summary, this study is the first to report the relationship between C10orf90 gene polymorphisms and CRC risk in Chinese people, which suggests that C10orf90 rs12412320 might play a crucial role in preventing CRC occurrence. It provides the foundation for the study on the mechanism of C10orf90 in CRC and supplies the basis for personalized treatment of CRC patients.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by Hainan Cancer Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
JS and KW wrote the manuscript, ZC and DZ processed and analyzed the results, LL and LG prepared Figure 1 and collected data, and SY designed the research ideas and plans. All the authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by 2020 Hainan Province Major Science and Technology Plan Project (ZDKJ202005) and supported by the specific research fund of The Innovation Platform for Academinicians of Hainan Province(YSPTZX202029).
Acknowledgments
The authors thank all participants and volunteers in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1192378/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y, et al. Incidence, mortality, survival, risk factor and screening of colorectal cancer: a comparison among China, Europe, and northern America. Cancer Lett (2021) 522:255–68. doi: 10.1016/j.canlet.2021.09.034
3. Huang RL, Liu Q, Wang YX, Zou JY, Hu LF, Wang W, et al. Awareness, attitude and barriers of colorectal cancer screening among high-risk populations in China: a cross-sectional study. BMJ Open (2021) 11(7):e045168. doi: 10.1136/bmjopen-2020-045168
4. Deng X, Gao F, Li N, Li Q, Zhou Y, Yang T, et al. Antitumor activity of NKG2D CAR-T cells against human colorectal cancer cells in vitro and in vivo. Am J Cancer Res (2019) 9(5):945–58.
5. Wang H, Wang P, Liu X, Li L, Xiao X, Liu P, et al. Factors predicting the colorectal adenoma detection rate in colonoscopic screening of a Chinese population: a prospective study. Medicine (2019) 98(15):e15103. doi: 10.1097/MD.0000000000015103
6. Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients (2019) 11(1):164. doi: 10.3390/nu11010164
7. Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, et al. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr Drug Targets (2021) 22(9):998–1009. doi: 10.2174/1389450121999201117115717
8. O'Sullivan DE, Sutherland RL, Town S, Chow K, Fan J, Forbes N, et al. Risk factors for early-onset colorectal cancer: a systematic review and meta-analysis. Clin Gastroenterol Hepatol (2022) 20(6):1229–1240.e1225. doi: 10.1016/j.cgh.2021.01.037
9. Rebuzzi F, Ulivi P, Tedaldi G. Genetic predisposition to colorectal cancer: how many and which genes to test? Int J Mol Sci (2023) 24(3):2137. doi: 10.3390/ijms24032137
10. Quistrebert J, Orlova M, Kerner G, Ton LT, Luong NT, Danh NT, et al. Genome-wide association study of resistance to mycobacterium tuberculosis infection identifies a locus at 10q26.2 in three distinct populations. PloS Genet (2021) 17(3):e1009392. doi: 10.1371/journal.pgen.1009392
11. Kenawy N, Kalirai H, Sacco JJ, Lake SL, Heegaard S, Larsen AC, et al. Conjunctival melanoma copy number alterations and correlation with mutation status, tumor features, and clinical outcome. Pigment Cell melanoma Res (2019) 32(4):564–75. doi: 10.1111/pcmr.12767
12. Zhang X, Zhang Q, Zhang J, Qiu L, Yan SS, Feng J, et al. FATS is a transcriptional target of p53 and associated with antitumor activity. Mol Cancer (2010) 9:244. doi: 10.1186/1476-4598-9-244
13. Yan S, Qiu L, Ma K, Zhang X, Zhao Y, Zhang J, et al. FATS is an E2-independent ubiquitin ligase that stabilizes p53 and promotes its activation in response to DNA damage. Oncogene (2014) 33(47):5424–33. doi: 10.1038/onc.2013.494
14. Zhang TM, Zhang J, Zhou DJ, Wang CL. [Expression of FATS in non-small cell lung cancer and its relationship with prognosis]. Zhonghua zhong liu za zhi [Chinese J oncology] (2019) 41(11):826–30. doi: 10.3760/cma.j.issn.0253-3766.2019.11.005
15. Zhang J, Wu N, Zhang T, Sun T, Su Y, Zhao J, et al. et al: the value of FATS expression in predicting sensitivity to radiotherapy in breast cancer. Oncotarget (2017) 8(24):38491–500. doi: 10.18632/oncotarget.16630
16. Song F, Zhang J, Qiu L, Zhao Y, Xing P, Lu J, et al. A functional genetic variant in fragile-site gene FATS modulates the risk of breast cancer in triparous women. BMC Cancer (2015) 15:559. doi: 10.1186/s12885-015-1570-9
17. Yoshino T, Argilés G, Oki E, Martinelli E, Taniguchi H, Arnold D, et al. Pan-Asian adapted ESMO clinical practice guidelines for the diagnosis treatment and follow-up of patients with localised colon cancer. Ann Oncol (2021) 32(12):1496–510. doi: 10.1016/j.annonc.2021.08.1752
18. Derakhshani A, Hashemzadeh S, Asadzadeh Z, Shadbad MA, Rasibonab F, Safarpour H, et al. et al: cytotoxic T-lymphocyte antigen-4 in colorectal cancer: another therapeutic side of capecitabine. Cancers (2021) 13(10):2414. doi: 10.3390/cancers13102414
19. Heinimann K. [Hereditary colorectal cancer: clinics, diagnostics and management]. Therapeutische Umschau Rev therapeutique (2018) 75(10):601–6. doi: 10.1024/0040-5930/a001046
20. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800
21. Fernandez-Rozadilla C, Timofeeva M, Chen Z, Law P, Thomas M, Schmit S, et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat Genet (2023) 55(1):89–99. doi: 10.1038/s41588-022-01222-9
22. Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet (2019) 51(1):76–87. doi: 10.1038/s41588-018-0286-6
23. Tian Y, Zhang J, Yan S, Qiu L, Li Z. FATS expression is associated with cisplatin sensitivity in non small cell lung cancer. Lung Cancer (Amsterdam Netherlands) (2012) 76(3):416–22. doi: 10.1016/j.lungcan.2011.11.009
24. Kim SE, Paik HY, Yoon H, Lee JE, Kim N, Sung MK. Sex- and gender-specific disparities in colorectal cancer risk. World J Gastroenterol (2015) 21(17):5167–75. doi: 10.3748/wjg.v21.i17.5167
25. Hultcrantz R. Aspects of colorectal cancer screening, methods, age and gender. J Internal Med (2021) 289(4):493–507. doi: 10.1111/joim.13171
26. Botteri E, Borroni E, Sloan EK, Bagnardi V, Bosetti C, Peveri G, et al. Smoking and colorectal cancer risk, overall and by molecular subtypes: a meta-analysis. Am J Gastroenterol (2020) 115(12):1940–9. doi: 10.14309/ajg.0000000000000803
27. Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K. Body fatness and cancer–viewpoint of the IARC working group. N Engl J Med (2016) 375(8):794–8. doi: 10.1056/NEJMsr1606602
28. Zhou X, Wang L, Xiao J, Sun J, Yu L, Zhang H, et al. Alcohol consumption, DNA methylation and colorectal cancer risk: results from pooled cohort studies and mendelian randomization analysis. Int J Cancer (2022) 151(1):83–94. doi: 10.1002/ijc.33945
29. Fu Y, Zhang Y, Cui J, Yang G, Peng S, Mi W, et al. SNP rs12982687 affects binding capacity of lncRNA UCA1 with miR-873-5p: involvement in smoking-triggered colorectal cancer progression. Cell Communication Signaling CCS (2020) 18(1):37. doi: 10.1186/s12964-020-0518-0
30. Song N, Shin A, Oh JH, Kim J. Effects of interactions between common genetic variants and alcohol consumption on colorectal cancer risk. Oncotarget (2018) 9(5):6391–401. doi: 10.18632/oncotarget.23997
31. Chen K, Collins G, Wang H, Toh JWT. Pathological features and prognostication in colorectal cancer. Curr Oncol (Toronto Ont) (2021) 28(6):5356–83. doi: 10.3390/curroncol28060447
32. Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol (2022) 7(3):262–74. doi: 10.1016/S2468-1253(21)00426-X
Keywords: colorectal cancer, C10orf90, gene polymorphisms, demographic characteristics, clinical features
Citation: Song J, Wang K, Chen Z, Zhong D, Li L, Guo L and Yu S (2023) Association study between C10orf90 gene polymorphisms and colorectal cancer. Front. Oncol. 13:1192378. doi: 10.3389/fonc.2023.1192378
Received: 23 March 2023; Accepted: 28 June 2023;
Published: 31 July 2023.
Edited by:
Jorge Melendez-Zajgla, National Institute of Genomic Medicine (INMEGEN), MexicoReviewed by:
Mansoor-Ali Vaali-Mohammed, King Saud University, Saudi ArabiaTianbo Jin, Northwest University, China
Antonella Argentiero, National Cancer Institute Foundation (IRCCS), Italy
Mónica Alejandra Rosales-Reynoso, Centro de Investigación Biomédica de Occidente (CIBO), Mexico
Copyright © 2023 Song, Wang, Chen, Zhong, Li, Guo and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyong Yu, eXVzaHV5b25nMjAyMkAxNjMuY29t
†These authors share first authorship
 Jian Song1†
Jian Song1† Kaixuan Wang
Kaixuan Wang Shuyong Yu
Shuyong Yu