- 1Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 2Department of Surgery, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 3Department of Biochemistry and Molecular Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 4SCHEQ Foundation, New York, NY, United States
- 5Department of Translational Genomics, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 6Hastings Center for Pulmonary Research, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 7Department of Mechanical and Aerospace Engineering, University of Florida, Gainesville, FL, United States
- 8College of Pharmacy and Pharmaceutical Sciences, Institute of Public Health, Florida A&M University, Tallahassee, FL, United States
Lung cancer is the leading cause of cancer death in the United States and worldwide, and a major source of cancer health disparities. Lung cancer cell lines provide key in vitro models for molecular studies of lung cancer development and progression, and for pre-clinical drug testing. To ensure health equity, it is imperative that cell lines representing different lung cancer histological types, carrying different cancer driver genes, and representing different genders, races, and ethnicities should be available. This is particularly relevant for cell lines from Black men, who experience the highest lung cancer mortality in the United States. Here, we undertook a review of the available lung cancer cell lines and their racial and ethnic origin. We noted a marked imbalance in the availability of cell lines from different races and ethnicities. Cell lines from Black patients were strongly underrepresented, and we identified no cell lines from Hispanic/Latin(x) (H/L), American Indian/American Native (AI/AN), or Native Hawaiian or other Pacific Islander (NHOPI) patients. The majority of cell lines were derived from White and Asian patients. Also missing are cell lines representing the cells-of-origin of the major lung cancer histological types, which can be used to model lung cancer development and to study the effects of environmental exposures on lung tissues. To our knowledge, the few available immortalized alveolar epithelial cell lines are all derived from White subjects, and the race and ethnicity of a handful of cell lines derived from bronchial epithelial cells are unknown. The lack of an appropriately diverse collection of lung cancer cell lines and lung cancer cell-of-origin lines severely limits racially and ethnically inclusive lung cancer research. It impedes the ability to develop inclusive models, screen comprehensively for effective compounds, pre-clinically test new drugs, and optimize precision medicine. It thereby hinders the development of therapies that can increase the survival of minority and underserved patients. The noted lack of cell lines from underrepresented groups should constitute a call to action to establish additional cell lines and ensure adequate representation of all population groups in this critical pre-clinical research resource.
Introduction
Lung cancer remains the leading cause of cancer death in the United States (1, 2) and in the world (3) and is a prominent source of cancer health disparities (4). In the United States, Black men have the highest rate of lung cancer mortality among all groups (5). Lung cancer deaths in the United States have steadily declined due in large part to a decrease in smoking rates, particularly within Black men and women (4, 6). As a result, the gap in lung cancer deaths between Black and White men is slowly closing (1). Yet Black men in the United States still show a 12% higher lung cancer incidence rate and a 15% higher lung cancer death rate compared to White men (4, 6). Many factors are thought to contribute to this disparity, including socioeconomic factors, such as a lower frequency of screening, lack of awareness of and access to molecular testing, lack of awareness and participation in clinical trials, mistrust of the medical profession, and lack of diversity in the biomedical workforce (7–9). Importantly, genetic differences between Black and White subjects likely also play a role (10–14), with further studies required to uncover additional associations (15). It has been determined that genetics can affect lung cancer risk (16, 17), for example through differences in nicotine and carcinogen uptake (18–24) or the strength of detoxification responses (10, 25–27). Genetic background/ancestry can also affect the nature of driver mutations acquired by tumors (28–32), tumor mutational burden (33), and patient response to therapy (34). Given the numerous possible effects of genetic background on lung cancer development, pathology, and treatment, it is vital that race/ethnicity be considered in lung cancer research (35).
There are many established model systems to study lung cancer in vitro or in vivo (36). Among these, lung cancer cell lines represent a versatile and relatively affordable resource that can be widely disseminated to the scientific community (36, 37). Cell lines can be used to gain molecular insights into the development and progression of lung cancer and to pre-clinically test prospective lead candidate drugs (36, 37). Given the disproportionate impact of lung cancer on Black individuals as documented in the United States (4–6), we investigated the availability of lung cancer cell lines from Black and other underrepresented population groups, in order to determine whether available cell lines adequately represent the diversity in histological type, gender, race, and ethnicity required for optimal lung cancer research. The current review summarizes our findings.
Lung cancer types
An important consideration for the use of cancer cell lines is that they must represent the diversity of cancer types for a given organ. In the case of lung cancer, the major histological types of lung cancer should be represented. Historically, four major histological types were designated: lung adenocarcinoma (LUAD), squamous cell carcinoma (LUSQ), small cell lung cancer (SCLC), and large cell carcinoma (LULCC) (Figure 1). Based on the 2015 World Health Organization reclassification of the 2004-designated lung cancer histological types, these four groups were reclassified into three major types: Lung adenocarcinoma, squamous cell carcinoma, and neuroendocrine tumors (40–42), the latter including small cell lung cancer and large cell carcinoma. Lung adenocarcinoma (LUAD), arising in the air sacs (alveoli) of the distal lung, is the most frequently occurring histological type and commonly presents in the following subtypes: lepidic, acinar, papillary, micropapillary, and solid (42). In addition, LUAD can present as invasive mucinous, colloid, fetal, enteric, and minimally invasive (42). Squamous cell lung cancer (LUSQ) is thought to arise in the airways, is the second most common major lung cancer type, and shows clearly present squamous morphologic patterns. LUSQ can be subclassified as keratinizing, nonkeratinizing, and basaloid. Within neuroendocrine tumors, the most common type is small cell lung cancer (SCLC), a very aggressive cancer that is thought to arise mainly from rare pulmonary neuroendocrine cells [though therapy-resistant lung adenocarcinoma can recur as SCLC, through genetic alterations and a possible stem cell intermediate (43, 44)]. Large cell lung carcinomas (LULCC) are poorly differentiated and when neuroendocrine morphology or staining patterns are seen, large cell lung cancers are referred to as large cell neuroendocrine carcinomas. Large cell carcinomas lacking neuroendocrine markers have been largely reclassified and assigned to other groups depending on immunohistochemical analyses, leaving only a small group of highly undifferentiated cancers designated as large cell carcinomas (~ 1%) (42).
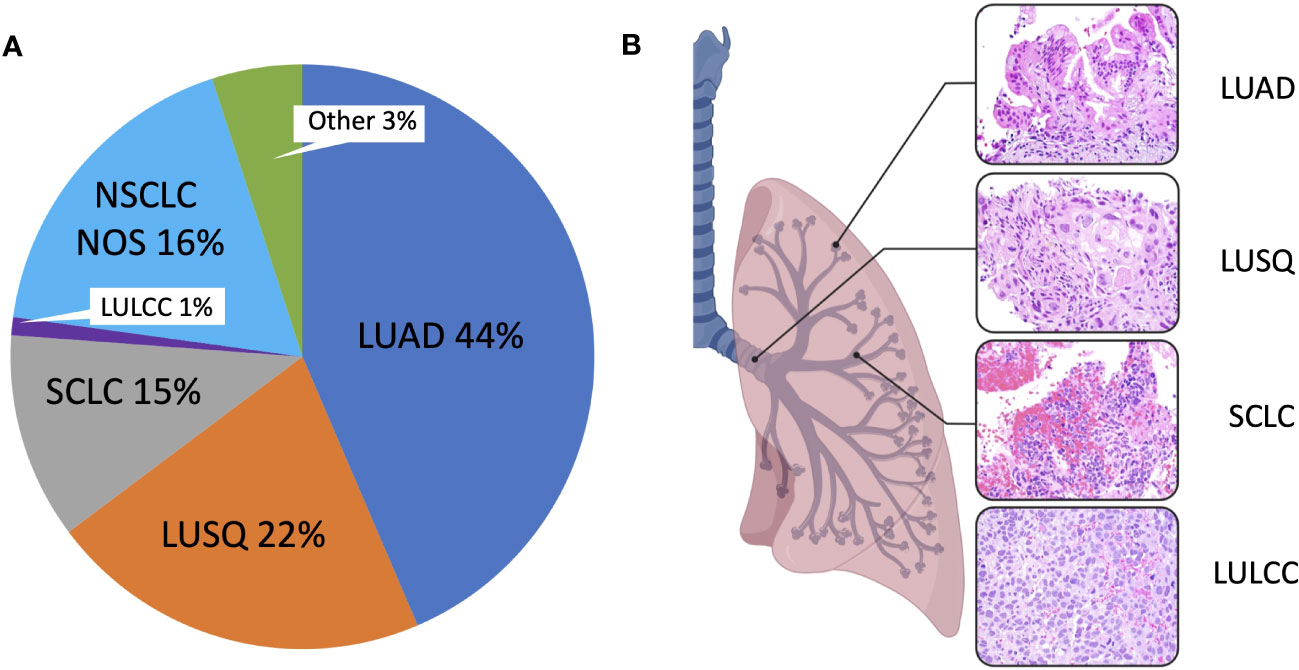
Figure 1 Major lung cancer histological subtypes. (A) Pie chart showing mortality data indicating the proportion of different histological subtypes. Mortality data was obtained from the Surveillance, Epidemiology, and End Results (SEER) Program based on 17 registries in different regions of the United States (www.seer.cancer.gov). SEER*Stat Database: Incidence-Based Mortality - SEER Research Data, 17 Registries, Nov 2021 Sub (2000-2019) - Linked To County Attributes - Time Dependent (1990-2019) Income/Rurality, 1969-2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2022, based on the November 2021 submission. Mortality was calculated via incidence-based mortality (IBM), a method to capture population-level mortality which can be attributable to particular tumor types or other variable reported to SEER registries. IBM calculations were done as described (38). ICD-O-3 morphology codes were grouped together to form the main histologic subtypes, as described (39). LUAD, Lung adenocarcinoma; LULCC, Lung large cell carcinoma; LUSQ, Lung squamous cell cancer; NOS, Lung cancer, not otherwise specified; Other, Other specified carcinoma, including but not limited to carcinoid carcinoma, adenosquamous carcinoma, salivary gland-type carcinomas; SCLC, Small cell lung cancer. (B) Hematoxylin and eosin-stained sections of different lung cancer types at 400x magnification.
We used data from the Surveillance, Epidemiology and End Results (SEER, https://seer.cancer.gov/) program, a large United States-based cancer registry that at present includes over 331 million subjects from 17 regions, to assess lung cancer mortality for different races/ethnicities for the main histological types (Figure 2). The data shows LUAD as the most common histological type across all gender and racial/ethnic categories. Black men and White women show the highest age-adjusted mortality rates for LUAD. Squamous cell lung cancer is the second most common histological type, with Black men and women showing the highest age-adjusted mortality rates for LUSQ. Age-adjusted mortality rates for SCLC are highest for White men and women, while for LULCC they are highest for Black men. Cell lines have been established from the most common lung cancer types (36), with lung adenocarcinoma cell lines predominating because cultures were relatively easy to establish. It should be noted that the histological classifications of cell lines are based on the WHO classification in use at the time the lines were established and may thus not fully match current designations.
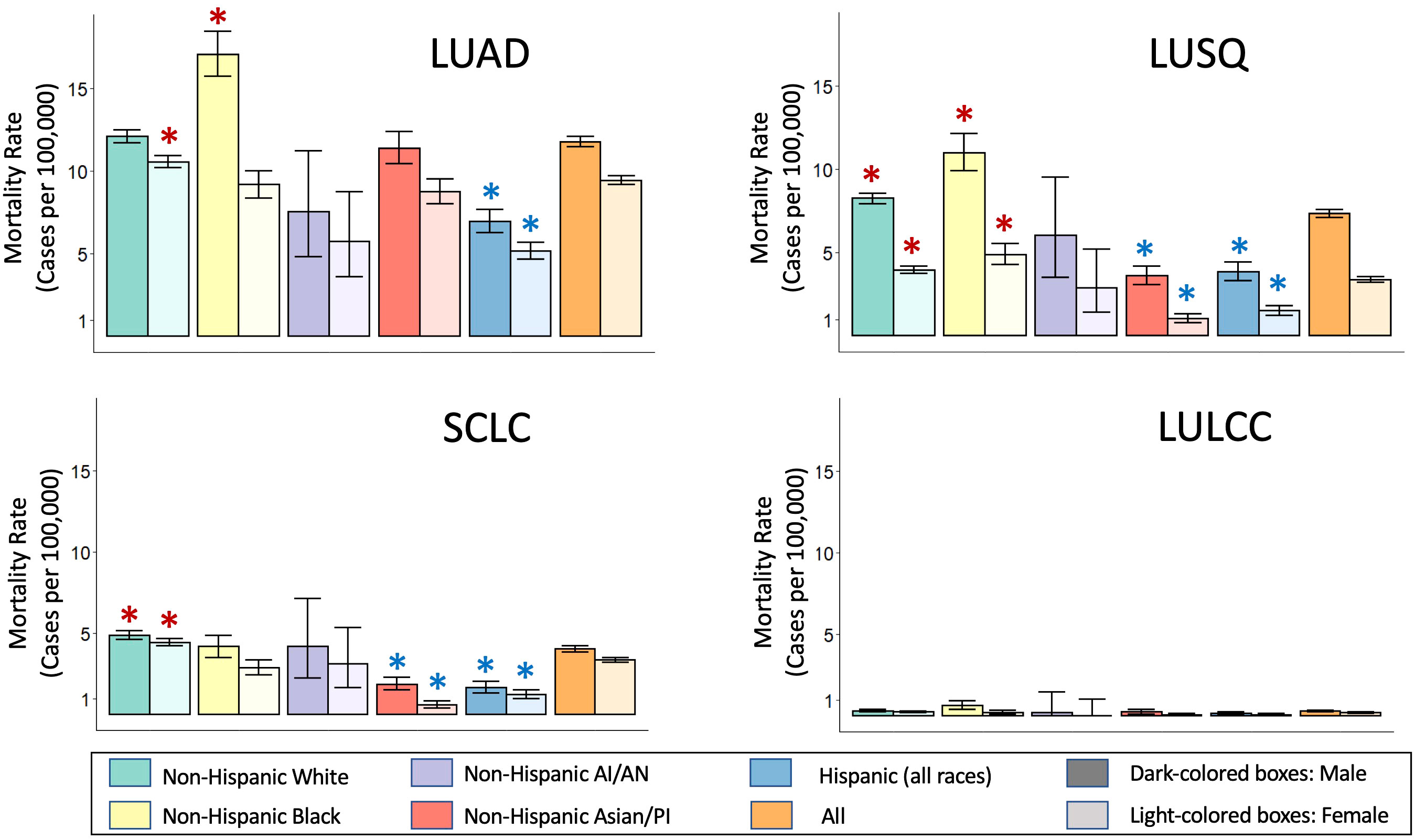
Figure 2 Race and ethnicity-specific mortality data for lung cancer histological subtypes. Mortality data was obtained from the Surveillance, Epidemiology, and End Results (SEER) Program based on 17 registries in different regions of the United States (www.seer.cancer.gov). SEER*Stat Database: Incidence-Based Mortality - SEER Research Data, 17 Registries, Nov 2021 Sub (2000-2019) - Linked To County Attributes - Time Dependent (1990-2019) Income/Rurality, 1969-2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2022, based on the November 2021 submission. Mortality rates are given for each group per 100,000 individuals in that group. Mortality rates were calculated via incidence-based mortality (IBM), a method to capture population-level mortality which can be attributable to particular tumor types or other variable reported to SEER registries. IBM calculations were done as described (38). ICD-O-3 morphology codes were grouped together to form the main histologic subtypes, as described (39). Rate ratio comparison for mortality between individual race/ethnic groups compared to the overall rate of the respective gender were requested as outputs from SEER*Stats, which utilizes the Tiwari method (45). Taking into account a Bonferroni correction for the 40 comparisons, made, we considered the rate ratio significantly different compared to the “All” rate of their respective gender (orange bars) for p-value<0.00125. *,Significantly higher than the All rates for that gender; *,Significantly lower that the All rates for that gender. LUAD, Lung adenocarcinoma; LULCC, Lung large cell carcinoma; LUSQ, Lung squamous cell cancer; SCLC, Small cell lung cancer.
Cell lines as model systems
Lung cancer cell lines allow the in vitro study of human lung cancer, and are especially important for facilitating research when tumor samples are difficult to obtain. SCLC is one such case; it is rarely surgically resected because patients usually present with metastases. In principle, cell lines can be propagated indefinitely and relatively cheaply and are easily disseminated, which allows different labs to study the same cells and compare their results, for example in drug screens. Another advantage of cancer cell lines is that they are pure populations of cells, lacking contaminating stroma and other cell types, thereby allowing detailed genetic and epigenetic studies. This lack of context also has its drawbacks, but these can be addressed using certain culture conditions and model systems as described in a later section.
The first cancer cell line to be cultured was the HeLa cell line, derived from Henrietta Lacks, a black woman with cervical cancer (46). Important ethical questions have been raised about the fact that the cells were obtained at the time without informed consent from the patient (47). The establishment of the HeLa cell line was a scientific breakthrough, and HeLa cells have been widely used in academic and biotech laboratories (47). The demonstrated ability to culture tumor cells from a human patient set the stage for subsequent work establishing cell lines from many kinds of cancer, including lung cancer cell lines.
Due in large part to intensive efforts by Drs. Gazdar, Minna, and Carney to optimize methods to derive cell cultures from patient lung tumors, a large number of cell lines were established (37, 48). With their collaborators, these investigators ultimately cultured more than 200 lung cancer cell lines of different histological types, initially at the National Cancer Institute (NCI-designated lung cancer cell lines), and later at UT Southwestern Medical Center at the Hamon Cancer Center (HCC-designated cell lines). Combined with the efforts of other investigators across the world, over 400 lung cancer cell lines have been reported (36). Many of these cell lines have been cultured for decades, raising concerns among some investigators that the lines might experience genetic drift. Fortunately, the genetic and epigenetic alterations seen in lung cancer cell lines have remained relatively stable over time (37). It has also been asked how well the obtained cell lines represent the tumors from which they are derived. A comparison between a large number of lung cancer cell lines and primary lung cancers has demonstrated that many key genetic and epigenetic changes seen in lung cancer tumors have also been observed in cell lines (37).
Cell line quality and authentication
Two important considerations when using cell lines for research are cell line quality and authenticity. The presence of contaminating microorganisms, particularly mycoplasma, and the cross-contamination with other cell lines can invalidate performed research (49, 50). Mycoplasma is a type of infectious prokaryote lacking a rigid cell wall. While primary cells can be contaminated, laboratory personnel can also be a source of infection (49). Infection can affect cell growth and physiology, thereby nullifying experimental results and making it imperative that cultures be routinely tested so that contaminated cultures be discarded (49, 51–54). If discarding is not an option because a cell line is rare or even irreplaceable, treatment with antibiotics may be considered (55). Authentication of cell lines is also of critical importance. A previous lack of cell line authentication has resulted in large numbers of publications based on incorrect cell types, including many cell lines found to be, in truth, HeLa cells (56). Thus, cell lines should be obtained from reliable sources and should be routinely authenticated through DNA fingerprinting, i.e., the use of short tandem repeats (STRs) (47, 51, 53, 54, 57) (see Table 1 for useful web sites). Journals and granting agencies can help minimize misidentification by requiring authors to authenticate cell lines used in publications (58, 59), such as required by the National Institutes of Health (https://grants.nih.gov/grants/guide/notice-files/not-od-15-103.html). Current recommendations are to test cell lines for mycoplasma and authenticity when they first reach a new laboratory, before publication, and every two months while in culture.
Representation of different races/ethnicities in current lung cancer cell line collections
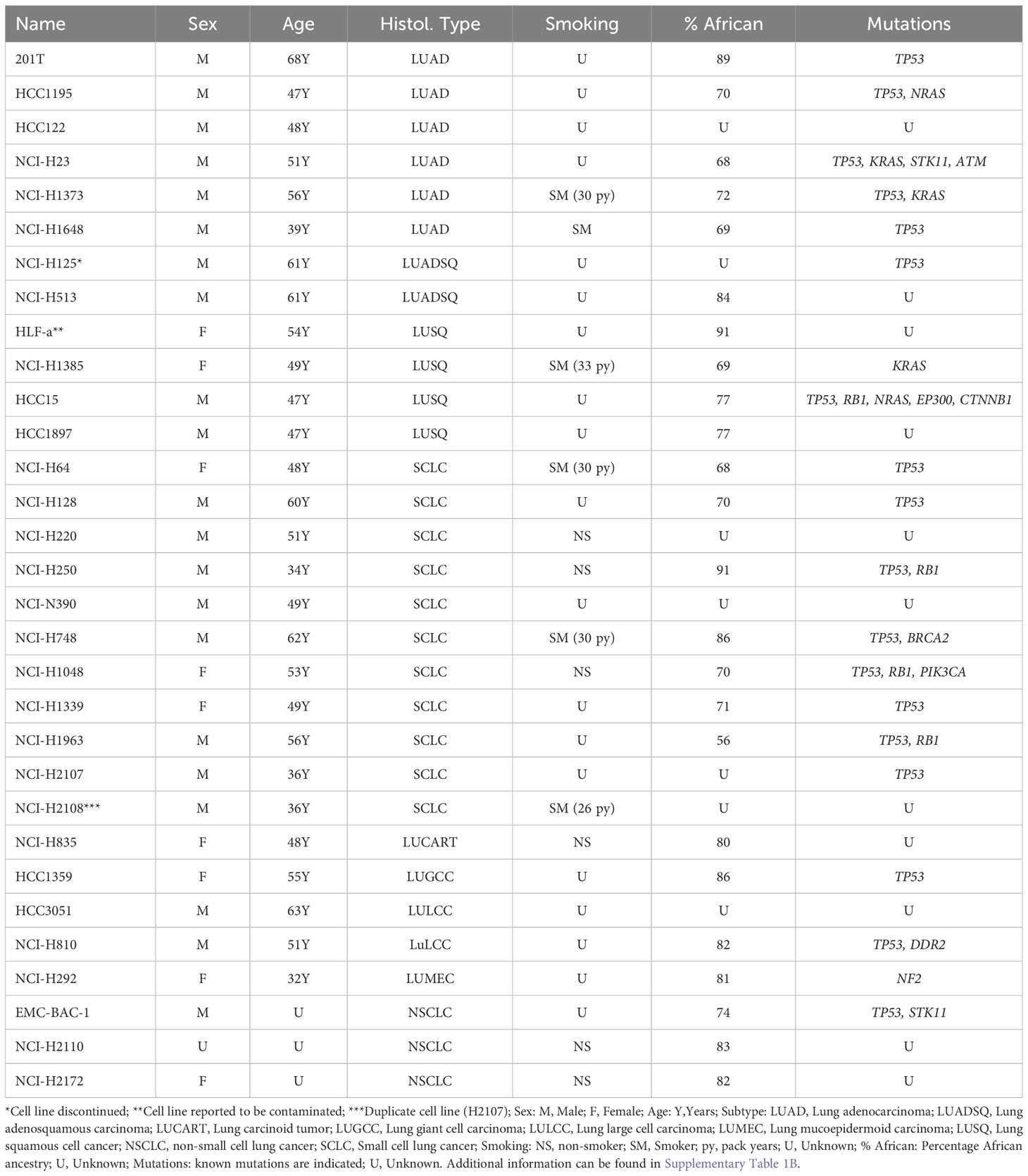
We investigated the availability of lung cancer cell lines representing different races/ethnicities using the resources listed in Table 2. We identified over 800 lung cancer cell lines (Supplementary Table 1). A substantial fraction of these are isogenic (derived from the same parental line) or derived from different sites of the same patient. We identified almost 200 lung cancer cell lines from White subjects (Supplementary Table 1A), 6.5-fold more than the 31 cell lines available from Black patients (Table 3, Supplementary Table 1B). One of the 31 cell lines appears to be a duplicate (NCI-H2108 lists identical patient age, gender, cancer histology, and cell line STR analysis to NCI-H2107). Of the 30 unique Black lung cancer cell lines, 6 were derived from lung adenocarcinomas, 4 from squamous cell cancers, 11 from SCLCs, and the remainder were from unspecified non-small cell lung cancers (3), adenosquamous carcinomas (2), large cell carcinomas (2), a carcinoid tumor, a giant cell carcinoma, and a mucinoepidermoid carcinoma. Ancestry information was available for 24 of these lines, and showed African ancestry, ranging from 56% to 91%.
We identified 390 cell lines from Asian lung cancer patients (Supplementary Table 1C), of which 20% appear to be non-unique (e.g. from different metastatic sites in the body of a given patient), or sister cell lines derived through manipulation of the original cell line.
We did not identify any cell lines representing H/L, AI/AN, or NHOPI individuals. It is possible that H/L ethnicity has not been properly documented for existing cell lines and thus, that such cell lines might be present in the current collection. However, while cell line race can be retrospectively examined using ancestry informative markers (single nucleotide polymorphisms that help infer ancestry admixtures (70–73)), H/L individuals in the Unites States represent an admixture population that may include White, Black, and AI/AN components and would be difficult to genetically identify. Going forward, ethnicity information would need to be documented at the time of sample collection.
We found almost 300 cell lines for which race/ethnicity is unknown (Supplementary Table 1D). Thus, there may be Black, AI/AN, and NHOPI cell lines among these unclassified lines and it may be worth determining their genetic ancestry (70–73).
We noted that the number of cell lines developed from men was over 2-fold higher than cell lines developed from women, and this excess was most prominent for the cell lines developed from Asian individuals (almost 7-fold) (Supplementary Table 1). This difference exceeds what might be expected based on the higher frequency of lung cancer detected in males, and indicates a disparity in the representation of female individuals in lung cancer cell lines. Overall, we conclude that there is a marked lack of cell lines from underrepresented populations and an underrepresentation of cell lines from women.
Cell lines representing the cells-of-origin of different types of lung cancer
In addition to cell lines derived from tumors, it is also important to establish cell lines derived from the cells-of-origin for the different lung cancer histological types. These cells can be useful for modeling the sequential development of the different lung cancer histological types and the effects of environmental exposures on lung cells from the airway or alveolar compartments. Genetic background can affect lung cancer predisposition as well as the metabolism and detoxification of tobacco smoke components (10, 18–27, 74). Thus, just as we need lung cancer cell lines from different races and ethnicities, we need cell-of-origin cell lines from different races and ethnicities to appropriately model lung cancer development. Normal lung cells derived from humans are not immortal and will undergo senescence when propagated in vitro (75). Immortalized cell lines must therefore be created using either viral genes such as Simian Virus 40 large T antigen (SV40LgT) (76, 77) or human papillomavirus E6+E7 genes (78), or overexpression/modification of human genes that allow cell cycle progression and prevent telomere shortening and the resulting senescence (79).
LUAD arises from alveolar epithelium, and to model human lung adenocarcinoma development in vitro, human immortalized alveolar epithelial cells are required. Four immortalized alveolar epithelial cell lines (hAECs) were established using SV40LgT antigen (80, 81). Race is only known for 3 of these cell lines, which were derived from White subjects (81). In addition, a polyclonal alveolar epithelial cell line of unknown race/ethnicity was established using a proprietary cocktail of 33 immortalization genes (82) and from it, a monoclonal cell line (Arlo) was recently derived (83). It will be important to develop additional immortalized alveolar epithelial cell lines for other racial/ethnic groups, given that LUAD is the most common lung cancer histological type in the United States for both genders and all races and ethnicities (Figure 2).
Human bronchial epithelial cells, the putative cells of origin of LUSQ, have been immortalized with SV40LgT, resulting in the BEAS-2B cell line (84), and by using overexpression of the telomerase gene in combination with either overexpression of G1 cell cycle kinase CDK4 or short hairpin RNA-based knockdown of cell cycle regulatory proteins p16INK4A and p14ARF. The latter yielded human bronchial epithelial cells (HBECs) and small airway epithelial cells (SAECs) (85, 86). BEAS-2B, HBEC, and SAEC cell lines can be useful to model the development of squamous cell lung cancer or determine the effects of environmental exposures on airway cells. To our knowledge, the race/ethnicity of the individuals from whom the cell lines were derived is unknown. Thus, ancestry tests of these lines would be useful, as would establishing more of these types of cell lines representing diverse races.
The availability of methods to establish immortalized alveolar and airway cells allows progress to be made in deriving additional cell lines from racially and ethnically diverse subjects. However, there is one important cell type for which no immortalized human cell lines have yet been established: pulmonary neuroendocrine (PNE) cells, the main cell-of-origin of SCLC (87). Immortalized PNE cells would be an important added tool to study the development of SCLC and may be especially relevant for studies of Black SCLC, as this type of cancer may arise at an earlier age in Black subjects than in other races (88, 89). However, PNE cells are rare (less than 1% of lung epithelial cells) making their isolation and immortalization challenging. One possible strategy is to derive these cells from induced pluripotent stem cells (iPSCs), a feat that was recently achieved (90).
Derivation of cell line types from induced pluripotent cells has also been used to obtain bronchial epithelial cells (91) and alveolar epithelial cells (92–94). The availability of racially/ethnically diverse iPSCs (95) provides an opportunity to derive diverse cell lines representing lung cancer cells-of-origin. However, iPSC-derived cell populations can consist of mixed cell types, and considerable time and expertise are required to differentiate them correctly (96, 97). Whether the epigenomes of such iPSC-derived cells fully match those of the corresponding adult differentiated cell types would also need to be determined. Using cell lines with the correct initial epigenome is particularly relevant in studies of the effect of environmental exposures (98). Epigenetic changes play a role in the development of all cancer types (99) and can be driven by environmental exposures such as tobacco smoke (100, 101). Using cell lines with epigenomes matching the natural cells-of-origin is also highly relevant for the study of disease-risk single nucleotide polymorphisms (SNPs) (98). Most risk SNPs, including those for lung cancer, lie in intergenic regions or introns, and likely affect risk by introducing changes in epigenetic regulatory elements (102). If cells differentiated from iPSCs do not epigenetically match their normal mature counterparts, regulatory elements may be missing or altered, thus affecting the correct interpretation of risk SNP epigenetic environments.
Applications of lung cancer cell lines and immortalized lung cell lines
Lung cancer cell lines and cell-of-origin cell lines can be used in a wide variety of ways to study lung cancer (36). In the simplest form, they can be grown on Petri dishes in two-dimensional culture or, in the case of classic SCLC cell lines, in suspension (48). Such in vitro cultures can be useful for the study of cancer driver and tumor suppressor genes, epigenetic changes in cancer cells, the effects of environmental exposures, and the investigation of lung cancer risk SNPs, among other topics. Cell lines provide a relatively pure population of cells compared to heterogeneous tumor or tissue samples that can contain variable amounts of contaminating blood cells and stroma. This simplification can greatly facilitate analyses and provides one powerful strategy to leverage cell-based models. However, it lacks the complexity arising from growth in three-dimensional space or from the interactions with other cell types, such as fibroblasts and blood vessels. Growth of pure cell lines with a defined medium in three dimensions can provide the next level of complexity, while the addition of fibroblasts, endothelial, and blood cells can further simulate in vivo characteristics. Even further advanced are three-dimensional models, so-called “organs-on-a-chip”, which may incorporate an air-liquid interface and/or the movement associated with breathing (103, 104). Organ-on-a-chip devices allow epithelial cells to be coated on a main channel and supportive cells or endothelial cells on a parallel secondary channel separated by a thin porous membrane (105). They can be used to study cancerous cells or cancer cells-of-origin, and should be considered for drug testing as the cellular microenvironment can affect cancer cells’ susceptibility to drugs (106, 107).
No matter how advanced an in vitro model is, it will not provide a natural tumor microenvironment identical to that found in vivo. To achieve the latter, implantation of cell lines into model organisms such as mice is required. To avoid rejection, immunocompromised (“nude”) mice or humanized mice need to be used. Such models, known as xenografts, can be made using human cancer cell lines, primary patient tumors, or even circulating tumor cells (108). Subcutaneous implantation is often used; while not fully mimicking the natural microenvironment, it allows easy monitoring of tumor size and thereby any therapeutic responses. However, if the cells used do not capture the racial and ethnic diversity of lung cancer patients, all models will fall short in moving lung cancer research forward for all population groups.
Discussion
Lung cancer cell lines and cell lines from lung cancer cells-of-origin are a key part of the research toolkit needed to advance knowledge on the development, progression, diagnosis, and treatment of lung cancer. However, in order to ensure that the knowledge gained, tools developed, and treatments devised are applicable to the population regardless of race or ethnicity, we need to ensure that cell lines representing all groups are available. In particular, cell lines representative of Black males should be at hand as Black males show the highest rates of lung cancer death. Here, we investigated the availability of lung cancer cell lines from underrepresented minority populations. We identified over 800 lung cancer cell lines, including ~200 unique lung cancer cell lines from White subjects and over 300 from Asian subjects. This contrasted with just 30 unique lung cancer cell lines available from Black patients. No lung cancer cell lines from H/L, AI/AN, or NHOPI individuals were identified, though some maybe present among the almost 300 lung cancer cell lines of unknown race/ethnicity. It is important to carry out ancestry analyses of existing cell lines to verify which race these lines best represent. In addition, a concerted effort should be made to generate more cell lines from women and underrepresented groups, and to document ethnicity at the time of tissue collection. Expanding the cell line repertoire is even more relevant for cell-of-origin lines, of which there are very few, and to our knowledge none from underrepresented groups.
It should be considered that certain racial/ethnic groups may have cultural objections to donating cells or tissues. Those desires should be respected, even if it means that population groups may not be represented in research. It is also important to keep in mind that broadly defined race/ethnicity groups do not capture the heterogenicity of admixed populations. For example, an analysis of Hispanic men in Florida showed that while lung cancer mortality rates were lower than those of White men, they were 50% higher in Puerto Rican than non-Puerto Rican men (109). Once cell lines from all groups willing to participate have been collected and represent all three major lung cancer types and cells-of-origin (from both men and women), thought should be given to key subpopulations that may merit disaggregation.
One short-term way to partially alleviate the current paucity of lung cancer cell lines representing different racial/ethnic groups is to use genome engineering to derive isogenic cell lines from the handful of underrepresented cell lines available. Cancer driver genes present in the cell lines can be replaced by other driver genes to generate cell lines in which the effects of different driver genes within a similar genomic context can be examined. This would expand the cell line repertoire available for molecular and drug development studies. However, to do this in a biologically meaningful way, the key cancer driver genes present in the different racial/ethnic populations of lung cancer patients must be identified. Unfortunately, cancer driver genes in underrepresented populations are under-studied. For example, in the public database The Cancer Genome Atlas (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), the number of sequenced lung cancer samples from White patients outnumbers that of Black patients by almost 9:1. Thus, data on driver mutations in underrepresented patients must also be expanded. Clearly, much work remains to be done. The first step is to highlight current shortcomings in knowledge and resources, and to disseminate information to lung cancer patients of all races and ethnicities about the need for cell lines representing lung cancer in their communities. Explaining how lung cancer cell lines and cell-of-origin lines can be used to improve research and develop new therapies for people of the patients’ own racial/ethnic backgrounds can help patients make an informed decision about whether to participate. In addition, it would be beneficial if the donations of tissues/cells were discussed with patients by researchers and/or clinicians from their own racial/ethnic group, supporting mutual trust and a better understanding of research goals (7–9). To this end, all races and ethnicities should be well-represented in the medical and biomedical research professions. Thus, we need to build not only the tools, but also foster the success of clinicians and biomedical researchers who can advocate for the establishment and implementation of those tools.
Author contributions
CL, EM, DIR, NSL, PEP, YH and IAO conceived of the study. CL identified and tabulated lung cancer cell lines. CL, AMN, MRC, DAV, MY, BR, WC, DIR, and MG drafted sections of the manuscript. JC extracted data from SEER and generated figures, all authors assisted with editing, and IAO oversaw and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Positionality statement
The authors represent the following racial/ethnic groups: White Hispanic/LaIn(x) (CL, JC, MRC, DAV, PEP, DIR, MG), Black non-Hispanic(EM,Jr, AMN, NSL), White non-Hispanic (IAO), and Asian (MY, BR, WC, YH).
Funding
This study was supported by grants U54CA233396, U54CA233444, and U54CA233465 from the National Institutes of Health (NIH)/National Cancer Institute (NCI), R21HL162405 from the NIH/National Heart, Lung, and Blood Institute (NHLBI), the Norris Comprehensive Cancer Center core grant, award number P30CA014089 from the NIH/NCI. IAO is supported in part by NIH/NHLBI grant R35HL135747, the California Tobacco-Related Disease Research Program (T31IP913 and T32IP5392), the Wright Foundation, and generous donations from Mrs. Judy Glick and Mrs. Conya Pembroke. MG is supported by the Keck School of Medicine Dean’s Fellowship. AMN is supported by a fellowship from the Hastings Center for Pulmonary Research.
Acknowledgments
The authors gratefully acknowledge all the members of the Florida-California Cancer Research Education and Engagement (CaRE2) Health Equity Center for their support as well as members of the Offringa and Dr. Crystal Marconett labs for helpful discussions.
Conflict of interest
EM sits on an advisory board for Takeda.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1187585/full#supplementary-material
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin (2023) 73(1):17–48. doi: 10.3322/caac.21763
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
4. DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin (2019) 69(3):211–33. doi: 10.3322/caac.21555
5. Cronin KA, Scott S, Firth AU, Sung H, Henley SJ, Sherman RL, et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer (2022) 128(24):4251–84. doi: 10.1002/cncr.34479
6. Giaquinto AN, Miller KD, Tossas KY, Winn RA, Jemal A, Siegel RL. Cancer statistics for African American/black people 2022. CA Cancer J Clin (2022) 72(3):202–29. doi: 10.3322/caac.21718
7. Byrd GS, Edwards CL, Kelkar VA, Phillips RG, Byrd JR, Pim-Pong DS, et al. Recruiting intergenerational African American males for biomedical research Studies: a major research challenge. J Natl Med Assoc (2011) 103(6):480–7. doi: 10.1016/S0027-9684(15)30361-8
8. Lin JJ, Mhango G, Wall MM, Lurslurchachai L, Bond KT, Nelson JE, et al. Cultural factors associated with racial disparities in lung cancer care. Ann Am Thorac Soc (2014) 11(4):489–95. doi: 10.1513/AnnalsATS.201402-055OC
9. Mitchell E, Alese OB, Yates C, Rivers BM, Blackstock W, Newman L, et al. Cancer healthcare disparities among African Americans in the United States. J Natl Med Assoc (2022) 114(3):236–50. doi: 10.1016/j.jnma.2022.01.004
10. Byun J, Han Y, Li Y, Xia J, Long E, Choi J, et al. Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer. Nat Genet (2022) 54(8):1167–77. doi: 10.1038/s41588-022-01115-x
11. Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med (2006) 354(4):333–42. doi: 10.1056/NEJMoa033250
12. Jones CC, Bush WS, Crawford DC, Wenzlaff AS, Schwartz AG, Wiencke JK, et al. Germline genetic variants and lung cancer survival in African Americans. Cancer Epidemiol Biomarkers Prev (2017) 26(8):1288–95. doi: 10.1158/1055-9965.EPI-16-0998
13. Mitchell KA, Shah E, Bowman ED, Zingone A, Nichols N, Pine SR, et al. Relationship between West African ancestry with lung cancer risk and survival in African Americans. Cancer Causes Control (2019) 30(11):1259–68. doi: 10.1007/s10552-019-01212-z
14. Stram DO, Park SL, Haiman CA, Murphy SE, Patel Y, Hecht SS, et al. Racial/ethnic differences in lung cancer incidence in the multiethnic cohort study: an update. J Natl Cancer Inst (2019) 111(8):811–9. doi: 10.1093/jnci/djy206
15. Ryan BM. Lung cancer health disparities. Carcinogenesis (2018) 39(6):741–51. doi: 10.1093/carcin/bgy047
16. Basu A, Kopulos L, Geissen N, Sukhal S, Smith SB. Analysis of the 30-pack-year smoking threshold in African Americans from an underserved lung cancer screening program. J Am Coll Radiol (2021) 18(1 Pt A):27–33. doi: 10.1016/j.jacr.2020.08.017
17. Etzel CJ, Kachroo S, Liu M, D'Amelio A, Dong Q, Cote ML, et al. Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev Res (Phila) (2008) 1(4):255–65. doi: 10.1158/1940-6207.CAPR-08-0082
18. Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, et al. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst (2010) 102(15):1199–205. doi: 10.1093/jnci/djq232
19. Blackman E, Ashing K, Gibbs D, Kuo YM, Andrews A, Ramakodi M, et al. The Cancer Prevention Project of Philadelphia: preliminary findings examining diversity among the African diaspora. Ethn Health (2021) 26(5):659–75. doi: 10.1080/13557858.2018.1548695
20. Hancock DB, Guo Y, Reginsson GW, Gaddis NC, Lutz SM, Sherva R, et al. Genome-wide association study across European and African American ancestries identifies a SNP in DNMT3B contributing to nicotine dependence. Mol Psychiatry (2018) 23(9):1911–9. doi: 10.1038/mp.2017.193
21. Hansen HM, Xiao Y, Rice T, Bracci PM, Wrensch MR, Sison JD, et al. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum Mol Genet (2010) 19(18):3652–61. doi: 10.1093/hmg/ddq268
22. Murphy SE, Park SL, Balbo S, Haiman CA, Hatsukami DK, Patel Y, et al. Tobacco biomarkers and genetic/epigenetic analysis to investigate ethnic/racial differences in lung cancer risk among smokers. NPJ Precis Oncol (2018) 2:17. doi: 10.1038/s41698-018-0057-y
23. Perez-Stable EJ, Herrera B, Jacob P 3rd, Benowitz NL. Nicotine metabolism and intake in black and white smokers. JAMA (1998) 280(2):152–6. doi: 10.1001/jama.280.2.152
24. Quach BC, Bray MJ, Gaddis NC, Liu M, Palviainen T, Minica CC, et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun (2020) 11(1):5562. doi: 10.1038/s41467-020-19265-z
25. Arauz RF, Byun JS, Tandon M, Sinha S, Kuhn S, Taylor S, et al. Whole-exome profiling of NSCLC among African Americans. J Thorac Oncol (2020) 15(12):1880–92. doi: 10.1016/j.jtho.2020.08.029
26. Boldry EJ, Patel YM, Kotapati S, Esades A, Park SL, Tiirikainen M, et al. Genetic determinants of 1,3-butadiene metabolism and detoxification in three populations of smokers with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev (2017) 26(7):1034–42. doi: 10.1158/1055-9965.EPI-16-0838
27. Taioli E, Ford J, Trachman J, Li Y, Demopoulos R, Garte S. Lung cancer risk and CYP1A1 genotype in African Americans. Carcinogenesis (1998) 19(5):813–7. doi: 10.1093/carcin/19.5.813
28. Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discovery (2021) 11(3):591–8. doi: 10.1158/2159-8290.CD-20-1165
29. Heath EI, Lynce F, Xiu J, Ellerbrock A, Reddy SK, Obeid E, et al. Racial disparities in the molecular landscape of cancer. Anticancer Res (2018) 38(4):2235–40. doi: 10.21873/anticanres.12466
30. Kytola V, Topaloglu U, Miller LD, Bitting RL, Goodman MM, D. Agostino RBJ, et al. Mutational landscapes of smoking-related cancers in Caucasians and African Americans: precision oncology perspectives at wake forest baptist comprehensive cancer center. Theranostics (2017) 7(11):2914–23. doi: 10.7150/thno.20355
31. Lusk CM, Watza D, Dyson G, Craig D, Ratliff V, Wenzlaff AS, et al. Profiling the mutational landscape in known driver genes and novel genes in African American non-small cell lung cancer patients. Clin Cancer Res (2019) 25(14):4300–8. doi: 10.1158/1078-0432.CCR-18-2439
32. Soh J, Toyooka S, Matsuo K, Yamamoto H, Wistuba II, Lam S, et al. Ethnicity affects EGFR and KRAS gene alterations of lung adenocarcinoma. Oncol Lett (2015) 10(3):1775–82. doi: 10.3892/ol.2015.3414
33. Schenk A, Lopez S, Kschischo M, McGranahan N. Germline ancestry influences the evolutionary disease course in lung adenocarcinomas. Evol Appl (2020) 13(7):1550–7. doi: 10.1111/eva.12964
34. Sausville LN, Jones CC, Aldrich MC, Blot WJ, Pozzi A, Williams SM. Genetic variation in the eicosanoid pathway is associated with non-small-cell lung cancer (NSCLC) survival. PloS One (2017) 12(7):e0180471. doi: 10.1371/journal.pone.0180471
35. Harrison S, Judd J, Chin S, Ragin C. Disparities in lung cancer treatment. Curr Oncol Rep (2022) 24(2):241–8. doi: 10.1007/s11912-022-01193-4
36. Gazdar AF, Hirsch FR, Minna JD. From mice to men and back: an assessment of preclinical model systems for the study of lung cancers. J Thorac Oncol (2016) 11(3):287–99. doi: 10.1016/j.jtho.2015.10.009
37. Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: Useless artifacts or invaluable tools for medical science? Lung Cancer (2010) 68(3):309–18. doi: 10.1016/j.lungcan.2009.12.005
38. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med (2020) 383(7):640–9. doi: 10.1056/NEJMoa1916623
39. Lewis DR, Check DP, Caporaso NE, Travis WD, Devesa SS. US lung cancer trends by histologic type. Cancer (2014) 120(18):2883–92. doi: 10.1002/cncr.28749
40. Rodak O, Peris-Diaz MD, Olbromski M, Podhorska-Okolow M, Dziegiel P. Current landscape of non-small cell lung cancer: epidemiology, histological classification, targeted therapies, and immunotherapy. Cancers (Basel) (2021) 13(18). doi: 10.3390/cancers13184705
41. Travis WD. The 2015 WHO classification of lung tumors. Pathologe (2014) 35(Suppl 2):188. doi: 10.1007/s00292-014-1974-3
42. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol (2015) 10(9):1243–60. doi: 10.1097/JTO.0000000000000630
43. Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol (2019) 14(10):1784–93. doi: 10.1016/j.jtho.2019.06.002
44. Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, et al. Multiomic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discovery (2021) 11(12):3028–47. doi: 10.1158/2159-8290.CD-20-1863
45. Tiwari RC, Clegg LX, Zou Z. Efficient interval estimation for age-adjusted cancer rates. Stat Methods Med Res (2006) 15(6):547–69. doi: 10.1177/0962280206070621
46. Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human Malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med (1953) 97(5):695–710. doi: 10.1084/jem.97.5.695
47. Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer (2002) 2(4):315–9. doi: 10.1038/nrc775
48. Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res (1985) 45(6):2913–23.
49. Drexler HG, Uphoff CC. Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology (2002) 39(2):75–90. doi: 10.1023/A:1022913015916
50. Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst (2010) 102(17):1310–21. doi: 10.1093/jnci/djq279
51. Dennert K, Kumar R. Traceability methods for cell line authentication and mycoplasma detection. SLAS Technol (2021) 26(6):630–6. doi: 10.1177/24726303211030290
52. Olarerin-George AO, Hogenesch JB. Assessing the prevalence of mycoplasma contamination in cell culture via a survey of NCBI's RNA-seq archive. Nucleic Acids Res (2015) 43(5):2535–42. doi: 10.1093/nar/gkv136
53. Uphoff CC, Drexler HG. Eradication of Mycoplasma contaminations from cell cultures. Curr Protoc Mol Biol (2014) 106:28 5 1– 5 12. doi: 10.1002/0471142727.mb2805s106
54. Young L, Sung J, Stacey G, Masters JR. Detection of Mycoplasma in cell cultures. Nat Protoc (2010) 5(5):929–34. doi: 10.1038/nprot.2010.43
55. Molla Kazemiha V, Shokrgozar MA, Arabestani MR, Shojaei Moghadam M, Azari S, Maleki S, et al. PCR-based detection and eradication of mycoplasmal infections from various mammalian cell lines: a local experience. Cytotechnology (2009) 61(3):117–24. doi: 10.1007/s10616-010-9252-6
56. Horbach S, Halffman W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PloS One (2017) 12(10):e0186281. doi: 10.1371/journal.pone.0186281
57. Eltonsy N, Gabisi V, Li X, Russe KB, Mills GB, Stemke-Hale K. Detection algorithm for the validation of human cell lines. Int J Cancer (2012) 131(6):E1024–30. doi: 10.1002/ijc.27533
58. Babic Z, Capes-Davis A, Martone ME, Bairoch A, Ozyurt IB, Gillespie TH, et al. Incidences of problematic cell lines are lower in papers that use RRIDs to identify cell lines. Elife (2019) 8. doi: 10.7554/eLife.41676
59. Souren NY, Fusenig NE, Heck S, Dirks WG, Capes-Davis A, Bianchini F, et al. Cell line authentication: a necessity for reproducible biomedical research. EMBO J (2022) 41(14):e111307. doi: 10.15252/embj.2022111307
60. Bepler G, Jaques G, Neumann K, Aumuller G, Gropp C, Havemann K. Establishment, growth properties, and morphological characteristics of permanent human small cell lung cancer cell lines. J Cancer Res Clin Oncol (1987) 113(1):31–40. doi: 10.1007/BF00389964
61. Bepler G, Koehler A, Kiefer P, Havemann K, Beisenherz K, Jaques G, et al. Characterization of the state of differentiation of six newly established human non-small-cell lung cancer cell lines. Differentiation (1988) 37(2):158–71. doi: 10.1111/j.1432-0436.1988.tb00806.x
62. Blanco R, Iwakawa R, Tang M, Kohno T, Angulo B, Pio R, et al. A gene-alteration profile of human lung cancer cell lines. Hum Mutat (2009) 30(8):1199–206. doi: 10.1002/humu.21028
63. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature (2019) 569(7757):503–8. doi: 10.1038/s41586-019-1186-3
64. Gottschling S, Jauch A, Kuner R, Herpel E, Mueller-Decker K, Schnabel PA, et al. Establishment and comparative characterization of novel squamous cell non-small cell lung cancer cell lines and their corresponding tumor tissue. Lung Cancer (2012) 75(1):45–57. doi: 10.1016/j.lungcan.2011.05.020
65. Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol (2015) 33(3):306–12. doi: 10.1038/nbt.3080
66. Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell (2020) 180(2):387–402 e16. doi: 10.1016/j.cell.2019.12.023
67. Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, et al. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl (1996) 24:32–91. doi: 10.1002/jcb.240630505
68. Sekido Y, Sato M, Usami N, Shigemitsu K, Mori S, Maeda O, et al. Establishment of a large cell lung cancer cell line (Y-ML-1B) producing granulocyte colony-stimulating factor. Cancer Genet Cytogenet (2002) 137(1):33–42. doi: 10.1016/S0165-4608(02)00536-8
69. Zheng C, Sun YH, Ye XL, Chen HQ, Ji HB. Establishment and characterization of primary lung cancer cell lines from Chinese population. Acta Pharmacol Sin (2011) 32(3):385–92. doi: 10.1038/aps.2010.214
70. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res (2009) 19(9):1655–64. doi: 10.1101/gr.094052.109
71. Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, et al. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PloS Genet (2012) 8(3):e1002554. doi: 10.1371/journal.pgen.1002554
72. Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat (2009) 30(1):69–78. doi: 10.1002/humu.20822
73. Phillips C, Salas A, Sanchez JJ, Fondevila M, Gomez-Tato A, Alvarez-Dios J, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet (2007) 1(3-4):273–80. doi: 10.1016/j.fsigen.2007.06.008
74. Zanetti KA, Wang Z, Aldrich M, Amos CI, Blot WJ, Bowman ED, et al. Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer (2016) 98:33–42. doi: 10.1016/j.lungcan.2016.05.008
75. Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res (1965) 37:614–36. doi: 10.1016/0014-4827(65)90211-9
76. Bryan TM, Reddel RR. SV40-induced immortalization of human cells. Crit Rev Oncog (1994) 5(4):331–57. doi: 10.1615/CritRevOncog.v5.i4.10
77. Jha KK, Banga S, Palejwala V, Ozer HL. SV40-mediated immortalization. Exp Cell Res (1998) 245(1):1–7. doi: 10.1006/excr.1998.4272
78. Halbert CL, Demers GW, Galloway DA. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol (1992) 66(4):2125–34. doi: 10.1128/jvi.66.4.2125-2134.1992
79. Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol (2000) 12(6):705–9. doi: 10.1016/S0955-0674(00)00155-1
80. Kemp SJ, Thorley AJ, Gorelik J, Seckl MJ, O'Hare MJ, Arcaro A, et al. Immortalization of human alveolar epithelial cells to investigate nanoparticle uptake. Am J Respir Cell Mol Biol (2008) 39(5):591–7. doi: 10.1165/rcmb.2007-0334OC
81. Tran E, Shi T, Li X, Chowdhury AY, Jiang D, Liu Y, et al. Development of human alveolar epithelial cell models to study distal lung biology and disease. iScience (2022) 25(2):103780. doi: 10.1016/j.isci.2022.103780
82. Kuehn A, Kletting S, de Souza Carvalho-Wodarz C, Repnik U, Griffiths G, Fischer U, et al. Human alveolar epithelial cells expressing tight junctions to model the air-blood barrier. ALTEX (2016) 33(3):251–60. doi: 10.14573/altex.1511131
83. Carius P, Jungmann A, Bechtel M, Grissmer A, Boese A, Gasparoni G, et al. A monoclonal human alveolar epithelial cell line ("Arlo") with pronounced barrier function for studying drug permeability and viral infections. Adv Sci (Weinh) (2023) 10(8):e2207301. doi: 10.1002/advs.202207301
84. Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res (1988) 48(7):1904–9.
85. Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res (2004) 64(24):9027–34. doi: 10.1158/0008-5472.CAN-04-3703
86. Smith JL, Lee LC, Read A, Li Q, Yu B, Lee CS, et al. One-step immortalization of primary human airway epithelial cells capable of oncogenic transformation. Cell Biosci (2016) 6:57. doi: 10.1186/s13578-016-0122-6
87. Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell (2011) 19(6):754–64. doi: 10.1016/j.ccr.2011.04.019
88. Tapan U, Furtado VF, Qureshi MM, Everett P, Suzuki K, Mak KS. Racial and other healthcare disparities in patients with extensive-stage SCLC. JTO Clin Res Rep (2021) 2(1):100109. doi: 10.1016/j.jtocrr.2020.100109
89. Thomas PL, Madubata CJ, Aldrich MC, Lee MM, Owonikoko TK, Minna JD, et al. A call to action: dismantling racial injustices in preclinical research and clinical care of black patients living with small cell lung cancer. Cancer Discovery (2021) 11(2):240–4. doi: 10.1158/2159-8290.CD-20-1592
90. Hor P, Punj V, Calvert BA, Castaldi A, Miller AJ, Carraro G, et al. Efficient generation and transcriptomic profiling of human iPSC-derived pulmonary neuroendocrine cells. iScience (2020) 23(5):101083. doi: 10.1016/j.isci.2020.101083
91. McIntyre BA, Alev C, Mechael R, Salci KR, Lee JB, Fiebig-Comyn A, et al. Expansive generation of functional airway epithelium from human embryonic stem cells. Stem Cells Transl Med (2014) 3(1):7–17. doi: 10.5966/sctm.2013-0119
92. Kanagaki S, Ikeo S, Suezawa T, Yamamoto Y, Seki M, Hirai T, et al. Directed induction of alveolar type I cells derived from pluripotent stem cells via Wnt signaling inhibition. Stem Cells (2021) 39(2):156–69. doi: 10.1002/stem.3302
93. Korogi Y, Gotoh S, Ikeo S, Yamamoto Y, Sone N, Tamai K, et al. In vitro disease modeling of Hermansky-Pudlak syndrome type 2 using human induced pluripotent stem cell-derived alveolar organoids. Stem Cell Rep (2019) 12(3):431–40. doi: 10.1016/j.stemcr.2019.01.014
94. Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods (2017) 14(11):1097–106. doi: 10.1038/nmeth.4448
95. Chang EA, Tomov ML, Suhr ST, Luo J, Olmsted ZT, Paluh JL, et al. Derivation of ethnically diverse human induced pluripotent stem cell lines. Sci Rep (2015) 5:15234. doi: 10.1038/srep15234
96. Meng G, Liu S, Rancourt DE. Synergistic effect of medium, matrix, and exogenous factors on the adhesion and growth of human pluripotent stem cells under defined, xeno-free conditions. Stem Cells Dev (2012) 21(11):2036–48. doi: 10.1089/scd.2011.0489
97. Volpato V, Webber C. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis Model Mech (2020) 13(1). doi: 10.1242/dmm.042317
98. Stueve TR, Marconett CN, Zhou B, Borok Z, Laird-Offringa IA. The importance of detailed epigenomic profiling of different cell types within organs. Epigenomics (2016) 8(6):817–29. doi: 10.2217/epi-2016-0005
99. Jones PA, Baylin SB. The epigenomics of cancer. Cell (2007) 128(4):683–92. doi: 10.1016/j.cell.2007.01.029
100. Stueve TR, Li WQ, Shi J, Marconett CN, Zhang T, Yang C, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet (2017) 26(15):3014–27. doi: 10.1093/hmg/ddx188
101. Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O'Hagan HM, et al. Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell (2017) 32(3):360–76 e6. doi: 10.1016/j.ccell.2017.08.006
102. Yang C, Stueve TR, Yan C, Rhie SK, Mullen DJ, Luo J, et al. Positional integration of lung adenocarcinoma susceptibility loci with primary human alveolar epithelial cell epigenomes. Epigenomics (2018) 10(9):1167–87. doi: 10.2217/epi-2018-0003
103. Agraval H, Sharma JR, Dholia N, Yadav UCS. Air-liquid interface culture model to study lung cancer-associated cellular and molecular changes. Methods Mol Biol (2022) 2413:133–44. doi: 10.1007/978-1-0716-1896-7_14
104. Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat BioMed Eng (2021) 5(8):815–29. doi: 10.1038/s41551-021-00718-9
105. Bai H, Ingber DE. What can an organ-on-a-chip teach us about human lung pathophysiology? Physiol (Bethesda) (2022) 37(5):0. doi: 10.1152/physiol.00012.2022
106. Breslin S, O'Driscoll L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget (2016) 7(29):45745–56. doi: 10.18632/oncotarget.9935
107. Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol (2014) 12(4):207–18. doi: 10.1089/adt.2014.573
108. Lallo A, Schenk MW, Frese KK, Blackhall F, Dive C. Circulating tumor cells and CDX models as a tool for preclinical drug development. Transl Lung Cancer Res (2017) 6(4):397–408. doi: 10.21037/tlcr.2017.08.01
Keywords: lung cancer, cell lines, underrepresented, diversity, cancer health disparities, lung adenocarcinoma, squamous cell lung cancer, in vitro models
Citation: Leon C, Manley E Jr., Neely AM, Castillo J, Ramos Correa M, Velarde DA, Yang M, Puente PE, Romero DI, Ren B, Chai W, Gladstone M, Lamango NS, Huang Y and Offringa IA (2023) Lack of racial and ethnic diversity in lung cancer cell lines contributes to lung cancer health disparities. Front. Oncol. 13:1187585. doi: 10.3389/fonc.2023.1187585
Received: 16 March 2023; Accepted: 10 October 2023;
Published: 01 November 2023.
Edited by:
Farnam Mohebi, University of California, Berkeley, United StatesReviewed by:
Giamila Fantuzzi, University of Illinois Chicago, United StatesQing Lin, Johns Hopkins University, United States
Copyright © 2023 Leon, Manley, Neely, Castillo, Ramos Correa, Velarde, Yang, Puente, Romero, Ren, Chai, Gladstone, Lamango, Huang and Offringa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ite A. Offringa, aWxhaXJkQHVzYy5lZHU=
†These authors share first authorship
 Christopher Leon
Christopher Leon Eugene Manley Jr.
Eugene Manley Jr. Aaron M. Neely
Aaron M. Neely Jonathan Castillo
Jonathan Castillo Michele Ramos Correa
Michele Ramos Correa Diego A. Velarde1,2,3
Diego A. Velarde1,2,3 Minxiao Yang
Minxiao Yang Pablo E. Puente
Pablo E. Puente Bing Ren
Bing Ren Wenxuan Chai
Wenxuan Chai Matthew Gladstone
Matthew Gladstone Nazarius S. Lamango
Nazarius S. Lamango Yong Huang
Yong Huang Ite A. Offringa
Ite A. Offringa