- Department of Hematology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
Background: Diffuse large B-cell lymphoma (DLBCL) is a kind of highly heterogeneous non-Hodgkin lymphoma, both in clinical and genetic terms. DLBCL is admittedly categorized into six subtypes by genetics, which contain MCD, BN2, EZB, N1, ST2, and A53. Dyslipidemia is relevant to a multitude of solid tumors and has recently been reported to be associated with hematologic malignancies. We aim to present a retrospective study investigating dyslipidemia in DLBCL based on the molecular subtypes.
Results: This study concluded that 259 patients with newly diagnosed DLBCL and their biopsy specimens were available for molecular typing. Results show that the incidence of dyslipidemia (87.0%, p <0.001) is higher in the EZB subtype than in others, especially hypertriglyceridemia (78.3%, p = 0.001) in the EZB subtype. Based on the pathological gene-sequencing, patients with BCL2 gene fusion mutation are significantly correlative with hyperlipidemia (76.5%, p = 0.006) and hypertriglyceridemia (88.2%, p = 0.002). Nevertheless, the occurrence of dyslipidemia has no remarkable influence on prognosis.
Conclusion: In summary, dyslipidemia correlates with genetic heterogeneity in DLBCL without having a significant influence on survival. This research first connects lipids and genetic subtypes in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is characterized by aggressiveness and heterogeneity epigenetically and genetically. Besides, DLBCL accounts for approximately 35% of non-Hodgkin lymphoma (NHL) and is the most common lymphoma in adults (1). In 2018, the definition of molecular typing was initially propounded. Shipp et al. classified DLBCL into clusters 1–5 by genetic abnormality (2). Staudt et al. implemented molecular typing by analyzing genetic alterations and proposed four prominent subtypes: MCD (featured as the co-occurrence of MYD88 L265p and CD79B mutations), BN2 (featured as BCL6 fusion mutations and NOTCH2 mutations), EZB (featured as EZH2 mutations and BCL2 fusion mutations), and N1 (featured as NOTCH1 mutations). Later, two other types were added, including ST2 (featured as SGK1 and TET2 mutations) and A53 (featured as TP53 aneuploid mutations) (3, 4). Researchers had considered the influence of MYC mutations and therefore divided the EZB subtype into MYC-positive EZB isoforms and MYC-negative EZB isoforms (4). While the prior simple algorithm of six subtypes is more frequently being applied in clinical. Dyslipidemia has been considered correlative to multitudes of solid tumors, like breast cancer, prostate cancer, thyroid cancer, colorectal cancer, and lung cancer (5–9). Recently, researchers have found that a portion of hematologic malignancies is relative to the incidence of dyslipidemia, for instance, chronic leukemia and acute promyelocytic leukemia (10, 11). Ma et al. recently found that cholesterol biosynthesis relative axis may have an oncogenic activation in DLBCL with BCL2-IGH translocations (12). While the correlation between dyslipidemia and DLBCL has not been reported, especially on the genetic aspect. Therefore, we aim to investigate dyslipidemia in DLBCL in the context of molecular typing and explore the correlation between dyslipidemia and gene mutations.
In this retrospective study, we analyzed 259 DLBCL patients with gene sequencing to assess dyslipidemia in newly diagnosed DLBCL.
Methods
Patients and sample collection
A total of 259 newly diagnosed DLBCL patients were selected with indispensable pathological issues to analyze molecular typing. All patients were admitted to the clinic center affiliated with the First Affiliated Hospital of Zhejiang University School of Medicine from April 2014 to November 2022. A total of 121 genes covering exons, fusion-relevant introns and alternative splicing areas are analyzed. Six typing methods were applied in this study, which contain MCD, BN2, EZB, N1, ST2, and A53. In terms of dyslipidemia, we collected lipid indices, including triglycerides (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C). Lipid profiles were stratified according to The Guidelines on Prevention and Treatment of Blood Lipid Abnormality in Chinese Adults (Version 2016) (13). Dyslipidemia is defined as a disease with an abnormality in lipids, usually with increased levels of TC and TG. Dyslipidemia is classified into different subtypes in clinical aspects, pathogenesis, and physical-chemistry aspects. According to the WHO criteria developed by Fredrickson, which discriminate by physical and chemical features. Dyslipidemia is categorized into six types (14). Due to the complexity of this classification, another standard is rather easier. Dyslipidemia is defined into four types: hypertriglyceridemia, hypercholesterolemia, hypoalbuminemia, and mixed hyperlipidemia (both hypertriglyceridemia and hypercholesterolemia). This method is now used more frequently in clinics, and in our study, this method was applied (15, 16).
All patients were initially treated with the first-line therapy recommended by the National Comprehensive Cancer Network (NCCN) (17). Patients with CD20 positivity were primely treated with rituximab in combination with cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) or R-CHOP-like regimens. Older patients were medicated with a dose-decreased R-CHOP regimen. The R-DA-EPOCH regimen was administered to patients with poor left ventricular functions.
Diagnosis and prognosis indicators related to DLBCL were collected and concluded as follows: B symptoms, Ann Arbor stage, international prognosis index (IPI), Eastern Cooperative Oncology Group stage (ECOG stage), and results of immunohistochemistry (IHC). We also collected demographic and other clinical data from medical records that concluded: age, sex, height, weight, body mass index (BMI), white blood count (WBC), hemoglobin count, platelet counts, albumin, alanine transaminase (ALT), serum creatinine, lactate dehydrogenase (LDH), ferritin, C-reactive protein (CRP), β microglobulin (β-MG), fibrinogen, prothrombin time (PT), activated prothrombin time (APTT), thrombin time (TT), International normalized ratio (INR), D-dimer, and cytokines including interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-6 (IL-6), interleukin-10 (IL-10), interleukin-17α (IL-17α), tumor necrosis factor-α (TNF-α), and interferon-γ (IFN-γ). We collected a total of 22,375 samples.
The study was approved by the institutional Ethics Committee and conducted in accordance with the Declaration of Helsinki.
Classification of abnormality
According to the Guidelines on Prevention and Treatment of Blood Lipid Abnormality in Chinese Adults (Version 2016), we define dyslipidemia as hypertriglyceridemia, hypercholesterolemia, hypoalbuminemia, and mixed hyperlipidemia, which meets both hypertriglyceridemia and hypercholesterolemia.
The upper limits of normal TG, TC, and LDL were 1.70 mmol/L (150 mg/dl), 5.86 mmol/L (226.6 mg/dl), and 3.29 mmol/L (127.2 mg/dl), respectively. The lower limit of normal HDL was 0.78 mmol/L (30.2 mg/dl).
Definition of molecular subtypes
Biopsy samples and tissue sections from pathological entities, such as swollen lymph nodes and gastrointestinal tissues, were saved for analysis at the institute. The testing platform was the Hiseq 4000 NGS platform (Illumina system), and the testing method was high-throughput sequencing, also called next-generation sequencing (NGS), contrary to the reference genome of GRCh37/hg19. This analysis covered 121 genes including exonic areas, intronic areas related to fusion mutations, and alternative splicing sites (3, 4). All these 121 genes are recorded in Figure 1 and Table S1. There were 2,876 cases of gene abnormalities occurring in 259 patients, including fusion mutations, copy number variation, gene amplification, missense variation, nonsense variation, frameshift insertion/deletion, and non-frameshift insertion/deletion. Besides, 2,790 protein-alternative mutations were found based on the analysis. The LymphGen algorithm was applied in the analysis for DLBCL subtype prediction with probability. The probability was analyzed in the Naïve Bayes algorithm [4]. All information was collected from the clinical center. All these gene abnormalities are listed in Table S2.
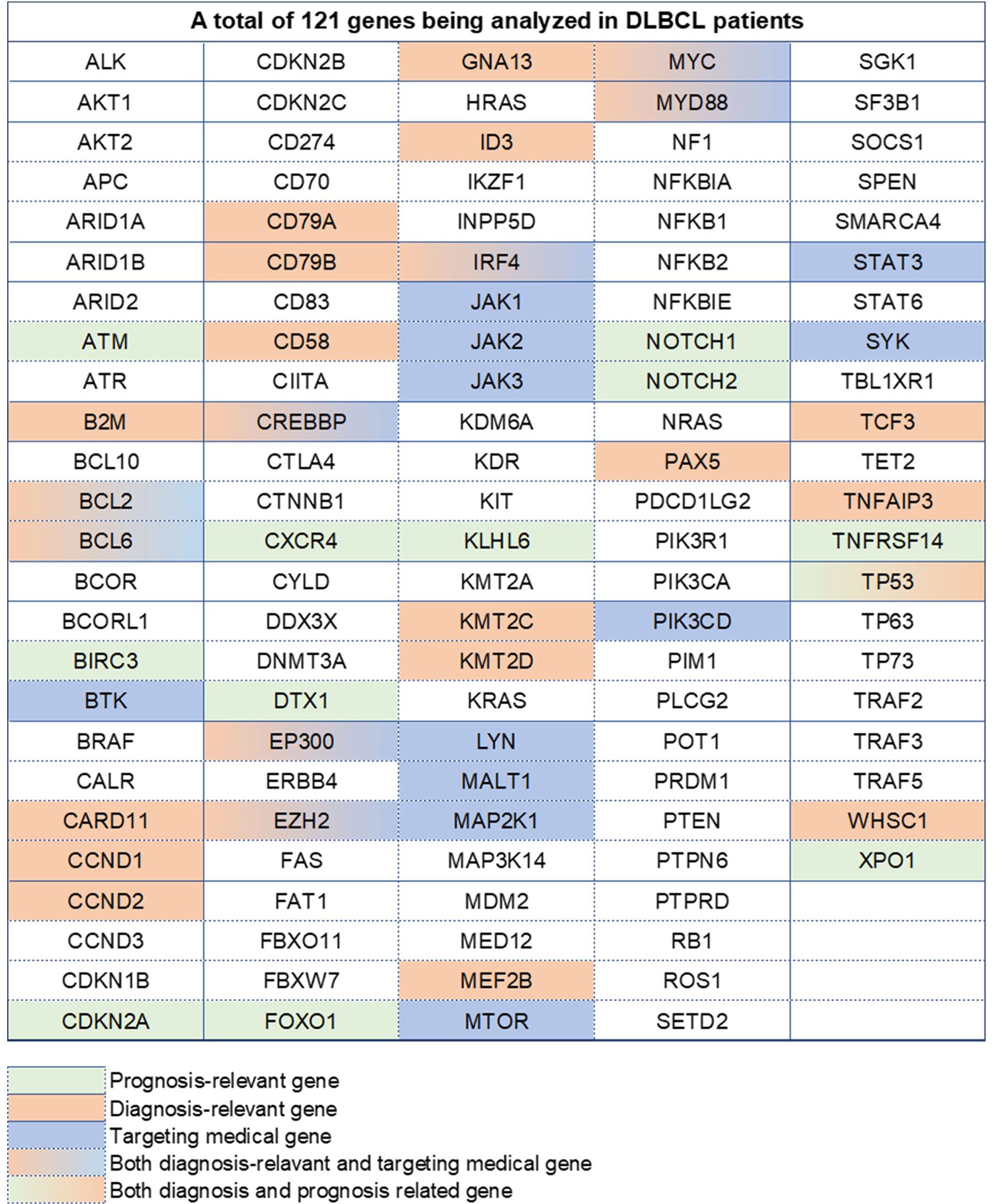
Figure 1 A total of 121 genes analyzed in DLBCL patients. Different colors represent different types, containing prognosis-relevant, diagnosis-relevant, and targeting medical-relevant genes.
Statistical analysis
Data are presented as medians, absolute ranges, or frequencies. The Kruskal–Wallis H test was used to compare the distribution of numerical variables. The χ2 test was used for qualitative variables. The relationships between clinical factors and dyslipidemia were assessed using univariable and multivariable logistic regression models. All multivariable analyses were adjusted for sex and age. The Kaplan–Meier method was used to estimate univariate survival curves, and the differences between curves were calculated via the log-rank test. Multivariable Cox proportional hazard regression models were used to assess the prognostic impact of hypertriglyceridemia with regard to overall survival (OS) and progress-free survival (PFS). Statistical analyses were performed using SPSS software (version 23.0) and GraphPad Prism (version 9.5). P-values below 0.05 were considered statistically significant.
Results
Patient characteristics
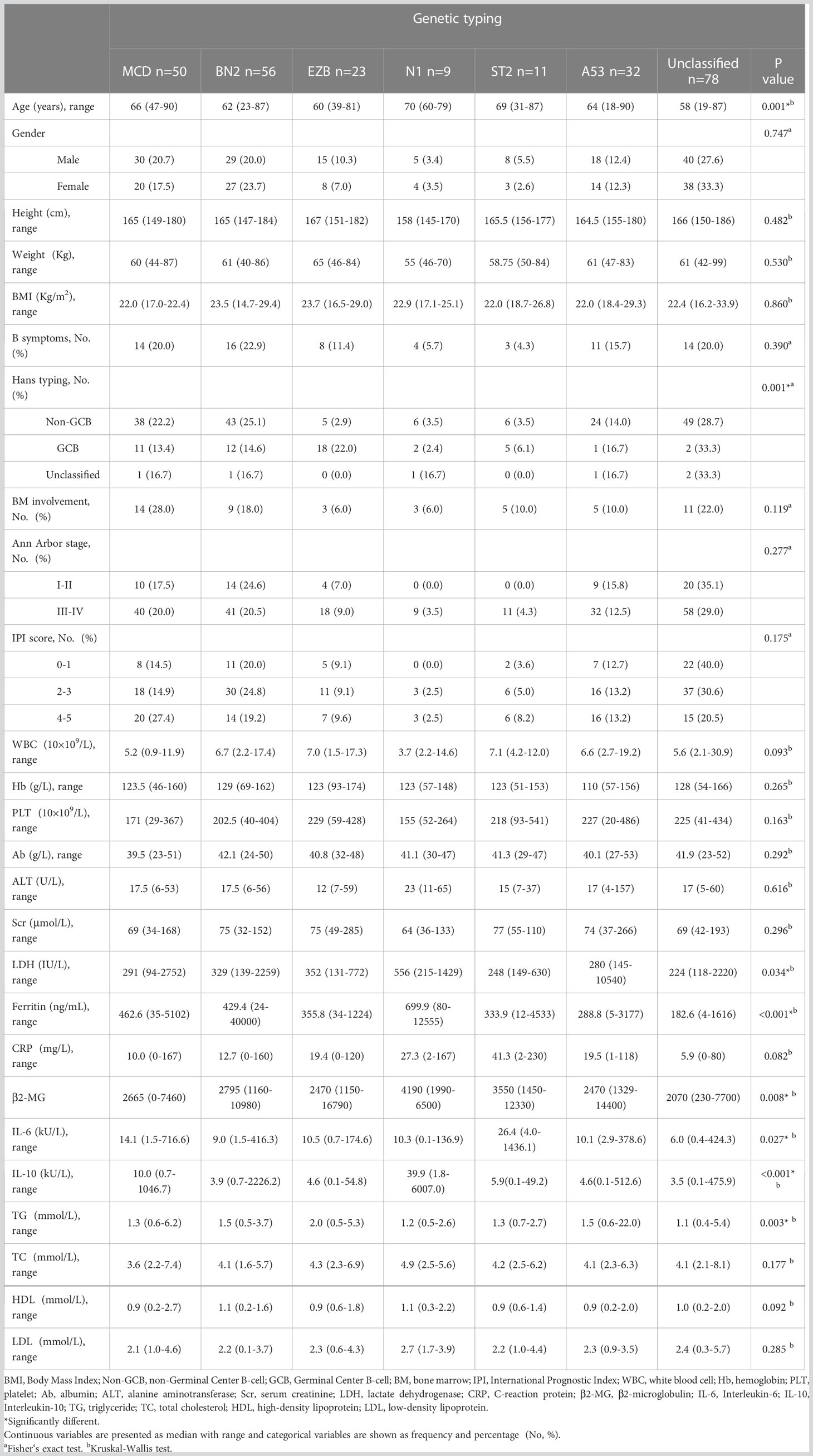
A total of 259 newly diagnosed DLBCL patients are categorized into six genetic subtypes, and the proportion of each type is 21.6% MCD subtype, 19.3% BN2 subtype, 8.9% EZB subtype, 3.5% N1 subtype, 4.2% ST2 subtype, and 12.4% A53 subtype, respectively (Figure 2). Besides, 30.1% of patients are unclassified, and this isoform is defined as a black control. To investigate the concrete genes influencing the lipids, we considered 60 familiar decisive molecular typing genes such as BCL2, BCL6, TP53, MYD88, MYC, NOTCH2, and EZH2. More details regarding genetic typing and gene translocation are shown in Figure 3.
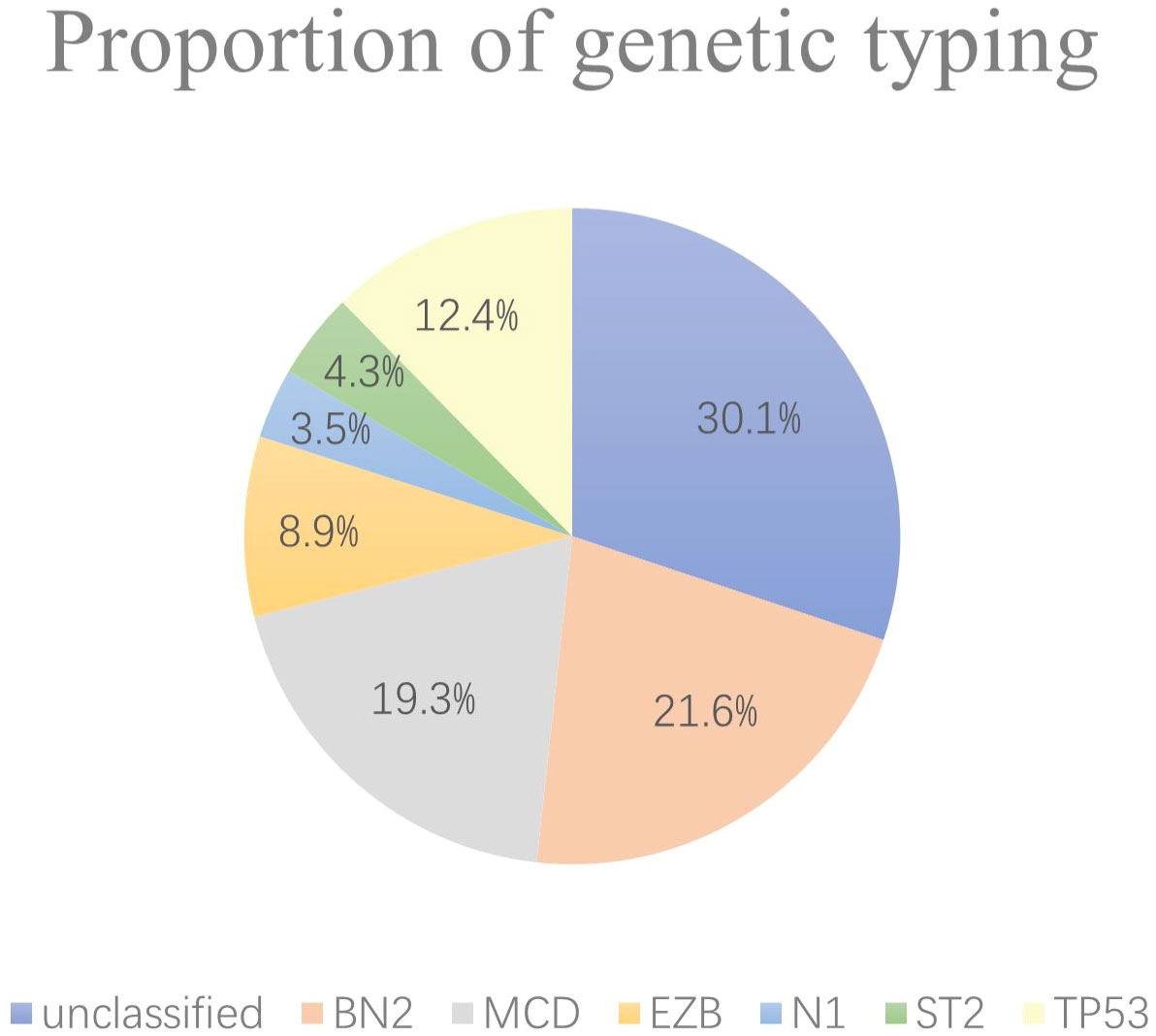
Figure 2 Proportion of genetic typing. In the six subgroups divided by genetics, the BN2 subtype, MCD subtype, and EZB subtype account for most, while the N1 and ST2 subtypes account for no more than 10% of the total.
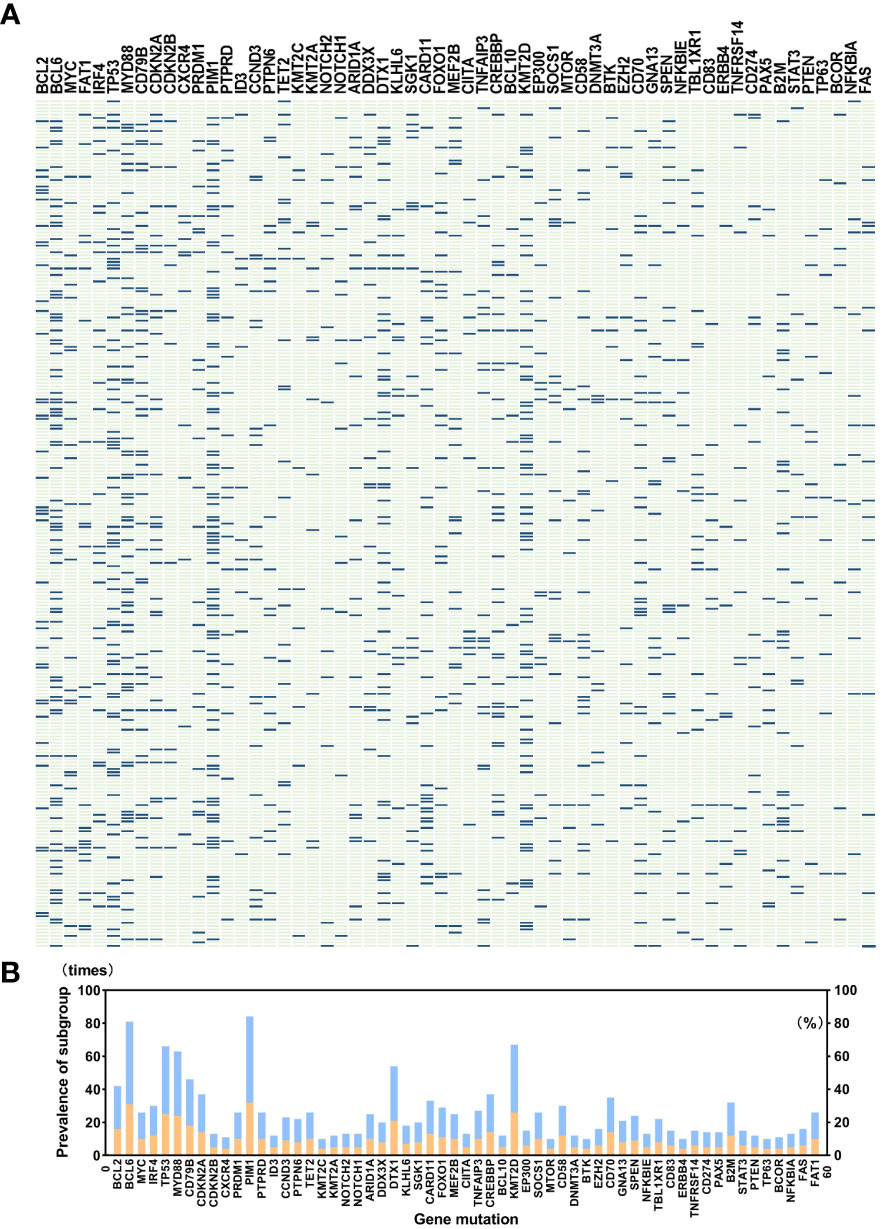
Figure 3 (A) shows gene mutations in detail in a total of 259 patients, and (B) indicates the prevalence of gene mutations. A total of 121 genes covering exon, fusion-relevant intron, and alternative splicing are analyzed, and we selected 60 types of gene mutations that occurred rather frequently or were molecular typing-related, like BCL2, BCL6, MYC, MYD88, CD79B, PIM1, KMT2D, DTX1, TET2, and SGK1. Panel (B) illustrates the prevalence of gene mutations; the blue band in the histogram represents mutation times, and the orange band shows percentage. Abnormalities of BCL6, TP53, PIM1, MYD88, and KMT2D occurred in more than 20% of patients, and PIM1 mutations appeared most common.
We recorded and analyzed the initial concentration of lipids before DLBCL patients received any treatment. Results show that TG in the EZB subtype was higher than in the unclassified subtype (Kruskal–Wallis H test, p = 0.001). The median concentration of TG in these two groups is 2.01 mmol/L (range 1.76–2.48 mmol/L) and 1.16 mmol/L (range 0.84–1.58 mmol/L), respectively. More importantly, results show that in the EZB subtype, the incidence of hypertriglyceridemia is 78.3% compared with 19.2% in the unclassified subtype, 36.0% in the MCD subtype, and 18.2% in the ST2 subtype (chi-square test and Fisher’s exact test, p <0.001, = 0.001, <0.001). In addition, dyslipidemia is discovered more commonly in the EZB subtype than in the unclassified subtype, with a significant statistical difference (87% vs 39.7%, p <0.001). The other three types of lipids (TC, HDL-C, and LDL-C), meanwhile, demonstrated no significant differences among the seven types.
According to the World Health Organization (WHO) classification of lymphoma, researchers put forward the concept of high-grade B-cell lymphoma based on three types of gene translocation: BCL2, BCL6, and MYC. Thus, we also analyzed the correlation between dyslipidemia and gene translocation. Based on pathological gene-sequencing, patients with BCL2 gene fusion mutation are significantly more likely to have dyslipidemia (chi-square test, 76.5%, p = 0.006) and hypertriglyceridemia (chi-square test, 88.2%, p = 0.002) than patients without rearrangement of these three genes.
Other demography and clinical characteristics are demonstrated in Table 1.
Other clinical factors about dyslipidemia
Binary logistic regression is used to analyze other clinical elements related to hypertriglyceridemia and dyslipidemia. In univariable analysis, molecular typing, BCL2 fusion translation, ferritin, and BMI were risk factors for hypertriglyceridemia (p <0.05). In terms of dyslipidemia, B symptoms, molecular typing, BCL2 fusion translation, ferritin, CRP, and IL-6 have influenced dyslipidemia (p <0.05).
In multivariable analysis, results indicate that being one of the EZB subtypes (p = 0.002, OR 34.524, 95% CI 3.700–322.120) is correlative to hypertriglyceridemia. Besides, the level of BMI affects triglycerides (p = 0.002, OR 1.183, 95% CI 1.064–1.315). As for dyslipidemia, the EZB subtype is significantly associated with dyslipidemia (p = 0.020, OR 14.931, 95% CI 1.532–145.477). The influence of ferritin level is familiar (p = 0.026, OR 1.001, 95% CI 1.000–1.001). The detailed results of the logistic regression model are shown in Table 2.
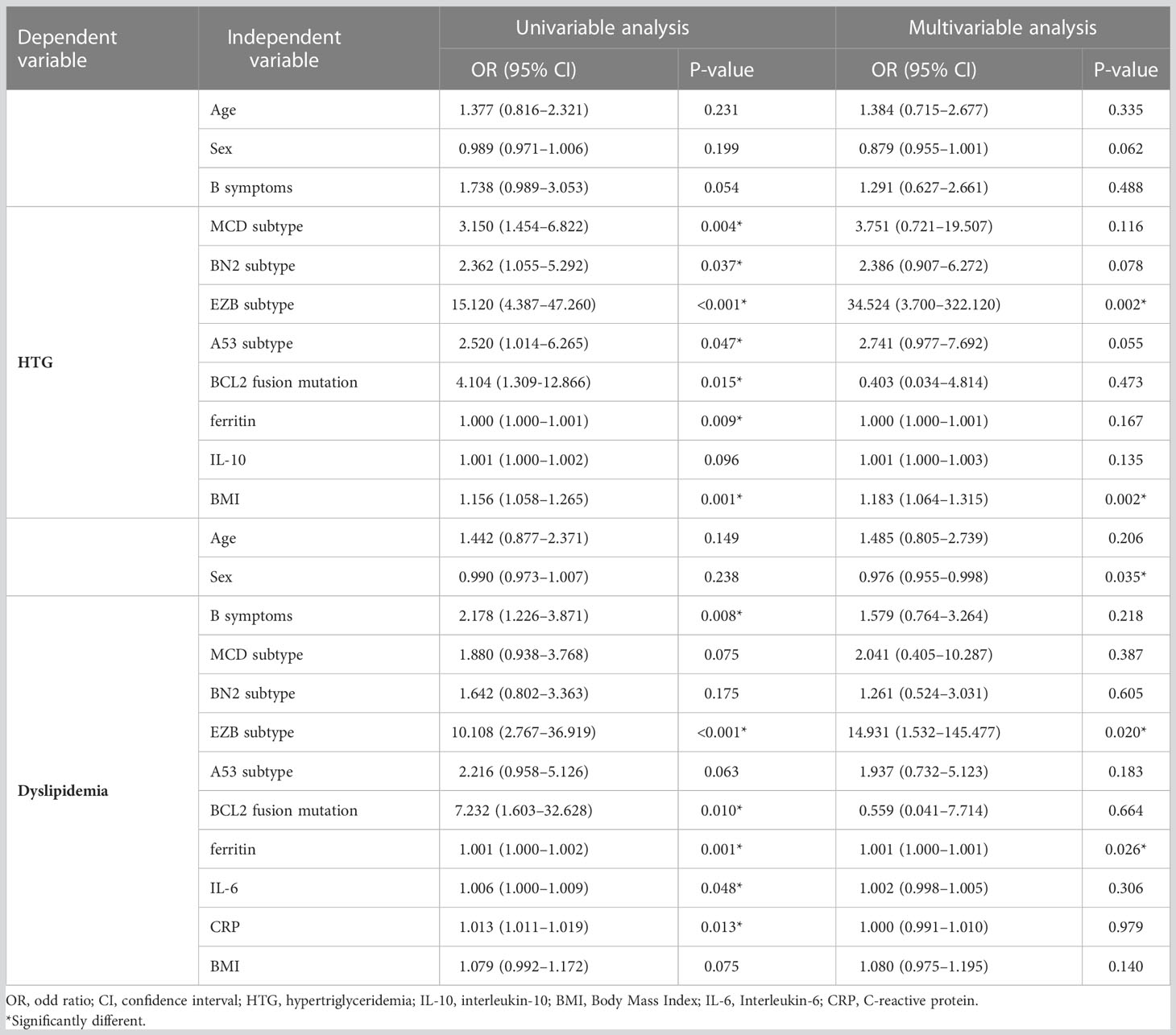
Table 2 Logistic regression models evaluating the associations between clinical variables and hypertriglyceridemia and dyslipidemia in DLBCL patients.
Relation between dyslipidemia and survival
The median follow-up time is 12.3 months (range 1.1–105.0 months) among the 235 surviving patients. A total of 21 patients died, three of whom died of complications, and 18 patients died of disease progression. Significantly, only two patients died during induction treatment. Moreover, up to the cut-off time, disease progress occurred in 57 patients, and in 35 patients, disease progress occurred at the first assessment during therapy. Three patients were lost to follow-up. The 2-year OS and 2-year PFS rates were 81.7% and 61.3%, respectively. We interpret the influence of lipids on survival. Neither hypertriglyceridemia nor dyslipidemia significantly affected OS and PFS in DLBCL patients (hypertriglyceridemia: OS hazard ratio HR 0.855, 95% CI 0.353–2.075, p = 0.7336; PFS HR 0.824, 95% CI 0.482–1.407, p = 0.4730. dyslipidemia: OS hazard ratio HR 0.637, 95% CI 0.270–1.504, p = 0.3012; PFS HR 0.692, 95% CI 0.408-1.176, p = 0.1490, Figure 4). OS and PFS in different genetic subtypes showed no significant statistical divergence (Figure 5). Interestingly, OS in the A53 subgroup showed differences. The 32 patients who are defined as the A53 subgroup were divided into the HTG group and the non-HTG group by the level of triglycerides, and they also divided into the dyslipidemia group and the non-dyslipidemia group. Although the small number of samples precludes achieving a significant difference in PFS, the OS showed significant results. The non-HTG group revealed better survival than the HTG group, while the non-dyslipidemia group showed worse survival than the dyslipidemia group (Figure 6). Relative parameters that significantly influenced survival were not discovered in either univariable or multivariable Cox regression models (Table S3).
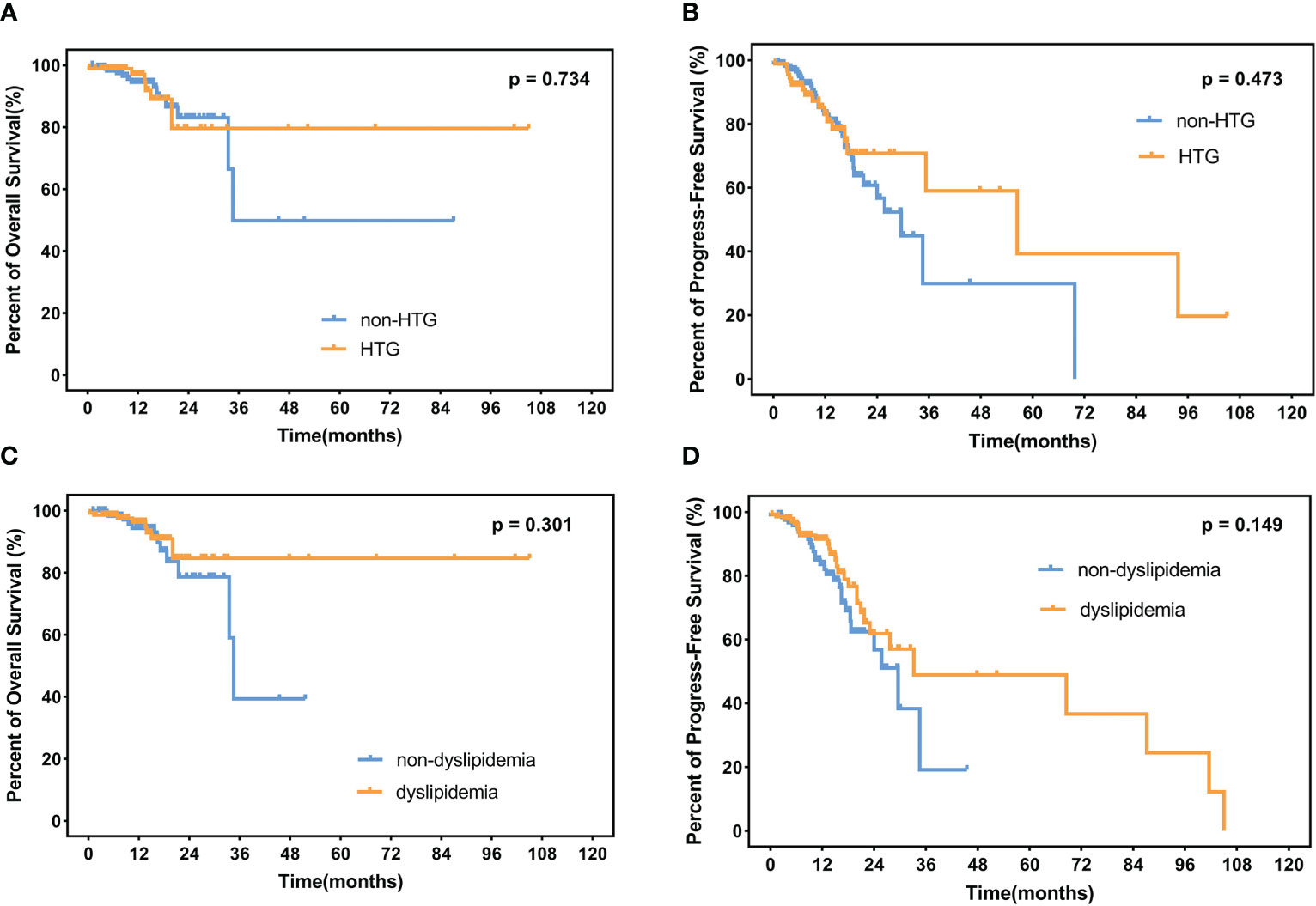
Figure 4 OS and PFS in patients. (A) Percent of OS and (B) progress-free survival in patients with or without HTG. (C) Percent of overall survival and (D) progress-free survival in dyslipidemia patients and non-dyslipidemia patients. OS, overall survival; PFS, progress-free survival; HTG, hypertriglyceridemia.
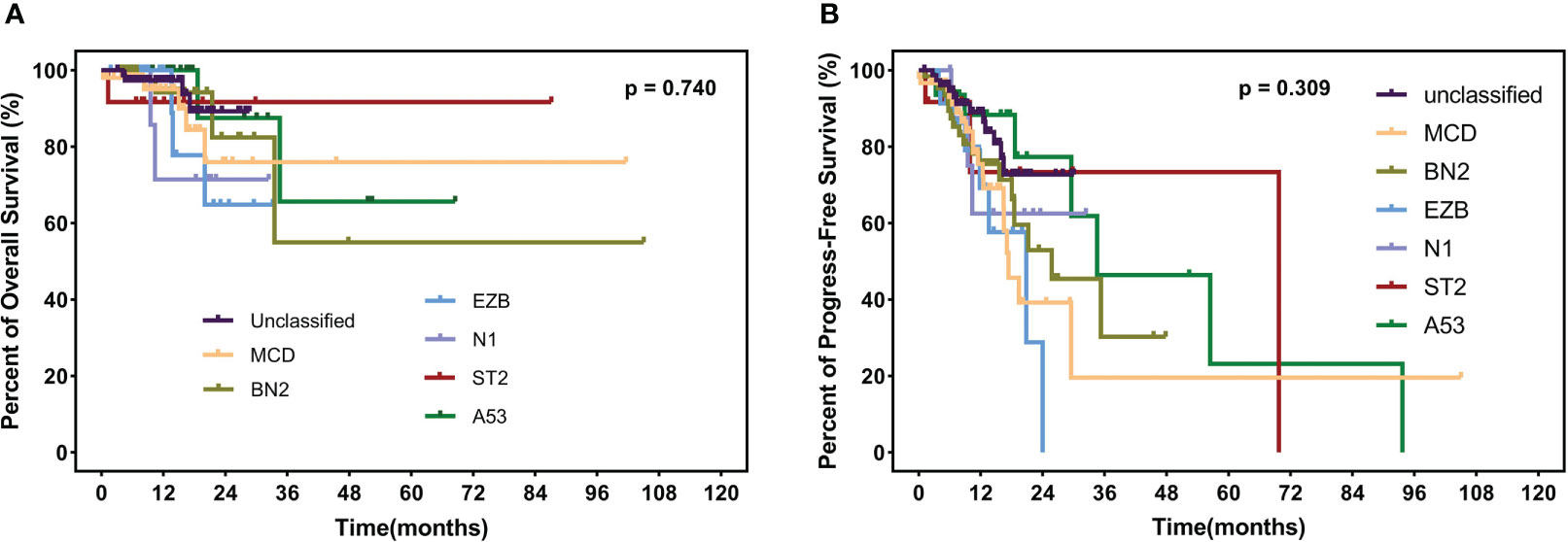
Figure 5 OS (A) and PFS (B) in different subtypes, including MCD, BN2, EZB, N1, ST2, A53, and unclassified subtypes. OS, overall survival; PFS, progress-free survival.
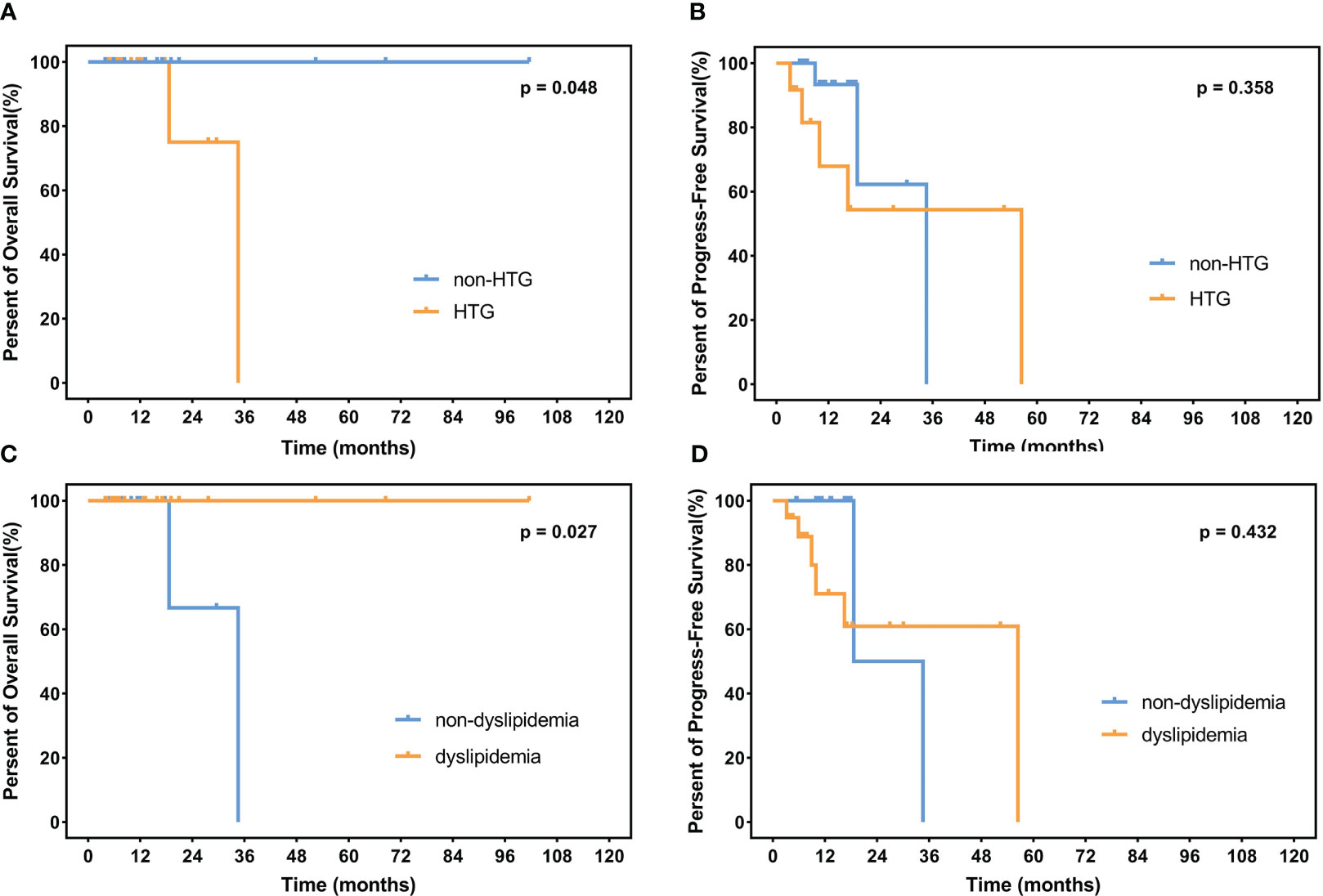
Figure 6 OS and PFS in the A53 subgroup. (A) illustrates OS in the HTG group and the non-HTG group with significant statistical differences. (B) shows PFS in these two groups without a significant statistical difference. (C, D) indicate OS and PFS in the dyslipidemia group and the non-dyslipidemia group, with a significant statistical difference only in OS. OS, overall survival; PFS, progress-free survival. HTG, hypertriglyceridemia.
Discussion
In our study, hypertriglyceridemia and dyslipidemia are associated with the molecular typing of DLBCL, and the most relevant is the EZB subtype. The other clinical factors also connect with dyslipidemia, including B symptoms, BCL2 fusion translation, ferritin, and BMI. We demonstrated the correlation of dyslipidemia with the genetic type of DLBCL at the genetic level. While the incidence of hypertriglyceridemia and dyslipidemia has not influenced OS and PFS. Research shows that prostate cancer patients with high levels of triglycerides have a higher risk of relapse. But in some malignancies, dyslipidemia is not associated with a poor prognosis, such as gastric cancer and breast cancer (18–20). We conjectured that patients who have dyslipidemia were provided therapy to lower lipids, and dyslipidemia is not an independent prognosis factor. Besides, Schmitz et al. reported different OS in MCD, BN2, EZB, and N1 subtypes (3). In our study, OS and PFS did not show statistical differences. We inferred that it might be related to the small size of the patients and not enough follow-up time.
Dyslipidemia is characterized by abnormal serum triglycerides, cholesterol, low-density lipoprotein, and high-density lipoprotein. The Frederickson model was raised to systematically categorize dyslipidemia into five phenotypes. Type 2 is characterized by elevated levels of LDL, and the rest of the types are characterized by increased levels of various triglyceride-rich lipoprotein subfractions (16). A more practical model of dyslipidemia defines it as hypertriglyceridemia, hypercholesterolemia, hypo-HDL, and hyper-LDL. The recommended levels of TG, LDL, and HDL are 2–9.9 mmol/L, 3.4–4.9 mmol/L, and 0.7–0.9 mmol/L (15). Exorbitant levels of lipids express an increased risk of pancreatitis (21). In our retrospective study, two patients had high levels of triglycerides (28.02 and 22.00 mmol/L, respectively). While there were no episodes of acute pancreatitis. Elevated plasma LDL-cholesterol levels are the 8th leading risk factor for death in 2019 worldwide (22). A large study conducted in China indicated that the most common dyslipidemia subtypes are hypo-HDL (20.4%) and HTG (13.8%) (23). The mechanism related to primary dyslipidemia has not been proved definitively. According to genome-wide association studies (GWASes), TMEM57, DOCK7, CELSR2, APOB, ABCG5, HMGCR, TRIB1, FADS2/S3, LDLR, NCAN, and TOMM40-APOE appear to be related to increased TC levels. Similarly, CELSR2-PSRC1-SORT1, PCSK9, NCAN-CILP2-PBX4, LDLR, and APOC1-APOE are associated with variations in LDL levels (24). D9N and N291S variants have been associated with elevated TG, while the S447X variant appears to be related to depressed TG. Moreover, APOA5-related abnormalities appear to be potentially correlative to TG (25, 26). Besides, pre-B-cell leukemia transcription factor 4 (PBX4/CILP2 locus) and B-cell CLL/lymphoma 3 genes are considered to be the association factors for dyslipidemia (27, 28). Secondary hyperlipidemia has been acknowledged as being associated with obesity, smoking, the metabolic syndrome, proinflammatory and prothrombotic biomarkers, and type 2 diabetes (29). Recently, the interaction between dyslipidemia-related genes and hematologic malignancy genes has been reported; for instance, the PML/RARα fusion protein and peroxisome proliferator-activated receptor-α have a synergistic effect on acute promyelocytic leukemia (30). Besides, the incidence of cardiovascular disease was higher in patients with DLBCL than in normal people (31).
Dyslipidemia has been reported to be associated with a multitude of solid tumors, such as breast cancer, prostate cancer, thyroid cancer, colorectal cancer, and lung cancer (5–9). In terms of hematologic malignancies, chronic lymphocyte leukemia and acute promyelocytic leukemia are reported to be associated with dyslipidemia and hypertriglyceridemia (11, 32). Whereas, the correlation between dyslipidemia and lymphoma or multiple myeloma has not been reported. More recently, studies aimed at lipids in DLBCL patients appeared to indicate a prognosis (33–35). Fundamental experiments investigated cholesterol and DLBCL. SYK inhibits cholesterol biosynthesis by modulating PI3K/AKT-dependent survival pathways (36). SOX9–DHCR24–cholesterol biosynthesis axis in IGH-BCL2 fusion translation positive DLBCL plays an oncogenic role via upregulating cholesterol synthesis (12, 37). Based on molecular typing, the EZB subtype is characterized by IGH-BCL2 fusion translation and the EZH2 mutation. Jiao et al. proposed the SOX9–DHCR24–cholesterol biosynthesis axis, which may interpret the association between the EZB subtype and dyslipidemia to some extent. SOX9 was researched more frequently in other solid tumors like colon cancer, hepatocellular carcinoma, and breast cancer. SOX9 plays an oncogenic role in GCB and IGH-BCL2-positive DLBCL. In other words, SOX9-positive DLBCLs are more likely to be categorized as EZB subtypes and are more likely to be associated with NF-κB signaling. SOX9 upregulates DHCR24 expression, which is important in the cholesterol biosynthesis pathway (38–40). Other studies indicate that BCL2 may influence lipid metabolism. Downregulation of BCL2-related transcription factors has been considered to affect carotid atherosclerosis and be associated with oxidized LDL trans-differentiation. Thus, we speculate that BCL2 fusion influences lipid metabolism (41).
The concept of genetic typing was raised to provide potential nosology for precision medicine strategies in DLBCL. According to the research containing 574 DLBCL biopsy samples identified genes, the BN2 subtype is dominated by NOTCH pathway aberrations and the NF-κB pathway. Besides, the MCD subtype is characterized by BCR and NF-κB pathway aberrations. EZB subtype is enriched for BCL2 translocation, EZH2 mutation, and REL amplification, involving Janus-associated kinase–signal transducers and activators of transcription (JAK-STAT) signaling and the PI3K pathway. BCR has been shown to directly promote cholesterol biosynthesis through intermediate kinases downstream of BCR to maintain cell membrane integrity and BCR signaling (3, 42–46). Aimed at this, targeting cellular cholesterol provides new precise treatment strategies. Thaxton et al. investigated functional lipoprotein nanoparticles in DLBCL via targeting both cellular cholesterol uptake and BCR-associated de novo cholesterol synthesis and achieved cellular cholesterol reduction and induced apoptosis in otherwise resistant ABC DLBCL cell lines (47).
We analyzed 259 DLBCL patients in the lipid aspect based on genetic typing and deduced the association between dyslipidemia and the EZB subtype without prognosis discrepancy. This study has some deficiencies. Firstly, it is a retrospective study with limitations in patient selection and analysis. Secondly, follow-up times are not long enough because there have been 121 types of gene sequencing analyses conducted in the clinical institution in recent years. Thirdly, the role of dynamic alterations in blood biomarkers during and after treatment was not taken into consideration during the analysis. Further prospective and multicenter research is needed to investigate dyslipidemia in DLBCL to instruct prognosis evaluation and treatment.
Conclusion
A total of 259 newly diagnosed DLBCL patients were categorized by genetic typing and analyzed for dyslipidemia. Significant differences were found in the EZB subtype; dyslipidemia and hypertriglyceridemia occurred in this isoform without influence on survival. In summary, the EZB subtype is exposed to higher risks of dyslipidemia and hypertriglyceridemia, which may guide clinical treatment strategies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Research Ethics Committee of the first affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent was not required to participate in this study in accordance with the national legislation and the institutional requirements
Author contributions
WX contributed to the conception and design of the article. YX, HS, YS, and YZ wrote the manuscript. XZ, JS, XL, DZ, CY, JHW, XH, JYW, JH, HM, WY, HT, and JJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by grants from the National Program on Key Research Project of China (2022YFC2502700).
Acknowledgments
We thank all people involved in this study at nine medical centers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1172623/full#supplementary-material
References
1. Rodriguez-Abreu D, Bordoni A, Zucca E. Epidemiology of hematological malignancies. Ann Oncol (2007) 18 Suppl 1:i3–8. doi: 10.1093/annonc/mdl443
2. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large b cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med (2018) 24(5):679–90. doi: 10.1038/s41591-018-0016-8
3. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse Large b-cell lymphoma. N Engl J Med (2018) 378(15):1396–407. doi: 10.1056/NEJMoa1801445
4. Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse Large b cell lymphoma with therapeutic implications. Cancer Cell (2020) 37(4):551–68.e14. doi: 10.1016/j.ccell.2020.03.015
5. Yao X, Tian Z. Dyslipidemia and colorectal cancer risk: a meta-analysis of prospective studies. Cancer Causes Control (2015) 26(2):257–68. doi: 10.1007/s10552-014-0507-y
6. Hao B, Yu M, Sang C, Bi B, Chen J. Dyslipidemia and non-small cell lung cancer risk in Chinese population: a case-control study. Lipids Health Dis (2018) 17(1):278. doi: 10.1186/s12944-018-0925-z
7. Song Y, Lee HS, Park G, Kang SW, Lee JW. Dyslipidemia risk in thyroid cancer patients: a nationwide population-based cohort study. Front Endocrinol (Lausanne) (2022) 13:893461. doi: 10.3389/fendo.2022.893461
8. Rice KR, Koch MO, Cheng L, Masterson TA. Dyslipidemia, statins and prostate cancer. Expert Rev Anticancer Ther (2012) 12(7):981–90. doi: 10.1586/era.12.75
9. Schairer C, Freedman DM, Gadalla SM, Pfeiffer RM. Lipid-lowering drugs, dyslipidemia, and breast cancer risk in a Medicare population. Breast Cancer Res Treat (2018) 169(3):607–14. doi: 10.1007/s10549-018-4680-7
10. Li K, Wang F, Yang ZN, Cui B, Li PP, Li ZY, et al. PML-RARalpha interaction with TRIB3 impedes PPARgamma/RXR function and triggers dyslipidemia in acute promyelocytic leukemia. Theranostics (2020) 10(22):10326–40. doi: 10.7150/thno.45924
11. Mozessohn L, Earle C, Spaner D, Cheng SY, Kumar M, Buckstein R. The association of dyslipidemia with chronic lymphocytic leukemia: a population-based study. J Natl Cancer Inst (2017) 109(3). doi: 10.1093/jnci/djw226
12. Shen Y, Zhou J, Nie K, Cheng S, Chen Z, Wang W, et al. Oncogenic role of the SOX9-DHCR24-cholesterol biosynthesis axis in IGH-BCL2+ diffuse large b-cell lymphomas. Blood (2022) 139(1):73–86. doi: 10.1182/blood.2021012327
13. Zhu JR, Gao RL, Zhao SP, Lu GP, Zhao D, Li JJ. Chinese guidelines for the management of dyslipidemia in adults. J Geriatr Cardiol (2018) 15(1):1–29. doi: 10.11909/j.issn.1671-5411.2018.01.011
14. Fredrickson DS, Levy RI, Lees RS. Fat transport in lipoproteins–an integrated approach to mechanisms and disorders. N Engl J Med (1967) 276(4):215–25. doi: 10.1056/NEJM196701262760406
15. Garg A, Garg V, Hegele RA, Lewis GF. Practical definitions of severe versus familial hypercholesterolaemia and hypertriglyceridaemia for adult clinical practice. Lancet Diabetes Endocrinol (2019) 7(11):880–6. doi: 10.1016/S2213-8587(19)30156-1
16. Berberich AJ, Hegele RA. A modern approach to dyslipidemia. Endocr Rev (2022) 43(4):611–53. doi: 10.1210/endrev/bnab037
17. Zelenetz AD, Gordon LI, Chang JE, Christian B, Abramson JS, Advani RH, et al. NCCN Guidelines(R) insights: b-cell lymphomas, version 5.2021. J Natl Compr Canc Netw (2021) 19(11):1218–30. doi: 10.6004/jnccn.2021.0054
18. Allott EH, Howard LE, Cooperberg MR, Kane CJ, Aronson WJ, Terris MK, et al. Serum lipid profile and risk of prostate cancer recurrence: results from the SEARCH database. Cancer Epidemiol Biomarkers Prev (2014) 23(11):2349–56. doi: 10.1158/1055-9965.EPI-14-0458
19. Pih GY, Gong EJ, Choi JY, Kim MJ, Ahn JY, Choe J, et al. Associations of serum lipid level with gastric cancer risk, pathology, and prognosis. Cancer Res Treat (2021) 53(2):445–56. doi: 10.4143/crt.2020.599
20. Goto W, Kashiwagi S, Kamei Y, Watanabe C, Aomatsu N, Ikeda K, et al. Relationship between serum lipid levels and the immune microenvironment in breast cancer patients: a retrospective study. BMC Cancer (2022) 22(1):167. doi: 10.1186/s12885-022-09234-8
21. Berglund L, Brunzell JD, Goldberg AC, Goldberg IJ, Sacks F, Murad MH, et al. Evaluation and treatment of hypertriglyceridemia: an endocrine society clinical practice guideline. J Clin Endocrinol Metab (2012) 97(9):2969–89. doi: 10.1210/jc.2011-3213
22. Pirillo A, Casula M, Olmastroni E, Norata GD, Catapano AL. Global epidemiology of dyslipidaemias. Nat Rev Cardiol (2021) 18(10):689–700. doi: 10.1038/s41569-021-00541-4
23. Zhang M, Deng Q, Wang L, Huang Z, Zhou M, Li Y, et al. Prevalence of dyslipidemia and achievement of low-density lipoprotein cholesterol targets in Chinese adults: a nationally representative survey of 163,641 adults. Int J Cardiol (2018) 260:196–203. doi: 10.1016/j.ijcard.2017.12.069
24. Jeemon P, Pettigrew K, Sainsbury C, Prabhakaran D, Padmanabhan S. Implications of discoveries from genome-wide association studies in current cardiovascular practice. World J Cardiol (2011) 3(7):230–47. doi: 10.4330/wjc.v3.i7.230
25. Hegele RA, Pollex RL. Hypertriglyceridemia: phenomics and genomics. Mol Cell Biochem (2009) 326(1-2):35–43. doi: 10.1007/s11010-008-0005-1
26. Khalil YA, Rabès JP, Boileau C, Varret M. APOE gene variants in primary dyslipidemia. Atherosclerosis (2021) 328:11–22. doi: 10.1016/j.atherosclerosis.2021.05.007
27. Vrablik M, Hubacek JA, Dlouha D, Lanska V, Rynekrova J, Zlatohlavek L, et al. Impact of variants within seven candidate genes on statin treatment efficacy. Physiol Res (2012) 61(6):609–17. doi: 10.33549/physiolres.932341
28. Miao L, Yin RX, Pan SL, Yang S, Yang DZ, Lin WX. BCL3-PVRL2-TOMM40 SNPs, gene-gene and gene-environment interactions on dyslipidemia. Sci Rep (2018) 8(1):6189. doi: 10.1038/s41598-018-24432-w
29. Yuan G, Al-Shali KZ, Hegele RA. Hypertriglyceridemia: its etiology, effects and treatment. CMAJ (2007) 176(8):1113–20. doi: 10.1503/cmaj.060963
30. Wu S, Li S, Jin P, Zhang Y, Chen L, Jin W, et al. Interplay between hypertriglyceridemia and acute promyelocytic leukemia mediated by the cooperation of peroxisome proliferator-activated receptor-alpha with the PML/RAR alpha fusion protein on super-enhancers. Haematologica (2022) 107(11):2589–600. doi: 10.3324/haematol.2021.280147
31. Ekberg S, Harrysson S, Jernberg T, Szummer K, Andersson PO, Jerkeman M, et al. Myocardial infarction in diffuse large b-cell lymphoma patients - a population-based matched cohort study. J Intern Med (2021) 290(5):1048–60. doi: 10.1111/joim.13303
32. Sun J, Lou Y, Zhu J, Shen H, Zhou D, Zhu L, et al. Hypertriglyceridemia in newly diagnosed acute promyelocytic leukemia. Front Oncol (2020) 10:577796. doi: 10.3389/fonc.2020.577796
33. Xu J, Wei Z, Zhang Y, Chen C, Li J, Liu P. A novel scoring system based on the level of HDL-c for predicting the prognosis of t-DLBCL patients: a single retrospective study. BioMed Res Int (2018) 2018:2891093. doi: 10.1155/2018/2891093
34. Gao R, Liang JH, Wang L, Zhu HY, Wu W, Cao L, et al. Low serum cholesterol levels predict inferior prognosis and improve NCCN-IPI scoring in diffuse large b cell lymphoma. Int J Cancer (2018) 143(8):1884–95. doi: 10.1002/ijc.31590
35. Zhang Y, Chen Q, Lu C, Yu L. Prognostic role of controlling nutritional status score in hematological malignancies. Hematology (2022) 27(1):653–8. doi: 10.1080/16078454.2022.2078040
36. Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D, et al. SYK inhibition modulates distinct PI3K/AKT- dependent survival pathways and cholesterol biosynthesis in diffuse large b cell lymphomas. Cancer Cell (2013) 23(6):826–38. doi: 10.1016/j.ccr.2013.05.002
37. Russo V. Cholesterol: a putative oncogenic driver for DLBCL. Blood (2022) 139(1):5–6. doi: 10.1182/blood.2021014300
38. Xia P, Chen J, Liu Y, Cui X, Wang C, Zong S, et al. MicroRNA-22-3p ameliorates alzheimer's disease by targeting SOX9 through the NF-kappaB signaling pathway in the hippocampus. J Neuroinflamm (2022) 19(1):180. doi: 10.1186/s12974-022-02548-1
39. Heckman CA, Mehew JW, Boxer LM. NF-kappaB activates bcl-2 expression in t(14;18) lymphoma cells. Oncogene (2002) 21(24):3898–908. doi: 10.1038/sj.onc.1205483
40. Buhrmann C, Brockmueller A, Mueller AL, Shayan P, Shakibaei M. Curcumin attenuates environment-derived osteoarthritis by Sox9/NF-kB signaling axis. Int J Mol Sci (2021) 22(14):7645. doi: 10.3390/ijms22147645
41. Rykaczewska U, Zhao Q, Saliba-Gustafsson P, Lengquist M, Kronqvist M, Bergman O, et al. Plaque evaluation by ultrasound and transcriptomics reveals BCLAF1 as a regulator of smooth muscle cell lipid transdifferentiation in atherosclerosis. Arterioscler Thromb Vasc Biol (2022) 42(5):659–76. doi: 10.1161/ATVBAHA.121.317018
42. Kuliszkiewicz-Janus M, Malecki R, Mohamed AS. Lipid changes occuring in the course of hematological cancers. Cell Mol Biol Lett (2008) 13(3):465–74. doi: 10.2478/s11658-008-0014-9
43. Lionakis MS, Dunleavy K, Roschewski M, Widemann BC, Butman JA, Schmitz R, et al. Inhibition of b cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell (2017) 31(6):833–43.e5. doi: 10.1016/j.ccell.2017.04.012
44. Phelan JD, Young RM, Webster DE, Roulland S, Wright GW, Kasbekar M, et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature (2018) 560(7718):387–91. doi: 10.1038/s41586-018-0290-0
45. Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active b-cell-receptor signalling in diffuse large b-cell lymphoma. Nature (2010) 463(7277):88–92. doi: 10.1038/nature08638
46. Wilson WH, Wright GW, Huang DW, Hodkinson B, Balasubramanian S, Fan Y, et al. Effect of ibrutinib with r-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell (2021) 39(12):1643–53.e3. doi: 10.1016/j.ccell.2021.10.006
47. Rink JS, Yang S, Cen O, Taxter T, McMahon KM, Misener S, et al. Rational targeting of cellular cholesterol in diffuse Large b-cell lymphoma (DLBCL) enabled by functional lipoprotein nanoparticles: a therapeutic strategy dependent on cell of origin. Mol Pharm (2017) 14(11):4042–51. doi: 10.1021/acs.molpharmaceut.7b00710
Keywords: diffuse large B-cell lymphoma, molecular typing, dyslipidemia, hypertriglyceridemia, BN2 subtype, MCD subtype, EZB subtype, A53 subtype
Citation: Xu Y, Shen H, Shi Y, Zhao Y, Zhen X, Sun J, Li X, Zhou D, Yang C, Wang J, Huang X, Wei J, Huang J, Meng H, Yu W, Tong H, Jin J and Xie W (2023) Dyslipidemia in diffuse large B-cell lymphoma based on the genetic subtypes: a single-center study of 259 Chinese patients. Front. Oncol. 13:1172623. doi: 10.3389/fonc.2023.1172623
Received: 23 February 2023; Accepted: 23 May 2023;
Published: 09 June 2023.
Edited by:
Gaël Roué, Josep Carreras Leukaemia Research Institute (IJC), SpainReviewed by:
Jue Wang, Huazhong University of Science and Technology, ChinaJoaquim Carreras, Tokai University, Japan
Copyright © 2023 Xu, Shen, Shi, Zhao, Zhen, Sun, Li, Zhou, Yang, Wang, Huang, Wei, Huang, Meng, Yu, Tong, Jin and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanzhuo Xie, eGlld2Fuemh1b0B6anUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yi Xu†
Yi Xu† Huafei Shen
Huafei Shen Jianai Sun
Jianai Sun Xianbo Huang
Xianbo Huang Jian Huang
Jian Huang Jie Jin
Jie Jin Wanzhuo Xie
Wanzhuo Xie