- Department of Gynecology, the Affiliated Hospital of Qingdao University, Qingdao, China
Regulated Cell Death (RCD) is a mode of cell death that occurs through drug or genetic intervention. The regulation of RCDs is one of the significant reasons for the long survival time of tumor cells and poor prognosis of patients. Long non-coding RNAs (lncRNAs) which are involved in the regulation of tumor biological processes, including RCDs occurring on tumor cells, are closely related to tumor progression. In this review, we describe the mechanisms of eight different RCDs which contain apoptosis, necroptosis, pyroptosis, NETosis, entosis, ferroptosis, autosis and cuproptosis. Meanwhile, their respective roles in the tumor are aggregated. In addition, we outline the literature that is related to the regulatory relationships between lncRNAs and RCDs in tumor cells, which is expected to provide new ideas for tumor diagnosis and treatment.
1 Introduction
Cell death is a common physiological process that can be found in nearly all organisms. According to the Nomenclature Committee on Cell Death in 2018, cell death can be divided into Accidental Cell Death (ACD) and Regulated Cell Death (RCD) (1). ACD is an inevitable cell death due to physical breakdown of the plasma membrane caused by drastic external stimuli, such as chemical, physical, or mechanical stimuli, and this process cannot be biologically controlled (2). Differently, cell death regulated by drug or genetic intervention is named RCD that relies on precise signaling cascades and intracellular molecular mechanisms (3). RCDs can be divided into apoptosis, necroptosis, lysosome-dependent cell death (LDCD), pyroptosis, NETotic cell death (NETosis), immunogenic cell death (ICD), entotic cell death (entosis), PARP-1-dependent cell death (parthanatos), ferroptosis, autophagy-dependent cell death (ADCD/autosis), oxeiptosis, pH-dependent cell death (alkaliptosis), cuproptosis etc. (1). Usually, one signal transduction pathway can regulate a variety of RCDs, and a certain RCD can be regulated by multiple signal pathways. Therefore, blocking some important molecules on the signal transduction pathways can inhibit the occurrence of cell death (4).
Cancer is a major global public health problem. Due to cancer incidence and mortality increasing rapidly all over the world, cancer becomes one of the leading causes of death worldwide (5). Cancer has a high mortality rate and poor prognosis, which is closely related to the unlimited proliferation ability of tumor cells (6). The ability of tumor cells to proliferate indefinitely depends on the inhibition of cell death in the cells, such as imbalance of pro-apoptotic versus anti-apoptotic systems, and loss of p53 gene function (7, 8).
Long non-coding RNAs (lncRNAs) are RNAs of more than 200 nucleotides in length that lack open reading frames and do not encode proteins. In recent years, the role of lncRNAs in regulating biological functions and cellular activities has gradually been discovered. The mechanisms of lncRNAs have been reported which include interaction with DNA, RNA, and proteins (9–11). LncRNAs are involved in the development and progression of many diseases, especially the progression of tumors. LncRNAs play complex and precise regulatory roles in biological processes such as tumor cell proliferation, migration, invasion and drug resistance, etc. (12, 13). More than one study has reported that multiple RCDs occurring within tumor cells can be regulated mediated by lncRNAs (14, 15).
Recently, many scholars have conducted in-depth studies on the generation mechanisms of lncRNAs in regulating different RCDs in tumor cells and have made remarkable progress. These have certain guiding significance for the diagnosis and treatment of tumors. In this paper, we focus on the regulatory relationships between apoptosis, necroptosis, pyroptosis, NETosis, entosis, ferroptosis, autosis, cuproptosis and lncRNAs in tumor cells in recent years, hoping to provide new ideas for tumor diagnosis and treatment.
2 Apoptosis
2.1 Mechanisms of apoptosis
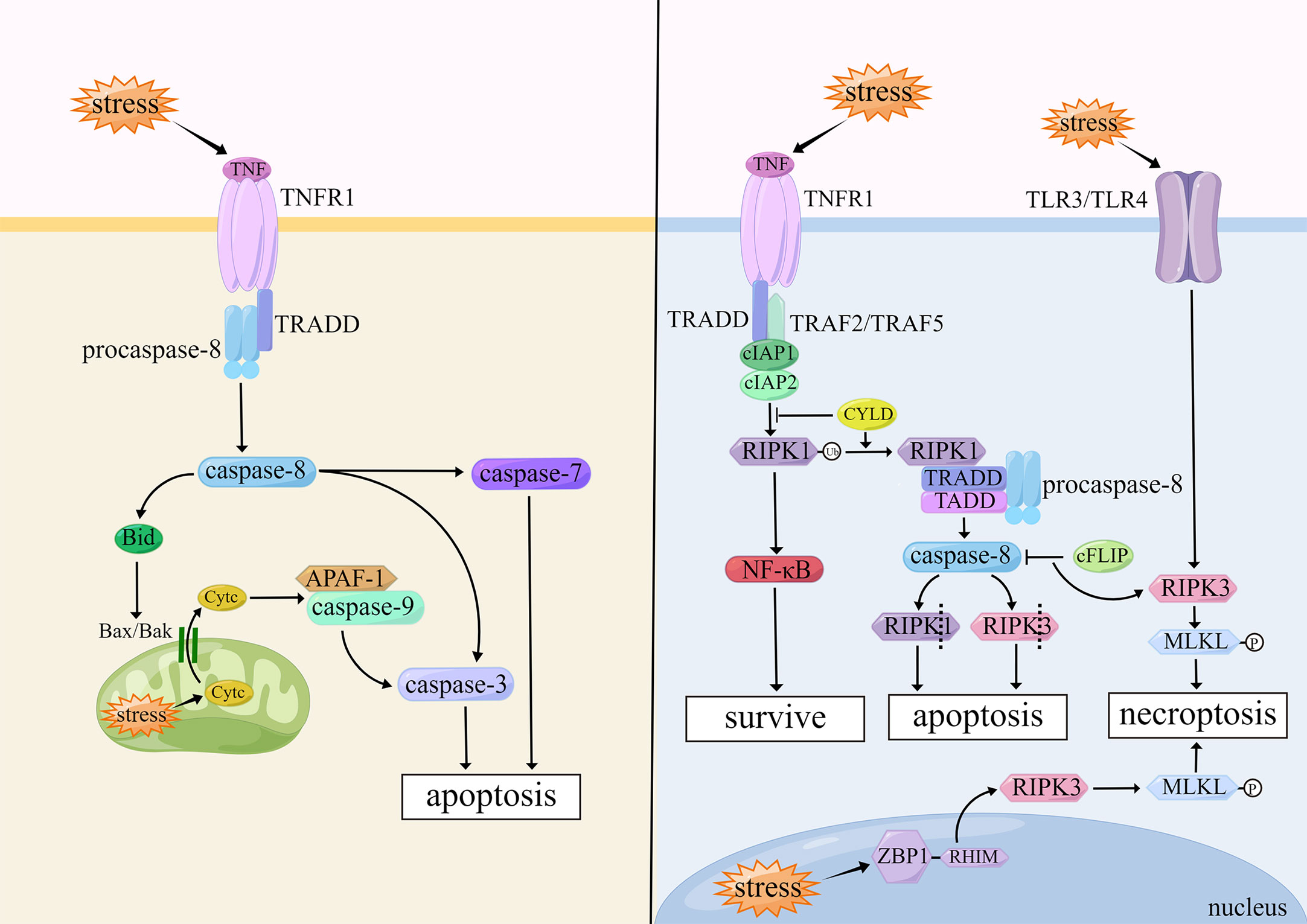
Apoptosis plays a significant role in tissue differentiation, organ development and aging, and clearance of injured or mutant cells. When cells undergo apoptosis, their morphology is mainly characterized by cell contraction, chromatin condensation, apoptotic body formation, and DNA fragmentation (1). The core mechanism of apoptosis is the activation of caspases, which are a large class of cysteine proteases that act in a cascade manner, of which caspase-3, -7, -8, and -9 play important roles in apoptosis (16). The two common activation pathways of apoptosis are the intrinsic pathway (also called mitochondrial pathway) and extrinsic pathway (also called death receptor pathway).
The initiation of the intrinsic pathway is caused by internal stimulus such as oxidative stress and genetic damage in cells. The intrinsic pathway is closely regulated by B cell lymphoma-2 (Bcl-2) family proteins, which are divided into two major classes: pro-apoptotic proteins (e.g., Bax, Bak, and Bid) and anti-apoptotic proteins (e.g., Bcl-2, Bcl-xL, and Bcl-w) (17). When the balance of Bcl-2 family proteins is broken, Bax and Bak form an opening in the outer mitochondrial membrane and induce changes in mitochondrial outer membrane permeability (MOMP) (18). Cytochrome c (Cytc) enters the cytoplasm through the mitochondrial intermembrane space and assists the pro-apoptotic factor, apoptotic protease activating factor 1 (APAF-1), in functioning to form apoptotic bodies (19). At the same time, when MOMP events occur in cells, caspase-9 binds to APAF-1 and is activated. And then caspase-9 further activates downstream caspase-3, ultimately triggering apoptosis (20).
The extrinsic pathway is activated by the combined action of different tumor necrosis factor (TNF) family members, including Fas receptor, tumor necrosis factor receptor (TNFR), TNF-related apoptosis-inducing ligand receptor (TRAILR), which are initiated by binding with their corresponding ligands. After binding to each other, these factors become type II membrane proteins with homotrimeric structures. They cleave the cell membrane to form soluble forms which can increase cell membrane permeability (21). The death-inducing signaling complex (DISC) is formed intracellularly, which includes the Fas-associated death domain (FADD), TNFR1-associated death domain (TRADD), and pro-caspase-8, after binding to the TNF family ligands. DISC can activate caspase-8 intracellularly and promote its maturation, which in turn activates downstream caspase-3 and caspase-7 and triggers apoptosis (22). In hepatocytes and fibroblasts, caspase-8 activates the pro-apoptotic protein Bid, and then induces Cytc release from mitochondria which leads to the initiation of the intrinsic pathway (23).
There is also a less common apoptosis within cells: the endoplasmic reticulum (ER) stress-induced apoptosis. When events occur in cells, such as hypoxia, oxidative damage, Ca2+ imbalance and viral infection, misfolded or unfolded proteins in the ER will increase, allowing the stress signal of the ER to be transmitted from the ER membrane to the nucleus, and promoting apoptosis (24). There are three apoptotic pathways induced by ER stress: IRE1/ASK1/JNK pathway, caspase-12 kinase pathway, and CHOP/GADD153 pathway. All three pathways play important roles in ER stress-induced apoptosis (25). (Figure 1)
2.2 Apoptosis in tumor cells
In tumor cells, the imbalance between the regulation of apoptosis and anti-apoptosis system often results in its antagonistic effects, promoting and suppressing cancer, and thus affecting cells survival. Anti-apoptosis, as an acquired feature of tumor cells, gives tumor cells survival advantages and promotes tumor evolution, growth and drug resistance (26). It has been reported that under apoptotic stress, cells release the growth factor FGF2, which rapidly up-regulates Bcl-2 and triggers the Bcl-2 dependent MEK-ERK pathway, thereby protecting neighboring cells from apoptosis (27). Similarly, apoptotic cells attract macrophages and polarize them into a regeneration-activated state to become tumor-associated macrophages (TAMs) in tumors. TAMs have the potential to promote tumor evolution and progression through multiple pathways. TAMs release anti-inflammatory mediators such as TGF-β1, IL-10 and PGE2, which promote tumor growth, activate angiogenesis, and promote tumor invasion and metastasis through proliferative signaling, regeneration and repair responses, and anti-tumor immune silencing of TAMs (28). For example, in aggressive non-Hodgkin lymphoma, apoptotic cells highly express genes encoding growth factors and angiogenesis, which can provide nutrients to the tumor microenvironment and promote TAMs accumulation, tumor growth and angiogenesis (29). However, how the balance between apoptosis and anti-apoptosis in tumor cells is disrupted, and how more elaborate mechanisms are regulated has yet to be elucidated, which require us to conduct further studies.
2.3 Apoptosis and lncRNAs in tumors
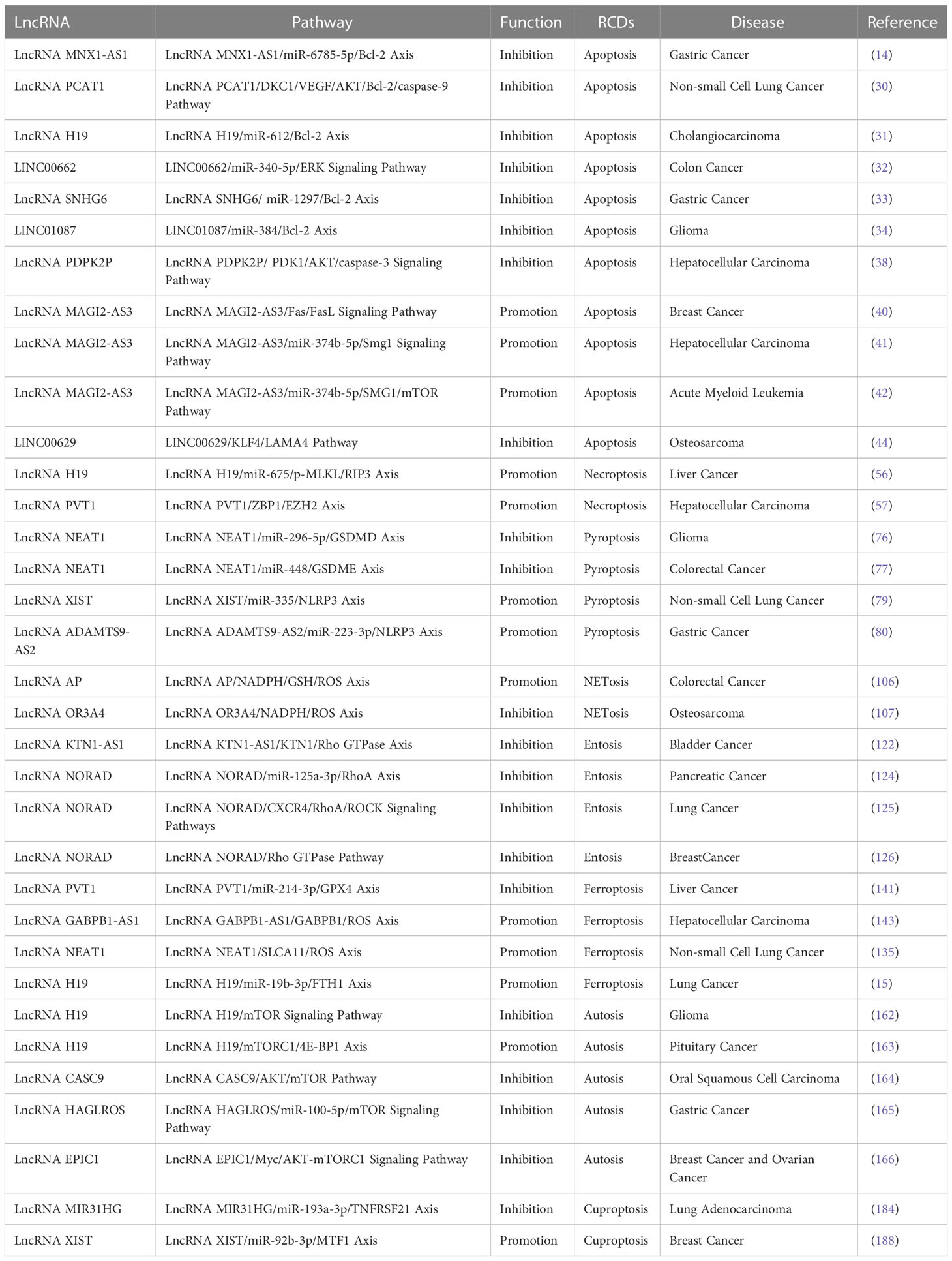
LncRNAs can induce or inhibit apoptosis of tumor cells by regulating key molecules of apoptosis. One study has suggested that lncRNA MNX1-AS1 up-regulates the expression of Bcl-2 by sponging miR-6785-5p in gastric cancer cells, which can promote proliferation, migration and invasion, and then inhibits apoptosis (14). LncRNA PCAT1 interacts with dyskerin pseudouridine synthase-1 (DKC1) to activate the vascular endothelial growth factor (VEGF)/protein kinase B (AKT)/Bcl-2/caspase-9 pathway and promote the transcription and translation of Bcl-2, contributing to enhancing abilities in proliferation and invasion of non-small cell lung cancer cells and inhibiting apoptosis of tumor cells (30). Subsequent experiments revealed that lncRNA H19, LINC00662, lncRNA SNHG6, and LINC01087 can act on different miRNAs in cholangiocarcinoma, colon cancer, gastric cancer and glioma, respectively. However, these all lncRNAs cause increased expression level of Bcl-2, inhibition of apoptosis, and tumor progression ultimately (31–34). Among them, lncRNA H19 plays a role in sepsis, myocardial injury, and vascular smooth muscle injury in addition to acting in cholangiocarcinoma (35–37). LncRNAs can induce or inhibit apoptosis by regulating apoptosis-related molecules in the caspase family. Experiments performed by Pan et al. indicated that lncRNA PDPK2P can interact with 1,3-phosphoinositide‐dependent protein kinase-1 (PDK1) and regulate the development of hepatocellular carcinoma through PDK1/AKT/caspase-3 signaling pathway (38). PDK1 plays a crucial part in regulating the phosphorylation of AKT, which is involved in apoptosis thereby changing the states of cells (39). LncRNA PDPK2P promotes AKT phosphorylation and inhibits the expression of caspase-3 by interacting with PDK1, and then inhibits the apoptotic pathway of hepatocellular carcinoma cells and promotes proliferation of tumor cells (38).
LncRNAs are also involved in the regulation of the extrinsic pathway of apoptosis. It has been reported that lncRNA MAGI2-AS3 can inhibit breast cancer cell growth by increasing the expression of Fas and Fas ligand and promoting apoptosis (40). LncRNA MAGI2-AS3 inhibits tumor cell proliferation in hepatocellular carcinoma cells as well (41). LncRNA MAGI2-AS3 binds to miR-374b-5p and inhibits its expression, thereby playing a positive regulation part on genitalia family member-1 (SMG1), and increasing the expression levels of SMG1, which in turn plays a tumor suppressor role. SMG1 is a potential tumor suppressor that can antagonize mammalian target of rapamycin (mTOR) to regulate leukemia cell growth in acute myeloid leukemia (42). However, conclusions from studies regarding the effect of SMG1 on apoptosis are not entirely consistent. In response to Smac mimetic compounds (SMC), an experimental small molecule, SMG1 inhibited SMC-mediated TNF-α-induced apoptosis to protect cells. Downregulation of SMG1 expression promotes SMC-mediated TNF-α-induced caspase-8 activation (43).
Wang et al. revealed that when osteosarcoma cells undergo ER stress, LINC00629 expression is increased by activating the KLF4/LAMA4 pathway, and ER stress-induced apoptosis is inhibited to promote tumorigenesis and metastasis (44). Taken together, lncRNAs play a significant role in the regulation of various pathways of apoptosis. (Table 1)
3 Necroptosis
3.1 Mechanisms of necroptosis
Necroptosis belongs to a nonapoptotic cell death independent of the caspase pathway. Necroptosis differs from necrosis. Necroptosis can be induced by stimulation with superfamily members of TNFR, pattern recognition receptor (PRR), T cell receptor, and chemotherapeutic agents; but necrosis is considered an “accidental” death that is not regulated by molecular events (45). Necroptosis is morphologically similar to necrosis, which is characterized by loss of integrity of the plasma membrane, swelling of the cytoplasm and organelles, chromosome condensation, and release of cellular contents (46).
Necroptosis is associated with activation of receptor-interacting protein-1 (RIP1), receptor-interacting protein-3 (RIP3), and mixed lineage kinase domain-like protein (MLKL), of which RIP3 and MLKL are considered biomarkers of necroptosis (47). Upon binding of TNF and TNFR1 on cell membranes, the complex I is formed, including receptor-interacting protein kinase 1 (RIPK1), TRADD, cellular inhibitor of apoptosis-1/2 (cIAP1/2), TNFR-related factor 2/5 (TRAF2/5), which in turn stimulates different signaling pathways and induces cell death (48). When RIPK1 is polyubiquitinated by cIAP1/2, the nuclear factor-κB (NF-κB) pathway is activated and cells survival predominate. However, when RIPK1 is deubiquitinated by cylindromatosis (CYLD), the NF-κB pathway is suppressed, forming the complex II consisting of RIPK1, TRADD, caspase-8, and FADD. Caspase-8 inactivates RIPK1 and RIPK3 by proteolytic cleavage, which leads to apoptosis. When caspase-8 is inhibited by cell FLICE-like inhibitory protein (cFLIP), RIPK3 phosphorylates its substrate, MLKL, to form p-MLKL. P-MLKL oligomerizes and translocates to the plasma membrane, ultimately leading to necroptosis (49). Toll-like receptor 3/4 (TLR3/TLR4) on the cell membrane can also initiate RIPK3-mediated necroptosis after lipopolysaccharide (LPS) stimulation (50).
Furthermore, there is an inside-out death pathway in cells. Z-DNA-binding protein-1 (ZBP1/DAI/DLM-1) in the nucleus is a nucleic acid sensor containing a RIP homotypic interaction motif (RHIM). Upon viral stimulation, RHIM can recruit and activate RIPK3, thereby initiating RIPK3- and MLKL-dependent necroptosis, which contributes to nuclear envelope breakdown, DNA leakage into the cytoplasm, and cell death eventually (51). (Figure 1)
3.2 Necroptosis in tumor cells
In tumors, foci of cell death (i.e., tumor necrosis) are often present due to conditions such as hypoxia and nutritional deficiencies (46). Necroptosis plays a very complex part in tumor cells. For one thing, down-regulating expression of necrosis factors allow tumor cells to resist necroptosis. Necroptosis-induced inflammatory reactions can also promote tumorigenesis and metastasis. For another, necroptosis can trigger strong immune responses in tumor cells, induce cell death, and then protect against tumor progression (45). MLKL, RIPK1, and RIPK3 have all been revealed to be down-regulated in breast cancer, rendering tumors unable to recruit immune cells. Consequently, immune surveillance is evaded and cancer-promoting effects are exerted (52). Accumulation of large amounts of RIPK3 in intestinal epithelial cells brings about severe necroptosis and promoting colon carcinogenesis (53). When tumor cells are glucose deficient, ZBP1 and its downstream pathways will be activated. RIPK3 expression increases, and necroptosis of tumor cells is induced, but cell metastasis is promoted as well (54). Additionally, in ovarian cancer, RIP1 has a dual role, both promoting cancer cell proliferation and reducing tumor resistance of drug thereby promoting the anticancer effects of chemotherapeutic agents such as cisplatin (55). However, in tumor cells, it still is an open question how the carcinogenic and anticancer effects of these necrosis factors are coordinated, and the mechanism behind it needs further study.
3.3 Necroptosis and lncRNAs in tumors
LncRNAs can regulate necroptosis through different pathways in tumor cells and improve or reduce tumor proliferation, invasion and migration. LncRNA H19 is the upstream of miR-675, and the expression levels of the two are positively correlated. MiR-675 can downregulate FADD and inhibit apoptosis; however, miR-675 has the ability to induce significant upregulation in p-MLKL and RIP3, which triggers necroptosis (56). A study collectively suggested that upregulation of lncRNA PVT1 is associated with elevated expression of necroptosis-related proteins, such as ZBP1, RIPK3, and p-MLKL (57). Enhancer of Zeste Homolog-2 (EZH2) negatively correlates with lncRNA PVT1 in hepatocellular carcinoma cells, and lncRNA PVT1 can increase methylation of the ZBP1 promoter and promote necroptosis by combining with DNA methyltransferase-1 through EZH2 (57, 58). However, in studies on breast cancer, promoter hypermethylation is found to lead to downregulation of ZBP1 expression, which promoted growth and migration of tumor cells (59).
Necroptosis-associated lncRNAs perform increasingly vital regulatory roles in many different tumors. In a key study by Luo et al., LINC00460, LINC02773, CHROMR, LINC01094, FLNB-AS1, ITFG1-AS1, LASTR, and LINC01638 are analyzed in TCGA database, which are highly expressed in gastric adenocarcinoma cells, while REPIN1-AS1, UBL7-AS1, PINK1-AS, and PVT1 are lowly expressed. These lncRNAs together regulate necroptosis in gastric adenocarcinoma cells (60). LncRNAs such as AP003392.3, AL928654.1, AL133371.2, AC007991.4, AC011445.1, and LINC00996 have been associated with necroptosis in ovarian cancer studies (61).
4 Pyroptosis
4.1 Mechanisms of pyroptosis
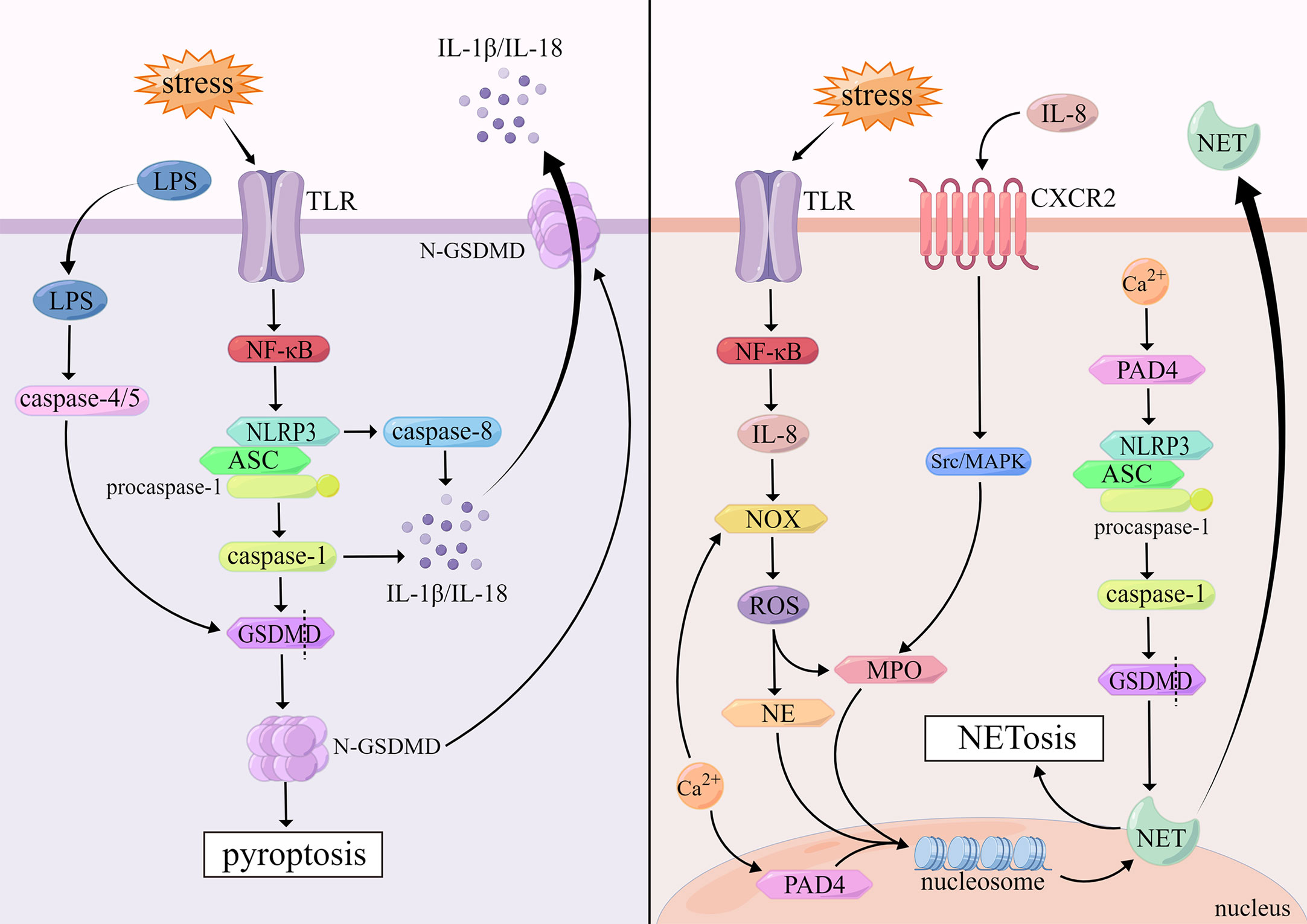
Pyroptosis is an inflammatory cell death mediated by the gasdermin superfamily that is triggered by the activation of certain inflammasomes and presents a form of DNA damage distinct from apoptosis (62). When pyroptosis occurs, the cytomorphology shows the following changes: the cell membrane forms transmembrane pores under inflammatory stimulation, and the cell permeability increases, resulting in cell swelling and osmotic lysis, intact nuclei but fragmented chromatin, and the release of intracellular pro-inflammatory components (63).
The gasdermin superfamily consists of gasdermin A/B/C/D/E (GSDMA/B/C/D/E) and DFNB59, of which GSDMD is of great importance to induce pyroptosis (64). Most inflammasomes contain three parts: nucleotide-binding domain, leucine-rich-repeat-containing receptors (NLRs), pro-caspase-1 and apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC) which consists of pyrin domain (PYD) and caspase activation and recruitment domain (CARD). The NLRP3 inflammasome, containing PYD, as an important cytosolic PRR, is activated upon TLR recognition of stimuli such as infection. NLRP3 recruits ASC via PYD-PYD domain interaction, and then activates pro-caspase-1 via CARD-CARD domain interaction (65). Activated caspase-1 can stimulate the activation of pro-IL-1β, pro-IL-18 and GSDMD cleavage N-terminal domain of GSDMD (N-GSDMD) appears oligomerization, inducing the formation of holes on the cell membrane, promoting cell membrane rupture, while releasing inflammatory factors such as IL-1β and IL-18, which leads to the occurrence of pyroptosis ultimately (66). It has been documented that caspase-8 can be activated by inflammasomes in caspase-1-deficient cells (67). Pyroptosis mediated by caspase-8 is independent of GSDMD, which belongs to a delayed, alternative form of cell death involving inflammasomes, accompanied by the release of massive mature IL-1 which has a protective function in the host (67).
There is also a pyroptosis pathway induced by caspase-4/5 in human cells. Caspase-4/5 can be activated by direct binding to the CARD at the N-terminus of LPS, which in turn cuts GSDMD into N-GSDMD. And then, N-GSDMD is oligomerized and transferred to the cell membrane to induce the assembly of NLRP3 Inflammasomes, and pyroptosis occurs finally (68). (Figure 2)
4.2 Pyroptosis in tumor cells
Pyroptosis is a double-edged sword in tumors. On the one hand, it can inhibit the occurrence and development of tumors; on the other hand, it can form a microenvironment suitable for the growth of tumors (69). For example, as an critical link in the development of pyroptosis, high expression of GSDMD in some tumors can improve the survival time of patients, while high expression of GSDMD often leads to poor prognosis in adrenocortical carcinoma, chromophobe renal cell carcinoma and other tumors (70). GSDMD expression is decreased in gastric cancer tissues, and S/G2 cell transition is accelerated by activating signal transducer and activator of transcription-3 (STAT3) and phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway to regulate cell cycle-related proteins and promote tumor proliferation in vitro and in vivo (71). Besides GSDMD, GSDME also plays different roles in tumor development. GSDME can enhance the phagocytosis of TAMs and the number and function of tumor lymphocytes, activating caspase-independent pyroptosis in target cells to inhibit tumors and enhance anti-tumor immunity (72). However, some studies run counter to the conclusions above. GSDME-mediated pyroptosis can promote the release of high-mobility group box-1 (HMGB1) to induce the proliferation of tumor cells and the development of colorectal cancer (73). Although the research on pyroptosis is getting deeper and deeper, the outcome brought about by pyroptosis on tumors still needs us to continue to explore.
4.3 Pyroptosis and lncRNAs in tumors
Recent advances have begun to clarify that molecules in the mechanism of pyroptosis (e.g., GSDM) are regulated by lncRNAs. Knockdown of lncRNA PVT1 significantly decreased the production and release of inflammatory factors, while the expression of GSDMD and caspase-1 is inhibited, which gives rise to inhibition of pyroptosis (74). LncRNA MALAT1 inhibits miR-141-3p, which in turn promotes the expression of GSDMD and induces pyroptosis (75). One study has demonstrated that lncRNA KCNQ1OT1, LINC01278, lncRNA MIRLET7BHG and lncRNA NEAT1 can act as upstream targets of miR-296-5p and regulate the occurrence and development of glioma (76). Among them, lncRNA NEAT1 can inhibit pyroptosis by regulating GSDMD (76). However, findings by Su et al. showed that down-regulated lncRNA NEAT1 suppresses pyroptosis through sponging miR-448 to regulate GSDME expression levels in ionizing radiation-induced colorectal cancer cells (77).
LncRNAs also regulate pyroptosis through other molecules in the pyroptosis mechanisms. For instance, lncRNA KCNQ1OT1 reduces pyroptosis by targeting miR-214-3p and caspase-1 (78). In non-small cell lung cancer, lncRNA XIST increases the expression of mitochondrial superoxide dismutase-2 through sponging miR-335; while knockdown of lncRNA XIST promotes reactive oxygen species (ROS) production and NLRP3 inflammasomes activation, resulting in triggering pyroptosis (79). Overexpressed lncRNA ADAMTS9-AS2 can downregulate miR-223-3p and activate NLRP3 inflammasomes in gastric cancer, which promotes the development of pyroptosis (80).
Wang et al. analyzed the database that lncRNA ZFPM2-AS1, lncRNA KDM4A-AS1, lncRNA LUCAT1, lncRNA NRAV, AL031985.3, AL049840.5, lncRNA MKLN1-AS, AC099850.3 and LINC01224 promote pyroptosis in hepatocellular carcinoma, while lncRNA CRYZL2P-SEC16B and AC008549.1 play a protective role and inhibit pyroptosis (81). AC006001.2, LINC02585, AL136162.1, AC005041.3, AL023583.1, and LINC02881 are associated with the development of ovarian cancer and involved in regulating pyroptosis in ovarian cancer (82). LncRNAs involved in the regulation of pyroptosis have been reported in different tumors such as breast cancer, prostate cancer, glioblastoma, head and neck squamous cell carcinoma, soft tissue sarcoma and so on, which are not repeated here (83–87).
5 NETosis
5.1 Mechanisms of NETosis
NETosis is a form of cell death that depends on neutrophil extracellular traps (NET) produced after neutrophil activation. NET is composed of DNA-histone complexes and cytotoxic proteins. NETs can not only capture invading pathogens, but also degrade them with NETs associated proteolytic enzymes (88, 89). Morphological changes in NETosis are characterized by chromatin decondensation accompanied by separation of the inner and outer layers of the nuclear membrane, fusion of the nucleoplasm and cytoplasm, cell size reduction, cell membrane rupture, NETs release, and ultimately cell death (90).
The mechanisms of neutrophil extracellular trapping net withering are divided into two types according to whether nicotinamide adenine dinucleotide phosphate (NADPH) is dependent or not. Cells attacked by viruses, bacteria and other pathogens activate TLR on the cell membrane, which initiates NF-κB signaling pathway, promotes the release of IL-8, and then induces NADPH-oxidase (NOX) activation and ROS production (91). ROS facilitates the release of antimicrobial proteins such as myeloperoxidase (MPO) and neutrophil elastase (NE) from neutrophils (92). These proteases induce chromatin decondensation in the nucleus, which is enhanced by MPO (93). Furthermore, IL-8 also activates downstream Src/MAPK signaling pathway through combining with CXC chemokine receptor-2 (CXCR2), which will promote the release of MPO (94). These proteins enter the nucleus, disrupt chromatin structure. The decondensed chromatin binds to proteins released by neutrophils to form NETs, and finally the cell membrane is destroyed, the cell ruptures, and intracellular NETs are released into the plasma (92). However, it remains unresolved how NETs are released from the nucleus into the cytoplasm through the nuclear envelope. One theory is that GSDMD mediates the nuclear envelope permeabilization, resulting in the release of NETs into the cytoplasm (95). Another perspective suggests that DNA is released from the nucleus into the cytoplasm via vesicles (96). In addition, neutrophil stimulation triggers the release of calcium stored in the ER. Upon intracellular calcium overload, NOX is also activated to stimulate the production of ROS, triggering NETosis that is not associated with infection (97).
NADPH-independent NETosis is mainly driven by peptidyl arginine deiminase-4 (PAD4) via citrullinated histones. Activation of PAD4 requires high concentrations of calcium (98). PAD4 is mainly localized in the nucleus of resting neutrophils, where it mediates citrullination of nucleosomal histone H3, resulting in reducing histone positive charge and the affinity between histones and negatively charged DNA, which brings about dissociation of histones from DNA and loss of chromatin structure, and induces NETs formation (96). In addition, PAD4 is involved in NLRP3-mediated ASC oligomerization and speckle formation in neutrophils, and then activates caspase-1 and its downstream molecule, GSDMD, touching off NETosis (99, 100). (Figure 2)
5.2 NETosis in tumor cells
NETs have been reported to exert carcinogenic and anticancer effects. Important components in NETs, such as MPO and protease, can inhibit tumor growth and metastasis and promote tumor cell death; however, some proteases in NETs can also degrade the extracellular matrix and promote tumor cell extravasation and metastasis (88). NETs also have the ability to awaken dormant tumor cells (101). There was a study showing that CXCR1 and CXCR2 agonists acting as major mediators of NETosis, interact with chemokines, CXC motif chemokine ligand 1/6/7/8 (CXCL1/6/7/8), induce NETs formation in tumors, and inhibit immune-mediated cytotoxicity (102). Studies using a nude mice model have suggested that CXCL1/2 knockdown significantly reduces tumor metastasis (103). NETs can directly alter the metabolic program of tumor cells to promote tumor growth as well, which is mainly due to NE released from NETs activating TLR4, increasing mitochondrial biogenesis related gene expression, increasing mitochondrial density, increasing ATP production, and accelerating tumor growth (104). A study has shown that the NETs content in the blood of patients with early stage of head and neck cancer is significantly higher than that of healthy people, while in the advanced stage of cancer, the NETs level is reduced (105). Therefore, NETs are of essential as cancer therapeutic targets to delve into the regulation of NETosis and the balance between NETs formation and destruction in tumor cells.
5.3 NETosis and lncRNAs in tumors
In recent years, there have not been many reports on the regulation of NETosis by lncRNAs, but studies have shown that lncRNAs are involved in regulating related molecules of NETosis. Pep-AP encoded by lncRNA AP can inhibit pentose phosphate pathway, reduce NADPH/NADP+ and glutathione (GSH) levels, promote ROS accumulation, induce redox imbalance, and thus inhibit colorectal cancer cell growth (106). In osteosarcoma, knockdown of lncRNA OR3A4 suppresses NADPH production and increases intracellular ROS, leading to ER stress and cell death (107). He et al. predicted five lncRNAs (AC079336.5, LINC00623, AC087752.4, AL645933.2, and LINC00426) to be associated with NETosis by database analysis, which affects cancer prognosis in head and neck squamous cell carcinoma (108).
6 Entosis
6.1 Mechanisms of entosis
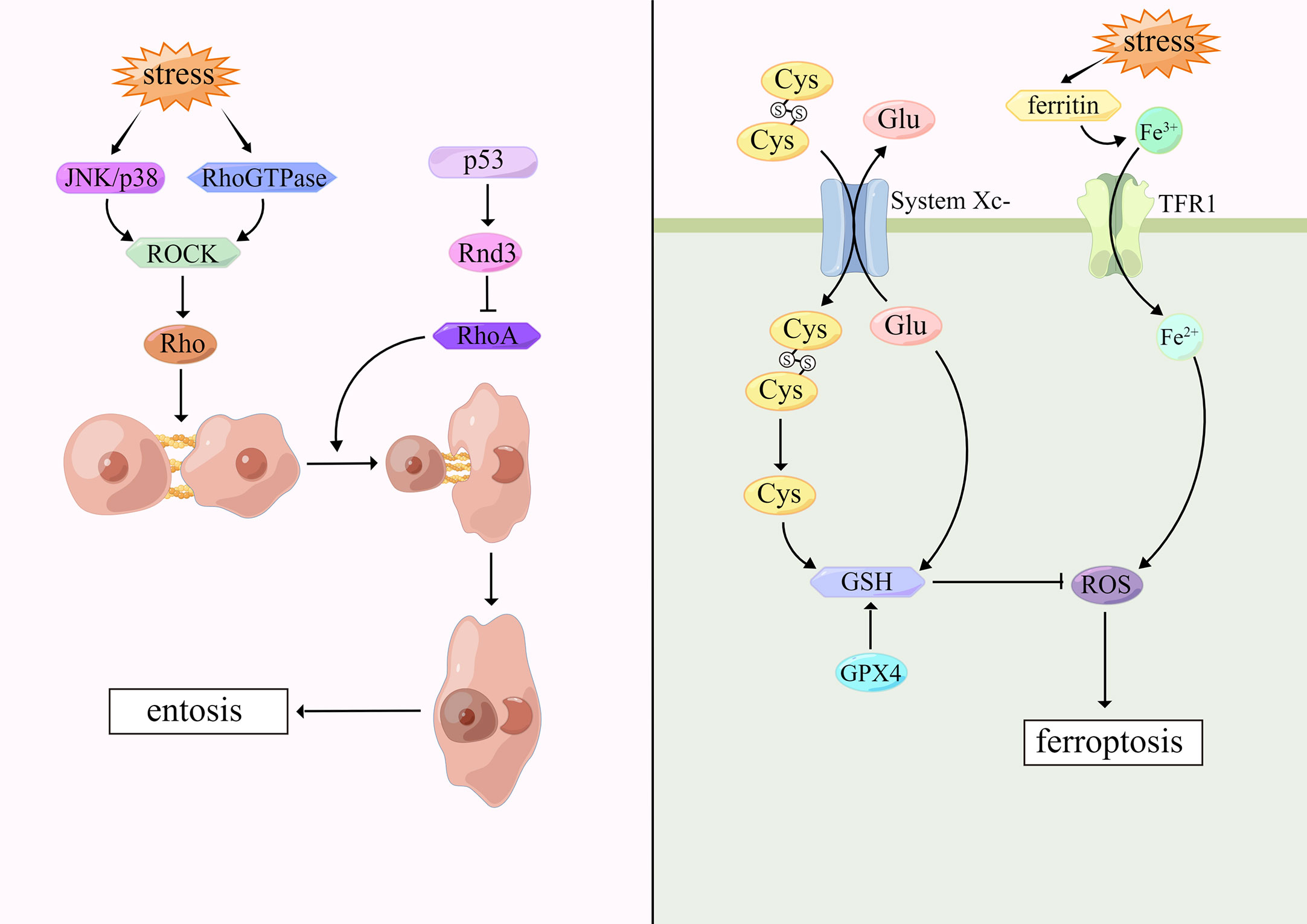
Entosis was first proposed in 2007 by Overholtzer and colleagues (109). This cell death pattern occurs in tumor cells and epithelial cells when cells are embedded in vacuoles of host cells and degraded by lysosomal enzymes of host cells. Under the microscope, internalized cells are surrounded by host cells which contain large vesicles, forming cell-in-cell (CIC) structures, but the cell membrane integrity of both cells is not disrupted. Host cells are stretched and form narrow cytoplasmic areas with crescent-shaped nuclei (110). Garanina et al. described five stages of entosis through electron microscopy. In the first stage, the internalized cells are round and the nuclei remain round. In the second stage, the internalized cells contract and produce short protrusions extending from the cell body to the entotic vacuole membrane. During the third stage, internalized cells further decreases with irregular nucleus shape and accumulation of cytoplasmic vacuoles. At the fourth stage, the shape of the internalized cells and nuclei is further deformed. The accumulation of cytoplasmic vacuoles increases and the nucleoli disappear. In the fifth stage, the entotic cells contain only residuals (111).
Cell adhesion and cytoskeletal rearrangement play a central part in entosis (4). Recent evidence suggests that cell-cell junctions are involved by both E-cadherin and α-catenin and regulated by Rho guanosine triphosphatases (Rho GTPases) (112). Aneuploid mitotic arrest occurs when cells are exposed to external stimuli that cause DNA damage (113). Cells detach from the extracellular matrix. Rho-GTP activates the downstream effector molecule Rho-associated coiled-coil containing protein kinase (ROCK), and then induces the activation of Rho protein, which promote actin-myosin interaction and increase contractility. Due to the action of cadherin, cells are tightly connected to their adjacent cells, so that cells are internalized by adjacent cells, and encapsulated by lysosomes of host cells. Finally, internalized cells degrade and die through the lysosomal pathway mediated by LC3-associated phagocytosis (114). However, it has been suggested that P53 locally inhibits RhoA signaling and myosin contraction at cell-cell junctions by targeting Rnd3, which leads to asymmetric RhoA activation and thus promotes entotic CIC formation (113). UV radiation is also one of the triggers of entosis. Upon cells stimulated, JNK/p38 signaling pathway is activated, which triggers ROCK-dependent entosis (115). Most interestingly, the present study has demonstrated that entosis occurs when breast tumor cells are deficient in glucose in which AMPK plays an important role (116). (Figure 3)
6.2 Entosis in tumor cells
In many cancers, tumor malignancy and poor prognosis are the link with entosis which even acts as an escape mechanism to evade adverse factors from other cells, contributing to treatment failure or cancer recurrence (117). In high-grade clear cell renal carcinomas, CIC structures tend to indicate high malignancy and metastasis (118). While Durgan, J et al. found that epithelial cells can phagocytose and kill abnormally dividing cells to inhibit tumor growth when entosis happens (119). For example, TRAIL-induced entosis in colorectal tumor cells results in the appearance of CIC structures and poor prognosis (120). Entosis can exert anticancer effects in novel methylselenoesters-induced pancreatic cancer cells (121). Entosis plays a dual role of cancer, but there is no complete and systematic study on the antagonistic mechanism of these two effects, which suggests new ideas for future studies on entosis.
6.3 Entosis and lncRNAs in tumors
Rho GTPases are important links in entosis. LncRNAs have been reported to modulate Rho GTPases-mediated signaling and affect the survival of cells (122, 123). In bladder cancer, lncRNA KTN1-AS1 promotes tumor development by regulating the KTN1/Rho GTPase axis (122). An elegant study out of Li et al. examined that lncRNA NORAD competitively sponges miR-125a-3p, bringing about dysregulation of RhoA and migration and invasion of pancreatic cancer cells (124). LncRNA NORAD has also been shown to promote lung cancer cell proliferation, invasion, and migration through CXCR4 and RhoA/ROCK signaling pathway (125). According to latest bioinformatics research, lncRNA NORAD can also promote breast cancer progression through the Rho GTPase pathway (126). However, the research about lncRNAs and entosis in other tumors is still elusive.
7 Ferroptosis
7.1 Mechanisms of ferroptosis
Ferroptosis was first proposed by Dixon in 2012 (127). Morphologically, ferroptosis is characterized by mitochondria shrunk, mitochondrial cristae reduced or disappeared, mitochondrial membrane density increased and mitochondrial outer membrane ruptured, while nuclear structure is intact, without nuclear fissures and chromatin marginalization, as opposed to cellular morphological changes that occur in apoptosis and necroptosis (128). Biochemically, ferroptosis is characterized by iron-dependent lipid peroxidation and increased intracellular ROS (129).
Ferroptosis is based on iron accumulation and lipid peroxidation. Typically, excess iron is present as ferritin. However, when cells are stimulated internally or externally, ferritin is degraded and Fe3+ is released in large amounts, which is reduced to Fe2+ by transferrin receptor-1 (TFR1) and released into the iron pool in the cytoplasm, increasing ROS and causing destructive effects (130, 131). On the cell membrane, SLC7A11 and SLC3A2 are critical subunits of glutamate-cystine reverse transporter (System Xc-) which can mediate glutamate transport out of the cells, cystine transport into the cells. Intracellular cystine is reduced to cysteine that synthesizes reduced GSH (132). Glutathione peroxidase 4 (GPX4) is a selenocysteine enzyme which can regulate the sensitivity of cells to ferroptosis. GPX4, which is activated by cystine via mTOR complex 1 (mTORC1) signaling pathway, can detoxify lipid peroxidation in the presence of GSH, thus reducing lipid ROS to inhibit ferroptosis (133, 134). When the antioxidant system, especially the System Xc-/GSH/GPX4-dependent antioxidant defense system, is inactivated, lipid ROS accumulates and ferroptosis occurs. Erastin is a commonly used ferroptosis-inducing factor, which suppresses SLC7A11 expression rendering System Xc-dysfunction, and then inhibits cystine uptake, reduces GSH production, and produces ferroptosis (135). (Figure 3)
7.2 Ferroptosis in tumor cells
Since the concept of “ferroptosis” has been proposed, there have been increasing studies on the role of ferroptosis in tumor suppression. Ferroptosis can be activated or resisted through different signaling pathways to regulate tumor growth and drug resistance, such as hippo signaling pathway, PI3K/AKT/mTOR signaling pathway and so on (136). PARP inhibitors have been approved for use in ovarian and breast cancers. It has been suggested that PARP inhibitors promote ferroptosis by suppressing SLC7A11 in a p53-dependent manner. In combination with erastin, the sensitivity of PARP inhibitors can be improved in the treatment of ovarian cancer, thereby exerting anticancer effects (137). Like PARP inhibitors, STAT3 inhibitors down-regulate SLC7A11 and GPX4 to trigger ferroptosis, inhibit gastric cancer progression and reduce chemoresistance (138). Ferroptosis, in addition to anticancer effects, can also produce inflammation-related immunosuppression in the tumor microenvironment, which facilitates tumor growth. HMGB1 can bind to receptor for advanced glycation end-products (RAGE or AGER) to promote inflammatory responses in macrophages, and inhibition of HMGB1 may have potential therapeutic effects on ferroptosis-related diseases (139). Furthermore, AGER in pancreatic cancer mediates the uptake of oncogenic protein KRAS by macrophages, which ultimately leads to macrophage polarization and stimulates tumor growth (140). Ferroptosis plays an anticancerous role in most tumors and is still a major hotspot in the study of cancer treatment.
7.3 Ferroptosis and lncRNAs in tumors
With the growing knowledge of ferroptosis, the regulation between lncRNAs and ferroptosis is continuously being investigated and developed. It has been shown that high level of lncRNA PVT1 directly interacts with miR-214-3p, hindering adsorption of GPX4, increasing GPX4 content and maintaining GSH in a reduced state, which inhibits ferroptosis and promotes tumor cell proliferation (141). Under the induction of erastin, miR-214 increases with GSH decreasing, lipid oxidation enhancing and ROS production increasing, causing ferroptosis in hepatocellular carcinoma (142). Erastin can also up-regulate lncRNA GABPB1-AS1 which downregulates the protein level of GABPB1 by blocking the translation of GABPB1 mRNA. Due to a series of biological effects mediated by GABPB1, intracellular ROS is increased which promotes ferroptosis and prolongs the survival time of liver cancer patients (143). There has been a study reporting that silencing of lncRNA NEAT1 can act in combination with erastin to down-regulate SLCA11 and GPX4, induce ROS increase, and promote the upregulation of Bax and caspase-3, inhibit the expression of Bcl-2, which motivates ferroptosis and apoptosis, and inhibits tumor proliferation, metastasis and invasion in non-small cell lung cancer (135). In addition to lncRNA NEAT1, lncRNA H19 can also trigger ferroptosis in lung cancer via the miR-19b-3p/FTH1 axis (15).
LncRNAs can promote or suppress ferroptosis by regulating the expression of miRNAs or mRNAs which affect key molecules of ferroptosis. There are still many studies on ferroptosis mediated by lncRNAs, which have been reported in different tumors, such as ovarian cancer, prostate cancer, bladder cancer, acute myeloid leukemia and so on (144–147). However, in benign tumors, the role of lncRNAs in regulating ferroptosis remains to be explored.
8 Autosis
8.1 Mechanisms of autosis
According to published guidelines, we can classify cell death into three types by the tightness of the relationship between autophagy and cell death: autophagy-associated cell death, autophagy-mediated cell death, and autophagy-dependent cell death (148). In 2013, Beth Levine named autophagy-dependent cell death “autosis” (149). We focus here on autophagy-dependent cell death. When autosis occurs, the morphological features of cells are characterized by enhanced adhesion between cells and extracellular matrix, disruption or disappearance of ER structure, swelling around the nucleus, and mild chromatin condensation (149).
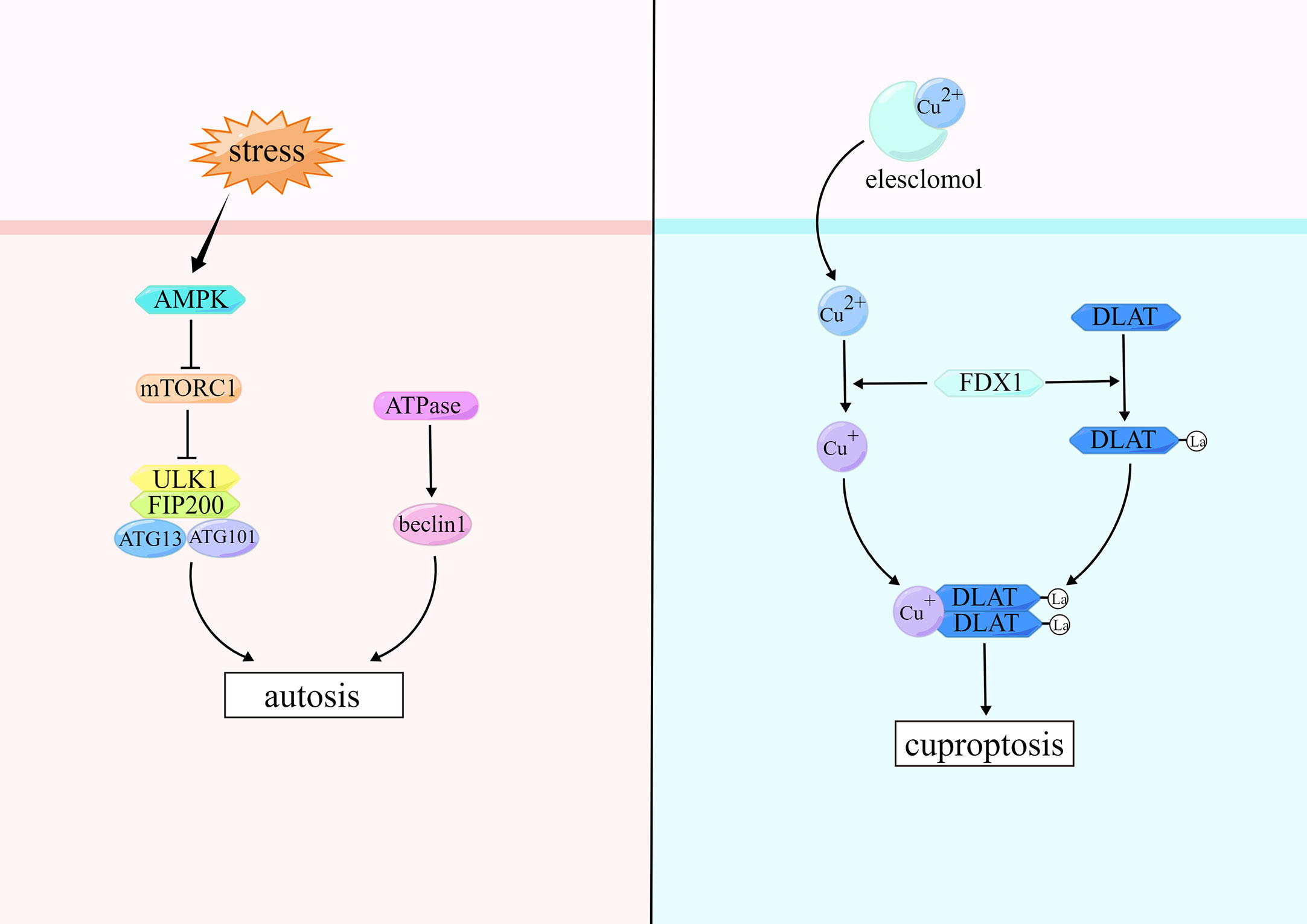
Autosis usually occurs as a result of stimulation by high doses of autophagy-inducing peptides, starvation, and permanent cerebral ischemia, and the key to initiation is the Unc-51-like autophagy activating kinase 1 (ULK1) complex (150). ULK1 complex which is the core complex in autosis consists of ULK1, ATG13, ATG101, and FAK family kinase-interacting protein of 200 kDa (FIP200) (151). The ULK1 kinase complex recruits the PI3K complex to phagocytes, which catalyzes the conversion of phosphatidylinositol to phosphatidylinositol 3-phosphate (PI3P) and promotes autophagosomes formation. The PI3K complex consists of PI3K, the autophagy protein beclin-1 (BECN1), vacuolar protein sorting-15, and other partners (150). PI3K can interact with a variety of regulatory proteins to form different complexes that will selectively participate in different stages of autosis and initiate downstream cascade enzymatic reactions (152). MTORC1 regulates the ULK1 complex to inhibit autosis (153). The upstream molecule of mTORC1 is AMP-activated protein kinase (AMPK), which inhibits mTORC1 and induces autosis (154, 155). Moreover, it has been shown that Na+, K+-adenosine triphosphatase (ATPase) can promote autosis by activating BECN1 (156). (Figure 4)
8.2 Autosis in tumor cells
It is well-known that autophagy exists as a pro-survival stress response in most cases. Moreover, the above three types of autophagy are not clearly divided in most studies, and most of the studies are on autophagy-mediated cell death. However, autosis still plays an important role in inducing tumor cell death, especially as a spare cell death program in apoptosis-deficient tumors (157). As early as more than a decade ago, Degenhardt et al. reported that autosis may suppress tumors by attenuating metabolic stress in apoptosis-deficient cells (158). In addition, autosis can act together with ferroptosis to exert anticancer effects in tumor cells. The AMPK signaling pathway activates BECN1, which promotes ferroptosis in tumor cells by directly blocking System Xc- and binding to SLC7A11 to improve anticancer therapy (159). The interaction with two pathways of GPX4 and mTOR can regulate autophagy-dependent ferroptosis in pancreatic cancer cells (160). Interestingly, autosis, in addition to its anticancer role, also plays a carcinogenic role in the early stage of tumors. Studies using a Drosophila melanogaster malignant tumour model have demonstrated that TNF and IL-6 mediated autophagy can modulate the tumor microenvironment and participate in tumor growth (161).
8.3 Autosis and lncRNAs in tumors
LncRNAs have been reported to affect autosis in tumor cells by regulating the ULK1 complex and mTOR signaling pathway. Zhao et al. found that lncRNA H19 can impact on glioma cell proliferation, migration, and autophagy by regulating the mTOR signaling pathway. Overexpression of lncRNA H19 inhibits mTOR phosphorylation and promotes ULK1 phosphorylation, thereby inhibiting the development of autosis and promoting tumor cell proliferation (162). LncRNA H19 has also been reported in pituitary tumor (163). LncRNA H19 suppresses mTORC1 function and blocks mTORC1-mediated phosphorylation of 4E-BP1 to inhibit pituitary tumor cell proliferation in vitro and in vivo (163). LncRNA CASC9 can promote tumor proliferation by inhibiting autosis and autophagy-mediated apoptosis through AKT/mTOR pathway in oral squamous cell carcinoma (164). There are two pathways by which lncRNA HAGLROS regulates the mTOR signaling pathway in gastric cancer. One is to increase the expression of mTOR mRNA and mTOR by sponging miR-100-5p; the other is that lncRNA HAGLROS interacts with mTORC1 components to activate the mTORC1 signaling pathway and inhibit autosis in gastric cancer cells (165). LncRNA EPIC1 can activate AKT-mTORC1 signaling pathway through regulating the expression of transcription factor Myc, which leads to rapamycin resistance and reduced autosis in tumor cells in breast and ovarian cancers (166). In addition to ULK1, two other proteins in the ULK1 complex have not been covered to be regulated by lncRNAs.
There are quite a few reports on the regulation of autophagy by lncRNAs, but there are few reports on the involvement of lncRNAs in the regulation of autophagy-dependent cell death, and there is still much room to be explored on the relationship between lncRNAs and autophagy-dependent cell death.
9 Cuproptosis
9.1 Mechanisms of cuproptosis
Cuproptosis belongs to RCDs discovered in 2022. Tsvetkov and colleagues showed that intracellular copper accumulation leads to aggregation of lipoylated proteins and destabilization of Fe-S cluster proteins in mitochondria, which in turn induces a unique type of cell death named cuproptosis (167). Changes in cell morphology have not been reported when cuproptosis occurs.
Elesclomol and copper ion complex (ES-Cu) can induce cuproptosis. ES in ES-Cu complexes can promote Cu2+ into cells. Ferredoxin 1 (FDX1) is a direct target of ES, and Cu2+ are reduced to Cu+ by FDX1 (168). FDX1 is a key component in the Fe-S cluster assembly pathway and also involved in lipoylation of the tricarboxylic acid (TCA) cycle proteins (169). TCA cycle proteins that can be lipoylated include line dihydrolipamide branched-chain transacylase E2, glycine cleavage system protein H, dihydrolipamide S-succinyltransferase, and dihydrolipamide S-acetyltransferase (DLAT), which are important components of the pyruvate dehydrogenase complex (170, 171). Cu+ reduced by FDX1 can combine directly with lipoylated DLAT and promote disulfide-bond-dependent DLAT oligomers formation. Meanwhile, FDX1-dependent Fe-S cluster proteins undergo degradation, causing cuproptosis in cells (167). (Figure 4)
9.2 Cuproptosis in tumor cells
As a key molecule of cuproptosis, serum copper has been reported to be more abundant in lung cancer, hepatocellular carcinoma, colorectal cancer, breast cancer, cervical cancer, oral cancer and other tumors compared with normal tissues, and copper is considered to be involved in tumor growth, metastasis, and drug resistance (172–178). A mitochondria-targeted, copper-depleting nanoparticle chelates copper in mitochondria, reduces oxygen consumption and oxidative phosphorylation, converts metabolism to glycolysis to reduce ATP production in triple-negative breast cancer cells, and ultimately inhibits tumor growth and improves survival rate (179). In addition, copper depletion activates AMP-activated protein kinase to inhibit mTORC1 pathway, and reduce oxidative phosphorylation which in turn weakens the ability of tumor invasion (180). The expression level of cuproptosis-related gene SLC31A1, i.e., copper transporter 1 (CTR1), is negatively correlated with overall survival. The study revealed that CTR1 is abnormally elevated in breast cancer and copper activates the PDK1/AKT pathway in a CTR1-dependent manner and promotes tumorigenesis (181). Apart from copper and SLC31A1, other cuproptosis-related genes, such as FDX1, ATP7B, and lipoyltransferase-1, are also involved in tumor development (182, 183). FDX1 which expression is significantly lower than normal tissues in a variety of cancer types, is positively correlated with immune cell infiltration and tumor mutation load, and has a clear correlation with tumor survival and prognosis. Therefore, FDX1 is expected to be a tumor biomarker and potential therapeutic target (169).
9.3 Cuproptosis and lncRNAs in tumors
After cuproptosis was reported, studies about lncRANs and cuproptosis-related genes have gradually increased, and have been reported in a variety of tumors. Mo et al. analyzed that AC008764.2, AL022323.1, lncRNA ELN-AS1 and LINC00578 are protective lncRNAs that promote cuproptosis in lung adenocarcinoma cells, while AL031667.3, AL606489.1 and lncRNA MIR31HG are considered as dangerous lncRNAs (184). Among them, lncRNA MIR31HG can inhibit cuproptosis and promote the proliferation, migration, and invasion of lung adenocarcinoma cells by down-regulating miR-193a-3p and increasing downstream TNFRSF21 expression (184). In osteosarcoma, AL645608.6, AL591767.1, lncRNA UNC5B-AS1, lncRNA CARD8-AS1, AC098487.1, AC005041, lncRNA TIPARP-AS1, lncRNA RUSC1-AS1, and LINC02315 play a role in regulating cuproptosis (185, 186). The studies on the regulatory relationship of lncRNAs on cuproptosis in breast cancer have also been reported. Jiang et al. found that lncRNAs such as lncRNAs GORAB-AS1, AC079922.2, AL589765.4, AC005696.4, lncRNA Cytor, lncRNA ZNF197-AS1, AC002398.1, AL451085.3, lncRNA YTHDF3-AS1, AC008771.1, and LINC02446 are associated with cuproptosis and can affect the prognosis of breast cancer and the sensitivity of immunotherapy (187). LncRNA XIST can sponge miR-92b-3p and regulate the cuproptosis-related gene MTF1 to influence the progression of breast cancer (188). In cervical cancer, AL441992.1, LINC01305, AL354833.2, lncRNA CNNM3-DT and lncRNA SCAT2 can promote cuproptosis to protect the body from tumor cells attack and improve tumor prognosis; while AL354733.3 and AC009902.2 can inhibit cuproptosis to facilitate tumor growth (189). In addition, there are many relevant reports on lncRNAs regulating cuproptosis in head and neck squamous cell carcinoma, gastric cancer, liver cancer, and colorectal cancer (190–193).
However, the regulatory relationship between lncRNAs and cuproptosis is only analyzed in the database, but there are fewer experimental studies about the two relationships. Therefore, it remains many doubts whether lncRNAs can regulate cuproptosis in tumor cells, which needs further exploration.
10 Discussion
LncRNAs have been the research focus in recent years, especially in the regulation of tumor biological function. LncRNAs can initiate RCDs mediated by different pathways which act on tumor cells for the purpose of promoting or inhibiting tumor proliferation, migration, invasion, prognosis, and drug resistance. The occurrence of a particular RCD is often regulated by many different lncRNAs in the same tumor. For example, lncRNA PDPK2p/PDK1/AKT/caspase-3 signaling pathway can inhibit apoptosis, while the lncRNA MAGI2-AS3/miR-374b-5p/Smg1 axis can promote apoptosis in liver cancer cells (38, 41). In cervical cancer, lncRNA FAM13A-AS1/miRNA-205-3p/DDI2 axis and lncRNA PTENP1/miR-106b/PTEN axis can jointly promote apoptosis and inhibit the progression of cervical cancer, while lncRNA HOXD-AS1 plays an anti-apoptosis role to promote the progression of cervical cancer (194–196). In addition to apoptosis, different lncRNAs have also been reported to regulate the same tumor in other RCDs such as necroptosis and ferroptosis. Homologous lncRNAs can play different roles in different tumors and initiate different RCDs. LncRNA H19 has been reported to regulate apoptosis, ferroptosis, and others (197). LncRNA NEAT1 regulates pyroptosis in glioma cells and colorectal cancer cells by targeting miR-296-5p or miR-448; while its high expression promotes ferroptosis in non-small cell lung cancer (76, 77, 135). Furthermore, lncRNA H19 and lncRNA NEAT1 act upon each other and respectively regulate apoptosis by targeting miR-675 and miR-204 in breast cancer (198). Although lncRNAs have been demonstrated successively as potential targets for tumor diagnosis and treatment, there is no mature means to use lncRNAs for clinical diagnosis and treatment, which is still a great gap waiting to be explored.
RCDs can occur on almost all cells and play indispensable roles in the normal growth and development of the human body. The signaling pathways of RCDs can interact with each other. As a very vital connection in the process of apoptosis, caspase family also play a part in necroptosis and pyroptosis; ROS is not only working in ferroptosis, but also participates in NETosis. In addition, some forms of cell death can also act depending on other forms of cell death. Apoptosis, which is dependent on caspase, can stimulate GSDM cleavage through caspase activation and alter GSDM expression in triggering pyroptosis (199). LINC00618 exerts apoptosis by upregulating levels of Bax and caspase-3, while LINC00618 induces ferroptosis by increasing ROS and decreasing SLC7A11. Findings used caspase inhibitors Z-VAD-FMK and erastin demonstrated that LINC00618-induced ferroptosis was dependent on apoptosis (147). However, the specific mechanism by which lncRNAs promote or inhibit different RCDs in tumor cells through targeting common key molecules that regulate multiple pathways remains unreported.
Finally, due to the limited available medical technology, people still do not conquer cancers. It is difficult to diagnose malignant tumors in time and provide effective and specific treatment for a long time, resulting in the prognosis of most malignant tumors is still not optimistic. Therefore, we need a target that is specific and sensitive, and lncRNAs may become this target. LncRNAs can improve tumor prognosis by triggering different RCDs, which provides us with new ideas for diagnosis and treatment.
Author contributions
Conceptualization, YL and YW; manuscript preparation, YW; manuscript revision, YL, YW, XW, YX and XY. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by grants from the Shinan District Science and Technology Program in Qingdao, Shandong Province, China, grant number 2022–2–006-YY.
Acknowledgments
We sincerely thank the grants and all those who contributed to this review. All figures here were made by Figdraw.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ (2018) 25(3):486–541. doi: 10.1038/s41418-017-0012-4
2. Aguilera A, Klemencic M, Sueldo DJ, Rzymski P, Giannuzzi L, Martin MV. Cell death in cyanobacteria: current understanding and recommendations for a consensus on its nomenclature. Front Microbiol (2021) 12:631654. doi: 10.3389/fmicb.2021.631654
3. Christgen S, Tweedell RE, Kanneganti TD. Programming inflammatory cell death for therapy. Pharmacol Ther (2022) 232:108010. doi: 10.1016/j.pharmthera.2021.108010
4. Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res (2019) 29(5):347–64. doi: 10.1038/s41422-019-0164-5
5. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
6. Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell (2020) 78(6):1045–54. doi: 10.1016/j.molcel.2020.05.014
7. Goolsby C, Paniagua M, Tallman M, Gartenhaus RB. Bcl-2 regulatory pathway is functional in chronic lymphocytic leukemia. Cytometry B Clin Cytom (2005) 63(1):36–46. doi: 10.1002/cyto.b.20034
8. Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, et al. DNA Repair processes are critical mediators of P53-dependent tumor suppression. Nat Med (2018) 24(7):947–53. doi: 10.1038/s41591-018-0043-5
9. Arab K, Karaulanov E, Musheev M, Trnka P, Schafer A, Grummt I, et al. Gadd45a binds r-loops and recruits Tet1 to cpg island promoters. Nat Genet (2019) 51(2):217–23. doi: 10.1038/s41588-018-0306-6
10. Entezari M, Taheriazam A, Orouei S, Fallah S, Sanaei A, Hejazi ES, et al. Lncrna-mirna axis in tumor progression and therapy response: an emphasis on molecular interactions and therapeutic interventions. BioMed Pharmacother (2022) 154:113609. doi: 10.1016/j.biopha.2022.113609
11. Shaath H, Vishnubalaji R, Elango R, Kardousha A, Islam Z, Qureshi R, et al. Long non-coding rna and rna-binding protein interactions in cancer: experimental and machine learning approaches. Semin Cancer Biol (2022) 86(Pt 3):325–45. doi: 10.1016/j.semcancer.2022.05.013
12. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. Lncrna-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) (2021) 41(2):109–20. doi: 10.1002/cac2.12108
13. Esposito R, Bosch N, Lanzos A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding rnas using crispr-Cas9 screening. Cancer Cell (2019) 35(4):545–57. doi: 10.1016/j.ccell.2019.01.019
14. Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang H, et al. Tead4 modulated lncrna Mnx1-As1 contributes to gastric cancer progression partly through suppressing Btg2 and activating Bcl2. Mol Cancer (2020) 19(1):6. doi: 10.1186/s12943-019-1104-1
15. Zhang R, Pan T, Xiang Y, Zhang M, Xie H, Liang Z, et al. Curcumenol triggered ferroptosis in lung cancer cells Via lncrna H19/Mir-19b-3p/Fth1 axis. Bioact Mater (2022) 13:23–36. doi: 10.1016/j.bioactmat.2021.11.013
16. Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res (2011) 30(1):87. doi: 10.1186/1756-9966-30-87
17. Osterlund EJ, Hirmiz N, Pemberton JM, Nougarede A, Liu Q, Leber B, et al. Efficacy and specificity of inhibitors of bcl-2 family protein interactions assessed by affinity measurements in live cells. Sci Adv (2022) 8(16):eabm7375. doi: 10.1126/sciadv.abm7375
18. Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol (2020) 17(7):395–417. doi: 10.1038/s41571-020-0341-y
19. Bhola PD, Letai A. Mitochondria-judges and executioners of cell death sentences. Mol Cell (2016) 61(5):695–704. doi: 10.1016/j.molcel.2016.02.019
20. Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and datp-dependent formation of apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell (1997) 91(4):479–89. doi: 10.1016/s0092-8674(00)80434-1
21. Razeghian E, Suksatan W, Sulaiman Rahman H, Bokov DO, Abdelbasset WK, Hassanzadeh A, et al. Harnessing trail-induced apoptosis pathway for cancer immunotherapy and associated challenges. Front Immunol (2021) 12:699746. doi: 10.3389/fimmu.2021.699746
22. Cui Z, Dabas H, Leonard BC, Shiah JV, Grandis JR, Johnson DE. Caspase-8 mutations associated with head and neck cancer differentially retain functional properties related to trail-induced apoptosis and cytokine induction. Cell Death Dis (2021) 12(8):775. doi: 10.1038/s41419-021-04066-z
23. Yin XM, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, et al. Bid-deficient mice are resistant to fas-induced hepatocellular apoptosis. Nature (1999) 400(6747):886–91. doi: 10.1038/23730
24. Bhat TA, Chaudhary AK, Kumar S, O'Malley J, Inigo JR, Kumar R, et al. Endoplasmic reticulum-mediated unfolded protein response and mitochondrial apoptosis in cancer. Biochim Biophys Acta Rev Cancer (2017) 1867(1):58–66. doi: 10.1016/j.bbcan.2016.12.002
25. Hu H, Tian M, Ding C, Yu S. The C/Ebp homologous protein (Chop) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol (2018) 9:3083. doi: 10.3389/fimmu.2018.03083
26. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
27. Bock FJ, Sedov E, Koren E, Koessinger AL, Cloix C, Zerbst D, et al. Apoptotic stress-induced fgf signalling promotes non-cell autonomous resistance to cell death. Nat Commun (2021) 12(1):6572. doi: 10.1038/s41467-021-26613-0
28. Morana O, Wood W, Gregory CD. The apoptosis paradox in cancer. Int J Mol Sci (2022) 23(3):1328. doi: 10.3390/ijms23031328
29. Ford CA, Petrova S, Pound JD, Voss JJ, Melville L, Paterson M, et al. Oncogenic properties of apoptotic tumor cells in aggressive b cell lymphoma. Curr Biol (2015) 25(5):577–88. doi: 10.1016/j.cub.2014.12.059
30. Liu SY, Zhao ZY, Qiao Z, Li SM, Zhang WN. Lncrna Pcat1 interacts with Dkc1 to regulate proliferation, invasion and apoptosis in nsclc cells Via the Vegf/Akt/Bcl2/Caspase9 pathway. Cell Transplant (2021) 30:963689720986071. doi: 10.1177/0963689720986071
31. Yu A, Zhao L, Kang Q, Li J, Chen K, Fu H. Transcription factor Hif1alpha promotes proliferation, migration, and invasion of cholangiocarcinoma Via long noncoding rna H19/Microrna-612/Bcl-2 axis. Transl Res (2020) 224:26–39. doi: 10.1016/j.trsl.2020.05.010
32. Cheng B, Rong A, Zhou Q, Li W. Lncrna Linc00662 promotes colon cancer tumor growth and metastasis by competitively binding with mir-340-5p to regulate Cldn8/Il22 Co-expression and activating erk signaling pathway. J Exp Clin Cancer Res (2020) 39(1):5. doi: 10.1186/s13046-019-1510-7
33. Mei J, Liu G, Li R, Xiao P, Yang D, Bai H, et al. Lncrna Snhg6 knockdown inhibits cisplatin resistance and progression of gastric cancer through mir-1297/Bcl-2 axis. Biosci Rep (2021) 41(12):BSR20211885. doi: 10.1042/BSR20211885
34. Tian YH, Jia LW, Liu ZF, Chen YH. Linc01087 inhibits glioma cell proliferation and migration, and increases cell apoptosis Via mir-384/Bcl-2 axis. Aging (Albany NY) (2021) 13(16):20808–19. doi: 10.18632/aging.203478
35. Li X, Luo S, Zhang J, Yuan Y, Jiang W, Zhu H, et al. Lncrna H19 alleviated myocardial I/Ri Via suppressing mir-877-3p/Bcl-2-Mediated mitochondrial apoptosis. Mol Ther Nucleic Acids (2019) 17:297–309. doi: 10.1016/j.omtn.2019.05.031
36. Hao X, Wei H. Lncrna H19 alleviates sepsis-induced acute lung injury by regulating the mir-107/Tgfbr3 axis. BMC Pulm Med (2022) 22(1):371. doi: 10.1186/s12890-022-02091-y
37. Zhang L, Cheng H, Yue Y, Li S, Zhang D, He R. H19 knockdown suppresses proliferation and induces apoptosis by regulating mir-148b/Wnt/Beta-Catenin in ox-ldl -stimulated vascular smooth muscle cells. J BioMed Sci (2018) 25(1):11. doi: 10.1186/s12929-018-0418-4
38. Pan W, Li W, Zhao J, Huang Z, Zhao J, Chen S, et al. Lncrna-Pdpk2p promotes hepatocellular carcinoma progression through the Pdk1/Akt/Caspase 3 pathway. Mol Oncol (2019) 13(10):2246–58. doi: 10.1002/1878-0261.12553
39. Jiang Q, Zheng N, Bu L, Zhang X, Zhang X, Wu Y, et al. Spop-mediated ubiquitination and degradation of Pdk1 suppresses akt kinase activity and oncogenic functions. Mol Cancer (2021) 20(1):100. doi: 10.1186/s12943-021-01397-5
40. Yang Y, Yang H, Xu M, Zhang H, Sun M, Mu P, et al. Long non-coding rna (Lncrna) Magi2-As3 inhibits breast cancer cell growth by targeting the Fas/Fasl signalling pathway. Hum Cell (2018) 31(3):232–41. doi: 10.1007/s13577-018-0206-1
41. Yin Z, Ma T, Yan J, Shi N, Zhang C, Lu X, et al. Lncrna Magi2-As3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the mir-374b-5p/Smg1 signaling pathway. J Cell Physiol (2019) 234(10):18825–36. doi: 10.1002/jcp.28521
42. Du Y, Lu F, Li P, Ye J, Ji M, Ma D, et al. Smg1 acts as a novel potential tumor suppressor with epigenetic inactivation in acute myeloid leukemia. Int J Mol Sci (2014) 15(9):17065–76. doi: 10.3390/ijms150917065
43. Cheung HH, St Jean M, Beug ST, Lejmi-Mrad R, LaCasse E, Baird SD, et al. Smg1 and nik regulate apoptosis induced by smac mimetic compounds. Cell Death Dis (2011) 2(4):e146. doi: 10.1038/cddis.2011.25
44. Wang Y, Zheng S, Han J, Li N, Ji R, Li X, et al. Linc00629 protects osteosarcoma cell from er stress-induced apoptosis and facilitates tumour progression by elevating Klf4 stability. J Exp Clin Cancer Res (2022) 41(1):354. doi: 10.1186/s13046-022-02569-x
45. Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan K, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer (2019) 18(1):100. doi: 10.1186/s12943-019-1029-8
46. Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer (2022) 8(1):21–7. doi: 10.1016/j.trecan.2021.09.003
47. Wang Y, Zhao M, He S, Luo Y, Zhao Y, Cheng J, et al. Necroptosis regulates tumor repopulation after radiotherapy Via Rip1/Rip3/Mlkl/Jnk/Il8 pathway. J Exp Clin Cancer Res (2019) 38(1):461. doi: 10.1186/s13046-019-1423-5
48. Feoktistova M, Makarov R, Yazdi AS, Panayotova-Dimitrova D. Ripk1 and tradd regulate tnf-induced signaling and ripoptosome formation. Int J Mol Sci (2021) 22(22):12459. doi: 10.3390/ijms222212459
49. Li X, Li F, Zhang X, Zhang H, Zhao Q, Li M, et al. Caspase-8 auto-cleavage regulates programmed cell death and collaborates with Ripk3/Mlkl to prevent lymphopenia. Cell Death Differ (2022) 29(8):1500–12. doi: 10.1038/s41418-022-00938-9
50. Kaiser WJ, Sridharan H, Huang C, Mandal P, Upton JW, Gough PJ, et al. Toll-like receptor 3-mediated necrosis Via trif, Rip3, and mlkl. J Biol Chem (2013) 288(43):31268–79. doi: 10.1074/jbc.M113.462341
51. Zhang T, Yin C, Boyd DF, Quarato G, Ingram JP, Shubina M, et al. Influenza virus z-rnas induce Zbp1-mediated necroptosis. Cell (2020) 180(6):1115–29.e13. doi: 10.1016/j.cell.2020.02.050
52. Stoll G, Ma Y, Yang H, Kepp O, Zitvogel L, Kroemer G. Pro-necrotic molecules impact local immunosurveillance in human breast cancer. Oncoimmunology (2017) 6(4):e1299302. doi: 10.1080/2162402X.2017.1299302
53. Xie Y, Zhao Y, Shi L, Li W, Chen K, Li M, et al. Gut epithelial Tsc1/Mtor controls Ripk3-dependent necroptosis in intestinal inflammation and cancer. J Clin Invest (2020) 130(4):2111–28. doi: 10.1172/JCI133264
54. Baik JY, Liu Z, Jiao D, Kwon HJ, Yan J, Kadigamuwa C, et al. Zbp1 not Ripk1 mediates tumor necroptosis in breast cancer. Nat Commun (2021) 12(1):2666. doi: 10.1038/s41467-021-23004-3
55. Zheng XL, Yang JJ, Wang YY, Li Q, Song YP, Su M, et al. Rip1 promotes proliferation through G2/M checkpoint progression and mediates cisplatin-induced apoptosis and necroptosis in human ovarian cancer cells. Acta Pharmacol Sin (2020) 41(9):1223–33. doi: 10.1038/s41401-019-0340-7
56. Harari-Steinfeld R, Gefen M, Simerzin A, Zorde-Khvalevsky E, Rivkin M, Ella E, et al. The lncrna H19-derived microrna-675 promotes liver necroptosis by targeting fadd. Cancers (Basel) (2021) 13(3):411. doi: 10.3390/cancers13030411
57. Qiannan D, Qianqian J, Jiahui S, Haowei F, Qian X. Lncrna Pvt1 mediates the progression of liver necroptosis Via Zbp1 promoter methylation under nonylphenol exposure. Sci Total Environ (2022) 844:157185. doi: 10.1016/j.scitotenv.2022.157185
58. Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. Lncrna Pvt1 promotes hepatitis b Virus−Positive liver cancer progression by disturbing histone methylation on the C−Myc promoter. Oncol Rep (2020) 43(2):718–26. doi: 10.3892/or.2019.7444
59. Gu W, Pan F, Singer RH. Blocking beta-catenin binding to the Zbp1 promoter represses Zbp1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci (2009) 122(Pt 11):1895–905. doi: 10.1242/jcs.045278
60. Luo L, Li L, Liu L, Feng Z, Zeng Q, Shu X, et al. A necroptosis-related lncrna-based signature to predict prognosis and probe molecular characteristics of stomach adenocarcinoma. Front Genet (2022) 13:833928. doi: 10.3389/fgene.2022.833928
61. He YB, Fang LW, Hu D, Chen SL, Shen SY, Chen KL, et al. Necroptosis-associated long noncoding rnas can predict prognosis and differentiate between cold and hot tumors in ovarian cancer. Front Oncol (2022) 12:967207. doi: 10.3389/fonc.2022.967207
62. Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther (2021) 6(1):128. doi: 10.1038/s41392-021-00507-5
63. Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF, Cheng KC, et al. Inflammation-related pyroptosis, a novel programmed cell death pathway, and its crosstalk with immune therapy in cancer treatment. Theranostics (2021) 11(18):8813–35. doi: 10.7150/thno.62521
64. He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin d is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res (2015) 25(12):1285–98. doi: 10.1038/cr.2015.139
65. Billingham LK, Stoolman JS, Vasan K, Rodriguez AE, Poor TA, Szibor M, et al. Mitochondrial electron transport chain is necessary for Nlrp3 inflammasome activation. Nat Immunol (2022) 23(5):692–704. doi: 10.1038/s41590-022-01185-3
66. Xia S, Zhang Z, Magupalli VG, Pablo JL, Dong Y, Vora SM, et al. Gasdermin d pore structure reveals preferential release of mature interleukin-1. Nature (2021) 593(7860):607–11. doi: 10.1038/s41586-021-03478-3
67. Schneider KS, Gross CJ, Dreier RF, Saller BS, Mishra R, Gorka O, et al. The inflammasome drives gsdmd-independent secondary pyroptosis and il-1 release in the absence of caspase-1 protease activity. Cell Rep (2017) 21(13):3846–59. doi: 10.1016/j.celrep.2017.12.018
68. Wright SS, Vasudevan SO, Rathinam VA. Mechanisms and consequences of noncanonical inflammasome-mediated pyroptosis. J Mol Biol (2022) 434(4):167245. doi: 10.1016/j.jmb.2021.167245
69. Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, et al. The role of pyroptosis in cancer: pro-cancer or pro-"Host"? Cell Death Dis (2019) 10(9):650. doi: 10.1038/s41419-019-1883-8
70. Qiu S, Hu Y, Dong S. Pan-cancer analysis reveals the expression, genetic alteration and prognosis of pyroptosis key gene gsdmd. Int Immunopharmacol (2021) 101(Pt A):108270. doi: 10.1016/j.intimp.2021.108270
71. Wang WJ, Chen D, Jiang MZ, Xu B, Li XW, Chu Y, et al. Downregulation of gasdermin d promotes gastric cancer proliferation by regulating cell cycle-related proteins. J Dig Dis (2018) 19(2):74–83. doi: 10.1111/1751-2980.12576
72. Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin e suppresses tumour growth by activating anti-tumour immunity. Nature (2020) 579(7799):415–20. doi: 10.1038/s41586-020-2071-9
73. Tan G, Huang C, Chen J, Zhi F. Hmgb1 released from gsdme-mediated pyroptotic epithelial cells participates in the tumorigenesis of colitis-associated colorectal cancer through the Erk1/2 pathway. J Hematol Oncol (2020) 13(1):149. doi: 10.1186/s13045-020-00985-0
74. Li C, Song H, Chen C, Chen S, Zhang Q, Liu D, et al. Lncrna Pvt1 knockdown ameliorates myocardial ischemia reperfusion damage Via suppressing gasdermin d-mediated pyroptosis in cardiomyocytes. Front Cardiovasc Med (2021) 8:747802. doi: 10.3389/fcvm.2021.747802
75. Wu A, Sun W, Mou F. Lncrna−Malat1 promotes high Glucose−Induced H9c2 cardiomyocyte pyroptosis by downregulating Mir−141−3p expression. Mol Med Rep (2021) 23(4):259. doi: 10.3892/mmr.2021.11898
76. Zi H, Tuo Z, He Q, Meng J, Hu Y, Li Y, et al. Comprehensive bioinformatics analysis of gasdermin family of glioma. Comput Intell Neurosci (2022) 2022:9046507. doi: 10.1155/2022/9046507
77. Su F, Duan J, Zhu J, Fu H, Zheng X, Ge C. Long Non−Coding rna nuclear paraspeckle assembly transcript 1 regulates ionizing Radiation−Induced pyroptosis Via Microrna−448/Gasdermin e in colorectal cancer cells. Int J Oncol (2021) 59(4):79. doi: 10.3892/ijo.2021.5259
78. Yang F, Qin Y, Lv J, Wang Y, Che H, Chen X, et al. Silencing long non-coding rna Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis (2018) 9(10):1000. doi: 10.1038/s41419-018-1029-4
79. Liu J, Yao L, Zhang M, Jiang J, Yang M, Wang Y. Downregulation of lncrna-xist inhibited development of non-small cell lung cancer by activating mir-335/Sod2/Ros signal pathway mediated pyroptotic cell death. Aging (Albany NY) (2019) 11(18):7830–46. doi: 10.18632/aging.102291
80. Ren N, Jiang T, Wang C, Xie S, Xing Y, Piao D, et al. Lncrna Adamts9-As2 inhibits gastric cancer (Gc) development and sensitizes chemoresistant gc cells to cisplatin by regulating mir-223-3p/Nlrp3 axis. Aging (Albany NY) (2020) 12(11):11025–41. doi: 10.18632/aging.103314
81. Wang T, Yang Y, Sun T, Qiu H, Wang J, Ding C, et al. The pyroptosis-related long noncoding rna signature predicts prognosis and indicates immunotherapeutic efficiency in hepatocellular carcinoma. Front Cell Dev Biol (2022) 10:779269. doi: 10.3389/fcell.2022.779269
82. Zhang Z, Xu Z, Yan Y. Role of a pyroptosis-related lncrna signature in risk stratification and immunotherapy of ovarian cancer. Front Med (Lausanne) (2021) 8:793515. doi: 10.3389/fmed.2021.793515
83. Ping L, Zhang K, Ou X, Qiu X, Xiao X. A novel pyroptosis-associated long non-coding rna signature predicts prognosis and tumor immune microenvironment of patients with breast cancer. Front Cell Dev Biol (2021) 9:727183. doi: 10.3389/fcell.2021.727183
84. Yu J, Tang R, Li J. Identification of pyroptosis-related lncrna signature and Ac005253.1 as a pyroptosis-related oncogene in prostate cancer. Front Oncol (2022) 12:991165. doi: 10.3389/fonc.2022.991165
85. Xing Z, Liu Z, Fu X, Zhou S, Liu L, Dang Q, et al. Clinical significance and immune landscape of a pyroptosis-derived lncrna signature for glioblastoma. Front Cell Dev Biol (2022) 10:805291. doi: 10.3389/fcell.2022.805291
86. Zhu W, Ye Z, Chen L, Liang H, Cai Q. A pyroptosis-related lncrna signature predicts prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int Immunopharmacol (2021) 101(Pt B):108268. doi: 10.1016/j.intimp.2021.108268
87. Lin Z, Xu Y, Zhang X, Wan J, Zheng T, Chen H, et al. Identification and validation of pyroptosis-related lncrna signature and its correlation with immune landscape in soft tissue sarcomas. Int J Gen Med (2021) 14:8263–79. doi: 10.2147/IJGM.S335073
88. Masucci MT, Minopoli M, Del Vecchio S, Carriero MV. The emerging role of neutrophil extracellular traps (Nets) in tumor progression and metastasis. Front Immunol (2020) 11:1749. doi: 10.3389/fimmu.2020.01749
89. Fousert E, Toes R, Desai J. Neutrophil extracellular traps (Nets) take the central stage in driving autoimmune responses. Cells (2020) 9(4):915. doi: 10.3390/cells9040915
90. Hamam HJ, Palaniyar N. Post-translational modifications in netosis and nets-mediated diseases. Biomolecules (2019) 9(8):369. doi: 10.3390/biom9080369
91. Munoz-Caro T, Gibson AJ, Conejeros I, Werling D, Taubert A, Hermosilla C. The role of Tlr2 and Tlr4 in recognition and uptake of the apicomplexan parasite eimeria bovis and their effects on net formation. Pathogens (2021) 10(2):118. doi: 10.3390/pathogens10020118
92. Tokuhiro T, Ishikawa A, Sato H, Takita S, Yoshikawa A, Anzai R, et al. Oxidized phospholipids and neutrophil elastase coordinately play critical roles in net formation. Front Cell Dev Biol (2021) 9:718586. doi: 10.3389/fcell.2021.718586
93. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol (2010) 191(3):677–91. doi: 10.1083/jcb.201006052
94. An Z, Li J, Yu J, Wang X, Gao H, Zhang W, et al. Neutrophil extracellular traps induced by il-8 aggravate atherosclerosis Via activation nf-kappab signaling in macrophages. Cell Cycle (2019) 18(21):2928–38. doi: 10.1080/15384101.2019.1662678
95. Sollberger G, Choidas A, Burn GL, Habenberger P, Di Lucrezia R, Kordes S, et al. Gasdermin d plays a vital role in the generation of neutrophil extracellular traps. Sci Immunol (2018) 3(26):eaar6689. doi: 10.1126/sciimmunol.aar6689
96. Thiam HR, Wong SL, Qiu R, Kittisopikul M, Vahabikashi A, Goldman AE, et al. Netosis proceeds by cytoskeleton and endomembrane disassembly and Pad4-mediated chromatin decondensation and nuclear envelope rupture. Proc Natl Acad Sci USA (2020) 117(13):7326–37. doi: 10.1073/pnas.1909546117
97. Inoue M, Enomoto M, Yoshimura M, Mizowaki T. Pharmacological inhibition of sodium-calcium exchange activates nadph oxidase and induces infection-independent netotic cell death. Redox Biol (2021) 43:101983. doi: 10.1016/j.redox.2021.101983
98. Liu X, Arfman T, Wichapong K, Reutelingsperger CPM, Voorberg J, Nicolaes GAF. Pad4 takes charge during neutrophil activation: impact of Pad4 mediated net formation on immune-mediated disease. J Thromb Haemost (2021) 19(7):1607–17. doi: 10.1111/jth.15313
99. Munzer P, Negro R, Fukui S, di Meglio L, Aymonnier K, Chu L, et al. Nlrp3 inflammasome assembly in neutrophils is supported by Pad4 and promotes netosis under sterile conditions. Front Immunol (2021) 12:683803. doi: 10.3389/fimmu.2021.683803
100. Yang S, Feng Y, Chen L, Wang Z, Chen J, Ni Q, et al. Disulfiram accelerates diabetic foot ulcer healing by blocking net formation via suppressing the Nlrp3/Caspase-1/Gsdmd pathway. Transl Res (2022) 254:115–27. doi: 10.1016/j.trsl.2022.10.008
101. Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science (2018) 361(6409):eaao4227. doi: 10.1126/science.aao4227
102. Teijeira A, Garasa S, Gato M, Alfaro C, Migueliz I, Cirella A, et al. Cxcr1 and Cxcr2 chemokine receptor agonists produced by tumors induce neutrophil extracellular traps that interfere with immune cytotoxicity. Immunity (2020) 52(5):856–71.e8. doi: 10.1016/j.immuni.2020.03.001
103. Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Transl Med (2016) 8(361):361ra138. doi: 10.1126/scitranslmed.aag1711
104. Yazdani HO, Roy E, Comerci AJ, van der Windt DJ, Zhang H, Huang H, et al. Neutrophil extracellular traps drive mitochondrial homeostasis in tumors to augment growth. Cancer Res (2019) 79(21):5626–39. doi: 10.1158/0008-5472.CAN-19-0800
105. Decker AS, Pylaeva E, Brenzel A, Spyra I, Droege F, Hussain T, et al. Prognostic role of blood netosis in the progression of head and neck cancer. Cells (2019) 8(9):105. doi: 10.3390/cells8090946
106. Wang X, Zhang H, Yin S, Yang Y, Yang H, Yang J, et al. Lncrna-encoded pep-ap attenuates the pentose phosphate pathway and sensitizes colorectal cancer cells to oxaliplatin. EMBO Rep (2022) 23(1):e53140. doi: 10.15252/embr.202153140
107. Wang X, Chen K, Zhao Z. Lncrna Or3a4 regulated the growth of osteosarcoma cells by modulating the mir-1207-5p/G6pd signaling. Onco Targets Ther (2020) 13:3117–28. doi: 10.2147/OTT.S234514
108. He X, Xiao Y, Liu S, Deng R, Li Z, Zhu X. Predicting the immune microenvironment and prognosis with a netosis-related lncrna signature in head and neck squamous cell carcinoma. BioMed Res Int (2022) 2022:3191474. doi: 10.1155/2022/3191474
109. Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-Cell invasion. Cell (2007) 131(5):966–79. doi: 10.1016/j.cell.2007.10.040
110. Borensztejn K, Tyrna P, Gawel AM, Dziuba I, Wojcik C, Bialy LP, et al. Classification of cell-in-Cell structures: different phenomena with similar appearance. Cells (2021) 10(10):2569. doi: 10.3390/cells10102569
111. Garanina AS, Kisurina-Evgenieva OP, Erokhina MV, Smirnova EA, Factor VM, Onishchenko GE. Consecutive entosis stages in human substrate-dependent cultured cells. Sci Rep (2017) 7(1):12555. doi: 10.1038/s41598-017-12867-6
112. Zeng RJ, Zheng CW, Chen WX, Xu LY, Li EM. Rho gtpases in cancer radiotherapy and metastasis. Cancer Metastasis Rev (2020) 39(4):1245–62. doi: 10.1007/s10555-020-09923-5
113. Liang J, Niu Z, Zhang B, Yu X, Zheng Y, Wang C, et al. P53-dependent elimination of aneuploid mitotic offspring by entosis. Cell Death Differ (2021) 28(2):799–813. doi: 10.1038/s41418-020-00645-3
114. Kianfar M, Balcerak A, Chmielarczyk M, Tarnowski L, Grzybowska EA. Cell death by entosis: triggers, molecular mechanisms and clinical significance. Int J Mol Sci (2022) 23(9):4985. doi: 10.3390/ijms23094985
115. Chen R, Ram A, Albeck JG, Overholtzer M. Entosis is induced by ultraviolet radiation. iScience (2021) 24(8):102902. doi: 10.1016/j.isci.2021.102902
116. Hamann JC, Surcel A, Chen R, Teragawa C, Albeck JG, Robinson DN, et al. Entosis is induced by glucose starvation. Cell Rep (2017) 20(1):201–10. doi: 10.1016/j.celrep.2017.06.037
117. Gutwillig A, Santana-Magal N, Farhat-Younis L, Rasoulouniriana D, Madi A, Luxenburg C, et al. Transient cell-in-Cell formation underlies tumor relapse and resistance to immunotherapy. Elife (2022) 11:e80315. doi: 10.7554/eLife.80315
118. Kong Y, Liang Y, Wang J. Foci of entotic nuclei in different grades of noninherited renal cell cancers. IUBMB Life (2015) 67(2):139–44. doi: 10.1002/iub.1354
119. Durgan J, Tseng YY, Hamann JC, Domart MC, Collinson L, Hall A, et al. Mitosis can drive cell cannibalism through entosis. Elife (2017) 6:e27134. doi: 10.7554/eLife.27134
120. Bozkurt E, Dussmann H, Salvucci M, Cavanagh BL, Van Schaeybroeck S, Longley DB, et al. Trail signaling promotes entosis in colorectal cancer. J Cell Biol (2021) 220(11):e202010030. doi: 10.1083/jcb.202010030
121. Khalkar P, Diaz-Argelich N, Antonio Palop J, Sanmartin C, Fernandes AP. Novel methylselenoesters induce programed cell death Via entosis in pancreatic cancer cells. Int J Mol Sci (2018) 19(10):2849. doi: 10.3390/ijms19102849
122. Hu X, Xiang L, He D, Zhu R, Fang J, Wang Z, et al. The long noncoding rna Ktn1-As1 promotes bladder cancer tumorigenesis Via Ktn1 cis-activation and the consequent initiation of rho gtpase-mediated signaling. Clin Sci (Lond) (2021) 135(3):555–74. doi: 10.1042/CS20200908
123. Saliani M, Mirzaiebadizi A, Mosaddeghzadeh N, Ahmadian MR. Rho gtpase-related long noncoding rnas in human cancers. Cancers (Basel) (2021) 13(21):5386. doi: 10.3390/cancers13215386
124. Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, et al. Long noncoding rna norad, a novel competing endogenous rna, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer (2017) 16(1):169. doi: 10.1186/s12943-017-0738-0
125. Wu Y, Shen QW, Niu YX, Chen XY, Liu HW, Shen XY. Lncnorad interference inhibits tumor growth and lung cancer cell proliferation, invasion and migration by down-regulating Cxcr4 to suppress Rhoa/Rock signaling pathway. Eur Rev Med Pharmacol Sci (2020) 24(10):5446–55. doi: 10.26355/eurrev_202005_21329
126. Nicknam A, Khojasteh Pour S, Hashemnejad MA, Hussen BM, Safarzadeh A, Eslami S, et al. Expression analysis of rho gtpase-related lncrnas in breast cancer. Pathol Res Pract (2023) 244:154429. doi: 10.1016/j.prp.2023.154429
127. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell (2012) 149(5):1060–72. doi: 10.1016/j.cell.2012.03.042
128. Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther (2020) 5(1):108. doi: 10.1038/s41392-020-00216-5
129. Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy (2021) 17(9):2054–81. doi: 10.1080/15548627.2020.1810918
130. Mancardi D, Mezzanotte M, Arrigo E, Barinotti A, Roetto A. Iron overload, oxidative stress, and ferroptosis in the failing heart and liver. Antioxid (Basel) (2021) 10(12):1864. doi: 10.3390/antiox10121864
131. Park E, Chung SW. Ros-mediated autophagy increases intracellular iron levels and ferroptosis by ferritin and transferrin receptor regulation. Cell Death Dis (2019) 10(11):822. doi: 10.1038/s41419-019-2064-5
132. Liu J, Xia X, Huang P. Xct: a critical molecule that links cancer metabolism to redox signaling. Mol Ther (2020) 28(11):2358–66. doi: 10.1016/j.ymthe.2020.08.021
133. Sha R, Xu Y, Yuan C, Sheng X, Wu Z, Peng J, et al. Predictive and prognostic impact of ferroptosis-related genes Acsl4 and Gpx4 on breast cancer treated with neoadjuvant chemotherapy. EBioMedicine (2021) 71:103560. doi: 10.1016/j.ebiom.2021.103560
134. Zhang Y, Swanda RV, Nie L, Liu X, Wang C, Lee H, et al. Mtorc1 couples Cyst(E)Ine availability with Gpx4 protein synthesis and ferroptosis regulation. Nat Commun (2021) 12(1):1589. doi: 10.1038/s41467-021-21841-w
135. Wu H, Liu A. Long non-coding rna Neat1 regulates ferroptosis sensitivity in non-Small-Cell lung cancer. J Int Med Res (2021) 49(3):300060521996183. doi: 10.1177/0300060521996183
136. Zhao L, Zhou X, Xie F, Zhang L, Yan H, Huang J, et al. Ferroptosis in cancer and cancer immunotherapy. Cancer Commun (Lond) (2022) 42(2):88–116. doi: 10.1002/cac2.12250
137. Hong T, Lei G, Chen X, Li H, Zhang X, Wu N, et al. Parp inhibition promotes ferroptosis Via repressing Slc7a11 and synergizes with ferroptosis inducers in brca-proficient ovarian cancer. Redox Biol (2021) 42:101928. doi: 10.1016/j.redox.2021.101928
138. Ouyang S, Li H, Lou L, Huang Q, Zhang Z, Mo J, et al. Inhibition of Stat3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol (2022) 52:102317. doi: 10.1016/j.redox.2022.102317
139. Wen Q, Liu J, Kang R, Zhou B, Tang D. The release and activity of Hmgb1 in ferroptosis. Biochem Biophys Res Commun (2019) 510(2):278–83. doi: 10.1016/j.bbrc.2019.01.090
140. Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, et al. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization Via release and uptake of oncogenic kras protein. Autophagy (2020) 16(11):2069–83. doi: 10.1080/15548627.2020.1714209
141. He GN, Bao NR, Wang S, Xi M, Zhang TH, Chen FS. Ketamine induces ferroptosis of liver cancer cells by targeting lncrna Pvt1/Mir-214-3p/Gpx4. Drug Des Devel Ther (2021) 15:3965–78. doi: 10.2147/DDDT.S332847
142. Bai T, Liang R, Zhu R, Wang W, Zhou L, Sun Y. Microrna-214-3p enhances erastin-induced ferroptosis by targeting Atf4 in hepatoma cells. J Cell Physiol (2020) 235(7-8):5637–48. doi: 10.1002/jcp.29496
143. Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, et al. Lncrna Gabpb1-As1 and Gabpb1 regulate oxidative stress during erastin-induced ferroptosis in Hepg2 hepatocellular carcinoma cells. Sci Rep (2019) 9(1):16185. doi: 10.1038/s41598-019-52837-8
144. Yuan J, Liu Z, Song R. Antisense lncrna as-Slc7a11 suppresses epithelial ovarian cancer progression mainly by targeting Slc7a11. Pharmazie (2017) 72(7):402–7. doi: 10.1691/ph.2017.7449
145. Zhang Y, Guo S, Wang S, Li X, Hou D, Li H, et al. Lncrna Oip5-As1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through mir-128-3p/Slc7a11 signaling. Ecotoxicol Environ Saf (2021) 220:112376. doi: 10.1016/j.ecoenv.2021.112376
146. Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, et al. Lncrna Rp11-89 facilitates tumorigenesis and ferroptosis resistance through Prom2-activated iron export by sponging mir-129-5p in bladder cancer. Cell Death Dis (2021) 12(11):1043. doi: 10.1038/s41419-021-04296-1
147. Wang Z, Chen X, Liu N, Shi Y, Liu Y, Ouyang L, et al. A nuclear long non-coding rna Linc00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol Ther (2021) 29(1):263–74. doi: 10.1016/j.ymthe.2020.09.024
148. Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th Edition)(1). Autophagy (2021) 17(1):1–382. doi: 10.1080/15548627.2020.1797280
149. Liu Y, Shoji-Kawata S, Sumpter RM Jr., Wei Y, Ginet V, Zhang L, et al. Autosis is a Na+,K+-Atpase-Regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA (2013) 110(51):20364–71. doi: 10.1073/pnas.1319661110
150. Nah J, Zablocki D, Sadoshima J. Autosis: a new target to prevent cell death. JACC Basic Transl Sci (2020) 5(8):857–69. doi: 10.1016/j.jacbts.2020.04.014
151. Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer (2020) 19(1):12. doi: 10.1186/s12943-020-1138-4
152. Xiang H, Zhang J, Lin C, Zhang L, Liu B, Ouyang L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm Sin B (2020) 10(4):569–81. doi: 10.1016/j.apsb.2019.10.003
153. Papinski D, Kraft C. Regulation of autophagy by signaling through the Atg1/Ulk1 complex. J Mol Biol (2016) 428(9 Pt A):1725–41. doi: 10.1016/j.jmb.2016.03.030
154. Wong PM, Puente C, Ganley IG, Jiang X. The Ulk1 complex: sensing nutrient signals for autophagy activation. Autophagy (2013) 9(2):124–37. doi: 10.4161/auto.23323
155. Kim J, Kundu M, Viollet B, Guan KL. Ampk and mtor regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol (2011) 13(2):132–41. doi: 10.1038/ncb2152
156. Fernandez AF, Liu Y, Ginet V, Shi M, Nah J, Zou Z, et al. Interaction between the autophagy protein beclin 1 and Na+,K+-atpase during starvation, exercise, and ischemia. JCI Insight (2020) 5(1):e133282. doi: 10.1172/jci.insight.133282
157. Linder B, Kogel D. Autophagy in cancer cell death. Biol (Basel) (2019) 8(4):82. doi: 10.3390/biology8040082
158. Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell (2006) 10(1):51–64. doi: 10.1016/j.ccr.2006.06.001
159. Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, et al. Ampk-mediated Becn1 phosphorylation promotes ferroptosis by directly blocking system X(C)(-) activity. Curr Biol (2018) 28(15):2388–99.e5. doi: 10.1016/j.cub.2018.05.094
160. Liu Y, Wang Y, Liu J, Kang R, Tang D. Interplay between mtor and Gpx4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther (2021) 28(1-2):55–63. doi: 10.1038/s41417-020-0182-y
161. Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM, et al. Microenvironmental autophagy promotes tumour growth. Nature (2017) 541(7637):417–20. doi: 10.1038/nature20815
162. Zhao W, Lin X, Han H, Zhang H, Li X, Jiang C, et al. Long noncoding rna H19 contributes to the proliferation and autophagy of glioma cells through Mtor/Ulk1 pathway. Neuroreport (2021) 32(5):352–8. doi: 10.1097/WNR.0000000000001602
163. Wu ZR, Yan L, Liu YT, Cao L, Guo YH, Zhang Y, et al. Inhibition of Mtorc1 by lncrna H19 Via disrupting 4e-Bp1/Raptor interaction in pituitary tumours. Nat Commun (2018) 9(1):4624. doi: 10.1038/s41467-018-06853-3
164. Yang Y, Chen D, Liu H, Yang K. Increased expression of lncrna Casc9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis Via the Akt/Mtor pathway in oral squamous cell carcinoma. Cell Death Dis (2019) 10(2):41. doi: 10.1038/s41419-018-1280-8
165. Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, et al. Stat3-induced lncrna haglros overexpression contributes to the malignant progression of gastric cancer cells Via mtor signal-mediated inhibition of autophagy. Mol Cancer (2018) 17(1):6. doi: 10.1186/s12943-017-0756-y
166. Wang Y, Zhang M, Wang Z, Guo W, Yang D. Myc-binding lncrna Epic1 promotes akt-Mtorc1 signaling and rapamycin resistance in breast and ovarian cancer. Mol Carcinog (2020) 59(10):1188–98. doi: 10.1002/mc.23248
167. Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated tca cycle proteins. Science (2022) 375(6586):1254–61. doi: 10.1126/science.abf0529
168. Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat Chem Biol (2019) 15(7):681–9. doi: 10.1038/s41589-019-0291-9
169. Xiao C, Yang L, Jin L, Lin W, Zhang F, Huang S, et al. Prognostic and immunological role of cuproptosis-related protein Fdx1 in pan-cancer. Front Genet (2022) 13:962028. doi: 10.3389/fgene.2022.962028
170. Solmonson A, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem (2018) 293(20):7522–30. doi: 10.1074/jbc.TM117.000259
171. Tang Q, Guo Y, Meng L, Chen X. Chemical tagging of protein lipoylation. Angew Chem Int Ed Engl (2021) 60(8):4028–33. doi: 10.1002/anie.202010981
172. Zhang L, Shao J, Tan SW, Ye HP, Shan XY. Association between serum Copper/Zinc ratio and lung cancer: a systematic review with meta-analysis. J Trace Elem Med Biol (2022) 74:127061. doi: 10.1016/j.jtemb.2022.127061
173. Fang AP, Chen PY, Wang XY, Liu ZY, Zhang DM, Luo Y, et al. Serum copper and zinc levels at diagnosis and hepatocellular carcinoma survival in the guangdong liver cancer cohort. Int J Cancer (2019) 144(11):2823–32. doi: 10.1002/ijc.31991
174. Baszuk P, Marciniak W, Derkacz R, Jakubowska A, Cybulski C, Gronwald J, et al. Blood copper levels and the occurrence of colorectal cancer in Poland. Biomedicines (2021) 9(11):1628. doi: 10.3390/biomedicines9111628
175. Yucel I, Arpaci F, Ozet A, Doner B, Karayilanoglu T, Sayar A, et al. Serum copper and zinc levels and Copper/Zinc ratio in patients with breast cancer. Biol Trace Elem Res (1994) 40(1):31–8. doi: 10.1007/BF02916818
176. Zhang M, Shi M, Zhao Y. Association between serum copper levels and cervical cancer risk: a meta-analysis. Biosci Rep (2018) 38(4):BSR20180161. doi: 10.1042/BSR20180161
177. Baharvand M, Manifar S, Akkafan R, Mortazavi H, Sabour S. Serum levels of ferritin, copper, and zinc in patients with oral cancer. BioMed J (2014) 37(5):331–6. doi: 10.4103/2319-4170.132888
178. Michalczyk K, Cymbaluk-Ploska A. The role of zinc and copper in gynecological malignancies. Nutrients (2020) 12(12):3732. doi: 10.3390/nu12123732
179. Cui L, Gouw AM, LaGory EL, Guo S, Attarwala N, Tang Y, et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat Biotechnol (2021) 39(3):357–67. doi: 10.1038/s41587-020-0707-9
180. Ramchandani D, Berisa M, Tavarez DA, Li Z, Miele M, Bai Y, et al. Copper depletion modulates mitochondrial oxidative phosphorylation to impair triple negative breast cancer metastasis. Nat Commun (2021) 12(1):7311. doi: 10.1038/s41467-021-27559-z
181. Guo J, Cheng J, Zheng N, Zhang X, Dai X, Zhang L, et al. Copper promotes tumorigenesis by activating the Pdk1-akt oncogenic pathway in a copper transporter 1 dependent manner. Adv Sci (Weinh) (2021) 8(18):e2004303. doi: 10.1002/advs.202004303
182. Yan C, Niu Y, Ma L, Tian L, Ma J. System analysis based on the cuproptosis-related genes identifies Lipt1 as a novel therapy target for liver hepatocellular carcinoma. J Transl Med (2022) 20(1):452. doi: 10.1186/s12967-022-03630-1
183. Yun Y, Wang Y, Yang E, Jing X. Cuproptosis-related gene - Slc31a1, Fdx1 and Atp7b - polymorphisms are associated with risk of lung cancer. Pharmgenomics Pers Med (2022) 15:733–42. doi: 10.2147/PGPM.S372824
184. Mo X, Hu D, Yang P, Li Y, Bashir S, Nai A, et al. A novel cuproptosis-related prognostic lncrna signature and lncrna Mir31hg/Mir-193a-3p/Tnfrsf21 regulatory axis in lung adenocarcinoma. Front Oncol (2022) 12:927706. doi: 10.3389/fonc.2022.927706
185. Liu B, Liu Z, Feng C, Li C, Zhang H, Li Z, et al. Identification of cuproptosis-related lncrna prognostic signature for osteosarcoma. Front Endocrinol (Lausanne) (2022) 13:987942. doi: 10.3389/fendo.2022.987942
186. Yang M, Zheng H, Xu K, Yuan Q, Aihaiti Y, Cai Y, et al. A novel signature to guide osteosarcoma prognosis and immune microenvironment: cuproptosis-related lncrna. Front Immunol (2022) 13:919231. doi: 10.3389/fimmu.2022.919231
187. Jiang ZR, Yang LH, Jin LZ, Yi LM, Bing PP, Zhou J, et al. Identification of novel cuproptosis-related lncrna signatures to predict the prognosis and immune microenvironment of breast cancer patients. Front Oncol (2022) 12:988680. doi: 10.3389/fonc.2022.988680
188. Jiang B, Zhu H, Feng W, Wan Z, Qi X, He R, et al. Database mining detected a cuproptosis-related prognostic signature and a related regulatory axis in breast cancer. Dis Markers (2022) 2022:9004830. doi: 10.1155/2022/9004830
189. Liu X, Zhou L, Gao M, Dong S, Hu Y, Hu C. Signature of seven cuproptosis-related lncrnas as a novel biomarker to predict prognosis and therapeutic response in cervical cancer. Front Genet (2022) 13:989646. doi: 10.3389/fgene.2022.989646
190. Yang L, Yu J, Tao L, Huang H, Gao Y, Yao J, et al. Cuproptosis-related lncrnas are biomarkers of prognosis and immune microenvironment in head and neck squamous cell carcinoma. Front Genet (2022) 13:947551. doi: 10.3389/fgene.2022.947551
191. Feng A, He L, Chen T, Xu M. A novel cuproptosis-related lncrna nomogram to improve the prognosis prediction of gastric cancer. Front Oncol (2022) 12:957966. doi: 10.3389/fonc.2022.957966
192. Chen S, Liu P, Zhao L, Han P, Liu J, Yang H, et al. A novel cuproptosis-related prognostic lncrna signature for predicting immune and drug therapy response in hepatocellular carcinoma. Front Immunol (2022) 13:954653. doi: 10.3389/fimmu.2022.954653
193. Hou D, Tan JN, Zhou SN, Yang X, Zhang ZH, Zhong GY, et al. A novel prognostic signature based on cuproptosis-related lncrna mining in colorectal cancer. Front Genet (2022) 13:969845. doi: 10.3389/fgene.2022.969845
194. Qiu Z, He L, Yu F, Lv H, Zhou Y. Lncrna Fam13a-As1 regulates proliferation and apoptosis of cervical cancer cells by targeting mirna-205-3p/Ddi2 axis. J Oncol (2022) 2022:8411919. doi: 10.1155/2022/8411919
195. Fan Y, Sheng W, Meng Y, Cao Y, Li R. Lncrna Ptenp1 inhibits cervical cancer progression by suppressing mir-106b. Artif Cells Nanomed Biotechnol (2020) 48(1):393–407. doi: 10.1080/21691401.2019.1709852
196. Liu H, Liu L, Liu Q, He F, Zhu H. Lncrna hoxd-As1 affects proliferation and apoptosis of cervical cancer cells by promoting Frrs1 expression Via transcription factor Elf1. Cell Cycle (2022) 21(4):416–26. doi: 10.1080/15384101.2021.2020962
197. Hashemi M, Moosavi MS, Abed HM, Dehghani M, Aalipour M, Heydari EA, et al. Long non-coding rna (Lncrna) H19 in human cancer: from proliferation and metastasis to therapy. Pharmacol Res (2022) 184:106418. doi: 10.1016/j.phrs.2022.106418
198. Muller V, Oliveira-Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncrna H19/Mir-675 and lncrna Neat1/Mir-204 in breast cancer. Mol Oncol (2019) 13(5):1137–49. doi: 10.1002/1878-0261.12472
Keywords: lncRNAs, apoptosis, necroptosis, pyroptosis, NEtosis, entosis, ferroptosis, cuproptosis
Citation: Wu Y, Wen X, Xia Y, Yu X and Lou Y (2023) LncRNAs and regulated cell death in tumor cells. Front. Oncol. 13:1170336. doi: 10.3389/fonc.2023.1170336
Received: 21 February 2023; Accepted: 17 May 2023;
Published: 29 May 2023.
Edited by:
Erik Arnold Bey, Wood Hudson Cancer Research Laboratory, United StatesCopyright © 2023 Wu, Wen, Xia, Yu and Lou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhui Lou, bG91eWhAcWR1LmVkdS5jbg==
 Yingying Wu
Yingying Wu Xiaoling Wen
Xiaoling Wen Yufang Xia
Yufang Xia Yanhui Lou
Yanhui Lou