- 1Department of Pathology, The Affiliated Huai’an No. 1 People’s Hospital of Nanjing Medical University, Huai’an, Jiangsu, China
- 2Key Laboratory of Epigenetics and Oncology, The Research Center for Preclinical Medicine, Southwest Medical University, Luzhou, Sichuan, China
- 3Department of Pathology, Taizhou People's Hospital of Nanjing University of Chinese Medicine, Jiangsu, China
- 4Department of Chemistry and Chemical Engineering, Hunan Institute of Science and Technology, Yueyang, Hunan, China
- 5NHC Key Laboratory of Cancer Proteomics, Department of Oncology, Xiangya Hospital, Central South University, Changsha, Hunan, China
Introduction: In the world, the incidence of breast cancer has surpassed that of lung cancer, and it has become the first malignant tumor among women. Triple-negative breast cancer (TNBC) shows an extremely heterogeneous malignancy toward high recurrence, metastasis, and mortality, but there is a lack of effective targeted therapy. It is urgent to develop novel molecular targets in the occurrence and therapeutics for TNBC, and novel therapeutic strategies to block the recurrence and metastasis of TNBC.
Methods: In this study, CTSL (cathepsin L) expression in tissues and adjacent tissues of TNBC patients was monitored by immunohistochemistry and western blots. The correlations between CTSL expressions and clinicopathological characteristics in the patient tissues for TNBC were analyzed. Cell proliferation, migration, and invasion assay were also performed when over-expressed or knocked-down CTSL.
Results: We found that the level of CTSL in TNBC is significantly higher than that in the matched adjacent tissues, and associated with differentiated degree, TNM Stage, tumor size, and lymph node metastatic status in TNBC patients. The high level of CTSL was correlated with a short RFS (p<0.001), OS (p<0.001), DMFS (p<0.001), PPS (p= 0.0025) in breast cancer from online databases; while in breast cancer with lymph node-positive, high level of CTSL was correlated with a short DMFS (p<0.001) and RFS (p<0.001). Moreover, in vitro experiments showed that CTSL overexpression promotes the abilities for proliferation, migration, and invasion in MCF-7 and MDA-MB-231 cell lines, while knocking-down CTSL decreases its characteristics in MDA-MB-231 cell lines.
Conclusion: CTSL might involve into the regulation of the proliferation, invasion, and metastasis of TNBC. Thus, CTSL would be a novel, potential therapeutic, and prognostic target of TNBC.
1 Introduction
Breast cancer (BC) is the most common malignancy in women worldwide. Relevant data show that there are about 2.3 million new cases of BC, and about 685,000 deaths worldwide in 2020 (1, 2). By 2040, the burden of breast cancer predicts to increase to more than 3 million new cases and more than 1 million deaths each year. While triple-negative BC (TNBC), a specific subtype of BC, accounts for 15% ~ 20% of all pathological types of breast cancer (3). This special subtype of BC is defined by the lack of the human receptors’ expression for estrogen (ER), and progesterone (PR) and the lack of amplification or expression for epidermal growth factor receptor 2 (HER2).
TNBC is the most difficult type of BC to treat at present, with the worst prognosis, the highest mortality, and the most prone to recurrence and metastasis. The treatment is almost only chemotherapy. Precision therapy is an important development direction for the future clinical treatment of TNBC patients. Although some progress has been made, there are still many difficulties. There is an urgent need to develop novel molecular targets in the occurrence and therapeutics for TNBC, and novel diagnostic and therapeutic strategies to block the recurrence and metastasis of TNBC.
Cathepsin L (CTSL, OMIM 116880), a member of the peptidase C1 family, is a lysosomal cysteine protease, which forms a dimer consisting of disulfide-linked heavy and light chains (4, 5). The cytogenetic location of the CTSL gene was mapped 9q21.33, which encodes a 333 amino acid length and 37564Da. In addition to playing important roles in intracellular catabolism, CTSL protein is involved in several pathological processes, such as myofibrillary necrosis (6, 7), and tumor progression/tumorigenesis (8–10), such as glioma cell migration and invasion by ionizing radiation-induced or X-ray-reduced manner (11, 12). CTSL was reported to proteolytically cleave the S1 subunit of spike protein on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or glycoprotein of Ebola virus (EboV) (13) for viral invasion into host cells (14–16). Gingival overgrowth and middle east respiratory syndrome (MERS) were also reported to associate with CTSL (17–19). CTSL is highly expressed in many malignant tumors, including gastrointestinal stromal tumors and metastatic bone tumors, identifying it as a new diagnostic or prognostic marker (20–22). Methyltransferase-like 3 (METTL3) enhanced the stability of CTSL mRNA m6A-IGF2BP2-dependent manners and promoted metastasis in cervical cancer cells (23). CTSL was reported to involve in the proliferation and invasion of breast cancer cells (24). In addition, studies in the breast cancer mouse model of PyMT have presented a strong influence of the CTSL/CTSB on lung metastasis and shown distinct effects on proteome composition in the metastatic lungs (25). However, it is unclear whether the regulation of CTSL expression is associated with metastasis, aggressiveness, and poor prognosis in TNBC patients.
In this research, we tested the CTSL role in TNBC and the correlations between CTSL expression and TNBC clinicopathology. The influence of CTSL on the biological function of the TNBC cell lines was analyzed by overexpressing and interfering with CTSL expression.
2 Materials and methods
2.1 Patient information and samples of TNBC
One hundred cases of paraffin-embedded breast cancer and paracancer tissues and thirty cases of fresh breast cancer and paracancer tissues were sampled from TNBC patients undergoing surgery in the Huai’an No. 1 People’s Hospital. The lymph node metastasis and received regional lymph node dissection were confirmed or suspected. All patients were diagnosed with TNBC by Immunohistochemistry (In the triple-negative breast cancer tissues, immunohistochemical staining of ER and PR in the nucleus and HER2 in the cell membrane were showed blue, which was interpreted as negative) and signed informed consent before surgery. The study protocol was approved by the Ethics Committee of Huai’an No.1 People’s Hospital.
2.2 Immunohistochemistry
We conducted immunohistochemistry (IHC) staining analysis to measure the protein expression of CTSL in TNBC tissues and adjacent normal breast tissues according to the standard immunoperoxidase staining procedure (26, 27). The Human TNBC tissues and adjacent normal breast tissues were embedded in paraffin and cut into 3mm thick slices, then placed on pre-coated slides. Placed them in an oven at 60°C for 1 h. Dewaxed and rehydrated: xylened for 3min, changed the solution again for 3min, then 50% xylene for 5min, 100% alcohol I-III for 1min each, 95% alcohol for 1 min, 75% alcohol for 1 min, ddH2O washed twice; Antigen retrieval was performed by heating the sample to 100°C in a 10 mM citric acid buffer; 1× PBS washed twice; ddH2O washed 3 times; 3%H2O2 incubated at room temperature for 10 min. Tissue sections were sealed with 5% BSA-1×PBST for 30min, incubated with CTSL antibody (1:100, Catalog No. ABIN1172740; Aachen, Germany), primary antibody at 4°C overnight, washed with 1×PBS twice, and incubated with secondary antibody at room temperature for 2h. The ABC (avidin-biotin-peroxidase complex) was incubated at room temperature for 2h, 1× PBS washed three times. The immunoreactivity was observed with DAB (diaminobenzidine), followed by reverse staining with hematoxylin. Dehydration and preservation: 75% ethanol for 30s; 100% ethanol for 30s; xylene for 30s; After sealed with neutral gum, covered with glass slides. The assessment was carried out by two independent pathologists in a double-blind method. Staining intensity and percentage of staining cells were scored respectively. The staining intensity score was 0 point: negative; 1 point: weak; 2 points: moderate; 3 points: strong. Staining cell percentage score 0: no cell staining; 1 point: staining cells <25%; 2 points: 25-50% cell staining; 3 points: 51-75% cells were stained; 4 points: >75% of the cells were stained. The final score for each tissue specimen is determined by multiplying the percentage of stained cells score by the staining strength score. The score of 0 was classified as negative, 1-4 as weak positive, 5-8 as positive, and 9-12 as strong positive. To distinguish the cut-off point of the CTSL expression level, receiver operating characteristic (ROC) curve analysis was used, and the score closest to the maximum Yoden index was used as the optimal cut-off value (27). Low CTSL expression was defined as samples with scores lower than cut-off values (score <6.3), while high CTSL expression was defined as samples with scores higher than cut-off values (score >6.3).
2.3 Western blotting
Firstly, the protein was extracted by 1×EBC lysate and 2xSDS, and the protein expression level was analyzed by Western Blot: 10 μl denatured protein samples were added to each well, electrophoresis with 10% SDS-PAGE, and protein separation completely was performed with 1 x running buffer at 100 V (28–30). The protein was transferred to the PVDF membrane at 100V for about 90 min. Then sealed the PVDF membrane at room temperature with 5% milk for 2h, and incubated the primary antibody with 2% milk at 4°C overnight. Primary antibodies included CTSL antibody (1:10000, Abcam Catalog No. ab200738), β-actin antibody (1:5000 Cell Signaling Technology), and HSP70 antibody (1:2000 Sigma USA). On the second day, PVDF (polyvinylidene fluoride) membrane was washed three times with 1×TBST, and the secondary antibody was incubated at room temperature for 1-2h. The secondary antibodies included anti-rabbit secondary antibodies (1:2000) and anti-mouse secondary antibodies (1:2000) which were purchased from Sigma (USA). The PVDF membrane was then cleaned with TBST. An imaging scanner was used to detect the strength of each cell membrane strip after TBST washing three times. Each experiment was repeated three times.
2.4 Online analysis for survivals
The Kaplan–Meier analysis for survivals was used to detect the prognosis of breast cancer patients including TNBC using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service&start=1) (31, 32).
2.5 CTSL overexpression and knocking-down assays
MCF-7 cells and MDA-MB-231 cells were purchased from the American Type Culture Collection (ATCC), DMEM from Thermo Fisher Scientific, and Fetal Bovine Serum (FBS) from Pan Biotech, Germany. MDA-MB-231 and MCF-7 breast cancer cell lines were cultured in an incubator containing 5% carbon dioxide at 37°C. When the cell density reached 60%, the 900 ng CTSL overexpression plasmid was transfected with opti-MEM medium and Lipofectamine 3000, and the control group was transfected with a 900 ng empty vector. Then the transfected cells were cultured for another 24h.
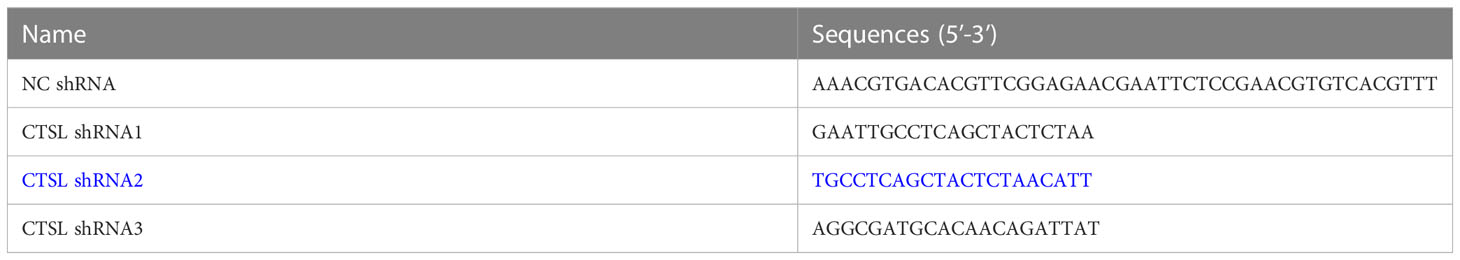
The virus packaging process includes: designing CTSL target sequences (Table 1), constructing CTSL interference vector (pLV-hU6-CTSL shRNA2 (blue in Table 1, human):-hef1a-mNeongreen-P2A-Puro), 293T cells are often used for lentivirus packaging production in experiments because they can stably and efficiently package viruses. Then 293T cells were transfected, and the supernatant of the cells was collected, the viruses were harvested, the impurities were removed by centrifugation, the viruses were concentrated and purified, and the virus quality was tested. The MDA-MB-231 cells in good condition were seeded into a 24-well plate and the cells were planned at a density of 5x104 cells/well. Then 500 μl DMEM medium is added to each well to ensure a fusion rate of 30-40% the next day. On the second day, the culture medium was replaced with DMEM containing 8µg/ml Polybrene, then the virus (MOI=20) was added to infect the MDA-MB-231 cells. The cell status was observed 12-24h after the virus infection. If the cells were in good condition, the DMEM medium was replaced within 24h. After 48h of virus infection, cells were observed under a fluorescence microscope to determine the efficiency of lentivirus infection.
2.6 Cell proliferation, migration, and invasion assays
6-well plates were applied to culture cell lines MDA-MB-231 and MCF-7 were cultured. When the cell density was about 60%, The cells were cleaned with PBS twice, digested with 0.25% trypsin, then added 1 ml serum-containing medium to stop digestion, centrifuged at 12000 rpm for 3 min, the supernatant was discarded, and 1ml serum-free medium was added to mix and counted with a bob-enumerating plate. On the CIM plate(monitoring cell growth), 13 μl serum and 17 μl serum-free medium, 100 μl cell suspension with a concentration of 2×104 cells/ml were added to the upper chamber, and 165μl serum-containing medium was added to the lower chamber. On the CIM plate (monitoring cell migration), 30 μl serum-free medium and 100μl cell suspension with a concentration of 2×104 cells/ml were added to the upper chamber, and 165 μl serum-containing medium was added to the lower chamber. CIM plate for monitoring cell invasion: First, apply glue (Matrigel matrix glue, purchased from BD Biosciences, Dilute with PBS at 1:40 ratio). After the upper chamber was gelled and the pores were dried for 1~2h, 30 μl serum-free medium and 100 μl cell suspension with a concentration of 2×104 cells/ml were added to the upper chamber. 165 μl medium with 10% FBS was added to the lower chamber (28). The experimental group and the control group were provided with three secondary Wells. The cell proliferation, migration, and invasion were monitored by the real-time cell analyzer (xCELLigence RTCA DP, Roche, Germany) (28, 33).
2.7 Statistical analysis
The data were analyzed by SPSS 20 and GraphPad Prism 9. Pairwise comparisons of normally distributed data were analyzed by Student’s t-test or for multi-group comparisons, one-way analysis of variance (ANOVA). Counting data were tested by chi-square test. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) are considered as statistical significances.
3 Results
3.1 CTSL expression in TNBC tissues is higher than that in the matched paracancerous tissues
To explore the expression of CTSL in TNBC, one hundred of Paraffin-embedded TNBC and paracancer tissues were collected for immunohistochemical, and thirty fresh TNBC tissues and the matched paracancer tissues were for western blotting analysis. The results showed high expression of CTSL in 22 cases of TNBC tissue, low expression in one case of TNBC tissue, and no significant difference in the expression of CTSL in six cases of TNBC tissues when compared with adjacent tissues. In one case, CTSL protein expression was not detected (Figures 1A, B). Therefore, the protein expression levels of CTSL were significantly higher in TNBC tissues than that in the matched adjacent tissues (Figure 1B, P<0.001). Meanwhile, the immunohistochemical results also showed that CTSL is more highly expressed in TNBC than that in the matched adjacent tissues (Figures 1C–E).
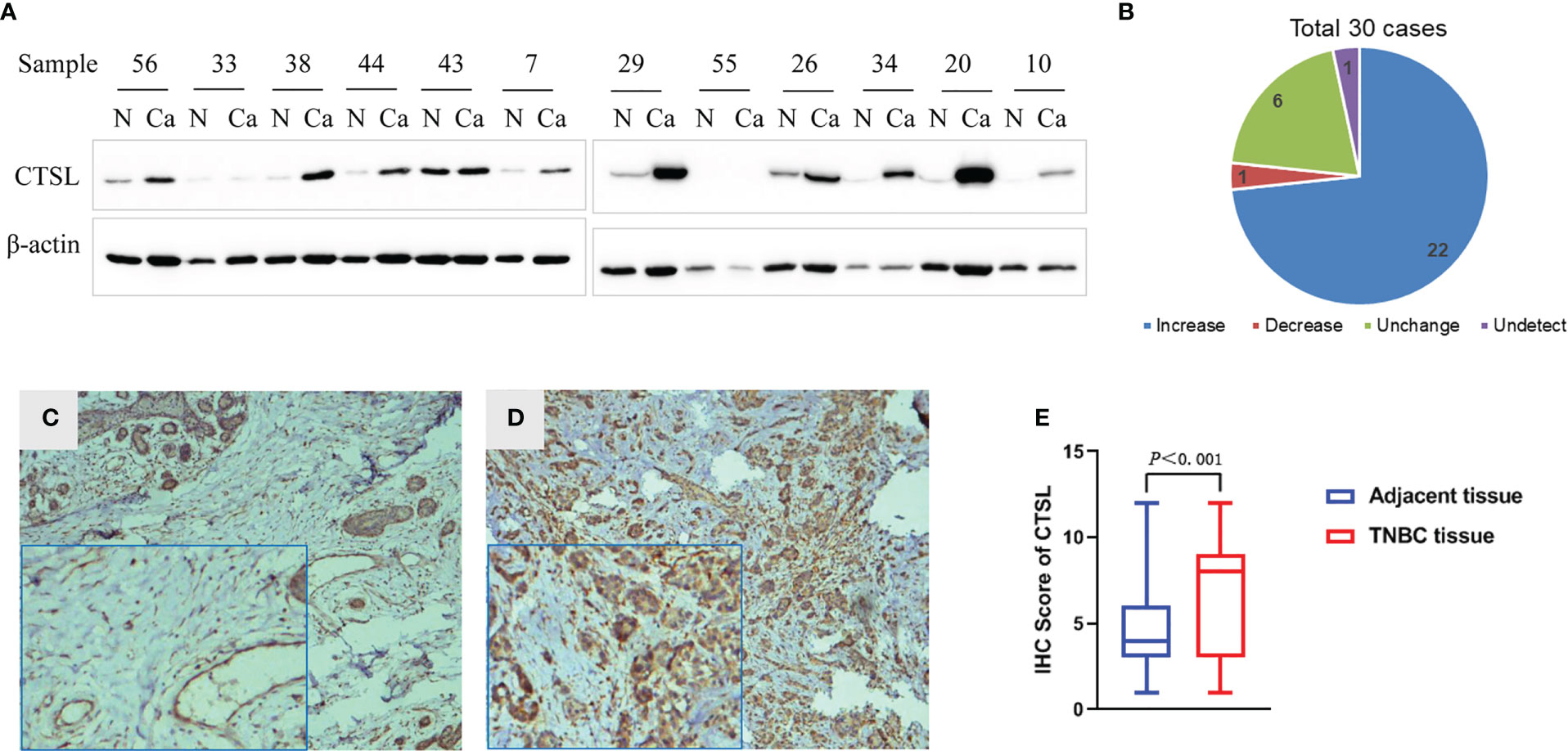
Figure 1 CTSL expression in TNBC by western blotting (WB) and immunohistochemistry (IHC). (A) Representative figures of CTSL expression in 30 cases of TNBC and the matched adjacent tissues by western blotting (WB), the expression of CTSL in TNBC tissues is mostly higher than that in the matched adjacent tissues. (B) Statistical analysis of CTSL expression in TNBC and the matched adjacent tissues of WB). (C) The CTSL expression is low in paracancer tissues than that of TNBC tissues (IHC 10×). (D) CTSL is more highly expressed in TNBC tissues than that of paracancer tissues (IHC 10×). Enlarged images are shown in the left corners of panels C and D, respectively. (E) Statistical analysis of CTSL expression in TNBC and the adjacent tissues (IHC).
3.2 Analysis of the correlations between CTSL expression and clinicopathological characteristics in patients for TNBC
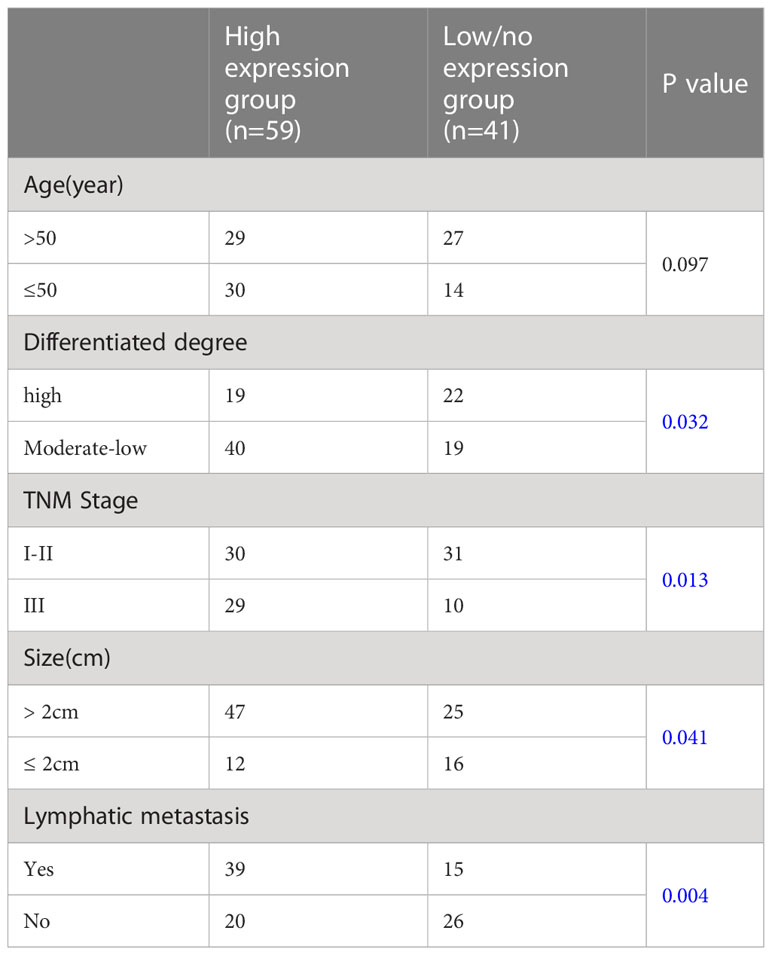
The information of one hundred patients with TNBC cancer was collected for analyzing the correlations between CTSL expression and clinicopathological data. In 100 cases of TNBC, 85% was positive for CTSL. We thus divided them into CTSL high expression group (n=59) and CTSL low/no expression group (n=41) based on the CTSL immunohistochemical score. Comparing the clinicopathological data, the results showed that the CTSL expression was associated with differentiated degree, tumor size, TNM stage, and lymph node metastasis in TNBC patients (Table 2, blue colors). Thus, these data demonstrated that CTSL might play clinicopathological characteristics roles including metastasis in TNBC patients.
3.3 CTSL expression is a poor prognostic marker for BC
We analyzed the correlations between CTSL expression and outcomes of survivals including recurrence-free (RFS), overall (OS), distant metastasis-free (DMFS), and post-progression (PPS) in BC by online databases. The results showed that a high level of CTSL is significantly correlated with a short RFS (Figure 2A, p<0.001), OS (Figure 2B, p<0.001), DMFS (Figure 2C, p<0.001), PPS (Figure 2D, p=0.0025) in breast cancer. Hence, CTSL expression could be a poor prognostic marker for survival in BC.
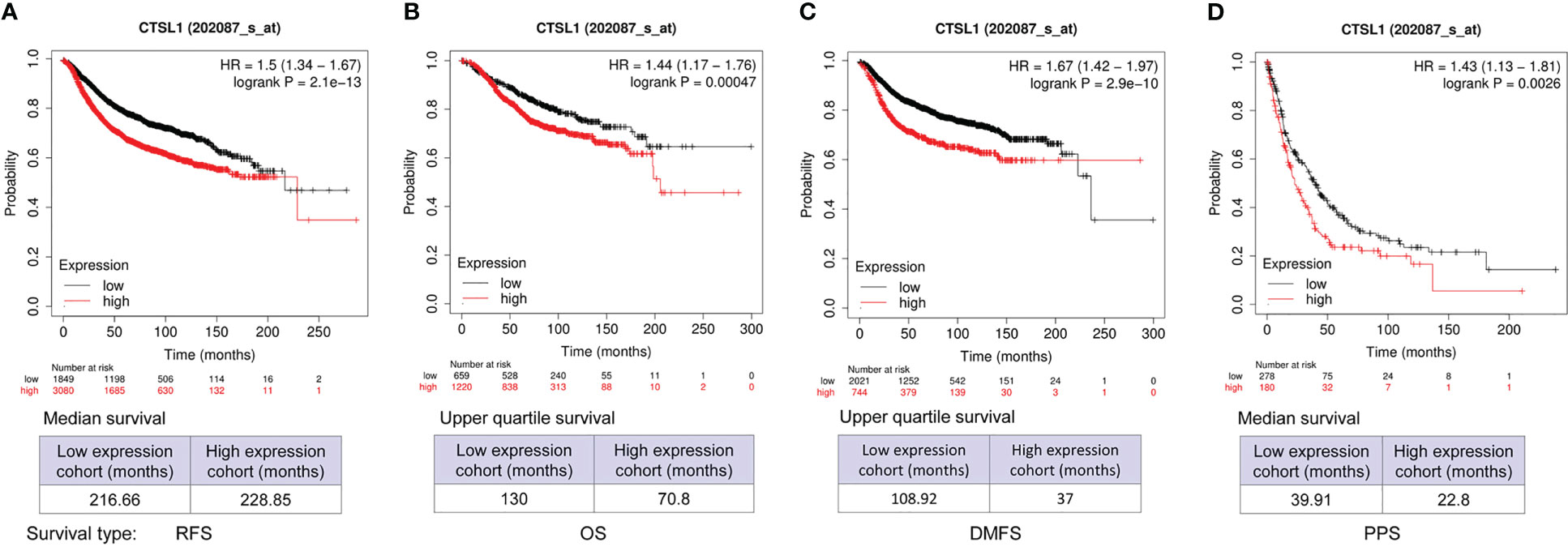
Figure 2 CTSL expression affects the survivals of BC patients for recurrence-free survival (RFS), overall survival (OS), and post-progression survival (PPS). (A) The clinical correlations between CTSL level and RFS (p<0.001). (B) The clinical correlations between CTSL level and OS (p<0.001). (C) The clinical correlations between CTSL level and DMS (p<0.001). (D) The correlations between CTSL level and PPS (p<0.001). The Kaplan–Meier analysis for survivals was used to detect the prognosis of breast cancer patients using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service&start=1).
3.4 Prognosis results of CTSL expression in TNBC
We further analyzed the correlations between CTSL expression and outcomes of survivals including RFS, OS, and DMFS in TNBC by Kaplan–Meier analysis. The results revealed that a high level of CTSL is correlated with a short RFS (Figure 3A, p=0.1), and OS (Figure 3B, p=0.11) in TNBC. While the results revealed that a high level of CTSL is correlated with a long DMFS (Figure 3C, p=0.071). Hence, CTSL expression might be a poor prognostic marker for RFS and OS in TNBC.
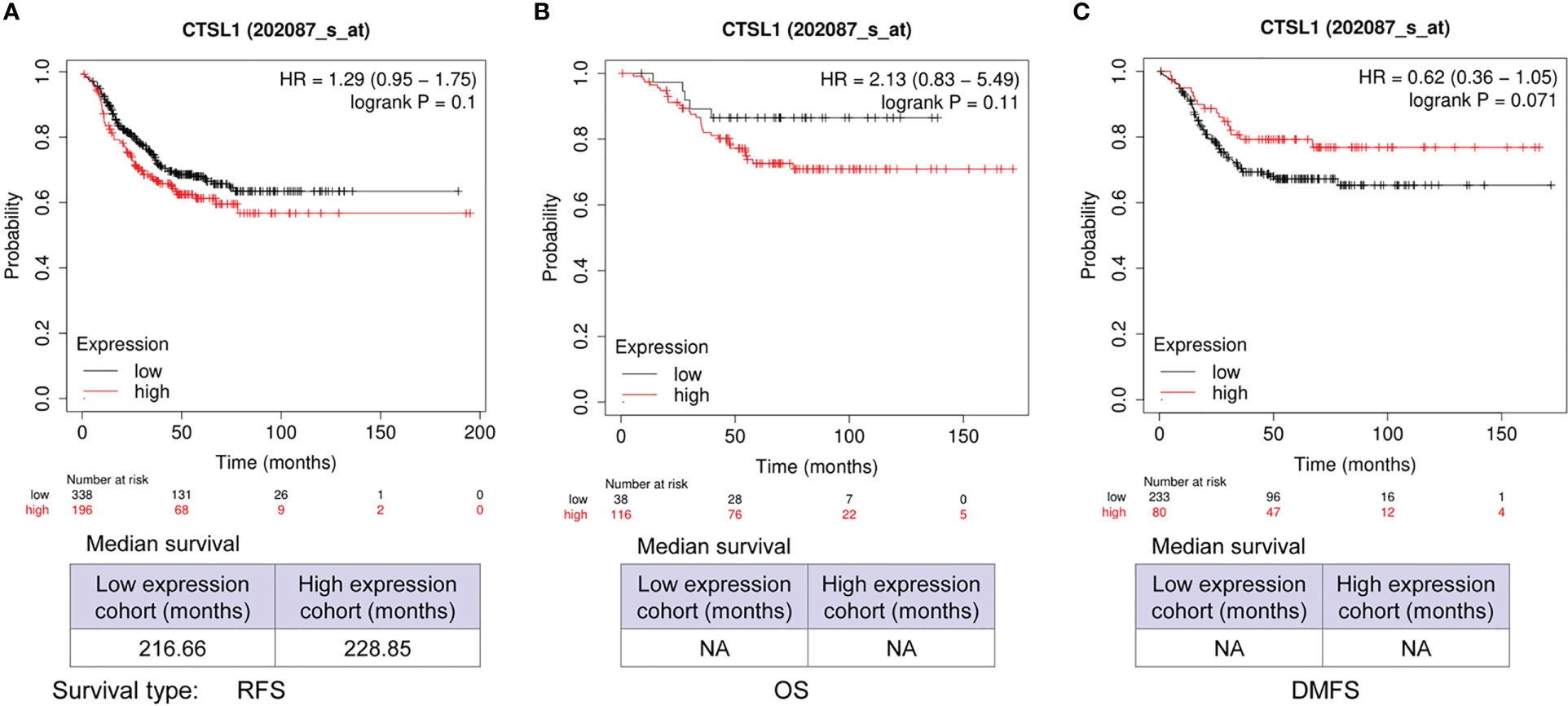
Figure 3 Prognosis analysis of CTSL expression in TNBC for recurrence-free survival (RFS), overall survival (OS), and post-progression survival (PPS). (A) The correlations between CTSL expression and RFS (p=0.1). (B) The correlations between CTSL expression and OS (p=0.11). (C) The clinical correlations between CTSL level and DMFS (p=0.071). The Kaplan–Meier analysis for survivals was used to detect the prognosis of TNBC patients using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service&start=1).
3.5 Prognosis results of CTSL expression in BC with lymph node-positive
We further analyzed the correlations between CTSL expression and outcomes of survivals of DMFS and RFS in breast cancer with Lymph node-positive. The results revealed that a high expression of CTSL is correlated with a short DMFS (Figure 4A, p<0.001) and RFS (Figure 4B, p<0.001). Hence, CTSL expression could be a poor prognostic marker in breast cancer with lymph node-positive.
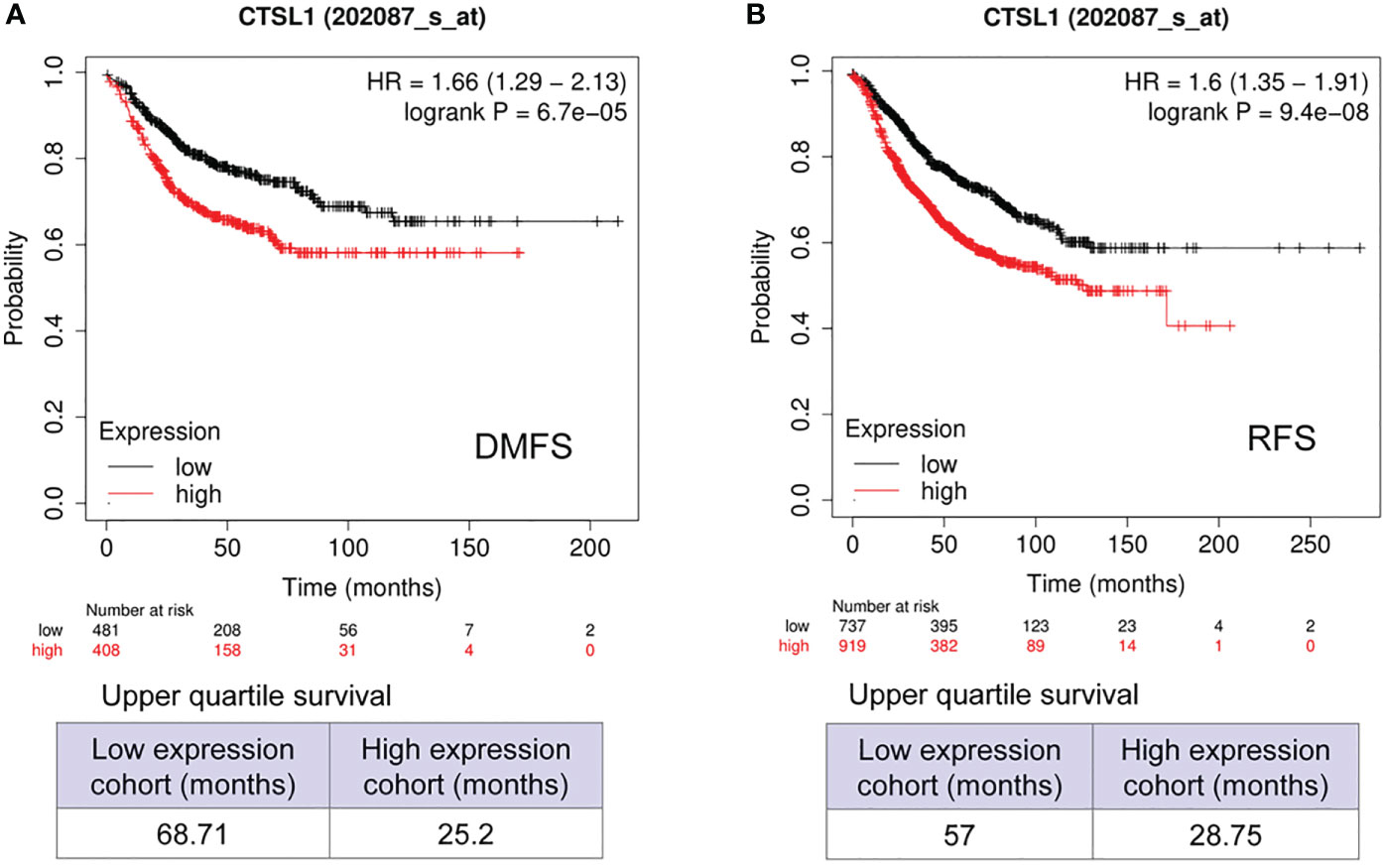
Figure 4 Prognosis analysis of CTSL expression in BC with Lymph node positive for distant metastasis-free survival (DMFS) and recurrence-free survival (RFS). (A) The correlations between CTSL level and DMFS (p<0.001). (B) The correlations between CTSL level and RFS (p<0.001). The Kaplan–Meier analysis for survivals was used to detect the prognosis of breast cancer patients using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service&start=1).
3.6 Overexpression of CTSL promotes cell proliferation, migration, and invasion of MDA-MB-231 and MCF-7
To further analyze the influence of CTSL expression on the behaviology of TNBC cells, we constructed a Flag-tagged CTSL overexpression vector, and transfected CTSL overexpression plasmid into MDA-MB-231 and MCF-7 respectively, which showed successful overexpression by western blotting with Flag antibody (Figures 5A, E). Then, we analyzed their proliferation, migration, and invasion capacities and the results revealed that overexpression of CTSL significantly enhanced the capacity of proliferation, migration, and invasion in MDA-MB-231 cell lines (Figures 5B–D) and MCF-7 cell lines (Figures 5F–H), respectively.
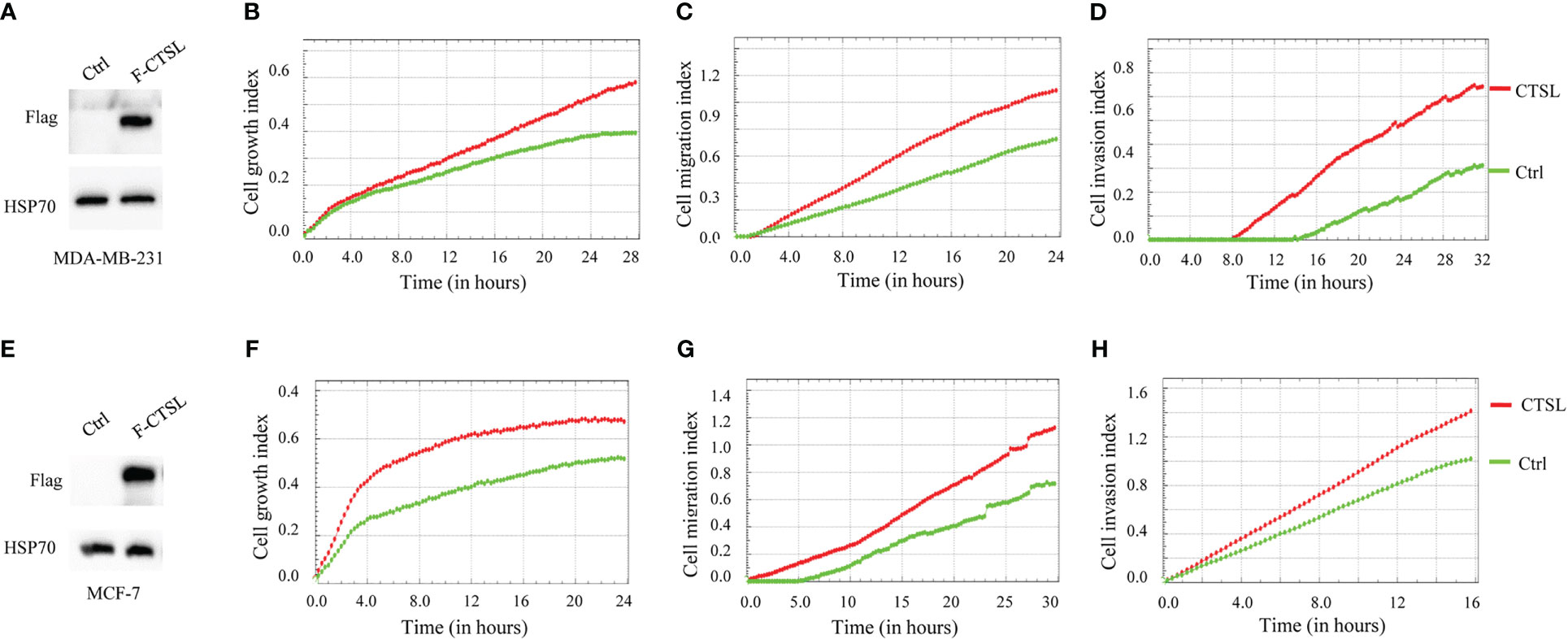
Figure 5 CTSL expression affects proliferation, migration, and invasion. (A) CTSL is successfully overexpressed in the indicated MDA-MB-231 cells. (B) Overexpression of CTSL increases the proliferation in the indicated MDA-MB-231 cells. (C) Overexpression of CTSL enhances the migration ability in MDA-MB-231 cells. (D) Overexpression of CTSL increases the invasion ability of MDA-MB-231 cells. (E) CTSL is successfully overexpressed in the indicated MCF-7 cells. (F) Overexpression of CTSL increased the proliferation in MCF-7 cells. (G) Overexpression of CTSL enhances the migration ability in MCF-7 cells. (H) Overexpression of CTSL enhances the invasion ability of MCF-7 cells. Flag antibody was used for the detection of CTSL overexpression. Breast cancer cell lines MDA-MB-231 and MCF-7 were used for CTSL overexpression and then to monitor the proliferation, migration, and invasion by the real-time cell analyzer (xCELLigence RTCA DP, Roche, Germany).
3.7 Downregulation of CTSL inhibits proliferation, migration, and invasion of MDA-MB-231 cell lines
On the other hand, we constructed three CTSL knocking-down vectors (Table 1, in blue color), and one of them (CTSL shRNA2) was knocked down well in TNBC cell lines MDA-MB-231 (Figure 6A). We then analyzed their proliferation and migration and invasion capacity. The results revealed that CTSL knocking-down could significantly inhibit the capacity of proliferation and migration and invasion in TNBC cancer cells MDA-MB-231 (Figures 6B–D).
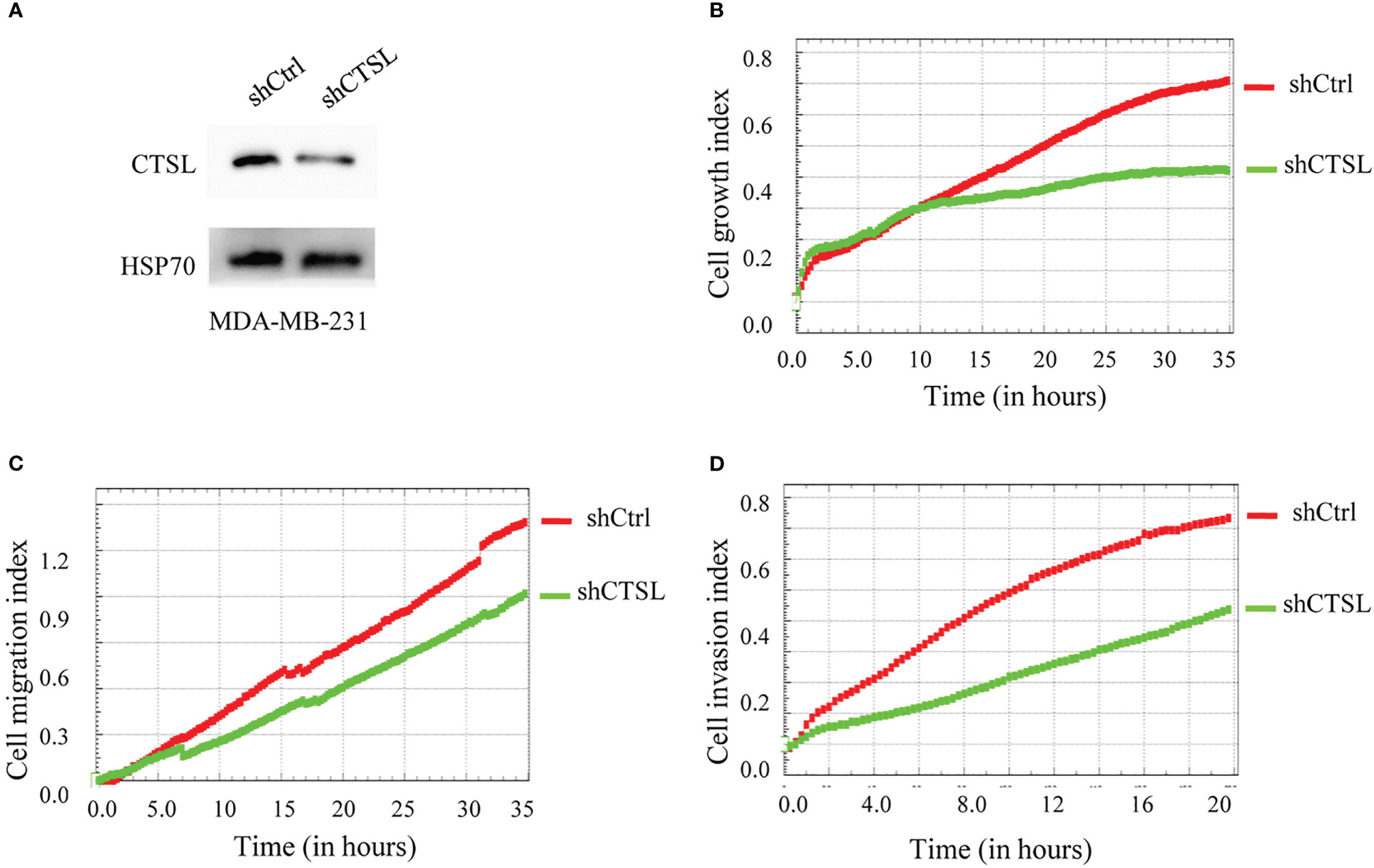
Figure 6 CTSL knocking-down affects the proliferation, migration, and invasion of MDA-MB-231 cells. (A) CTSL is successfully knockdown in MDA-MB-231 cells by western blots (CTSL shRNA2). (B): knockdown CTSL decreases the proliferation in MDA-MB-231 cells. (C) knockdown CTSL decreases the migration ability of MDA-MB-231 cells. (D) knockdown CTSL decreases the invasion ability in MDA-MB-231 cells. Breast cancer cell line MDA-MB-231 was used for CTSL knocking-down and then to monitor the proliferation, migration, and invasion by the real-time cell analyzer (xCELLigence RTCA DP, Roche, Germany).
4 Discussion
Proteases were reported to involve several stages of tumorigenesis and progression and act as prognostic markers (34–36). CTSL, a member of the papain superfamily of cysteine protease, is involved in the proliferation, invasion, and metastasis of various cancers. For example, relevant studies have found the expression of CTSL level in glioblastoma multiforme (GBM) tissues is higher than that in healthy brain tissues (37), which is a proteolytic enzyme as a potential therapeutic target in this tumor disease. CTSL was reported to involve the growth and invasion of human ovarian cancer cells (38). Down-regulation of CTSL could significantly inhibit the growth and invasion of ovarian cancer cells and sensitized these cells to chemotherapy (38, 39). CTSL enzyme activity or expression was significantly higher in many cancers, including highest in kidney and testicular tumors, then higher in the lung, adrenal, bladder, breast, colon, ovary, prostate, and thyroid cancers than in normal controls (21, 40). This suggests that the upregulation of CTSL expression may be closely related to tumor progression.
Thus, in addition to CTSL acting as one of the SARS-CoV-2 receptors, there is increasing evidence that proteolytic enzymes play important roles in cancers. Several CTSL inhibitors have been developed and used for clinical trials for cancer treatment (41, 42). CTSL protein levels were also reported to be downregulated by the treatment of cordycepin, a potential anti-cancer small molecule, in MDA-MB-231 (27). However, no too much progress has been made in TNBC (43). Identification of CTSL as a new potential therapeutic and prognostic target for TNBC is of great significance for TNBC patients.
In this study, we demonstrated that CTSL level is more highly expressed in TNBC tissues than that of paracancer tissues by western blotting and IHC, which was significantly associated with differentiated degree, TNM stage, tumor size, and lymphatic metastasis. We scored the IHC results of CTSL in paraffin breast cancer tissues to distinguish CTSL expression levels, where the score closest to the maximum point of the Yoden index was selected as the optimal threshold through receiver operating characteristic (ROC) curve analysis. Samples with scores below the critical value (score <6.3) were defined as low CTSL expressions, while samples with scores above the critical value (score >6.3) were defined as high CTSL expressions. We think that the optimal threshold value obtained by ROC is more objective, even though some studies considered an IHC score higher than 4 as a high expression (44). We further found that high CTSL expression predicted poor prognosis in TNBC patients, and CTSL expression in BC with Lymph node-positive revealed that a high expression of CTSL is correlated with a short DMFS and RFS. Moreover, overexpression of CTSL could significantly enhance the proliferation, migration, and invasion in the cells of MDA-MB-231 and MCF-7, while knockdown CTSL could significantly inhibit in MDA-MB-231 cells. These results imply that CTSL may be an effective biomarker for TNBC. Of course, it will be much better if have the in vivo data to support the conclusion as a biomarker. And we will do it in the future. Mechanistically, CTSL/CTSB was reported to regulate TGF-β production/signaling, which was required for the activation of fibroblasts and their promotion of invasive growth in human melanoma cells (45). Inflammatory BC (IBC) is the most aggressive and lethal form of BC, showing a poor prognosis and high infiltration of tumor-associated macrophages. El-Nadi et al. reported that CTSL and CD14+ monocytes-derived IL-10 play vital roles in the pathogenesis and their targeting might have therapeutic significances in IBC (46). Overall, CTSL which promoted the proliferation, migration, and invasion of MDA-MB-231 and MCF-7 may be associated with biological processes and signaling pathways, but it is still unclear for the potential mechanisms/pathways, which need to be further studied. Besides, CTSL is reported to contribute to tumor angiogenesis and its inhibition may have therapeutic significances in BC patients (47). The potential mechanisms/pathways BC angiogenesis need to be further studied.
In conclusion, CTSL is highly expressed in TNBC tissues and correlated with clinical progression. CTSL promotes the proliferation and migration/invasion in TNBC cells. High CTSL expression predicted poor prognosis in TNBC patients.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The work was approved by the Ethical Committee of Southwest Medical University and Huai’an People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
LZ, YZha, and YZhu collected the samples and did IHC. JY, CW, JC, TL, XL, and PZ did cell culture, western blotting, RT-PCR, assays of cell growth, migration and invasion, and data analysis. JF, SS, and LZ designed and supervised the project. LZ and JF wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81672887 and 82073263), the Joint Innovation Special Project of Science and Technology Plan of Sichuan Province (grant nos. 2022YFS0623-C4 and 2022YFS0623-C3), the Project Fund for Young Innovative Talents of Huai’an No. 1 Hospital Affiliated to Nanjing Medical University (grant nos. QC202209), and the Primary Research & Development Plan of Hunan Province (2020SK2071).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast (2022) 66:15–23. doi: 10.1016/j.breast.2022.08.010
2. Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the national comprehensive cancer network. Cancer (2012) 118:5463–72. doi: 10.1002/cncr.27581
4. Fujishima A, Imai Y, Nomura T, Fujisawa Y, Yamamoto Y, Sugawara T. The crystal structure of human cathepsin l complexed with e-64. FEBS Lett (1997) 407:47–50. doi: 10.1016/S0014-5793(97)00216-0
5. Chauhan SS, Popescu NC, Ray D, Fujisawa Y, Yamamoto Y, Sugawara T. Cloning, genomic organization, and chromosomal localization of human cathepsin l. J Biol Chem (1993) 268:1039–45. doi: 10.1016/S0021-9258(18)54038-2
6. Okitani A, Matsukura U, Kato H, Fujimaki M. Purification and some properties of a myofibrillar protein-degrading protease, cathepsin l, from rabbit skeletal muscle. J Biochem (1980) 87:1133–43.
7. Nonaka I, Ishiura S, Arahata K, Ishibashi-Ueda H, Maruyama T, Ii K. Progression in nemaline myopathy. Acta Neuropathol (1989) 78:484–91. doi: 10.1007/BF00687709
8. Thomssen C, Schmitt M, Goretzki L, Oppelt P, Pache L, Dettmar P, et al. Prognostic value of the cysteine proteases cathepsins b and cathepsin l in human breast cancer. Clin Cancer Res (1995) 1:741–6.
9. Herszènyi L, Plebani M, Carraro P, De Paoli M, Roveroni G, Cardin R, et al. The role of cysteine and serine proteases in colorectal carcinoma. Cancer (1999) 86:1135–42. doi: 10.1002/(SICI)1097-0142(19991001)86:7<1135::AID-CNCR6>3.0.CO;2-2
10. Sheahan K, Shuja S, Murnane MJ. Cysteine protease activities and tumor development in human colorectal carcinoma. Cancer Res (1989) 49:3809–14.
11. Qian F, Xu H, Zhang Y, Li L, Yu R. Methionine deprivation inhibits glioma growth through downregulation of CTSL. Am J Cancer Res (2022) 12:5004–18.
12. Fei Y, Xiong Y, Shen X, Zhao Y, Zhu Y, Wang L, et al. Cathepsin l promotes ionizing radiation-induced U251 glioma cell migration and invasion through regulating the GSK-3beta/CUX1 pathway. Cell Signal (2018) 44:62–71. doi: 10.1016/j.cellsig.2018.01.012
13. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science (2005) 308:1643–5. doi: 10.1126/science.1110656
14. Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin l to infect ACE2-expressing cells. J Biol Chem (2006) 281:3198–203. doi: 10.1074/jbc.M508381200
15. Bosch BJ, Bartelink W, Rottier PJ. Cathepsin l functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol (2008) 82:8887–90. doi: 10.1128/JVI.00415-08
16. Zhao MM, Yang WL, Yang FY, Zhang L, Huang WJ, Hou W, et al. Cathepsin l plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther (2021) 6:134. doi: 10.1038/s41392-021-00558-8
17. Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin l prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA (2005) 102:11876–81. doi: 10.1073/pnas.0505577102
18. Matsuyama S, Shirato K, Kawase M, Terada Y, Kawachi K, Fukushi S, et al. Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J Virol (2018) 92:e00683-18. doi: 10.1128/JVI.00683-18
19. Nishimura F, Naruishi H, Naruishi K, Yamada T, Sasaki J, Peters C, et al. Cathepsin-l, a key molecule in the pathogenesis of drug-induced and I-cell disease-mediated gingival overgrowth: a study with cathepsin-l-deficient mice. Am J Pathol (2002) 161:2047–52. doi: 10.1016/S0002-9440(10)64483-5
20. Miyamoto K, Iwadate M, Yanagisawa Y, Ito E, Imai J, Yamamoto M, et al. Cathepsin l is highly expressed in gastrointestinal stromal tumors. Int J Oncol (2011) 39:1109–15. doi: 10.3892/ijo.2011.1127
21. Chauhan SS, Goldstein LJ, Gottesman MM. Expression of cathepsin l in human tumors. Cancer Res (1991) 51:1478–81.
22. Park IC, Lee SY, Jeon DG, Lee JS, Hwang CS, Hwang BG, et al. Enhanced expression of cathepsin l in metastatic bone tumors. J Korean Med Sci (1996) 11:144–8. doi: 10.3346/jkms.1996.11.2.144
23. Liu P, Ju M, Zheng X, Jiang Y, Yu X, Pan B, et al. Methyltransferase-like 3 promotes cervical cancer metastasis by enhancing cathepsin l mRNA stability in an N6-methyladenosine-dependent manner. Cancer Sci (2023) 114:837–54. doi: 10.1111/cas.15658
24. Parigiani MA, Ketscher A, Timme S, Bronsert P, Schlimpert M, Kammerer B, et al. Conditional gene targeting reveals cell type-specific roles of the lysosomal protease cathepsin l in mammary tumor progression. Cancers (Basel) (2020) 12:2004. doi: 10.3390/cancers12082004
25. Sigloch FC, Tholen M, Gomez-Auli A, Biniossek ML, Reinheckel T, Schilling O. Proteomic analysis of lung metastases in a murine breast cancer model reveals divergent influence of CTSB and CTSL overexpression. J Cancer (2017) 8:4065–74. doi: 10.7150/jca.21401
26. Wang K, Deng H, Song B, He J, Liu S, Fu J, et al. The correlation between immune invasion and SARS-COV-2 entry protein ADAM17 in cancer patients by bioinformatic analysis. Front Immunol (2022) 13:923516. doi: 10.3389/fimmu.2022.923516
27. Zhang L, Wei C, Li D, He J, Liu S, Deng H, et al. COVID-19 receptor and malignant cancers: association of CTSL expression with susceptibility to SARS-CoV-2. Int J Biol Sci (2022) 18:2362–71. doi: 10.7150/ijbs.70172
28. Fu J, Qin L, He T, Qin J, Hong J, Wong J, et al. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res (2011) 21:275–89. doi: 10.1038/cr.2010.118
29. Tan Q, Fu J, Liu Z, Deng H, Zhang L, He J, et al. Impacts of transmembrane serine protease 4 expression on susceptibility to severe acute respiratory syndrome coronavirus 2. Chin Med J (Engl) (2023) 136:860–2. doi: 10.1097/CM9.0000000000002443
30. Du J, Fu J, Zhang W, Zhang L, Chen H, Cheng J, et al. Effect of DPP4/CD26 expression on SARS−CoV−2 susceptibility, immune response, adenosine (derivatives m(6)(2)A and CD) regulations on patients with cancer and healthy individuals. Int J Oncol (2023) 62:41. doi: 10.3892/ijo.2023.5489
31. Fu J, Zhou B, Zhang L, Balaji KS, Wei C, Liu X, et al. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol Biol Rep (2020) 47:4383–92. doi: 10.1007/s11033-020-05478-4
32. Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J (2021) 19:4101–9. doi: 10.1016/j.csbj.2021.07.014
33. Wei C, Liu Y, Liu X, Cheng J, Fu J, Xiao X, et al. The speckle-type POZ protein (SPOP) inhibits breast cancer malignancy by destabilizing TWIST1. Cell Death Discovery (2022) 8:389. doi: 10.1038/s41420-022-01182-3
35. Pulz LH, Strefezzi RF. Proteases as prognostic markers in human and canine cancers. Vet Comp Oncol (2017) 15:669–83. doi: 10.1111/vco.12223
36. Sullivan S, Tosetto M, Kevans D, Coss A, Wang L, O'Donoghue D, et al. Localization of nuclear cathepsin l and its association with disease progression and poor outcome in colorectal cancer. Int J Cancer (2009) 125:54–61. doi: 10.1002/ijc.24275
37. Dong Q, Li Q, Duan L, Wang H, Yan Y, Yin H, et al. Expressions and significances of CTSL, the target of COVID-19 on GBM. J Cancer Res Clin Oncol (2022) 148:599–608. doi: 10.1007/s00432-021-03843-9
38. Zhang L, Wei L, Shen G, He B, Gong W, Min N, et al. Cathepsin l is involved in proliferation and invasion of ovarian cancer cells. Mol Med Rep (2015) 11:468–74. doi: 10.3892/mmr.2014.2706
39. Zhang H, Zhang L, Wei L, Gao X, Tang LI, Gong W, et al. Knockdown of cathepsin l sensitizes ovarian cancer cells to chemotherapy. Oncol Lett (2016) 11:4235–9. doi: 10.3892/ol.2016.4494
40. Shuja S, Murnane MJ. Marked increases in cathepsin b and l activities distinguish papillary carcinoma of the thyroid from normal thyroid or thyroid with non-neoplastic disease. Int J Cancer (1996) 66:420–6. doi: 10.1002/(SICI)1097-0215(19960516)66:4<420::AID-IJC2>3.0.CO;2-Y
41. Li Y, Ai X, Zou C, Liu Y, Ma L, Men J, et al. Discovery of a novel and selective cathepsin l inhibitor with anti-metastatic ability in vitro and in vivo against breast cancer cells. Bioorg Chem (2021) 115:105256. doi: 10.1016/j.bioorg.2021.105256
42. Sudhan DR, Siemann DW. Cathepsin l targeting in cancer treatment. Pharmacol Ther (2015) 155:105–16. doi: 10.1016/j.pharmthera.2015.08.007
43. Wang Z, Xiang Z, Zhu T, Chen J, Zhong MZ, Huang J, et al. Cathepsin l interacts with CDK2-AP1 as a potential predictor of prognosis in patients with breast cancer. Oncol Lett (2020) 19:167–76. doi: 10.3892/ol.2019.11067
44. Sui H, Shi C, Yan Z, Wu M. Overexpression of cathepsin l is associated with chemoresistance and invasion of epithelial ovarian cancer. Oncotarget (2016) 7:45995–6001. doi: 10.18632/oncotarget.10276
45. Yin M, Soikkeli J, Jahkola T, Virolainen S, Saksela O, Hölttä E. TGF-beta signaling, activated stromal fibroblasts, and cysteine cathepsins b and l drive the invasive growth of human melanoma cells. Am J Pathol (2012) 181:2202–16. doi: 10.1016/j.ajpath.2012.08.027
46. El-Nadi M, Hassan H, Saleh ME, Nassar E, Ismail YM, Amer M, et al. Induction of heparanase via IL-10 correlates with a high infiltration of CD163+ M2-type tumor-associated macrophages in inflammatory breast carcinomas. Matrix Biol Plus (2020) 6-7:100030. doi: 10.1016/j.mbplus.2020.100030
Keywords: the CTSL gene, triple-negative breast cancer (TNBC), prognostics, cell proliferation, migration, invasion
Citation: Zhang L, Zhao Y, Yang J, Zhu Y, Li T, Liu X, Zhang P, Cheng J, Sun S, Wei C and Fu J (2023) CTSL, a prognostic marker of breast cancer, that promotes proliferation, migration, and invasion in cells in triple-negative breast cancer. Front. Oncol. 13:1158087. doi: 10.3389/fonc.2023.1158087
Received: 03 February 2023; Accepted: 14 June 2023;
Published: 29 June 2023.
Edited by:
Armando Pérez Torres, National Autonomous University of Mexico, MexicoCopyright © 2023 Zhang, Zhao, Yang, Zhu, Li, Liu, Zhang, Cheng, Sun, Wei and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junjiang Fu, ZnVqdW5qaWFuZ0Bzd211LmVkdS5jbg==; Lianmei Zhang, emhhbmdsaWFubWVpMjAwOEAxNjMuY29t; Chunli Wei, d2VpY2h1bmxpMjAxNUBzd211LmVkdS5jbg==; Suan Sun, aGF5eXNzYUAxNjMuY29t
 Lianmei Zhang1,2,3*
Lianmei Zhang1,2,3* Yaning Zhu
Yaning Zhu Ting Li
Ting Li Pengfei Zhang
Pengfei Zhang Jingliang Cheng
Jingliang Cheng Chunli Wei
Chunli Wei Junjiang Fu
Junjiang Fu