
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Oncol. , 23 August 2023
Sec. Cancer Epidemiology and Prevention
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1155123
Background: The nuanced relationship between inflammatory bowel disease (IBD) and pancreatic cancer is noticed in recent years. However, the underlying causal effects of these two diseases are still unclear.
Methods: The two-sample mendelian randomization (MR) was conducted to explore the causal effect of IBD condition on pancreatic cancer. Methods of Wald ratio, inverse variance weighted (IVW), MR-Egger, weighted median, and weighted mode were used to investigate the causal relationship between IBD and pancreatic cancer. Besides, Cochrane’s Q test, MR-Egger, and leave-one-out method were further conducted to detect heterogeneity, stability, and pleiotropy of MR results.
Results: In the MR analysis, we found Crohn’s disease had a significant causal effect on pancreatic cancer. Specifically, Crohn’s disease would increase 11.1% the risk of pancreatic cancer by the IVW method (p= 0.022), 33.8% by MR Egger (p= 0.015), by 35.3% by the Weighted model (p= 0.005). Regarding ulcerative colitis, there was no statistically significant causal effect observed on pancreatic cancer (p>0.05). Additionally, the pleiotropic test and Leave-one-out analysis both proved the validity and reliability of the present two-sample MR analyses.
Conclusion: This study indicates that IBD, particularly Crohn’s disease, is causality associated with increased risk of pancreatic cancer. Our results may help public health managers to make better follow-up surveillance of IBD patients.
The prevalence of inflammatory bowel disease (IBD), classified as Crohn’s disease (CD) and ulcerative colitis (UC), is increased substantially in many regions during the past decades, especially in Western societies such as Europe and North America (1, 2). Results of the IBD statistical report, in 2017, there were 6.8 million cases of IBD worldwide. The age-standardized prevalence rate increased from 79.5 per 100,000 population in 1990 to 84.3 per 100,000 population in 2017 (1). As one typical chronic inflammatory disease of the gastrointestinal tract, an increased cancer risk has also been determined in IBD patients during long-term follow-up (3, 4). Also, IBD treatments, such as immunosuppressant medications, may independently increase the risk of cancer in IBD (5–9).
In recent years, numerous studies were settled to assess the association between IBD and varied cancer risks. Compelling evidence has proved the positive association between IBD and future increased cancer risks. Especially, colorectal cancer is now a well-known risk in IBD patients during follow-up (3, 10–12). Moreover, with a deeper understanding of this chronic inflammation disease, relationships between extraintestinal cancers and IBD also raised clinicians’ interest and concerns (13). For instance, in one prospective observational cohort study within almost half a million populations in European, Wu et al. determined an increased risk of digestive, non-melanoma skin cancer, and male genital cancers in IBD patients (14). Similarly, results from Korean (7) and Chinese (4) cohorts also noticed a higher cancer risk in IBD patients, when compared with the background population.
Moreover, as one of the leading causes of cancer mortality globally with increasing prevalence and mortality, pancreatic cancer is considered to have an unfavorable clinical outcome (15, 16). Nevertheless, pancreatic cancer was frequently mixed with other upper gastrointestinal cancers during the calculations (14, 17, 18). To date, limited studies have evaluated the relationship between IBD and pancreatic cancer (7, 19). Notably, Everhov et al. conducted a large-scale population-based study to explore the association between IBD and pancreatic cancer. The results revealed that the 20-year cumulative incidence was 0.34% (95%CI: 0.30-0.38) vs 0.29% (95%CI: 0.28-0.30) and the overall hazard ratio (HR) for pancreatic cancer was increased overall in IBD population (Crohn’s disease: HR= 1.44; ulcerative colitis: HR=1.35, respectively) (19). However, a recent meta-analysis did not find any significant difference in the development of pancreatic cancer among the IBD population and the non-IBD population (20). Some scholars even assumed that the increased pancreatic risk might be due to misdiagnosis of periampullary cancer (13), as the IBD patients with primary sclerosing cholangitis had remarkably higher pancreatic cancer risk (HR=7.55) (19).
There is still a lack of robust evidence to conclude the association between IBD and pancreatic cancer, as previous studies were mainly based on clinical cohort data. Although some potential associations between IBD and pancreatic cancer were suggested, none of them could explain the exact causal relationship. Notably, gene prediction based on genome-wide association study (GWAS) data seems to be able to help us solve this puzzle. The application of Mendelian randomization (MR) methods (21–24) can assist us in predicting causality at the genetic level by correlating GWAS data of IBD and pancreatic cancer. The findings from MR analysis might provide us with insightful perspectives to deeply understand the genetic association between IBD and pancreatic cancer.
To fill this research gap, we aim to investigate the causal effect of IBD and pancreatic cancer, which can provide more robust evidence for clinical IBD management and help the active surveillance, early diagnosis, and precise clinical decision for high-risk pancreatic cancer subpopulation.
The classification of IBD incorporates two sub-diseases, named ulcerative colitis and Crohn’s disease. The GWAS data for IBD in this study was derived from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC, an organization aimed at identifying genetic risk factors for IBD and their clinical features as well as assessing the underlying interaction between genetic risks and the disease phenotypes). Specifically, the GWAS data of ulcerative colitis (GWAS id: ieu-a-970) was derived from the summary data of Liu et al. (25) and further download from the “MR-base”, a platform for MR developed by the “MRC Integrative Epidemiology Unit” at the University of Bristol (26) (https://gwas.mrcieu.ac.uk/datasets/ieu-a-970/). It contained 13,768 European-descent IBD cases and 33,977 European-descent health control with 156,116 single-nucleotide polymorphisms (SNPs). Similarly, the GWAS data for Crohn’s disease (GWAS id: ieu-a-12) was also extracted from the summary data of Liu et al. (25) and further download from the “MR-base” (https://gwas.mrcieu.ac.uk/datasets/ieu-a-12/). The GWAS data of Crohn’s disease contained 17,897 European-descent IBD cases and 33,977 European-descent health control with 124,888 SNPs. The data on pancreatic cancer was primarily derived from the Pancreatic Cancer Cohort Consortium release 1 (PanScan 1), including a large-scale GWAS analysis of 3,835 people from 12 prospective cohorts plus one hospital-based case-control study (27), and we downloaded the GWAS data from the same platform (https://gwas.mrcieu.ac.uk/datasets/ieu-a-822/). Ulcerative colitis and Crohn’s disease were defined as the exposure factor and pancreatic cancer was determined as the primary outcome.
Our study satisfied the assumptions of MR analysis. First and foremost, the SNPs selected for MR analysis must be strongly associated with ulcerative colitis and Crohn’s disease. In the present study, to ensure satisfy the assumption, only SNPs in IBD whose p-values were below the genome-wide significance level (5 × 10−8) were included for analysis. To ensure the robust association between instrumental variables and exposure factors, we excluded the weak instrumental variables with F values (formula: (R2/(R2-1)) *((N-K-1)/K)) <10 (28). Detailly, N represents the sample size of the exposure dataset, K is the number of SNPs, and R2 is the proportion of variation explained by IVs in the exposure dataset. Secondly, the chosen instrumental variables must meet an independence test. Genetic distance refers to the length of the region, regarding the linkage disequilibrium. Therefore, 1000 Genomes project European samples data were used as the reference panel to calculate the linkage disequilibrium between the SNPs (29). Namely, the SNP linkage disequilibrium value (r2) was set to 0.001 and the genetic distance was set to 10000 kb. We removed the SNP with r2 greater than 0.001 with the most significant SNPs to reduce the linkage disequilibrium impact and keep the independence of selected instrument variables. The SNPs characteristics of IBD from the European population were collected, including the number of SNPs, chromosome location, effective allele, effective allele frequency, effect value, standard error, and associated P-value of the effective allele.
Besides, Each SNP was checked at PhenoScanner V2 (30), which was a database of human genotype-phenotype associations (http://www.phenoscanner.medschl.cam.ac.uk/). Based on the PhenoScanner, the SNPs significantly associated with the potential confounders, like significantly existing in other tissues or diseases, in the present study were subsequently excluded. The mentioned instrumental variables screening guarantees the validity of our study’s findings. The schematic diagram for the Mendelian randomization analysis was presented (Figure 1).
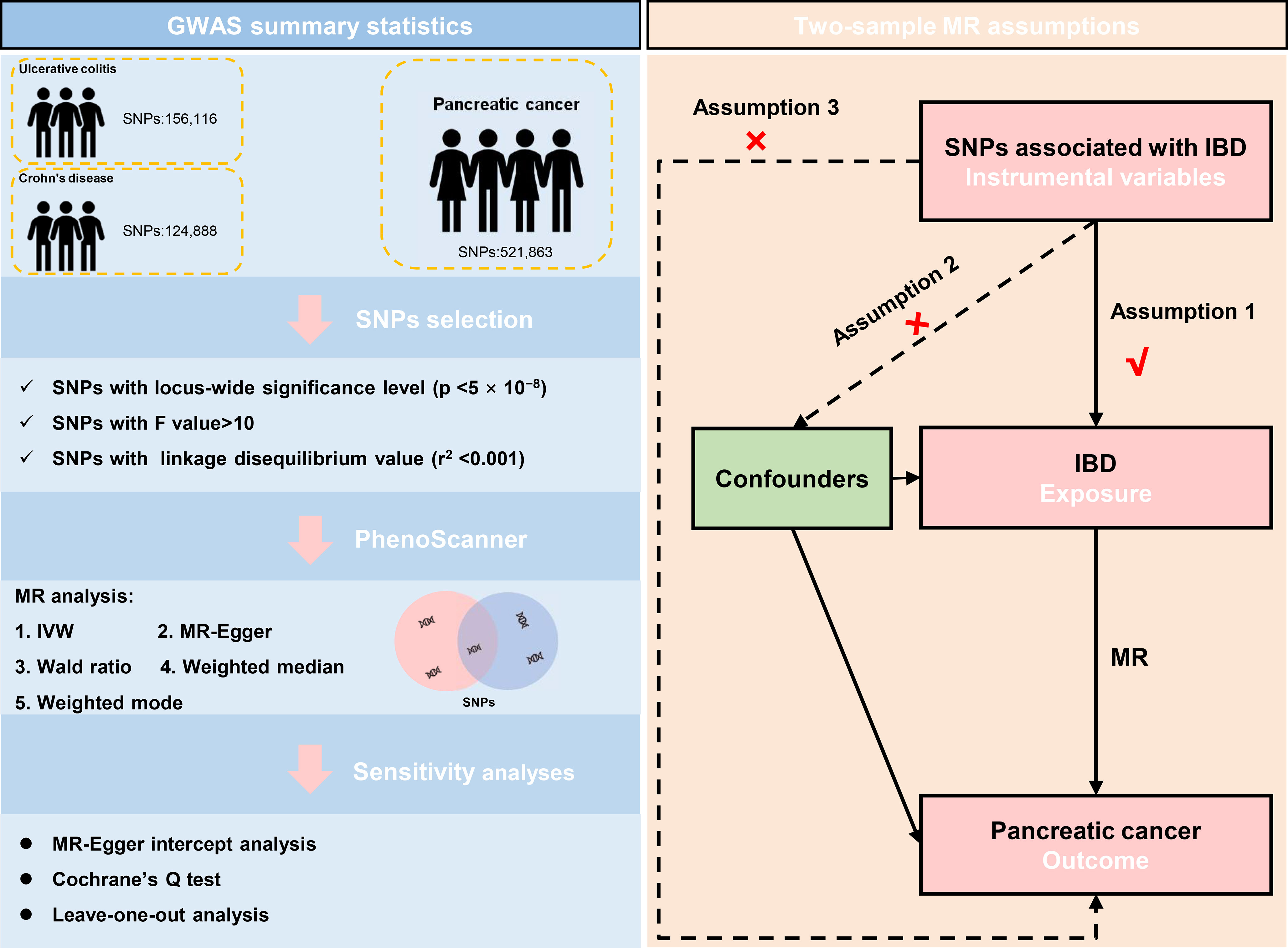
Figure 1 The data selection and the assumptions for the Mendelian randomization analysis in the present study. SNP, single-nucleotide polymorphism; IVW, inverse variance weighted; IBD, inflammatory bowel disease; MR, Mendelian randomization.
Four methods, including Inverse variance weighting (IVW), MR-Egger, weighted median, and weighted mode, were performed to assess the causal effect between exposures and outcomes. Different methods have different sensitivities to various issues, accommodate different scenarios, and vary in their statistical efficiency. As the primary analysis method, IVW is a time-honored method for combining the Wald ratio estimates of all relevant instrumental variables. This strategy is analogous to using weighted linear regression to probe the ties between the instrumental factors and the result. The instrumental variables’ intercept is restricted to zero. IVW can obtain unbiased estimates of the status without horizontal pleiotropy. Under the premise of Instrument Strength Independent of Direct Effect (InSIDE) (31), the MR-Egger method can primarily demonstrate the dosage relationship between instrumental variables and outcomes while accounting for some pleiotropy. The class one error rate can be lowered using the weighted median method, which also permits the possibility of invalidity for some specific genetic variants. Even if a certain instrumental variable does not satisfy the requirements of the MR technique for causal inference, the weighted mode approach remains valid when the vast majority of instrumental variables with identical causal estimates are valid. If the results of these methods are inconsistent, we give priority to IVW as the main result.
Furthermore, pleiotropy (refers to a genetic variant with numerous independent phenotypic effects), which might affect the causal effects, was assessed by the method of MR Egger intercept. To verify the conformity of each SNP, the heterogeneity test was performed utilizing MR Egger and IVW methods to calculate Cochran Q statistics and find the heterogeneity among genetic variants (32). Meanwhile, the leave-one-out analysis was conducted by excluding the genetic variants one by one. Then, the causal relationship would be rational and stable if the result of the leave-one-out analysis was in line with the global IVW analysis. To determine a more rigorous interpretation of the causal relationship between IBD and pancreatic cancer, Bonferroni-corrected was used in this study, according to the number of study exposures (0.05/2, 0.025). A nominal causal effect was determined when the p-value was between 0.05 and the corrected value of 0.025.
This study was followed by the STROBE-MR guideline for designation (33). All of the statistical analyses were performed by R (version 4.2.0, https://www.rproject.org/) with the “TwoSampleMR” package.
From the same European ancestry, there were 103454 cases involved in the present study, including 47,745 cases (13,768 patients and 33,977 health control) with 156,116 SNPs in the ulcerative colitis group, 51,874 cases (17,897 patients and 33,977 health control) with 124,888 SNPs in Crohn’s disease group, and 3835 cases (1,896 patients and 1,939 health control) with 521,863 SNPs in pancreatic cancer group, respectively. Moreover, there were 80 instrumental variables in ulcerative colitis and 122 instrumental variables in Crohn’s disease selected to conduct the MR analysis (Table 1). The detailed information of each selected SNP was summarized in Supplementary Files (Supplementary Files).
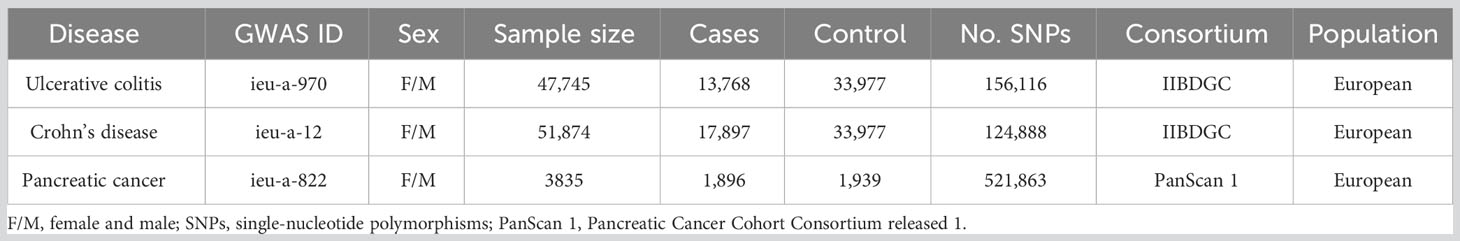
Table 1 The list of Genome-wide summary association studies (GWAS) included in the Mendelian randomization (MR) study.
The two-sample MR analysis was conducted to investigate the causal effect of IBD on pancreatic cancer. Specifically, ulcerative colitis was not observed to have causal effect on pancreatic cancer (Figure 2A) across the IVW (OR= 0.946, 95% CI= 0.830-1.079, p= 0.409), Weighted median (OR= 0.910, 95% CI= 0.753-1.101, p= 0.332), Weighted mode (OR= 0.902, 95% CI= 0.682-1.194, p= 0.474), and MR-Egger (OR= 0.951, 95% CI= 0.648-1.397, p= 0.799) methods (Table 2). Alternatively, there was a significant causality between Crohn’s disease and pancreatic cancer, namely, Crohn’s disease would increase the risk of pancreatic cancer (Figure 2B).
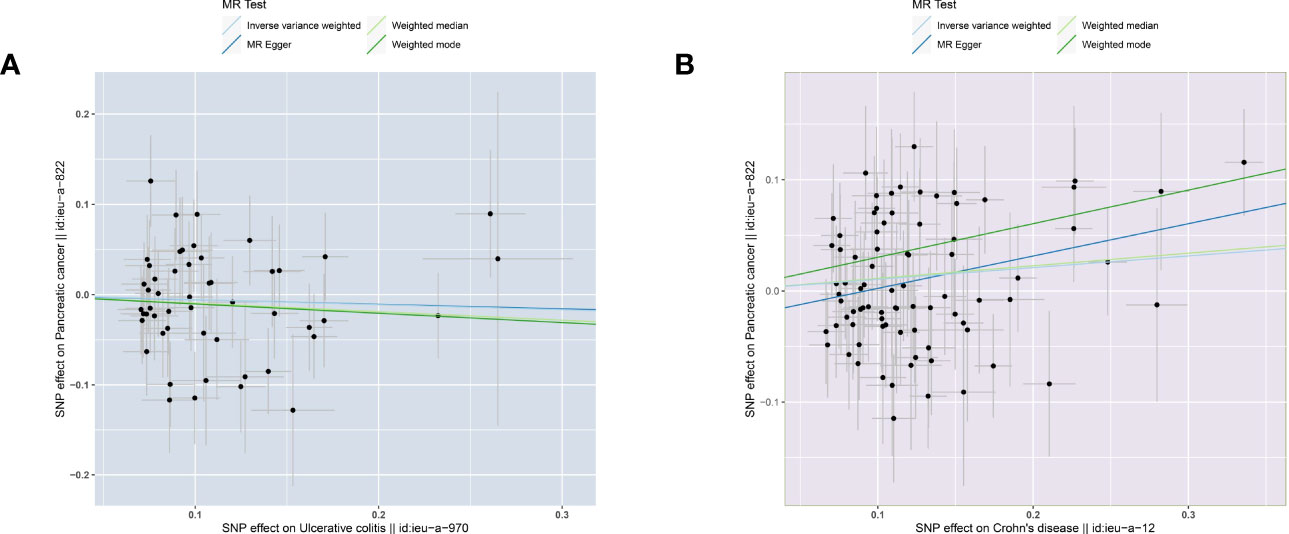
Figure 2 Scatter plots of causal estimates from genetically predicted IBD on pancreatic cancer. (A) ulcerative colitis. (B) Crohn’s disease.
According to the MR analysis, Crohn’s disease could increase approximately 11.1% risk of pancreatic cancer by the IVW method (OR= 1.111, 95% CI= 1.015-1.213, p= 0.022), by 33.8% by MR Egger (OR= 1.338, 95% CI= 1.064-1.683, p= 0.015), by 35.3% by the Weighted model (OR= 1.353, 95% CI =1.100-1.662, p= 0.005) methods (Table 3). Each SNP singly estimated the causal effect of IBD on pancreatic cancer by using the Wald ratio method, which was represented in two forest plots (Figures 3A, C). Moreover, the leave-one-out method revealed that the MR results remained stable after excluding the instrumental variables one by one. (Figures 3B, D).
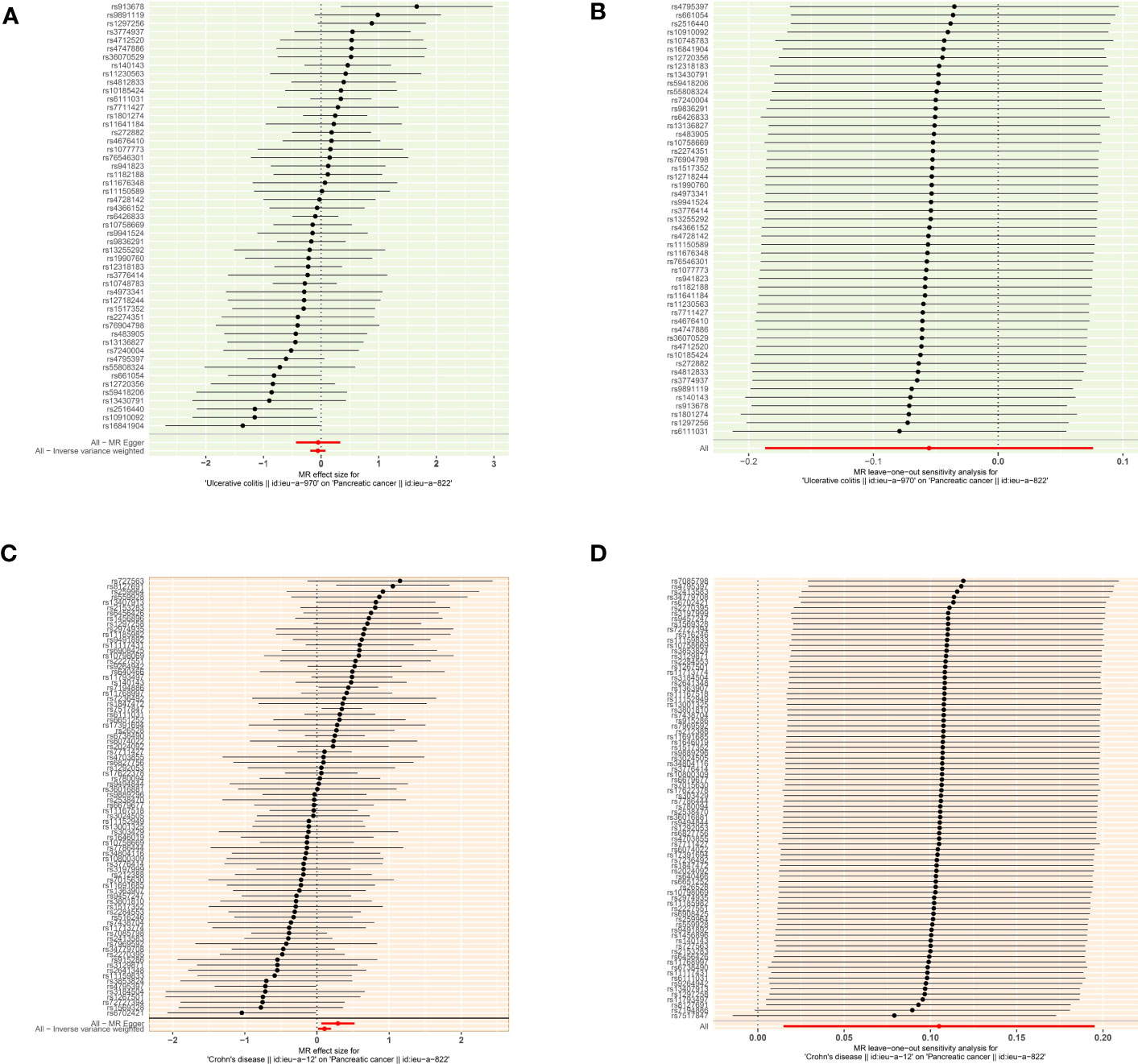
Figure 3 The causal effect of exposure on the outcome and leave-one-out sensitivity analysis for each SNP (A) The causal effect of exposure on the outcome is estimated using each SNP for ulcerative colitis and pancreatic cancer. (B) Leave-one-out sensitivity analysis for each SNP for ulcerative colitis and pancreatic cancer. (C) The causal effect of exposure on the outcome is estimated using each SNP for ulcerative colitis and pancreatic cancer. (D) Leave-one-out sensitivity analysis for each SNP for Crohn’s disease and pancreatic cancer.
The MR-Egger analysis showed that there was no horizontal pleiotropy or outliers in MR analysis for IBD and pancreatic cancer (Egger intercept= -0.001, Standard error= 0.022, p= 0.978 in ulcerative colitis, and Egger intercept= -0.026, Standard error= 0.015, p= 0.087 in Crohn’s disease, respectively). The funnel plot was adopted to show the distribution balance of single SNP effects (Figures 4A, B).
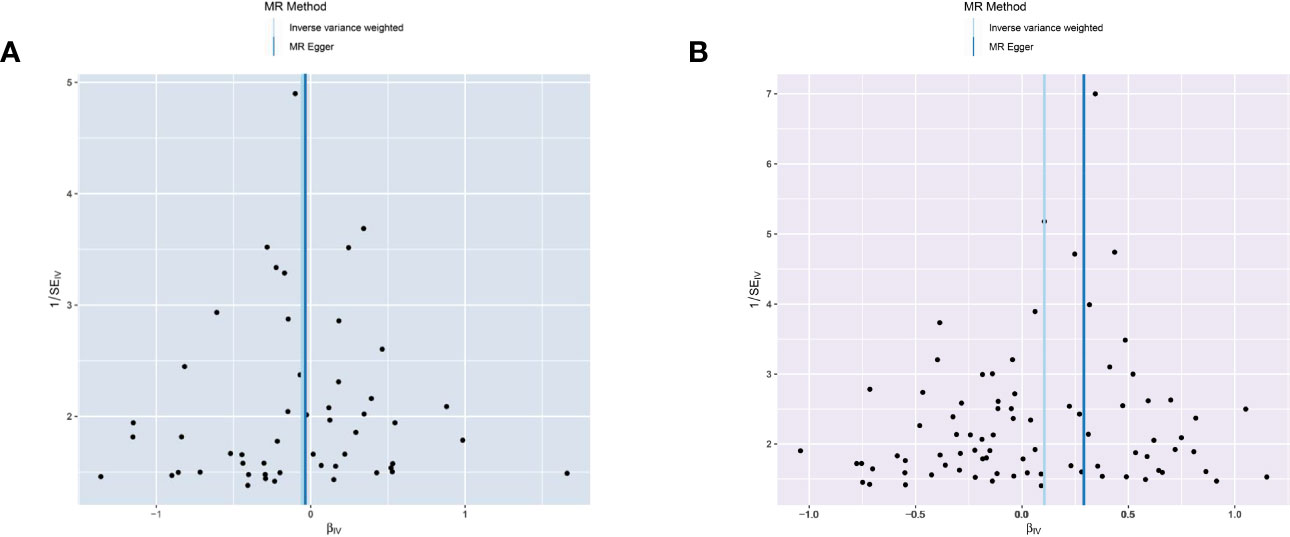
Figure 4 Funnel plot to assess the heterogeneity in causal estimates from genetically predicted IBD on pancreatic cancer. (A) ulcerative colitis. (B) Crohn’s disease.
The plots indicated that the effect of each SNP and distribution were in equilibrium. Additionally, to evaluate the heterogeneity in MR analysis, Cochrane’s Q test was also applied. In Crohn’s disease and pancreatic cancer group, the Q was 82 in MR Egger and 85 in inverse variance weighted, respectively (p>0.05). Similarly, in the ulcerative colitis and pancreatic cancer group, the Q was 54 in MR Egger and 54 in inverse variance weighted, respectively (p>0.05). Therefore, the heterogeneity test found limited evidence of heterogeneity. Only Crohn’s disease was observed to achieve a significant causal effect after using the Bonferroni-corrected test.’
To the best of our knowledge, we conducted the preliminary MR analysis on evaluating the causality between IBD and pancreatic cancer in the European population, based on the large-scale GWAS databases. We identified the positive association between Crohn’s disease and pancreatic cancer. The findings might provide noteworthy value for future preclinical studies on this topic.
To date, the association between IBD and pancreatic cancer raised wide concerns (19, 34). Notably, evidence from the Scandinavian register-based cohort study, Everhov et al. determined a statistically significant increased risk of pancreatic cancer in IBD patients. Interestingly, the long-term cumulative incidence was similar between the case and control groups (0.34% vs. 0.29%). Meanwhile, results from a Korean study confirmed an increased pancreatic cancer risk (OR=8.6, 95%CI: 1.0-31.0) in women with Crohn’s disease (7). Herein, some scholars even suggested that patients should not be nervous about this potential risk, while the primary sclerosing cholangitis or misdiagnosis of periampullary cancer would be contributed to this increased risk instead (13, 35). Nonetheless, the exact causal effect of IBD and pancreatic cancer remains unclear and could not be evaluated by observational studies, as the confounders could not be comprehensively adjusted.
In the present study, we noticed that Crohn’s disease but not Ulcerative colitis could significantly increase the risk of pancreatic cancer (IVW analysis: OR= 1.111, p=0.022) via the MR analysis. This divergence was consistent with recent clinical observational studies in different ethnic populations (7, 36). Apart from the IVW analysis, the other two MR analysis methods, including MR Egger (OR=1.338, p=0.015) and Weighted mode (OR= 1.353, p=0.005), also supported that Crohn’s disease caused by genetic factors can elevate the risk of pancreatic cancer. Reviewing the previous study on evaluating Crohn’s disease and pancreatic cancer, the majority of studies yielded that Crohn’s disease could increase pancreatic cancer risk. Focused on the cancer risk in Crohn’s disease patients with more detailed cancer sites, Hemminki et al. found an increased risk for many subsequent cancers, including pancreatic cancer. Interestingly, the highest standardized incidence ratio (SIR) frequently appeared in patients with more active surveillance (Follow-up interval<1 year, SIR=6.15) and elderly patients (>64 years, SIR=3.3). Most recently, in one large population-based study from the UK Biobank prospective database, Wu et al. demonstrated IBD patients had a higher incidence of digestive cancer and worse cancer-specific mortality, compared with reference controls. Moreover, the prevalence of each site-specific cancer was higher in ulcerative colitis patients than that in Crohn’s disease patients. As for pancreatic cancer, however, it was included in “Other digestive cancers” during analysis and there was no remarkable increased cancer risk observed in the results, after adjusting for thirteen confounders (14). In our study, no significant increased pancreatic cancer risk was determined in ulcerative colitis patients. Although previous studies also discovered this discrepancy (7, 19), we could not conclude this phenomenon as ulcerative colitis patients showed higher incidence in the majority of cancer sites. Therefore, with more comprehensive genetic data or adjusting more confounders, future works could be settled to further investigate the causality between ulcerative colitis and cancer sites that present relatively lower incidence in the background population.
Currently, the indications of screening for pancreatic cancer are restricted and not recommended for the general population of asymptomatic adults (37). Only when individuals who have a family history of pancreatic cancer, have a pathogenic variant in one of the high-risk pancreatic cancer susceptibility genes, or individuals at high risk owing to a personal history of pancreatic disease might consider screening. Thus, there is a lack of evidence for applying pancreatic cancer screening for individuals with comorbidities like diabetes mellitus, and chronic pancreatitis, even though these diseases are clinically observed to be associated with the risk of pancreatic cancer. Therefore, whether physicians need to inform patients about the increased risk of pancreatic cancer and subsequent annual screening is still a controversial topic (35). With the continually updated GWAS data, the common variants in several genomic regions as significantly associated with pancreatic cancer risk could be identified (16, 27). This milestone could help the development of pancreatic cancer screening with high-accuracy and non-invasive strategies.
There are some strengths of the present study that need to be mentioned. First, we evaluated the causality between IBD and pancreatic cancer via different MR methods. Besides, the design and SNPs evaluation of the study is followed by the guidelines of MR analysis (22). The evidence of MR sits at the interface of experimental and observational studies, which could provide more robust evidence for investigating the causality between two events. Thus, with the heterogeneity and horizontal pleiotropy-sensitive analysis, the findings drawn in this study were reasonable.
Indeed, there are still some limitations that need to be addressed in the following works. First, while the study population in this work eliminates the race discrepancy, whether the findings could be generalized to other countries with different races remain unclear. Thus, GWAS studies from different regions could provide a more comprehensive analysis of the association between IBD and pancreatic cancer. Second, as the IBD patients were collected from different medical centers, the differences in diagnosis methods, information acquisition, and data processing could lead to calculation bias. Third, the GWAS data of outcome was solely derived from the PanScan 1 project, which is the pancreatic cancer data that we can obtain to the greatest extent. If possible, we will continue exploring other pancreatic cancer GWAS datasets to understand the relationship between IBD and pancreatic cancer risk fully. Future data with more SNPs included for these diseases could help validate the results we determined.
In summary, based on the large-scale MR analysis, our results revealed Crohn’s disease instead of ulcerative colitis had a positive causal effect on pancreatic cancer. Future clinical guidelines and recommendations could consider to make better follow-up management and screening of IBD patients and minimize the risk of secondary malignancies, especially in terms of pancreatic cancer.
The R packages mentioned in the manuscript are available and can be downloaded on the website (https://github.com/MRCIEU/TwoSampleMR). Besides, the code and data for conducting this study have been uploaded on the website (https://github.com/Minyu96/IBD-and-PC).
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
This research has been conducted using published studies and consortia providing publicly available summary statistics. Besides, no individual-level data was used in this study. Therefore, the ethical approval was waived by the West China Hospital. The patient consent has already been obtained in the primary research of the IIBDGC and PanSan1 program, so they are not required in our study.
Conceptualization, YP. Data curation, RL and JJ. Formal analysis, JJ and ZW. Funding acquisition, XH and XP. Investigation, ZL, XH, and XP. Methodology, YM, RL, and ZW. Project administration, XH and XP. Software, JJ and XH. Supervision, XH and XP. Validation, ZL, RL, and YP. Visualization, YM and XP. Writing – original draft, YM, ZL, RL, JJ, ZW, YP, and XP. Writing – review & editing, YM, ZL, RL, JJ, ZW, YP, and XH. All authors contributed to the article and approved the submitted version.
The work was supported by the National Key Research and Development Program of China (2021YFE0206600), and the National Natural Science Foundation of China (82172842 and 81672386). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
We thank all the investigators of the International Inflammatory Bowel Disease Genetics Consortium, PanSan1, and IEU OpenGWAS project for exposing the data publicly.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1155123/full#supplementary-material
1. Alatab S, Sepanlou SG, Ikuta K, Vahedi H, Bisignano C, Safiri S, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol (2020) 5(1):17–30. doi: 10.1016/S2468-1253(19)30333-4
2. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol (2015) 12(12):720–7. doi: 10.1038/nrgastro.2015.150
3. Nadeem MS, Kumar V, Al-Abbasi FA, Kamal MA, Anwar F. Risk of colorectal cancer in inflammatory bowel diseases. Semin Cancer Biol (2020) 64:51–60. doi: 10.1016/j.semcancer.2019.05.001
4. Zhang H, Zhang M, Chen X, Guo M, Zhou R, Lv H, et al. Risk of Malignancy in patients with inflammatory bowel disease: A population-based cohort study from China. Int J Cancer (2022) 150(11):1770–8. doi: 10.1002/ijc.33932
5. Katsanos KH, Tatsioni A, Pedersen N, Shuhaibar M, Ramirez VH, Politi P, et al. Cancer in inflammatory bowel disease 15 years after diagnosis in a population-based European Collaborative follow-up study. J Crohns Colitis (2011) 5(5):430–42. doi: 10.1016/j.crohns.2011.04.013
6. Axelrad JE, Lichtiger S, Yajnik V. Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J Gastroenterol (2016) 22(20):4794–801. doi: 10.3748/wjg.v22.i20.4794
7. Jung YS, Han M, Park S, Kim WH, Cheon JH. Cancer risk in the early stages of inflammatory bowel disease in Korean patients: A nationwide population-based study. J Crohns Colitis (2017) 11(8):954–62. doi: 10.1093/ecco-jcc/jjx040
8. Yao D, Dong M, Dai C, Wu S. Inflammation and inflammatory cytokine contribute to the initiation and development of ulcerative colitis and its associated cancer. Inflammation Bowel Dis (2019) 25(10):1595–602. doi: 10.1093/ibd/izz149
9. Biancone L, Armuzzi A, Scribano ML, Castiglione F, D’Incà R, Orlando A, et al. Cancer risk in inflammatory bowel disease: A 6-year prospective multicenter nested case-control IG-IBD study. Inflammation Bowel Dis (2020) 26(3):450–9. doi: 10.1093/ibd/izz155
10. Bopanna S, Ananthakrishnan AN, Kedia S, Yajnik V, Ahuja V. Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol (2017) 2(4):269–76. doi: 10.1016/S2468-1253(17)30004-3
11. Liang L, Lin R, Xie Y, Lin H, Shao F, Rui W, et al. The role of cyclophilins in inflammatory bowel disease and colorectal cancer. Int J Biol Sci (2021) 17(10):2548–60. doi: 10.7150/ijbs.58671
12. Porter RJ, Arends MJ, Churchhouse AMD, Din S. Inflammatory bowel disease-associated colorectal cancer: translational risks from mechanisms to medicines. J Crohns Colitis (2021) 15(12):2131–41. doi: 10.1093/ecco-jcc/jjab102
13. Mala A, Foteinogiannopoulou K, Koutroubakis IE. Solid extraintestinal Malignancies in patients with inflammatory bowel disease. World J Gastrointest Oncol (2021) 13(12):1956–80. doi: 10.4251/wjgo.v13.i12.1956
14. Wu S, Xie S, Yuan C, Yang Z, Liu S, Zhang Q, et al. Inflammatory bowel disease and long-term risk of cancer: A prospective cohort study among half a million adults in UK biobank. Inflammation Bowel Dis (2022) 29 (3):384–95. doi: 10.1093/ibd/izac096
15. Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol (2016) 22(44):9694–705. doi: 10.3748/wjg.v22.i44.9694
16. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol (2021) 18(7):493–502. doi: 10.1038/s41575-021-00457-x
17. Pedersen N, Duricova D, Elkjaer M, Gamborg M, Munkholm P, Jess T. Risk of extra-intestinal cancer in inflammatory bowel disease: meta-analysis of population-based cohort studies. Am J Gastroenterol (2010) 105(7):1480–7. doi: 10.1038/ajg.2009.760
18. Scharl S, Barthel C, Rossel JB, Biedermann L, Misselwitz B, Schoepfer AM, et al. Malignancies in inflammatory bowel disease: frequency, incidence and risk factors-results from the swiss IBD cohort study. Am J Gastroenterol (2019) 114(1):116–26. doi: 10.1038/s41395-018-0360-9
19. Everhov ÅH, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, et al. Inflammatory bowel disease and pancreatic cancer: a Scandinavian register-based cohort study 1969-2017. Aliment Pharmacol Ther (2020) 52(1):143–54. doi: 10.1111/apt.15785
20. Lo B, Zhao M, Vind I, Burisch J. The risk of extraintestinal cancer in inflammatory bowel disease: A systematic review and meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol (2021) 19(6):1117–38.e19. doi: 10.1016/j.cgh.2020.08.015
21. Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol (2016) 27(11):3253–65. doi: 10.1681/ASN.2016010098
22. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. Bmj (2018) 362:k601. doi: 10.1136/bmj.k601
23. Carter AR, Sanderson E, Hammerton G, Richmond RC, Davey Smith G, Heron J, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol (2021) 36(5):465–78. doi: 10.1007/s10654-021-00757-1
24. Zhang X, Zhao H, Man J, Yin X, Zhang T, Yang X, et al. Investigating causal associations of diet-derived circulating antioxidants with the risk of digestive system cancers: A mendelian randomization study. Nutrients (2022) 14(15):3237. doi: 10.3390/nu14153237
25. Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet (2015) 47(9):979–86. doi: 10.1038/ng.3359
26. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife (2018) 7:e34408. doi: 10.7554/eLife.34408
27. Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet (2009) 41(9):986–90. doi: 10.1038/ng.429
28. Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol (2011) 40(3):755–64. doi: 10.1093/ije/dyr036
29. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature (2010) 467(7319):1061–73. doi: 10.1038/nature09534
30. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics (2016) 32(20):3207–9. doi: 10.1093/bioinformatics/btw373
31. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol (2017) 32(5):377–89. doi: 10.1007/s10654-017-0255-x
32. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol (2015) 44(2):512–25. doi: 10.1093/ije/dyv080
33. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
34. Massironi S, Fanetti I, Viganò C, Pirola L, Fichera M, Cristoferi L, et al. Systematic review-pancreatic involvement in inflammatory bowel disease. Aliment Pharmacol Ther (2022) 55(12):1478–91. doi: 10.1111/apt.16949
35. Holmer AK, Singh S. Editorial: pancreatic cancer risk in inflammatory bowel diseases-it’s all relative. Aliment Pharmacol Ther (2020) 52(3):550–1. doi: 10.1111/apt.15877
36. Biancone L, Armuzzi A, Scribano ML, D’Inca R, Castiglione F, Papi C, et al. Inflammatory bowel disease phenotype as risk factor for cancer in a prospective multicentre nested case-control IG-IBD study. J Crohns Colitis (2016) 10(8):913–24. doi: 10.1093/ecco-jcc/jjw048
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, risk factor, pancreatic cancer, mendelian randomization, GWAS
Citation: Min Y, Liu Z, Li R, Jin J, Wei Z, Pei Y, Hu X and Peng X (2023) Association between inflammatory bowel disease and pancreatic cancer: results from the two-sample Mendelian randomization study. Front. Oncol. 13:1155123. doi: 10.3389/fonc.2023.1155123
Received: 31 January 2023; Accepted: 14 August 2023;
Published: 23 August 2023.
Edited by:
Lu Zhang, Hong Kong Baptist University, Hong Kong SAR, ChinaReviewed by:
Gabriela Fonseca-Camarillo, Instituto Nacional de Cardiología Ignacio Chávez, MexicoCopyright © 2023 Min, Liu, Li, Jin, Wei, Pei, Hu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingchen Peng, cHh4MjAxNEAxNjMuY29t; Xiaolin Hu, aHV4aWFvbGluQHdjaHNjdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.