- 1Department of Experimental Medicine, University of Rome Tor Vergata, Rome, Italy
- 2Department of Biology, University of Rome Tor Vergata, Rome, Italy
- 3Clinical Infectious Diseases, Department of System Medicine, University of Rome Tor Vergata, Rome, Italy
HBV is one of the most widespread hepatitis viruses worldwide, and a correlation between chronic infection and liver cancer has been clearly reported. The carcinogenic capacity of HBV has been reported for other solid tumors, but the largest number of studies focus on its possible lymphomagenic role. To update the correlation between HBV infection and the occurrence of lymphatic or hematologic malignancies, the most recent evidence from epidemiological and in vitro studies has been reported. In the context of hematological malignancies, the strongest epidemiological correlations are with the emergence of lymphomas, in particular non-Hodgkin’s lymphoma (NHL) (HR 2.10 [95% CI 1.34-3.31], p=0.001) and, more specifically, all NHL B subtypes (HR 2.14 [95% CI 1.61-2.07], p<0.001). Questionable and unconfirmed associations are reported between HBV and NHL T subtypes (HR 1.11 [95% CI 0.88-1.40], p=0.40) and leukemia. The presence of HBV DNA in peripheral blood mononuclear cells has been reported by numerous studies, and its integration in the exonic regions of some genes is considered a possible source of carcinogenesis. Some in vitro studies have shown the ability of HBV to infect, albeit not productively, both lymphomonocytes and bone marrow stem cells, whose differentiation is halted by the virus. As demonstrated in animal models, HBV infection of blood cells and the persistence of HBV DNA in peripheral lymphomonocytes and bone marrow stem cells suggests that these cellular compartments may act as HBV reservoirs, allowing replication to resume later in the immunocompromised patients (such as liver transplant recipients) or in subjects discontinuing effective antiviral therapy. The pathogenetic mechanisms at the basis of HBV carcinogenic potential are not known, and more in-depth studies are needed, considering that a clear correlation between chronic HBV infection and hematological malignancies could benefit both antiviral drugs and vaccines.
Introduction
Several viruses, often causing chronic or persistent infections, are correlated with the occurrence of malignancy. Usually, the immune response, with its articulated actions, represents the best control against the emergence of neoplastic cells; however, the persistence of viruses with oncogenic characteristics represents the driver for immune evasion, facilitating poor control over malignancy emergence.
Epstein−Barr virus, human papillomavirus, human herpesvirus-8, Merkel cell polyomavirus, and hepatitis B viruses (HBV) and hepatitis C virus (HCV) are commonly considered viruses with oncogenic properties. The correlation between these infections and the emergence of cancer has been clearly demonstrated; although, the development of malignancy is not common among infected subjects (1, 2). Indeed, multiple additional factors, such as chronic inflammation, environmental mutagens, or immunosuppression, are required for cancer development; therefore, some virus-related tumors are more common in immunocompromised patients (solid organ or bone marrow transplant recipients, human immunodeficiency virus [HIV]-infected patients) and in patients with other chronic inflammatory conditions.
HBV is one of the most widespread hepatitis viruses worldwide, with 296 million infected subjects, including over 6 million children under the age of 5 (3). The correlation between chronic HBV infection and liver cancer is widely known, and it has been reported that more than 80% of hepatocellular carcinoma (HCC) diagnoses are related to HBV infection (4). However, HCC is not the only malignancy correlated with HBV infection, as a few other non-hepatic malignancies have been associated with the presence of chronic HBV infection. A recent study on 496,732 Chinese participants (5) assessed the associations between chronic HBV infection and the risk of all cancer types, and the authors found an increased risk of developing several non-liver cancers, including stomach, oral, colorectal, and pancreatic cancers, and lymphomas.
The positive correlation between chronic HBV infection and the emergence of lymphomas has been repeatedly reported in the literature. However, although several meta-analyses reported a positive correlation (6), others did not (7); therefore, the topic remains still dibated.
The purpose of this review is to provide information on HBV lymphotropism and its potential impact on lymphomagenesis. With this aim, the epidemiological correlates between HBV infection and the occurrence of lymphatic or hematological tumors have been updated, and the results of in vitro studies of HBV infection of hematopoietic and lymphatic cells have been reported, suggesting possible patterns of persistent infection and viral reactivation. Finally, some considerations on possible implications between HBV infection of blood cells and the possibility of new targets for eradication treatments are reported.
Correlation between HBV infection and hematological disorders: Data from epidemiological studies
The role of HBV in the field of oncogenesis is mainly related to the occurrence of HCC, and some studies have evaluated the protective effect of HBV vaccination and antiviral drugs on HCC emergence (8, 9). The association between HBV and HCC is directly related to HBV’s hepatotropic characteristics and its ability to infect and replicate inside hepatocyte cells. However, it is now known (10–12) that HBV can also infect other cells from extrahepatic sites, such as lymph nodes, spleen, thyroid, kidneys, and pancreas, and its influence on extrahepatic tumor emergence has been studied. HBV replicates in hematopoietic and lymphoid cells and their progenitors (10, 13), making this compartment an additional reservoir of infection and suggesting an implication of the virus in lymphomagenesis (Table 1).
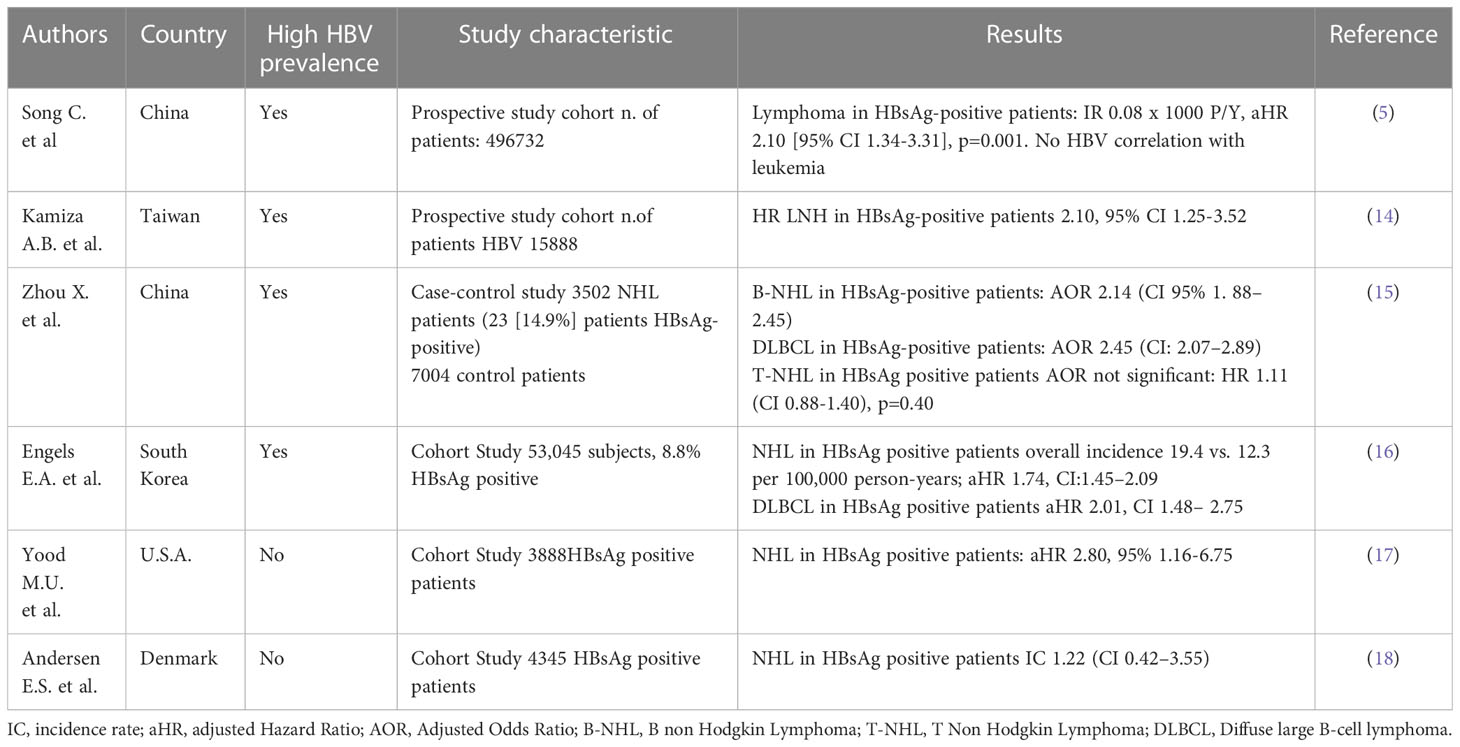
Table 1 Summary of studies addressing the correlation between HBV infection and hematological disorders.
Some recent studies have attempted to evaluate the impact of active HBV infection on the onset of all types of extrahepatic cancer (5). In a prospective cohort study by Ci Song et al. found a significant correlation for the occurrence of lymphoma, but not leukemia in HBV patients. Recent studies have confirmed the absence of a role for HBV in the pathogenesis of leukemia, such as the case–control study by Kamiza et al. in Taiwan (14) and the South Korean cohort study by Engels (16), which also distinguished different subtypes of leukemia (including multiple myeloma [MM]; lymphoid leukemia; myeloid/monocytic leukemia).
Lymphomas are the hematological tumors most epidemiologically associated with HBV infection (5, 14). Not all varieties of lymphoma were associated with active HBV infection, and Kamiza et al. (14) showed a two fold increase of risk for non-Hodgkin’s lymphoma (NHL) in the HBV-positive population.
A recent Chinese paper by Zhou et al. (15) investigated the impact of HBV on the onset of NHL, specifying the different subtypes of the onco-hematological pathology, given that the frequency of the major NHL subtypes varies substantially by geographic region (19). In this study, B-cell NHL (B-NHL) and T-cell NHL (T-NHL) were distinguished, and all further subtypes were considered. Diffuse large B-cell lymphoma (DLBCL) was the most common (n=1224 patients) and thus was further distinguished into germinal center-type lymphoma (DLBCL-GCB n=356) or non-GCB-DLBCL (n=663). Interestingly, 17% of patients with B-NHL were HBsAg-positive, while 9.4% of T-NHL patients were positive. Indeed, the association between HBV infection and the occurrence of B-NHL was statistically significant in multivariate analysis (adjusted for age, sex, and year of diagnosis) and a doubled risk of occurrence of B-NHL has been described yet. However, the association between HBV and the onset of T-NHL was not significant. The majority of B-NHL subtypes were found to be associated with HBV infection: a more than doubled risk has been identified in DLBCL, follicular lymphoma (FL), Small lymphocytic lymphoma/chronic lymphocytic leukemia (SLL/CLL) and precursor B lumphoblastic lymphoma. No differences were found between GCB-type and non-GCB-type patients. Furthermore, only two subtypes of T-NHL were found to be significantly associated with HBV infection: angioimmunoblastic T-cell lymphoma (AITL) and anaplastic large cell lymphoma (ALCL). Among other cohort studies (8, 16–18), only the Danish DANVIR study did not find statistical significance in the association between NHL and HBV infection and, consequently, with the different subtypes of NHL. This can be related to HBV epidemiological data of Denmark, which is a country with low HBV prevalence, with main sexual transmission and limited chronic HBV infection.
The influence of anti-HBV antiviral treatments and vaccinations on lymphomagenesis was also studied. Both variables are absent in most of the above cited study cohorts. One of the most relevant studies on this issue is that of Huang et al. (20) on a Taiwanese study cohort, which covered the 17-year period from 1997 to 2013 and compared the incidence of NHL in HBV-positive and HBV-negative groups. History of anti-HBV treatment, including interferon, adefovir, entecavir, lamivudine and tenofovir, was recorded, as well as the diagnosis of NHL based on The National Health Insurance Research Database (NHIRD). The study showed that only 3 of 4,069 (0.07%) patients developed NHL in the follow-up period, significantly less than the 107 of 34,559 (0.31%) patients without anti-HBV treatment (p = 0.0076) in the HBV cohort, but it was only a prevalence analysis with a lack of clinical and virological data at the time of diagnosis and during follow-up. In the same study, a significantly lower incidence of NHL was found in the group born after July 1984 (introduction of universal vaccination in Taiwan) as compared to the group born before 1984 (0.74 [95% CI 0.41-1.34] vs. 1.85 [95% CI 1.02-3.24], respectively, p=0.032) between the ages of 12.5 and 20 years but was not confirmed for the group aged 21 to 29.5 years. Therefore, the authors concluded that the definite impact of HBV vaccination in the population older than 21 years needs a much longer follow-up and should be adjusted for increasing incidence of NHL in the general population, even under the assumption that vaccination has a protective effect on developing NHL in the population younger than 20 years.
In conclusion, most cohort studies conducted in high HBV-prevalence countries have demonstrated a correlation between HBV infection and NHL, while the impact of HBV on the onset of leukemia, Hodgkin’s lymphoma and other hematological disorders is controversial. New studies are needed, including prospective clinical and virological studies that involve control groups, which are often lacking in retrospective and prospective epidemiological studies. Moreover, longer follow-up studies would allow us to better evaluate the influence of vaccination status and antiviral therapy on cancer development.
HBV DNA integration in immune cells as a mechanism mediating lymphomagenesis
The integration of HBV DNA into the genome of human hepatocytes is a recognized mechanism contributing to the onset of HCC (21). HBV DNA integration can promote the neoplastic transformation of the involved hepatocytes by several mechanisms, such as the direct dysregulation of oncogenes and onco-suppressors, the production of chimeric viral-human RNAs and proteins with transactivating properties, and the promotion of overall genome instability, further enhancing the accumulation of chromosomal aberrations (22–28). Overall, these mechanisms can trigger the clonal selection of hepatocytes with enhanced survival and proliferation, promoting their neoplastic transformation (23, 25, 29–31).
In a similar fashion, some recent studies have also supported the role of HBV DNA integration into the genome of immune cells in driving the pathogenesis of immunoproliferative disorders such as NHL and leukemia (32) (Figure 1). This hypothesis is in keeping with previous studies demonstrating the occurrence of HBV DNA integration into the genome of peripheral blood mononuclear cells (PBMCs) from patients with chronic HBV infection (33–36) and is also in line with data from the woodchuck model identifying multiple hepadnaviral integrations not only in PBMCs but also in lymphatic organs, including bone marrow, spleen, and lymph nodes (37).
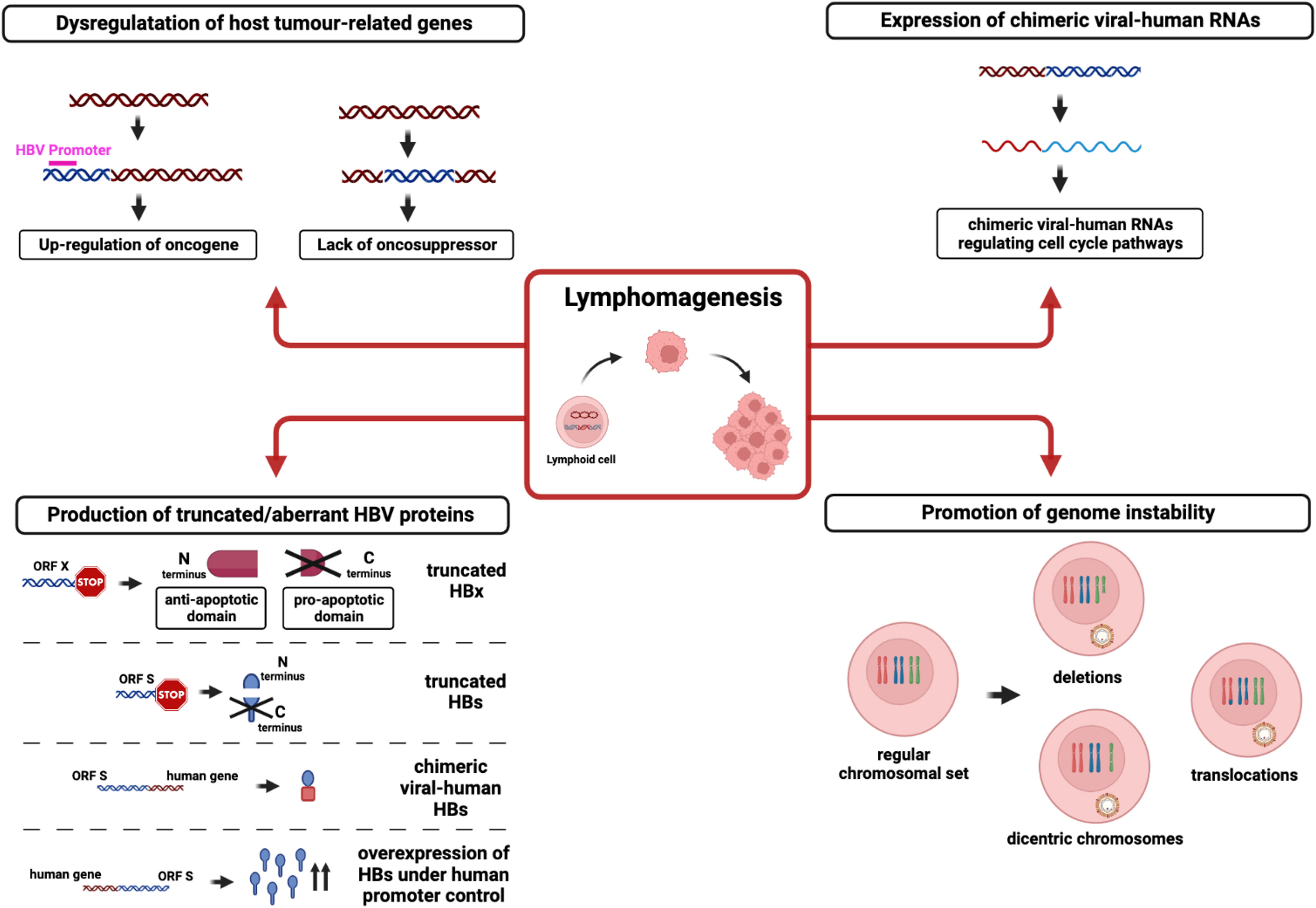
Figure 1 Mechanisms involved in lymphomagenesis caused by HBV DNA integration into the host genome. The figure shows the potential pathways by which HBV integration in lymphoid cell genome leads to lymphomagenesis: i) HBV DNA (blue) integration into the infected cells’ genome (red) can alter host tumor-related genes by up-regulating oncogene or by interrupting oncosuppressor activity; ii) HBV integration can generate chimeric viral-human RNAs with a role in regulating cell cycle pathways; iii) integrated HBV DNA can lead to the production of truncated/aberrant HBV proteins, (such as truncated HBx or HBs) as well as chimeric viral-human proteins; iv) HBV integration can promote genomic instability by altering the regular chromosomal set.
In this regard, a recent study revealed that HBV DNA integration represents a common phenomenon in NHL tissues of patients with HBV infection (38). Indeed, multiple HBV DNA integration events have been observed in half of the HBsAg-positive patients with NHL analyzed in this study. Notably, HBV DNA integration was found in the exonic regions (crucial for mRNA synthesis) of four specific genes (FAT2, SsTX, ITGA10, and CD63), determining their altered expression and potentially perturbing relevant intracellular pathways. Similarly, HBV DNA integration was also recurrently found in seven coding genes (ANKS1B, CAPZB, CTNNA3, EGFLAM, FHOD3, HDAC4, and OPCML) potentially involved in mechanisms underlying carcinogenesis. Of note, six of these seven genes, preferentially targeted by HBV DNA integration, showed increased expression in NHL with respect to normal lymphatic tissues, suggesting that HBV DNA integration could promote lymphomagenesis mostly by cis-activating pro-oncogenes (38).
In line with these data, another study has recently shown that HBV DNA integration profiles in PBMCs are superimposable to those observed in tumor liver tissues, since they mostly involve genes regulating cell survival and proliferation in both body compartments. This study also provided evidence of HBV DNA integration within an extrahepatic lymphoproliferative tumor in a patient with chronic hepatitis B. Overall findings further reinforce the role of HBV DNA integration in PBMCs in paving the way toward lymphomagenesis (39).
Beyond PBMCs, HBV DNA integration has also been detected in the bone marrow hematological stem cells (HSCs) of chronically infected HBV patients (40). These integrations in the precursors of immune cells could promote uncontrolled cell proliferation and, in turn, could represent a mechanism favoring the onset of different types of lymphoproliferative disorders.
Overall, further studies are necessary to better explore the functional consequences of HBV DNA integration on immune cell function, malignant cell surveillance, and oncogenic transformation to finally elucidate the role of HBV DNA integration in immune cells and their precursors in hematological malignancies. Similarly, the limited knowledge of the kinetics, frequencies and specific mechanisms underlying HBV DNA integration in immune cells calls for further investigations to unveil these relevant issues.
Evidence of HBV infection in hematopoietic stem cells and lymphoid cells in in vitro studies and animal models
Early studies after the discovery of HBV included poor information on the capability of HBV to infect, persist and propagate in cells other than hepatocytes, mainly due to limitations of the techniques employed at that time (32). Few data are present in the literature on in vitro models of HBV infection, especially in blood mononuclear cells. However, over the years, several studies have demonstrated the presence of viral genomes or other replicative intermediates in cells such as B and T lymphocytes and monocytes (41–48) (Figure 2). Nevertheless, the mechanism of HBV entry into immune cells is still unknown, as it is unlikely that the sodium taurocholate cotransporting polypeptide (NTCP) receptor mediating viral entry into human hepatocytes is involved (32).
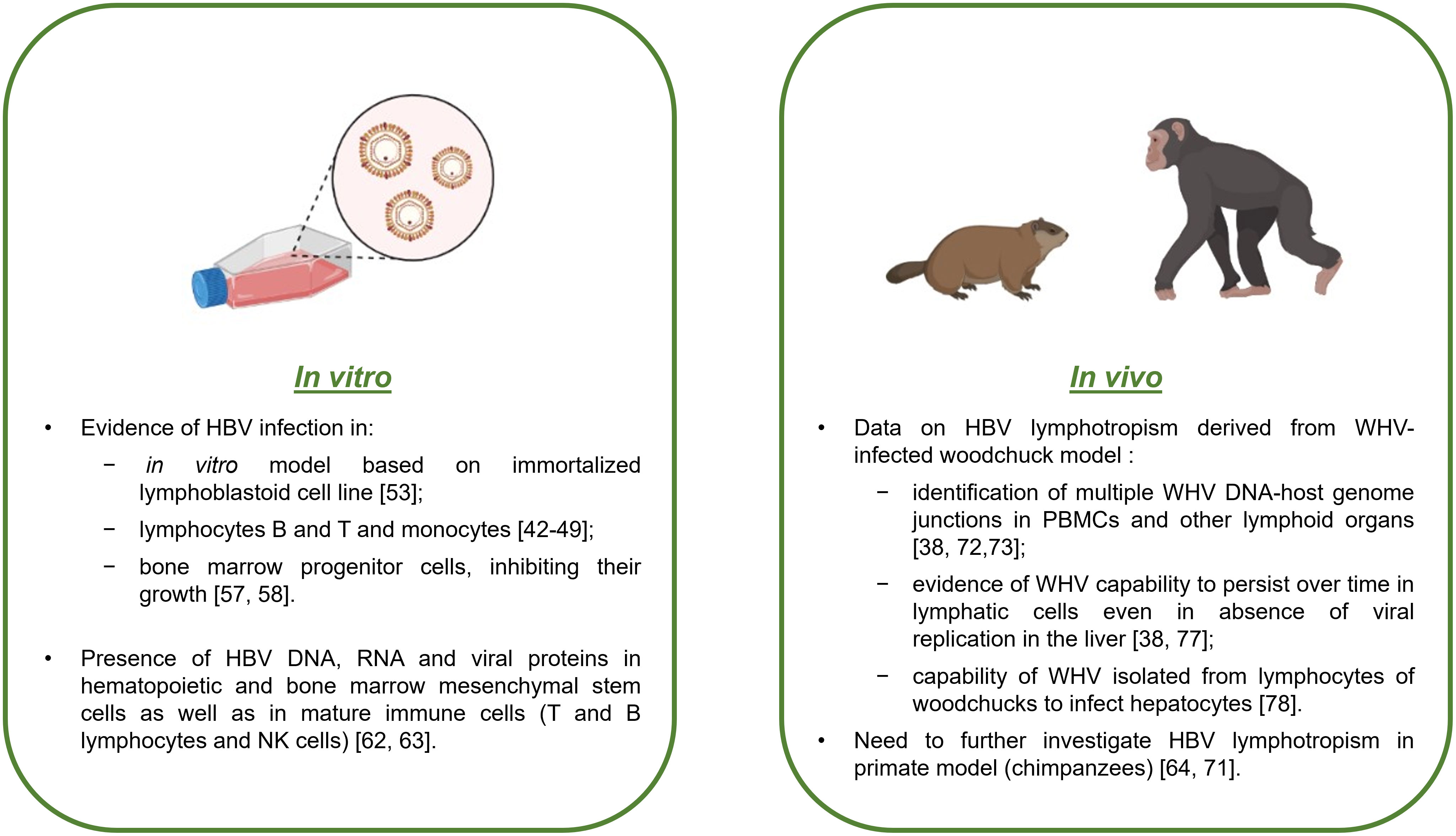
Figure 2 Evidence of HBV infection in lymphoid cells from in vitro and in vivo studies. Schematic summary with the main results of HBV infection in lymphoid cells based on cell cultures and animal models.
Although hepatocytes represent an eligible in vitro model for studying HBV infection (49–51), primary human hepatocytes are difficult to obtain, and consequently, it is difficult to study HBV infection in this kind of cell culture (51). However, since the end of the 1980s, it has been possible to create an in vitro model of an immortalized lymphoblastoid cell line sensitive to HBV infection, which is very useful for understanding the mechanisms of viral infection, replication, and propagation (52) (Figure 2). Early in vitro studies on HBV infection in PBMCs demonstrated that HBV was able to bind to the cell surface but was not able to completely infect or replicate inside PBMCs by evaluating the presence of cccDNA with the classical PCR method (51). From a methodological aspect, when analyzing HBV infection in in vitro models of PBMCs, it is important to be sure that the positivity of viral DNA effectively comes from the infected cells and not from viral particles or virus-free DNA fragments. Therefore, limited digestion of cell surfaces with trypsin and DNase and deep washing prior to nucleic acid extraction become crucial (33, 53, 54). The final confirmation of HBV lymphotropism is provided by the capability of immune cells to produce infectious virus (32).
One of the first findings resulting from in vitro studies on HBV infection in lymphatic cells was the capability of HBV to infect bone marrow progenitor cells and inhibit their growth (48, 55) (Figure 2). More specifically, only viral particles containing HBV DNA were infectious and capable of causing cell growth inhibition, unlike sub-viral particles or heat-inactivated virions, which had no effect on progenitor stem cells (48, 55).
Many studies have tested the susceptibility of healthy volunteer-derived hematopoietic and mesenchymal stem cells to HBV infection after exposure to HBV-positive sera (40–42, 48, 56–58). Positive results for intracellular HBV DNA and/or RNA and HBsAg were found in CD34+ hematopoietic stem cells (precursors of immune cells, myeloid and erythroid cell linages) (56, 57), as well as in bone marrow mesenchymal stem cells reactive to CD105 and CD90. Notably, the latter also represent hepatocyte progenitors (58).
Moreover, HBV lymphotropism was also demonstrated in mature immune cells (Figure 2). In particular, HBV mRNAs were detected in both B- and T-cell subsets with a higher degree in B lymphocytes (59). Another study revealed the presence of viral genome and proteins in NK cells (60) (Figure 2).
Regarding in vivo models, many studies have demonstrated the presence of HBV infection in extrahepatic sites, such as PBMCs, spleen, pancreas, and kidney, in animal models (46, 61–67). Notably, various animals can be used for studying HBV infection, but chimpanzees represent the most suitable model, as they can be infected by human HBV and present many useful characteristics, offering the opportunity to study different aspects of viral infection and its consequences (67) (Figure 2). Nonetheless, their use for research purposes is limited by handling due to the large size of the animals, ethical issues, and high costs (67). HBV DNA integration into the genome of PBMCs of chronically infected HBV chimpanzees was demonstrated (61).
Woodchucks represent another interesting model, where it was possible to detect woodchuck hepatitis virus (WHV) in immune cells (32) (Figure 2). In particular, the study’s results showed that immune cells support productive replication of WHV and that the derived viral progeny is infectious to hepatocytes and to immune cells in in vitro and in vivo models, where it can cause hepatitis (32). In the woodchuck model, multiple WHV DNA-host genome junctions were identified in PBMCs and other lymphoid organs, regardless of whether HCC had developed (37, 68, 69) (Figure 2). However, further studies are needed to understand the role of these WHV DNA integration events in PBMCs in lymphoproliferative disorders, such as NHL and chronic lymphocytic leukemia (34, 70, 71). Another important aspect, demonstrated in the woodchuck model, was the capability of WHV to persist in lymphatic cells without involving the liver (37, 72), representing a crucial point for understanding how this mechanism could be involved in viral persistence and likely consequent reactivation (Figure 2). Furthermore, it was demonstrated that in infected woodchucks, lymphotropic WHV was also able to infect hepatocytes and that the viral genomic sequences revealed in both types of cells were the same (73), underlining how lymphotropism could be a natural property of wild-type WHV and not a consequence of the development of lymphotropic variants (Figure 2).
HBV infection in lymphoid cells as a novel route of HBV persistence, a source of HBV reactivation and a site for HBV genetic evolution
There is evidence that HBV infection in PBMCs can contribute to viral persistence even in the absence of viral replication in the liver (Figure 3). In this regard, a previous study in patients undergoing liver transplantation detected HBV DNA in the PBMCs of 7 out of 11 patients with no evidence of HBV replicative markers in the liver graft (74). Moreover, further studies have shown that PBMC-specific variants (not detected in the liver or serum before transplantation) can become the predominant viral variant after liver transplantation, thus contributing to the infection of the liver graft (75–78). Notably, some of these PBMC-specific variants can be characterized by the presence of immune-escape mutations (known to promote HBV evasion from neutralizing antibodies), thus contributing to the failure of immune-based therapy, typically used to prevent HBV reactivation after liver transplantation (75–78) (Figure 3). Overall findings support the role of PBMCs as a critical reservoir capable of fueling the persistence and spread of HBV infection.
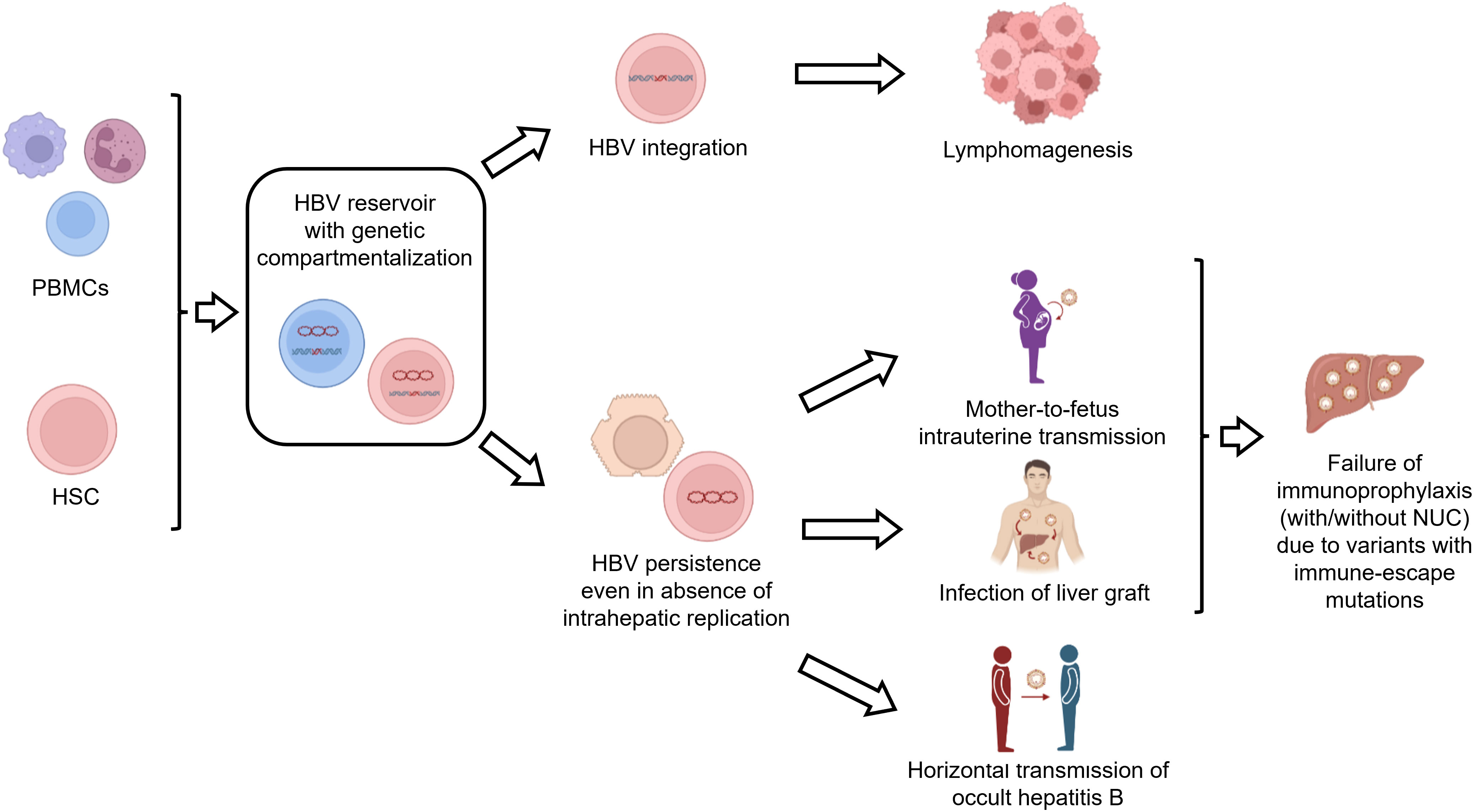
Figure 3 Consequences of HBV infection in lymphoid cells. The figure summarizes the implications of HBV infection in lymphoid cells. HBV-infected lymphoid cells can act as a reservoir of HBV infection. In particular, HBV persistence in lymphoid cells can contribute to i) the infection of the liver graft after liver transplantation, ii) intrauterine HBV transmission and iii) horizontal transmission of occult HBV infection. Furthermore, integrated HBV DNA can promote lymphomagenesis. Finally, the compartmentalization of viral variants with immune-escape mutations in PBMCs can contribute to the failure of vaccination or immunoglobulin treatment.
Furthermore, HBV-infected PBMCs can serve as a source of intrauterine infection (79–81) (Figure 3). In this regard, a previous study has shown that HBV DNA-positive maternal PBMCs can cross the placental barrier and enter the fetal circulation, thus increasing the risk of vertical HBV transmission (81). Notably, these results raise the possibility of using HBV DNA detection in PBMCs as a biomarker of HBV intrauterine infection (81). Again, immune-escape mutations, detected in maternal HBV-infected PBMCs, have been shown to contribute to vertical HBV transmission despite immunoprophylaxis at birth (80). These findings support that HBV infection in PBMCs can lead to a genetic compartmentalization of viral variants playing an important role in promoting vertical HBV transmission (Figure 3).
HBV-infected PBMCs can also contribute to the horizontal transmission of HBV infection (Figure 3). Indeed, using the woodchuck model of hepadnaviral infection, previous studies have found that HBV-infected PBMCs can act as an important reservoir of transmission-competent occult hepadnaviral infection that invariably infects the lymphatic system through parenteral or vertical routes but rarely infects the liver (82, 83). This is in keeping with further studies conducted in humans showing the horizontal transmission (between family members) of occult HBV infection, characterized by a unique genotype and immune-escape mutations within the PBMC compartment (84, 85), again corroborating the role of genetic compartmentalization in PBMCs in mechanisms modulating HBV persistence and spreading.
In a similar direction, unique HBV strains, detected only in PBMCs, have been detected in HIV-infected patients, leading to HBV reactivation and fulminant hepatic failure (75, 86).
Overall findings support the role of PBMCs as a reservoir of HBV infection, critical to fueling human-to-human transmission by different routes and promoting HBV persistence. HBV genetic compartmentalization in PBMCs can further enhance HBV pathobiological properties. The role of HBV-infected PBMCs as a predictive biomarker of liver reinfection after transplantation and of increased risk of vertical transmission deserves further investigation for optimized management of patients with HBV infection.
Potential implications of HBV lymphotropism for antiviral treatment and for HBV cure
The ideal treatment goal of anti-HBV treatment is the achievement of an HBV functional cure, defined as sustained HBsAg loss along with undetectable serum HBV DNA off-therapy, reflecting the silencing of cccDNA transcriptional activity (87–89). Indeed, HBsAg loss has been associated with a significant decrease in the risk of developing end-stage liver diseases, such as HCC and cirrhosis (90–92). Unfortunately, this goal is rarely achieved by currently available treatment options, including nucleos(t)ide analogs (NUCs) and peg-interferon alpha, with very limited or no impact on the cccDNA pool and its transcriptional activity.
Recently, experimental and clinical research in the field of HBV treatment has been devoted to the identification of novel drug molecules capable of promoting HBV functional cure by dampening viral antigen production and enhancing the strength of anti-HBV immune responses (88). In particular, promising data for the achievement of HBV functional cure derive from novel immunotherapies based on checkpoint inhibitors (i.e. PD-1 inhibitors) and therapeutic vaccination, aimed to restore T-cell and B-cell mediated anti-HBV immune response of chronically infected patients and from the RIG-I or Toll-like receptor (TLR) agonists (such as TLR-7 and TLR-8 agonists), aimed to potentiate the activation of innate immunity to counteract HBV infection (93, 94). In this light, the achievement of an HBV functional cure cannot disregard the role of PBMCs as an extrahepatic reservoir of HBV infection that could contribute to persistent HBV production and jeopardize the success of novel therapeutic approaches.
Notably, accumulating evidence demonstrates that liver macrophages play a double-faced role in HBV infection and pathogenesis. Indeed, on one hand M1 macrophages are known to favor HBV clearance by direct antiviral effect and by producing cytokines with trans-activating effect on adaptive immunity, on the other hand M2 macrophages and their phenotype modulation by HBV has been recently demonstrated to favor viral persistence and its related immunopathogenesis (95–97). Furthermore, according to literature, M1 macrophages suppresses early HCC tumorigenesis by eliminating cancer cells while M2 macrophages promote cancer cells proliferation and invasion by suppressing the adaptive immune system. In this light, this differential role should be taken into account for the design of novel immunotherapies, based on macrophages, aimed at achieving HBV functional cure as well as at hindering HBV-related tumorigenesis, including not only hepatocarcinogenesis but also lymphomagenesis.
Moreover, it has been recently shown that HBV DNA integration can represent an important source of HBsAg even when the transcriptional activity of cccDNA has been completely silenced. In this regard, the occurrence of HBV DNA integration in PBMCs suggests that this HBV reservoir can participate along with the liver in the continuous production and release of HBsAg in the patient’s serum. This can contribute to the elevated burden of viral antigens and the subsequent exhaustion of the anti-HBV immune response typically observed in chronic HBV infection (thus far recognized as a barrier to HBV cure) (98, 99). Furthermore, HBsAg production from PBMC-integrated HBV DNA can challenge the role of HBsAg loss as a surrogate marker of HBV functional cure.
Overall findings support the importance of deepening the role of lymphoid cells as HBV reservoir and finely characterizing the intermediates of HBV replication in this compartment. In particular, the evaluation of the efficacy of these novel drugs should include the monitoring of HBV replication in lymphoid cells to verify whether the drug can induce cccDNA silencing in all compartments where the virus hides and replicates.
Conclusions
Numerous studies have confirmed the possibility of finding HBV DNA inside cellular compartments outside the liver, such as the pancreas and kidney, with several data on cells of the hematopoietic system, such as circulating PBMCs, NK cells and bone marrow precursor stem cells. Little is known about the mechanisms of HBV entry into hematological cells; nevertheless, the role of PBMCs as an extrahepatic reservoir of HBV infection deserves further investigation, since it may contribute to persistent HBV production, potentially jeopardizing the success of the current and novel therapeutic approaches, aimed at achieving HBV functional cure.
Furthermore, it is possible that the presence of HBV in immune cell precursors can promote uncontrolled cell proliferation and, possibly, malignancy.
Data correlating HBV with other hematologic malignancies have been reported; however, they are not always confirmed and are often supported by limited studies. In particular, among hematological disorders so far investigated, a stronger association has been reported for B-cell NHL.
There are no clear pathogenetic studies investigating the connections between chronic HBV infection and the onset of lymphatic and/or hematological malignancy, and it is possible that HBV acts more as an inflammatory driver than as a direct carcinogen.
Nevertheless, it is known that HBV can frequently integrate its DNA into the genome of the infected cells. HBV DNA integration is recognized as a mechanism promoting neoplastic transformation by several mechanisms, including the direct dysregulation of oncogenes and onco-suppressors, the production of chimeric viral-human RNAs and proteins with transactivating properties, and the promotion of overall genome instability. Notably, HBV DNA integration represents a common phenomenon in NHL tissues, molecular data show its potential impact in carcinogenesis, and epidemiological evidence seems to confirm a possible correlation.
In conclusion, although many epidemiological and laboratory studies have been produced in recent years, this remains a field of study due to the possible benefits of therapeutic or prophylactic strategies involving antivirals and vaccines.
Author contributions
LS and VS: study conception and design. VS, VM, RS, MI, LP, and SD: literature revision. LS, VS, VM, RS, MI, LP, and SD: draft manuscript preparation. LS and VS: final manuscript revision. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zur Hausen H. The search for infectious causes of human cancers: Where and why. Virology (2009) 392:1–10. doi: 10.1016/j.virol.2009.06.001
2. Mui UN, Haley CT, Tyring SK. Viral oncology: Molecular biology and pathogenesis. J Clin Med (2017) 6:111. doi: 10.3390/jcm6120111
3. Fast facts on global hepatitis b . Available at: https://www.cdc.gov/globalhealth/immunization/diseases/hepatitis-b/data/fast-facts.html.
4. Arbuthnot P, Kew M. Hepatitis b virus and hepatocellular carcinoma. Int J Exp Pathol (2001) 82(2):77–100. doi: 10.1111/j.1365-2613.2001.iep178.x
5. Song C, Lv J, Liu Y, Chen JG, Ge Z, Zhu Z, et al. Associations between hepatitis b virus infection and risk of all cancer types. JAMA Netw Open (2019) 2(6):e195718. doi: 10.1001/jamanetworkopen.2019.5718
6. Yi HZ, Chen JJ, Cen H, Yan W, Tan XH. Association between infection of hepatitis b virus and onset risk of b-cell non-hodgkin's lymphoma: a systematic review and a meta-analysis. Med Oncol (2014) 31(8):84. doi: 10.1007/s12032-014-0084-7
7. Mullen CJR, Volesky KD, Greenwald ZR, El-Zein M, Franco EL. Is Hodgkin lymphoma associated with hepatitis b and c viruses? a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev (2021) 30(12):2167–75. doi: 10.1158/1055-9965.EPI-21-0548
8. Huang Z-H, Lu G-Y, Qiu L-X, Zhong G-H, Huang Y, Yao X-M, et al. Risk of hepatocellular carcinoma in antiviral treatment-naïve chronic hepatitis b patients treated with entecavir or tenofovir disoproxil fumarate: a network meta-analysis. BMC Cancer (2022) 22:287. doi: 10.1186/s12885-022-09413-7
9. Zhang X, Guan L, Tian H, Zeng Z, Chen J, Huang D, et al. Risk factors and prevention of viral hepatitis-related hepatocellular carcinoma. Front Oncol (2021) 11:686962. doi: 10.3389/fonc.2021.686962
10. Yoffe B, Burns DK, Bhatt HS, Combes B. Extrahepatic hepatitis b virus DNA sequences in patients with acute hepatitis b infection. Hepatology (1990) 12:187–92. doi: 10.1002/hep.1840120202
11. Inoue J, Ueno Y, Kogure T, Nagasaki F, Kimura O, Obara N, et al. Analysis of the full-length genome of hepatitis b virus in the serum and cerebrospinal fluid of a patient with acute hepatitis b and transverse myelitis. J Clin Virol (2008) 41:301–4. doi: 10.1016/j.jcv.2008.01.002
12. Marcucci F, Spada E, Mele A, Caserta CA, Pulsoni A. The association of hepatitis b virus infection with b-cell non-Hodgkin lymphoma - a review. Am J Blood Res (2012) 2:18–28.
13. Pontisso P, Vidalino L, Quarta S, Gatta A. Biological and clinical implications of HBV infection in peripheral blood mononuclear cells. Autoimmun Rev (2008) 8:13–7. doi: 10.1016/j.autrev.2008.07.016
14. Kamiza AB, Su F-H, Wang W-C, Sung F-C, Chang S-N, Yeh C-C. Chronic hepatitis infection is associated with extrahepatic cancer development: A nationwide population-based study in Taiwan. BMC Cancer (2016) 16:861. doi: 10.1186/s12885-016-2918-5
15. Zhou X, Pan H, Yang P, Ye P, Cao H, Zhou H. Both chronic HBV infection and naturally acquired HBV immunity confer increased risks of b-cell non-Hodgkin lymphoma. BMC Cancer (2019) 19:477. doi: 10.1186/s12885-019-5718-x
16. Engels EA, Cho ER, Jee SH. Hepatitis b virus infection and risk of non-Hodgkin lymphoma in south Korea: a cohort study. Lancet Oncol (2010) 11:827–34. doi: 10.1016/S1470-2045(10)70167-4
17. Ulcickas Yood M, Quesenberry CP, Guo D, Caldwell C, Wells K, Shan J, et al. Incidence of non-hodgkin’s lymphoma among individuals with chronic hepatitis b virus infection. Hepatology (2007) 46:107–12. doi: 10.1002/hep.21642
18. Andersen ES, Omland LH, Jepsen P, Krarup H, Christensen PB, Obel N, et al. Risk of all-type cancer, hepatocellular carcinoma, non-Hodgkin lymphoma and pancreatic cancer in patients infected with hepatitis b virus. J Viral Hepat (2015) 22:828–34. doi: 10.1111/jvh.12391
19. Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet (2017) 390:298–310. doi: 10.1016/S0140-6736(16)32407-2
20. Huang C-E, Yang Y-H, Chen Y-Y, Chang J-J, Chen K-J, Lu C-H, et al. The impact of hepatitis b virus infection and vaccination on the development of non-Hodgkin lymphoma. J Viral Hepat (2017) 24:885–94. doi: 10.1111/jvh.12713
21. Salpini R, D’Anna S, Benedetti L, Piermatteo L, Gill U, Svicher V, et al. Hepatitis b virus DNA integration as a novel biomarker of hepatitis b virus-mediated pathogenetic properties and a barrier to the current strategies for hepatitis b virus cure. Front Microbiol (2022) 13:972687. doi: 10.3389/fmicb.2022.972687
22. Feitelson MA, Lee J. Hepatitis b virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett (2007) 252:157–70. doi: 10.1016/j.canlet.2006.11.010
23. Zhao L-H, Liu X, Yan H-X, Li W-Y, Zeng X, Yang Y, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun (2016) 7:12992. doi: 10.1038/ncomms12992
24. Álvarez EG, Demeulemeester J, Otero P, Jolly C, García-Souto D, Pequeño-Valtierra A, et al. Aberrant integration of hepatitis b virus DNA promotes major restructuring of human hepatocellular carcinoma genome architecture. Nat Commun (2021) 12:6910. doi: 10.1038/s41467-021-26805-8
25. Bousali M, Papatheodoridis G, Paraskevis D, Karamitros T. Hepatitis b virus DNA integration, chronic infections and hepatocellular carcinoma. Microorganisms (2021) 9:1787. doi: 10.3390/microorganisms9081787
26. Péneau C, Imbeaud S, la Bella T, Hirsch TZ, Caruso S, Calderaro J, et al. Hepatitis b virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut (2022) 71:616–26. doi: 10.1136/gutjnl-2020-323153
27. Ramirez R, van Buuren N, Gamelin L, Soulette C, May L, Han D, et al. Targeted long-read sequencing reveals comprehensive architecture, burden, and transcriptional signatures from hepatitis b virus-associated integrations and translocations in hepatocellular carcinoma cell lines. J Virol (2021) 95(19):e0029921. doi: 10.1128/JVI.00299-21
28. Yu M-C, Lee C-W, Lee Y-S, Lian J-H, Tsai C-L, Liu Y-P, et al. Prediction of early-stage hepatocellular carcinoma using OncoScan chromosomal copy number aberration data. World J Gastroenterol (2017) 23:7818–29. doi: 10.3748/wjg.v23.i44.7818
29. Ding D, Lou X, Hua D, Yu W, Li L, Wang J, et al. Recurrent targeted genes of hepatitis b virus in the liver cancer genomes identified by a next-generation sequencing–based approach. PLoS Genet (2012) 8:e1003065. doi: 10.1371/journal.pgen.1003065
30. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet (2012) 44:760–4. doi: 10.1038/ng.2291
31. Lin SY, Zhang A, Lian J, Wang J, Chang T-T, Lin Y-J, et al. Recurrent HBV integration targets as potential drivers in hepatocellular carcinoma. Cells (2021) 10:1294. doi: 10.3390/cells10061294
32. Coffin CS, Mulrooney-Cousins PM, Michalak TI. Hepadnaviral lymphotropism and its relevance to HBV persistence and pathogenesis. Front Microbiol (2021) 12:695384. doi: 10.3389/fmicb.2021.695384
33. Umeda M, Marusawa H, Seno H, Katsurada A, Nabeshima M, Egawa H, et al. Hepatitis b virus infection in lymphatic tissues in inactive hepatitis b carriers. J Hepatol (2005) 42:806–12. doi: 10.1016/j.jhep.2005.01.016
34. Wang P, Wang X, Cong S, Ma H, Zhang X. Mutation analyses of integrated HBV genome in hepatitis b patients. J Genet Genomics (2008) 35:85–90. doi: 10.1016/S1673-8527(08)60013-2
35. Laskus T, Radkowski M, Wang L-F, Nowicki M, Rakela J. Detection and sequence analysis of hepatitis b virus integration in peripheral blood mononuclear cells. J Virol (1999) 73:1235–8. doi: 10.1128/JVI.73.2.1235-1238.1999
36. Murakami Y, Minami M, Daimon Y, Okanoue T. Hepatitis b virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis b virus surface antigen. J Med Virol (2004) 72:203–14. doi: 10.1002/jmv.10547
37. Mulrooney-Cousins PM, Chauhan R, Churchill ND, Michalak TI. Primary seronegative but molecularly evident hepadnaviral infection engages liver and induces hepatocarcinoma in the woodchuck model of hepatitis b. PLoS Pathog (2014) 10:e1004332. doi: 10.1371/journal.ppat.1004332
38. Li M, Shen Y, Chen Y, Gao H, Zhou J, Wang Q, et al. Characterization of hepatitis b virus infection and viral DNA integration in non-Hodgkin lymphoma. Int J Cancer (2020) 147:2199–209. doi: 10.1002/ijc.33027
39. Lau KCK, Burak KW, Coffin CS. Impact of hepatitis b virus genetic variation, integration, and lymphotropism in antiviral treatment and oncogenesis. Microorganisms (2020) 8:1470. doi: 10.3390/microorganisms8101470
40. Shi Y, Lan Y, Cao F, Teng Y, Li L, Wang F, et al. Infected hematopoietic stem cells and with integrated HBV DNA generate defective T cells in chronic HBV infection patients. J Viral Hepat (2014) 21(7):e39-47. doi: 10.1111/jvh.12236
41. Romet-Lemonne J-L, McLane MF, Elfassi E, Haseltine WA, Azocar J, Essex M. Hepatitis b virus infection in cultured human lymphoblastoid cells. Sci (1979) (1983) 221:667–9. doi: 10.1126/science.6867736
42. Elfassi E, Romet-Lemonne JL, Essex M, Frances-McLane M, Haseltine WA. Evidence of extrachromosomal forms of hepatitis b viral DNA in a bone marrow culture obtained from a patient recently infected with hepatitis b virus. Proc Natl Acad Sci (1984) 81:3526–8. doi: 10.1073/pnas.81.11.3526
43. Pontisso P, Poon MC, Tiollais P, Brechot C. Detection of hepatitis b virus DNA in mononuclear blood cells. BMJ (1984) 288:1563–6. doi: 10.1136/bmj.288.6430.1563
44. Laure F, Zagury D, Saimot AG, Gallo RC, Hahn BH, Brechot C. Hepatitis b virus DNA sequences in lymphoid cells from patients with AIDS and AIDS-related complex. Sci (1979) (1985) 229:561–3. doi: 10.1126/science.2410981
45. Pasquinelli C, Lauré F, Chatenoud L, Beaurin G, Gazengel C, Bismuth H, et al. Hepatitis b virus DNA in mononuclear blood cells. J Hepatol (1986) 3:95–103. doi: 10.1016/S0168-8278(86)80152-0
46. Korba BE, Wells F, Tennant BC, Yoakum GH, Purcell RH, Gerin JL. Hepadnavirus infection of peripheral blood lymphocytes in vivo: Woodchuck and chimpanzee models of viral hepatitis. J Virol (1986) 58:1–8. doi: 10.1128/jvi.58.1.1-8.1986
47. Noonan CA, Yoffe B, Mansell PW, Melnick JL, Hollinger FB. Extrachromosomal sequences of hepatitis b virus DNA in peripheral blood mononuclear cells of acquired immune deficiency syndrome patients. Proc Natl Acad Sci (1986) 83:5698–702. doi: 10.1073/pnas.83.15.5698
48. Zeldis JB, Mugishima H, Steinberg HN, Nir E, Gale RP. In vitro hepatitis b virus infection of human bone marrow cells. J Clin Invest (1986) 78:411–7. doi: 10.1172/JCI112591
49. Gripon P, Diot C, Thézé N, Fourel I, Loreal O, Brechot C, et al. Hepatitis b virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol (1988) 62:4136–43. doi: 10.1128/jvi.62.11.4136-4143.1988
50. Galle PR, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. In vitro experimental infection of primary human hepatocytes with hepatitis b virus. Gastroenterology (1994) 106:664–73. doi: 10.1016/0016-5085(94)90700-5
51. Köck J, Theilmann L, Galle P, Schlicht H. Hepatitis b virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology (1996) 23:405–13. doi: 10.1002/hep.510230303
52. Colucci G, Lyons P, Beazer Y, Waksal SD. Production of hepatitis b virus-infected human b-cell hybridomas: Transmission of the viral genome to normal lymphocytes in cocultures. Virology (1988) 164:238–44. doi: 10.1016/0042-6822(88)90641-1
53. Michalak TI, Pasquinelli C, Guilhot S, Chisari FV. Hepatitis b virus persistence after recovery from acute viral hepatitis. J Clin Invest (1994) 93:230–9. doi: 10.1172/JCI116950
54. Lew Y-Y, Michalak TI. In vitro and In vivo infectivity and pathogenicity of the lymphoid cell-derived woodchuck hepatitis virus. J Virol (2001) 75:1770–82. doi: 10.1128/JVI.75.4.1770-1782.2001
55. Steinberg HS, Bouffard P, Trépo C, Zeldis JB. In vitro inhibition of hemopoietic cell line growth by hepatitis b virus. J Virol (1990) 64:2577–81. doi: 10.1128/jvi.64.6.2577-2581.1990
56. Huang Y, Yan Q, Fan R, Song S, Ren H, Li Y, et al. Hepatitis b virus replication in CD34+ hematopoietic stem cells from umbilical cord blood. Med Sci Monitor (2016) 22:1673–81. doi: 10.12659/MSM.898680
57. Ma R, Xing Q, Shao L, Wang D, Hao Q, Li X, et al. Hepatitis b virus infection and replication in human bone marrow mesenchymal stem cells. Virol J (2011) 8:486. doi: 10.1186/1743-422X-8-486
58. Aurich I, Mueller LP, Aurich H, Luetzkendorf J, Tisljar K, Dollinger MM, et al. Functional integration of hepatocytes derived from human mesenchymal stem cells into mouse livers. Gut (2007) 56:405–15. doi: 10.1136/gut.2005.090050
59. Stoll-Becker S, Repp R, Glebe D, Schaefer S, Kreuder J, Kann M, et al. Transcription of hepatitis b virus in peripheral blood mononuclear cells from persistently infected patients. J Virol (1997) 71:5399–407. doi: 10.1128/jvi.71.7.5399-5407.1997
60. Chemin I, Vermot-Desroches C, Baginski I, Saurin JC, Laurent F, Zoulim F, et al. Selective detection of human hepatitis b virus surface and core antigens in peripheral blood mononuclear cell subsets by flow cytometry. J Viral Hepat (1994) 1:39–44. doi: 10.1111/j.1365-2893.1994.tb00060.x
61. Halpern R, Covey L. Community support for adolescent parents and their children: The parent-to-parent program in Vermont. J Prim Prev (1983) 3:160–73. doi: 10.1007/BF01325437
62. Jilbert AR, Freiman JS, Gowans EJ, Holmes M, Cossart YE, Burrell CJ. Duck hepatitis b virus DNA in liver, spleen, and pancreas: Analysis by in situ and southern blot hybridization. Virology (1987) 158:330–8. doi: 10.1016/0042-6822(87)90205-4
63. Lieberman HM, Tung WW, Shafritz DA. Splenic replication of hepatitis b virus in the chimpanzee chronic carrier. J Med Virol (1987) 21:347–59. doi: 10.1002/jmv.1890210407
64. Korba BE, Cote PJ, Gerin JL. Mitogen-induced replication of woodchuck hepatitis virus in cultured peripheral blood lymphocytes. Sci (1979) (1988) 241:1213–6. doi: 10.1126/science.3261887
65. Walter Ogston C, Schechter EM, Humes CA, Pranikoff MB. Extrahepatic replication of woodchuck hepatitis virus in chronic infection. Virology (1989) 169:9–14. doi: 10.1016/0042-6822(89)90035-4
66. Hosoda K, Omata M, Uchiumi K, Imazeki F, Yokosuka O, Ito Y, et al. Extrahepatic replication of duck hepatitis b virus: More than expected. Hepatology (1990) 11:44–8. doi: 10.1002/hep.1840110109
67. Walter E, Teubner K, Blum HE, Offensperger WB, Offensperger S, Gerok W. Duck hepatitis b virus infection of non-hepatocytes. Liver (1991) 11:53–62. doi: 10.1111/j.1600-0676.1991.tb00491.x
68. Toyoda H, Kumada T, Kaneoka Y, Murakami Y. Impact of hepatitis b virus (HBV) X gene integration in liver tissue on hepatocellular carcinoma development in serologically HBV-negative chronic hepatitis c patients. J Hepatol (2008) 48:43–50. doi: 10.1016/j.jhep.2007.08.016
69. Zhang X, You X, Li N, Zhang W, Gagos S, Wang Q, et al. Involvement of hepatitis b virus X gene (HBx) integration in hepatocarcinogenesis via a recombination of HBx/ alu core sequence/subtelomeric DNA. FEBS Lett (2012) 586:3215–21. doi: 10.1016/j.febslet.2012.06.039
70. Nath A, Agarwal R, Malhotra P, Varma S. Prevalence of hepatitis b virus infection in non-Hodgkin lymphoma: A systematic review and meta-analysis. Intern Med J (2010) 40:633–41. doi: 10.1111/j.1445-5994.2009.02060.x
71. Becker N, Schnitzler P, Boffetta P, Brennan P, Foretova L, Maynadié M, et al. Hepatitis b virus infection and risk of lymphoma: results of a serological analysis within the European case–control study epilymph. J Cancer Res Clin Oncol (2012) 138:1993–2001. doi: 10.1007/s00432-012-1279-y
72. Coffin CS, Michalak TI. Persistence of infectious hepadnavirus in the offspring of woodchuck mothers recovered from viral hepatitis. J Clin Invest (1999) 104:203–12. doi: 10.1172/JCI5048
73. Mulrooney-Cousins PM, Michalak TI. Repeated passage of wild-type woodchuck hepatitis virus in lymphoid cells does not generate cell type-specific variants or alter virus infectivity. J Virol (2008) 82:7540–50. doi: 10.1128/JVI.00405-08
74. Fèray C, Zigneco AL, Samule D, Bismuth A, Reynes M, Tiollais P, et al. Persistent hepatitis b virus infection of mononuclear blood cells without concomitant liver infection. Transplantation (1990) 49:1155–7. doi: 10.1097/00007890-199006000-00025
75. Cassini R, de Mitri MS, Gibellini D, Urbinati L, Bagaglio S, Morsica G, et al. A novel stop codon mutation within the hepatitis b surface gene is detected in the liver but not in the peripheral blood mononuclear cells of HIV-infected individuals with occult HBV infection. J Viral Hepat (2013) 20:42–9. doi: 10.1111/j.1365-2893.2012.01623.x
76. Coffin CS, Osiowy C, Gao S, Nishikawa S, van der Meer F, van Marle G. Hepatitis b virus (HBV) variants fluctuate in paired plasma and peripheral blood mononuclear cells among patient cohorts during different chronic hepatitis b (CHB) disease phases. J Viral Hepat (2015) 22:416–26. doi: 10.1111/jvh.12308
77. Brind A, Jiang J, Samuel D, Gigou M, Feray C, Bréchot C, et al. Evidence for selection of hepatitis b mutants after liver transplantation through peripheral blood mononuclear cell infection. J Hepatol (1997) 26:228–35. doi: 10.1016/S0168-8278(97)80035-9
78. Lee Z, Nishikawa S, Gao S, Eksteen JB, Czub M, Gill MJ, et al. Detection of hepatitis b virus (HBV) genomes and HBV drug resistant variants by deep sequencing analysis of HBV genomes in immune cell subsets of HBV mono-infected and/or human immunodeficiency virus type-1 (HIV-1) and HBV Co-infected individuals. PLoS One (2015) 10:e0137568. doi: 10.1371/journal.pone.0137568
79. Bai G, Wang Y, Zhang L, Tang Y, Fu F. The study on the role of hepatitis b virus X protein and apoptosis in HBV intrauterine infection. Arch Gynecol Obstet (2012) 285:943–9. doi: 10.1007/s00404-011-2096-2
80. Shao Q, Zhao X, Yao Li MD. Role of peripheral blood mononuclear cell transportation from mother to baby in HBV intrauterine infection. Arch Gynecol Obstet (2013) 288:1257–61. doi: 10.1007/s00404-013-2893-x
81. Xu Y-Y, Liu H-H, Zhong Y-W, Liu C, Wang Y, Jia L-L, et al. Peripheral blood mononuclear cell traffic plays a crucial role in mother-to-Infant transmission of hepatitis b virus. Int J Biol Sci (2015) 11:266–73. doi: 10.7150/ijbs.10813
82. Michalak TI. Occult persistence and lymphotropism of hepadnaviral infection: insights from the woodchuck viral hepatitis model. Immunol Rev (2000) 174:98–111. doi: 10.1034/j.1600-0528.2002.017406.x
83. Michalak TI, Mulrooney PM, Coffin CS. Low doses of hepadnavirus induce infection of the lymphatic system that does not engage the liver. J Virol (2004) 78:1730–8. doi: 10.1128/JVI.78.4.1730-1738.2004
84. Datta S, Panigrahi R, Biswas A, Chandra PK, Banerjee A, Mahapatra PK, et al. Genetic characterization of hepatitis b virus in peripheral blood leukocytes: Evidence for selection and compartmentalization of viral variants with the immune escape G145R mutation. J Virol (2009) 83:9983–92. doi: 10.1128/JVI.01905-08
85. Chakravarty R, Neogi M, Roychowdhury S, Panda CK. Presence of hepatitis b surface antigen mutant G145R DNA in the peripheral blood leukocytes of the family members of an asymptomatic carrier and evidence of its horizontal transmission. Virus Res (2002) 90:133–41. doi: 10.1016/S0168-1702(02)00147-8
86. Bagaglio S, Albarello L, Biswas P, Uberti-Foppa C, Fortis C, Morsica G. Virological pattern of hepatitis b infection in an HIV-positive man with fatal fulminant hepatitis b: A case report. J Med Case Rep (2009) 3:110. doi: 10.1186/1752-1947-3-110
87. Lampertico P, Agarwal K, Berg T, Buti M, Janssen HLA, Papatheodoridis G, et al. EASL 2017 clinical practice guidelines on the management of hepatitis b virus infection. J Hepatol (2017) 67:370–98. doi: 10.1016/j.jhep.2017.03.021
88. Degasperi E, Anolli MP, Lampertico P. Towards a functional cure for hepatitis b virus: A 2022 update on new antiviral strategies. Viruses (2022) 14:2404. doi: 10.3390/v14112404
89. Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis b cure: From discovery to regulatory approval. Hepatology (2017) 66:1296–313. doi: 10.1002/hep.29323
90. Yip TC-F, Lok AS-F. How do we determine whether a functional cure for HBV infection has been achieved? Clin Gastroenterol Hepatol (2020) 18:548–50. doi: 10.1016/j.cgh.2019.08.033
91. Vittal A, Sharma D, Hu A, Majeed NA, Terry N, Auh S, et al. Systematic review with meta-analysis: the impact of functional cure on clinical outcomes in patients with chronic hepatitis b. Aliment Pharmacol Ther (2022) 55:8–25. doi: 10.1111/apt.16659
92. Liu F, Wang X-W, Chen L, Hu P, Ren H, Hu H-D. Systematic review with meta-analysis: development of hepatocellular carcinoma in chronic hepatitis b patients with hepatitis b surface antigen seroclearance. Aliment Pharmacol Ther (2016) 43:1253–61. doi: 10.1111/apt.13634
93. Wang D, Fu B, Wei H. Advances in immunotherapy for hepatitis b. Pathogens (2022) 11(10):1116. doi: 10.3390/pathogens11101116
94. Hoogeveen RC, Boonstra A. Checkpoint inhibitors and therapeutic vaccines for the treatment of chronic HBV infection. Front Immunol (2020) 11:401. doi: 10.3389/fimmu.2020.00401
95. Li Y, Li S, Duan X, Yang C, Xu M, Chen L. Macrophage phenotypes and hepatitis b virus infection. J Clin Transl Hepatol (2020) 8(4):424–31. doi: 10.14218/JCTH.2020.00046
96. Faure-Dupuy S, Durantel D, Lucifora J. Liver macrophages: Friend or foe during hepatitis B infection? Liver Int (2018) 38(10):1718–29. doi: 10.1111/liv.13884
97. Li J, Yu M, Zong R, Fan C, Ren F, Wu W, et al. Deacetylation of Notch1 by SIRT1 contributes to HBsAg- and HBeAg-mediated M2 macrophage polarization. Am J Physiol Gastrointest Liver Physiol (2022) 322(4):G459–71. doi: 10.1152/ajpgi.00338.2021
98. Loggi E, Bihl FK, Cursaro C, Granieri C, Galli S, Brodosi L, et al. Virus-specific immune response in HBeAg-negative chronic hepatitis b: Relationship with clinical profile and HBsAg serum levels. PLoS One (2013) 8:e65327. doi: 10.1371/journal.pone.0065327
Keywords: hepatitis B virus, HBV, carcinogenesis, lymphotropism, lymphomagenesis, tumors
Citation: Svicher V, Salpini R, D’Anna S, Piermatteo L, Iannetta M, Malagnino V and Sarmati L (2023) New insights into hepatitis B virus lymphotropism: Implications for HBV-related lymphomagenesis. Front. Oncol. 13:1143258. doi: 10.3389/fonc.2023.1143258
Received: 12 January 2023; Accepted: 02 March 2023;
Published: 15 March 2023.
Edited by:
Giuseppe Gentile, Department of Translational and Precision Medicine, Sapienza University of Rome, ItalyReviewed by:
Kiana Shahzamani, Lorestan University of Medical Sciences, IranGiovanni Battista Gaeta, University of Campania Luigi Vanvitelli, Italy
Copyright © 2023 Svicher, Salpini, D’Anna, Piermatteo, Iannetta, Malagnino and Sarmati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Loredana Sarmati, c2FybWF0aUBtZWQudW5pcm9tYTIuaXQ=
 Valentina Svicher
Valentina Svicher Romina Salpini1
Romina Salpini1 Lorenzo Piermatteo
Lorenzo Piermatteo Marco Iannetta
Marco Iannetta Loredana Sarmati
Loredana Sarmati