
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 04 July 2023
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1142017
Objective: Ghrelin is a protein that regulate appetite and energy balance in the human body, which is encoded by the ghrelin prepropeptide gene (GHRL). GHRL is linked with carcinogenesis and immune regulation. However, the correlation of GHRL to prognosis and tumor-infiltrating lymphocytes in gastric cancer (GC) remains unclear.
Methods: In this study, we assessed the transcriptional expression, prognosis, and different clinicopathological features about GHRL and the correlation between GHRL and tumor infiltration immune cells in GC patients based on the data published in the following databases: TIMER, GEPIA, GEO, STRING, UALCAN, TISIDB, and Kaplan–Meier Plotter. Furthermore, R software analysis for GC Correa’ cascade was also provided. Finally, GHRL expression in GC tissues was assayed using quantitative real-time polymerase chain reaction and immunohistochemistry.
Results: We found that GHRL expression in GC samples was lower than in normal samples and verified by quantitative PCR (qPCR) and immunohistochemistry. However, sample type, cancer stage, and worse survival were correlated to high GHRL expression. We also found that the expression of GHRL in dysplasia was significantly lower than that in CNAG and in GC. High GHRL expression was connected with immunomodulators, chemokines, and infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in GC.
Conclusions: GHRL is a prognostic biomarker for GC patients, and it is correlated with progression of precancerous lesions in GC. It might lead to poor prognosis by regulating tumor immune microenvironment. Studies are important to explore therapeutic targeting GHRL in the future.
Gastric cancer (GC) is the fifth most common cancer and the third most common cause of cancer death globally (1). GC patients are usually at late stages when diagnosed, thus losing the opportunity for surgery and with the poor prognosis (2). Currently, immunotherapy has become an effective treatment in GC (1, 3–7). However, not all GC patients benefit from immunotherapy because of the different immune microenvironment of tumor (8, 9); it is urgent to seek immune-related biomarkers in GC.
Ghrelin was discovered and named by Kojima in 1999 (10), which was a 28-amino acid polypeptide secreted by the stomach and released into the blood, acting on the growth hormone secretagogue receptor (GHSR), thereby promoting the release of growth hormone. Ghrelin exists in the human body in two forms, active (acylated-ghrelin) and inactive (deacylated ghrelin). Ghrelin was encoded by the ghrelin prepropeptide gene (GHRL) (11). Dixit et al. found that ghrelin and its receptor (GHSR) were expressed in T cells and monocytes; ghrelin inhibited the expression of proinflammatory cytokines tumor necrosis factor alpha (TNF-a) and interleukin (IL)-6 and inhibited inflammatory reactions (12). It has been confirmed that the stomach was indeed not the only source of ghrelin (13); hypothalamus, hippocampus, pituitary gland, cortex, small intestine, and pancreas can also secrete a small amount of ghrelin (14–16), GHRL plays important roles in the development of tumors, and aberrant GHRL expression has been found in various cancers, such as breast cancer (17), lung cancer (18), bladder cancer (19), and adrenal cancer (20). At present, there were some researchers who found that ghrelin affects cancer cell proliferation, migration, and invasion through different signaling pathways (21–24). However, the possible mechanisms of GHRL about tumor development, the GHRL expression in different stages of GC Correa’ cascade, and immune engagement in GC has not been well understood before.
In this study, the GHRL expression and its connection to prognosis in GC were presented via diverse databases including the Gene Expression Profiling Interaction Analysis2.0 (GEPIA2), UALCAN, STRING, Kaplan–Meier (KM) plotter, and TISIDB datasets. Furthermore, we used R software to further analyze the dynamic changes in GHRL in different pathological stages of GC Correa’ cascade (25); the Tumor Immune Estimation Resource2.0 (TIMER2.0) was used to investigate the relationship of GHRL with immune-related cells in the tumor microenvironments. In addition, GHRL expression in GC tissues was assayed using quantitative real-time polymerase chain reaction and immunohistochemistry. This study uncovered the critical involvement of GHRL in GC, Correa’ cascade, and the possible mechanisms via which GHRL may regulate tumor-infiltrating immune cells.
We followed the methods of Junchang Zhang et al., 2022 (26). Furthermore, GEO analysis were also involved in this study for comprehensive bioinformatics analysis.
The Tumor Immune Estimation Resource (TIMER2.0) is a platform to analyze immune infiltration in various cancers (https://timer.cistrome.org/)1 (27). We investigated GHRL expression in GC and the relationship between the expression of GHRL and TILs. Furthermore, the relationship between GHRL expression with gene markers of TILs, including markers of CD8+/CD4+ T cells, B cells, monocytes, TAMs, M1 macrophages, M2 macrophages, natural killer (NK) cells, neutrophils, and dendritic cells (DCs) has been analyzed.
The Gene Expression Profiling Interactive Analysis2.0 (GEPIA2) database (http://gepia.cancer-pku.cn/index.html) 2is a platform that obtains the data from TCGA and The Genotype–Tissue Expression (GTEx) databases (28). In this study, we used the GEPIA data source to analyze the expression and survival of GHRL in GC.
In this study, we used the UALCAN website (29) (https://ualcan.path.uab.edu/)3 to examine the GHRL expression level from major clinical features such as tissue type (healthy/tumor), Helicobacter pylori status, and GC stage (stages 1–4).
To further validate the expression of GHRL in GC Correa’ cascade, GEO microarray series (GSE55696, GSE130823, GSE87666, GSE116312, GSE106656, GSE160116, GSE5081, and GSE78523) containing STAD tumor and non-tumor samples were obtained from the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/)4.
The prognostic significance of GHRL in GC, including overall survival (OS) and post-progression survival (PPS), was investigated using the KM plotter (30) (http://kmplot.com/analysis/)5.
TISIDB (http://cis.hku.hk/TISIDB/index.php)6 is an online platform that combines various heterogeneous data to find tumor and immune system interactions (31). In this research, TISIDB was used to investigate the association of GHRL with TILs, immunostimulators, immunoinhibitors, chemokines, and receptors in GC.
STRING (http://string-db.org)7 is an online database for the prediction of protein interaction relationships (32). The GHRL-related genes were displayed in the STRING website. Then, we performed enrichment analysis using Metascape (https://metascape.org/)8.
In this study, paired GC tissues and adjacent non-cancer tissues were acquired from the First Affiliated Hospital of Nanchang University. Nine pairs of fresh GC tissues and adjacent non-cancer tissues were cryopreserved and were used for the quantitative analysis of the expression of GHRL by quantitative real-time PCR (qRT-PCR) and immunohistochemistry. All patients signed an informed consent form. The Ethics Committee approved the study of the First Affiliated Hospital of Nanchang University (Ethical Application Ref: 2021006).
The total RNA of GC tissues and adjacent non-cancer tissues was extracted using RNA extraction kit (Yeasen Biotechnology, China). cDNA synthesis was performed using the Hifair III 1st Strand cDNA Synthesis SuperMix (Yeasen Biotechnology, China). qRT-PCR was performed using the SYBR Primix Ex Taq™ II (Tiangen Biotechnology, Germany) on ABI-7500 instrument (Applied Biosystems, United States). Actin was used as an internal reference gene, and the 2−△△Ct method was used to compare the expression of GHRL. Primers used for qRT-PCR were as follows: ACTIN, 5′-CTCCACCCTGGCCTCGCTGT-3′ (F), 5′-GCTGTCACCTTCGTTCC-3′(R); GHRL, 5′-TACTACTCTCCACGCCC-3′ (F), 5′-AGGGGCCATCCACAGTCTTC-3′ (R).
Immunohistochemistry (IHC) was performed to investigate the expression of GHRL. IHC staining of these specimens were conducted as previously described (33). Anti-GHRL (Santa Cruz Biotechnology, Dallas, TX, USA, sc-293422) were used for IHC staining.
HR and p-values were described using log-rank test. Spearman’s correlation coefficient was used to analyze the connection of GHRL expression with immune infiltration levels, immunostimulators, immunoinhibitors, chemokines, and receptors. Statistical significance was defined as p < 0.05.
According to the GEPIA database, we discovered that GHRL expression level was increased in diffuse large b-cell lymphoma and acute myeloid leukemia but was decreased in stomach adenocarcinoma (STAD) compared with healthy tissues. Figures 1A, B show the findings.
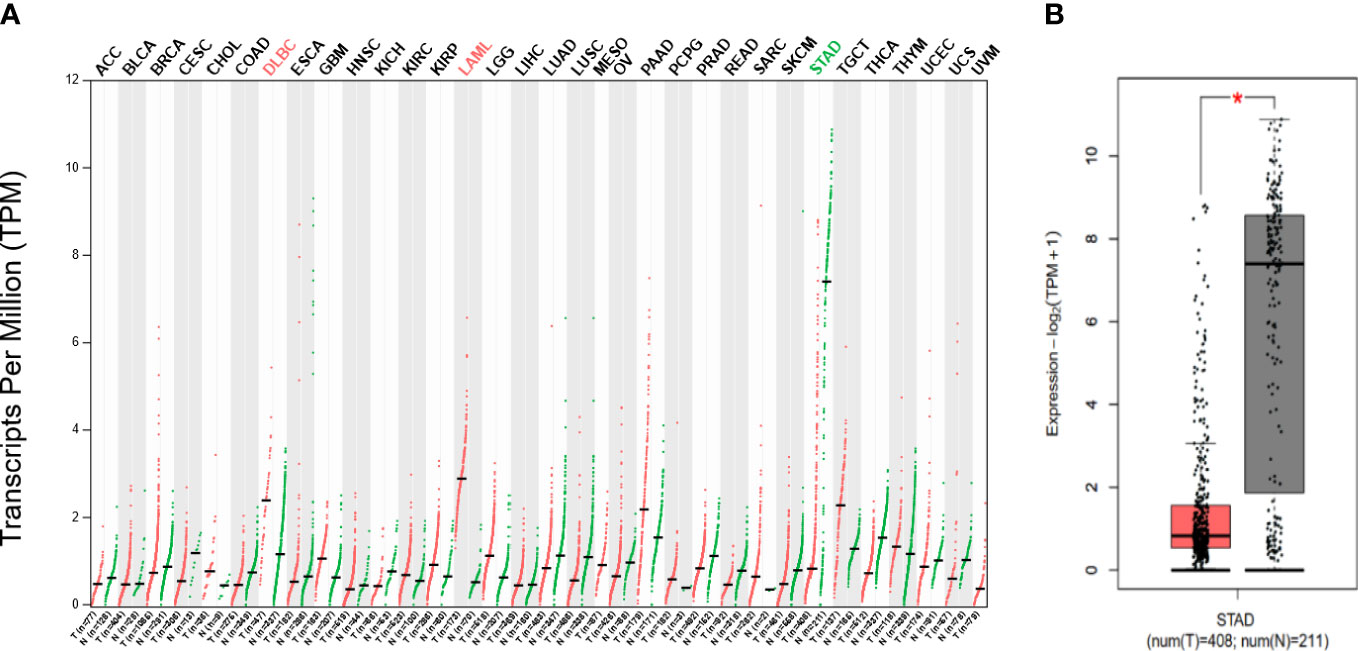
Figure 1 (A) Difference in GHRL expression in pan-carcinoma (GEPIA), low expression in STAD at RNA level (B). Red mark represents that GHRL expression in tumor is higher than adjacent issue; green mark represents that GHRL expression in tumor is lower than that in adjacent issue. *p < 0.05.
We examined GHRL expression in relation to sample type (healthy/primary tumor), tumor stage (stages 1–4), and H. pylori status by applying UALCAN. As shown in Figure 2A, GC samples had lower GHRL expression than healthy samples (p = 0.0036). However, there was no significant difference between H. pylori-positive and H. pylori-negative tumors (Figure 2B). A study of tumor stages revealed that GHRL in the middle and late-stage cancers was significantly higher expressed than in the early stages, suggesting a potential function for GHRL in cancer development and migration (Figure 2C).
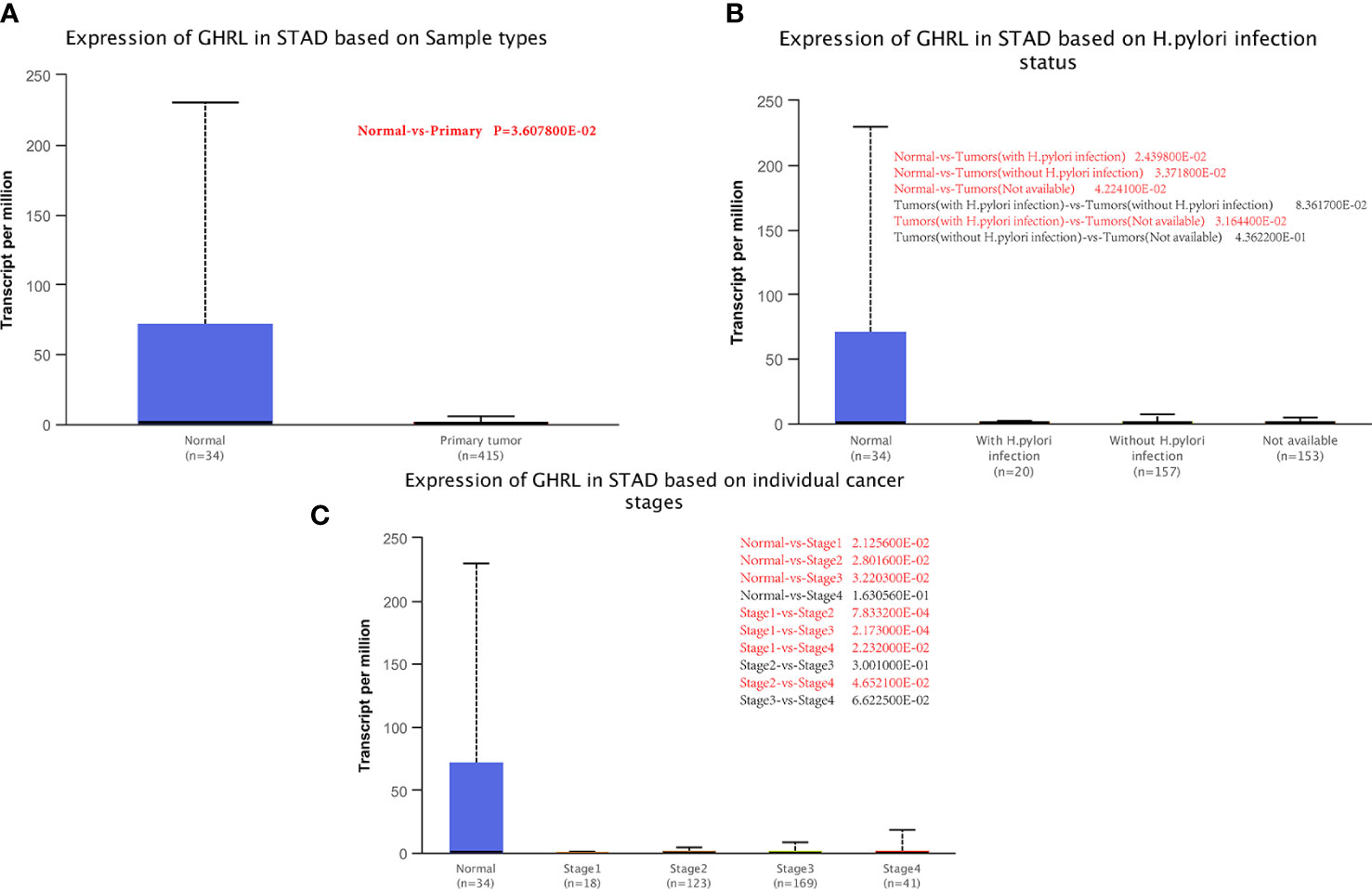
Figure 2 Correlation between GHRL mRNA expression level and clinicopathological parameters of GC through the UALCAN database. (A) Sample type (normal/primary tumor). (B) (H) pylori status. (C) Cancer stage (stage 1–4). N, normal; P, primary tumor; S1, stage 1; S2, stage 2; S3, stage 3; S4, stage 4; STAD, stomach adenocarcinoma. Red mark represents a statistically significant difference between groups.
To further analyze the dynamic changes of GHRL in different pathological stages of GC Correa’ cascade, we used R software to analyze the GEO dataset. We found that the expression of GHRL in GC was higher than that in dysplasia (LGIN+HGIN), and the expression of GHRL in dysplasia was significantly lower than that in CNAG (Figure 3A). However, there was no significant difference in GHRL expression among CAG, CNAG, and NC (Figure 3B).
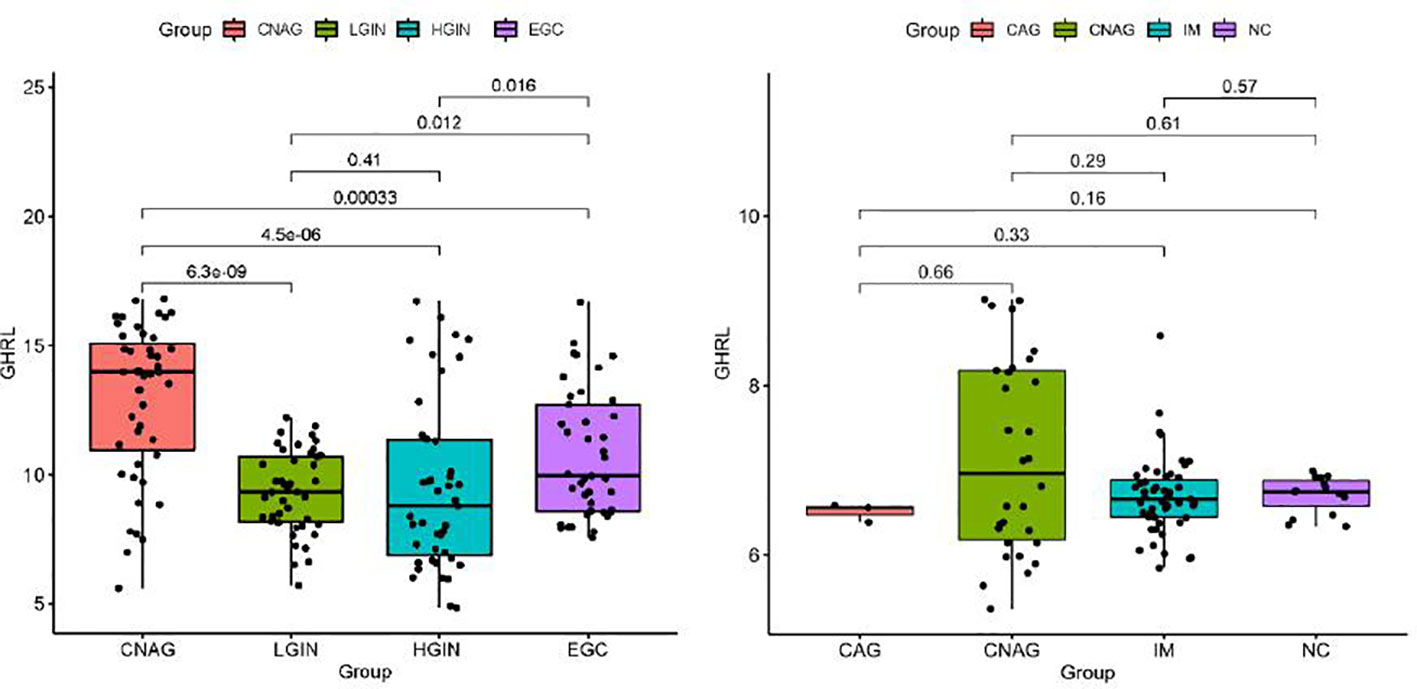
Figure 3 The GHRL expression in different stages of GC Correa’ cascade. CNAG, chronic non-atrophic gastritis; LGIN, low-grade intraepithelial neoplasia; HGIN, high-grade intraepithelial neoplasia; EGC, early GC; CAG, chronic atrophic gastritis; IM, intestinal metaplasia; NC, normal control.
In this research, we evaluate whether GHRL can be used as a prognostic biomarker in GC via Kaplan–Meier survival curves. This study showed that increased GHRL expression was linked with poorer overall survival (OS) and disease-free survival (DFS) in GC (Figure 4). To study the significance and possible molecular mechanisms of GHRL expression in tumor development, we found the correlation between the GHRL expression and clinical–pathological features of GC in the KM plotter. High GHRL expression was linked to poorer OS in female patients and poorer PPS in male patients. Specifically, increased GHRL expression was associated with poorer PPS in stage 3 (PPS HR = 0.61, p= 0.046) of GC patients (Table 1). We also discovered that OS and PPS at stage T3 (OS, HR = 0.62, p = 0.0077) and N1 + 2 + 3 (OS, HR = 0.75, p = 0.042) were related to GHRL expression. Surely, we found that PPS of intestinal GC based on Lauren classification was related to high GHRL expression. These findings suggested that the prognostic significance of GHRL in GC patients was determined by their clinical features.
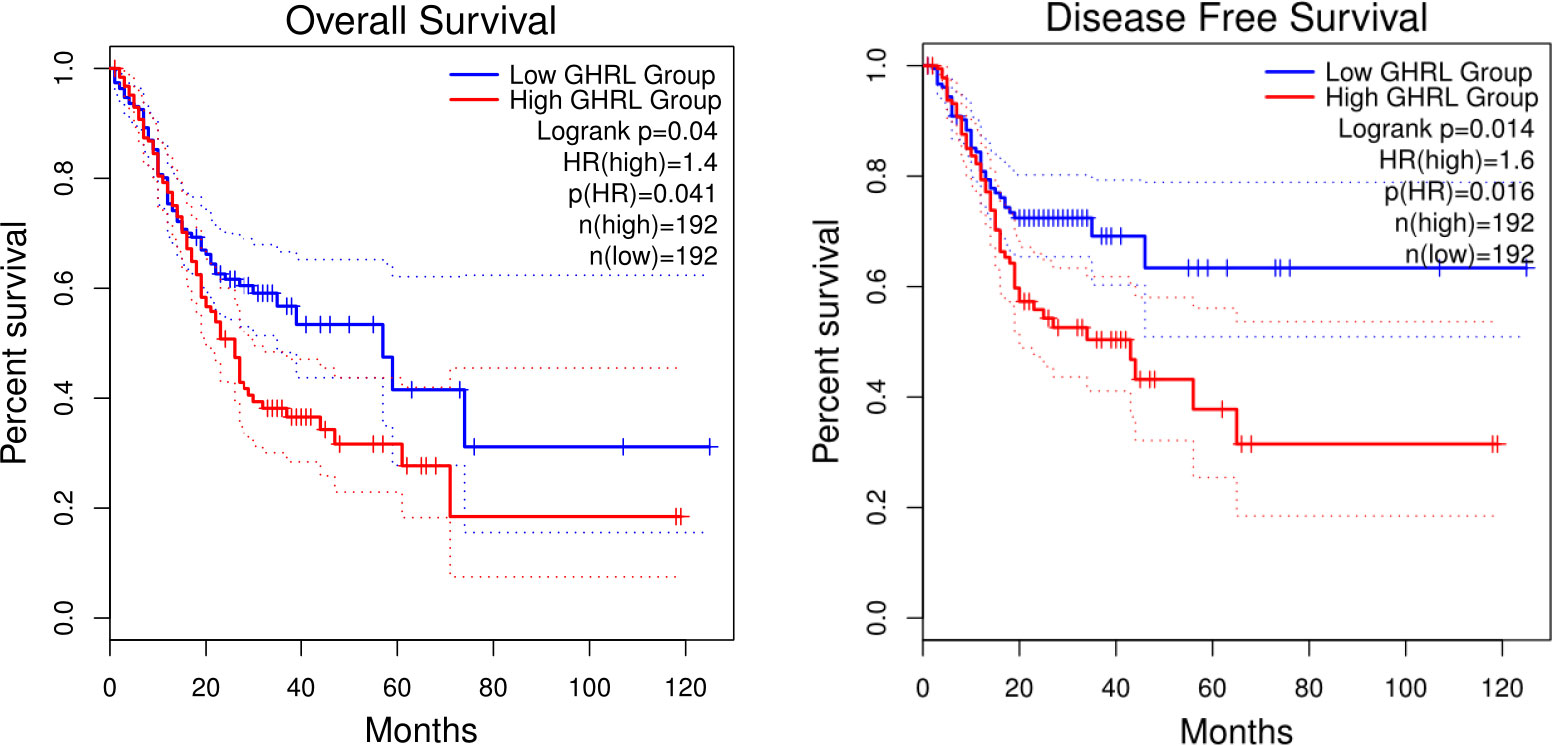
Figure 4 Kaplan–Meier survival curves comparing the high and low expression of GHRL in GC in GEPIA databases.
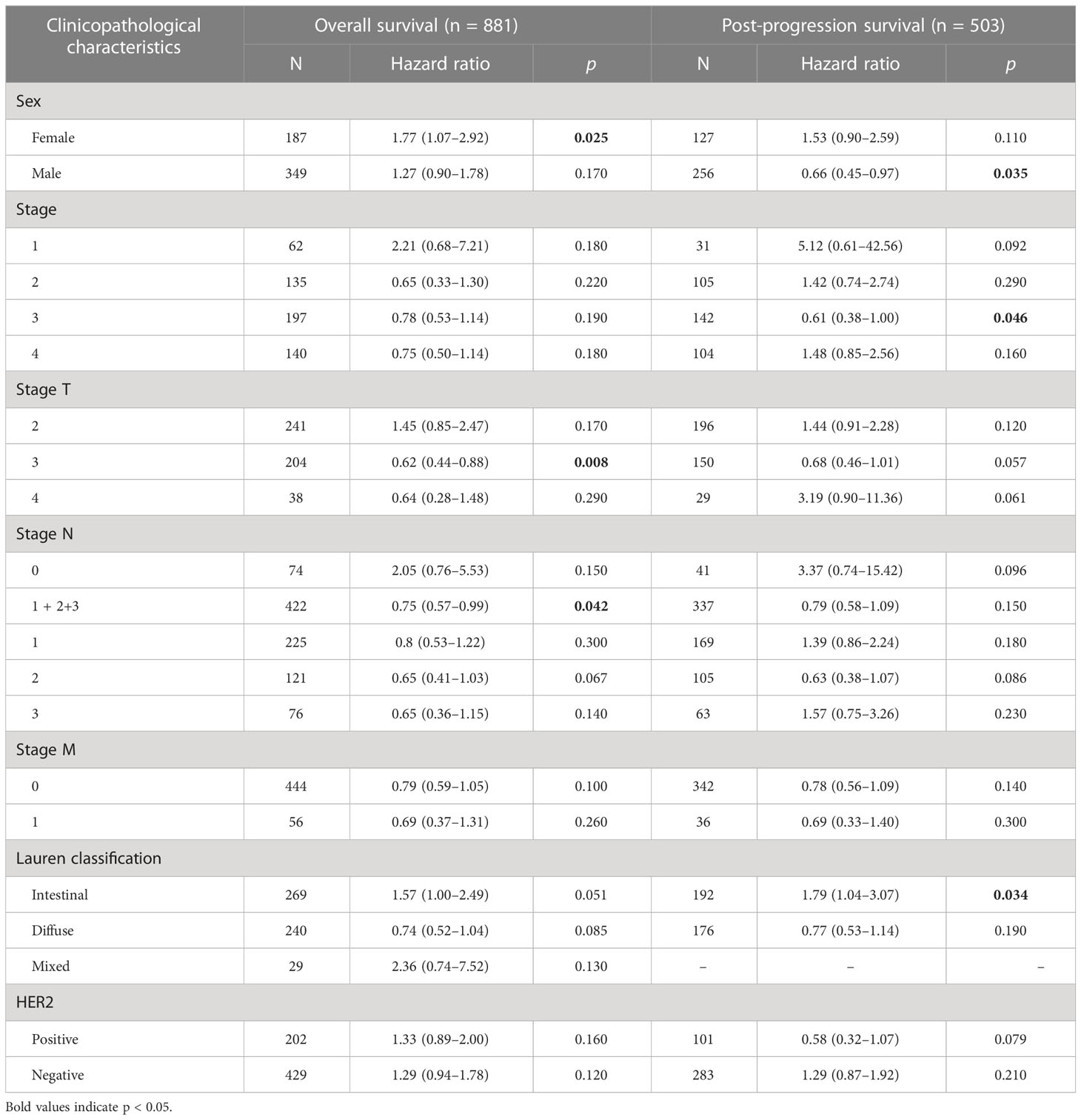
Table 1 Correlation of GHRL mRNA expression and clinical prognosis in gastric cancer with different clinicopathological factors by Kaplan–Meier plotter.
Immune infiltration is an important factor associated with tumor development (34). In this study, TISIDB and TIMER platforms were used to assess GHRL expression in connection with immune cell infiltration levels in GC. GHRL expression was adversely correlated with the purity of STAD (rho = −0.236, p <3.22e−6). Our results also discovered that GHRL had a correlation with the abundance of TILs (Figure 5A). High expression level of GHRL was positively correlated with infiltrating degree of B cells (rho = 0.217), CD8+ T cells (rho = 0.239), CD4+ T cells (rho = 0.345), macrophages (rho = 0.27), neutrophils (rho = 0.183), and dendritic cells (rho = 0.238) (Figure 5B). These results indicate that GHRL plays an important function in the immune infiltration of GC. TIMER and GEPIA databases were used to study the association between GHRL and different biomarkers of TILs (CD8+ T cells, B cells, T cells, NK cells, monocytes, DCs, TAMs, M1macrophages, M2 macrophages, neutrophils, and related subtypes) in STAD. We found that GHRL was associated with the majority of TILs markers in STAD. GHRL was linked to the majority of immune marker sets of TILs in STAD (Table 2). Indeed, GHRL had a significant association with the majority of marker sets of CD8+ T cells, B cells, monocytes, M2 macrophages, DCs, Th2, Th17, Treg, and T-cell exhaustion in STAD (Table 2), including CD8A, CD8B, CD19, CD86, CD163, BDCA-1(CD1C), GATA3, STAT6, STAT3, FOXP3, CCR8, CTLA4, and LAG3 (p<0.01). Furthermore, according to the GEPIA database, we further assessed the connection of GHRL expression with the markers of M1 macrophages, M2 macrophages, monocytes, TAMs, and T-cell exhaustion (Table 3). Therefore, GHRL may regulate T-cell exhaustion and macrophage polarization in GC.
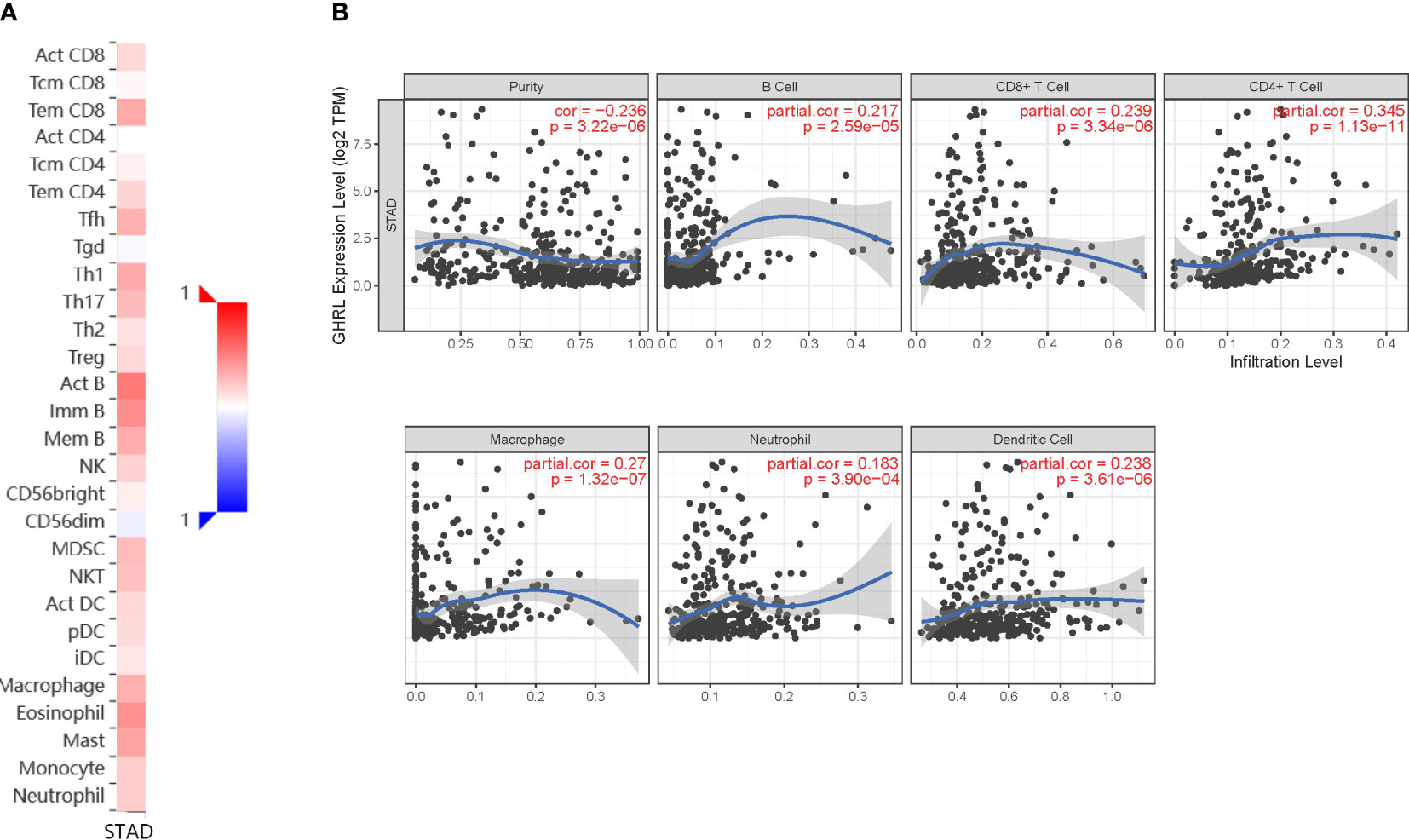
Figure 5 Correlation of GHRL expression with immune infiltration in GC. (A) Correlation between the expression of GHRL and the abundance of TILs in GC available at TISIDB database. (B) Correlation of GHRL expression with infiltration levels of B cell, CD8 + T cell, CD4 + T cell, macrophage, neutrophil, and dendritic cell in GC available at TIMER2.0 database. TILs, tumor-infiltrating lymphocytes.
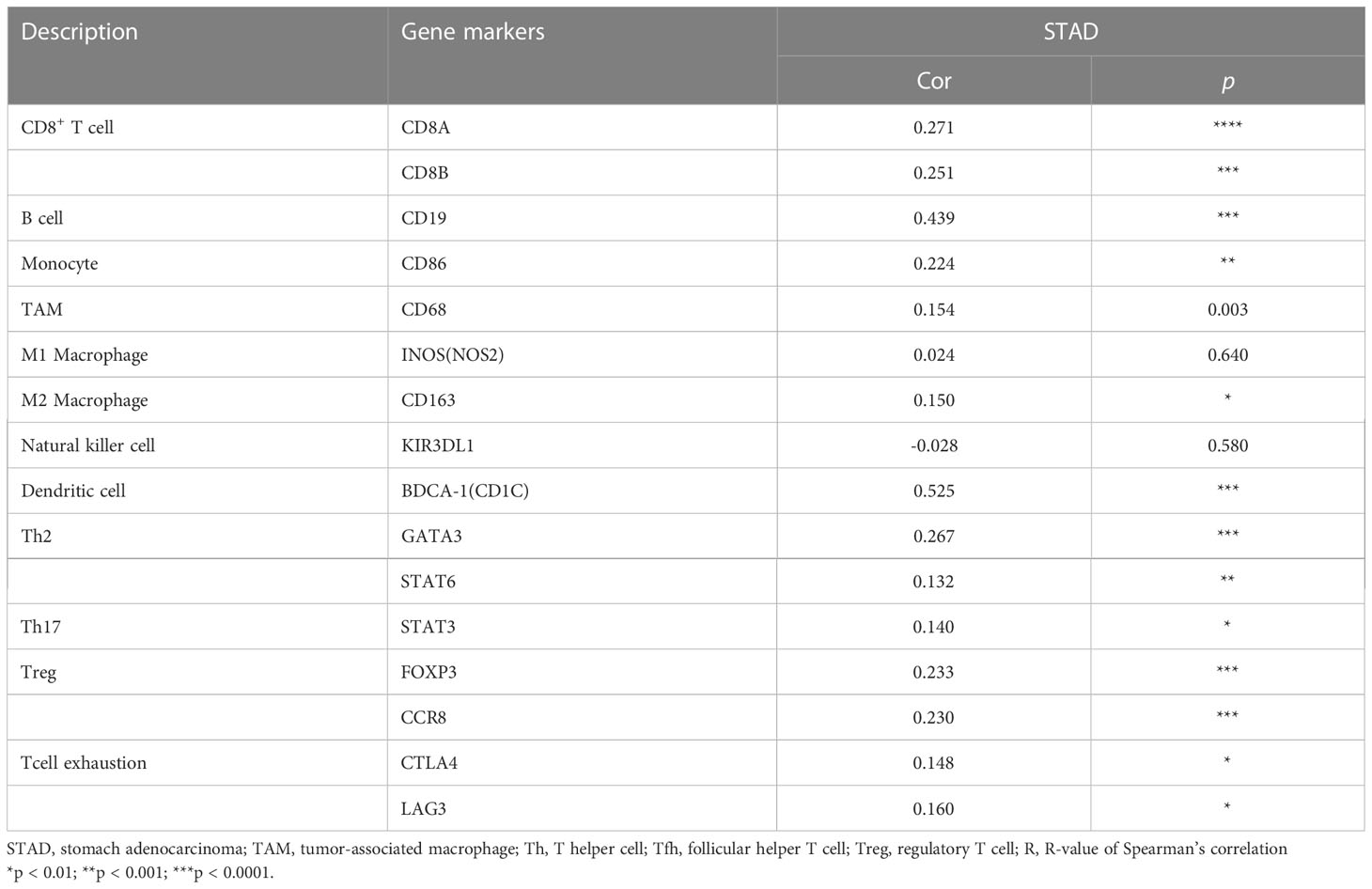
Table 2 Correlation analysis between GHRL and related genes and markers of immune cells in Tumor Immune Estimation Resource (TIMER2.0).
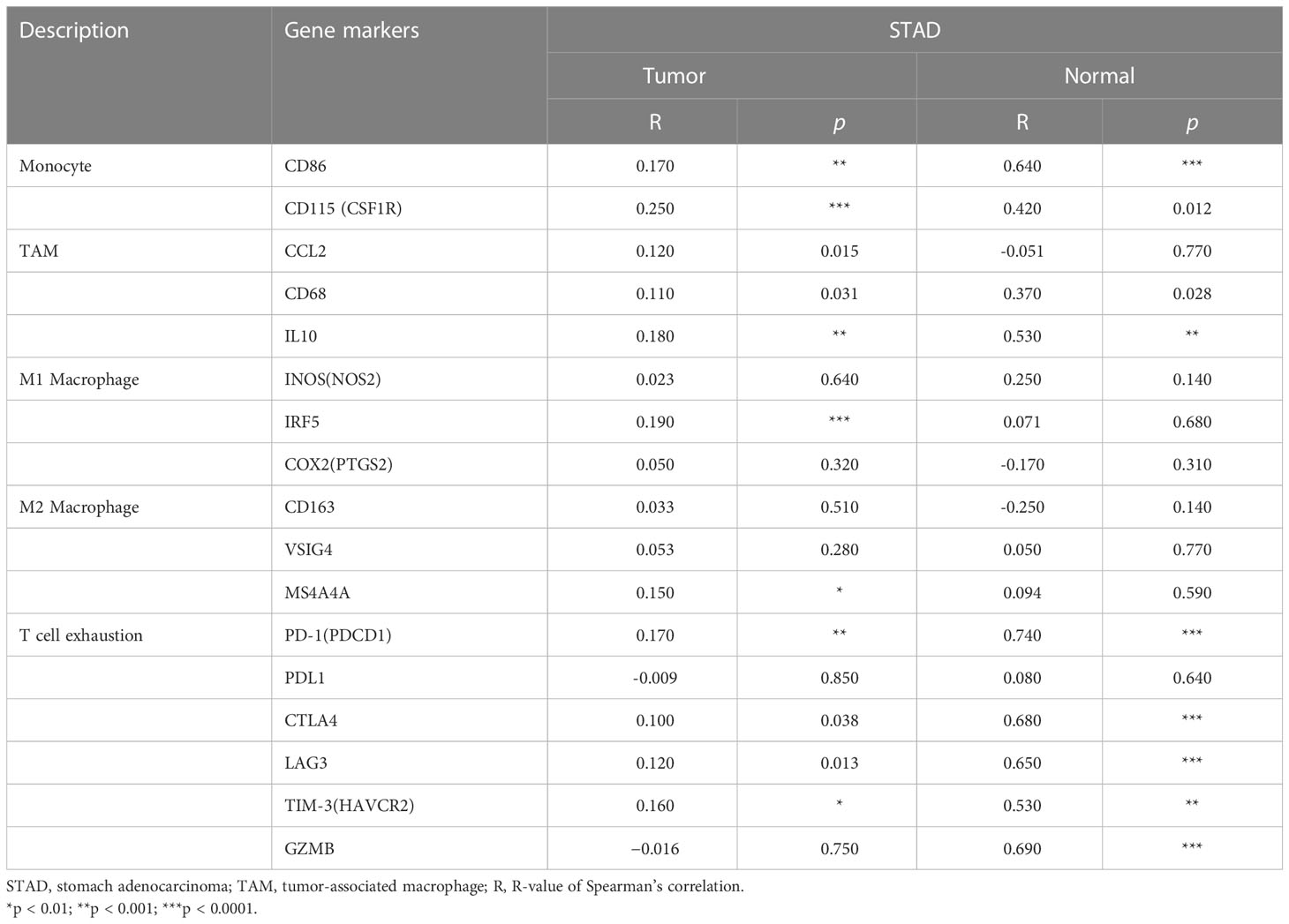
Table 3 Correlation analysis between GHRL and related genes and markers of immune cells in Gene Expression Profiling Interaction Analysis (GEPIA).
This research indicated that GHRL was significantly connected with immunoinhibitors (p < 0.0001), such as ADORA2A (rho = 0.347), BTLA (rho = 0.417), CD244 (rho = 0.262), and CSF1R (rho = 0.279) (Figure 6A). The expression of GHRL was also closely associated with immunostimulators (p < 0.0001), including CD27 (rho = 0.447), CD28 (rho = 0.409), CD40LG (rho = 0.466), CD48 (rho = 0.414), CXCR4 (rho = 0.375), IL6R (rho = 0.342), LTA (rho = 0.324), TMIGD2 (rho = 0.309), TNFRSF13C (rho = 0.335), and TNFRSF17 (rho = 0.438) (Figure 6B). These results suggested that GHRL is engaged in the regulation of immune interaction and may modulate tumor immune escape.
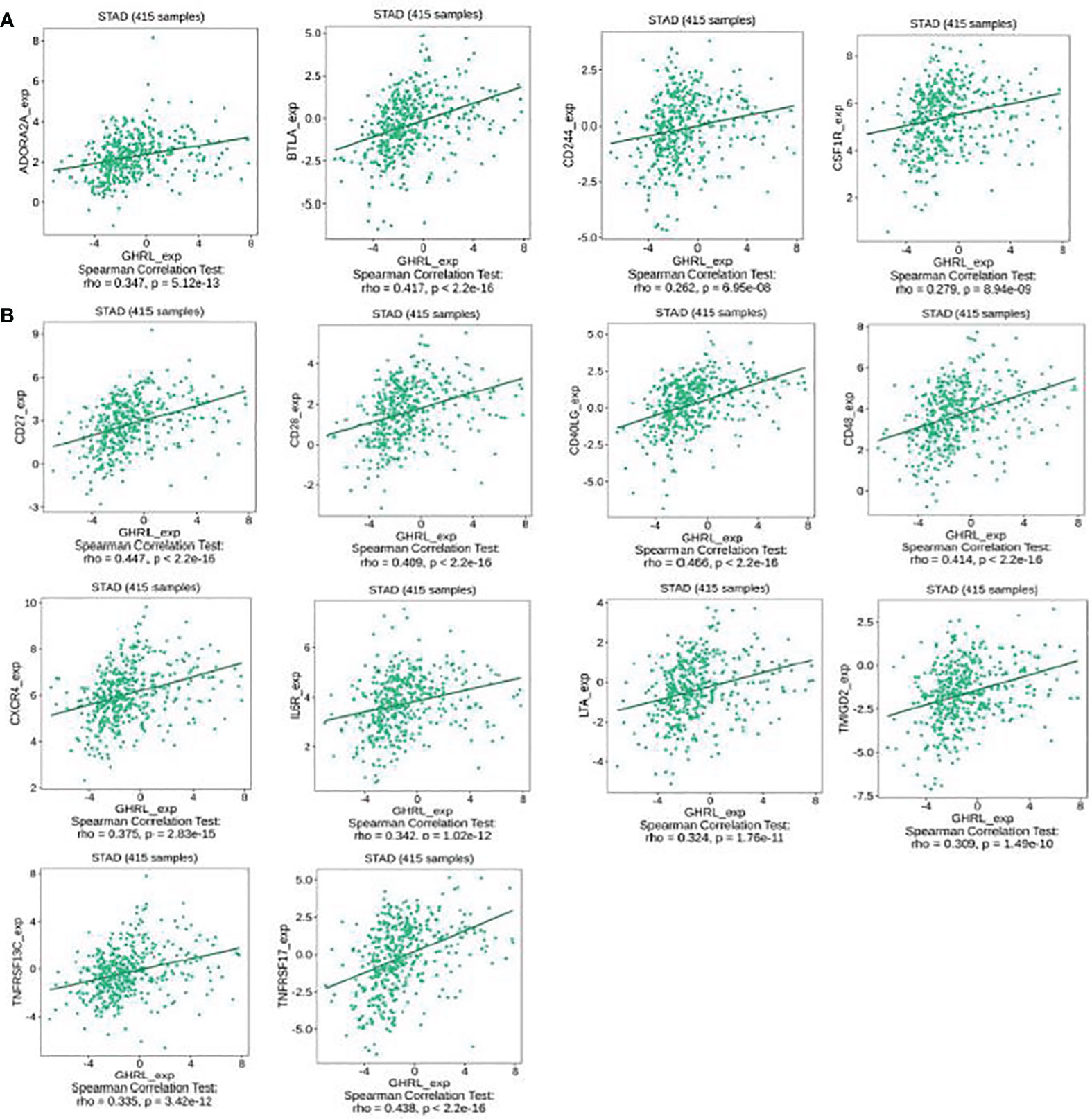
Figure 6 The expression of GHRL is associated with immunomodulators in GC. (A) Correlation between GHRL expression and immunoinhibitors in GC available at TISIDB database. (B) Correlation between GHRL expression and immunostimulators in GC available at TISIDB database.
This research also implicated the association between GHRL expression with chemokines. For instance, GHRL expression was significantly linked to CCL14 (rho = 0.316), CCL19 (rho = 0.438), CCL21 (rho = 0.365), CCL22 (rho = 0.319), CXCL14 (rho = 0.308), CXCL17 (rho = 0.404), CCR9 (rho = 0.375), and TNFRSF13B (rho = 0.518) (Figure 7A). Meanwhile, we demonstrated that GHRL expression was also correlated with chemokine receptors (p < 0.001), including CCR2 (rho = 0.355), CCR4 (rho = 0.427), CCR6 (rho = 0.395), CCR7 (rho = 0.483), CXCR4 (rho = 0.375), CXCR5 (rho = 0.503), and CX3CR1 (rho = 0.407) (Figure 7B). These results demonstrated that GHRL may function as an immunoregulatory factor in GC.
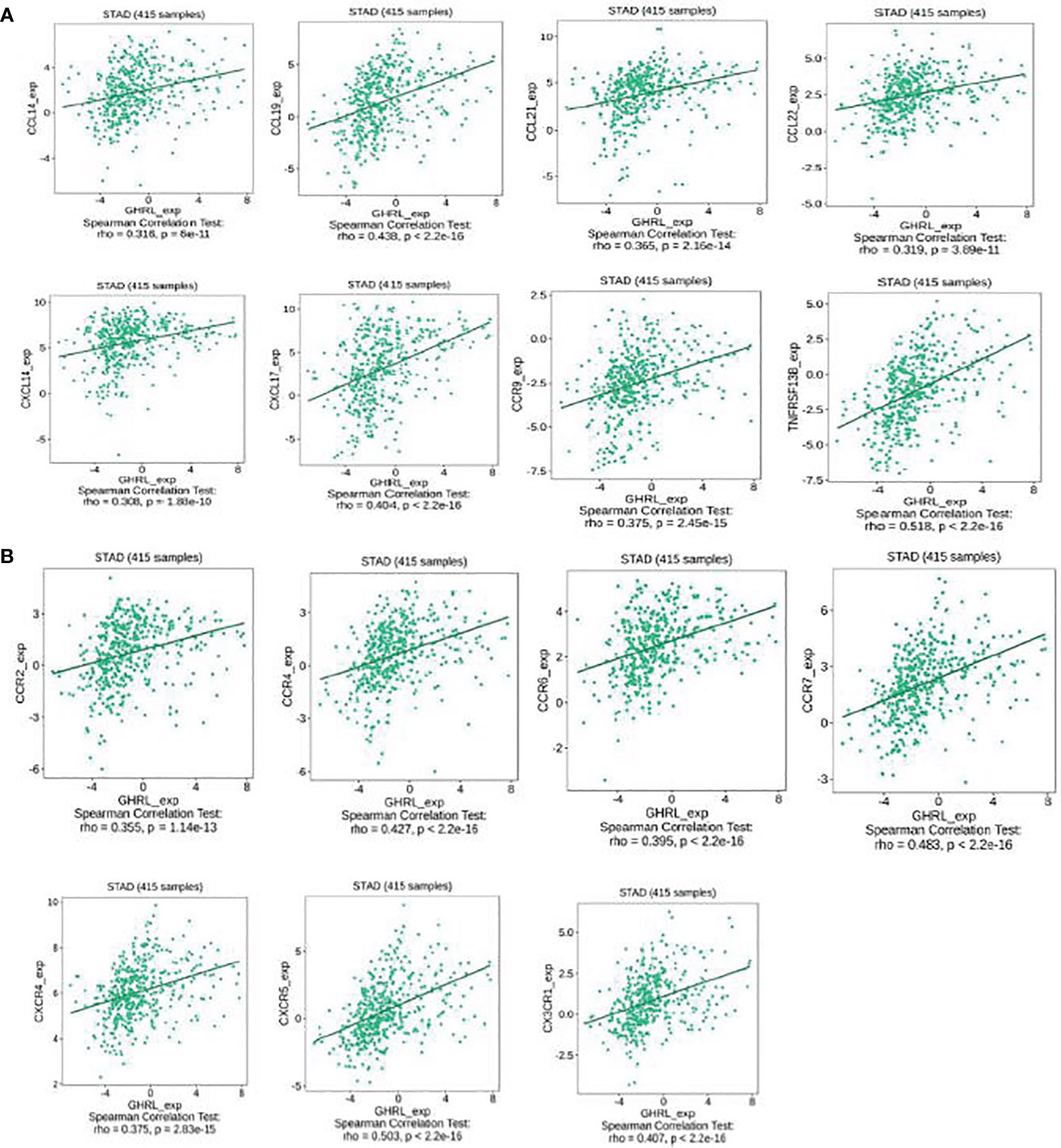
Figure 7 Correlation between the expression of GHRL and chemokines in GC. (A) Correlation between GHRL expression and chemokines in GC available at TISIDB database. (B) Correlation between GHRL expression and chemokine receptors in GC available at TISIDB database.
A total of 11 genes (GHRL, GHRH, GHSR, CCK, GPR39, MBOAT4, UCN, LEP, INS, GH1, and IGF1) were filtered into the target genes PPI network complex, containing 11 nodes and 38 edges (Figure 8A). The enrichment analysis indicated that co-expressed genes of GHRL were highly enriched in synthesis, secretion, and deacylation of Ghrelin, positive regulation of growth, positive regulation of insulin-like growth factor receptor signaling pathway, GPCR ligand binding (Figure 8B), eating disorders, bulimia nervosa, neuroendocrine tumors, and gastric ulcer (Figure 8C).
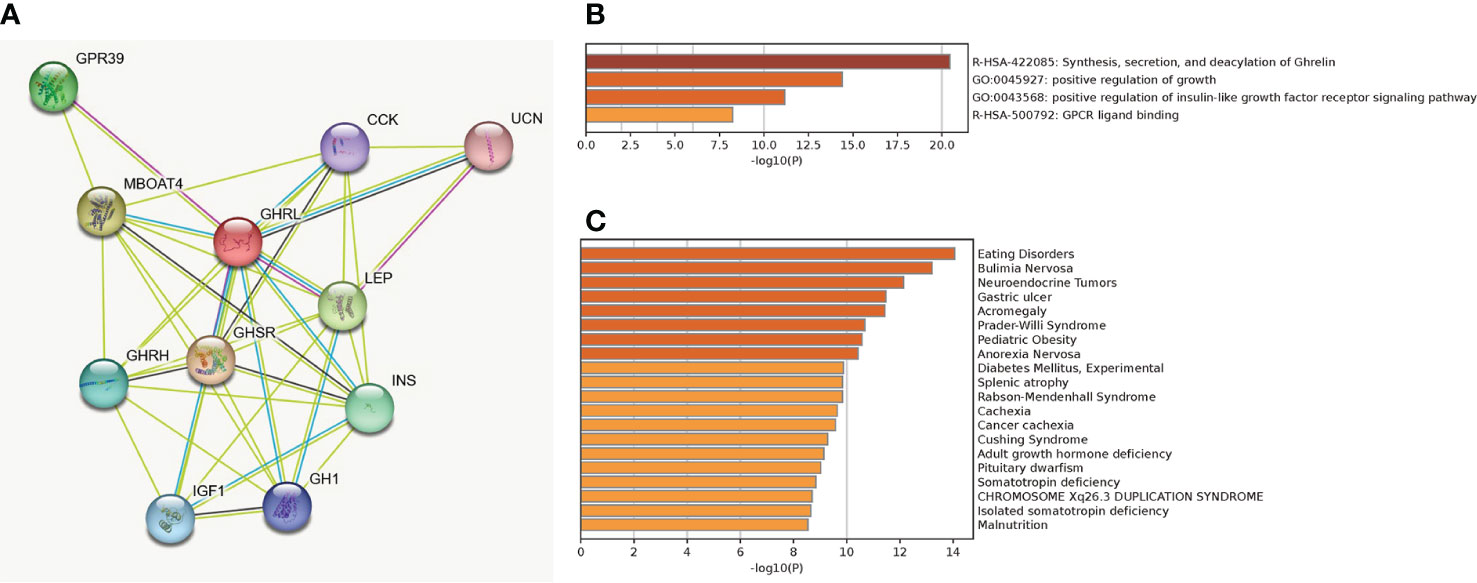
Figure 8 PPI network based on string database (A), biological process enrichment analysis of GHRL-related genes (B), and enrichment analysis on human disease (C).
In order to characterize GHRL expression in GC tissues, qRT-PCR was performed, and the results showed that GHRL expression was significantly lower in GC tissues than adjacent noncancerous tissues (Figure 9A). In addition, we conducted an IHC method to determine that GHRL expression was lower in GC tissues than those in adjacent non-cancerous tissues (Figure 9B).
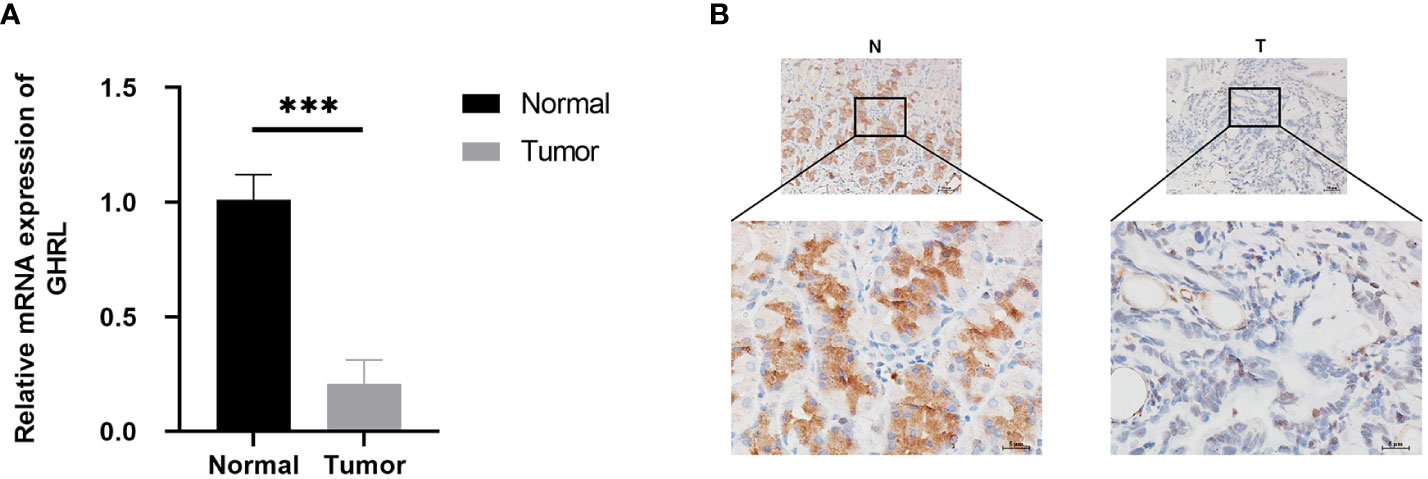
Figure 9 GHRL expression in nine pairs of GC tissues performed by qRT-PCR (A) and IHC (B), ***p < 0.0001.
In the present study, a comprehensive bioinformatics investigation was performed to analyze the clinical significance and expression level of GHRL in GC. Our results revealed that poor prognosis was consistent with high expression of GHRL in GC. Furthermore, our data also indicated that GHRL expression had a close association with the infiltration degrees of different immune cells, immunostimulators, immunoinhibitors, chemokines, and receptors in GC. Therefore, our research revealed new insights in understanding the function of GHRL, and it may be a prognostic biomarker linked to immune infiltration of GC.
Ghrelin, a widely distributed peptide hormone, participates in a series of cancer progression (35); GHRL plays a vital role in carcinogenic potential, which was correlated with breast cancer, lung cancer, bladder cancer, and adrenal cancer (17–20). There is an increasing evidence implicating an immunoregulatory role for ghrelin. Ghrelin mainly acts on the innate and adaptive immune systems to suppress inflammation and induce an anti-inflammatory profile (36). However, the possible function of GHRL in regulating tumor immunity and its clinical significance in GC are still unknown.
Therefore, we evaluated GHRL expression in GC using databases including GEPIA, TIMER, GEO, and UALCAN. We discovered that GHRL was clearly decreased in GC compared with normal samples. These results showed that the level of GHRL expression may serve as a potential diagnostic biomarker in GC. Furthermore, to confirm whether GHRL can be used as a prognostic biomarker, we used the KM plotter database to analyze the correlation between GHRL expression and OS and PPS. However, the results indicated that the higher GHRL expression correlated with worse OS and poor PPS of GC. In addition, high GHRL expression had a significant correlation with a worse prognosis of GC in stages T3 and N1 + 2 + 3 for worse OS and in stage 3 for worse PPS. These observations support our hypothesis that GHRL may act as an anti-oncogene in GC. For low expression of GHRL in GC and high expression of GHRL linked with poor prognosis in GC, previous studies have also found similar seemingly contradictory results, and they made no explanation and experimental verification (37–39). Our explanation may be that the higher is the malignancy, the more oncogene GHRL is needed in GC. In addition, previous studies found that the expression of GHRL was decreased in intestinal GC compared with diffuse histotype via transcriptome analysis on GC specimens (40). In addition, we first discovered that the expression of GHRL in dysplasia was significantly lower than that in CNAG and in GC, so we speculated that ghrelin may be associated with the progression of precancerous lesions. GHRL played different roles in different pathological stages. Previously, a large number of scholars studied the relationship among ghrelin, gastric mucosal atrophy, and H. pylori status, but the results were controversial (41–46). Our study first discovered that GHRL was not related to gastric mucosal atrophy and H. pylori status via bioinformatics investigation, probably because of the small sample size of GEO database on gastric mucosal atrophy; therefore, large sample clinical studies and fundamental experiment may be needed for validation in the future.
Additionally, this study discovered that GHRL was related to immune infiltration in GC. In the tumor microenvironment, it has been demonstrated that immune cell infiltration played vital roles in the development and progression of cancer (47, 48). GHRL played a role in inflammatory and immune responses (49). Recent studies have revealed that GHRL was involved in the regulation of metabolism, energy balance, and the immune, cardiovascular, and reproductive systems (50–52). However, whether GHRL expression was linked to immune infiltration in GC remains unknown. Therefore, we systematically examined the association between GHRL expression and the degree of immune infiltration in GC. In our study, this was the first time that GHRL regulating immune infiltration in GC was identified. Our study showed that GHRL expression had a correlation with TILs including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. At the same time, decreased GHRL expression was associated with immunostimulators, immunoinhibitors, chemokines, and receptors. In addition, this study also demonstrated the association between GHRL expression and the TIL marker genes of GC. Indeed, GHRL expression had an association with M1 macrophage markers, like IRF5, and M2 macrophage markers, such as MS4A4A and CD163, correlated with GHRL expression. These findings indicate that GHRL has a potential function to regulate macrophage polarization.
Furthermore, our results revealed that GHRL expression was significantly correlated to cell response to chemokines according to the TISIDB databases. These results reflected that it may be a direction for enhancing immunotherapy effectiveness by targeting GHRL. Above all, GHRL played a key function in recruiting and modulating TILs in GC, and it is worth to continue investigating the molecular mechanism and function of GHRL in modulating the tumor microenvironment.
Unfortunately, we are yet to conduct experimental studies validating the function of GHRL in the development of GC and the molecular mechanism of GHRL in GC immunity, but in the future, we guarantee that experiments will be performed to further validate the projected results.
The high GHRL expression is closely correlated with poor prognosis. GHRL may be associated with the progression of precancerous lesions in GC. The low GHRL expression enhanced immune infiltration degree including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in GC. Moreover, the expression of GHRL contributes to the regulation of CD8+ T cells, B cells, monocytes, M2 macrophages, DCs, Th2, Th17, Treg, and T-cell exhaustion. Therefore, this study suggests that GHRL may serve as a useful biomarker and therapeutic target for patients with GC.
A preprint has previously been published on https://doi.org/10.21203/rs.3.rs-1790354/v1.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by The Ethics Committee approved the study of the First Affiliated Hospital of Nanchang University (Ethical Application Ref: 2021006). The patients/participants provided their written informed consent to participate in this study.
YX designed the experiment and supervised the study. DL and JW contributed to formal analysis. JW wrote the manuscript. YX reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Nos. 82270593 and 81970502) and the Science and Technology Project of Jiangxi Province (No. 20201ZDG02007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
2. Ouyang S, Li H, Lou L, Huang Q, Zhang Z, Mo J, et al. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol (2022) 52:102317. doi: 10.1016/j.redox.2022.102317
3. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med (2018) 24(9):1449–58. doi: 10.1038/s41591-018-0101-z
4. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med (2015) 373(1):23–34. doi: 10.1056/NEJMoa1504030
5. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-Positive non-Small-Cell lung cancer. N Engl J Med (2016) 375(19):1823–33. doi: 10.1056/NEJMoa1606774
6. Wang D, Lin J, Yang X, Long J, Bai Y, Yang X, et al. Combination regimens with PD-1/PD-L1 immune checkpoint inhibitors for gastrointestinal malignancies. J Hematol Oncol (2019) 12(1):42. doi: 10.1186/s13045-019-0730-9
7. Lote H, Cafferkey C, Chau I. PD-1 and PD-L1 blockade in gastrointestinal malignancies. Cancer Treat Rev (2015) 41(10):893–903. doi: 10.1016/j.ctrv.2015.09.004
8. Tirino G, Pompella L, Petrillo A, Laterza MM, Pappalardo A, Caterino M, et al. What's new in gastric cancer: the therapeutic implications of molecular classifications and future perspectives. Int J Mol Sci (2018) 19(9). doi: 10.3390/ijms19092659
9. Roviello G, Catalano M, Iannone LF, Marano L, Brugia M, Rossi G, et al. Current status and future perspectives in HER2 positive advanced gastric cancer. Clin Transl Oncol (2022) 24(6):981–96. doi: 10.1007/s12094-021-02760-0
10. Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature (1999) 402(6762):656–60. doi: 10.1038/45230
11. Kotta AS, Kelling AS, Corleto KA, Sun Y, Giles ED. Ghrelin and cancer: examining the roles of the ghrelin axis in tumor growth and progression. Biomolecules (2022) 12(4). doi: 10.3390/biom12040483
12. Dixit VD, Schaffer EM, Pyle RS, Collins GD, Sakthivel SK, Palaniappan R, et al. Ghrelin inhibits leptin- and activation-induced proinflammatory cytokine expression by human monocytes and T cells. J Clin Invest (2004) 114(1):57–66. doi: 10.1172/JCI200421134
13. Hosoda H, Kojima M, Mizushima T, Shimizu S, Kangawa K. Structural divergence of human ghrelin. identification of multiple ghrelin-derived molecules produced by post-translational processing. J Biol Chem (2003) 278(1):64–70. doi: 10.1074/jbc.M205366200
14. Higgins SC, Gueorguiev M, Korbonits M. Ghrelin, the peripheral hunger hormone. Ann Med (2007) 39(2):116–36. doi: 10.1080/07853890601149179
15. Hou Z, Miao Y, Gao L, Pan H, Zhu S. Ghrelin-containing neuron in cerebral cortex and hypothalamus linked with the DVC of brainstem in rat. Regul Pept (2006) 134(2-3):126–31. doi: 10.1016/j.regpep.2006.02.005
16. Date Y, Kojima M, Hosoda H, Sawaguchi A, Mondal MS, Suganuma T, et al. Ghrelin, a novel growth hormone-releasing acylated peptide, is synthesized in a distinct endocrine cell type in the gastrointestinal tracts of rats and humans. Endocrinology (2000) 141(11):4255–61. doi: 10.1210/endo.141.11.7757
17. Ahmed AK, Asad M, Ahmed M, Asdaq S, Mohzari Y, Alrashed A, et al. A case control study on serum levels of potential biomarkers in Male breast cancer patients. Int J Environ Res Public Health (2021) 18(9). doi: 10.3390/ijerph18094852
18. Ghe C, Cassoni P, Catapano F, Marrocco T, Deghenghi R, Ghigo E, et al. The antiproliferative effect of synthetic peptidyl GH secretagogues in human CALU-1 lung carcinoma cells. Endocrinology (2002) 143(2):484–91. doi: 10.1210/endo.143.2.8654
19. Karimi M, Heshmati M, Fattahi S, Bagheri N, Alibeigi FM, Taheri F, et al. The relation between the ghrelin receptor and FOXP3 in bladder cancer. Biotech Histochem (2021) 96(4):287–95. doi: 10.1080/10520295.2020.1799074
20. Komarowska H, Malinska A, Komekbai Z, Brominska B, Bednarek-Rajewska K, Ruchala M, et al. Immunohistochemical analysis of ghrelin expression in various types of adrenal tumors. Folia Histochem Cytobiol (2021) 59(2):86–94. doi: 10.5603/FHC.a2021.0009
21. Hu XL, Zhu YJ, Hu CH, You L, Wu J, He XY, et al. Ghrelin affects gastric cancer progression by activating AMPK signaling pathway. Biochem Genet (2021) 59(3):652–67. doi: 10.1007/s10528-020-10022-x
22. Li H, Zhang X, Feng L. Ghrelin regulates cyclooxygenase-2 expression and promotes gastric cancer cell progression. Comput Math Methods Med (2021) 2021:5576808. doi: 10.1155/2021/5576808
23. Zhu J, Yao J, Huang R, Wang Y, Jia M, Huang Y. Ghrelin promotes human non-small cell lung cancer A549 cell proliferation through PI3K/Akt/mTOR/P70S6K and ERK signaling pathways. Biochem Biophys Res Commun (2018) 498(3):616–20. doi: 10.1016/j.bbrc.2018.03.031
24. Tian C, Zhang L, Hu D, Ji J. Ghrelin induces gastric cancer cell proliferation, migration, and invasion through GHS-R/NF-kappaB signaling pathway. Mol Cell Biochem (2013) 382(1-2):163–72. doi: 10.1007/s11010-013-1731-6
26. Zhang J, Wang H, Yuan C, Wu J, Xu J, Chen S, et al. ITGAL as a prognostic biomarker correlated with immune infiltrates in gastric cancer. Front Cell Dev Biol (2022) 10:808212. doi: 10.3389/fcell.2022.808212
27. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res (2020) 48(W1):W509–14. doi: 10.1093/nar/gkaa407
28. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
29. Chandrashekar DS, Bashel B, Balasubramanya S, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
30. Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat (2016) 160(3):439–46. doi: 10.1007/s10549-016-4013-7
31. Ru B, Wong CN, Tong Y, Zhong JY, Zhong S, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics (2019) 35(20):4200–2. doi: 10.1093/bioinformatics/btz210
32. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res (2017) 45(D1):D362–8. doi: 10.1093/nar/gkw937
33. Lin Y, Liang Z, He L, Yang M, Liu D, Gu HF, et al. Gut ghrelin regulates hepatic glucose production and insulin signaling via a gut-brain-liver pathway. Cell Commun Signal (2019) 17(1):8. doi: 10.1186/s12964-019-0321-y
34. Paijens ST, Vledder A, de Bruyn M, Nijman HW. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol (2021) 18(4):842–59. doi: 10.1038/s41423-020-00565-9
35. Leng Y, Zhao C, Yan G, Xu S, Yang Y, Gong T, et al. Ghrelin enhances cisplatin sensitivity in HO-8910 PM human ovarian cancer cells. J Ovarian Res (2021) 14(1):162. doi: 10.1186/s13048-021-00907-9
36. Pereira J, Da SF, de Moraes-Vieira P. The impact of ghrelin in metabolic diseases: an immune perspective. J Diabetes Res (2017) 4527980. doi: 10.1155/2017/4527980
37. Wu X, Wu Y, Ye B, Wu F, Wang P. High expression of ghrelin and obestatin prepropeptide in tumor tissues predicted adverse overall survival in gastric carcinoma patients. Med (Baltimore) (2020) 99(26):e20635. doi: 10.1097/MD.0000000000020635
38. Liu H, Qu Y, Zhou H, Zheng Z, Zhao J, Zhang J. Bioinformatic analysis of potential hub genes in gastric adenocarcinoma. Sci Prog (2021) 104(1):368504211004260. doi: 10.1177/00368504211004260
39. Hussein NA, Rashad MM, Abdou AS, Hussein AM, Mohamed HM. Gene profiling of SEC13, SMAD7, GHRL, long non-coding RNA GHRLOS, HIF-1α in gastric cancer patients. Sci Rep (2022) 12(1):6555. doi: 10.1038/s41598-022-10402-w
40. Carino A, Graziosi L, Marchianò S, Biagioli M, Marino E, Sepe V, et al. Analysis of gastric cancer transcriptome allows the identification of histotype specific molecular signatures with prognostic potential. Front Oncol (2021) 11:663771. doi: 10.3389/fonc.2021.663771
41. Isomoto H, Nakazato M, Ueno H, Date Y, Nishi Y, Mukae H, et al. Low plasma ghrelin levels in patients with helicobacter pylori-associated gastritis. Am J Med (2004) 117(6):429–32. doi: 10.1016/j.amjmed.2004.01.030
42. Gokcel A, Gumurdulu Y, Kayaselcuk F, Serin E, Ozer B, Ozsahin AK, et al. Helicobacter pylori has no effect on plasma ghrelin levels. Eur J Endocrinol (2003) 148(4):423–6. doi: 10.1530/eje.0.1480423
43. Nwokolo CU, Freshwater DA, O'Hare P, Randeva HS. Plasma ghrelin following cure of helicobacter pylori. Gut (2003) 52(5):637–40. doi: 10.1136/gut.52.5.637
44. Ikeda K, Chiba T, Sugai T, Kangawa K, Hosoda H, Suzuki K, et al. Correlation between plasma or mucosal ghrelin levels and chronic gastritis. Hepatogastroenterology (2011) 58(110-111):1622–7. doi: 10.5754/hge10271
45. Gao XY, Kuang HY, Liu XM, Ma ZB, Nie HJ, Guo H. Plasma obestatin levels in men with chronic atrophic gastritis. Peptides (2008) 29(10):1749–54. doi: 10.1016/j.peptides.2008.05.027
46. Eun BS, Hoon LJ, Soo PY, Ok KS, Young CJ, Yong AJ, et al. Decrease of serum total ghrelin in extensive atrophic gastritis: comparison with pepsinogens in histological reference. Scand J Gastroenterol (2016) 51(2):137–44. doi: 10.3109/00365521.2015.1083049
47. Liu Z, Wu X, Tian Y, Zhang W, Qiao S, Xu W, et al. H. pylori infection induces CXCL8 expression and promotes gastric cancer progress through downregulating KLF4. Mol Carcinog (2021) 60(8):524–37. doi: 10.1002/mc.23309
48. Xu R, Wu J, Zhang X, Zou X, Li C, Wang H, et al. Modified bu-zhong-yi-qi decoction synergies with 5 fluorouracile to inhibits gastric cancer progress via PD-1/PD- L1-dependent T cell immunization. Pharmacol Res (2020) 152:104623. doi: 10.1016/j.phrs.2019.104623
49. Skibola DR, Smith MT, Bracci PM, Hubbard AE, Agana L, Chi S, et al. Polymorphisms in ghrelin and neuropeptide y genes are associated with non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev (2005) 14(5):1251–6. doi: 10.1158/1055-9965.EPI-04-0895
50. Menzies M, Seim I, Josh P, Nagaraj SH, Lees M, Walpole C, et al. Cloning and tissue distribution of novel splice variants of the ovine ghrelin gene. BMC Vet Res (2014) 10:211. doi: 10.1186/s12917-014-0211-x
51. Safari R, Hoseinifar SH, Van Doan H, Dadar M. The effects of dietary myrtle (Myrtus communis) on skin mucus immune parameters and mRNA levels of growth, antioxidant and immune related genes in zebrafish (Danio rerio). Fish Shellfish Immunol (2017) 66:264–9. doi: 10.1016/j.fsi.2017.05.007
52. Pech-Pool S, Berumen LC, Martinez-Moreno CG, García-Alcocer G, Carranza M, Luna M, et al. Thyrotropin-releasing hormone (TRH) and somatostatin (SST), but not growth hormone-releasing hormone (GHRH) nor ghrelin (GHRL), regulate expression and release of immune growth hormone (GH) from chicken bursal b-lymphocyte cultures. Int J Mol Sci (2020) 21(4). doi: 10.3390/ijms21041436
Keywords: GHRL, gastric cancer, prognosis, tumor immune microenvironment, Correa’ cascade
Citation: Wang J, Liu D and Xie Y (2023) GHRL as a prognostic biomarker correlated with immune infiltrates and progression of precancerous lesions in gastric cancer. Front. Oncol. 13:1142017. doi: 10.3389/fonc.2023.1142017
Received: 15 February 2023; Accepted: 12 June 2023;
Published: 04 July 2023.
Edited by:
Yuming Jiang, Stanford University, United StatesReviewed by:
Baohong Liu, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2023 Wang, Liu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Xie, eGlleW9uZ190ZmFob25jdUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.