
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 03 April 2023
Sec. Cancer Molecular Targets and Therapeutics
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1140452
Gastric cancer remains the fourth most frequently diagnosed malignancy and the fifth leading cause of cancer-related mortality worldwide owning to the lack of efficient drugs and targets for therapy. Accumulating evidence indicates that UPS, which consists of E1, E2, and E3 enzymes and proteasome, plays an important role in the GC tumorigenesis. The imbalance of UPS impairs the protein homeostasis network during development of GC. Therefore, modulating these enzymes and proteasome may be a promising strategy for GC target therapy. Besides, PROTAC, a strategy using UPS to degrade the target protein, is an emerging tool for drug development. Thus far, more and more PROTAC drugs enter clinical trials for cancer therapy. Here, we will analyze the abnormal expression enzymes in UPS and summarize the E3 enzymes which can be developed in PROTAC so that it can contribute to the development of UPS modulator and PROTAC technology for GC therapy.
According to the global cancer statistics, one million new gastric cancer (GC) cases and about 769,000 deaths were estimated in 2020, ranking fourth for mortality and fifth for incidence among all types (1). At present, the clinical treatments of GC mainly include surgical resection, chemotherapy, radiotherapy, and molecular targeted therapy (2, 3). Molecular targeted therapy has achieved major success in recent years with the understanding of molecular mechanisms in cancer. However, only scant targets are used to develop drugs in GC such as VEGFR-2, HER2, PD-1, etc. (4). Moreover, most of traditional small molecule inhibitors affect active site of the targets to inhibit its function. Due to the limit of drugs targets and related technology, the development of targeted therapy for GC is still limited. Thus, investigating more effective therapeutic targets and developing novel technologies for GC treatment are highly expected. Ubiquitin-proteasome system (UPS) is one of the main pathways for protein degradation in mammals, which regulate various cellular biological processes through changing the protein levels, such as cell signal transduction, cell cycle, transcription, DNA damage and repair, etc. (5, 6). Through the coordination between ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin-ligase E3 in UPS, target proteins modified by ubiquitin are transferred to the proteasome for degradation. Therefore, UPS is essential for maintaining the normal levels of intracellular proteins by removing damaged organelles and misfolded proteins (7). During GC development, it is commonly observed that dysfunction of UPS due to the abnormal changes of E1, E2, and E3 enzymes causes the imbalanced accumulation of large numbers of proteins. Fortunately, it has been validated by clinical success of many UPS modulators, emphasizing the therapeutic potential of this pathway (8). Besides, PROTAC (proteolysis-targeting chimeras) is the novel drug development technology using UPS to degrade target proteins, which plays an important role in the treatment of prostate cancer and breast cancer (9). Hence, to develop targeted drugs by using UPS has bright prospects for further GC treatment. This review will elaborate the research progresses of UPS modulators and PROTAC to provide a novel perspective for targeted therapy of GC.
Compared with other cancers, such as breast cancer (HER2) and lung cancer (EGFR, ALK, ROS1), there are still no effective molecular targets for GC which lacks the dominant driver genes and epigenetics targets. Thus, researchers are turning their attention to other fields. UPS, as an important system for degradation of intracellular proteins, has attracted more attention for GC treatment. Accumulating evidence indicates that abnormal expression of E1, E2, and E3 enzymes is involved in the GC tumorigenesis, resulting in imbalance of intracellular protein homeostasis. UBE2C, UBE2T, UbcH10 are significantly upregulated in GC, which correlate with poor differentiation, high T classification, and poor prognosis (10–12). A fraction of E3 enzymes, such as MDM2, MKRN1, Cullin1, and Hakai, are overexpressed and have established oncogenic roles in gastric carcinogenesis (13–16). Numerous E3 enzymes, including FBXW7 and CHIP, have been shown to function as typical tumor suppressors in GC owing to frequent inactivating mutations or downregulated expression in GC (17). Besides, the proteasome is the primary site for protein degradation, and its activity affects UPS efficiency (18). Mechanically, abnormal UPS could lead to gastric cancer by regulating the epithelial-mesenchymal transition (EMT) through degradation of E-cadherin and N-cadherin, affecting the cell cycle through regulation of cell cycle proteins p21/p27 and cyclin D, impacting apoptosis by regulating the expression of BAX and Bcl-2 proteins. Furthermore, abnormal UPS influences the PI3K/AKT, Hippo pathway, p53 pathway, TGF-β signaling, STAT3 signaling, Wnt/β-catenin pathway, NF-κB pathway, autophagy, and so on by regulation of corresponding protein in these signaling pathway such as p53, β-catenin, SHP-1, etc. (Figure 1). Consequently, UPS can be considered as promising targets and it is a worth strategy by modulating the abnormality of UPS during the progression of GC for developing therapeutic drugs.
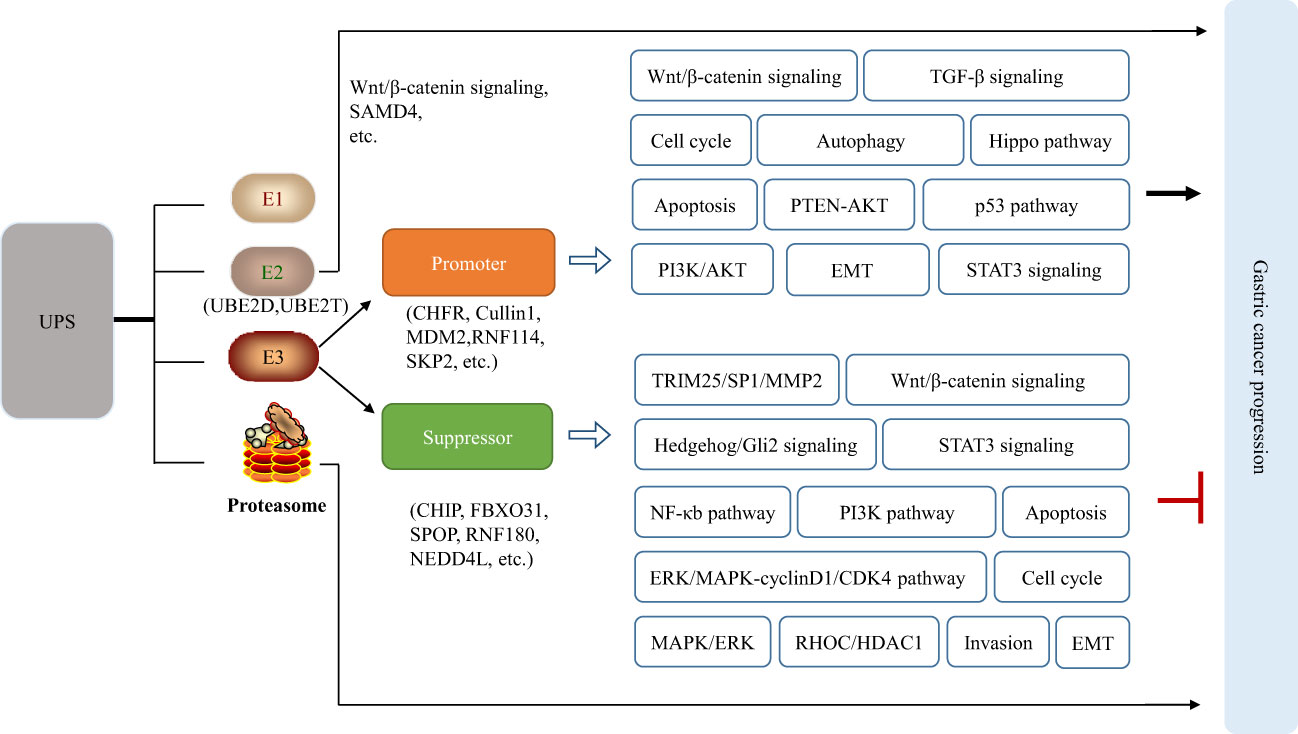
Figure 1 Abnormal UPS affects the normal signaling pathways in cells, leading to or inhibiting the occurrence of gastric cancer.
UPS is a complex structure involving the proteasome; and the E1, E2, and E3 enzymes (19). Derangements of UPS leads to alterations in protein homeostasis and causes many human diseases, particularly cancer (20). Here, the recent advances of UPS modulators by targeting proteasome and enzymes and how they pave the way towards GC treatment are discussed in detail below (with the working model seen in Figure 2).
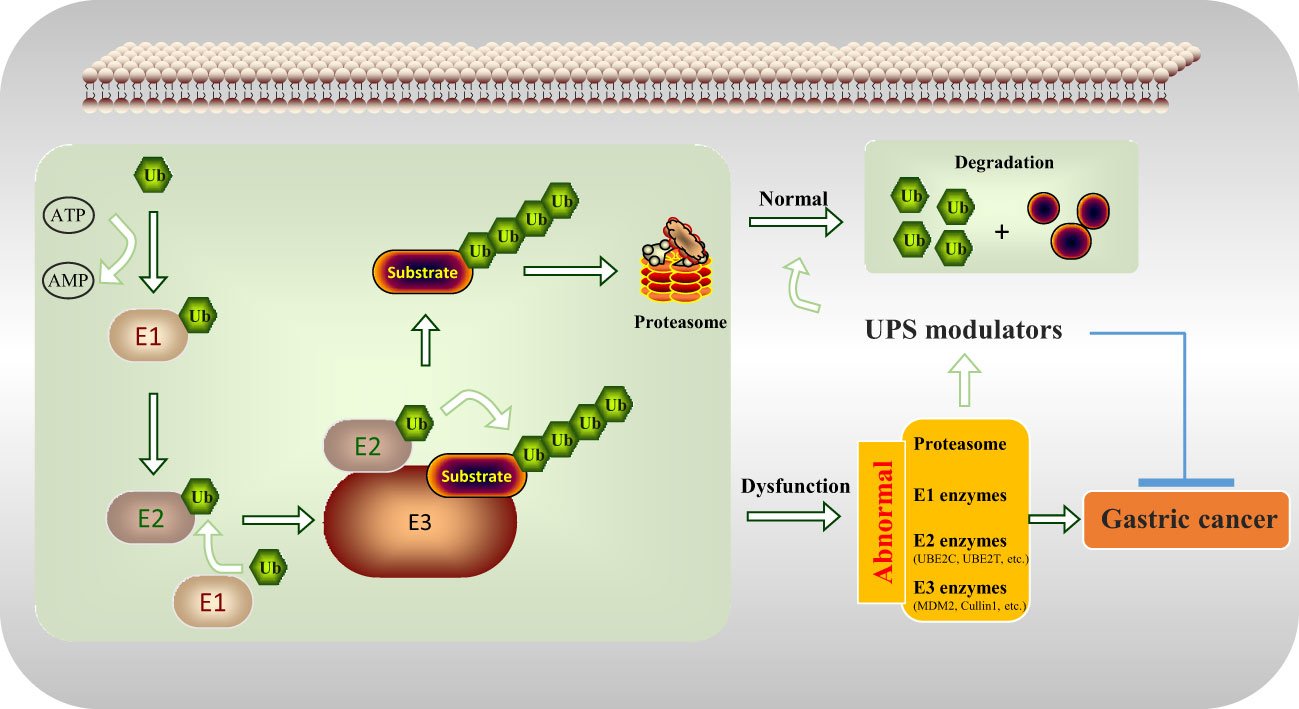
Figure 2 UPS modulators inhibit gastric tumorigenesis by regulating the abnormal components of the UPS including proteasome, E1 enzymes, E2 enzymes (UBE2C, UBE2T, etc.), and E3 enzymes (MDM2, Cullin1, etc.).
The proteasome is a large multi-protein complex designed to degrade proteins specifically marked by ubiquitination (21). In 2003, the FDA approved bortezomib as the first proteasome inhibitor for treating multiple myeloma that hugely increase the survival time of patients with multiple myeloma, which has provided ample evidence that targeting the proteasome is a viable approach for the treatment of human cancer (22, 23). For GC, bortezomib suppresses proliferation in vitro and in vivo, and is more effective in GC cells with lower NF-κB activation than others (24). In addition, proteasome inhibitor MG132 can effectively reverse the multidrug resistance by promoting drug-induced apoptosis of GC cells and inhibiting the expression of p-glycoprotein, confirming the hypothesis that proteasome inhibitors may be effective chemotherapeutics for GC with multidrug resistance (25). Unfortunately, the results of the phase II clinical trial show that bortezomib as a single inhibitor is inactive in advanced or metastatic GC therapy (26, 27). Therefore, future studies of proteasome inhibitor should focus on the combination of other targeted drugs in GC therapy.
Ubiquitin-activating enzyme E1 is the first enzyme in UPS, which mediates the activation of ubiquitin. In mammals, only two E1 enzymes including UAE and UBA6 have been discovered (28). At present, a variety of E1 enzyme inhibitors have been reported, such as PYZD-4409 and TAK-243 that inhibit the activity of UAE. PYZD-4409 not only inhibits the growth of primary acute myeloid leukemia cells in vitro, but also delays tumor growth in mouse models of leukemia in vivo (29). In addition, TAK-243, as a new class of drugs inhibiting UAE, can induce the death of various cancer cells and attenuate the growth of xenograft models of many types of tumors (8). To date, however, there is no report on E1 enzyme inhibitors for GC due to the low specificity of inhibitors and less E1 enzymes.
Ubiquitin-conjugating enzyme E2 as the key enzyme performs the second step of the ubiquitination reaction (30). The human genome encodes around 40 different E2 enzymes (31). Accumulating evidence suggests that E2 enzymes are crucial in the occurrence and development of cancer (32). Based on the indispensability and diversity of E2 enzymes in ubiquitination, more and more inhibitors are designed for E2 than E1 enzymes. For example, the silence of UBE2D1 reduces the ubiquitination of SMAD4, inhibiting the migration of GC cells (33). Moreover, UbcH10 promotes the growth of GC cells and may represent a potential biomarker for GC (12). Besides, the novel UBE2T inhibitor controls the overactivation of Wnt/β-catenin signaling and the progression of GC by blocking RACK1 ubiquitination (34). Thus, it is attractive to develop inhibitors targeting E2 enzymes such as UBED1, UbcH10, and UBE2T for GC treatment.
In the series of enzymatic cascades, ubiquitin-ligase enzyme E3 determines the specific recognition of target proteins and plays a key role in the functioning of the UPS (35). Compared to efforts against the E1 and E2 enzymes or proteasome, it is considered a better therapeutic target through targeting E3 enzymes for GC drug development because the E3 enzymes confer substrate specificity. Up to now, more than 600 types of E3 enzymes have been discovered in humans (36). In accordance with the roles, E3 ubiquitin ligases can be divided into two categories: tumor promoter and tumor suppressor based on their target proteins. Although many types of E3 enzymes are reported, the related research is rare in GC development. At present, about 66 kinds of E3 enzymes are involved in GC, of which 40 types exerted an oncogenic function for promoting cancer progression and 26 types play tumor-suppressive functions (Tables 1, 2). Subsequently, representative E3 enzymes that play key roles in the development of GC will be summarized separately and analyzed the possibility as the drug targets.
The growth and development of GC can be inhibited by suppressing E3 enzymes which are the tumor promoting factor. The upregulation of MDM2 and the accompanying inactivation of p53 pathway play an important role in diffuse gastric cancer (119). MiR-410 inhibits gastric cancer cells proliferation, migration, and invasion by targeting the MDM2 gene (120). Nutlin-3, which is the MDM2 inhibitor, has anti-tumor effects in GC cells in vitro and in vivo (121). Besides, SKP2, also named FBXL1, is an F-box typed E3 ligase, which is an overexpressed and modulated malignant phenotype via p27 proteolysis in GC (122). Downregulation of SKP2 inhibits the growth and metastasis of GC cells (123). Moreover, UHRF1 (Ubiquitin-Like PHD And RING Finger Domain-Containing Protein 1) expression is significantly higher in GC and is an independent and significant predictor of GC prognosis (124, 125). The knockdown of UHRF1 suppresses the growth, migration, invasion, and apoptosis of GC cells via an ROS-associated pathway (72). To date, there is no E3 enzyme inhibitor for clinical trials in GC treatment. However, the above studies show that the development of drugs that target the promoting function of E3 enzymes is valuable for GC therapy, though researching and finding the antagonist to inhibit promoter maybe the promising therapy for GC.
Different from a tumor promoter, the expression of a tumor suppressor needs to be enhanced to inhibit the occurrence of GC. RNF43 acts as a negative regulator of the Wnt signaling pathway by mediating the ubiquitination, endocytosis, and subsequent degradation of Wnt receptor complex component Frizzled. Research shows that RNF43 that inhibits cell proliferation is significantly downregulated in the gastric carcinoma, and its expression is positively correlated with p53 and negatively correlated with Ki67 and Lgr5 protein (107). RNF43 is related to the development of GC and attenuates the stemness of GC stem-like cells through the Wnt-β/catenin signaling pathway (108). The loss of endogenous RNF43 function enhances the growth of GC (126). FBXW7 is another important E3 ligase which negatively regulates GC progression. Low levels of FBXW7 protein in primary GC contributes to malignant potential and poor prognosis (127). In vitro studies found that FBXW7 inhibits GC progression by inducing apoptosis and growth arrest (94). In addition, CHIP is the ubiquitin ligase which contains a tetratricopeptide repeat and a U-box, and can significantly reduce the migration and invasion of GC cells though inhibiting the NF-κB signaling pathway (84). CHIP overexpression impedes the growth of xenografts in nude mice and inhibits endothelial cell growth and tube formation (128). Based on current research, enhancing the function of E3 enzymes can inhibit the development of GC. For example, eprenetapopt, as the first targeting drug of p53 as suppressor gene, binding the cysteine residue of mutant p53 and converting to the wild-type conformation, can restore p53 function. Therefore, by restoring the function of the inactivated mutant E3 enzymes to exert tumor suppressor roles, it is an interesting research field for GC treatment.
Traditional small molecule drugs and antibodies play a critical role for diseases treatment by activating or inhibiting the function of the target protein, which is defined as “occupation-driven” mode (129). This mode requires higher concentration of inhibitors or monoclonal antibodies to occupy the activity site of the target so that the transduction of downstream signaling pathways is blocked (130, 131). As an emerging new technology, PROTAC is different from the “occupation-driven” mode through the UPS system to ubiquitinate and degrade the target proteins (132–134). A PROTAC includes three key parts: E3 ubiquitin ligase ligand, POI (protein of interest) ligand, and linker (135, 136). As shown in Figure 3, in this technology, the specific target protein is recognized by the POI ligand, and the E3 ubiquitin ligase ligand is used to recruit the specific E3 enzyme of the target protein, so that the target protein and its E3 enzyme are spatially bound together via a flexible chemical linker to promote the degradation of the target protein (137–139). Based on this principle, a variety of PROTAC drugs have been designed and synthesized. For example, ARV-110 is the first oral bioavailable PROTAC small molecule drug that enters clinical trials in the field of PROTAC in world, which can selectively target degradation androgen receptor (AR) to treat prostate cancer (132). In the GC process, there is an abnormal expression of protein. It is attractive by looking for E3 enzymes of these proteins to design effective PROTAC drugs for GC therapy.
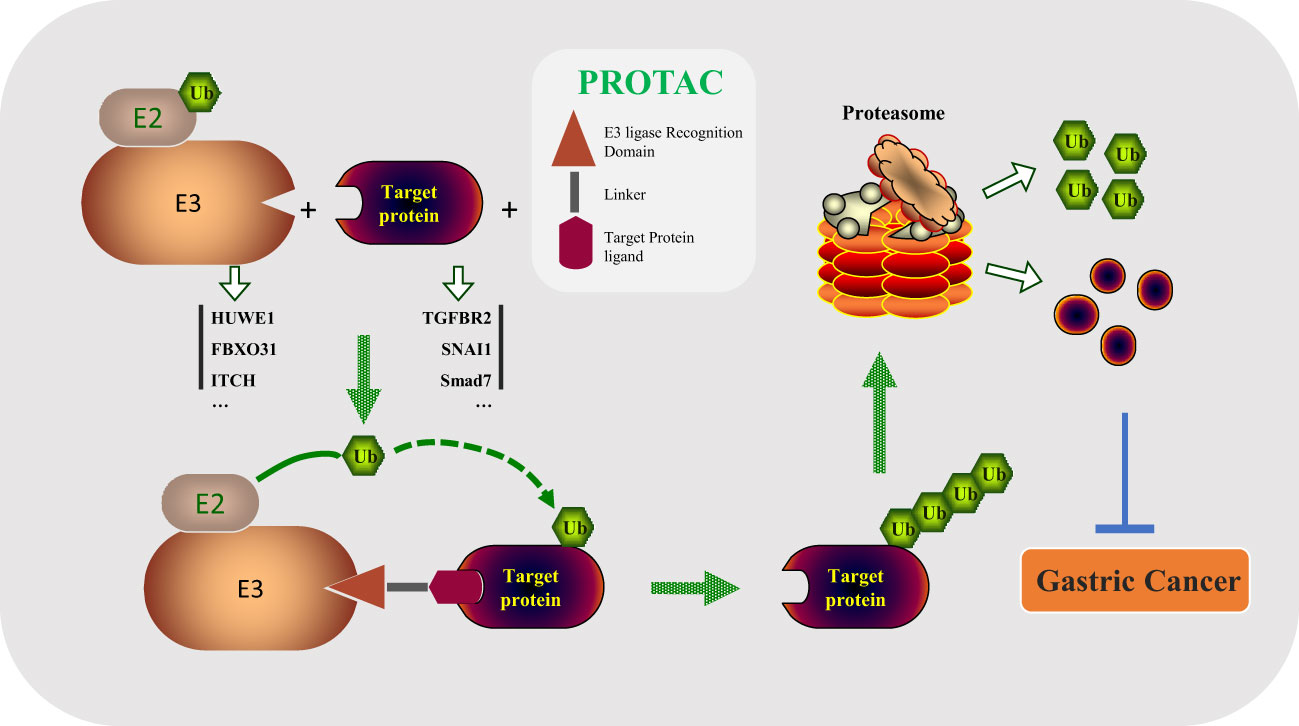
Figure 3 PROTAC drugs degrade important oncogenic substrates in the process of tumorigenesis by linking the E3 enzyme and substrate (HUWE1/TGFBR2, FBXO31/SNAI1, ITCH/Smad7, etc.), preventing the occurrence of gastric cancer.
PROTAC as the novel therapeutic technology offers numerous advantages over traditional inhibition strategies (140, 141). Firstly, the dose of traditional small molecules drugs is high, which may result in toxic side effects (142). However, in PROTAC technology, by catalyzing the degradation of the target protein, lower drug concentration could achieve good degradation efficiency to overcome on-target drug toxicity (143, 144). Secondly, the UPS is the main protein degradation system and E3 ubiquitin ligases are widely expressed in a variety of cells (145). The PROTAC molecule only needs to connect the target protein and E3 enzymes together; subsequently, protein is degraded through the proteasome (146). Therefore, PROTAC technologies have broad applications for different targets. Thirdly, PROTAC degrades the protein to the basal level within a few minutes (137). However, the re-synthesis rate of most proteins is very slow so that the cell still needs the time to restore the physiological protein level; even PROTAC is completely cleared, thereby PROTAC could prolong the action time (147).
In general, the use of PROTAC drugs is associated with several disadvantages, such as worse membrane permeability and bad oral availability (148). In addition, the molecular weight of PROTAC is higher compared with traditional small molecule drugs because of its triplet form; as a result, its solubility may be poor (140). Besides, the production of PROTAC is more difficult and costly and the potential toxicity of PROTAC is longer than traditional small molecule drugs (149). Hence, the researchers need still pay attention to solve these problems for PROTAC drug development.
With GC as heterogeneous cancer, is difficult to design effective drugs using traditional targets. Patients are prone to drug resistance in the current molecular targeted drug treatment. However, PROTAC technology as a means of precise treatment could partially solve the above problems. Thus, based on the characteristics of PROTAC technology, it is a promising strategy to investigate new substrate proteins and their corresponding E3 enzyme in GC, which will provide a basis for the development of PROTAC drugs. At present, E3 enzymes are often employed for designing PROTAC including VHL, CRBN, and IAPs that belong to the RING family (9, 150, 151). Besides, cancer-related proteins such as AR, ER, BRD4, CDK4, and CDK6 are the top five studied targets for degradation (152). To date, about 10 PROTAC drugs have already entered the clinical development stages and about 110 are in pre-clinical projects worldwide. Among them, ARV-110, ARV-471, and CFT7455 that are currently the fastest clinically progressing PROTAC drugs have entered clinical phase II trials (132, 153, 154). ARV-110 is a CRBN-based PROTAC, which is designed to degrade AR for prostate cancer treatment (155). ARV-471, by degrading ER for treating breast cancer, is also a CRBN-based PROTAC (156). Moreover, CFT7455 is designed based on CRBN E3 ligases for multiple myeloma through decreasing the level of IKZF1/3 protein (157). Therefore, PROTAC technology provides the potential for development of cancer-targeted therapy drugs.
However, the development of PROTAC drugs is relatively slow in GC. According to research, ARV-825 as the PROTAC drug effectively inhibits the growth of GC cells and elevates the apoptosis through downregulation of c-MYC and PLK1, suggesting that it may be a better therapeutic strategy for GC (158), while the clinical application of ARV-825 needs to explore continually. For GC, there are more than 40 types of E3 enzymes that have been reported, but it is the focus topic how to select appropriate E3 enzymes and their targets to develop PROTAC drugs. Three main points need to be considered when choosing E3 ligase. a. Choose the E3 ligase whose main function is to induce protein degradation. b. The substrate protein of E3 enzymes should play an important role in GC development. c. The tissue and cell-level distribution of E3 enzymes should be fully considered. Here, we summarize the E3 enzymes and corresponding substrates that have been studied in GC (Tables 1, 2) so as to provide the base for the development of PROTAC drugs in the future. In addition to the selection of E3 ligase and specific target protein, it is considered to select the corresponding ligand and appropriate length linker, which is the problem that restricts PROTAC technology (159). At present, most of the ligands are the thalidomide and its derivatives to recruit CRBN in clinic (157, 160). With the development of PTOTAC, a variety of E3 enzymes including AhR, clAP1, clAP2, CRBN, DCAF11, DCAF15, DCAF16, IAP, MDM2, RNF114, RNF4, VHL, and XIAP have developed corresponding ligands (147). Besides, Linker is a structure connecting the two ligands of PROTAC drugs, the length of which have an important influence on the biological activity of PROTAC drugs (137). However, there are no general rules for linker design (161). With current challenges solved, more and more E3 enzymes that are mentioned in Tables 1 and 2 will be used to develop PROTAC drugs in the future for GC treatment. PROTAC may become another important disease treatment drug after small molecule inhibitors and monoclonal antibodies.
The occurrence and development of GC is often accompanied by the disorder of the UPS system, which is manifested as the abnormal expression of the E1, E2, and E3 enzymes. Targeting these abnormally expressed enzymes or proteasomes is a promising strategy for GC treatment. At present, these E3 ligases are worth considering as targets including MDM2, SKP2, UHRF1, RNF43, FBXW7, and CHIP E3 enzymes widely studied in GC. However, it is easier to inhibit the function of oncogenes than to restore the roles of tumor suppressor, so E3 enzymes as oncogenes maybe more suitable. However, there are no E3 enzyme modulators for clinical trials in GC treatment to date. Considering the extensive and complex activities regulated by ubiquitination, blocking or activating E3 ligases for GC therapy may adversely affect other normal biological process. Therefore, it remains a considerable challenge to explore and solve these problems. Moreover, PROTAC is the novel technology to develop drugs for degrading intracellular important oncogenic proteins that could accomplish precise treatment of GC through the UPS. So far, only a fraction of E3 enzymes has been shown to be suitable for PROTAC. It also needs to identify more available E3 ligases and explore the mechanisms to develop PROTAC. It is believed that with the progress of basic research and clinical trials, these problems can be solved finally. Hence, targeting the Ubiquitin-proteasome system for gastric cancer is a promising strategy to supply a gap due to lack of sufficient drug selection via the UPS system in GC therapy. To develop targeted drugs by using UPS has bright prospects for GC treatment in further studies.
ZL and KL conceived and designed this review. HA collected the references. HY and JZ wrote the original manuscript and made the figures. ZL, JM and KL revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Medical science and technology Project of Henan Province (Grant Nos. 232102311077 and LHGJ20220198, China).
We acknowledge and appreciate our colleagues for their suggestions and assistance for this review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
GC, gastric cancer; PROTAC, proteolysis-targeting chimeras; UPS, ubiquitin-proteasome system
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology (2011) 141:476–85, 485 e1-11. doi: 10.1053/j.gastro.2011.04.042
3. Barbour AP, Walpole ET, Mai GT, Barnes EH, Watson DI, Ackland SP, et al. Preoperative cisplatin, fluorouracil, and docetaxel with or without radiotherapy after poor early response to cisplatin and fluorouracil for resectable oesophageal adenocarcinoma (AGITG DOCTOR): results from a multicentre, randomised controlled phase II trial. Ann Oncol (2020) 31:236–45. doi: 10.1016/j.annonc.2019.10.019
4. Cetin B, Gumusay O, Cengiz M, Ozet A. Advances of molecular targeted therapy in gastric cancer. J Gastrointest Cancer (2016) 47:125–34. doi: 10.1007/s12029-016-9806-8
5. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol (2006) 22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503
6. Ulrich H D, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol (2010) 11:479–89. doi: 10.1038/nrm2921
7. Ding F, Xiao H, Wang M, Xie X, Hu F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front Biosci (Landmark Ed) (2014) 19:886–95. doi: 10.2741/4254
8. Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat Med (2018) 24:186–93. doi: 10.1038/nm.4474
9. Li X, Song Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J Hematol Oncol (2020) 13:50. doi: 10.1186/s13045-020-00885-3
10. Wang Y, Huang F, Liu M, Zhao Q. UBE2C mRNA expression controlled by miR-300 and HuR determines its oncogenic role in gastric cancer. Biochem Biophys Res Commun (2021) 534:597–603. doi: 10.1016/j.bbrc.2020.11.034
11. Yu H, Xiang P, Pan Q, Huang Y, Xie N. Ubiquitin-conjugating enzyme E2T is an independent prognostic factor and promotes gastric cancer progression. Tumour Biol (2016) 37:11723–32. doi: 10.1007/s13277-016-5020-3
12. Yang M, Qu Y, Shi R, Wu X, Su C, Hu Z, et al. Ubiquitin-conjugating enzyme UbcH10 promotes gastric cancer growth and is a potential biomarker for gastric cancer. Oncol Rep (2016) 36:779–86. doi: 10.3892/or.2016.4906
13. Feng C, Xian Q, Liu S. Micro RNA-518 inhibits gastric cancer cell growth by inducing apoptosis via targeting MDM2. BioMed Pharmacother (2018) 97:1595–602. doi: 10.1016/j.biopha.2017.11.091
14. Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst (2012) 104:1660–72. doi: 10.1093/jnci/djs424
15. Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan Y, et al. Overexpression of Cullin1 is associated with poor prognosis of patients with gastric cancer. Hum Pathol (2011) 42:375–83. doi: 10.1016/j.humpath.2010.09.003
16. Figueroa A, Kotani H, Toda Y, Mazan-Mamczarz K, Mueller EC, Otto A, et al. Novel roles of hakai in cell proliferation and oncogenesis. Mol Biol Cell (2009) 20:3533–42. doi: 10.1091/mbc.e08-08-0845
17. Hou Y C, Deng JY. Role of E3 ubiquitin ligases in gastric cancer. World J Gastroenterol (2015) 21:786–93. doi: 10.3748/wjg.v21.i3.786
18. Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol (2012) 199:583–8. doi: 10.1083/jcb.201210077
19. Kim IS, Jo WM. Effects of a proteasome inhibitor on cardiomyocytes in a pressure-overload hypertrophy rat model: An animal study. Korean J Thorac Cardiovasc Surg (2017) 50:144–52. doi: 10.5090/kjtcs.2017.50.3.144
20. Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr Pharm Des (2013) 19:3215–25. doi: 10.2174/13816128113199990300
21. Gomes FP, Park R, Viana AG, Fernandez-Costa C, Topper E, Kaya A, et al. Protein signatures of seminal plasma from bulls with contrasting frozen-thawed sperm viability. Sci Rep (2020) 10:14661. doi: 10.1038/s41598-020-71015-9
23. Buac D, Shen M, Schmitt S, Kona FR, Deshmukh R, Zhang Z, et al. From bortezomib to other inhibitors of the proteasome and beyond. Curr Pharm Des (2013) 19:4025–38. doi: 10.2174/1381612811319220012
24. Nakata W, Hayakawa Y, Nakagawa H, Sakamoto K, Kinoshita H, Takahashi R, et al. Anti-tumor activity of the proteasome inhibitor bortezomib in gastric cancer. Int J Oncol (2011) 39:1529–36. doi: 10.1016/S0016-5085(11)62794-7
25. Wang H, Wang X, Li Y, Liao A, Fu B, Pan H, et al. The proteasome inhibitor bortezomib reverses p-glycoprotein-mediated leukemia multi-drug resistance through the NF-kappaB pathway. Pharmazie (2012) 67:187–92. doi: 10.1691/ph.2012.1585
26. Shah MA, Power DG, Kindler HL, Holen KD, Kemeny MM, Ilson DH, et al. A multicenter, phase II study of bortezomib (PS-341) in patients with unresectable or metastatic gastric and gastroesophageal junction adenocarcinoma. Invest New Drugs (2011) 29:1475–81. doi: 10.1007/s10637-010-9474-7
27. Ocean AJ, Christos P, Sparano JA, Shah MA, Yantiss RK, Cheng J, et al. Phase II trial of bortezomib alone or in combination with irinotecan in patients with adenocarcinoma of the gastroesophageal junction or stomach. Invest New Drugs (2014) 32:542–8. doi: 10.1007/s10637-014-0070-0
28. Huang Y, Xiao Y, Zhang X, Huang X, Li Y. The emerging roles of tripartite motif proteins (TRIMs) in acute lung injury. J Immunol Res (2021) 2021:1007126. doi: 10.1155/2021/1007126
29. Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood (2010) 115:2251–9. doi: 10.1182/blood-2009-07-231191
30. Wang S, Zhang G, Zhang Y, Song Q, Chen Z, Wang J, et al. Comparative studies of mitochondrial proteomics reveal an intimate protein network of male sterility in wheat (Triticum aestivum l. ). J Exp Bot (2015) 66:6191–203. doi: 10.1093/jxb/erv322
31. Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res (2016) 26:423–40. doi: 10.1038/cr.2016.35
32. Chen X, Wang L, Su X, Luo SY, Tang X, Huang Y. Identification of potential target genes and crucial pathways in small cell lung cancer based on bioinformatic strategy and human samples. PloS One (2020) 15:e0242194. doi: 10.1371/journal.pone.0242194
33. Xie H, He Y, Wu Y, Lu Q. Silencing of UBE2D1 inhibited cell migration in gastric cancer, decreasing ubiquitination of SMAD4. Infect Agent Cancer (2021) 16:63. doi: 10.1186/s13027-021-00402-2
34. Yu Z, Jiang X, Qin L, Deng H, Wang J, Ren W, et al. A novel UBE2T inhibitor suppresses wnt/beta-catenin signaling hyperactivation and gastric cancer progression by blocking RACK1 ubiquitination. Oncogene (2021) 40:1027–42. doi: 10.1038/s41388-020-01572-w
35. Qian S, Zhan X, Lu M, Li N, Long Y, Li X, et al. Quantitative analysis of ubiquitinated proteins in human pituitary and pituitary adenoma tissues. Front Endocrinol (Lausanne) (2019) 10:328. doi: 10.3389/fendo.2019.00328
36. Zheng N, Shabek N. Ubiquitin ligases: Structure, function, and regulation. Annu Rev Biochem (2017) 86:129–57. doi: 10.1146/annurev-biochem-060815-014922
37. Yang S, He F, Dai M, Pan J, Wang J, Ye B. CHFR promotes the migration of human gastric cancer cells by inducing epithelial-to-mesenchymal transition in a HDAC1-dependent manner. Onco Targets Ther (2019) 12:1075–84. doi: 10.2147/OTT.S191016
38. Kashima L, Idogawa M, Mita H, Shitashige M, Yamada T, Ogi K, et al. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J Biol Chem (2012) 287:12975–84. doi: 10.1074/jbc.M111.321828
39. Zheng Q, Zhao LY, Kong Y, Nan KJ, Yao Y, Liao ZJ. CDK-associated cullin 1 can promote cell proliferation and inhibit cisplatin-induced apoptosis in the AGS gastric cancer cell line. World J Surg Oncol (2013) 11:5. doi: 10.1186/1477-7819-11-5
40. Kamran M, Long ZJ, Xu D, Lv SS, Liu B, Wang CL, et al. Aurora kinase a regulates survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis (2017) 6:e298. doi: 10.1038/oncsis.2016.80
41. Sun C, Tao Y, Gao Y, Xia Y, Liu Y, Wang G, et al. F-box protein 11 promotes the growth and metastasis of gastric cancer via PI3K/AKT pathway-mediated EMT. BioMed Pharmacother (2018) 98:416–23. doi: 10.1016/j.biopha.2017.12.088
42. Sun X, Wang T, Guan ZR, Zhang C, Chen Y, Jin J, et al. FBXO2, a novel marker for metastasis in human gastric cancer. Biochem Biophys Res Commun (2018) 495:2158–64. doi: 10.1016/j.bbrc.2017.12.097
43. Zhang L, Hou Y, Wang M, Wu B, Li N. A study on the functions of ubiquitin metabolic system related gene FBG2 in gastric cancer cell line. J Exp Clin Cancer Res (2009) 28:78. doi: 10.1186/1756-9966-28-78
44. Yao Y, Liu Z, Huang S, Huang C, Cao Y, Li L, et al. The E3 ubiquitin ligase, FBXW5, promotes the migration and invasion of gastric cancer through the dysregulation of the hippo pathway. Cell Death Discovery (2022) 8:79. doi: 10.1038/s41420-022-00868-y
45. Mishra L, Katuri V, Evans S. The role of PRAJA and ELF in TGF-beta signaling and gastric cancer. Cancer Biol Ther (2005) 4:694–9. doi: 10.4161/cbt.4.7.2015
46. Jia J, Yang Y, Yan T, Chen T, Lu G. Down-regulation of RFWD3 inhibits cancer cells proliferation and migration in gastric carcinoma. Gen Physiol Biophys (2020) 39:363–71. doi: 10.4149/gpb_2020009
47. Feng Z, Li L, Zeng Q, Zhang Y, Tu Y, Chen W, et al. RNF114 silencing inhibits the proliferation and metastasis of gastric cancer. J Cancer (2022) 13:565–78. doi: 10.7150/jca.62033
48. Li R, Gu Z, Zhang X, Yu J, Feng J, Lou Y, et al. RNF115 deletion inhibits autophagosome maturation and growth of gastric cancer. Cell Death Dis (2020) 11:810. doi: 10.1038/s41419-020-03011-w
49. Migita K, Matsumoto S, Wakatsuki K, Kunishige T, Nakade H, Miyao S, et al. RNF126 as a marker of prognosis and proliferation of gastric cancer. Anticancer Res (2020) 40:1367–74. doi: 10.21873/anticanres.14078
50. Qiu D, Wang Q, Wang Z, Chen J, Yan D, Zhou Y, et al. RNF185 modulates JWA ubiquitination and promotes gastric cancer metastasis. Biochim Biophys Acta Mol Basis Dis (2018) 1864:1552–61. doi: 10.1016/j.bbadis.2018.02.013
51. Zhang J, Sun Z, Han Y, Yao R, Yue L, Xu Y, et al. Rnf2 knockdown reduces cell viability and promotes cell cycle arrest in gastric cancer cells. Oncol Lett (2017) 13:3817–22. doi: 10.3892/ol.2017.5868
52. Yue F, Peng K, Zhang L, Zhang J. Circ_0004104 accelerates the progression of gastric cancer by regulating the miR-539-3p/RNF2 axis. Dig Dis Sci (2021) 66:4290–301. doi: 10.1007/s10620-020-06802-5
53. Zhu F, Yi G, Liu X, Zhu F, Zhao A, Wang A, et al. Ring finger protein 31-mediated atypical ubiquitination stabilizes forkhead box P3 and thereby stimulates regulatory T-cell function. J Biol Chem (2018) 293:20099–111. doi: 10.1074/jbc.RA118.005802
54. Zhang J, Wu H, Yi B, Zhou J, Wei L, Chen Y, et al. RING finger protein 38 induces gastric cancer cell growth by decreasing the stability of the protein tyrosine phosphatase SHP-1. FEBS Lett (2018) 592:3092–100. doi: 10.1002/1873-3468.13225
55. Huang Z, Cai Y, Yang C, Chen Z, Sun H, Xu Y, et al. Knockdown of RNF6 inhibits gastric cancer cell growth by suppressing STAT3 signaling. Onco Targets Ther (2018) 11:6579–87. doi: 10.2147/OTT.S174846
56. Das L, Kokate SB, Dixit P, Rath S, Rout N, Singh SP, et al. Membrane-bound beta-catenin degradation is enhanced by ETS2-mediated Siah1 induction in helicobacter pylori-infected gastric cancer cells. Oncogenesis (2017) 6:e327. doi: 10.1038/oncsis.2017.26
57. Kokate SB, Dixit P, Poirah I, Roy AD, Chakraborty D, Rout N, et al. Testin and filamin-c downregulation by acetylated Siah2 increases invasiveness of helicobacter pylori-infected gastric cancer cells. Int J Biochem Cell Biol (2018) 103:14–24. doi: 10.1016/j.biocel.2018.07.012
58. Wen Y, Wang K, Yang K. Inhibiting the role of Skp2 suppresses cell proliferation and tumorigenesis of human gastric cancer cells via the upregulation of p27kip1. Mol Med Rep (2016) 14:3917–24. doi: 10.3892/mmr.2016.5676
59. Wang F, Ruan L, Yang J, Zhao Q, Wei W. TRIM14 promotes the migration and invasion of gastric cancer by regulating epithelialtomesenchymal transition via activation of AKT signaling regulated by miR1955p. Oncol Rep (2018) 40:3273–84. doi: 10.3892/or.2018.6750
60. Zhou W, Chen H, Ruan Y, Zeng X, Liu F. High expression of TRIM15 is associated with tumor invasion and predicts poor prognosis in patients with gastric cancer. J Invest Surg (2021) 34:853–61. doi: 10.1080/08941939.2019.1705443
61. Yao Y, Liu Z, Guo H, Huang S, Zhong M, Deng J, et al. Elevated TRIM23 expression predicts poor prognosis in Chinese gastric cancer. Pathol Res Pract (2018) 214:2062–8. doi: 10.1016/j.prp.2018.10.010
62. Fang Z, Deng J, Zhang L, Xiang X, Yu F, Chen J, et al. TRIM24 promotes the aggression of gastric cancer via the wnt/beta-catenin signaling pathway. Oncol Lett (2017) 13:1797–806. doi: 10.3892/ol.2017.5604
63. Qiu F, Xiong JP, Deng J, Xiang XJ. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol (2015) 8:5053–61.
64. Wang J, Fang Y, Liu T. TRIM32 promotes the growth of gastric cancer cells through enhancing AKT activity and glucose transportation. BioMed Res Int (2020) 2020:4027627. doi: 10.1155/2020/4027627
65. Wang C, Xu J, Fu H, Zhang Y, Zhang X, Yang D, et al. TRIM32 promotes cell proliferation and invasion by activating beta-catenin signalling in gastric cancer. J Cell Mol Med (2018) 22:5020–8. doi: 10.1111/jcmm.13784
66. Fu T, Ji K, Jin L, Zhang J, Wu X, Ji X, et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-kappaB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer (2021) 24:45–59. doi: 10.1007/s10120-020-01096-y
67. Kashimoto K, Komatsu S, Ichikawa D, Arita T, Konishi H, Nagata H, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci (2012) 103:2021–6. doi: 10.1111/j.1349-7006.2012.02407.x
68. Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH, et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology (2014) 147:1043–54. doi: 10.1053/j.gastro.2014.07.021
69. Mao J, Liang Z, Zhang B, Yang H, Li X, Fu H, et al. UBR2 enriched in p53 deficient mouse bone marrow mesenchymal stem cell-exosome promoted gastric cancer progression via wnt/beta-catenin pathway. Stem Cells (2017) 35:2267–79. doi: 10.1002/stem.2702
70. Yang M, Jiang N, Cao QW, Ma MQ, Sun Q. The E3 ligase UBR5 regulates gastric cancer cell growth by destabilizing the tumor suppressor GKN1. Biochem Biophys Res Commun (2016) 478:1624–9. doi: 10.1016/j.bbrc.2016.08.170
71. Ding F, Zhu X, Song X, Yuan P, Ren L, Chai C, et al. UBR5 oncogene as an indicator of poor prognosis in gastric cancer. Exp Ther Med (2020) 20:7. doi: 10.3892/etm.2020.9135
72. Zhang H, Song Y, Yang C, Wu X. UHRF1 mediates cell migration and invasion of gastric cancer. Biosci Rep (2019) 26(Suppl1):113–819. doi: 10.1042/BSR20181065
73. He Y, Zhou J, Wan Q. The E3 ligase HUWE1 mediates TGFBR2 ubiquitination and promotes gastric cancer cell proliferation, migration, and invasion. Invest New Drugs (2021) 39:713–23. doi: 10.1007/s10637-020-01041-x
74. Kim SS, Yoo NJ, Jeong EG, Kim MS, Lee SH. Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS (2008) 116:779–84. doi: 10.1111/j.1600-0463.2008.00999.x
75. Ji CD, Wang YX, Xiang DF, Liu Q, Zhou ZH, Qian F, et al. Kir2.1 interaction with Stk38 promotes invasion and metastasis of human gastric cancer by enhancing MEKK2-MEK1/2-ERK1/2 signaling. Cancer Res (2018) 78:3041–53. doi: 10.1158/0008-5472.CAN-17-3776
76. Tao Y, Sun C, Zhang T, Song Y. SMURF1 promotes the proliferation, migration and invasion of gastric cancer cells. Oncol Rep (2017) 38:1806–14. doi: 10.3892/or.2017.5825
77. Zhang Y, Xu J, Fu H, Wei Z, Yang D, Yan R. UBE3C promotes proliferation and inhibits apoptosis by activating the beta-catenin signaling via degradation of AXIN1 in gastric cancer. Carcinogenesis (2021) 42:285–93. doi: 10.1093/carcin/bgaa098
78. Li Q, Li Z, Wei S, Wang W, Chen Z, Zhang L, et al. Overexpression of miR-584-5p inhibits proliferation and induces apoptosis by targeting WW domain-containing E3 ubiquitin protein ligase 1 in gastric cancer. J Exp Clin Cancer Res (2017) 36:59. doi: 10.1186/s13046-017-0532-2
79. Zhang L, Wu Z, Ma Z, Liu H, Wu Y, Zhang Q. WWP1 as a potential tumor oncogene regulates PTEN-akt signaling pathway in human gastric carcinoma. Tumour Biol (2015) 36:787–98. doi: 10.1007/s13277-014-2696-0
80. Wang K, Liu J, Zhao X, Li H, Luo G, Yu Y, et al. WWP2 regulates proliferation of gastric cancer cells in a PTEN-dependent manner. Biochem Biophys Res Commun (2020) 521:652–9. doi: 10.1016/j.bbrc.2019.10.179
81. Fan Y, Qu X, Ma Y, Qu J, Liu Y, Hu X. Cbl-b accelerates trypsin-induced cell detachment through ubiquitination and degradation of proline-rich tyrosine kinase 2. Tumour Biol (2014) 35:11129–35. doi: 10.1007/s13277-014-2296-z
82. Che X, Zhang Y, Qu X, Guo T, Ma Y, Li C, et al. The E3 ubiquitin ligase cbl-b inhibits tumor growth in multidrug-resistant gastric and breast cancer cells. Neoplasma (2017) 64:887–92. doi: 10.4149/neo_2017_610
83. Fan Y, Che X, Hou K, Zhang M, Wen T, Qu X, et al. MiR-940 promotes the proliferation and migration of gastric cancer cells through up-regulation of programmed death ligand-1 expression. Exp Cell Res (2018) 373:180–7. doi: 10.1016/j.yexcr.2018.10.011
84. Liu F, Zhou J, Zhou P, Chen W, Guo F. The ubiquitin ligase CHIP inactivates NF-kappaB signaling and impairs the ability of migration and invasion in gastric cancer cells. Int J Oncol (2015) 46:2096–106. doi: 10.3892/ijo.2015.2893
85. Dai H, Chen H, Xu J, Zhou J, Shan Z, Yang H, et al. The ubiquitin ligase CHIP modulates cellular behaviors of gastric cancer cells by regulating TRAF2. Cancer Cell Int (2019) 19:132. doi: 10.1186/s12935-019-0832-z
86. Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, et al. Loss of COP1 expression determines poor prognosisin patients with gastric cancer. Oncol Rep (2013) 30:1971–5. doi: 10.3892/or.2013.2664
87. Hsu TS, Mo ST, Hsu PN, Lai MZ. C-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis (2018) 9:135. doi: 10.1038/s41419-017-0165-6
88. Wu P, Wang F, Wang Y, Men H, Zhu X, He G, et al. Significance of FBX8 in progression of gastric cancer. Exp Mol Pathol (2015) 98:360–6. doi: 10.1016/j.yexmp.2015.03.015
89. Li LQ, Pan D, Chen H, Zhang L, Xie WJ. F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1. FEBS Lett (2016) 590:445–52. doi: 10.1002/1873-3468.12071
90. Wu W, Ding H, Cao J, Zhang W. FBXL5 inhibits metastasis of gastric cancer through suppressing Snail1. Cell Physiol Biochem (2015) 35:1764–72. doi: 10.1159/000373988
91. Cen G, Ding HH, Liu B, Wu WD. FBXL5 targets cortactin for ubiquitination-mediated destruction to regulate gastric cancer cell migration. Tumour Biol (2014) 35:8633–8. doi: 10.1007/s13277-014-2104-9
92. Jiang Y, Liu X, Shen R, Gu X, Qian W. Fbxo21 regulates the epithelial-to-mesenchymal transition through ubiquitination of Nr2f2 in gastric cancer. J Cancer (2021) 12:1421–30. doi: 10.7150/jca.49674
93. Zou S, Ma C, Yang F, Xu X, Jia J, Liu Z. FBXO31 suppresses gastric cancer EMT by targeting Snail1 for proteasomal degradation. Mol Cancer Res (2018) 16:286–95. doi: 10.1158/1541-7786.MCR-17-0432
94. Li H, Wang Z, Zhang W, Qian K, Xu W, Zhang S. Fbxw7 regulates tumor apoptosis, growth arrest and the epithelial-to-mesenchymal transition in part through the RhoA signaling pathway in gastric cancer. Cancer Lett (2016) 370:39–55. doi: 10.1016/j.canlet.2015.10.006
95. Li C, Deng C, Pan G, Wang X, Zhang K, Dong Z, et al. Lycorine hydrochloride inhibits cell proliferation and induces apoptosis through promoting FBXW7-MCL1 axis in gastric cancer. J Exp Clin Cancer Res (2020) 39:230. doi: 10.1186/s13046-020-01743-3
96. Deng T, Shen P, Li A, Zhang Z, Yang H, Deng X, et al. CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in gastric cancer. Theranostics (2021) 11:8112–28. doi: 10.7150/thno.54961
97. Kuai X, Li L, Chen R, Wang K, Chen M, Cui B, et al. SCF(FBXW7)/GSK3beta-mediated GFI1 degradation suppresses proliferation of gastric cancer cells. Cancer Res (2019) 79:4387–98. doi: 10.1158/0008-5472.CAN-18-4032
98. Yin J, Ji Z, Hong Y, Song Z, Hu N, Zhuang M, et al. Sh-MARCH8 inhibits tumorigenesis via PI3K pathway in gastric cancer. Cell Physiol Biochem (2018) 49:306–21. doi: 10.1159/000492882
99. Liu Z, Xiang S, Guo X, Zhou J, Liao L, Kou J, et al. MKRN2 inhibits the proliferation of gastric cancer by downregulating PKM2. Aging (Albany NY) (2022) 13. doi: 10.18632/aging.203643
100. Zhao Z, Zhu L, Xing Y, Zhang Z. Praja2 suppresses the growth of gastric cancer by ubiquitylation of KSR1 and inhibiting MEK-ERK signal pathways. Aging (Albany NY) (2022) 14(4):2004–13. doi: 10.18632/aging.202356
101. Chen X, Wang Y, Zang W, Du Y, Li M, Zhao G. miR-194 targets RBX1 gene to modulate proliferation and migration of gastric cancer cells. Tumour Biol (2015) 36:2393–401. doi: 10.1007/s13277-014-2849-1
102. Xu Y, Feng Y, Sun Z, Li Q. RNF168 promotes RHOC degradation by ubiquitination to restrain gastric cancer progression via decreasing HDAC1 expression. Biochem Biophys Res Commun (2021) 557:135–42. doi: 10.1016/j.bbrc.2021.03.123
103. Xie XM, Deng JY, Hou YC, Cui JL, Wu WP, Ying GG, et al. Evaluating the clinical feasibility: The direct bisulfite genomic sequencing for examination of methylated status of E3 ubiquitin ligase RNF180 DNA promoter to predict the survival of gastric cancer. Cancer biomark (2015) 15:259–65. doi: 10.3233/CBM-150466
104. Wu Z, Liu H, Sun W, Du Y, He W, Guo S, et al. RNF180 mediates STAT3 activity by regulating the expression of RhoC via the proteasomal pathway in gastric cancer cells. Cell Death Dis (2020) 11:881. doi: 10.1038/s41419-020-03096-3
105. Cheung KF, Lam CN, Wu K, Ng EK, Chong WW, Cheng AS, et al. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer (2012) 118:947–59. doi: 10.1002/cncr.26189
106. Wang S, Wang X, Gao Y, Peng Y, Dong N, Xie Q, et al. RN181 is a tumour suppressor in gastric cancer by regulation of the ERK/MAPK-cyclin D1/CDK4 pathway. J Pathol (2019) 248:204–16. doi: 10.1002/path.5246
107. Niu L, Qin HZ, Xi HQ, Wei B, Xia SY, Chen L. RNF43 inhibits cancer cell proliferation and could be a potential prognostic factor for human gastric carcinoma. Cell Physiol Biochem (2015) 36:1835–46. doi: 10.1159/000430154
108. Gao Y, Cai A, Xi H, Li J, Xu W, Zhang Y, et al. Ring finger protein 43 associates with gastric cancer progression and attenuates the stemness of gastric cancer stem-like cells via the wnt-beta/catenin signaling pathway. Stem Cell Res Ther (2017) 8:98. doi: 10.1186/s13287-017-0548-8
109. Zeng C, Wang Y, Lu Q, Chen J, Zhang J, Liu T, et al. SPOP suppresses tumorigenesis by regulating Hedgehog/Gli2 signaling pathway in gastric cancer. J Exp Clin Cancer Res (2014) 33:75. doi: 10.1186/s13046-014-0075-8
110. Chen W, Lu C, Hong J. TRIM15 exerts anti-tumor effects through suppressing cancer cell invasion in gastric adenocarcinoma. Med Sci Monit (2018) 24:8033–41. doi: 10.12659/MSM.911142
111. Chen JJ, Ren YL, Shu CJ, Zhang Y, Chen MJ, Xu J, et al. JP3, an antiangiogenic peptide, inhibits growth and metastasis of gastric cancer through TRIM25/SP1/MMP2 axis. J Exp Clin Cancer Res (2020) 39:118. doi: 10.1186/s13046-020-01617-8
112. Sugiura T. The cellular level of TRIM31, an RBCC protein overexpressed in gastric cancer, is regulated by multiple mechanisms including the ubiquitin-proteasome system. Cell Biol Int (2011) 35:657–61. doi: 10.1042/CBI20100772
113. Sugiura T, Miyamoto K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J Cell Biochem (2008) 105:1081–91. doi: 10.1002/jcb.21908
114. Qin H, Cai A, Xi H, Yuan J, Chen L. ZnRF3 induces apoptosis of gastric cancer cells by antagonizing wnt and hedgehog signaling. Panminerva Med (2015) 57:167–75. doi: 10.1007/s12013-015-0607-7
115. Zhou Y, Lan J, Wang W, Shi Q, Lan Y, Cheng Z, et al. ZNRF3 acts as a tumour suppressor by the wnt signalling pathway in human gastric adenocarcinoma. J Mol Histol (2013) 44:555–63. doi: 10.1007/s10735-013-9504-9
116. Chen YL, Li DP, Jiang HY, Yang Y, Xu LL, Zhang SC, et al. Overexpression of HACE1 in gastric cancer inhibits tumor aggressiveness by impeding cell proliferation and migration. Cancer Med (2018) 7:2472–84. doi: 10.1002/cam4.1496
117. Gen Y, Yasui K, Kitaichi T, Iwai N, Terasaki K, Dohi O, et al. ASPP2 suppresses invasion and TGF-beta1-induced epithelial-mesenchymal transition by inhibiting Smad7 degradation mediated by E3 ubiquitin ligase ITCH in gastric cancer. Cancer Lett (2017) 398:52–61. doi: 10.1016/j.canlet.2017.04.002
118. Jiang X, Zhang S, Yin Z, Sheng Y, Yan Q, Sun R, et al. The correlation between NEDD4L and HIF-1alpha levels as a gastric cancer prognostic marker. Int J Med Sci (2019) 16:1517–24. doi: 10.7150/ijms.34646
119. Gunther T, Schneider-Stock R, Hackel C, Kasper HU, Pross M, Hackelsberger A, et al. Mdm2 gene amplification in gastric cancer correlation with expression of Mdm2 protein and p53 alterations. Mod Pathol (2000) 13:621–6. doi: 10.1038/modpathol.3880107
120. Shen J, Niu W, Zhou M, Zhang H, Ma J, Wang L, et al. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PloS One (2014) 9:e104510. doi: 10.1371/journal.pone.0104510
121. Endo S, Yamato K, Hirai S, Moriwaki T, Fukuda K, Suzuki H, et al. Potent in vitro and in vivo antitumor effects of MDM2 inhibitor nutlin-3 in gastric cancer cells. Cancer Sci (2011) 102:605–13. doi: 10.1111/j.1349-7006.2010.01821.x
122. Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, et al. Clinical and biological significance of s-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res (2002) 62:3819–25.
123. Wei Z, Jiang X, Liu F, Qiao H, Zhou B, Zhai B, et al. Downregulation of Skp2 inhibits the growth and metastasis of gastric cancer cells in vitro and in vivo. Tumour Biol (2013) 34:181–92. doi: 10.1007/s13277-012-0527-8
124. Zhou L, Shang Y, Jin Z, Zhang W, Lv C, Zhao X, et al. UHRF1 promotes proliferation of gastric cancer via mediating tumor suppressor gene hypermethylation. Cancer Biol Ther (2015) 16:1241–51. doi: 10.1080/15384047.2015.1056411
125. Ge M, Gui Z, Wang X, Yan F. Analysis of the UHRF1 expression in serum and tissue for gastric cancer detection. Biomarkers (2015) 20:183–8. doi: 10.3109/1354750X.2015.1061599
126. Niu L, Qin HZ, Xi HQ, Wei B, Xia SY, Chen L. Loss of endogenous RNF43 function enhances proliferation and tumour growth of intestinal and gastric cells. Carcinogenesis (2019) 40:551–9. doi: 10.1093/carcin/bgy152
127. Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T, et al. p53-altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res (2009) 69:3788–94. doi: 10.1158/0008-5472.CAN-08-2846
128. Wang S, Wu X, Zhang J, Chen Y, Xu J, Xia X, et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut (2013) 62:496–508. doi: 10.1136/gutjnl-2011-301522
129. Moon S, Lee BH. Chemically induced cellular proteolysis: An emerging therapeutic strategy for undruggable targets. Mol Cells (2018) 41:933–42. doi: 10.14348/molcells.2018.0372
130. Zeng S, Huang W, Zheng X, Liyan C, Zhang Z, Wang J, et al. Proteolysis targeting chimera (PROTAC) in drug discovery paradigm: Recent progress and future challenges. Eur J Med Chem (2021) 210:112981. doi: 10.1016/j.ejmech.2020.112981
131. An S, Fu L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine (2018) 36:553–62. doi: 10.1016/j.ebiom.2018.09.005
132. Qi SM, Dong J, Xu ZY, Cheng XD, Zhang WD, Qin JJ. PROTAC: An effective targeted protein degradation strategy for cancer therapy. Front Pharmacol (2021) 12:692574. doi: 10.3389/fphar.2021.692574
133. Liu Z, Wang P, Chen H, Wold EA, Tian B, Brasier AR, et al. Drug discovery targeting bromodomain-containing protein 4. J Med Chem (2017) 60:4533–58. doi: 10.1021/acs.jmedchem.6b01761
134. Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther (2021) 6:201. doi: 10.1038/s41392-021-00572-w
135. Zheng M, Huo J, Gu X, Wang Y, Wu C, Zhang Q, et al. Rational design and synthesis of novel dual PROTACs for simultaneous degradation of EGFR and PARP. J Med Chem (2021) 64:7839–52. doi: 10.1021/acs.jmedchem.1c00649
136. Yu F, Cai M, Shao L, et al. Targeting protein kinases degradation by PROTACs. Front Chem (2021) 9:679120. doi: 10.3389/fchem.2021.679120
137. Wang Y, Jiang X, Feng F, Liu W, Sun H. Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B (2020) 10:207–38. doi: 10.1016/j.apsb.2019.08.001
138. Chen S, Liu Y.Zhou H. Advances in the development ubiquitin-specific peptidase (USP) inhibitors. Int J Mol Sci (2021) 22(9):4546. doi: 10.3390/ijms22094546
139. Bhaduri U, Merla G. Ubiquitination, biotech startups, and the future of TRIM family proteins: A TRIM-endous opportunity. Cells (2021) 10(5):1015. doi: 10.3390/cells10051015
140. Weng G, Shen C, Cao D, Gao J, Dong X, He Q, et al. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res (2021) 49:D1381–7. doi: 10.1093/nar/gkaa807
141. Veggiani G, Gerpe MCR, Sidhu SS, Zhang W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol Ther (2019) 199:139–54. doi: 10.1016/j.pharmthera.2019.03.003
142. Salehi B, Selamoglu Z, KSM, Pezzani R, Redaelli M, Cho WC, et al. Liposomal cytarabine as cancer therapy: From chemistry to medicine. Biomolecules (2019) 9(12):773. doi: 10.3390/biom9120773
143. Cromm P M, Crews CM. Targeted protein degradation: from chemical biology to drug discovery. Cell Chem Biol (2017) 24:1181–90. doi: 10.1016/j.chembiol.2017.05.024
144. He Y, Zhang X, Chang J, Kim HN, Zhang P, Wang Y, et al. Using proteolysis-targeting chimera technology to reduce navitoclax platelet toxicity and improve its senolytic activity. Nat Commun (2020) 11:1996. doi: 10.1038/s41467-020-15838-0
145. Fan C L, Liu TB. The vacuolar morphogenesis protein Vam6-like protein Vlp1 is required for pathogenicity of cryptococcus neoformans. J Fungi (Basel) (2021) 7(6):418. doi: 10.3390/jof7060418
146. Song X, Zhong H, Qu X, Yang L, Jiang B. Two novel strategies to overcome the resistance to ALK tyrosine kinase inhibitor drugs: Macrocyclic inhibitors and proteolysis-targeting chimeras. MedComm (2020) (2021) 2:341–50. doi: 10.1002/mco2.42
147. Khan S, He Y, Zhang X, Yuan Y, Pu S, Kong Q, et al. PROteolysis TArgeting chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene (2020) 39:4909–24. doi: 10.1038/s41388-020-1336-y
148. Guo WH, Qi X, Yu X, Liu Y, Chung CI, Bai F, et al. Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun (2020) 11:4268. doi: 10.1038/s41467-020-17997-6
149. Pettersson M, Crews CM. PROteolysis TArgeting chimeras (PROTACs) - past, present and future. Drug Discovery Today Technol (2019) 31:15–27. doi: 10.1016/j.ddtec.2019.01.002
150. Yin S, Liu L.Gan W. The roles of post-translational modifications on mTOR signaling. Int J Mol Sci (2021) 22(4):1784. doi: 10.3390/ijms22041784
151. Zhang Z, Chang X, Zhang C, Zeng S, Liang M, Ma Z, et al. Identification of probe-quality degraders for Poly(ADP-ribose) polymerase-1 (PARP-1). J Enzyme Inhib Med Chem (2020) 35:1606–15. doi: 10.1080/14756366.2020.1804382
152. Poirier JT, George J, Owonikoko TK, Berns A, Brambilla E, Byers LA, et al. New approaches to SCLC therapy: From the laboratory to the clinic. J Thorac Oncol (2020) 15:520–40. doi: 10.1016/j.jtho.2020.01.016
153. Wang C, Zhang Y, Wu Y, Xing D. Developments of CRBN-based PROTACs as potential therapeutic agents. Eur J Med Chem (2021) 225:113749. doi: 10.1016/j.ejmech.2021.113749
154. Berdeja J, Ailawadhi S, Horwitz SM, Matous JV, Mehta-Shah N, Martin T, et al. A phase 1 study of CFT7455, a novel degrader of IKZF1/3, in multiple myeloma and non-Hodgkin lymphoma. Blood (2021) 138:1675–5. doi: 10.1182/blood-2021-153575
155. Maneiro MA, Forte N, Shchepinova MM, Kounde CS, Chudasama V, Baker JR, et al. Antibody-PROTAC conjugates enable HER2-dependent targeted protein degradation of BRD4. ACS Chem Biol (2020) 15:1306–12. doi: 10.1021/acschembio.0c00285
156. Shah VV, Duncan AD, Jiang S, Stratton SA, Allton KL, Yam C, et al. Mammary-specific expression of Trim24 establishes a mouse model of human metaplastic breast cancer. Nat Commun (2021) 12:5389. doi: 10.1038/s41467-021-25650-z
157. Cheng J, Guo J, North BJ, Tao K, Zhou P, Wei W. The emerging role for cullin 4 family of E3 ligases in tumorigenesis. Biochim Biophys Acta Rev Cancer (2019) 1871:138–59. doi: 10.1016/j.bbcan.2018.11.007
158. Liao X, Qian X, Zhang Z, Tao Y, Li Z, Zhang Q, et al. ARV-825 demonstrates antitumor activity in gastric cancer via MYC-targets and G2M-checkpoint signaling pathways. Front Oncol (2021) 11:753119. doi: 10.3389/fonc.2021.753119
159. Lebraud H, Wright DJ, Johnson CN, Heightman TD. Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. ACS Cent Sci (2016) 2:927–34. doi: 10.1021/acscentsci.6b00280
160. Yang K, Zhao Y, Nie X, Wu H, Wang B, Almodovar-Rivera CM, et al. A cell-based target engagement assay for the identification of cereblon E3 ubiquitin ligase ligands and their application in HDAC6 degraders. Cell Chem Biol (2020) 27:866–876.e8. doi: 10.1016/j.chembiol.2020.04.008
Keywords: UPS modulator, PROTAC, E3 ligase, gastric cancer, therapy
Citation: Yang H, Ai H, Zhang J, Ma J, Liu K and Li Z (2023) UPS: Opportunities and challenges for gastric cancer treatment. Front. Oncol. 13:1140452. doi: 10.3389/fonc.2023.1140452
Received: 09 January 2023; Accepted: 20 March 2023;
Published: 03 April 2023.
Edited by:
Massimo Broggini, Mario Negri Institute for Pharmacological Research (IRCCS), ItalyReviewed by:
Lin-Feng Chen Chen, University of Illinois at Urbana-Champaign, United StatesCopyright © 2023 Yang, Ai, Zhang, Ma, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Li, emx5eWxpemhpMDMxNkB6enUuZWR1LmNu; Kangdong Liu, a2RsaXVAenp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.