
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 30 March 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1134744
This article is part of the Research Topic Advances in Molecular Classification and Targeting of Solid Tumors View all 7 articles
Background: Opioids are widely used for patients with solid tumors during surgery and for cancer pain relief. We conducted a pan-cancer genomic analysis to investigate the prognostic features of Mu opioid receptor (MOR) mRNA expression across 18 primary solid cancers.
Methods: All the data of cancer with MOR mRNA were retrieved from cBioPortal for Cancer Genomics. Logistic regression was used to determine the associations between MOR mRNA expression and clinicopathological features. Log-rank test and Cox regression was used for survival analysis. Subgroup analysis and propensity score matching were also carried out.
Results: 7,274 patients, including 1,112 patients with positive MOR mRNA expression, were included for data analyses. Positive MOR mRNA expression was associated with more advanced stage of T (adjusted Odds ratio [OR], 1.176; 95% confidence interval [CI], 1.022-1.354; P=0.024), M (adjusted OR, 1.548; 95% CI, 1.095-2.189; P=0.013) except N (adjusted OR, 1.145; 95% CI, 0.975-1.346; P=0.101), and worse prognosis for overall survival (Hazard ratio [HR] 1.347, 95% CI 1.200-1.512, P<0.001), progression-free survival (HR 1.359, 95% CI 1.220-1.513, P<0.001), disease-free survival (HR 1.269, 95% CI 1.016-1.585, P<0.001) and disease-specific survival (HR 1.474, 95% CI 1.284-1.693, P<0.001). Patients with positive MOR mRNA expression tended to be classified as tumor microenvironment immune types II, representing low PD-L1 and low CD8A expression.
Conclusion: MOR mRNA overexpression is associated with poor prognosis and poor response to PD-L1 therapy.
Cancer is still a common cause of death globally although study into molecular mechanisms of cancer cell biology and treatments including immunotherapy are well advancing. However, surgery is still the frontline treatment of solid tumors (1). Sadly, cancer reoccurrence followings surgery is the main cause of death. This may be due to many factors including the malignant nature of disease, surgical stress and inflammatory responses. But beyond these, anesthetics/techniques used during the perioperative period may also contribute to cancer reoccurrence and patients’ death (2–4). In addition, opioids are widely used for cancer patients during surgery and beyond such as intraoperative anesthesia, postoperative analgesia, and advanced cancer pain relief. Opioids work through acting on opioid receptors expressed in the central and peripheral neurons, and then reducing pain transduction to the central nervous system (5). There are three sub-types of opioid receptors, namely Mu opioid receptors (MOR), Delta opioid receptors and Kappa opioid receptors (6). MOR is the primary receptor for endogenous opioids like endorphin and enkephalins, as well as exogenous opioids like morphine and fentanyl. It is a prototypical G protein-coupled receptor that plays an important role in regulating pain, reward, and addictive behaviors (7).
Retrospective studies suggested that increased opioid use during cancer surgery may be related to cancer recurrence (8). Subsequent studies found that high MOR expression indicated poor prognosis in a variety of cancers including lung cancer, hepatocellular carcinoma and esophageal carcinoma (9–11). Furthermore, in vivo and in vitro experimental data also suggested that MOR was involved in tumor proliferation and metastasis (11). MOR may also regulate the immune system through mediating immune suppression (12).
The relationship between MOR and various solid cancers on long term surgical outcomes is limited. In this study, MOR mRNA expression across 18 solid cancers was analyzed based on the integrative pan-cancer TCGA database (13). The significant role of MOR mRNA expression in clinicopathological characteristics and prognosis were reported together along with its immunogenic features in the current study.
The public and de-identified data of primary solid tumors from TCGA database by cBioPortal for Cancer Genomics (https://www.cbioportal.org, accessed in March 25, 2020), including genomic, demographic, clinicopathological and prognostic data were retrieved (14). The genomic data consisted of mRNA-Seq expression data, which was generated using Illumina HiSeq V2 platform. The mRNA-Seq data were processed and normalized using RNA-Seq by expectation maximization, and transformed into log 2 values for analysis (15). Our pan-cancer analysis only focused on Mu Opioid Receptor mRNA expression in 18 common primary solid tumors (Table S1) and any patients whose MOR mRNA expression was not available were excluded.
The MOR mRNA-Seq expression data (symbol: OPRM1; gene ID: 4988) of 18 common solid cancer types were retrieved from the TCGA database. Initially, we divided all patients into positive versus negative subgroups by median MOR mRNA expression values of each cancer type (16). Then we found MOR mRNA expression was at a low level, and most median values were zero. Finally, we defined patients with zero MOR expression as negative. Demographic data included age, gender and race. Age was classified as young (under 60 years old), old (over 60 years old) and unknown subgroups (the median of age was 60 years old). Gender was classified as male, female and unknown. Race was classified as Caucasian, African and others. Clinicopathological data were the American Joint Committee on Cancer (AJCC) Tumor-node-metastasis (TNM) stages and histological grade. AJCC pathologic TNM stage was classified as stage I, stage II, stage III, stage IV and others. AJCC pathologic T stage was classified as T1, T2, T3, T4 and others. N stage was classified as N0, N1, N2, N3 and others. M stage was classified as M0, M1 and others. Histological grade was classified as low grade, high grade and others. In this study, when investigating the prognostic features of MOR, we focused on overall survival (OS), progression-free survival (PFS), disease-free survival (DFS) and disease-specific survival (DSS).
We also analyzed the association between MOR mRNA expression and tumor microenvironmental immune types (TMIT) and assessed the immunogenic features of the MOR. According to previous studies (17), the TMIT classification was divided into high or low expressions based on the expression of PD-L1 and CD8A. The type I represents high PD-L1 and high CD8A expression, type II with low PD-L1 and low CD8A expression, type III as high PD-L1 and low CD8A expression, and type IV of low PD-L1 and high CD8A expression. To explain, TMIT type II implies decreased infiltration of CD8+ T cells in the tumor microenvironment and decreased expression of PD-L1 in cancer cells, which represents a poor response to PD-L1 therapy.
Continuous variables with normal distribution were described as mean and standard deviation. Continuous variables with skewed distribution were described as median, first quartile and third quartile. Categorical variables were described as frequency and percentage. Pearson’s Chi-squared test or Fisher’s exact test was used to detect the statistical significance for categorical variables of demographic and clinicopathological features between MOR subgroups, as well as the association between TMIT and MOR mRNA expression. Independent Student’s t-test was used to detect a statistical significance for continuous variables with normal distribution and homogeneity of variances between MOR subgroups; Otherwise, Kruskal-Wallis test was used. Binomial logistic regression models were used to detect associations between MOR mRNA expression and binary clinicopathological features. Multinomial logistic regression models were used to detect the associations between MOR mRNA expression and polyfactorial clinicopathological features. The greater odds ratio (OR) value indicates the more advanced stage and grade of cancer. The log-rank test and Kaplan-Meier estimator were used to screen significant prognostic factors that were associated with survival outcomes. Glioblastoma multiforme (GBM) and skin cutaneous melanoma (SKCM) were excluded from the disease-free survival analysis, because no disease-free survival data was available for GBM and SKCM. After adjustment of Cox regression model, the hazard ratio (HR) for each prognostic factor was calculated. The greater HR value suggested the greater risk of death. To eliminate potential disequilibrium caused by confounding factors, subgroup analysis and propensity score matching (PSM) were done. All statistical analysis was done by R statistical software (version 3.6.2, released on February 29, 2020). A Two-sided P value < 0.05 was considered to be of statistical significance.
There were 7,274 patients with 18 common solid cancer types included into this study; Of those, 6126 patients were with negative MOR mRNA expression and 1,112 patients with positive MOR mRNA expression (Table 1). There were 3,630 young patients with age up to 60 years old, 3,612 male patients, and 5, 222 Caucasian patients (Table 1). The majority of patients belonged to stage I (1755), N0 stage (3247) and M0 stage (3872) (Table 1). Generally, the positive rates of MOR mRNA expression varied from 2.7% to 50.2% across different cancer types (Table S1). The clinicopathological data were missing in some cancer types, and these cancer types were excluded in subsequent analysis. Compared with normal tissues, MOR mRNA was generally expressed in solid cancers, but the mRNA expression level was relatively low (Figure S1). The colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) had similar embryological and histological features and they were classified as one group (colon and rectal adenocarcinoma, COREAD) in the following analysis.
We explored the association between clinicopathological features (including AJCC TNM stage, histological grade, T stage, N stage and M stage) and mRNA expression of MOR across 18 major solid cancer types. Patients with positive MOR mRNA expression tended to have more advanced AJCC TNM stage, T stage, N stage and M stage when compared with patients with negative MOR mRNA expression (Figure 1). There was no visible relationship between MOR expression and histological grade. In the following analysis, we combined N1, N2 and N3 together.
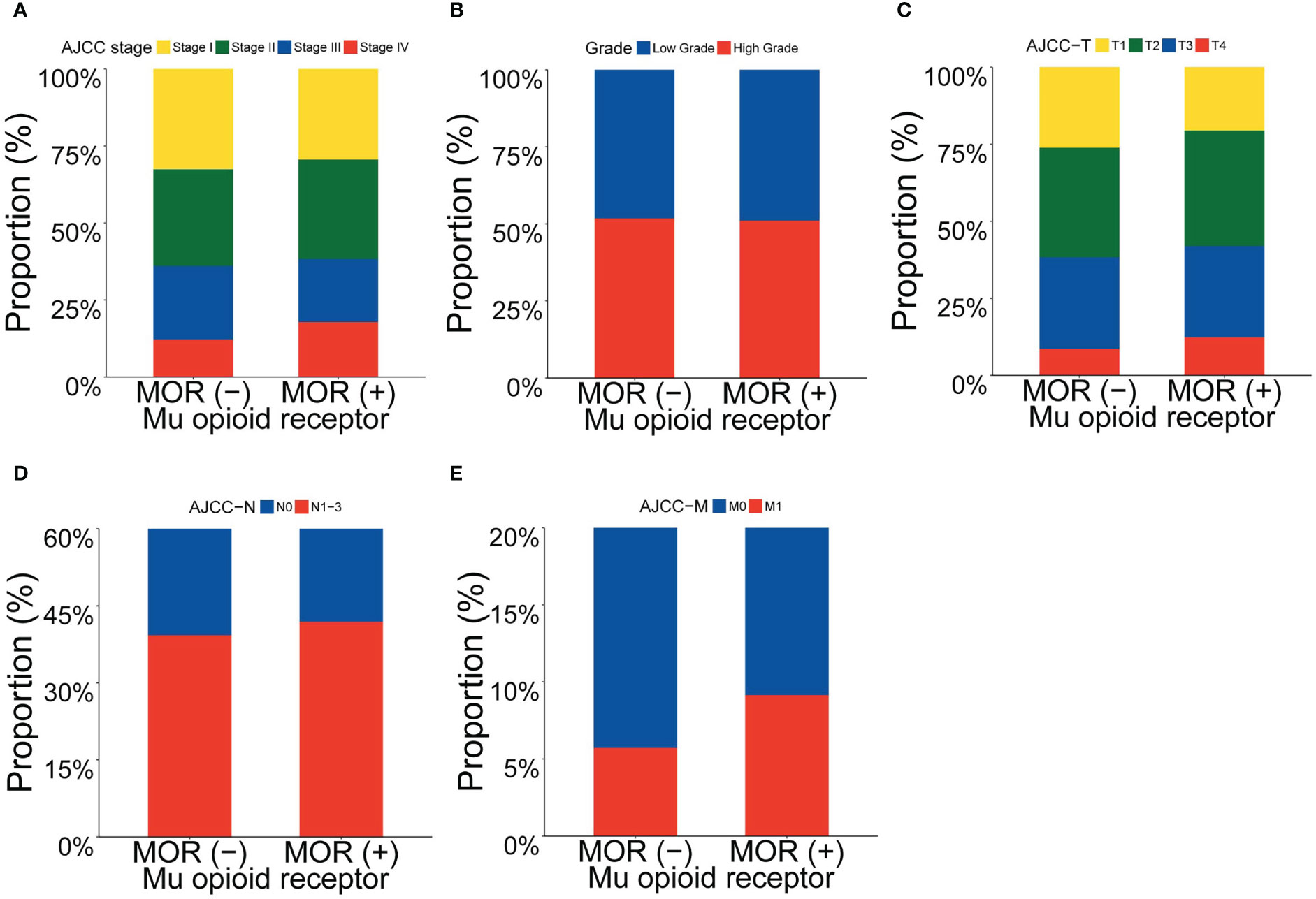
Figure 1 Proportion of clinicopathological features according to MOR mRNA expression. Footnotes: Proportion of (A) AJCC stage, (B) grade, (C) AJCC T stage, (D) AJCC N stage and (E) AJCC M stage.
After adjustment of multivariate logistic regression analysis, as shown in Table 2, we found that the association between T stage and MOR mRNA expression remained statistically significant (adjusted OR, 1.176; 95% confidence interval [CI], 1.022-1.354; P=0.024), as well as the association between M stage and MOR mRNA expression (adjusted OR, 1.548; 95% CI, 1.095-2.189; P=0.013), but association between N stage and MOR was not significant statistically (adjusted OR, 1.145; 95% CI, 0.975-1.346; P=0.101). In the sensitivity analysis, we removed one cancer type at each time and then re-analyze the association between clinicopathological features and MOR mRNA expression. We found that the association between T stage, M stage and MOR mRNA expression was stable (Figure S2). Subgroup analysis showed that there was an association trend between clinicopathological features and MOR mRNA expression (most adjusted OR>1.0), but the statistical difference is not significant (most P>0.05), (Figure S3).
There were 43 patients (0.6%) with missing OS data, 43 patients (0.6%) with missing PFS data, 3529 patients (48.5%) with missing DFS data, and 275 patients (3.8%) with missing DSS data. These patients were excluded from the corresponding survival analysis. The median follow-up periods were 23.87 months (13.12-46.45 months) for OS, 19.79 months (10.00-39.45 months) for PFS, 23.74 months (13.71-43.73 months) for DFS, and 23.87 months (13.25-46.32 months) for DSS.
The prognosis of patients with positive MOR mRNA expression was worse than patients with negative MOR expression (P<0.001 for OS, PFS, DFS and DSS) (Figure 2). The poor prognosis of positive MOR mRNA expression was observed in most cancer types (Figures S4-S7). Multivariate Cox regression models identified positive MOR mRNA expression as a significant prognostic factor in all cancer types (HR 1.347, 95% CI 1.200-1.512, P<0.001 for OS; HR 1.359, 95% CI 1.220-1.513, P<0.001 for PFS; HR 1.269, 95% CI 1.016-1.585, P<0.001 for DFS; HR 1.474, 95% CI 1.284-1.693, P<0.001 for DSS) (Figure S8). Sensitivity analysis confirmed the prognostic significance of MOR mRNA expression (Figure S8). After removing liver hepatocellular carcinoma (LIHC) from the study cohort, the effect of positive MOR mRNA expression on DFS turned more remarkable (HR 1.539, 95% CI 1.218−1.944, P<0.001) (Figure S8). We applied PSM to generate balanced data from the raw data, in which the demographics differences were eliminated between the two groups (Table S2). The adverse effect of positive MOR expression on prognosis still existed (P<0.001 for OS, PFS, DFS and DSS) with the balanced data (Figure S9).
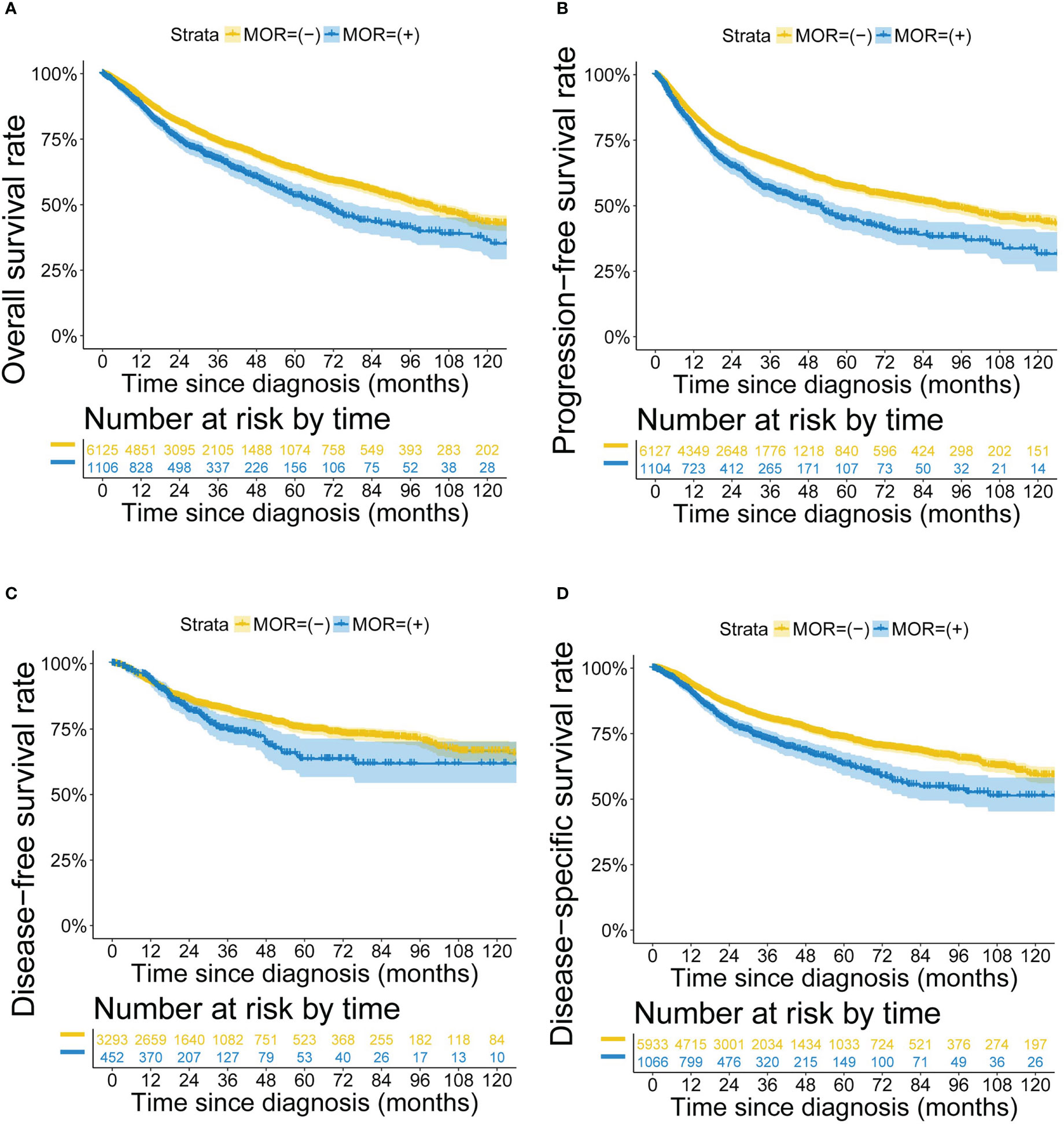
Figure 2 Kaplan-Meier survival curves according to MOR mRNA expression. Footnotes: Kaplan-Meier curves for (A) OS, (B) PFS, (C) DFS and (D) DSS.
We grouped all the patients into four TMIT groups according to the expression levels of CD8A and PD-L1. Among the included patients, 35.0% were classified as TMIT I, with high PD-L1 expression and high CD8A expression. High PD-L1 expression indicated a favorable response to the PD-L1 therapies. High CD8A expression represented a high proportion of CD8+ CTLs in the tumor microenvironment. The proportions of TMIT II (low PD-L1/low CD8A), III (high PD-L1/low CD8A), and IV (low PD-L1/high CD8A) were 35.0%, 15.0% and 15.0%, respectively (Table S3).
We analyzed the relationship between MOR mRNA expression and TMIT. As patients with positive MOR mRNA expression were likely to be classified as TMIT II, while patients with negative MOR expression were likely to be classified as TMIT I (Figure 3A). Patients of TMIT II had higher MOR mRNA expression, while patients of TMIT I had lower MOR expression (Figures 3B, D and Figure S10). This pattern remained in breast invasive carcinoma (BLCA), cervical squamous cell carcinoma (CESC), head and neck squamous cell carcinoma (HNSC), lung squamous cell carcinoma (LUSC). The expression levels of PD-L1 and CD8A were positively correlated overall and in most cancer types (Figure 3C and Figure S11).
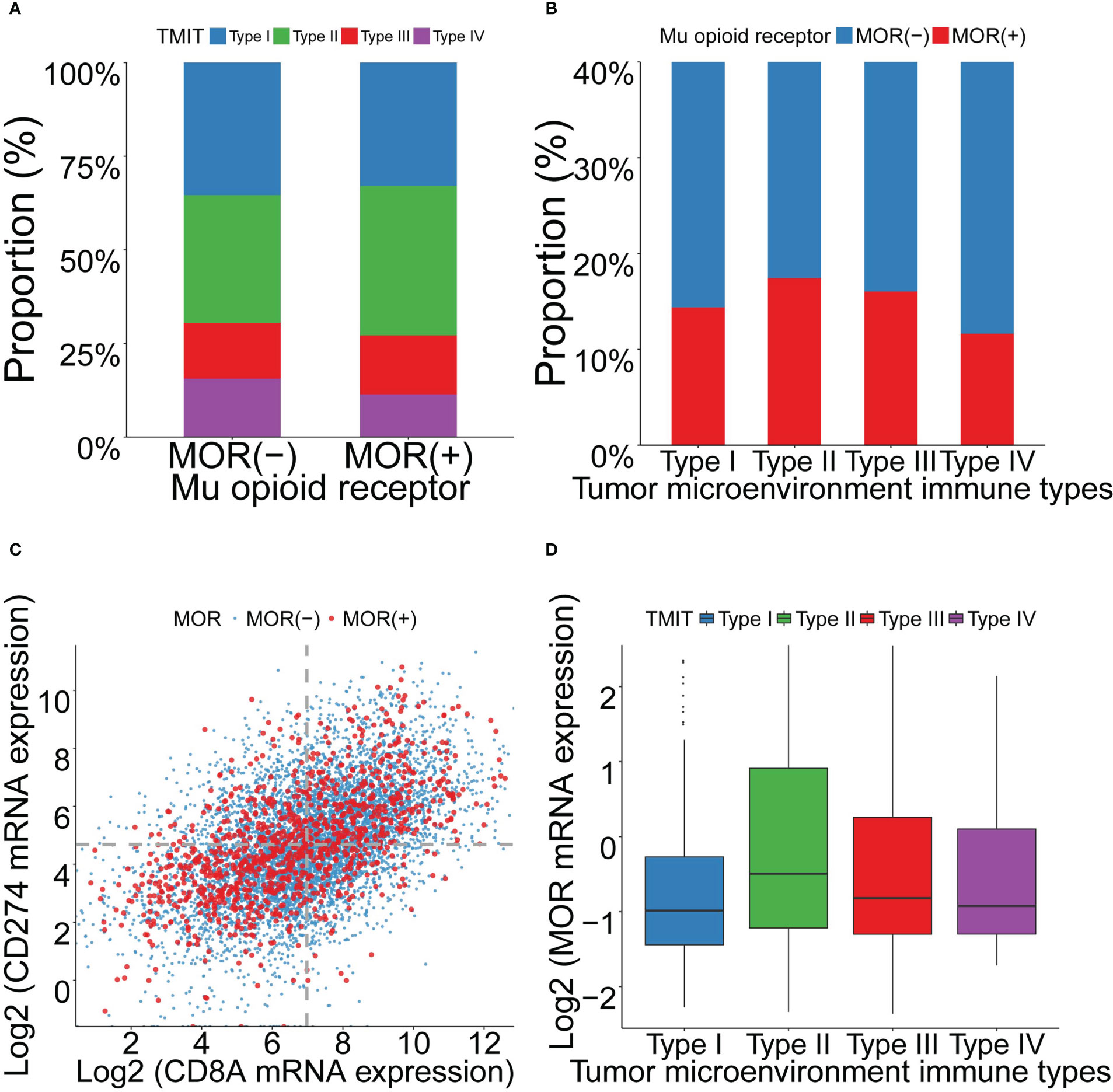
Figure 3 Association between MOR mRNA expression and TMITs, as well as correlation between PD-L1 and CD8A expression across 18 cancer types. Footnotes: (A) Distribution of TMITs according to MOR expression. (B) Distribution of MOR expression according to TMITs. (C) Correlation between mRNA expression level of PD-L1 and CD8A. (D) The mRNA expression level of MOR according to TMITs.
To summarize, MOR mRNA overexpression in solid tumors was associated with advanced cancer and poor survival, which may be related to the tumor microenvironment (Figure 4). In the tumor microenvironment, increased MOR expression indicated decreased PD-L1 expression on cancer cells and decreased CD8+ T cell infiltrations, demonstrating poor response to PD-L1 therapy (Figure 4).
In this study, we performed a pan-cancer analysis and found the prognostic features of MOR mRNA expression across 18 common solid cancer types. Firstly, we found that patients with positive MOR mRNA expression was associated with more advanced T and M stage. Secondly, MOR mRNA was identified as a prognostic biomarker in all cancer types. Positive MOR mRNA expression indicated worse prognosis for OS, PFS, DFS and DSS. Thirdly, patients with positive MOR mRNA expression tended to be classified as TMIT II, with low proportion of CD8+ CTLs and low PD-L1 expression, which implied poor response to PD-L1 therapies.
Opioids exert analgesic effects through acting on opioid receptors (mainly MOR) on neurons. Our study found that, out of a total of 7,274 patients, 1,112 were positive for MOR mRNA expression on the transcript level. When it comes to protein expression levels, a large number of laboratory studies have shown that cancer cells also expressed MOR, which were activated by opioids (18, 19). In line with our data, previous studies reported that MOR was upregulated in many cancers, and highly expressed MOR in cancers were correlated with cancer progression and recurrence (11, 20–22). A study of lung cancer also demonstrated the direct effect of opiates on cancer progression (23). The underlying molecular mechanisms remain elusive but it may be involved with the target receptors of opioids and downstream signaling pathways, induced by microRNAs’ modification (24). MOR overexpression promoted hepatocellular carcinoma cell proliferation and metastasis ability through EMT signaling pathway (11). MOR regulated self-renewal of hepatocellular carcinoma stem cells and acted as a potential therapeutic target via MOR-NFAT signaling pathway (19). Our study provides a potential explanation from the perspective of tumor immunology (25). Patients with refractory advanced cancer often receive anti-PD1/PDL1 therapy (26). However, not all solid tumors are sensitive to immunotherapy. The use of opioids is detrimental to survival outcomes for cancer patients receiving anti-PD-1/PD-L1 therapies (27). Two multicenter retrospective studies about solid cancers revealed that opioids used during immunotherapy were associated with a higher risk of early progression (28, 29), and shorter OS (30). Opioids may regulate immune cells in the tumor microenvironment, and then affect the tumor’s response to immunotherapy, including impairing T cell function and upregulating immunosuppressor cells (31), during which process MOR may serve as a potential target (29). For example, morphine-3-glucuronide upregulated PD-L1 expression through the PI3K signaling pathway, leading to the immune escape of non-small cell lung cancer cells (32). MOR may be used as a new biomarker for anti-PD1/PDL1 therapy sensitivity or not in patients with solid tumors (33).
Whether anesthesia influences cancer remains a key question in the field of anesthesia. Research on the relationship between opioid use and cancer outcomes is emerging (34, 35). MOR expression is associated with opioid use. Intraoperative opioid use increases MOR expression in cancer tissues (36, 37), and also exacerbates shorter survival (38). Contrarily, sufentanil consumption was higher in the MOR high expression group (21, 39). Generally, opioid administration may promote cancer progression, recurrence and reduce survival. For patients with non-small-cell lung carcinoma, increased doses of opioids during the postoperative period were associated with a higher 5-year recurrence rate (8). The consumption of opioids is increasing to manage chronic cancer pain in Western countries (40). While greater opioid requirement for advanced cancer pain was independently associated with reduced survival in advanced non-small cell lung cancer (41). However, a Danish population-based study found no association between opioid prescriptions and recurrence in breast cancer (42). There was also no association between postoperative opioid consumption and cancer progression or all-cause mortality in surgical patients with colorectal cancer according to another retrospective cohort study (43).
The impacts of opioids on cancer outcome vary from cancers to cancers. Certain types of cancer are sensitive to opioids, in which opioid consumption may lead to worse survival of patients. Our study may explain such differences. Our data showed that the correlation between MOR mRNA expression and poor prognosis is strong in BRCA, LIHC, lung adenocarcinoma (LUAD) and LUSC, which was consistent with previous studies (10, 11, 42). In contrast, the predictive outcome of MOR also exists in other cancer types although relatively weak (44). Interestingly, prostate adenocarcinoma (PRAD) patients with MOR mRNA expression have a better prognosis, which is rarely reported yet (45) but warrants further study.
Among patients of solid cancers, increased MOR mRNA expression was associated with reduced survival, also accompanied by advanced-stage cancer. In the tumor microenvironment, increased MOR expression indicated less PD-L1 expression in cancer cells, as well as less CD8+ T cell infiltrations, which responded poorly to immunotherapy. Our research is of practical significance. Although opioid-based analgesia is necessary for controlling cancer pain, the long-term opioid use may activate oncogenic pathways and lead to a worse prognosis (46). Therefore, use alternative analgesics including local and regional block for cancer relief is urgently needed to avoid its side effects including addiction (47); use intrathecal opioid pump or multimodal analgesia to reduce the use of opioids are strongly recommended (48, 49).
There were still some limitations in our study. Firstly, some patients’ clinicopathological data is missing in the TCGA database and therefore, our study may not reflect patient population. Secondly, it is a retrospective study and causal relationship of MOR and cancer outcomes is still unknown. Thirdly, the data were transcriptome in our study, further transcriptome, proteome or even epigenetic data analysis are also needed in future studies.
In conclusion, this is the first pan-cancer study revealing the prognostic role of MOR mRNA expression across 18 cancers. Our data showed that most tumors commonly express MOR mRNA. MOR mRNA was a prognostic biomarker cross all cancer types studied. Our work may indicate MOR mRNA overexpression in solid cancer represented poor prognosis and responded poorly to PD-L1 therapy although warrants future study.
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database.
WS, SZ, MC and ZQ were responsible for the conception and design of the study, drafting and writing of the article, acquisition and analysis of data. WS and ZQ were responsible for the interpretation of the data and drawing the figures. All authors contributed to the article and approved the submitted version.
This work was supported by Guangdong Province Hospital Pharmaceutical Research Fund, Xinchen Comfortable Medical Special Fund (grant numbers 2020XC19, 2020XC21), and Shantou Health Science and Technology Project (grant numbers [2021-4th-No.2], [2021-4th-No.3]).
The authors thanked TCGA database and cBioPortal for Cancer Genomics for their open source and maintenance of the database and Professor Daqing Ma, MD, PhD, FRCA, MAE, Imperial College London, London, UK, for his critical comments during manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1134744/full#supplementary-material
1. Sun W, Zhou H, Cheng M, Zhuang S, Qiu Z. Association between socioeconomic status and one-month mortality after surgery in 20 primary solid tumors: A pan-cancer analysis. J Cancer (2020) 11(18):5449–55. doi: 10.7150/jca.46088
2. Benzonana LL, Perry NJ, Watts HR, Yang B, Perry IA, Coombes C, et al. Isoflurane, a commonly used volatile anesthetic, enhances renal cancer growth and malignant potential via the hypoxia-inducible factor cellular signaling pathway in vitro. Anesthesiology (2013) 119(3):593–605. doi: 10.1097/ALN.0b013e31829e47fd
3. Huang H, Benzonana LL, Zhao H, Watts HR, Perry NJ, Bevan C, et al. Prostate cancer cell malignancy via modulation of HIF-1alpha pathway with isoflurane and propofol alone and in combination. Br J Cancer (2014) 111(7):1338–49. doi: 10.1038/bjc.2014.426
4. Luo X, Zhao H, Hennah L, Ning J, Liu J, Tu H, et al. Impact of isoflurane on malignant capability of ovarian cancer in vitro. Br J Anaesthesia (2015) 114(5):831–9. doi: 10.1093/bja/aeu408
5. Al-Hasani R, Bruchas MR. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology (2011) 115(6):1363–81. doi: 10.1097/ALN.0b013e318238bba6
6. Chen D, Chen Y, Yan Y, Pan J, Xing W, Li Q, et al. Down-regulation of the tumour suppressor kappa-opioid receptor predicts poor prognosis in hepatocellular carcinoma patients. BMC cancer (2017) 17(1):553. doi: 10.1186/s12885-017-3541-9
7. Lutz PE, Almeida D, Filliol D, Jollant F, Kieffer BL, Turecki G. Increased functional coupling of the mu opioid receptor in the anterior insula of depressed individuals. Neuropsychopharmacology (2021) 46(5):920–7. doi: 10.1038/s41386-021-00974-y
8. Maher DP, Wong W, White PF, McKenna R Jr., Rosner H, Shamloo B, et al. Association of increased postoperative opioid administration with non-small-cell lung cancer recurrence: A retrospective analysis. Br J Anaesthesia (2014) 113 Suppl 1:i88–94. doi: 10.1093/bja/aeu192
9. Zhang YF, Xu QX, Liao LD, Xu XE, Wu JY, Wu ZY, et al. Association of mu-opioid receptor expression with lymph node metastasis in esophageal squamous cell carcinoma. Dis Esophagus (2015) 28(2):196–203. doi: 10.1111/dote.12165
10. Singleton PA, Mirzapoiazova T, Hasina R, Salgia R, Moss J. Increased mu-opioid receptor expression in metastatic lung cancer. Br J Anaesthesia (2014) 113 Suppl 1:i103–8. doi: 10.1093/bja/aeu165
11. Chen DT, Pan JH, Chen YH, Xing W, Yan Y, Yuan YF, et al. The mu-opioid receptor is a molecular marker for poor prognosis in hepatocellular carcinoma and represents a potential therapeutic target. Br J Anaesthesia (2019) 122(6):e157–e67. doi: 10.1016/j.bja.2018.09.030
12. Eisenstein TK. The role of opioid receptors in immune system function. Front Immunol (2019) 10:2904. doi: 10.3389/fimmu.2019.02904
13. Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet (2013) 45(10):1113–20. doi: 10.1038/ng.2764
14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
15. Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf (2011) 12:323. doi: 10.1186/1471-2105-12-323
16. Lv JW, Zheng ZQ, Wang ZX, Zhou GQ, Chen L, Mao YP, et al. Pan-cancer genomic analyses reveal prognostic and immunogenic features of the tumor melatonergic microenvironment across 14 solid cancer types. J Pineal Res (2019) 66(3):e12557. doi: 10.1111/jpi.12557
17. Ock CY, Keam B, Kim S, Lee JS, Kim M, Kim TM, et al. Pan-cancer immunogenomic perspective on the tumor microenvironment based on PD-L1 and CD8 T-cell infiltration. Clin Cancer Res (2016) 22(9):2261–70. doi: 10.1158/1078-0432.CCR-15-2834
18. Ramirez MF, Gorur A, Cata JP. Opioids and cancer prognosis: A summary of the clinical evidence. Neurosci Letters (2021) 746:135661. doi: 10.1016/j.neulet.2021.135661
19. Li Y, Li G, Tao T, Kang X, Liu C, Zhang X, et al. The mu-opioid receptor (MOR) promotes tumor initiation in hepatocellular carcinoma. Cancer Letters (2019) 453:1–9. doi: 10.1016/j.canlet.2019.03.038
20. Lennon FE, Moss J, Singleton PA. The mu-opioid receptor in cancer progression: Is there a direct effect? Anesthesiology (2012) 116(4):940–5. doi: 10.1097/ALN.0b013e31824b9512
21. Zhang H, Sun M, Zhou D, Gorur A, Sun Z, Zeng W, et al. Increased mu-opioid receptor expression is associated with reduced disease-free and overall survival in laryngeal squamous cell carcinoma. Br J Anaesthesia (2020) 125(5):722–9. doi: 10.1016/j.bja.2020.07.051
22. Gorur A, Patino M, Takahashi H, Corrales G, Pickering CR, Gleber-Netto FO, et al. Mu-opioid receptor activation promotes in vitro and in vivo tumor growth in head and neck squamous cell carcinoma. Life Sci (2021) 278:119541. doi: 10.1016/j.lfs.2021.119541
23. Mathew B, Lennon FE, Siegler J, Mirzapoiazova T, Mambetsariev N, Sammani S, et al. The novel role of the mu opioid receptor in lung cancer progression: A laboratory investigation. Anesth Analgesia (2011) 112(3):558–67. doi: 10.1213/ANE.0b013e31820568af
24. Ishikawa M, Iwasaki M, Sakamoto A, Ma D. Anesthetics may modulate cancer surgical outcome: A possible role of miRNAs regulation. BMC Anesthesiol (2021) 21(1):71. doi: 10.1186/s12871-021-01294-w
25. Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer (2021) 21(6):345–59. doi: 10.1038/s41568-021-00347-z
26. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat Rev Immunol (2020) 20(11):651–68. doi: 10.1038/s41577-020-0306-5
27. Ju M, Gao Z, Liu X, Zhou H, Wang R, Zheng C, et al. The negative impact of opioids on cancer patients treated with immune checkpoint inhibitors: A systematic review and meta-analysis. J Cancer Res Clin Oncol (2022). doi: 10.1007/s00432-022-04513-0
28. Cortellini A, Tucci M, Adamo V, Stucci LS, Russo A, Tanda ET, et al. Integrated analysis of concomitant medications and oncological outcomes from PD-1/PD-L1 checkpoint inhibitors in clinical practice. J Immunother Cancer (2020) 8(2):e001361. doi: 10.1136/jitc-2020-001361
29. Botticelli A, Cirillo A, Pomati G, Cerbelli B, Scagnoli S, Roberto M, et al. The role of opioids in cancer response to immunotherapy. J Transl Med (2021) 19(1):119. doi: 10.1186/s12967-021-02784-8
30. Miura K, Sano Y, Niho S, Kawasumi K, Mochizuki N, Yoh K, et al. Impact of concomitant medication on clinical outcomes in patients with advanced non-small cell lung cancer treated with immune checkpoint inhibitors: A retrospective study. Thorac Cancer (2021) 12(13):1983–94. doi: 10.1111/1759-7714.14001
31. Prasetya RA, Metselaar-Albers M, Engels F. Concomitant use of analgesics and immune checkpoint inhibitors in non-small cell lung cancer: A pharmacodynamics perspective. Eur J Pharmacol (2021) 906:174284. doi: 10.1016/j.ejphar.2021.174284
32. Wang K, Wang J, Liu T, Yu W, Dong N, Zhang C, et al. Morphine-3-glucuronide upregulates PD-L1 expression via TLR4 and promotes the immune escape of non-small cell lung cancer. Cancer Biol Med (2021) 18(1):155–71. doi: 10.20892/j.issn.2095-3941.2020.0442
33. Banchereau R, Leng N, Zill O, Sokol E, Liu G, Pavlick D, et al. Molecular determinants of response to PD-L1 blockade across tumor types. Nat Commun (2021) 12(1):3969. doi: 10.1038/s41467-021-24112-w
34. Perry NJS, Buggy D, Ma D. Can anesthesia influence cancer outcomes after surgery? JAMA Surg (2019) 154(4):279–80. doi: 10.1001/jamasurg.2018.4619
35. Lu H, Zhang H, Weng ML, Zhang J, Jiang N, Cata JP, et al. Morphine promotes tumorigenesis and cetuximab resistance via EGFR signaling activation in human colorectal cancer. J Cell Physiol (2021) 236(6):4445–54. doi: 10.1002/jcp.30161
36. Levins KJ, Prendeville S, Conlon S, Buggy DJ. The effect of anesthetic technique on µ-opioid receptor expression and immune cell infiltration in breast cancer. J Anesthesia (2018) 32(6):792–6. doi: 10.1007/s00540-018-2554-0
37. Steele GL, Dudek AZ, Gilmore GE, Richter SA, Olson DA, Eklund JP, et al. Impact of pain, opioids, and the mu-opioid receptor on progression and survival in patients with newly diagnosed stage IV pancreatic cancer. Am J Clin Oncol (2020) 43(8):591–7. doi: 10.1097/COC.0000000000000714
38. Boland JW. Effect of opioids on survival in patients with cancer. Cancers (2022) 14(22):5720. doi: 10.3390/cancers14225720
39. Zhang H, Qu M, Gorur A, Sun Z, Cata JP, Chen W, et al. Association of mu-opioid Receptor(MOR) expression and opioids requirement with survival in patients with stage I-III pancreatic ductal adenocarcinoma. Front Oncol (2021) 11:686877. doi: 10.3389/fonc.2021.686877
40. Sjogren P, Kaasa S. The role of opioids in cancer progression: Emerging experimental and clinical implications. Ann Oncol (2016) 27(11):1978–80. doi: 10.1093/annonc/mdw407
41. Zylla D, Kuskowski MA, Gupta K, Gupta P. Association of opioid requirement and cancer pain with survival in advanced non-small cell lung cancer. Br J Anaesthesia (2014) 113(Suppl 1):i109–6. doi: 10.1093/bja/aeu351
42. Cronin-Fenton DP, Heide-Jorgensen U, Ahern TP, Lash TL, Christiansen PM, Ejlertsen B, et al. Opioids and breast cancer recurrence: A Danish population-based cohort study. Cancer (2015) 121(19):3507–14. doi: 10.1002/cncr.29532
43. Wu HL, Tai YH, Chang WK, Chang KY, Tsou MY, Cherng YG, et al. Does postoperative morphine consumption for acute surgical pain impact oncologic outcomes after colorectal cancer resection?: A retrospective cohort study. Medicine (2019) 98(18):e15442. doi: 10.1097/MD.0000000000015442
44. Falk W, Magnuson A, Eintrei C, Henningsson R, Myrelid P, Matthiessen P, et al. Comparison between epidural and intravenous analgesia effects on disease-free survival after colorectal cancer surgery: A randomised multicentre controlled trial. Br J Anaesthesia (2021) 127(1):65–74. doi: 10.1016/j.bja.2021.04.002
45. Zylla D, Gourley BL, Vang D, Jackson S, Boatman S, Lindgren B, et al. Opioid requirement, opioid receptor expression, and clinical outcomes in patients with advanced prostate cancer. Cancer (2013) 119(23):4103–10. doi: 10.1002/cncr.28345
46. Tripolt S, Neubauer HA, Knab VM, Elmer DP, Aberger F, Moriggl R, et al. Opioids drive breast cancer metastasis through the delta-opioid receptor and oncogenic STAT3. Neoplasia (New York NY) (2021) 23(2):270–9. doi: 10.1016/j.neo.2020.12.011
47. Yang J, Bauer BA, Wahner-Roedler DL, Chon TY, Xiao L. The modified WHO analgesic ladder: Is it appropriate for chronic non-cancer pain? J Pain Res (2020) 13:411–7. doi: 10.2147/JPR.S244173
48. Wang R, Wang Q, Jiang S, Chen C, Zheng J, Liu H, et al. Spontaneous ventilation video-assisted thoracoscopic surgery for non-small-cell lung cancer patients with poor lung function: Short- and long-term outcomes. Front Surg (2022) 9:800082. doi: 10.3389/fsurg.2022.800082
Keywords: Mu opioid receptor, pan-cancer analysis, prognostic features, solid cancer, opioid
Citation: Sun W, Zhuang S, Cheng M and Qiu Z (2023) Mu opioid receptor mRNA overexpression predicts poor prognosis among 18 common solid cancers: A pan-cancer analysis. Front. Oncol. 13:1134744. doi: 10.3389/fonc.2023.1134744
Received: 30 December 2022; Accepted: 21 March 2023;
Published: 30 March 2023.
Edited by:
Manoj Pandey, Banaras Hindu University, IndiaReviewed by:
Li Zhang, University of Minnesota Twin Cities, United StatesCopyright © 2023 Sun, Zhuang, Cheng and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeting Qiu, cWl1enRAc3R1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.