- 1Department of Medical Oncology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 24 + 4 Medical Doctor (MD) Program, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Geneplus-Beijing, Beijing, China
- 4Molecular Pathology Research Center, Department of Pathology, Peking Union Medical College Hospital, and Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 5Department of International Medical Services, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Background: HER2-targeted therapy provides survival benefits to HER2-mutant non-small cell lung cancer (NSCLC). A better understanding of the clinical and genomic characterization of treatment-naïve HER2-positive NSCLC, as well as the efficacy of and resistance to HER2-targeted therapy in HER2-altered NSCLC, could promote further improvement of HER2 targeted therapy.
Methods: HER2-altered NSCLC patients was retrospectively included and their genomic profiles were performed by next-generation sequencing. The clinical outcomes included overall response rate, disease control rate and progression-free survival.
Results: Among 176 treatment-naïve patients with HER2 alterations, 64.8% harbored HER2 mutations with/without HER2 amplification, and 35.2% carried HER2 amplification only. Molecular characterization was correlated with tumor stage that late-stage NSCLC with HER2 oncogenic mutations showed a higher prevalence of TP53 mutations and a higher tumor mutation burden. However, this correlation was not found in patients with HER2 amplification only. Twenty-one patients with HER2 alterations treated with pyrotinib or afatinib were retrospectively enrolled. Pyrotinib yielded a longer median progression-free survival than afatinib (5.9 [95% CI, 3.8-13.0] vs. 4.0 months [95% CI, 1.9-6.3], P = 0.06) in these patients. Analysis of the genomic profiles before and after anti-HER2 targeted therapies identified de novo HER2 copy number gain and G518W mutation, as well as mutations involving DNA damage repair signaling, SWI–SNF complex, and epigenetic regulations as potential resistance mechanisms.
Conclusion: HER2-mutant NSCLC had different molecular features from HER2-amplified NSCLC, and its genomic profile was dependent of tumor stage. Pyrotinib had superior therapeutic effects than afatinib in HER2-altered NSCLC, although larger cohorts are warranted to validate it. HER2-dependent and -independent resistance mechanisms to afatinib and pyrotinib were unveiled.
1 Introduction
Human epidermal growth factor receptor 2 (HER2), a member of the ERBB family of tyrosine kinase receptors, has been suggested to be a therapeutic target of non-small cell lung cancer (NSCLC) in recent years. Its oncogenic mutations occur in 2-4% of NSCLC and are almost mutually exclusive with activating mutations in other oncogenic drivers. Patients with HER2 mutations were common in women, nonsmokers, and adenocarcinoma (1–4). Although exon 20 insertions are the most common mutations and emerging as a clinically validated therapeutic target, point mutations and other insertions have also been suggested to be actionable (5–7). The heterogeneity of HER2 mutations offers therapeutic challenges. Compared with HER2 mutations, HER2 amplification has been reported as a distinct entity and therapeutic target that might be less relevant in lung carcinogenesis (1, 8). However, the co-alteration profiles of HER2 mutations and amplification in treatment-naïve NSCLC remain largely unknown.
Molecular testing based on next-generation sequencing (NGS) is not routinely conducted for early-stage NSCLC experiencing curative-intent pulmonary resection in many medical centers. However, early-stage lung cancers have a high risk of recurrence, with 5-year disease-free survival ranging from 36% to 82% (9). In the event of recurrent disease, tumor tissue samples for molecular testing are unavailable in many cases. The absence of molecular profiles brings challenges to better guide treatment strategy. Therefore, elucidating the molecular characteristics of HER2 alterations, especially in treatment-naive NSCLC patients at different stages, is essential to refine the therapeutic strategy.
Recently, several studies supported the clinical response of anti-HER2 drugs in HER2-positive breast cancer, gastric adenocarcinoma, and gastroesophageal adenocarcinoma, which promotes the exploration of anti-HER2 agents in NSCLC. Pan-HER tyrosine kinase inhibitors (TKIs) such as afatinib, pyrotinib, dacomitinib, neratinib, and poziotinib have been evaluated in HER2-positive NSCLC patients, with several of them showing effectiveness (10, 11). Afatinib is a selective inhibitor of the ERBB protein family, which irreversibly blocks the signaling of these proteins (12). Several studies have reported the clinical response in HER2-positive NSCLC patients with an objective response rate (ORR) of 7.7-33% and a median progression-free survival (mPFS) of 3.2-3.7 months (11, 13). However, increasing evidence suggests the disappointing effectiveness of afatinib in HER2-mutated NSCLC. In a phase II trial that enrolled eighteen patients with HER2-positive advanced NSCLC, no patient achieved an objective response and harbored a mPFS of 2.76 months (14). Xu et al. reported that chemotherapy could benefit more than afatinib to HER2-mutated advanced lung cancer, especially in those with A775_G776insYVMA (15). Therefore, the clinical application of afatinib in HER2-positive NSCLC needs further exploration. Pyrotinib, another irreversible pan-HER TKI, has been suggested to be a beneficial therapeutic agent in HER2-mutated NSCLC (6, 16). In patient-derived organoids and xenografts harboring HER2 A775_G776YVMA advanced NSCLC, pyrotinib showed a superior antitumor activity than afatinib or T-DM1 (17). Several studies demonstrated the clinical response of pyrotinib in HER2-positive metastasis NSCLC with an ORR of 19.2-53.3% and a mPFS of 5.6-6.8 months (6, 11). Although afatinib and pyrotinib offer clinical benefits for HER2-mutated NSCLC, resistance to targeted therapy is observed in the clinical setting and brings a significant challenge in cancer management. Uncovering the mechanism underlying the acquisition of HER2 TKI resistance is warranted to overcome the resistance.
In this retrospective study, we aimed to dissect the molecular characteristics of HER2 alterations in treatment-naïve NSCLC, especially the differences of molecular characteristics between early- and late-stage NSCLC with HER2 alterations, assess the efficacy of afatinib and pyrotinib in HER2-positive NSCLC patients, and uncover the potential resistance mechanism to them.
2 Methods
2.1 Patients and cohort
A total of 176 treatment-naïve NSCLC patients with HER2 alterations receiving commercial NGS of 1,021 cancer-related gene panel (Geneplus-Beijing, Beijing, China) between August 2016 and March 2022 were retrospectively enrolled. Twenty-one patients with HER2 alterations who received targeted therapy at Peking Union Medical College Hospital between March 2018 and April 2022 were retrospectively enrolled to investigate the efficacy and resistance mechanism to anti-HER2 TKIs. Written informed consent was obtained from each patient. All procedures conformed to the principles of the Helsinki Declaration with approval by institutional review boards of Peking Union Medical College Hospital.
2.2 Data collection and response assessment
The clinicopathological data, including age, sex, tumor histology, smoking status, and treatment information, were collected from the patient medical record. The outcome measures are PFS, ORR and disease control rate (DCR) assessed by standard radiologic criteria. PFS was defined as the interval from initial anti-HER2 targeted therapy to progression or death from any cause. ORR was defined as the proportion of patients with a partial response (PR) or complete response (CR). DCR was defined as the proportion of patients with a stable disease or PR or CR.
2.3 DNA extraction and targeted next-generation sequencing
DNA was isolated from tissue samples, peripheral blood, cerebrospinal fluid and hydrothorax using commercial kits (Qiagen, Hilden, Germany). The KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) was used to prepare indexed Illumina NGS libraries. Custom-designed 1,021 or 59 cancer-related gene panels were used to hybridize the DNA libraries, and their selected regions and genes are listed in Supplementary Tables 1 and 2. The hybridized libraries were sequenced using a 100-bp paired-end configuration on a DNBSEQ-T7RS sequencer (MGI Tech, Shenzhen, China).
After the removal of terminal adaptor sequences and low-quality reads with FASTP (18), the remaining reads were mapped to the reference human genome (hg19) and aligned using the Burrows-Wheel Aligner (version 0.7.12-r1039) with default parameters. GATK (3.4–46-gbc02625) and MuTect2 (1.1.4) were used to call somatic single nucleotide variants and small insertions and deletions. Contra (2.0.8) was used to identify copy number variations (19). NCsv (in-house algorithm 0.2.3) was employed to detect structural variants (20, 21). All candidate variants were manually confirmed by using the integrative genomics viewer browser. Variants were filtered to exclude clonal hematopoietic mutations with an inhouse database of clonal hematopoiesis variants of >10000 pan-cancer patients and healthy individuals (22), germline mutations in dbSNP, as well as variants that occur at a population frequency of >1% in the Exome Sequencing Project.
2.4 Tumor mutation burden (TMB) and PD-L1 expression evaluation
The TMB was determined as the number of somatic nonsynonymous single nucleotide variants and small insertions/deletions per mega-base in the coding region (with VAF ≥ 0.03) (23). Immunohistochemistry with the PD-L1 IHC 22C3 pharmDx assay (Agilent Technologies, Santa Clara, CA, USA) was performed to evaluate PD-L1 expression of tumor tissues.
2.5 Statistical analyses
Statistical analyses were performed using SPSS version 19.0 (SPSS Company, Chicago, IL). The Mann-Whiney U test and Student’s t-test were used for non-normally and normally distributed continuous variables, respectively. The comparison of categorical variables was conducted with Pearson’s χ2 test or Fisher’s exact tests. All statistical tests were performed with two-sided methods, and P < 0.05 was considered to indicate statistical significance.
3 Results
3.1 The molecular characteristics of treatment-naïve NSCLC patients with HER2 alterations
A total of 176 treatment-naïve NSCLC patients with HER2 alterations were enrolled to analyze the co-alteration profile of HER2 alterations. The median age at diagnosis was 59.5 years old (range, 25-80), with 56.8% (100/176) males and 30.1% (53/176) smokers. Most patients had stage IV disease (54.0%, 95/176), followed by stage I (21.6%, 38/176), stage III (16.4%, 29/176), and stage II (8.0%, 14/176). Via clinicopathological examination, 86.4% (152/176) patients had adenocarcinoma, with 7.4% (13/176) squamous cell carcinoma, 1.7% (3/176) adenosquamous carcinoma and 4.5% (8/176) unclear tumor histology. Of all 176 tumor tissues used for 1,021 gene panel sequencing, 123 (69.9%) samples were from primary lesions, 33 (18.7%) from metastases lesions and 20 (11.4%) were of unknown origin (Table 1).
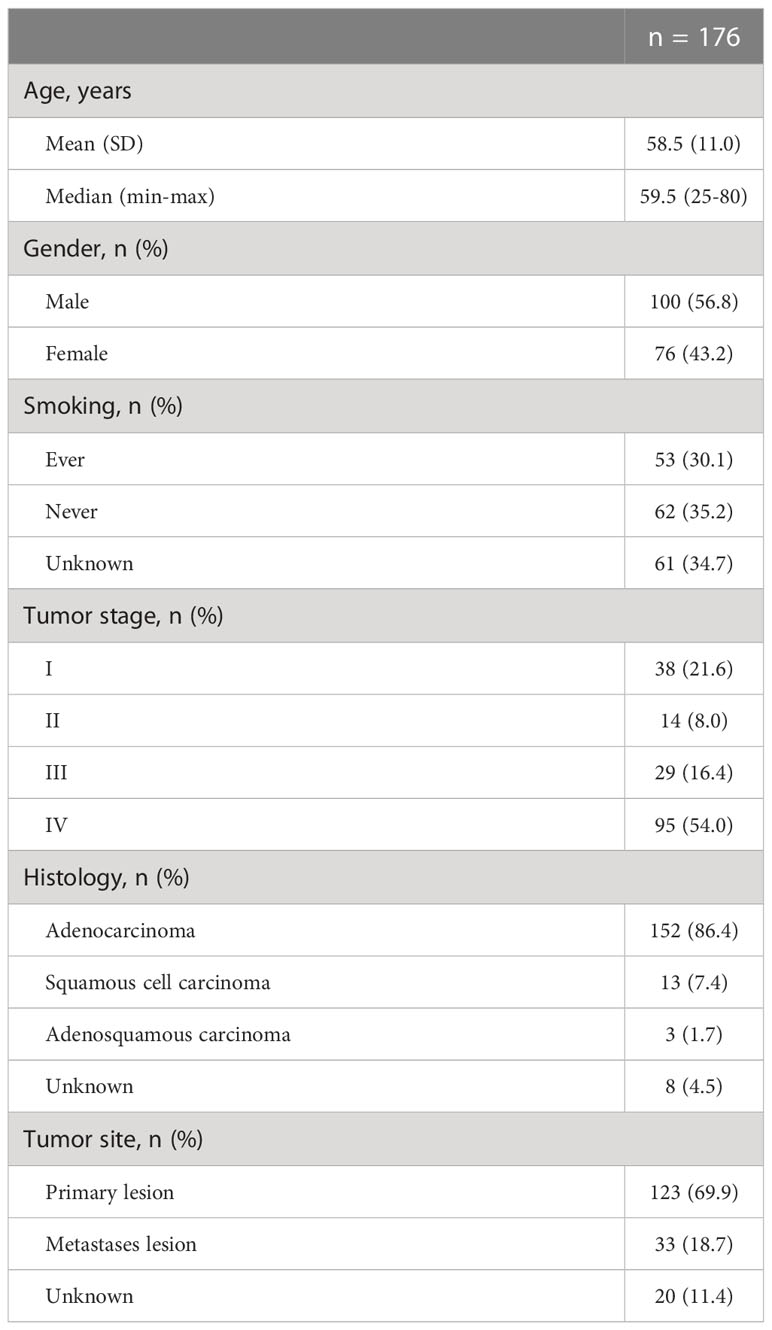
Table 1 Demographic and baseline characteristics of 176 treatment-naïve NSCLC patients with HER2 alterations.
According to the sequencing data, a total of 1,945 somatic alterations were identified, with a mean of 11.1 alterations per sample. Among these alterations, 1,361 were substitution/small insertions and deletions, with 514 gene amplifications, 65 truncations, and 5 fusion/rearrangements. HER2 mutations were detected in 114 (64.8%) patients, including 17 patients with HER2 co-amplification. HER2 amplification only was found in 62 (35.2%) patients (Figure 1A). A total of 32 types of HER2 mutations were identified, including 22 (68.6%) oncogenic types, which are documented as pathogenic in the COSMIC database (Figure 1B). The tyrosine kinase domain (TKD) of HER2 was detected as the most frequently mutated region (75.4%), followed by the furin-like domain (19.3%) and others (13.1%). Across all HER2 oncogenic mutations, Y772_A775dup was the most common mutation (46.5%), followed by S310F/Y (15.8%) and G776delinsVC/IC/LC/SVC/VV (14.9%) (Figure 1B). A comparison of the clinical features between oncogenic TKD and non-TKD mutations indicated that patients with oncogenic non-TKD mutations were older than those with oncogenic TKD mutations (Table 2). Furthermore, patients with oncogenic non-TKD mutations had a significantly higher prevalence of EGFR, KEAP1, and RBM10 mutations and EGFR amplification than those with oncogenic TKD mutations (EGFR mutation: 51.9% vs. 2.4%, P < 0.001; KEAP1 mutation: 14.8% vs. 1.2%, P = 0.01; RBM10 mutation: 18.5% vs. 0.0%, P = 0.001; EGFR amplification: 11.1 vs. 1.2%, P = 0.04).
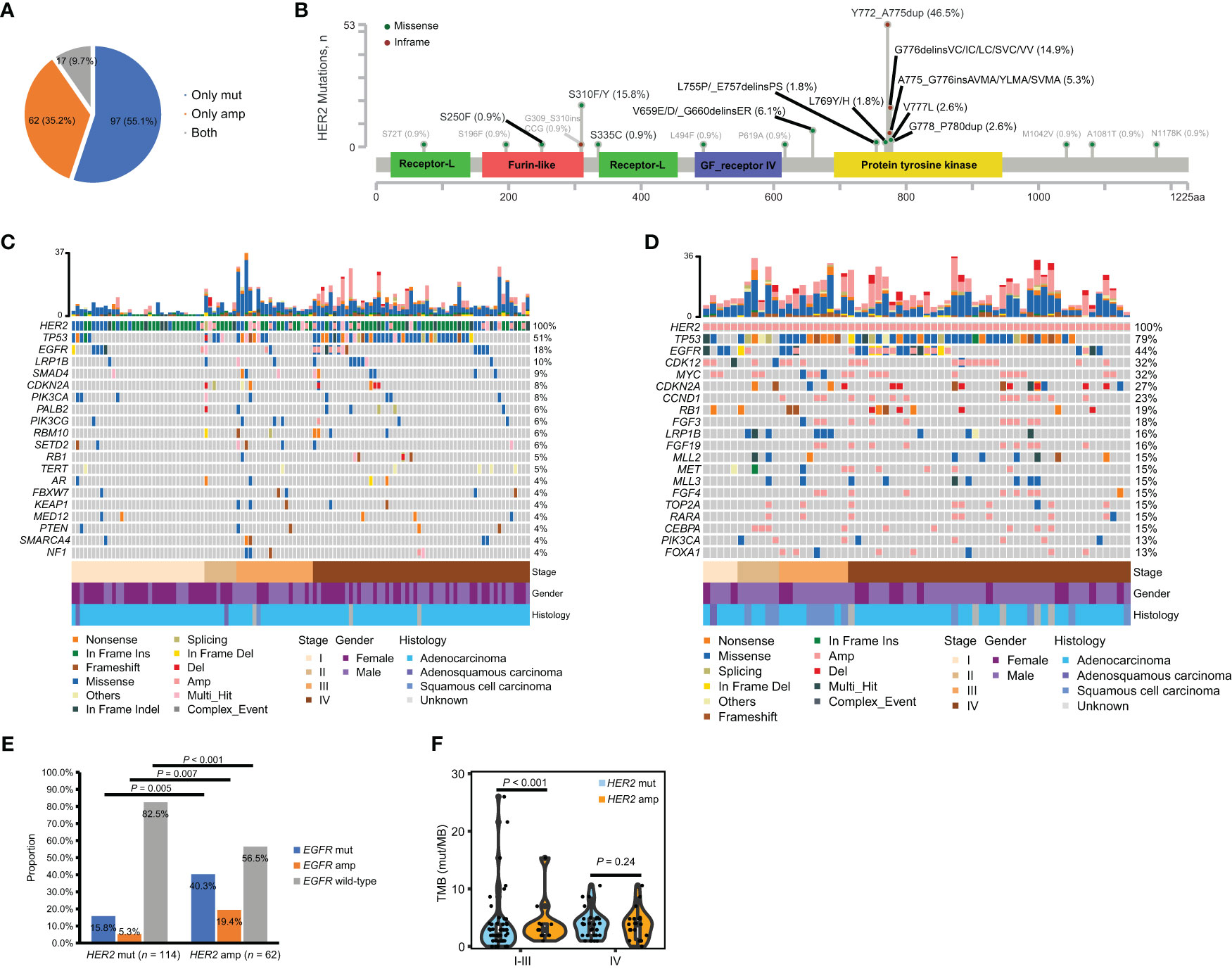
Figure 1 Incidence and co-mutation profiles among treatment-naïve HER2-altered NSCLC patients from the geneplus dataset. (A) Pie plot showing the incidence of HER2 mutation only (n = 97), HER2 amplification only (n = 62) or both HER2 mutation and amplification (n = 17) within our cohort. (B) Oncogenic (black) and non-oncogenic mutations (gray) in HER2-mutant patients were localized to HER2 protein. (C, D) Heatmap displaying the mutation landscapes of patients with HER2 mutations (n = 114) (C) and with HER2 amplification only (n = 62) (D). Each column represents data from a single patient. The number of somatic mutations of each patient and the mutation frequency of each gene are shown in the top and right respectively. The bottom heatmaps indicate key patient characteristics. (E) The proportion of EGFR mutation, amplification and EGFR wild-type in patients with HER2 mutations (n = 114) and with HER2 amplification only (n = 62). (F) Difference of TMB between patients with HER2 mutations (n = 106) and with HER2 amplification only (n = 60) in early- and late-stage disease respectively. Amp, amplification; mut, mutation; TMB, tumor mutation burden.
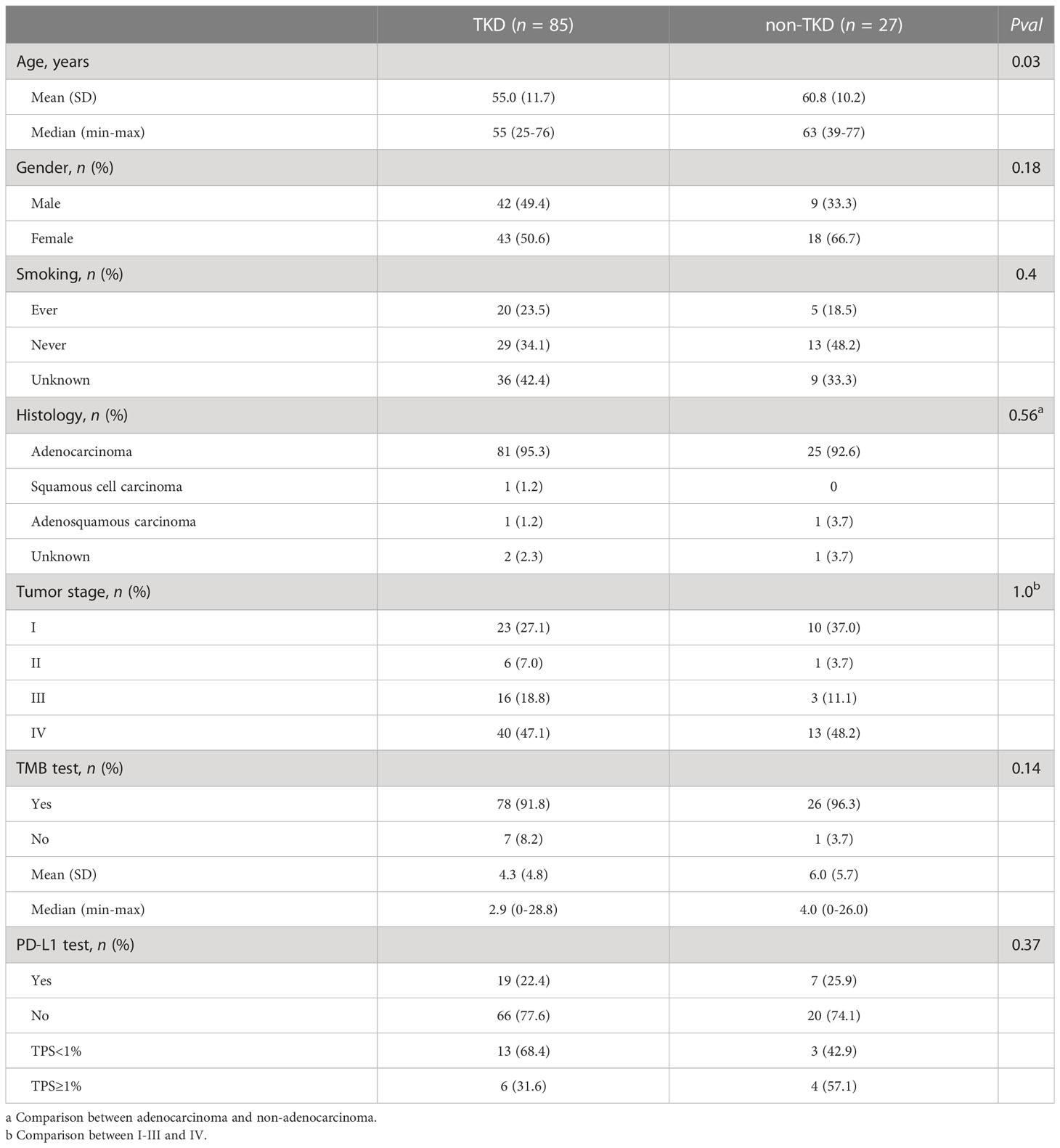
Table 2 Baseline clinical and molecular characteristics between treatment-naïve patients with HER2 oncogenic TKD and non-TKD mutations.
Given the evidence that HER2 mutations and amplification exhibit distinct entities in carcinogenesis (1, 8), we elucidated the co-alteration profiles of HER2 mutations and amplification respectively. The most frequent concomitant alterations of HER2 mutations with/without amplification were TP53 (50.9%), EGFR (17.5%), LRP1B (9.6%), SMAD4 (8.7%), PIK3CA (7.9%), CDKN2A (7.9%) and SMARCA4 (7.0%) (Figure 1C). The most common concomitant alterations of HER2 amplification only were TP53 (79.0%), EGFR (43.6%), CDK12 (32.3%), MYC (32.3%), CDKN2A (27.4%), CCND1 (22.6%) and RB1 (19.4%) (Figure 1D). Of note, patients with HER2 amplification only had a significantly higher prevalence of EGFR mutation (40.3% vs. 15.8%, P = 0.005) and EGFR amplification (19.4% vs. 5.2%, P = 0.007), as well as lower prevalence of EGFR wild-type (56.5% vs. 82.5%, P < 0.001) than those with HER2 mutations (Figure 1E). We further compared TMB between patients with HER2 mutations and with HER2 amplification only in early- and late-stage disease respectively. TMB data was available for 106 patients with HER2 mutations and 60 patients with HER2 amplification only. Our results showed that patients with HER2 amplification only had a significantly higher TMB than those with HER2 mutation in early-stage but not in late-stage disease (Figure 1F). Collectively, the molecular characteristics of HER2 mutations and amplification differed.
3.2 The molecular characteristics of early- and late-stage NSCLC with HER2 alterations
Among 114 patients with HER2 mutations, 60 of them (52.6%) were early stage (stage I-III). Compared with late NSCLC, patients with early NSCLC were younger (Supplementary Table 3). To compare the mutational profiles between early- and late-stage NSCLC with HER2 mutation and to avoid age as a potentially confounding variable, we conducted a propensity-matched comparison manually. As a result, a total of 100 matched patients were obtained, including 50 out of the 60 patients from the early-stage group and 50 out of 54 patients from the late-stage group. Their baseline characteristics are depicted in Table 3. The matched early- and late-stage patients had similar HER2 mutation types (Figure 2A). Remarkably, the result of the co-alteration comparison between matched early- and late-stage patients indicated that TP53 mutation was more frequently observed in patients with late-stage disease (78.0% vs. 30.0%, P < 0.001) (Figure 2B). Moreover, patients with late-stage NSCLC had significantly higher TMB than those with early-stage (P = 0.03) (Table 3). No significant correlations were found between PD-L1 expression and tumor stage in patients with HER2 mutations.
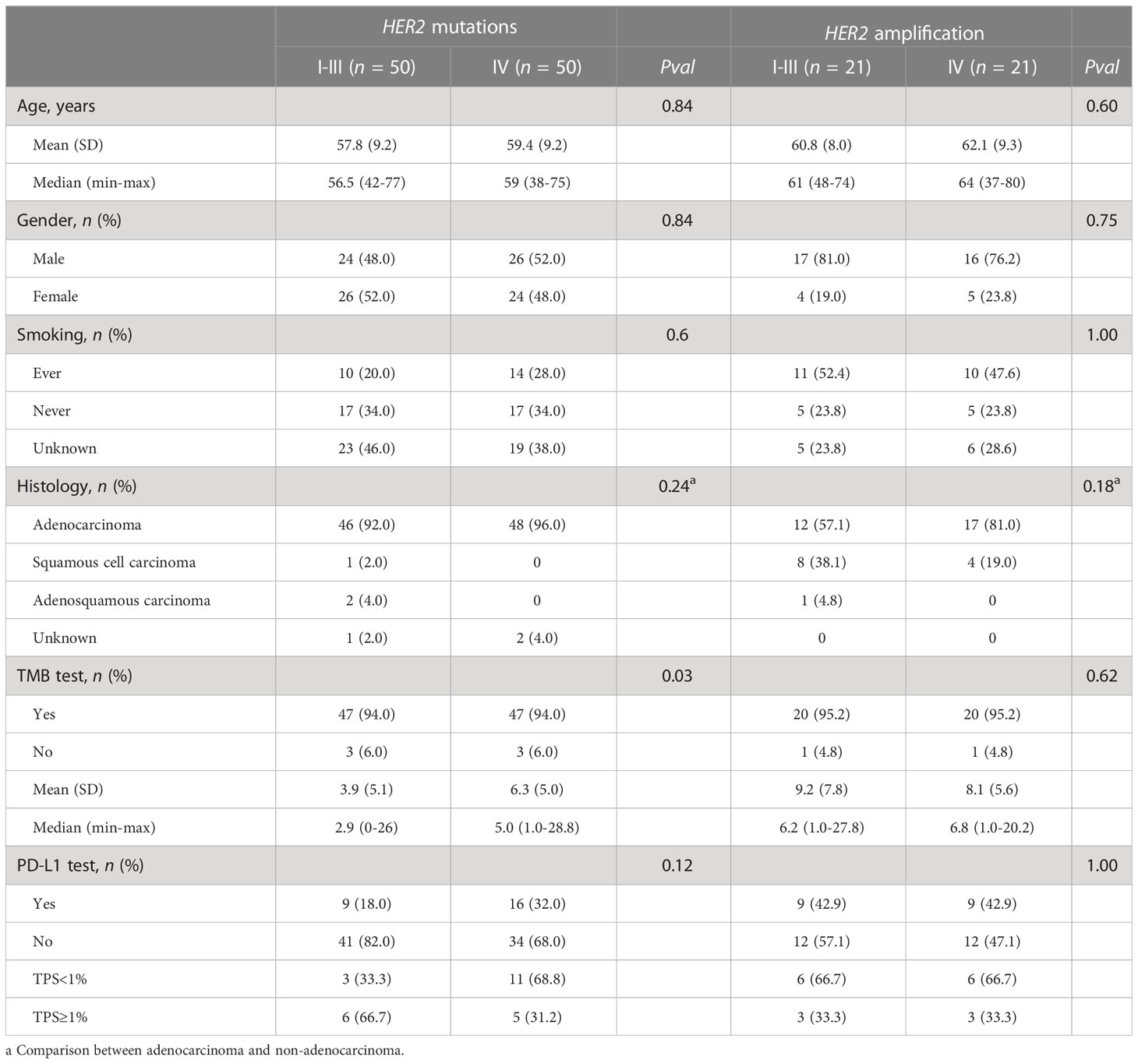
Table 3 Baseline characteristics by stage of NSCLC among matched patients with HER2 mutations or amplification.
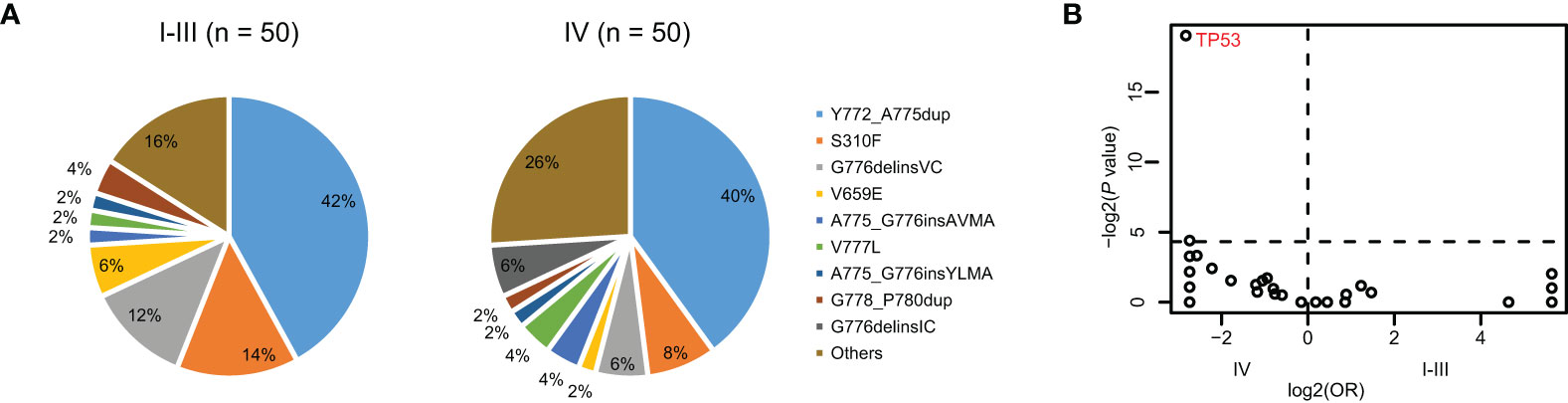
Figure 2 The differences of co-alteration profile between early- and late-stage NSCLC with HER2 mutation. (A) Pie plot showing the incidence of HER2 mutation types within early- (n = 50) and late-stage NSCLC (n = 50) with HER2 mutation. (B) Volcano plots showing the difference of co-occurring alterations between early- and late-stage NSCLC with HER2 mutation.
Across 62 patients with HER2 amplification only, 21 (33.9%) and 41 (66.1%) patients were at early and late stage respectively. Patients with late-stage NSCLC had a higher proportion of adenocarcinoma than those with early stage (Supplementary Table 3). Similarly, a manual propensity-matched comparison was performed to compare the difference in genomic alterations between early- and late-stage NSCLC with HER2 amplification only. We identified 21 out of the 41 patients from the late-stage group that matched with 21 patients with early-stage NSCLC, and their clinical characteristics are displayed in Table 3. No statistical differences were detected when comparing the frequency of co-alterations, TMB value, and PD-L1 expression level between the two matched groups. Therefore, molecular characteristics and PD-L1 expression of HER2-amplified NSCLC tumors were independent of tumor stages.
3.3 The efficacy of afatinib and pyrotinib in NSCLC patients with HER2 alterations
To assess the efficacy of targeted therapy in NSCLC patients with HER2 alterations, we retrospectively enrolled patients with HER2 alterations who received anti-HER2 targeted therapy in our center. Twenty-one patients were eligible, and their clinical and molecular characteristics are presented in Table 4. The median age was 56 years (range, 39-76), and 52.4% (11/21) were female. Most patients had stage IV disease (81.0%, 17/21) and adenocarcinoma (95.2%, 20/21). Via genetic testing, most patients carried HER2 exon 20 insertion (76.2%, 16/21), one carried HER2 V659E mutation, one carried HER2 S310X (exact sequence unknown) mutation, two carried HER2 amplification only, and one carried HER2 alteration with unclear sequence.
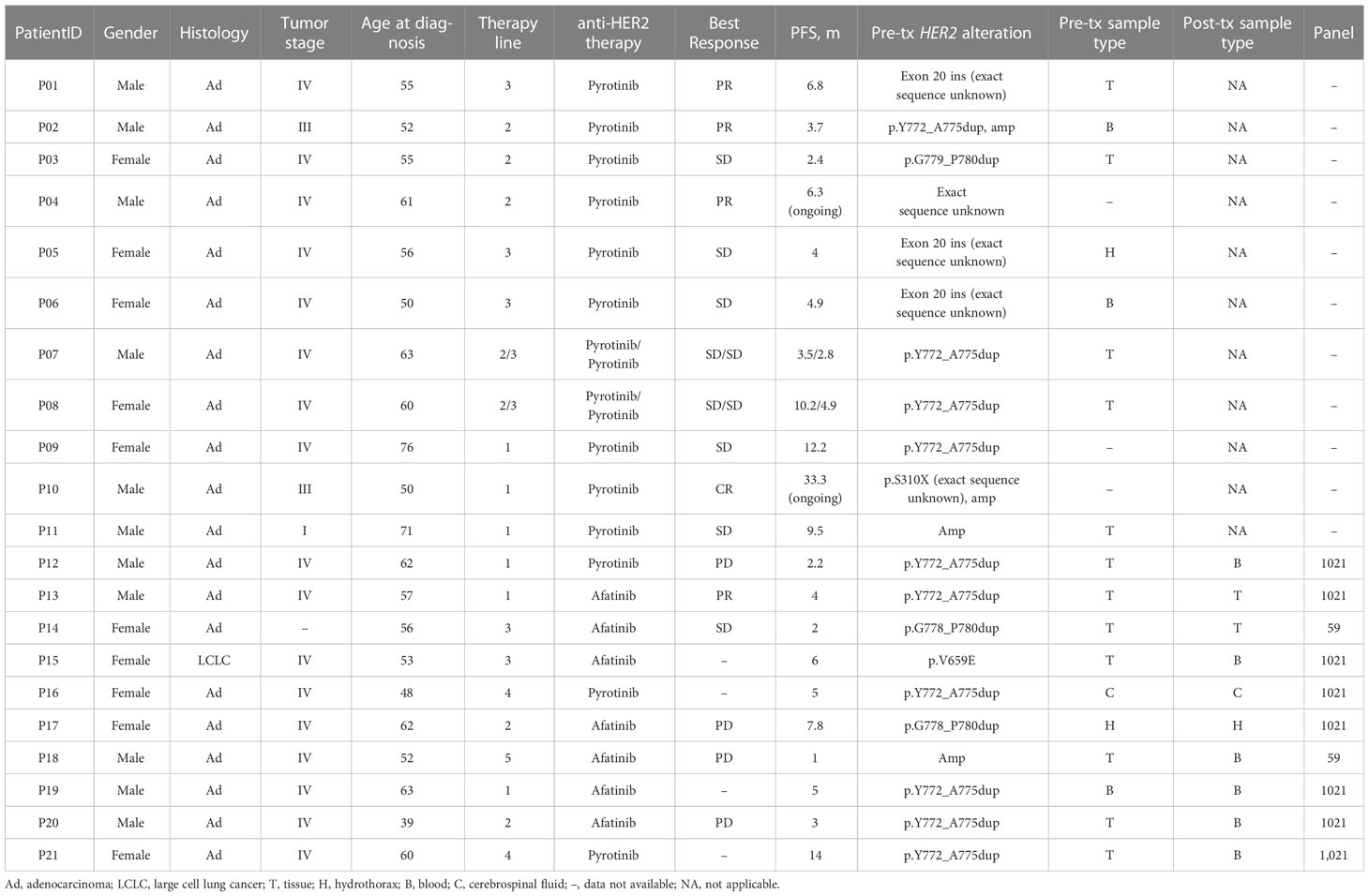
Table 4 Clinical and molecular features of patients with HER2 alterations treated with anti-targeted therapy.
Among these 21 patients, 14 (66.7%) patients received pyrotinib and 7 (33.3%) patients received afatinib. Two patients received pyrotinib as both second- and third-line treatment, of whom the clinical efficacy was only assessed on the second-line treatment. At the last follow-up date of October 2022, two patients were still on pyrotinib treatment and did not reach PFS, 18 patients developed disease progression, and one had died from the disease. The median line of afatinib and pyrotinib treatment was 2 (range, 1-5) and 2 (range, 1-4) respectively. The clinical response to targeted therapy was evaluated according to RECIST v1.1. As a result, pyrotinib yielded an ORR of 33.3% (4/12) and a DCR of 91.67% (11/12), and afatinib produced an ORR of 20% (1/5) and a DCR of 20% (1/5) (Figure 3A). The total mPFS was 5.0 months (95% CI, 3.8-10.1). Of note, patients who received pyrotinb treatment had a mPFS of 5.9 months (95% CI, 3.8-13.0), which is longer than those who received afatinib (4.0 months [95% CI, 1.9-6.3]), although statistical significance was not reached (P = 0.06) (Figure 3B). However, due to the limited number of patients enrolled in this analysis, this result is preliminary.
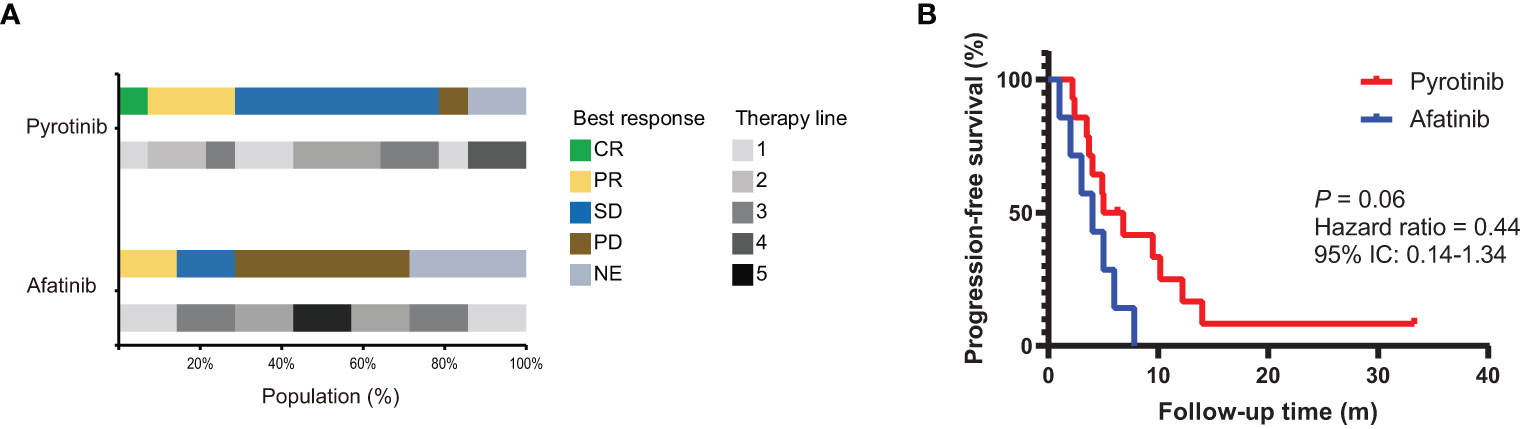
Figure 3 The efficacy of afatinib and pyrotinib in NSCLC patients with HER2 alterations. (A) The proportion of best clinical response and therapy line for HER2-altered NSCLC patients treated with afatinib (n = 7) or pyrotinib (n = 14). (B) Kaplan–Meier estimates of progression-free survival of HER2-altered NSCLC patients treated with afatinib or pyrotinib.
3.4 Potential resistance to afatinib and pyrotinib in NSCLC patients with HER2 alterations
Pre- and post-treatment genomic landscapes were dissected to uncover the potential resistance to afatinib and pyrotinib in ten eligible patients. Among them, nine patients carried HER2 mutations, and one carried HER2 amplification (Figure 4). All patients still harbored HER2 alterations after afatinib or pyrotinib treatment. We first assessed the HER2-dependent resistance to afatinib or pyrotinib. For patient P14 harboring HER2 G778_p780dup mutations and received afatinib as second-line treatment for 2 months, HER2 amplification was identified at disease progression. For patient P18, carrying HER2 amplification and received fifth-line afatinib treatment for one month, HER2 G518W oncogenic mutation and PIK3CA mutation were found upon progression disease (PD) (Figure 4).
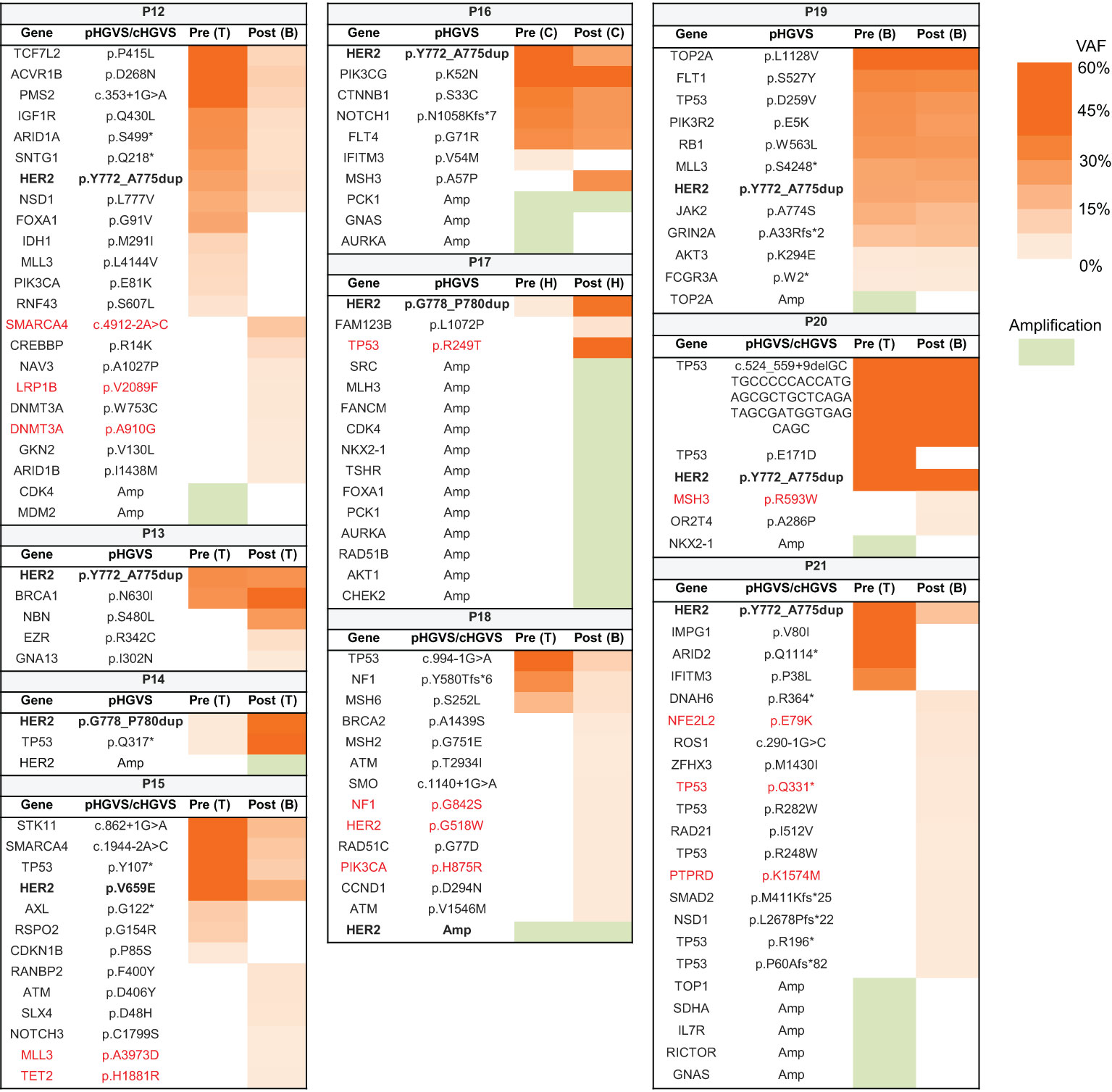
Figure 4 Potential resistance mechanism to afatinib and pyrotinib in lung cancer patients with HER2 alterations. Heat map showing the variants and their allele frequencies in patients with HER2 alterations before and after treatment. The acquired oncogenic mutations after treatment are marked with red color. B, blood; C, cerebrospinal fluid; H, hydrothorax; T, tissue; VAF, variant allele frequency.
To explore HER2-independent resistance to afatinib and pyrotinib, we analyzed the acquired alterations from eight patients who did not have HER2 secondary alterations at PD. Mutations reported by COSMIC datasets as oncogenic mutations were elaborately analyzed. Ten oncogenic mutations were identified in five patients. TP53 participating in DNA damage repair (DDR) pathway, its oncogenic mutation was identified in two patients (P17 and P21) who received second-line afatinib and fourth-line pyrotinib respectively. Other alterations of DDR pathway mediated by MSH2 oncogenic mutation was found in patient P20, who received second-line afatinib. Of note, two of these three patients harbored high genomic instability, with acquired 12 copy number alterations in P17 and acquired 13 mutations in P21, likely due to the deficiency of DDR pathway. SMARCA4 gene, encoding a part of the SWI-SNF chromatin-remodeling complex, its loss of function mutation was detected in patient P12 treated with first-line pyrotinib, indicating that compromise of the SWI–SNF complex potentially confers resistance to pyrotinib. Additionally, mutations occurred in genes involved in epigenetic regulation (LRP1B, DNMT3A, MLL3, and TET2) were found in two patients (P12 and P15), suggesting epigenetic dysregulating might also play a role in resistance to HER2 targeted therapy.
4 Discussion
With growing evidence that anti-HER2 targeted therapy provides clinical efficacies in HER2-altered NSCLC, a comprehensive understanding of the clinical and molecular characteristics of HER2-altered NSCLC tumors is urgently needed. Our retrospective cohort of 176 treatment-naïve NSCLC patients with HER2 alterations was elaborately analyzed to understand the molecular characteristics of HER2-altered NSCLC tumors. HER2 lacks ligand binding domain and activates downstream signaling by binding to other ligand-bound epidermal growth factors to form a heterodimer. HER2 oncogenic mutations are located in both TKD and non-TKD, which play roles in receptor dimerization. Our results showed that patients with oncogenic non-TKD mutations were older, and had a higher prevalence of EGFR, KEAP1, and RBM10 mutations and EGFR amplification than those with oncogenic TKD mutation, indicating that HER2 oncogenic non-TKD mutations exhibit weaker oncogenic features than HER2 oncogenic TKD mutations. Previous studies have suggested that HER2 mutations and amplification are distinct entities and therapeutic targets owing to no or low overlap (8, 24). In our study, 9.7% (17/176) patients had concurrent HER2 mutation and amplification, which is consistent with the previous study (24). HER2 oncogenic mutations are almost mutually exclusive with other driver mutations, indicating a powerful function in lung carcinogenesis. Patients with HER2 amplification only had a higher prevalence of EGFR mutations and amplification, supporting that HER2 amplification has a distinct entity and may be less relevant in lung carcinogenesis. Moreover, we found that patients with HER2 amplification only had higher TMB than those with HER2 mutations in early-stage disease. The elevated TMB in early-stage HER2-amplified NSCLC suggests a weaker oncogenic feature of HER2 amplification as well. Furthermore, previous studies reported similar results that TMB is higher in NSCLC patients with HER2 amplification than those with HER2 mutations in mixed cohorts consisting of early and advanced disease, which is thought to be associated with the higher prevalence of male smokers in HER2-amplified group (25, 26). The comparable TMB between HER2-mutated and HER2-amplified NSCLC in late-stage disease was likely due to the increased tumor mutation load of late-stage HER2-mutated tumors, as indicated by our results that in HER2-mutated NSCLC, patients with late-stage disease had higher TMB than those with early-stage disease.
Co-alteration profiles of early- and late-stage NSCLC with HER2 mutations or amplification only were studied. TP53 mutation and high TMB are known prognostic factors of poor survival in NSCLC (27–29). Compared with HER2-mutant patients with early-stage NSCLC, those with late-stage NSCLC harbored higher frequency of TP53 mutation and higher TMB. This result is congruent with the aggressive tumor biology of advanced NSCLC. However, the co-alteration profiles and TMB were similar in HER2-amplified patients with early- or late-stage NSCLC, providing evidence that HER2 mutations and amplification may induce different lung carcinogenesis procedures.
Chemotherapy is the standard treatment for patients with HER2-mutant or -amplified NSCLC but showed unsatisfactory efficacies. Currently, several studies focused on the clinical response of HER2 TKIs in HER2-positive NSCLC, including afatinib, dacomitinib, neratinib, poziotinib and pyrotinib (11). Among them, afatinib has shown controversial clinical benefits results, with an ORR of 0%-33% and a mPFS of 2.76-3.7 months (11, 13–15). Our cohort enrolled seven patients received afatinib treatment, and only one with HER2 Y772_A775dup achieved a partial response. The mPFS afatinib yielded was 4.0 months, which is comparable with previous reports (11, 13). Increasing evidences suggested pyrotinib had superior antitumor activity over afatinib with an ORR of 19.2-53.3% and a mPFS of 5.6-6.8 months (6, 11). Indeed, our retrospective study included 14 patients received pyrotinib and suggested an ORR of 33.3% and a mPFS of 5.9 months. The absence of statistical significance of mPFS between these two groups may be explained by the limited number of patients. In our cohort, pyrotinib produced a mPFS of 5.9 months, which is slightly shorter than Zhou C’s study but similar to Song Z’s study (7, 13). Differences in the sample size and between retrospective and prospective cohorts may lead to this discrepancy. Of great interest, patient P10 with IIIB adenocarcinoma received first-line pyrotinib treatment, and had a long-term complete remission of 33.3 months. This patient carried HER2 S310X mutation, which is reminiscent of previous results that patients with non-exon 20 HER2 mutations have superior clinical benefits from pyrotinib than those with exon 20 HER2 mutations (6). However, due to the low incidence of non-exon 20 HER2 mutations, the efficacy of pyrotinib in large cohorts with non-exon 20 HER2 mutations requires further investigation. Additionally, trastuzumab deruxtecan, a novel HER2 antibody-drug conjugate, has shown promising therapeutic effects in HER2-mutant NSCLC with an ORR of 61.9% and a mPFS of 14.0 months in a phase II trial (30). We propose that pyrotinib and HER2-targeted antibody-drug conjugates may provide promising treatment options for patients with HER2-mutant NSCLC.
We investigated the potential resistance mechanism to afatinib and pyrotinib in 10 HER2-positive NSCLC by comparing pre- and post-treatment genomic profiles. HER2 amplification was identified from one patient, consistent with previous reports that HER2 amplification conferred resistance to anti-HER2 targeted therapy in HER2-mutant NSCLC (6, 31). In another patient, HER2 G518W oncogenic mutation combined with PIK3CA mutation was identified at PD. Since a preclinical study suggested that dual PI3K/Akt/mTOR and HER2 blockade could conquer the resistance of anti-HER2 therapy (32), we propose that combing anti-HER2 agent with mTOR inhibitor might be a potential treatment option to reinforce the antitumor activities in this subset of patients. Three patients carried oncogenic mutations involved in the DDR pathway at PD. Since mutations in the DDR pathway were associated with improved survival for immune checkpoint inhibitors in NSCLC (33–35), a combination of pyrotinib and immune checkpoint inhibitors may vanquish the resistance. SMARCA4 oncogenic mutation was identified in one patient who received pyrotinib. As an inhibitor of the pro-oncogenic transcriptional coactivators, SMARCA4 is associated with increased metastatic incidence and leads to afatinib resistance in a patient with EGFR-mutant NSCLC (36, 37). We speculated that SMRACA4 mutation might also play a role in HER2-TKI resistance. Additionally, mutations functioning in epigenetic regulating might also lead to HER2-TKI resistance. However, in vivo, in vitro and clinical evidences are needed to determine the biological impact of these mutations to anti-HER2 targeted therapy and to study the strategies to overcome these resistances.
This study has certain limitations. First, survival data were unavailable to analyze the prognostic value of HER2 alteration. Second, comparison with afatinib and pyrotinib to HER2-mutant NSCLC should be validated in a larger cohort, especially a prospective cohort. Third, the number of patients used to evaluate the resistance mechanism to afatinib and pyrotinib was limited, and sample types before and after treatment were different. Since circulating tumor DNA is easier to obtain, it is recommended to use blood sample profiling to investigate resistance to anti-HER2 TKIs in a larger NSCLC population.
5 Conclusion
Our study displayed the molecular characteristics in treatment-naïve HER2-altered NSCLC. Compared with afatinib, pyrotinib showed better efficacy in patients with HER2 alterations. Both HER2-dependent and -independent resistance mechanisms were found in patients treated with HER2 TKIs. Further larger cohorts, especially prospective cohorts, are warranted to validate the clinical activity and resistance mechanism of pyrotinib in NSCLC patients with HER2 alterations.
Data availability statement
The data presented in the study are deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences that are publicly accessible at https://ngdc.cncb.ac.cn/gvm, accession number: GVM000466.
Ethics statement
The studies involving human participants were reviewed and approved by the ethics board of Peking Union Medical College Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
YW, YX, and RC contributed to conception and design of the study. YW, YH, TL, and YX collected the data and organized the database. YX and YH performed data analysis and interpretation. YX and YH contributed to manuscript writing. All authors contributed to manuscript revision, read and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National High Level Hospital Clinical Research Funding (2022-PUMCH-A-213) and the Beijing Municipal Science and Technology Commission (Z211100003021034).
Conflict of interest
YX and RC are current employees of Geneplus-Beijing Ltd. All financial interests are unrelated to this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JD declared a shared parent affiliation with the authors YH, TL, YL, HT, RG, YW to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1121708/full#supplementary-material
References
1. Mazieres J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, et al. Lung cancer that harbors an Her2 mutation: Epidemiologic characteristics and therapeutic perspectives. J Clin Oncol (2013) 31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095
2. Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of Erbb2 (Her2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res (2012) 18(18):4910–8. doi: 10.1158/1078-0432.CCR-12-0912
3. Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the Her2 kinase domain in lung adenocarcinomas. Cancer Res (2005) 65(5):1642–6. doi: 10.1158/0008-5472.CAN-04-4235
4. Tomizawa K, Suda K, Onozato R, Kosaka T, Endoh H, Sekido Y, et al. Prognostic and predictive implications of Her2/Erbb2/Neu gene mutations in lung cancers. Lung Cancer (2011) 74(1):139–44. doi: 10.1016/j.lungcan.2011.01.014
5. Wei XW, Gao X, Zhang XC, Yang JJ, Chen ZH, Wu YL, et al. Mutational landscape and characteristics of Erbb2 in non-small cell lung cancer. Thorac Cancer (2020) 11(6):1512–21. doi: 10.1111/1759-7714.13419
6. Song Z, Li Y, Chen S, Ying S, Xu S, Huang J, et al. Efficacy and safety of pyrotinib in advanced lung adenocarcinoma with Her2 mutations: A multicenter, single-arm, phase ii trial. BMC Med (2022) 20(1):42. doi: 10.1186/s12916-022-02245-z
7. Zhou C, Li X, Wang Q, Gao G, Zhang Y, Chen J, et al. Pyrotinib in Her2-mutant advanced lung adenocarcinoma after platinum-based chemotherapy: A multicenter, open-label, single-arm, phase ii study. J Clin Oncol (2020) 38(24):2753–61. doi: 10.1200/JCO.20.00297
8. Li BT, Ross DS, Aisner DL, Chaft JE, Hsu M, Kako SL, et al. Her2 amplification and Her2 mutation are distinct molecular targets in lung cancers. J Thorac Oncol (2016) 11(3):414–9. doi: 10.1016/j.jtho.2015.10.025
9. Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, et al. The iaslc lung cancer staging project: Proposals for revision of the tnm stage groupings in the forthcoming (Eighth) edition of the tnm classification for lung cancer. J Thorac Oncol (2016) 11(1):39–51. doi: 10.1016/j.jtho.2015.09.009
10. Rolfo C, Russo A. Her2 mutations in non-small cell lung cancer: A herculean effort to hit the target. Cancer Discovery (2020) 10(5):643–5. doi: 10.1158/2159-8290.CD-20-0225
11. Ren S, Wang J, Ying J, Mitsudomi T, Lee DH, Wang Z, et al. Consensus for Her2 alterations testing in non-Small-Cell lung cancer. ESMO Open (2022) 7(1):100395. doi: 10.1016/j.esmoop.2022.100395
12. Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. Bibw2992, an irreversible Egfr/Her2 inhibitor highly effective in preclinical lung cancer models. Oncogene (2008) 27(34):4702–11. doi: 10.1038/onc.2008.109
13. Song Z, Lv D, Chen S, Huang J, Wang L, Xu S, et al. Efficacy and resistance of afatinib in Chinese non-small cell lung cancer patients with Her2 alterations: A multicenter retrospective study. Front Oncol (2021) 11:657283. doi: 10.3389/fonc.2021.657283
14. Fan Y, Chen J, Zhou C, Wang H, Shu Y, Zhang J, et al. Afatinib in patients with advanced non-small cell lung cancer harboring Her2 mutations, previously treated with chemotherapy: A phase ii trial. Lung Cancer (2020) 147:209–13. doi: 10.1016/j.lungcan.2020.07.017
15. Xu F, Yang G, Xu H, Yang L, Qiu W, Wang Y. Treatment outcome and clinical characteristics of Her2 mutated advanced non-small cell lung cancer patients in China. Thorac Cancer (2020) 11(3):679–85. doi: 10.1111/1759-7714.13317
16. Song Z, Lv D, Chen SQ, Huang J, Li Y, Ying S, et al. Pyrotinib in patients with Her2-amplified advanced non-small cell lung cancer: A prospective, multicenter, single-arm trial. Clin Cancer Res (2022) 28(3):461–7. doi: 10.1158/1078-0432.CCR-21-2936
17. Wang Y, Jiang T, Qin Z, Jiang J, Wang Q, Yang S, et al. Her2 exon 20 insertions in non-Small-Cell lung cancer are sensitive to the irreversible pan-her receptor tyrosine kinase inhibitor pyrotinib. Ann Oncol (2019) 30(3):447–55. doi: 10.1093/annonc/mdy542
18. Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-One fastq preprocessor. Bioinformatics (2018) 34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
19. Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. Contra: Copy number analysis for targeted resequencing. Bioinformatics (2012) 28(10):1307–13. doi: 10.1093/bioinformatics/bts146
20. Zhang J-T, Liu S-Y, Gao W, Liu S-YM, Yan H-H, Ji L, et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non–small cell lung cancer. Cancer Discov (2022) 12(7):1690–701. doi: 10.1158/2159-8290.Cd-21-1486
21. Ai X, Cui J, Zhang J, Chen R, Lin W, Xie C, et al. Clonal architecture of egfr mutation predicts the efficacy of egfr-tyrosine kinase inhibitors in advanced nsclc: A prospective multicenter study (Nct03059641). Clin Cancer Res (2021) 27(3):704–12. doi: 10.1158/1078-0432.CCR-20-3063
22. Zhang Y, Yao Y, Xu Y, Li L, Gong Y, Zhang K, et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun (2021) 12(1):11. doi: 10.1038/s41467-020-20162-8
23. Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun (2018) 9(1):5361. doi: 10.1038/s41467-018-07767-w
24. Chen R, Zhao J, Lin G, Liu L, Chen L, Hu X, et al. Jces 01.28 ngs sequencing based liquid / tissue biopsy identified coexistence of Her2 amplification and mutation in advanced nsclc patients. J Thorac Oncol (2017) 12(11):S1740. doi: 10.1016/j.jtho.2017.09.315
25. Wang Q, Mao Z, Li W, Wang S, Wang L, Chen L, et al. Characteristics of the immunogenicity and tumor immune microenvironment in Her2-amplified lung adenocarcinoma. Front Immunol (2022) 13:1042072. doi: 10.3389/fimmu.2022.1042072
26. Dudnik E, Bshara E, Grubstein A, Fridel L, Shochat T, Roisman LC, et al. Rare targetable drivers (Rtds) in non-small cell lung cancer (Nsclc): Outcomes with immune check-point inhibitors (Icpi). Lung Cancer (2018) 124:117–24. doi: 10.1016/j.lungcan.2018.07.044
27. Molina-Vila MA, Bertran-Alamillo J, Gasco A, Mayo-de-las-Casas C, Sanchez-Ronco M, Pujantell-Pastor L, et al. Nondisruptive P53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res (2014) 20(17):4647–59. doi: 10.1158/1078-0432.CCR-13-2391
28. Li XM, Li WF, Lin JT, Yan HH, Tu HY, Chen HJ, et al. Predictive and prognostic potential of Tp53 in patients with advanced non-Small-Cell lung cancer treated with egfr-tki: Analysis of a phase iii randomized clinical trial (Ctong 0901). Clin Lung Cancer (2021) 22(2):100–9.e3. doi: 10.1016/j.cllc.2020.11.001
29. Owada-Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, et al. Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: Brief report. J Thorac Oncol (2018) 13(8):1217–21. doi: 10.1016/j.jtho.2018.04.003
30. Smit E, Nakagawa K, Nagasaka M, Felip E, Goto Y, Li B, et al. Ma11.03 trastuzumab deruxtecan in Her2-mutated metastatic non-small cell lung cancer (Nsclc): Interim results of destiny-Lung01. J Thorac Oncol (2021) 16(3, Supplement):S173. doi: 10.1016/j.jtho.2021.01.248
31. Chuang JC, Stehr H, Liang Y, Das M, Huang J, Diehn M, et al. Erbb2-mutated metastatic non-small cell lung cancer: Response and resistance to targeted therapies. J Thorac Oncol (2017) 12(5):833–42. doi: 10.1016/j.jtho.2017.01.023
32. Garcia-Garcia C, Ibrahim YH, Serra V, Calvo MT, Guzman M, Grueso J, et al. Dual Mtorc1/2 and Her2 blockade results in antitumor activity in preclinical models of breast cancer resistant to anti-Her2 therapy. Clin Cancer Res (2012) 18(9):2603–12. doi: 10.1158/1078-0432.CCR-11-2750
33. Zhou Z, Ding Z, Yuan J, Shen S, Jian H, Tan Q, et al. Homologous recombination deficiency (Hrd) can predict the therapeutic outcomes of immuno-neoadjuvant therapy in nsclc patients. J Hematol Oncol (2022) 15(1):62. doi: 10.1186/s13045-022-01283-7
34. Xiong A, Nie W, Zhou Y, Li C, Gu K, Zhang D, et al. Comutations in ddr pathways predict atezolizumab response in non-small cell lung cancer patients. Front Immunol (2021) 12:708558. doi: 10.3389/fimmu.2021.708558
35. Ricciuti B, Recondo G, Spurr LF, Li YY, Lamberti G, Venkatraman D, et al. Impact of DNA damage response and repair (Ddr) gene mutations on efficacy of pd-(L)1 immune checkpoint inhibition in non-small cell lung cancer. Clin Cancer Res (2020) 26(15):4135–42. doi: 10.1158/1078-0432.CCR-19-3529
36. Kunimasa K, Hirotsu Y, Miyashita Y, Goto T, Amemiya K, Mochizuki H, et al. Multiregional sequence revealed Smarca4 R1192c mutant clones acquired egfr C797s mutation in the metastatic site of an egfr-mutated nsclc patient. Lung Cancer (2020) 148:28–32. doi: 10.1016/j.lungcan.2020.07.035
Keywords: HER2, non-small cell lung cancer, HER2-TKI, resistance mechanism, targeted sequencing
Citation: Han Y, Xiong Y, Lu T, Chen R, Liu Y, Tang H, Geng R and Wang Y (2023) Genomic landscape and efficacy of HER2-targeted therapy in patients with HER2-mutant non-small cell lung cancer. Front. Oncol. 13:1121708. doi: 10.3389/fonc.2023.1121708
Received: 12 December 2022; Accepted: 22 March 2023;
Published: 03 April 2023.
Edited by:
Jill Kolesar, University of Kentucky, United StatesReviewed by:
Jianchun Duan, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaChristine Fillmore Brainson, University of Kentucky, United States
Xiaomin Niu, Shanghai Jiao Tong University, China
Copyright © 2023 Han, Xiong, Lu, Chen, Liu, Tang, Geng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingyi Wang, d2FuZ3lpbmd5aUBwdW1jaC5jbg==
†These authors have contributed equally to this work and share first authorship
 Yanjie Han
Yanjie Han Yuanyuan Xiong
Yuanyuan Xiong Tao Lu4
Tao Lu4 Rongrong Chen
Rongrong Chen Hui Tang
Hui Tang Ruixuan Geng
Ruixuan Geng