
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 03 February 2023
Sec. Genitourinary Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1116648
This article is part of the Research Topic The Mechanism of Tumor Evolution and Microenvironmental Changes of Genitourinary Oncology in Clinical Diagnosis and Treatment View all 14 articles
Background: Xp11.2 translocation renal cell carcinoma (Xp11.2 tRCC) is a group of rare and highly heterogeneous renal cell carcinoma (RCC). The translocation involving TFE3 and different fusion partners lead to overexpression of the chimeric protein. The purpose of this study is to explore the clinicopathological features of Xp11.2 tRCC with four common fusion subtypes.
Methods: We screened out 40 Xp11.2 tRCC patients from January 2007 to August 2021 in our institution. The diagnosis was initially confirmed by TFE3 immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) assay and their fusion partners were verified by RNA sequencing. Then the 40 cases were divided into two groups (DBHS family and non-DBHS family group) and a clinical comparison among the four common fusion subtypes was performed.
Results: Among the 40 cases, 11 cases with SFPQ-TFE3 gene fusion and 7 cases with NONO-TFE3 gene fusion were classified in DBHS group, the remaining cases with ASPL-TFE3 (11 cases) or PRCC-TFE3 (11 cases) gene fusion were classified in non-DBHS group. Lymph node (LN) metastasis (P=0.027) and distant metastasis (P=0.009) were more common seen in non-DBHS family group than DBHS family group and cases in DBHS family group have better progressive-free survival (PFS) (P=0.02). In addition, ASPL-TFE3 fusion was associated with worse outcome (P=0.03) while NONO-TFE3 fusion (P=0.04) predicted a better prognosis.
Conclusions: Different fusion partner genes may play a functional role in various morphology, molecular and biological features of Xp11.2 tRCCs. The impact of fusion partners on clinical characteristics of Xp11.2 tRCCs deserves further exploration.
Xp11.2 translocation renal cell carcinoma (Xp11.2 tRCC) is a rare and distinct subtype of RCC, classified in Microphthalmia (MiT) transcription factor family translocation renal cell carcinomas (RCC) in 2016 (1). The most notable feature of Xp11.2 tRCC is chromosome translocations involving TFE3 gene, resulting in fusion with various gene partners (2). Since the discovery of ASPL-TFE3 as the first gene fusion in Xp11.2 tRCC (3), the number of fusion partners has expanded with the development of next-generation sequencing (NGS) technologies. The relatively common fusion genes included ASPSCR1 (ASPL), PRCC, NONO (p54nrb), SFPQ (PSF) and other rare fusion genes such as CLTC, RBM10, MED15, SETD1B, ZC3H4, LUC7L3, KHSRP, PARP14, DVL2, GRIPAP1 were occasionally reported (3–13). The diversity of the fusion partners drastically affects biological behaviors and chromosome structures, which in turn leads to the clinical heterogeneity of Xp11.2 tRCC (14). However, due to the rarity of Xp11.2 tRCC, more cases with definite fusion types are needed to compare the clinical characteristics of Xp11.2 tRCC.
The impact of fusion genes on Xp11.2 tRCC is multifaceted. The common fusion partners of Xp11.2 tRCC, SFPQ and NONO, are both members of the drosophila behavior/human splicing (DBHS) protein family and participate in almost all steps of gene regulation (15–18). The protein products encoded by DBHS family are functionally conserved and largely considered as nuclear factors (15). Previous study has suggested SFPQ and NONO may play an important role in nuclear localization of TFE3 during tumor progression(2). In addition, the inversion of the TFE3 and NONO results in an equivocal split signal distance in fluorescence in situ hybridization (FISH), which makes it difficult to diagnose Xp11.2 with this special fusion type (19). On the other hand, another common fusion partner, ASPL, was proved to be associated with aggressive behavior and poor prognosis compared with other fusion genes (12, 14). PRCC can bind to MAD2B (a mitotic checkpoint protein) directly and regulate mitosis, but this interaction can be impaired by the translocation of PRCC and TFE3 (20). As most molecularly confirmed Xp11.2 tRCC cases was described in small series, so far, there are few reports about systematic clinical comparison of Xp11.2 tRCC with common fusion types.
In the present study, we identified 40 cases of Xp11.2 tRCC with four common fusion types (ASPL, PRCC, NONO and SFPQ) by RNA sequencing and described their typical morphological and molecular features. Furthermore, we divided the 40 cases into two groups (DBHS family and non-DBHS family group) and compared their clinicopathological characteristics and prognosis.
In this study, 40 cases with suspicious morphological features of Xp11.2 tRCC were retrieved from the diagnostic files in Nanjing Drum Tower Hospital between January 2007 to August 2021. The hematoxylin & eosin (H&E) slides were reviewed independently by two specialist uropathologists and the diagnosis of these cases was based on preliminary TFE3 immunohistochemistry (IHC) or FISH assay. Some of these cases have been reported in the previous literature and their fusion partners have been confirmed by reverse transcriptase polymerase chain reaction (RT-PCR) (21, 22). The available clinicopathological features and follow-up data were recorded. The TNM stage and nuclear grade were classified by the AJCC 2017 TNM Staging System and WHO/ISUP grading system, respectively.
The 4-μm-thick sections were prepared from 10% FFPE tissue blocks for TFE3 IHC staining. All slides were exposed to 3% H2O2 for 10 minutes at room temperature to block endogenous peroxidase activity. TFE3 (HPA023881, Sigma, USA), cathepsin K (ab19027, Abcam, Cambridge, UK), CD10 (ab227640, Abcam, Cambridge, UK), CA-IX (ab107257, Abcam, Cambridge, UK), Vimentin (ab8978, Abcam, Cambridge, UK), CD117(ab32363, Abcam, Cambridge, UK), CK7 (ab181598, Abcam, Cambridge, UK) antibody were incubated with tumor sections in a humidified chamber at 4°C overnight, then the anti-mouse or anti-rabbit peroxidase-conjugated secondary antibody (EnVision™ Detection Kit, DAKO, Denmark) were used with the sections at 37°C for 30 minutes.
The result was evaluated in a semiquantitative manner by multiplying the staining intensity (0 = no staining, 1 = mild staining, 2 =moderate staining, and 3 = strong staining) by the percentage of immunoreactive tumor cells (0–100). The final immunostaining result was calculated as following: negative (0), score <25; weak positive (1+), score 26–100; moderate positive (2+), score 101–200; strong positive (3+), score 201–300.
FISH assay was performed on 3-μm -thick FFPE tissue sections with TFE3 positive immunostaining. The commercial dual-color break-apart FISH probes (LBP, Guangzhou, China) were used to detect TFE3 gene arrangement. The telomere and centromeric sides were labeled with 5-ROX-dUTP (red) and fluorescein-12-dUTP (green), respectively. Briefly, the FFPE sections were deparaffinized and permeabilized after a series of treatments, then the probes were applied to the tumor region. All the slides containing the tissue DNA probes denatured at 85°C in a in situ thermocycler for 5 minutes and hybridized at 37°C overnight. After washing in 2×SSC for 10 minutes and in 0.1% NP-40/2×SSC for 5 minutes at room temperature, the slides were air dried and 5 μL of 4′,6-diamidino-2-phenylindole (DAPI) was used to counterstain the nuclei. The detail FISH protocol has been reported previously. Routinely, at least 100 non-overlapping nuclei were counted under Olympus BX51TRF fluorescence microscope (Olympus, Tokyo, Japan) at ×1000 magnification. The signals separated by a distance >2 signal diameter was considered to be split. For cases with suspicious NONO-TFE3 fusion (equivocal FISH pattern), RNA sequencing was performed to verify the result. When >10% of the nuclei showed evidence of split signals, the result was considered to be positive.
The 40 cases with positive FISH results were analyzed by RNA sequencing. Total RNA from FFPE samples was extracted using RNeasy kit (QIAGEN). RNase H was used to depleted ribosomal RNA and KAPA Stranded RNA-seq Kit with RiboErase (HMR) (KAPA Biosystems) was used to library preparation. Library concentration and library quality was accessed by KAPA Library Quantification Kit (KAPA Biosystems) and Agilent High Sensitivity DNA kit on Bioanalyzer 2100 (Agilent Technologies), respectively. Then the products were sequenced on Illumina HiSeq next-generation sequencing (NGS) platforms (Illumina).
Statistical analyses were conducted by SPSS software version 26.0 (SPSS, Inc., Chicago, IL, USA) and figures were depicted by GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA). Student’ s t test or Mann–Whitney U test or was performed to compare continuous data. Pearson chi-square test or Fisher’ s exact test was performed to compare categorical data. Kaplan– Meier method was used to compare the survival data, and statistical comparisons between the two groups were evaluated with Log-rank test. A two-sided P < 0.05 was considered statistically significant.
The detailed clinicopathologic features of the 40 Xp11.2 tRCC patients are shown in Table 1. Xp11.2 tRCC most commonly occurred in young adults, at a median age at diagnosis of 35.5 years and a mean of 37.3 years, ranging from 7 to 70 years. The incidence rate was slightly higher in female than that in male, with a male: female ratio of 1:1.4. The median tumor size was 4.5cm and the mean size was 5.5cm, respectively. Regional lymph node metastasis was found in 9 patients (22.5%) at diagnosis and distant metastasis had developed in 13 patients (32.5%) at the last follow‐up. 31 patients (77.5%) were at earlier pT stage (T1-T2) while 9 patients (22.5%) were at an advanced stage (T3-T4) at the time of diagnosis. Higher nuclear grades (WHO/ISUP grade: 3-4) was observed in more than half the patients (60%). 10 (25.0%) patients died at the end of follow-up and 4 patients (10%) were alive with disease.
Xp11.2 tRCC showed variable morphological characteristics according to different fusion types. The typical feature of Xp11.2 tRCC was the presence of papillary, glandular, nested, or tubular architectures with clear or eosinophilic cytoplasm, psammoma bodies or calcification were occasionally seen. Papillary architecture was the most common morphology in the 40 cases. Nested architecture and pseudorosettes were seen in 4 SFPQ-TFE3 cases and 5 ASPL-TFE3 cases while none of cases with NONO-TFE3 and PRCC-TFE3 showed this architecture. The most distinctive feature of PRCC-TFE3 cases was the compact (Closely arranged tumor cells with less voluminous cytoplasm and few psammoma bodies) architecture (7/11,63.6%), which was quite different from the other subtypes. Psammoma bodies appeared the most in ASPL-TFE3 cases (8/11, 72.7%) while they were rarely observed in NONO-TFE3 (1/7, 14.3%) and PRCC-TFE3 cases (0/11, 0%). The histologic features of Xp11.2 tRCC with four main fusion types were shown in Figure 1. In term of IHC profiles, all the cases showed TFE3 nuclear positivity and the majority of Xp11.2 tRCC cases (90%) showed moderate (++) to strong (+++) positivity. Cathepsin K were seen predominantly in PRCC-TFE3 cases (7/11.63.6%), which is useful to distinguish PRCC-TFE3 cases from the other types. CD10 was positive in most of the cases (34/40, 85%) and half of the cases (22/40, 55%) showed vimentin positivity. The positivity of CA-IX, CD117 and CK7 is uncommon. The morphological and IHC profiles of 40 cases were shown in Table 2. In FISH results, 33 cases showed typical translocation signals and 7 cases with NONO-TFE3 fusion showed equivocal signals (split but adjacent signals). The representative IHC and FISH features were shown in Figures 2, 3. Gene fusions were verified by RNA sequencing, of which the gene fusions involve ASPL-TFE3 gene fusion (11 cases), PRCC-TFE3 gene fusion (11 cases), SFPQ-TFE3 gene fusion (11 case) and NONO-TFE3 gene fusion (7 cases). The representative sequencing results were shown in Table S1.
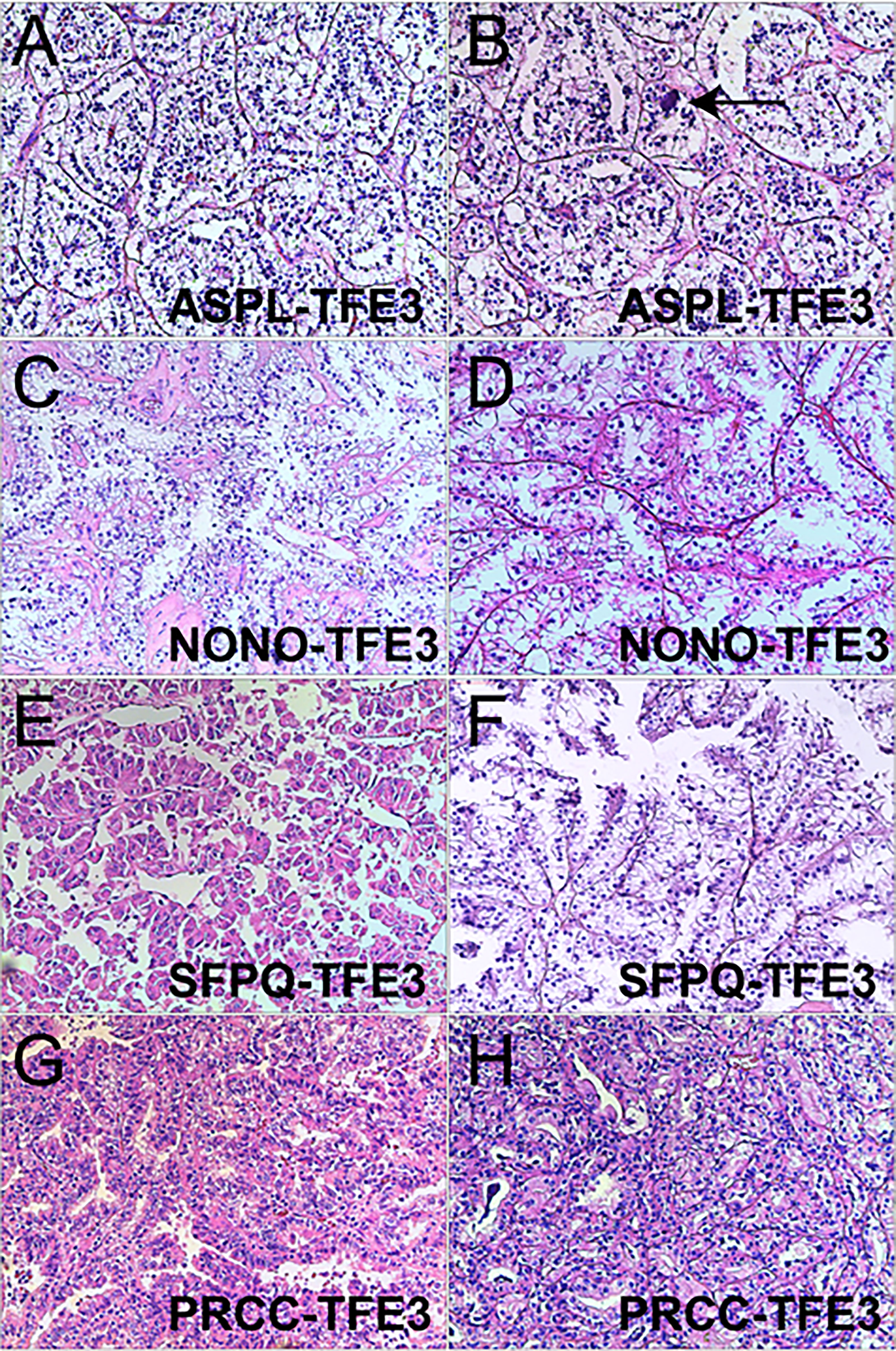
Figure 1 Representative images of morphologic features in Xp11.2 tRCCs with four common fusion subtypes. (A, B) ASPL-TFE3 cases showed nested architecture, clear to eosinophilic cells with voluminous cytoplasm and round nuclei, Psammoma bodies were frequently seen (arrowhead). (C, D) NONO-TFE3 cases showed papillary architecture with clear to flocculent eosinophilic cytoplasm. (E, F) SFPQ-TFE3 cases showed papillary architecture. (G, H) PRCC-TFE3 showed compact architecture with less voluminous cytoplasm. Original magnification: × 100 (A–H). H&E: hematoxylin and eosin.
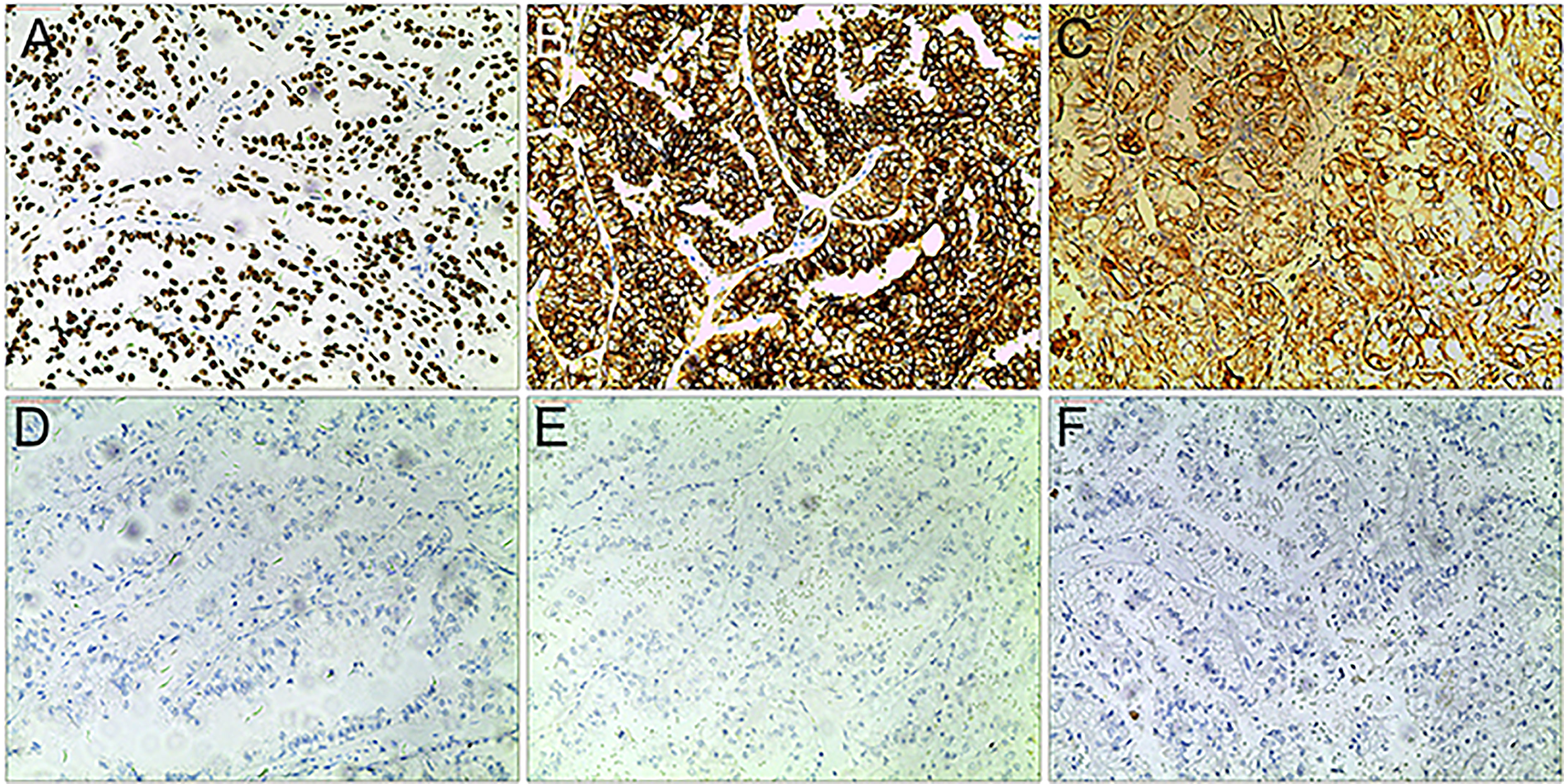
Figure 2 Representative IHC images of Xp11.2 tRCCs. (A–C) positive results of TFE3, CD10 and Vimentin. (D–F) Negative results of cathepsin K, CD117 and CA-IX. Original magnification: × 100 (A–F).
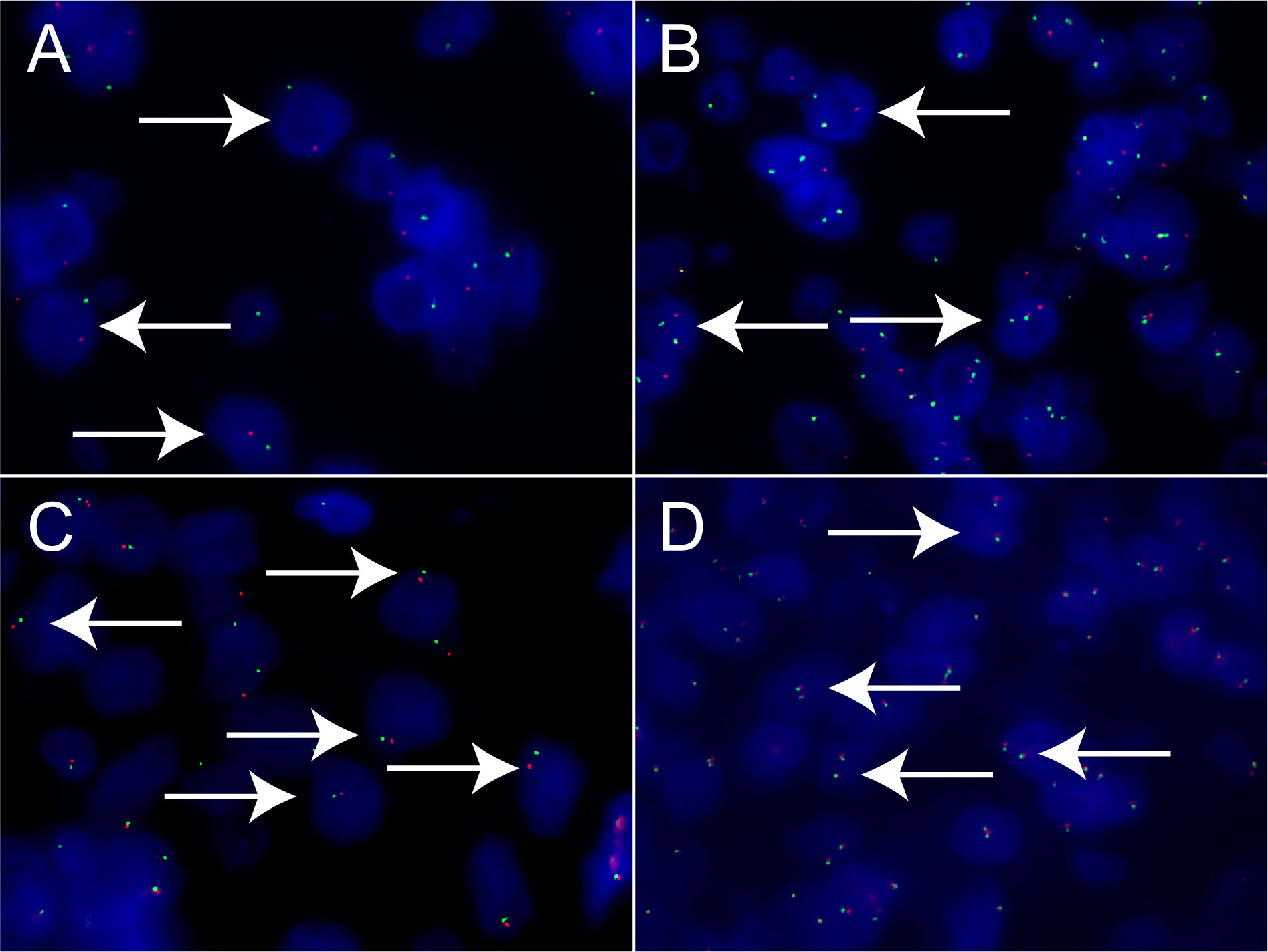
Figure 3 Results of TFE3 break-apart FISH in Xp11.2 tRCC. (A, B) Typical signal patterns in a male case (1R1G) and a female case (1R1G1F). (C, D) Equivocal signal patterns of NONO-TFE3 in a male case (1R1G) and a female case (1R1G1F). Original magnification: × 1000 (A–D). R, red; G, green; F, fusion; FISH, fluorescence in situ hybridization.
Among all the 40 cases, 11 cases with SFPQ-TFE3 gene fusion and 7 cases with NONO-TFE3 gene fusion were classified in DBHS group, the remaining cases with ASPL-TFE3 or PRCC-TFE3 gene fusion were classified in non-DBHS group (Table 3). No statistical difference was seen in the baseline clinicopathologic characteristics, including age, gender, laterality, tumor size, pT stage and AJCC stage (P >0.05) between DBHS family group and non-DBHS family group. However, there was a significant difference in pN stage (P = 0.027) at surgery and M stage at the last follow‐up (P = 0.009). Furthermore, although no statistical difference was found in nuclear grade and follow-up time (all P >0.05), cases in non-DBHS family group tend to have a worse outcome (P =0.013).
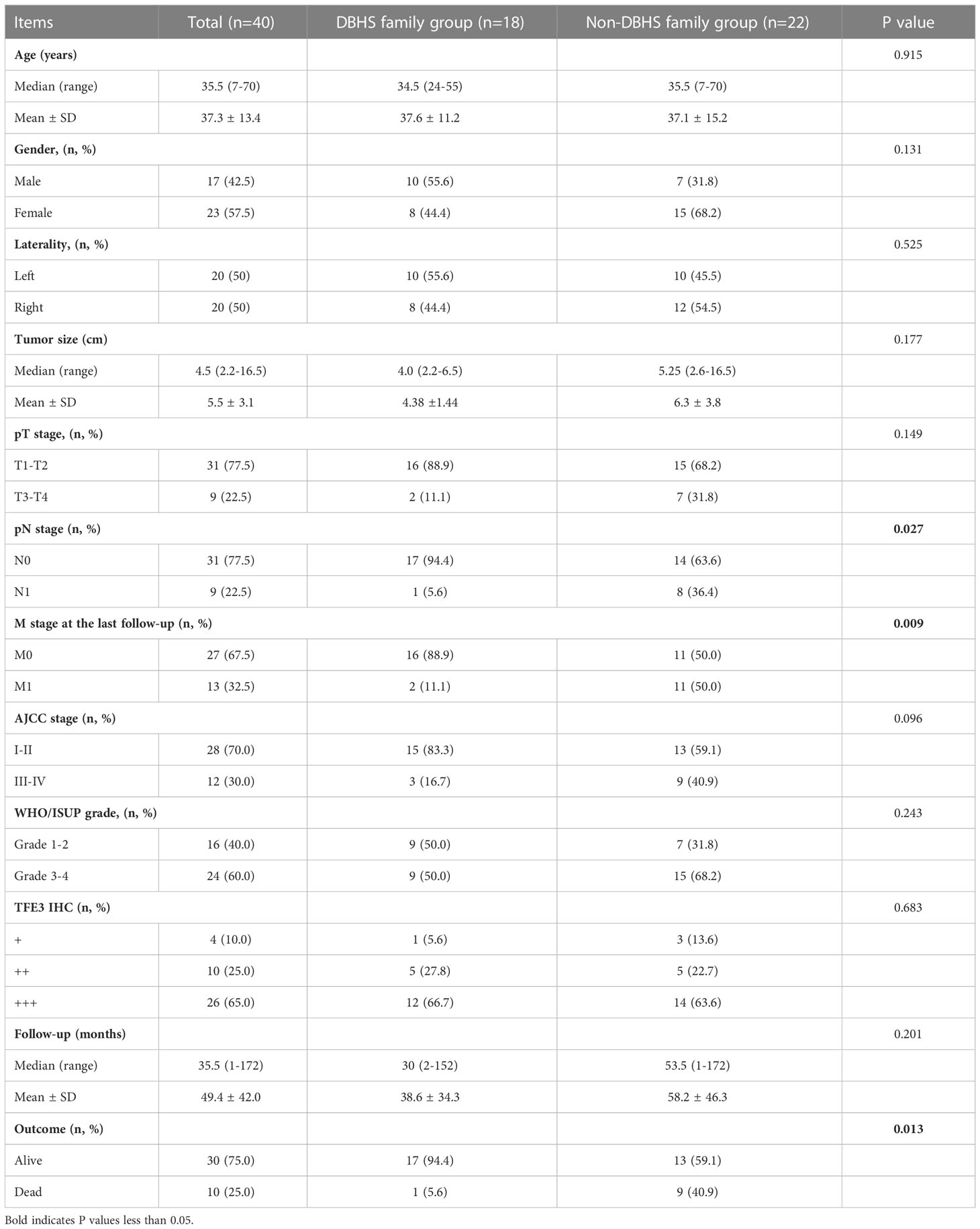
Table 3 Comparison of clinicopathological features between DBHS family group and non-DBHS family group.
The follow-up time of the 40 Xp11.2 tRCC cases with the four main fusion types was compared. The median follow-up time was 35.5 months (range, 1–172 months). Survival analysis showed that cases in DBHS family group have better progressive-free survival (PFS) (median PFS: not reached vs. 48 months, P = 0.023, Figure 4A) and overall survival (OS) (median OS: not reached vs. 75 months, P = 0.115, Figure 4B) compared with those in non-DBHS family group. In the subgroup analysis for the four different fusion types, patients with ASPL-TFE3 fusion were associated with poor PFS compared with other subtypes (median PFS: 18 months vs. 135 months, P = 0.026, Figure 4C) even though there was no significant difference in OS (median OS: 62 months vs. 148 months, P = 0.379, Figure 4D). In contrast, patients with NONO-TFE3 fusion showed better PFS (median PFS: not reached vs. 55 months, P = 0.040, Figure 4E) while there remained no significant difference in OS (median OS: not reached vs. 75 months, P = 0.077, Figure 4F). The difference in follow-up between TFE3-SFPQ group and the other subtypes were not compared due to the short follow-up period of TFE3-SFPQ group. Survival analysis indicated that cases with ASPL-TFE3 fusion have the worst PFS among the other three subtypes with sufficiently long follow-up (median PFS: 18 months vs. not reached vs. 135 months, P = 0.04, Figure 5A) although there was no difference in OS (P >0.05, Figure 5B). No statistically significant difference was seen in PFS and OS between PRCC and non-PRCC group (P >0.05, Figures 5C–D).
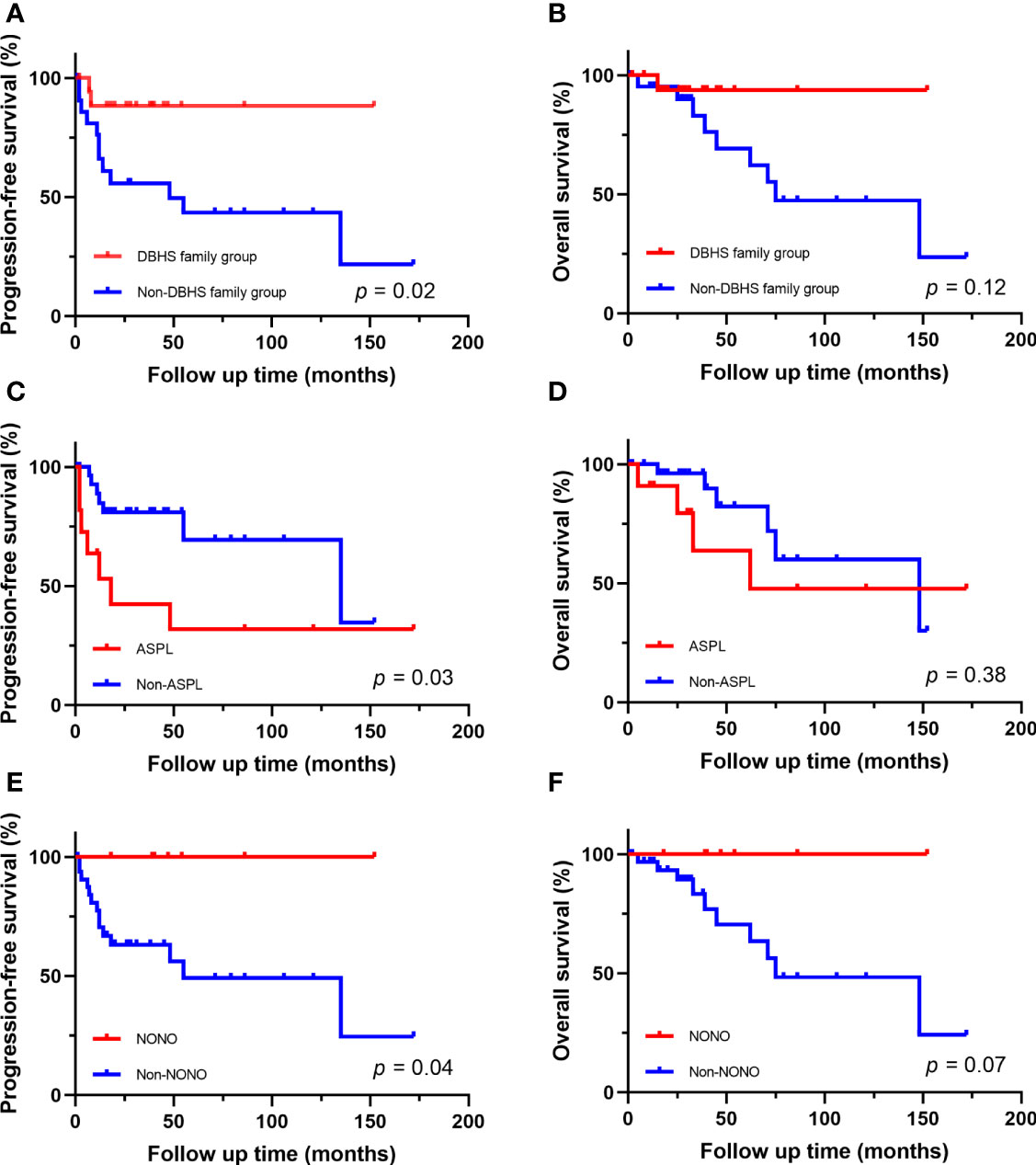
Figure 4 Survival analysis of different fusion partners. (A, B) PFS and OS for patients in DBHS famliy group and non-DBHS famliy group. (C, D) PFS and OS for patients with ASPL-TFE3 fusion and non-ASPL fusion. (E, F) PFS and OS for patients with NONO-TFE3 fusion and non-NONO fusion.
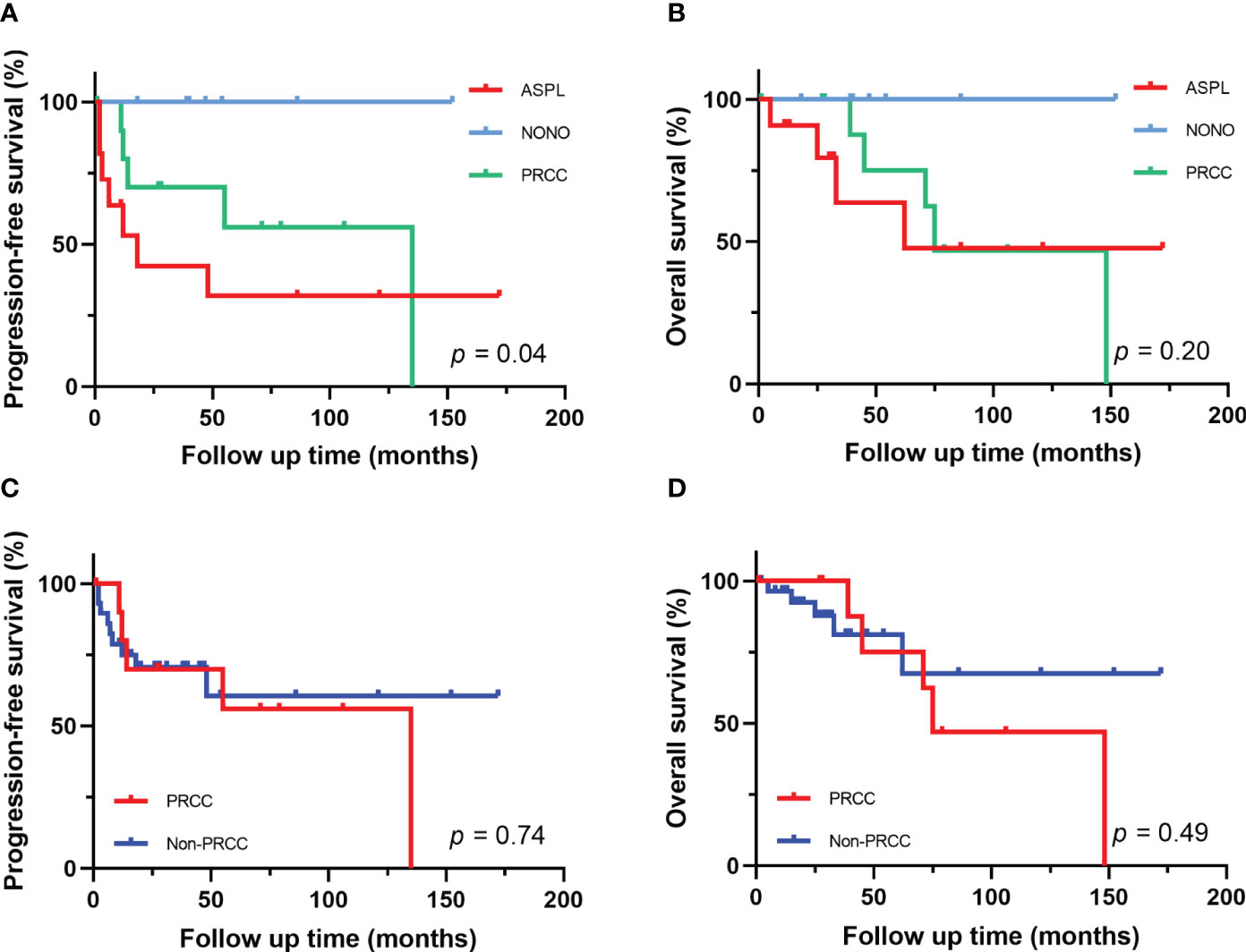
Figure 5 Survival analysis of different fusion partners. (A, B) PFS and OS for patients with different fusion subtypes. (C, D) PFS and OS for patients with PRCC-TFE3 fusion and non-PRCC fusion.
In recent years, with an in-depth understanding of the genomic spectrum of RCCs, Xp11.2 tRCC has received increasing attention. Apart from morphological and molecular features different from conventional RCCs, Xp11.2 tRCC itself is a group of highly heterogeneous tumors and various fusion partners has proven to be the source of this heterogeneity (12, 14, 23, 24). ASPL, PRCC and SFPQ were regarded as relatively common fusion partners initially while the pericentric inversion of NONO and TFE3 is also not rare nowadays. Therefore, in this study, we retrospectively investigated the clinicopathological features of 40 Xp11.2 tRCC cases with the above four fusion types in our cohort by IHC, FISH and RNA sequencing. The initial diagnosis of Xp11.2 tRCC was based on IHC and FISH analysis and then the fusion types were verified by RNA sequencing.
Xp11.2 tRCCs with different fusion subtypes have respective histological characteristics. As regards morphology, cases with ASPL-TFE3 fusion could present with various features including alveolar, papillary or nested architecture, which is not much different from other subtypes, but psammoma bodies were most common seen in ASPL-TFE3 cases among the four subtypes (25, 26). Papillary architecture frequently occurred in SFPQ-TFE3 subtype while pseudorosette-like architecture was occasionally described (27). NONO-TFE3 cases could also show a papillary architecture but it was more like the appearance of secretory endometrioid(7). PRCC-TFE3 cases tended to show a compact architecture and psammoma bodies were rare (28). In addition, recent literature reported the distinctive morphology of MED15-TFE3 cases. This subtype often showed a multicystic architecture without solid composition, resembling the feature of multilocular cystic renal cell neoplasm of low malignant potential (MCRN-LMP) (29). In regard to IHC, moderate (++) to strong (+++) TFE3 nuclear positivity is the primary clue to the initial diagnosis of Xp11.2 tRCC and cathepsin K positivity seems to be relevant to PRCC-TFE3 subtype. CA-IX and CK7 were always negative in Xp11.2 tRCC, which could help to exclude clear cell RCC (ccRCC) and papillary RCC (PRCC) (30). TFE3 break-apart FISH was the most effective method to detect TFE3 rearrangement in clinical practice (31), while equivocal or false-negative split signal pattern could be observed in several special fusion subtypes such as NONO(19) and RBM10(6). Hence, suspicious Xp11.2 tRCC cases with negative FISH results should be confirmed by further sequencing.
The fusion gene partners are likely to have a functional role in the oncogenesis of Xp11.2 tRCC and the underlying mechanisms may influence clinical behavior(2). SFPQ and NONO, belonging to DBHS family, are both pre-mRNA splicing factors and associated with tumorigenesis of multiple cancers such as prostate cancer and breast cancer (16–18, 32). Due to the homology of SFPQ and NONO gene, we classified Xp11.2 tRCC harboring SFPQ-TFE3 and NONO-TFE3 as a group and compared the characteristics of this group with non-DBHS family group (cases with PRCC-TFE3 and ASPL-TFE3 fusion). In the DBHS family group, only one patient (5.6%) was found lymph node (LN) metastasis in surgery, while 8 patients (36.4%) in non-DBHS family group had LN metastasis. This phenomenon suggested that cases in DBHS family group tend to show more lymph node metastasis in the early stages of disease. More importantly, half of the cases (50%) in non-DBHS family group showed local recurrence or distant metastasis at the last follow‐up, which demonstrated that cases in non-DBHS family group were likely to display a more aggressive and invasive behavior compared with cases in DBHS family group. It is worth mentioning that previous literature reported that Xp11.2 tRCC with ASPL-TFE3 fusion are more prone to present at advanced stage than cases with PRCC-TFE3 (14), however, in our cohort, although positive lymph node status at surgery is more common in ASPL-TFE3 cases (6/11, 54.5%), there was no statistically significant difference in pM stage at the last follow‐up between ASPL-TFE3 and PRCC-TFE3 cases. In addition, cases 25 presented with LN and vertebral metastasis after 10 years, which indicated follow-up period need to be long enough to estimate metastasis status and outcome.
The prognosis of Xp11.2 tRCC could be affected by a variety of factors. Above all, the impact of age on prognosis is prominent in Xp11.2 tRCC. Pediatric patients tended to show an indolent course whether the presence of lymph node metastasis (33). Case 3 in our cohort is a 7-year-old boy and showed no evidence of disease after 172-month follow-up, which seemed to support this view. Recent study has demonstrated that pediatric patients with Xp11.2 tRCC have a lower burden of genetic alteration compared with adult patients (34) and this could be a probable explanation of this phenomenon. On the contrary, Xp11.2 tRCC was more aggressive in adults, older age and distant metastasis were two predictors of poor prognosis (14, 35). Beyond that, the correlation between fusion subtypes and outcomes are currently being explored. In our study, cases with ASPL-TFE3 fusion showed a worse PFS compared with non-ASPL group, which supported that ASPL-TFE3 fusion may represent a more adverse prognosis in previous studies (12, 14, 36). Apart from fusion subtypes, copy number alterations (CNA) and chromosomal amplification could also affect the prognosis of Xp11.2 tRCC (23, 24). Patients with CNA burden had worse survival outcomes and 22q loss was an independent adverse prognostic marker (12, 37).In addition, a recent proteogenomic study revealed that deletions of 3p could lead to decreased OS via trans- effect or cis-effect (36). Overall, genetic alteration is an important cause of the clinical heterogeneity in Xp11.2 tRCC.
The effective treatment strategy for Xp11.2 tRCC is still unclear. Several clinical investigations have indicated that Xp11.2 tRCC patients had a poor response to immune checkpoint inhibitors (ICIs) and tyrosine kinase inhibitors (TKIs) (38, 39). However, the response to immunotherapy and vascular endothelial growth factor receptor (VEGFR)-targeted therapy seemed to vary according to the fusion subtypes. A previous case report demonstrated an 18-year-old male Xp11.2 tRCC patient with ASPL-TFE3 fusion had a favorable response to sorafenib (40). Recent transcriptomic analysis also revealed tumors with ASPL-TFE3 fusion are more likely to benefit from antiangiogenic treatments compared with the other subtypes and ICI plus TKI combination therapy may be a better choice (12). Besides, due to the activation of RPS6KB1 in Xp11.2 tRCC, Trilaciclib, an FDA-approved CDK5 inhibitor, was expected to become a potential therapeutic drug (36).
There are some limitations to our research. Firstly, although we have collected as much data of Xp11.2 tRCC cases as possible to fulfill the research goal, the sample size still needs to be further expanded. Secondly, our follow-up data only suggested there was a significant difference in PFS, therefore the patients require a longer follow-up time to validate the results.
In summary, Xp11.2 tRCC is a group of highly heterogeneous tumors. Of the four common fusion subtypes, cases in non-DBHS family group more frequently developed LN and distant metastases than cases in DBHS family group. Tumors with ASPL-TFE3 fusion tend to have a worse outcome while those with NONO-TFE3 fusion exhibit a relatively good prognosis. Novel therapeutic approaches and targets for Xp11.2 tRCC with different fusion subtypes remain to be explored.
The original contributions presented in the study are included in the article and Supplementary Material. The Illumina HiSeq next generation sequencing was performed by GloriousMed. The datasets presented in this article are not readily available because patient confidentiality and participant privacy, thus requests to access the datasets should be directed to aW5mb0BnbG9yaW91c21lZC5jb20=.
The studies involving human participants were reviewed and approved by Institutional Review Board of the Nanjing Drum Tower Hospital. The patients/participants provided their written informed consent to participate in this study.
WGu and YZ performed study concept and design; YZ and XP performed developpment of methodology and writing, review and revision of the paper; WGu provided acquisition, analysis and interpretation of data, and statistical analysis; WGa and HG provided technical and material support. All authors read and approved the final paper.
This research was supported by the Project of Invigorating Health Care through Science, Technology and Education, Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKB2016014) and Beijing Ronghe Medical Development Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank Xiaogong Li, Gutian Zhang, Xiaozhi Zhao, Changwei Ji (Department of Urology, Nanjing Drum Tower Hospital) for providing patient information. We also thank Jun Yang and Ming Chen (Department of Pathology, Nanjing Drum Tower Hospital) for providing technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1116648/full#supplementary-material
1. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and Male genital organs-part a: Renal, penile, and testicular tumours. Eur Urol (2016) 70(1):93–105. doi: 10.1016/j.eururo.2016.02.029
2. Kauffman EC, Ricketts CJ, Rais-Bahrami S, Yang Y, Merino MJ, Bottaro DP, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol (2014) 11(8):465–75. doi: 10.1038/nrurol.2014.162
3. Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol (2001) 159(1):179–92. doi: 10.1016/s0002-9440(10)61684-7
4. Argani P, Antonescu CR, Couturier J, Fournet JC, Sciot R, Debiec-Rychter M, et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am J Surg Pathol (2002) 26(12):1553–66. doi: 10.1097/00000478-200212000-00003
5. Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, Ladanyi M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol (2003) 27(6):750–61. doi: 10.1097/00000478-200306000-00005
6. Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR. RBM10-TFE3 renal cell carcinoma: A potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am J Surg Pathol (2017) 41(5):655–62. doi: 10.1097/pas.0000000000000835
7. Argani P, Zhong M, Reuter VE, Fallon JT, Epstein JI, Netto GJ, et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol (2016) 40(6):723–37. doi: 10.1097/pas.0000000000000631
8. Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene (1997) 15(18):2233–9. doi: 10.1038/sj.onc.1201394
9. Classe M, Malouf GG, Su X, Yao H, Thompson EJ, Doss DJ, et al. Incidence, clinicopathological features and fusion transcript landscape of translocation renal cell carcinomas. Histopathology (2017) 70(7):1089–97. doi: 10.1111/his.13167
10. Huang W, Goldfischer M, Babayeva S, Mao Y, Volyanskyy K, Dimitrova N, et al. Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer (2015) 54(8):500–5. doi: 10.1002/gcc.22261
11. Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res (2014) 20(15):4129–40. doi: 10.1158/1078-0432.CCR-13-3036
12. Sun G, Chen J, Liang J, Yin X, Zhang M, Yao J, et al. Integrated exome and RNA sequencing of TFE3-translocation renal cell carcinoma. Nat Commun (2021) 12(1):5262. doi: 10.1038/s41467-021-25618-z
14. Ellis CL, Eble JN, Subhawong AP, Martignoni G, Zhong M, Ladanyi M, et al. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age, and stage. Mod Pathol (2014) 27(6):875–86. doi: 10.1038/modpathol.2013.208
15. Knott GJ, Bond CS, Fox AH, The DBHS proteins SFPQ. NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res (2016) 44(9):3989–4004. doi: 10.1093/nar/gkw271
16. de Silva HC, Lin MZ, Phillips L, Martin JL, Baxter RC. IGFBP-3 interacts with NONO and SFPQ in PARP-dependent DNA damage repair in triple-negative breast cancer. Cell Mol Life Sci (2019) 76(10):2015–30. doi: 10.1007/s00018-019-03033-4
17. Feng P, Li L, Deng T, Liu Y, Ling N, Qiu S, et al. NONO and tumorigenesis: More than splicing. J Cell Mol Med (2020) 24(8):4368–76. doi: 10.1111/jcmm.15141
18. Petti E, Buemi V, Zappone A, Schillaci O, Broccia PV, Dinami R, et al. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat Commun (2019) 10(1):1001. doi: 10.1038/s41467-019-08863-1
19. Xia QY, Wang Z, Chen N, Gan HL, Teng XD, Shi SS, et al. Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod Pathol (2017) 30(3):416–26. doi: 10.1038/modpathol.2016.204
20. Weterman MA, van Groningen JJ, Tertoolen L, van Kessel AG. Impairment of MAD2B-PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc Natl Acad Sci U.S.A. (2001) 98(24):13808–13. doi: 10.1073/pnas.241304198
21. Chen X, Yang Y, Gan W, Xu L, Ye Q, Guo H. Newly designed break-apart and ASPL-TFE3 dual-fusion FISH assay are useful in diagnosing Xp11.2 translocation renal cell carcinoma and ASPL-TFE3 renal cell carcinoma: a STARD-compliant article. Med (Baltimore) (2015) 94(19):e873. doi: 10.1097/MD.0000000000000873
22. Xiong L, Chen X, Liu N, Wang Z, Miao B, Gan W, et al. PRCC-TFE3 dual-fusion FISH assay: A new method for identifying PRCC-TFE3 renal cell carcinoma in paraffin-embedded tissue. PloS One (2017) 12(9):e0185337. doi: 10.1371/journal.pone.0185337
23. Macher-Goeppinger S, Roth W, Wagener N, Hohenfellner M, Penzel R, Haferkamp A, et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod Pathol (2012) 25(2):308–15. doi: 10.1038/modpathol.2011.169
24. Malouf GG, Monzon FA, Couturier J, Molinie V, Escudier B, Camparo P, et al. Genomic heterogeneity of translocation renal cell carcinoma. Clin Cancer Res (2013) 19(17):4673–84. doi: 10.1158/1078-0432.CCR-12-3825
25. Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, Chan DY, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol (2007) 31(8):1149–60. doi: 10.1097/PAS.0b013e318031ffff
26. Gandhi JS, Malik F, Amin MB, Argani P, Bahrami A. MiT family translocation renal cell carcinomas: A 15th anniversary update. Histol Histopathol (2020) 35(2):125–36. doi: 10.14670/HH-18-159
27. Wang XT, Xia QY, Ni H, Ye SB, Li R, Wang X, et al. SFPQ/PSF-TFE3 renal cell carcinoma: a clinicopathologic study emphasizing extended morphology and reviewing the differences between SFPQ-TFE3 RCC and the corresponding mesenchymal neoplasm despite an identical gene fusion. Hum Pathol (2017) 63:190–200. doi: 10.1016/j.humpath.2017.02.022
28. Wang XT, Xia QY, Zhou XJ, Rao Q. Xp11 translocation renal cell carcinoma and the mesenchymal counterparts: An evolving concept with novel insights on clinicopathologic features, prognosis, treatment, and classification. Crit Rev Oncog (2017) 22(5-6):481–97. doi: 10.1615/CritRevOncog.2017020558
29. Song Y, Yin X, Xia Q, Zheng L, Yao J, Zeng H, et al. Xp11 translocation renal cell carcinoma with morphological features mimicking multilocular cystic renal neoplasm of low malignant potential: a series of six cases with molecular analysis. J Clin Pathol (2021) 74(3):171–6. doi: 10.1136/jclinpath-2020-206681
30. Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol (2015) 32(2):103–13. doi: 10.1053/j.semdp.2015.02.003
31. Rao Q, Williamson SR, Zhang S, Eble JN, Grignon DJ, Wang M, et al. TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol (2013) 37(6):804–15. doi: 10.1097/PAS.0b013e31827e17cb
32. Takayama KI. Splicing factors have an essential role in prostate cancer progression and androgen receptor signaling. Biomolecules (2019) 9(4). doi: 10.3390/biom9040131
33. Geller JI, Argani P, Adeniran A, Hampton E, De Marzo A, Hicks J, et al. Translocation renal cell carcinoma: lack of negative impact due to lymph node spread. Cancer (2008) 112(7):1607–16. doi: 10.1002/cncr.23331
34. Marcon J, DiNatale RG, Sanchez A, Kotecha RR, Gupta S, Kuo F, et al. Comprehensive genomic analysis of translocation renal cell carcinoma reveals copy-number variations as drivers of disease progression. Clin Cancer Res (2020) 26(14):3629–40. doi: 10.1158/1078-0432.CCR-19-3283
35. Calio A, Segala D, Munari E, Brunelli M, Martignoni G. MiT family translocation renal cell carcinoma: from the early descriptions to the current knowledge. Cancers (2019) 11(8). doi: 10.3390/cancers11081110
36. Qu Y, Wu X, Anwaier A, Feng J, Xu W, Pei X, et al. Proteogenomic characterization of MiT family translocation renal cell carcinoma. Nat Commun (2022) 13(1):7494. doi: 10.1038/s41467-022-34460-w
37. Pan CC, Sung MT, Huang HY, Yeh KT. High chromosomal copy number alterations in Xp11 translocation renal cell carcinomas detected by array comparative genomic hybridization are associated with aggressive behavior. Am J Surg Pathol (2013) 37(7):1116–9. doi: 10.1097/PAS.0b013e318293d872
38. McKay RR, Bosse D, Xie W, Wankowicz SAM, Flaifel A, Brandao R, et al. The clinical activity of PD-1/PD-L1 inhibitors in metastatic non-clear cell renal cell carcinoma. Cancer Immunol Res (2018) 6(7):758–65. doi: 10.1158/2326-6066.CIR-17-0475
39. Boileve A, Carlo MI, Barthelemy P, Oudard S, Borchiellini D, Voss MH, et al. Immune checkpoint inhibitors in MITF family translocation renal cell carcinomas and genetic correlates of exceptional responders. J Immunother Cancer (2018) 6(1):159. doi: 10.1186/s40425-018-0482-z
Keywords: Xp11.2 translocation renal cell carcinoma, TFE3, FISH, DBHS family, prognosis
Citation: Guo W, Zhu Y, Pu X, Guo H and Gan W (2023) Clinical and pathological heterogeneity of four common fusion subtypes in Xp11.2 translocation renal cell carcinoma. Front. Oncol. 13:1116648. doi: 10.3389/fonc.2023.1116648
Received: 05 December 2022; Accepted: 16 January 2023;
Published: 03 February 2023.
Edited by:
Wen-Hao Xu, Fudan University, ChinaReviewed by:
Ali Amin, Rhode Island Hospital, United StatesCopyright © 2023 Guo, Zhu, Pu, Guo and Gan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weidong Gan, Z3dkQG5qdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.