
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 20 February 2023
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1094131
This article is part of the Research Topic Gynecological and Gastrointestinal Cancers: Recent Advances in Molecular Diagnosis and Targeted Therapy View all 6 articles
Introduction: Growing evidence shows that long non-coding RNA small nucleolar RNA host genes (lncRNA SNHGs) enact an pivotal regulatory roles in the shorter survival outcome of colorectal cancer (CRC). However, no research has systematically evaluated the correlation among lncRNA SNHGs expression and survival outcome of CRC. This research indented to screen whether exist potential prognostic effect of lncRNA SNHGs in CRC patientss using comprehensive review and meta-analysis.
Methods: Systematic searches were performed from the six relevant databases from inception to October 20, 2022. The quality of published papers was evaluated in details. We pooled the hazard ratios (HR) with 95% confidence interval (CI) through direct or indirect collection of effect sizes, and odds ratios (OR) with 95% CI by collecting effect sizes within articles. Detailed downstream signaling pathways of lncRNA SNHGs were summarized in detail
Results: 25 eligible publications including 2,342 patients were finally included to appraise the association of lncRNA SNHGs with prognosis of CRC. Elevated lncRNA SNHGs expression was revealed in colorectal tumor tissues. High lncSNHG expression means bad survival prognosis in CRC patients (HR=1.635, 95% CI: 1.405–1.864, P<0.001). Additionally, high lncRNA SNHGs expression was inclined to later TNM stage (OR=1.635, 95% CI: 1.405–1.864, P<0.001), distant lymph node invasion, distant organ metastasis, larger tumor diameter and poor pathological grade. Begg's funnel plot test using the Stata 12.0 software suggested that no significant heterogeneity was found.
Conclusion: Elevated lncRNA SNHGs expression was revealed to be positively correlated to discontented CRC clinical outcome and lncRNA SNHG may act as a potential clinical prognostic index for CRC patients.
Colorectal cancer (CRC) seriously threatens human life and health, well-being and happiness quotient. According to 2021 cancer statistics, the annual incidence and mortality of CRC worldwide was ranked second and third in the world, respectively (1). With the development of medicine, various treatment options, for instance, radiotherapy, chemotherapy, biological targeted therapy and molecular biological therapy have been adopted in CRC therapy (2-4). Nevertheless, the five-year and ten-year survival rate of CRC patientss is unsatisfactory as before (5). While optimizing the strategies of combined radiotherapy and chemotherapy and targeted therapy and immunotherapy, researchers are also trying to hunt for splendid therapeutic targets to ameliorate the survival and prognosis of CRC cases.
Over the past ten years, mounting investigations have focalized long non-coding RNAs (lncRNA), a sort of small molecules of >200 nucleotide units without protein-coding functions, which enact a crucial part in the stride of CRC (6, 7). lncRNAs can affect the biological property of tumor cells at the cellular function level by affecting intracellular transduction pathways, such as cell proliferation, drug resistance, immune evasion and apoptosis (8, 9). Hence, lncRNAs could be used as underlying therapeutic targets and effective prognostic markers for tumor therapy.Small nucleolar RNA host genes (SNHGs), is a long non-coding RNA family, have been shown to be up-regulated in CRC, mounting publications suggests that lncRNA SNHG play increasingly prominent motivation in the biological properties of CRC cells (10, 11), for example, some researchers have uncovered that high SNHG expression could drive the growth, distant organ migration, invasion, and inhibition of apoptosis in CRC cells (12-15). Till date, there is no literature that evaluates the correlation between the lncRNA SNHG family and CRC prognosis. Results of some studies are inconsistent, and a single study has insufficient data. At the same time, we executed a comprehensively systematic assessment and meta-analysis to figure out the relation of the lncRNA SNHG family and CRC prognosis.
A systematical search of databases of PubMed (Medline), Embase, ISI Web of Knowledge, Springer, the Cochrane Library, Scopus, BioMed Central, ScienceDirect, Wanfang, Weipu, and China National Knowledge Internet was conducted from inception to November 1, 2022. Appropriate publications was searched in accordance with the detailed Mesh, including (“lnc SNHG” OR “long non-coding RNA SNHG” OR “Small nucleolar RNA host genes”) AND (“prognosis” OR “prognostic” OR “survival” OR “outcome”) based on PRISMA (The Preferred Reporting Items for Systematic Reviews and Meta-Analyses).
Studies were selected by two independent researchers. Inclusion criteria were as follows: (1) investigation of the association between SNHG expression and survival outcome, as well as clinical prognosis of CRC patients; (2) classification of patients into high and low expression groups in accordance with primary literature; (3) detection of SNHG expression level using validated techniques; (4) sufficient data to calculate odds ratio (OR) or hazard ratio (HR); and (5) studies written in English. Exclusion criteria were: (1) no investigation on the relationship between SNHG expression level and CRC prognosis different from a mere exploration of the involved molecular biological mechanisms; (2) reviews and meta-analyses, letters, animal studies, and conference proceedings; (3) insufficient data for prognostic analysis; and (4) duplicate studies.
For inclusion in this meta-analysis, two researches independently scored the selected studies using the Newcastle-Ottawa quality assessment scale (NOS) to assess the quality and suitability of the selected studies (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) (16). The NOS items for selection of cohorts, comparability among included studies, and outcome were included. Study selection flow diagram is shown in Figure 1.
For all included literatures, two researchers independently extracted the following index in detail: name of first author, article release year, country where the patient belongs, number of cases, expression level of SNHG, cutoff value, presentation of HR value, follow-up month, HR with 95% CI for overall survival (OS) and reference-free survival (RFS); OR with 95% CI for dichotomous data included TNM staging, distant lymph node metastasis (LNM), organ metastasis, pathological score, tumor size etc. If the literature provides both multivariate and univariate analyses methods, then we extract the values of multivariate analysis. In situations where an article simply presents survival curves without providing HR and its 95% CI, the HR was extracted by extracting survival curves using the Engauge Digitizer v10.4 software (17). In the event where the extracted data of the two investigators were inconsistent, a third investigator was asked to evaluate the data to decide whether to extract the data or not. Additionally, we seek out The Cancer Genome Atlas (TCGA) database of Interactive Analysis of Gene Expression Profile, and explored the GEPIA2 website to further explore SNHG expression in colorectal tumor tissues and adjacent tissues.
Review Manager V 5.4 Software and Stata SE14.0 software were utilized to implement the statistical analysis in this study. patientss included in the study were spanided into high-expression and low-expression groups on the ground of reports in the original literature. Pooling OR besides 95% CI to investigate the correlation both lncSNHGs expression and the clinical prognostic parameters of CRC patientss, such as TNM stage, LNM, DM, histological grade, tumor size, depth of invasion, etc. Pooling HR with 95% CI was implemented to explore the pertinence of lncSNHGs expression levels with OS progression-free survival (PFS) and DFS in cancer patients. The heterogeneity amid incorporated studies was appraised by I2 and P value. If I2 was >50% and P<0.05, we assumed there exist markedly strengthened heterogeneity in this outcome and a random-effects model was performed. At the same time, we tried to found the provenience of heterogeneity through classification analysis in view of follow-up visits, cut-off index, HR calculation procedures, and sample size. Sensitivity analyses were performed to find individual studies with large heterogeneity in OS outcomes. Clinical and statistical bias was detected using Begg's test.
Basic characteristics of enrolled documents according to our search using the keywords, 263 articles were screened in the initial trial, excluding 74 articles that were repeatedly published. One-hundred and thirty-seven papers did not assess the interrelation between lncRNA SNHGs expression and CRC prognosis, 14 papers were based on animal experiments, 6 papers did not classify lncRNA SNHGs into high and low expression patients, and 7 papers had insufficient data. This study finally included 25 suitable papers, including 2,342 CRC cases published between 2016 and 2022, with the number of patients ranging between 30–338 (14, 18–31). All the patients were from China, and were diagnosed as CRC by pathology or histology. High and low lncRNA SNHG expression was verified using the real-time quantitative reverse transcription reaction. Twenty-two papers included survival data of patients, and three papers only provided clinical pathological characteristics of patients (Table 1). The NOS score among covered investigations ranged between 6–9 based on overall evaluation of literature quality in detail (Table 2).
Twenty-two researches comprising 2,134 patientss were obtained to revealed the relativity between SNHG expression and the survival outcome of CRC patients. Twelve papers directly reported HR values and 95% CI; 10 papers simply provided survival curves and were extracted via the Engauge Software (indirect extraction). Pooled HRs demonstrated that high SNHG expression predicts poor cancer prognosis (HR = 1.635; 95% CI, 1.405–1.864; P<0.0001) (Figure 2). We took into consideration different cut-off index (mean or median), follow-up month, and HR estimation method (Directly and Indirectly). The classification analysis was carried out to reveal the marked heterogeneity between different groups based on HR estimation method (Directly and Indirectly), follow-up month (<60 months and not less than 60 months), number of patients (>100 and <100), cut-off value (mean and median), NOS score (9 and <9), and analysis method (univariate and multivariate analyses). A positive association was expose between increasing SNHG expression and short OS in the multivariate analysis (HR = 1.527; 95% CI, 1.276–1.779), univariate analysis (HR = 2.186; 95% CI, 1.616–2.756), HRs withdraw straight from studies (HR = 1.527; 95% CI, 1.276–1.777), or explicitly abstracted from the survival curve (HR = 2.224; 95% CI, 1.639–2.808), cancer with >100 (HR = 2.300; 95% CI, 1.729–2.871) and <100 (HR = 1.505; 95% CI, 1.254–1.757), not less than 60 months of follow-up (HR = 1.907; 95% CI, 1.546–2.267) and <60 months (HR = 1.448; 95% CI, 1.149–1.746) (Table 3). Three studies comprising 301 patients, 2 studies comprising 173 patientss, and 1 study with 141 patientss were obtained to evaluate the connection between SNHG expression and cancer survival outcome, including PFS, DFS, and recurrence free survival (RFS). Pooled HRs showed that increasing SNHG expression reminds worse PFS (HR = 2.35; 95% CI, 1.46–3.78) (Figure 3A), DFS (HR = 1.85; 95% CI, 1.27–2.71) (Figure 3B), and RFS (HR = 2.70; 95% CI, 1.17–6.23) (Figure 3C).
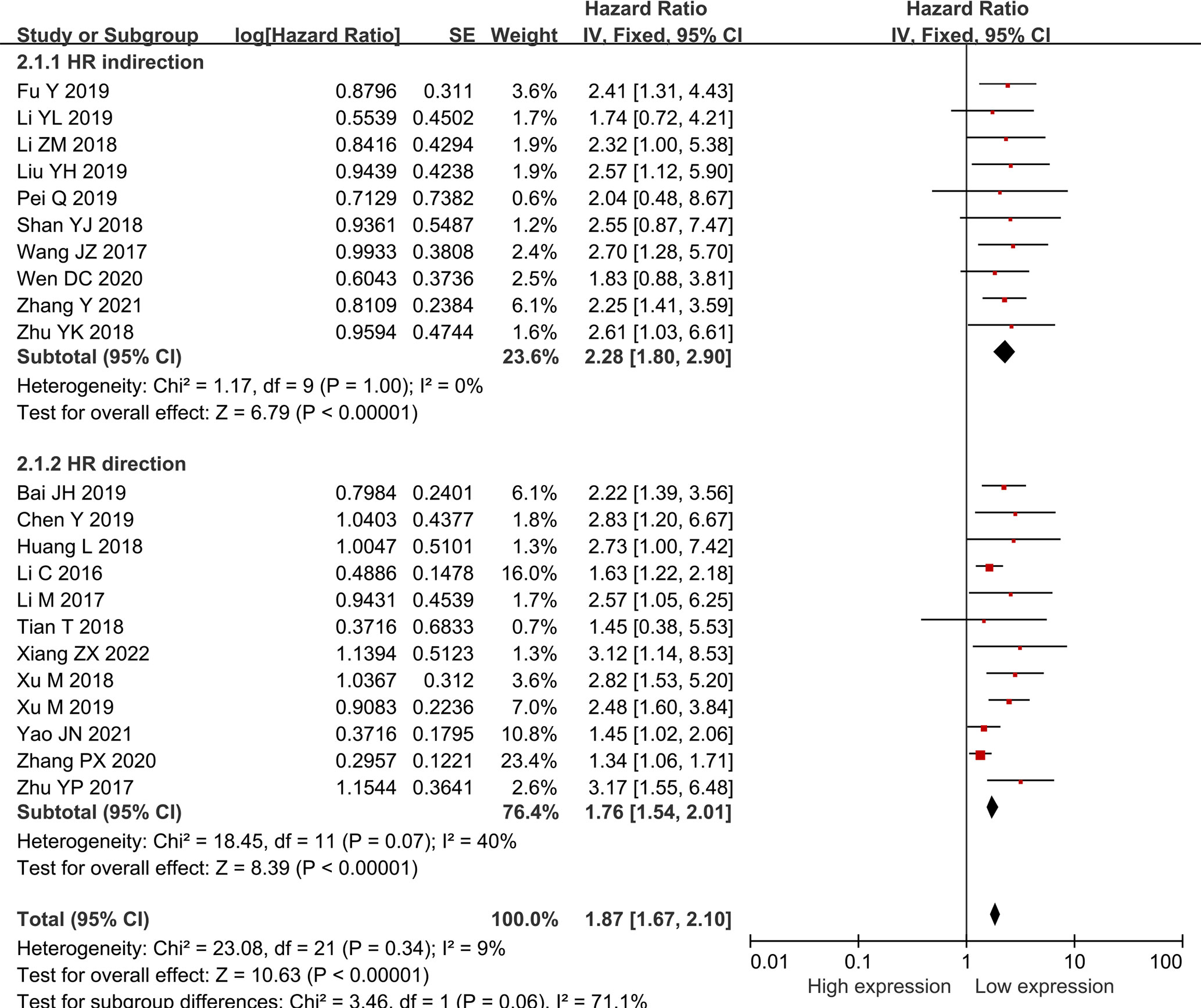
Figure 2 Forest plot showing the relationship between small nucleolar RNA host gene expression and overall survival (OS) in colorectal cancers.
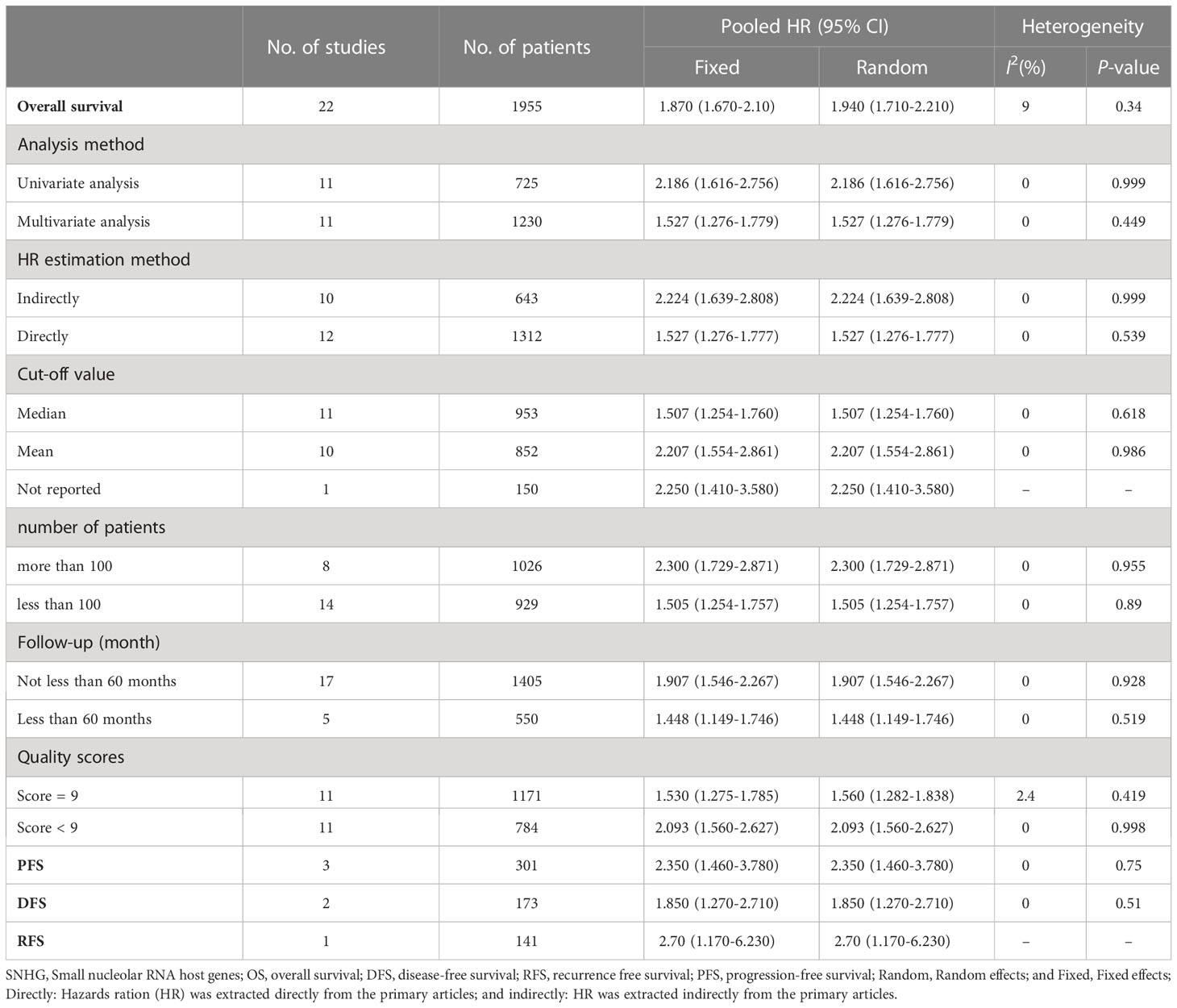
Table 3 Subgroup analysis of lncRNA SNHGs expression and overall survival in colorectal cancer patients.
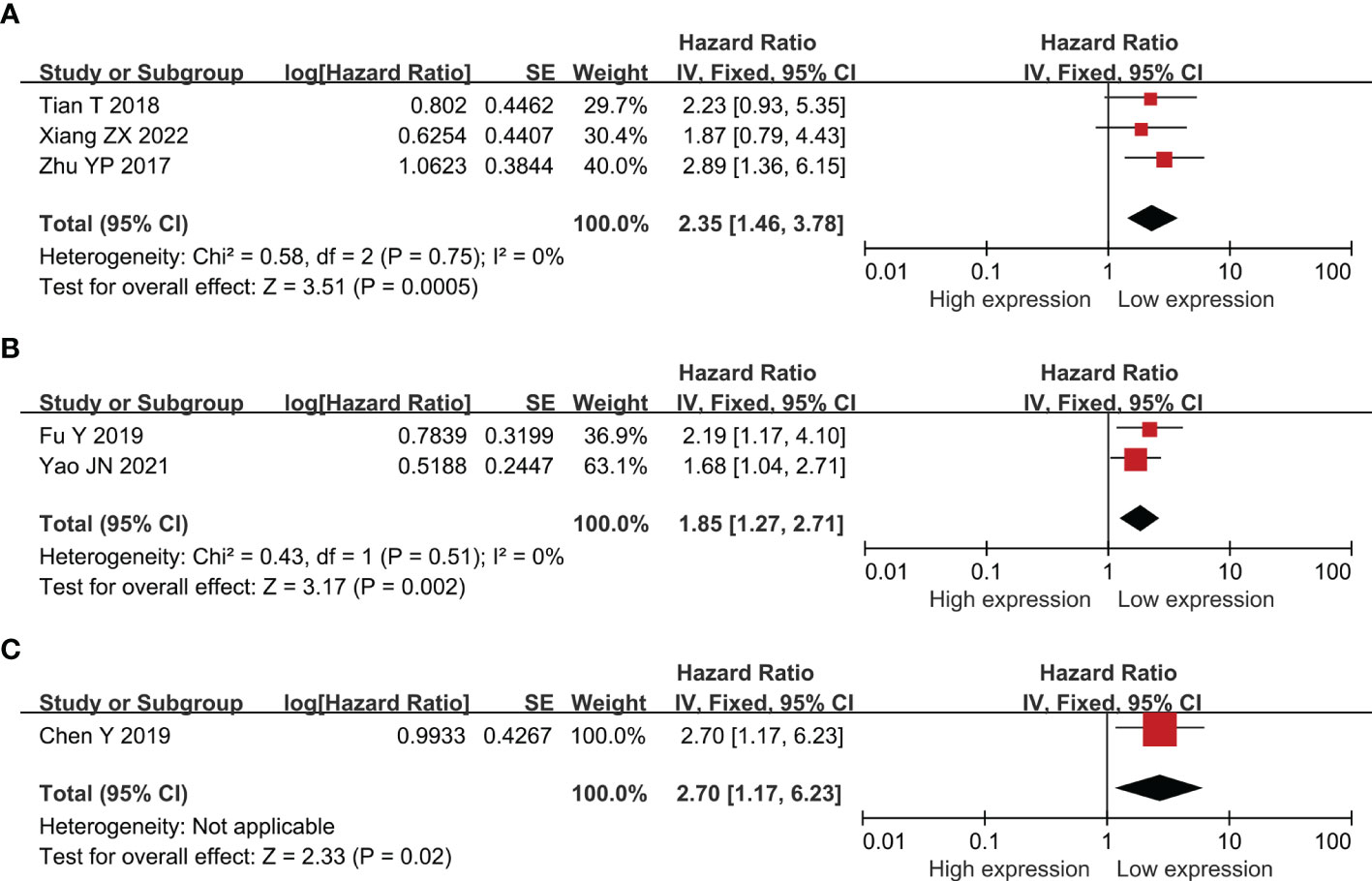
Figure 3 Forest plot showing the relationship between small nucleolar RNA host gene (SNHG) expression and progression-free survival (PFS) (A); disease-free survival (DFS) (B); and reference-free survival (RFS) in colorectal cancers (C).
Twenty publications with 1,873 patientss were obtained to inquire the dependency between lncRNA SNHGs expression and clinical stage of CRC patientss. The publications showed that increasing SNHG expression predicts an poor TNM stage (OR = 1.750; 95% CI, 1.481–2.067; P=0.0001) (Figure 4). A classification analysis redouble revealed that high SNHG expression means a terminal TNM stage in a group that used mean value as the cut-off value (OR = 1.656; 95% CI, 1.315–2.085; P=0.0001) and median value as the cut-off value (OR = 1.859; 95% CI, 1.460–2.368; P=0.0001). At the same time, enhanced SNHG expression indicates advanced TNM stage in both high NOS score (OR = 1.841; 95% CI, 1.490–2.274; P=0.0001) and low (OR = 1.609; 95% CI, 1.226–2.110; P=0.0001) NOS score groups. A fixed effects model was exerted to derive from a small heterogeneity level (I2 = 19.2, P=0.216) (Table 4).
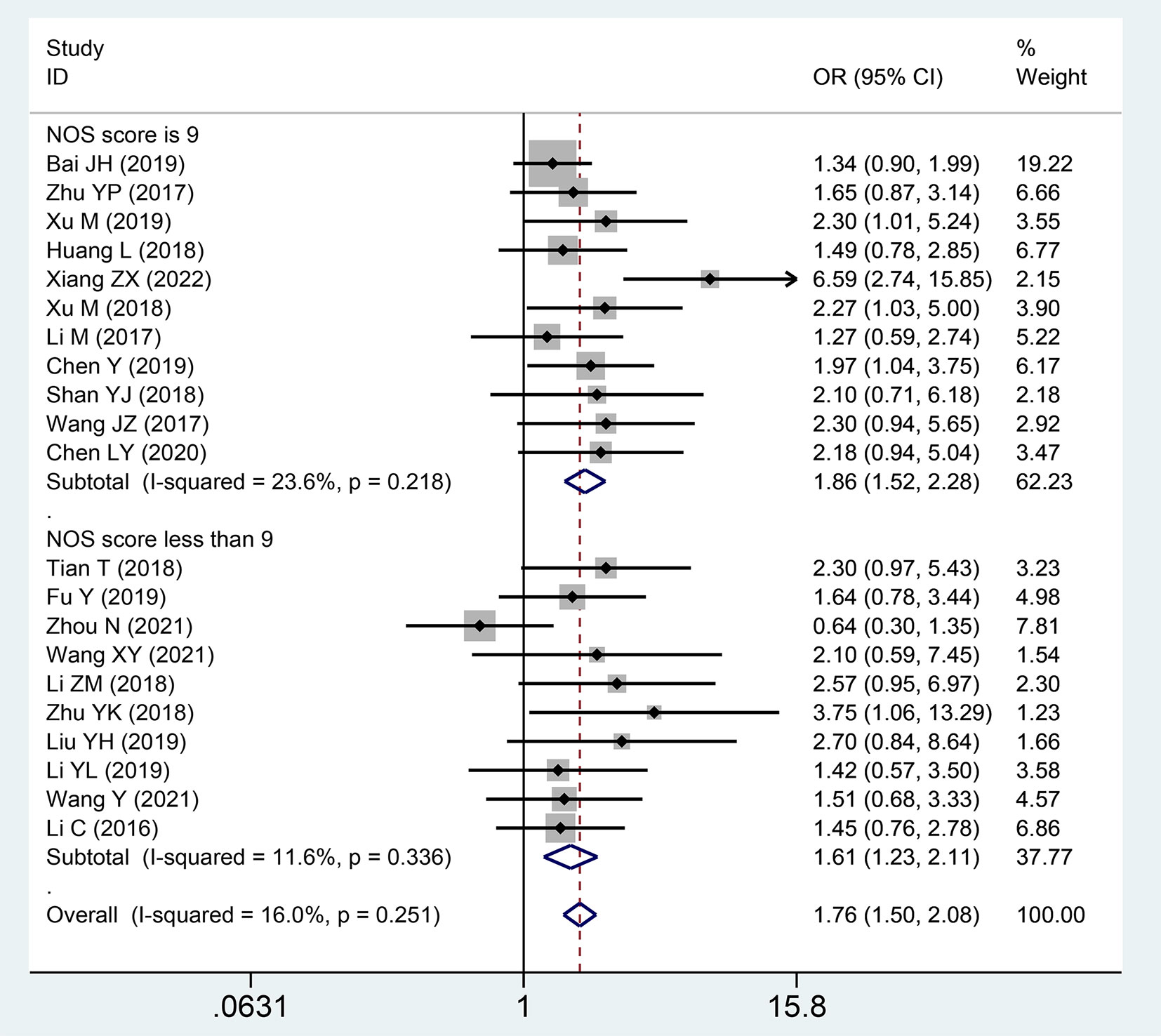
Figure 4 Forest plot showing the relationship between small nucleolar RNA host gene (SNHG) expression and TNM stage in colorectal cancers.
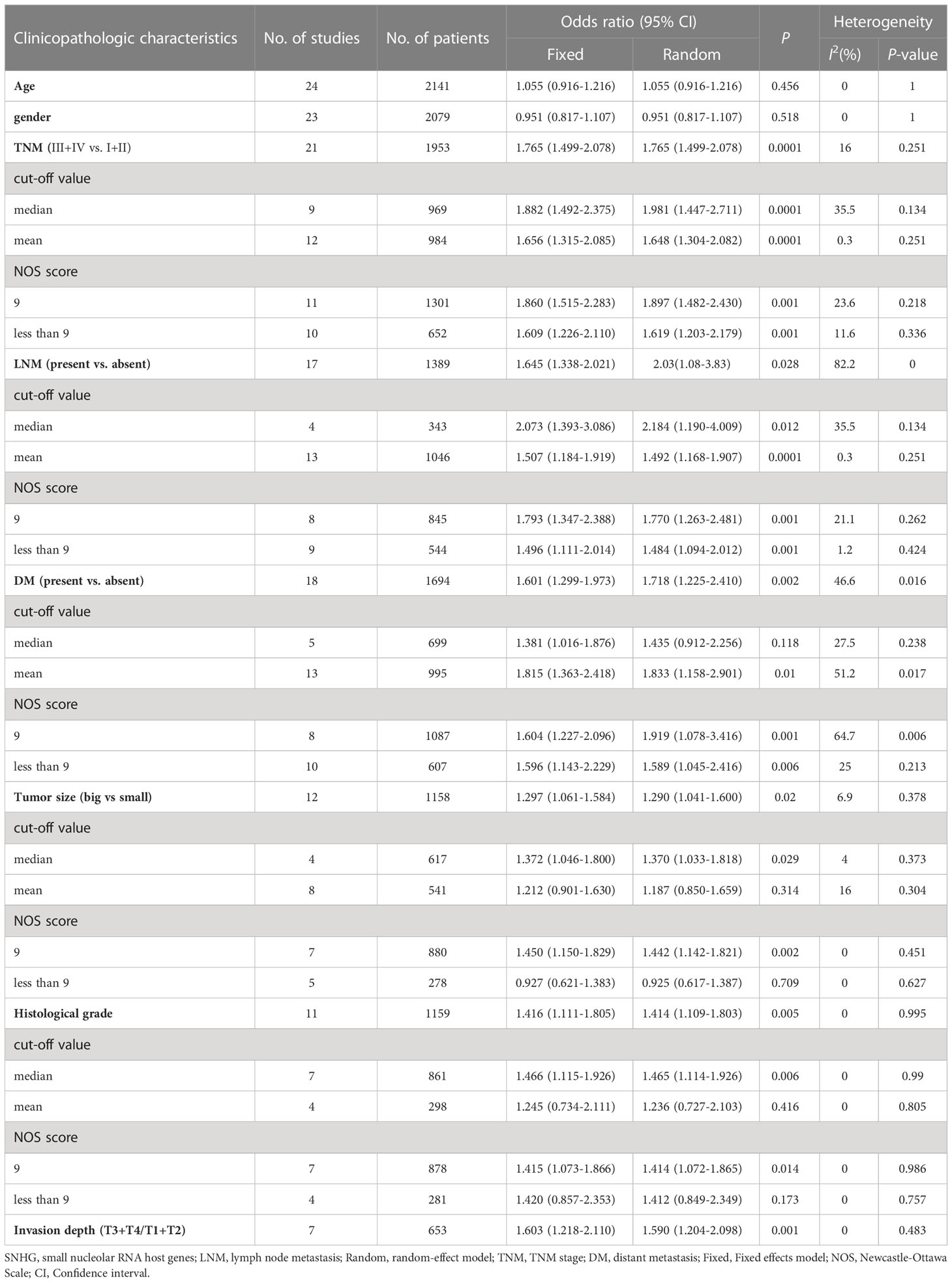
Table 4 Pool effects of clinicopathological characteristics in colorectal cancer patients with abnormal lncRNA SNHGs expression.
Seventeen publications with 1,389 patientss were incorporated to inspect the pertinence between the lncRNA SNHGs expression and LNM of CRC patientss. These studies showed that high SNHG expression was noteworthy relativity to lymph node invasion (OR = 1.645; 95% CI, 1.338–2.021; P=0.0001) (Figure 5). In a subgroup analysis, we detected that elevated lncRNA SNHGs expression demonstrated significant association with lymph node metastasis in a group that used mean value as the cut-off index (OR = 1.507; 95% CI, 1.184–1.919; P=0.0001) and median value as the cut-off index (OR = 2.073; 95% CI, 1.393–3.086; P=0.0001). A remarkable relevance was observed in both high (OR = 1.793; 95% CI, 1.347–2.388; P=0.0001) and low (OR = 1.496; 95% CI, 1.111–2.014; P=0.0001) NOS score groups. A fixed effects model was adopted for small heterogeneity level (I2 = 8.3, P=0.357) (Table 4).
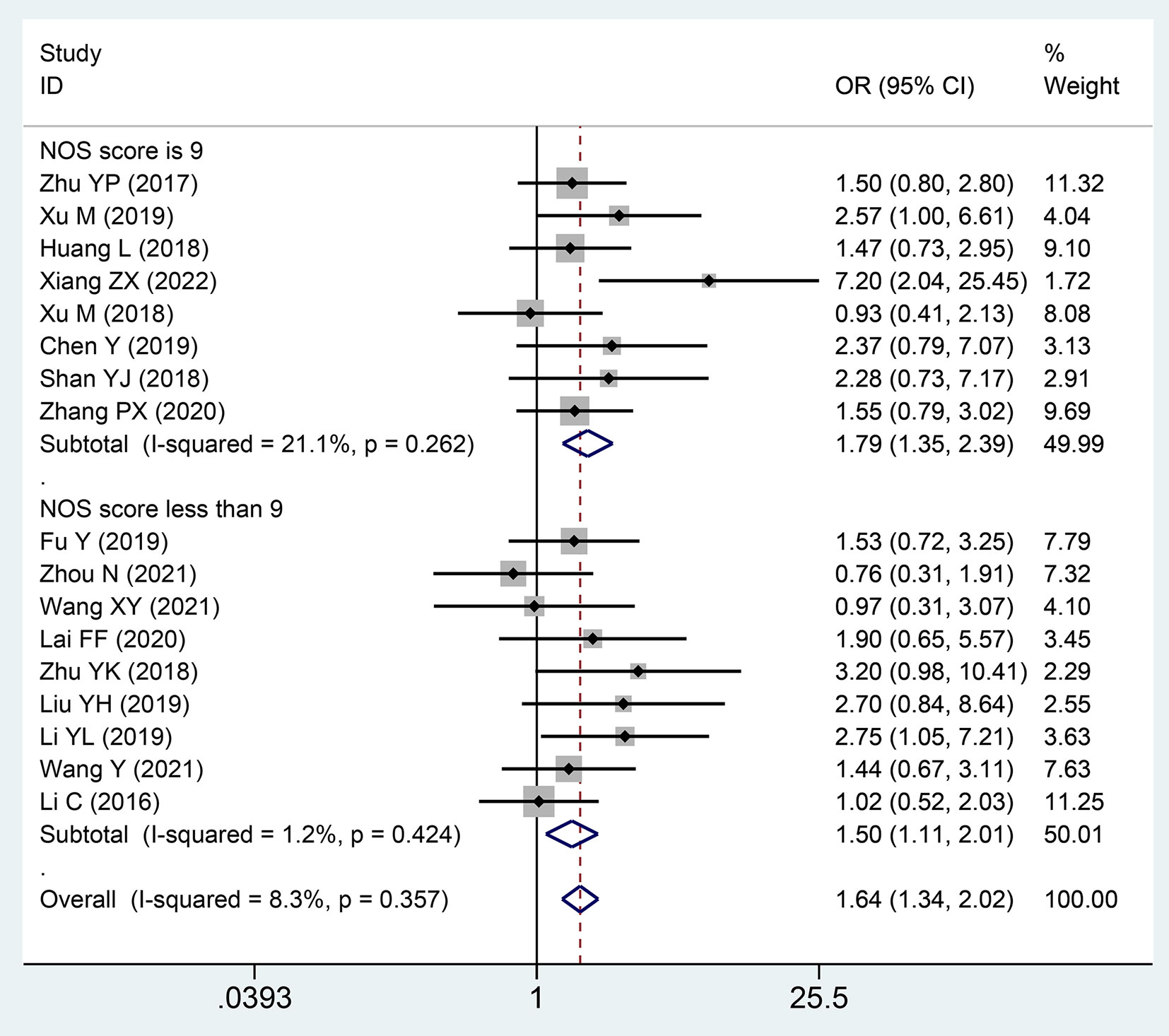
Figure 5 Forest plot showing the relationship between small nucleolar RNA host gene (SNHG) expression and lymph node metastasis (LNM) stage in colorectal cancers.
A total of 18 studies with 1,694 patients described the association between the correlation of the expression level of lncRNA SNHGs and distant metastasis in patients with CRC. A significantly positive correlation was revealed between increased lncRNA SNHGs expression and earlier distant metastases (OR = 1.601; 95% CI, 1.299–1.973; P=0.0001) (Figure 6). A subgroup analysis further found a positive association between elevated SNHG expression and earlier distant metastases in the group that used mean value as the cut-off value (OR = 1.815; 95% CI, 1.363–2.418; P=0.0001) and median value as the cut-off value (OR = 1.381; 95% CI, 1.016–1.876; P=0.0001). A similar positive relationship was observed in groups with high (OR = 1.604; 95% CI, 1.227–2.096; P=0.0001) and low (OR = 1.596; 95% CI, 1.143–2.229; P=0.0001) NOS scores. A fixed effects model was adopted due to small heterogeneity level (I2 = 46.6, P=0.016) (Table 4).
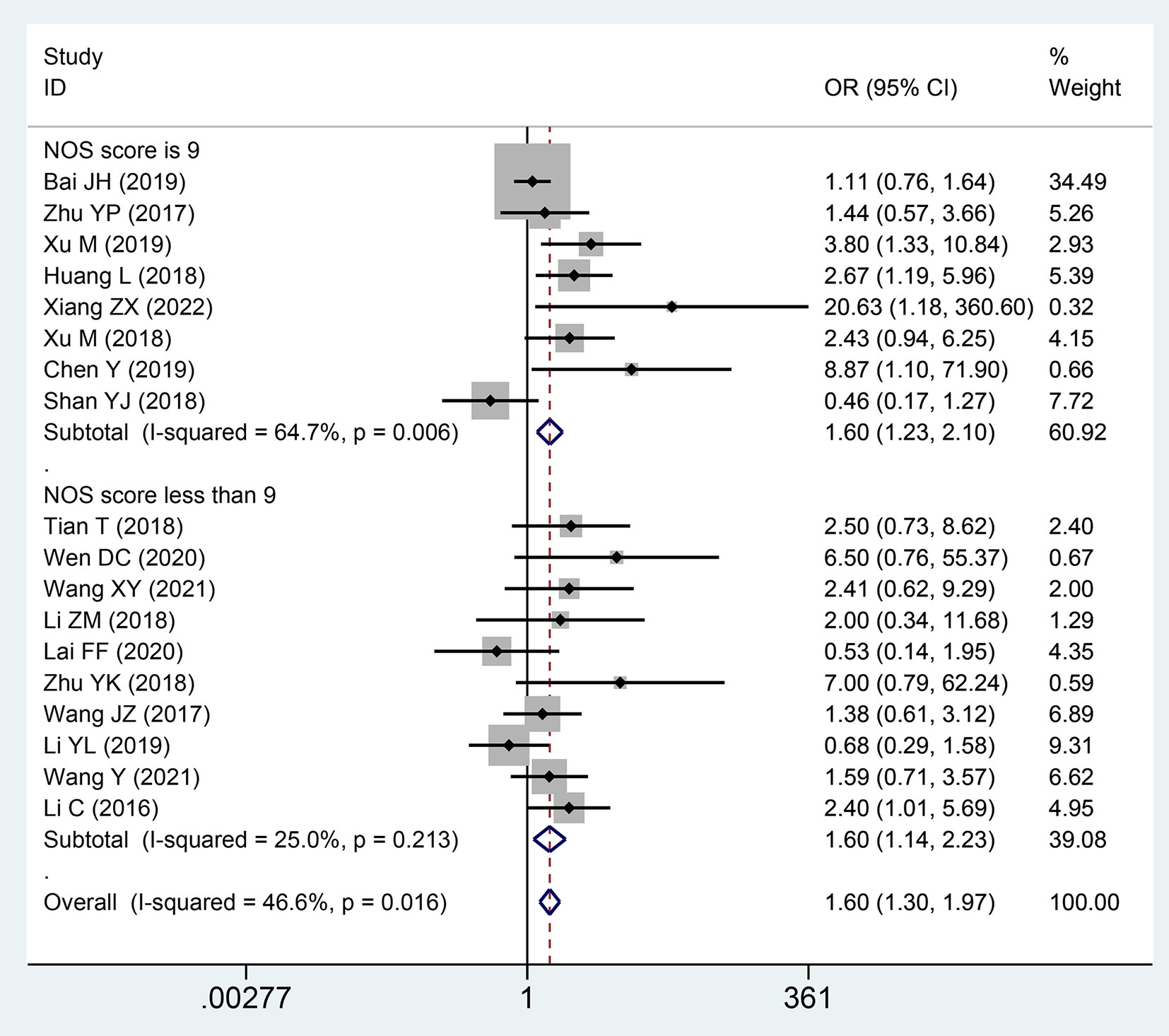
Figure 6 Forest plot showing the relationship between small nucleolar RNA host gene (SNHG) expression and distant metastasis (DM) stage in colorectal cancers.
Twelve publications comprising 1,158 patients were included to assess the relationship between the expression level of lncRNA SNHGs and tumor size in CRC patients. A positive association was observed between high lncRNA SNHGs expression and bigger tumor size (OR = 1.297; 95% CI, 1.061–1.584; P=0.011) (Figure 7). A subgroup analysis was based on a cut-off value and NOS score, and significantly positive correlations were also confirmed in groups that used median value as the cut-off value (OR = 1.372; 95% CI, 1.046–1.800; P=0.0001) and those with high NOS score (OR = 1.450; 95% CI, 1.150–1.829; P=0.0001). Meanwhile, no significance was observed in groups that used mean value as the cut-off value (OR = 1.212; 95% CI, 0.901–1.630; P=0.0001) and those with low NOS score (OR = 0.927; 95% CI, 0621–1.383; P=0.0001). A fixed effects model was adopted due to small heterogeneity level (I2 = 46.6, P=0.016) (Table 4).
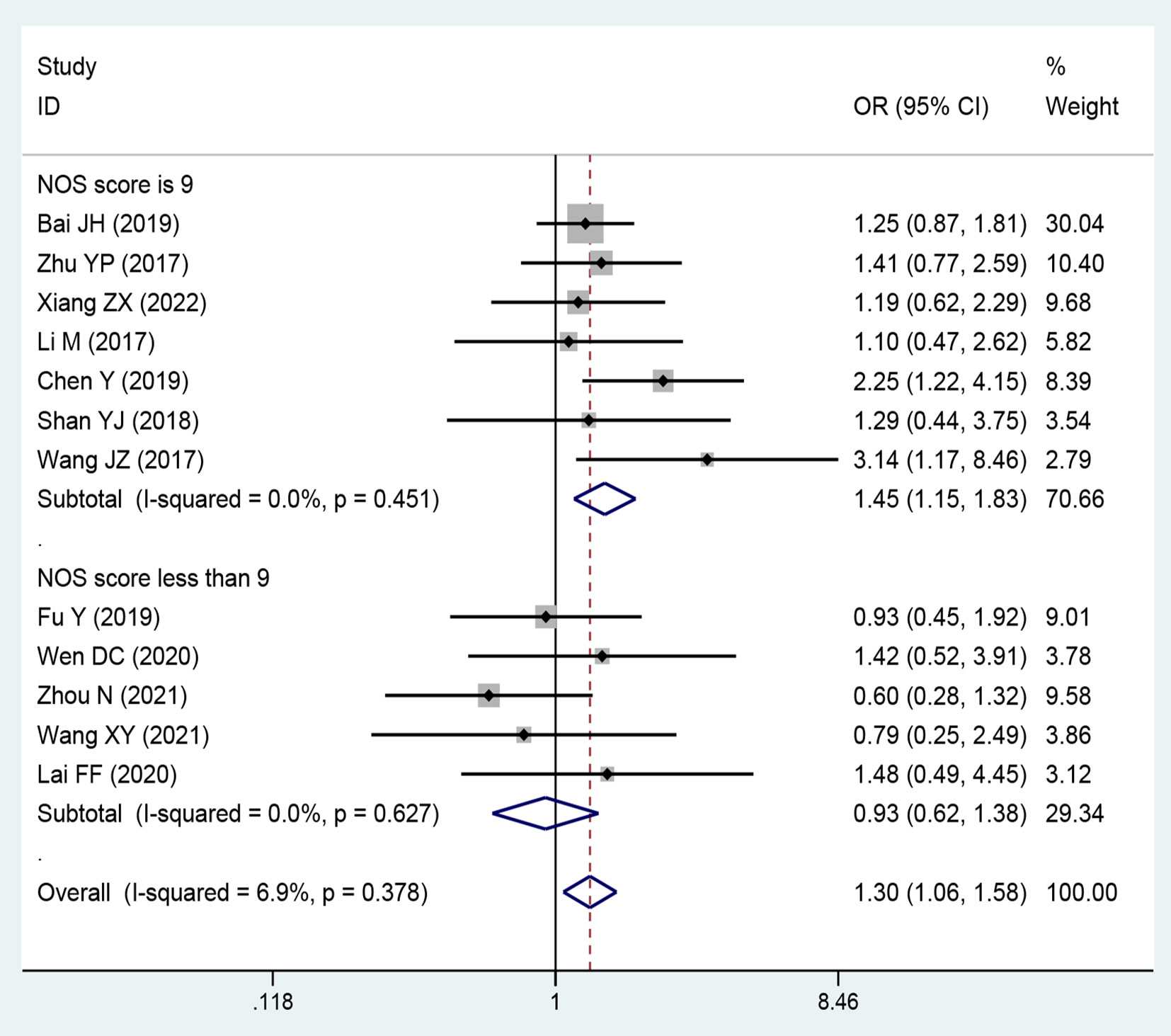
Figure 7 Forest plot showing the relationship between small nucleolar RNA host gene (SNHG) expression and tumor size stage in colorectal cancers.
The relationship between lncRNA SNHGs expression and other clinicopathological indicators, including histological grade, age, and gender were also assessed. A significantly positive correlation was observed between high SNHG expression and depth of invasion (HR = 1.603; 95% CI, 1.218–2.110; P=0.001) and bad histological grade (HR = 1.416; 95% CI, 1.111–1.805; P=0.005). Meanwhile, no significance was observed between lncRNA SNHGs expression and age (OR = 1.055; 95% CI, 0.916–1.216; P=0.456) and gender (OR = 0.951; 95% CI, 0.817–1.107; P=0.61) (Table 4).
Sensitivity analyses were performed to explore methodological heterogeneity in these studies, and to explore whether single study data significantly affected the overall outcome. We did not find a study data that affected the overall outcome significantly (Figure 8). A potential publication bias was explored using the Begg’s test, and unobvious publication bias was found in the included studies (Figure 9).
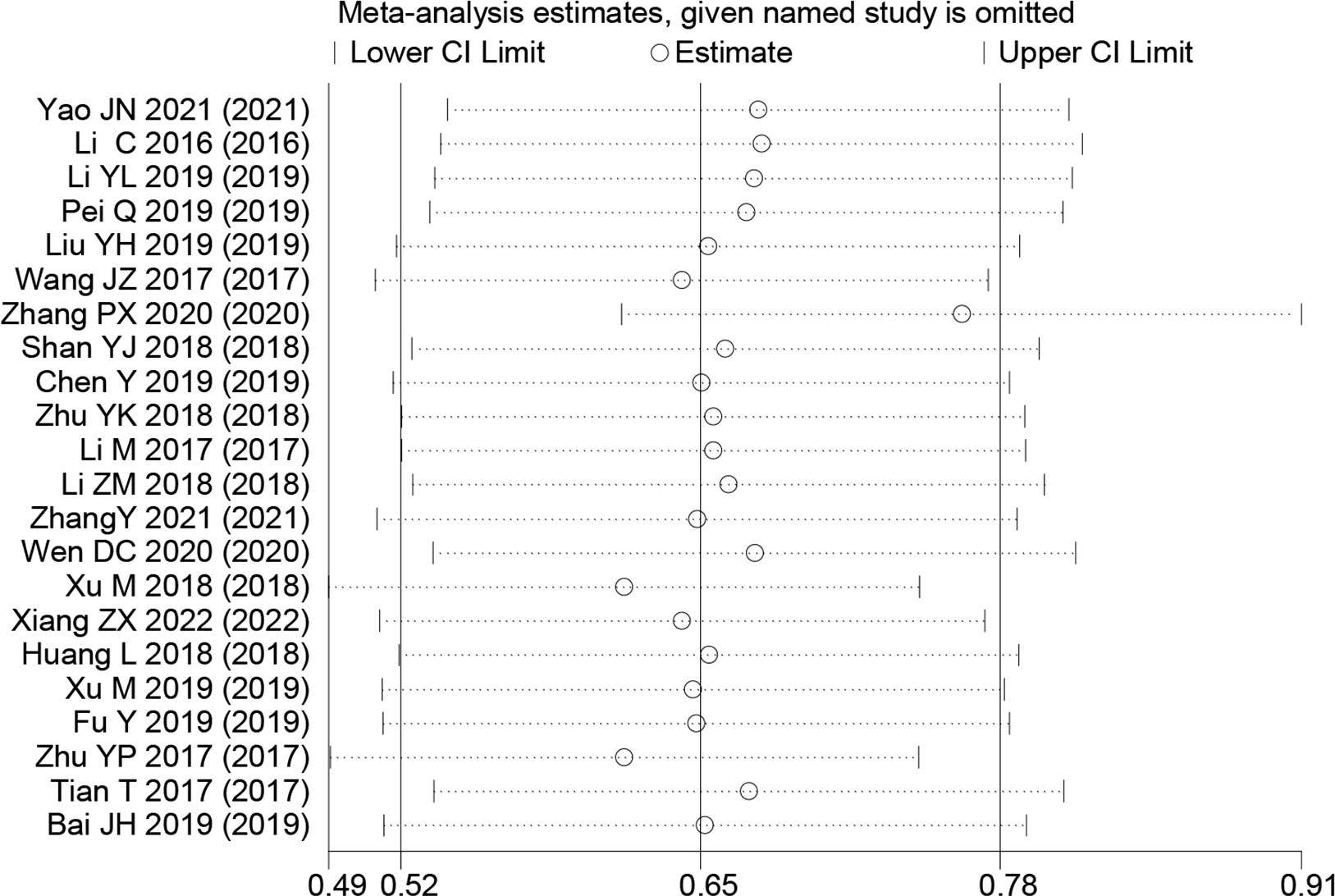
Figure 8 Sensitivity analysis for small nucleolar RNA host gene expression with overall survival in colorectal cancers. HR, hazard ratio; CI, confidence interval.
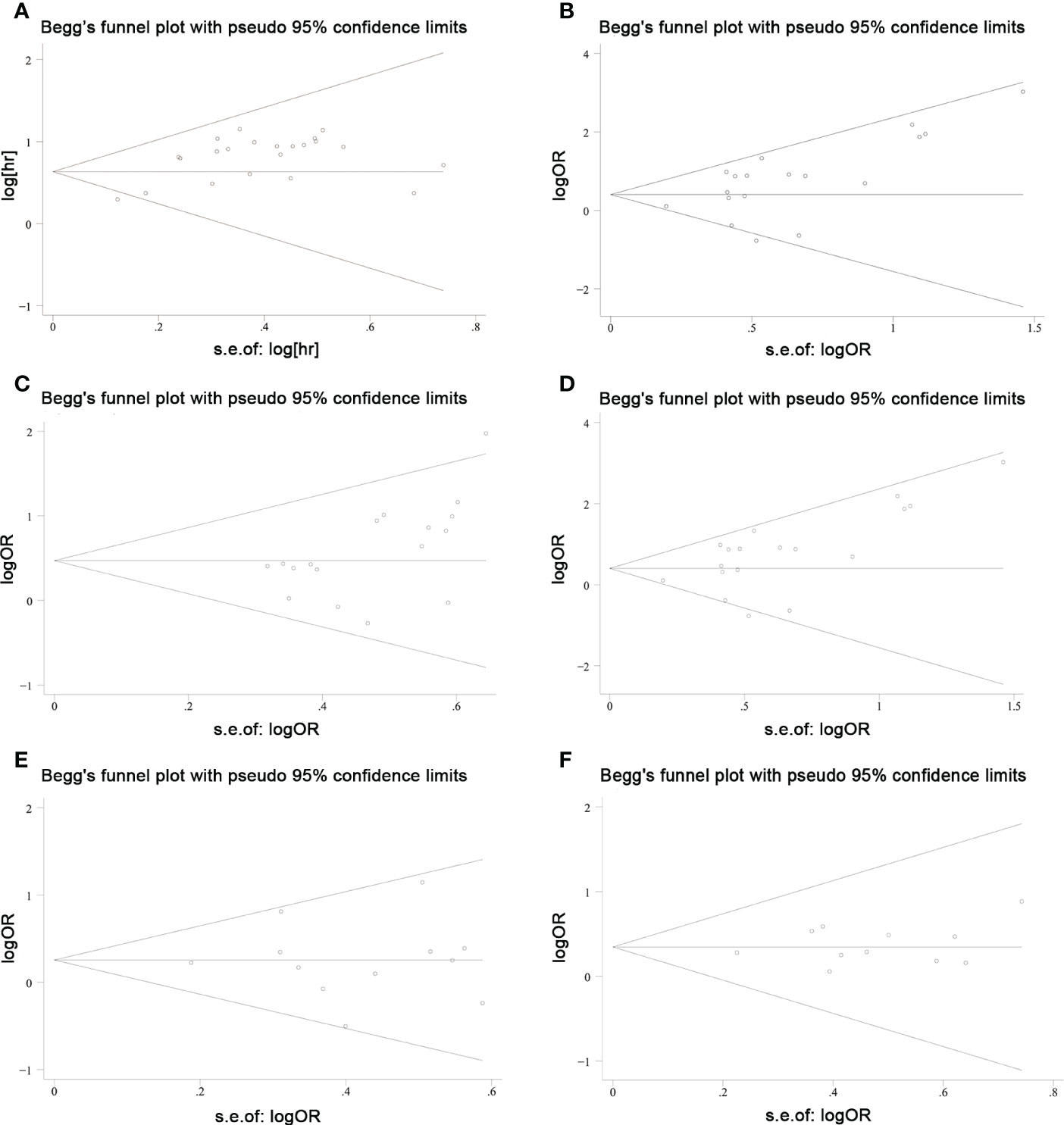
Figure 9 Publication bias was explored using Begg’s test in colorectal cancers. (A) overall survival; (B) TNM stage; (C) lymph node metastasis; (D) distant metastasis; (E) tumor size; and (F) histological grade.
Based on TCGA-COAD and TCGA-READ datasets of TCGA database (https://portal.gdc.cancer.gov/repository?facetTab=cases) and the GEPIA website (http://gepia.cancer-pku.cn/), we analyzed the expression of SNHGs in tumor tissues and adjacent tissues. The results showed that the expression level of SNHGs in tumor tissues was higher than that in adjacent tissues. This result is consistent with the results of this meta-analysis. (Figure 10).
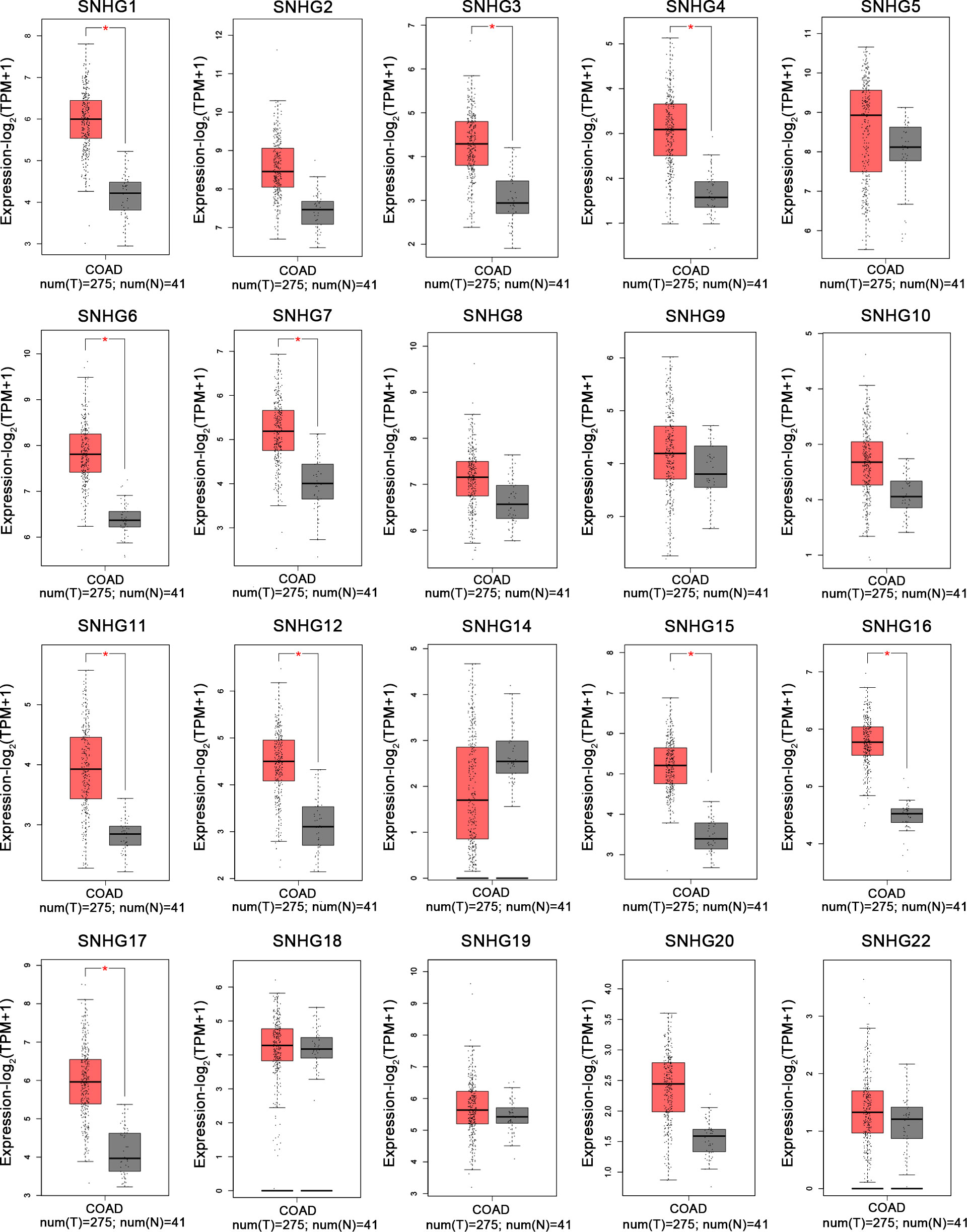
Figure 10 Expression levels of SNHG1-SNHG22 in colonic tumor and normal tissues in the GEPIA cohort by merging SNHG expression data (n=316).
In the past ten years, increasing lncRNA SNHGs have been confirmed to regulate the cell biological behavior of CRC cells, for instance, both proliferation, chemotherapy resistance and immune escape (32). Many researchers tried to control the expression quantity of lncRNA SNHGs to achieve the impact of treating CRC. Mounting evidence attempts to seek the relativity between lncRNA SNHGs expression and survival outcome of CRC (33). It is gratifying that most studies have proclaimed that high expression of lncRNA SNHGs is memorably bound up positively with poor prognosis of CRC (23, 34). However, due to insufficient data in a single study and inconsistent research conclusions of several researches, so far, no research has systematically probed the relations between outlier expression of SNHG and CRC prognosis. In this paper, we summarized and explored the intervention of lncRNA SNHGs in colorectal malignancies. In this study, 25 original studies were included, and lncRNAs were highly expressed in CRC. This finding could also be obviously supported by TCGA and GEPIA (Figure 10). Pooling HR with 95% CI showed high expression of lncSNHG, thereby indicating poor cancer prognosis, OS, PFS, and DFS. We further explored the prognostic relevance of lncSNHGs through subgroup analysis. The results stated clearly that increased lncRNA SNHGs expression emphatically predicted unsatisfactory colorectal prognosis in various subgroup such as HR direct extraction group (Detail in Table 3). Additionally, some studies also tried to explore the correlation between lncRNA SNHGs expression and other prognostic indicators, including PFS, DFS, and RFS of CRC patients. Pooled HR with 95% CI make clear that high lncRNA SNHGs expression was positively interrelated with worse PFS, DFS and RFS. This study also seek the relativity between lncRNA SNHGs expression and clinical pathological parameters. The results showed that high lncRNA SNHGs predicted advanced clinical stage, deeper invasion, worse differentiation grade, and larger tumor size. Overall, lncRNA SNHGs was positively correlated with poor CRC prognosis, and lncRNA SNHGs may be an excellent indicator of survival prognosis and a aussichtsreich therapeutic target for CRC. Increasing researchers are attempting inquired the underlying biological mechanisms of lncRNA SNHGs in CRC cells at the molecular level (Table 5 and Figure 11). First, SNHG could directly bind on downstream target proteins, thereby affecting the tumor cell growth, metastasis and apoptosis of through some signal cascades. For example, Huang et al. demonstrated that lncSNHG15 is preeminently associated with liver metastasis of CRC, but the specific mechanism has not yet been discovered (35). Li et al. reported that SNHG 20 could induce cell growth, distant metastasis and inhibit cell apoptosis of CRC cells by modulating P21 and cyclin A1 (19). Wang et al. reported that lncSNHG6 may drive the migration, growth and make inroads on CRC cells via TGF-β/Smad signaling pathway activation by targeting UPF1 (36). Shan et al. speculated that lncSNHG7 induces the migration and proliferation of CRC cells by enhanced N-acetylgalactosaminyltransferase 1 expression and promoting epithelial-mesenchymal transition (EMT) markers (E-cadherin and vimentin) by binding to miR-216b (25). Wang et al. revealed that lnSNHG12 promotes growth by upregulating cell cycle-related proteins and stifled apoptosis via decrease in caspase 3 expression (23).
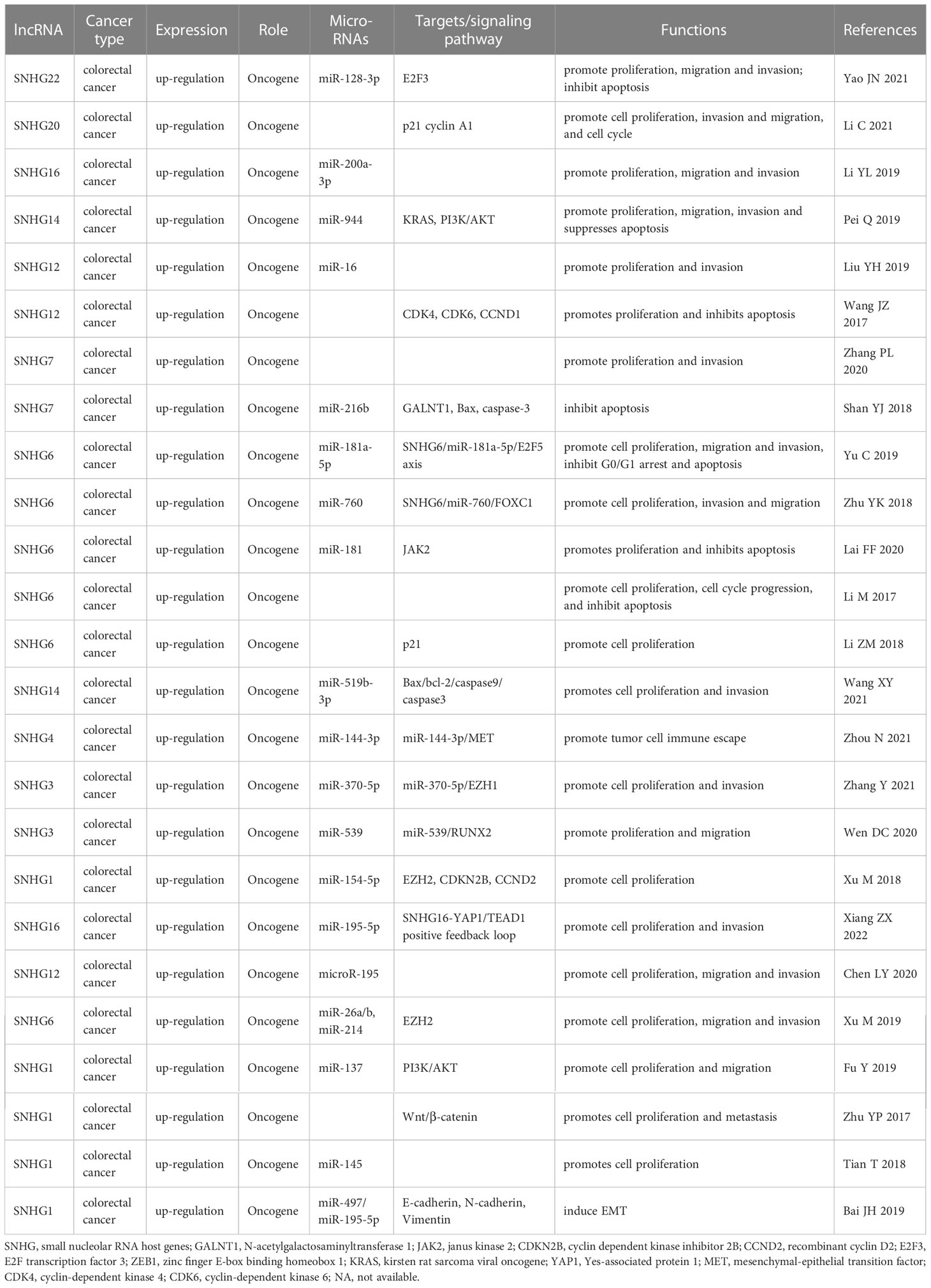
Table 5 Transition of cell phenotype and related molecular mechanisms with abnormal lncRNA SNHGs expression in colorectal cancers.
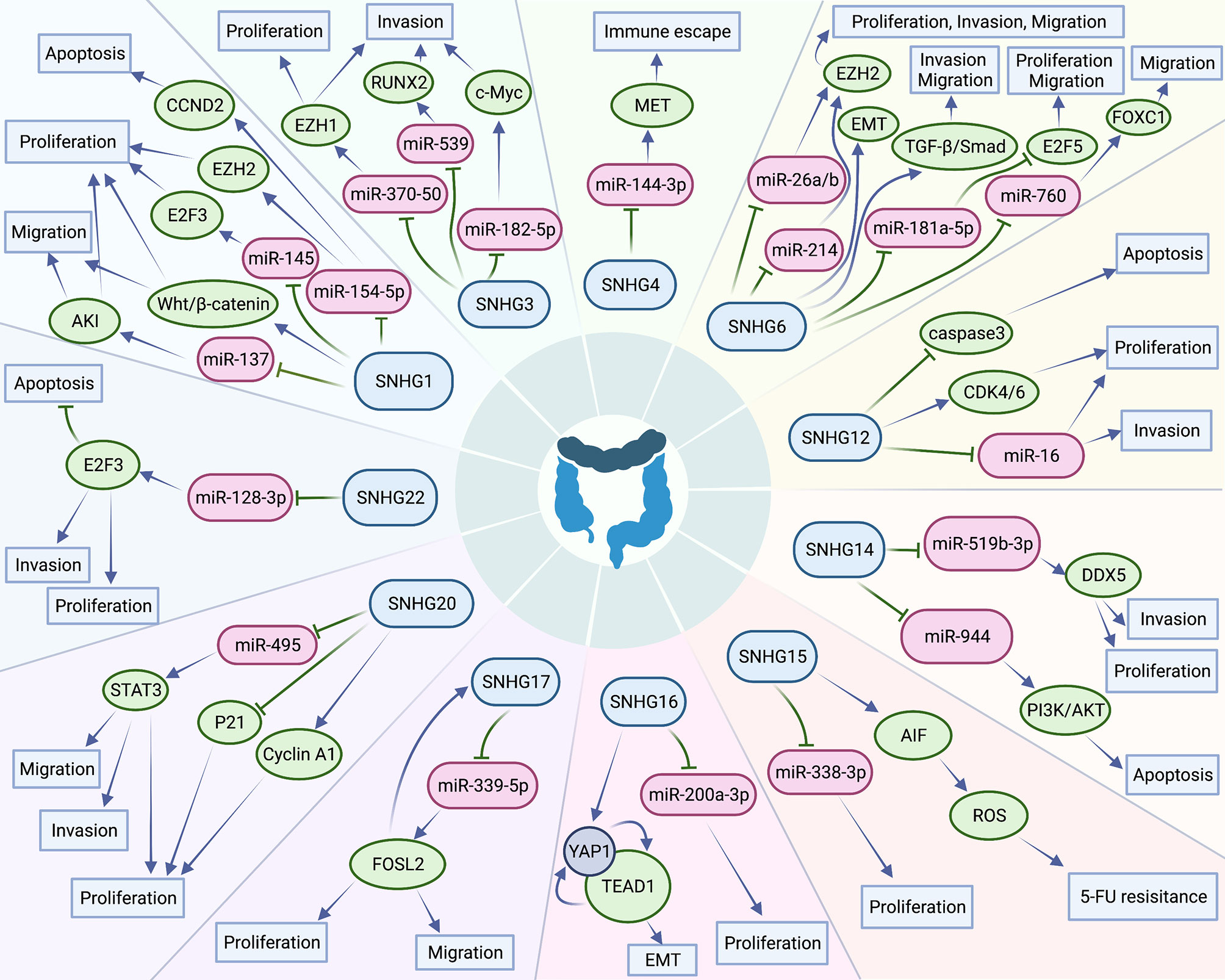
Figure 11 Small nucleolar RNA host genes involved in a series of cellular biological roles in colorectal cancers.
MicroRNAs have been proved to play an critical part in the progress of CRC (37). Different expression of microRNAs could surpassingly promote growth, local invasion, and distant organ migration of CRC cells (38). Additionally, lncSNHGs could work as competitive endogenous RNA (ceRNA) (Figure 10, thereby influencing the biological characteristics of CRC tumor cells by affecting the downstream signal axis through sponge adsorption of microRNAs. Bai et al. indicated that lncSNHG1 may functions as an oncogene in order to drive CRC cell violation and transference via EMT modification by cooperating with miR-195-5p/miR-497, downregulating E-cadherin, and upregulating N-cadherin and vimentin (12). Zhang et al. uncovered that lncSNHG3 facilitates callus grow and violation of CRC cells by upregulating the enhancer of zeste homolog 1 (EZH1) and downregulating miR-375-5p (32). Similarly, Wen et al. revealed that lncSNHG3 induces the cell multiplication and migration of CRC cells by acting as ceRNAs, sponing miR-539, and increasing the expression of runt-related transcription factor 2 expression (33). Zhen et al. suggested that lncSNHG8 can function as a ceRNA, thereby promoting the multiplication, invasion, and migration of CRC cells by directly sponging with miR-663 (39). Xu et al. implied that lncSNHG6 function as an oncogene and could promote the cell cycle, enhance the invasion and migration ability of CRC cells by upregulating EZH1 expression via co binding and downregulation of miR-26a/b and miR-214 (40).
Some studies also revealed that lncSNHG could participate in the immune escape of tumor cells. For example, Zhou et al. revealed that lncSNHG4 could induce the immune escape of CRC cells via PD-1/PD-L1 activation and inducing CD4+ T cell apoptosis by sponging on miR-144-3p (13).
Several lncSNHG may induce chemotherapy resistance of CRC and promote cancer progression. For example, Ghasemi et al. validated that lncSNHG6 could drive the proliferation and drug resistance of CRC cells by upregulating the RAS and MAPK/AKT pathway (41). Saeinasab al. reported that lncSNHG15 may promote colon cancer and increases the resistance of CRC cells to 5-fluorouracil by interacting with apoptosis induced factor (42).
This study has some limitations. First, all the patients included were Asias; therefore, our conclusions can only represent the Asian population. Secondly, many original literatures only provided survival curve without HR value, which causes methodological bias to some extent. Finally, the number of patients in a single study is small, which can affect the overall results to some extent.
This is study is the first to explore the correlation between lncRNA SNHGs expression level and the prognosis of CRC. We revealed that high expression of lncRNA SNHGs is significantly related to poor CRC prognosis, and the expression level of lncRNA SNHGs may be used as a significant prognostic marker and potential therapeutic target of CRC.
High lncRNA SNHG expression implies worse prognosis for CRC, especially SNHG1. Additionally, lncRNA SNHG may function as potential therapeutic target for CRC. Therefore, an original study with higher quality is required to further support the consequence of this study.
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
PL designed the study; PL and YL searched databases and performed literature screening; PL and JD extracted and analyzed the data; JM and WS evaluated the quality of included literature; PL, JD, YL and JM contributed to writing the manuscript. Final draft was approved by all the authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
2. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. Jama. (2021) 325(7):669–85. doi: 10.1001/jama.2021.0106
3. Piawah S, Venook AP. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer. (2019) 125(23):4139–47. doi: 10.1002/cncr.32163
4. Johdi NA, Sukor NF. Colorectal cancer immunotherapy: Options and strategies. Front Immunol (2020) 11:1624. doi: 10.3389/fimmu.2020.01624
5. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (London England) (2019) 394(10207):1467–80. doi: 10.1016/s0140-6736(19)32319-0
6. Bridges MC, Daulagala AC, Kourtidis A. Lnccation: Lncrna localization and function. J Cell Biol (2021) 220(2). doi: 10.1083/jcb.202009045
7. Peng WX, Koirala P, Mo YY. Lncrna-mediated regulation of cell signaling in cancer. Oncogene. (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
8. Li J, Meng H, Bai Y, Wang K. Regulation of lncrna and its role in cancer metastasis. Oncol Res (2016) 23(5):205–17. doi: 10.3727/096504016x14549667334007
9. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. Lncrna-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (London England) (2021) 41(2):109–20. doi: 10.1002/cac2.12108
10. Yang Z, Li Q, Zheng X, Xie L. Long noncoding rna small nucleolar host gene: A potential therapeutic target in urological cancers. Front Oncol (2021) 11:638721. doi: 10.3389/fonc.2021.638721
11. Chu Q, Gu X, Zheng Q, Guo Z, Shan D, Wang J, et al. Long noncoding rna Snhg4: A novel target in human diseases. Cancer Cell Int (2021) 21(1):583. doi: 10.1186/s12935-021-02292-1
12. Bai J, Xu J, Zhao J, Zhang R. Lncrna Snhg1 cooperated with mir-497/Mir-195-5p to modify epithelial-mesenchymal transition underlying colorectal cancer exacerbation. J Cell Physiol (2020) 235(2):1453–68. doi: 10.1002/jcp.29065
13. Zhou N, Chen Y, Yang L, Xu T, Wang F, Chen L, et al. Lncrna Snhg4 promotes malignant biological behaviors and immune escape of colorectal cancer cells by regulating the mir-144-3p/Met axis. Am J Trans Res (2021) 13(10):11144–61.
14. Liu Y, Zhou J, Wang S, Song Y, Zhou J, Ren F. Long non-coding rna Snhg12 promotes proliferation and invasion of colorectal cancer cells by acting as a molecular sponge of microrna-16. Exp Ther Med (2019) 18(2):1212–20. doi: 10.3892/etm.2019.7650
15. Fang SX, Chen C, Guo Q, Ke XX, Lu HL, Xu G. High Lncsnhg15 expression may predict poor cancer prognosis: A meta-analysis based on the prisma and the bio-informatics analysis. Bioscience Rep (2020) 40(7). doi: 10.1042/bsr20194468
16. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol (2010) 25(9):603–5. doi: 10.1007/s10654-010-9491-z
17. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-Event data into meta-analysis. Trials. (2007) 8:16. doi: 10.1186/1745-6215-8-16
18. Yao J, Wang C, Dong X, Zhang Y, Li Y, Zhou H, et al. Lncrna Snhg22 sponges Mir-128-3p to promote the progression of colorectal cancer by upregulating E2f3. Int J Oncol (2021) 59(3). doi: 10.3892/ijo.2021.5251
19. Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding rna Snhg20 predicts poor prognosis in colorectal cancer. BMC canc (2016) 16:655. doi: 10.1186/s12885-016-2719-x
20. Wang Y, Fu J, Yang L, Liang Z. Long Non-Coding rna Snhg20 promotes colorectal cancer cell proliferation, migration and invasion Via Mir-495/Stat3 axis. Mol Med Rep (2021) 23(1). doi: 10.3892/mmr.2020.11669
21. Li Y, Lu Y, Chen Y. Long non-coding rna Snhg16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer Via sponging mir-200a-3p. Bioscience Rep (2019) 39(5). doi: 10.1042/bsr20182498
22. Pei Q, Liu GS, Li HP, Zhang Y, Xu XC, Gao H, et al. Long noncoding rna Snhg14 accelerates cell proliferation, migration, invasion and suppresses apoptosis in colorectal cancer cells by targeting mir-944/Kras axis through Pi3k/Akt pathway. Eur Rev Med Pharmacol Sci (2019) 23(22):9871–81. doi: 10.26355/eurrev_201911_19551
23. Wang JZ, Xu CL, Wu H, Shen SJ. Lncrna Snhg12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res = Rev Bras pesquisas medicas e biologicas (2017) 50(3):e6079. doi: 10.1590/1414-431x20176079
24. Zhang P, Shi L, Song L, Long Y, Yuan K, Ding W, et al. Lncrna crnde and lncrna Snhg7 are promising biomarkers for prognosis in synchronous colorectal liver metastasis following hepatectomy. Cancer Manage Res (2020) 12:1681–92. doi: 10.2147/cmar.S233147
25. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. Lncrna Snhg7 sponges mir-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating Galnt1. Cell Death disease (2018) 9(7):722. doi: 10.1038/s41419-018-0759-7
26. Yu C, Sun J, Leng X, Yang J. Long noncoding rna Snhg6 functions as a competing endogenous rna by sponging mir-181a-5p to regulate E2f5 expression in colorectal cancer. Cancer Manage Res (2019) 11:611–24. doi: 10.2147/cmar.S182719
27. Zhu Y, Xing Y, Chi F, Sun W, Zhang Z, Piao D. Long noncoding rna Snhg6 promotes the progression of colorectal cancer through sponging mir-760 and activation of Foxc1. OncoTargets Ther (2018) 11:5743–52. doi: 10.2147/ott.S170246
28. Lai F, Deng W, Fu C, Wu P, Cao M, Tan S. Long non-coding rna Snhg6 increases Jak2 expression by targeting the mir-181 family to promote colorectal cancer cell proliferation. J Gene Med (2020) 22(12):e3262. doi: 10.1002/jgm.3262
29. Li M, Bian Z, Yao S, Zhang J, Jin G, Wang X, et al. Up-regulated expression of Snhg6 predicts poor prognosis in colorectal cancer. Pathology Res practice (2018) 214(5):784–9. doi: 10.1016/j.prp.2017.12.014
30. Li Z, Qiu R, Qiu X, Tian T. Snhg6 promotes tumor growth Via repression of P21 in colorectal cancer. Cell Physiol Biochem (2018) 49(2):463–78. doi: 10.1159/000492986
31. Wang X, Yang P, Zhang D, Lu M, Zhang C, Sun Y. Lncrna Snhg14 promotes cell proliferation and invasion in colorectal cancer through modulating mir-519b-3p/Ddx5 axis. J Canc (2021) 12(16):4958–70. doi: 10.7150/jca.55495
32. Zhang Y, Li L, Lu KX, Yu LB, Meng J, Liu CY. Lncrna Snhg3 is responsible for the deterioration of colorectal carcinoma through regulating the mir-370-5p/Ezh1 axis. Eur Rev Med Pharmacol Sci (2021) 25(19):6131–7. doi: 10.26355/eurrev_202110_26891
33. Dacheng W, Songhe L, Weidong J, Shutao Z, Jingjing L, Jiaming Z. Lncrna Snhg3 promotes the growth and metastasis of colorectal cancer by regulating mir-539/Runx2 axis. Biomed pharmacother = Biomed pharmacotherapie (2020) 125:110039. doi: 10.1016/j.biopha.2020.110039
34. Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, et al. The long noncoding rna Snhg1 regulates colorectal cancer cell growth through interactions with Ezh2 and mir-154-5p. Mol canc (2018) 17(1):141. doi: 10.1186/s12943-018-0894-x
35. Huang L, Lin H, Kang L, Huang P, Huang J, Cai J, et al. Aberrant expression of long noncoding rna Snhg15 correlates with liver metastasis and poor survival in colorectal cancer. J Cell Physiol (2019) 234(5):7032–9. doi: 10.1002/jcp.27456
36. Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, et al. Lncrna Snhg6 promotes proliferation, invasion and migration in colorectal cancer cells by activating tgf-B/Smad signaling pathway Via targeting Upf1 and inducing emt Via regulation of Zeb1. Int J Med Sci (2019) 16(1):51–9. doi: 10.7150/ijms.27359
37. Mei H, Wen Y. Micrornas for diagnosis and treatment of colorectal cancer. Endocrine Metab Immune Disord Drug targets (2021) 21(1):47–55. doi: 10.2174/1871530320999200818134339
38. Wang H. Micrornas and apoptosis in colorectal cancer. Int J Mol Sci (2020) 21(15). doi: 10.3390/ijms21155353
39. Zhen Y, Ye Y, Wang H, Xia Z, Wang B, Yi W, et al. Knockdown of Snhg8 repressed the growth, migration, and invasion of colorectal cancer cells by directly sponging with mir-663. Biomed pharmacother = Biomed pharmacotherapie (2019) 116:109000. doi: 10.1016/j.biopha.2019.109000
40. Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, et al. Lncrna Snhg6 regulates Ezh2 expression by sponging mir-26a/B and mir-214 in colorectal cancer. J Hematol Oncol (2019) 12(1):3. doi: 10.1186/s13045-018-0690-5
41. Ghasemi T, Khalaj-Kondori M, Hosseinpour Feizi MA, Asadi P. Aberrant expression of lncrnas Snhg6, Trpm2-As1, Mir4435-2hg, and hypomethylation of Trpm2-As1 promoter in colorectal cancer. Cell Biol Int (2021) 45(12):2464–78. doi: 10.1002/cbin.11692
42. Saeinasab M, Bahrami AR, González J, Marchese FP, Martinez D, Mowla SJ, et al. Snhg15 is a bifunctional myc-regulated noncoding locus encoding a lncrna that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with aif. J Exp Clin Cancer Res CR (2019) 38(1):172. doi: 10.1186/s13046-019-1169-0
Keywords: lncRNA, SNHGs, colorectal cancer, prognosis, meta-analysis, bioinformatics analysis
Citation: Luo P, Du J, Li Y, Ma J and Shi W (2023) Association between small nucleolar RNA host gene expression and survival outcome of colorectal cancer patients: A meta-analysis based on PRISMA and bioinformatics analysis. Front. Oncol. 13:1094131. doi: 10.3389/fonc.2023.1094131
Received: 15 November 2022; Accepted: 16 January 2023;
Published: 20 February 2023.
Edited by:
Jin Zhang, Shenzhen University, ChinaReviewed by:
Jin Wenke, Southwest Jiaotong University, ChinaCopyright © 2023 Luo, Du, Li, Ma and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei Luo, MjcyNzYzNTE1NEBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.