
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Oncol., 26 January 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1092870
This article is part of the Research TopicTowards a New 3Rs Era in Experimental ResearchView all 37 articles
Background: Research on tumor organoids has developed rapidly over the past 20 years, but a systematic analysis of current research trends is lacking. Researchers in the field need relevant references and knowledge of current research hot spots. Bibliometric analysis and visualization is a systematic method of acquiring an in-depth understanding of the status of research on tumor organoids.
Methods: CiteSpace, VOSviewer and the Bibliometric Online Analysis Platform from the Web of Science Core Collection were used to analyze and predict publishing trends and research hot spots worldwide in the field of tumor organoids.
Results: A total of 3,666 publications on tumor organoids were retrieved, and 2,939 eligible articles were included in the final analysis. The number of publications has grown significantly, with the United States of America as the leading country for research on tumor organoids. Among journals, Cancers published the largest number of articles. Harvard Medical School published the highest number of articles among all institutions. The Chinese Academy of Sciences was ranked highest among all contributing institutions on the importance of their publications. A trend in multi-disciplinary collaboration was observed in studies on tumor organoids. Keywords indicated that the current research largely concentrated on optimizing the construction of organoid models to use for medication development and screening in the clinical setting, and to provide patients with individualized treatment for gastric cancer and colorectal cancer, which are newly emerging research hotspots. Gastric and colorectal cancers were the top two tumors that have received increasing attention and have become the focal points of recent studies.
Conclusion: This study analyzed 2,939 publications covering the topic of tumor organoids. Although optimizing the construction of organoid models has always been a hot topic in this field, the application of tumor organoids to the development of medications and screenings will foster individualized treatment for patients, which is another emerging hot spot in this field of research.
The International Agency for Research on Cancer reported 19.3 million new cases of cancer and approximately 10 million deaths due to cancer worldwide in 2020. Approximately 28.4 million new cases of cancer are expected worldwide in 2040, which is an increase of 47%. Cancer has become a key problem endangering global public health (1). In recent years, the treatment of patients with cancer has evolved from interventions based on tumor types to those based on the tumor’s molecular characteristics or microenvironment. This approach, known as precision medicine or individualized treatment, has saved many patients with advanced cancer. At present, commonly used tumor models, such as the 2D cell culture (2), genetically engineered mouse models (3) and the human-derived tumor xenograft models (4), although essential for tumor research, are difficult to simulate perfectly the actual state of tumors in vivo, resulting in the failures of many clinical trials. Organoids are derived from the stem cells of 3D cultures, which can reproduce the structural and functional characteristics of a native organ and simulate the development of diseases of human organs in culture dishes (5). Tumor organoids are in vitro models established by surgical resection or tissue biopsy to obtain tumor tissue in patients, followed by mincing and enzymatic hydrolysis of tumor tissue and the 3D culture of the tumor cells in it (6). Compared with traditional tumor models, tumor organoids not only reflect the genetic characteristics and tissue structure heterogeneity of a patient’s tumor tissues, but they also maintain gene stability during self-renewal and throughout long-term expansion. Given these reasons, they can be used to study cancers caused by infection or gene mutation, and have unique advantages for clinical drug development and guiding individualized treatment (7).
Although the research on tumor organoids has developed rapidly in recent years, a scientific and systematic analysis of their status and trends is lacking. Therefore, we used the Web of Science Core Collection (WoSCC) as our data source to perform a quantitative analysis of the literature in the field of tumor organoid research, in order to understand and determine the research hot spots and frontiers, and to provide a relevant reference for scientific researchers in the field.
All of the data in this study were derived from the WoSCC. The search terms were: Tumor or Neoplasm (TS) = (“Cancer” or “Tumor” or “Neoplasm” or “Neoplasia” or “Malignant Neoplasm” or “Malignancy” or “Malignant Neoplasms” or “Benign Neoplasms” or “carcinoma”) AND (“Organoid” or “Organoids”) AND Language = English. The search period was from January 1, 2011 to December 31, 2021 and the required language was English. A total of 3,666 articles were retrieved and 727 were excluded from the analysis because either they were not research reports or they were review articles. Thus, 2,939 articles were included in the final sample for the subsequent metrological analysis, as shown in Figure 1. All data in the text were extracted on September 14, 2022 (the same day) to avoid deviations due to daily updates of the database.
The full records and cited references of the eligible publications were downloaded from the WoSCC database and saved in TXT format; then they were imported into the CiteSpace software V5.8.R3 SE, 64 bits. CiteSpace is visual analysis software based on the JAVA environment developed by Chen Chaomei. The software is designed to analyze information in the scientific literature. The user can detect the development of rules and the distribution of scientific knowledge in a field through visualization, and identify key points in the development of a field of study based on the existing data, especially turning points and key points in the field of knowledge (8). VOSviewer is visualization software developed by the Science and Technology Research Center in Leiden, the Netherlands. The user can also build a visual network map based on information in the scientific literature and acquire a comprehensive understanding of trends in the scientific structure and dynamic development of a field (9). The full records and cited references of these publications were downloaded from the WoSCC database, saved as a tab-delimited file and imported into the Bibliometrics Online Analysis Platform1. We chose the “total literature analysis” option to examine trends in publications of different countries and the “partnership analysis” option to explore collaborations between countries and regions.
In this bibliometric analysis, the quantity and quality of the research results were evaluated using indicators, such as the number of articles published, the frequency of citations of the article and the impact factor, which reflect the level of research and academic status of a region, institution or author in a certain field. The number of papers published is an important indicator for evaluating the capacity for scientific research output. Frequency of citations in bibliometric analysis, refers to the number of times a published paper is cited in other articles, which reflect the value of the paper to a field and the degree of attention it has received. The H-index refers to rankings of articles published using relevant statistical measures, based on an articles’ frequency of citations (from high to low). Articles with the lowest H-index have the least amount of citations, which we used as a measure to assess the number of scientific research outputs and the quality of the research in this bibliometric analysis. The journal’s impact factor and other metrics were retrieved from the 2021 edition of Journal Citation Reports. These are important indicators of the academic level and quality of the journals and the articles.
To clarify the rate of the development of research on tumor organoids, global trends in the volume of published research articles from 2011 to 2021 were plotted, as shown in Figure 2. Only 129 articles were published worldwide from 2011 to 2014. With the global shift to a rapid growth of research on tumor organoids before and after 2017, as of 2021, 912 studies have been published worldwide, accounting for 31.03% of the total number of articles published from 2011 to 2021, indicating that research on tumor organoids has become a focus of global enquiry. Given the upward trend line in Figure 2, the global number of publications in 2022 is expected to be significantly higher than the number of articles published last year.
Between 2011 and 2021, there were 72 countries worldwide that published research on tumor organoids, as shown in Figure 3. Indicators of the top ten countries with the highest number of articles are shown in Table 1. The highest and most significant number of articles were published in the United States of America (USA) (1,320, 44.91%) and China (447, 15.21%), followed by Germany (329, 11.19%), Japan (297, 10.10%) and the Netherlands (289, 9.83%). The H-indices of the USA, the Netherlands and Germany were 93, 67 and 48, respectively, ranking them among the top three countries with the highest H-indices. Articles from the USA, the Netherlands and England with 44,814, 25,368 and 10,089 citations, respectively, were ranked among the top three countries with the highest total number of citations, and articles from the Netherlands, England and Switzerland, with an average of 87.78, 42.93 and 36.17 citations per paper respectively, were ranked among the top three countries for the highest average number of citations per article.
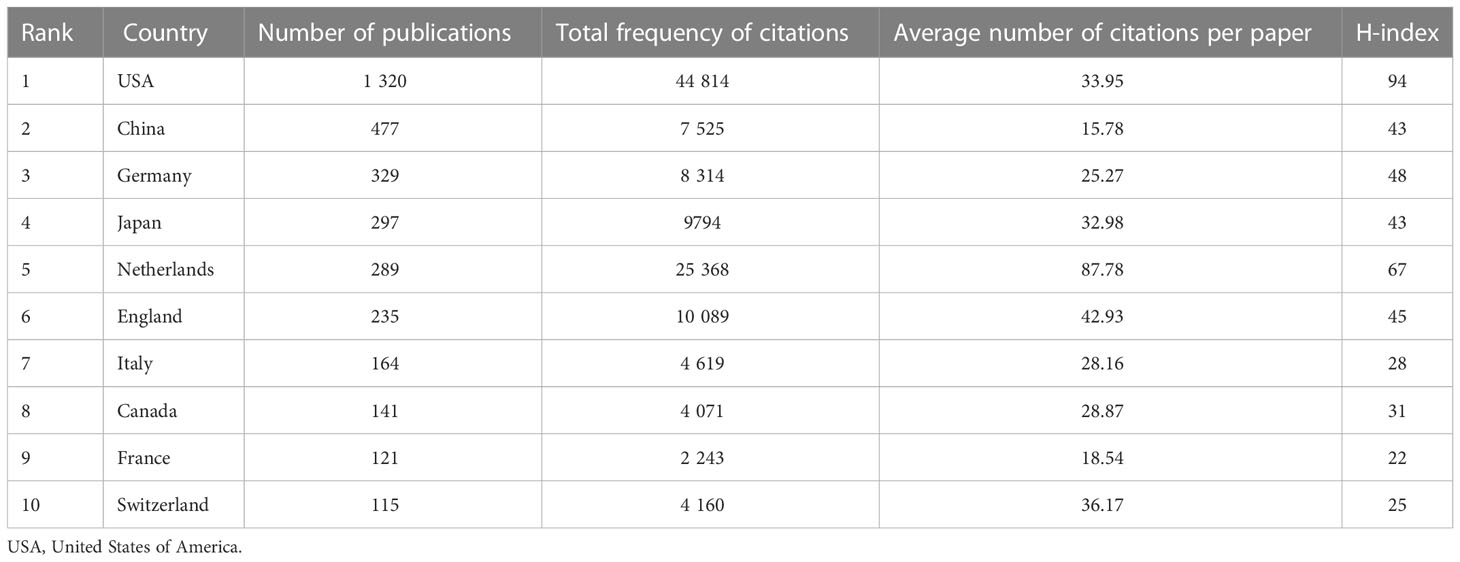
Table 1 Publication metrics of articles on tumor organoids of the top ten countries by number of publications.
All relevant information was exported from the WoSCC database and saved as a tab-delimited file. International cooperation between the countries publishing relevant research was analyzed using the Bibliometric Online Analysis Platform1, as shown in Figure 4A, with the USA having the highest frequency of international cooperation, followed by Germany and the Netherlands. China had close cooperative relationships with the USA, the Netherlands and Singapore. The mediation centrality of the publishing countries was analyzed using CiteSpace software, as shown in Figure 4B. Mediation centrality refers to the ratio of the shortest path passing through a certain point and connecting these two points in the network to the total number of shortest path lines between these two points, which is an indicator used to characterize the importance of the node. The numerical value indicates that the node is in a key position in the network and has influence. The information in Figure 4B was exported to report the rankings of centrality of the top five nations’ publications in Table 2. The USA (0.38), Germany (0.21), the England (0.21), France (0.19) and Italy (0.19) exhibited high degrees of centrality, which is represented by purple in Figure 4B. The above results indicate that these countries play an important role in tumor organoid research, and the academic influence of their research results influence countries worldwide. The reliability, quality and innovation of the relevant research from these countries are far ahead of other countries.
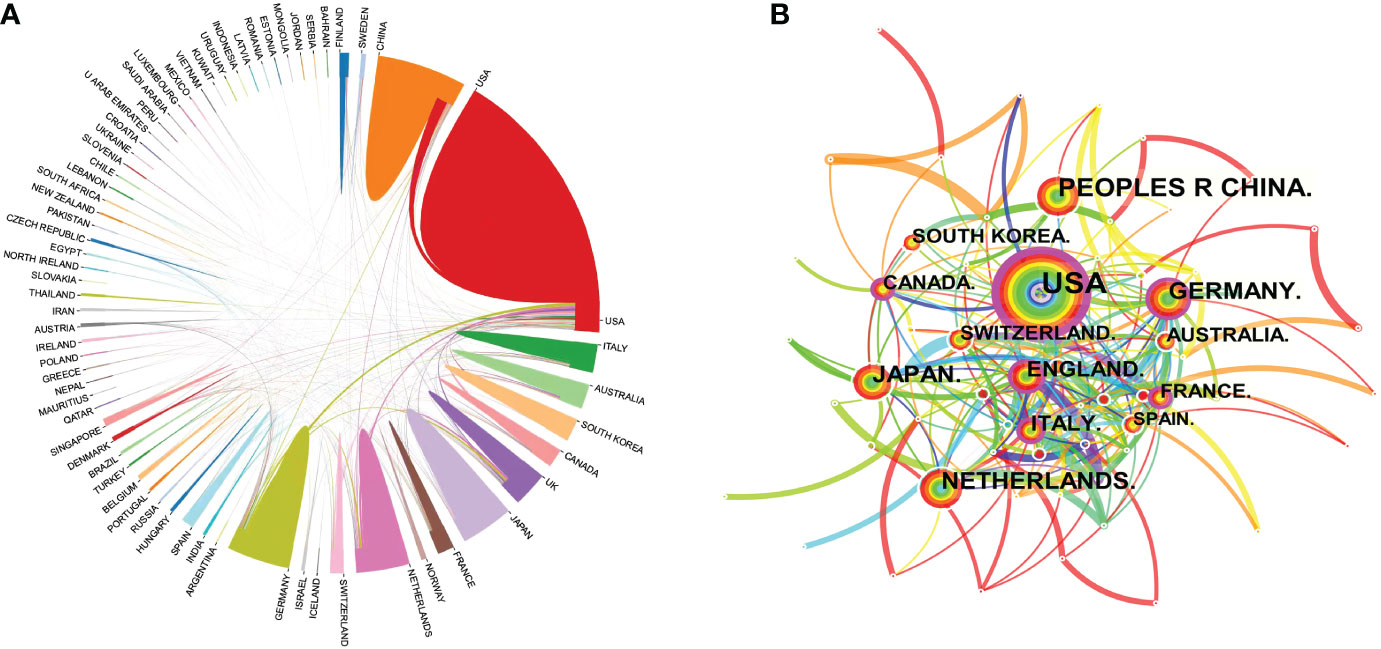
Figure 4 Map of the co-occurrence of tumor organoid research by nation. Figure 4 (A) illustrates the international cooperation among countries. These countries are represented by different colors; the links represent international cooperation and links between countries represent connections between countries. Figure 4 (B) shows the centrality of the articles. Each node represents a country, and the node size represents the number of articles published by the country. The larger the node, the more articles that were published. The purple color in the outermost ring represents a central node or the central location of the cooperation between the institution and other institutions.
Academic cooperation between institutions is crucial for strengthening exchanges between scholars and for disseminating advanced experiences. CiteSpace software was used in this study to conduct network co-occurrence analysis of the publishing institution, as shown in Figure 5. The information in Figure 5 was exported to report the rankings of centrality and the number of publications among the top five institutions in Table 3. Harvard Medical School (111), the University Medical Center of Utrecht (102), Memorial Sloan Kettering Cancer Center (88), Johns Hopkins University (61) and Vanderbilt University (54) were the top five research institutions in terms of their numbers of published articles. The Chinese Academy of Sciences (0.32), Dana Farber Cancer Institute (0.30), the University of Nebraska Medical Center (0.24), Columbia University (0.23) and Memorial Sloan Kettering Cancer Center (0.22) were the top five research institutions in terms of intermediary centrality. These institutions occupy core positions in the field of tumor organoid research and engage in cooperative academic research with most institutions.
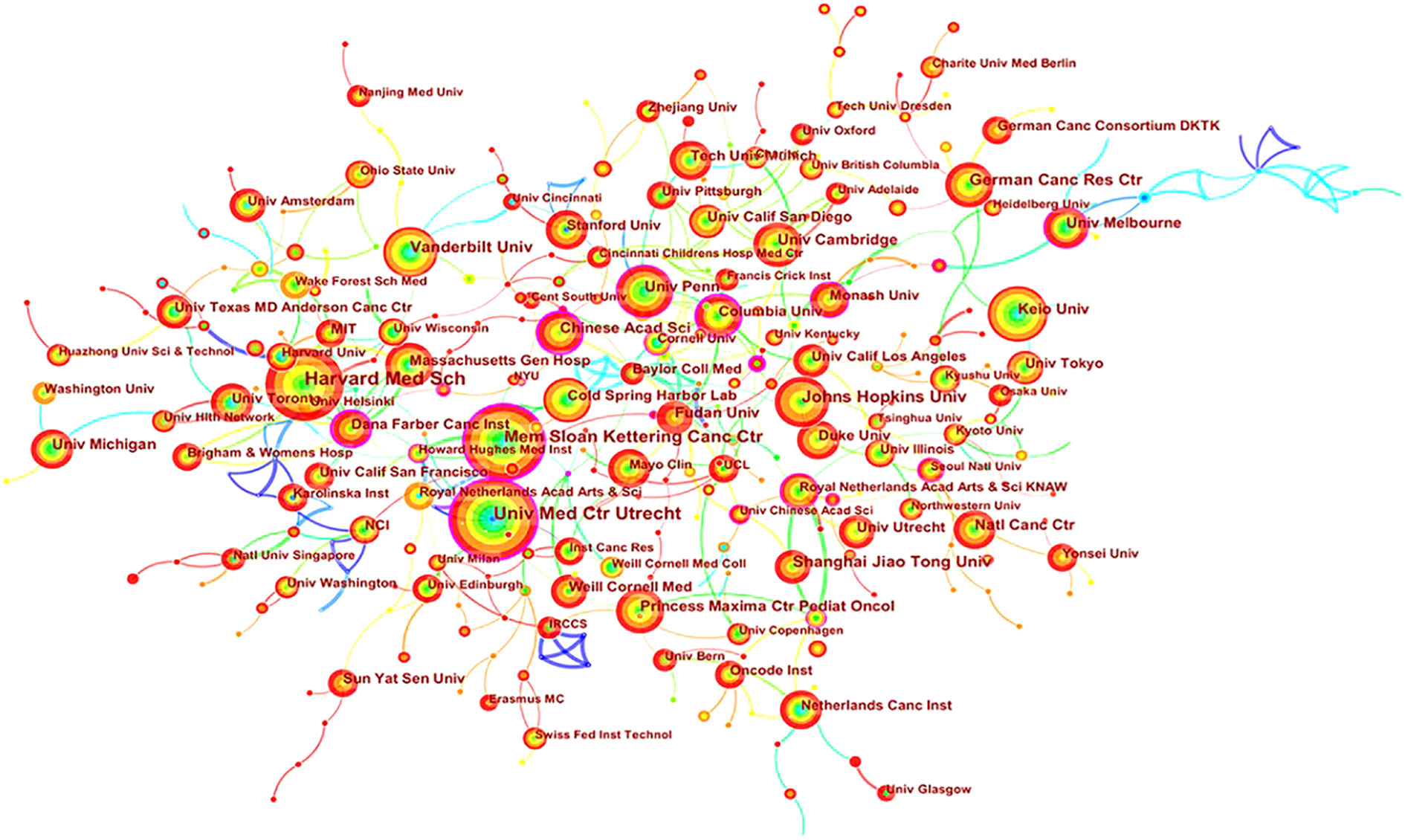
Figure 5 Map of cooperation in tumor organoid research. The nodes in Figure 5 represent different institutions, and the size of the nodes represents the number of articles published. The purple ring in the outermost circle indicates a central node, which has a central position in the cooperation between this institution and other institutions. The wired lines indicate cooperation among institutions.
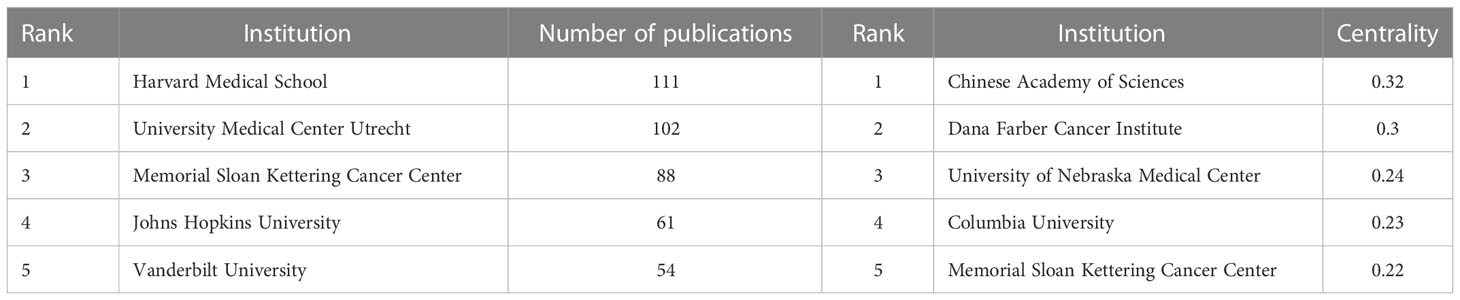
Table 3 Centrality rankings and number of publications of the top five institutions in the field of tumor organoid research.
A total of 20,139 scholars published articles in the field of tumor organoid research worldwide. The indicators of the top ten scholars as shown in Table 4. Four articles were published by the top ten scholars; five scholars were from the USA and three were from the Netherlands, Japan and England. Clevers H (103) at the University Medical Center of Utrecht, Sato T (34) at the Keio University School of Medicine and Chen Y (32) at Memorial Sloan Kettering Cancer Center were identified as the top three authors with the highest number of published articles. Clevers H also ranked first in the total number of citations received, the average frequency of citations per paper and the H-index. These authors have influenced tumor organoid research worldwide.
Based on the cooperative network analysis of the authors with high numbers of published articles using the VOSviewer, we further defined “core authors” as those who had at least 5 publications, which included 316 authors in the analysis. We found some collaborative research teams had a high publication volume, as shown in Figure 6. Clevers H, Sato T, Drost J, Chen Y, Sansom OJ, Braker N and Van Der collaborated closely, but most of the inter-author collaborations were limited to intra-team collaborations, with little international collaboration, such as several small cooperative networks in the periphery (e.g., Hippo Y, Onuma K and Inoue M., and Jun P, Meyer TF and Bartfeld S). If cooperative research between authors can be strengthened, especially between authors from different countries or institutions, exchanges and innovations will likely improve significantly.
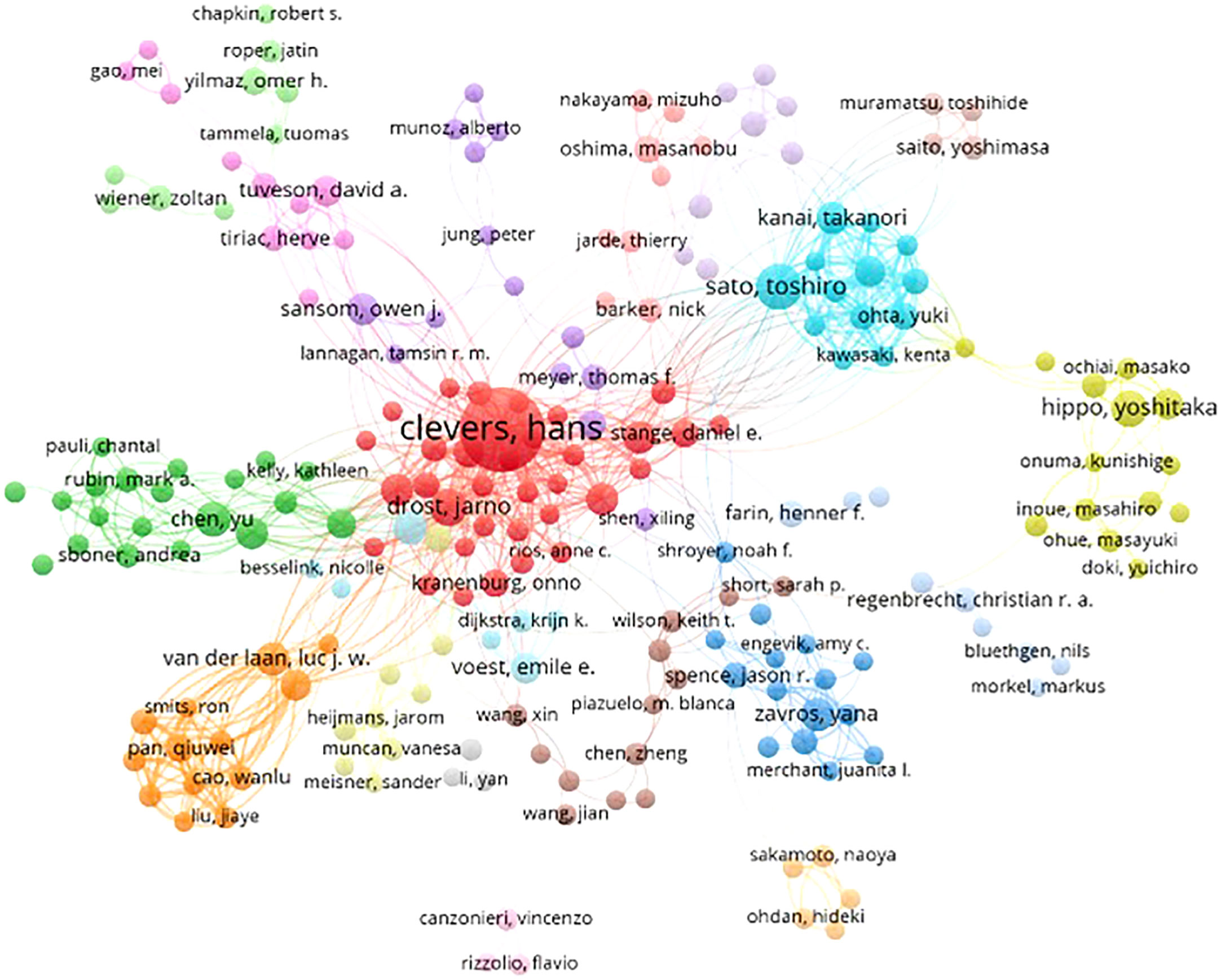
Figure 6 Collaborative networks among authors with a high publication volume of tumor organoid research.
Journals play a crucial role in promoting international cooperation and progress in the level of scientific research. Between 2011 and 2021, 752 journals worldwide published academic papers on tumor organoid research, of which the top ten journals published 5 papers on the topic as shown in Table 5. Cancers published the most research results, with 130 articles, accounting for 4.42% of the total publication volume, followed by Scientific Reports and Nature Communications, which published 85 and 74 articles, respectively, accounting for 2.89% and 2.51% of the total volume of publications. Gastroenterology ranked first in both citation frequency and the H-index. Among the top ten journals, eight journals belonged to partition Q1, 2 belonged to partition Q2 and 1 to partition Q3, with an impact factor ranging from 1.696 to 23.937, and Gastroenterology having the highest impact factor. The analysis shows that these international journals were the academic authorities on the findings of tumor organoid research, which were most likely to be of concern to scholars in various countries.
The 2,939 articles in this study were classified into 95 research directions. It should be noted that the sum of the percentages of all the studies’ directions exceeded 100% because the same article could be classified into multiple research directions. Changing trends in research directions in recent years is shown in Figure 7. The most articles published were in the direction of Oncology with a total of 802, accounting for 27.25% of the total number of articles published. These were followed by Cell Biology and Multidisciplinary Sciences, with 599 and 362 articles, respectively, followed by Biochemistry Molecular Biology and Gastroenterology Hepatology. Experimental Medicine Research, Genetics Heredity, Pathology, Cell Tissue Engineering and Pharmacology Pharmacy, with more than 100 articles. Thus, this study of tumor organoids showed the developing characteristics of multidisciplinary intersection. Given the changes in the number of articles published in each research direction over time, the top five research directions of the articles indicated an increasing trend in fluctuations before 2016. All research directions reached a state of rapid growth after 2016, with Oncology developing most rapidly.
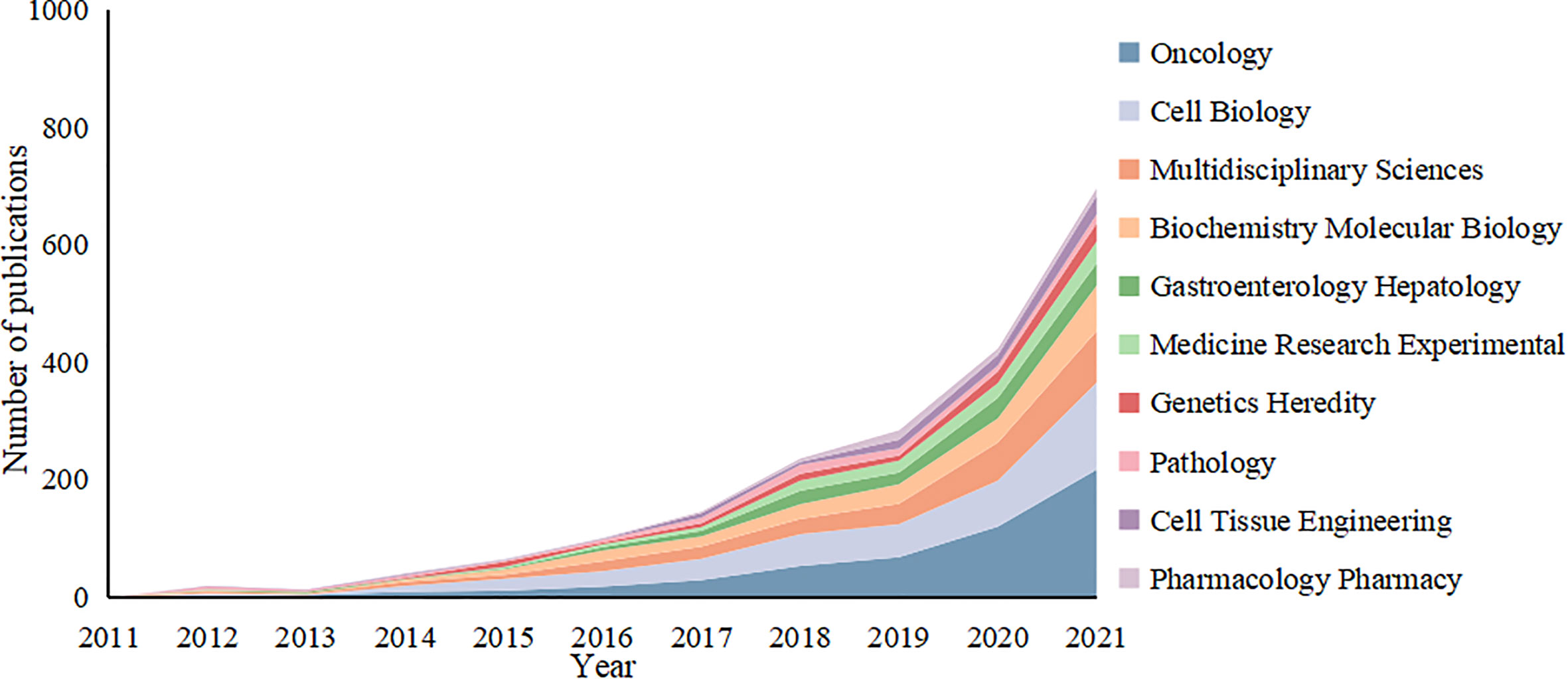
Figure 7 Variation curve of the number of published papers by main research direction of the tumor organoid research.
The total frequencies of citations of the top ten articles were derived from the citation reports of the WoSCC. These articles were cited more than 600 times, of which 9 were from the team led by Clevers H (Table 6). In 2011, the Clevers H team successfully constructed a small intestinal crypt-villus structure in vitro without mesenchymal conditions from a single LGR5 stem cell (10). The team then added different growth factors or inhibitors to different organoids, based on previous studies, so that the replication potential of the adult stem cells was not limited in vitro. This improvement promoted the use of organoid models as a tool for investigating diseases with increasing complexity (11–13). The team also constructed a variety of tumor organoid models, such as models of colorectal, prostate and pancreatic cancers (14–19). These constructed models had a better fit with real tumor growth and development, and the organoid tumor models that were constructed using the patient’s own tumor cells facilitated a more individualized approach to the treatment of tumors. These studies have opened up a new path for acquiring a more in-depth understanding of the mechanism of tumorigenesis and development, and clinical drug development and screening.
Table 7 shows the top twenty most frequently occurring keywords analyzed by CiteSpace. Nodes were set as keywords with a time interval of 2011–2021 and yearly time slices. We found that “stem cells,” “cancer,” “in vitro,” “colorectal cancer,” “model,” “culture,” “differentiation” and “growth” were the most frequently occurring keywords, and “3D culture,” “tumor model,” “beta catenin,” “inflammatory bowel disease,” “small intestine,” “P53” and “cancer metabolism” were the most central keywords.
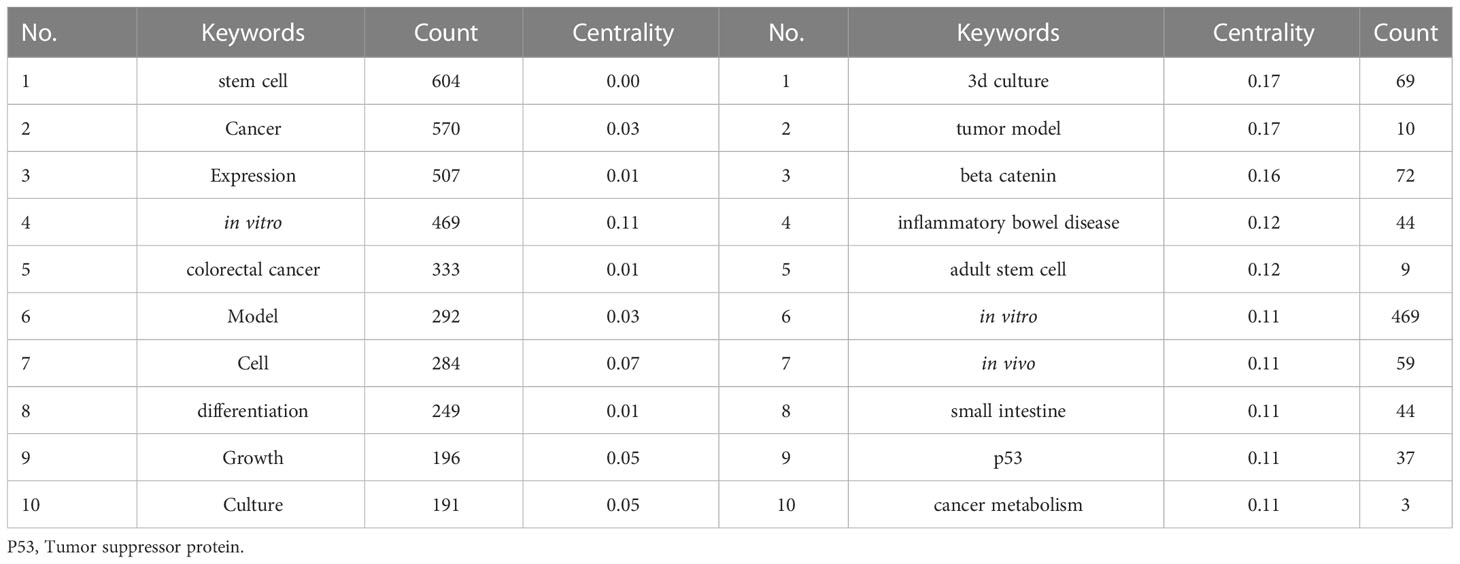
Table 7 The top twenty most frequently occurring keywords and their centrality in research related to tumor organoids.
A K-means cluster analysis of the keywords was performed using CiteSpace, and 19 clusters were obtained (Table 8; Supplementary Figure 1). The keyword co-occurrence network in the field of research on tumor organoids is shown in Supplementary Figure 1. The main research directions were divided into the following categories: #0 (cell), #1 (stem cell), #2 (generation), #3 (expression), #4 (cerebral organoid), #5 (extracellular matrix), #8 (in vitro), #9 (landscape), #13 (in vitro) and #14 (culture) the culture and characteristics of the tumor organoid models. Categories: #6 (gastric cancer), #7 (rectal cancer), #10 (inflammatory bowel disease), #12 (colorectal cancer) and #17 (cancer) were classified as diseases currently used in tumor organoids, and categories #11 (precision medicine), #15 (personalized medicine), #16 (progression) and #18 (photo thermal medicine) were classified as current research and trends in tumor organoids. These 19 clusters summarize the process of development of tumor organoid research, and highlight research hot spots.
Although the 3D culture was proposed more than 100 years ago, it was not until 2009 that the first organoid model of a single stem cell source was established, and Clevers’ laboratory demonstrated that a single LGR5 stem cell could successfully construct small intestinal crypt-villous structures in vitro in the absence of interstitial space (20, 21). In recent years, the application of 3D culture models in cancer research has developed rapidly, and studies of almost every tumor have solved a wide variety of research problems. In contrast to the widely used 2D cell culture and animal models, organoids are an important preclinical model for cancer research with distinct advantages. The 2D cell culture is simpler, cheaper and allows more direct genetic manipulation than the 3D culture models permits; however many cell lines cannot mimic the real situation of tumors accurately due to the lack of spatial organization and the tumor’s micro-environment. Increasing the dimension of the extracellular matrix from 2D to 3D significantly improved cell proliferation, differentiation and survival, and the success rate of patient-derived tumor organoid cultures (22, 23). The patient-derived xenograft model is similar to the original tumor in terms of its histological expression and biological behavior, such as protein expression, tumor biomarker status and genetic status, which cannot be achieved by tumor organoid models (24). However, patient-derived xenograft model transplantation is difficult to perform (25) and time consuming (26), and its cost is relatively high. In addition, genetically engineered mouse models can be used to assess in vivo phenotypes and incorporate the complexity of the syngeneic tumor microenvironment. However, the complexities of time, cost and genetic manipulation are significant drawbacks of these models, compared to those of the organoid models (24). The advantages and disadvantages of the organoid model compared with other models are summarized in Supplementary Figure 2.
Non-clinical testing was used to determine the potential risk of drugs and their efficacy in humans. However, the most commonly used 2D in vitro cell culture system lacked heterogeneity and could not accurately describe or mimic the rich environment in vivo and the complex process of disease formation. Significant flaws were also found in animal studies, with species differences causing most drugs to exhibit different drug sensitivities in different in vitro models (27). The 3D cell culture models have been developed to reflect drug sensitivity in humans more accurately. Compared to the 2D cell culture systems, the 3D cell culture models can replicate the complex micro-ecological environment of human organs can be used for drug safety assessments and combination studies and they can provide key pharmacokinetic and biomarker data (28–30). The intestinal epithelium is an important site for drug absorption and metabolism and plays an important role in the oral bioavailability of drugs. In particular, intestinal epithelial cells serve as gatekeepers for drug and nutrient absorption (31). The data from cell and animal models related to intestinal nutrient absorption, metabolism and oral bioavailability of drugs have poor accuracy due to tumor heterogeneity and species differences. In contrast, intestinal organoids are used to examine different aspects of bioavailability, including drug distribution, individual differences, and high-throughput screening (32). Furthermore, the establishment of organoid models for the liver, kidney and blood-brain barrier meet the basic requirements for evaluating the different pharmacological aspects of drugs (33). In addition to efficacy, drug metabolism and toxicological mechanisms are also important to assess. Simplified culture models of primary human hepatocytes or liver cell lines that are currently used differ considerably from in vivo physiology in terms of drug toxicity, often resulting in failed drug conversion (34). Therefore, the determination of toxicological properties relies mainly on animals. However, due to species differences between humans and animals, the results might not be consistent with the real situation (35). At present, a reproducible human liver organoid model can detect bile acid uptake and excretion in modified human liver organoids in real time by knocking out bile acid transporter genes. The assay platform can be used for large-scale compound screening and has been shown to be genotype-specific sensitive to bosentan-induced cholestasis. This powerful assay will facilitate diagnoses, functional studies, drug development and individualized therapies (36).
Traditional cancer treatments (chemotherapy and radiation therapy) are ineffective or have a positive effect in only a subset of patients due to the heterogeneity of malignancies. To ensure the effectiveness of the treatment, it is necessary to consider the patient’s personal characteristics (37). Precision medicine has been propelled forward by current large-scale tumor sequencing efforts and numerous therapeutic targets that have been identified as a result. Despite the reports of many successful cases based on DNA sequencing, the need for effective treatment remains significant for the majority of patients with cancer (38). However, genetic changes in a small percentage of coding regions do not adequately describe the complexities of cancer progression. Although DNA sequencing provides information about cancer drivers that are relatively well-preserved during cancer progression, it does not include the effects of other regulatory factors, such as epigenetic changes or non-coding regions, which are more dynamic and their relevance is more difficult to understand (39, 40). Therefore, a dynamic and versatile model system is needed to analyze tumor biology from multiple dimensions accurately and to reflect the behavior of the original tumor in the patient. The ability of tumor organoids to retain the original tumor characteristics makes them unique in studies at the individual level of patients with cancer, and they have been proposed as a model for precision medicine. For example, a study established a bank of living tumor samples from patients with advanced rectal cancer who received adjuvant chemo-radiotherapy. The combined clinical trial data confirmed that the chemo-radiotherapy response of the patients was a very close match with the organoid response of the rectal cancer, with an accuracy of 84.43%, a sensitivity of 78.01% and a specificity of 91.97% (41). Organoids have also successfully predicted the sensitivity of colorectal cancer (42), gastric cancer (43), ovarian cancer (44) and prostate cancer (45) to drugs. It appears at the present time that extracting samples from patient tumor tissues, creating and culturing organoids, exposing patient-derived tumor organoids to various drugs to find the best drug or drug combination to treat patients, revealing potential weaknesses of individual treatments based on gene mutation profiles and treatment responses of organoids and determining the next treatment route when first-line treatment is ineffective will become the ultimate mode of treatment for cancer (46, 47)1.
According to the latest literature and further analysis of keywords, the chemopreventive and therapeutic effects of antioxidant polyphenols on tumors were found to have received increasing attention from the scientific community in recent years. In an examination of antioxidant polyphenols, the complex technology of organoids as a new anti-cancer strategy has displayed unique advantages. Polyphenols that largely exist in plants serve as reactive oxygen species scavengers with prominent antioxidant activity, which exert the inhibitory effects on inflammation by manipulating the inflammation-related signaling pathways and suppressing the release of inflammatory mediators (48, 49). Not only contributing to treating non-neoplastic diseases, such as natural inducers from saffron targeting Nrf2/vitagene pathway to suppress oxidative stress and neuroinflammation and consequently inhibiting cognitive dysfunction (50), polyphenols could also play their roles in preventing and treating tumors by regulating cell apoptosis, autophagy, cell-cycle progression, inflammation, invasion, and metastasis through the activation of tumor suppressor genes and the inhibition of oncogenes (51, 52). Professor Scuto M’s team demonstrated that, polyphenols with low concentrations could suppress oxidative stress and inflammation, and induced the apoptotic of brain cancer cells by activating Nrf2/vitagene pathway (53), in addition, the supplementation of dietary polyphenols and vitamin D can also manipulate oxidative stress, inflammation and autophagy dysfunction in tumor cells through Nrf2/vitagene pathway, the low concentrated polyphenols and vitamin D could serve as cytotoxic agents of pro-oxidants, in the form of molecular activation through reactive oxygen species production and several survival pathways (e. g., glutathione) to induce apoptosis and cell-cycle arrest in tumor cells, for the chemoprevention and treatment of tumors in the presence of metal ions. Furthermore, the study has fully investigated a range of potential interventions in tumor organoid models depending on plant polyphenols and vitamin D, which is required to be introduced into clinical practice combined to powerful and novel technique through future interventions incorporating redox mesums of oxidative stress (54). The establishment of tumor model aims to approach 100% simulate the practical condition of the true tumor in the patient, so as to contribute to preclinical research and clinical application. Patient-derived organoids as a stable biology could facilitate gene operation, which is suitable for high-throughput sequencing analysis, and can reproduce the development from native tissue to the whole process of tumor occurrence (55). Currently some studies have taken tumor organoids as a model to explore the effect of polyphenols in tumor treatment, the results indicated that the low-dose polyphenols combined with chemotherapeutic drugs could not only optimize the efficacy, but also alleviate cytotoxicity and prevent the drug resistance (56). Another study with patient-derived organoids model revealed that the oligomeric proanthocyanidins combined to the curcumin attenuated the expression of cyclin D1, PCNA, and HSPA 5, which could serve as a method that preferentially targets cancer cells without affecting normal cells (57). The latest development of organoid models provides an excellent preclinical platform for the research of tumors, as well as makes the personalization of tumor therapy, evaluation of drug efficacy easier, contributing to the more in-depth exploration the mechanisms of cancer.
In summary, organoids mimic the structural and functional properties of native tumors, and compared to traditional tumor models, tumor organoids maintain gene stability while self-renewing and expanding over the long-term, and reflecting the genetic characteristics and tissue structure heterogeneity of the patients’ tumor tissues. In recent years, the study of tumor organoids has steadily increased, and effective organoid cultures have made it possible to screen individualized drugs within the time line of clinical treatment and apply them to translational medicine and individualized treatment. The use of organoids as precise and high-throughput preclinical tools for precision medicine is an unavoidable future trend. CiteSpace and VOSview software were used in this paper to analyze the bibliometrics and visualize the data derived from tumor organoid research.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
YS is responsible for conception design, article writing and proofreading. WS and ZX are responsible for literature collection, analysis and summary. CY and ZT are responsible for project guidance. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant number 81360531), the Natural Science Foundation of Guangxi Zhuang Autonomous Region, China (grant number 2020GXNSFAA238030), the Innovation Project of Guangxi Graduate Education of Guangxi University of Chinese Medicine, China (grant number YCSZ2022018), and the Top Talents Program of Guangxi University of Chinese Medicine, China (grant number: 2022C001).
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evauluate in this article, or claim that made by its manufactured, is nopt guaranteed or endorsed by the publisher.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1092870/full#supplementary-material
Web of Science Core Collection, WoSCC; United States of America, USA.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Kuenzi BM, Park J, Fong SH, Sanchez KS, Lee J, Kreisberg JF, et al. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell (2020) 38(5):672–84. doi: 10.1016/j.ccell.2020.09.014
3. Weber J, Rad R. Engineering CRISPR mouse models of cancer. Curr Opin Genet Dev (2019) 54:88–96. doi: 10.1016/j.gde.2019.04.001
4. Hansel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet (2020) 52(9):878–83. doi: 10.1038/s41588-020-0672-8
5. Yi SA, Zhang Y, Rathnam C, Pongkulapa T, Lee KB. Bioengineering approaches for the advanced organoid research. Adv Mater (2021) 33(45):e2007949. doi: 10.1002/adma.202007949
6. LeSavage BL, Suhar RA, Broguiere N, Lutolf MP, Heilshorn SC. Next-generation cancer organoids. Nat Mater (2022) 21(2):143–59. doi: 10.1038/s41563-021-01057-5
7. Bock C, Boutros M, Camp JG, Clarke L, Clevers H, Knoblich JA, et al. The organoid cell atlas. Nat Biotechnol (2021) 39(1):13–7. doi: 10.1038/s41587-020-00762-x
8. Synnestvedt MB, Chen C, Holmes JH. CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu Symp Proc (2005), 724–8.
9. van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics (2010) 84(2):523–38. doi: 10.1007/s11192-009-0146-3
10. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature (2011) 469(7330):415–8. doi: 10.1038/nature09637
11. Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell (2015) 160(1-2):324–38. doi: 10.1016/j.cell.2014.12.021
12. Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature (2015) 525(7568):251–5. doi: 10.1038/nature14966
13. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and barrett’s epithelium. Gastroenterology (2011) 141(5):1762–72. doi: 10.1053/j.gastro.2011.07.050
14. Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science (2013) 340(6137):1190–4. doi: 10.1126/science.1234852
15. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell (2014) 159(1):176–87. doi: 10.1016/j.cell.2014.08.016
16. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell (2015) 161(4):933–45. doi: 10.1016/j.cell.2015.03.053
17. Clevers H. Modeling development and disease with organoids. Cell (2016) 165(7):1586–97. doi: 10.1016/j.cell.2016.05.082
18. Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med (2017) 214(3):579–96. doi: 10.1084/jem.20162024
19. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (2018) 359(6378):920–6. doi: 10.1126/science.aao2774
20. Wilson HV. A new method by which sponges may be artificially reared. Science (1907) 25(649):912–5. doi: 10.1126/science.25.649.912
21. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature (2009) 459(7244):262–5. doi: 10.1038/nature07935
22. Duval K D, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiol (Bethesda) (2017) 32(4):266–77. doi: 10.1152/physiol.00036.2016
23. Sarin V, Yu K, Ferguson ID, Gugliemini O, Nix MA, Hann B, et al. Evaluating the efficacy of multiple myeloma cell lines as models for patient tumors via transcriptomic correlation analysis. Leukemia (2020) 34(10):2754–65. doi: 10.1038/s41375-020-0785-1
24. Wood LD, Ewald AJ. Organoids in cancer research: a review for pathologist-scientists. J Pathol (2021) 254(4):395–404. doi: 10.1002/path.5684
25. Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res (2013) 73(15):4885–97. doi: 10.1158/0008-5472.CAN-12-4081
26. Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discovery (2014) 4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001
27. Fontoura JC, Viezzer C, Dos Santos FG, Ligabue RA, Weinlich R, Puga RD, et al. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater Sci Eng C Mater Biol Appl (2020) 107:110264. doi: 10.1016/j.msec.2019.110264
28. Dame K, Ribeiro AJ. Microengineered systems with iPSC-derived cardiac and hepatic cells to evaluate drug adverse effects. Exp Biol Med (Maywood) (2021) 246(3):317–31. doi: 10.1177/1535370220959598
29. Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discovery Today (2013) 18(5-6):240–9. doi: 10.1016/j.drudis.2012.10.003
30. Mittal R, Woo FW, Castro CS, Cohen MA, Karanxha J, Mittal J, et al. Organ-on-chip models: Implications in drug discovery and clinical applications. J Cell Physiol (2019) 234(6):8352–80. doi: 10.1002/jcp.27729
31. Macedo MH, Araújo F, Martínez E, Barrias C, Sarmento B. iPSC-derived enterocyte-like cells for drug absorption and metabolism studies. Trends Mol Med (2018) 24(8):696–708. doi: 10.1016/j.molmed.2018.06.001
32. Zietek T, Boomgaarden W, Rath E. Drug screening, oral bioavailability and regulatory aspects: A need for human organoids. Pharmaceutics (2021) 13(8):1280. doi: 10.3390/pharmaceutics13081280
33. Wang H, Brown PC, Chow ECY, Ewart L, Ferguson SS, Fitzpatrick S, et al. 3D cell culture models: Drug pharmacokinetics, safety assessment, and regulatory consideration. Clin Transl Sci (2021) 14(5):1659–80. doi: 10.1111/cts.13066
34. Gupta R, Schrooders Y, Hauser D, van Herwijnen M, Albrecht W, Ter Braak B, et al. Comparing in vitro human liver models to in vivo human liver using RNA-seq. Arch Toxicol (2021) 95(2):573–89. doi: 10.1007/s00204-020-02937-6
35. Hedrich WD, Panzica-Kelly JM, Chen SJ, Strassle B, Hasson C, Lecureux L, et al. Development and characterization of rat duodenal organoids for ADME and toxicology applications. Toxicology (2020) 446:152614. doi: 10.1016/j.tox.2020.152614
36. Shinozawa T, Kimura M, Cai Y, Saiki N, Yoneyama Y, Ouchi R, et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology (2021) 160(3):831–46. doi: 10.1053/j.gastro.2020.10.002
37. Supplitt S, Karpinski P, Sasiadek M, Laczmanska I. Current achievements and applications of transcriptomics in personalized cancer medicine. Int J Mol Sci (2021) 22(3):1422. doi: 10.3390/ijms22031422
38. Cobain EF, Wu YM, Vats P, Chugh R, Worden F, Smith DC, et al. Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol (2021) 7(4):525–33. doi: 10.1001/jamaoncol.2020.7987
39. Perri F, Longo F, Giuliano M, Sabbatino F, Favia G, Ionna F, et al. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit Rev Oncol Hematol (2017) 111:166–72. doi: 10.1016/j.critrevonc.2017.01.020
40. van Galen P. Decoding the noncoding cancer genome. Cancer Discovery (2020) 10(5):646–7. doi: 10.1158/2159-8290.CD-20-0285
41. Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell (2020) 26(1):17–26. doi: 10.1016/j.stem.2019.10.010
42. Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discovery (2020) 10(8):1129–39. doi: 10.1158/2159-8290.CD-20-0187
43. Seidlitz T, Koo BK, Stange DE. Gastric organoids-an in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ (2021) 28(1):68–83. doi: 10.1038/s41418-020-00662-2
44. Tao M, Wu X. The role of patient-derived ovarian cancer organoids in the study of PARP inhibitors sensitivity and resistance: from genomic analysis to functional testing. J Exp Clin Cancer Res (2021) 40(1):338. doi: 10.1186/s13046-021-02139-7
45. Pamarthy S, Sabaawy HE. Patient derived organoids in prostate cancer: improving therapeutic efficacy in precision medicine. Mol Cancer (2021) 20(1):125. doi: 10.1186/s12943-021-01426-3
46. Veninga V, Voest EE. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell (2021) 39(9):1190–201. doi: 10.1016/j.ccell.2021.07.020
47. Yao J, Yang M, Atteh L, Liu P, Mao Y, Meng W, et al. A pancreas tumor derived organoid study: from drug screen to precision medicine. Cancer Cell Int (2021) 21(1):398. doi: 10.1186/s12935-021-02044-1
48. Kopustinskiene DM, Jakstas V, Savickas A, Bernatoniene J. Flavonoids as anticancer agents. Nutrients (2020) 12(2):457. doi: 10.3390/nu12020457
49. Kim HH, Ha SE, Vetrivel P, Bhosale PB, Kim SM, Kim GS, et al. Potential antioxidant and anti-inflammatory function of gynura procumbens polyphenols ligand. Int J Mol Sci (2021) 22(16):8716. doi: 10.3390/ijms22168716
50. Scuto M, Modafferi S, Rampulla F, Zimbone V, Tomasello M, Spano S, et al. Redox modulation of stress resilience by crocus sativus l. for potential neuroprotective and anti-neuroinflammatory applications in brain disorders: From molecular basis to therapy. Mech Ageing Dev (2022) 205:111686. doi: 10.1016/j.mad.2022.111686
51. Patra S, Pradhan B, Nayak R, Behera C, Das S, Patra SK, et al. Dietary polyphenols in chemoprevention and synergistic effect in cancer: Clinical evidences and molecular mechanisms of action. Phytomedicine (2021) 90:153554. doi: 10.1016/j.phymed.2021.153554
52. Arrigoni R, Ballini A, Santacroce L, Cantore S, Inchingolo A, Inchingolo F, et al. Another look at dietary polyphenols: Challenges in cancer prevention and treatment. Curr Med Chem (2022) 29(6):1061–82. doi: 10.2174/0929867328666210810154732
53. Scuto M, Ontario ML, Salinaro AT, Caligiuri I, Rampulla F, Zimbone V, et al. Redox modulation by plant polyphenols targeting vitagenes for chemoprevention and therapy: Relevance to novel anti-cancer interventions and mini-brain organoid technology. Free Radic Biol Med (2022) 179:59–75. doi: 10.1016/j.freeradbiomed.2021.12.267
54. Scuto M, Trovato SA, Caligiuri I, Ontario ML, Greco V, Sciuto N, et al. Redox modulation of vitagenes via plant polyphenols and vitamin d: Novel insights for chemoprevention and therapeutic interventions based on organoid technology. Mech Ageing Dev (2021) 199:111551. doi: 10.1016/j.mad.2021.111551
55. Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U.S.A. (2015) 112(43):13308–11. doi: 10.1073/pnas.1516689112
56. Ye HS, Gao HF, Li H, Nie JH, Li TT, Lu MD, et al. Higher efficacy of resveratrol against advanced breast cancer organoids: A comparison with that of clinically relevant drugs. Phytother Res (2022) 36(8):3313–24. doi: 10.1002/ptr.7515
Keywords: tumor organoids, bibliometric analysis, drug screening, precise medicine, antioxidant polyphenols, CiteSpace, VOSviewer
Citation: Shuoxin Y, Shuping W, Xinyue Z, Tao Z and Yuanneng C (2023) Progress of research on tumor organoids: A bibliometric analysis of relevant publications from 2011 to 2021. Front. Oncol. 13:1092870. doi: 10.3389/fonc.2023.1092870
Received: 08 November 2022; Accepted: 04 January 2023;
Published: 26 January 2023.
Edited by:
Michael Paul Kim, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Maria Concetta Scuto, University of Catania, ItalyCopyright © 2023 Shuoxin, Shuping, Xinyue, Tao and Yuanneng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Yuanneng, Y3luNjA2NjhAYWxpeXVuLmNvbQ==; Zhang Tao, emhhbmd0YW80MUBhbGl5dW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.