- 1The Department of Surgery, the First Dongguan Affiliated Hospital of Guangdong Medical University, Guangdong, China
- 2The Department of Surgery, Guangzhou Medical University, Guangdong, China
Background: It has been reported that ING3 inhibits the progression of various cancers. However, some studies have shown that it promotes the development of prostate cancer. The purpose of this study was to investigate whether ING3 expression is associated with the prognosis of patients with cancer.
Materials and methods: PubMed, Cochrane Database, Embase, Medline, ScienceDirect, Scopus and Web of Science were searched until September 2022. The hazard ratio (HR)/odds ratio (OR) and 95% confidence interval (95% CI) were calculated using Stata 17 software. We used the Newcastle-Ottawa Scale (NOS) to assess the risk of bias.
Result: Seven studies involving 2371 patients with five types of cancer were included. The results showed that high expression of ING3 was negatively associated with a more advanced TNM stage (III-IV vs. I-II) (OR=0.61, 95% CI: 0.43-0.86), lymph node metastasis (OR=0.67, 95% CI: 0.49-0.90) and disease-free survival (HR=0.63, 95% CI: 0.37-0.88). However, ING3 expression was not associated with overall survival (HR=0.77, 95% CI: 0.41-1.12), tumor size (OR=0.67, 95% CI: 0.33-1.37), tumor differentiation (OR=0.86, 95% CI: 0.36-2.09) and gender (OR=1.14, 95% CI: 0.78-1.66).
Conclusion: This study showed that the expression of ING3 was associated with better prognosis, suggesting that ING3 may be a potential biomarker for cancer prognosis.
Systematic review registration: https://www.crd.york.ac.uk/prospero/, identifier (CRD42022306354).
Introduction
The ING family, including ING1, ING2, ING3, ING4 and ING5, is a group of type II tumor suppressors. The ING family is involved in DNA repair, chromatin remodeling, cell cycle control, senescence and apoptosis through multiple mechanisms (1–4). The highly conserved plant homeodomain (PHD) is considered to be the main domain of ING protein (5, 6). PHD domains play a key role in the function of epigenetic regulatory proteins, which are known as chromatin remodeling factors and perform a function in gene expression (7). The epigenetic regulatory protein is added at lysine 4 (H3K4me3) after binding to histone H3 with 3 methyl groups through the PHD domains. H3K4me3 is a biomarker of active transcription, in this way ING proteins participate in cell growth regulation (8, 9).
ING3 encodes a 418 amino acid protein located on chromosome 7q31 with a nuclear localization sequence and a conserved carboxy-terminal PHD, the latter of which is thought to play an important role in inhibiting tumorigenic processes (10–13). ING3 has long been considered as a tumor suppressor gene because of its biological functions in limiting cell growth, controlling cell cycle arrest, and initiating apoptosis in a P53-dependent pathway (14). However, Nabbi et al. found that ING3 promotes cell proliferation and functions as an oncogene in prostate cancer (15). In addition, according to another study, ING3 is necessary for the growth of breast and ovarian cancer cells (16). Unlike ING1 and ING2, which can act as AR corepressors to inhibit androgen signaling, ING3 can act as an AR coactivator and thus play a role in prostate cancer (17, 18).
Due to the contradictory findings above, the predictive function of ING3 remains unknown in most investigations. Therefore, the purpose of this study was to determine whether ING3 expression is associated with cancer prognosis. In this study, we performed a systematic review and meta-analysis to summarize the results of global studies predicting the clinical outcomes of ING3 expression in cancer patients.
Materials and methods
This study was designed and implemented according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) criteria (19). This study aimed to determine whether ING3 expression is associated with cancer prognosis. The study was registered in the International Systematic Review Prospective Register. (PROSPERO 2022 CRD42022306354).
Selection criteria
Inclusion and exclusion criteria were based on Population, Intervention, Comparison, Outcome (PICO) criteria.
The following criteria were used to determine the eligibility for candidate studies: (1) All included patients were diagnosed with cancer by established means. (2) A well-established molecular approach, such as PCR, was used to determine the expression level of ING3 in the included patients. (3) The included literature provided data that could be used for prognostic analysis such as HR and 95% CI for overall survival (OS)/disease-free survival (DFS), or data that could be used to estimate HR and 95% CI, such as Kaplan-Meier survival curves. (4) The study provided an association between ING3 expression and clinicopathological characteristics, such as gender, lymph node metastasis, TNM stage, tumor size, and tumor differentiation.
Articles that met the following criteria were excluded: (1) Clinicopathological characteristics (such as gender, lymph node metastasis, TNM stage, etc.) were not provided in the article. (2) Non-clinical experiments, such as in vivo or in vitro experiments. (3) Articles were review articles, case reports, letters, clinical guidelines, etc. (4) The article did not provide or could not extract HR and 95% CI. (5) For studies with multiple publications, the study that provided the most data was selected.
All articles that met the inclusion criteria and did not meet the exclusion criteria were read in full. Articles that had been read in full text and met the criteria underwent data extraction. All publications were independently screened by two authors (Li, Xu), and any disagreements were resolved after discussion between the two authors or by consulting a third reviewer (Chen).
Literature search
We conducted a literature search on the relationship between ING3 expression and patient prognosis up to September 2022. The following databases were searched: PubMed, Cochrane database, Embase, Medline, ScienceDirect, Scopus, and Web of Science. The ClinicalTrials was also searched for any related trials. No filters were used in the search process. We also searched Google Scholar to see if there was any grey literature.
The following keywords were used for the search: "tumour*", " tumor*", " malignan*", " carcinom*", "neoplas*", "cancer*", "ING 3", "ING3", "inhibitor of growth 3", "inhibitor of growth protein 3", "ING protein" 3".
Taking PubMed as an example, the specific search strategies were as follows:
#1 ING3[Title/Abstract] OR ING 3[Title/Abstract] OR inhibitor of growth 3[Title/Abstract] OR Inhibitor of growth protein 3[Title/Abstract] OR ING protein 3[Title/Abstract]
#2 "Neoplasms"[Mesh]
#3 tumour*[Title/Abstract] OR tumor*[Title/Abstract] OR malignan*[Title/Abstract] OR carcinom*[Title/Abstract] OR neoplas*[Title/Abstract] OR cancer*[Title/Abstract]
#4 #2 OR #3
#5 #1 And #4
We used Endnote X9 software to exclude duplicate documents. We first excluded articles with the same author, year, title, journal, volume, issue and page, and then gradually narrowed the criteria until only articles with the same title were excluded. Each time we double-checked the excluded articles, we believed that by doing so we could avoid excluding any articles relevant to this study.
Data extraction
The following data were extracted from the study: author, year, tumor type, sample size, country, follow-up time, lymph node metastasis, TNM stage, tumor size, tumor differentiation, the cut-off value of hazard ratios (HRs) and 95% confidence intervals (CIs) of ING3 for overall survival (OS) and disease-free survival (DFS). All data were extracted separately by two authors (Li, Xu).
Quality assessment
The Newcastle-Ottawa Scale (NOS) was used by two authors (Li, Xu) to assess the quality of each study (20), and any disagreements were resolved through mutual discussion or consultation with a third reviewer (Chen). Object selection, comparability and outcome were the most important evaluation items. Quality evaluation scores ranged from 0 (lowest) to 9 (highest); studies with NOS scores greater than 6 were regarded as high quality.
Statistical analysis
HR is the best performance indicator for measuring time-to-event outcomes (21). The ING3 expression was treated as a binary variable for the sake of this study (high expression vs low expression). To obtain an overall estimate for the DFS and OS analyses, the HRs and 95% CIs were calculated for each study. HRs and 95% CIs were extracted directly from the included studies or transformed from Kaplan-Meier survival curves using the digitizing program Engauge Digitizer (22). We used two endpoints in this study. The primary endpoint was the relationship between ING3 expression and overall survival (OS) and disease-free survival (DFS). The secondary endpoint was the relationship between ING3 expression and clinicopathological features, such as gender, lymph node metastasis, TNM stage, tumor size, and tumor differentiation. Due to conflicting findings in prostate cancer, to explore whether ING3 plays a different role in prostate cancer, we decided to perform a subgroup analysis by cancer type (prostate cancer vs. other) before the study started. To detect statistical heterogeneity between various studies, the Higgin's I2 statistics and the Cochran's Q test were used (23). Heterogeneity was considered statistically significant at I2>50%. When I2>50%, a random-effects model was used to evaluate the pooled results; otherwise, a fixed-effects model was used to analyze the pooled results. The combined HRs and 95% CIs from each included study were used to assess the effect of ING3 expression on prognosis. In general, a combined HR > 1 is considered to indicate a significant association with poor prognosis, and a combined HR < 1 indicates a better prognosis. A sensitivity analysis was performed to assess the stability of the combined data and to pinpoint the cause of any heterogeneity. If the results are consistent, they are considered robust; otherwise, they will be regarded with caution. Begg's and Egger's linear regression tests were used to investigate publication bias. All the above analyses were performed using STATA 17.0 and Review Manager 5.4.
Result
Study selection
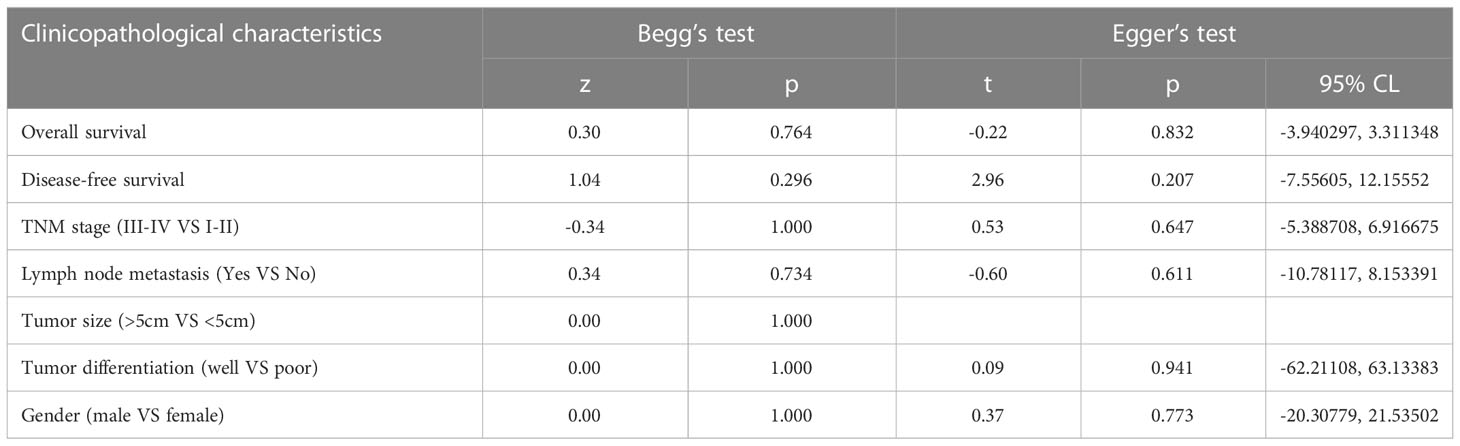
As shown in the literature search flow chart (Figure 1), up to September 2022, there were 357 literature from PubMed, Cochrane Database, Embase, Medline, ScienceDirect, Scopus and Web of Science. No relevant studies were found on the trial registration website (clinicaltrials.gov) or Google Scholar. Among these studies, 241 were identified as duplicates and subsequently excluded. 102 publications were excluded after reading the titles and abstracts. Five publications were excluded after reading the full text, including three that did not provide relevant outcomes (12, 24, 25) and two that did not fully meet the inclusion criteria (26, 27). Finally, seven publications were included in the study based on the inclusion and exclusion criteria (14, 15, 28–32).
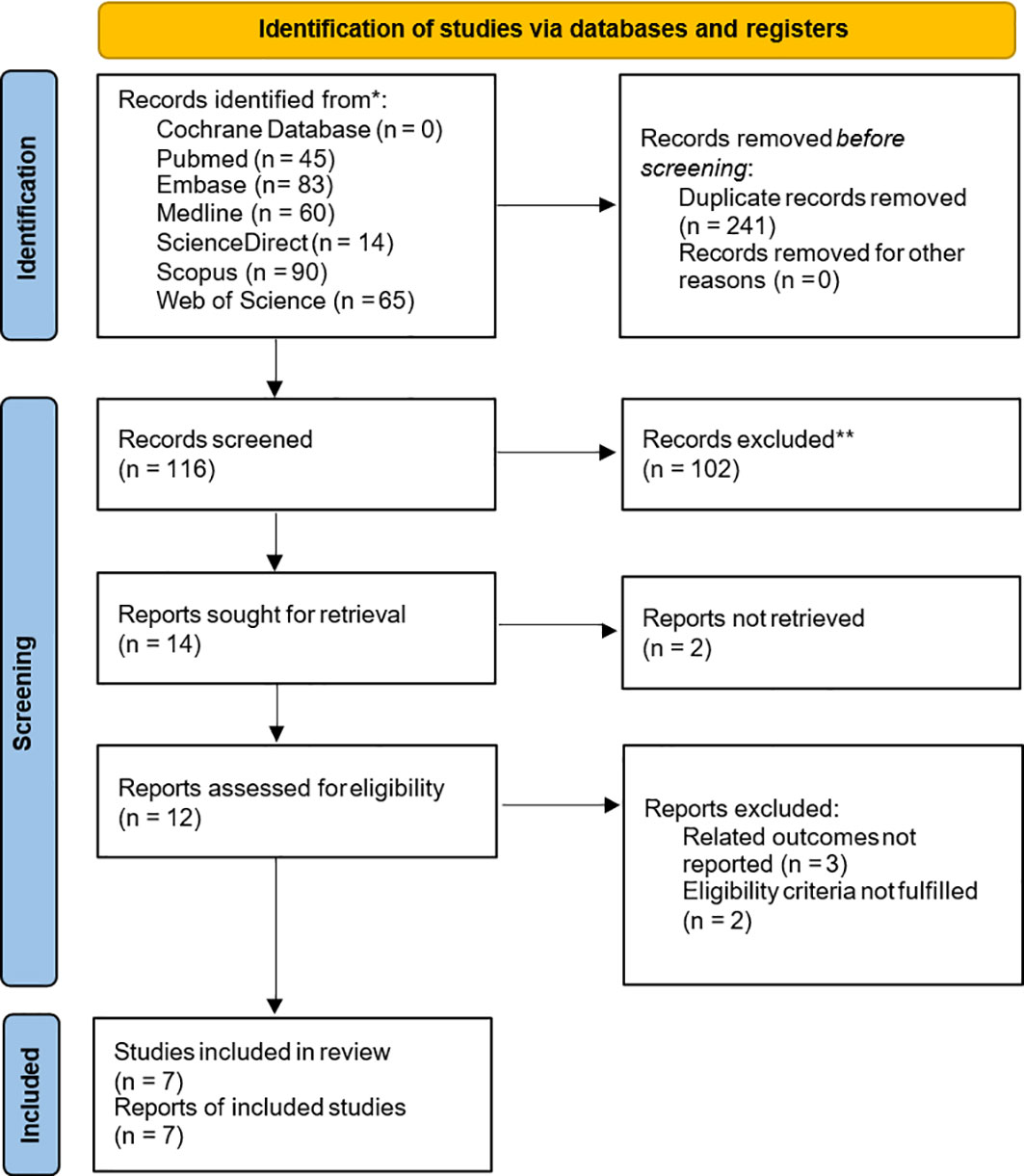
Figure 1 Flowchart of literature search and study selection. Flowchart of literature search and study selection. *Consider, if feasible to do so, reporting the number of records identified from each database or register searched (rather than the total number across all databases/registers). **If automation tools were used, indicate how many records were excluded by a human and how many were excluded by automation tools.
Characteristics of the included studies and quality evaluation
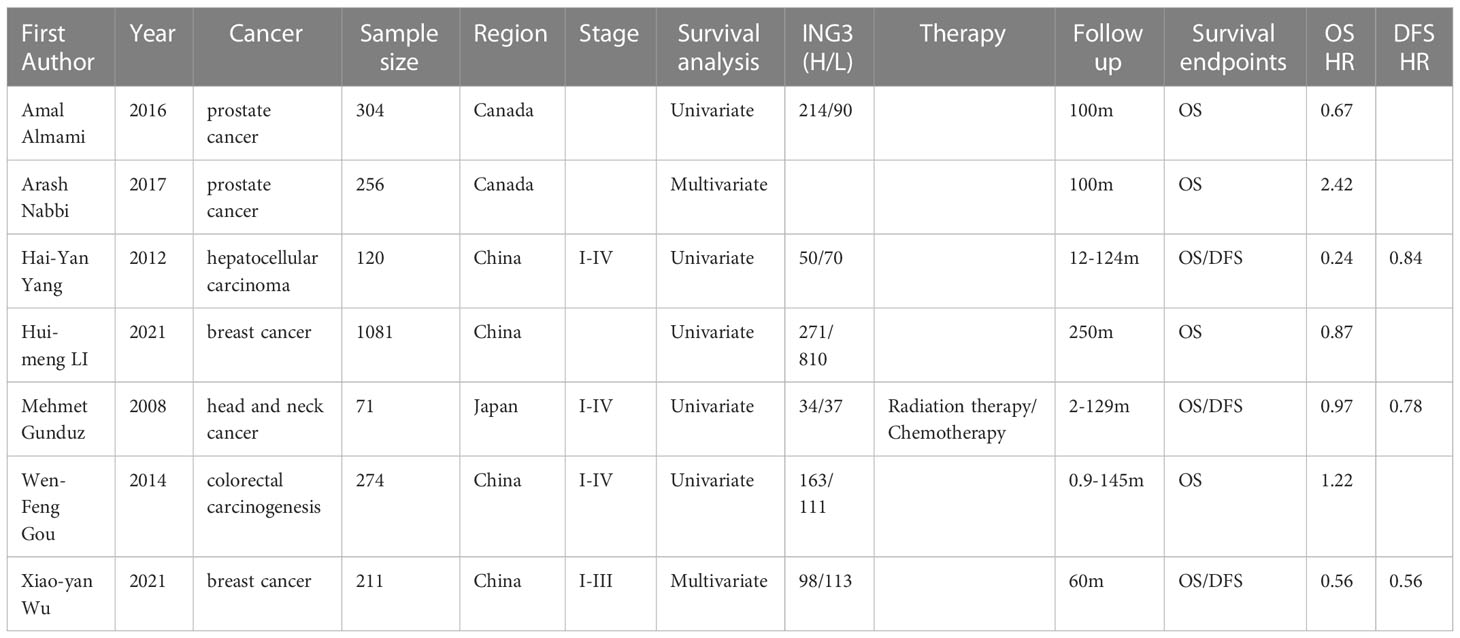
Among the eligible studies, two were related to prostate cancer (15, 30), two were related to breast cancer (28, 29), and the remaining three were related to hepatocellular carcinoma (31), head and neck cancer (32), and colorectal cancer (14). The majority of studies (n = 4) were conducted in China, followed by Canada (n = 2) and Japan (n = 1). Three studies provided HRs and 95% CIs in the article, and for the remaining studies, we derived HRs and 95% CIs from Kaplan-Meier curves. The systematic review and meta-analysis comprised 2317 patients from seven selected publications, with sample sizes ranging from 71 to 1081. Table 1 summarizes the characteristics of the seven published studies. All patients with malignancy were diagnosed based on histology. Tissue samples were obtained from tumors and adjacent normal tissues, and ING3 expression was evaluated by qRT-PCR.
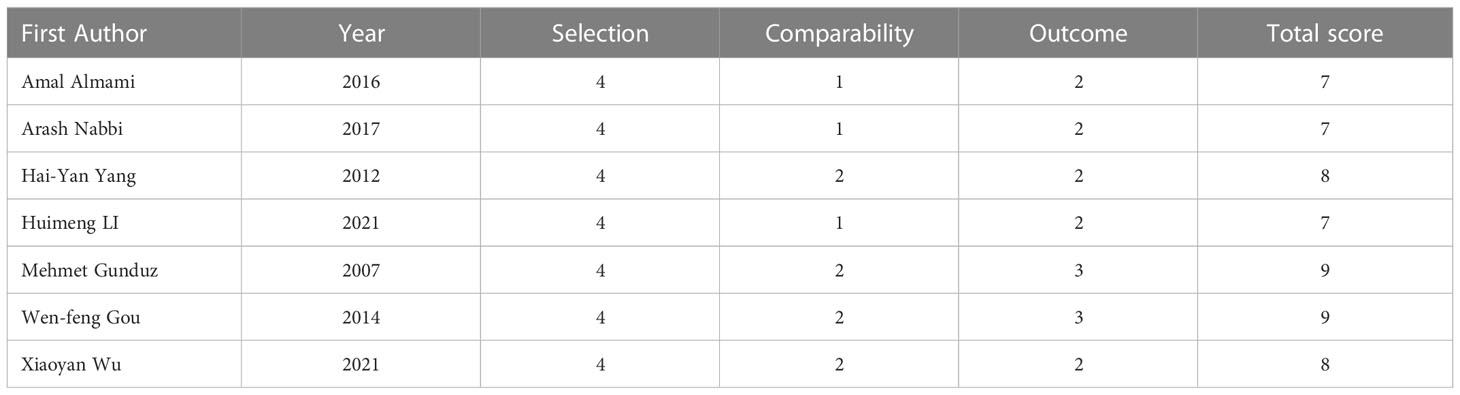
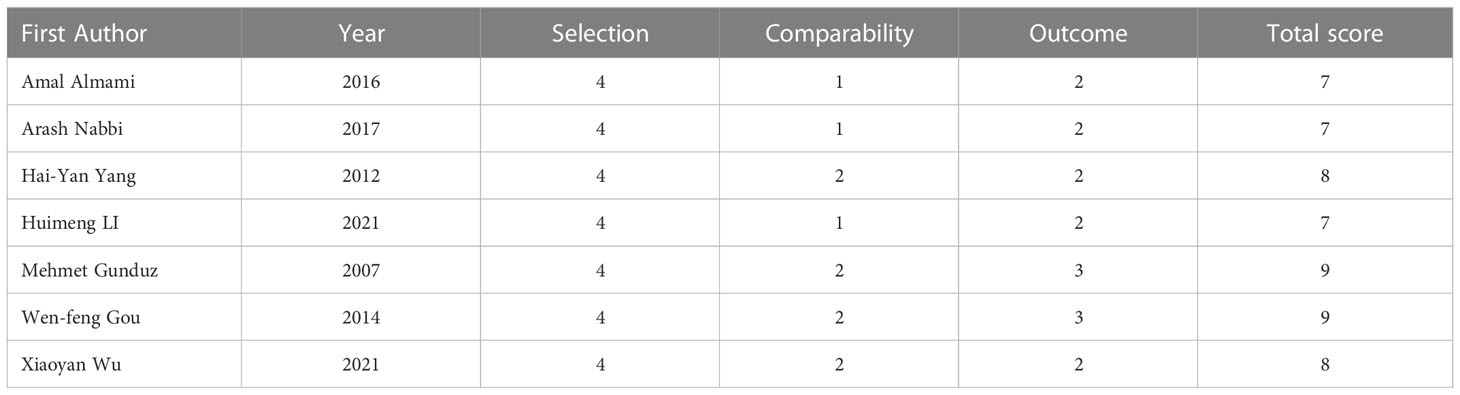
All included articles were assessed using the NOS scale, with scores ranging from 7 to 9. Three publications had a NOS score of 7, two publications had a NOS score of 8, and two publications had a NOS score of 9, for a total of seven publications with a median score of 8 (Table 2).
Primary endpoint: Relationship of ING3 to DFS and OS
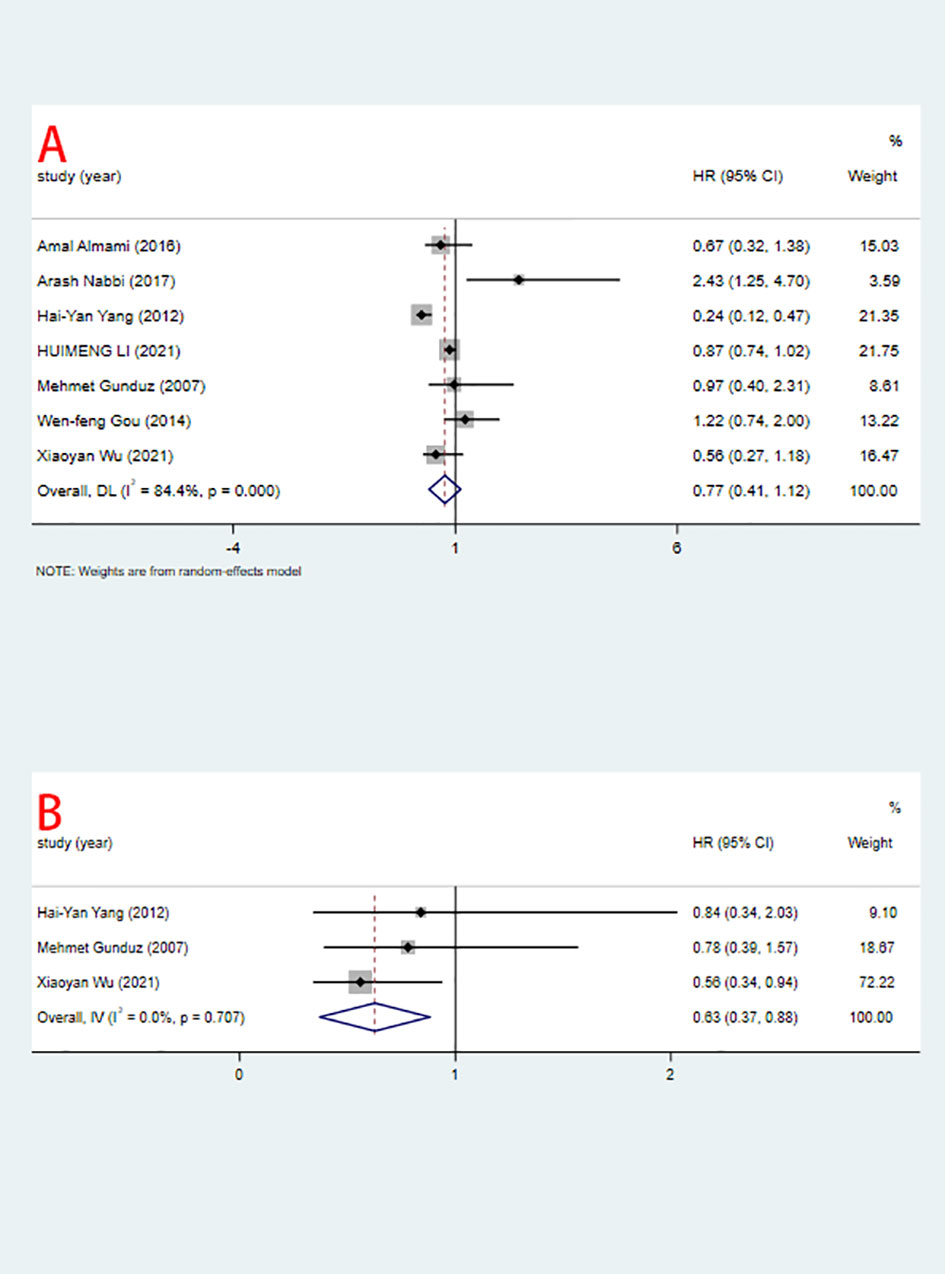
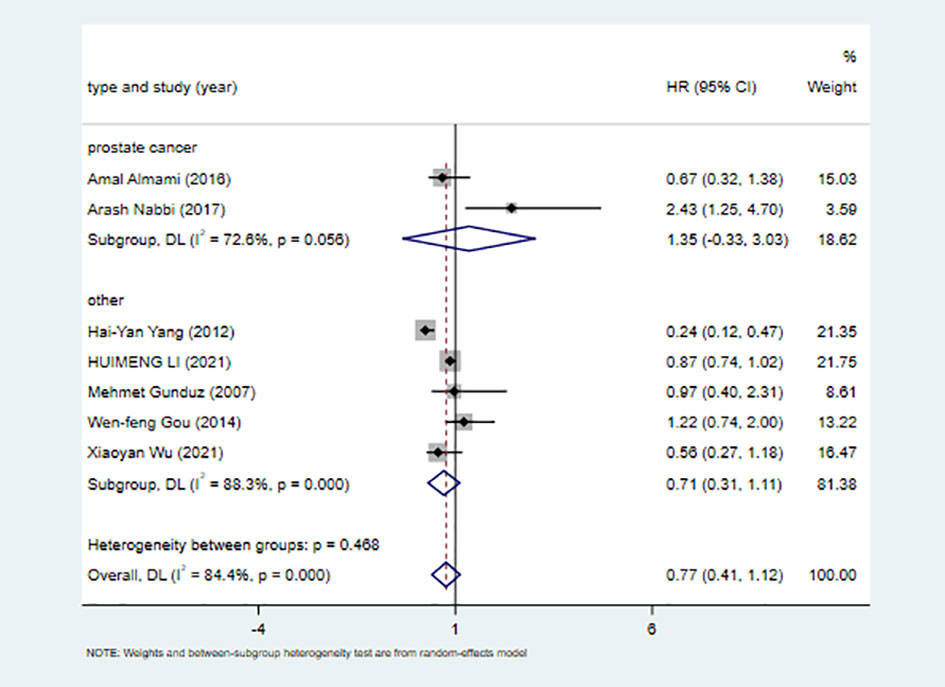
Seven studies including 2371 patients reported HRs for OS based on ING3 expression levels. As shown in Figure 2A and Table 3, given the significant degree of heterogeneity (I2=84.4%), a random-effects model was used. In most studies, ING3 expression seemed to be associated with a better prognosis; however, since their 95% CI crossed the null effect line (HR=1), these results lacked statistical significance. In one of these studies, high expression of ING3 indicated poorer overall survival, while in another study, high expression of ING3 indicated a better prognosis. In the remaining studies, no relationship was found between ING3 expression and patient prognosis, as the range of 95% CI crossed 1. The combined data showed that ING3 expression was not associated with prognosis in these patients, and the pooled HR for OS was 0.77 (95% CI: 0.41–1.12, P = 0).
Three studies including 402 patients reported HRs for DFS. As shown in Figure 2B and Table 3. Due to the low heterogeneity (I2=0%), a fixed-effects model was used for the analysis. The combined data showed that high expression of ING3 indicated a better prognosis, with a pooled HR for DFS of 0.63 (95% CI: 0.37-0.88, P = 0). This implies that in individuals with malignancies, increased expression of ING3 may lead to improved clinical outcomes, while low expression of ING3 corresponds to a poor prognosis.
Secondary endpoints: Relationship of ING3 to clinicopathological features
In the seven included studies, the correlation between ING3 expression and clinicopathological features is shown in Figure 3 and Table 3. We found that high expression of ING3 was negatively associated with TNM stage 3-4 (OR=0.61, 95% CI: 0.43-0.86) and lymph node metastasis (OR=0.67, 95% CI: 0.49-0.90). This means that in patients with low ING3 expression, tumors are more likely to progress to advanced stages or have lymph node metastases. Usually, this means a poor prognosis and leads to a poor clinical outcome. In the analysis of tumor size (OR=0.67, 95% CI: 0.33-1.37), tumor differentiation (OR=0.86, 95% CI: 0.36-2.09) and gender (OR=1.14, 95% CI: 0.78-1.66), they were all found to be unrelated to ING3 expression.
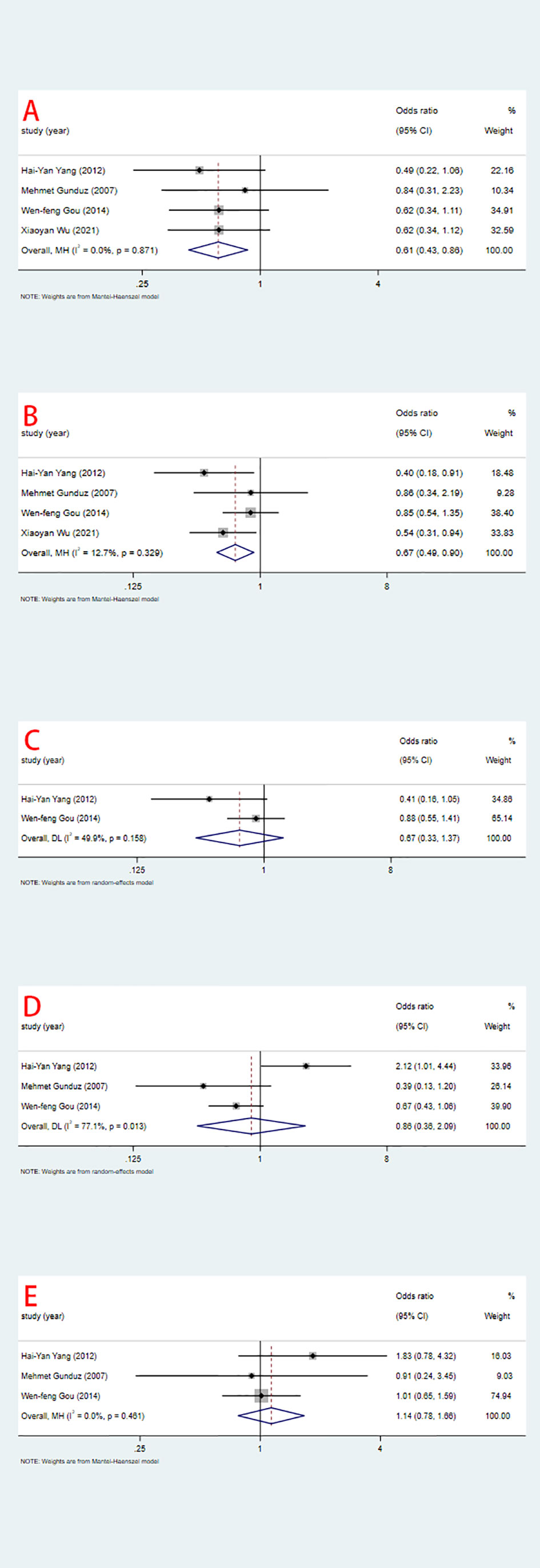
Figure 3 Relationship between ING3 expression and TNM stage (A), lymph node metastasis (B), tumor size (C), tumor differentiation (D) and gender (E).
Subgroup analysis
To investigate whether ING3 plays a different role in prostate cancer, we performed a subgroup analysis by cancer type. The results of the subgroup analysis are shown in Figure 4 and Table 4. We found a similar result in different types of cancer. There was significant heterogeneity in both subgroups, and overall survival was not associated with ING3 expression in either prostate cancer or other cancers.

Table 4 Subgroup analysis for the relationship between high ING3 expression and survival of cancer patients.
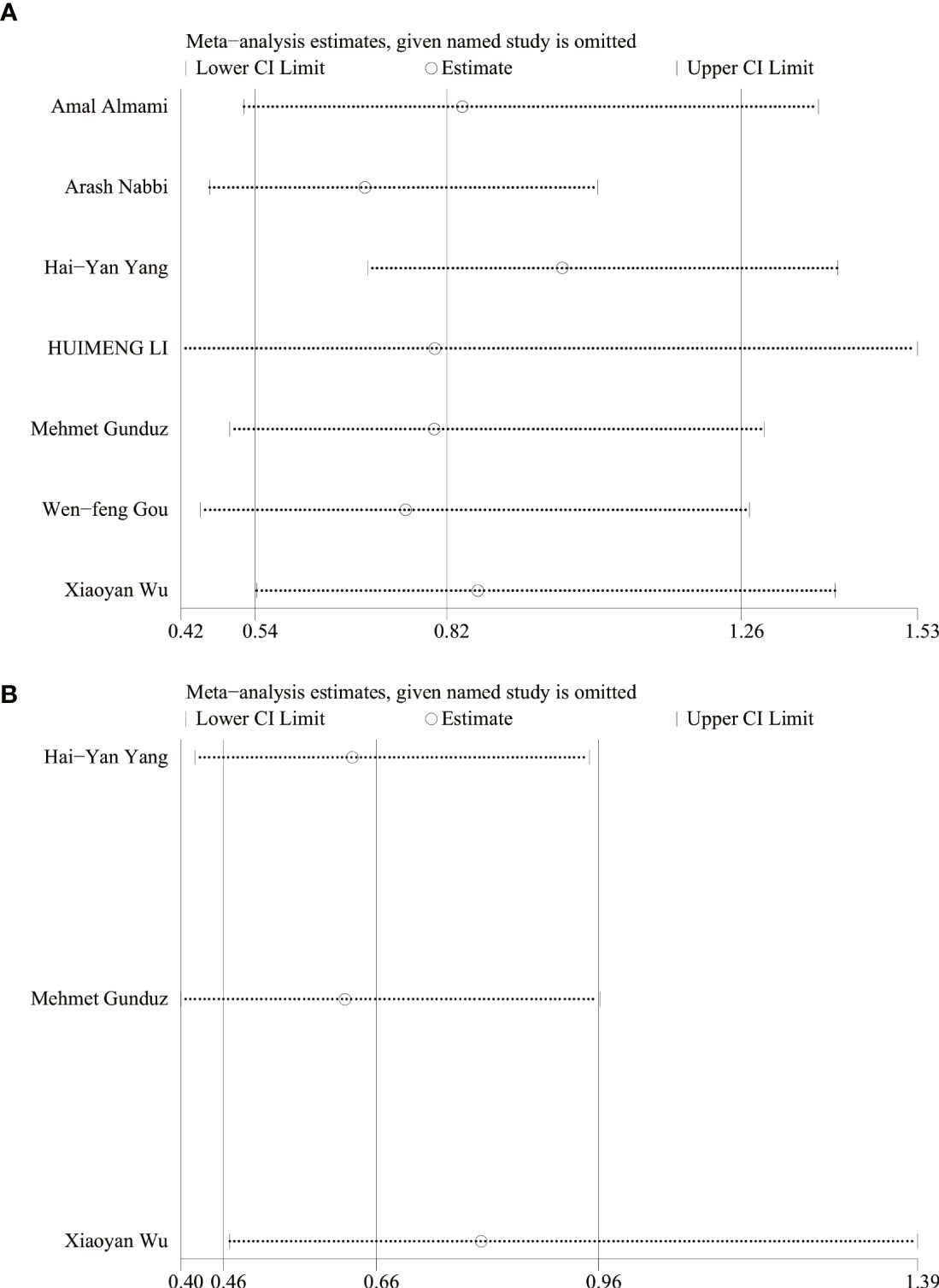
Analysis of sensitivity
Sensitivity analysis was performed using STATA 17 software. The purpose of the sensitivity analysis was to determine whether any single study affected the overall results. We found that removing studies one by one did not significantly change the results, suggesting that the overall results were not influenced by individual studies. The HRs and 95% CIs varied in the range from 0.70 (95% CI: 0.47–1.05) to 0.99 (95% CI: 0.70–1.41) for overall survival (Figure 5A), and from 0.61 (95% CI: 0.40–0.96) to 0.80 (95% CI: 0.46–1.39) for disease-free survival (Figure 5B). The ORs and 95% CIs varied in the range from 0.58 (95% CI: 0.40–0.85) to 0.64 (95% CI: 0.44–0.95) for TNM stage (Figure 6A), from 0.54 (95% CI: 0.36–0.82) to 0.73 (95% CI: 0.51–1.05) for lymph node metastasis (Figure 6B), from 0.54 (95% CI: 0.36– 0.82) to 0.73 (95% CI: 0.51–1.05) for lymph node metastasis (Figure 6B), from 0.41 (95% CI: 0.16–1.05) to 0.88 (95% CI: 0.55–1.40) for tumor size (Figure 6C), from 0.62 (95% CI: 0.41– 0.95) to 1.15 (95% CI: 0.37–3.52) for tumor differentiation (Figure 6D), from 1.00 (95% CI: 0.65–1.54) to 1.50 (95% CI: 0.73–3.06) for gender (Figure 6E).
Figure 6 Sensitivity analysis for relationship between ING3 expression and TNM stage (A), lymph node metastasis (B), tumor size (C), tumor differentiation (D) and gender (E).
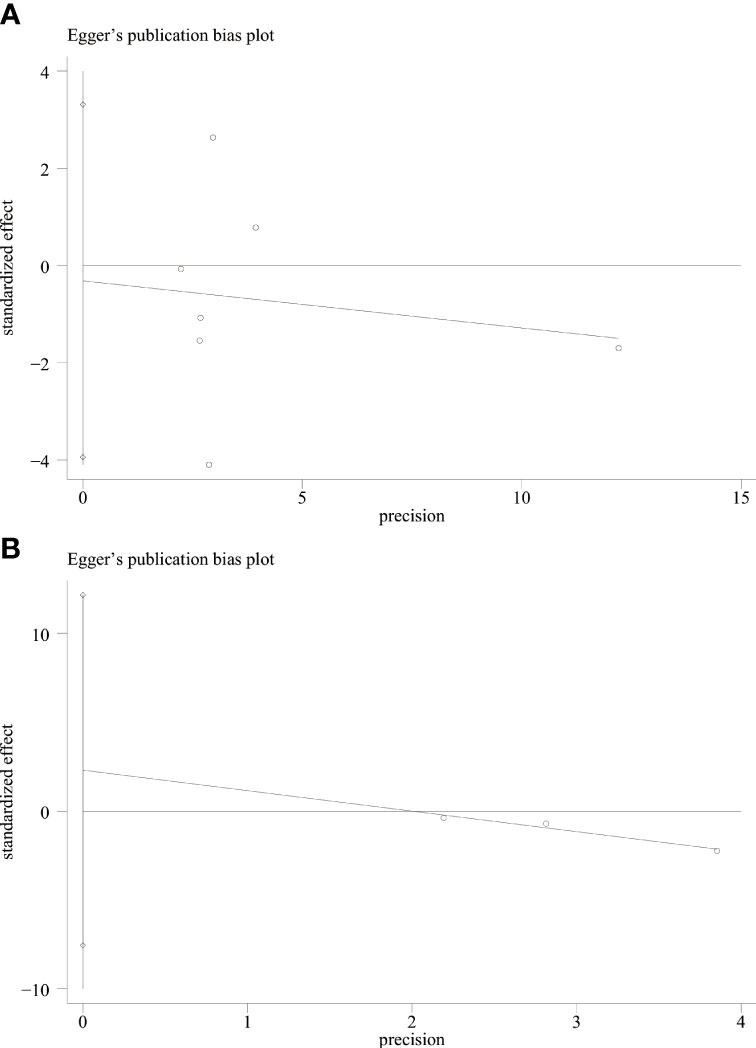
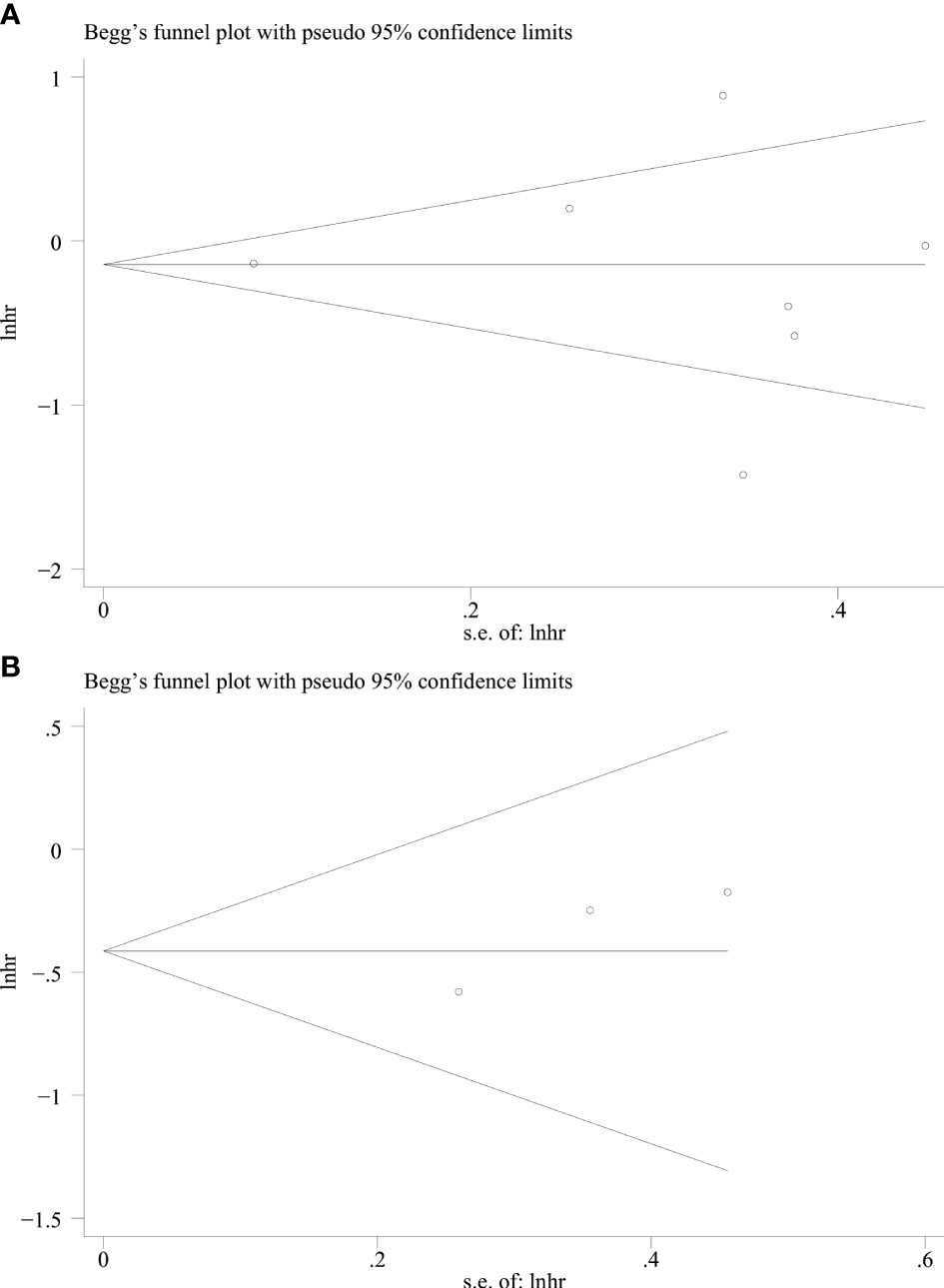
Publication bias test
We used Begg’s and Egger’s linear regression tests to detect any publication bias. The results of the Egger’s test are shown in (Figure 7). No asymmetry was observed in the funnel plot in the assessment of ING3 expression and OS (Figure 8A), DFS (Figure 8B), TNM stage (Figure 9A), lymph node metastasis (Figure 9B), tumor size (Figure 9C), tumor differentiation (Figure 9D), and gender (Figure 9E). No publication bias was observed in assessing ING3 expression and OS, DFS, TNM stage, lymph node metastasis, tumor size, tumor differentiation, and gender according to Begg’s test (P = 0.764, P = 0.296, P = 1.000, P = 0.734, P = 1.000, P = 1.000, and P = 1.000, respectively). A similar conclusion can be derived from Egger’s test (P=0.832, P=0.207, P=0.647, P=0.611, P=0.941, and P=0.773, respectively). Table 5 shows the detailed data for Begg’s test and Egger’s test.
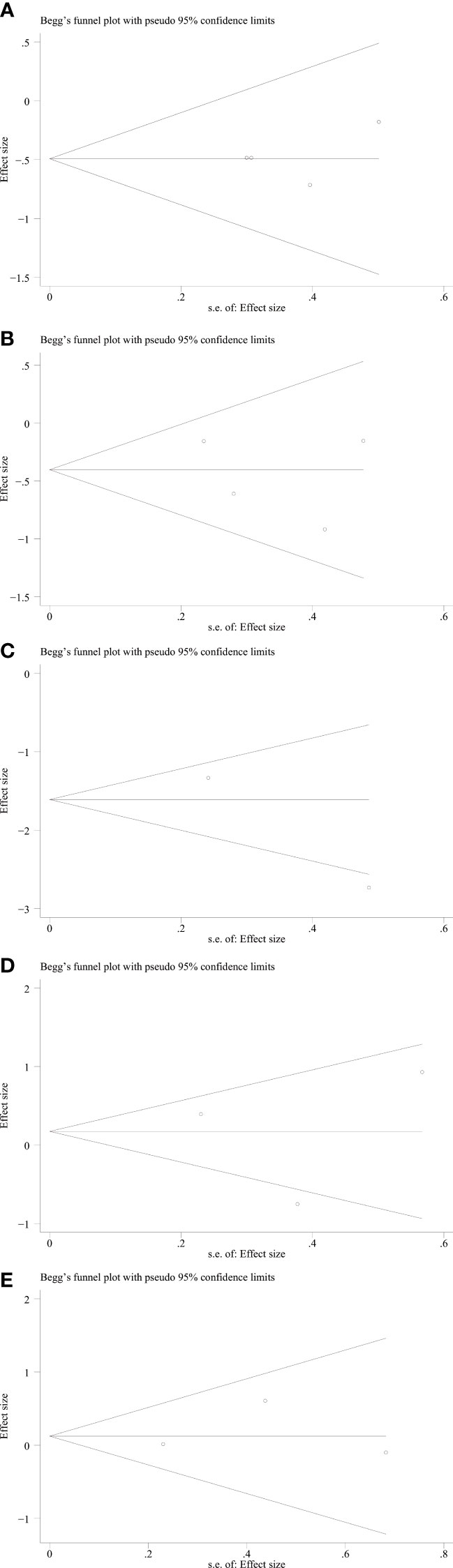
Figure 9 Begg’s funnel plot for relationship between ING3 expression and TNM stage (A), lymph node metastasis (B), tumor size (C), tumor differentiation (D) and gender (E).
Discussion
Cancer is becoming a growing public health problem worldwide. In 2021, the United States is anticipated to have 1,898,160 new cancer cases and 608,570 cancer deaths, making cancer the second leading cause of death in the country (33). Although the prognosis of cancer patients has improved substantially due to advances in medical therapy, the prognosis of the majority of patients remains dismal. One explanation is that most tumors have already progressed or metastasized by the time they are detected, which makes treatment extremely challenging. Another explanation is that oncologists may overestimate cancer patients’ prognoses, resulting in delayed or even missed conversations regarding patients’ goals and ultimately poor end-of-life care (34).
Cancer biomarkers can be classified into three types: predictive biomarkers, prognostic biomarkers and diagnostic biomarkers. Prognostic biomarkers can guide individualized treatment, assess efficacy, and optimize follow-up (35). Several prognostic biomarkers have been discovered, such as CA19-9 (36), MIR4435-2HG (37), AFP (38), and CEA (39). In recent years, with the development of technology, especially the advent of liquid biopsy (40), circulating tumor DNA (ctDNA) has become a new biomarker for the diagnosis of cancer (41, 42). However, the specificity and sensitivity of prognostic biomarkers remain low (43), and few biomarkers have entered clinical practice (35). Hence, there is an urgent need to discover prognostic biomarkers that might assist doctors in early cancer diagnosis and treatment (44, 45).
ING3 is the most unique member of the ING family. Unlike the high similarity exhibited between ING1, ING2, ING4 and ING5, ING3 has the least similarity to the others (46). Previous studies have reported that ING3 expression varies in different types of cancer. Almost all in vitro studies have shown that ING3 has the ability to inhibit growth (47), however, it was found that ING3 can function as an oncogene in prostate cancer (30). Nabbi et al. reported that ING3 may act as a coactivator of androgen receptor in prostate cancer (15). Since androgens can stimulate prostate cancer cell growth, this may lead to prostate cancer progression. Due to the controversial findings of existing studies, we conducted this meta-analysis and systematic review to investigate what role ING3 actually plays in cancer.
This systematic review and meta-analysis included seven studies involving 2,371 patients and five types of cancer. Our findings showed that high expression of ING3 was negatively associated with disease-free survival (HR=0.63, 95% CI: 0.37-0.88), a more advanced TNM stage (III-IV vs. I-II) (OR=0.61, 95% CI: 0.43-0.86) and lymph node metastasis (OR=0.67, 95% CI: 0.49-0.90). In other words, reduced expression of ING3 may lead to faster cancer progression, greater lymph node metastasis, and worse disease-free survival, all of which usually lead to poor prognosis. In the analysis of the relationship between ING3 expression and OS, we found no relationship between ING3 expression and OS, and a significant heterogeneity was observed. We assumed that the heterogeneity might come from the different types of cancer included in the study. We also found that ING3 expression did not correlate with clinicopathological data such as tumor size, tumor differentiation and gender. In the subgroup analysis, we found no significant difference between prostate cancer and other cancers. However, given the small number of included studies, this finding should be interpreted with caution. Sensitivity analysis showed that the pooled results were not affected by any single study, and after removing individual studies, the conclusions remained consistent with the main study findings. Begg’s test and Egger’s test showed no significant publication bias.
After considering all the results together, we conclude that ING3 expression predicts a better prognosis for cancer patients and can be used as a prognostic biomarker. ING3 may inhibit cancer progression through its PHD domains. PHD domains play a key role in the function of epigenetic regulatory proteins, which are known as chromatin remodeling factors and perform a function in the expression of genes (7). Malakhov et al. found that knockdown of the PHD domain resulted in epithelial-to-mesenchymal transition (EMT) and cellular senescence in LNCaP prostate cancer cell lines, suggesting that ING3 could limit the diffusion of cancer cells (48). ING3 may inhibit other types of cancer in the same way. In addition, the nucleolar translocation sequence (NTS) can aid in the translocation of ING proteins when DNA is damaged, and when it is mutated, it leads to a decrease in the level of apoptosis (49). Furthermore, ING3 expression was found to be associated with the expression levels of p300, p21 and acetylated p53. The interaction between ING3 and p300 can lead to the upregulation of acetylated p53 and induce cell cycle arrest and senescence associated with p53 (50). Although ING3 can act as a coactivator of the androgen receptor and thus contribute to the development of prostate cancer, it can still inhibit the progression of other types of cancer because they are rarely affected by androgens.
However, this study still has limitations. First, several HRs and 95% Cls were calculated using Kaplan-Meier survival curves, which may deviate from the actual situation. In addition, the number of included studies was small, and the number of studies was even smaller when it came to each type of cancer, so the results for some cancers lacked statistical significance. Finally, our study included different tumors and different treatments, due to the different pathogenesis of different cancers, which may lead to great heterogeneity.
Cancer biomarkers are key to the discovery and development of new cancer therapies. They are also an important factor in clinical practice as they can be used for risk assessment, diagnosis, prognosis and evaluation of efficacy. In addition, biomarkers are key to the success of precision medicine. The use of biomarkers that can identify and predict therapeutic response is essential in precision medicine (51). Overall, the findings of this study provide positive support for more research evaluating the expression levels of ING3 in cancer patients and may have practical clinical applications in treatment decision-making or early malignancy detection.
Conclusion
This meta-analysis investigated the relationship between high expression of ING3 and the prognosis of various cancers. We found that high expression of ING3 was negatively associated with more advanced TNM stage, lymph node metastasis, and disease-free survival. This suggests that ING3 may be related to cancer prognosis and can be used as a prognostic biomarker. However, more studies are still needed to confirm these findings.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
ZL developed the search strategy for the review and assisted with the method description. ZL, SX, and LC completed the article screening and completed the data extraction. ZL and LC created the PRISMA diagram. ZL and SX created the tables. ZL performed the meta-analysis. SH and XK helped interpret the results and correct grammatical errors in the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Guangdong Medical University Innovation Project (ZCDS004) and 2021 Guangdong University Student Innovation and Entrepreneurship Project (S202110571034).
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ING, inhibitor of growth; PHD, plant homeodomain; HR, hazard ratio; OR, odds ratio; OS, overall survival; DFS, disease-free survival; NTS, translocation sequence; EMT, epithelial-to-mesenchymal transition.
References
1. Hu G-W, Yan X-W, Qin Y-J, Nie H-T. Molecular cloning and expression analysis of inhibitor of growth protein 3 (ING3) in the Manila clam, ruditapes philippinarum. Mol Biol Rep (2014) 41(6):3583–90. doi: 10.1007/s11033-014-3221-7
2. Gong W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int J Biochem Cell Biol (2005) 37(5):1054–65. doi: 10.1016/j.biocel.2004.09.008
3. Coles AH, Jones SN. The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol (2009) 218(1):45–57. doi: 10.1002/jcp.21583
4. Russell M, Berardi P, Gong W, Riabowol K. Grow-ING, age-ING and die-ING: ING proteins link cancer, senescence and apoptosis. Exp Cell Res (2006) 312(7):951–61. doi: 10.1016/j.yexcr.2006.01.020
5. Ludwig S, Klitzsch A, Baniahmad A. The ING tumor suppressors in cellular senescence and chromatin. Cell Biosci (2011) 1(1):25. doi: 10.1186/2045-3701-1-25
6. Gou W-F, Yang X-F, Shen D-F, Zhao S, Sun H-Z, Luo J-S, et al. Immunohistochemical profile of ING3 protein in normal and cancerous tissues. Oncol Lett (2017) 13(3):1631–6. doi: 10.3892/ol.2017.5632
7. Sanchez R, Zhou M-M. The PHD finger: A versatile epigenome reader. Trends Biochem Sci (2011) 36(7):364–72. doi: 10.1016/j.tibs.2011.03.005
8. Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature (2007) 450(7172):1106–10. doi: 10.1038/nature06431
9. Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell (2007) 131(1):58–69. doi: 10.1016/j.cell.2007.08.016
10. Zhou R, Rotte A, Li G, Chen X, Chen G, Bhandaru M. Nuclear localization of ING3 is required to suppress melanoma cell migration, invasion and angiogenesis. Biochem Biophys Res Commun (2020) 527(2):418–24. doi: 10.1016/j.bbrc.2020.04.056
11. Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, et al. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene (2003) 22(3):343–50. doi: 10.1038/sj.onc.1206115
12. Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, et al. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene (2002) 21(28):4462–70. doi: 10.1038/sj.onc.1205540
13. Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Imai H, Minami N. ING3 is essential for asymmetric cell division during mouse oocyte maturation. PloS One (2013) 8(9):e74749. doi: 10.1371/journal.pone.0074749
14. Gou WF, Sun HZ, Zhao S, Niu ZF, Mao XY, Takano Y, et al. Downregulated inhibitor of growth 3 (ING3) expression during colorectal carcinogenesis. Indian J Med Res (2014) 139(APR):561–7.
15. Nabbi A, McClurg UL, Thalappilly S, Almami A, Mobahat M, Bismar TA, et al. ING3 promotes prostate cancer growth by activating the androgen receptor. BMC Med (2017) 15(1):103. doi: 10.1186/s12916-017-0854-0
16. McClurg UL, Nabbi A, Ricordel C, Korolchuk S, McCracken S, Heer R, et al. Human ex vivo prostate tissue model system identifies ING3 as an oncoprotein. Br J Cancer (2018) 118(5):713–26. doi: 10.1038/bjc.2017.447
17. Melekhova A, Baniahmad A. ING tumour suppressors and ING splice variants as coregulators of the androgen receptor signalling in prostate cancer. Cells (2021) 10(10):2599. doi: 10.3390/cells10102599
18. Esmaeili M, Pungsrinont T, Schaefer A, Baniahmad A. A novel crosstalk between the tumor suppressors ING1 and ING2 regulates androgen receptor signaling. J Mol Med (Berl) (2016) 94(10):1167–79. doi: 10.1007/s00109-016-1440-1
19. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PloS Med (2009) 6(7):e1000097.
20. Wong WCW, Cheung CSK, Hart GJ. Development of a quality assessment tool for systematic reviews of observational studies (QATSO) of HIV prevalence in men having sex with men and associated risk behaviours. Emerg Themes Epidemiol (2008) 5:23. doi: 10.1186/1742-7622-5-23
21. Riley RD, Moons KGM, Snell KIE, Ensor J, Hooft L, Altman DG, et al. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ (2019) 364:k4597. doi: 10.1136/bmj.k4597
22. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16
23. Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (2003) 327(7414):557–60. doi: 10.1136/bmj.327.7414.557
24. Lu M, Chen F, Wang Q, Wang K, Pan Q, Zhang X. Downregulation of inhibitor of growth 3 is correlated with tumorigenesis and progression of hepatocellular carcinoma. Oncol Lett (2012) 4(1):47–52. doi: 10.3892/ol.2012.685
25. Wang Y, Dai DL, Martinka M, Li G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin Cancer Res (2007) 13(14):4111–6. doi: 10.1158/1078-0432.CCR-07-0408
26. Martinez-Vargas YDC, TJd S-F, DHIPd O, Goncalo RIC, Queiroz LMG. ING3 and ING4 immunoexpression and their relation to the development of benign odontogenic lesions. Braz Dental J (2021) 32(4):74–82. doi: 10.1590/0103-6440202104279
27. Cengiz B, Gunduz M, Gunduz E, Ohuchida M, Shimizu K, Nagai N, et al. Frequent deletion and down-regulation of ING3 in head and neck cancer. J Hard Tissue Biol (2005) 14(2):298–9. doi: 10.2485/jhtb.14.298
28. Li H, Zhang H, Tan X, Liu D, Guo R, Wang M, et al. Overexpression of ING3 is associated with attenuation of migration and invasion in breast cancer. Exp Ther Med (2021) 22(1):699. doi: 10.3892/etm.2021.10131
29. Wu X, Chen C, Luo B, Yan D, Yan H, Chen F, et al. Nuclear ING3 expression is correlated with a good prognosis of breast cancer. Front Oncol (2020) 10. doi: 10.21203/rs.3.rs-33232/v1
30. Almami A, Hegazy SA, Nabbi A, Alshalalfa M, Salman A, Abou-Ouf H, et al. ING3 is associated with increased cell invasion and lethal outcome in ERG-negative prostate cancer patients. Tumor Biol (2016) 37(7):9731–8. doi: 10.1007/s13277-016-4802-y
31. Yang HY, Liu HL, Tian LT, Song RP, Song X, Yin DL, et al. Expression and prognostic value of ING3 in human primary hepatocellular carcinoma. Exp Biol Med (2012) 237(4):352–61. doi: 10.1258/ebm.2011.011346
32. Gunduz M, Beder LB, Gunduz E, Nagatsuka H, Fukushima K, Pehlivan D, et al. Downregulation of ING3 mRNA expression predicts poor prognosis in head and neck cancer. Cancer sci (2008) 99(3):531–8. doi: 10.1111/j.1349-7006.2007.00708.x
33. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
34. Parikh RB, Manz CR, Nelson MN, Evans CN, Regli SH, O'Connor N, et al. Clinician perspectives on machine learning prognostic algorithms in the routine care of patients with cancer: A qualitative study. Support Care Cancer (2022) 30(5):4363–72. doi: 10.1007/s00520-021-06774-w
35. Li Z, Liu J, Zhang X, Fang L, Zhang C, Zhang Z, et al. Prognostic significance of cyclin D1 expression in renal cell carcinoma: A systematic review and meta-analysis. Pathol Oncol Res (2020) 26(3):1401–9. doi: 10.1007/s12253-019-00776-0
36. Zhong W, Yu Z, Zhan J, Yu T, Lin Y, Xia Z-S, et al. Association of serum levels of CEA, CA199, CA125, CYFRA21-1 and CA72-4 and disease characteristics in colorectal cancer. Pathol Oncol Res (2015) 21(1):83–95. doi: 10.1007/s12253-014-9791-9
37. Zhong C, Xie Z, Zeng L-H, Yuan C, Duan S. MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Front Immunol (2022) 13:855078. doi: 10.3389/fimmu.2022.855078
38. Toyoda H, Kumada T, Tada T, Sone Y, Kaneoka Y, Maeda A. Tumor markers for hepatocellular carcinoma: Simple and significant predictors of outcome in patients with HCC. Liver Cancer (2015) 4(2):126–36. doi: 10.1159/000367735
39. Tsai P-L, Su W-J, Leung W-H, Lai C-T, Liu C-K. Neutrophil-lymphocyte ratio and CEA level as prognostic and predictive factors in colorectal cancer: A systematic review and meta-analysis. J Cancer Res Ther (2016) 12(2):582–9.
40. Romero A, Serna-Blasco R, Calvo V, Provencio M. Use of liquid biopsy in the care of patients with non-small cell lung cancer. Curr Treat Options Oncol (2021) 22(10):86. doi: 10.1007/s11864-021-00882-9
41. Tjensvoll K, Nordgård O, Smaaland R. Circulating tumor cells in pancreatic cancer patients: methods of detection and clinical implications. Int J Cancer (2014) 134(1):1–8. doi: 10.1002/ijc.28134
42. Chaudhuri AA, Binkley MS, Osmundson EC, Alizadeh AA, Diehn M. Predicting radiotherapy responses and treatment outcomes through analysis of circulating tumor DNA. Semin Radiat Oncol (2015) 25(4):305–12. doi: 10.1016/j.semradonc.2015.05.001
43. Pezaro C, Woo HH, Davis ID. Prostate cancer: measuring PSA. Intern Med J (2014) 44(5):433–40. doi: 10.1111/imj.12407
44. Nedelcu T, Kubista B, Koller A, Sulzbacher I, Mosberger I, Arrich F, et al. Livin and bcl-2 expression in high-grade osteosarcoma. J Cancer Res Clin Oncol (2008) 134(2):237–44.
45. Partridge AH, Rumble RB, Carey LA, Come SE, Davidson NE, Di Leo A, et al. Chemotherapy and targeted therapy for women with human epidermal growth factor receptor 2-negative (or unknown) advanced breast cancer: American society of clinical oncology clinical practice guideline. J Clin Oncol (2014) 32(29):3307–29. doi: 10.1200/JCO.2014.56.7479
46. He GHY, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol (2005) 22(1):104–16. doi: 10.1093/molbev/msh256
47. Taheri M, Hussen BM, Najafi S, Abak A, Ghafouri-Fard S, Samsami M, et al. Molecular mechanisms of inhibitor of growth (ING) family members in health and malignancy. Cancer Cell Int (2022) 22(1):272. doi: 10.1186/s12935-022-02693-w
48. Melekhova A, Leeder M, Pungsrinont T, Schmäche T, Kallenbach J, Ehsani M, et al. A novel splice variant of the inhibitor of growth 3 lacks the plant homeodomain and regulates epithelial-mesenchymal transition in prostate cancer cells. Biomolecules (2021) 11(8):1152. doi: 10.3390/biom11081152
49. Scott M, Boisvert FM, Vieyra D, Johnston RN, Bazett-Jones DP, Riabowol K. UV Induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res (2001) 29(10):2052–8. doi: 10.1093/nar/29.10.2052
50. Li X, Zhang Q, Zhang M, Luo Y, Fu Y. Downregulation of nuclear ING3 expression and translocalization to cytoplasm promotes tumorigenesis and progression in head and neck squamous cell carcinoma (HNSCC). Histol Histopathol (2020) 35(7):681–90.
Keywords: cancer, gene, biomarker, prognosis, meta-analysis
Citation: Li Z, Xu S, Chen L, Huang S, Kuerban X and Li T (2023) Prognostic significance of ING3 expression in patients with cancer: A systematic review and meta-analysis. Front. Oncol. 13:1090860. doi: 10.3389/fonc.2023.1090860
Received: 06 November 2022; Accepted: 19 January 2023;
Published: 09 February 2023.
Edited by:
Xin Zhou, Nanjing Medical University, ChinaReviewed by:
Zeyan Li, Shandong University, ChinaSridhar Malkaram, West Virginia State University, United States
Copyright © 2023 Li, Xu, Chen, Huang, Kuerban and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianyu Li, RGF2aWRsZWUtZ2RAdG9tLmNvbQ==
 Zehan Li
Zehan Li Shengchao Xu2
Shengchao Xu2 Shuqi Huang
Shuqi Huang Xieyida Kuerban
Xieyida Kuerban Tianyu Li
Tianyu Li