- 1Pancreatic Endocrinology Ward, Department of General Surgery, Shengjing Hospital of China Medical University, Shenyang, China
- 2Department of Gastroenterology, The Fourth Affiliated Hospital of China Medical University, Shenyang, China
- 3Beijing ChosenMed Clinical Laboratory Co. Ltd., Beijing, China
- 4Computer Network Information Center, Chinese Academy of Sciences, Beijing, China
- 5University of the Chinese Academy of Sciences, Beijing, China
SMARCA4 (BRG1)-deficient undifferentiated carcinoma is a rare and highly aggressive malignancy. It has been reported to occur in a multiple range of organs. However, to the best of our knowledge, SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder has not yet been reported. Here, we describe a case of SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder. Through comprehensive genetic analysis, we hypothesized that in addition to SMARCA4 (BRG1) deficiency, other genetic changes might also be involved in the tumorigenesis of undifferentiated gallbladder cancer in this patient, particularly somatic mutations in the CTNNB1, KRAS, PIK3CA, TP53, CREBBP, and FANCI genes. To the best of our knowledge, this is the first report of SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder with genetic analysis.
Background
Gallbladder carcinoma (GBC) is a rare malignancy. Nearly 98% of GBCs are adenocarcinomas. Other rare histological types include mucinous, squamous, adenosquamous, small-cell, and undifferentiated carcinomas (1). Undifferentiated carcinoma of gallbladder accounts for 3.4% of all GBCs. This malignancy is more aggressive and frequently presents a larger tumor size compared to GBC (5.0 cm versus 3.0 cm). Its prognosis is extremely poor, with a median overall survival of 7.3 months.
The switch/sucrose non-fermenting (SWI/SNF) complex is a family of ATP-dependent chromatin remodeling proteins that play a crucial role in regulating gene transcription. SWI/SNF complex consists of 12–15 subunits encoded by 29 genes, including SMARCA2 (BRM), SMARCA4 (BRG1), SMARCC1, SMARCC2, SMARCB1 (INI1), ARID1A, and PBRM1. SWI/SNF complex exerts a crucial role in the differentiation, cell adhesion, motility of cancer cells and induces abnormal activation of the hedgehog signaling pathway (2).
SMARCA4 (BRG1) is one of the catalytic subunits of SWI/SNF complex. Inactivation of SMARCA4 (BRG1) has recently been suggested to be involved in the pathogenesis of some undifferentiated carcinomas. Previous studies have reported that SMARCA4 (BRG1)-deficient undifferentiated carcinomas could occur in the lung, ovary, gastrointestinal tract, uterus, and other organs (3–9). However, to the best of our knowledge, SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder has not yet been reported.
Here, we report the first case of SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder and analyze comprehensively its genetic alterations. We also discussed the potential targeted therapy.
Case presentation
On June 30, 2022, a 65-year-old female patient was admitted to the hospital due to right upper abdominal pain for more than one month and aggravation for half a month. A whole abdominal enhanced computer tomography (CT) scan showed a space-occupying lesion in the gallbladder, with enlargement of multiple peripheral and retroperitoneal lymph nodes, suspected a malignant lesion (Figure 1A). On July 5, 2022, the patient underwent radical resection of the gallbladder, which was approximately 5.0 cm in diameter (Figure 1B). Upon partially opening the gallbladder, it was discovered that a mass took up a significant portion of it. The mass exhibited a yellow-white color and had a brittle texture upon being cut. Further inspection revealed that it had infiltrated the gallbladder’s muscle layer. The space-occupying lesion was approximately 4.5 cm × 3.0 cm × 3.0 cm in size (Figure 1C). Postoperative pathology revealed that the tumor cells presented obvious atypia, diffuse patchy infiltration, growth, poor cell adhesion, chromatin vacuolated, nucleoli obvious, mitotic figures were easy to see, necrosis and more lymphocyte infiltration were seen (Figure 2A). Notably, there were no cancer cells detected in the stump of the cystic duct, but retroperitoneal lymph node metastasis was observed. Immunohistochemistry (IHC) showed that the tumor cells were positive for cytokeratin, synaptophysin, SMARCB1 (INI1), and negative for SMARCA4 (BRG1), CK7, CK19, p63, p40, vimentin (Figure 2B). IHC for PD-L1 (DAKO 22C3) showed that the tumor proportion score (TPS) was <1%, and the combined positive score (CPS) was 50. The tumor had metastasized to the retroperitoneal lymph nodes as well as lymph node groups 7, 8, 9, 12, and 13. The final diagnosis was SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder (stage II-III).
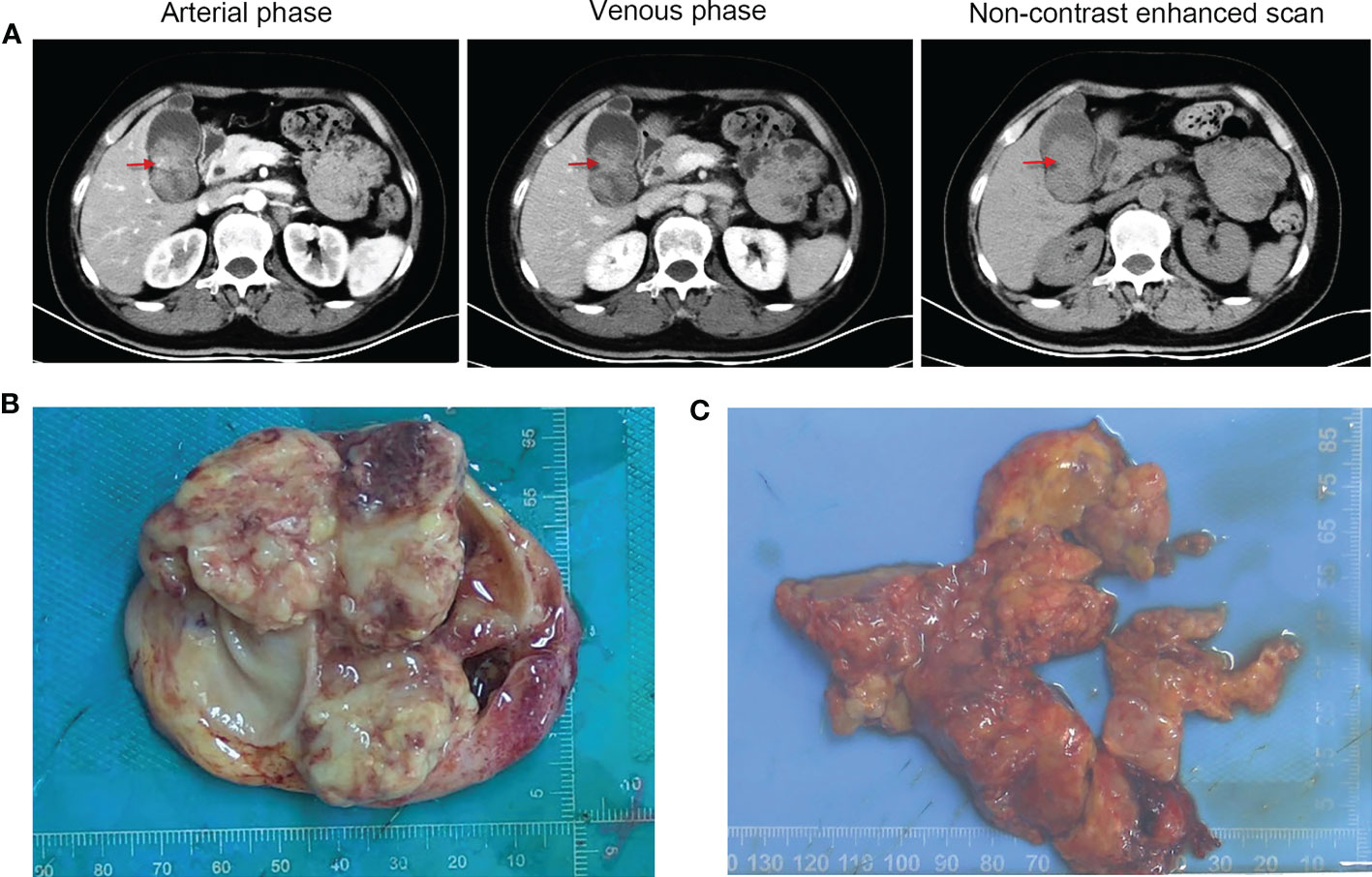
Figure 1 Radiographic and macroscopic images of the tumor. (A) Computed tomography images of the patient (red arrows refer to the space-occupying lesion). (B) The resected gallbladder was approximately 5.0 cm in diameter. (C) The irregular mass in the gallbladder was approximately 4.5 cm × 3.0 cm × 3.0 cm in size.
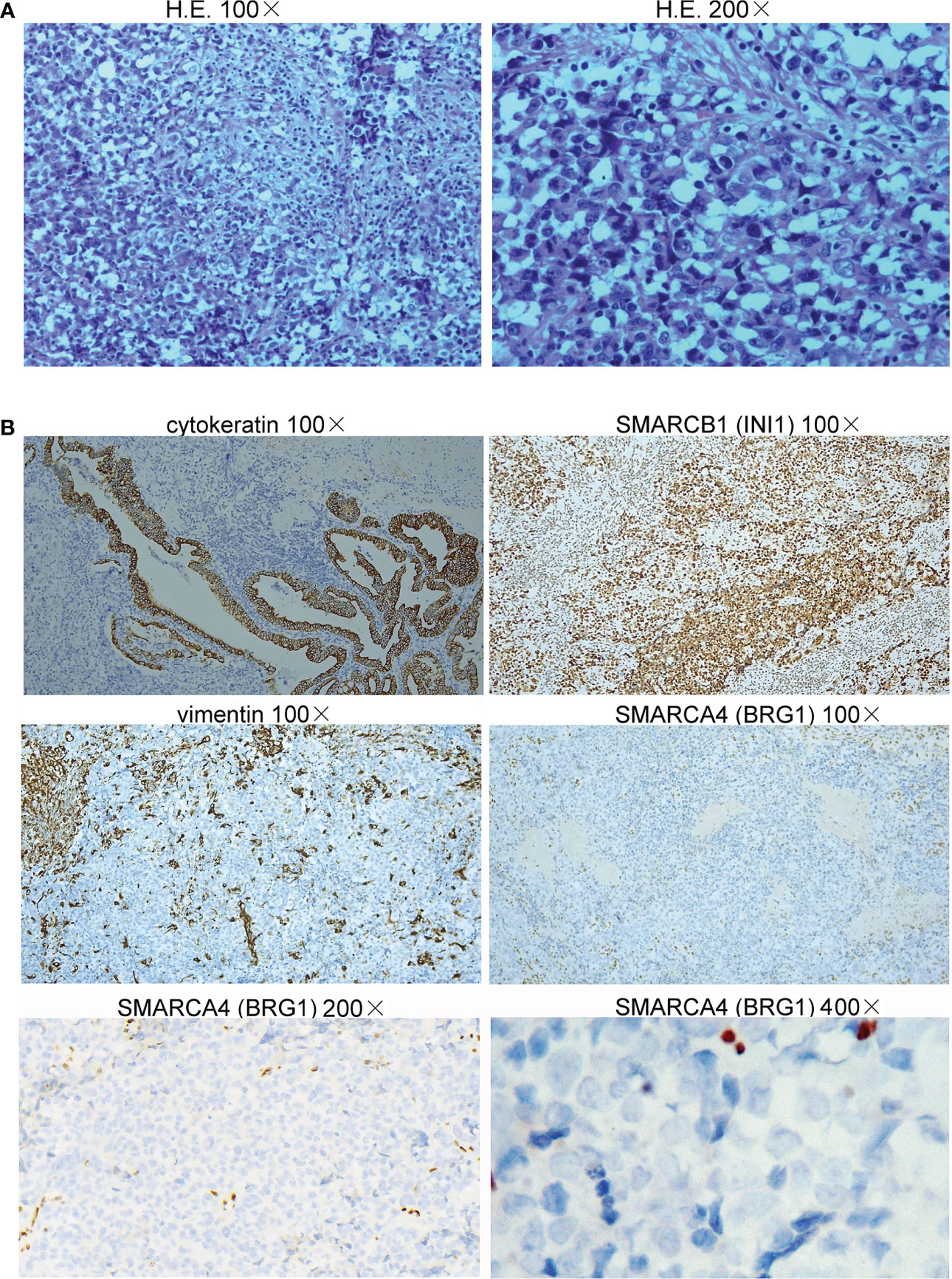
Figure 2 Histopathology of the undifferentiated carcinoma of the gallbladder. (A) The tumor cells presented obvious atypia, diffuse patchy infiltration, growth, and poor cell adhesion (H&E staining, 100× and 200×). (B) Immunohistochemical staining showed that the neoplastic cells were positive for cytokeratin and SMARCB1 (INI1) but negative for vimentin (100×) and SMARCA4 (BRG1) (100×, 200×, and 400×).
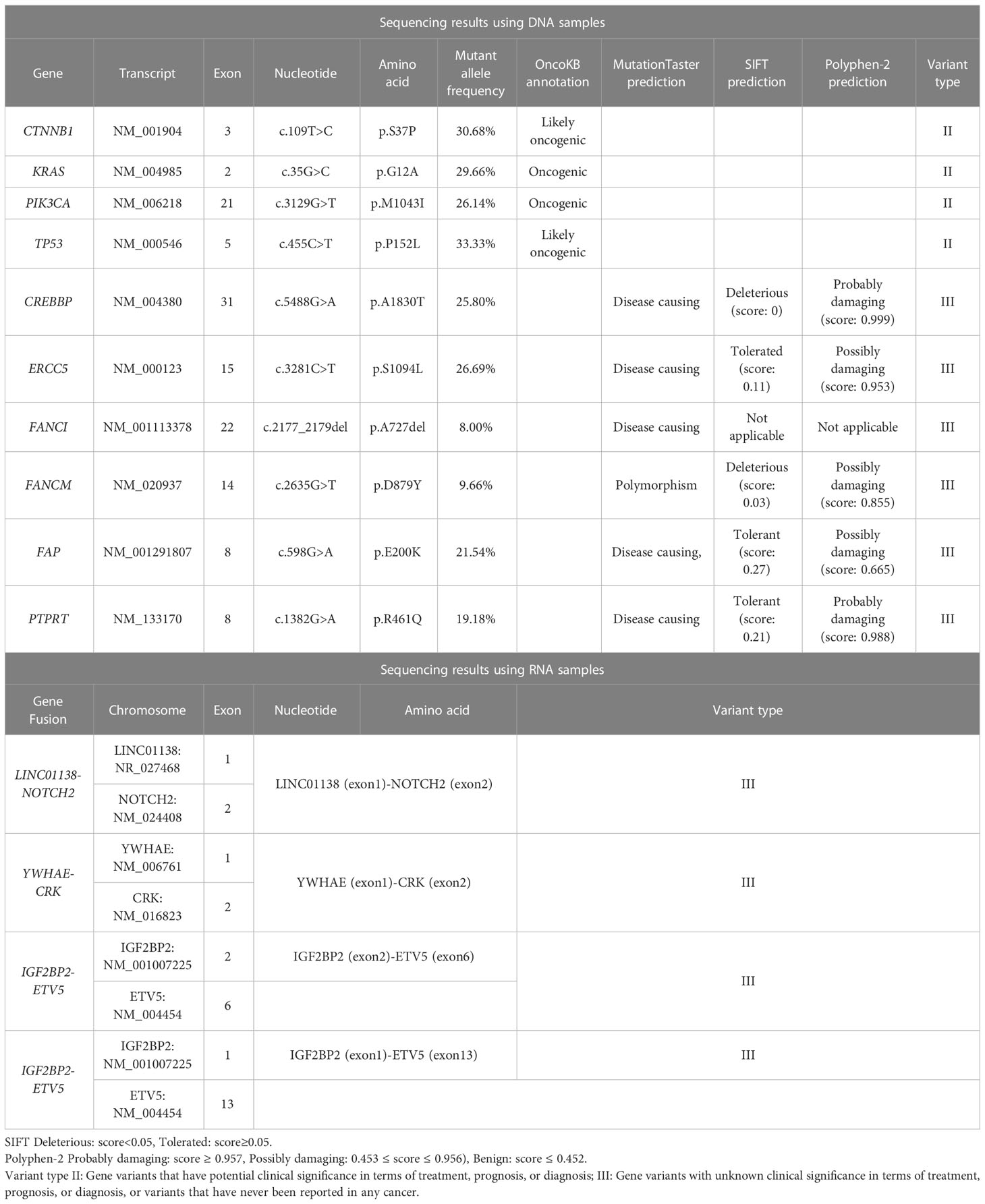
On July 14, 2022, DNA and RNA were extracted from the resected formalin fixed and paraffin embedded tumor tissues of the patient, and were subjected to next-generation sequencing (NGS) using a 1123-gene panel and a 102-gene panel, respectively (ChosenMed Technology [Beijing] Co. Ltd, Beijing, China). The sequencing results using DNA samples revealed that the patient harbored somatic mutations in the CTNNB1, KRAS, PIK3CA, TP53, CREBBP, ERCC5, FANCI, FANCM, FAP and PTPRT genes. The sequencing results using RNA samples revealed that the patient harbored four gene fusions: LINC01138-NOTCH2, YWHAE-CRK, IGF2BP2-ETV5, and IGF2BP2-ETV5. In addition, analysis of the NGS data revealed microsatellite-stability (MSS) status and medium tumor mutational burden (M-TMB, 8.22 muts/Mb). The genomic profile of the patient is demonstrated in Table 1. The patient had no obvious discomfort after the operation and was discharged from the hospital one month later. The patient died five months after surgery on December 14, 2022.
Discussion
Gallbladder cancer (GBC) is an uncommon but highly fatal malignancy, and undifferentiated GBC is extremely rare. Some research had revealed the genomics profile of GBC. The common somatic mutation genes were TP53 (64%-73%), CDKN2A (11.0%-25.0%), ERBB2 (9.3%), and PIK3CA (10%-20%) in GBC (10, 11). The SMARCA4 alteration frequency was 7.0% in a 60 GBC patients study (11). A case reported an SWI/SNF-deficient undifferentiated/rhabdoid carcinoma of the gallbladder carrying a POLE mutation in a 30-year-old woman (12).
The incidence of SMRCA4 somatic mutations in NSCLC is 8% (407/4813); the major co-mutated genes were TP53 (56%), KEAP1 (41%), STK11 (39%), and KRAS (36%) in SMARCA4-mutant NSCLC. Patients with SMARCA4 co-mutations with STK11 or KEAP1 had a worse prognosis than those with single mutations, and patients with triple mutations had the worst prognosis (13).
In addition, SMARCA4 (BRG1)-deficient in other undifferentiated cancers has been reported sporadically, such as colon (14), gastrointestinal tract (15), and ovary (16). The SMARCA4 (BRG1)-deficient in undifferentiated gallbladder carcinoma has not yet been reported. This case first reported a patient with SMARCA4 (BRG1)-deficient undifferentiated gallbladder carcinoma. GBC is a multifactorial disease whose occurrence is related to chronic inflammation of the gallbladder, dietary factors and female gender. Multiple genetic alterations are involved in this malignancy, the most common being KRAS, TP53, and erbB-2 (neu/HER-2) genes (17). In the patient, mutations in the KRAS (p.G12A) and TP53 (p.P152L) genes were also detected, but no aberrations in erbB-2.
As one of the most prevalent mutations of KRAS, the mutation p.G12A in the KRAS has the potential capability to activate MEK/ERK- and PI3K/AKT-signaling pathways in cancer cells and is an oncogenic mutation (18).
TP53 is a tumor suppressor in the DNA damage pathway. According to the annotation in the OncoKB database, TP53 p.P152L may lead to loss of protein function and is probably oncogenic.
In addition, we also detected two type II variations of CTNNB1 (beta-catenin) and PIK3CA genes, as well as six type III variations of CREBBP, ERCC5, FANCI, FANCM, FAP, and PTPRT genes.
The gene CTNNB1 encodes beta-catenin protein, which acts as an intracellular signal converter in the Wnt signaling pathway. Aberrant CTNNB1 activates proto oncogenes and cyclins, driving the occurrence, progress, survival, and recurrence of cancer. CTNNB1 is recurrently mutated in multiple cancer types (19). According to the annotation in the OncoKB database, CTNNB1 p.S37P mutation may be pathogenic.
PIK3CA gene encodes the catalytic subunit of class I phosphatidylinositol-3-kinase (PI3K). It plays an important role in the cell growth, proliferation, migration and survival through the PI3K/AKT/mTOR signaling pathway. Mutated PIK3CA gene leads to the continuously activated PI3K/Akt signaling pathway in various cancers (20–23). p.M1043I is located in the PI3K/PI4K domain of the PIK3CA protein. According to the annotation in the OncoKB database, PIK3CA p.M1043I alteration is an oncogenic variation, which can increase the level of AKT phosphorylation and activate the downstream signaling pathway (24, 25).
CREBBP is a tumor suppressor and transcriptional co-activator, and it is frequently inactivated in hematologic malignancies (26, 27). The association between CREBBP and carcinoma has not yet been reported. MutationTaster, SIFT and Polyphen-2 consistently predicted that CREBBP p.A1830T is harmful.
ERCC5 is also a tumor suppressor and serves as a DNA endonuclease involved in the nucleotide-excision repair (NER) pathway (28). Germline mutations of ERCC5 gene are associated with several disorders with defective DNA repair, which are susceptible to develop certain cancers (29). Although abnormal expression of ERCC5 has been detected in breast and ovarian cancers (30), somatic mutations of this gene have not yet been reported to be oncogenic drivers. ERCC5 p.S1094L is a novel variation. MutationTaster, SIFT and Polyphen-2 predicted this mutation is disease causing, tolerated, and possibly damaging, respectively.
FANCI is an important component of the Fanconi anemia (FA) pathway, which is a DNA damage response (DDR) pathway (31). Among the identified 22 FA proteins (FANCA-FANCW), FANCI is an evolutionarily relevant partner of FANCD2. These two proteins form a protein complex FANCI-FANCD2 (ID2), which is a critical step in the activation of the FA pathway (32). A study showed that mutated FANCI may be a candidate ovarian cancer-predisposing gene (33). The non-frameshift deletion (p.A727del) in FANCI is a novel alteration, which is predicted to be disease causing by MutationTaster software.
FANCM is another essential member of the FA pathway. It is a tumor suppressor gene encoding a conserved and structure-specific DNA translocase. Loss-of-function mutations in FANCM are associated with predisposition to breast and ovarian cancer (34, 35). FANCM p.D879Y is a novel mutation. MutationTaster predicted that this variation is a polymorphism, whereas SIFT and Polyphen-2 predicted that it is deleterious and possibly damaging, respectively.
Fibroblast activation protein alpha (FAP) is a type II integral serine protease expressed specifically by activated fibroblasts. FAP can promote tumor growth, invasion, metastasis, and immunosuppression (36). MutationTaster, SIFT and Polyphen-2 predicted that the FAP p.E200K variation is disease causing, tolerant, and possibly damaging, respectively.
The PTPRT gene encodes a receptor-type tyrosine-protein phosphatase T enzyme. It is a tumor suppressor gene involved in signal transduction and cellular adhesion. PTPRT inhibits cell proliferation through the STAT3 pathway (37–39). MutationTaster, SIFT, and Polyphen-2 predicted that the PTPRT p.R461Q variation is disease causing, tolerant, and probably damaging, respectively.
Meanwhile, we identified four gene fusions through sequencing RNA samples, i.e., LINC01138-NOTCH2, YWHAE-CRK, IGF2BP2-ETV5, and IGF2BP2-ETV5. The association between these four gene fusions and malignancy has not yet been reported.
Regretfully, we didn’t detect the SMARCA4 (BRG1) deletion by NGS. The possible reason is that NGS can accurately detect small insertion/deletion mutations, point mutations and exonic copy-number changes. However, NGS cannot accurately detect large deletions. We speculate that the patient harbors a large SMARCA4 (BRG1) deletion. In addition, the alterations may occur in the regulatory regions of the SMARCA4 gene, such as the promoter region, CpG islands, enhancers, and so on, resulting in a loss of gene expression. Epigenetic modifications involving chemical alterations to chromatin or DNA molecules may also contribute to gene expression deficiency. These modifications include DNA methylation, histone modifications, and non-coding RNAs. For instance, DNA methylation refers to adding methyl groups to DNA molecules, often leading to gene silencing and suppressed expression.
Based on the above genetic analysis, we speculate that, in addition to SMARCA4 (BRG1) deficiency, other genetic alterations might also participate in the occurrence and progress of the undifferentiated carcinoma of the gallbladder in this patient, particularly the mutations in the CTNNB1, KRAS, PIK3CA, TP53, CREBBP, and FANCI genes (Table 1).
Conventional treatment is usually ineffective for SMARCA4 (BRG1)-deficient tumors. Based on the antagonism of SWI/SNF and polycomb repressive complex 2 (PRC2), SWI/SNF deletion leads to the loss of inhibition of EZH2 methyltransferase, which in turn results in PRC2-mediated tumorigenesis (40). Targeted therapy with EZH2 inhibitors might be effective (41). Clinical trials on SMARCB1 (INI1)-deficient rhabdoid tumors and epithelioid sarcomas have shown that EZH2 inhibitors have lasting anti-proliferative effects (42). In a phase II clinical trial of epithelioid sarcoma, 15% of patients achieved clinical remission, of which 67% achieved remission for at least six months (43). Therefore, the US Food and Drug Administration accelerated the approval of the first EZH2 inhibitor tazemetostat in January 2020 for treating locally advanced or metastatic epithelioid sarcoma patients. In vitro and in vivo studies have suggested that tazemetostat has the potential anti-tumor effects on SMARCA4 (BRG1)- and SMARCA2 (BRM)-deficient small-cell carcinomas of the ovary of hypercalcaemic type (SCCOHT) (44), which brings hope for the targeted therapy of other types of SMARCA4 (BRG1)-deficient undifferentiated tumors. In addition to EZH2 inhibitors, other possible targeted therapies include histone deacetylase inhibitors and DNA methyltransferase inhibitors. Furthermore, another potential therapy is immune checkpoint inhibitor (ICI) therapy. The patient gave up the therapy after discharge from the hospital because of personal reasons.
To the best of our knowledge, this is the first case of SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder with genetic analysis. This case might contribute to the understanding of SWI/SNF-deficient carcinoma of gallbladder.
Data availability statement
The sample data of this patient for this study is included in the article/Supplementary Material.
Ethics statement
The work was approved by the Ethics Committee of Shengjing Hospital of China Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the participant/patient(s) for the publication of this case report.
Author contributions
XM: conceptualization. JM: postoperative care. TY: Collection data. NM: writing the draft. BN: Editing the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors wish to thank the patient for her participation.
Conflict of interest
BN, NM, and TY are employees of Beijing ChosenMed Clinical Laboratory.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1086266/full#supplementary-material
References
1. Roa JC, Tapia O, Cakir A, Basturk O, Dursun N, Akdemir D, et al. Squamous cell and adenosquamous carcinomas of the gallbladder: clinicopathological analysis of 34 cases identified in 606 carcinomas. Modern Pathol (2011) 24(8):1069–78. doi: 10.1038/modpathol.2011.68
2. Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev pathology. (2015) 10:145–71. doi: 10.1146/annurev-pathol-012414-040445
3. Agaimy A, Fuchs F, Moskalev EA, Sirbu H, Hartmann A, Haller F. SMARCA4-deficient pulmonary adenocarcinoma: clinicopathological, immunohistochemical, and molecular characteristics of a novel aggressive neoplasm with a consistent TTF1(neg)/CK7(pos)/HepPar-1(pos) immunophenotype. Virchows Archiv an Int J pathology. (2017) 471(5):599–609. doi: 10.1007/s00428-017-2148-5
4. Karnezis AN, Wang Y, Ramos P, Hendricks WP, Oliva E, D'Angelo E, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J pathology. (2016) 238(3):389–400. doi: 10.1002/path.4633
5. Agaimy A, Daum O, Markl B, Lichtmannegger I, Michal M, Hartmann A. SWI/SNF complex-deficient Undifferentiated/Rhabdoid carcinomas of the gastrointestinal tract: a series of 13 cases highlighting mutually exclusive loss of SMARCA4 and SMARCA2 and frequent Co-inactivation of SMARCB1 and SMARCA2. Am J Surg pathology. (2016) 40(4):544–53. doi: 10.1097/PAS.0000000000000554
6. Tessier-Cloutier B, Schaeffer DF, Bacani J, Marginean CE, Kalloger S, Kobel M, et al. Loss of switch/sucrose non-fermenting complex protein expression in undifferentiated gastrointestinal and pancreatic carcinomas. Histopathology (2020) 77(1):46–54. doi: 10.1111/his.14096
7. Ramalingam P, Croce S, McCluggage WG. Loss of expression of SMARCA4 (BRG1), SMARCA2 (BRM) and SMARCB1 (INI1) in undifferentiated carcinoma of the endometrium is not uncommon and is not always associated with rhabdoid morphology. Histopathology (2017) 70(3):359–66. doi: 10.1111/his.13091
8. Agaimy A, Jain D, Uddin N, Rooper LM, Bishop JA. SMARCA4-deficient sinonasal carcinoma: a series of 10 cases expanding the genetic spectrum of SWI/SNF-driven sinonasal malignancies. Am J Surg pathology. (2020) 44(5):703–10. doi: 10.1097/PAS.0000000000001428
9. Agaimy A, Bertz S, Cheng L, Hes O, Junker K, Keck B, et al. Loss of expression of the SWI/SNF complex is a frequent event in undifferentiated/dedifferentiated urothelial carcinoma of the urinary tract. Virchows Archiv an Int J pathology. (2016) 469(3):321–30. doi: 10.1007/s00428-016-1977-y
10. de Bitter TJJ, de Reuver PR, de Savornin Lohman EAJ, Kroeze LI, Vink-Börger ME, van Vliet S, et al. Comprehensive clinicopathological and genomic profiling of gallbladder cancer reveals actionable targets in half of patients. NPJ Precis Oncol (2022) 6(1):83. doi: 10.1038/s41698-022-00327-y
11. Lin J, Dong K, Bai Y, Zhao S, Dong Y, Shi J, et al. Precision oncology for gallbladder cancer: insights from genetic alterations and clinical practice. Ann Transl Med (2019) 7(18):467. doi: 10.21037/atm.2019.08.67
12. Gerber TS, Agaimy A, Hartmann A, Habekost M, Roth W, Stenzinger A, et al. SWI/SNF-deficient undifferentiated/rhabdoid carcinoma of the gallbladder carrying a POLE mutation in a 30-year-old woman: a case report. Diagn Pathol (2021) 16(1):52. doi: 10.1186/s13000-021-01112-4
13. Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, et al. The genomic landscape of SMARCA4 alterations and associations with outcomes in patients with lung CancerSMARCA4 alterations in lung cancer. Clin Cancer Res (2020) 26(21):5701–8. doi: 10.1158/1078-0432.CCR-20-1825
14. Duan H, Gao W, Wang L, Cao F, Teng L. Undifferentiated colonic neoplasm with SMARCA4 germline gene mutation and loss of SMARCA4 protein expression: a case report and literature review. Diagn Pathol (2021) 16(1):1–5. doi: 10.1186/s13000-021-01091-6
15. Zhu P, Li X, Liu J, Du X, Su H, Wang J. SMARCA4-deficient undifferentiated carcinoma of the gastrointestinal tract: a clinicopathological and immunohistochemical study of nine cases. Zhonghua Bing li xue za zhi= Chin J Pathology. (2022) 51(9):868–74. doi: 10.3760/cma.j.cn112151-20220226-00130
16. Jelinic P, Schlappe BA, Conlon N, Tseng J, Olvera N, Dao F, et al. Concomitant loss of SMARCA2 and SMARCA4 expression in small cell carcinoma of the ovary, hypercalcemic type. Modern Pathology. (2016) 29(1):60–6. doi: 10.1038/modpathol.2015.129
17. Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J gastroenterology. (2017) 23(22):3978–98. doi: 10.3748/wjg.v23.i22.3978
18. Weissbach S, Heredia-Guerrero SC, Barnsteiner S, Grosshans L, Bodem J, Starz H, et al. Exon-4 mutations in KRAS affect MEK/ERK and PI3K/AKT signaling in human multiple myeloma cell lines. Cancers (2020) 12(2):455. doi: 10.3390/cancers12020455
19. Kurnit KC, Kim GN, Fellman BM, Urbauer DL, Mills GB, Zhang W, et al. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Modern Pathol (2017) 30(7):1032–41. doi: 10.1038/modpathol.2017.15
20. Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res (2012) 18(14):3791–802. doi: 10.1158/1078-0432.CCR-11-2342
21. Abubaker J, Bavi PP, Al-Harbi S, Siraj AK, Al-Dayel F, Uddin S, et al. PIK3CA mutations are mutually exclusive with PTEN loss in diffuse large b-cell lymphoma. Leukemia (2007) 21(11):2368–70. doi: 10.1038/sj.leu.2404873
22. Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, et al. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica (2010) 95(5):819–28. doi: 10.3324/haematol.2009.013797
23. Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res (2008) 68(17):6913–21. doi: 10.1158/0008-5472.CAN-07-5084
24. Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci United States America. (2007) 104(13):5569–74. doi: 10.1073/pnas.0701005104
25. Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell (2018) 33(3):450–62 e10. doi: 10.1016/j.ccell.2018.01.021
26. Juskevicius D, Jucker D, Klingbiel D, Mamot C, Dirnhofer S, Tzankov A. Mutations of CREBBP and SOCS1 are independent prognostic factors in diffuse large b cell lymphoma: mutational analysis of the SAKK 38/07 prospective clinical trial cohort. J Hematol Oncol (2017) 10(1):70. doi: 10.1186/s13045-017-0438-7
27. Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci United States America. (2015) 112(10):E1116–25. doi: 10.1073/pnas.1501199112
28. Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, et al. Allowing transactivation of nuclear receptors: implications for cockayne syndrome in XP-G/CS patients. Mol Cell (2007) 26(2):231–43. doi: 10.1016/j.molcel.2007.03.013
29. Jaeken J, Klocker H, Schwaiger H, Bellmann R, Hirsch-Kauffmann M, Schweiger M. Clinical and biochemical studies in three patients with severe early infantile cockayne syndrome. Hum Genet (1989) 83(4):339–46. doi: 10.1007/BF00291378
30. Bartolucci R, Wei J, Sanchez JJ, Perez-Roca L, Chaib I, Puma F, et al. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung cancer. (2009) 10(1):47–52. doi: 10.3816/CLC.2009.n.007
31. Ceccaldi R, Sarangi P, D'Andrea AD. The fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol (2016) 17(6):337–49. doi: 10.1038/nrm.2016.48
32. Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the fanconi anemia DNA repair pathway. J Biol Chem (2010) 285(41):31525–36. doi: 10.1074/jbc.M110.138206
33. Fierheller CT, Guitton-Sert L, Alenezi WM, Revil T, Oros KK, Gao Y, et al. A functionally impaired missense variant identified in French Canadian families implicates FANCI as a candidate ovarian cancer-predisposing gene. Genome Med (2021) 13(1):186. doi: 10.1186/s13073-021-00998-5
34. Catucci I, Osorio A, Arver B, Neidhardt G, Bogliolo M, Zanardi F, et al. Individuals with FANCM biallelic mutations do not develop fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med (2018) 20(4):452–7. doi: 10.1038/gim.2017.123
35. Fouquet B, Pawlikowska P, Caburet S, Guigon C, Makinen M, Tanner L, et al. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. eLife (2017) 6:e30490. doi: 10.7554/eLife.30490
36. Wang H, Wu Q, Liu Z, Luo X, Fan Y, Liu Y, et al. Downregulation of FAP suppresses cell proliferation and metastasis through PTEN/PI3K/AKT and ras-ERK signaling in oral squamous cell carcinoma. Cell Death disease. (2014) 5:e1155. doi: 10.1038/cddis.2014.122
37. Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. (2011) 11(1):35–49. doi: 10.1038/nrc2980
38. Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z. Tumor-derived extracellular mutations of PTPRT /PTPrho are defective in cell adhesion. Mol Cancer Res MCR. (2008) 6(7):1106–13. doi: 10.1158/1541-7786.MCR-07-2123
39. Lui VW, Peyser ND, Ng PK, Hritz J, Zeng Y, Lu Y, et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci United States America. (2014) 111(3):1114–9. doi: 10.1073/pnas.1319551111
40. Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell (2010) 18(4):316–28. doi: 10.1016/j.ccr.2010.09.006
41. Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol (2020) 17(7):435–48. doi: 10.1038/s41571-020-0357-3
42. Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci United States America. (2013) 110(19):7922–7. doi: 10.1073/pnas.1303800110
43. Gounder M, Schoffski P, Jones RL, Agulnik M, Cote GM, Villalobos VM, et al. Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet Oncol (2020) 21(11):1423–32. doi: 10.1016/S1470-2045(20)30451-4
44. Chan-Penebre E, Armstrong K, Drew A, Grassian AR, Feldman I, Knutson SK, et al. Selective killing of SMARCA2- and SMARCA4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: In vitro and In vivo preclinical models. Mol Cancer Ther (2017) 16(5):850–60. doi: 10.1158/1535-7163.MCT-16-0678
Keywords: SMARCA4, BRG1, SWI/SNF, gallbladder carcinoma, case report
Citation: Meng X, Ma J, Meng N, Yun T and Niu B (2023) Case Report: SMARCA4 (BRG1)-deficient undifferentiated carcinoma of gallbladder with genetic analysis. Front. Oncol. 13:1086266. doi: 10.3389/fonc.2023.1086266
Received: 09 February 2023; Accepted: 26 May 2023;
Published: 30 June 2023.
Edited by:
Satvinder Singh Mudan, The London Clinic, United KingdomReviewed by:
Patricia N. Tonin, McGill University, CanadaBingcheng Wu, National University Hospital, Singapore
Copyright © 2023 Meng, Ma, Meng, Yun and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangpeng Meng, bXhwY211QDE2My5jb20=
†These authors have contributed equally to this work
 Xiangpeng Meng
Xiangpeng Meng Jia Ma2†
Jia Ma2† Nan Meng
Nan Meng Beifang Niu
Beifang Niu