
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 14 February 2023
Sec. Cancer Genetics
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1068110
This article is part of the Research Topic Case Reports in Cancer Genetics : 2022 View all 16 articles
Heterozygous, loss-of-function germline variants in ATM have been associated with an increased lifetime risk of breast, pancreas, prostate, stomach, ovarian, colorectal, and melanoma cancers. We conducted a retrospective review of thirty-one unrelated patients found to be heterozygous for a germline pathogenic variant in ATM and identified a significant proportion of patients in this cohort with cancers not currently associated with the ATM hereditary cancer syndrome, including carcinomas of the gallbladder, uterus, duodenum, kidney, and lung as well as a vascular sarcoma. A comprehensive review of the literature found 25 relevant studies where 171 individuals with a germline deleterious ATM variant have been diagnosed with the same or similar cancers. The combined data from these studies were then used to estimate the prevalence of germline ATM pathogenic variants in these cancers, which ranged between 0.45% and 2.2%. Analysis of tumor sequencing performed in large cohorts demonstrated that the frequency of deleterious somatic ATM alterations in these atypical cancers equaled or exceeded the alteration frequency in breast cancer and occurred at a significantly higher rate than in other DNA-damage response tumor suppressors, namely BRCA1 and CHEK2. Furthermore, multi-gene analysis of somatic alterations in these atypical cancers demonstrated significant co-occurrence of pathogenic alterations in ATM with BRCA1 and CHEK2, while there was significant mutual exclusivity between pathogenic alterations in ATM and TP53. This indicates that germline ATM pathogenic variants may play a role in cancer initiation and progression in these atypical ATM malignancies, potentially influencing these cancers to be driven toward DNA-damage repair deficiency and away from loss of TP53. As such, these findings provide evidence for broadening of the ATM-cancer susceptibility syndrome phenotype to improve the recognition of affected patients and provide more efficacious, germline-directed therapies.
ATM (OMIM 607585) encodes the PI3K-related serine/threonine protein kinase comprising 3,056 amino acids, ataxia-telangiectasia mutated. This protein kinase primarily resides in the nucleus of dividing cells and has long been recognized as a key upstream modulator of the response to DNA double-stranded breaks, as well as oxidative and other genotoxic stresses. A critical function of ATM involves the recruitment and cooperation with DNA-damage sensing proteins such as BRCA1 in the face of double-strand breaks (1). ATM also plays a critical role in the DNA damage response through the phosphorylation of downstream substrates including CHK2 and p53, which then stimulate the cell-cycle checkpoint arrest and cellular apoptosis pathways, respectively (1, 2).
Biallelic pathogenic loss-of-function variants in ATM have long been associated with the genomic instability syndrome, ataxia-telangiectasia. Manifestations of the disorder include cerebellar ataxia, oculocutaneous telangiectasia, immunodeficiency, radiosensitivity, premature aging, and a predisposition to cancer development, primarily of lymphoid origin (3).
It was later recognized that women who are heterozygous for pathogenic loss-of-function variants in ATM have an increased risk for breast cancer, leading ATM to become regarded as a moderate penetrance breast cancer susceptibility gene, conferring a 2.3-fold increased risk for breast cancer compared to the general population (4, 5).
The risk of cancer among individuals heterozygous for ATM pathogenic variants has since been demonstrated for a broader range of malignancies. ATM pathogenic variant carriers appear to be at moderate-to-high increased risk for pancreatic, prostate, and gastric cancers and at low-to-moderate increased risk for ovarian and colorectal cancer as well as melanoma (6–9).
The association of ATM with cancer susceptibility has directly influenced clinical practice, from the creation of gene panels for molecular testing to cancer surveillance guidance and treatment recommendations. Here we present six novel cases of individuals with carcinomas of the gallbladder, uterus, duodenum, kidney, and lung as well as a sarcoma, all harboring germline ATM pathogenic variants, which suggests further expansion of the ATM-associated cancer susceptibility phenotype.
We retrospectively reviewed patients seen in our hereditary cancer clinic between March 2017 and December 2021 who had undergone germline testing with next-generation, multi-gene sequencing. We then identified all patients found to harbor germline likely pathogenic or pathogenic variants in ATM producing a loss-of-function using the criteria of the American College of Medical Genetics standards and guidelines for sequence variant interpretation (10). A comprehensive chart review through the electronic medical record was conducted to collect relevant clinical data including patient demographics, personal and family cancer history, as well as complete genetic testing results. Evidence substantiating the pathogenicity of the germline ATM variants among the six patients with atypical ATM-associated cancers was collected (Supplementary Table 1).
Multi-gene hereditary cancer syndrome testing was performed in commercial clinical laboratories accredited by the College of American Pathologists and certifyed by the Clinical Laboratory Improvement Amendment. Laboratories used included Ambry Genetics (Aliso Viejo, CA), GeneDx (Gaithersburg, MD), Myriad (Salt Lake City, UT), Sema4 (Stamford, CT), or Invitae (San Francisco, CA). One patient had sequencing performed at the medical institution where he had previously received care (Memorial Sloan Kettering, New York, NY). The number of analyzed genes ranged from 25-84, each of which was associated with hereditary cancers in peer-reviewed scientific literature (Supplementary Table 2).
A comprehensive review of the peer-reviewed literature through MEDLINE was conducted using PubMed for articles published between 1970 and July 2022. Search keywords included “pathogenic,” “germline,” “ATM,” “variant,” “hereditary,” “cancer,” “gallbladder,” “uterus,” “duodenum,” “kidney,” “lung,” “epithelioid hemangioendothelioma,” and then later expanded to include “biliary tract,” “small bowel,” “ampulla,” and “sarcoma.” Keywords were connected by the Boolean functions AND and OR.
All cross-sectional studies involving thirty or more patients affected with cancer of the biliary tract, uterus, small intestine/ampulla, kidney, lung, or sarcoma, who had undergone hereditary cancer predisposition testing with a multigene panel, exome, or genome sequencing were considered for inclusion. Studies using redundant patient information drawn from large datasets were excluded from the review. The study text and supplemental information were reviewed for patient characteristics, total germline pathogenic variants detected, and specific germline ATM variants identified.
The prevalence for each cancer type was estimated by dividing the number of individuals with a germline ATM pathogenic variant by the total sample size. To estimate the prevalence among those with hereditary cancer susceptibility, the number of individuals with a germline ATM pathogenic variant was divided by the number of those testing positive for any pathogenic variant in a hereditary cancer gene.
De-identified genomic sequencing data from The Cancer Genome Atlas and other large-scale, cancer-specific sequencing studies were accessed and queried online through the cBioPortal for Cancer Genomics at https://www.cbioportal.org/ (11, 12). Patient cohorts were primarily selected from the TCGA pan-cancer atlas, and the accession numbers for each tumor-specific analysis were recorded (Supplementary Methods). Somatic alterations included were loss-of-function single nucleotide variants (SNVs), indels, and copy number variants (CNVs) classified as pathogenic or likely pathogenic. Statistical analysis to compare deleterious alteration counts of ATM, BRCA1, and CHEK2 within each tumor type was performed with Microsoft Excel version 16.64 (Microsoft, WA, USA) using the Z-test for independent proportions. A p-value equal to or less than 0.05 was considered significant.
Pathogenic alterations in the relevant cancer types accessed through cBioPortal as above were jointly analyzed to determine the relationship between variants in ATM, BRCA1, CHEK2, and TP53. A Log2 odds ratio was used to calculate how strongly the presence or absence of alterations in one gene was associated with the presence or absence of alterations in a second gene within the selected samples. A q-value derived from the Benjamini-Hochberg FDR correction procedure equal to or less than 0.05 was considered significant.
All studies involving human participants were approved by the University of Miami institutional research board (IRB #20081166) and are in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
We reviewed the patients seen in our hereditary cancer clinic who had undergone comprehensive multigene panel testing and were found to be heterozygous for a germline pathogenic variant in ATM. A total of thirty-one patients met these criteria, none of which had a pathogenic variant in any other hereditary cancer gene. Twenty-five of these patients had a personal or family history of cancer that is consistent with the currently described spectrum of ATM-associated malignancies. Notably, the remaining six patients (19.4%) had a personal history of cancers not currently associated with ATM hereditary cancer susceptibility. Only one of these six patients was identified to harbor variants of uncertain significance in other hereditary cancer genes (Supplementary Table 3). To evaluate for genotype-phenotype correlations, we compared the pathogenic ATM variants in the 6 atypical cancer cases to the 25 typical cancer cases, though no clear differences were evident (Figure 1).
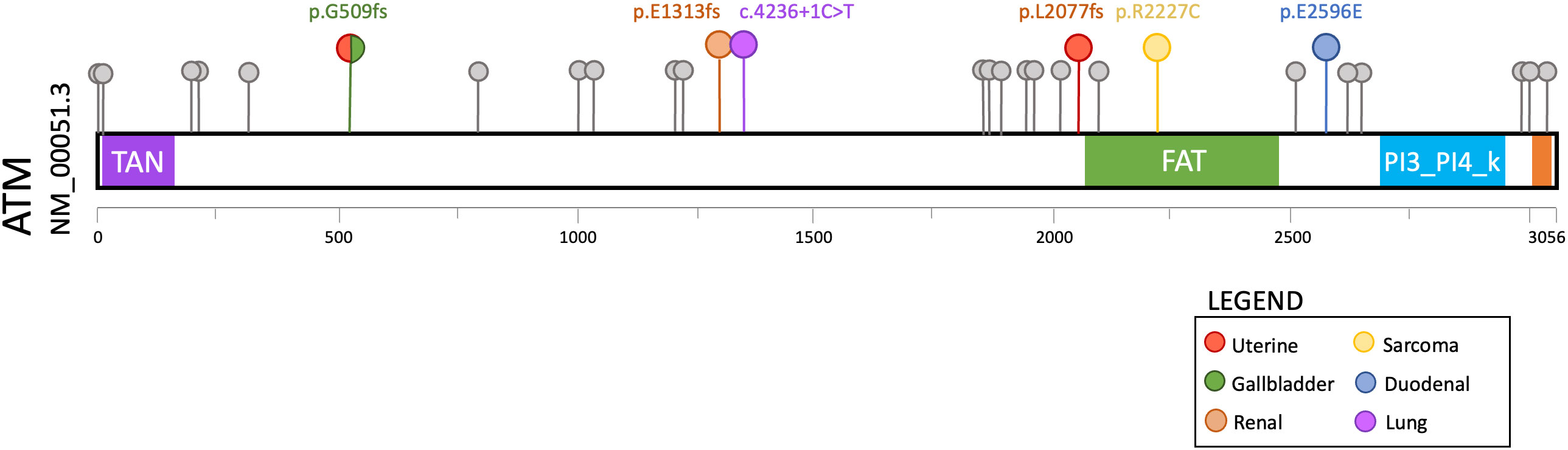
Figure 1 Lollipop plot of germline ATM pathogenic variants detected in 31 consecutive patients with a personal or family history of cancer who underwent hereditary cancer susceptibility testing. The six variants associated with cancers that are atypical for ATM hereditary cancer susceptibility are highlighted in color (see legend).
The personal and family history of cancer among the six patients with malignancies atypical for ATM-associated hereditary cancer susceptibility were then evaluated in detail (Table 1). We observed that two of the six patients had a personal history of known ATM-associated cancers. Patient 1 had a history of breast adenocarcinoma, and Patient 6 had a history of melanoma. All six patients had at least one 1st or 2nd-degree relative with ATM-associated cancer including breast, pancreatic, prostate, gastric, and colorectal adenocarcinomas. Four of the six patients meet current NCCN criteria for germline genetic testing based on either their personal history of cancer or family history of cancer (Supplementary Table 4).
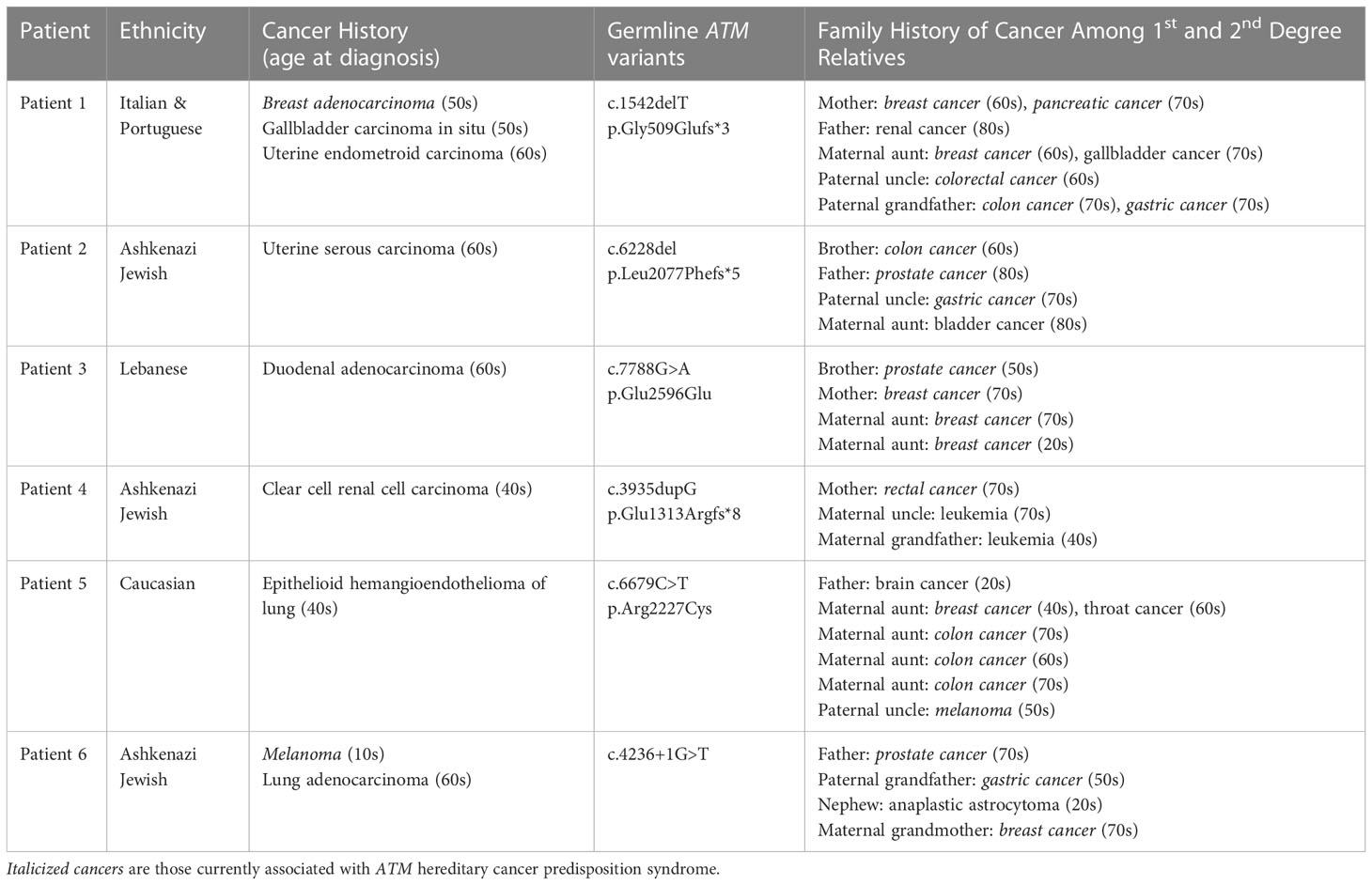
Table 1 Clinical summary of the six patients identified to harbor a pathogenic germline variant in ATM and a personal history of cancer not associated with ATM hereditary cancer syndrome.
To better characterize the association between germline ATM pathogenic variants and the atypical cancers seen in our six patients, we performed a comprehensive review of all cross-sectional studies where germline sequencing was performed in patients diagnosed with carcinomas of the biliary tract, uterus, small bowel and ampulla, kidney, and lung, as well as sarcomas. We identified twenty-five broadly relevant, cross-sectional studies where next-generation sequencing of cancer susceptibility genes was performed (Supplementary Table 5, 13–37). A total of 171 unique patients harboring a germline pathogenic variant in ATM and diagnosed with one of the atypical ATM cancers were identified. Of these, we found that fewer than 20 cases of germline ATM pathogenic variants have been associated with biliary tract carcinoma, uterine carcinoma, small bowel carcinoma, or sarcoma, while we identified 95 cases of lung adenocarcinoma and 32 cases of renal cell carcinoma associated with germline ATM pathogenic variants.
We then estimated the prevalence of germline ATM pathogenic variants for each relevant cancer type by compiling data from each reviewed study (Figure 2A). The prevalence was calculated to be as low as 0.45% for sarcoma, 0.57% for uterine carcinoma, 0.6% for biliary tract carcinoma, 0.63% for lung carcinoma, and 0.87% for renal cell carcinoma. In the case of small intestine & ampullary carcinoma, the prevalence reached 2.2% albeit with a broad range due to the limited sample size. We then calculated the prevalence of germline ATM pathogenic variants among individuals diagnosed with an atypical ATM cancer who tested positive for hereditary cancer predisposition (Figure 2B). We found that among sarcoma and renal cell carcinoma patients diagnosed with a hereditary cancer susceptibility syndrome, the prevalence of a germline ATM pathogenic variant was 2.4% and 6.5%, respectively. Meanwhile, patients with biliary tract, uterine, small bowel & ampullary, as well as lung carcinomas who tested positive for a hereditary cancer susceptibility syndrome had a germline ATM pathogenic variant in 9% - 12.7% of cases.
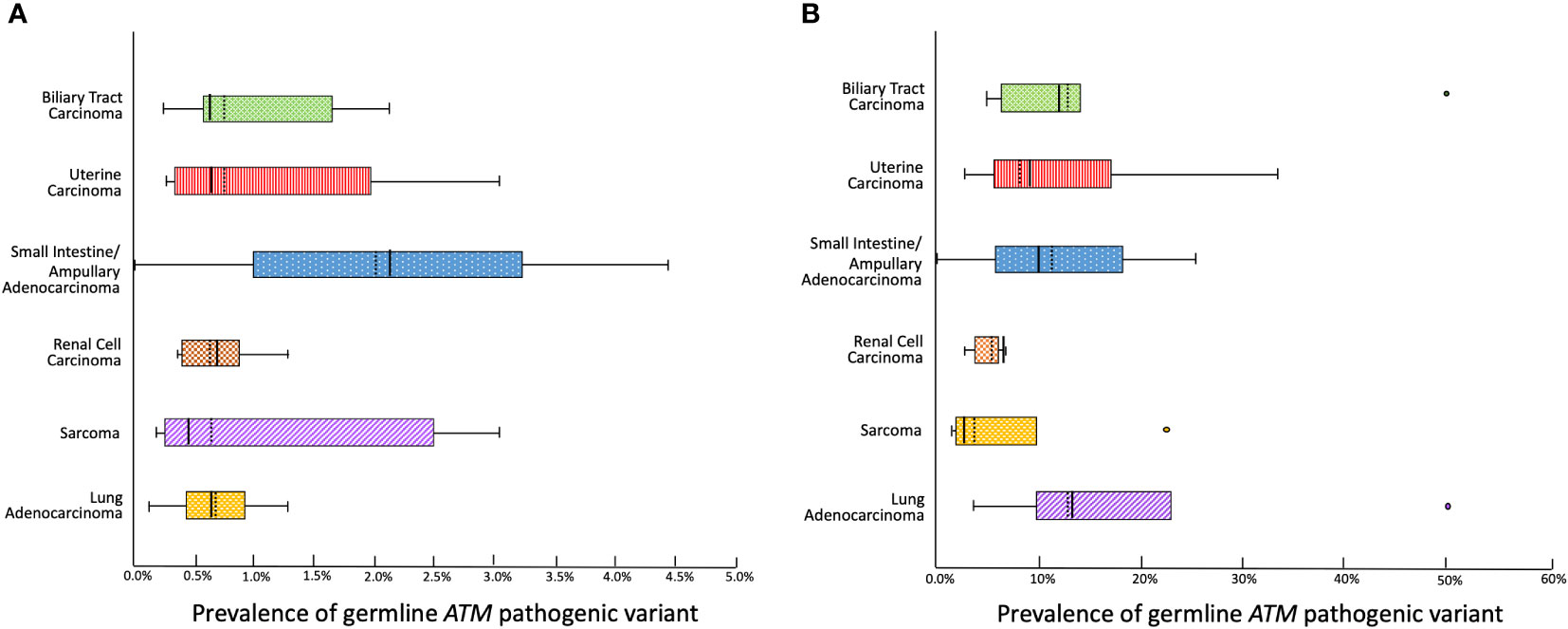
Figure 2 Box and whiskers plot depicting (A) The prevalence of a germline pathogenic ATM variant among individuals affected with each cancer type based on pooled data from a comprehensive review of cross-sectional studies. (B) The prevalence of a germline pathogenic ATM variant among individuals affected with each cancer who test positive for hereditary cancer predisposition (mean depicted with a solid vertical line, median depicted with a dashed vertical line).
These data provide evidence that germline ATM pathogenic variants exhibit a low penetrance for the atypical cancers investigated herein. Nevertheless, these germline ATM pathogenic variants were associated with a significant proportion of the known causes for hereditary cancer susceptibility in these cancers.
Next, we explored available tumor sequencing data from large cohorts that closely match the tumor type and subtype described in our patient cases with the aim of further clarifying the biological relevance of ATM loss-of-function variants in the carcinogenesis of gallbladder, uterus, duodenum, kidney, lung, and sarcomas. We queried data available through cBioPortal and determined the frequency of somatic deleterious ATM variants and compared this alteration frequency to that of BRCA1 and CHEK2, as these are DNA-damage response tumor suppressors also implicated in hereditary cancer, but not established to drive oncogenesis in the cancer types being investigated.
ATM variants were detected in as many as 12% of uterine carcinomas, 8% of gallbladder carcinomas, 7.8% of small bowel carcinomas, and 5% of lung adenocarcinomas, whereas BRCA1 and CHEK2 variants were found in less than 3% of uterine carcinomas, less than 2% of lung adenocarcinomas, less than 1% of gallbladder carcinomas, and none were found in small bowel carcinomas (Figure 3). Clear cell renal cancer had the fewest ATM pathogenic alterations, reported at a frequency of 2.3%, though still at a significantly higher frequency than that of BRCA1 and CHEK2. In the case of sarcoma, the ATM alteration frequency was 2.8%, which was significantly greater than BRCA1 but not CHEK2. We then compared these alteration frequencies to those seen in breast cancer, where all three genes have been implicated in carcinogenesis, and found pathogenic variants in ATM only occurred in 2.5% of cases and with a similar frequency to BRCA1, though more frequently than CHEK2. These data provide complementary evidence that ATM may contribute to cancer risk in each of these cancer types.
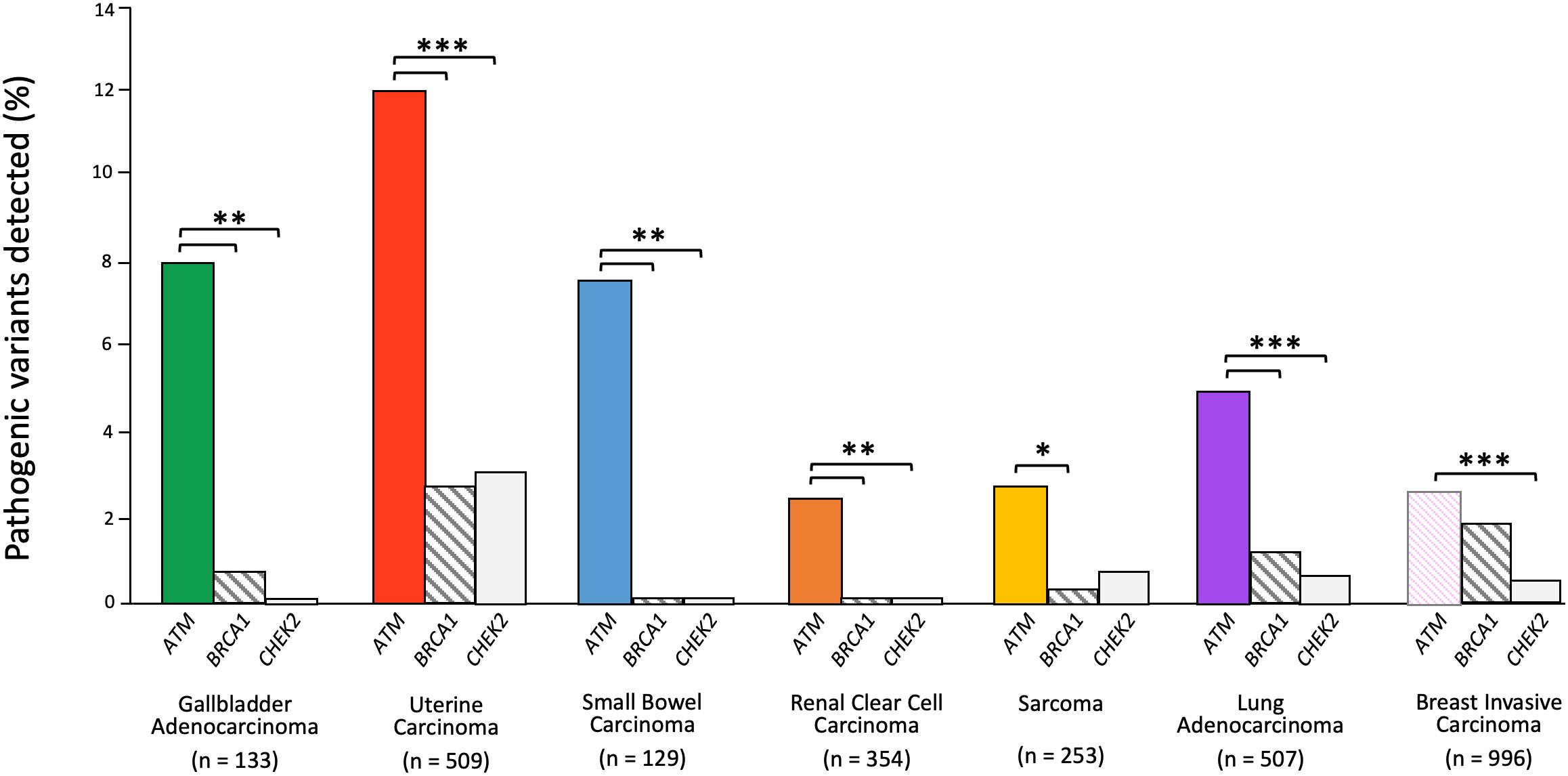
Figure 3 ATM somatic alteration frequency compared to BRCA1 and CHEK2 alterations in relevant cancers. Z-test for independent proportions was used to identify significant differences (* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, all other comparisons not significant). Tumor sequencing data was accessed through cBioPortal.
To better understand the influence of germline and somatic ATM variants on carcinogenesis in these atypical ATM cancers, we pooled the cancer sequencing data to examine the relationship between ATM alterations and the alteration of other genes in the DNA damage response and cell cycle pathways (Figure 4). We found a significant co-occurrence of pathogenic alterations in ATM with pathogenic alterations in BRCA1 and CHEK2 (Log2 OR 2.7, 2.5, p<0.01). In addition, we saw significant mutual exclusivity between ATM and TP53 (Log2 OR -0.67, p<0.05). This suggests that the presence of germline ATM pathogenic variants may influence associated cancers to be driven toward DNA-damage repair deficiency, and away from loss of TP53, the most ubiquitous driver of cancer (38).
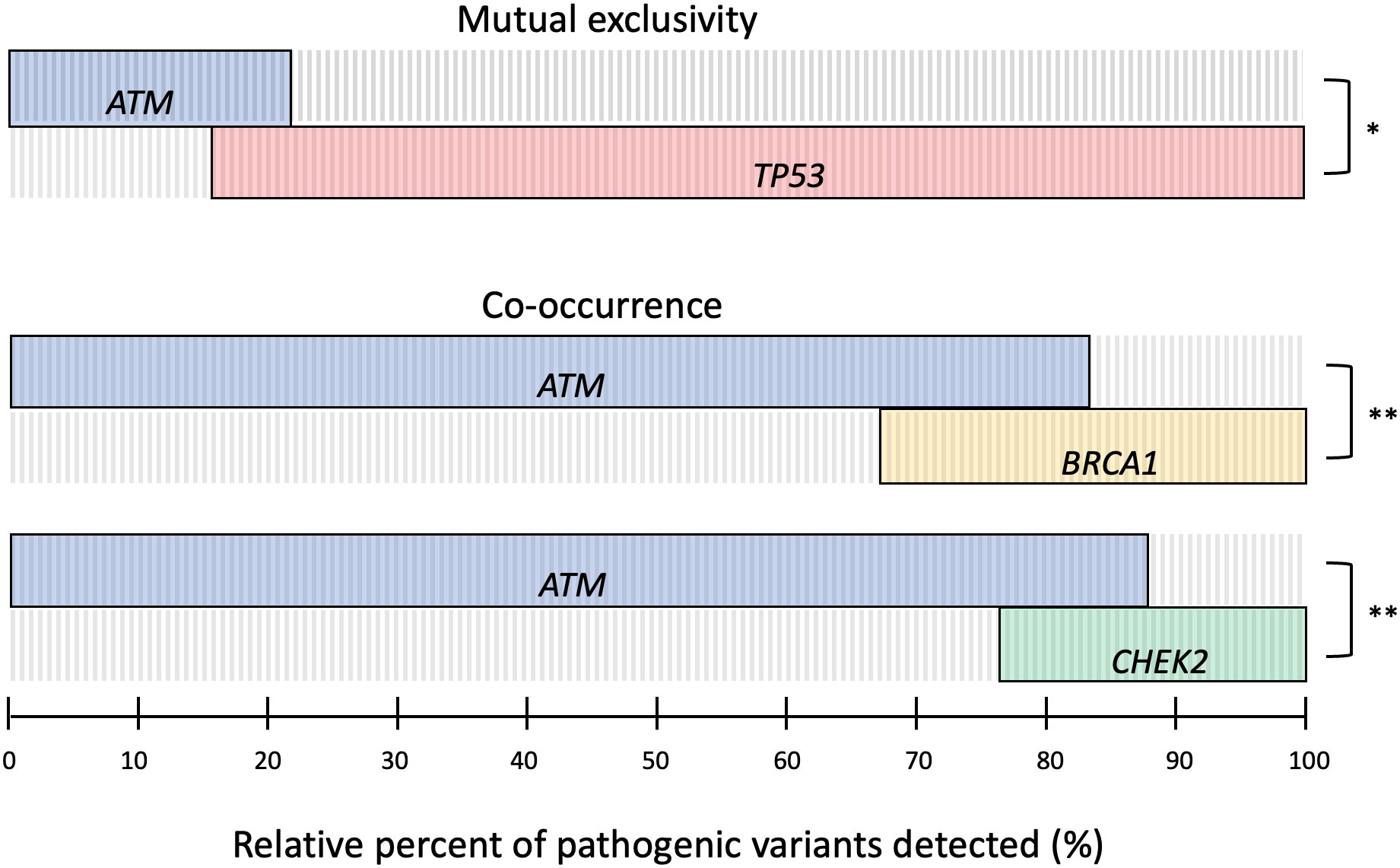
Figure 4 Oncoprint plot of somatic pathogenic alterations in ATM, TP53, BRCA, and CHEK2 across relevant tumor sequencing data to determine the relationship between alterations. Log2 odds ratio was used to identify mutual exclusivity and co-occurrence, with significant differences defined by Benjamini-Hochberg FDR correction (* q ≤ 0.05, ** q ≤ 0.01, *** q ≤ 0.001).
Here we describe six unrelated patients found to be heterozygous for germline, loss-of-function variants in ATM and diagnosed with at least one malignancy that is not currently associated with the ATM cancer susceptibility syndrome, including gallbladder carcinoma, uterine carcinoma, renal cell carcinoma, duodenal adenocarcinoma, lung adenocarcinoma, and a vascular sarcoma. These atypical cancers were diagnosed in a significant proportion of patients with ATM-associated hereditary cancer susceptibility in our cohort. Nevertheless, our review of the literature indicates that the prevalence of germline ATM pathogenic variants among individuals diagnosed with these atypical cancers appears to be less than 1% in most cases.
Exploration of somatic alterations in The Cancer Genome Atlas (TCGA) datasets demonstrated there was a high frequency of pathogenic alterations in ATM relative to other tumor suppressors associated with hereditary cancer syndromes, suggesting that ATM loss may contribute to oncogenesis within these cancer types. Furthermore, our analysis reveals that alterations in ATM tend to co-occur with pathogenic alterations in BRCA1 and CHEK2 while being mutually exclusive of TP53. This latter finding underscores the shared functional pathway between ATM and TP53 and reinforces prior work showing that DNA damage creates selection pressure for the inactivation of p53 that can be abrogated through the loss of ATM (39, 40). More broadly, these pathway interactions illustrate how germline ATM pathogenic variants may influence the acquisition of somatic alterations throughout cancer initiation and progression.
The cases reported in this series exemplify how the tumor spectra associated with cancer susceptibility syndromes are broadening through the clinical use of pan-cancer panels. The germline pathogenic variants in ATM detected in all six of our patients were discovered through comprehensive hereditary cancer panels. This testing approach is supported by studies demonstrating significantly higher detection rates compared to panels oriented specifically to the genes that are most relevant to the personal or family history of cancer (41–43). The heterogeneity in cancers documented among our patients with germline ATM variants and their families further emphasizes the value of pan-cancer panels, and certainly the importance of including ATM in multi-gene panels testing for hereditary cancer predisposition.
Moreover, our cases illustrate the way NCCN Guidelines may significantly limit the detection of hereditary cancer susceptibility, as many cancer types, particularly those with a low prevalence, do not currently qualify individuals for genetic testing. In our series, neither Patient 4 nor Patient 6 met guidelines for hereditary cancer testing based on their personal or family history of cancer, illustrating circumstances in which ATM germline variants may go undetected. This was underscored in a recent study where germline genetic testing in an unselected pan-cancer patient population found that over half of the patients who tested positive for pathogenic variants were not eligible for hereditary cancer testing based on current guidelines (44). Improving the detection of ATM-associated hereditary cancer is clinically meaningful as it can enable cascade genetic testing for family members who are at risk, facilitate appropriate cancer surveillance for those affected, and prompt the use of precision-based therapies.
Although no specific ATM genotype-phenotype correlations were discerned in this study, we did observe that the ATM c.1564_1565delGA variant was present in two patients with uterine cancer, and the ATM p.V2716A variant was present in two patients with lung adenocarcinoma. In addition, we found the germline ATM variant c.6679C>T identified in Patient 5 with epithelioid hemangioendothelioma was also seen in a case of endometrioid uterine cancer, and the germline ATM variant c.4236+1G seen in Patient 6 with lung adenocarcinoma was also present in a case of renal cell carcinoma. Meanwhile, none of the pathogenic germline ATM variants identified in our six patients overlapped among the somatic loss-of-function ATM variants present in the cancer sequencing datasets. Larger sample sizes will be needed to systematically investigate whether specific germline ATM variants confer a greater risk for atypical ATM cancers.
A limitation of this study is that we cannot exclude the possibility that our patients’ malignancies were incidental to, rather than caused by the pathogenic variant in ATM, as the incidence of heterozygosity for pathogenic variants in ATM has been estimated in gnomAD for European populations to be 0.48% (45). A segregation analysis of our patients’ families may have strengthened the association between atypical cancers and pathogenic variants in ATM, as this would have revealed whether the gallbladder carcinoma in the maternal aunt of Patient 1, the leukemias in the maternal uncle and grandfather of Patient 4, or the anaplastic astrocytoma in the paternal nephew of Patient 6 were also associated with pathogenic variants in ATM. Moreover, we were not able to confirm whether ATM loss of heterozygosity occurred in our six patients, which could have provided further evidence that ATM is driving carcinogenesis based on the two-hit hypothesis (46). Yet, this does not appear to be a reliable finding as the frequency of biallelic two-hit events across all cancers where a germline pathogenic variant is present has been estimated at 38.3%, with a recent study finding that ATM somatic second hits occurred in 11.3% in a wide spectrum of cancers including breast, pancreatic, bladder, uterine and lung cancers, among others (15, 47). A case-control analysis for each of the atypical-ATM cancer types could have also provided compelling evidence of an association. This has been previously performed in two cohorts of patients with lung adenocarcinoma, demonstrating a significant enrichment for germline pathogenic variants in ATM in cases compared to controls (48, 49). Unfortunately, the feasibility of case-control studies is limited by the rarity of many of these cancers, as well as the low penetrance of ATM in association with these atypical cancers.
Nevertheless, our finding of relatively frequent somatic ATM alterations across many of the atypical cancer types has been corroborated by an analysis showing that ATM was among the most frequently mutated genes in the DNA-damage response pathway across 33 distinct types of cancers (50). This evidence has also been bolstered by a recent study demonstrating a high prevalence of germline pathogenic variants in ATM relative to other hereditary cancer genes across a spectrum of cancers lacking testing guidelines, including bladder, brain, esophagus, and head and neck cancers (15). Furthermore, our identification that pathogenic ATM alterations in these atypical cancers are associated with the co-occurrence of alterations in BRCA1 and CHEK2, and mutual exclusivity with TP53, suggests that cancers with a germline pathogenic variant in ATM may depend on specific molecular alterations that confer targetable vulnerabilities. This has already been documented in patients with metastatic prostate cancer harboring a germline pathogenic variant in ATM sensitizing to PARP inhibition, as well as in patients with pancreatic cancer where monoallelic pathogenic variants in ATM produce susceptibility to combined therapies that synergistically target the DNA-damage response pathway (51, 52).
In conclusion, this study combines detailed clinical phenotyping with a comprehensive review of relevant germline ATM reports and large-scale tumor sequencing data to propose that germline pathogenic variants in ATM may be associated with the development of cancers of the gallbladder, duodenum, uterus, kidney, and lung as well as sarcoma cancers. Consideration should be given to a broadening of the ATM-cancer susceptibility syndrome phenotype within hereditary cancer testing guidelines, as this has the potential to improve the detection of affected patients and facilitate the use of more effective cancer treatments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by University of Miami institutional research board (IRB #20081166). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
NB: Conceptualization, methodology, data curation, formal analysis, project administration, visualization, writing–original draft, and writing–review and editing. RS-S: Conceptualization, investigation, resources, data curation, writing–review and editing. MH: Investigation, resources, data interpretation, writing–review and editing. DP: Investigation, resources, and writing–review and editing. DS: Investigation, resources, writing–review and editing. MT: Conceptualization, methodology, formal analysis, investigation, writing–original draft, and writing–review and editing, visualization, supervision. All authors contributed to the article and approved the submitted version.
Institutional support was received for open-access publication fees.
We thank Thilani Samarakoon and Cameron Riopelle for their help with the data analysis and graphical visualizations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1068110/full#supplementary-material
1. Choi M, Kipps T, Kurzrock R. ATM Mutations in cancer: Therapeutic implications. Mol Cancer Ther (2016) 15:1781–91. doi: 10.1158/1535-7163.mct-15-0945
2. Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Bio (2013) 14:197–210. doi: 10.1038/nrm3546
3. Rothblum-Oviatt C, Wright J, Lefton-Greif MA, McGrath-Morrow SA, Crawford TO, Lederman HM. Ataxia telangiectasia: A review. Orphanet J Rare Dis (2016) 11:159. doi: 10.1186/s13023-016-0543-7
4. Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. Jnci J Natl Cancer Inst (2005) 97:813–22. doi: 10.1093/jnci/dji141
5. Marabelli M, Cheng S, Parmigiani G. Penetrance of ATM gene mutations in breast cancer: A meta-analysis of different measures of risk. Genet Epidemiol (2016) 40:425–31. doi: 10.1002/gepi.21971
6. Hall MJ, Bernhisel R, Hughes E, Larson K, Rosenthal ET, Singh NA, et al. Germline pathogenic variants in the ataxia telangiectasia mutated (ATM) gene are associated with high and moderate risks for multiple cancers. Cancer Prev Res (2021) 14:433–40. doi: 10.1158/1940-6207.capr-20-0448
7. Chaffee KG, Oberg AL, McWilliams RR, Majithia N, Allen BA, Kidd J, et al. Prevalence of germline mutations in cancer genes among pancreatic cancer patients with positive family history. Genet Med Off J Am Coll Med Genet (2018) 20:119–27. doi: 10.1038/gim.2017.85
8. Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol (2016) 3:464. doi: 10.1001/jamaoncol.2016.5194
9. Goldstein AM, Xiao Y, Sampson J, Zhu B, Rotunno M, Bennett H, et al. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum Mol Genet (2017) 26:4886–95. doi: 10.1093/hmg/ddx368
10. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med (2015) 17:405–23. doi: 10.1038/gim.2015.30
11. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal (2013) 6:pl1. doi: 10.1126/scisignal.2004088
12. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery (2012) 2:401–4. doi: 10.1158/2159-8290.cd-12-0095
13. Maynard H, Stadler ZK, Berger MF, Solit DB, Ly M, Lowery MA, et al. Germline alterations in patients with biliary tract cancers: A spectrum of significant and previously underappreciated findings. Cancer (2020) 126:1995–2002. doi: 10.1002/cncr.32740
14. Wardell CP, Fujita M, Yamada T, Simbolo M, Fassan M, Karlic R, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol (2018) 68:959–69. doi: 10.1016/j.jhep.2018.01.009
15. Yap TA, Ashok A, Stoll J, Mauer E, Nepomuceno VM, Blackwell KL, et al. Prevalence of germline findings among tumors from cancer types lacking hereditary testing guidelines. JAMA Netw Open (2022) 5:e2213070. doi: 10.1001/jamanetworkopen.2022.13070
16. Lin J, Dong K, Bai Y, Zhao S, Dong Y, Shi J, et al. Precision oncology for gallbladder cancer: insights from genetic alterations and clinical practice. Ann Transl Med (2019) 7:467–7. doi: 10.21037/atm.2019.08.67
17. Bertelsen B, Tuxen IV, Yde CW, Gabrielaite M, Torp MH, Kinalis S, et al. High frequency of pathogenic germline variants within homologous recombination repair in patients with advanced cancer. NPJ Genom Med (2019) 4:13. doi: 10.1038/s41525-019-0087-6
18. Karpel HC, Chern J-Y, J. MS, A. JS, Pothuri B. Utility of germline multi-gene panel testing in patients with endometrial cancer. Gynecol Oncol (2022) 165:546–51. doi: 10.1016/j.ygyno.2022.04.003
19. Ring KL, Bruegl AS, Allen BA, Elkin EP, Singh N, Hartman A-R, et al. Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Modern Pathol (2016) 29:1381–9. doi: 10.1038/modpathol.2016.135
20. Kondrashova O, Shamsani J, O’Mara TA, Newell F, Reed AEM, Lakhani SR, et al. Tumor signature analysis implicates hereditary cancer genes in endometrial cancer development. Cancers (2021) 13:1762. doi: 10.3390/cancers13081762
21. Long B, Lilyquist J, Weaver A, Hu C, Gnanaolivu R, Lee KY, et al. Cancer susceptibility gene mutations in type I and II endometrial cancer. Gynecol Oncol (2019) 152:20–5. doi: 10.1016/j.ygyno.2018.10.019
22. Latham A, Shia J, Patel Z, Reidy-Lagunes DL, Segal NH, Yaeger R, et al. Characterization and clinical outcomes of DNA mismatch repair–deficient small bowel adenocarcinoma. Clin Cancer Res (2021) 27:1429–37. doi: 10.1158/1078-0432.ccr-20-2892
23. Kumari N, Singh RK, Mishra SK, L R, Mohindra S, Krishnani N. Prevalence and spectrum of pathogenic germline variants in intestinal and pancreatobiliary type of ampullary cancer. Pathol - Res Pract (2021) 217:153309. doi: 10.1016/j.prp.2020.153309
24. Wong W, Lowery MA, Berger MF, Kemel Y, Taylor B, Zehir A, et al. Ampullary cancer: Evaluation of somatic and germline genetic alterations and association with clinical outcomes. Cancer (2019) 125:1441–8. doi: 10.1002/cncr.31951
25. Kong W, Yang T, Wen X, Mu Z, Zhao C, Han S, et al. Germline mutation landscape and associated clinical characteristics in Chinese patients with renal cell carcinoma. Front Oncol (2021) 11:737547. doi: 10.3389/fonc.2021.737547
26. Truong H, Sheikh R, Kotecha R, Kemel Y, Reisz PA, Lenis AT, et al. Germline variants identified in patients with early-onset renal cell carcinoma referred for germline genetic testing. Eur Urol Oncol (2021) 4:993–1000. doi: 10.1016/j.euo.2021.09.005
27. Carlo MI, Mukherjee S, Mandelker D, Vijai J, Kemel Y, Zhang L, et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol (2018) 4:1228. doi: 10.1001/jamaoncol.2018.1986
28. Santos M, Lanillos J, Roldan-Romero JM, Caleiras E, Montero-Conde C, Cascón A, et al. Prevalence of pathogenic germline variants in patients with metastatic renal cell carcinoma. Genet Med (2021) 23:698–704. doi: 10.1038/s41436-020-01062-0
29. Alaiwi SA, Nassar AH, Adib E, Groha SM, Akl EW, McGregor BA, et al. Trans-ethnic variation in germline variants of patients with renal cell carcinoma. Cell Rep (2021) 34:108926. doi: 10.1016/j.celrep.2021.108926
30. Yngvadottir B, Andreou A, Bassaganyas L, Larionov A, Cornish AJ, Chubb D, et al. Frequency of pathogenic germline variants in cancer susceptibility genes in 1336 renal cell carcinoma cases. Hum Mol Genet (2022) 31:ddac089. doi: 10.1093/hmg/ddac089
31. Chan SH, Lim WK, Ishak NDB, Li S-T, Goh WL, Tan GS, et al. Germline mutations in cancer predisposition genes are frequent in sporadic sarcomas. Sci Rep-uk (2017) 7:10660. doi: 10.1038/s41598-017-10333-x
32. Gillani R, Camp SY, Han S, Jones JK, Chu H, O’Brien S, et al. Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes. Am J Hum Genet (2022) 109:1026–37. doi: 10.1016/j.ajhg.2022.04.007
33. Mirabello L, Zhu B, Koster R, Karlins E, Dean M, Yeager M, et al. Frequency of pathogenic germline variants in cancer-susceptibility genes in patients with osteosarcoma. JAMA Oncol (2020) 6:724–34. doi: 10.1001/jamaoncol.2020.0197
34. Kim J, Light N, Subasri V, Young EL, Wegman-Ostrosky T, Barkauskas DA, et al. Pathogenic germline variants in cancer susceptibility genes in children and young adults with rhabdomyosarcoma. Jco Precis Oncol (2021) 5:75–87. doi: 10.1200/po.20.00218
35. Parry EM, Gable DL, Stanley SE, Khalil SE, Antonescu V, Florea L, et al. Germline mutations in DNA repair genes in lung adenocarcinoma. J Thorac Oncol (2017) 12:1673–8. doi: 10.1016/j.jtho.2017.08.011
36. Mukherjee S, Bandlamudi C, Hellmann MD, Kemel Y, Drill E, Rizvi H, et al. Germline pathogenic variants impact clinicopathology of advanced lung cancer. Cancer Epidemiol Biomarkers Prev (2022) 31:1450–9. doi: 10.1158/1055-9965.epi-21-1287
37. Tian P, Cheng X, Zhao Z, Zhang Y, Bao C, Wang Y, et al. Spectrum of pathogenic germline mutations in Chinese lung cancer patients through next-generation sequencing. Pathol Oncol Res (2020) 26:109–14. doi: 10.1007/s12253-019-00771-5
38. Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature (2020) 578:82–93. doi: 10.1038/s41586-020-1969-6
39. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Bio (2010) 11:220–8. doi: 10.1038/nrm2858
40. Pettitt AR, Sherrington PD, Stewart G, Cawley JC, Taylor AMR, Stankovic T. p53 dysfunction in b-cell chronic lymphocytic leukemia: Inactivation of ATM as an alternative toTP53 mutation. Blood (2001) 98:814–22. doi: 10.1182/blood.v98.3.814
41. Rosenthal ET, Bernhisel R, Brown K, Kidd J, Manley S. Clinical testing with a panel of 25 genes associated with increased cancer risk results in a significant increase in clinically significant findings across a broad range of cancer histories. Cancer Genet-ny (2017) 218:58–68. doi: 10.1016/j.cancergen.2017.09.003
42. LaDuca H, Polley EC, Yussuf A, Hoang L, Gutierrez S, Hart SN, et al. A clinical guide to hereditary cancer panel testing: Evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet Med (2020) 22:407–15. doi: 10.1038/s41436-019-0633-8
43. Cavaillé M, Uhrhammer N, Privat M, Ponelle-Chachuat F, Gay-Bellile M, Lepage M, et al. Feedback of extended panel sequencing in 1530 patients referred for suspicion of hereditary predisposition to adult cancers. Clin Genet (2021) 99:166–75. doi: 10.1111/cge.13864
44. Perkins AT, Haslem D, Goldsberry J, Shortt K, Sittig L, Raghunath S, et al. Universal germline testing of unselected cancer patients detects pathogenic variants missed by standard guidelines without increasing healthcare costs. Cancers (2021) 13:5612. doi: 10.3390/cancers13225612
45. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature (2020) 581:434–43. doi: 10.1038/s41586-020-2308-7
46. Knudson AG. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci (1971) 68:820–3. doi: 10.1073/pnas.68.4.820
47. Huang K, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, et al. Pathogenic germline variants in 10,389 adult cancers. Cell (2018) 173:355–370.e14. doi: 10.1016/j.cell.2018.03.039
48. Selvan ME, Zauderer MG, Rudin CM, Jones S, Mukherjee S, Offit K, et al. Inherited rare, deleterious variants in ATM increase lung adenocarcinoma risk. J Thorac Oncol (2020) 15:1871–9. doi: 10.1016/j.jtho.2020.08.017
49. Sang J, Zhang T, Kim J, Li M, Pesatori AC, Consonni D, et al. Rare germline deleterious variants increase susceptibility for lung cancer. Hum Mol Genet (2022) 31:3558–65. doi: 10.1093/hmg/ddac123
50. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep (2018) 23:239–254.e6. doi: 10.1016/j.celrep.2018.03.076
51. Gout J, Perkhofer L, Morawe M, Arnold F, Ihle M, Biber S, et al. Synergistic targeting and resistance to PARP inhibition in DNA damage repair-deficient pancreatic cancer. Gut (2021) 70:743–60. doi: 10.1136/gutjnl-2019-319970
Keywords: ATM, ataxia-telangiectasia mutated (ATM), germline, pathogenic variant (PV), hereditary cancer, cancer susceptibility association
Citation: Borja NA, Silva-Smith R, Huang M, Parekh DJ, Sussman D and Tekin M (2023) Atypical ATMs: Broadening the phenotypic spectrum of ATM-associated hereditary cancer. Front. Oncol. 13:1068110. doi: 10.3389/fonc.2023.1068110
Received: 12 October 2022; Accepted: 03 February 2023;
Published: 14 February 2023.
Edited by:
Athina Markou, National and Kapodistrian University of Athens, GreeceReviewed by:
Emilio Cordova, National Institute of Genomic Medicine (INMEGEN), MexicoCopyright © 2023 Borja, Silva-Smith, Huang, Parekh, Sussman and Tekin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mustafa Tekin, bXRla2luQG1lZC5taWFtaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.