- 1Department of Hepatobiliary and Pancreatic Surgery, Second Affiliated Hospital of Jilin University, Jilin University, Changchun, China
- 2Department of General Affairs, First Hospital of Jilin University (the Eastern Division), Jilin University, Changchun, China
- 3Department of Pediatric Neurology, First Hospital of Jilin University, Jilin University, Changchun, China
The COMMD proteins are a highly conserved protein family with ten members that play a crucial role in a variety of biological activities, including copper metabolism, endosomal sorting, ion transport, and other processes. Recent research have demonstrated that the COMMD proteins are closely associated with a wide range of disorders, such as hepatitis, myocardial ischemia, cerebral ischemia, HIV infection, and cancer. Among these, the role of COMMD proteins in tumors has been thoroughly explored; they promote or inhibit cancers such as lung cancer, liver cancer, gastric cancer, and prostate cancer. COMMD proteins can influence tumor proliferation, invasion, metastasis, and tumor angiogenesis, which are strongly related to the prognosis of tumors and are possible therapeutic targets for treating tumors. In terms of molecular mechanism, COMMD proteins in tumor cells regulate the oncogenes of NF-κB, HIF, c-MYC, and others, and are related to signaling pathways including apoptosis, autophagy, and ferroptosis. For the clinical diagnosis and therapy of malignancies, additional research into the involvement of COMMD proteins in cancer is beneficial.
1 Introduction
The protein family of copper metabolism MURR1 domain-containing (COMMD) is highly conserved in eukaryotic multicellular organisms and participates in numerous cellular processes, such as copper metabolism, endosomal sorting, ion transport, and transcription factor regulation (1). The COMMD family consists of ten members (Figure 1), of which COMMD1 (also known as MURR1) has been the subject of most research since its discovery by Nabetani et al. (2). Exon 2 deletion of the COMMD1 gene was identified in Bedlington Terriers with copper toxicosis in a pervious study (3), demonstrating for the first time that COMMD1 regulates copper metabolism. Copper toxicosis is an autosomal recessive illness characterized by copper secretory abnormalities and hepatic copper accumulation, hepatitis, and even cirrhosis. Subsequently, additional members of the COMMD protein family with comparable structure and a highly conserved COMM domain at the carboxy-terminal region were discovered; the conserved COMM domain participates in protein-protein interaction and nuclear transport, and is ubiquitination targets (4, 5). In contrast, the amino terminal domain is highly variable and practically nonexistent in COMMD6 (4). COMMD proteins are extensively expressed in a range of human tissues, with expression difference between each COMMD member (6) indicating different roles of each COMMD member in particular tissues. In recent years, the majority of research on the function of COMMD proteins has been on COMMD1; little is known about the function of the other COMMD proteins (7). This review focuses on the biological roles of the COMMD protein family and the particularity of COMMD1, and involves as far as possible the research results about other members of the COMMD protein family. Given the functional diversity of COMMD proteins, the COMMD protein family is implicated in numerous disease processes, such as hepatitis (8, 9), myocardial ischemia (10–12), cerebral ischemia (13), HIV infection (14, 15), and malignancies (4). COMMD1 deficiency induces or worsens hepatitis in mouse and dogs, and the mechanism might involve an increase in intracellular hepatic copper and hepatic lipid accumulation (8, 9). Heart ischemia is frequently accompanied by Cu depletion (16). Cu concentration is decreased in the ischemic zone of a mouse model of myocardial ischemia, but COMMD1 protein levels are up. COMMD1-knockdown can decrease the outflow of myocardial Cu, safeguard myocardial function, and diminish the size of infarct focus. However, the cause of COMMD1 rise due to myocardial ischemia requires additional study (10, 12). Although the majority of molecular mechanism research have focused on COMMD1, nearly all COMMD protein family members regulate tumor progression. For example, COMMD1 (17), COMMD4 (18), COMMD8 (19), and COMMD9 (20) regulate the proliferation, invasion, and metastasis of lung cancer. COMMD2 (21), COMMD3 (22), COMMD7 (23), COMMD8 (24), and COMMD10 (25) promote or inhibit hepatocellular carcinoma (HCC). Additionally, COMMD1 (26) and COMMD10 (27) are linked to drug resistance in tumors. After briefly explaining the biological functions of the COMMD protein family, this review then comprehensively detailed the specific roles of COMMD proteins in tumors of diverse systems in order to serve as a resource for future tumor research and clinical diagnosis and therapy.
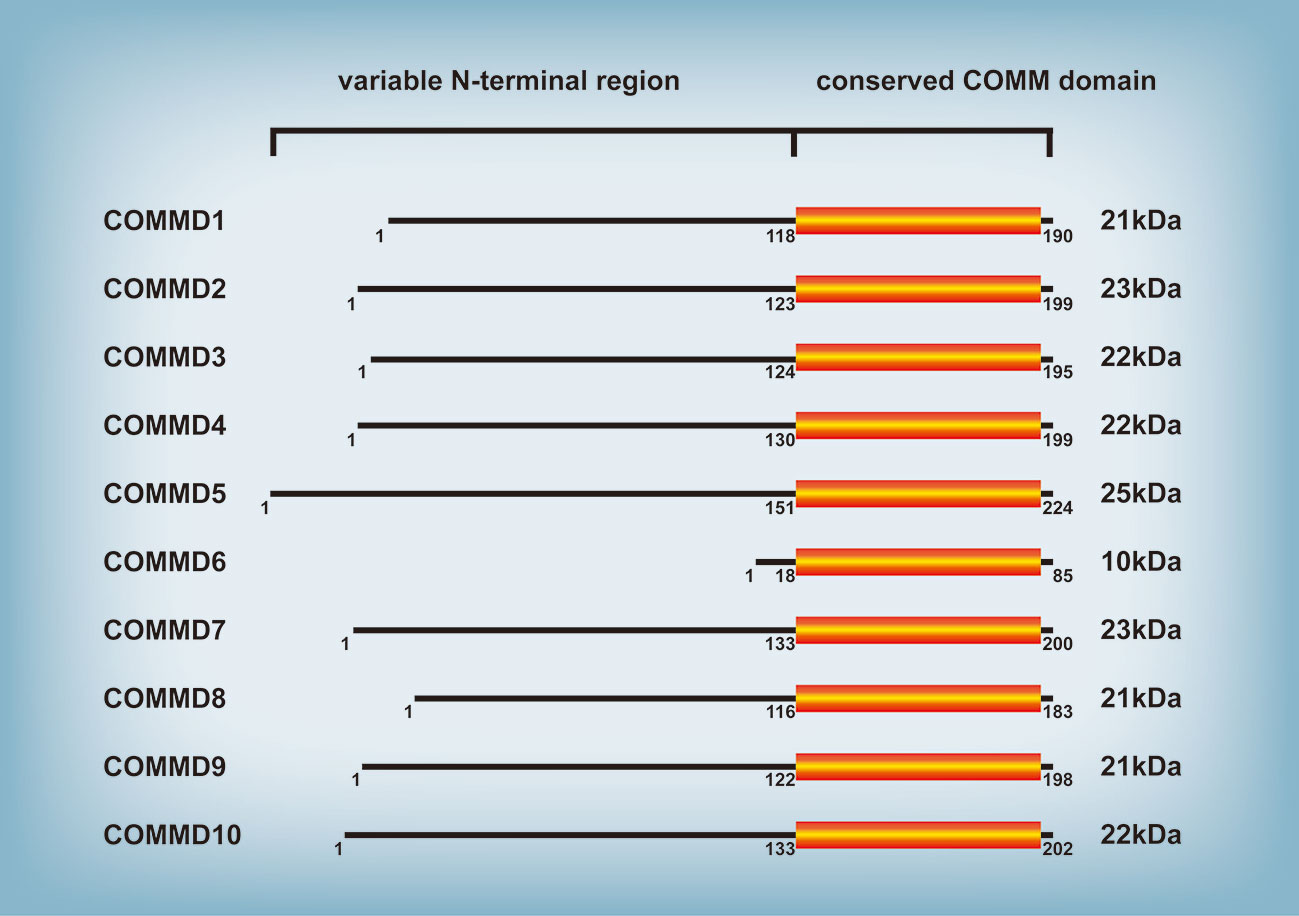
Figure 1 COMMD family of proteins. All ten members of the COMMD protein family share a conserved COMM domain. In contrast, the amino terminal domain is highly variable among different COMMD proteins.
2 The primary biological functions of COMMD protein family
2.1 Regulating copper metabolism
Copper is an important trace element that plays a significant role in many biological processes, including cell respiration, cell proliferation, antioxidant defense, neurotransmitter synthesis and angiogenesis. Wilson’s disease, Alzheimer’s disease, and Parkinson’s disease have been associated with an abnormal copper metabolism (28, 29). Normal life activities rely heavily on copper homeostasis being maintained. The liver plays a crucial role in controlling systemic copper homeostasis. Copper is absorbed in the intestine to fulfill the body’s needs, and excreted through the bile to prevent toxicity. Cu transporting ATPase B (Atp7b) in hepatocytes assists secreting excess copper, which will be excreted with bile (28). In humans, mutations in the Atp7b gene are responsible for Wilson’s disease, a disorder that produces hepatic copper overload comparable to copper toxicosis in dogs. Exon 2 deletion of COMMD1 in Bedlington Terriers results in the loss of COMMD1 protein expression, which ultimately results in aberrant biliary copper excretion and copper toxicosis (30). However, the mechanism by which COMMD1 regulates biliary copper excretion remains unknown. Studies have demonstrated that COMMD1 could bind to mutant Atp7b and promoted its degradation via the proteasomal pathway (31). Researchers speculated that COMMD1 is involved in the quality regulation of Atp7b through hydrolyzing misfolded and dysfunctional proteins caused by mutations. Failure to properly regulate Atp7b in Bedlington Terriers due to COMMD1 deficiency may be the underlying cause of copper toxicosis (32). Copper is efficiently secreted due to the bidirectional translocation of Atp7b between trans-Golgi network and cytosolic vesicles. Miyayama et al. discovered that COMMD1-knockdown decreased the expression of Atp7b; concurrently, the recycling of Atp7b from cytosolic vesicles to the trans-Golgi network is hindered, resulting in the accumulation of intracellular copper (33). The majority of dietary copper is absorbed in the small intestine, and Atp7a transports copper from small intestine cells to the portal vein. A mutation in the Atp7a gene could cause Menkes’ disease, which is mostly characterized by copper deficiency (28). COMMD1 could bind to both wild-type and mutant Atp7a, increasing Atp7a’s stability and ameliorating the copper exporting capacities of Atp7a mutants (34). In contrast, another research published in the same year shown that COMMD1 triggered the degradation of wild-type and mutant Atp7a through proteasomal pathway (31). The role of COMMD1 in the stability of Atp7a requires additional study. COMMD1 can also regulate the intracellular transport of Atp7a. COMMD1 forms the COMMD/CCDC22/CCDC93 (CCC) complex with CCDC22, CCDC93, and C16orf62, which is necessary for the proper sorting and relocalization of Atp7a (4, 35). COMMD1 can directly bind copper in addition to interacting with Atp7a and Atp7b. However, the importance of this binding effect on copper homeostasis is unexplored (36). There is no indication that COMMD1 gene mutation induces human copper metabolic-related disorders such as Wilson’s disease, while numerous pieces of evidence indicate that COMMD1 is involved in the regulation of copper metabolism. Also, the role of COMMD2-10 in regulating copper metabolism is rarely explored. It has been observed that the deletion of any one of COMMD1, COMMD6, or COMMD9 leads to a deficiency in the transport of Atp7b from cytosolic vesicles to the plasma membrane in mouse hepatocytes, and a hepatic copper accumulation in conditions of a high-copper diet (37). The function of the COMMD protein family in copper homeostasis requires additional investigation.
2.2 Assisting in endosomal sorting of transmembrane receptors
Receptor endocytosis and subsequent sorting is an important way to maintain cell homeostasis. Early endosomes internalize transmembrane proteins and their associated macromolecules, some of which degrade through the lysosomal pathway while others are transported to trans-Golgi network or plasma membrane for reuse by retromer, retriever, WASH complex, CCC complex, and SNX proteins (38). Phosphatidylinositol 4, 5-bisphosphate (PtdIns (4, 5) P2) is an important membrane-anchoring molecule, functioning in vesicular trafficking and modulation of transporter activity. The specific binding of COMMD1 to PtdIns(4, 5)P2 makes COMMD1 recruited to the endocytotic membranes, where it functions in endosomal sorting (39). Subsequent investigations demonstrated that COMMD1’s endosomal sorting role is performed through the formation of the CCC complex. Depletion of COMMD1 or CCC complex inhibits the transfer of Atp7a from endosomal vesicles to the plasma membrane. CCC complex binds to the WASH complex member FAM21, and CCC complex, WASH complex, and retromer are all implicated in the regulation of endosomal trafficking of Atp7a (35). The low-density lipoprotein receptor (LDLR) plays significant role in eliminating low-density lipoprotein (LDL). CCC and WASH complexes are both involved in the endosomal sorting of LDLR to maintain the homeostasis of circulating cholesterol. Similar to COMMD1, COMMD6-knockout and COMMD9 in mouse hepatocytes also destabilizes the CCC complex, resulting in reduced LDLR levels on the cell surface and elevated plasma cholesterol (40, 41). Another research conducted in the same year indicated that COMMD9 is also a component of the CCC complex and is essential for the endosomal sorting of Notch receptors. Given that almost all members of the COMMD protein family can connect with the CCC complex component CCDC22 (40), it is hypothesized that the various COMMD proteins associated with the CCC complex aid in the specific cargos selection (42). Unlike Atp7a, the endosomal cargo recycling of α5β1-integrin is retromer-independent. Retriever interacts with cargo adaptor SNX17 and combines CCC and WASH complex to facilitate cell surface recycling of α5β1-integrin (43). In conclusion, CCC complex and related proteins play an important role in the endosomal sorting of transmembrane receptors.
2.3 Regulating ion transport
ENaC is an amiloride-sensitive Na+ channel, and is comprised of three homologous subunits (α, β, and γ; or δ, β, and γ), which plays a significant role in Na+ and body fluid volume homeostasis (44). COMMD1 interacts with Nedd4-2 to increase the ubiquitination and promote the degradation of ENaC (α, β, and γ) (45). In addition, COMMD1 was also found able to bind to δENaC, therefore promoting δENaC into an intracellular recycling pool and reducing δENaC on the cell surface while enhancing ENaC ubiquitination. However, it is unknown if Nedd4-2 is involved in the regulation of δENaC (46). Additionally, COMMD2-10 interacts with ENaC, and later investigations on COMMD3 and COMMD9 have shown that they both lower ENaC expression on the cell surface and amiloride-sensitive current in mammalian epithelia. The COMMD protein family appears to play a negative regulatory role on ENaC (47). However, COMMD10-knockdown in Fischer rat thyroid epithelia resulted in increased Nedd4-2 and decreased ENaC current. In conclusion, COMMD1 inhibits Na+ transport by promoting ENaC ubiquitination and endocytosis; nevertheless, the regulatory mechanism of COMMD on ENaC proteins and its subunits need more investigation. In epithelial tissue, the cystic fibrosis transmembrane conductance regulator (CFTR) is a vast polytopic cAMP-regulated Cl- channel. Overexpression of COMMD1 in Hela cells increased the amount of CFTR on the cell surface, while COMMD1-knockdown induced ubiquitination and probably proteasomal degradation of CFTR (48). The Na-K-Cl co-transporter 1(NKCC1), a member of the cation-Cl cotransport family, interacts with COMMD1 to ubiquitinate itself, hence enhancing its own expression in the basolateral membrane. One potential reason is that COMMD1 improves the biological activity of NKCC1 by promoting non-classical ubiquitination, which stabilizes rather than degrades the target protein (4, 49).
2.4 Regulating NF-κB-mediated transcription
The NF-κB family is an essential transcription factor that regulates the expression of several genes and participates in a variety of physiological processes, such as immune response, inflammation, and cell survival. RelA(p65), RelB, cREL, NF-κB1(p50/p105), and NF-κB2(p52/p100) occur as homodimers or heterodimers in cells as five members of the NF-κB family. Furthermore, the NF-κB family members in inactive status bind to inhibitor of NF-κB (IκB) (7, 50). In the canonical NF-κB pathway, PAMPs/DAMPs (such as LPS and CpG DNA), proinflammatory cytokines, and other stimulations, activate the receptors on the cell membrane. IκB is subsequently phosphorylated by IκB kinase (IKK) and degraded by proteasomes. RelA/p50 dimers that are no longer inhibited by IκB translocate to the nucleus to bind DNA and mediate transcription (50). In the majority of instances, all ten COMMD proteins may bind selectively to distinct subunits of NF-κB and inhibit the activity of NF-κB (6, 51)(Figure 2). COMMD1 is the most well investigated regulator of the NF-κB signaling pathway. COMMD1 interacts with ECSSOCS1, a multimeric E3 ubiquitin ligase complex composed of Elongins B and C, cullin2, and SOCS1, in order to increase RelA binding to SOCS1, resulting in the ubiquitination and proteasomal degradation of RelA, hence inhibiting NF-κB-mediated transcription (52). RelA Ser468 phosphorylation induced by IKK is required for COMMD1-dependent ubiquitination of RelA (53, 54). The phosphorylation of RelA at Ser468 increases its binding to general control non-repressed protein 5(GCN5), which forms a complex with COMMD1 and ECSSOCS1, increasing RelA ubiquitination and degradation (54). Notably, COMMD1 continues to occupy the promoter site even after RelA is removed, indicating that COMMD1 may inhibit the expression of NF-κB target genes by this method (53). In addition, COMMD1 overexpression inhibits NF-κB-mediated transcription by decreasing the binding time of RelA to chromosomes (6) and IκB ubiquitination (51, 55). Acetylated COMMD1 in the cytoplasm induced the nucleolus translocation of RelA to mediate apoptosis by ubiquitylating RelA (56). Acetylated COMMD1 ubiquitinates RelA. Ubiquitinated RelA translocates to the nucleus to mediate apoptosis. CCDC22, a highly conserved protein associated with X-linked intellectual disability, binds to COMMD proteins in conjunction to regulate the NF-κB signaling pathway. Cullin1 interacts with the CCDC22-COMMD8 complex to enhance IκB ubiquitination and NF-κB-mediated transcription. In contrast, the CCDC22-COMMD1 complex binds Cullin2 in order to promote RelA ubiquitination and inhibits the NF-κB pathway. Given that CCDC22 deficit inhibits NF-κB activation, the CCDC22-COMMD8 complex may serve as the primary regulator (57). Reportedly, COMMD7 enhances the activation of the NF-κB signaling pathway in HCC, which will be described in depth in the next section of this review (58).
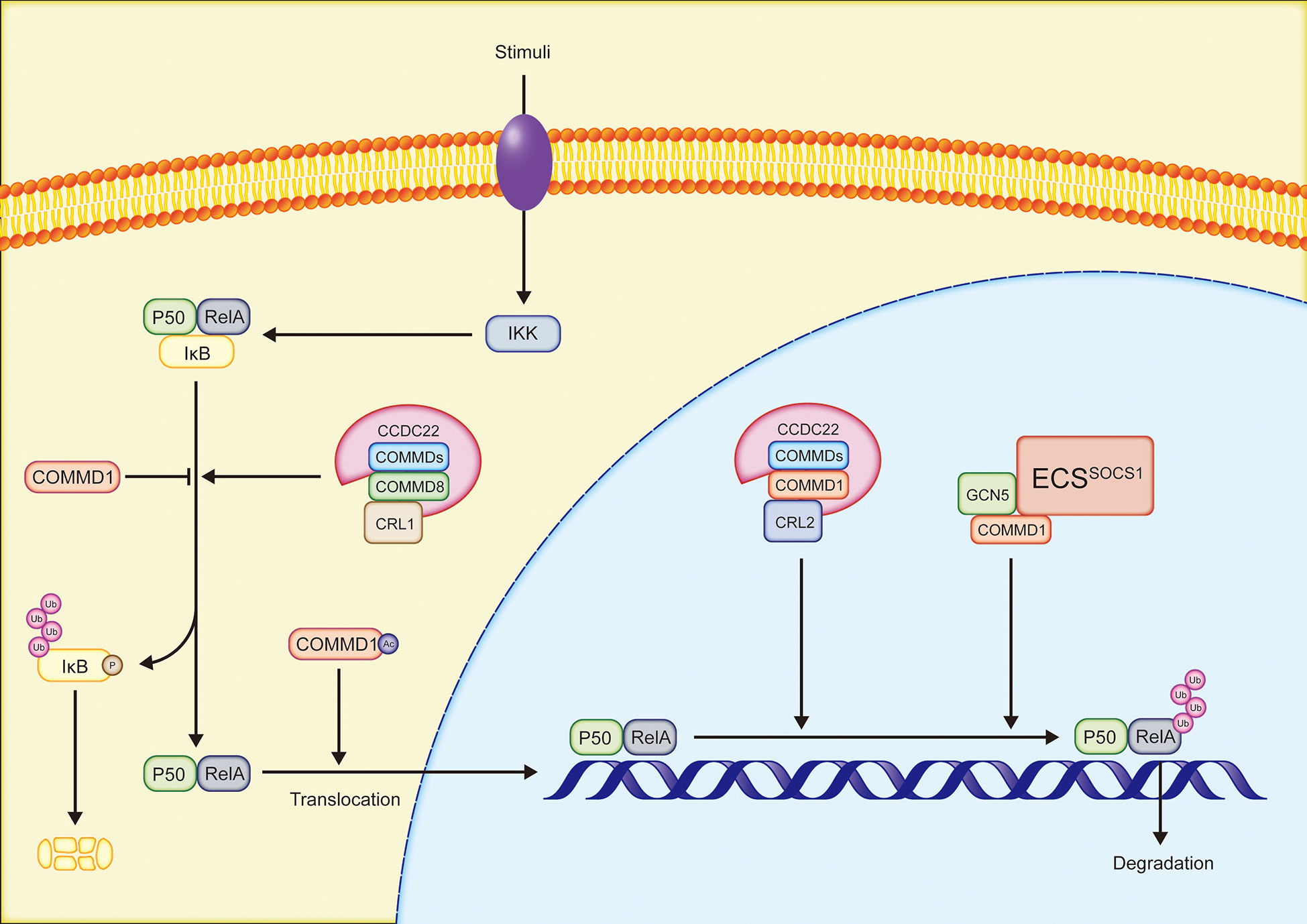
Figure 2 Simplified overview of the regulation of NF-κB pathway by COMMD proteins: COMMD, copper metabolism MURR1 domain-containing; CCDC22, coiled-coil domain-containing protein 22; CRL, Cullin-RING ligase; IKK, IκB kinase complex; GCN5, general control non-repressed protein 5.
2.5 Other biological functions of the COMMD protein family
Superoxide dismutase 1(SOD1), a member of the Cu,Zn superoxide dismutase family, catalyzes the production of molecular oxygen and hydrogen peroxide from superoxide anions, therefore acting as an antioxidant. Homodimer formation is the last stage in SOD1 maturation, and COMMD1 may decrease the amount of SOD1 homodimers, hence inhibiting SOD1 maturation and activity (59). Also regulated by COMMD1 is the hypoxia inducible factor 1 (HIF-1). Overexpression of COMMD1 promotes the degradation of HIF-1α, while COMMD1 deficiency raises the stability of HIF-1α, increasing the transcription of HIF-1α target genes, and resulting in developmental defects and embryonic lethality (60). On the one hand, COMMD1 competitively inhibits Heat Shock Protein 90 (HSP90), which binds to and stabilizes HIF-1α. On the other hand, after binding to HIF-1α, COMMD1 cooperates with Heat Shock Protein 70(HSP70) to induce the ubiquitin-independent degradation of HIF-1 (61). In addition, COMMD1 may directly inhibit the formation of HIF-α and HIF-β to heterodimers with transcriptional activity (62). In a recent investigation on hepatocellular cancer, COMMD10 was also discovered to bind and inhibit HIF-1α (63), and it has to be determined if additional members of the COMMD protein family have comparable effects. Additionally, the COMMD prtotein family may engage in cell cycle control. Overexpression or silencing of COMMD1 in HEK293T cells regulates the cell cycle and cell proliferation via altering the level of p21 Cip1 (64). The interaction between Lamin A and COMMD1 indicates that COMMD1 may potentially be associated with aging and laminopathies (65). Many investigations have shown that the COMMD protein family is broadly involved in physiological processes, including copper metabolism, endosomal sorting, ion transport, transcriptional control, and others; nevertheless, the probable molecular mechanism remains unexplored.
3 The COMMD protein family and tumors
3.1 Lung cancer
Lung cancer is a leading cause of cancer-related mortality, with roughly 85% of cases attributable to smoking (which is also affected by other environmental factors). Lung cancer is separated histologically into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC); NSCLC includes adenocarcinoma (ADC), squamous cell carcinoma (SCC), and large cell carcinoma (undifferentiated NSCLC), accounting for over 80% of lung cancer cases and causing over 1.2 million deaths annually. Despite the widespread use of surgery, radiotherapy, chemotherapy, targeted therapy, and other treatments for the treatment of lung cancer, the prognosis of patients remains dismal, and the 5-year overall survival rate is just 15% (66, 67). By upregulating genes associated to cell proliferation, metastasis, angiogenesis, and anti-apoptosis, NF-κB may contribute to oncogenesis. It is believed that aberrant activation of the NF-κB signaling pathway promotes tumor survival and medication resistance by upregulating anti-apoptotic proteins and genes (67, 68). Given the obvious inhibitory effect of the COMMD protein family on NF-κB, it is plausible to hypothesize that COMMD proteins play a crucial role in oncogenesis, progression, and death. CIGB-552 is a synthetic anti-tumor peptide that stimulates the ubiquitination of RelA, a subunit of NF-κB, in lung cancer cells, therefore decreasing the anti-apoptotic action of NF-κB and promoting the death of tumor cells. The protein-protein interaction between COMMD1 and CIGB-552 and the accumulation of COMMD1 were also observed in lung cancer cells influenced by CIGB-552. Further research revealed that the COMMD1 knockout inhibited CIGB-552-induced NF-κB degradation and cell death. Therefore, COMMD1 upregulation produce antitumor effects through inhibiting the NF-κB signaling pathway (68). HIF-1 is another COMMD1-regulated transcription factor, and is involved in energy metabolism, cell growth, and angiogenesis. Similar to NF-κB, CIGB-552 promotes COMMD1 accumulation and inhibits HIF-1 activity in H460 cells (69), hence exerting anti-inflammatory and anti-angiogenic actions. CIGB-552 and cisplatin performed a synergistic impact in inhibiting tumor cell growth, inducing apoptosis and oxidative stress response, and overcoming cisplatin resistance in a recent research involving lung cancer cells and mice models (70). It is worth noting that COMMD1 promoted the repair of DNA double-strand breaks (DSBs) in lung cancer cells. The research also revealed that the upregulation of COMMD1 increased the proliferation of NSCLC cells and was associated with a bad prognosis for patients. Even COMMD1-knockdown might increase the radiation sensitivity of NSCLC cells (17). As a potential novel anticancer treatment target, the function of COMMD1 in NSCLC requires additional investigation. In addition to COMMD1, additional COMMD protein family members have showed promise as prospective lung cancer therapeutic targets. Compared to healthy tissues and cells, the gene and protein expression levels of COMMD4 is upregulated in NSCLC, and patients with high COMMD4 expression are more likely to have a poor prognosis. COMMD4 depletion significantly inhibits NSCLC cell growth, induces mitotic catastrophe and death, and increases NSCLC sensibility to irradiation and camptothecin. Because irradiation and camptothecin might produce DNA DSBs, COMMD4 activity is crucial for protecting NSCLC cells from DNA damage (18). COMMD8 is up-regulated in NSCLC, and COMMD8 promoted cell proliferation, migration, glycolysis and inhibited cell apoptosis in NSCLC. In addition, research have shown that COMMD8 might be activated in NSCLC through activating MALAT1/miR-613 axis and LINC00657/miR-26b-5p axis, playing a carcinogenic role (19, 71). In NSCLC cells, DRTF1 and E2F1 join to create a heterodimer in order to enhance G1/S transition and inhibit P53 activity. The high expression of COMMD9 in NSCLC increases cell proliferation and invasion. COMMD9-knockdown results in G1/S arrest, induces autophagy, inhibits the activation of TFDP1/E2F1 and enhances P53 signaling pathway, playing an antitumor effect (20).
3.2 Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the most prevalent type of primary liver cancer and the third biggest cause of cancer-related mortality globally. In recent years, several studies have shown that the COMMD protein family plays a crucial role in the development and incidence of HCC. Analysis of bioinformatic data revealed that COMMD2 was highly expressed in HCC and promoted the proliferation and migration of HCC cells, making it a potential oncogene. A high expression of COMMD2 is associated with poor prognosis, higher histological grade, more advanced clinical stage, lymph node metastasis and the TP53 mutation status in HCC patients. Additionally, COMMD2 impacts the overall survival of patients with HCC by enhancing tumor immune infiltration (21). Similarly with COMMD2, COMMD3, COMMD7, and COMMD8 are overexpressed in HCC, increasing cell proliferation, migration, and invasion (22, 24, 58). In HCC patients, COMMD3 upregulation is related with advanced TNM stage, poor overall survival, and vascular invasion. In null mice with subcutaneous xenograft tumors, silencing COMMD3 inhibited tumor development and expression of HIF1α, VEGF, and CD34. In HCC, COMMD3 seemed to operate as an upstream regulator of HIF1α (22). Several investigations have elaborated on the involvement of COMMD7 in HCC, despite the fact that COMMD7 inhibits NF-κB activation in HEK293T cells (72). Instead, COMMD7 stimulated NF-κB in HCC cells. A positive feedback loop was formed when COMMD7 silencing induces HCC cell apoptosis by inhibiting NF-κB, and inhibited NF-κB further lowerd COMMD7 transcription. Researchers suggested that the inconsistent action of COMMD7 on NF-κB might be due to the different cell types or cytokine stimulation (73, 74). COMMD7 overexpression alone activated NF-κB signaling pathway by increasing PIAS4-mediated NEMO sumoylation in Nanog+ HCSCs, but the overexpression of COMMD1 and COMMD7 together ended NF-κB signaling (58). Moreover, overexpression of COMMD7 in HCC might increase cell proliferation and invasion by promoting ROS accumulation and activating NF-κB, and downstream CXCL10 (23, 75). COMMD10, unlike COMMD7, inhibits HCC cell proliferation by inhibits the NF-κB signaling pathway and enhances cell death by activating the signaling pathway of Bcl‐2/Bax/caspase-9/3. On the on hand, COMMD10 binds to the Rel homology domain of p65 in HCC cells and inhibits the nuclear translocation of NF-κB. On the other hand, COMMD10 reduces the ubiquitination and degradation of IκB, hence reducing the action of NF-κB (25). Another research revealed that COMMD10 increased radiosensitivity in HCC cells by decreasing intracellular Cu, inhibiting HIF1α, and stimulating ferroptosis (63). COMMD10 exhibits an anticancer impact in HCC and is related with increased survival.
3.3 Other cancers
Similar to the effect in lung cancer, COMMD1 exerts an anti-tumor effect in malignant tumors, including prostate cancer (76), diffuse large B-cell lymphoma (77), head and neck squamous-cell carcinoma (HNSCC) (78), and neuroblastoma (79). Downregulation of COMMD1 was related with a poor prognosis for diffuse large B-cell lymphoma, according to bioinformatics study (77). COMMD1 decreases tumor cell activity by inhibiting the NF-κB signaling pathway in prostate cancer and neuroblastoma (76, 79). COMMD1 increases its stability by forming a complex with DRR1 and F-actin in the nucleus of neuroblastoma. Subsequently, DRR1 and COMMD1 inhibits cyclinD1 expression, the G1/S transition, and neuroblastoma cell proliferation (79). miR-205 enhances inflammatory and stemness properties in stemness-enriched HNSCC cells and promotes tumorigenesis and tumor progression via downregulating COMMD1. COMMD1 downregulation also enhances the activation of NF-κB in HNSCC cells, which in turn promotes the release of inflammatory factors into the tumor microenvironment and further increases miR-205 expression (78). HIF-1, a transcription factor, is similarly regulated by COMMD1 in tumor cells. In HT29 and U2OS cells, reduced COMMD1 expression inhibits many HIF-responsive genes, including VEGFA, TGFA, HK2, and GLUT1. Notably, COMMD1-deficient tumor cells did not exhibit an increase of protein level in HIF-1α or HIF-2α. Further research demonstrated that COMMD1 bond to HIF-1α in a competitive manner and prevented the formation of the HIF-1α/β heterodimer (62), hence inhibiting the DNA binding and transcriptional activity of HIF-1α. Furthermore, COMMD1 is associated to drug resistance in ovarian cancer and multiple myeloma (26, 80). Ovarian cancer cells A2780 are more susceptible to cisplatin when nuclear COMMD1 expression is increased. The mechanism might include COMMD1’s regulation of the G2/M checkpoint, DNA repair, and apoptosis (26). In Bortezomib-resistant multiple myeloma, however, COMMD1 expression was shown to be elevated. By inhibiting SCF complex, COMMD1-knockdown could overcome Bortezomib resistance (80). By activating COMMD1, CIGB-552 reduced the growth of breast cancer cells MCF-7 and colon cancer cells HT-29 in addition to lung cancer cells (81). Gene rearrangement is often linked with an aggressive phenotype and poor prognosis in prostate cancer. The researchers discovered that COMMD3: BMI1 fusion expression was significantly elevated in metastatic prostate cancer. COMMD3 activates oncogenes such as c-MYC, which promotes prostate cancer cell proliferation, migration, and invasion; COMMD3 expression is positively connected with tumor recurrence and decreased survival rate (82). COMMD5, also known as hypertension-related, calcium-regulated gene (HCaRG), is strongly expressed in renal proximal tubules, where it regulates cell proliferation and differentiation, and accelerate the repair of renal tubules. However, excessive proliferation and poor differentiation are significant tumor progression indicators. In renal cancer cells, COMMD5 decreases proliferation, improves differentiation, accelerates autophagic cell death, and lowers VEGF production through downregulating HIF-1α, ultimately inhibiting tumor angiogenesis. The overexpression of ErbB receptors (including EGFR, ErbB2, ErbB3, and other receptors) is related with the development and progression of cancer, and have been observed in several epithelial-derived human malignancies. In renal cell cancer, COMMD5 inhibits the methylation of EGFR and ErbB3 promoters. COMMD5 significantly reduces the transcription and translation of EGFR and ErbB3 by inhibiting the methylation of EGFR and ErbB3 promoters, ultimately performing an antitumor function (83, 84). Expression of COMMD5 is reduced in gastric cancer. Rosiglitazone, a synthetic PPAR agonist, was associated by COMMD5 up-regulation in inhibiting gastric carcinogenesis, according to studies. However, the precise function of COMMD5 in gastric cancer requires additional investigation (85). In pancreatic ductal adenocarcinoma (PDAC) (86) and acute myeloid leukemia (87), high COMMD7 expression is associated with a poor prognosis. COMMD7 is positively linked with PDAC histological differentiation, lymph node metastasis, and TNM staging. By downregulating cyclin D1, inhibiting MMP-2 secretion, and activating the ERK1/2 and apoptosis signaling pathways, inhibition of COMMD7 may reduce tumor growth and invasion (86). Additionally, COMMD7 is connected with medication resistance. In gastric cancer, the activation of the Linc00852/miR-514a-5p/COMMD7 axis enhances cisplatin resistance (88). Loss of ETV6 function is associated with the development of hematological cancers. Using a functional genome-wide shRNA screen, researchers discovered that COMMD9-knockdown in leukemic cells might inhibit ETV6 transcriptional activity (89). However, the underlying mechanism by which COMMD9 regulates ETV6 remains unknown. Formin-like2 (FMNL2) is a member of the Formins family, which is upregulated in colorectal cancer and increases tumor cell motility, invasion, and metastasis (90). Additional research demonstrated that FMNL2 enhanced the ubiquitin-mediated proteasome degradation of COMMD10, hence reducing the nuclear translocation of NF-κB subunit p65 and promoting tumor progression. COMMD10 downregulation is often correlated with worse clinical outcomes. Consequently, COMMD10 may be employed as an independent survival predictive marker for CRC patients (91). Bioinformatics analysis revealed that the expression of COMMD10 in lung squamous cell carcinoma, breast invasive carcinoma, and renal clear cell carcinoma is distinct from that of their corresponding normal tissues. Moreover, high COMMD10 expression in renal clear cell carcinoma is predictive with improved patient survival (27).
4 Conclusions
COMMD proteins serve various biological functions in eucaryon. It is hypothesized that the action of COMMD1 on copper metabolism is connected to its regulation of the stability as well as intracellular transport of copper-transporting ATPases Atp7a and Atp7b (31, 33, 35). In addition, COMMD1 may directly bind to Cu (II), although the importance of this binding for copper metabolism regulation is unknown (36). In the hepatocyte COMMD-specific knockout mouse model, the absence of COMMD1, COMMD6, or COMMD9 similarly impaired the endosomal recycling of Atp7b, according to a research published last year (37). However, the regulation on copper metabolism by COMMD protein family members other than COMMD1 remains poorly known. COMMD proteins may also form CCC complexes with CCDC22, CCDC93, and C16orf62, and participate in endosomal sorting of many transmembrane proteins including Atp7a (35), LDLR (41), and α5β1-integrin (43). It has to be determined if the COMMD protein family and CCC complex are involved in the protein trafficking of other proteins. COMMD may also regulate ion transport by influencing the ubiquitination and surface expression of ENaC (45), CFTR (48), and NKCC1 (92) on the cell membrane. COMMD1 increases the ubiquitination of ENaC (45), CFTR (48), NKCC1 (92). COMMD1 promotes the ubiquitination of ENaC (45), NKCC1 (92) and Nf-κB (52), but inhibits the ubiquitination of CFTR (48) and IκB (51).Therefore, the precise mechanism of COMMD protein-mediated ubiquitination needs further investigation. In addition, the action of COMMD proteins in regulating NF-κB, HIF, and other transcription factors enables it to play a crucial role in inflammation, cancer, and other disease progression.
Almost all COMMD proteins are implicated in regulating cancer signaling (Table 1). They regulate the proliferation, differentiation, invasion, and influence metastasis of tumor cells, tumor angiogenesis, chemotherapeutic drug resistance, and prognosis of cancer. Current research indicates that the great majority of COMMD proteins, such as COMMD2 (21), COMMD3 (22), COMMD4 (18), COMMD7 (7), COMMD8 (24) and COMMD9 (20), are up-regulated in tumor cells and promote tumor proliferation, invasion, and metastasis. COMMD1 (68, 76), COMMD5 (83), and COMMD10 (25, 91) have antitumor effects in general. In a recent study, researchers used the GEPIA database to evaluate COMMD6 mRNA expression in 31 human cancers with matching normal tissue species. COMMD6 was discovered to be highly expressed in 20 types of tumors, such as colon cancer and brain lower grade glioma (LGG), and to be poorly expressed in 11 other types of tumors, including adrenocortical carcinoma (ACC) and pheochromocytoma. Further research indicated that high expression of COMMD6 is associated with shorter survival in HNSC, cholangiocarcinoma, and ACC patients, but it correlates with longer survival in LGG, uveal melanoma, testicular germ cell tumors, thyroid carcinoma, and uterine corpus endometrial carcinoma patients (93). Depending on the kind of COMMD protein and the type of tumor, it is evident that the function of COMMD protein differs somewhat. Even if there are only dozens of pieces of experimental researches on the function of COMMD protein in tumors, we can still determine the general rule regarding the involvement of COMMD protein in tumors from these restricted data. Often, the same COMMD protein has a similar role in various tumors. For instance, COMMD1 inhibits the development of cancer in a number of malignancies. Inhibition of COMMD1 expression stimulated the growth of tumor cells in lung cancer (69), neuroblastoma (79), head and neck squamous cell carcinoma (78), and prostate cancer (76). The mechanism may be associated with COMMD1’s deactivation of HIF-1 and NF-κB. In contrast, the high expression of COMMD7 stimulated the proliferation and invasion of hepatocellular carcinoma (23) and pancreatic ductal adenocarcinoma (86). Various COMMD proteins serve different roles for the same tumor. COMMD1 (68) functioned as an anti-cancer agent in lung cancer, whereas COMMD4 (18), COMMD8 (19), and COMMD9 (20) increased the growth of lung cancer cells. Due to a dearth of relevant research, it remains unclear if distinct COMMD proteins have synergistic or antagonistic effects on the same tumor. Recent research has shown the relationship between COMMD1 and COMMD7. Overexpression of COMMD1 and COMMD7 concurrently inhibited NF-κB signaling in Nanog+ HCSCs (58). It is hypothesized that the antagonistic action of COMMD1 on COMMD7 is achieved through regulating the NF-κB signaling pathway. However, it remains unknown if additional COMMD proteins interact with each other in tumor cells. In terms of molecular mechanism, COMMD proteins in tumor cells regulate the oncogenes expression of NF-κB, HIF (68), C-YMC (82), and others. In lung cancer (68), prostate cancer (76), and neuroblastoma (79), for example, COMMD1 exerts anti-tumor actions via inhibiting NF-κB. Further investigation is required to determine if additional oncogenes are regulated by COMMD proteins. In addition, COMMD proteins are engaged in the regulation of cell cycle (20) in tumors and is strongly associated with apoptosis (18), autophagy, ferroptosis (63) and other signaling pathways, influencing the proliferation and death of tumor cells. Given the broad significance of COMMD proteins in tumor-related signaling pathways, COMMD is a viable therapeutic target for tumors. CIGB-552 is a synthetic anti-tumor peptide that functions as an anti-tumor agent in lung cancer and colorectal cancer by activating COMMD1 (69, 94). Several noncoding RNAs, such as Linc00852 (88), Linc657 (71), MALAT1 (19), and MNX1-AS1 (24), have been reported to influence the incidence, development of tumor, and the death of tumor cells by influencing COMMD proteins expression. Prospects for the discovery of novel anticancer medicines that target the COMMD proteins and their upstream pathways are vast. In addition, since COMMD proteins are expressed in a range of tumor tissues and there are considerable variations between normal and malignant tissues, COMMD has high predictive value that may be used for clinical staging and prognostic assessment of cancers.
Although many pieces of research have established that the COMMD proteins are closely associated with cancer, the actual molecular mechanism is still not well understood, and the great majority of studies have focused on the COMMD as a regulation for oncogenes of NF-κB and HIF. Copper homeostasis may be employed as a novel cancer therapeutic target, according to studies. Cu levels are greatly elevated in several malignant tumors, such as breast cancer, ovarian cancer, and gastric cancer. Cu can also enhance cancer proliferation, angiogenesis, and metastasis (28). Although COMMD proteins play a critical role in the regulation of copper metabolism, few studies have shown that interference with copper homeostasis by COMMD proteins affect tumor cell activity. Cu improved the radioresistance of HCC cells in a recent research, but COMMD10 increased their radiosensitivity by decreasing intracellular copper levels and inducing ferroptosis (63). This discovery offers fresh insights into the relationship between the COMMD protein family and tumors. In addition, mounting data demonstrates that ENaC (95), CFTR (96), SOD1 (97) and other signaling molecules play crucial roles in tumor cell proliferation, migration, and apoptosis, and this review notes that these molecules are similarly regulated by COMMD proteins. To be investigated further is the existence of a complex protein-protein interaction network between COMMD proteins and tumor phenotypes.
In conclusion, the COMMD proteins family has a broad variety of biological roles and is involved in tumor proliferation, invasion, metastasis, angiogenesis, and drug resistance, among other functions. Additionally, the COMMD protein may be exploited as a therapeutic target and a biomarker for the prognosis and staging of malignancies. Additional research on the function of the COMMD proteins in cancer is beneficial for the clinical diagnosis and treatment of malignancies.
Author contributions
GY drafted the manuscript. CZ, LW, HF, ZL, XZ and XY discussed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Jilin Provincial Health Special Project(2020SCZT017), and the Project of Hepatobiliary and Pancreatic Disease Translational Medicine Platform Construction (2017F009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Healy MD, Hospenthal MK, Hall RJ, Chandra M, Chilton M, Tillu V, et al. Structural insights into the architecture and membrane interactions of the conserved COMMD proteins. Elife (2018) 7:e35898. doi: 10.7554/eLife.35898
2. Nabetani A, Hatada I, Morisaki H, Oshimura M, Mukai T. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol Cell Biol (1997) 17(2):789–98. doi: 10.1128/mcb.17.2.789
3. van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet (2002) 11(2):165–73. doi: 10.1093/hmg/11.2.165
4. Riera-Romo M. COMMD1: A multifunctional regulatory protein. J Cell Biochem (2018) 119(1):34–51. doi: 10.1002/jcb.26151
5. Maine GN, Mao X, Muller PA, Komarck CM, Klomp LW, Burstein E. COMMD1 expression is controlled by critical residues that determine XIAP binding. Biochem J (2009) 417(2):601–9. doi: 10.1042/bj20080854
6. Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, et al. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem (2005) 280(23):22222–32. doi: 10.1074/jbc.M501928200
7. Bartuzi P, Hofker MH, van de Sluis B. Tuning NF-κB activity: A touch of COMMD proteins. Biochim Biophys Acta (2013) 1832(12):2315–21. doi: 10.1016/j.bbadis.2013.09.014
8. Favier RP, Spee B, Schotanus BA, van den Ingh TS, Fieten H, Brinkhof B, et al. COMMD1-deficient dogs accumulate copper in hepatocytes and provide a good model for chronic hepatitis and fibrosis. PloS One (2012) 7(8):e42158. doi: 10.1371/journal.pone.0042158
9. Bartuzi P, Wijshake T, Dekker DC, Fedoseienko A, Kloosterhuis NJ, Youssef SA, et al. A cell-type-specific role for murine Commd1 in liver inflammation. Biochim Biophys Acta (2014) 1842(11):2257–65. doi: 10.1016/j.bbadis.2014.06.035
10. Li K, Li C, Xiao Y, Wang T, James Kang Y. The loss of copper is associated with the increase in copper metabolism MURR domain 1 in ischemic hearts of mice. Exp Biol Med (Maywood) (2018) 243(9):780–5. doi: 10.1177/1535370218773055
11. Li C, Peng H, Kang YJ. Cardiomyocyte-specific COMMD1 deletion suppresses ischemia-induced myocardial apoptosis. Cardiovasc Toxicol (2021) 21(7):572–81. doi: 10.1007/s12012-021-09650-5
12. Li C, Wang T, Xiao Y, Li K, Meng X, James Kang Y. COMMD1 upregulation is involved in copper efflux from ischemic hearts. Exp Biol Med (Maywood) (2021) 246(5):607–16. doi: 10.1177/1535370220969844
13. Guo Y, Zhou J, Li X, Xiao Y, Zhang J, Yang Y, et al. The association of suppressed hypoxia-inducible factor-1 transactivation of angiogenesis with defective recovery from cerebral ischemic injury in aged rats. Front Aging Neurosci (2021) 13:648115. doi: 10.3389/fnagi.2021.648115
14. Taura M, Kudo E, Kariya R, Goto H, Matsuda K, Hattori S, et al. COMMD1/Murr1 reinforces HIV-1 latent infection through IκB-α stabilization. J Virol (2015) 89(5):2643–58. doi: 10.1128/jvi.03105-14
15. Kudo E, Taura M, Suico MA, Goto H, Kai H, Okada S. Transcriptional regulation of HIV-1 host factor COMMD1 by the sp family. Int J Mol Med (2018) 41(4):2366–74. doi: 10.3892/ijmm.2018.3386
16. Klevay LM. Copper and ischemic heart disease. Biol Trace Elem Res (1983) 5(4-5):245–55. doi: 10.1007/bf02987211
17. Suraweera A, Duijf PHG, Jekimovs C, Schrobback K, Liu C, Adams MN, et al. COMMD1, from the repair of DNA double strand breaks, to a novel anti-cancer therapeutic target. Cancers (2021) 13(4):830. doi: 10.3390/cancers13040830
18. Suraweera A, Duff A, Adams MN, Jekimovs C, Duijf PHG, Liu C, et al. Defining COMMD4 as an anti-cancer therapeutic target and prognostic factor in non-small cell lung cancer. Br J Cancer (2020) 123(4):591–603. doi: 10.1038/s41416-020-0899-2
19. Wang S, Wang T, Liu D, Kong H. LncRNA MALAT1 aggravates the progression of non-small cell lung cancer by stimulating the expression of COMMD8 via targeting miR-613. Cancer Manag Res (2020) 12:10735–47. doi: 10.2147/cmar.S263538
20. Zhan W, Wang W, Han T, Xie C, Zhang T, Gan M, et al. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell Signal (2017) 30:59–66. doi: 10.1016/j.cellsig.2016.11.016
21. Fang W, Gan Y, Zhang L, Xiong J. COMMD2 upregulation mediated by an ncRNA axis correlates with an unfavorable prognosis and tumor immune infiltration in liver hepatocellular carcinoma. Front Oncol (2022) 12:853026. doi: 10.3389/fonc.2022.853026
22. Cheng W, Cheng Z, Zhang C, Weng L, Xing D, Zhang M. Investigating the association between COMMD3 expression and the prognosis of hepatocellular carcinoma. J Cancer (2022) 13(6):1871–81. doi: 10.7150/jca.62454
23. You N, Li J, Huang X, Wu K, Tang Y, Wang L, et al. COMMD7 activates CXCL10 production by regulating NF-κB and the production of reactive oxygen species. Mol Med Rep (2018) 17(5):6784–8. doi: 10.3892/mmr.2018.8706
24. Ji D, Wang Y, Sun B, Yang J, Luo X. Long non-coding RNA MNX1-AS1 promotes hepatocellular carcinoma proliferation and invasion through targeting miR-218-5p/COMMD8 axis. Biochem Biophys Res Commun (2019) 513(3):669–74. doi: 10.1016/j.bbrc.2019.04.012
25. Yang M, Wu X, Li L, Li S, Li N, Mao M, et al. COMMD10 inhibits tumor progression and induces apoptosis by blocking NF-κB signal and values up BCLC staging in predicting overall survival in hepatocellular carcinoma. Clin Transl Med (2021) 11(5):e403. doi: 10.1002/ctm2.403
26. Fedoseienko A, Wieringa HW, Wisman GB, Duiker E, Reyners AK, Hofker MH, et al. Nuclear COMMD1 is associated with cisplatin sensitivity in ovarian cancer. PloS One (2016) 11(10):e0165385. doi: 10.1371/journal.pone.0165385
27. Fan Y, Zhang L, Sun Y, Yang M, Wang X, Wu X, et al. Expression profile and bioinformatics analysis of COMMD10 in BALB/C mice and human. Cancer Gene Ther (2020) 27(3-4):216–25. doi: 10.1038/s41417-019-0087-9
28. Li Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life (2020) 72(9):1900–8. doi: 10.1002/iub.2341
29. Scheiber I, Dringen R, Mercer JF. Copper: Effects of deficiency and overload. Met Ions Life Sci (2013) 13:359–87. doi: 10.1007/978-94-007-7500-8_11
30. Klomp AE, van de Sluis B, Klomp LW, Wijmenga C. The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J Hepatol (2003) 39(5):703–9. doi: 10.1016/s0168-8278(03)00380-5
31. Materia S, Cater MA, Klomp LW, Mercer JF, La Fontaine S. Clusterin and COMMD1 independently regulate degradation of the mammalian copper ATPases ATP7A and ATP7B. J Biol Chem (2012) 287(4):2485–99. doi: 10.1074/jbc.M111.302216
32. de Bie P, van de Sluis B, Burstein E, van de Berghe PV, Muller P, Berger R, et al. Distinct wilson’s disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology (2007) 133(4):1316–26. doi: 10.1053/j.gastro.2007.07.020
33. Miyayama T, Hiraoka D, Kawaji F, Nakamura E, Suzuki N, Ogra Y. COMM-domain-containing 1 in stability and recruitment of the copper-transporting ATPase in a mouse hepatoma cell line. Biochem J (2010) 429(1):53–61. doi: 10.1042/bj20100223
34. Vonk WI, de Bie P, Wichers CG, van den Berghe PV, van der Plaats R, Berger R, et al. The copper-transporting capacity of ATP7A mutants associated with menkes disease is ameliorated by COMMD1 as a result of improved protein expression. Cell Mol Life Sci (2012) 69(1):149–63. doi: 10.1007/s00018-011-0743-1
35. Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol Biol Cell (2015) 26(1):91–103. doi: 10.1091/mbc.E14-06-1073
36. Narindrasorasak S, Kulkarni P, Deschamps P, She YM, Sarkar B. Characterization and copper binding properties of human COMMD1 (MURR1). Biochemistry (2007) 46(11):3116–28. doi: 10.1021/bi0620656
37. Singla A, Chen Q, Suzuki K, Song J, Fedoseienko A, Wijers M, et al. Regulation of murine copper homeostasis by members of the COMMD protein family. Dis Model Mech (2021) 14(1):dmm045963. doi: 10.1242/dmm.045963
38. Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD. Endosomal receptor trafficking: Retromer and beyond. Traffic (2018) 19(8):578–90. doi: 10.1111/tra.12574
39. Burkhead JL, Morgan CT, Shinde U, Haddock G, Lutsenko S. COMMD1 forms oligomeric complexes targeted to the endocytic membranes via specific interactions with phosphatidylinositol 4,5-bisphosphate. J Biol Chem (2009) 284(1):696–707. doi: 10.1074/jbc.M804766200
40. Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, et al. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat Commun (2016) 7:10961. doi: 10.1038/ncomms10961
41. Fedoseienko A, Wijers M, Wolters JC, Dekker D, Smit M, Huijkman N, et al. The COMMD family regulates plasma LDL levels and attenuates atherosclerosis through stabilizing the CCC complex in endosomal LDLR trafficking. Circ Res (2018) 122(12):1648–60. doi: 10.1161/circresaha.117.312004
42. Li H, Koo Y, Mao X, Sifuentes-Dominguez L, Morris LL, Jia D, et al. Endosomal sorting of notch receptors through COMMD9-dependent pathways modulates notch signaling. J Cell Biol (2015) 211(3):605–17. doi: 10.1083/jcb.201505108
43. McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, et al. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol (2017) 19(10):1214–25. doi: 10.1038/ncb3610
44. Pitzer AL, Van Beusecum JP, Kleyman TR, Kirabo A. ENaC in salt-sensitive hypertension: Kidney and beyond. Curr Hypertens Rep (2020) 22(9):69. doi: 10.1007/s11906-020-01067-9
45. Ke Y, Butt AG, Swart M, Liu YF, McDonald FJ. COMMD1 downregulates the epithelial sodium channel through Nedd4-2. Am J Physiol Renal Physiol (2010) 298(6):F1445–56. doi: 10.1152/ajprenal.00257.2009
46. Chang T, Ke Y, Ly K, McDonald FJ. COMMD1 regulates the delta epithelial sodium channel (δENaC) through trafficking and ubiquitination. Biochem Biophys Res Commun (2011) 411(3):506–11. doi: 10.1016/j.bbrc.2011.06.149
47. Liu YF, Swart M, Ke Y, Ly K, McDonald FJ. Functional interaction of COMMD3 and COMMD9 with the epithelial sodium channel. Am J Physiol Renal Physiol (2013) 305(1):F80–9. doi: 10.1152/ajprenal.00158.2013
48. Drévillon L, Tanguy G, Hinzpeter A, Arous N, de Becdelièvre A, Aissat A, et al. COMMD1-mediated ubiquitination regulates CFTR trafficking. PloS One (2011) 6(3):e18334. doi: 10.1371/journal.pone.0018334
49. Smith L, Litman P, Liedtke CM. COMMD1 interacts with the COOH terminus of NKCC1 in calu-3 airway epithelial cells to modulate NKCC1 ubiquitination. Am J Physiol Cell Physiol (2013) 305(2):C133–46. doi: 10.1152/ajpcell.00394.2012
50. Liu D, Zhong Z, Karin M. NF-κB: A double-edged sword controlling inflammation. Biomedicines (2022) 10(6):1250. doi: 10.3390/biomedicines10061250
51. de Bie P, van de Sluis B, Burstein E, Duran KJ, Berger R, Duckett CS, et al. Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem J (2006) 398(1):63–71. doi: 10.1042/bj20051664
52. Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J (2007) 26(2):436–47. doi: 10.1038/sj.emboj.7601489
53. Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep (2009) 10(4):381–6. doi: 10.1038/embor.2009.10
54. Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, et al. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev (2009) 23(7):849–61. doi: 10.1101/gad.1748409
55. Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature (2003) 426(6968):853–7. doi: 10.1038/nature02171
56. O’Hara A, Simpson J, Morin P, Loveridge CJ, Williams AC, Novo SM, et al. p300-mediated acetylation of COMMD1 regulates its stability, and the ubiquitylation and nucleolar translocation of the RelA NF-κB subunit. J Cell Sci (2014) 127(Pt 17):3659–65. doi: 10.1242/jcs.149328
57. Starokadomskyy P, Gluck N, Li H, Chen B, Wallis M, Maine GN, et al. CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling. J Clin Invest (2013) 123(5):2244–56. doi: 10.1172/jci66466
58. Zheng L, You N, Huang X, Gu H, Wu K, Mi N, et al. COMMD7 regulates NF-κB signaling pathway in hepatocellular carcinoma stem-like cells. Mol Ther Oncolyt (2019) 12:112–23. doi: 10.1016/j.omto.2018.12.006
59. Vonk WI, Wijmenga C, Berger R, van de Sluis B, Klomp LW. Cu,Zn superoxide dismutase maturation and activity are regulated by COMMD1. J Biol Chem (2010) 285(37):28991–9000. doi: 10.1074/jbc.M110.101477
60. van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, et al. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol Cell Biol (2007) 27(11):4142–56. doi: 10.1128/mcb.01932-06
61. van de Sluis B, Groot AJ, Vermeulen J, van der Wall E, van Diest PJ, Wijmenga C, et al. COMMD1 promotes pVHL and O2-independent proteolysis of HIF-1alpha via HSP90/70. PloS One (2009) 4(10):e7332. doi: 10.1371/journal.pone.0007332
62. van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, et al. COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J Clin Invest (2010) 120(6):2119–30. doi: 10.1172/jci40583
63. Yang M, Wu X, Hu J, Wang Y, Wang Y, Zhang L, et al. COMMD10 inhibits HIF1α/CP loop to enhance ferroptosis and radiosensitivity by disrupting Cu-fe balance in hepatocellular carcinoma. J Hepatol (2022) 76(5):1138–50. doi: 10.1016/j.jhep.2022.01.009
64. Jiang Z, Yuan Y, Zheng H, Cui H, Sun X, Zhao W, et al. COMMD1 regulates cell proliferation and cell cycle progression by modulating p21 Cip1 levels. Biosci Biotechnol Biochem (2019) 83(5):845–50. doi: 10.1080/09168451.2019.1569497
65. Jiang Z, Chen W, Zhou J, Peng Q, Zheng H, Yuan Y, et al. Identification of COMMD1 as a novel lamin a binding partner. Mol Med Rep (2019) 20(2):1790–6. doi: 10.3892/mmr.2019.10419
66. Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: Biology and treatment options. Biochim Biophys Acta (2015) 1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002
67. Rasmi RR, Sakthivel KM, Guruvayoorappan C. NF-κB inhibitors in treatment and prevention of lung cancer. BioMed Pharmacother (2020) 130:110569. doi: 10.1016/j.biopha.2020.110569
68. Fernández Massó JR, Oliva Argüelles B, Tejeda Y, Astrada S, Garay H, Reyes O, et al. The antitumor peptide CIGB-552 increases COMMD1 and inhibits growth of human lung cancer cells. J Amino Acids (2013) 2013:251398. doi: 10.1155/2013/251398
69. Daghero H, Fernández Massó JR, Astrada S, Guerra Vallespí M, Bollati-Fogolín M. The anticancer peptide CIGB-552 exerts anti-inflammatory and anti-angiogenic effects through COMMD1. Molecules (2020) 26(1):152. doi: 10.3390/molecules26010152
70. Gomez Rodriguez Y, Oliva Arguelles B, Riera-Romo M, Fernandez-De-Cossio J, Garay HE, Fernandez Masso J, et al. Synergic effect of anticancer peptide CIGB-552 and cisplatin in lung cancer models. Mol Biol Rep (2022) 49(4):3197–212. doi: 10.1007/s11033-022-07152-3
71. Zhang R, Niu Z, Pei H, Peng Z. Long noncoding RNA LINC00657 induced by SP1 contributes to the non-small cell lung cancer progression through targeting miR-26b-5p/COMMD8 axis. J Cell Physiol (2020) 235(4):3340–9. doi: 10.1002/jcp.29222
72. Esposito E, Napolitano G, Pescatore A, Calculli G, Incoronato MR, Leonardi A, et al. COMMD7 as a novel NEMO interacting protein involved in the termination of NF-κB signaling. J Cell Physiol (2016) 231(1):152–61. doi: 10.1002/jcp.25066
73. Zheng L, Liang P, Li J, Huang XB, Liu SC, Zhao HZ, et al. ShRNA-targeted COMMD7 suppresses hepatocellular carcinoma growth. PloS One (2012) 7(9):e45412. doi: 10.1371/journal.pone.0045412
74. Zheng L, Deng CL, Wang L, Huang XB, You N, Tang YC, et al. COMMD7 is correlated with a novel NF-κB positive feedback loop in hepatocellular carcinoma. Oncotarget (2016) 7(22):32774–84. doi: 10.18632/oncotarget.9047
75. You N, Li J, Huang X, Wu K, Tang Y, Wang L, et al. COMMD7 promotes hepatocellular carcinoma through regulating CXCL10. BioMed Pharmacother (2017) 88:653–7. doi: 10.1016/j.biopha.2017.01.046
76. Zoubeidi A, Ettinger S, Beraldi E, Hadaschik B, Zardan A, Klomp LW, et al. Clusterin facilitates COMMD1 and I-kappaB degradation to enhance NF-kappaB activity in prostate cancer cells. Mol Cancer Res (2010) 8(1):119–30. doi: 10.1158/1541-7786.Mcr-09-0277
77. Taskinen M, Louhimo R, Koivula S, Chen P, Rantanen V, Holte H, et al. Deregulation of COMMD1 is associated with poor prognosis in diffuse large b-cell lymphoma. PloS One (2014) 9(3):e91031. doi: 10.1371/journal.pone.0091031
78. Yeh DW, Chen YS, Lai CY, Liu YL, Lu CH, Lo JF, et al. Downregulation of COMMD1 by miR-205 promotes a positive feedback loop for amplifying inflammatory- and stemness-associated properties of cancer cells. Cell Death Differ (2016) 23(5):841–52. doi: 10.1038/cdd.2015.147
79. Mu P, Akashi T, Lu F, Kishida S, Kadomatsu K. A novel nuclear complex of DRR1, f-actin and COMMD1 involved in NF-κB degradation and cell growth suppression in neuroblastoma. Oncogene (2017) 36(41):5745–56. doi: 10.1038/onc.2017.181
80. Malek E, Abdel-Malek MA, Jagannathan S, Vad N, Karns R, Jegga AG, et al. Pharmacogenomics and chemical library screens reveal a novel SCF(SKP2) inhibitor that overcomes bortezomib resistance in multiple myeloma. Leukemia (2017) 31(3):645–53. doi: 10.1038/leu.2016.258
81. Astrada S, Fernández Massó JR, Vallespí MG, Bollati-Fogolín M. Cell penetrating capacity and internalization mechanisms used by the synthetic peptide CIGB-552 and its relationship with tumor cell line sensitivity. Molecules (2018) 23(4):801. doi: 10.3390/molecules23040801
82. Umbreen S, Banday MM, Jamroze A, Mansini AP, Ganaie AA, Ferrari MG, et al. COMMD3:BMI1 fusion and COMMD3 protein regulate c-MYC transcription: Novel therapeutic target for metastatic prostate cancer. Mol Cancer Ther (2019) 18(11):2111–23. doi: 10.1158/1535-7163.Mct-19-0150
83. Matsuda H, Campion CG, Fujiwara K, Ikeda J, Cossette S, Verissimo T, et al. HCaRG/COMMD5 inhibits ErbB receptor-driven renal cell carcinoma. Oncotarget (2017) 8(41):69559–76. doi: 10.18632/oncotarget.18012
84. Ikeda J, Matsuda H, Ogasawara M, Ishii Y, Yamaguchi K, Takahashi S, et al. COMMD5 inhibits malignant behavior of renal cancer cells. Anticancer Res (2021) 41(6):2805–15. doi: 10.21873/anticanres.15061
85. Chen BL, Yu J, Zeng ZR, Chu WK, Wong CY, Cheng YY, et al. Rosiglitazone suppresses gastric carcinogenesis by up-regulating HCaRG expression. Oncol Rep (2008) 20(5):1093–7. doi: 10.3892/or_00000114
86. You N, Li J, Gong Z, Huang X, Wang W, Wang L, et al. COMMD7 functions as molecular target in pancreatic ductal adenocarcinoma. Mol Carcinog (2017) 56(2):607–24. doi: 10.1002/mc.22520
87. Li K, Chen L, Zhang H, Wang L, Sha K, Du X, et al. High expression of COMMD7 is an adverse prognostic factor in acute myeloid leukemia. Aging (2021) 13(8):11988–2006. doi: 10.18632/aging.202901
88. Cao S, Fu B, Cai J, Zhang D, Wang C, Wu H. Linc00852 from cisplatin-resistant gastric cancer cell-derived exosomes regulates COMMD7 to promote cisplatin resistance of recipient cells through microRNA-514a-5p. Cell Biol Toxicol (2022). doi: 10.1007/s10565-021-09685-y
89. Neveu B, Richer C, Cassart P, Caron M, Jimenez-Cortes C, St-Onge P, et al. Identification of new ETV6 modulators through a high-throughput functional screening. iScience (2022) 25(3):103858. doi: 10.1016/j.isci.2022.103858
90. Liang L, Li X, Zhang X, Lv Z, He G, Zhao W, et al. MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology (2013) 144(3):624–35.e4. doi: 10.1053/j.gastro.2012.11.033
91. Yang SS, Li XM, Yang M, Ren XL, Hu JL, Zhu XH, et al. FMNL2 destabilises COMMD10 to activate NF-κB pathway in invasion and metastasis of colorectal cancer. Br J Cancer (2017) 117(8):1164–75. doi: 10.1038/bjc.2017.260
92. McDonald FJ. COMMD1 and ion transport proteins: What is the COMMection? focus on “COMMD1 interacts with the COOH terminus of NKCC1 in calu-3 airway epithelial cells to modulate NKCC1 ubiquitination”. Am J Physiol Cell Physiol (2013) 305(2):C129–30. doi: 10.1152/ajpcell.00128.2013
93. Yang M, Huang W, Sun Y, Liang H, Chen M, Wu X, et al. Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis. Br J Cancer (2019) 121(8):699–709. doi: 10.1038/s41416-019-0571-x
94. Vallespí MG, Pimentel G, Cabrales-Rico A, Garza J, Oliva B, Mendoza O, et al. Antitumor efficacy, pharmacokinetic and biodistribution studies of the anticancer peptide CIGB-552 in mouse models. J Pept Sci (2014) 20(11):850–9. doi: 10.1002/psc.2676
95. Liu C, Zhu LL, Xu SG, Ji HL, Li XM. ENaC/DEG in tumor development and progression. J Cancer (2016) 7(13):1888–91. doi: 10.7150/jca.15693
96. Amaral MD, Quaresma MC, Pankonien I. What role does CFTR play in development, differentiation, regeneration and cancer? Int J Mol Sci (2020) 21(9):3133. doi: 10.3390/ijms21093133
Keywords: COMMD, cancer, copper homeostasis, endosomal sorting, ion transport, NF-κB
Citation: You G, Zhou C, Wang L, Liu Z, Fang H, Yao X and Zhang X (2023) COMMD proteins function and their regulating roles in tumors. Front. Oncol. 13:1067234. doi: 10.3389/fonc.2023.1067234
Received: 11 October 2022; Accepted: 12 January 2023;
Published: 23 January 2023.
Edited by:
Massimo Broggini, Mario Negri Institute for Pharmacological Research (IRCCS), ItalyCopyright © 2023 You, Zhou, Wang, Liu, Fang, Yao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxao Yao, eWFveHhAamx1LmVkdS5jbg==; Xuewen Zhang, Wmhhbmd4d0BqbHUuZWR1LmNu
 Guangqiang You
Guangqiang You Chen Zhou2
Chen Zhou2 Lei Wang
Lei Wang Zefeng Liu
Zefeng Liu He Fang
He Fang Xuewen Zhang
Xuewen Zhang