
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 20 January 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1063930
This article is part of the Research Topic 365 Days of Progress In Molecular and Cellular Oncology View all 4 articles
The heterogeneous species of tRNA-derived fragments (tRFs) with specific biological functions was recently identified. Distinct roles of tRFs in tumor development and viral infection, mediated through transcriptional and post-transcriptional regulation, has been demonstrated. In this review, we briefly summarize the current literatures on the classification of tRFs and the effects of tRNA modification on tRF biogenesis. Moreover, we highlight the tRF repertoire of biological roles such as gene silencing, and regulation of translation, cell apoptosis, and epigenetics. We also summarize the biological roles of various tRFs in cancer development and viral infection, their potential value as diagnostic and prognostic biomarkers for different types of cancers, and their potential use in cancer therapy.
Over the past few decades, non-coding RNAs including tRNA-derived fragments (tRFs) (also named as tRNA-derived small RNAs (tsRNAs) have been verified to play crucial roles in the pathophysiological processes of cancer (1–3). Challenging the older paradigm that tRFs are merely random products of tRNAs, tRF complementarity to specific locations of pre-tRNAs or mature tRNAs demonstrates that tRFs are purposeful products of tRNAs (4). The biogenesis of tRFs is strictly controlled by a set of precise ribonucleases that produce 14–50 nucleotide-long small RNAs (5).
Evidence indicates that tRFs participate in biological processes such as gene destabilization (6, 7), mRNA processing (8), translation (9), and epigenetic regulation (10). An increasing number of studies verify that tRFs play indispensable roles in diverse diseases including cancer and viral infection (11). Here, we outline tRF biogenesis, classification, and the role of tRNA modifications on the tRF production. We also delineate the major biological functions of tRFs and the roles of tRFs in various types of cancers and viral infections. In addition, we summarize the possible usage of tRFs for cancer diagnostic and prognostic biomarkers, and as therapeutic targets. Lastly, we explore the mechanism of tRF biology that may be involved in chemoresistance.
In the 1970s, researchers discovered tRFs by examining the urine of cancer patients (12). However, they did not realize that these small RNAs had biological functions. After a few decades, researchers found that tRFs are not useless debris derived from tRNAs, as they have precise sequence structures and are involved in various biological processes (13). Pre-tRNAs are produced by RNA polymerase III (RNA Pol III) located in the eukaryotic nucleus. During the maturation process of pre-tRNAs, 5′-leader and 3′-poly U nucleotides are removed by endoribonuclease P (RNase P) and ribonuclease Z (RNase Z)/cytoplasmic homolog ribonuclease Z2 (ELAC2), respectively (14, 15). The 3′-CCA tail is attached to the 3′-acceptor stem of tRNAs with the assistance of a specific tRNA nucleotide transferase (16). During the process of enzymatic splicing and chemical modifications of pre-tRNAs and mature tRNAs, tRFs are created (17). Based on the disparate cleavage sites, tRFs are classified into various distinct categories (18) (Figure 1): 1) 3′ U tRFs (tRF-1s), which are 16–27 nucleotides long, and are released by RNase Z or ELAC2 in the 3′-trailer sequences of pre-tRNA (19); 2) 5′-tRFs start from the 5′-termini of parental tRNAs and end at the D-loop or around the anticodon-loop; 3) tRF-3s span the 3′-termini and progress to the TΨC loop of parental tRNAs and are 18 nucleotides or 22 nucleotides in length; 4) i-tRFs (also known as tRF-2s) originate from the internal region of mature tRNAs spanning anticodons and contain D-loop and T-loop sequences, with the sequences being variable in length (13). Numerous studies have verified that the vast majority of tRFs are exclusively produced by Dicer (20, 21), while the specific ribonuclease involved in the cutting process of i-tRFs needs further elucidation; 5) tRNA halves (tiRNAs or tRHs), including 5′-tiRNAs and 3′-tiRNAs (30–40 nucleotides in length) correspond to half of a mature tRNA (22, 23). Substantial evidence has demonstrated that most tiRNAs are produced by angiogenin (ANG) under stress, such as ischemia, oxidative injury, ultraviolet exposure, arsenite exposure, or infection diseases (13, 24–27). In non-stress conditions, ANG is confined to the nucleus and exists in an inhibited state associated with the ribonuclease inhibitor RNH1. Once exposed to stress stimuli, ANG dissociates from RNH1 and translocates to the cytoplasm for tRNA processing. Sex hormone-dependent tRNA-derived RNAs are found to be abnormally expressed in non-stress conditions such as breast cancers (BCAs) with estrogen receptors (ERs) or prostate cancers (PCAs) with androgen receptors (ARs) (28). It is worth noting that ANG-dependent tiRNAs may be limited to a few tRNAs. A study identified that ANG specifically produces tiRNAGly, tiRNAGlu, tiRNALys, tiRNAVal, tiRNAHis, tiRNAAsp, and tiRNASec (22). Surprisingly, the small RNA sequences from ANG-knockout cells revealed that only the abundance of tiRNAHis and tiRNAAsp changed, suggesting that there are other ribonucleases associated with the generation of tiRNAs (29). In the ciliate Tetrahymena and Saccharomyces cerevisiae, tiRNAs are produced from the cleavage of RNase T2 family members named RNT2 and Rny1p, respectively (30, 31). All these findings have indicated the sophistication and complexity of tRFs biogenesis.
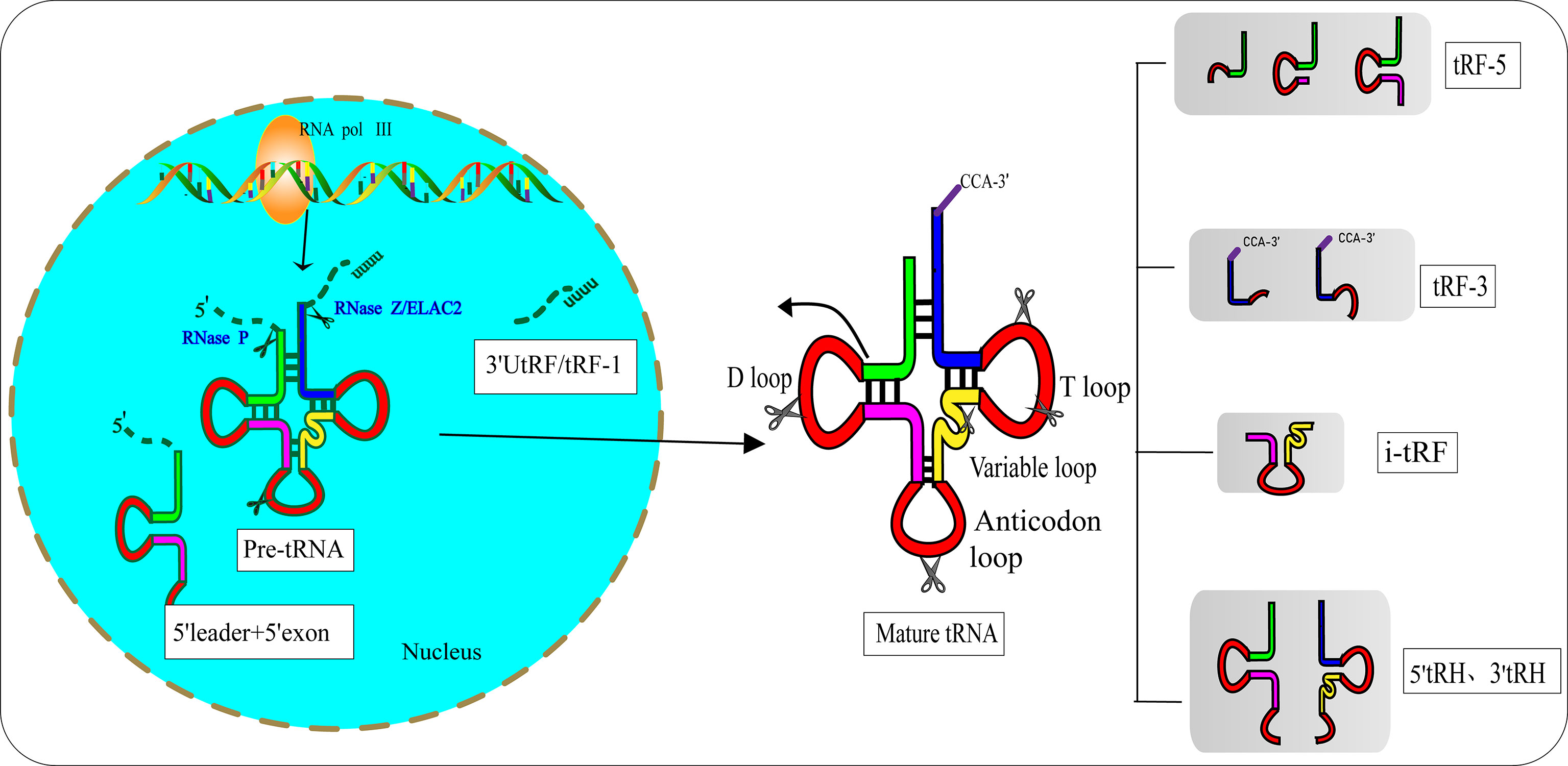
Figure 1 Biogenesis and classification of tRNA-derived fragments (tRFs). Pre-tRNA is transcribed by RNA Pol III in the nucleus and undergoes 5′-leader, 3′-polyU removal and 3′ CCA addition, based on disparate cleavage sites on pre-tRNA or mature tRNA. tRFs can be divided into different types including 3′U tRFs (tRF-1s), 5′-tRFs, 3′-tRFs, i′-tRFs, 5′-tRNA halves (5′-tRHs) and 3′-tRHs.
Hanada’s team discovered an atypical type of tRF that accumulate in mice with spinal motor neuron degenerative disease (32). These novel tRFs were derived from pre-tRNAtyr after the aberrant removal of introns. RNA sequencing indicated that these novel tRFs encompass 5′ leader sequences starting with PPP-nucleotide and followed by 5′ exon sequences (Figure 1).
Modified nucleotides account for 17% of all residues, and tRNAs are the most extensively modified RNAs in eukaryotes (33). Some modifications are essential for translation efficiency and the accuracy of translation through ensuring the correct wobble base pairing (34, 35), and for maintaining the stability and folding of tRNAs (36). Recently, numerous studies have shown the significance of tRNA modification in the production of tRFs (37–40).
5-methylcytosine modification is one of the important determiners in the stability of tRNAs, and can prevent tRNAs from ANG-mediated cleavage. As a result, the loss of modification leads to the degradation of tRNAs (40). Blanco et al. identified that NSun2-mediated cytosine-5 RNA methylation at the variable loop can lead to an increased affinity of tRNA and ANG, which gives rise to an accumulation of 5′-tRFs and the subsequent attenuation of protein translation rates (41). Similarly, Tuorto et al. revealed a quantitative loss of cytosine-C5 tRNA methylation in mice with DNMT2 and NSUN2 deficiencies, which led to a substantial decrease in abundance of tRNAAsp-GTC and tRNAGly-GCC and reduced efficiency of overall protein synthesis (42). However, specific tRFs were not revealed by the authors. Analogously, Chen et al. developed a mouse strain with demethylase α-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) knockdown and found that ALKBH3 potently and selectively demethylated the m1A and m3C residues on tRNA. However, the expression level of the most targeted tRNAs, aside from tRNAGlyGCC, were not significantly changed in HeLa cells with ALKBH3 deletion (38). However, tRNAGlyGCC had low levels in epididymis, testis and lung samples from mice with ALKBH3 knockdown.
Pseudouridylation also plays critical roles in regulating the abundance and species of tRFs (43). A family of pseudouridine synthases (PUS7) were reported to catalyze pseudouridylation of RNAs (44). Guzzi’s team indicated that the deficiency of PUS7 results in reduced levels of ~18 nucleotide-long 5′-tRFs but increased levels of 5′-tiRNAs, suggesting that the pseuduridylated modification on tRNAs are correlated with their differentiated endonuclease affinity and subsequent processing products (43).
Queuosine modification is uniquely detected on eukaryotic tRNA anticodon-loop containing G34U35N36 sequences (45). In the process of queuosine modification, guanine is substituted by queuine with the assistance of queuine tRNA-ribosyl transferase catalytic subunit 1 (QTRT1) (46). Wang et al. revealed that queuosine modification significantly protects tRNAHis and tRNAAsn from ANG-mediated cleavage (37). These data provide new insights into how tRNA modifications affect small RNA pools. However, it is unclear how these modifications enhance tRNA stability. Thus, a detailed understanding of the biogenesis of tRFs requires more research.
tRFs are ubiquitous in all domains of organisms and are associated with the pathophysiological processes of various diseases (26, 47, 48). The biological functions of tRFs have been reported in recent years. Here, we summarize the main biological roles of tRFs such as in gene silencing, RNA processing, and translational, apoptotic, and epigenetic regulation in different types of diseases.
Researchers previously treated tRFs as a distinctive type of microRNA (miRNA). It is logical to speculate that tRFs function similarly to miRNAs. For example, Green’s group detailed that tRF-3003a (derived from tRNACysGCA) confers gene silencing of Janus Kinase 3 (JAK3) by ‘seed sequence’ complementarity in osteoarthritis chondrocytes (49). They further verified that tRF-3003a associated with AGO2 and GW182, and forms RNA-induced silencing complex by performing AGO2 and GW182 RNA immunoprecipitation assays. Another study found that the C-terminus of AGO2 was indispensable for functionality, but the N-terminus was not indispensable for interacting with GW182 (50). The effects of GW182 were suggested to be quite important in AGO2-mediated gene silencing. However, more research is needed to uncover the details of tRFs in AGO-dependent gene silencing.
Several studies have indicated that disparate tRFs show distinct affinities with various AGO subtypes. An earlier meta-analysis revealed that several tRFs have a stronger affinity to AGO1, AGO3 and AGO4 in comparison with AGO2, indicating that tRFs possess other mechanisms of action beyond binding to AGO2, unlike miRNAs (51). A tRF named CU1276 suppresses the endogenous expression of Replication Protein1 (RPA1) by sequence complementarity, while the RNA-induced silencing complex is composed of AGO3, AGO4 and AGO1 rather than AGO2 (4, 52). Likewise, Zhong et al. identified that Gly-tRF (5′-tRF, with the length of 29–34 nucleotides) is upregulated in both ethanol-fed mice and alcoholic fatty liver disease patients. Further research has indicated that alcohol consumption can result in the activation of oxidative stress and the subsequent upregulation of Gly-tRFs (53). Gly-tRFs interact with AGO3, but not with other types of AGOs, to silence sirtuin1 (Sirt1) expression by targeting its 3′-UTR. This leads to the disruption of lipid metabolism pathways and liver injury (Figure 2A). However, the potential mechanism of discrepant affinity of tRFs to AGO needs further study.
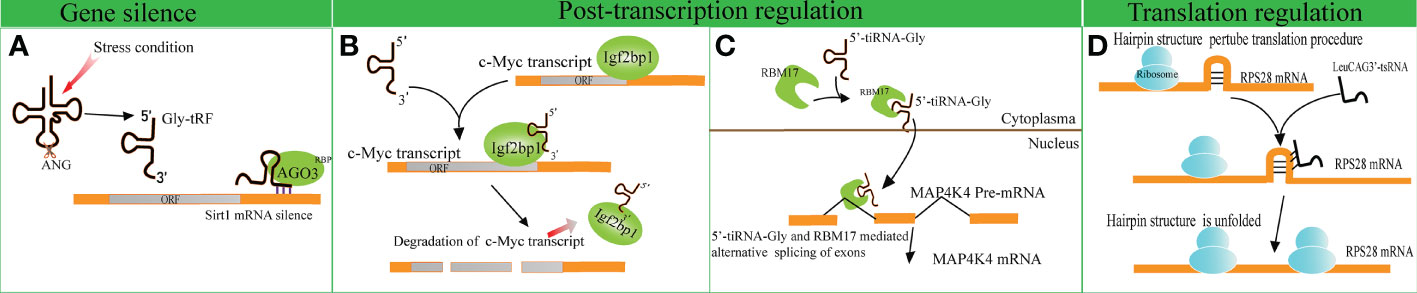
Figure 2 Gene expression regulation by tRNA-derived fragments (tRFs). (A) Gly-tRFs inhibit the expression of Sirt1 by associating with AGO3 to target Sirt1 mRNA 3′-UTR. (B) 5′-tsRNAs sequester RBP IGF2BP1 from the c-Myc mRNA and affects the stability of the target mRNA. (C) tiRNA-Gly promotes translocation of RBM17 from the cytoplasm to the nucleus, and induces RBM17-dependent splicing of MAP4K4 mRNA. (D) LeuCAG3’-tsRNA promotes the translation efficiency of ribosomal protein S28 (RPS28) mRNA by unfolding the hairpin structure of RPS28.
In comparison to the studies described above, other studies support that tRFs play a role in gene silencing by interacting with RNA binding proteins (RBPs). A previous study verified that a series of i-tRFs (derived from tRNAGluYTC, tRNAAspGTC, tRNAGlyTCC, and tRNATyrGTA) competitively bind to Y-box binding protein 1 (YBX1), known to stabilize oncogenic transcripts. The upregulation of i-tRFs sequesters YBX1 away from oncogenic mRNAs, leading to the degradation of oncogenic transcripts. Krishna et al. reported that a series of 5′-tsRNAs (including tsRNA-GlnCTG, tsRNA-GlyGCC, tsRNA-GluTTC, and tsRNA-ValCCC) modulate the states of mouse embryonic stem cells by influencing the abundance of stemness-marker c-Myc (7). Based on RNA pulldown assays and mass spectrometry, it has been verified that 5′-tsRNAs preferentially bind to RBP IGF2BP1. IGF2BP1 is known to maintain the stability of c-Myc mRNA by interacting with the coding region instability determinant. This association results in the instability of c-Myc mRNA and lowers c-Myc expression (Figure 2B). The study provides new insights in gene silencing at the post-transcriptional level.
A recent study identified that tiRNA-Gly promotes the proliferation and migration of papillary thyroid cancer cells by binding to RNA binding motif protein 17 (RBM17) (8), a spliceosome protein that can selectively splice mRNAs. The study revealed that the interaction of tiRNA-Gly and RBM17 suppresses ubiquitin-dependent degradation of RBM17, and facilitates the translocation of RBM17 from the cytoplasm to the nucleus. RBM17 mediates alternative exon splicing of Mitogen-Activated Protein 4 Kinase 4 (MAP4K4) pre-mRNA. Increased tiRNA-Gly induced higher levels of truncated MAP4K4 mRNA and lower levels of long variant MAP4K4 mRNA. MAP4K4 was known as a protein kinase to activate the MAPK pathway (54), though it was revealed that the two variants of MAP4K4 substantially phosphorylated the downstream proteins of the MAPK pathway, and the truncated variant showed a stronger effect than that of the long variant (Figure 2C). These data revealed the significance of alternative splicing in altering signal pathways mediated by tiRNA-Gly. This study provides novel insights into the role of tiRNAs in post-transcriptional gene expression regulation.
It has been demonstrated that the overall translation speed can be decreased by about 10% by tRFs (25). However, the underlying mechanism of the inhibitory effect of tRFs on protein translation is unclear. Ivanovet et al. reported that 5′-tiRNAAla and 5′-tiRNACys promotes the synthesis of stress granules in a phospho-eIF2a-independent way, and disturbs the formation of the translation initiation complex by replacing the eukaryotic initiation factors from m7G-capped mRNAs (55). Further studies have clarified that the G-quadruplex-like structure (G4-motif) at the 5′-end of 5′-tiRNAs contribute to the formation of intermolecular RNA G-quadruplexes (9). These complexes can interact with the cold shock domain of translational silencer protein YBX1 (also referred as YB-1), which facilitates the assembly of stress granules and strengthens the resistance against stress (56). Mechanistic studies have indicated that RNA G-quadruplexes are necessary for replacing translational initiation factors (eIF4G/A) from mRNAs. However, YBX1 protein is not indispensable for interfering with translation-initiation complexes, but it is required in facilitating the assembly of stress granules (57). In addition, another study provided deep insights into how pseudouridylated tRFs affect translational initiation in embryogenesis and hematopoietic lineage manifestation (43). It was identified that tRF-5s are abundant in human embryonic stem cells (43). Pseudouridylation of tRF-5 at the U8 position (referred to as mTOGs) inhibits translation initiation by competitively binding to polyA binding protein-1 (PABPC1), eIF4G/A, and eIF4E.
tRFs not only disrupt translation initiation but also enhance translation efficiency. For example, the overexpression of LeuCAG3′-tsRNA significantly promotes the biogenesis of 18S rRNA (58). LeuCAG3′-tsRNA promotes the translation efficiency of RPS28 mRNA by interacting with the coding sequence and unfolding the hairpin structure of RPS28 mRNA. (Figure 2D).
Moreover, research has verified that the pre-tRNA trailer derived tRFs sequesters La/SSB in the cytoplasm and leads to the inhibition of HCV internal ribosome entry site-mediated translation (59). Certain aspects of tRF-5 play a critical role in translation silencing. For example, a conserved “GG” dinucleotide structure in tRF-5 contributes to translation inhibition regardless of the shortened length of RF-5 (60). These results pave the way for tRFs in translation regulation by interacting with RBPs or mRNAs.
Studies have disclosed the potential role of tRFs in regulating cell apoptosis. Saikia et al. verified that a series of tiRNAs perturb the formation of the apoptosome by interfering with the interaction of apoptotic protease activating factor-1 (APAF1) and cytochrome c, resulting in increased survival of mouse embryonic fibroblasts. The affinity of cytochrome c for disparate tiRNAs varies considerably, and it was shown that cytochrome c showed significantly lower affinity to tiRNAArg than to tiRNAAla. However, how cytochrome c recognizes specific tRNA targets needs further research (61).
A recent study revealed that tRF-21-VBY9PYKHD (i-tRF, derived from tRNAGlyGCC) is involved in cell apoptosis regulation (62). Inflammatory cytokine-induced tRF-21 was downregulated in pancreatic ductal adenocarcinoma (PDAC), and overexpression of tRF-21 significantly enhanced apoptosis and inhibited growth of PDAC cells. Further research revealed that tRF-21 knockdown promotes the phosphorylation of heterogeneous nuclear ribonucleoprotein L (hnRNP L) and the formation of hnRNP L and dead-box helicase 17 (DDX17) complexes. These complexes play crucial roles in splicing Caspase 9 into Caspase 9b (with anti-apoptotic specificity) and mH2A1.2 (with pro-invasive specificity), while upregulation of tRF-21 exerts the opposite effect (Figure 3A). These studies reveal novel apoptosis-related mechanisms for the treatment of cancers.
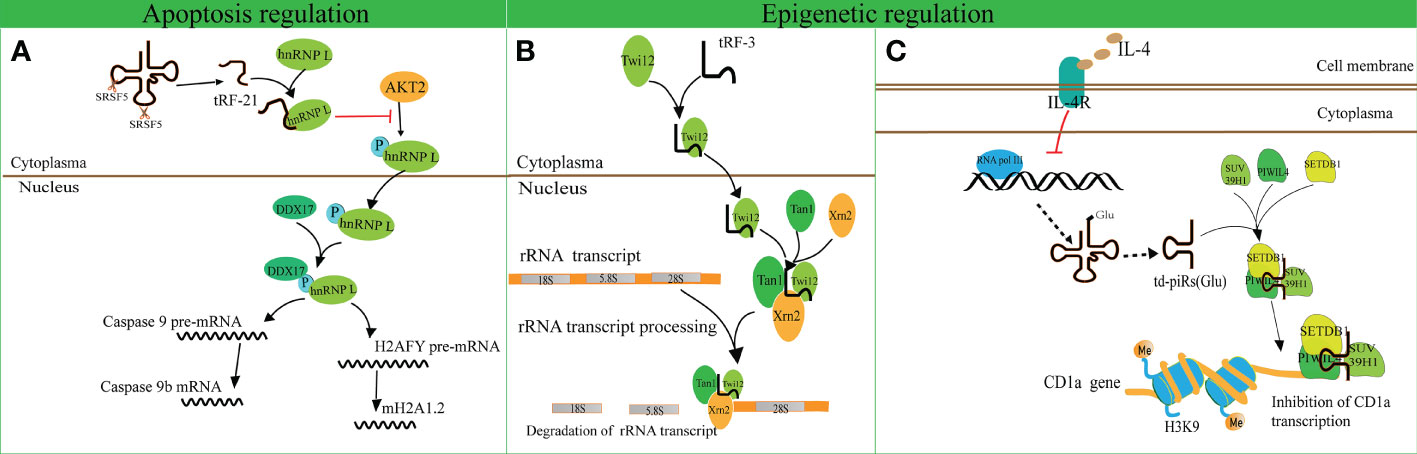
Figure 3 Apoptosis and epigenetic regulation by tRNA-derived fragments (tRFs). (A) The interaction of tRF-21-VBY9PYKHD and hnRNP L inhibits the phosphorylation of hnRNP L mediated by AKT2 and promotes the formation of the hnRNP L and DDX17 complex. This complex splices Caspase 9 and mH2A1 pre-mRNAs into Caspase 9b mRNA and mH2A1.2 mRNA. (B) tRF-3 translocates into the nucleus with the assistance of Twi12. The Twi12-tRF-3 complex binds to the exonuclease Xrn2 and Tan1 to form a complex, which plays important roles in rRNA processing. (C) IL-4 decreases the production of tRNAGlu followed by the downregulation of td-piR, which assembles with PIWIL4 and recruits SETDB1, SUV39H1, and HP1β to the promoter of CD1a mRNA and facilitates methylation of H3K9 histone followed by inhibition of CD1a transcription.
Transposons are genetic sequences that can translocate their sites within a genome (63). The mobilizable peculiarity of transposons is beneficial to the diversity of life and strengthens the adaptation to stress conditions (64). However, substantial evidence has shown that various cancers are significantly correlated with the transcriptional activity of transposons (65, 66). To constrain the possible harmful effects of transposons, eukaryotes have developed various mechanisms such as DNA methylation, chromatin modification, as well as RNA silencing mediated by piRNA-Piwi complexes to keep these genetic elements in a quiescent state (67). The question naturally arises of how the genome protects itself from being destructed when most of the epigenetic marks and piRNAs disappear during epigenetic reprogramming, like during embryonic development prior to implantation. It was verified that a novel class of tRF-3s (derived from mature tRNAsLysUUU) 18–22 nucleotides long was discovered to be enriched in SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) knockout mouse embryonic stem cells (68). SETDB1 induces histone H3K9 trimethylation and plays a passive role in the transcription of long terminal repeat-retrotransposons, named endogenous retroviruses. The 18 nucleotide-long tRF-3 interferes with retroviral cDNA synthesis by displacing tRNAs from the primer binding site located in the long terminal repeat retrotransposon. The 22 nucleotide-long tRF-3 leads to the gene silencing of endogenous retrovirus mRNA through sequence complementarity to the primer binding site and results in reduced retrotransposon integration. MERVL is a retroelement that functions to drive the transcription of specific genes (69). GlyGCC 5′-tRF suppresses MERVL-mediated gene transcription by binding to heterogeneous nuclear ribonucleoproteins F and H (hnRNP F/H) to form complexes, which play a crucial role in the biogenesis of several classes of small non-coding RNAs including U7 snRNAs. The stability and utility of U7 snRNAs rely on Cajal bodies. U7 snRNAs promote the production of histone proteins by interacting with histone downstream elements. As a result, the biogenesis of histone partially halts the post-transcriptional expression of MERV-mediated genes (10).
tRF-3 has a length of 26–31 nucleotides and has been found to interact with ribonucleoproteins AGO/Piwi and participate in epigenetic regulation (70). Couvillion et al. challenged the conventional wisdom that AGO/Piwi typically induces mRNA degradation and represses translation through RNA-induced silencing complex formation (70). They revealed that tRF-3 associates with the Tetrahymena thermophila AGO/Piwi protein Twi12 and promotes its nuclear translocation, while Twi12 plays essential roles in ribosomal RNA processing by assembling with Xrn2 and Tan1 proteins (Figure 3B). This study unveiled the roles of tRF-3s in nuclear translocation of Twi12 and possible mechanisms of epigenetic regulation. Simultaneously, it was speculated that the modified bases on tRF-3s attenuated the effects of sequence complementarity to target genes. This study may help to broaden the roles of tRF-3 and differentiate these roles from those of tRF-5.
Another study identified that tRFGlu derives td-piR(Glu) with a 2′-O-methylation and 3′-terminus, and is highly enriched in monocytes in comparison to dendritic cells (71). In addition, interleukin-4 (IL-4) decreases the production of tRNAGlu and its by-product td-piR(Glu) by regulating the activity of polymerase III. td-piR(Glu) functions as an IL-4-mediated signaling molecule by promoting H3K9 histone methylation, binding to PIWIL4 protein and recruiting SETDB1 and heterochromatin protein 1β (HP1β). Suppressor of variegation 3-9 homolog 1 (SUV39H1) is also recruited to the promoter of CD1A, which results in the suppression of CD1A transcriptional activity (71). These results suggest that td-piR(Glu) participates in chromatin remodeling in immune cells (Figure 3C).
Mounting evidence indicates that the dysregulation of tRFs are key players of various malignant tumors.
Upregulated oncogenic signaling and downregulated anti-cancer signaling triggers the initiation and progression of cancers. tRF-19-3L7L73JD (i-tRF, derived from tRNAValAAC) is downregulated in the plasma of pre-operative gastric cancer patients, while overexpression of tRF-19-3L7L73JD attenuates viability of gastric cancer cells by promoting cell apoptosis (72). Lee et al. demonstrated that tRF-1001 is abundantly expressed in PCA cells and the knockdown of tRF-1001 affects cell cycle distribution and reduces cell proliferation (19). In another study, 5′ tRFHisGTG (derived from tRFHisGTG) was upregulated in colorectal cancer (CRC) tissues and positively correlated with tumor size. Moreover, the overexpression of 5′ tRFHisGTG promoted cancer cell division by targeting large tumor suppressor 2 (LATS2), which functions in the tumor-suppressive Hippo signaling pathway (73). Analogously, tRF-Val was found to be upregulated in GC cell lines and tissues. Functionally, tRF-Val promoted proliferation of GC cells in vivo and in vitro by destabilizing the eukaryotic translation elongation gene, elongation factor 1-alpha 1 (EEF1A1), a regulator that mediates p53 ubiquitination by enhancing the effects of E3 ubiquitin ligase (74). This study suggests the substantial potential of tRFs in cell proliferation regulation.
Migration and invasion allow cancer cells to spread to distant tissues or organs from the primary tumor site. Accumulating studies have shown that dysregulated tRFs are correlated with the invasion and metastasis of tumors. For example, Zhang et al. discovered that tRF-03357 is more abundant in ovarian cancer cells, and overexpression of tRF-03357 significantly inhibits the migration and invasion of ovarian cancer cells (75). Li et al. revealed that a cluster of 5′-tiRNAs regulate the metastatic and invasive abilities of CRC cells, and among the detected 5′- tiRNAs, 5′-tiRNAVal (derived from tRNAVal) was verified to be positively correlated with lymph node and distant metastasis in vivo and in vitro (76). Meanwhile, another study revealed that tRF-20-MEJB5Y13 promotes the migration and invasion of CRC cells (77). Dong et al. discovered that the overexpression of tRF-24-V29K9UV3IU hinders the migratory capacity of gastric cells, and a bioinformatics analysis revealed that tRF-24-V29K9UV3IU influences signaling pathways involved in cancer metastasis (78). However, the detailed mechanism of these phenotypes requires further study.
Malignancy, characterized by an attenuation of cancer cell apoptosis, can also be regulated by certain tRFs. For example, overexpression of tRF-315 (derived from tRNAlys) inhibits the apoptosis of PCA cells by perturbing the expression of growth arrest and DNA damage 45a (GADD45a), which plays a vital role in sustaining BAX mRNA stability and facilitating the expression of the apoptotic factor BAX (79). These studies shed light on novel apoptosis-promoting mechanisms and could be relevant in the treatment of PCA.
A growing number of studies have identified dysregulated tRFs in cells or tissues associated with viral infection, and have revealed the function of tRFs in viral replication (11, 80–83). 5′-tRF-GlyCCC and 5′-tRF-LysCTT were discovered to be upregulated in A549 cells upon respiratory syncytial virus (RSV) infection, and overexpression of 5′-tRF-GlyCCC and 5′-tRF-LysCTT significantly promoted RSV replication (81). However, the mechanism underlying this observation was not specified in the study. Ruggero et al. demonstrated the specific function of tRF-3019 in promoting human T-celll leukemia virus type1 (HTLV-1) replication in CD4+ T cells (11). It was revealed that tRF-3019 exhibited perfect base pairing to the primer binding site of HTLV-1, and served as a primer in guiding the reverse-transcriptional activity of HTLV-1. A study by Deng et al. revealed a novel targeting mechanism of tRFs in regulating viral replication (80). For example, tRF5-GluCTC was highly abundant in airway epithelial cells upon RSV infection (80). Contrary to typical microRNAs, tRF5-GluCTC utilizes a novel gene silencing mechanism. It was reported that the interaction of the 3′-portion of and 3′-UTR of apolipoprotein E receptor 2 (APOER2) mRNA resulted in reduced expression of APOER2. APoER2 is an antiviral protein whose inhibition leads to RSV replication. Given the role of tRF5-GluCTC in promoting RSV replication (84), Choi et al. examined whether tRF5-GluCTC silences target gene expression in miRNA machinery (83). Both AGO1 and AGO4 was found to contribute to gene silencing of tRF5-GluCTC, while AGO2 and AGO3 were not involved in tRF5-GluCTC-induced gene expression activity. These observations signify the possible development of novel therapeutics against viral infection by exploiting the function of tRFs.
Numerous studies have elucidated the abnormal expression of tRFs in various kinds of tumors and body fluids in cancer patients. There is enormous potential for tRFs to function as novel biomarkers for diagnosis, tracking the prognosis of cancer, as therapeutic targets, as well as in improving chemotherapy resistance (Figure 4 and Table 1).
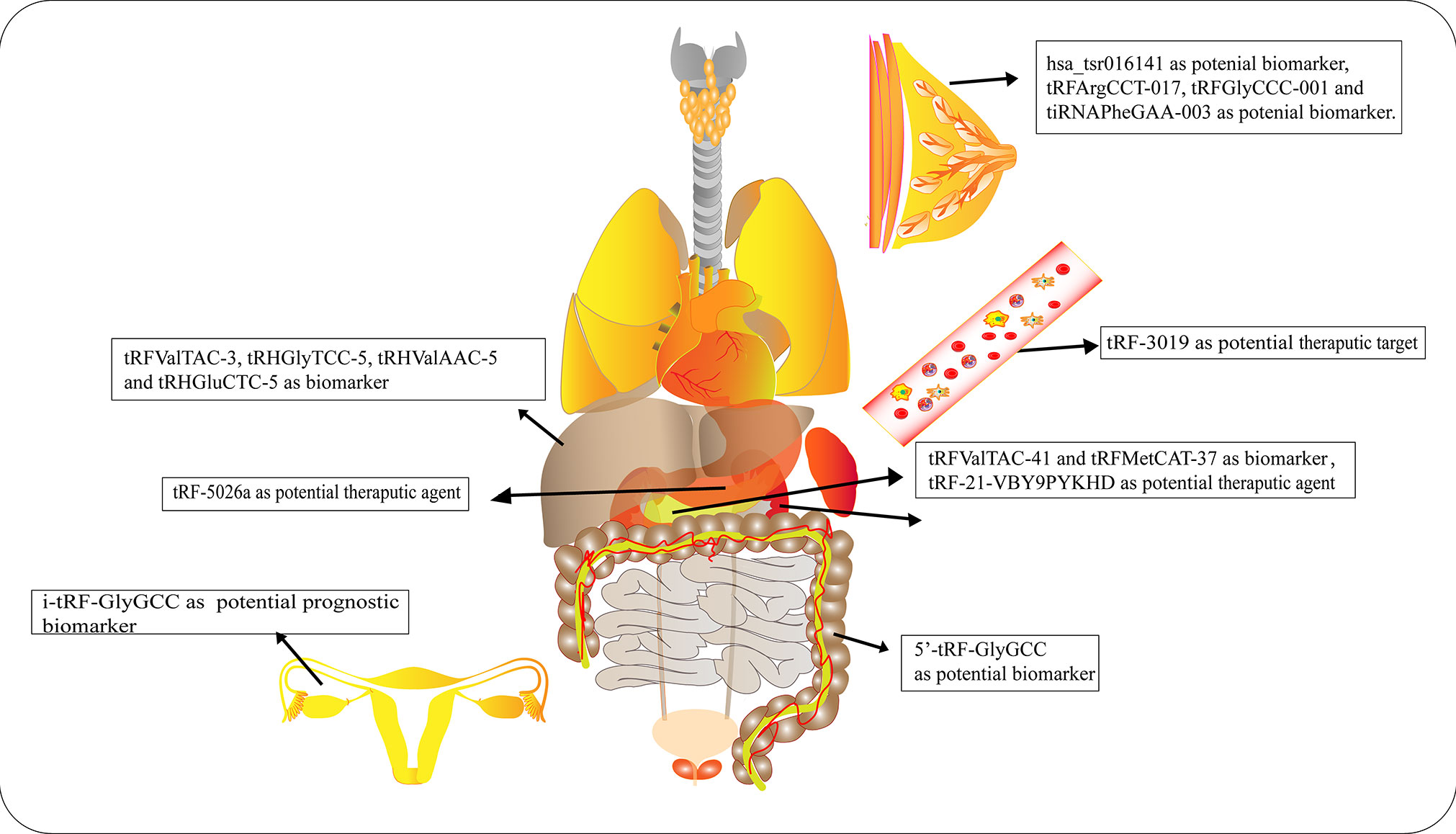
Figure 4 Roles of tRNA-derived fragments (tRFs) in various cancers. Different types of tRFs could serve as various biomarkers or therapeutic targets for distinct cancers or potential therapeutic agents.
Early diagnosis and treatment are considered critical factors relating to the improved prognosis of cancer patients. Therefore, it is crucial to develop specific biomarkers to significantly improve the diagnostic efficiency for cancer patients. The fact that various types of tRFs have been detected in body fluids, such as blood, urine, saliva and sperm (92–94), has rendered tRFs promising biomarkers. Recent studies have affirmed the values of tRFs as diagnostic and prognostic markers. Panoutsopoulou et al. revealed that i-tRF-GlyGCC is abundant in ovarian cancer compared to healthy controls. A Kaplan-Meier survival analysis confirmed the diagnostic value of elevated i-tRF-GlyGCC in predicting poor overall survival (OS) and worse progression-free survival of patients (90). Another study verified that the diagnostic values of tRFArgCCT-017, tRFGlyCCC-001, and tiRNAPheGAA-003 in BCA within an area under the curve were 0.683, 0.656, and 0.666, respectively (86). Meanwhile, elevated tRFArgCCT-017 or tiRNAPhe-GAA-003 levels are correlated with worse OS and disease-free survival rates in BCA patients (86). Wu et al. revealed that the plasma levels of 5′-tRF-GlyGCC increase with the progression and metastasis of CRC (87). The combination of 5′-tRF-GlyGCC and carcinoembryonic antigen, and carbohydrate antigen 19-9 (CA19-9) improves the AUC to 0.926. Gu et al. reported that the serum level of hsa_tsr016141 is correlated with metastasis and the cancer stage of gastric cancer, and can improve the diagnostic efficiency when combined with carcinoembryonic antigen and CA19-9 (85).
Xue et al. found that the combination of tRFValTAC-41 or tRFMetCAT-37 with CA19-9 can increase the diagnostic value (AUC = 0.947 and 0.949, respectively) in PDAC compared to CA19-9 alone (AUC = 0.906), and confirmed the clinical significance of tsRNA-ValTAC-41in predicting poor OS (89). In addition, high expression of tsRNA-5001a was revealed to be associated with increased risk of postoperative recurrences and poor OS in lung adenocarcinoma (3). Moreover, several studies have revealed that tRFs can be selectively exported in extracellular vesicles (95). Zhu et al. verified that 5′-tRH-GlyTCC, 5′-tRH-ValAAC, 5′-tRH-GluCTC, and 3′-tRF-ValTAC exhibit higher levels in plasma exosomes in patients with liver cancer (91). Intriguingly, it was reported that 5′-tiRNAs can form homodimers or heterodimers to prevent endonucleolytic cleavage, thus enhancing the stability of 5′-tiRNAs in extracellular vesicles (96). The presented studies indicate the immense potential of tRFs as novel forms of bio-liquid-based markers in diagnosing and evaluating prognosis.
In the context of tRFs with suppressive effects on tumor development, synthesized tRF mimics can be introduced into cancer cells or tissues to treat cancers. Pan et al. identified that inflammatory cytokine-induced tRF-21-VBY9PYKHD was downregulated in PDAC cells, and overexpression of tRF-21 significantly enhanced apoptosis and inhibited growth of PDAC cells (62). Mice treated with tRF-21 agomir showed a reduction in tumor volume and had longer survival times. Likewise, Zhu et al. transfected gastric cancer cells with tRF-5026a mimics and injected these cells subcutaneously into nude mice, successfully attenuating the tumor growth in vivo (88). Han’s group transfected CRC cells with tRF3008A (derived from tRNAVal) mimics and subcutaneously or intravenously injected these cells into mice to identify the effects of tRF3008A (97). tRF3008A had suppressive effects on the proliferation and migration of CRC. The results indicate the potential value of tRFs as therapeutic agents (98). In the context of oncogenic tRFs, the inhibition of certain tRFs may have therapeutic effects on cancer (99). Yang et al. demonstrated that upregulation of AS-tDR-007333 significantly promoted the growth and migration of non-small cell lung cancer cells (99). A mechanistic study revealed that AS-tDR-007333 and HSPB1 synergistically enhance transcription of mediator complex subunit 29 (MED29) by modifying histone modifications on MED29 promoter regions. The therapeutic efficacy of AS-tDR-007333 was evaluated in vivo. Reduced levels of AS-tDR-007333 significantly inhibited growth of NSCLC tumors (99). However, it should be noted that several endogenous tRFs harbor modifications that may confer improved stability compared to synthesized tRFs.
Chemotherapy resistance is a challenge in cancer treatment and affects patient survival directly (100). It is therefore imperative to ascertain the potential mechanisms of cancer chemoresistance. We speculate on the potential methods that tRFs could be involved in cancer chemoresistance. For example, Cui et al. determined that tDR-0009 (derived from tRNAGlyGCC-1-1) and tDR-7336 (derived from tRNAGlyGCC-1-2) were significantly upregulated in hypoxic BCA cells, and a bioinformatics analysis revealed the two upregulated tRFs were mainly involved in cellular response to IL-6 in TNBC (101). IL-6 was reported to promote transcription of HIF1A by activating STAT3 signaling, and enhancing cisplatin resistance of ovarian cancer cells both in vitro and vivo by upregulation of HIF1A (102). As a transcription factor, HIF-1α induces paclitaxel and cisplatin resistance of BCA cells by increasing transcription of apoptosis-resistant Bcl-2, as well as ATP-binding cassette (ABC) transport proteins, P-glycoproteins (P-gps) and multidrug-resistant protein 1s (MRP1s) (103). On the basis of the above studies, we speculate that specific tRFs may act as new classes of regulators in cancer chemoresistance (Figure 5A).
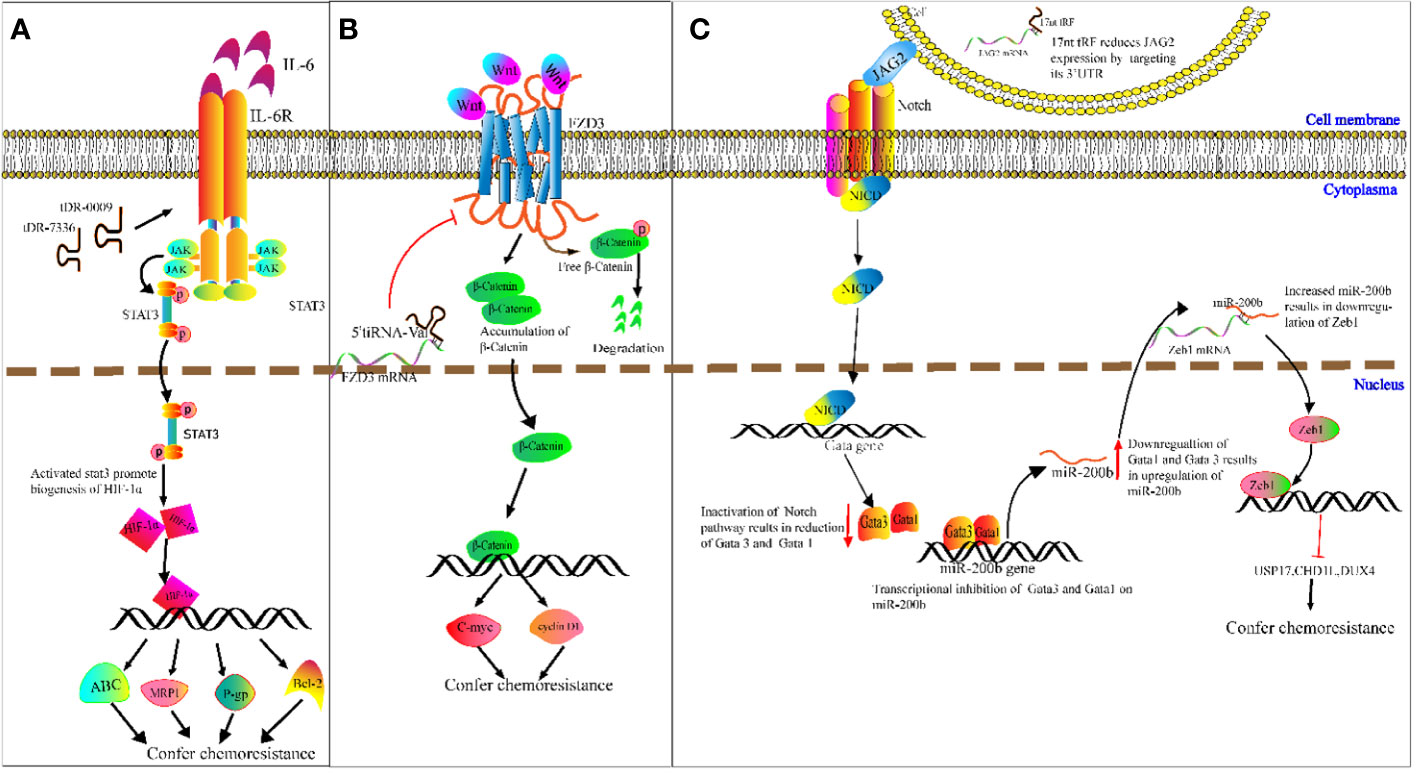
Figure 5 Molecular mechanism underlying tRNA-derived fragment (tRFs) regulation of carcinogenesis and chemoresistance. (A) DR-0009 and tDR-7336 activate the JAK/STAT3 pathway. Activated STAT3 promotes the biogenesis of HIF-1α. HIF-1α upregulates the expression of Bcl-2, ABC transporters, P-gp and MRP1. (B) 5′ tiRNAVal suppresses FZD3/Wnt/β-Catenin signaling. Inhibition of Wnt signals results in the degradation of free β-Catenin followed by downregulation of c-Myc and cyclin D1. (C) The 17nt-tRF/miR-1280 suppresses Notch/JAG2 transmembrane signal transduction by downregulating JAG2. This then leads to transcriptional repression of Gata1 and Gata3 and upregulation of miR-200b. miR-200b downregulates ZEB1 by targeting its 3′-UTR, followed by reduced expression of USP17, CHD1L and DUX4.
Meanwhile, Mo et al. demonstrated that overexpressed 5′ tiRNAVal suppresses cell proliferation and migration of BCA cells by targeting frizzled-3 (Fz-3) (104). As a vital component of the Wnt/β-catenin pathway, Fz-3 upregulation promotes expression of c-Myc, while Fz-3 inhibition results in degradation of free β-catenin (104) (105). This results in the subsequent attenuation of c-Myc expression (Figure 5B). c-Myc has been reported to confer antiestrogen resistance in BCA cells (106, 107).
Moreover, Huang’s team demonstrated that the 17 nucleotide-long tRF/miR-1280 suppresses proliferation and metastasis activity by destabilizing jagged canonical Notch ligand 2 (JAG2) (108). JAG2 is a membrane-bound ligand, and the binding of JAG2 to notch receptors results in proteolysis of the Notch intracellular domain, which translocates into the nucleus and binds to the promoter of GATA1 and GATA3 genes. GATA1 and GATA3 proteins exert transcriptional inhibition of miR-200b. As a result, the inhibition of Notch/JAG2 signaling reduces GATA1 and GATA3 expression, upregulates miR-200b expression, and subsequently increases expression of ZEB1 (target gene of miR-200b) (108). While ZEB1 was verified in CRC cells to inhibit transcription of ubiquitin-specific peptidase 17 (USP17), chromodomain helicase DNA-binding protein 1-like (CHD1L), and double homeobox 4 (DUX4), knockdown of ZEB1 improved CRC cell response to cisplatin (109). For example, 17 nucleotide-long tRF/miR-1280 mediated suppression of the Notch/JAG2 pathway and results in reduced levels of ZEB1. This may improve cisplatin sensitivity in CRC (Figure 5C). The regulatory mechanism that tRFs may play in chemoresistance is complicated, and is unlikely to be limited to the mechanism mentioned above. However, it is likely that tRFs may participate in this elaborate network.
According to existing literature, there are more than 500 tRNAs used to transport amino acids during translation (110). Therefore, it is rational to speculate that the number of existing tRF species may exceed the current estimates. The advanced technology of RNA sequencing contributes to the detection of diverse types of tRFs. tRFs have been verified to play roles in regulating the proliferation, metastasis, invasiveness and chemoresistance of cancer cells. Their specialties in cancer tissues and plasma makes it possible for scientists to develop novel screening, diagnostic and prognostic “liquid biopsy” biomarkers, as well as treatment targets for cancers. Nevertheless, as we have only scratched the surface of the biological roles of tRFs, the availability and biological significance of the various tRFs still require further investigation.
Firstly, the clinical applications of tRFs require in-depth research. Because of the extensively modified residues on tRNAs (111), tRFs inevitably contain these modifications, and may lead to inaccurate cDNA production. Our group previously combined tRF pretreatment and qRT-PCR to quantify tRFs (88), and we also applied hairpin structure primers to achieve accurate cDNA synthesis. The conventional methods used to detect tRFs mainly include high-throughput sequencing and Northern blotting. As these methods are not suitable for large-scale clinical testing, more advanced methods are needed for clinical applications.
Secondly, as tRF research in the context of chemotherapy resistance is in its infancy, it is still challenging to fully comprehend the potential mechanisms of the role of tRFs in chemoresistance. Therefore, the depth and breadth of research on the carcinogenesis of tRFs should be further expanded, especially with chemotherapeutic resistance.
In conclusion, despite the limited knowledge about tRFs, it is obvious that tRFs exist ubiquitously in all domains of organisms and play formidable roles in different pathophysiological processes. However, in-depth studies are required before clinical applications can be performed.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
JG designed the study. SZ and JG wrote the manuscript. XY and YX contributed to the literature search. All authors revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors are thankful for the financial support from the Zhejiang Provincial Natural Science Foundation of China (no. LGF21H200004), the National Natural Science Foundation of China (no. 81974316), the Ningbo Municipal Bureau of Science and Technology (nos. 2021Z133 and 2022Z130), and the K.C. Wong Magna Fund from Ningbo University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wang S, Luo Z, Yuan L, Lin X, Tang Y, Yin L, et al. tRNA-derived small RNAs: Novel insights into the pathogenesis and treatment of cardiovascular diseases. J Cardiovasc Trans Res (2022). doi: 10.1007/s12265-022-10322-0
2. Wang Y, Weng Q, Ge J, Zhang X, Guo J, Ye G. tRNA-derived small RNAs: Mechanisms and potential roles in cancers. Genes diseases. (2022) 9(6):1431–42. doi: 10.1016/j.gendis.2021.12.009
3. Hu F, Niu Y, Mao X, Cui J, Wu X, Simone CB 2nd, et al. tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Trans Lung Cancer Res (2021) 10(10):3957–72. doi: 10.21037/tlcr-21-829
4. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in b cell lymphoma. Proc Natl Acad Sci United States America. (2013) 110(4):1404–9. doi: 10.1073/pnas.1206761110
5. Weng Q, Wang Y, Xie Y, Yu X, Zhang S, Ge J, et al. Extracellular vesicles-associated tRNA-derived fragments (tRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases. J Mol Med (Berlin Germany). (2022) 100(5):679–95. doi: 10.1007/s00109-022-02189-0
6. Green JA, Ansari MY, Ball HC, Haqqi TM. tRNA-derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1β stimulated chondrocytes. Osteoarthritis cartilage. (2020) 28(8):1102–10. doi: 10.1016/j.joca.2020.04.014
7. Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE, et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep (2019) 20(7):e47789. doi: 10.15252/embr.201947789
8. Han L, Lai H, Yang Y, Hu J, Li Z, Ma B, et al. A 5’-tRNA halve, tiRNA-gly promotes cell proliferation and migration via binding to RBM17 and inducing alternative splicing in papillary thyroid cancer. J Exp Clin Cancer research: CR. (2021) 40(1):222. doi: 10.1186/s13046-021-02024-3
9. Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun (2017) 8(1):1127. doi: 10.1038/s41467-017-01278-w
10. Boskovic A, Bing XY, Kaymak E, Rando OJ. Control of noncoding RNA production and histone levels by a 5’ tRNA fragment. Genes Dev (2020) 34(1-2):118–31. doi: 10.1101/gad.332783.119
11. Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J virology. (2014) 88(7):3612–22. doi: 10.1128/jvi.02823-13
12. Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. (1979) 44(6):2120–3. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6
13. Yu X, Xie Y, Zhang S, Song X, Xiao B, Yan Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics. (2021) 11(1):461–69. doi: 10.7150/thno.51963
14. Jarrous N, Reiner R. Human RNase p: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res (2007) 35(11):3519–24. doi: 10.1093/nar/gkm071
15. Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase z processes mitochondrial and nuclear pre-tRNA 3’ ends. vivo. Nucleic Acids Res (2004) 32(1):255–62. doi: 10.1093/nar/gkh182
16. Liapi E, van Bilsen M, Verjans R, Schroen B. tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function. Biochim Biophys Acta Mol Cell Res (2020) 1867(3):118465. doi: 10.1016/j.bbamcr.2019.03.012
17. Martinez G. tRNA-derived small RNAs: New players in genome protection against retrotransposons. RNA Biol (2018) 15(2):170–75. doi: 10.1080/15476286.2017.1403000
18. Xie Y, Yao L, Yu X, Ruan Y, Li Z, Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther (2020) 5(1):109. doi: 10.1038/s41392-020-00217-4
19. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev (2009) 23(22):2639–49. doi: 10.1101/gad.1837609
20. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA (New York NY). (2009) 15(12):2147–60. doi: 10.1261/rna.1738409
21. Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev (2005) 19(4):489–501. doi: 10.1101/gad.1248505
22. Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS letters. (2009) 583(2):437–42. doi: 10.1016/j.febslet.2008.12.043
23. Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem (2010) 285(14):10959–68. doi: 10.1074/jbc.M109.077560
24. Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol (2020) 235(2):683–90. doi: 10.1002/jcp.29057
25. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol (2009) 185(1):35–42. doi: 10.1083/jcb.200811106
26. Zong T, Yang Y, Lin X, Jiang S, Zhao H, Liu M, et al. 5’-tiRNA-Cys-GCA regulates VSMC proliferation and phenotypic transition by targeting STAT4 in aortic dissection. Mol Ther Nucleic Acids (2021) 26:295–306. doi: 10.1016/j.omtn.2021.07.013
27. Elkordy A, Rashad S, Shehabeldeen H, Mishima E, Niizuma K, Abe T, et al. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res (2019) 1714:8–17. doi: 10.1016/j.brainres.2019.02.019
28. Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci United States America. (2015) 112(29):E3816–25. doi: 10.1073/pnas.1510077112
29. Su Z, Kuscu C, Malik A, Shibata E, Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem (2019) 294(45):16930–41. doi: 10.1074/jbc.RA119.009272
30. Andersen KL, Collins K. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate tetrahymena. Mol Biol Cell (2012) 23(1):36–44. doi: 10.1091/mbc.E11-08-0689
31. Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in saccharomyces cerevisiae. J Cell Biol (2009) 185(1):43–50. doi: 10.1083/jcb.200811119
32. Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. (2013) 495(7442):474–80. doi: 10.1038/nature11923
33. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
34. Ranjan N, Rodnina MV. tRNA wobble modifications and protein homeostasis. Translation (Austin). (2016) 4(1):e1143076. doi: 10.1080/21690731.2016.1143076
35. Tuorto F, Lyko F. Genome recoding by tRNA modifications. Open Biol (2016) 6(12):160287. doi: 10.1098/rsob.160287
36. Lorenz C, Mörl M, Lünse CE. tRNA modifications: Impact on structure and thermal adaptation. Biomolecules. (2017) 7(2):35. doi: 10.3390/biom7020035
37. Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA (New York NY). (2018) 24(10):1305–13. doi: 10.1261/rna.067033.118
38. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res (2019) 47(5):2533–45. doi: 10.1093/nar/gky1250
39. Lyons SM, Fay MM, Ivanov P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS letters. (2018) 592(17):2828–44. doi: 10.1002/1873-3468.13205
40. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA Methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev (2010) 24(15):1590–5. doi: 10.1101/gad.586710
41. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J (2014) 33(18):2020–39. doi: 10.15252/embj.201489282
42. Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, et al. RNA Cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol (2012) 19(9):900–5. doi: 10.1038/nsmb.2357
43. Guzzi N, Cieإla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. (2018) 173(5):1204–16 e26. doi: 10.1016/j.cell.2018.03.008
44. Hamma T, Ferré-D’Amaré AR. Pseudouridine synthases. Chem Biol (2006) 13(11):1125–35. doi: 10.1016/j.chembiol.2006.09.009
45. Donovan J, Rath S, Kolet-Mandrikov D, Korennykh A. Rapid RNase l-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA (New York NY). (2017) 23(11):1660–71. doi: 10.1261/rna.062000.117
46. Fergus C, Barnes D, Alqasem MA, Kelly VP. The queuine micronutrient: charting a course from microbe to man. Nutrients. (2015) 7(4):2897–929. doi: 10.3390/nu7042897
47. Wu W, Lee I, Spratt H, Fang X, Bao X. tRNA-derived fragments in alzheimer’s disease: Implications for new disease biomarkers and neuropathological mechanisms. J Alzheimer’s disease: JAD. (2021) 79(2):793–806. doi: 10.3233/jad-200917
48. Ma C, Du J, Ma X. tRNA-derived fragment tRF-1020 ameliorates diabetes-induced retinal microvascular complications. J Cell Mol Med (2022) 26(20):5257–66. doi: 10.1111/jcmm.17555
49. Green JA, Ansari MY, Ball HC, Haqqi TM. tRNA-derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1خ² stimulated chondrocytes. Osteoarthritis cartilage. (2020) 28(8):1102–10. doi: 10.1016/j.joca.2020.04.014
50. Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The c-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA (New York NY). (2009) 15(5):804–13. doi: 10.1261/rna.1229409
51. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol (2014) 12:78. doi: 10.1186/s12915-014-0078-0
52. Hefel A, Honda M, Cronin N, Harrell K, Patel P, Spies M, et al. RPA complexes in caenorhabditis elegans meiosis; unique roles in replication, meiotic recombination and apoptosis. Nucleic Acids Res (2021) 49(4):2005–26. doi: 10.1093/nar/gkaa1293
53. Zhong F, Hu Z, Jiang K, Lei B, Wu Z, Yuan G, et al. Complement C3 activation regulates the production of tRNA-derived fragments gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res (2019) 29(7):548–61. doi: 10.1038/s41422-019-0175-2
54. Gao X, Chen G, Gao C, Zhang DH, Kuan SF, Stabile LP, et al. MAP4K4 is a novel MAPK/ERK pathway regulator required for lung adenocarcinoma maintenance. Mol Oncol (2017) 11(6):628–39. doi: 10.1002/1878-0261.12055
55. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell (2011) 43(4):613–23. doi: 10.1016/j.molcel.2011.06.022
56. Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res (2016) 44(14):6949–60. doi: 10.1093/nar/gkw418
57. Ivanov P, O’Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-Quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci United States America. (2014) 111(51):18201–6. doi: 10.1073/pnas.1407361111
58. Kim HK, Xu J, Chu K, Park H, Jang H, Li P, et al. A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell Rep (2019) 29(12):3816–24 e4. doi: 10.1016/j.celrep.2019.11.062
59. Cho H, Lee W, Kim GW, Lee SH, Moon JS, Kim M, et al. Regulation of La/SSB-dependent viral gene expression by pre-tRNA 3’ trailer-derived tRNA fragments. Nucleic Acids Res (2019) 47(18):9888–901. doi: 10.1093/nar/gkz732
60. Sobala A, Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol (2013) 10(4):553–63. doi: 10.4161/rna.24285
61. Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol (2014) 34(13):2450–63. doi: 10.1128/mcb.00136-14
62. Pan L, Huang X, Liu ZX, Ye Y, Li R, Zhang J, et al. Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses pancreatic ductal adenocarcinoma progression. J Clin Invest. (2021) 131(22):e148130. doi: 10.1172/jci148130
63. Yu T, Huang X, Dou S, Tang X, Luo S, Theurkauf WE, et al. A benchmark and an algorithm for detecting germline transposon insertions and measuring de novo transposon insertion frequencies. Nucleic Acids Res (2021) 49(8):e44. doi: 10.1093/nar/gkab010
64. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. (2001) 409(6822):860–921. doi: 10.1038/35057062
65. Burns KH. Our conflict with transposable elements and its implications for human disease. Annu Rev Pathol (2020) 15:51–70. doi: 10.1146/annurev-pathmechdis-012419-032633
66. Chenais B. Transposable elements in cancer and other human diseases. Curr Cancer Drug Targets. (2015) 15(3):227–42. doi: 10.2174/1568009615666150317122506
67. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol (2011) 12(4):246–58. doi: 10.1038/nrm3089
68. Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon control by tRNA-derived small RNAs. Cell. (2017) 170(1):61–71 e11. doi: 10.1016/j.cell.2017.06.013
69. Yang F, Huang X, Zang R, Chen J, Fidalgo M, Sanchez-Priego C, et al. DUX-miR-344-ZMYM2-Mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell (2020) 26(2):234–50 e7. doi: 10.1016/j.stem.2020.01.004
70. Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A tetrahymena piwi bound to mature tRNA 3’ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell (2012) 48(4):509–20. doi: 10.1016/j.molcel.2012.09.010
71. Zhang X, He X, Liu C, Liu J, Hu Q, Pan T, et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on Monocytes/Dendritic cells. J Immunol (2016) 196(4):1591–603. doi: 10.4049/jimmunol.1500805
72. Shen Y, Xie Y, Yu X, Zhang S, Wen Q, Ye G, et al. Clinical diagnostic values of transfer RNA-derived fragment tRF-19-3L7L73JD and its effects on the growth of gastric cancer cells. J Cancer. (2021) 12(11):3230–38. doi: 10.7150/jca.51567
73. Tao EW, Wang HL, Cheng WY, Liu QQ, Chen YX, Gao QY. A specific tRNA half, 5’tiRNA-His-GTG, responds to hypoxia via the HIF1خ±/ANG axis and promotes colorectal cancer progression by regulating LATS2. J Exp Clin Cancer research: CR. (2021) 40(1):67. doi: 10.1186/s13046-021-01836-7
74. Cui H, Li H, Wu H, Du F, Xie X, Zeng S, et al. A novel 3’tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death disease. (2022) 13(5):471. doi: 10.1038/s41419-022-04930-6
75. Zhang M, Li F, Wang J, He W, Li Y, Li H, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther (2019) 12:6371–83. doi: 10.2147/ott.s206861
76. Li S, Shi X, Chen M, Xu N, Sun D, Bai R, et al. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J cancer. (2019) 145(5):1395–407. doi: 10.1002/ijc.32245
77. Luan N, Mu Y, Mu J, Chen Y, Ye X, Zhou Q, et al. Dicer1 promotes colon cancer cell invasion and migration through modulation of tRF-20-MEJB5Y13 expression under hypoxia. Front Genet (2021) 12:638244. doi: 10.3389/fgene.2021.638244
78. Dong X, Fan X, He X, Chen S, Huang W, Gao J, et al. Comprehensively identifying the key tRNA-derived fragments and investigating their function in gastric cancer processes. Onco Targets Ther (2020) 13:10931–43. doi: 10.2147/ott.s266130
79. Yang C, Lee M, Song G, Lim W. tRNA(Lys)-derived fragment alleviates cisplatin-induced apoptosis in prostate cancer cells. Pharmaceutics. (2021) 13(1):55. doi: 10.3390/pharmaceutics13010055
80. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol therapy: J Am Soc Gene Ther (2015) 23(10):1622–9. doi: 10.1038/mt.2015.124
81. Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS, et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen virology. (2017) 98(7):1600–10. doi: 10.1099/jgv.0.000852
82. Suhasini AN, Sirdeshmukh R. Onconase action on tRNA(Lys3), the primer for HIV-1 reverse transcription. Biochem Biophys Res Commun (2007) 363(2):304–9. doi: 10.1016/j.bbrc.2007.08.157
83. Choi EJ, Ren J, Zhang K, Wu W, Lee YS, Lee I, et al. The importance of AGO 1 and 4 in post-transcriptional gene regulatory function of tRF5-GluCTC, an respiratory syncytial virus-induced tRNA-derived RNA fragment. Int J Mol Sci (2020) 21(22):8766. doi: 10.3390/ijms21228766
84. Choi EJ, Wu W, Zhang K, Lee I, Kim IH, Lee YS, et al. ELAC2, an enzyme for tRNA maturation, plays a role in the cleavage of a mature tRNA to produce a tRNA-derived RNA fragment during respiratory syncytial virus infection. Front Mol biosciences. (2020) 7:609732. doi: 10.3389/fmolb.2020.609732
85. Gu X, Ma S, Liang B, Ju S. Serum hsa_tsr016141 as a kind of tRNA-derived fragments is a novel biomarker in gastric cancer. Front Oncol (2021) 11:679366. doi: 10.3389/fonc.2021.679366
86. Wang J, Ma G, Ge H, Han X, Mao X, Wang X, et al. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ breast cancer (2021) 7(1):4. doi: 10.1038/s41523-020-00211-7
87. Wu Y, Yang X, Jiang G, Zhang H, Ge L, Chen F, et al. 5’-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med (2021) 13(1):20. doi: 10.1186/s13073-021-00833-x
88. Zhu L, Li Z, Yu X, Ruan Y, Shen Y, Shao Y, et al. The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem Cell Res Ther (2021) 12(1):418. doi: 10.1186/s13287-021-02497-1
89. Xue M, Shi M, Xie J, Zhang J, Jiang L, Deng X, et al. Serum tRNA-derived small RNAs as potential novel diagnostic biomarkers for pancreatic ductal adenocarcinoma. Am J Cancer Res (2021) 11(3):837–48.
90. Panoutsopoulou K, Dreyer T, Dorn J, Obermayr E, Mahner S, Gorp TV, et al. tRNA(GlyGCC)-derived internal fragment (i-tRF-GlyGCC) in ovarian cancer treatment outcome and progression. Cancers. (2021) 14(1):24. doi: 10.3390/cancers14010024
91. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol cancer. (2019) 18(1):74. doi: 10.1186/s12943-019-1000-8
92. Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep (2017) 7:44061. doi: 10.1038/srep44061
93. Guo Y, Bai D, Liu W, Liu Y, Zhang Y, Kou X, et al. Altered sperm tsRNAs in aged male contribute to anxiety-like behavior in offspring. Aging Cell (2021) 20(9):e13466. doi: 10.1111/acel.13466
94. Li F, Kaczor-Urbanowicz KE, Sun J, Majem B, Lo HC, Kim Y, et al. Characterization of human salivary extracellular RNA by next-generation sequencing. Clinical chemistry. (2015) 23(10):1622–29. doi: 10.1373/clinchem.2017.285072
95. Chiou NT, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep (2018) 25(12):3356–70 e4. doi: 10.1016/j.celrep.2018.11.073
96. Tosar JP, Gأmbaro F, Darrأ© L, Pantano S, Westhof E, Cayota A. Dimerization confers increased stability to nucleases in 5’ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res (2018) 46(17):9081–93. doi: 10.1093/nar/gky495
97. Han Y, Peng Y, Liu S, Wang X, Cai C, Guo C, et al. tRF3008A suppresses the progression and metastasis of colorectal cancer by destabilizing FOXK1 in an AGO-dependent manner. J Exp Clin Cancer research: CR. (2022) 41(1):32. doi: 10.1186/s13046-021-02190-4
98. Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Sci (New York NY). (2016) 351(6271):397–400. doi: 10.1126/science.aad7977
99. Yang W, Gao K, Qian Y, Huang Y, Xiang Q, Chen C, et al. A novel tRNA-derived fragment AS-tDR-007333 promotes the malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes. J Hematol Oncol (2022) 15(1):53. doi: 10.1186/s13045-022-01270-y
100. Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao Z, et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene. (2016) 35(10):1302–13. doi: 10.1038/onc.2015.189
101. Cui Y, Huang Y, Wu X, Zheng M, Xia Y, Fu Z, et al. Hypoxia-induced tRNA-derived fragments, novel regulatory factor for doxorubicin resistance in triple-negative breast cancer. J Cell Physiol (2019) 234(6):8740–51. doi: 10.1002/jcp.27533
102. Xu S, Yu C, Ma X, Li Y, Shen Y, Chen Y, et al. IL-6 promotes nuclear translocation of HIF-1α to aggravate chemoresistance of ovarian cancer cells. Eur J Pharmacol (2021) 894:173817. doi: 10.1016/j.ejphar.2020.173817
103. Wang Y, Niu XL, Qu Y, Wu J, Zhu YQ, Sun WJ, et al. Autocrine production of interleukin-6 confers cisplatin and paclitaxel resistance in ovarian cancer cells. Cancer letters. (2010) 295(1):110–23. doi: 10.1016/j.canlet.2010.02.019
104. Mo D, Jiang P, Yang Y, Mao X, Tan X, Tang X, et al. A tRNA fragment, 5’-tiRNA(Val), suppresses the Wnt/خ²-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer letters. (2019) 457:60–73. doi: 10.1016/j.canlet.2019.05.007
105. Alshaer W, Alqudah DA, Wehaibi S, Abuarqoub D, Zihlif M, Hatmal MM, et al. Downregulation of STAT3, خ²-catenin, and notch-1 by single and combinations of siRNA treatment enhance chemosensitivity of wild type and doxorubicin resistant MCF7 breast cancer cells to doxorubicin. Int J Mol Sci (2019) 20(15):3696. doi: 10.3390/ijms20153696
106. Venditti M, Iwasiow B, Orr FW, Shiu RP. C-myc gene expression alone is sufficient to confer resistance to antiestrogen in human breast cancer cells. Int J cancer. (2002) 99(1):35–42. doi: 10.1002/ijc.10269
107. Yin S, Cheryan VT, Xu L, Rishi AK, Reddy KB. Myc mediates cancer stem-like cells and EMT changes in triple negative breast cancers cells. PloS One (2017) 12(8):e0183578. doi: 10.1371/journal.pone.0183578
108. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res (2017) 77(12):3194–206. doi: 10.1158/0008-5472.Can-16-3146
109. Wang M, He SF, Liu LL, Sun XX, Yang F, Ge Q, et al. Potential role of ZEB1 as a DNA repair regulator in colorectal cancer cells revealed by cancer-associated promoter profiling. Oncol Rep (2017) 38(4):1941–48. doi: 10.3892/or.2017.5888
110. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol (2018) 19(1):45–58. doi: 10.1038/nrm.2017.77
Keywords: tRNA-derived fragments, cancer, biological role, clinical value, chemotherapy resistance
Citation: Zhang S, Yu X, Xie Y, Ye G and Guo J (2023) tRNA derived fragments:A novel player in gene regulation and applications in cancer. Front. Oncol. 13:1063930. doi: 10.3389/fonc.2023.1063930
Received: 07 October 2022; Accepted: 04 January 2023;
Published: 20 January 2023.
Edited by:
Srikala Raghavan, Institute for Stem Cell Science and Regenerative Medicine (inStem), IndiaReviewed by:
Mailin Gan, Sichuan Agricultural University, ChinaCopyright © 2023 Zhang, Yu, Xie, Ye and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junming Guo, Z3VvanVubWluZ0BuYnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.