- 1Center for Pancreatic Cancer Research, School of Medicine, The South China University of Technology, Guangzhou, China
- 2Department of Pathology, School of Medicine, The South China University of Technology, Guangzhou, China
Pancreatic cancer (PC) is the most lethal human cancer, with less than 10% 5-year survival. Pancreatic premalignancy is a genetic and epigenomic disease and is linked to PC initiation. Pancreatic premalignant lesions include pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasm (IPMN), and mucinous cystic neoplasm (MCN), with pancreatic acinar-to-ductal metaplasia (ADM) as the major source of pancreatic premalignant lesions. Emerging evidence reveals that an epigenetic dysregulation is an early event in pancreatic tumorigenesis. The molecular mechanisms of epigenetic inheritance include chromatin remodeling; modifications in histone, DNA, and RNA; non-coding RNA expression; and alternative splicing of RNA. Changes in those epigenetic modifications contribute to the most notable alterations in chromatin structure and promoter accessibility, thus leading to the silence of tumor suppressor genes and/or activation of oncogenes. The expression profiles of various epigenetic molecules provide a promising opportunity for biomarker development for early diagnosis of PC and novel targeted treatment strategies. However, how the alterations in epigenetic regulatory machinery regulate epigenetic reprogramming in pancreatic premalignant lesions and the different stages of their initiation needs further investigation. This review will summarize the current knowledge of epigenetic reprogramming in pancreatic premalignant initiation and progression, and its clinical applications as detection and diagnostic biomarkers and therapeutic targets in PC.
1. Introduction
Pancreatic cancer (PC) is one of the most lethal diseases, with an incidence nearly equal to the mortality; e.g., there are 495,773 new cases and 466,003 deaths worldwide in 2020 (1). The 5-year survival rate of PC patients is less than 10%, the lowest among common cancers (2). The poor patient survival is mainly due to the fact that PC is asymptomatic in the early stages and difficult for early diagnosis. Although improvements in diagnosis, surgical procedure, and adjuvant therapy have contributed to a sustained decrease in the overall mortality rate, the death rate in PC has increased over the past decades (3). The development of minimally invasive biomarker assays for early diagnosis and effective clinical management of PC is urgently needed to reduce high morbidity and mortality.
The PC develops through the accumulation of a series of genomic and epigenomic alterations and progresses into invasive carcinoma. The process can take up to 15–20 years to develop from the occurrence of the first initiating mutational event. Understanding and discovering early PC and its precursor lesions are fundamentally important to effectively control this deadly disease and improve the outcome of patients. It is generally believed that pancreatic acinar-to-ductal metaplasia (ADM) is the major source of pancreatic premalignant lesions (4). ADM is a reversible process during injury or acute pancreatitis and becomes irreversible with chronic pancreatitis or Kras mutations (5). Pancreatic premalignant lesions include pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasm (IPMN), and mucinous cystic neoplasm (MCN), with PanIN being the most frequent (4, 6). PC is well investigated at the genetic level, while the molecular basis of its precursor, particularly epigenetics, remains to be explored.
Epigenetics studies the inheritance of phenotypes that occur without a corresponding change in DNA sequence. The molecular mechanisms of epigenetic inheritance include DNA methylation, RNA methylation, non-coding RNA, alternative splicing, histone modifications, chromatin remodeling, and phosphorylation (7–11). Epigenetic changes contribute to the silencing of tumor suppressor genes and the activation of oncogenes. Aberrant epigenetic alterations link to many genes with important roles in PC initiation and progression (12, 13). Epigenetic dysregulation is an early event in pancreatic tumorigenesis. The cancer-associated epigenetic changes occur hours after pancreatic injury at the beginning of ADM and before the appearance of widespread PanIN lesions and involve an “acinar-to-neoplasia” chromatin remodeling that leads to the early malfunction of genes that initiate PC (14). PC initiation results from a complex interaction of genetic and epigenetic damage that triggers changes in cell identity and tissue state and leads to a neoplastic cell fate, i.e., from normal lineage-specifying to cancer-defining (14, 15).
In this review, we first present an overview of the basic molecular mechanisms of epigenetic reprogramming in pancreatic premalignancy and describe the potential use of epigenetic reprogramming for early PC detection and targeted intervention.
2. DNA methylation in pancreatic premalignancy
Epigenetic changes could inhibit tumor suppressor genes and activate oncogenes. DNA methylation is a predominant epigenetic modification and differs in pancreatic neoplasia vs. normal pancreas (8, 16, 17). DNA methylation is catalyzed by DNA methyltransferases (DNMTs), which are encoded by a unique family of genes, DNMT1, DNMT2, DNMT3, and MGMT (18, 19) (Table 1; Figure 1).
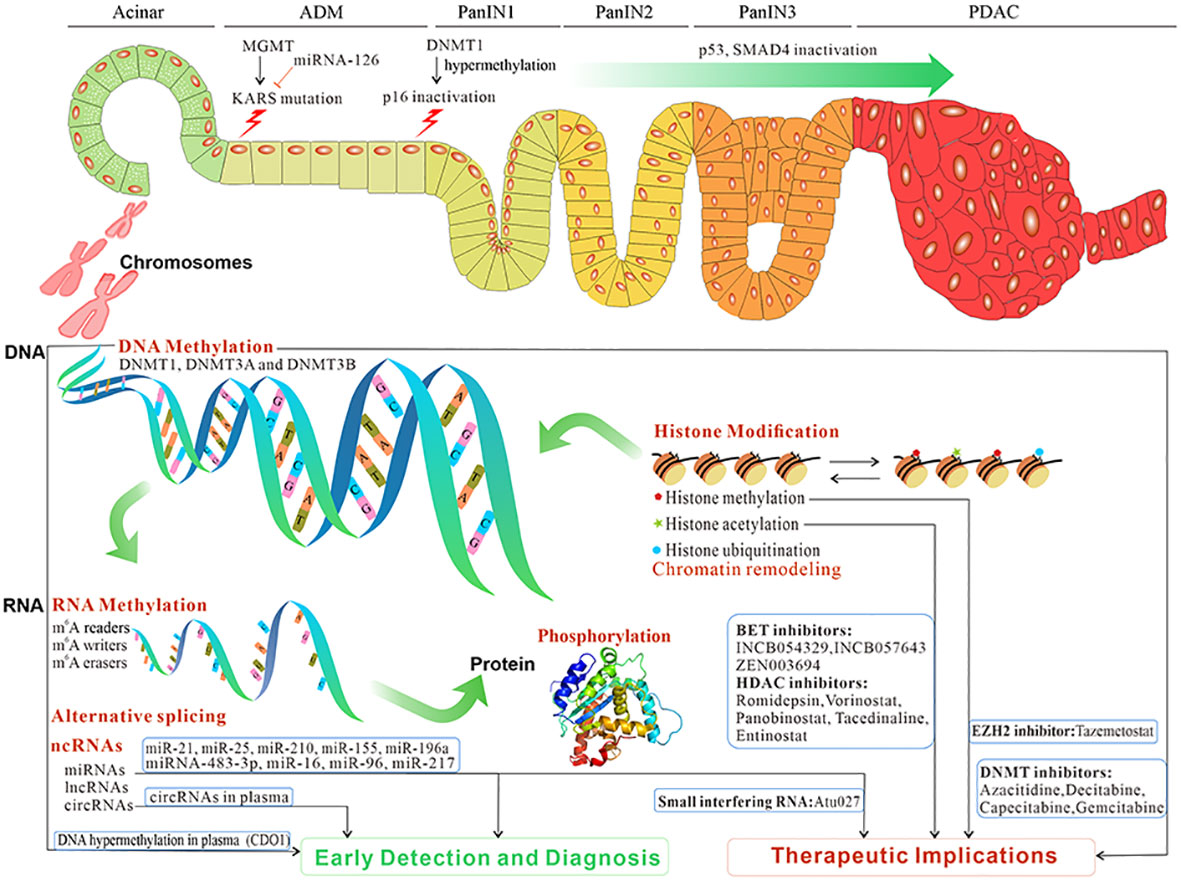
Figure 1 The overview and clinical implications of epigenetic reprogramming in pancreatic premalignancy and PC progression. Epigenetic reprogramming contributes to ADM, and its progression to PanIN and then PC. The initiation and progression of pancreatic premalignancy occur after epigenetic reprogramming such as DNA methylation, RNA methylation, microRNAs, lncRNAs, circRNAs, alternative splicing, histone modification, and phosphorylation. Those epigenetic changes potentially constitute biomarkers for early detection and diagnosis and targeted treatment in PC (data from ClinicalTrials.gov). PC, pancreatic cancer; ADM, acinar-to-ductal metaplasia; PanIN, pancreatic intraepithelial neoplasia.
2.1. Expression of DNA methyltransferases
DNMT1 is the most abundant and primarily a DNA maintenance methyltransferase and is responsible for copying methylation patterns after DNA replication (20). DNMT1, DNMT3A, and DNMT3B mRNA levels significantly increase from normal ducts to PanINs to PC (74). Similar to the mRNA results, DNMT1 protein expression increases from normal ducts with or without inflammation to pre-cancerous lesions (PanINs or IPMNs) to PC (64).
DNMT1 protein overexpression is involved in multistage tumorigenesis of the pancreas. PC patients with high levels of DNMT1 protein expression have a poorer outcome than those with low levels of DNMT1 expression (64, 75). Higher DNMT1 expression correlates with advanced stages of the disease (64). DNMT1 is oncogenic in promoting malignant phenotype and stemness in PC. These effects are achieved via promoter hypermethylation of tumor suppressor genes, e.g., cyclin-dependent kinase inhibitors, tumor suppressor microRNAs, and suppressors of epithelial–mesenchymal transition (EMT) (21).
DNMT2 methylates specifically cytosine 38 in the anticodon loop. DNMT2 is also involved in DNA recombination, DNA damage recognition, and mutation repair. The levels of DNMT3A, DNMT3B, and DNMT3L are usually elevated in cancer tissues and cells, in part due to the hypermethylation of promoter CpG-rich regions of tumor suppressor genes (22). DNMT3A and DNMT3B function as de novo methyltransferases (20). Patients having PC with higher levels of DNMT3A and/or DNMT3B expression have a poorer prognosis than those with a lower-level expression (74). In addition, the expressions of DNMT1 and DNMT3a in PC tissues are all higher than those in adjacent normal tissues (23, 76). However, the potential changes in their expression and function and underlying mechanisms in pancreatic premalignancy remain to be defined.
O6-Methylguanine DNA methyltransferase (MGMT) is another important DNMT and DNA-repair protein that removes mutagen from the O6 position of guanine. O6-Methylguanine often mispairs with thymine during replication and results in G-to-A point mutations if the adduct is not removed (18). MGMT methylation is associated with mutations of KRAS, TP53, and SMAD4 in PC (77). There is approximately 25% loss of MGMT in PC (78) and pancreatic endocrine tumors (79).
2.2. DNA methylation of tumor genes
DNA methylation of CpG islands in the promoter regions of tumor-related genes is associated with the silencing of those genes (12). Aberrant DNA methylation is associated with chromosomal instability, while deamination of 5-methylcytosine to thymine increases gene mutagenicity (80). Aberrant methylation of the ppENK gene increases progressively from PanIN-1A and PanIN-3 to PC. Aberrant methylation of p16 gene is also increased from PanIN-1A to PanIN-3. The methylation of p16 and ppENK genes increases with PanIN grades, suggesting that methylation of these genes is an intermediate or late event during pancreatic carcinogenesis (8). Moreover, the majority of IPMNs exhibit hypermethylation of CpG islands. Hypermethylation of p16 and ppENK occurs significantly higher in high-grade IPMNs than in low-grade IPMNs. The methylation of multiple CpG islands is one of the important pathways linked to the development and progression of IPMNs (16).
The aberrant methylation is identified in the majority of the earliest PanIN-1A lesions. NPTX2 increases in methylation from PanIN-1 to PanIN-2, while SARP2, Reprimo, and LHX1 increase in methylation from PanIN-2 to PanIN-3. The aberrant hypermethylation of CpG islands occurs in the early stages of PanINs, and its prevalence progressively increases during tumor progression (17). WNK2 is a cytoplasmic serine–threonine kinase. WNK2 interferes with the activation of the MEK1/ERK1/2 MAP kinase pathway and inhibits cell proliferation. WNK2 hypermethylation is higher in PanIN lesions than in surrounding tissues (81).
3. RNA methylation in pancreatic premalignancy
N6-Methyladenosine (m6A) is the most common modification that exists in mRNAs and long non-coding RNA (lncRNAs) (11). The m6A modification system is comprised of m6A writers (WTAP, METTL3, and METTL14), m6A erasers (ALKBH5 and FTO), and m6A readers (YTH domain-containing family proteins, IGF2BP1, IGF2BP2, and HNRNPA2B1) (11, 82–84). m6A has critical roles in tumorigenesis, and alterations in the m6A modification system may provide new therapeutic strategies (Table 1; Figures 1, 2).
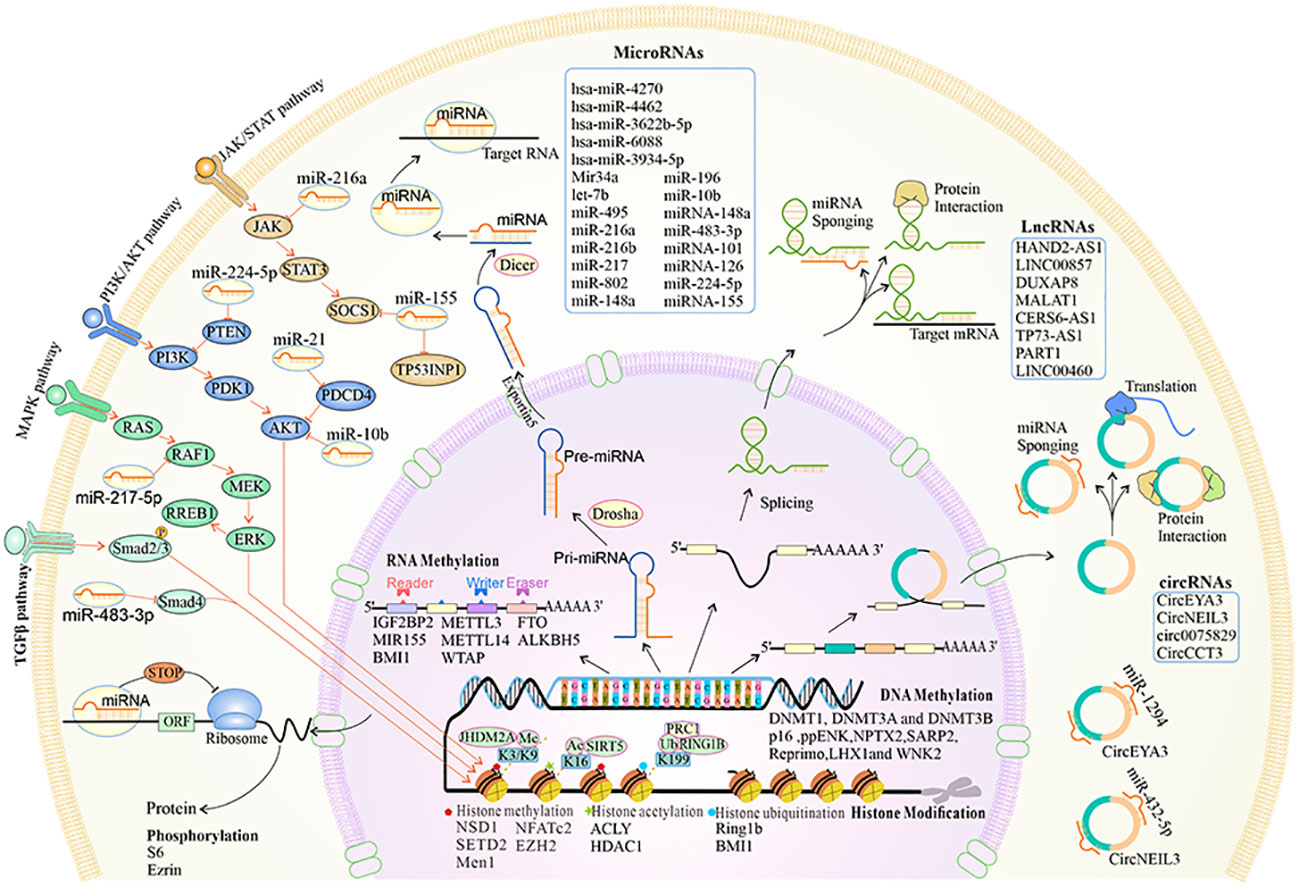
Figure 2 The overview of the biogenesis, functions, and mechanisms of epigenetic reprogramming in pancreatic premalignancy. At the chromatin level, histone methylation, acetylation and deacetylation, and histone ubiquitination occur. DNA and RNA can be modified by methylation. Alternative splicing leads to different isoforms of mRNA and proteins. MiRNAs can regulate different key downstream gene targets that mediate cellular signaling pathways. The lncRNAs are cell type-specific and cell state-specific. The lncRNAs can act as miRNA sponges, regulate mRNA stability, and interact with proteins. The circRNAs are transcribed by the RNA polymerase and can be processed through alternative splicing of the pre-mRNA. The circRNAs can act as miRNA sponges, interact with proteins, and translate into proteins.
3.1. m6A writers
METTL3 mRNA and protein are significantly higher in PC than in adjacent tissues. The increased expression of METTL3 is associated with advanced stages and decreased survival (85). Moreover, the m6A RNA methylation of IGF2BP2, IGF2BP3, METTL3, FTO, YTHDC1, and YTHDF1 is increased, while that of ALKBH5 METTL16, RBM15, and ZC3H13 is downregulated (86). Overexpression of METTL3 causes m6A modification and an excessive maturation of miR-25-3p. The upregulation of miR-25-3p inhibits PHLPP2 and activates AKT-p70S6K oncogenic signaling, forming a malignant METTL3–miR-25-3p–PHLPP2–AKT axis and promoting PC development and progression (13).
Leukemia inhibitory factor receptor antisense RNA 1 (LIFR-AS1) is a novel cancer-related lncRNA. METTL3 induces m6A hypermethylation on the 3′-UTR of LIFR-AS1 and enhances its stability. LIFR-AS1 interacts directly with miR-150-5p and indirectly increases VEGFA expression, promoting pancreatic tumorigenesis (24). The nucleobindin 1 (NUCB1), a calcium-binding protein, controls the unfolded protein response. METTL3 mediates m6A modification on NUCB1 5′-UTR via YTHDF2 and downregulates NUCB1, thus promoting PC cell proliferation (87).
METTL14 is an enzyme that modulates m6A methylation. METTL14 overexpression significantly promotes pancreatic tumorigenesis by directly targeting the mRNA of p53 effector related to PMP-22 (25), MPKα, ERK1/2, and mTOR signaling pathways (88), cytidine deaminase, an enzyme inactivating gemcitabine (89), and Cdc2-like kinases 1/SRSF5/cyclin L2 pathways (24). WTAP plays an important role in transcriptional and post-transcriptional regulation. WTAP is an independent prognostic indicator for PC. The WTAP promotes malignancy and suppresses gemcitabine chemo-sensitivity in PC (26). The pseudogene WTAPP1 RNA significantly increases in PC and is associated with poor prognosis in patients. Decreasing WTAPP1 RNA significantly suppresses the growth and metastasis of PC cells (90). However, their involvement in pancreatic premalignancy is unclear.
3.2. m6A erasers
Fat mass and obesity-associated (FTO) protein is a critical demethylase, or m6A eraser, in regulating mRNA stability by removing m6A residues in mRNA. The role of FTO in cancer cell homeostasis remains unclear. FTO is required for PC cell proliferation, and its knockdown causes a compromised DNA synthesis and PC cell proliferation but an increase in apoptosis. FTO interacts with MYC and bHLH transcription factors and enhances their stability by decreasing m6A levels. Reduced FTO expression results in elevated levels of m6A RNA modification in PC. Moreover, FTO demethylates the m6A modification of Praja ring finger ubiquitin ligase 2 (PJA2), reduces its mRNA degradation, suppresses Wnt signaling, and ultimately inhibits the malignant phenotype of PC cells (27). This novel mechanism for the regulation of PC malignancy by FTO could be a therapeutic target of PC (91).
Alkylation repair homolog protein 5 (ALKBH5) is an m6A demethylase. Its overexpression globally downregulates RNA m6A levels. ALKBH5 is generally downregulated in PC cells (92). P53-mediated activation of ALKBH5 transcription regulates the m6A modifications in a feedback loop (93).
ALKBH5 substrates include those mRNAs encoding mitochondrial iron importers SLC25A37 and SLC25A28 and ubiquitin ligase FBXL5. Downregulation of FBXL5 in tumor samples correlates with shorter patient survival. ALKBH5 overexpression incurs FBXL5-mediated degradation of and a significant reduction in iron regulatory protein IRP2 and the modulator of EMT, SNAI1. ALKBH5 overexpression leads to a significant reduction in intracellular iron levels as well as cell migration and invasion, which could be rescued by FBXL5 knockdown (28). Wnt inhibitory factor 1 (WIF-1) is also ALKBH5 target gene. Increased ALKBH5 decreases WIF-1 RNA methylation and transactivates WIF-1, thus inhibiting Wnt signaling and PC (94). ALKBH5 activates PER1 by m6A demethylation in an m6A-YTHDF2-dependent manner. Upregulated PER1 reactivates ATM-CHK2-P53/CDC25C signaling and inhibits PC cell growth (93).
ALKBH5 can also demethylate lncRNA KCNK15-AS1. Both ALKBH5 and KCNK15-AS1 are lower in PC tissues than in adjacent normal tissues. KCNK15-AS1 inhibits the malignancy of PC cells, while ALKBH5 downregulation and then KCNK15-AS1 downregulation promote PC cells (92). KCNK15-AS1 overexpression suppresses cell proliferation and EMT while promoting cell apoptosis in PC (92). ALKBH5 induces m6A demethylation of KCNK15-AS1 mRNA and then KCNK15-AS1 upregulation. Interestingly, KCNK15-AS1 recruits MDM2 proto-oncogene (MDM2) to promote the ubiquitination of RE1 silencing transcription factor (REST), thus transcriptionally upregulating phosphatase and tensin homolog (PTEN) and attenuating the AKT pathway (95).
3.3. m6A readers
The m6A modifications are recognized by m6A-binding proteins, such as YTHDF1/2/3, IGF2BP1/2, and HNRNPA2/B1. These proteins are regarded as “m6A readers” (84). Abnormal modifications in the m6A levels are closely related to tumorigenesis. Specifically, IGF2BP2 is a member of the IGF2BP protein family. IGF2BP2 has the capacity of binding to many transcripts. The expression of IGF2BP2 is upregulation with a poor prognosis in PC. IGF2BP2 protein is gradually upregulated from normal pancreas, PanIN to PC in the KrasG12D mouse model. IGF2BP2 promotes aerobic glycolysis and cellular proliferation via directly binding to and stabilizing Glut1 mRNA (29, 96). The hnRNP A2/B1 is upregulated in the PanIN and pancreatic cancer tissues (97). Moreover, hnRNP A2/B1 increases in PC tissues and cell lines (98).
4. Non-coding RNA in pancreatic premalignancy
Non-coding RNAs (ncRNAs) include circular RNAs (circRNAs), lncRNAs, and microRNAs (miRNAs) (Table 1; Figure 2).
4.1. MicroRNAs
MicroRNAs are small non-coding RNAs of 20–22 nucleotides. Growing evidence suggests that miRNAs interfere with cellular functions in pancreatic premalignancy and malignancy (41). The direct targeting of miRNAs provides a novel approach to the development of targeted intervention of early PC.
4.1.1. MicroRNAs in ADM
ADM in an inflammatory environment is a precursor of PC. From the miRNA–mRNA networks in the serum, hsa-miR-24-3p, hsa-miR-149-3p, hsa-miR-6785-5p, hsa-miR-4728-5p, hsa-miR-6808-5p, hsa-miR-6779-5p, hsa-miR-6799-5p, hsa-miR-6086, hsa-miR-4722-5p, and hsa-miR-4433a-3p are significantly upregulated, while hsa-miR-5100 is downregulated. hsa-miR-4270, hsa-miR-4462, hsa-miR-3622b-5p, hsa-miR-6088, and hsa-miR-3934-5p are also involved in ADM (30). In Ptf1a+/Cre and Kras+/LSL-G12D mice, pancreas-specific loss of Mir34a leads to an accelerated formation of pre-neoplastic lesions and a faster PC development (31).
As a critical component of the miRNA processing machinery, Dicer is an enzyme containing RNase III. Dicer essentially maintains acinar cell identity. Acinar cells without Dicer exhibit increased plasticity, as evidenced by loss of polarity and initiation of ADM and EMT. Moreover, homozygous Dicer deletion accelerates the formation of ADM but not PanIN, whereas heterozygous Dicer deletion accelerates PanIN initiation, suggesting complex roles for Dicer in the regulation of normal and neoplastic pancreatic epithelial identity (99). Let-7b and miR-495 repress HNF6 and express in developing acini. Let-7b and miR-495 expressions are downregulated in Dicer-deficient acini and pancreatitis-induced ADM. Moreover, suppressing let-7b and miR-495 causes similar effects in Dicer-deficient acini and metaplastic cells, i.e., induction of HNF6 and other hepatic genes and suppression of acinar differentiation. This gene network of Let-7b, miR-495, and their targets establish and maintain pancreatic acinar cell differentiation (32).
MiR217HG encodes miR-216a, miR-216b, and miR-217, which are enriched in pancreatic acinar cells. In germline knockout mice of miR-216a, miR-216b, or miR-217, the acini from each of the three miRNA-knockout mice produce many more ducts than those from control mice. There is an increased expression in ductal genes and reduced expression in acinar cells after acinar transdifferentiation. In caerulein-induced acute pancreatitis, miR-216a- and miR-216b-KO mice have more pancreatic duct glands and delayed recovery (33).
MiR-802 is highly expressed in pancreatic acinar cells and silenced during the early stages of pancreatic injury or cellular transformation. Knockout of miR-802 promotes ADM formation in Ptf1a+/Cre;Kras+/LSL-G12D mice. Downregulation of miR-802 activates Sox9 and enhances the expression levels of ductal identity genes but attenuates the expression of acinar identity genes. MiR-802 also inhibits PC development by suppressing oncogenic Kras-induced ADM (34).
4.1.2. MicroRNAs in PanIN
MicroRNA deregulation is a very early event in the progression of PC. MiRNA-21, miRNA-221, miRNA-222, and let-7a are upregulated in PanIN lesions, with peak expression occurring in PanIN-2 and PanIN-3 (100, 101). MiR-148a and miR-217 expression levels are downregulated in PanIN-2 and PanIN-3 and PC, whereas the level of miR-196 is significantly elevated in PC and PanIN-2 and PanIN-3 as compared with normal pancreatic parenchyma. Moreover, miR-10b is highly expressed in PanIN-2 and PanIN-3. These markers could be used as biomarkers to distinguish PC and its precursors from benign lesions (35). MiRNA-196 is most specifically overexpressed miRNA in PanIN-3, which has potential use as a marker for PanIN-3 lesions (102).
The expression of miRNA-148a is significantly downregulated in PanIN-1B, PanIN-2, and PanIN-3 lesions as compared with normal duct and PanIN-1A. The methylation level in the coding region of miRNA-148a is significantly higher in PC samples than in chronic pancreatitis samples. MiRNA-148a downregulation occurs at an early stage of PC initiation and progression (103). MiRNA-148a targets the mRNA of DNMT3B. The downregulation of miRNA-148a occurs early in premalignancy lesions, leading to DNMT3B upregulation and the inactivation of tumor suppressor genes (36). MiR-155 is significantly overexpressed in both PanIN-2 and PanIN-3, while miR-21 is highly expressed only in PanIN-3 (104). MiR-196b only is expressed in PanIN-3 tissue (102).
MiR-483-3p is overexpressed in PanIN and PC. Moreover, miR-483-3p expression levels correlate with increases in the PanIN grade. Circulating miR-483-3p levels are significantly increased in the serum and serum exosomes of PC patients. Interestingly, serum levels of miR-483-3p can distinguish patients with early-stage PC (≤2 cm) from healthy controls. MiR-483-3p expression negatively correlates with Smad4 expression in PC and PanIN tissues (37).
4.1.3. MicroRNAs in IPMN
The expression of miRNA-101 is significantly lower and EZH2 is dramatically higher in malignant IPMN than in benign IPMN. The loss of miRNA-101 could be a trigger for the tumorigenesis of IPMN by upregulation of EZH2 (38). Downregulation of miRNA-126 is detected in PC as compared to low malignant pancreatic benign cystic tumors. MiRNA-126 directly targets KRAS transcript through its binding site within 3′-UTR (39).
In high-risk IPMNs, oncogenic targets are upregulated, e.g., miR-126 (IRS-1), miR-130a (ATG2B and MEOX2), and miR-342-3p (DNMT1) (105). MiR-21 and miR-155 are higher in invasive IPMNs than in non-invasive IPMNs and normal tissues. Conversely, miR-101 is lower in invasive IPMNs than in non-invasive IPMNs and normal tissues. High levels of miR-21 are associated with worse overall survival. Patients with high-miR-21 expression also have a shorter median disease-free survival (106). MiR-10a-5p and miR-221-3p are upregulated while miR-148a-3p is downregulated in invasive IPMN as compared with non-invasive IPMN. Moreover, miR-10a-5p is highly expressed in invasive IPMN (107). MiR-99a, miR-99b, miR-100, miR-126, miR-130a, and miR-342-3p are downregulated in high-risk IPMNs. The alteration of those miRNAs may provide novel insights into miRNA-mediated formation and progression to pancreatic malignancy. Whether they could be promising potential biomarkers to distinguish between benign and invasive IPMN warrants further studies.
4.1.4. MicroRNAs in MCN
The expression of miR-17-3p, miR-21, and miR-221 is significantly higher in mucinous than in non-mucinous cysts (108). MiR-224-5p expression is significantly higher while PTEN expression is lower in pancreatic mucinous cystadenocarcinoma cells than in normal tissues. Overexpression of miR-224-5p promotes cell proliferation and invasion. PTEN is the direct target gene of miR-224-5p and negatively correlates with each other (41).
4.2. Long non-coding RNAs
LncRNAs are RNA molecules and usually consist of more than 200 nucleotides without protein-coding potential (109).
4.2.1. LncRNAs in pancreatic premalignancy
Neat1 is a large intergenic non-coding RNA (lincRNA) and is p53-regulated with a key role in suppressing transformation and tumorigenesis. Neat1 deficiency promotes the development of ADM, PanIN, and PC in KrasG12D mice (110). LncRNA Ppp3ca and lincRNA1611 expressions are significantly high in PC cells and tissues. Ppp3ca is low in normal pancreatic tissues and PanIN-I and PanIN-II tissues, while it is upregulated in PanIN-III and PC tissues (111).
LncRNA HAND2-AS1 is a suppressor in multiple cancer (42, 112, 113). HAND2-AS1 could regulate cytokine–cytokine receptor interaction, calcium signaling pathway, PI3K–Akt signaling pathway, and actin cytoskeleton (42). LncRNA HAND2-AS1 and CTD-2033D15.2 are gradually downregulated from the normal main pancreatic duct to low-grade IPMN, high-grade IPMN, and invasive IPMN, while LncRNA-TFG is gradually upregulated. LncRNAs promote tumorigenesis of IPMN and could serve as early diagnostic biomarkers and therapeutic targets in PC (114). Moreover, lncRNAs ADARB2-AS1, PANDA, ANRIL, LINC00472, MEG3, PVT1, GLIS3-AS1, and UCA1h in plasma from patients have high accuracy in discriminating between malignant and benign IPMNs (115).
4.2.2. LncRNAs in PC
N6-methyladenosine of LINC00857 is highly upregulated and enhances its RNA stability (43). The LINC00857 competes with endogenous RNA for sponging miR-150-5p, then upregulates its target E2F3, promotes proliferation, and inhibits apoptosis in PC cells. DUXAP8 is upregulated whereas miR-448 is downregulated in PC tissue and cells. DUXAP8 directly targets miR-448, which directly binds to WTAP. DUXAP8 sponges miR-448 to modulate the malignancy of PC cells (44).
LncRNA MALAT1 is highly expressed in PC tissues, while the large tumor suppressor 1 (LATS1) expression is downregulated and YAP1 is upregulated. MALAT1 influences proliferation, migration, and invasion in PC by regulating Hippo-YAP signaling (45). MALAT1 also interacts with RNA binding protein HuR, dramatically enhances the regulation of TIA-1, and has further effects on inhibiting autophagy (46).
CERS6 antisense RNA 1 (CERS6-AS1) is highly increased in PC. CERS6-AS1 competitively binds to miR-15a-5p as a molecular sponge in PC, increasing the expression of fibroblast growth factor receptor 1 (FGFR1), which also is a direct target of miR-15a-5p (47). Additionally, CERS6-AS1 exerts as a molecular sponge for miR-217-5p. MiR-217-5p suppresses cell proliferation and metastasis by targeting YWHAG, which targets RAF1, promotes its phosphorylation, and activates RAF1-mediated ERK signaling (116). Moreover, CERS6-AS1 sponges miR-15a-5p and miR-6838-5p to regulate HMGA1, which involves cell proliferation and migration in PC (117).
The expression of lncRNA TP73-AS1 is upregulated and miRNA-128-3p is downregulated in PC. TP73-AS1 promotes PC progression through binding to miR-128-3p and upregulating GOLM1 (48). High expression of TP73-AS1 correlates with poor overall survival. The TP73-AS1 positively regulates BDH2 expression by sponging miR-141 (49). The TP73-AS1 led to high MMP14 expression through miR-200a sponging, which significantly enhances migration and invasion in PC (118).
LncRNA PART1 expression is significantly upregulated in PC and correlates with tumor size, clinical stage, and poor overall survival. PART1 serves as a molecular sponge of miR-122. PART1 knockdown significantly reduces cell proliferation and invasion of PC (50). LncRNA ELFN1-AS1 is highly upregulated in PC. The knockdown of ELFN1-AS1 significantly increases cancer cell apoptosis and growth arrest (119).
The expression level of LINC00460 is increased in PC. Moreover, miR-320b directly targets LINC00460, whose knockdown leads to a downregulation in ARF1 expression. LINC00460 modulates the miR-320b/ARF1 axis, which inhibits tumor cell proliferation, migration, and invasion (51). LINC00460 directly targets and attenuates the tumor suppressor miR-491-5p and accelerates PC progression (120).
4.3. Circular RNAs
Alternative splicing of both exons and introns of the pre-mRNA can generate circRNAs. The circRNAs usually act as miRNA sponges because of their multiple miRNA response elements in the sequence. Certain circRNAs can also bind to proteins with RNA-binding sites. They can also encode for and translate into various proteins.
There are several circRNAs that are known to be important in PC development and progression. CircEYA3 is highly elevated in PC and associated with a poorer prognosis. CircEYA3 works as an endogenous miR-1294 sponge to upregulate c-Myc expression and promotes PC progression (52). CircNEIL3 is increased in PC and promotes PC progression. CircNEIL3 regulates ADAR1 expression by sponging miR-432-5p and induces the editing of glioma-associated oncogene 1 mRNA, thus affecting the cell cycle and promoting EMT in PC (121). The expression of circ_0075829 is also upregulated and regulates the LAMTOR3/p-ERK signaling pathway via sponging miR-1287-5p in PC. Knockdown of circ_0075829 significantly suppresses the malignancy of PC cells (53). The expression of CircCCT3 is significantly increased in PC tissues and cell lines. Patients with high expression of CircCCT3 have significantly poorer survival than those with low CircCCT3 expression. Moreover, CircCCT3 facilitates VEGFA and VEGFR2 expression as a sponge for miR-613 and promotes the malignancy of PC (54).
5. Alternative splicing in pancreatic premalignancy
Alternative splicing has an important role in tumorigenesis, and the splicing-derived isoforms are potentially powerful diagnostic, therapeutic, and prognostic factors. Alternative splicing produces multiple actin regulators, hMENA protein isoforms with hMENA11a and hMENAΔv6 (7). hMENA is absent in normal pancreas and low-grade PanINs and very weak in PanIN-3, while high hMENA protein levels are present in PC. However, the hMENA11a isoform is only strongly expressed in 26% of PC cases. In the absence of hMENA11a, increased expression of hMENA and hMENAΔv6 is key to SMAD2-mediated TGF-β1 signaling and EMT. The absence of hMENA11a significantly correlates with poor prognosis. The hMENA splicing and associative pathways are promising targets for the prognostic biomarkers and treatment in PC (55).
Tissue factor (TF) is a membrane glycoprotein and an enzymatic cofactor of the serine protease FVII/FVIIa and triggers the blood coagulation cascade. The murine alternatively spliced tissue factor (masTF) lacks exon 5 and has a distinct 93-amino-acid C-terminus. The intensity of masTF expression increases from early PanINs to PC phenotype, suggesting that masTF could promote PC growth (56).
The alternative splicing of FGFR-2 produces two variants: IIIb and IIIc. The expression of FGFR-2 IIIb correlates with a poor prognosis and may promote migration and invasion in PC (57). FGFR-2 IIIc is highly expressed in PC tissues and cell lines, which correlates with increased proliferation, migration, and invasion. High FGFR-2 IIIc levels in PC contribute to cancer stemness and could be a novel target for PC therapy (58).
6. Histone modification in pancreatic premalignancy
Epigenetic regulation can effectively change gene expression through histone post-translational modification and chromatin remodeling (122) (Table 1; Figures 1, 2). The factors that regulate epigenetics undergo certain expression changes in ADM, leading to the silence of genes specific to acinar cells and upregulation of the genes specific to ductal cells.
6.1. Histone methylation
NSD1 and SETD2 genes encode two histone H3K36 methyltransferases and are altered in approximately 10% of PC cases. NSD1 protein expression gradually increases in PanIN, IPMN, and MCN, and plays an important role in PC. Also, NSD1 expression is significantly higher in metastatic PC than in normal pancreas and primary cancer. The expression of SETD2 significantly decreases in metastatic PC and PanIN as compared to that in the normal pancreas (59). Moreover, the expression of SETD2 is downregulated in human PC and mouse models. The decreased expression of SETD2 mainly silences Fbxw7 through epigenetics to relieve its inhibitory effect on c-Myc (123).
The multiple endocrine neoplasia syndrome type 1 (MEN1) is a component of the macromolecular SET domain histone methyltransferase complex, which mediates the methylation of histone 3 on lysine 4 (60). The expression of Men1 is significantly reduced in PC (124). The absence of Men1 in the mouse pancreas impairs caerulein-induced pancreas regeneration and accelerates Kras-induced tumor formation (61). The tumor-promoting effect of Men1 deletion is related to the overexpression of the transcription factor Jund. Men1 inhibits the transcription of Jund, and Men1 deletion leads to Jund upregulation (125).
NFATc2 binds to the p15INK4b promoter site, induces the H3K9 trimethylation and heterochromatin formation, allows docking of heterochromatin protein HP1γ to the complex, and promotes PC cell growth (62). NFATc2 expression increases in PanIN lesions. Enhancer of zeste 2 homolog 2 (EZH2) is a histone methyltransferase, silences Nfatc1 epigenetically by histone methylation, and promotes acinar cell redifferentiation. However, NFATC1 is required for EZH2-mediated transcriptional activation of KRAS signaling in PC cells (126).
6.2. Histone acetylation
Histone acetylation is a chromatin modification and plays important roles in gene regulation, DNA replication, and DNA damage response. Pancreatic acinar cells with Kras mutations acquire histone H4 acetylation prior to the phenotype of premalignant lesions (127). Acetyl-CoA is upregulated in Kras-mutant acinar cells, and the mevalonate pathway supports ADM formation. Specific knockout of the acetyl-CoA-producing enzyme ATP-citrate lyase in the pancreas suppresses ADM and carcinogenesis. The acetyl-CoA regulates histone acetylation and facilitates cell plasticity and proliferation (63). In acute or chronic pancreatitis, inhibition of histone deacetylase (HDAC) can decrease the formation of ADM and inflammatory cell infiltration (128), suggesting that it is feasible to inhibit ADM development by modulations of epigenetics. Also, PC patients having a higher expression of HDAC1 have lower survival than those having a lower expression. High expression of HDAC1 correlates with advanced stages of and reflects the malignancy of PC (64).
6.3. Histone ubiquitination
The expression levels of ubiquitination significantly increase in the nucleus and cytoplasm in premalignant atypical acinar cells and cells of malignant adenocarcinoma in situ (129). Ring1b catalyzes histone modification H2AK119ub and silences acinar cell transcription factors, which is a key mechanism for triggering acinar cell dedifferentiation and PC development (66). Ring1b expression increases in PC (130), and knockout of acinar-specific Ring1b attenuates the formation of ADM induced by Kras (65).
B cell-specific Moloney murine leukemia virus insertion site 1 (BMI1) belongs to the polycomb group of proteins, including polycomb repressive complex 1, which consists of BMI1, RING1, and RING2 and acts as an E3-ubiquitin ligase. Upregulation of BMI1 in early PanINs may be a protective response to Kras-driven oxidative stress. BMI1 is highly expressed in human and murine PC (131, 132). BMI1-knockout mice have an increased expression of the Ink4a/Arf locus and a decreased β-cell mass and impaired glucose tolerance. The depletion of Bmi1 greatly decreases the levels of H2AK119Ub at the Chk2 locus. In pancreatic carcinogenesis, the role of Bmi1 is independent of the Ink4a/Arf expression, because its downregulation abrogates PanIN formation even in Ink4a-null animals (67). In addition, high expression of H2AK119Ub1 is significantly associated with poor prognosis and metastasis (66).
6.4. Chromatin remodeling
Chromatin remodeling affects gene transcription by changing the binding of transcription factors to chromosomes. BRG1 is the ATPase subunit of the SWI/SNF chromatin remodeling complex and is often inactivated in PC (68). In the presence of Kras oncogenic mutations, duct-specific BRG1 deletion leads to the formation of IPMN (69, 133). BRG1 binds to the Sox9 promoter region, regulates its expression, and recruits Pdx-1 through chromatin conformation changes (134). ARID1A, another component of the SWI/SNF complex, has also been frequently mutated in PC (70). ARID1A deletion changes the cancer precursor spectrum from PanIN to IPMN and accelerates the onset of invasive cancer (71).
7. Phosphorylation in pancreatic premalignancy
S6 ribosomal protein (S6) phosphorylation is involved in PC cell proliferation. As compared to that in normal cells, the expression of S6 phosphorylation is significantly upregulated in IPMN and is associated with glucose uptake increase in malignant cells of IPMN (9). Therefore, its expression level can be detected in pancreatic juice as a biomarker of malignancy of IPMN.
pSmad3C gradually downregulates from low-grade dysplasia and high-grade dysplasia to malignant IPMN, whereas pSmad3L upregulates. There are inverse relationships between the expression of pSmad3C and that of c-Myc, and between pSmad3C and Ki-67. There are positive relationships between the expression of pSmad3L and that of c-Myc, and between pSmad3L and Ki-67 (135).
As a member of the ezrin and moesin protein family, Ezrin acts as a cross-linker between the cell membrane and actin cytoskeleton. Ezrin is activated by threonine and tyrosine phosphorylation (72). The expression of phosphorylated ezrin (p-ezrin, tyr354) is higher significantly in IPMN than in invasive carcinoma. Moreover, p-ezrin (tyr353) is highly expressed in all grades of PanINs. The p-ezrin (tyr354) expression is associated with invasion in IPMNs, while p-ezrin (tyr353) expression plays an important role in early PanIN development (73).
8. Epigenetic biomarkers for early PC detection and diagnosis
The discovery of specific biomarkers is essential for the detection of PC premalignancy and diagnosis of PC at its early stages (136). Currently, only the serum carbohydrate antigen 19-9 (CA19-9) is a Food and Drug Administration (FDA)-approved marker for clinical treatment efficacy in PC (137). However, CA9-9 has limitations, including ineffectiveness, low sensitivity, and low specificity. Other biomarkers such as carcinoembryonic antigen (CEA) and cancer antigen 125 (CA125) are ineffective as early PC biomarkers (138). Evidently, epigenetic biomarkers have many potential advantages including stability in serum and non-invasive, economical, and convenient detection (Table 2; Figures 1, 3).
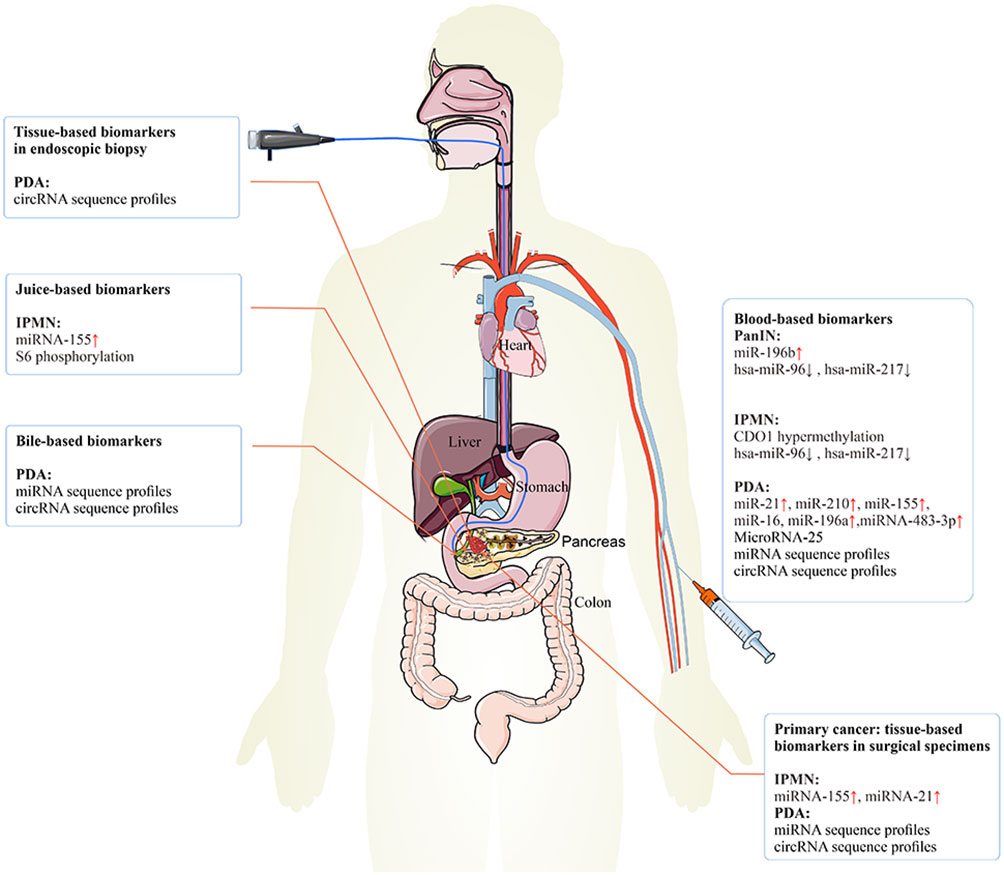
Figure 3 The epigenetic biomarkers for early PC detection. The epigenetic biomarkers or regulators can be exploited for clinical detection and diagnosis of early PC. Tissue-based biomarkers in endoscopic biopsy specimens may be used clinically to improve the prediction of the risk of PC. Tissue-based biomarkers in surgical specimens may have diagnosis value in IPMN and PC. Blood-based biomarkers may have diagnosis value in PanIN, IPMN, and PC. Juice-based biomarkers may have diagnosis value in IPMN. Bile-based biomarkers may have diagnosis value in PC. PC, pancreatic cancer; IPMN, intraductal papillary mucinous neoplasm; PanIN, pancreatic intraepithelial neoplasia.
8.1. DNA methylation biomarkers
DNA methylation changes during the development of cancer. The methylation usually occurs in the regulatory region of the gene and leads to the inactivation of the gene. Small amounts of DNA are released into the blood and can be detected in a blood sample. The DNA methylation changes can be tumor-specific and potentially usable tumor biomarkers. The alteration in hypermethylation of DNA from plasma is a diagnostic biomarker for PC (NCT02079363). Hypermethylation of cysteine dioxygenase 1 (CDO1) promoter is specific to IPMN and may increase with IPMN tumor progression, suggesting that CDO1 is a potential diagnostic marker of pancreatic cystic diseases (139).
8.2. MiRNAs and circRNAs as biomarkers
8.2.1. Plasma-associated miRNAs
Plasma miRNA profiling could be a sensitive and specific blood-based biomarker assay for PC in the clinic. MiR-155, miR-196a, miR-21, and miR-210 are implicated in PC development. Analyses of plasma levels for this panel of four miRNAs have shown certain sensitivity and specificity (140). The combination of CA19-9, miR-16, and miR-196a in plasma is more effective for distinguishing PC from non-PC (normal or chronic pancreatitis), especially in early tumor screening (141). MiRNA-483-3p and miRNA-21 from blood plasma could be PC biomarkers. MiR-483-3p expression is significantly increased in PC patients than in IPMN patients (142).
8.2.2. Tissue-associated miRNAs
MiRNA-155 and miRNA-21 are also significantly overexpressed in IPMN lesions (40). Locked nucleic acid in situ hybridization has verified that the expression of miRNA-155 is much higher in IPMNs than in normal ducts and that the expression of miRNA-21 is much higher in IPMNs than in normal ducts. The specificity of miRNA-155 upregulation as a biomarker of IPMNs is observed in 60% of IPMN cases in pancreatic juice samples compared to 0% of healthy controls (143).
MiR-196b is overexpressed selectively in PanIN-3 and PC lesions as compared with lower-grade PanINs and normal pancreatic ducts. These samples are isolated by laser capture microdissection. The expression of miR-196b is restricted in PanIN-3 lesions and could be an early diagnostic marker (102). The expression of hsa-miR-96 and hsa-miR-217 is significantly downregulated with increasing grades of PanINs and IPMNs as compared with normal pancreatic ductal epithelium in formalin-fixed paraffin-embedded specimens. They may serve as potential early biomarkers (144).
8.2.2.1. Ongoing clinical trials (selected data from ClinicalTrials.gov)
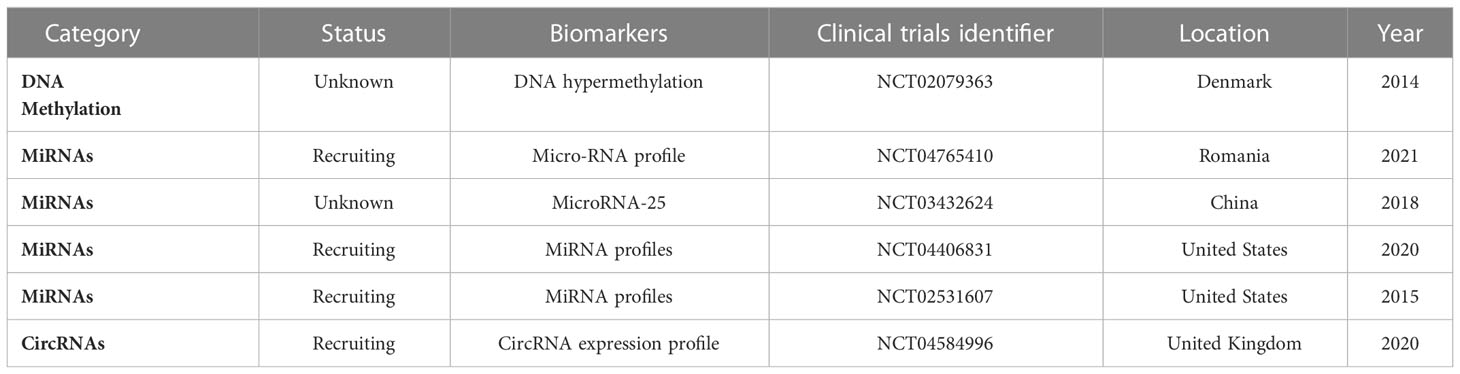
There are clinical trials on epigenetic biomarkers for the early detection of PC (Table 2). For example, NCT04765410 analyzes miRNA profile (84 miRNAs) using qRT-PCR array in every participant to identify the correlation between microRNA expression in PC tissue and tumor aggressive behavior, treatment response, and patient survival. Samples are obtained from the participants through fine-needle aspiration during endoscopic ultrasonography. NCT03432624 tries to improve the efficacy of PC diagnosis by combined detection of microRNA, conventional tumor markers, and imaging. NCT04406831 determines whether the detection of certain miRNAs present in the blood of PC patients is important to the early diagnosis of the disease and whether miRNA detection in PC patients helps predict treatment response and produces prognostic information.
In the NCT02531607 trial, the patients are assigned to control, non-carcinoma, carcinoma non-pancreatic, and pancreatic ductal adenocarcinoma. Comparative efficacy of biomarkers, brush cytology, CA19-9, and biliary and blood biomarkers including lipidomics, miRNAs, and proteomics is determined for the diagnosis of pancreatic malignancy.
In the NCT04584996 trial, paired samples of PC tissue and associated normal pancreatic tissue will be collected after pancreatic surgery. The expression levels of circRNAs will be determined, and significantly dysregulated candidate circRNAs will be selected. Candidate circRNA expression in blood plasma samples as a clinically diagnostic biomarker in PC and superior sensitivity than serum CA19-9 will be evaluated. Moreover, the expression of candidate circRNAs in patient tissue, blood, bile, and biopsy samples will be explored as biomarkers for PC diagnosis. Finally, bioinformatic data will be reviewed to determine the abilities of miRNAs to bind candidate circRNAs.
9. Therapeutic implications of epigenetic regulators
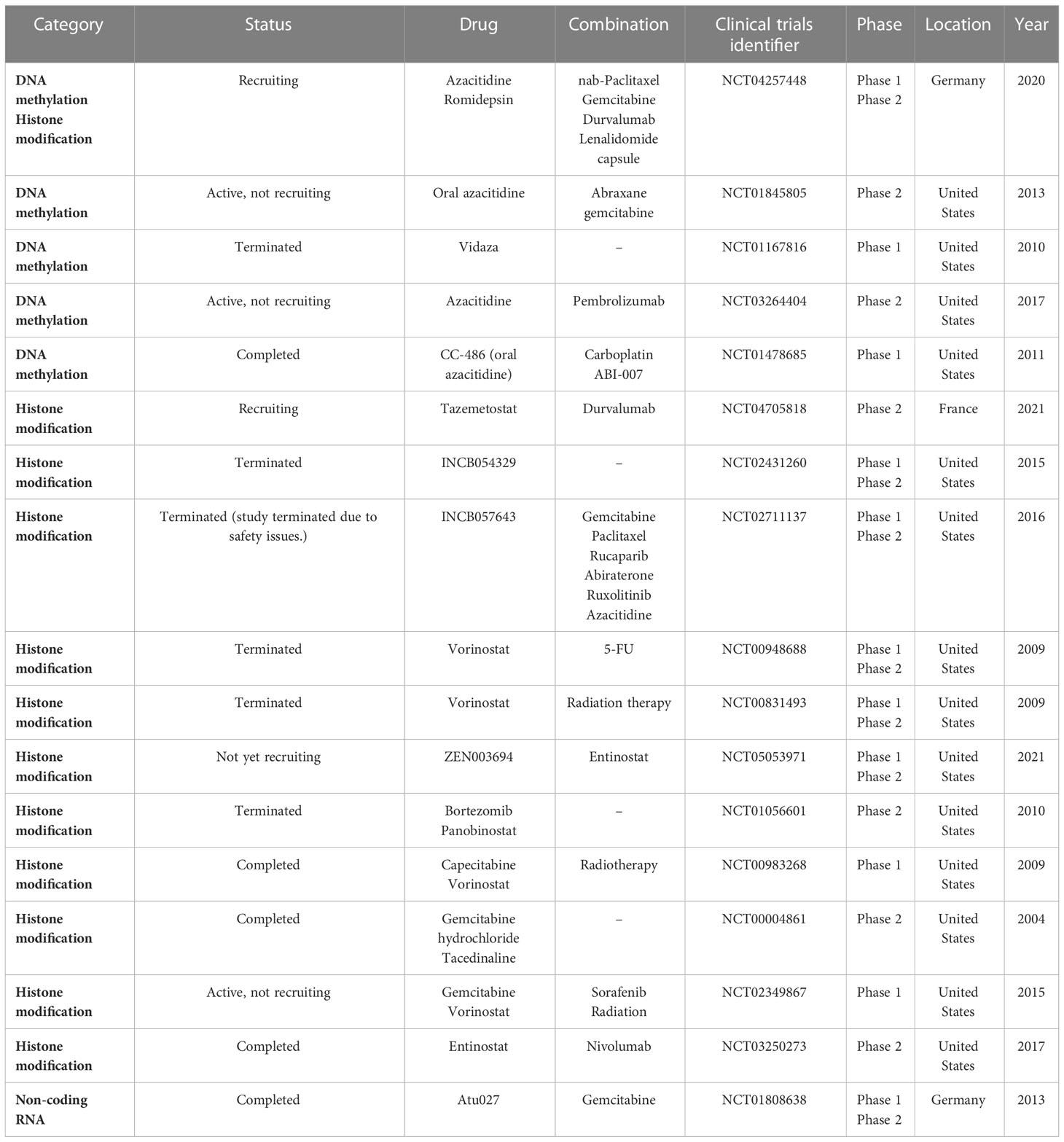
Unlike genetic changes, epigenetic alterations can be easily reversed by pharmacological manipulations. Numerous studies have focused on understanding the key epigenetic regulators that regulate these epigenetic processes and developing molecule inhibitors targeting these regulators. Increasing clinical trials are conducted on epigenetic alterations as valuable targets to develop cancer therapies (Table 3; Figure 1).
Many clinical trials on epigenetic regulators are being carried out for treating PC according to the data from ClinicalTrials.gov. The DNMT inhibitors 5-azacytidine and decitabine can trap DNMT proteins during the S-phase and degrade the trapped DNMTs and hypomethylated CpG islands of tumor suppressor genes (145, 146). Azacitidine and decitabine are used for hematological cancers in clinics.
There are clinical trials on the epigenetic treatment of PC (Table 3). For example, NCT04257448 aims to determine the safety and tolerability of HDAC inhibition by romidepsin and DNMT inhibition by azacitidine or both agents, in combination with gemcitabine/nab-paclitaxel in patients with advanced PC. Similarly, a phase II trial (NCT01845805) is ongoing to improve outcomes in patients with resected PC and high recurrence risk using oral azacitidine therapy. This trial is for patients who have resected PC and have completed adjuvant therapy or who are unable to receive adjuvant therapy due to an increased CA19-9 or advanced disease. Moreover, the phase I trial of 5-azacitidine plus gemcitabine (NCT01167816) is for patients having advanced PC. The primary objective is to define the maximum tolerated dose (MTD) of gemcitabine and azacitidine in patients with unresectable and previously untreated PC and to determine the effect of azacitidine therapy on DNA methylation in peripheral blood cells.
In the NCT02847000 trial, patients with PC who have stopped responding to one or more chemotherapy drugs are asked to take part in this study on decitabine therapy that decreases DNMT1 protein level. The study hopes to find out whether decitabine will have an effect on PC. Decitabine is also being given with another drug, tetrahydrouridine (THU), to improve the exposure of the PC cells to decitabine. Phase II trials (NCT04705818) are conducted in parallel to assess the efficacy of durvalumab when prescribed with tazemetostat, separately, in advanced PC.
Because preclinical data in a PC mouse model demonstrates an improvement in survival with the combined use of a hypomethylating agent and immune therapy, NCT03264404 phase II study is designed to determine the effectiveness of combining immune therapy, pembrolizumab, with a hypomethylating agent, azacitidine, for locally advanced or metastatic PC. Another phase I study (NCT01478685) is to use CC-486 alone or in combination with ABI-007 or carboplatin in patients with relapsed or refractory solid tumors.
The bromodomain and extra-terminal domain (BET) binds to acetylated histone lysine residues within chromatin and interacts with gene promoters/enhancers (147). After binding, BET proteins recruit transcription complexes, thereby promoting gene transcription. INCB054329 and INCB057643 are new oral BET inhibitors that are tested in phase I and II clinical trials for patients with advanced cancer (NCT02431260 and NCT02711137) (148).
Vorinostat (Zolinza) is an HDAC inhibitor. In this first study (NCT00948688), vorinostat will be given along with radiation therapy (RT) and 5-FU to determine whether vorinostat combined with radiation and 5-FU may improve PC treatment. The phase I and II trials (NCT00831493) determine the MTD of vorinostat plus radiation therapy in patients with locally advanced PC. Another phase I trial (NCT00983268) studies the side effects and optimal dose of vorinostat when used in combination with capecitabine and radiation therapy for patients with non-metastatic PC. In the NCT02349867 phase I study, concurrent chemoradiation uses a regimen of sorafenib and vorinostat with gemcitabine and radiation followed by chemotherapy in patients with PC to find the recommended phase II dose of the concurrent chemoradiation combination. The U.S. FDA has approved vorinostat (Merck & Co., Inc.) to treat cutaneous manifestations in patients with progressive, persistent, or recurrent cutaneous T-cell lymphoma disease or following two systemic therapies (149).
Phase II (NCT05053971) will determine the MTD of entinostat and BET bromodomain inhibitor ZEN-3694 (ZEN003694) in combination in patients having advanced and refractory PC, and the overall response rate of entinostat and ZEN003694 in advanced/progressive PC. Similarly, the phase II clinical trial (NCT03250273) of combined use of entinostat and nivolumab is for patients having metastatic or previously treated unresectable PC.
Panobinostat (LH589) produces diverse effects on endothelial cells and suppresses tumor angiogenesis. Bortezomib (Velcade) triggers cell death in PC cells. A phase II study (NCT01056601) of panobinostat in combination with bortezomib focuses on Patients with PC progressing on gemcitabine monotherapy or gemcitabine in combination. Moreover, a phase II trial (NCT00004861) compares the effectiveness of gemcitabine with or without tacedinaline (CI-994) in treating patients with advanced PC.
Finally, Atu027 is a liposomal formulation of siRNA targeting protein kinase N3 and can suppress cancer growth. In vitro Atu027 suppresses PKN3 function in endothelial cells and impairs their tube formation (150). Disease stabilization has been achieved in 41% of patients at end of treatment (151). In the NCT01808638 clinical trial, combined use of Atu027 and gemcitabine appeared to be safe and well tolerated. In patients with advanced PC, twice-weekly Atu027 significantly improved outcomes (152).
10. Conclusion and future perspectives
Effective management of PC urgently needs early detection through the discovery of highly sensitive and specific biomarkers for pancreatic premalignancy and novel interventional strategies with precise therapeutic targets. PanIN, MCN, and IPMN are the main pancreatic premalignant lesions. PanINs are the most frequent and too small to detect clinically. Identification and validation of precursor biomarkers are fundamentally important. Epigenetics are important regulatory mechanisms in pancreatic premalignant, including cellular proliferation and apoptosis, making them appealing candidates for diagnostic, therapeutic, and prognostic biomarkers. The expression profiles of various epigenetics molecules offer an exciting and promising opportunity for biomarker development for early diagnosis of PC. In this review, we describe several epigenetic biomarkers with the potential for early PC detection and diagnosis, including methylation of CDO1, miR-21, miR-25, miR-155, and miR-483-3p. However, there is no sufficient evidence to support the clinical use of these candidate biomarkers because of their mediocre performance and lack of thorough validation. Nonetheless, the performance of some investigational biomarkers could be improved through combination with existing tumor biomarkers (e.g., serum CA19-9) and radiological features (137, 138).
DNA methylation is a major epigenetic program regulating gene transcription. Aberrant hypermethylation of CpG islands in the promoters/enhancers of tumor suppressor genes and hypomethylation of oncogenes are key to early tumorigenesis (153, 154). m6A is the most abundant reversible methylation in mammalian mRNA and plays crucial roles in tumorigenesis. Histone post-translational modification and chromatin remodeling are the most complicated types of epigenetic programs and are crucial in PC development and progression (122).
Likewise, miRNAs in pancreatic premalignancy regulate cell proliferation, growth, metastasis, invasion, and apoptosis (41, 43). The direct targeting of miRNAs provides a novel insight into the development of diagnostic biomarkers and targeted therapy of PC. Currently, only the microRNA-25 and miRNA profiles have been in clinical trials, while not a single miRNA biomarker is applied in the clinic for the detection and diagnosis of PC or premalignant lesions. Investigation into the regulation of miRNAs within pancreatic premalignant lesions using an effective and credible approach remains a major challenge. Both lncRNAs and circRNAs are emerging and promising molecules that need further systematical studies for their full potential as biomarkers in pancreatic premalignant lesions.
Preclinical studies and clinical trials have shown that epigenetic alterations are potentially reversible through the use of DNMT, EZH2, BET, and HDAC inhibitors. However, it remains to determine whether PC patients benefit from receiving epigenetic inhibitors with improved survival, and none of the available epigenetic regulators that entered clinical trials has passed phase II. Nevertheless, the potential of targeting epigenetic regulators should be systematically investigated, particularly in combination with chemotherapy, radiotherapy, immunotherapy, and targeted therapy to increase therapeutic efficacy and avoid toxicity.
With improving knowledge and understanding of the molecular basis on the initiation and progression of pancreatic premalignancy and with potential identification and validation of novel diagnostic biomarkers and therapeutic targets, it becomes possible not only for early detection and diagnosis of PC but also for screening of patients with a high risk for developing a malignant transformation and clinical management of these patients before the development of high-grade dysplasia or invasive PC.
Author contributions
WZ, TJ, and KX reviewed the literature for the article and wrote, reviewed, and/or edited the manuscript before its submission. All authors contributed to the article and approved the submitted version.
Acknowledgments
We are grateful to the members of our laboratory for their critical reading of the manuscript and constructive thoughts. The work is partly supported by the National Natural Science Foundation of China (#82072632) and the Guangzhou Municipality Bureau of Science and Technology, Guangzhou, China (#202102010033).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–249. doi: 10.3322/caac.21660
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
4. Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol (2001) 25:579–86. doi: 10.1097/00000478-200105000-00003
5. Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol (2017) 14:296–304. doi: 10.1038/nrgastro.2017.12
6. Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2000) 6:2969–72.
7. Di Modugno F, Iapicca P, Boudreau A, Mottolese M, Terrenato I, Perracchio L, et al. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc Natl Acad Sci U.S.A. (2012) 109:19280–5. doi: 10.1073/pnas.1214394109
8. Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol (2002) 160:1573–81. doi: 10.1016/S0002-9440(10)61104-2
9. Hirashita T, Hirashita Y, Iwashita Y, Endo Y, Kiyonaga M, Matsumoto S, et al. S6 ribosomal protein phosphorylation is associated with malignancy of intraductal papillary mucinous neoplasm of the pancreas. Ann Gastroenterol Surg (2020) 4:571–9. doi: 10.1002/ags3.12367
10. Kouzarides T. Chromatin modifications and their function. Cell (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
11. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev (2015) 29:1343–55. doi: 10.1101/gad.262766.115
12. Peng D-F, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, et al. DNA Methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis (2006) 27:1160–8. doi: 10.1093/carcin/bgi361
13. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun (2019) 10:1858. doi: 10.1038/s41467-019-09712-x
14. Alonso-Curbelo D, Ho Y-J, Burdziak C, Maag JLV, Morris JP, Chandwani R, et al. A gene–environment-induced epigenetic program initiates tumorigenesis. Nature (2021) 590:642–8. doi: 10.1038/s41586-020-03147-x
15. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell (2013) 155:934–47. doi: 10.1016/j.cell.2013.09.053
16. Sato N, Ueki T, Fukushima N, Iacobuzio–Donahue CA, Hruban RH, Goggins M, et al. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology (2002) 123:365–72. doi: 10.1053/gast.2002.34160
17. Sato N, Fukushima N, Hruban RH, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol Off J U S Can Acad Pathol Inc (2008) 21:238–44. doi: 10.1038/modpathol.3800991
18. Patra SK. Ras regulation of DNA-methylation and cancer. Exp Cell Res (2008) 314:1193–201. doi: 10.1016/j.yexcr.2008.01.012
19. Robertson KD. DNA Methylation, methyltransferases, and cancer. Oncogene (2001) 20:3139–55. doi: 10.1038/sj.onc.1204341
20. Miremadi A, Oestergaard MZ, Pharoah PDP, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet (2007) 16:R28–49. doi: 10.1093/hmg/ddm021
21. Wong KK. DNMT1 as a therapeutic target in pancreatic cancer: mechanisms and clinical implications. Cell Oncol (2020) 43:779–92. doi: 10.1007/s13402-020-00526-4
22. Subramaniam D, Thombre R, Dhar A, Anant S. DNA Methyltransferases: A novel target for prevention and therapy. Front Oncol (2014) 4. doi: 10.3389/fonc.2014.00080
23. Clarke AR, Cummings MC, Harrison DJ. Interaction between murine germline mutations in p53 and APC predisposes to pancreatic neoplasia but not to increased intestinal malignancy. Oncogene (1995) 11:1913–20.
24. Chen J-Q, Tao Y-P, Hong Y-G, Li H-F, Huang Z-P, Xu X-F, et al. M6A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell Cycle (2021) 20:2507–18. doi: 10.1080/15384101.2021.1991122
25. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer (2020) 19:130. doi: 10.1186/s12943-020-01249-8
26. Li B-Q, Liang Z-Y, Seery S, Liu Q-F, You L, Zhang T-P, et al. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing fak mRNA in pancreatic cancer. Cancer Lett (2019) 451:48–57. doi: 10.1016/j.canlet.2019.02.043
27. Zeng J, Zhang H, Tan Y, Wang Z, Li Y, Yang X. m6A demethylase FTO suppresses pancreatic cancer tumorigenesis by demethylating PJA2 and inhibiting wnt signaling. Mol Ther Nucleic Acids (2021) 25:277–92. doi: 10.1016/j.omtn.2021.06.005
28. Huang R, Yang L, Zhang Z, Liu X, Fei Y, Tong W-M, et al. RNA m6A demethylase ALKBH5 protects against pancreatic ductal adenocarcinoma via targeting regulators of iron metabolism. Front Cell Dev Biol (2021) 9:724282. doi: 10.3389/fcell.2021.724282
29. Huang S, Wu Z, Cheng Y, Wei W, Hao L. Insulin-like growth factor 2 mRNA binding protein 2 promotes aerobic glycolysis and cell proliferation in pancreatic ductal adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochim Biophys Sin (2019) 51:743–52. doi: 10.1093/abbs/gmz048
30. Sheng L-P, Han C-Q, Nie C, Xu T, Zhang K, Li X-J, et al. Identification of potential serum exosomal microRNAs involved in acinar-ductal metaplasia that is a precursor of pancreatic cancer associated with chronic pancreatitis. Med (Baltimore) (2021) 100:e25753. doi: 10.1097/MD.0000000000025753
31. Hidalgo-Sastre A, Lubeseder-Martellato C, Engleitner T, Steiger K, Zhong S, Desztics J, et al. Mir34a constrains pancreatic carcinogenesis. Sci Rep (2020) 10:9654. doi: 10.1038/s41598-020-66561-1
32. Prévot P, Augereau C, Simion A, Van den Steen G, Dauguet N, Lemaigre FP, et al. Let-7b and miR-495 stimulate differentiation and prevent metaplasia of pancreatic acinar cells by repressing HNF6. Gastroenterology (2013) 145:668–678.e3. doi: 10.1053/j.gastro.2013.05.016
33. Sutaria DS, Jiang J, Azevedo-Pouly AC, Wright L, Bray JA, Fredenburg K, et al. Knockout of acinar enriched microRNAs in mice promote duct formation but not pancreatic cancer. Sci Rep (2019) 9:11147. doi: 10.1038/s41598-019-47566-x
34. Ge W, Goga A, He Y, Silva PN, Hirt CK, Herrmanns K, et al. miR-802 suppresses acinar-to-Ductal reprogramming during early pancreatitis and pancreatic carcinogenesis. Gastroenterology (2021) 162:269–284. doi: 10.1053/j.gastro.2021.09.029
35. Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet (2013) 206:217–21. doi: 10.1016/j.cancergen.2013.05.020
36. Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA (2008) 14:872–7. doi: 10.1261/rna.972008
37. Shao H, Zhang Y, Yan J, Ban X, Fan X, Chang X, et al. Upregulated MicroRNA-483-3p is an early event in pancreatic ductal adenocarcinoma (PDAC) and as a powerful liquid biopsy biomarker in PDAC. OncoTargets Ther (2021) 14:2163–75. doi: 10.2147/OTT.S288936
38. Nakahara O, Takamori H, Iwatsuki M, Baba Y, Sakamoto Y, Tanaka H, et al. Carcinogenesis of intraductal papillary mucinous neoplasm of the pancreas: Loss of MicroRNA-101 promotes overexpression of histone methyltransferase EZH2. Ann Surg Oncol (2012) 19:565–71. doi: 10.1245/s10434-011-2068-6
39. Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PloS One (2012) 7:e32068. doi: 10.1371/journal.pone.0032068
40. Frampton AE, Gall TM, Giovannetti E, Stebbing J, Castellano L, Jiao LR, et al. Distinct miRNA profiles are associated with malignant transformation of pancreatic cystic tumors revealing potential biomarkers for clinical use. Expert Rev Mol Diagn (2013) 13:325–9. doi: 10.1586/erm.13.18
41. Peng X, Guo C, Wu Y, Ying M, Chang R, Song L, et al. miR−224−5p regulates the proliferation, migration and invasion of pancreatic mucinous cystadenocarcinoma by targeting PTEN. Mol Med Rep (2021) 23:1–10. doi: 10.3892/mmr.2021.11985
42. He J, Zhao H, Deng D, Wang Y, Zhang X, Zhao H, et al. Screening of significant biomarkers related with prognosis of liver cancer by lncRNA-associated ceRNAs analysis. J Cell Physiol (2020) 235:2464–77. doi: 10.1002/jcp.29151
43. Meng X, Deng Y, He S, Niu L, Zhu H. m(6)A-mediated upregulation of LINC00857 promotes pancreatic cancer tumorigenesis by regulating the miR-150-5p/E2F3 axis. Front Oncol (2021) 11:629947. doi: 10.3389/fonc.2021.629947
44. Li J-R, Liu L, Luo H, Chen Z-G, Wang J-H, Li N-F. Long noncoding RNA DUXAP8 promotes pancreatic carcinoma cell migration and invasion Via pathway by miR-448/WTAP/Fak signaling axis. Pancreas (2021) 50:317–26. doi: 10.1097/MPA.0000000000001751
45. Zhou Y, Shan T, Ding W, Hua Z, Shen Y, Lu Z, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating hippo-YAP signaling. J Cell Physiol (2018) 233:5805–14. doi: 10.1002/jcp.26357
46. Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther (2016) 15:2232–43. doi: 10.1158/1535-7163.MCT-16-0008
47. Yun Z, Meng F, Li S, Zhang P. Long non-coding RNA CERS6-AS1 facilitates the oncogenicity of pancreatic ductal adenocarcinoma by regulating the microRNA-15a-5p/FGFR1 axis. Aging (2021) 13:6041–54. doi: 10.18632/aging.202540
48. Wang B, Sun X, Huang K-J, Zhou L-S, Qiu Z-J. Long non-coding RNA TP73-AS1 promotes pancreatic cancer growth and metastasis through miRNA-128-3p/GOLM1 axis. World J Gastroenterol (2021) 27:1993–2014. doi: 10.3748/wjg.v27.i17.1993
49. Cui XP, Wang CX, Wang ZY, Li J, Tan YW, Gu ST, et al. LncRNA TP73-AS1 sponges miR-141-3p to promote the migration and invasion of pancreatic cancer cells through the up-regulation of BDH2. Biosci Rep (2019) 39(3):BSR20181937. doi: 10.1042/BSR20181937
50. Hu X, Zhang L, Tian J, Ma J. Long non-coding RNA PART1 predicts a poor prognosis and promotes the malignant progression of pancreatic cancer by sponging miR-122. World J Surg Oncol (2021) 19:1–11. doi: 10.1186/s12957-021-02232-3
51. Cheng J, Lou Y, Jiang K. Downregulation of long non-coding RNA LINC00460 inhibits the proliferation, migration and invasion, and promotes apoptosis of pancreatic cancer cells via modulation of the miR-320b/ARF1 axis. Bioengineered (2021) 12:96–107. doi: 10.1080/21655979.2020.1863035
52. Rong Z, Shi S, Tan Z, Xu J, Meng Q, Hua J, et al. Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol Cancer (2021) 20:106. doi: 10.1186/s12943-021-01400-z
53. Zhang X, Xue C, Cui X, Zhou Z, Fu Y, Yin X, et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J Cell Mol Med (2020) 24:14596–607. doi: 10.1111/jcmm.16089
54. Hou JP, Men XB, Yang LY, Han EK, Han CQ, Liu LB. CircCCT3 acts as a sponge of miR-613 to promote tumor growth of pancreatic cancer through regulating VEGFA/VEGFR2 signaling. Balk Med J (2021) 38:229–38. doi: 10.5152/balkanmedj.2021.21145
55. Melchionna R, Iapicca P, Di Modugno F, Trono P, Sperduti I, Fassan M, et al. The pattern of hMENA isoforms is regulated by TGF-β1 in pancreatic cancer and may predict patient outcome. Oncoimmunology (2016) 5:e1221556. doi: 10.1080/2162402X.2016.1221556
56. Godby RC, van den Berg YW, Srinivasan R, Sturm R, Hui DY, Konieczny SF, et al. Nonproteolytic properties of murine alternatively spliced tissue factor: Implications for integrin-mediated signaling in murine models. Mol Med (2012) 18:771–9. doi: 10.2119/molmed.2011.00416
57. Nomura S, Yoshitomi H, Takano S, Shida T, Kobayashi S, Ohtsuka M, et al. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br J Cancer (2008) 99:305–13. doi: 10.1038/sj.bjc.6604473
58. Ishiwata T, Matsuda Y, Yamamoto T, Uchida E, Korc M, Naito Z. Enhanced expression of fibroblast growth factor receptor 2 IIIc promotes human pancreatic cancer cell proliferation. Am J Pathol (2012) 180:1928–41. doi: 10.1016/j.ajpath.2012.01.020
59. Ettel M, Zhao L, Schechter S, Shi J. Expression and prognostic value of NSD1 and SETD2 in pancreatic ductal adenocarcinoma and its precursor lesions. Pathol (Phila) (2019) 51:392–8. doi: 10.1016/j.pathol.2019.02.005
60. Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci (2005) 102:749–54. doi: 10.1073/pnas.0408836102
61. Wasylishen AR, Sun C, Chau GP, Qi Y, Su X, Kim MP, et al. Men1 maintains exocrine pancreas homeostasis in response to inflammation and oncogenic stress. Proc Natl Acad Sci U.S.A. (2020) 117:6622–9. doi: 10.1073/pnas.1920017117
62. Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J, et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology (2012) 142:388-98.e1-7. doi: 10.1053/j.gastro.2011.11.001
63. Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, et al. Acetyl-CoA metabolism supports multistep pancreatic tumorigenesis. Cancer Discovery (2019) 9:416–35. doi: 10.1158/2159-8290.CD-18-0567
64. Wang W, Gao J, Man X-H, Li Z-S, Gong Y-F. Significance of DNA methyltransferase-1 and histone deacetylase-1 in pancreatic cancer. Oncol Rep (2009) 21:1439–47. doi: 10.3892/or_00000372
65. Benitz S, Straub T, Mahajan UM, Mutter J, Czemmel S, Unruh T, et al. Ring1b-dependent epigenetic remodelling is an essential prerequisite for pancreatic carcinogenesis. Gut (2019) 68:2007–18. doi: 10.1136/gutjnl-2018-317208
66. Chen S, Chen J, Zhan Q, Zhu Y, Chen H, Deng X, et al. H2AK119Ub1 and H3K27Me3 in molecular staging for survival prediction of patients with pancreatic ductal adenocarcinoma. Oncotarget (2014) 5:10421–33. doi: 10.18632/oncotarget.2126
67. Bednar F, Schofield HK, Collins MA, Yan W, Zhang Y, Shyam N, et al. Bmi1 is required for the initiation of pancreatic cancer through an Ink4a-independent mechanism. Carcinogenesis (2015) 36:730–8. doi: 10.1093/carcin/bgv058
68. Marquez SB, Thompson K, Lu L, Reisman D. Mechanism of BRG1 silencing in primary cancers. Oncotarget (2016) 7:56153–69. doi: 10.18632/oncotarget.10593
69. von Figura G, Morris JP, Wright CVE, Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic kras-mediated pancreatic neoplastic initiation. Gut (2014) 63:656–64. doi: 10.1136/gutjnl-2012-304287
70. Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U.S.A. (2012) 109:E252–9. doi: 10.1073/pnas.1114817109
71. Wang W, Friedland SC, Guo B, O’Dell MR, Alexander WB, Whitney-Miller CL, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut (2019) 68:1245–58. doi: 10.1136/gutjnl-2017-315541
72. Sato N, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. A gene family consisting of ezrin, radixin and moesin. its specific localization at actin filament/plasma membrane association sites. J Cell Sci (1992) 103(Pt 1):131–43. doi: 10.1242/jcs.103.1.131
73. Oda Y, Aishima S, Morimatsu K, Hayashi A, Shindo K, Fujino M, et al. Differential ezrin and phosphorylated ezrin expression profiles between pancreatic intraepithelial neoplasia, intraductal papillary mucinous neoplasm, and invasive ductal carcinoma of the pancreas. Hum Pathol (2013) 44:1487–98. doi: 10.1016/j.humpath.2012.12.001
74. Zhang J-J, Zhu Y, Zhu Y, Wu J-L, Liang W-B, Zhu R, et al. Association of increased DNA methyltransferase expression with carcinogenesis and poor prognosis in pancreatic ductal adenocarcinoma. Clin Transl Oncol (2012) 14:116–24. doi: 10.1007/s12094-012-0770-x
75. Peng D-F, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci (2005) 96:403–8. doi: 10.1111/j.1349-7006.2005.00071.x
76. He S, Wang F, Yang L, Guo C, Wan R, Ke A, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PloS One (2011) 6:e27684. doi: 10.1371/journal.pone.0027684
77. Masetti M, Acquaviva G, Visani M, Tallini G, Fornelli A, Ragazzi M, et al. Long-term survivors of pancreatic adenocarcinoma show low rates of genetic alterations in KRAS, TP53 and SMAD4. Cancer biomark Sect Dis Markers (2018) 21:323–34. doi: 10.3233/CBM-170464
78. Bergmann F, Aulmann S, Sipos B, Kloor M, von Heydebreck A, Schweipert J, et al. Acinar cell carcinomas of the pancreas: a molecular analysis in a series of 57 cases. Virchows Arch (2014) 465:661–72. doi: 10.1007/s00428-014-1657-8
79. Gilbert JA, Adhikari LJ, Lloyd RV, Halfdanarson TR, Muders MH, Ames MM. Molecular markers for novel therapeutic strategies in pancreatic endocrine tumors. Pancreas (2013) 42:411–21. doi: 10.1097/MPA.0b013e31826cb243
81. Dutruel C, Bergmann F, Rooman I, Zucknick M, Weichenhan D, Geiselhart L, et al. Early epigenetic downregulation of WNK2 kinase during pancreatic ductal adenocarcinoma development. Oncogene (2014) 33:3401–10. doi: 10.1038/onc.2013.312
82. Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24:177–89. doi: 10.1038/cr.2014.3
83. Zhang H, Wu H, Zheng J, Yu P, Xu L, Jiang P, et al. Transforming growth factor beta1 signal is crucial for dedifferentiation of cancer cells to cancer stem cells in osteosarcoma. Stem Cells (2013) 31:433–46. doi: 10.1002/stem.1298
84. Chen X-Y, Zhang J, Zhu J-S. The role of m6A RNA methylation in human cancer. Mol Cancer (2019) 18:103. doi: 10.1186/s12943-019-1033-z
85. Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, et al. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol - Res Pract (2019) 215:152666. doi: 10.1016/j.prp.2019.152666
86. Guo Y, Wang R, Li J, Song Y, Min J, Zhao T, et al. Comprehensive analysis of m6A RNA methylation regulators and the immune microenvironment to aid immunotherapy in pancreatic cancer. Front Immunol (2021) 12:769425. doi: 10.3389/fimmu.2021.769425
87. Hua Y-Q, Zhang K, Sheng J, Ning Z-Y, Li Y, Shi W-D, et al. NUCB1 suppresses growth and shows additive effects with gemcitabine in pancreatic ductal adenocarcinoma via the unfolded protein response. Front Cell Dev Biol (2021) 9:641836. doi: 10.3389/fcell.2021.641836
88. Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, et al. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol (2020) 122:105731. doi: 10.1016/j.biocel.2020.105731
89. Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li Z, et al. m6A methyltransferase METTL14-mediated upregulation of cytidine deaminase promoting gemcitabine resistance in pancreatic cancer. Front Oncol (2021) 11:696371. doi: 10.3389/fonc.2021.696371
90. Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, et al. N6-methyladenosine–mediated upregulation of WTAPP1 promotes WTAP translation and wnt signaling to facilitate pancreatic cancer progression. Cancer Res (2021) 81:5268–83. doi: 10.1158/0008-5472.CAN-21-0494
91. Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett (2019) 17:2473–8. doi: 10.3892/ol.2018.9873
92. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 48:838–46. doi: 10.1159/000491915
93. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA Demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer (2020) 19:91. doi: 10.1186/s12943-020-01158-w
94. Tang B, Yang YH, Kang M, Wang YS, Wang Y, Bi Y, et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating wnt signaling. Mol Cancer (2020) 19:3. doi: 10.1186/s12943-019-1128-6
95. He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C, et al. ALKBH5-mediated m6A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis (2021) 12:1121. doi: 10.1038/s41419-021-04401-4
96. Wang C, Gu Y, Zhang E, Zhang K, Qin N, Dai J, et al. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene (2019) 38:1611–24. doi: 10.1038/s41388-018-0548-x
97. Wang L, Liu H-L, Li Y, Yuan P. Proteomic analysis of pancreatic intraepithelial neoplasia and pancreatic carcinoma in rat models. World J Gastroenterol (2011) 17:1434–41. doi: 10.3748/wjg.v17.i11.1434
98. Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett (2002) 183:215–20. doi: 10.1016/S0304-3835(02)00168-4
99. Wang YJ, McAllister F, Bailey JM, Scott S-G, Hendley AM, Leach SD, et al. Dicer is required for maintenance of adult pancreatic acinar cell identity and plays a role in kras-driven pancreatic neoplasia. PloS One (2014) 9:e113127. doi: 10.1371/journal.pone.0113127
100. du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem (2010) 56:603–12. doi: 10.1373/clinchem.2009.137364
101. Zhu Z, Gao W, Qian Z, Miao Y. Genetic variation of miRNA sequence in pancreatic cancer. Acta Biochim Biophys Sin (2009) 41:407–13. doi: 10.1093/abbs/gmp023
102. Yu J, Li A, Hong S-M, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res (2012) 18:981–92. doi: 10.1158/1078-0432.CCR-11-2347
103. Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, et al. The silencing of MicroRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem (2010) 56:1107–18. doi: 10.1373/clinchem.2010.144709
104. Ryu JK, Hong S-M, Karikari CA, Hruban RH, Goggins MG, Maitra A. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology (2010) 10:66–73. doi: 10.1159/000231984
105. Permuth-Wey J, Chen YA, Fisher K, McCarthy S, Qu X, Lloyd MC, et al. A genome-wide investigation of MicroRNA expression identifies biologically-meaningful MicroRNAs that distinguish between high-risk and low-risk intraductal papillary mucinous neoplasms of the pancreas. PloS One (2015) 10:e0116869. doi: 10.1371/journal.pone.0116869
106. Caponi S, Funel N, Frampton AE, Mosca F, Santarpia L, van der Velde AG, et al. The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol (2013) 24:734–41. doi: 10.1093/annonc/mds513
107. Kuratomi N, Takano S, Fukasawa M, Maekawa S, Kadokura M, Shindo H, et al. MiR-10a in pancreatic juice as a biomarker for invasive intraductal papillary mucinous neoplasm by miRNA sequencing. Int J Mol Sci (2021) 22:3221. doi: 10.3390/ijms22063221
108. Ryu JK, Matthaei H, Dal Molin M, Hong S-M, Canto MI, Schulick RD, et al. Elevated microRNA miR-21 levels in pancreatic cyst fluid are predictive of mucinous precursor lesions of ductal adenocarcinoma. Pancreatol Off J Int Assoc Pancreatol IAP Al (2011) 11:343–50. doi: 10.1159/000329183
109. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17:47–62. doi: 10.1038/nrg.2015.10
110. Mello SS, Sinow C, Raj N, Mazur PK, Bieging-Rolett K, Broz DK, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev (2017) 31:1095–108. doi: 10.1101/gad.284661.116
111. Wang Q, Jiang H, Ping C, Shen R, Liu T, Li J, et al. Exploring the wnt pathway-associated LncRNAs and genes involved in pancreatic carcinogenesis driven by Tp53 mutation. Pharm Res (2015) 32:793–805. doi: 10.1007/s11095-013-1269-z
112. Yan Y, Li S, Wang S, Rubegni P, Tognetti L, Zhang J, et al. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem (2019) 120:9564–71. doi: 10.1002/jcb.28233
113. Zhou J, Lin J, Zhang H, Zhu F, Xie R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun (2018) 503:1848–53. doi: 10.1016/j.bbrc.2018.07.125
114. Ding J, Li Y, Zhang Y, Fan B, Li Q, Zhang J, et al. Identification of key lncRNAs in the tumorigenesis of intraductal pancreatic mucinous neoplasm by coexpression network analysis. Cancer Med (2020) 9:3840–51. doi: 10.1002/cam4.2927
115. Permuth JB, Chen D-T, Yoder SJ, Li J, Smith AT, Choi JW, et al. Linc-ing circulating long non-coding RNAs to the diagnosis and malignant prediction of intraductal papillary mucinous neoplasms of the pancreas. Sci Rep (2017) 7:10484. doi: 10.1038/s41598-017-09754-5
116. Xu J, Wang J, He Z, Chen P, Jiang X, Chen Y, et al. LncRNA CERS6-AS1 promotes proliferation and metastasis through the upregulation of YWHAG and activation of ERK signaling in pancreatic cancer. Cell Death Dis (2021) 12:648. doi: 10.1038/s41419-021-03921-3
117. Shen R, Wang X, Wang S, Zhu D, Li M. Long noncoding RNA CERS6-AS1 accelerates the proliferation and migration of pancreatic cancer cells by sequestering MicroRNA-15a-5p and MicroRNA-6838-5p and modulating HMGA1. Pancreas (2021) 50:617–24. doi: 10.1097/MPA.0000000000001806
118. Miao H, Lu J, Guo Y, Qiu H, Zhang Y, Yao X, et al. LncRNA TP73-AS1 enhances the malignant properties of pancreatic ductal adenocarcinoma by increasing MMP14 expression through miRNA -200a sponging. J Cell Mol Med (2021) 25:3654–64. doi: 10.1111/jcmm.16425
119. Ma G, Li G, Gou A, Xiao Z, Xu Y, Song S, et al. Long non-coding RNA ELFN1-AS1 in the pathogenesis of pancreatic cancer. Ann Transl Med (2021) 9:877. doi: 10.21037/atm-21-2376
120. Wu J, Sun S, Liao W, Chen E, Wang X, Song Y, et al. LINC00460 promotes pancreatic cancer progression by sponging miR-491-5p. J Gene Med (2021) 23:e3333. doi: 10.1002/jgm.3333
121. Shen P, Yang T, Chen Q, Yuan H, Wu P, Cai B, et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and a-to-I RNA-editing. Mol Cancer (2021) 20:51. doi: 10.1186/s12943-021-01333-7
122. Barrero MJ, Boué S, Izpisúa Belmonte JC. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell (2010) 7:565–70. doi: 10.1016/j.stem.2010.10.009
123. Niu NN, Lu P, Yang YL, He RZ, Zhang L, Shi JJ, et al. Loss of Setd2 promotes kras-induced acinar-to-ductal metaplasia and epithelia-mesenchymal transition during pancreatic carcinogenesis. Gut (2020) 69:715–26. doi: 10.1136/gutjnl-2019-318362
124. Cavallari I, Silic-Benussi M, Rende F, Martines A, Fogar P, Basso D, et al. Decreased expression and promoter methylation of the menin tumor suppressor in pancreatic ductal adenocarcinoma. Genes Chromosomes Cancer (2009) 48:383–96. doi: 10.1002/gcc.20650
125. Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell (1999) 96:143–52. doi: 10.1016/S0092-8674(00)80967-8
126. Shen X, Chang LG, Hu MY, Yan D, Zhou LN, Ma Y, et al. KrasG12D-LOH promotes malignant biological behavior and energy metabolism of pancreatic ductal adenocarcinoma cells through the mTOR signaling pathway. Neoplasma (2018) 65:81–8. doi: 10.4149/neo_2018_170224N142
127. Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab (2014) 20:306–19. doi: 10.1016/j.cmet.2014.06.004
128. Bombardo M, Saponara E, Malagola E, Chen R, Seleznik GM, Haumaitre C, et al. Class I histone deacetylase inhibition improves pancreatitis outcome by limiting leukocyte recruitment and acinar-to-ductal metaplasia. Br J Pharmacol (2017) 174:3865–80. doi: 10.1111/bph.13984
129. Tóth S, Vastagh C, Pálfia Z, Réz G. Ubiquitin cytochemical changes during azaserine-initiated pancreatic carcinogenesis. Acta Biol Hung (2001) 52:383–91. doi: 10.1556/ABiol.52.2001.4.3
130. Benitz S, Regel I, Reinhard T, Popp A, Schäffer I, Raulefs S, et al. Polycomb repressor complex 1 promotes gene silencing through H2AK119 mono-ubiquitination in acinar-to-ductal metaplasia and pancreatic cancer cells. Oncotarget (2015) 7:11424–33. doi: 10.18632/oncotarget.6717
131. Martínez-Romero C, Rooman I, Skoudy A, Guerra C, Molero X, González A, et al. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. J Pathol (2009) 219:205–13. doi: 10.1002/path.2585
132. Song W, Tao K, Li H, Jin C, Song Z, Li J, et al. Bmi-1 is related to proliferation, survival and poor prognosis in pancreatic cancer. Cancer Sci (2010) 101:1754–60. doi: 10.1111/j.1349-7006.2010.01577.x
133. von Figura G, Fukuda A, Roy N, Liku ME, Morris JP, Kim GE, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol (2014) 16:255–67. doi: 10.1038/ncb2916
134. Tsuda M, Fukuda A, Roy N, Hiramatsu Y, Leonhardt L, Kakiuchi N, et al. The BRG1/SOX9 axis is critical for acinar cell-derived pancreatic tumorigenesis. J Clin Invest (2018) 128:3475–89. doi: 10.1172/JCI94287
135. Hori Y, Ikeura T, Yamaguchi T, Yoshida K, Matsuzaki K, Ishida M, et al. Role of phosphorylated Smad3 signal components in intraductal papillary mucinous neoplasm of pancreas. Hepatobiliary Pancreat Dis Int HBPD Int (2020) 19:581–9. doi: 10.1016/j.hbpd.2020.05.007
136. Wray CJ, Ahmad SA, Matthews JB, Lowy AM. Surgery for pancreatic cancer: Recent controversies and current practice. Gastroenterology (2005) 128:1626–41. doi: 10.1053/j.gastro.2005.03.035
137. Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a biomarker for pancreatic cancer–a comprehensive review. Indian J Surg Oncol (2011) 2:88–100. doi: 10.1007/s13193-011-0042-1
138. Winter JM, Yeo CJ, Brody JR. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J Surg Oncol (2013) 107:15–22. doi: 10.1002/jso.23192
139. Fujiyama Y, Kumamoto Y, Nishizawa N, Nakamoto S, Harada H, Yokota K, et al. Promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in intraductal papillary mucinous neoplasm (IPMN). Ann Surg Oncol (2020) 27:4007–16. doi: 10.1245/s10434-020-08291-2
140. Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila Pa) (2009) 2:807–13. doi: 10.1158/1940-6207.CAPR-09-0094
141. Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer (2012) 131:683–91. doi: 10.1002/ijc.26422
142. Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol (2014) 46:539–47. doi: 10.3892/ijo.2014.2743
143. Habbe N, Koorstra J-BM, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther (2009) 8:340–6. doi: 10.4161/cbt.8.4.7338
144. Chang X, Yu C, Li J, Yu S, Chen J. Hsa-miR-96 and hsa-miR-217 expression down-regulates with increasing dysplasia in pancreatic intraepithelial neoplasias and intraductal papillary mucinous neoplasms. Int J Med Sci (2017) 14:412–8. doi: 10.7150/ijms.18641
145. Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol (2010) 28:1069–78. doi: 10.1038/nbt.1678
146. Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood (2016) 127:42–52. doi: 10.1182/blood-2015-07-604512
147. Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discovery (2014) 13:337–56. doi: 10.1038/nrd4286
148. Falchook G, Rosen S, LoRusso P, Watts J, Gupta S, Coombs CC, et al. Development of 2 bromodomain and extraterminal inhibitors with distinct pharmacokinetic and pharmacodynamic profiles for the treatment of advanced malignancies. Clin Cancer Res Off J Am Assoc Cancer Res (2020) 26:1247–57. doi: 10.1158/1078-0432.CCR-18-4071
149. Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA Approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncol. (2007) 12:1247–52. doi: 10.1634/theoncologist.12-10-1247
150. Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res (2008) 68:9788–98. doi: 10.1158/0008-5472.CAN-08-2428
151. Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol Off J Am Soc Clin Oncol (2014) 32:4141–8. doi: 10.1200/JCO.2013.55.0376
152. Schultheis B, Strumberg D, Kuhlmann J, Wolf M, Link K, Seufferlein T, et al. Safety, efficacy and pharcacokinetics of targeted therapy with the liposomal RNA interference therapeutic Atu027 combined with gemcitabine in patients with pancreatic adenocarcinoma. A Randomized Phase Ib/IIa Study. Cancers (2020) 12:E3130. doi: 10.3390/cancers12113130
153. Kim M, Costello J. DNA Methylation: an epigenetic mark of cellular memory. Exp Mol Med (2017) 49:e322. doi: 10.1038/emm.2017.10
Keywords: pancreatic premalignancy, detection, therapy, epigenetics, pancreatic cancer
Citation: Zhang W, Jiang T and Xie K (2023) Epigenetic reprogramming in pancreatic premalignancy and clinical implications. Front. Oncol. 13:1024151. doi: 10.3389/fonc.2023.1024151
Received: 21 August 2022; Accepted: 01 February 2023;
Published: 16 February 2023.
Edited by:
Nilabja Sikdar, Indian Statistical Institute, Kolkata, IndiaReviewed by:
Marlise Guerrero Schimpf, University of Miami, United StatesAntonella De Cola, University of Cambridge, United Kingdom
Yunfeng Cheng, Fudan University, China
Copyright © 2023 Zhang, Jiang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keping Xie, c2N1dG1lZGljaW5lQHNjdXQuZWR1LmNu
 Wei Zhang
Wei Zhang Tingting Jiang1,2
Tingting Jiang1,2 Keping Xie
Keping Xie