- 1People’s Hospital of Henan University, Henan Provincial People’s Hospital, Microbiome Laboratory, Zhengzhou, Henan, China
- 2Intensive Care Unit, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China
- 3Department of Surgery of Spine and Spinal Cord, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, Zhengzhou, Henan, China
- 4Microbiome Laboratory, Henan Provincial People’s Hospital, Henan University People’s Hospital, Zhengzhou, China
- 5Department of Urology, Lanzhou University Second Hospital, Lanzhou, Gansu, China
- 6Department of Neurosurgery, Henan Provincial People’s Hospital, People’s Hospital of Henan University, People’s Hospital of Zhengzhou University, Zhengzhou, Henan, China
Nuclear pore complex (NPC) is a major transport pivot for nucleocytoplasmic molecule exchange. Nucleoporin 205 (NUP205)—a main component of NPC—plays a key regulatory role in tumor cell proliferation; however, few reports document its effect on the pathological progression of lower-grade glioma (LGG). Therefore, we conducted an integrated analysis using 906 samples from multiple public databases to explore the effects of NUP205 on the prognosis, clinicopathological characteristics, regulatory mechanism, and tumor immune microenvironment (TIME) formation in LGG. First, multiple methods consistently showed that the mRNA and protein expression levels of NUP205 were higher in LGG tumor tissue than in normal brain tissue. This increased expression was mainly noted in the higher WHO Grade, IDH-wild type, and 1p19q non-codeleted type. Second, various survival analysis methods showed that the highly expressed NUP205 was an independent risk indicator that led to reduced survival time of patients with LGG. Third, GSEA analysis showed that NUP205 regulated the pathological progress of LGG via the cell cycle, notch signaling pathway, and aminoacyl-tRNA biosynthesis. Ultimately, immune correlation analysis suggested that high NUP205 expression was positively correlated with the infiltration of multiple immune cells, particularly M2 macrophages, and was positively correlated with eight immune checkpoints, particularly PD-L1. Collectively, this study documented the pathogenicity of NUP205 in LGG for the first time, expanding our understanding of its molecular function. Furthermore, this study highlighted the potential value of NUP205 as a target of anti-LGG immunotherapy.
1 Introduction
Glioma, which originates from neural stem cells or progenitor cells, is the most common primary malignancy of the intracranial brain parenchyma (1). According to the World Health Organization (WHO) 2016 classification system, gliomas are generally divided into two types: WHO Grade I to III, considered as lower-grade gliomas (LGGs), and WHO Grade IV, considered as glioblastomas (GBMs) (2). LGG accounts for approximately 43.2% of all gliomas and has a median survival rate of 7 years (3, 4). However, due to the invasive growth characteristic of LGG cells, LGGs exhibit a high recurrence rate, and 70% of LGGs eventually progress to high-grade malignancies within 5–10 years (5). The current mainstay treatment for patients with LGG is surgical resection and adjunct radiotherapy combined with chemotherapy; however, their therapeutic effects do not meet ideal expectations (6). Fortunately, cancer research has expanded from solely studying the molecular event changes of tumor cells to studying the overall abnormality of the tumor immune microenvironment (TIME), allowing the advent of new immunotherapies that provide precise, individualized cancer treatment (7). However, current immunotherapeutic strategies are limited to only a subset of cancer patients, as only 20% of patients respond to therapy (8). Thus, finding more targets to mediate the immune response in the LGG TIME is crucial for prolonging the life of patients with LGG.
Abnormal export of pathogenic mRNAs from the nucleus increases the malignant biological behavior of cancer cells, which accelerates their malignant progression (9). The nuclear pore complex (NPC) is the major transport pivot for nucleocytoplasmic molecule exchange, including mRNAs. By controlling mRNA export from the nucleus, the NPC regulates vital activities of cells, such as cell growth, differentiation, apoptosis, and immune response (9–11). Recently, an increasing body of evidence showed that the NPC is closely related to tumorigenesis and regulates immune response in the tumor microenvironment (TME). For example, overexpression of NUP37 activates the PI3K/AKT/mTOR signaling pathway promoting proliferation, migration, and invasion of gastric cancer cells, and also gives rise to an immunosuppressive microenvironment in gliomas (12, 13). NUP205, as a core subunit of the NPC, is responsible for the gate control function and long-term maintenance of NPC, which is of great significance to the normal progress of cell life activities (14, 15). Thus, many scientists have studied the role of abnormal NUP205 in cancers and found that NUP205 is also closely related to tumorigenesis. For example, the high expression of NUP205 increases the proliferation ability of hepatocellular carcinoma cells, leading to poor prognosis in these patients (16, 17). The expression of NUP205 in nasopharyngeal carcinoma tumor tissue was significantly upregulated compared with that in adjacent tissue, which was an important factor leading to the proliferation of nasopharyngeal carcinoma cells (18). NUP205 has also been reported to be an oncogene in colorectal cancer, bladder cancer, and lung cancer (19–21). However, the role of NUP205 in the pathological process of LGG remains elusive, especially in the TIME of LGG. Therefore, this study, for the first time, revealed the relationship between NUP205 and the prognosis of LGG patients, as well as the TIME of LGG.
This study attempts to reveal the function of NUP205 in the malignant pathological progression of LGG through mutual validation from multiple databases. Our study is the first to document high NUP205 expression in LGG using public databases, and the results were confirmed by our basic experiments. We found that aberrantly expressed NUP205 had a profound impact on the prognosis of patients with LGG by data analysis of 903 cases from two datasets, which suggested that NUP205 could be a potential biomarker for LGG. At the same time, we explored the mechanism of the overexpression of NUP205 in LGG by documenting the relationship between NUP205 and its DNA methylation. Ultimately, we found that NUP205 might play an important role in the immunosuppressive microenvironment of LGG, which could provide a theoretical basis for the use of NUP205 as an immunotherapeutic target.
2 Materials and methods
2.1 Data collection and tissue preparation
In this study, Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/), an online bioinformatics analysis tool (22), was used to explore the mRNA expression of NUP205 in LGG. We collected data from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database to verify the results of the GEPIA database (GSE12657: 13 LGG tissues vs 5 control tissues; GSE21354: 10 LGG tissues vs 4 normal brain tissues; GSE70231: 24 LGG tissues vs 6 normal brain tissues) (23–25). In the database of The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) (26), we collected RNA sequencing data with the corresponding clinical information of 503 patients with LGG and the DNA methylation sequencing data of 511 patients with LGG, which were used to explore the effects of NUP205 on the prognosis, clinicopathological characteristics, regulatory mechanism, and TIME in LGG. The detailed patient information of the TCGA RNA-seq database is presented in Supplementary Table S1. Finally, we collected the RNA sequencing data of 403 LGG tissue samples in the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) database to verify the results obtained from the TCGA RNA-seq database (27). The detailed patient information of the CGGA RNA-seq database is presented in Supplementary Table S2.
Five brain tissue samples from epilepsy patients and five tumor tissue samples from patients with LGG were obtained from the operating room of Henan Provincial People’s Hospital and used to determine the change of NUP205 mRNA expression in LGG; the histopathology and IDH mutation status of these five glioma patients is shown in Supplementary Table S3. Then, three additional brain tissue samples from epilepsy patients and three tumor tissue samples from patients with LGG were used to detect the change in protein expression of NUP205, CD163, and CD274. Finally, we collected three glioma tissues of WHO Grade III and three glioma tissues of WHO Grade II and compared the protein expression of NUP205 between them. All participating patients provided informed consent, and the project was approved by the ethics committee of Henan Provincial People’s Hospital (Ethics approval number: 2020107).
2.2 Culture of human astrocyte cells and glioma cells
Human astrocyte (HA) and glioma cell lines (SHG44, T98, and LN229) were purchased from the Cell Bank of the Chinese Academy of Sciences and Qingqi Cell Bank (Shanghai, China), respectively. The cells in both groups were grown in Dulbecco’s Modified Eagle Medium culture (Procell, China) with 10% fetal bovine serum (Gibco, USA) and 1% penicillin (Procell, China) and incubated at 37°C and 5% carbon dioxide. After cells were grown to confluence in 60-mm dishes, total RNA was extracted by RT-qPCR to compare the expression levels of NUP205 in normal astrocytes and glioma cells. To demonstrate whether NUP205 mRNA expression in glioma cells is regulated by DNA methylation, we added 100 µM ademetionine disulfate tosylate (SAM) (Topscience, China) and treated them with SHG44, T98, and LN229 for 10 h. After culturing with SAM, glioma cells were used to identify changes in NUP205 expression after DNA hypermethylation.
2.3 Extraction of total RNA and RT-qPCR
According to the protocol of Total RNA Kit I (Omega, USA), we obtained total RNA from cells and tissue samples, and then determined total RNA concentration using a NanoDrop machine (Thermo, USA). Total RNA was reverse transcribed to synthesize cDNA according to the instructions of the NovoScript Plus All-in-one 1st Strand cDNA Synthesis SuperMix kit (Novoprotein, China). cDNA was then amplified by the StepOne Plus Real-Time PCR System (Thermo, USA) according to the instructions of NovoStart® SYBR qPCR SuperMix Plus (Novoprotein, China). The internal-reference gene, 18S, was selected to standardize the NUP205 expression. The specific primer sequences of 18S and NUP205 were as follows: 18S forward: 5’-GTAACCCGTTGAACCCCATT-3’ and 18S reverse: 5’-CCATCCAATCGGTAGTAGCG-3’. NUP205 forward: 5’-CATCACCCAGAAGGAGCAAG-3’ and NUP205 reverse: 5’-GGAGTCCCAGAATCACCACA-3’.
2.4 Immunohistochemistry
Immunohistochemistry (IHC) staining was performed following a standard procedure. After brain tissue samples were sliced into 5.0-µm paraffin sections, the sections were deparaffinized in xylene and dehydrated in graded concentrations of ethanol. After EDTA (ZSGB-BIO, China) antigen retrieval and quenching of endogenous peroxidase, 10% bovine serum albumin solution was used to block non-specific antigens for 1 h at 30 °C. These sections were incubated with 1:200 Anti-NUP205 (Proteintech, China), 1:250 Anti-CD163 (Invitrogen, USA), or 1:250 Anti-CD274 (Proteintech, China) overnight at 4 °C, respectively. Subsequently, the incubation of secondary antibodies and DAB color development were completed following the instructions of the Mouse/Rabbit enhanced polymer detection system (ZSGB-BIO, China). Finally, the stained area was photographed under a 200× microscope, and the results were calculated by ImagePro-Plus software (version 6.0).
2.5 Western blot
An appropriate amount of brain tissue was homogenized with RIPA lysate (EpiZyme, China) and a protease inhibitor (EpiZyme, China). The brain tissue was then split on ice for 30 min, and the protein supernatant was centrifuged at 12000 rpm for 30 min at 4 °C. The protein concentration was detected using a BCA kit (Biosharp, China). Briefly, the protein was boiled at 100 °C for 10 min in 4× loading buffer (Solarbio, China), separated by SDS-PAGE electrophoresis, and then transferred to a PVDF membrane (Bio-Rad, USA). After the membrane was sealed with 5% evaporated milk, NUP205 (1:1000; Proteintech, China) and β-actin (1:1000; Bioss, China) primary antibodies were added overnight at 4 °C. Then, the membrane was incubated in goat anti-rabbit IgG H&L antibody (1:2000; Bioss, China) at 37 °C for 1 h. Finally, the protein blots were developed with a chemiluminescence reagent kit (Beyotime Biotechnology), and ImagePro-Plus software (version 6.0) was used for the quantitative analysis.
2.6 Meta-analysis of NUP205 in LGG
In this study, a meta-analysis was used to explore the effect of NUP205 on the OS of patients with LGG. First, after searching PubMed databases, we found a few studies on the relationship between NUP205 and the OS of patients with LGG. Therefore, we had to assemble 924 samples from three datasets (TCGA RNA-seq database: 503 LGG samples, CGGA RNA-seq database: 403 LGG samples, and GSE43378: 18 LGG samples) to perform a meta-analysis. Subsequently, we adopted the Cox analysis model on the three datasets separately to calculate the hazard ratio (HR) of NUP205 on the prognosis of patients with LGG using R software (version 4.0.3). Meanwhile, the Q test (I2 statistics) was performed to evaluate the heterogeneity of the three datasets. Ultimately, we selected a random effect model according to I2 > 50% and p < 0.05 to calculate combined HR and a 95% confidence interval (95% CI).
2.7 Correlation analysis between NUP205 and TIME in LGG
Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a common platform for analyzing the relationship between immune cell infiltrates and multiple variables based on the TCGA samples (28). Therefore, we utilized the TIMER database to analyze the relationship between NUP205 and TIME in LGG. First, we explored the correlation between the expression of NUP205 and six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells). Second, Kaplan–Meier analysis was used to reveal the effects of NUP205 expression and the six immune infiltrates on the OS of patients with LGG. Finally, we analyzed the correlation between the types of somatic copy number alteration (SCNA) of NUP205 and the six immune infiltrates.
The TIMER database cannot analyze the relationship between target genes and immune cell subtypes. For this reason, we used data from the TCGA RNA-seq database to further understand the relationship between the expression of NUP205 and various immune cell subtypes. The “CIBERSORT” package on R software (version 4.0.3) was performed to calculate and visualize the effects of NUP205 expression on infiltrates of various immune-cell subtypes. Spearman analysis was used to reveal the correlation between NUP205 expression and markers of various immune-cell subtypes. Immune checkpoint therapy has now become a hotspot for antitumor therapy, and thus, we used Pearson analysis on R software (version 4.0.3) to calculate the correlation between NUP205 expression and the 8 famous immune checkpoints (CD274, PDCD1, HAVCR2, CD96, KLRB1, IDO1, CD276, LAG3) based on the TCGA database.
2.8 Gene set enrichment analysis of the high NUP205 phenotype in LGG
Gene set enrichment analysis (GSEA) is a common tool used to arrange the position of target genes into a preconstructed functional gene set to predict their function (29). Therefore, we used GSEA software (version 4.0) to predict the cell signaling pathways through which NUP205 regulates LGG. First, according to the median value of NUP205 expression level in LGG, we divided the data from the TCGA RNA-Seq database into high NUP205 phenotype and low NUP205 phenotype. Then, we permuted the high NUP205 phenotype 1000 times, and hundreds of cell signaling pathways emerged. Finally, according to nominal (NOM) p < 0.05 and false discovery rate (FDR) Q < 0.25, we selected the cell signaling pathways where a high NUP205 phenotype might influence LGG.
2.9 Statistical analysis
The results of RT-qPCR, IHC staining, and western blotting were calculated by unpaired t-tests on GraphPad Prism 9 (San Diego, CA, USA) software. The chi-squared test was used to compare NUP205 expression between normal brain and LGG tissue, as well as to analyze the relationship between the expression of NUP205 and several clinical features. Kaplan–Meier analysis was used to document the correlation between NUP205 expression and the OS of patients with LGG. Receiver operating characteristic (ROC) curves were used to calculate the diagnostic value of NUP205 in patients with LGG. Cox regression analysis was used to determine whether the high expression of NUP205 was a risk factor for patients with LGG. Pearson analysis was used to search for the co-expressed genes of NUP205. All results were derived by R software (version 4.0.3), and p < 0.05 was considered statistically significant.
3 Results
3.1 Significantly increased NUP205 in the transcriptome and proteome of LGG
The association between the clinical progression of cancer and the high expression of pathogenic genes prompted us to report the expression of NUP205 in patients with LGG. First, the results of both the GEPIA database and chi-squared test based on the GEO databases (GSE12657, GSE21354, GSE70231) showed that NUP205 mRNA expression was higher in LGG than in normal brain tissue (Figures 1A–D respectively). These results were subsequently verified with RT-qPCR analysis (Figures 1E, F). Since the protein serves as the last unit to perform certain biological functions, we performed IHC staining on NUP205 and showed significantly increased protein expression in the LGG tissue compared to the controls (Figure 1G). Taken together, these results show that NUP205 had abnormally high expression in LGG compared with the normal brain, implying that NUP205 might play an important role in the pathological progression of LGG.
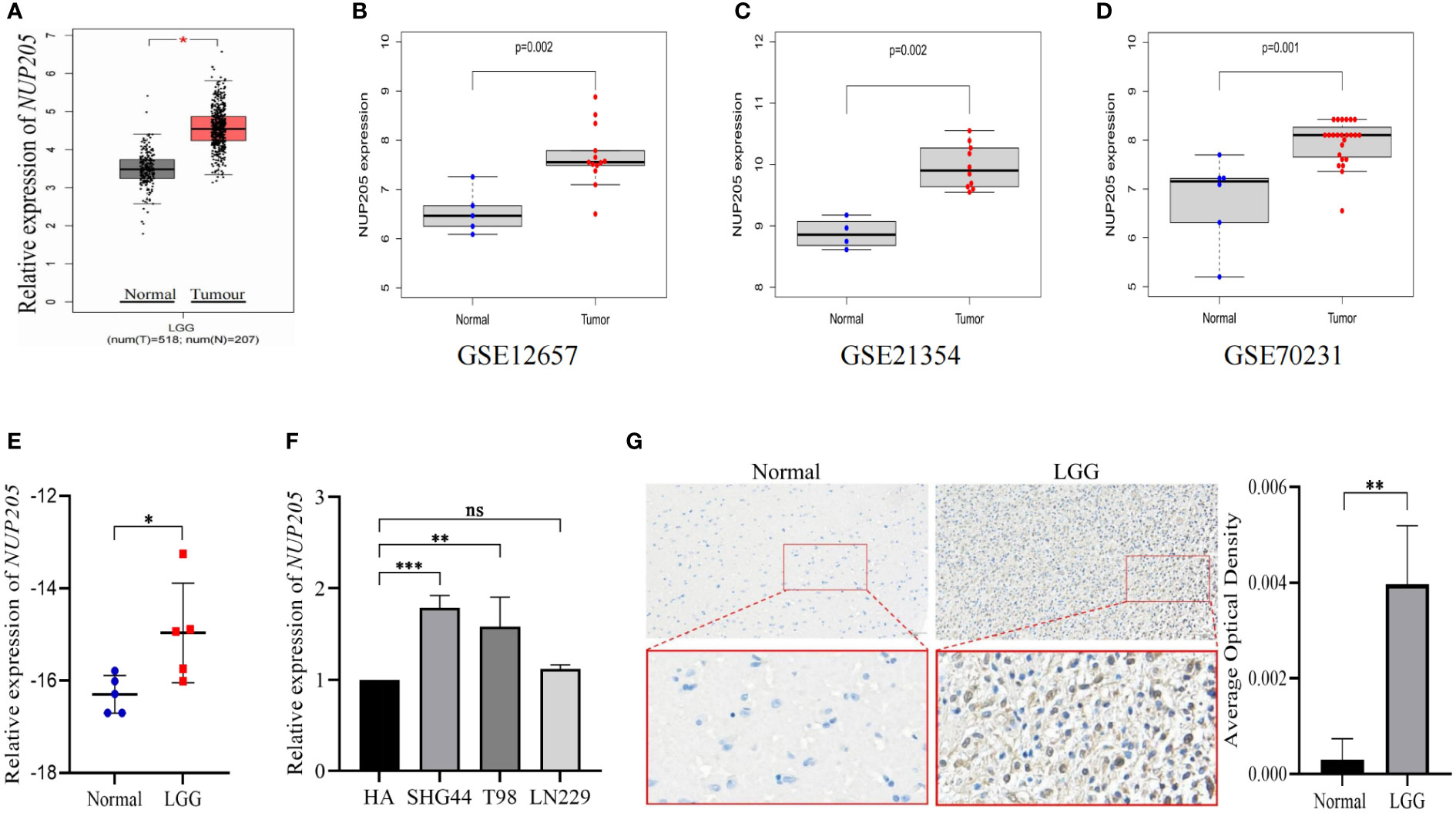
Figure 1 The mRNA and protein expression of NUP205 was increased in LGG. The results of (A) GEPIA, (B) GSE12657, (C) GSE21354, and (D) GSE70231 database showed that the mRNA expression of NUP205 increased in LGG tumor tissues. (E) The results of RT-qPCR showed that the mRNA expression of NUP205 was increased in LGG tissue. (F) The results of RT-qPCR showed that the mRNA expression of NUP205 was increased in glioma cells. (G) The results of IHC staining showed that the protein expression of NUP205 was increased in LGG tissue. ns, no statistically significant; *: p<0.05, **: p<0.01, ***: p<0.001. p<0.05 was considered statistically significant.
3.2 Relationship between NUP205 expression and clinical characteristics in LGG
It is well known that high pathogenic gene expression is often concomitant with malignant clinical characteristics in cancer. This highlights the importance of exploring the relationship between NUP205 expression and the clinical characteristics of patients with LGG. First, we found that the mRNA expression of NUP205 increased with WHO Grade based on the TCGA RNA-seq and CGGA RNA-seq databases (Figure 2A). In addition, the results of western blotting showed that the protein expression of NUP205 in the WHO Grade III tumor tissue was significantly higher than that of WHO Grade II tissue (Figure 2B). Second, based on the TCGA and CGGA databases, we found that the expression of NUP205 in chemotherapy-type patients was significantly higher than that of non-chemotherapy types (Figure 2C). Third, analysis of the histological types of LGG from the TCGA RNA-seq database showed that NUP205 expression is highest in anaplastic astrocytoma (AA) and lowest in astrocytoma (A) (Figure 2D). Fourth, we found that NUP205 expression was higher in the radiotherapy type and IDH-wild type LGG compared to the non-radiotherapy type and IDH-mutated type based on the TCGA RNA-seq database (Figures 2E, F). Finally, we found that the expression of NUP205 was higher in the 1p19q non-codeleted type than in the 1p19q codeleted type LGG based on the CGGA RNA-seq database (Figure 2G). In short, the above results suggested that the high expression of NUP205 always appeared in the malignant clinical subtypes of LGG, implying its effect on poor prognosis in patients with LGG.
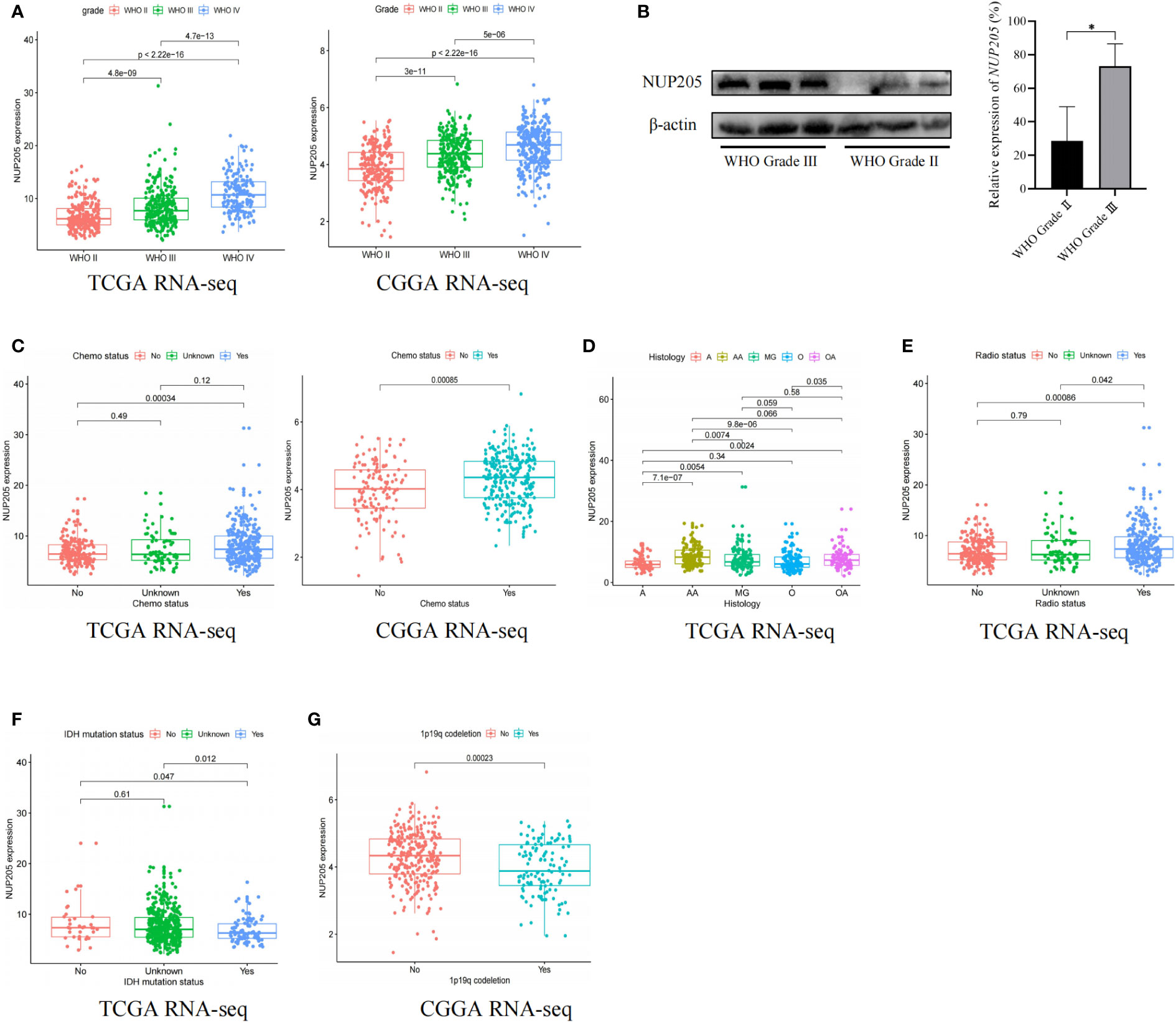
Figure 2 The relationship between the expression of NUP205 and clinical features in LGG. (A) WHO grade, (B) Western blot results showed that the protein expression of NUP205 was significantly higher in WHO grade III than in WHO grade II. (C) Chemotherapy status, (D) Histology, (E) Radiotherapy status, (F) IDH-mutation status, (G) 1p19q codeletion status. *p<0.05. p<0.05 was considered statistically significant.
3.3 High NUP205 expression led to poor prognosis in patients with LGG and was an independent risk factor for LGG
To find more evidence to support the correlation between NUP205 and the prognosis of patients, we conducted Kaplan–Meier analysis, ROC curves analysis, univariate and multivariate analysis, and meta-analysis based on the TCGA and CGGA databases. First, Kaplan–Meier analysis showed that LGG patients of WHO Grades II & III with high NUP205 expression had shorter OS (Figure 3A and Supplemental Figure S1A). For the LGG patients of WHO Grade II, the Kaplan–Meier analysis had similar results in the CGGA database (Supplemental Figure S1B), while there was no statistical significance in the TCGA database (Figure 3B). LGG patients of WHO Grade III with high NUP205 expression were found to have significantly lower OS than those with low expression (Figure 3C and Supplemental Figure S1C). These results suggest that high NUP205 expression might lead to a poor prognosis in LGG patients, whether they are of WHO Grade II or III. Subsequently, to explore the diagnosis value of NUP205 for patients with LGG, we constructed ROC curves and found that NUP205 can be used as a biomarker for LGG diagnosis (Figure 3D and Supplemental Figure S1D). In addition, the results of univariate and multivariate analysis showed that high NUP205 expression was HR > 1 (p < 0.05) in LGG, which suggests its role as an independent risk factor for patients with LGG (Figures 3E, F, and Supplemental Figures S1E, F). Finally, the results of the meta-analysis showed that the HR and 95% CI were 1.41 and 0.94–2.10, respectively (Figure 3G). In summary, all evidence shows that NUP205 is a pathogenetic gene for LGG and leads to a poor prognosis in patients with LGG.
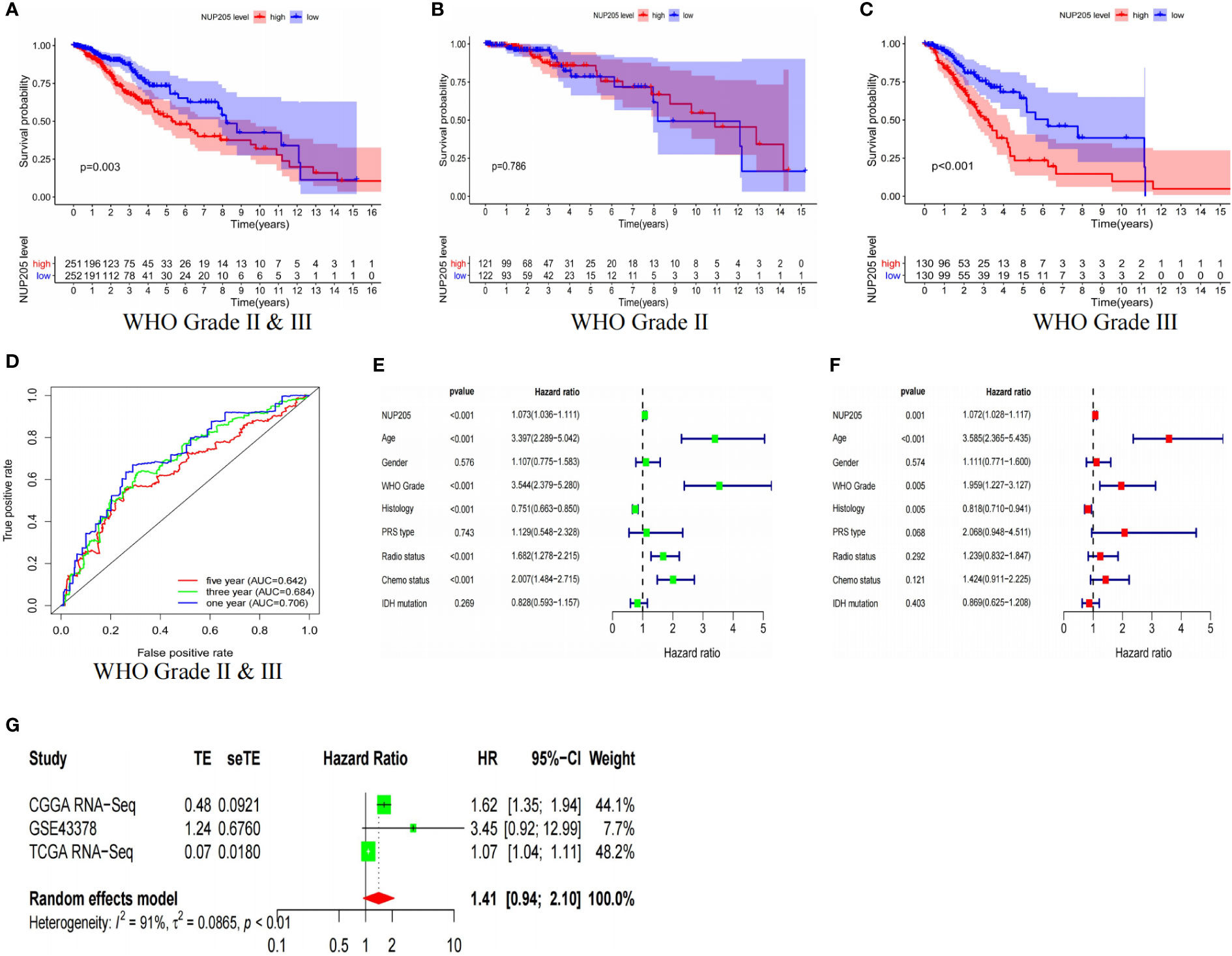
Figure 3 The results based on TCGA RNA-seq database showed that the high expression of NUP205 leads to poor prognosis of LGG patients. The result of (A) Kaplan-Meier analysis for LGG patients of WHO Grade II & III, (B) Kaplan-Meier analysis for LGG patients of WHO Grade II, (C) Kaplan-Meier analysis for LGG patients of WHO Grade III, (D) ROC curve, (E) Univariate analysis, (F) Multivariate analysis. (G) Meta analysis. p<0.05 was considered statistically significant.
3.4 DNA methylation of NUP205 was negatively correlated with its mRNA expression and influenced the prognosis of patients with LGG
Previous studies reported that DNA methylation could negatively regulate the expression of pathogenic genes in LGG (30), therefore, we tried to reveal the mechanism through which DNA methylation regulates NUP205 expression. First, using the TCGA database to extract DNA methylation data, we screened ten methylation sites that may regulate NUP205 expression (Figure 4A). Subsequently, Kaplan–Meier analysis was used to analyze whether these methylation sites were related to the prognosis of LGG patients. We found that LGG patients with hypermethylation of cg25119219 had longer OS than those with hypomethylation of cg25119219 (Figure 4B). Finally, SAM, a drug used to promote DNA methylation, was cultured with glioma cells (SHG44, T98, and LN229). The RT-qPCR results showed that NUP205 expression significantly decreased in glioma cells cultured with SAM (Figure 4C). Therefore, we speculated that the abnormally high expression of NUP205 might result from its DNA demethylation, and cg25119219 could be used as a biomarker for LGG patients.
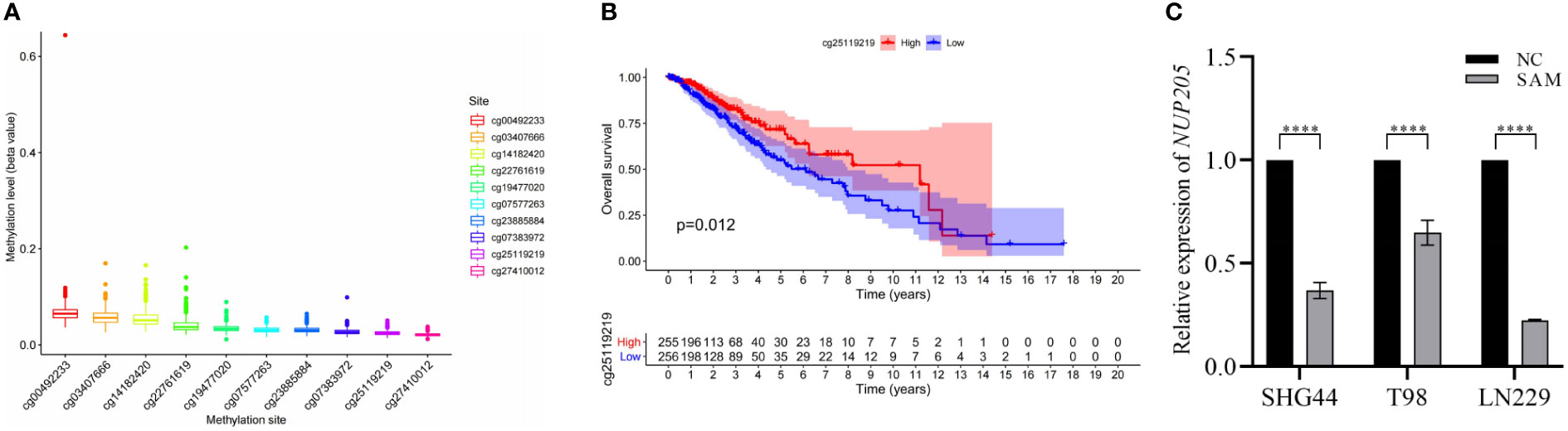
Figure 4 The relationship between the mRNA expression of NUP205 and its DNA methylation in LGG. (A) DNA methylation sites of NUP205. (B) The result of Kaplan-Meier analysis showed that hypermethylation of cg25119219 led to reduce the overall survival of LGG patients. (C) The result of RT-qPCR showed that the expression of NUP205 was decreased in glioma cells treated with SAM. ns, no statistically significant; ****: p<0.0001. p<0.05 was considered statistically significant.
3.5 Co-expression analysis of NUP205 and GSEA analysis of NUP205 in LGG
The co-expression of pathogenic genes may exert similar functions to synergistically regulate cancer progression (31). Therefore, to better understand the role that NUP205 plays in the pathological progression of LGG, we performed co-expression analyses of NUP205 based on the TCGA database. We picked out the five most positively correlated genes (CASP2, NCAPG2, PAXIP1, BAZ1B, ZNF800) and the five most negatively correlated genes (MAPK3, AVPI1, HDAC11, FKBP8, ALDH2) (Figures 5A, B). By reviewing previous studies, we found that CASP2, NCAPG2, BAZ1B, and ZNF800, the genes most positively related to NUP205, were reported as carcinogenic genes in cancer, especially CASP2, NCAPG2, and BAZ1B, which were found to be carcinogenic genes in glioma (32–35). However, the genes most negatively associated with NUP205 were reported to be tumor suppressor genes, such as AVPI1, HDAC11, FKBP8, and ALDH2 (36–39). Therefore, the results of the co-expression analyses also supported our view that NUP205 is a pathogenic gene of LGG.
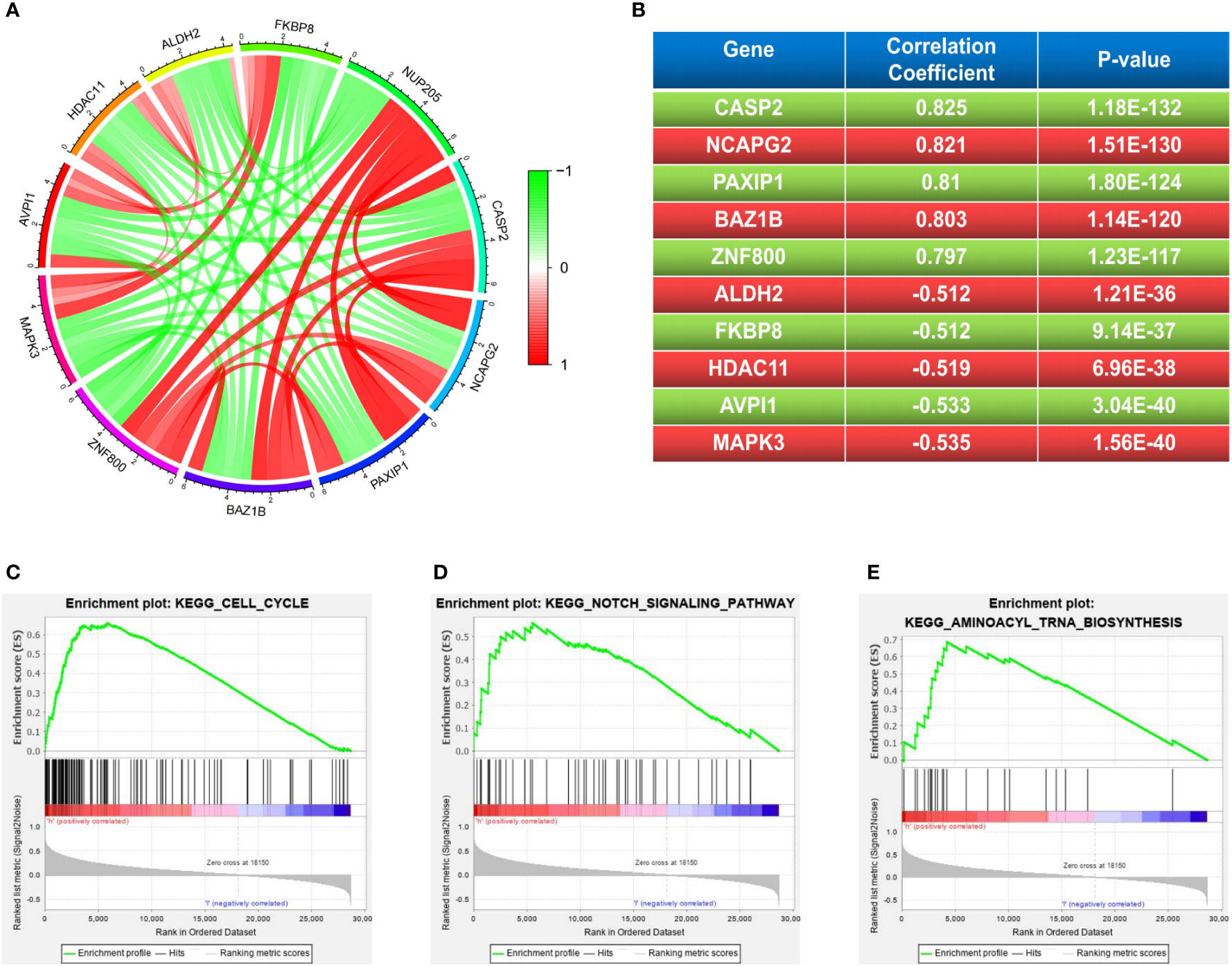
Figure 5 (A, B) The 5 most positive and negative genes related to NUP205 in LGG. The results of GSEA analysis suggested that NUP205 participated in the pathological process of LGG via (C) cell cycle, (D) notch signaling pathway, (E) aminoacyl-tRNA biosynthesis. p<0.05 was considered statistically significant.
To better understand the molecular mechanism of NUP205 in the pathological progression of LGG, we used GSEA analysis to explore the cell signaling pathways that NUP205 might act through in LGG. Our results showed that high NUP205 expression was significantly enriched in the cell cycle, notch signaling pathway, and aminoacyl-tRNA biosynthesis (Figures 5C–E and Supplementary Table S4). Previous studies have documented the effect of the aforementioned pathways in LGG progression, such as malignant proliferation of tumor cells and immune evasion (40, 41). Therefore, we speculated that the high expression of NUP205 might promote the malignant biological behavior of LGG and induce the formation of the LGG immunosuppressive microenvironment through these pathways.
3.6 Association of NUP205 with immune cell infiltration and immune checkpoints in LGG
Previous studies have shown that pathogenic genes can shape the immunosuppressive microenvironment of cancer (42). Also, the results of GSEA suggested that NUP205 might promote the immunosuppressive microenvironment of LGG. Thus, these hints inspired us to explore the role of NUP205 in the TIME of LGG.
The TIMER database was used to explore the relationship between NUP205 expression and immune infiltration in LGG, and we found that NUP205 expression positively correlated with infiltration of six types of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) (Figure 6A). Subsequently, the Kaplan–Meier analysis showed that both high infiltration of the six immune cells and high expression of NUP205 led to a shorter OS in patients with LGG (Figure 6B). Finally, we found that arm-level gain and arm-level deletion of NUP205 in LGG led to higher infiltration levels of 5 immune cells (B cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) (Figure S2A). Taken together, the above results suggest that high NUP205 expression is positively correlated with the infiltration of immune cells, thereby leading to poor prognosis in LGG patients.
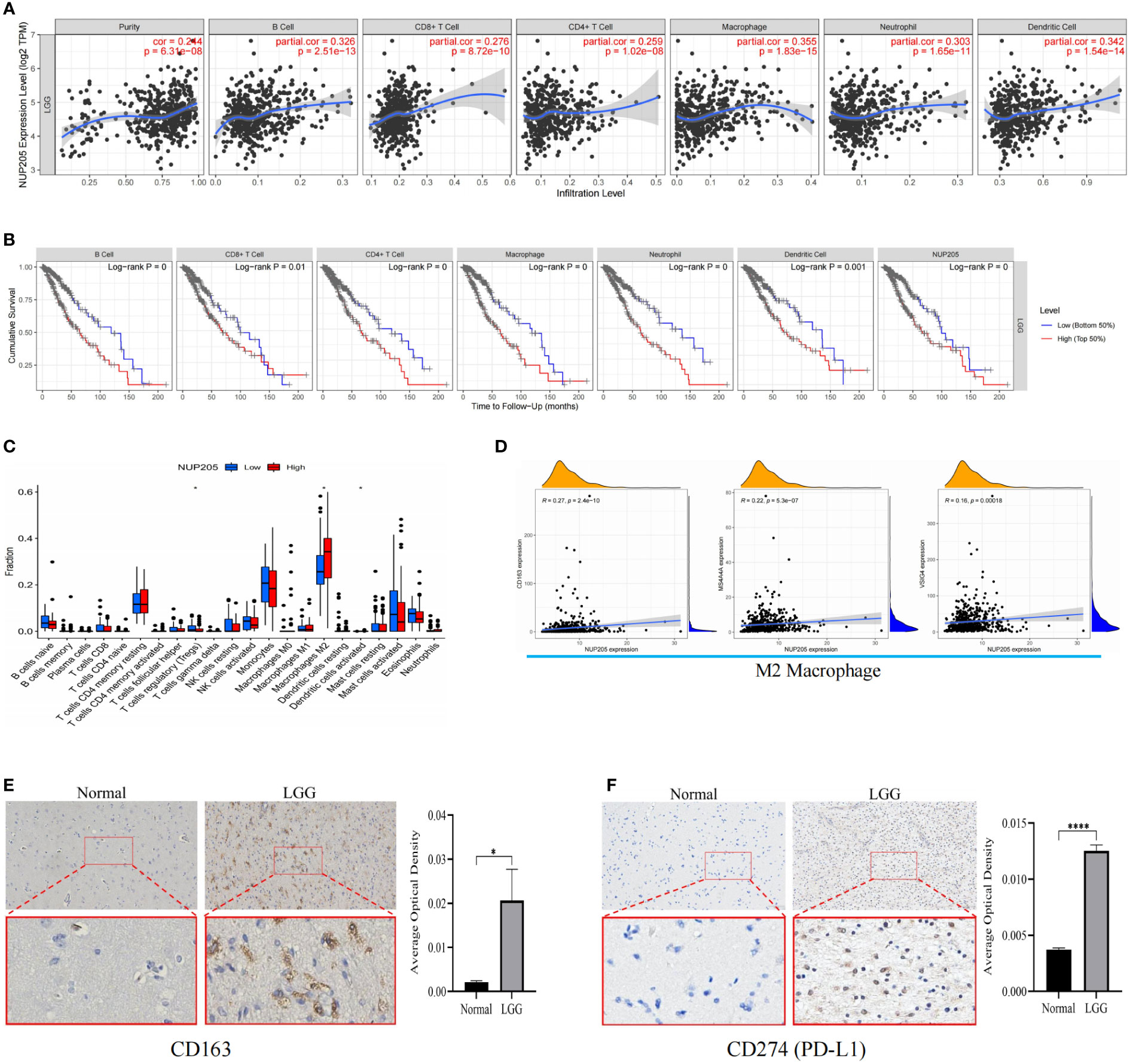
Figure 6 (A) The results of TIMER database showed that NUP205 expression was positively correlated with six immune infiltrations (B cell, CD4 + T cell, CD8 + T cell, neutrophil, macrophage, and dendritic cell). (B) The results of Kaplan-Meier analysis showed that the high level of six immune infiltration and high expression of NUP205 lead to poor prognosis of LGG patients. (C) The results of “CIBERSTART” analysis. (D) The results of Pearson analysis showed that NUP205 was positively correlated with the markers of M2 macrophages (CD163, VSIG4, MS4A4A). (E) The IHC staining of CD163 showed that the protein expression of CD163 in LGG tumor tissue was higher than that in control brain tissue. (F) The IHC staining of PD-L1 showed that the protein expression of PD-L1 in LGG tumor tissue was higher than that in control brain tissue. ns, no statistically significant; *: p<0.05, ****: p<0.0001. p<0.05 was considered statistically significant.
Since the TIMER database lacks analysis between the target gene and the immune cell subtypes, we used the “CIBERSTART” analysis to explore the relationship between NUP205 expression and the infiltration levels of 22 immune cell subtypes based on the TCGA database. We found that the NUP205 expression in multiple immune cell subtypes was markedly increased in LGG, particularly M2 macrophages (Figure 6C). Surprisingly, the correlation analysis also showed that NUP205 expression was positively correlated with markers of M2 macrophages (CD163, VSIG4, MS4A4A) and negatively correlated with those of M1 macrophages (Figure 6D and Table 1). Finally, we performed IHC staining for CD163 (a M2 macrophage marker) and found that CD163 expression, like NUP205, was significantly increased in LGG compared to control brain tissue (Figure 6E). In summary, these results show that the expression of NUP205 is positively correlated with M2 macrophages, suggesting that NUP205 might be related to the infiltration of M2 macrophages in the TIME of LGG.
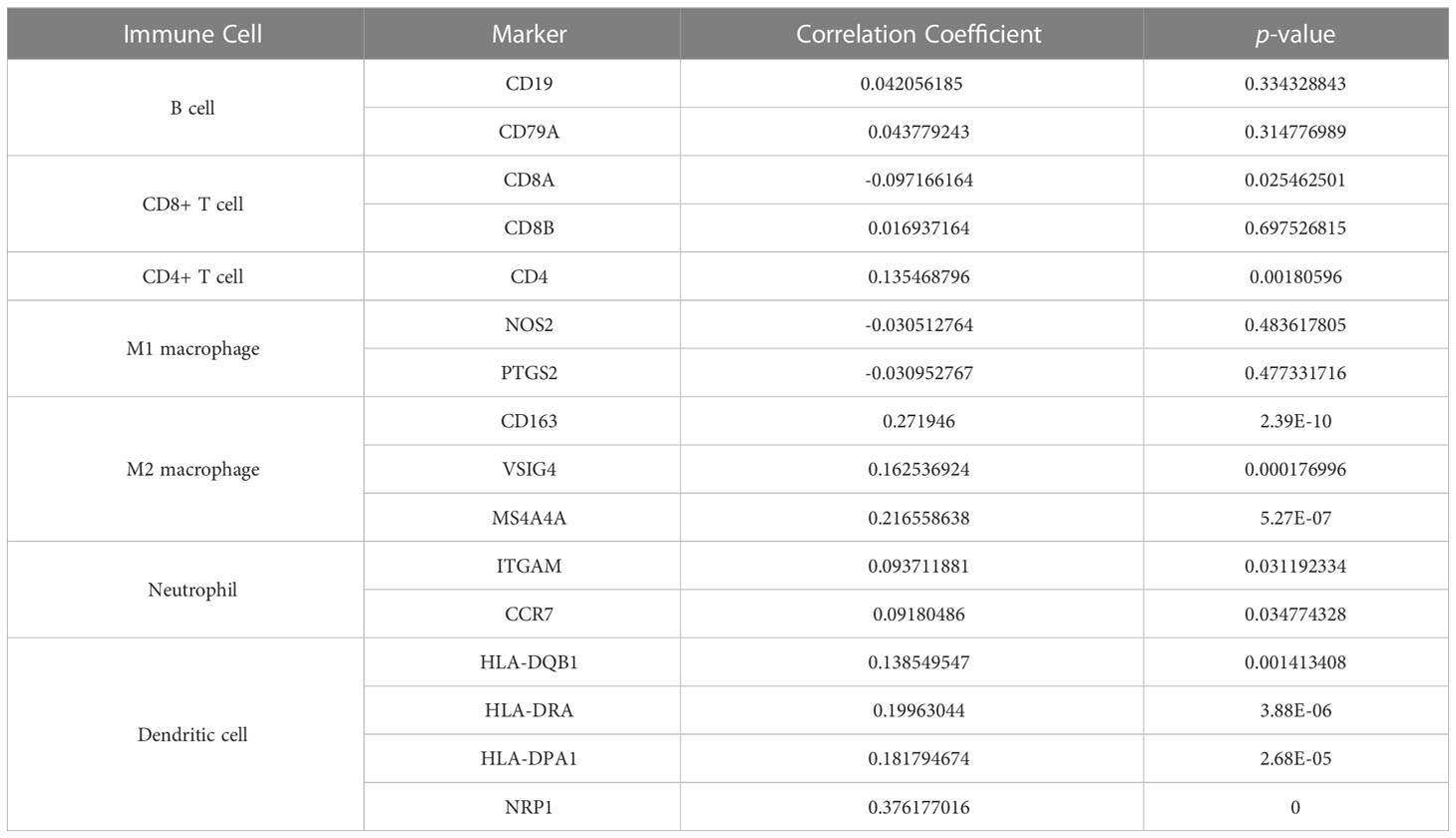
Table 1 Results of correlation analysis between NUP205 and multiple immune-cell markers based on TCGA database.
Immune checkpoint treatment has become a hotspot in cancer treatment. Therefore, we explored the relationship between NUP205 and 8 famous immune checkpoints in LGG by Spearman analysis based on the TCGA database. We found that NUP205 expression positively correlated with the eight immune checkpoints (CD274, PDCD1, HAVCR2, CD96, KLRB1, IDO1, CD276, LAG3) (Table 2). Surprisingly, CD274 (PD-L1), the most famous immune checkpoint marker, expression was significantly increased in LGG, similar to that of NUP205 (Figure 6F). In conclusion, NUP205 might be a potential target for LGG immunotherapy because it is positively correlated with the expression of various immune checkpoints, especially PD-L1.
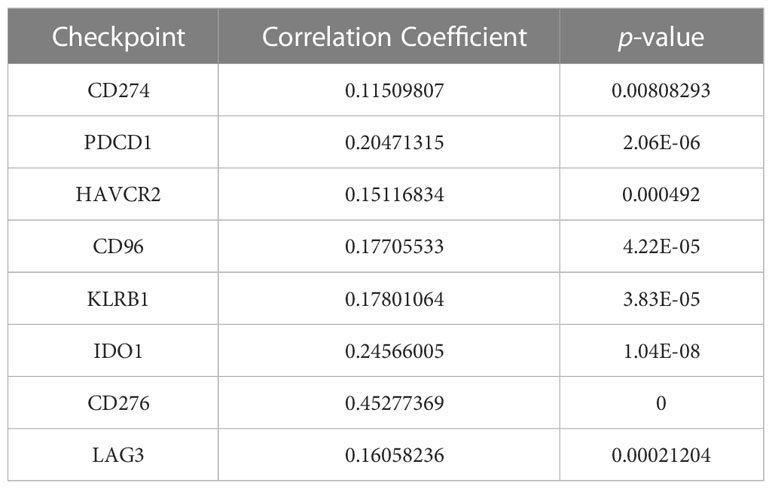
Table 2 Results of correlation between NUP205 and multiple immune-checkpoints based on TCGA database.
4 Discussion
NPC can not only regulate transport of nucleocytoplasmic molecules, but can also control DNA damage repair and chromatin translocation and silencing, making it relevant to the pathological progression of cancers (43). Since NUP205 preserves the integrity of NPC, recent studies have focused on revealing its role in various malignancies. For example, LncRNA HOTAIR can upregulate the expression of NUP205 to increase the growth, migration, and invasion of papillary thyroid carcinoma cells via absorbing the role of miR-488-5p (44). The overexpression of NUP205 can accelerate the cell cycle, increasing the proliferation of acute myoid leukemia cells (45). Unfortunately, few reports on the role of NUP205 in the malignant process of LGG exist. Existing studies have all reported that NUP205 is an oncogene in several cancers, leading us to believe that NUP205 may also be a pathogenic gene in LGG.
In this study, we attempted to reveal the influence of NUP205 expression on the pathological progression of LGG. First, we found that the mRNA expression of NUP205 was significantly increased in LGG tissue using the GEPIA and GEO databases. Our own experimental data verified that both mRNA and protein expression of NUP205 are increased. These different perspective analyses made our findings more credible. Previous studies have shown that the oncogene is present in a high expression state and promotes the pathological progression of LGG. For example, the oncogene METTL21B is highly expressed in LGG tissue and leads to a poor prognosis in patients (46). These findings are highly consistent with our own findings, thereby further proving that NUP205 is a pathogenic gene in LGG.
To solidify our hypothesis that NUP205 is in fact a pathogenic gene in the pathological process of LGG, we performed a correlation analysis between NUP205 and the clinical characteristics of patients with LGG. First, we found that the mRNA and protein expression of NUP205 in WHO Grade III gliomas was significantly higher than that in WHO Grade II gliomas, and that the prognosis of glioma patients worsens with the improvement of WHO Grade (47). Second, we found that NUP205 expression was higher in chemotherapy type, IDH wild type, and 1p19q no-codeleted type of LGG, and these clinical features are associated with a worse prognosis (48). After the above discussion, we speculated that highly expressed NUP205 was closely related to the poor prognosis of patients with LGG. To further support our hypothesis, the Kaplan–Meier analysis showed that patients with LGG with high NUP205 expression had a poor prognosis, and its increased expression can promote malignant progression of LGG. Previous studies reported similar findings. For example, high NUP205 expression leads to poor prognosis in patients with hepatocellular carcinoma (17). In addition, Cox regression and meta-analysis further confirmed that NUP205 is an independent risk factor for LGG, and an indicator of poor prognosis in patients with LGG, respectively. Together, these results demonstrate that NUP205 is indeed a pathogenic gene for LGG and, as such, may serve as a biomarker for predicting the prognosis of patients with LGG and a potential therapeutic target for the treatment of LGG.
Next, we aimed to explore the reason behind the increased expression of NUP205 in LGG. The changes in aberrant DNA methylation status often occur during the pathological progression of LGG, and as such, DNA hypomethylation of the target gene can significantly increase its expression level (49). Therefore, we sought to reveal the reason behind the high NUP205 expression in LGG from the perspective of DNA methylation. We found that NUP205 expression decreased in glioma cells after treatment with promethylating drugs compared to untreated LGG cells. The expression of NUP205 in these glioma cells was higher than that in normal human astrocytes. This illustrated that NUP205 expression was negatively regulated by DNA methylation. Previous studies have reported that the methylation sites of pathogenic genes, such as DNA hypomethylation of EMILIN2 (50), could serve as markers for predicting the prognosis of patients with LGG. Thus, in this study, we screened ten methylation sites that could affect NUP205 expression, and by using the Kaplan–Meier analysis method, and we found that the hypomethylation of cg125119219 was closely related to poor prognosis in LGG patients. In short, we found that NUP205 expression might be negatively regulated by its DNA methylation level in LGG, and that cg125119219 could be used as a biomarker for predicting prognosis in patients with LGG.
Co-expression analysis can be applied to determine the function of target genes by looking up their co-expressed genes (51). Our results suggest that many genes that positively correlate with NUP205 play a role in the carcinogenesis of glioma. For instance, overexpression of NCAPG2 can regulate the activation of Wnt/β-catenin pathway to promote proliferation, migration, and invasion of GBM cells, and knockdown of NCAPG2 inhibited tumorigenesis of GBM in vivo (33). Also, the high expression of BAZ1B can also increase proliferation, migration, invasion, and inhibition of apoptosis in GBM cells (34). In our study, the NCAPG2 and BAZ1B genes were positively related to NUP205 and have been reported as oncogenes for glioma. In addition, HDAC11, which was negatively correlated with the expression of NUP205, was found to have low expression in glioma tissue and was associated with a better prognosis in glioma patients (37). Interestingly, AVPI1, FKBP8, and ALDH2, which were most negatively correlated with NUP205 in LGG, were all reported to be tumor suppressor genes in cancer (36, 38, 39). This discovery further strengthens our previous view that NUP205 is a pathogenic gene in LGG.
Exploring the molecular mechanism of NUP205 in LGG will improve our understanding of the malignant progression of the disease. As such, we performed a GSEA analysis and found that NUP205 influenced LGG progression via the cell cycle, notch signaling pathway, and aminoacyl-tRNA biosynthesis. Many studies have reported that activation of these cell signaling pathways is associated with the malignant progression of cancers. For example, knockdown of MRPL42 increased the G1 and G2/M phases and decreased the S phase in the cell cycle of glioma cells, suggesting its role in accelerating the malignant advance of gliomas (52). In addition, aminoacyl-tRNA biosynthesis is upregulated in gastric cancer, leading to a poor prognosis seen in patients (53). Interestingly, the activation of the notch signaling pathway is not only able to maintain glioma cells in an aggressive and proliferative state, but is also closely associated with the formation of the TIME of multiple cancers (54, 55). Briefly, NUP205 may play an important role in LGG malignant progression, including the growth, invasion, and formation of the immunosuppressive microenvironment.
Crosstalk between glioma and immune cells in the TME drives immune cells to reprogram, thereby promoting the transition of TIME to an immunosuppressive microenvironment. Ultimately, cancer cells escape immune cell surveillance, evading immune cell-mediated destruction, and thereby leading to poor prognosis in glioma patients (56, 57). Previous studies have reported that pathogenic genes can participate in regulating the above process (58). Our study showed that NUP205 acts as a pathogenic gene and may be involved in regulating the immunosuppressive microenvironment of LGG. To prove this, we found that NUP205 expression positively correlated with the level of six immune-cell infiltrations in LGG TIME. Previous studies have also shown that higher levels of immune cell infiltration in cancer led to worse prognosis in patients (59), which strongly supports our finding that the high level of six immune cell infiltrations led to a short overall survival of patients with LGG. Thus, we speculated that high NUP205 expression may be involved in the formation of the LGG TIME, which may be one of the important reasons leading to poor prognosis in LGG patients. More importantly, we also found that the expression of NUP205 was positively correlated with the infiltration of M2 macrophages and negatively correlated with the infiltration of M1 macrophages. Two types of polarization of macrophages exist: classically activated macrophages (M1 macrophages) and alternatively activated macrophages (M2 macrophages) (60). Previous studies reported that tumor-associated macrophages (TAMs) are the most commonly infiltrating immune cells in glioma, and that polarization of TAMs to M2 macrophages led to a poorer prognosis in glioma patients (61). Thus, this finding further strengthened our view that NUP205 may play an important role in the formation of the TIME in LGG.
The emergence of immunotherapy has provided a new tool for cancer treatment, causing more scientists and researchers to focus on immune-targeted therapy (62). For example, therapeutic anti-CTLA-4 monoclonal antibody drugs have shown remarkable clinical efficacy in treating melanoma (63). In addition, anti-PD-L1 antibody therapy showed good clinical efficacy and superior tolerance in urothelial carcinoma and non-small cell lung cancer (64). Therefore, in the present study, we sought to explore the expression relationship between NUP205 and multiple immune checkpoint genes and found a positive correlation between NUP205 and 8 well-known immune-checkpoint genes (CD274, PDCD1, HAVCR2, CD96, KLRB1, IDO1, CD276, LAG3). We also found that protein expression of NUP205 and PD-L1 (CD274) was indeed positively correlated by IHC staining. Based on the findings that NUP205 was positively correlated with multiple immune checkpoints in LGG, and that NUP205 might participate in the formation of the TIME in LGG, we can infer that NUP205 is a potential immunotherapy target for LGG.
5 Conclusions
This study was the first to identify highly expressed NUP205 as a pathogenic gene and to document its role in the malignant progression and poor prognosis of LGG. More importantly, we found that highly expressed NUP205 might participate in the formation of the TIME in LGG, leading to a poor prognosis in LGG patients. In summary, this study not only broadened our general understanding of the molecular function of NUP205 but also proved it as an immunotherapeutic target that may improve the prognosis of patients with LGG.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by the ethics committee of Henan Provincial People’s Hospital (Ethics approval number: 2020107). The patients/participants provided their written informed consent to participate in this study.
Author contributions
WL, CH, and QZ contributed equally to this work. YG and RQ designed the research. WL and CH revised and wrote the manuscript. QZ and XC performed the experiment. ZL, HW, and PL collected clinical samples. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Tackling key problems of science and technology in Henan Province (Grant ID: 222102310002), Henan Postdoctoral Fund (Grant ID: 2021), The Medical Science and technology research plan in Henan Province (Grant ID: LHGJ20210013), and the Project of Action for Postgraduate Training Innovation and Quality Improvement of Henan University (Grant ID: SYLYC2022174). The funding source was not involved in collecting, analyzing and interpreting the data.
Acknowledgments
We appreciate the support of Henan Provincial People’s Hospital.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1007198/full#supplementary-material
Supplementary Figure 1 | The results based on CGGA RNA-seq database showed that the high expression of NUP205 leads to poor prognosis of LGG patients. The results of (A) Kaplan-Meier analysis for LGG patients of WHO Grade II & III, (B) Kaplan-Meier analysis for LGG patients of WHO Grade II, (C) Kaplan-Meier analysis for LGG patients of WHO Grade III, (D) ROC curve, (E) Univariate analysis, (F) Multivariate analysis. p<0.05 was considered statistically significant.
Supplementary Figure 2 | (A) In LGG, arm-level gain and arm-level deletion of NUP205 led to higher infiltration levels of 5 immune cells (B) cell, CD8 + T cell, neutrophil, macrophage, and dendritic cell).
References
1. Zhang L, Liu Z, Li J, Huang T, Wang Y, Chang L, et al. Genomic analysis of primary and recurrent gliomas reveals clinical outcome related molecular features. Sci Rep (2019) 9:16058. doi: 10.1038/s41598-019-52515-9
2. Louis D, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee W, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol (2016) 131:803–20. doi: 10.1007/s00401-016-1545-1
3. Tian W, Yan G, Chen K, Han X, Zhang W, Sun L, et al. Development and validation of a novel prognostic model for lower-grade glioma based on enhancer RNA-regulated prognostic genes. Front Oncol (2022) 12:714338. doi: 10.3389/fonc.2022.714338
4. Naidoo M, Jones L, Conboy B, Hamarneh W, D'Souza D, Anthony K, et al. Duchenne muscular dystrophy gene expression is an independent prognostic marker for IDH mutant low-grade glioma. Sci Rep (2022) 12:3200. doi: 10.1038/s41598-022-07223-2
5. Dixit K, Raizer J. Newer strategies for the management of low-grade gliomas. Oncology (2017) 31:680–2.
6. Nicholson J, Fine H. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discovery (2021) 11:575–90. doi: 10.1158/2159-8290.Cd-20-1474
7. Friedrich M, Bunse L, Wick W, Platten M. Perspectives of immunotherapy in isocitrate dehydrogenase-mutant gliomas. Curr Opin Oncol (2018) 30:368–74. doi: 10.1097/cco.0000000000000478
8. Rameshbabu S, Labadie B, Argulian A, Patnaik A. Targeting innate immunity in cancer therapy. Vaccines (2021) 9:138. doi: 10.3390/vaccines9020138
9. Borden K. The nuclear pore complex and mRNA export in cancer. Cancers (2020) 13:42. doi: 10.3390/cancers13010042
10. Bonnet A, Palancade B. Regulation of mRNA trafficking by nuclear pore complexes. Genes (2014) 5:767–91. doi: 10.3390/genes5030767
11. Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol (2017) 18:73–89. doi: 10.1038/nrm.2016.147
12. Zhang J, Lv W, Liu Y, Fu W, Chen B, Ma Q, et al. Nucleoporin 37 promotes the cell proliferation, migration, and invasion of gastric cancer through activating the PI3K/AKT/mTOR signaling pathway. In Vitro Cell Dev Biol Anim (2021) 57:987–97. doi: 10.1007/s11626-021-00627-w
13. He Y, Li J, Shen L, Zhou H, Fei W, Zhang G, et al. Pan-cancer analysis reveals NUP37 as a prognostic biomarker correlated with the immunosuppressive microenvironment in glioma. Aging (2022) 14:1033–47. doi: 10.18632/aging.203862
14. Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol (2010) 189:1129–42. doi: 10.1083/jcb.200912045
15. Krull S, Thyberg J, Björkroth B, Rackwitz H, Cordes V. Nucleoporins as components of the nuclear pore complex core structure and tpr as the architectural element of the nuclear basket. Mol Biol Cell (2004) 15:4261–77. doi: 10.1091/mbc.e04-03-0165
16. Deng Z, Huang K, Liu D, Luo N, Liu T, Han L, et al. Key candidate prognostic biomarkers correlated with immune infiltration in hepatocellular carcinoma. J Hepatocell Carcinoma (2021) 8:1607–22. doi: 10.2147/jhc.S337067
17. Xiong D, Feng Z, Lai Z, Qin Y, Liu L, Fu H, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis (2019) 10:658. doi: 10.1038/s41419-019-1890-9
18. Yue W, Wang Y, Li W, Wang Z. LINC00887 regulates the proliferation of nasopharyngeal carcinoma via targeting miRNA-203b-3p to upregulate NUP205. Eur Rev Med Pharmacol Sci (2020) 24:8863–70. doi: 10.26355/eurrev_202009_22826
19. Ji L, Fu J, Hao J, Ji Y, Wang H, Wang Z, et al. Proteomics analysis of tissue small extracellular vesicles reveals protein panels for the reoccurrence prediction of colorectal cancer. J Proteomics (2021) 249:104347. doi: 10.1016/j.jprot.2021.104347
20. Zhang C, Gou X, He W, Yang H, Yin H. A glycolysis-based 4-mRNA signature correlates with the prognosis and cell cycle process in patients with bladder cancer. Cancer Cell Int (2020) 20:177. doi: 10.1186/s12935-020-01255-2
21. Fujitomo T, Daigo Y, Matsuda K, Ueda K, Nakamura Y. Critical function for nuclear envelope protein TMEM209 in human pulmonary carcinogenesis. Cancer Res (2012) 72:4110–8. doi: 10.1158/0008-5472.Can-12-0159
22. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–W102. doi: 10.1093/nar/gkx247
23. Wu Y, Peng Z, Gu S, Wang H, Xiang W. A risk score signature consisting of six immune genes predicts overall survival in patients with lower-grade gliomas. Comput Math Methods Med (2022) 2022:2558548. doi: 10.1155/2022/2558548
24. Liu Z, Yao Z, Li C, Lu Y, Gao C. Gene expression profiling in human high-grade astrocytomas. Comp Funct Genomics (2011) 2011:245137. doi: 10.1155/2011/245137
25. Rickman D, Bobek M, Misek D, Kuick R, Blaivas M, Kurnit D, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res (2001) 61:6885–91.
26. Zhang Z, Li H, Jiang S, Li R, Li W, Chen H, et al. A survey and evaluation of web-based tools/databases for variant analysis of TCGA data. Brief Bioinform (2019) 20:1524–41. doi: 10.1093/bib/bby023
27. Zhao Z, Zhang K, Wang Q, Li G, Zeng F, Zhang Y, et al. Chinese Glioma genome atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinf (2021) 19:1–12. doi: 10.1016/j.gpb.2020.10.005
28. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu J, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77:e108–e10. doi: 10.1158/0008-5472.Can-17-0307
29. Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U.S.A. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
30. Tan Y, Zhang S, Xiao Q, Wang J, Zhao K, Liu W, et al. Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics (2020) 112:4808–16. doi: 10.1016/j.ygeno.2020.08.035
31. Pidò S, Ceddia G, Masseroli M. Computational analysis of fused co-expression networks for the identification of candidate cancer gene biomarkers. NPJ Syst Biol Appl (2021) 7:17. doi: 10.1038/s41540-021-00175-9
32. Mall R, Bynigeri R, Karki R, Malireddi R, Sharma B, Kanneganti T. Pancancer transcriptomic profiling identifies key PANoptosis markers as therapeutic targets for oncology. NAR Cancer (2022) 4:zcac033. doi: 10.1093/narcan/zcac033
33. Wu J, Li L, Jiang G, Zhan H, Zhu X, Yang W. NCAPG2 facilitates glioblastoma cells' malignancy and xenograft tumor growth via HBO1 activation by phosphorylation. Cell Tissue Res (2021) 383:693–706. doi: 10.1007/s00441-020-03281-y
34. Yang L, Du C, Chen H, Diao Z. Downregulation of williams syndrome transcription factor (WSTF) suppresses glioblastoma cell growth and invasion by inhibiting PI3K/AKT signal pathway. Eur J Histochem (2021) 65:3255. doi: 10.4081/ejh.2021.3255
35. Zhuo E, Cai C, Liu W, Li K, Zhao W. Downregulated microRNA-140-5p expression regulates apoptosis, migration and invasion of lung cancer cells by targeting zinc finger protein 800. Oncol Lett (2020) 20:390. doi: 10.3892/ol.2020.12253
36. Motwani J, Rodger E, Stockwell P, Baguley B, Macaulay E, Eccles M. Genome-wide DNA methylation and RNA expression differences correlate with invasiveness in melanoma cell lines. Epigenomics (2021) 13:577–98. doi: 10.2217/epi-2020-0440
37. Li J, Yan X, Liang C, Chen H, Liu M, Wu Z, et al. Comprehensive analysis of the differential expression and prognostic value of histone deacetylases in glioma. Front Cell Dev Biol (2022) 10:840759. doi: 10.3389/fcell.2022.840759
38. Hsu F, Chou Y, Chiang M, Li F, Yeh C, Lee W, et al. Signal peptide peptidase promotes tumor progression via facilitating FKBP8 degradation. Oncogene (2019) 38:1688–701. doi: 10.1038/s41388-018-0539-y
39. Yao S, Yin X, Chen T, Chen W, Zuo H, Bi Z, et al. ALDH2 is a prognostic biomarker and related with immune infiltrates in HCC. Am J Cancer Res (2021) 11:5319–37.
40. Besson A, Yong V. Mitogenic signaling and the relationship to cell cycle regulation in astrocytomas. J Neurooncol (2001) 51:245–64. doi: 10.1023/a:1010657030494
41. Chang W, Lai A. Aberrations in notch-hedgehog signalling reveal cancer stem cells harbouring conserved oncogenic properties associated with hypoxia and immunoevasion. Br J Cancer (2019) 121:666–78. doi: 10.1038/s41416-019-0572-9
42. Wang Z, He L, Li W, Xu C, Zhang J, Wang D, et al. GDF15 induces immunosuppression via CD48 on regulatory T cells in hepatocellular carcinoma. J Immunother Cancer (2021) 9:e002787. doi: 10.1136/jitc-2021-002787
43. Simon D, Rout M. Cancer and the nuclear pore complex. Adv Exp Med Biol (2014) 773:285–307. doi: 10.1007/978-1-4899-8032-8_13
44. Xia F, Xia W, Yu X. LncRNA HOTAIR influences the growth, migration, and invasion of papillary thyroid carcinoma via affection on the miR-488-5p/NUP205 axis. Technol Cancer Res Treat (2020) 19:1533033820962125. doi: 10.1177/1533033820962125
45. Bao X, Zhang L, Song W. LncRNA SNHG1 overexpression regulates the proliferation of acute myeloid leukemia cells through miR-488-5p/NUP205 axis. Eur Rev Med Pharmacol Sci (2019) 23:5896–903. doi: 10.26355/eurrev_201907_18334
46. Shu X, Li X, Xiang X, Wang Q, Wu Q. METTL21B is a prognostic biomarker and potential therapeutic target in low-grade gliomas. Aging (2021) 13:20661–83. doi: 10.18632/aging.203454
47. Davis M. Epidemiology and overview of gliomas. Semin Oncol Nurs (2018) 34:420–9. doi: 10.1016/j.soncn.2018.10.001
48. Chen R, Smith-Cohn M, Cohen A, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics (2017) 14:284–97. doi: 10.1007/s13311-017-0519-x
49. Ding W, Chen G, Shi T. Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis. Epigenetics (2019) 14:67–80. doi: 10.1080/15592294.2019.1568178
50. Wang L, Cui W, Zhang Z, Tan Z, Lv Q, Chen S, et al. Expression, methylation and prognostic feature of EMILIN2 in low-Grade-Glioma. Brain Res Bull (2021) 175:26–36. doi: 10.1016/j.brainresbull.2021.07.013
51. Savino A, Provero P, Poli V. Differential Co-expression analyses allow the identification of critical signalling pathways altered during tumour transformation and progression. Int J Mol Sci (2020) 21:9461. doi: 10.3390/ijms21249461
52. Hao C, Duan H, Li H, Wang H, Liu Y, Fan Y, et al. Knockdown of MRPL42 suppresses glioma cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Rep (2018) 38:bsr20171456. doi: 10.1042/bsr20171456
53. Gao X, Guo R, Li Y, Kang G, Wu Y, Cheng J, et al. Contribution of upregulated aminoacyl-tRNA biosynthesis to metabolic dysregulation in gastric cancer. J Gastroenterol Hepatol (2021) 36:3113–26. doi: 10.1111/jgh.15592
54. Stockhausen M, Kristoffersen K, Poulsen H. Notch signaling and brain tumors. Adv Exp Med Biol (2012) 727:289–304. doi: 10.1007/978-1-4614-0899-4_22
55. Janghorban M, Xin L, Rosen J, Zhang X. Notch signaling as a regulator of the tumor immune response: To target or not to target? Front Immunol (2018) 9:1649. doi: 10.3389/fimmu.2018.01649
56. Zindl C, Chaplin D. Immunology. Tumor Immune evasion. Sci (2010) 328:697–8. doi: 10.1126/science.1190310
57. Hinshaw D, Shevde L. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79:4557–66. doi: 10.1158/0008-5472.Can-18-3962
58. Paliouras A, Monteverde T, Garofalo M. Oncogene-induced regulation of microRNA expression: Implications for cancer initiation, progression and therapy. Cancer Lett (2018) 421:152–60. doi: 10.1016/j.canlet.2018.02.029
59. Bruni D, Angell H, Galon J. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer (2020) 20:662–80. doi: 10.1038/s41568-020-0285-7
60. Gordon S, Taylor P. Monocyte and macrophage heterogeneity. Nat Rev Immunol (2005) 5:953–64. doi: 10.1038/nri1733
61. Xu J, Zhang J, Zhang Z, Gao Z, Qi Y, Qiu W, et al. Hypoxic glioma-derived exosomes promote M2-like macrophage polarization by enhancing autophagy induction. Cell Death Dis (2021) 12:373. doi: 10.1038/s41419-021-03664-1
62. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol (2020) 17:807–21. doi: 10.1038/s41423-020-0488-6
63. Simpson T, Li F, Montalvo-Ortiz W, Sepulveda M, Bergerhoff K, Arce F, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med (2013) 210:1695–710. doi: 10.1084/jem.20130579
Keywords: NUP205, lower-grade glioma, prognosis, pathogenic gene, immunotherapeutic target
Citation: Liang W, Hu C, Zhu Q, Cheng X, Gao S, Liu Z, Wang H, Li P, Gao Y and Qian R (2023) Exploring the relationship between abnormally high expression of NUP205 and the clinicopathological characteristics, immune microenvironment, and prognostic value of lower-grade glioma. Front. Oncol. 13:1007198. doi: 10.3389/fonc.2023.1007198
Received: 02 August 2022; Accepted: 29 March 2023;
Published: 22 May 2023.
Edited by:
Adam J. Dupuy, The University of Iowa, United StatesReviewed by:
Yu Zeng, Xiangya Hospital, Central South University, ChinaZahid Nawaz, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2023 Liang, Hu, Zhu, Cheng, Gao, Liu, Wang, Li, Gao and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanzheng Gao, eWFuemhlbmdnYW9obkAxNjMuY29t; Rongjun Qian, ZG9jdG9ycWlhbnJvbmdqdW5AMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Wenjia Liang1†
Wenjia Liang1† Zhendong Liu
Zhendong Liu Yanzheng Gao
Yanzheng Gao