
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol., 29 September 2022
Sec. Neuro-Oncology and Neurosurgical Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.997506
This article is part of the Research TopicChordoma: Advances in Biology and Clinical ManagementView all 14 articles
 Franco Rubino1
Franco Rubino1 Christopher Alvarez-Breckenridge1
Christopher Alvarez-Breckenridge1 Kadir Akdemir1
Kadir Akdemir1 Anthony P. Conley2
Anthony P. Conley2 Andrew J. Bishop3
Andrew J. Bishop3 Wei-Lien Wang4
Wei-Lien Wang4 Alexander J. Lazar4
Alexander J. Lazar4 Laurence D. Rhines1
Laurence D. Rhines1 Franco DeMonte1
Franco DeMonte1 Shaan M. Raza1*
Shaan M. Raza1*Introduction and objective: Despite the improvements in management and treatment of chordomas over time, the risk of disease recurrence remains high. Consequently, there is a push to develop effective systemic therapeutics for newly diagnosed and recurrent disease. In order to tailor treatment for individual chordoma patients and develop effective surveillance strategies, suitable clinical biomarkers need to be identified. The objective of this study was to systematically review all prognostic biomarkers for chordomas reported to date in order to classify them according to localization, study design and statistical analysis.
Methods: Using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, we systematically reviewed published studies reporting biomarkers that correlated with clinical outcomes. We included time-to-event studies that evaluated biomarkers in skull base or spine chordomas. To be included in our review, the study must have analyzed the outcomes with univariate and/or multivariate methods (log-rank test or a Cox-regression model).
Results: We included 68 studies, of which only 5 were prospective studies. Overall, 103 biomarkers were analyzed in 3183 patients. According to FDA classification, 85 were molecular biomarkers (82.5%) mainly located in nucleus and cytoplasm (48% and 27%, respectively). Thirty-four studies analyzed biomarkers with Cox-regression model. Within these studies, 32 biomarkers (31%) and 22 biomarkers (21%) were independent prognostic factors for PFS and OS, respectively.
Conclusion: Our analysis identified a list of 13 biomarkers correlating with tumor control rates and survival. The future point will be gathering all these results to guide the clinical validation for a chordoma biomarker panel. Our identified biomarkers have strengths and weaknesses according to FDA’s guidelines, some are affordable, have a low-invasive collection method and can be easily measured in any health care setting (RDW and D-dimer), but others molecular biomarkers need specialized assay techniques (microRNAs, PD-1 pathway markers, CDKs and somatic chromosome deletions were more chordoma-specific). A focused list of biomarkers that correlate with local recurrence, metastatic spread and survival might be a cornerstone to determine the need of adjuvant therapies.
Chordomas are malignant bone tumors, of possible embryologic origin, arising anywhere along the neuroaxis (1), that are prone to both local recurrence and systemic relapse. Successful treatment of chordomas requires an experienced multidisciplinary team. The median overall survival of patients with chordoma is between 6.3 and 7.7 years (1, 2) with a 5-year overall survival around 70% in most series (2–4). For newly diagnosed patients, primary treatment consists of radical but safe resection; importantly, the extent of resection directly impacts outcomes in patients with skull base and spine chordomas. Over time, treatment of newly diagnosed chordomas has also evolved to incorporate adjuvant radiation, commonly with particle therapy (carbon-ion or proton-ion), particularly for skull base chordomas (5–8). While outcomes have improved over time, the rates of both local recurrence and distant metastases remain high. Ultimately, it is widely accepted that chordoma behaves across a biological spectrum – ranging from an indolent to a more aggressive phenotype with poorer survival and increased risk of local recurrence and metastatic disease. Unfortunately, there is still an unmet clinical need to develop effective systemic therapeutic agents that can be incorporated into the treatment paradigm for newly diagnosed and recurrent disease. To this end, clinical biomarkers that can be used to better tailor treatment and implement surveillance strategies for patients with chordoma are needed.
To meet this need for clinical biomarkers, several molecular studies have been conducted to identify therapeutic targets and prognostic markers in order to predict outcome (9–12). While many of the identified biomarkers have been validated in the research laboratory setting, the challenge lies in creating a biomarker panel that can be used in the clinical setting to guide real-time decision-making (13, 14). According to the US Food and Drug Administration (FDA), a biomarker is defined as a measurable characteristic that serves as an indicator of normal or pathological biological processes or responses to an exposure or intervention. Additionally, biomarkers can include molecular, histologic, radiographic or physiologic features (15). To be clinically utilized, there are certain critical features that any biomarker should include: 1) it should be non-invasive, easily measured, affordable, and produce rapid results; 2) it should be accessible from readily available sources, such as blood or urine; 3) it should have high sensitivity to allow early detection and have a high specificity in diseased samples; 4) the expression level of the biomarker should aid in risk stratification and possess prognostic value in terms of clinical outcomes; and 5) the biomarker should provide insight into the underlying disease mechanism (16). Specifically, a prognostic biomarker identifies the likelihood of a clinical event, disease recurrence, or progression in patients who have a medical condition of interest (17).
The objective of the present study was to review all prognostic biomarkers reported to date for chordomas in order to classify them according to localization, clinical utility, study design and statistical analysis.
The current systematic review was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (18). Studies were classified according to their design as prospective or retrospective. The level of evidence of each study was determined according to Oxford Centre for Evidence-Based Medicine (OCEBM) guidelines (19, 20). A systematic search was conducted in the PubMed/Medline and Google Scholar databases using the following terms: “chordoma”, “biomarker”, “overall survival”, “progression free survival”, “prognosis” and “biological marker”. Any discrepancies were resolved through consensus. The search strategy utilized is reported in Figure 1. Inclusion criteria were as follows: i) the study must have been a prospective clinical trial or a retrospective case-control/cross sectional study published in English with no start date restriction and until February 2022 (when analyses for this review were performed); ii) the studies had to have reported chordomas confirmed by pathology (either in the skull base, mobile spine or sacrum); iii) the studies must have been time-to-event studies, which determined Progression Free Survival (PFS), and/or Overall Survival (OS); iv) the outcomes must have been analyzed by employing Kaplan-Meier (K-M) analysis with log-rank test or a Cox proportional hazards analysis (Cox model); v) the study must have included a full biomarker description including the brand name of the assay used, the source/matrix, the measurement technique and the assay technology used; vi) studies must not have had overlapping patients.
Studies using other biomarker categories, such as susceptibility/risk, diagnostic, monitoring, predictive, pharmacodynamic/response or safety were excluded.
The extracted variables of each study include the following: Type of study, number of patients, localization of the primary tumor (skull base, mobile spine and/or sacrum), localization of the biomarker (cellular [nuclear, cytoplasm or cell membrane] or extracellular [blood or extracellular matrix]), assay technique for measurement, prognostic variable analyzed, non-parametric statistical approach, level of significance (p-value), and study design (prospective or retrospective). For classification purposes, we divide the biomarker as ‘positive’ or ‘negative’, according to its correlation with PFS or OS.
The primary goal was to determine whether there was significant correlation between the biomarker and prognostic variable analyzed, either using Log-rank univariate analysis or Cox regression multivariate analysis. All statistical analyses were performed using Microsoft Excel 365 (Microsoft, Washington, USA) and GraphPad Prism 9.0 statistical software package (La Jolla, California, USA). Descriptive statistics was the main statistical technique. Categorical variables are presented as proportions and the continuous variables are presented with mean and standard deviation or median and interquartile range.
Sixty-eight studies met predetermined eligibility criteria and were included for data abstraction (Figure 1). Fourteen studies used the same cohort of patients with different biomarkers, so a total number of 1263 patients are duplicated (21–33). Two studies were multicenter, which includes different populations, increasing the strength of the study (21, 34). With regard to study design, 5 studies were prospective (7.3%) (26, 35–38) and 63 studies were retrospective (92.7%) (21–25, 27–34, 39–87).
Data for a total of 3183 patients (not duplicated) were analyzed using different prognostic biomarkers. We excluded data from patients that was collected from the same hospital and in the same time frame. Of these, 3150 cases were adult patients (99%) and 33 were pediatric patients (1%); there were 1595 (50%) skull base chordomas, 1165 (36.6%) sacrococcygeal chordomas and 423 (13.4%) chordomas in mobile spine segments. A total of 103 biomarkers were analyzed in the 68 studies included in our analysis. Using the FDA classification system (15) we divided the biomarkers into 4 groups: molecular (82.5%; n=85), physiological (9.7%; n=10), histological (6.8%; n=7) and radiographic biomarkers (1%; n=1) (82).
Localization for each biomarker was different. A total of 40% were located in the nucleus (n=41), 22% in the cytoplasm (n=23), 14% in the cellular membrane (n=14), 7% in the extracellular matrix (n=7), and 16% from blood samples (n=17). The most common detection technique used was immunohistochemistry (IHC) in 66% of the studies (n=45) followed by polymerase chain reaction (PCR), serum protein analysis, and DNA sequencing in 20% (n=14), 9% (n=6) and 5% (n=3), respectively.
Three studies did not consider the expression of individual biomarkers, but instead they described a score using a group of biomarkers. One study describes an “miRNA-score”, which measures the nuclear concentration of different microRNAs (miR-1290; miR-574-3p; miR-1; miR-1237-3p; miR-155; miR-140-3p) using PCR-based techniques (42). Another study describes a “Systemic Inflammatory Index” (SII), which is obtained by multiplying blood platelet count and neutrophil count and dividing by lymphocyte blood counts (P x N/L) (41), and the last study describes an “Immunoscore” based on the expression of CD3+ and CD8+ cells in the tumor specimen, using IHC (58).
The clinical outcomes commonly evaluated in the selected studies were Progression free survival (PFS) and overall survival (OS). Only those biomarkers that demonstrated significant correlation with clinical outcomes in non-parametric tests were considered. Significant correlation of biomarkers with OS, PFS, or both, appeared in 35% (n=24), 23% (n=16) and 40% (n=27) of the studies, respectively. One study found that the lack of 1p36 LOH correlates with improved OS and PFS but did not report the HR and/or the CI (60). Because of this, we did not include their results in the following figures and analysis. Ultimately, 82 biomarkers were demonstrated to have statistical correlation in PFS and/or OS.
Elevated expression of 15 biomarkers was found to be associated with improved PFS and/or OS in chordoma patients (23, 24, 30, 31, 39, 55, 58, 62, 69, 76, 78, 84, 88). Among them, five biomarkers correlated with improved OS (elevated CD8+/Foxp3+ ratio, H-TERT promoter mutations, and high expression of SNF5 protein, c-MET receptor, and miRNA-1), eight biomarkers correlated with improved PFS (high Immunoscore, elevated PD1+ Tumor-Infiltrating Lymphocytes (TIL), high expression of PHLPP1 protein, Raf-1 and ERK proteins, elevated MMP-9/RECK protein ratio, and the high nuclear expression of miRNA-1290 and mi-RNA-1237-3p) and, 2 biomarkers were linked with both PFS and OS (elevated PD-L1+ TIL and high preoperative serum albumin level).
Increased expression of the remaining biomarkers (82%, n=67) correlated with poorer PFS and/or OS outcomes. Among them, the Ki-67 Level Index (LI) was the most frequent biomarker analyzed across the collection of studies (n=7, 10.3%). A high Ki-67 LI correlated with a shorter OS or PFS (even on COX regression analysis) (27, 33, 56).
Thirty-four studies analyzed the correlation between biomarkers and clinical outcomes using Cox multivariate regression and found that 35 biomarkers (43%) and 22 biomarkers (27%) were independent prognostic factors for PFS and OS, respectively. Although these biomarkers demonstrated a p-value less than 0.05, the confidence intervals (95% CI) varied greatly, partly due to limited sample sizes. To mention some examples, in one study, high Brachyury gene expression or increased number of alterations in chromosome 2p (CNA 2p) correlated with improved PFS in a sample of 37 patients, but the CI of both is wide and include the value 1 (45). Furthermore, other biomarkers like VEGFA and E-cadherin had a CI of almost 1 in a sample size of 151 patients (27).
Forest plots in Figures 2 and 3, help to clearly differentiate those studies with more statistical strength. Overall, 13 biomarkers (19%) had significant correlation with both clinical outcomes (Table 1).
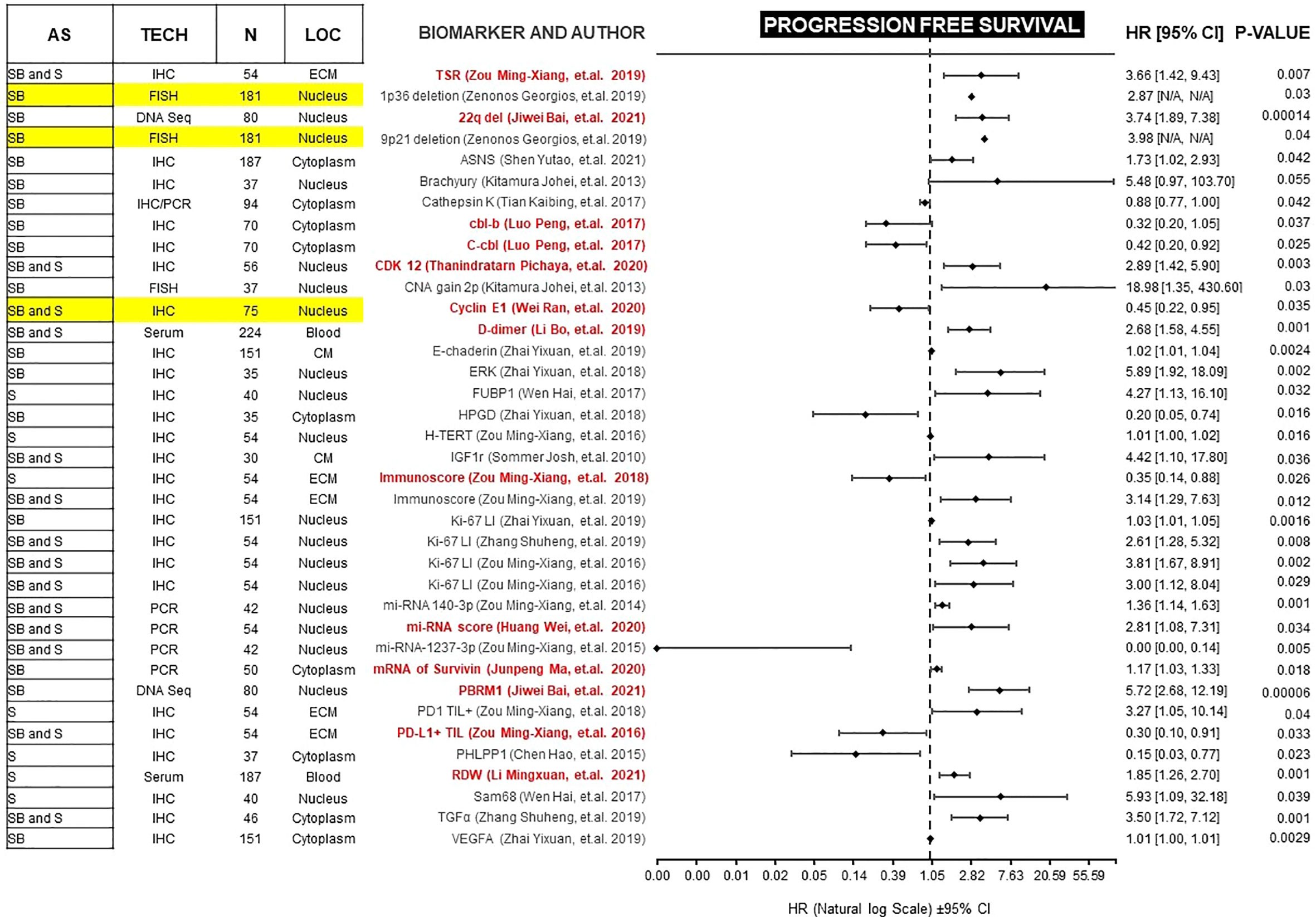
Figure 2 Forest plot showing biomarkers associated with PFS benefit on multivariate analysis. Those biomarkers with benefit in PFS and OS are marked in red. Prospective studies are marked in yellow. AS (Anatomic Site), TECH (Assay technique used), N (number of patients), LOC (Biomarker localization), SB (Skull base), S (Spine), IHC (Immunohistochemistry), PCR (Polymerase chain reaction), DNA Seq (DNA sequencing), FISH (fluorescence in situ hybridization), ECM (extracellular matrix), CM (Cell Membrane), TSR (Tissue-stroma ratio), PD-L1 TIL (Programmed death ligand 1, Tumor infiltrating lymphocytes), CDK (Cyclin-dependent kinase), TGFα (Tumor Growth Factor α), RDW (Red cell distribution width), CNA (copy number alterations), N/A (Not available).
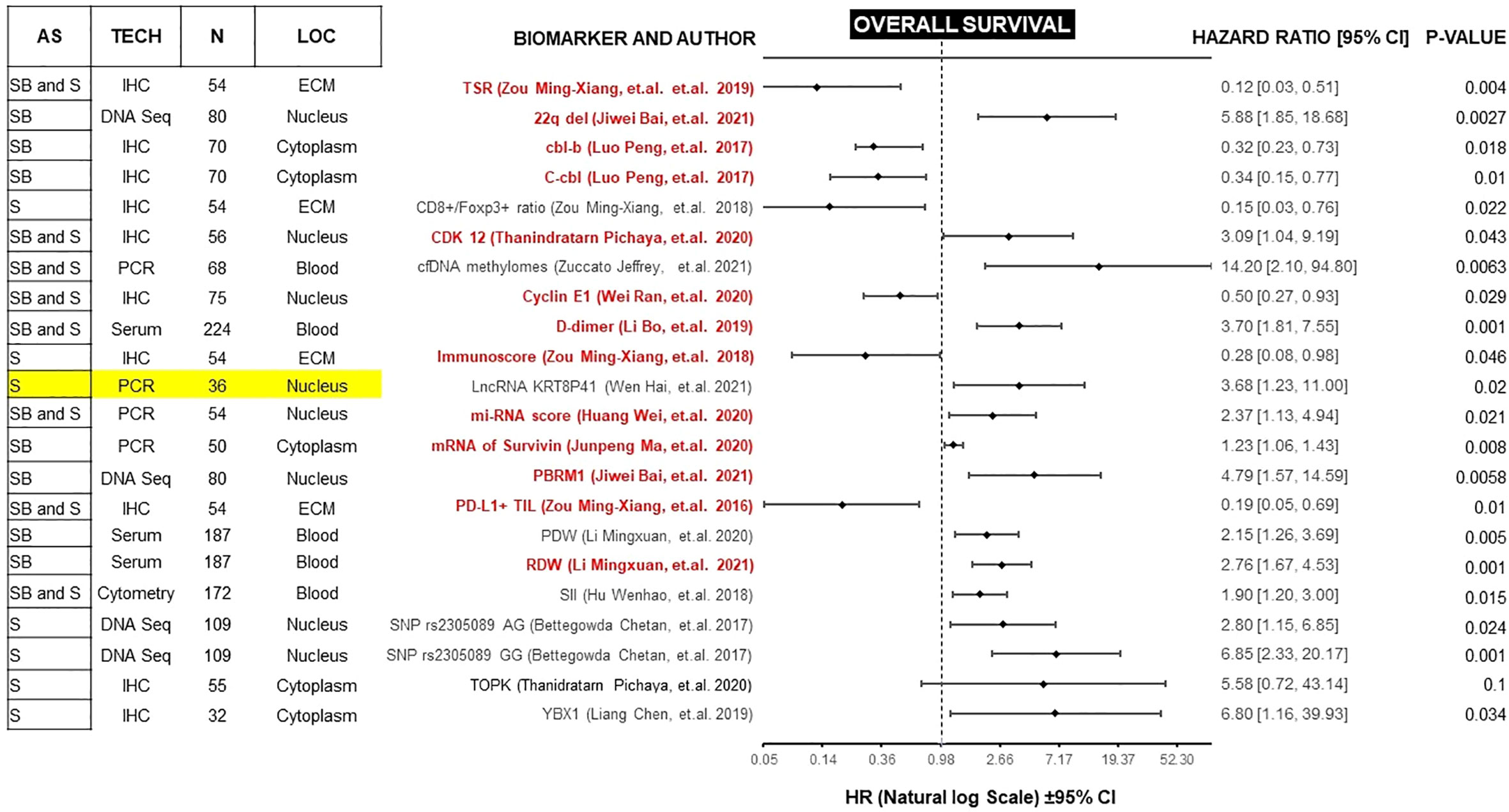
Figure 3 Forest plot showing biomarkers associated with OS benefit on multivariate analysis. Those biomarkers with benefit in PFS and OS are marked in red. Prospective studies are marked in yellow. AS (Anatomic Site), TECH (Assay technique used), N (number of patients), LOC (Biomarker localization), SB (Skull base), S (Spine), IHC (Immunohistochemistry), PCR (Polymerase chain reaction), DNA Seq (DNA sequencing), FISH (fluorescence in situ hybridization), ECM (extracellular matrix), CM (Cell Membrane), TSR (Tissue-stroma ratio), PD-L1 TIL (Programmed death ligand 1, Tumor infiltrating lymphocytes), RDW (Red cell distribution width), PDW (Platelet distribution width), LncRNA (Long non coding RNA).
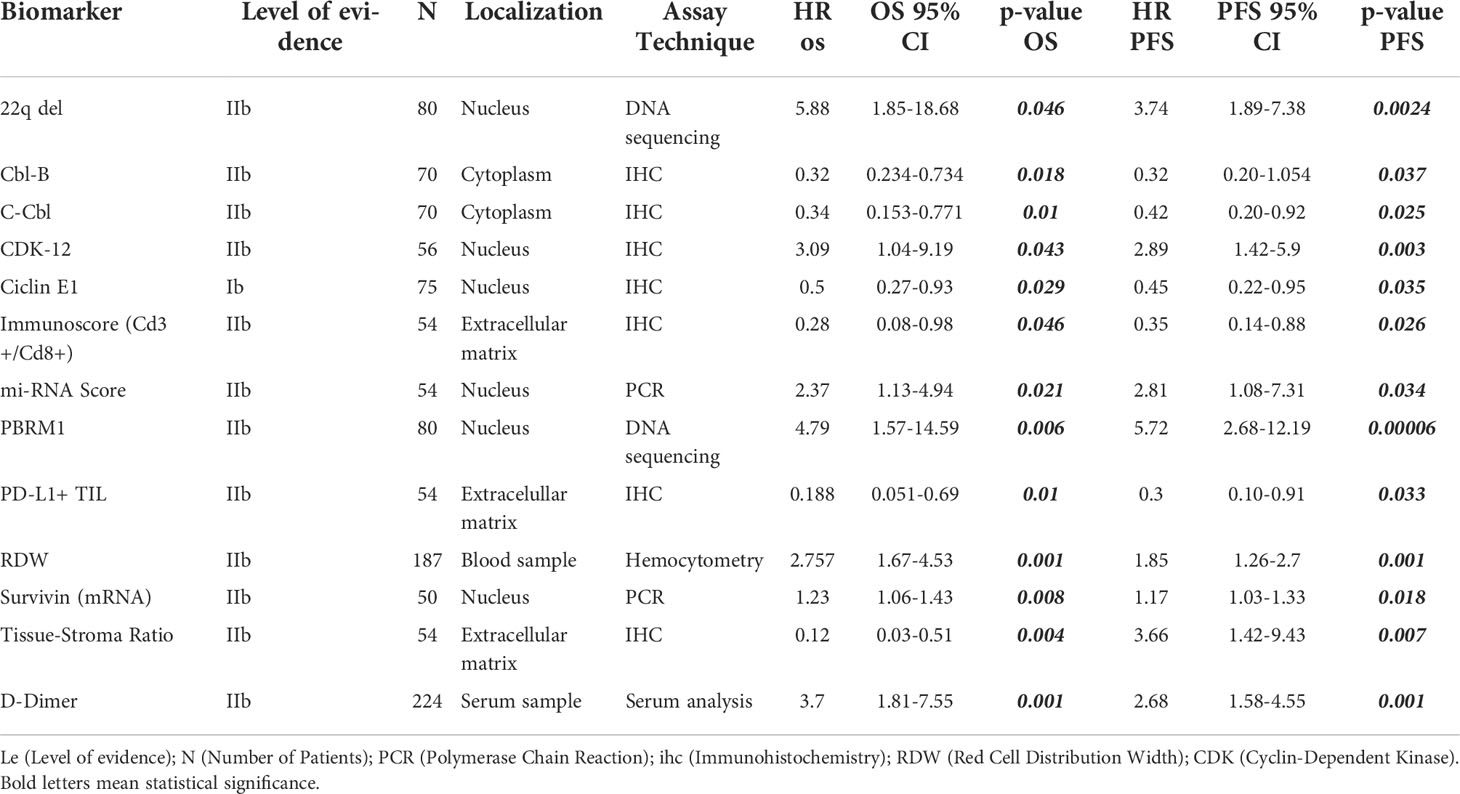
Table 1 List of reported biomarkers with independent prognostic impact in PFS and OS (with Cox multivariate analysis).
There is increasing recognition of the need for predictive biomarkers to assist in the management of chordomas. To the best of our knowledge, this is the first comprehensive review that summarizes prognostic biomarkers in chordoma that have been reported to date. The mainstay of treatment for localized chordomas is extensive resection with negative margins, but local recurrence and systemic metastases are common. Clinically reliable prognostic biomarkers that could aid in determining whether a particular chordoma is likely to recur would impact clinical decision-making, particularly with regard to the need for adjuvant therapy, the early incorporation of systemic therapy agents, and surveillance strategies.
PFS is a surrogate for OS (89). However, OS in diseases with more than 12 months of survival after disease progression/recurrence (like chordomas) is not a reliable endpoint, and a very large number of patients is required to power studies to detect statistically significant differences in solely in OS (90). Therefore, we focused on studies of biomarkers with a significant correlation for both endpoints. Because most of these studies are retrospective, potential confounders must be unmasked and multivariate analysis is an effective method of ranking various prognostic factors (91, 92). We found 13 biomarkers which are independently prognostic for both PFS and OS with statistical significance (Table 1). In each case, we compared the Hazard ratio (HR) and the Confidence Interval (CI). As a rule, the width of the CI correlates inversely with the precision of the biomarker. In other words, some biomarkers have a significant p-value, but the CI could be wide or includes the value of 1 (Figures 2 and 3). This situation decreases the precision of the biomarker when compared with others and is therefore an important consideration. We further discuss some of the 13 biomarkers below.
The expression of Programmed Death-1 (PD-1) receptor, its ligand, and the immune microenvironment in chordoma have been widely studied (30, 58). The PD-1/PD-L1 pathway is responsible for modulating T-cell activation (anti-tumor response) and exhaustion in cancer (93). Tumor cells express PD-L1 as an adaptive immune mechanism to escape this anti-tumor environment with a high number of CD8+ cells (Immunoscore) (58) or a high CD8+/Foxp3 ratio (30), consistently analyzed in our review. The blockade of this immune checkpoint signal has the potential clinical benefit of increasing the sensitivity of chordoma cells to T-cell mediated lysis. In this sense, PD-L1+ is currently a prognostic and therapeutic biomarker (10, 94). FDA-approved monoclonal antibodies currently used to block the PD-1/PD-L1 pathway include Pembrolizumab, Nivolumab, and Avelumab. Additionally, Nivolumab is currently being evaluated amongst rare CNS cancers including chordoma in 3 open clinical trials (NCT03173950, NCT02989636, NCT03623854). Even though the PD-1/PD-L1 pathway appears to be a very promising therapeutic and prognostic marker, the clones of anti-mouse PD-1 antibodies used are not consistent, and validation of them is needed to reach a wide reproducibility.
MicroRNA are small noncoding RNAs that play important roles in transcriptional gene regulation. Different miRNAs have been implicated in several cancers, including chordomas (30, 31, 42, 69, 88). Multiple proteins are involved in chordomagenesis, therefore it is unlikely that a single miRNA would provide optimal information regarding prognosis. Rather than focusing on a single miRNA, Huang et al. identified 6 prognostic miRNAs and developed a miRNA score (miRscore) based on integrated data. Using this approach, the miRscore was able to predict clinical outcome in chordoma (42). However, the main limitation of using this biomarker in clinical practice is the affordability along with ease and consistency of measurement using RT-PCR. An alternative approach is miRNA-seq but that is also hampered by technical complications limiting broad adoption (95).
Survivin is an apoptosis inhibitor protein widely expressed in tumors and non-terminally differentiated tissues (96). Ma et al. evaluated the expression of survivin in primary versus recurrent chordomas and demonstrated that survivin expression is elevated in recurrent tumors. Additionally, they found survivin levels were increased in recurrent tumors collected from the same patient, and expression of survivin was found to be an independent prognostic factor for progression (44). However, the lab technique used to quantify survivin expression was quantitative RT-PCR which is not practical assay from a clinical testing standpoint, although the use of survivin as a biomarker offers the advantage of measuring a single molecule as compared with the use of miRscore which measures 6 different miRNAs (42).
The Cyclin-dependent kinases (CDKs) are serine/threonine nuclear protein kinases involved in DNA transcription and cell cycle progression. In chordoma, CDK overexpression induces migration of tumor cells, proliferation, and tumor growth. In particular, the CDKN2A/p16INK4a/CDK4-6/RB pathway is a well-known cascade involved in tumor cell proliferation across multiple cancer types (97). The CDKN2A gene, and its protein product p16INK4a, have an inhibitory role over CDK4 and 6, decreasing cell proliferation. Mutations in CDKN2A gene have been studied as a prognostic biomarker in independent studies (38, 43, 50), but none have found a significant correlation with clinical outcomes. Only Sommer et al. (50) found that CDKN2A gene expression level correlated with a higher PFS (p=.03); however, this correlation was not significant in multivariate analysis. CDK4 expression was found to be significantly associated with OS in a study by Yakkioui et al. (83), however, this was only in univariate analysis (p=.021). These data suggest that this pathway has an unclear prognostic role in chordoma PFS or OS. Despite these uncertain results, the pathway is currently being evaluated for chordoma therapy. For example, palbociclib is an approved CDK4/6 inhibitor for chordomas with CDKN2A gene loss (98) and is currently being evaluated in a clinical trial (NCT03110744).
CDK12 overexpression increases the phosphorylation of antiapoptotic proteins, including Survivin and MCL-1 and correlates with shorter overall survival in breast cancer, ovarian cancer and gastric cancer (99, 100). Pichaya et al. measured the expression of CDK12 in tumor tissue in a cohort of 56 spine chordoma patients using immunohistochemistry, and grouped chordomas as those with low CDK-12 expression (less than 75% of the cells with positive nuclear staining) and those with high CDK-12 expression (more than 75% of the cells with positive nuclear staining) (52). The statistical analysis demonstrated a negative effect of this CDK in prognosis, however the results need to be interpreted with caution because the wide CI indicates lack of precision and little knowledge about the effect. While a study with a larger sample is required, the strength of this biomarker lies in the relatively easy assay technique, affordability and it resides upstream of other biomarkers implicated in poor clinical outcomes.
Somatic chromosome deletions have been studied as a type of genomic driver event in chordomas. Bai et al. conducted whole-genome sequencing of 80 skull base chordomas and evaluated the prognostic value of 17 somatic chromosomal events including gains of chromosomes 1q, 7p, and 7q, and deletions of 1p, 3, 4, 9, 10, 13q, 14q, 18, and 22q (43). Of these, the hemizygous deletion of 22q, where SMARCB1 gene is located, demonstrated a strong association with clinical outcomes. The homozygous deletion of SMARCB1 is a hallmark of poorly differentiated chordoma (101), but this finding suggests that even partial inactivation of SMARCB1 in conventional chordoma could be used as a prognostic biomarker. Nevertheless, the wide CI in this study highlights the need for validation with a larger sample. Although genome sequencing is an expensive and highly specialized assay technique, chromosomal deletion can be assessed with Fluorescence in situ hybridization (FISH) or spectral karyotyping (SKY) (102). In this regard, it is worth mentioning that Zenonos et al. present one of the few prospective studies with a strong statistical design (36). Using FISH, they found that skull base chordoma patients with a threshold percentage of cells with 1p36 and 9p21 deletions (more than 15% for 1p36 and more than 4% for 9p21) have a shorter PFS. Despite being a prospective study with thorough statistical design and promising HR, we did not include in Table 1 because the study did not include OS and CIs in the analysis.
The last two biomarkers were assessed in blood samples and are the most promising in terms of affordability, limited invasiveness, reproducibility, assay technique, and required sample size (22, 46). Other studies have reported that increased levels of RDW or D-dimer are observed in many types of cancers and act as useful markers of adverse prognosis (103–106). In chordoma patients, a preoperative measurement above the cut-off values of 12.7 for RDW (in a spine chordoma population) and 840 μg/L for D-dimer (in a skull base chordoma population) were associated with a poor prognosis. However, these biomarkers do not provide an insight about the underlying disease mechanism, and they lack specificity (106–108).
After analyzing all of the available information gathered for this review, Table 1 presents a set of biomarkers that have been demonstrated to correlate with both PFS and OS. While additional studies are needed to validate a narrower list of biomarkers with the strongest prognostic value, this proposed panel serves as a starting point. Given the techniques employed and the already available testing, there are biomarkers that can be readily incorporated into routine clinical setting. For example, preoperative D-dimer can be readily integrated as a potential prognostic biomarker due to its low-invasiveness, accessible measurement technique, and affordability. Also, the D-dimer study collected a significant cohort of patients (n=224) when compared with other biomarker studies and reported results using multivariate analysis with narrow CI (PFS, HR 2.68 [1.58, 4.55], p=.001; OS, 3.70 [1.81, 7.55], p=.001).
Furthermore, our inferential statistical analysis of the data has limitations because chordoma has intrinsic low incidence. Most of the studies report a limited patient cohort, making it difficult to assess the multivariate analysis of each biomarker. It is worth mentioning that none of the biomarkers met all the criteria mentioned in the introduction of this manuscript. They need to be validated in prospective studies with a larger sample size, and across multiple institutions. Affordability is one of the most important factors to consider for a biomarker that can be tested ubiquitously. The less expensive techniques used were serum protein analysis or hemocytometry, followed by immunohistochemistry and finally, some of the studies used expensive measurement techniques such as DNA sequencing (21, 43, 62), DNA microarrays (60, 86) and FISH (36, 45, 74). Also, when the lab technique is expensive or difficult to use, the studies are concentrated among the same centers, resulting in limited widespread integration. In order to use biomarker information to evaluate chordoma patients and enable clinical decision-making, high-quality prospective studies to evaluate the known potential biomarkers with strong biological underpinnings in chordomas are warranted. While retrospective studies remain useful in identifying potential new biomarkers, prospective trials with larger cohorts are essential for the eventual clinical translation of biomarkers.
In this study we gathered all the prognostic biomarker studies reported to date according to PRISMA guidelines. More than a hundred biomarkers have been analyzed. The information is assorted, heterogenous, and lacks consistency in terms of markers used and tests performed. There is an increasing need to gather all these results to guide future validation studies to determine its clinical utility. A comprehensive understanding of this information can eventually help determine adjuvant treatments and surveillance strategies for patients with the worst prognosis. Further work is needed to validate a narrower list of biomarkers that correlate with overall survival, local recurrence, and metastatic spread with the strongest significance. Taken collectively, we encourage the comprehensive centers that evaluate and treat chordoma patients to collaboratively work to create a panel of prognostic biomarkers that can then be applied universally.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The authors confirm contribution to the paper as follows:
Study conception and design: FR and SR. Data collection: FR and SR. Analysis and interpretation of results: FR, CA-B, AC, AB, W-LW, AL, LR, FD, and SR. Draft manuscript preparation: FR, CA-B, KA, LR, FD, and SR. All authors reviewed the results and approved the final version of the manuscript.
We thank Preeti Ramadoss, PhD for her editorial assistance in preparing this publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses; FDA ,Food and Drug Administrations; PFS, Progression Free Survival; OS, Overall Survival; PD-1, programmed cell death protein 1; CDK, cyclin dependent kinases.
1. Ulici V, Hart J. Chordoma. Arch Pathol Lab Med (2022) 146(3):386–95. doi: 10.5858/arpa.2020-0258-RA
2. Smoll NR, Gautschi OP, Radovanovic I, Schaller K, Weber DC. Incidence and relative survival of chordomas: The standardized mortality ratio and the impact of chordomas on a population. Cancer (2013) 119(11):2029–37. doi: 10.1002/cncr.28032
3. Jones PS, Aghi MK, Muzikansky A, Shih HA, Barker FG 2nd, Curry WT. Outcomes and patterns of care in adult skull base chordomas from the surveillance, epidemiology, and end results (SEER) database. J Clin Neurosci (2014) 21(9):1490–6. doi: 10.1016/j.jocn.2014.02.008
4. Scampa M, Tessitore E, Dominguez DE, Hannouche D, Buchs NC, Kalbermatten DF, et al. Sacral chordoma: A population-based analysis of epidemiology and survival outcomes. Anticancer Res (2022) 42(2):929.LP–37. doi: 10.21873/anticanres.15552
5. Takahashi S, Kawase T, Yoshida K, Hasegawa A, Mizoe J-E. Skull base chordomas: Efficacy of surgery followed by carbon ion radiotherapy. Acta Neurochirurgica (2009) 151(7):759–69. doi: 10.1007/s00701-009-0383-5
6. Bugoci DM, Girvigian MR, Chen JCT, Miller MM, Rahimian J. Photon-based fractionated stereotactic radiotherapy for postoperative treatment of skull base chordomas. Am J Clin Oncol (2013) 36(4):404–10. doi: 10.1097/COC.0b013e318248dc6f
7. Yu E, Koffer PP, DiPetrillo TA, Kinsella TJ. Incidence, treatment, and survival patterns for sacral chordoma in the united states, 1974-2011. Front Oncol (2016) 6:203–. doi: 10.3389/fonc.2016.00203
8. Chambers KJ, Lin DT, Meier J, Remenschneider A, Herr M, Gray ST. Incidence and survival patterns of cranial chordoma in the united states. Laryngoscope (2014) 124(5):1097–102. doi: 10.1002/lary.24420
9. Liu T, Shen JK, Choy E, Zhang Y, Mankin HJ, Hornicek FJ, et al. CDK4 expression in chordoma: A potential therapeutic target. J Orthopaedic Res (2018) 36(6):1581–9. doi: 10.1002/jor.23819
10. Fujii R, Friedman ER, Richards J, Tsang KY, Heery CR, Schlom J, et al. Enhanced killing of chordoma cells by antibody-dependent cell-mediated cytotoxicity employing the novel anti-PD-L1 antibody avelumab. Oncotarget (2016) 7(23):33498–511. doi: 10.18632/oncotarget.9256
11. Stacchiotti S, Tamborini E, Lo Vullo S, Bozzi F, Messina A, Morosi C, et al. Phase II study on lapatinib in advanced EGFR-positive chordoma. Ann Oncol (2013) 24(7):1931–6. doi: 10.1093/annonc/mdt117
12. Houessinon A, Boone M, Constans J-M, Toussaint P, Chauffert B. Sustained response of a clivus chordoma to erlotinib after imatinib failure. Case Rep Oncol (2015) 8:25–9. doi: 10.1159/000371843
13. Romeo S, Hogendoorn PCW. Brachyury and chordoma: the chondroid-chordoid dilemma resolved? J Pathol (2006) 209(2):143–6. doi: 10.1002/path.1987
14. Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol (2006) 209(2):157–65. doi: 10.1002/path.1969
15. FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource [Internet]. (US: Silver Spring (MD): Food and Drug Administration) (2016).
16. Group BDW. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther (2001) 69(3):89–95. doi: 10.1067/mcp.2001.113989
17. Oldenhuis CNAM, Oosting SF, Gietema JA, de Vries EGE. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer (Oxford England: 1990). (2008) 44(7):946–53. doi: 10.1016/j.ejca.2008.03.006
18. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol (2009) 21:6(7). doi: 10.1371/journal.pmed.1000097
19. Richards D. GRADING – levels of evidence. Evidence-Based Dentistry (2009) 10(1):24–5. doi: 10.1038/sj.ebd.6400636
20. OCEBM Levels of Evidence Working Group. The 2011 Oxford levels of evidence. (UK: University of Oxford) (2011). Available at: http://www.cebm.net/index.aspx?o=5653.
21. Bettegowda C, Yip S, Lo S-FL, Fisher CG, Boriani S, Rhines LD, et al. Spinal column chordoma: prognostic significance of clinical variables and T (brachyury) gene SNP rs2305089 for local recurrence and overall survival. Neuro-Oncology (2017) 19(3):405–13. doi: 10.1093/neuonc/now156
22. Li M, Shen Y, Xiong Y, Bai J, Wang S, Li C, et al. High red cell distribution width independently predicts adverse survival in patients with newly diagnosed skull base chordoma. OncoTargets Ther (2021) 14:5435. doi: 10.2147/OTT.S335454
23. Li M, Bai J, Wang S, Zhai Y, Zhang S, Li C, et al. Prognostic value of cumulative score based on preoperative fibrinogen and albumin level in skull base chordoma. OncoTargets Ther (2020) 13:8337. doi: 10.2147/OTT.S257779
24. Li M, Zhai Y, Bai J, Wang S, Gao H, Li C, et al. SNF5 as a prognostic factor in skull base chordoma. J Neuro-Oncol (2018) 137(1):139–46. doi: 10.1007/s11060-017-2706-3
25. Thanindratarn P, Dean DC, Nelson SD, Hornicek FJ, Duan Z. T-LAK cell-originated protein kinase (TOPK) is a novel prognostic and therapeutic target in chordoma. Cell Proliferation (2020) 53(10):e12901–e. doi: 10.1111/cpr.12901
26. Wen H, Fu Y, Zhu Y, Tao S, Shang X, Li Z, et al. Long non-coding RNA KRT8P41/miR-193a-3p/FUBP1 axis modulates the proliferation and invasion of chordoma cells. J Bone Oncol (2021) 31:100392–. doi: 10.1016/j.jbo.2021.100392
27. Zhai Y, Bai J, Li M, Wang S, Li C, Wei X, et al. A nomogram to predict the progression-free survival of clival chordoma. J Neurosurg (2019) 134(1):144–52. doi: 10.3171/2019.10.JNS192414
28. Zhai Y, Bai J, Gao H, Wang S, Li M, Gui S, et al. Clinical features and prognostic factors of children and adolescents with clival chordomas. World Neurosurg (2017) 98:323–8. doi: 10.1016/j.wneu.2016.11.015
29. Zhang K, Chen H, Wu G, Chen K, Yang H. High expression of SPHK1 in sacral chordoma and association with patients’ poor prognosis. Med Oncol (2014) 31(11):1–5. doi: 10.1007/s12032-014-0247-6
30. Zou M-X, Guo K-M, Lv G-H, Huang W, Li J, Wang X-B, et al. Clinicopathologic implications of CD8+/Foxp3+ ratio and miR-574-3p/PD-L1 axis in spinal chordoma patients. Cancer Immunol Immunother (2018) 67(2):209–24. doi: 10.1007/s00262-017-2080-1
31. Zou M-X, Huang W, Wang X-b, Li J, Lv G-H, Wang B, et al. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J (2015) 24(8):1738–46. doi: 10.1007/s00586-015-3927-9
32. Zou M-X, Huang W, Wang X-B, Lv G-H, Li J, Deng Y-W. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol (2014) 7(8):4877.
33. Zou M-X, Lv G-H, Li J, She X-L, Jiang Y. Upregulated human telomerase reverse transcriptase (hTERT) expression is associated with spinal chordoma growth, invasion and poor prognosis. Am J Trans Res (2016) 8(2):516.
34. Zuccato JA, Patil V, Mansouri S, Liu JC, Nassiri F, Mamatjan Y, et al. DNA Methylation based prognostic subtypes of chordoma tumors in tissue and plasma. Neuro-Oncology (2021) 24(3):442–54. doi: 10.1093/neuonc/noab235
35. Wei R, Dean DC, Thanindratarn P, Hornicek FJ, Guo W, Duan Z. Prognostic significance of cyclin E1 expression in patients with chordoma: A clinicopathological and immunohistochemical study. Frontiers Oncology (2020) 10:596330. doi: 10.3389/fonc.2020.596330
36. Zenonos GA, Fernandez-Miranda JC, Mukherjee D, Chang Y-F, Panayidou K, Snyderman CH, et al. Prospective validation of a molecular prognostication panel for clival chordoma. J Neurosurg (2018) 130(5):1528–37. doi: 10.3171/2018.3.JNS172321
37. Lebellec L, Bertucci F, Tresch-Bruneel E, Bompas E, Toiron Y, Camoin L, et al. Circulating vascular endothelial growth factor (VEGF) as predictive factor of progression-free survival in patients with advanced chordoma receiving sorafenib: an analysis from a phase II trial of the french sarcoma group (GSF/GETO). Oncotarget (2016) 7(45):73984–94. doi: 10.18632/oncotarget.12172
38. Yang C, Sun J, Yong L, Liang C, Liu T, Xu Y, et al. Deficiency of PTEN and CDKN2A Tumor-suppressor genes in conventional and chondroid chordomas: Molecular characteristics and clinical relevance. OncoTargets Ther (2020) 13:4649. doi: 10.2147/OTT.S252990
39. Chen H, Zhang K, Wu G, Song D, Chen K, Yang H. Low expression of PHLPP1 in sacral chordoma and its association with poor prognosis. Int J Clin Exp Pathol (2015) 8(11):14741–. doi: 10.1007/s12032-014-0247-6
40. Wen H, Li P, Ma H, Zheng J, Yu Y, Lv G. High expression of Sam68 in sacral chordomas is associated with worse clinical outcomes. OncoTargets Therapy (2017) 10:4691–700. doi: 10.2147/OTT.S147446.
41. Hu W, Yu J, Huang Y, Hu F, Zhang X, Wang Y. Lymphocyte-related inflammation and immune-based scores predict prognosis of chordoma patients after radical resection. Trans Oncol (2018) 11(2):444–9. doi: 10.1016/j.tranon.2018.01.010
42. Huang W, Yan Y-G, Wang W-J, Ouyang Z-H, Li X-L, Zhang T-L, et al. Development and validation of a 6-miRNA prognostic signature in spinal chordoma. Front Oncol (2020) 10:2298–. doi: 10.3389/fonc.2020.556902
43. Bai J, Shi J, Li C, Wang S, Zhang T, Hua X, et al. Whole genome sequencing of skull-base chordoma reveals genomic alterations associated with recurrence and chordoma-specific survival. Nature Communications (2021) 12(1):757. doi: 10.1038/s41467-021-21026-5
44. Ma J, Tian K, Du J, Wu Z, Wang L, Zhang J. High expression of survivin independently correlates with tumor progression and mortality in patients with skull base chordomas. J Neurosurg (2020) 132(1):140–9. doi: 10.3171/2018.8.JNS181580
45. Kitamura Y, Sasaki H, Kimura T, Miwa T, Takahashi S, Kawase T, et al. Molecular and clinical risk factors for recurrence of skull base chordomas: Gain on chromosome 2p, expression of brachyury, and lack of irradiation negatively correlate with patient prognosis. J Neuropathol Exp Neurol (2013) 72(9):814–21. doi: 10.1093/jnen/72.9.814
46. Li B, Zhang H, Zhou P, Yang J, Wei H, Yang X, et al. Prognostic significance of pretreatment plasma d-dimer levels in patients with spinal chordoma: A retrospective cohort study. Eur Spine J (2019) 28(6):1480–90. doi: 10.1007/s00586-018-05872-4
47. Li M, Bai J, Wang S, Zhai Y, Zhang S, Li C, et al. Mean platelet volume and platelet distribution width serve as prognostic biomarkers in skull base chordoma: A retrospective study. BMC Cancer (2020) 20(1):988. doi: 10.1186/s12885-020-07497-7
48. Liang CA-O, Ma Y, Yong L, Yang C, Wang P, Liu X, et al. Y-box binding protein-1 promotes tumorigenesis and progression via the epidermal growth factor receptor/AKT pathway in spinal chordoma Cancer Science (2019) 110(1):166–79. doi: 10.1111/cas.13875
49. Luo P, Wang X, Zhou J, Li L, Jing Z. C-cbl and cbl-b expression in skull base chordomas is associated with tumor progression and poor prognosis. Human Pathology (2018) 74:129–34. doi: 10.1016/j.humpath.2017.12.019
50. Sommer J, Itani DM, Homlar KC, Keedy VL, Halpern JL, Holt GE, et al. Methylthioadenosine phosphorylase and activated insulin-like growth factor-1 receptor/insulin receptor: Potential therapeutic targets in chordoma. J Pathol (2010) 220(5):608–17. doi: 10.1002/path.2679
51. Shen Y, Li M, Xiong Y, Gui S, Bai J, Zhang Y, et al. Proteomics analysis identified ASNS as a novel biomarker for predicting recurrence of skull base chordoma. Frontiers in oncology (Switzerland). (2021) 2234–943X.
52. Thanindratarn P, Dean DC, Feng W, Wei R, Nelson SD, Hornicek FJ, et al. Cyclin-dependent kinase 12 (CDK12) in chordoma: Prognostic and therapeutic value. Eur Spine J (2020) 29(12):3214–28. doi: 10.1007/s00586-020-06543-z
53. Tian K, Ma J, Wang L, Wang K, Li D, Hao S, et al. Expression of cathepsin K in skull base chordoma. (US: World Neurosurgery) (2017) 1878–8769.
54. Wen H, Ma H, Li P, Zheng J, Yu Y, Lv G. Expression of far upstream element-binding protein 1 correlates with c-myc expression in sacral chordomas and is associated with tumor progression and poor prognosis. Biochem Biophys Res Commun (2017) 491(4):1047–54. doi: 10.1016/j.bbrc.2017.08.008
55. Zhai Y, Bai J, Wang S, Li M, Wang F, Li C, et al. Aberrant expression of extracellular signal-regulated kinase and 15-hydroxyprostaglandin dehydrogenase indicates radiation resistance and poor prognosis for patients with clival chordomas. World Neurosurg (2018) 115:e146–e51. doi: 10.1016/j.wneu.2018.03.216
56. Zhang S, Bai J, Li M, Zhai Y, Wang S, Liu Q, et al. Predictive value of transforming growth factor-α and ki-67 for the prognosis of skull base chordoma. World Neurosurg (2019) 129:e199–206. doi: 10.1016/j.wneu.2019.05.110
57. Zou MX, Peng AB, Lv GH, Wang XB, Li J, She XL, et al. Expression of programmed death-1 ligand (PD-L1) in tumor-infiltrating lymphocytes is associated with favorable spinal chordoma prognosis. American J Translational Res (2016) 8(7):3274–87.
58. Zou M-X, Zheng B-W, Liu F-S, Wang X-B, Hu J-R, Huang W, et al. The relationship between tumor-stroma ratio, the immune microenvironment, and survival in patients with spinal chordoma. Neurosurgery (2019) 85(6):E1095–E110. doi: 10.1093/neuros/nyz333
59. Zou MX, Lv GH, Wang XB, Huang W, Li J, Jiang Y, et al. Clinical impact of the immune microenvironment in spinal chordoma: Immunoscore as an independent favorable prognostic factor. Neurosurgery (2019) 84(6):E318–33. doi: 10.1093/neuros/nyy274
60. Longoni M, Orzan F, Stroppi M, Boari N, Mortini P, Riva P. Evaluation of 1p36 markers and clinical outcome in a skull base chordoma study. Neuro-Oncology (2008) 10(1):52–60. doi: 10.1215/15228517-2007-048
61. Akhavan-Sigari R, Gaab Mr Fau - Rohde V, Rohde V Fau - Abili M, Abili M Fau - Ostertag H, Ostertag H. Prognostic significance of immunohistochemical expression of VEGFR2 and iNOS in spinal chordoma. European Spine J (2014) 23(11):2416–22. doi: 10.1007/s00586-014-3417-5
62. Bettegowda C, Yip S, Jiang B, Wang W-L, Clarke MJ, Lazary A, et al. Prognostic significance of human telomerase reverse transcriptase promoter region mutations C228T and C250T for overall survival in spinal chordomas. Neuro-Oncology (2019) 21(8):1005–15. doi: 10.1093/neuonc/noz066
63. Boari N, Gagliardi F, Cavalli A, Gemma M, Ferrari L, Riva P, et al. Skull base chordomas: Clinical outcome in a consecutive series of 45 patients with long-term follow-up and evaluation of clinical and biological prognostic factors. J Neurosurg, (2016) 125(2):450–60. doi: 10.3171/2015.6.JNS142370
64. Chen C, Yang Hl Fau - Chen K-W, Chen Kw Fau - Wang G-L, Wang Gl Fau - Lu J, Lu J Fau - Yuan Q, Yuan Q Fau - Gu Y-P, et al. High expression of survivin in sacral chordoma. Medical oncology (Northwood, London, England), (2013) 30(2):529. doi: 10.1007/s12032-013-0529-4.
65. Chen H, Garbutt CC, Spentzos D, Choy E, Hornicek FJ, Duan Z. Expression and therapeutic potential of SOX9 in chordoma. Clinical cancer research: An official journal of the American Association for Cancer Research (US). (2017); pp. 1557–3265.
66. Chen KW, Yang Hl Fau - Lu J, Lu J Fau - Wang G-L, Wang Gl Fau - Ji Y-M, Ji Ym Fau - Wu G-Z, Wu Gz Fau - Zhu L-F, et al. Expression of vascular endothelial growth factor and matrix metalloproteinase-9 in sacral chordoma. J Neuro-Oncology, (2011) 101(3):357–63. doi: 10.1007/s11060-010-0263-0.
67. de Castro CV, Guimaraes G, Aguiar S Jr, Aguiar S Jr, Lopes A, Lopes A, et al. Tyrosine kinase receptor expression in chordomas: phosphorylated AKT correlates inversely with outcome. Human Pathology (2013) 44(9):1747–55. doi: 10.1016/j.humpath.2012.11.024
68. Ding Y, Bui MM, Wang Q, Sun X, Zhang M, Niu X, et al. ADAM10 is a potential novel prognostic biomarker for sacral chordoma. Annals of clinical and laboratory science (US). (2019) pp. 1550–8080.
69. Duan Z, Shen J, Yang X, Yang P, Osaka E, Choy E, et al. Prognostic significance of miRNA-1 (miR-1) expression in patients with chordoma. J Orthopaedic Res (2014) 32(5):695–701. doi: 10.1002/jor.22589
70. Tauziéde-Espariat A, Bresson D, Bresson D, Polivka M, Polivka M, Bouazza S, et al. Prognostic and therapeutic markers in chordomas: A study of 287 tumors. Journal of neuropathology and experimental neurology (2016) 75(2):111–20. doi: 10.1093/jnen/nlv010
71. Feng Y, Zhang Q, Wang Z, Yan B, Wei W, Li P. Overexpression of the BMP4/SMAD signaling pathway in skull base chordomas is associated with poor prognosis. International J Clin E Pathol (2015) 8(7):8268–75.
72. Gulluoglu S, Tuysuz EC, Sahin M, Yaltirik CK, Kuskucu A, Ozkan F, et al. The role of TNF-α in chordoma progression and inflammatory pathways. Cellular Oncology (Dordrecht) (2019) 42(5):663–77. doi: 10.1007/s13402-019-00454-y
73. Hobusch GM, Bodner F, Walzer S, Marculescu R, Funovics PT, Sulzbacher I, et al. C-reactive protein as a prognostic factor in patients with chordoma of lumbar spine and sacrum–a single center pilot study. World J Surgical Oncology (2016) 14:111. doi: 10.1186/s12957-016-0875-8
74. Horbinski C, Oakley GJ, Cieply K, Mantha GS, Nikiforova MN, Dacic S, et al. The prognostic value of ki-67, p53, epidermal growth factor receptor, 1p36, 9p21, 10q23, and 17p13 in skull base chordomas. Arch Pathol Lab Med (2010) 134(8):1170–6. doi: 10.5858/2009-0380-OA.1
75. Morimoto Y, Tamura R, Ohara K, Kosugi K, Oishi Y, Kuranari Y, et al. Prognostic significance of VEGF receptors expression on the tumor cells in skull base chordoma. J Neuro-Oncology (2019) 144(1):65–77. doi: 10.1007/s11060-019-03221-z
76. Naka T, Kuester D, Boltze C, Scheil-Bertram S, Samii A, Herold C, et al. Expression of hepatocyte growth factor and c-MET in skull base chordoma. Cancer: Interdiscip Int J Am Cancer Society (2008) 112(1):104–10. doi: 10.1002/cncr.23141
77. Otani R, Mukasa A, Shin M, Omata M, Takayanagi S, Tanaka S, et al. Brachyury gene copy number gain and activation of the PI3K/Akt pathway: Association with upregulation of oncogenic brachyury expression in skull base chordoma. J Neurosurg (2018) 128(5):1428–37. doi: 10.3171/2016.12.JNS161444
78. Rahmah NN, Sakai K, Nakayama J, Hongo K. Reversion-inducing cysteine-rich protein with kazal motifs and matrix metalloproteinase-9 are prognostic markers in skull base chordomas. Neurosurg review (2010) 33(2):167–73. doi: 10.1007/s10143-009-0228-y
79. Saad AG, Collins MH. Prognostic value of MIB-1, e-cadherin, and CD44 in pediatric chordomas. Pediatric Developmental Pathology (2005) 8(3):362–368. doi: 10.1007/s10024-005-1127-z
80. Tosuner Z, Bozkurt Su Fau - Kiliç T, Kiliç T Fau - Yilmaz B, Yilmaz B. The role of EGFR, hepatocyte growth factor receptor (c-met), c-ErbB2 (HER2-neu) and clinicopathological parameters in the pathogenesis and prognosis of chordoma. Turk Patoloji Dergisi (2017) 33(2):112–20. doi: 10.5146/tjpath.2016.01378
81. Wang B, Zhang K, Chen H, Lu J, Wu G, Yang H, et al. miR-1290 inhibits chordoma cell proliferation and invasion by targeting Robo1. Translational Cancer Res (2019) 8(2):542–51. doi: 10.21037/tcr.2019.03.13
82. Wei W, Wang K, Liu Z, Tian K, Wang L, Du J, et al. Radiomic signature: A novel magnetic resonance imaging-based prognostic biomarker in patients with skull base chordoma. Radiother Oncol (2019) 141:239–46. doi: 10.1016/j.radonc.2019.10.002
83. Yakkioui Y, Temel Y, Creytens D, Jahanshahi A, Fleischeuer R, Santegoeds RGC, et al. A comparison of cell-cycle markers in skull base and sacral chordomas. World Neurosurg (2014) 82(1-2):e311–e8. doi: 10.1016/j.wneu.2013.01.131
84. Zhang K, Chen H, Zhang B, Sun J, Lu J, Chen K, et al. Overexpression of raf-1 and ERK1/2 in sacral chordoma and association with tumor recurrence. Int J Clin Exp Pathol (2015) 8(1):608.
85. Zhou M, Chen K Fau - Yang H, Yang H Fau - Wang G, Wang G Fau - Lu J, Lu J Fau - Ji Y, Ji Y Fau - Wu C, et al. Expression of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in sacral chordoma. J Neuro-Oncology (2014) 116(1):77–82. doi: 10.1007/s11060-013-1274-4
86. Zhu GG, Ramirez D, Chen W, Lu C, Wang L, Frosina D, et al. Chromosome 3p loss of heterozygosity and reduced expression of H3K36me3 correlate with longer relapse-free survival in sacral conventional chordoma. Hum Pathol (2020) 104:73–83. doi: 10.1016/j.humpath.2020.07.002
87. Li Z, Lv T, Lv T, Liu Y, Liu Y, Huang X, et al. PARP1 is a novel independent prognostic factor for the poor prognosis of chordoma. Cancer biomarkers: Section A of Disease markers (US). (2016) pp. 1875–8592.
88. Wang Y, Chen K, Chen H, Zhang K, Lu J, Mao H, et al. Low expression of miRNA-1290 associated with local invasion and recurrence in sacral chordoma. Int J Clin Exp Pathol (2017) 10(11):10934.
89. Miksad RA, Zietemann V, Gothe R, Schwarzer R, Conrads-Frank A, Schnell-Inderst P, et al. Progression-free survival as a surrogate endpoint in advanced breast cancer. Int J Technol Assess Health Care (2008) 24(04):371–83. doi: 10.1017/S0266462308080495
90. Broglio KR, Berry DA. Detecting an overall survival benefit that is derived from progression-free survival. JNCI: J Natl Cancer Inst (2009) 101(23):1642–9. doi: 10.1093/jnci/djp369
91. Randolph KA, Myers LL. Basic statistics in multivariate analysis. (USA: Oxford University Press) (2013).
92. Hortobagyi GN, Smith TL, Legha SS, Swenerton KD, Gehan EA, Yap HY, et al. Multivariate analysis of prognostic factors in metastatic breast cancer. J Clin Oncol (1983) 1(12):776–86. doi: 10.1200/JCO.1983.1.12.776
93. Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res (2020) 10(3):727–42.
94. Gill CM, Fowkes M, Shrivastava RK. Emerging therapeutic targets in chordomas: A review of the literature in the genomic era. Clin Neurosurg: Oxford Univ Press (2020) p:E118–E23. doi: 10.1093/neuros/nyz342
95. Potla P, Ali SA, Kapoor M. A bioinformatics approach to microRNA-sequencing analysis. Osteoarthritis Cartilage Open (2021) 3(1):100131. doi: 10.1016/j.ocarto.2020.100131
96. Li F, Yang J Fau - Ramnath N, Ramnath N Fau - Javle MM, Javle Mm Fau - Tan D, Tan D. Nuclear or cytoplasmic expression of survivin: what is the significance. International J Cancer (US). (2005) pp. 0020–7136.
97. Barber SM, Sadrameli SS, Lee JJ, Fridley JS, Teh BS, Oyelese AA, et al. Chordoma-current understanding and modern treatment paradigms. J Clin Med (2021) 10(5):1054. doi: 10.3390/jcm10051054
98. Von Witzleben A, Goerttler LT, Marienfeld R, Barth H, Lechel A, Mellert K, et al. Preclinical characterization of novel chordoma cell systems and their targeting by pharmocological inhibitors of the CDK4/6 cell-cycle pathway. Cancer Res (2015) 75(18):3823–31. doi: 10.1158/0008-5472.CAN-14-3270
99. Ekumi KM, Paculova H, Lenasi T, Pospichalova V, Bösken CA, Rybarikova J, et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res (2015) 43(5):2575–89. doi: 10.1093/nar/gkv101
100. Naidoo K, Wai PT, Maguire SL, Daley F, Haider S, Kriplani D, et al. Evaluation of CDK12 protein expression as a potential novel biomarker for DNA damage response-targeted therapies in breast cancer. Mol Cancer Ther (2018) 17(1):306–15. doi: 10.1158/1535-7163.MCT-17-0760
101. Mobley BC, McKenney Jk, Bangs CD, Bangs Cd, Callahan K, Callahan K, et al. Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathologica (Germany) (2010) pp. 1432–0533.
102. Song Q, Peng M, Chu Y, Huang S. Techniques for detecting chromosomal aberrations in myelodysplastic syndromes. Oncotarget (2017) 8(37):62716–29. doi: 10.18632/oncotarget.17698
103. Chen Y, Yu H, Wu C, Li J, Jiao S, Hu Y, et al. Prognostic value of plasma d-dimer levels in patients with small-cell lung cancer. Biomed Pharmacother = Biomed Pharmacother (2016) 81:210–7. doi: 10.1016/j.biopha.2016.02.030
104. Kilic M, Yoldas O, Keskek M, Ertan T, Tez M, Gocmen E, et al. Prognostic value of plasma d-dimer levels in patients with colorectal cancer. Colorectal Dis (2008) 10(3):238–41. doi: 10.1111/j.1463-1318.2007.01374.x
105. Hu L, Li M, Ding Y, Pu L, Liu J, Xie J, et al. Prognostic value of RDW in cancers: A systematic review and meta-analysis. Oncotarget (2017) 8(9):16027–35. doi: 10.18632/oncotarget.13784
106. Furtado S, Dunogué B, Jourdi G, Chaigne B, Chibah A, Legendre P, et al. High d-dimer plasma concentration in systemic sclerosis patients: Prevalence and association with vascular complications. J Scleroderma Related Disord (2021) 6(2):178–86. doi: 10.1177/2397198320957558
107. He Q, Ding J, He S, Yu Y, Chen X, Li D, et al. The predictive value of procalcitonin combined with c-reactive protein and d dimer in moderately severe and severe acute pancreatitis. Eur J Gastroenterol Hepatol (2022) 34(7):744–50. doi: 10.1097/MEG.0000000000002376
Keywords: Chordoma, prognostic biomarkers, molecular, skull base, spine, systematic review, multivariate analysis
Citation: Rubino F, Alvarez-Breckenridge C, Akdemir K, Conley AP, Bishop AJ, Wang W-L, Lazar AJ, Rhines LD, DeMonte F and Raza SM (2022) Prognostic molecular biomarkers in chordomas: A systematic review and identification of clinically usable biomarker panels. Front. Oncol. 12:997506. doi: 10.3389/fonc.2022.997506
Received: 19 July 2022; Accepted: 12 September 2022;
Published: 29 September 2022.
Edited by:
Sebastien Froelich, Assistance Publique Hopitaux De Paris, FranceReviewed by:
Johannes Wach, University Hospital Bonn, GermanyCopyright © 2022 Rubino, Alvarez-Breckenridge, Akdemir, Conley, Bishop, Wang, Lazar, Rhines, DeMonte and Raza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaan M. Raza, c21yYXphQG1kYW5kZXJzb24ub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.