
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 17 October 2022
Sec. Thoracic Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.992596
This article is part of the Research Topic365 Days of Progress In Thoracic OncologyView all 6 articles
 Changqing Dong1†
Changqing Dong1† Wanwan Cheng2†
Wanwan Cheng2† Meiling Zhang3†
Meiling Zhang3† Si Li4
Si Li4 Lele Zhao4
Lele Zhao4 Dongsheng Chen4
Dongsheng Chen4 Yong Qin4
Yong Qin4 Mingzhe Xiao4
Mingzhe Xiao4 Shencun Fang2*
Shencun Fang2*Background: To evaluate the potential treatment for patients with non-small cell lung cancer (NSCLC) and rare malignant pulmonary lymphangitis carcinomatosis (PLC), our study provided a genomic profile and clinical outcome of this group of patients.
Methods: We retrospectively reviewed patients with NSCLC who developed PLC. The genomic alterations, tumor mutation burden (TMB), and microsatellite instability (MSI) based on DNA-based next-generation sequencing were reviewed and compared in a Chinese population with lung adenocarcinomas (Chinese-LUAD cohort). Clinical outcomes after exploratory anlotinib treatment and factors influencing survival are summarized.
Results: A total of 564 patients with stage IV NSCLC were reviewed, and 39 patients with PLC were included. Genomic profiling of 17 adenocarcinoma patients with PLC (PLC-LUAD cohort) revealed TP53, EGFR, and LRP1B as the three most frequently altered genes. EGFR was less mutated in PLC-LUAD than Chinese-LUAD cohort of 778 patients (35.3% vs. 60.9%, P = 0.043). BRIP1 was mutated more often in the PLC-LUAD cohort (11.8% vs. 1.8%, P= 0.043). Two patients presented with high tumor mutational burden (TMB-H, 10 mutations/MB). Combing alterations in the patient with squamous cell carcinoma, the most altered pathways of PLC included cell cycle/DNA damage, chromatin modification, the RTK/Ras/MAPK pathway and VEGF signaling changes. Fourteen of the participants received anlotinib treatment. The ORR and DCR were 57.1% and 92.9%, respectively. Patients achieved a median progression-free survival of 4.9 months and a median overall survival of 7 months. The adverse effects were manageable. In patients with adenocarcinoma, the mPFS (5.3 months vs. 2.6 months) and mOS (9.9 months vs. 4.5 months) were prolonged in patients receiving anlotinib treatment compared to those receiving other treatment strategies (P < 0.05).
Conclusion: Patients with PLC in NSCLC demonstrated distinct genetic alterations. The results improve our understanding of the plausible genetic underpinnings of tumorigenesis in PLC and potential treatment strategies. Exploratory anlotinib treatment achieved considerable benefits and demonstrated manageable safety.
Pulmonary lymphangitic carcinomatosis (PLC) is caused by lymphatic spreading of metastatic cancer cells in the lungs. Among malignant tumors, PLC accounts for 6%–8% of intrathoracic metastases (1). Patients harboring this rare type of cancer often exhibit a poor prognosis due to delayed diagnosis, poor health conditions, and resistance to various therapies (2). Therefore, improving the understanding of PLC and exploring efficient treatments are urgent and important for these patients.
Lung cancer is the second most commonly reported primary tumor and is associated with the development of PLC (2). Recently, targeted therapy and immunotherapy have brought significant improvements in the survival rates of patients with NSCLC (3), but observations of the clinical and molecular characteristics of lung cancer with malignant PLC are still scarce. Genomic profiling of gene alterations could provide a comprehensive view of a patient’s targetable mutations. Simultaneously, alterations to the pathways involved in PLC might also indicate the mechanisms involved in the development of PLC and potential therapeutic strategies.
Researchers have reported few evaluations of the efficacy and safety of regimens for PLC due to its relatively low prevalence and dispersed locations of primary tumors. Isolated studies included the assessment of the remission of PLC and disease control with hormonal therapy (4), chemotherapy (5) (6), antiangiogenic drugs, including apatinib (7), regorafenib (8), bevacizumab (9), cetuximab (10), and gefitinib (11, 12), and immunotherapy (13). These highlighted potential treatment strategies for patients who develop PLC and studies with detailed descriptions are warranted to guide the management of patients. Anlotinib has recently been demonstrated to suppress lymphangiogenesis and lymphatic metastasis in mouse models of lung adenocarcinoma (14). A summary of its efficacy and safety in clinical practice would provide an important reference for patients considering anlotinib.
In this study, we retrospectively reviewed patients with NSCLC who developed PLC and illustrated the full picture of the patients’ clinical characteristics, genomic alterations, and clinical outcomes. The results of our study may provide a comprehensive understanding of NSCLC with PLC and offer potential treatment options for this group of patients. Our preliminary results were presented at the annual meeting of the American Society of Clinical Oncology in 2021 (15).
We retrospectively reviewed patients who were treated at the Department of Respiratory Medicine of Nanjing Chest Hospital (The Affiliated Brain Hospital of Nanjing Medical University) from January 2018 to May 2021 (Figure 1). The inclusion criteria were metastatic or recurrent NSCLC (Stage IV) with a complete diagnosis report and confirmed metastasis of PLC.
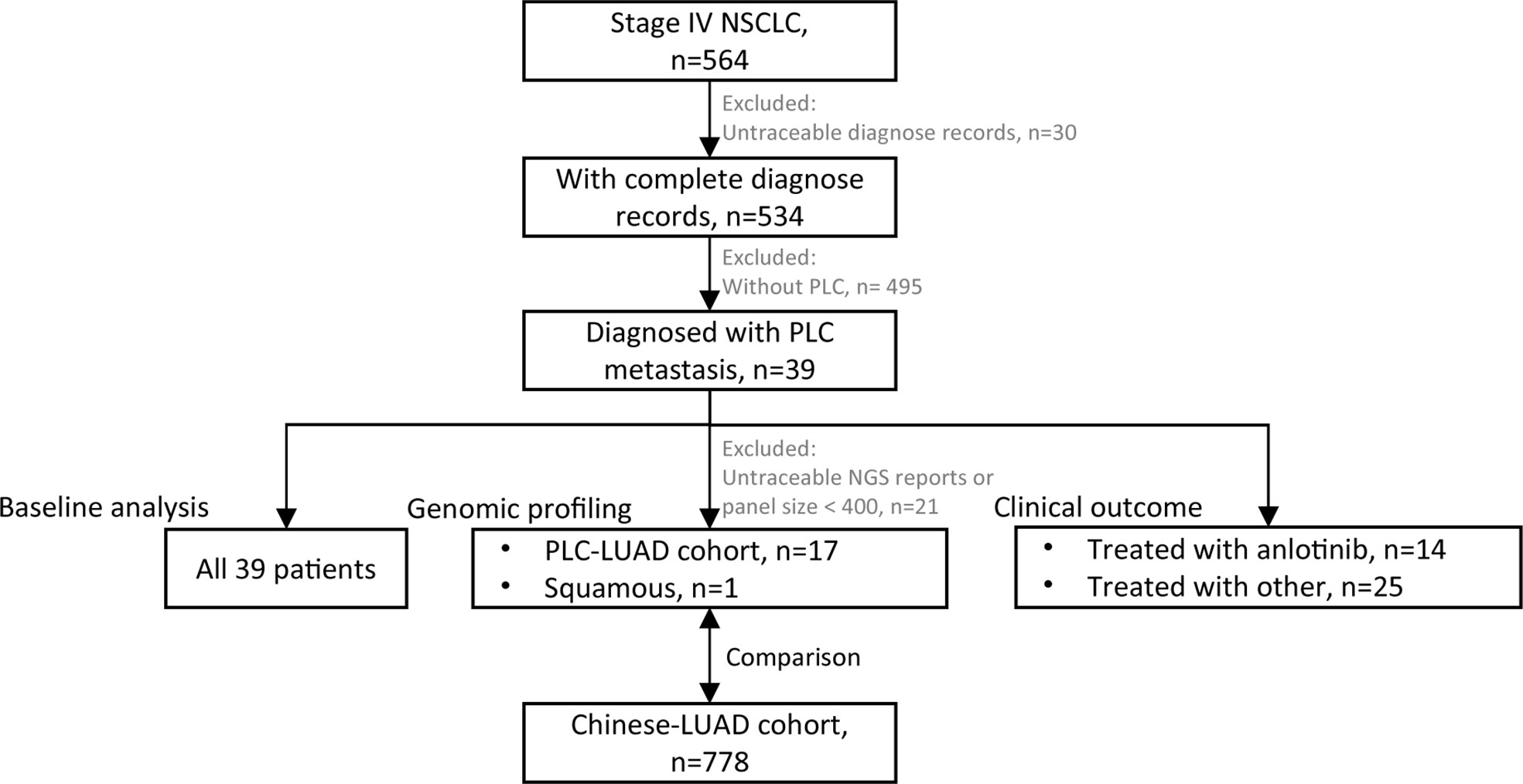
Figure 1 Flowchart of patient selection. 39 patients were enrolled for baseline analysis; 18 patients (17 of PLC-LUAD cohort, 1 squamous) with traceable DNA-based next generation sequencing (NGS, panel size > 400 genes) reports were enrolled for genomic profiling, and that of PLC-LUAD was compared to lung adenocarcinoma in Chinese population (Chinese-LUAD cohort); 14 patients treated with anlotinib and 25 with other treatment strategies were enrolled for clinical outcome evaluation.
The following information on the enrolled patients was collected for baseline analysis: age, sex, Karnofsky performance scale (KPS) score, smoking history, histological subtype, sites of metastatic disease, site of PLC, and mutation status of EGFR. The Research Ethics Committee of Nanjing Chest Hospital approved this study.
Among all patients, 18 patients previously diagnosed with DNA-based next-generation sequencing (NGS) with a panel size of > 400 genes (see Supplementary Table S1 for a list of the panels and genes) were enrolled for genomic analysis. The samples included 11 tumor tissues from biopsy specimens, seven plasma specimens, and one cerebrospinal fluid specimen, all of which were obtained before treatment. To illustrate the common genomic profile, 274 intersecting genes were identified (Table S1). Alteration types [single nucleotide variation (SNV)/insertion and deletion (InDel)/copy number variation (CNV)/intron/rearrangement] and the frequencies of the intersecting 274 genes, TMB (tumor mutation burden, nonsynonymous), and microsatellite instability (MSI) were reviewed.
Alterations found in 778 patients with lung adenocarcinoma in a Chinese population (Chinese-LUAD cohort) were extracted and compared to the alterations found in the PLC-LUAD cohort. From January 1, 2019, to December 31, 2020, tumor or plasma samples collected from the 778 patients with lung adenocarcinoma were analyzed with a panel-based NGS assay (539 genes) at the College of American Pathologists-certified laboratory in Jiangsu Simcere Diagnostics Co., Ltd. Genomic DNA (gDNA) of the sample tissues was extracted using a tissue sample DNA extraction kit (Kai Shuo). Genomic DNA from leucocytes and cell-free DNA (cfDNA) in plasma were extracted with a MagMAXTM DNA Multi-Sample Ultra kit (Thermo Fisher). Following the manufacturer’s instructions, extraction procedures were performed. The probe hybridization capture method was used for gene library construction. Commercial reagents and customized probes were used for library construction and hybridization capture. In brief, 15–200 ng of gDNA was sheared into 200–350 bp fragments with fragmentation enzymes. Base calls obtained with the Illumina NovaSeq6000 were converted to FASTQ files. The fastp tool (v.2.20.0) was used for adapter trimming and filtering low-quality bases (16). Duplicate reads obtained by PCR were excluded using Dedup with Error Correct. SNVs/InDels were called and annotated via VarDict (v.1.5.7) and InterVar (17, 18), and then, the variants were filtered against common SNPs in public databases, including the 1,000 Genome Project (Aug 2015) and Exome Aggregation Consortium Browser28 (v.0.3). Next, CNVs and fusions were analyzed with a CNV kit (dx1.1) and factera (v1.4.4) (19) (20), respectively. Alterations in SNVs, InDels, CNVs, introns, and rearrangements were extracted from the Chinese-LUAD cohort and compared with the LUAD-PLC cohort.
Fourteen patients received anlotinib-based treatment (the Anlotinib group), and the other 25 patients received other stratifications based on clinical practice (the Other group) (baseline comparison in Table S2). The response, survival, and safety data for patients treated with anlotinib were assessed. Anlotinib was administered orally at a dose of 12 mg daily. Responses to anlotinib were evaluated after 3 to 4 weeks of initial administration and during the regular follow-up visits (every 1–2 months), according to the Response Evaluation Criteria in Solid Tumors (RECIST, v1.1). Next, the progression-free survival (PFS) rate, overall survival (OS) rate, and objective response rate (ORR) were collected. The toxicities were assessed according to the Common Terminology Criteria for Adverse Events. PFS and OS were collected for the Other group.
Fisher’s Exact Test and Wilcoxon rank sum test were used for comparisons of categorical and continuous variables, respectively. Survival data were presented in Kaplan-Meier curves and compared using the log-rank test. The impact of factors on PFS and OS rates were evaluated using univariable COX regression analysis. All the statistical analyses were conducted with R version 4.0.3 (http://www.r-project.org). A two-sided P < 0.05 was considered statistically significant.
A total of 564 patients with stage IV NSCLC were reviewed, and 39 patients were eventually enrolled with confirmed metastasis of PLC (Figure 1). The patients’ baseline characteristics are summarized in Table 1. The median age was 63 years, with a range of 30–80 years. Twenty-six (66.7%) patients were male. The median (range) KPS was 70 (range 40–90). Twenty-one (53.8%) patients presented with a history of smoking, and 35 (89.7%) were diagnosed with adenocarcinoma. The most frequent metastatic sites were the bones (25; 64.1%) and lungs (18; 46.2%). PLC lesions were predominantly observed in the right lung (34; 87.3%).
Eighteen patients were enrolled for genomic profiling (Table 1), and the landscape of 17 patients in PLC-LUAD is given in Figure 2. Overall, 80 alterations were discovered in PLC-LUAD cohort. TP53 (10/17; 58.8%), EGFR (6/17; 35.3%), and LRP1B (3/17; 17.6%) were the most commonly altered genes. A total of 76.5% of patients harbored driver gene mutations, including those in EGFR (35.3%), KRAS (11.8%), NTRK3 (5.9%), BRAF (5.9%), ERBB2 (5.9%), and MET (5.9%), as well as fusions in ALK (5.9%) and ROS1 (5.9%). Compared to the alterations in the Chinese-LUAD cohort (Figure S1), mutations in EGFR were less often observed in patients with PLC (35.3% vs. 60.9%; P = 0.043). Mutations of the BRIP1 gene were more commonly observed in PLC (11.8% vs. 1.8%, P = 0.043). The remaining most frequently altered genes in the PLC-LUAD cohort were involved in the cell cycle/DNA damage pathway (64.7%), chromatin modifying pathway (17.6%), RTK/Ras/MAPK pathway (11.8%), and VEGF signaling pathway (11.8%). One patient with squamous cell carcinoma (Patient 33) harbored alterations in TP53, LRP1B, ATM and VEGFA, with a TMB of 2.87 mutations/MB and MSI-S.
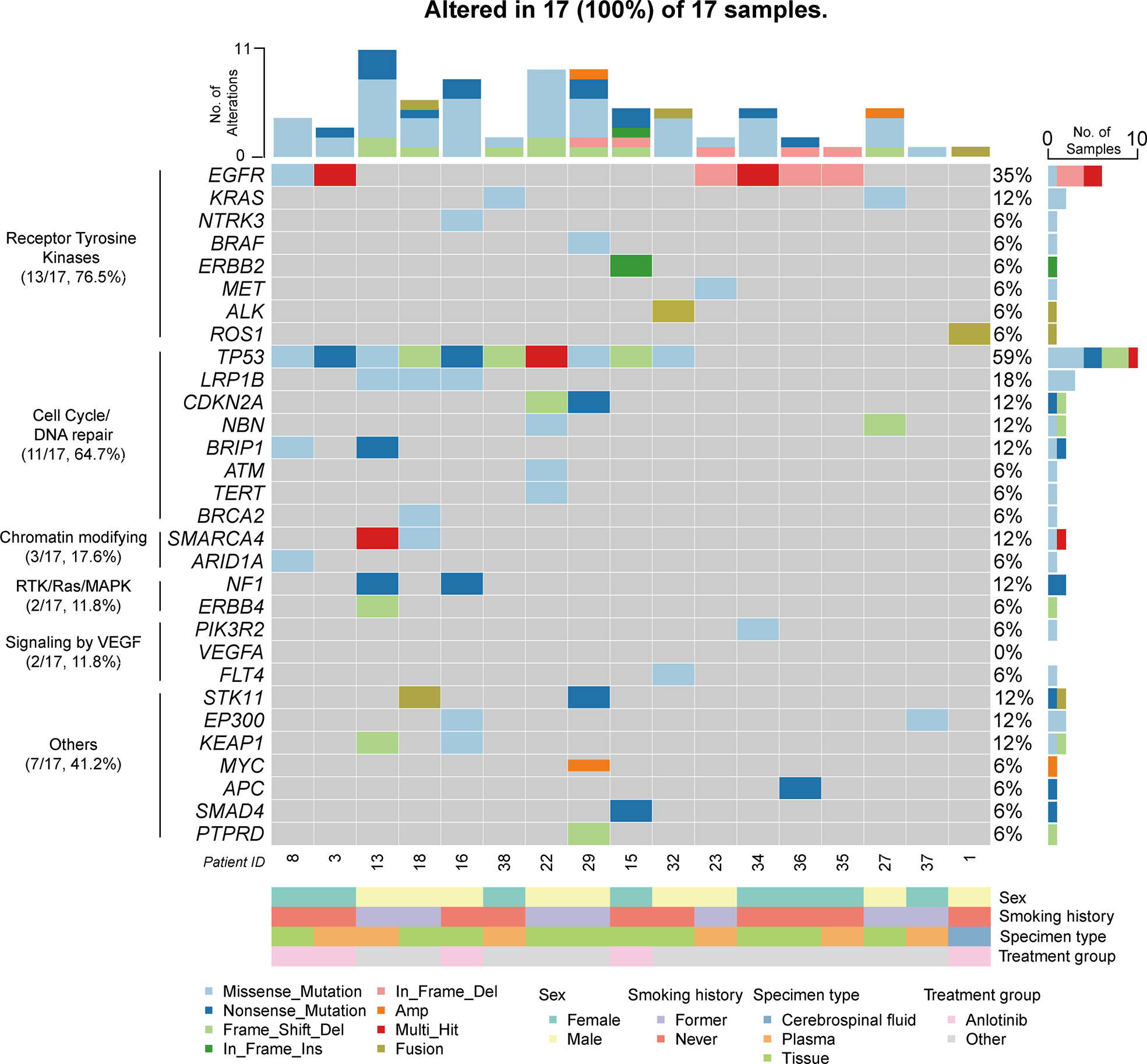
Figure 2 Genomic profiling of PLC-LUAD cohort (17 patients). Alterations presented here included single nucleotide variation/insertion and deletion/copy number variation/fusion. Top 30 altered genes were illustrated.
The mean and median TMB of the PLC-LUAD cohort were 5.2 mutations/MB and 3.8 mutations/MB, respectively. Two patients (patients 13 and 16; 11.8%) demonstrated a TMB ≥ 10 mutations/MB (which was TMB-H). MSI was available in 15 patients, and none of these were considered microsatellite instability-high (MSI-H). The frequencies of TMB-H and MSI-H were comparable to those of the Chinese-LUAD cohort (TMB-H, 10%; MSI-H, 0.3%; P > 0.5).
Detailed baseline and clinical outcomes in the Anlotinib group are shown in Table 2. In all patients, the objective response rate (ORR) was 57.1% (8/14), and the disease control rate was 92.9% (13/14). The median PFS (mPFS) was 4.9 months, and the median OS (mOS) was 7 months (Figures 3 A, B). More specifically, patients with adenocarcinoma and squamous cell carcinoma had a mPFS of 5.3 months and 1.5 months, respectively (Figure S2A). The mOS of these patients were 9.9 months and 3.3 months, respectively (Figure S2B). The results of a univariable Cox regression analysis in 12 patients with adenocarcinoma (Table 3) revealed that patients with a favorable KPS (≥ 60) demonstrated longer mOS [13.2 months vs. 5.2 months, P = 0.013; (Table 3; Figure 4A)].
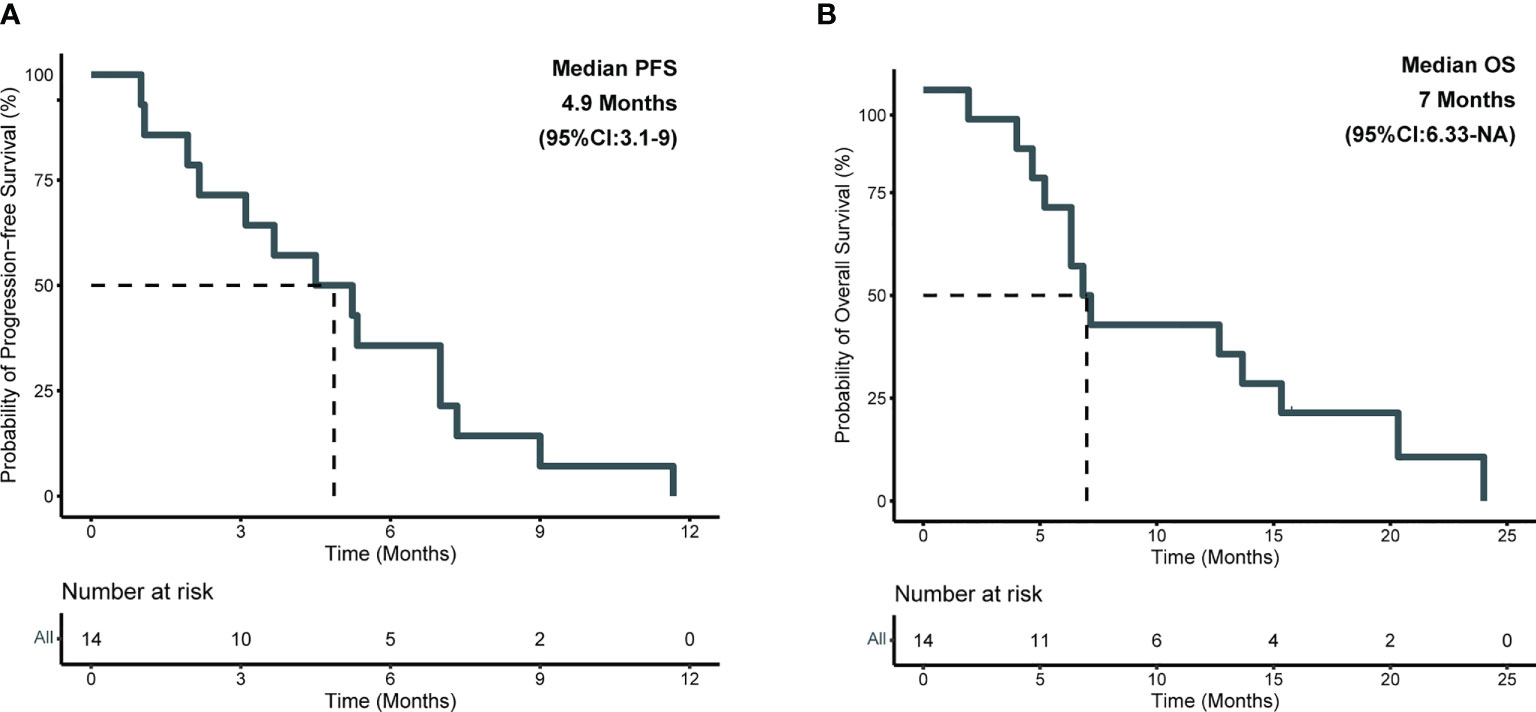
Figure 3 Kaplan-Meier survival analysis in anlotinib group. (A) Plots of progression-free survival (PFS); (B) Plots of overall survival (OS).
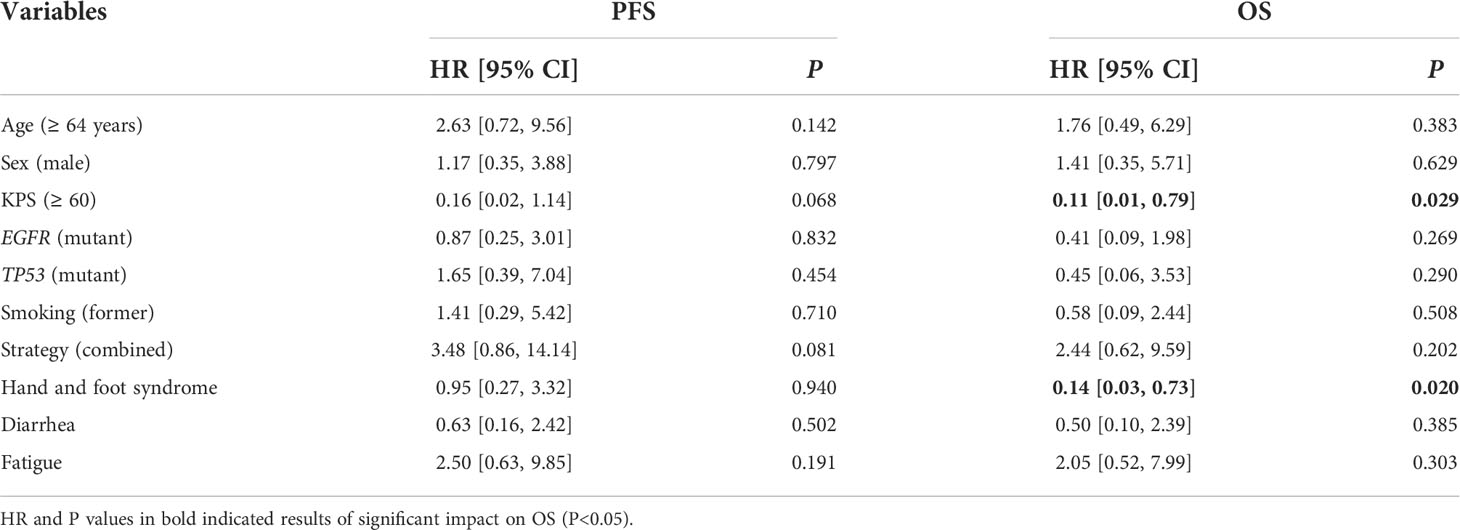
Table 3 Results of univariable COX regression analysis of risk factors for PFS and OS in 12 patients with adenocarcinoma treated with anlotinib.
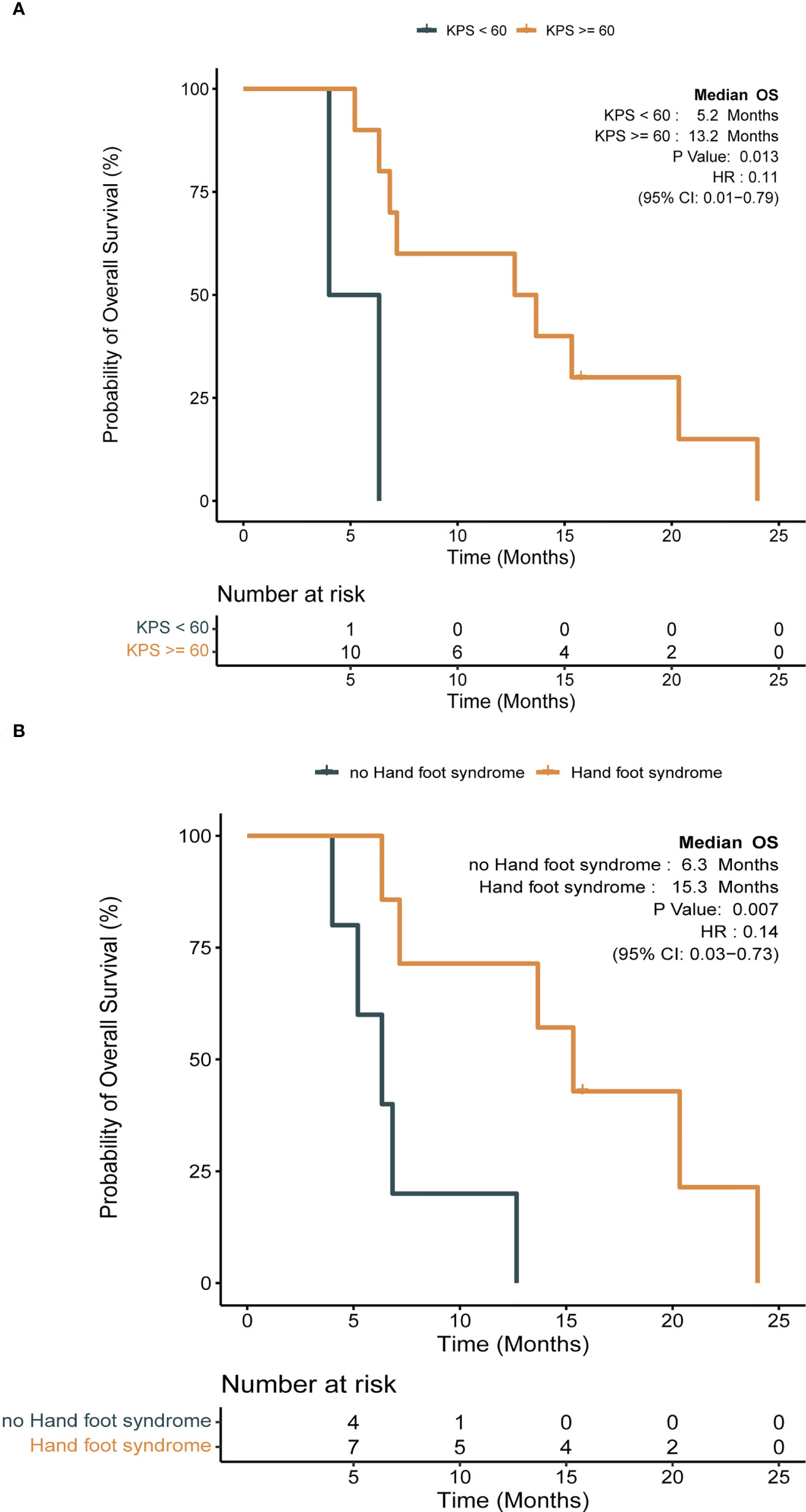
Figure 4 Impact of clinical factors on survival with Kaplan-Meier survival analysis in anlotinib group (log-rank test, P < 0.05). (A) Impact of Karnofsky performance scale (KPS) on OS; (B) impact of hand and foot syndrome (HFS) on OS.
Among 14 patients, the most frequently reported AEs were hypertension (10; 71.4%), hand foot syndrome (7; 50.0%) (HFS), fatigue (6; 42.9%), diarrhea (5; 35.7%), and hoarseness (3; 21.4%). An event of grade 3 was reported in 1 patient (7.1%) who presented with hypertension, and he continued treatment by reducing the dosage to 10 mg/day. Patients who presented with HFS during anlotinib treatment tended to demonstrate superior mOS [15.3 months vs. 6.3 months, P = 0.007; (Table 3; Figure 4B)].
Compared to the Other group of patients with adenocarcinoma, those in the Anlotinib group showed prolonged mPFS and mOS (mPFS, 5.3 months vs. 2.6 months, P = 0.032; mOS, 9.9 months vs. 4.5 months, P = 0.005; Figure S3). There were four patients with squamous cell carcinoma in total (Anlotinib group, n = 2; Other group, n = 2). The PFS was 1 and 1.93 months in the Anlotinib group and 1.4 and 3.3 months in the Other group; the OS was 1.93 and 4.67 months (Anlotinib group) and 5.5 and 6 months (Other group), respectively (Table 2; Supplementary Data of Clinical and Treatment). Comparisons in patients with squamous cell carcinoma were not performed due to limited sample size.
A 79-year-old female nonsmoker (patient 12) was diagnosed with lung adenocarcinoma, and chest computed tomography (CT) revealed a mass in the left inferior lobe with diffuse carcinomatous lymphangitis in both lungs (Figure 5A). A microscopic image that could be used for making a pathological diagnosis of PLC is presented Figure 5B. DNA sequencing of primary tumor specimens revealed a TP53 mutation. Then, the patient was treated with pemetrexed plus carboplatin chemotherapy as the first-line treatment. One week after chemotherapy, the patient developed refractory respiratory failure, accompanied by rapid progression of PLC in both lungs (Figure 5C). Then, anlotinib was initiated as salvage treatment. Dyspnea was significantly relieved after 3 days of treatment. The PLC in both lungs virtually disappeared after 1 month of treatment with anlotinib (Figure 5D), and the efficacy was evaluated as a partial response (PR). Furthermore, a good response in PLC was maintained for more than 7 months.
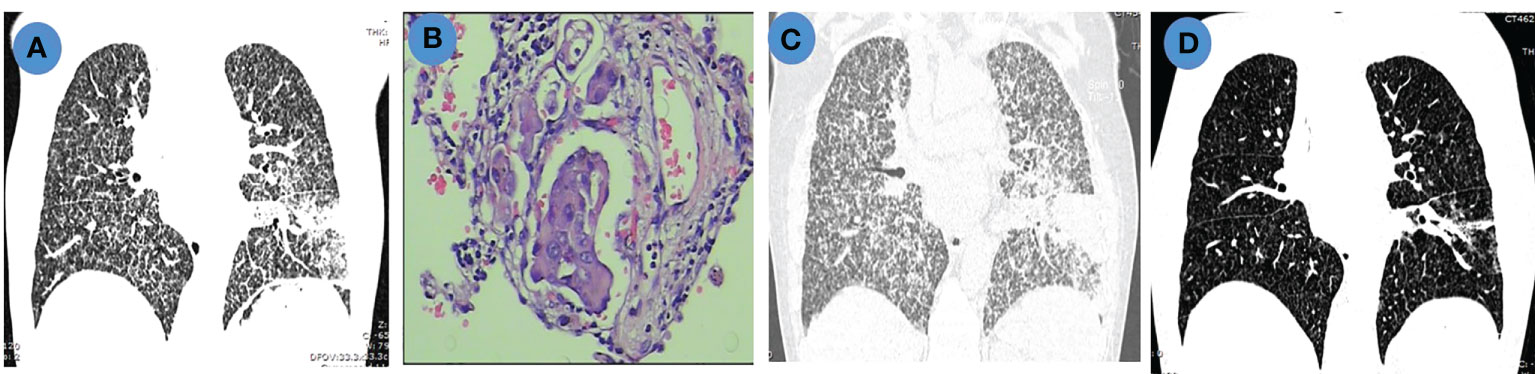
Figure 5 Chest imaging and pathological features in a patient with PLC. (A). Chest CT scan shows pulmonary mass with diffuse PLC before chemotherapy; (B). Hematoxylin and eosin (H&E) staining (200×) of lymphatic tumor embolus; (C). Chest CT scan shows progressive disease (PD) after chemotherapy; (D). Chest CT scan shows partial remission (PR) after anlotinib treatment.
PLC is a rare type of intrathoracic metastasis of malignant tumors that leads to a poor prognosis. Our study aimed to provide full clinical and genomic profiles for patients with NSCLC and PLC, and we evaluated the outcomes of anlotinib treatment in clinical practice.
In our study, 7.3% (39/534) of patients with metastatic NSCLC were diagnosed with PLC. This confirms its low incidence, as reported previously (2). The molecular features of these patients have not been reviewed previously. Our genomic profiling revealed that most patients harbored driver gene alterations (76.5%; Figure 2), and 11.8% of the patients presented with TMB-H. These patients might benefit from targeted therapy and immunotherapy. Among the mutations in the driver genes, EGFR mutations were observed less often in the PLC-LUAD cohort, leaving fewer chances to benefit from EGFR TKIs in these patients. BRCA1 interacting protein C-terminal helicase 1 (BRIP1) mutations occurred more frequently in the cohort in our study compared with the Chinese-LUAD cohort. BRIP1 is a homologous recombination-related gene, and previous reports included BRIP1 as a risk gene for ovarian cancer (21). Additionally, BRIP1 promotes cell proliferation and invasion in breast cancer (22). Next, in small-cell lung cancer, a patient harboring a germline mutation of BRIP1 achieved a notable disease response to DNA repair-targeted therapies (23). These may present an opportunity for therapeutic interventions, similar to other HR defects for patients with BRIP1 mutations. Additionally, BRIP1 tends to be coexpressed with PD-L1 in lung adenocarcinoma (24). It mainly participates in the immune response and cell activation. Additionally, this might provide complementary biomarkers for predicting responses to PD-1/PD-L1 inhibitors.
Of note, VEGF signaling (through the VEGFA, FLT4, and PIK3R2 genes) was one of the most frequently altered pathways in the PLC-LUAD cohort (11.8%, 5% in the Chinese-LUAD cohort, P = 0.22), and the patients with squamous cell carcinoma also harbored mutations in VEGFA. VEGF signaling is best known for its involvement in the development and maintenance of the blood and lymphatic vascular systems (25). Specifically, growth factor-C (VEGFC), whose receptor is encoded by FLT4, promotes the intralymphatic spread of metastases in the lung, thus inducing lymphangitic carcinomatosis (25–27). These results could provide important explanations for the development of PLC in our patients and, more importantly, suggest potential therapeutic approaches. Similarly, in a study of lung adenocarcinoma mouse models (14), anlotinib inhibited the growth and migration of human lymphatic endothelial cells and lymphangiogenesis in vitro and in vivo, probably through the inactivation of VEGFR-3 phosphorylation (28). This study indicated that anlotinib may be a promising treatment for PLC; however, its clinical outcomes still require systematic evaluation.
In this study, anlotinib had a mPFS of 4.9 months and a mOS of 7 months (Figure 3). More specifically, patients with adenocarcinoma achieved a mPFS of 5.3 months and a mOS of 9.9 months (Figure S2). Compared to the median survival of 2 months between symptom identification and death in the review by Klimek et al. (2), anlotinib showed preliminary improvements in survival benefits. This was also indicated by the improvements in the mPFS and mOS in the Anlotinib group compared to those in the Other group (Figure S3). Moreover, after reviewing previously reported cases (11–13, 29, 30), we calculated an ORR of 50% in gefitinib-treated patients with EGFR mutations (12), which was 100% in our study (Table 2). Apatinib-based treatment was reported to demonstrate a DCR of 69.2%, mPFS of 5.2 months, and mOS of 10.2 months (7) when not distinguishing patients’ baseline characteristics, treatment lines and strategies. A more detailed description might be important when evaluating the efficacy and treatment adoption for patients. In our results, a superior survival benefit was observed in patients with favorable KPS (mOS, 13.2 months), which was not inferior to patients treated with anlotinib with third-line treatment in the ALTER0303 study (PS, 0-1; mPFS, 5.4 months; mOS, 9.6 months) (31). This highlighted the benefit of anlotinib. Furthermore, we observed significantly longer OS (15.3 vs. 6.3 months, P = 0.007) in patients with HFS, which was in line with the ALTER0303 study (32). Inhibition of the VEGF pathway is widely discussed as the cause of HFS (33, 34), but the pathophysiology needs to be clarified. In summary, anlotinib might a potential regimen for treating PLC, especially in those patients with favorable KPS. The occurrence of HFS during treatment might predict superior efficacy.
Certainly, this study has intrinsic limitations. In this retrospective analysis, the samples collected and sequencing platforms were heterogeneous, which might introduce information bias on the results of genomic alterations. Furthermore, due to the lack of sequencing results in patients without PLC, we compared the profile of the PLC cohort with the whole population (LUAD cohort). This might have merged the difference between patients with and without PLC. Additionally, a larger sample size study might illustrate the alteration landscape more accurately. The limited number of evaluated patients and missing date due to the retrospective design also made it challenging to provide a comprehensive assessment of the clinical outcome of anlotinib and other strategies. This needs further verification.
In conclusion, our study provided preliminary clinical and genomic profiles for NSCLC with PLC. The landscape of common driver genes revealed that most patients were candidates for targeted therapy. Analysis of the TMB and distinct mutations shed light on potential immunotherapy approaches and might simultaneously enhance the understanding of the tumorigenesis of PLC. Exploratory anlotinib treatment achieved considerable benefits and manageable safety. Further studies are warranted to validate these findings.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by The Research Ethics Committee of Nanjing Chest Hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Conception and design: SF. Administrative support: MX, YQ. Provision of study materials or patients: CD, WC, MZ. Data analysis and interpretation: SL, LZ. All authors contributed to the article and approved the submitted version.
This study was supported by the “Six one projects” in Jiangsu Province (LGY2019006).
We thank Qin Zhang, Yaqing Wu, Yangyang Yu, and Chuang Qi from Jiangsu Simcere Diagnostics for their kindly assistance.
Authors SL, LZ, DC, YQ and MX were employed by the company Jiangsu Simcere Diagnostics Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.992596/full#supplementary-material
Supplementary Figure 1 | Genomic profiling of Chinese-LUAD cohort (778 patients). Alterations presented here included single nucleotide variation/insertion and deletion/copy number variation, etc.. Genes were listed according to the 30 top altered genes in PLC-LUAD cohort.
Supplementary Figure 2 | Kaplan-Meier survival analysis in patients with adenocarcinoma and squamous treated by anlotinib. (A) Plots of progression-free survival (PFS); (B) Plots of overall survival (OS).
Supplementary Figure 3 | Kaplan-Meier survival analysis and comparison of survival between group of anlotinib and other in patients with adenocarcinoma (log-rank test, P < 0.05). (A) Plots of progression-free survival (PFS); (B) Plots of overall survival (OS).
1. Bruce DM, Heys SD, Eremin O. Lymphangitis carcinomatosa: A literature review. J R Coll Surg Edinb (1996) 41:7–13.
2. Klimek M. Pulmonary lymphangitis carcinomatosis: Stematic review and meta-analysis of case reports, 1970-2018. Postgraduate Med (2019) 131:309–18. doi: 10.1080/00325481.2019.1595982
3. Yang C-Y, Yang JC-H, Yang P-C. Precision management of advanced non-small cell lung cancer. Annu Rev Med (2020) 71:117–36. doi: 10.1146/annurev-med-051718-013524
4. Fukuda M, Takashima H, Fuse H, Hirano S. [Prostatic cancer with multiple pulmonary metastases treated successfully with hormonal therapy: A case report]. Hinyokika Kiyo (2002) 48:499–502.
5. Watanabe T, Kawabata H, Takase I, Murata A, Nakajima H. [A case of advanced gastric cancer with lymphangitis carcinomatosa of the lung, successfully treated with paclitaxel and TS-1]. Gan To Kagaku Ryoho (2003) 30:849–53.
6. Fumet J-D, Wickre M, Jacquot J-P, Bizollon M-H, Melis A, Vanoli A, et al. Successfully treatment by eribulin in visceral crisis: A case of lymphangitic carcinomatosis from metastatic breast cancer. BMC Cancer (2018) 18:839. doi: 10.1186/s12885-018-4725-7
7. Yang P. P46 experience in the treatment of 13 cases of pulmonary lymphangitic carcinomatosis with apatinib. J Thorac Oncol (2018) 13:S178. doi: 10.1016/j.jtho.2018.07.085
8. Takeuchi N, Koike K, Yoshida S, Fujiwara M. Efficacy of regorafenib in acute pulmonary carcinomatous lymphangitis as a manifestation of rectal cancer: A case report. Oncol Lett (2019) 18:6469–6474. doi: 10.3892/ol.2019.11043
9. Aman J, Thunnissen E, Paul MA, van Nieuw Amerongen GP, Vonk-Noordegraaf A. Successful treatment of diffuse pulmonary lymphangiomatosis with bevacizumab. Ann Intern Med (2012) 156:839. doi: 10.7326/0003-4819-156-11-201206050-00016
10. Toshima H, Ikusue T, Hisamatsu A, Kobayashi K, Shimada K. Two cases of lymphangitic carcinomatosis as the primary symptom of colorectal carcinoma that achieved complete remission using combination anti-EGFR antibody therapy. OTT (2019) 12:2089–93. doi: 10.2147/OTT.S194224
11. Fujiwara K, Kiura K, Ueoka H, Tabata M, Hamasaki S, Tanimoto M. Dramatic effect of ZD1839 ('Iressa’) in a patient with advanced non-small-cell lung cancer and poor performance status. Lung Cancer (2003) 40:73–6. doi: 10.1016/s0169-5002(03)00028-x
12. Naito T, Hasegawa H, Asada K, Suda T, Chida K. Lymphangitis carcinomatosis as a potential predictor for a response to gefitinib. Clin Oncol (R Coll Radiol) (2006) 18:573–4. doi: 10.1016/j.clon.2006.05.004
13. Yamasaki M, Funaishi K, Kawamoto K, Matsumoto Y, Matsumoto N, Taniwaki M, et al. Platinum-doublet chemotherapy followed by pembrolizumab therapy for lung cancer with lymphangitis carcinomatosa mimicking interstitial pneumonitis: A case report. Med (Baltimore) (2019) 98:e16834. doi: 10.1097/MD.0000000000016834
14. Qin T, Liu Z, Wang J, Xia J, Liu S, Jia Y, et al. Anlotinib suppresses lymphangiogenesis and lymphatic metastasis in lung adenocarcinoma through a process potentially involving VEGFR-3 signaling. Cancer Biology & Medicine 2020 17:15. doi: 10.20892/j.issn.2095-3941.2020.0024
15. Fang S, Cheng W, Zhang Y, Zhang H, Li S, Xiao M. Successful treatment of pulmonary lymphangitic carcinomatosis by anlotinib in 14 Chinese patients with advanced non-small cell lung cancer. JCO (2021) 39:e21064–4. doi: 10.1200/JCO.2021.39.15_suppl.e21064
16. Hwang K-B, Lee I-H, Li H, Won D-G, Hernandez-Ferrer C, Negron JA, et al. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci Rep (2019) 9:3219. doi: 10.1038/s41598-019-39108-2
17. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res (2016) 44:e108. doi: 10.1093/nar/gkw227
18. Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet (2017) 100:267–80. doi: 10.1016/j.ajhg.2017.01.004
19. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PloS Comput Biol (2016) 12:e1004873. doi: 10.1371/journal.pcbi.1004873
20. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics (2014) 30:3390–3. doi: 10.1093/bioinformatics/btu549
21. Suszynska M, Ratajska M, Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: Mutation prevalence and precise risk estimates based on a pooled analysis of ~30,000 cases. J Ovarian Res (2020) 13:50. doi: 10.1186/s13048-020-00654-3
22. Rizeq B, Sif S, Nasrallah GK, Ouhtit A. Novel role of BRCA1 interacting c-terminal helicase 1 (BRIP1) in breast tumour cell invasion. J Cell Mol Med (2020) 24:11477–88. doi: 10.1111/jcmm.15761
23. Tlemsani C, Takahashi N, Pongor L, Rajapakse VN, Tyagi M, Wen X, et al. Whole-exome sequencing reveals germline-mutated small cell lung cancer subtype with favorable response to DNA repair-targeted therapies. Sci Transl Med (2021) 13:eabc7488. doi: 10.1126/scitranslmed.abc7488
24. Wu Y, Lin L, Liu X. Identification of PDL1-related biomarkers to select lung adenocarcinoma patients for PD1/PDL1 inhibitors. Dis Markers (2020) 2020:7291586. doi: 10.1155/2020/7291586
25. Karaman S, Leppänen V-M, Alitalo K. Vascular endothelial growth factor signaling in development and disease. Development (2018) 145:dev151019. doi: 10.1242/dev.151019
26. Das S, Ladell DS, Podgrabinska S, Ponomarev V, Nagi C, Fallon JT, et al. Vascular endothelial growth factor-c induces lymphangitic carcinomatosis, an extremely aggressive form of lung metastases. Cancer Res (2010) 70:1814–24. doi: 10.1158/0008-5472.CAN-09-3675
27. Matsumoto M, Roufail S, Inder R, Caesar C, Karnezis T, Shayan R, et al. Signaling for lymphangiogenesis via VEGFR-3 is required for the early events of metastasis. Clin Exp Metastasis (2013) 30:819–32. doi: 10.1007/s10585-013-9581-x
28. Otrock ZK, Makarem JA, Shamseddine AI. Vascular endothelial growth factor family of ligands and receptors: Review. Blood Cells Molecules Dis (2007) 38:258–68. doi: 10.1016/j.bcmd.2006.12.003
29. Suzuki E, Tanahashi M, Yukiue H, Yoshii N, Shitara M, Fujino T, et al. [A patient with lung adenocarcinoma, lymphangitis carcinomatosa, and multiple bone metastases who achieved long-term survival after successful treatment with carboplatin, paclitaxel, and bevacizumab]. Gan To Kagaku Ryoho (2016) 43:617–20.
30. Natsume M, Honda T, Haruyama T, Ishihara M, Fukasawa Y, Sakamoto T, et al. A case of lung adenocarcinoma with marked improvement of pulmonary lymphangitic carcinomatosis by adding bevacizumab to cisplatin and pemetrexed. Case Rep Oncol (2017) 10:1065–9. doi: 10.1159/000484662
31. Han B, Li K, Wang Q, Zhang L, Shi J, Wang Z, et al. Effect of anlotinib as a third-line or further treatment on overall survival of patients with advanced non–small cell lung cancer: The ALTER 0303 phase 3 randomized clinical trial. JAMA Oncol (2018) 4:1569. doi: 10.1001/jamaoncol.2018.3039
32. Nan X, Xie C, Zhu Q, Zhang J, Fu S, Han X, et al. Hand-foot syndrome and survival in patients with advanced non-small-cell lung cancer receiving anlotinib: a subgroup analysis of data from the ALTER 0303 study. Int J Clin Oncol (2020) 25:1492–8. doi: 10.1007/s10147-020-01683-0
33. Azad NS, Aragon-Ching JB, Dahut WL, Gutierrez M, Figg WD, Jain L, et al. Hand-foot skin reaction increases with cumulative sorafenib dose and with combination anti-vascular endothelial growth factor therapy. Clin Cancer Res (2009) 15:1411–6. doi: 10.1158/1078-0432.CCR-08-1141
Keywords: pulmonary lymphangitic carcinomatosis (PLC), non-small cell lung cancer (NSCLC), genomic profiling, anlotinib, survival
Citation: Dong C, Cheng W, Zhang M, Li S, Zhao L, Chen D, Qin Y, Xiao M and Fang S (2022) Genomic profiling of non-small cell lung cancer with the rare pulmonary lymphangitic carcinomatosis and clinical outcome of the exploratory anlotinib treatment. Front. Oncol. 12:992596. doi: 10.3389/fonc.2022.992596
Received: 12 July 2022; Accepted: 30 September 2022;
Published: 17 October 2022.
Edited by:
Luciano Mutti, Temple University, United StatesCopyright © 2022 Dong, Cheng, Zhang, Li, Zhao, Chen, Qin, Xiao and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shencun Fang, RmFuZzE5ODRAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.