- Department of Obstetrics and Gynecology, Tangdu Hospital, The Fourth Military Medical University, Xi`an, China
Purpose: High-grade serous ovarian cancer (HGSOC) remains the most lethal female cancer due to metastasis. CircRNAs are recently identified to be modified by N6-methyladenosine (m6A) in many cells. However, the significance of m6A-modified circular RNAs (circRNAs) has not been elucidated in HGSOC peritoneal metastasis. Here, we aimed to investigate the participation and potential functions of m6A-modified circRNAs in HGSCO peritoneal metastasis.
Methods: Cancerous tissues were collected from the in situ and the peritoneal metastasis lesions of HGSCO patients. M6A-tagged circRNAs were identified by m6A-modified RNA immunoprecipitation sequencing (m6A-RIP-seq). Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to predict the potential functions of the m6A-modified circRNAs.
Results: For the m6A-modified circRNAs, 259 were upregulated and 227 were downregulated in the peritoneal metastasis than in the situ lesions of HGSCO patients. For the m6A peaks, 1541 were upregulated and 1293 were downregulated in the peritoneal metastasis than in the in situ lesions of HGSCO patients. For the differential expressed circRNAs, 1911(19.6%) were upregulated and 2883(29.6%) were downregulated in the peritoneal metastasis than in the in situ lesions of HGSCO patients. The upregulated m6A-modified circRNAs were associated with the HIF-1 signaling. The downregulated m6A-modified circRNAs were associated with the MAPK signaling.
Conclusions: This work firstly identified the transcriptome-wide map of m6A-modified circRNAs in peritoneal metastasis of HGSCO. Our findings provided novel evidences about the participation of m6A-modified circRNAs via HIF-1 and MAPK signaling and a new insight in molecular target of HGSCO peritoneal metastasis.
Introduction
Ovarian cancer (OC) is the 7th most commonly diagnosed gynecologic malignancy worldwide, accounting for 2.5% of all female cancers (1, 2). In 2020, there were about 313,959 new cases and 207,252 deaths of OC worldwide in the world (3, 4). OC consists of subtypes with distinct biological and molecular properties, with epithelial ovarian cancer accounting for more than 95% and non-epithelial ovarian cancer accounting for 5%. OC is a heterogeneous disease that is histologically classified into 5 major subtypes: high-grade serous, low-grade serous, clear cell carcinoma, endometrioid carcinoma, and mucinous ovarian carcinoma (3, 4). High-grade serous ovarian cancer (HGSOC) is the most common histological subtype of OC and accounts for 67.5% of all OC subtypes (1, 2), and remains the most lethal female cancer with an estimated 21,410 new cases and 13,770 deaths expected in the United States in 2021 (5).
HGSOC is typically diagnosed at advanced stages owing to the deficiency of specific symptoms at early stages, which is characterized by the involvement of bilateral ovaries, aggressive behavior and low survival without effective molecular targeted therapy. To date, regardless of the continuous improvement and development of surgical and systematic management, the mortality of these patients has not been improved under the standard regimens with approximately 70% of patients having relapse and metastasis after treatment. Consequently, identifying the mechanisms that drive HGSOC metastasis will translate into clinical managements that would improve their outcomes (1, 5–8).
Currently, significant efforts have been made for personalized treatment to improve prognosis of HGSOC patients, including application of genomic technologies to identify the expression profile, copy-number changes, mutations, and epigenetic modifications of the genes. RNA sequencing (RNA-seq) is one of the key genomic technologies applied to reveal molecular pathogenesis in HGSOC (5–8).
Circular RNA (circRNA) is one type of noncoding RNA (ncRNAs) with a closed loop structure without 5′-3′polarity and poly A tail, which were firstly identified abundant in viruses by RNA-seq, and predominantly regulates the gene expressions at the transcriptional and post-transcriptional levels (9–12). Moreover, it has been reported that circRNAs are closely associated with HGSOC after circRNAs high-throughput sequencing (13).
Over 160 different chemical modifications in RNAs have been identified in all living organisms Among these RNA modifications, N6-methyladenosine (m6A), methylated at the N6 position of adenosine, is identified as the most universal epigenetic modification on ncRNAs in eukaryotes and affects the circRNA expressions (14–16). M6A modification is predominantly enriched near the stop codons, within the internal long exons and at the 3′untranslated regions (14, 17). The m6A methylation can be catalyzed by the methyltransferases (“writers”) such as METTL3/14/16, removed by demethylases (“erasers”) FTO or ALKBH5, and interacts with the m6A binding proteins (“readers”), such as YTHDF1/2/3 and IGF2BP1/2/3 (14, 18–20). The circRNAs displayed cell-type-specific M6A-methylation patterns in many human cells (14, 18, 20). The m6A-modifed circRNAs are expected to promote protein translation in a cap-independent pattern (14, 21). However, the m6A-modification of circRNAs in HGSOC remains unknown.
Therefore, we hypothesized that m6A-modified circRNAs might be related to the HGSOC metastasis by regulating the genes/signaling pathways associated with HGSOC development. Here, we conducted a genome-wide screening of m6A-modified circRNAs in the tissues of HGSOC patients with peritoneal metastasis to identify the expression profiling of the m6A-modified circRNAs. This study may provide new therapeutic targets for HGSOC patients.
Materials and methods
Reagents
TRIzol Reagent was bought from Invitrogen-Gibco (Carlsbad, USA). NEBNext rRNA Depletion Kit and NEBNext Ultra II Directional RNA Library Prep Kit New England Biolabs, Inc. (Ipswich, USA).GenSeqTM m6A-MeRIP Kit was bought from GenSeq Inc. (Cyberjaya, Malaysia). Anti-N6-methyladenosine (m6A) antibody and Rnase R were purchased from Merck Millipore-Sigma-Aldrich (Beijing, China).
Patients, informed consents and ethics statement
All study protocols were approved by the Ethics Committee of Tangdu Hospital, the Fourth Military Medical University and performed following the principles of the Helsinki Declaration. Each included patient provided a written informed consent. Cancerous tissues were collected from the in situ and the peritoneal metastasis lesions of 3 HGSCO patients during the surgery treatment, and were applied for the initial high throughput screening of m6A- methylated RNA immunoprecipitation sequencing (MeRIP-seq) (22, 23).
RNA extraction and quality control
Total RNA (10 mg) was obtained from the cancerous tissues by using the TRIzol Reagent following the manufacturer’s protocols. The extracted RNAs were purified with Rnase R (RNR07250, Epicentre) digestion to remove linear transcripts.
NEBNext rRNA Depletion Kit was used to remove rRNAs from the total RNAs. The RNA concentration was measured at 260/280 nm (1.8 to 2.1) by using NanoDrop ND-2000. Total RNA quality was evaluated with the band intensity ratio of 18S/28S ribosomal after electrophoresis in an ethidium bromide-containing 1% agarose gel (22, 23).
RNA-seq analysis of circRNAs
After quality filtering, paired-end reads were collected from Illumina Hiseq Sequence. The reads were aligned to the reference genome (UCSC RN5) with STAR software. The CIRI software was applied to detect and annotate circRNAs. Raw junction reads were normalized against per million reads mapped to the genome by using log2 scaled (14).
Library preparation and MeRIP-Seq for maps of m6A-methylated RNAs
To immunoprecipitate the m6A-methylated RNAs, the anti-N6-methyladenosine (m6A) antibody was cultured with the fragmented total RNAs extracted from the cancerous tissues of HGSCO patients after rRNA depletion with the GenSeqTM m6A-MeRIP Kit following the manufacturer’s instructions. The bound RNAs were eluted from the protein A/G magnetic beads after incubation.
The input RNAs and m6A-immunoprecipitated (m6A-IP) RNAs were reverse-transcribed to cDNA for the construction of the RNA-seq library by using NEBNext Ultra II Directional RNA Library Prep Kit. The library quality was weighed by using a BioAnalyzer 2100 system (Agilent Technologies, Inc., Santa Clara, USA). Then the qualified m6A-IP and input samples were subjected to 150 bp paired-end deep sequencing on Illumina HiSeq sequencer to obtain the high-resolution reads of m6A-methylated RNAs.
The RNA-seq of circRNAs and the m6A RNA-Seq were carried out by the Cloud-seq Biotech Inc. (Shanghai, China).
Bioinformatics analysis
Paired-end reads were collected from Illumina HiSeq sequencer with the quality control of Q30. After 3’ adaptor-trimming, the low-quality reads were removed with the cutadapt software (v1.9.3). First, the clean reads of input libraries were aligned to the reference genome (UCSC HG38) by using the STAR software. Based on the STAR alignment results, the circRNAs were identified by using the DCC software. Then, the clean reads of all libraries were aligned to the reference genome with the Hisat2 software (v2.0.4). The MACS software was applied to identify the methylated sites on circRNAs (peaks) and the diffReps was further used to identify the differentially methylated sites. The overlapping sites between the peaks, identified by both software based on the circRNA exons, were selected by the home-made scripts.
GO and KEGG analysis
The comprehensive function annotations of the source genes of differentially m6A methylated circRNAs were achieved by Gene Ontology (GO), as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses based on the online software DAVID.
Statistical analysis
Statistical analysis was performed with SPSS 25.0 (IBM, Chicago, USA) and GraphPad Prism software 7.0 (GraphPad Software, San Diego, USA). The single factor ANOVA was used for statistical analyses, p < 0.05 was considered statistically significant (24).
Results
Abundance of m6A-methylated circRNAs and circRNAs involve in peritoneal metastasis of HGSCO patients
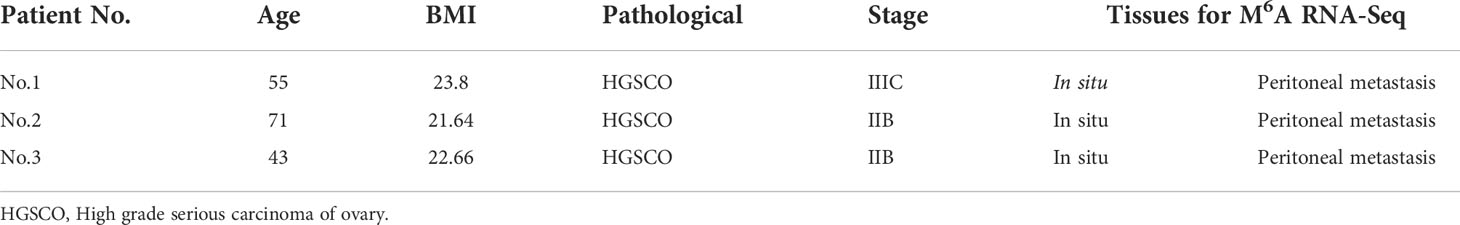
A genome-wide profiling of m6A-modified circRNAs was performed in cancerous specimens of three biological replicates from the in situ lesions (n = 3) and the peritoneal metastasis lesions (n = 3) from HGSCO patients. Patient information is presented in Table 1. The MeRIP-Seq data had been submitted to gene expression omnibus (accession number, GSEXXX). As shown in the Figures 1A, B, the m6A abundance in the peritoneal metastasis lesions (1070 + 259 for the m6A-modified circRNAs and 2089 + 1541 for the m6A peaks) was found to be slightly increased than the in situ lesions (1070 + 227 for the m6A-modified circRNAs and 2089 + 1293 for the m6A peaks) of the HGSCO patients, which were further visualized by the Heatmap (Figure 1C) and Bar plots (Figure 1D).
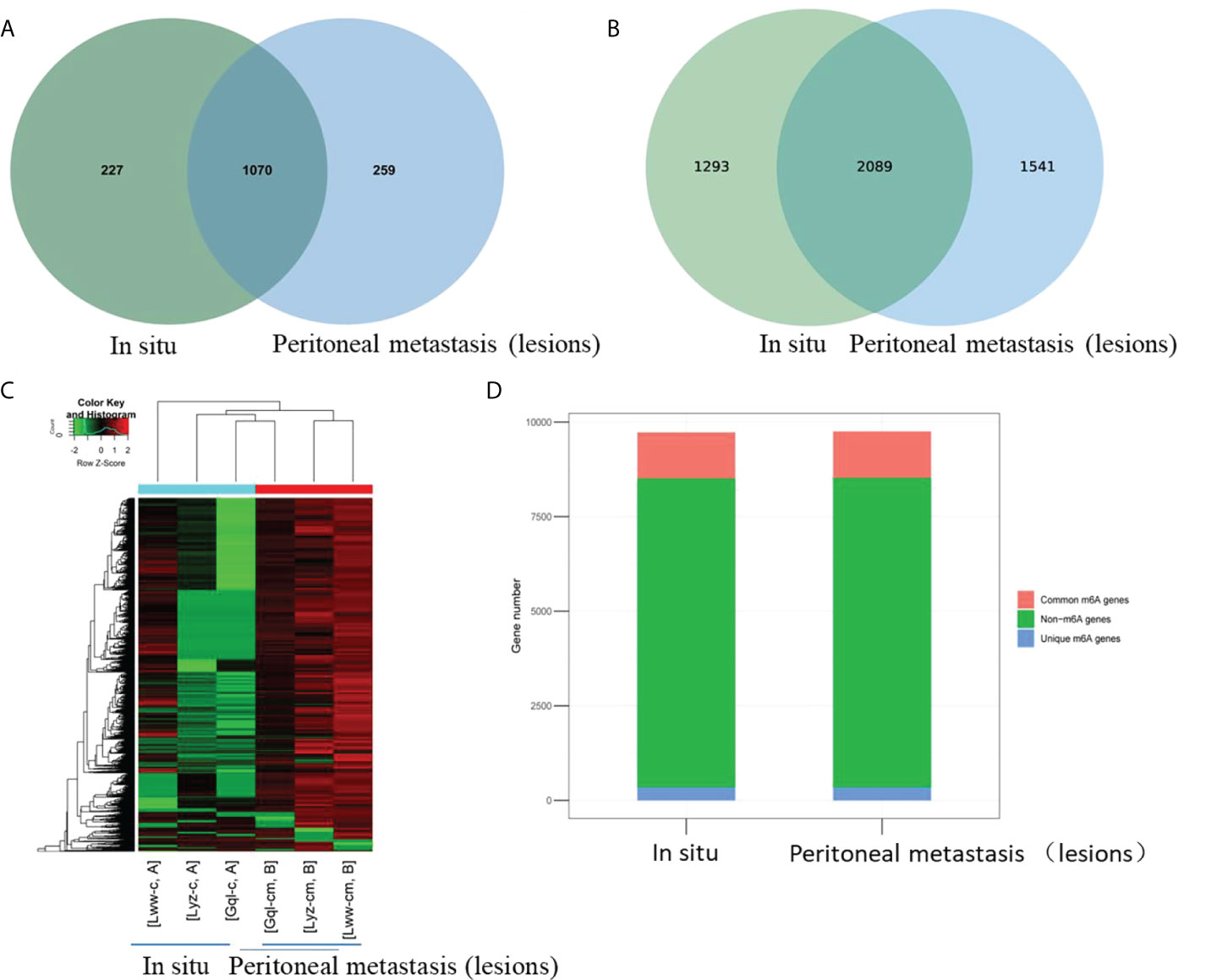
Figure 1 Overview of m6A-modified circRNAs of the in situ and the peritoneal metastasis lesions in the HGSCO patients. Vnn diagrams presenting the overlapped (A) m6A-modified circRNAs and (B) m6A peaks of the in situ and the peritoneal metastasis lesions in HGSCO patients. (C) Heatmap of the m6A-modified circRNAs. (D) Bar plots of the m6A peaks pear circRNAs.
To explore whether m6A methylation would influence circRNA expression levels in HGSCO patients, RNA-seq analysis of circRNAs was further performed, and 4934 (50.7%) overlapping circRNAs were identified. CircRNA expression was decreased in the peritoneal metastasis lesions than in the situ lesions. These findings suggest that m6A may downregulate the circRNA expressions in peritoneal metastasis lesions of HGSCO patients (Figure 2).
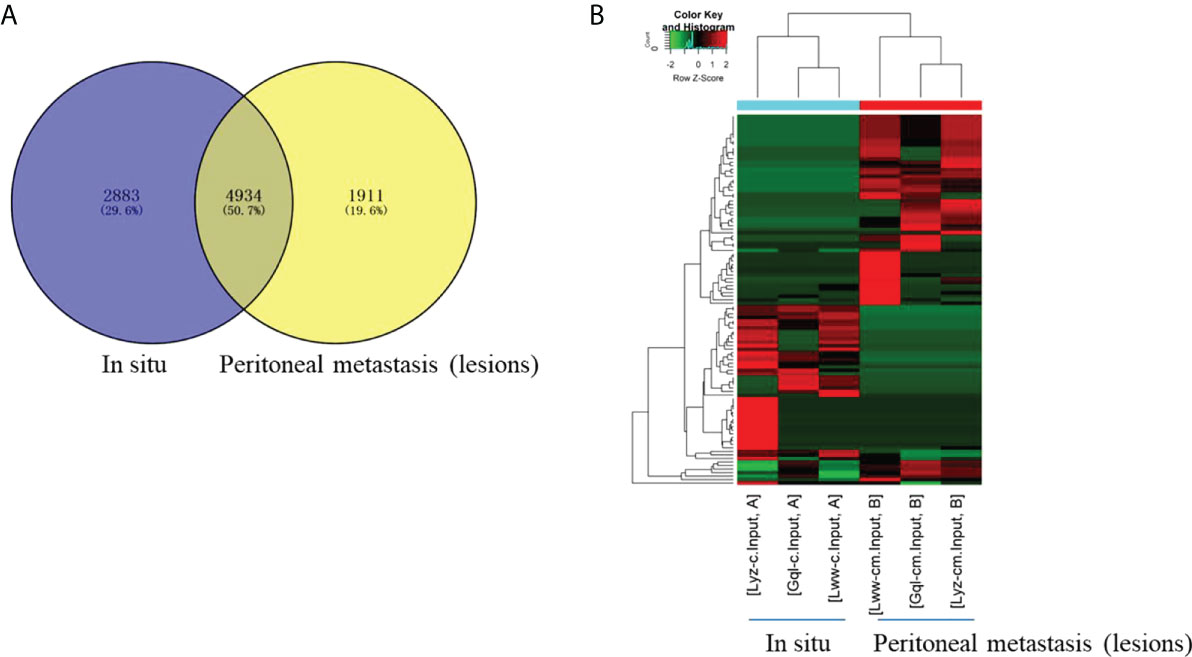
Figure 2 The differentially expressed circRNAs between the in situ and the peritoneal metastasis lesions of the HGSCO patients. (A) Vnn diagram and (B) Heatmap presenting the differential expressed circRNAs between the in situ and the peritoneal metastasis lesions of the HGSCO patients.
Distribution of circRNAs in the peritoneal metastasis lesions from HGSCO patients
To characterize the chromosomal and genomic distributions of all differently m6A-modified circRNA peaks and differentially expressed circRNAs of the peritoneal metastasis lesions than the in situ lesions from the HGSCO patients, the distributions of the down-regulated or upregulated m6A-modified circRNA peaks and the differentially expressed circRNAs on each chromosome was calculated and plotted. The upregulated m6A-modified circRNA peaks were mostly located in the chromosome 1, 2, 5 and 6 (Figure 3A), while the downregulated m6A-modified circRNA peaks were mostly located in chromosome 1, 2, 8 and 22 (Figure 3A). The upregulated differentially expressed circRNAs were mostly located in the chromosome 2, 3, and 22 (Figure 3B), while the downregulated m6A-modified circRNA peaks were mostly located in chromosome 1, 2 and 3 (Figure 3B). Genomic distribution of differently m6A-modified circRNA peaks (Figure 3C) and differently expressed circRNAs were mostly (over 65%) in the sense overlapping region (Figure 3D). Moreover, single m6A-modified circRNA peaks per gene was slightly decreased in the peritoneal metastasis lesions than the in situ lesions from the HGSCO patients, which was the predominant number of peaks per gene for both the peritoneal metastasis lesions and the in situ lesions from the HGSCO patients (Figure 3E), indicating that m6A methylation was abundant in circRNAs originated from single exons in both the peritoneal metastasis and the in situ lesions with a slight downregulation in the peritoneal metastasis lesion.
Figure 3 Chromosomal and genomic distributions of circRNAs. (A) Chromosomal distribution of differently m6A-modified circRNA peaks. (B) Chromosomal distribution of differently expressed circRNAs. (C) Genomic distribution of differently m6A-modified circRNA peaks. (D) Genomic distribution of differently expressed circRNAs. (E) The m6A peak numbers per circRNA.
Functional Annotation of the differently m6A-modified circRNAs
To explore the physiological and pathological significance of m6A modification in peritoneal metastasis of HGSCO patients, GO enrichment analysis and KEGG pathway analysis were conducted with the significantly changed m6A peaks. GO enrichment analysis was applied to predict the key functions of the target genes. Pathway analysis was accomplished to identify the significant pathways of the differential genes based on the KEGG.
In the GO analysis (Figure 4), the parent genes of circRNAs with upregulated m6A peaks were enriched in the localization and organelle organization, while downregulated m6A peaks were enriched in the cellular macromolecule metabolic process, the macromolecule modification, the protein modification process and the cellular protein modification process in the biological process (BP) analysis. The parent genes of circRNAs with upregulated m6A peaks were enriched in the intracellular, organelle and cytoplasm, while downregulated m6A peaks were enriched in the intracellular, organelle, cytoplasm and membrane-bounded organelle in the cellular component (CC) analysis. The parent genes of circRNAs with upregulated m6A peaks were enriched in the adenyl ribonucleotide binding and the adenyl nucleotide binding, while downregulated m6A peaks were enriched in the catalytic activity in the molecular functions (MF) analysis.
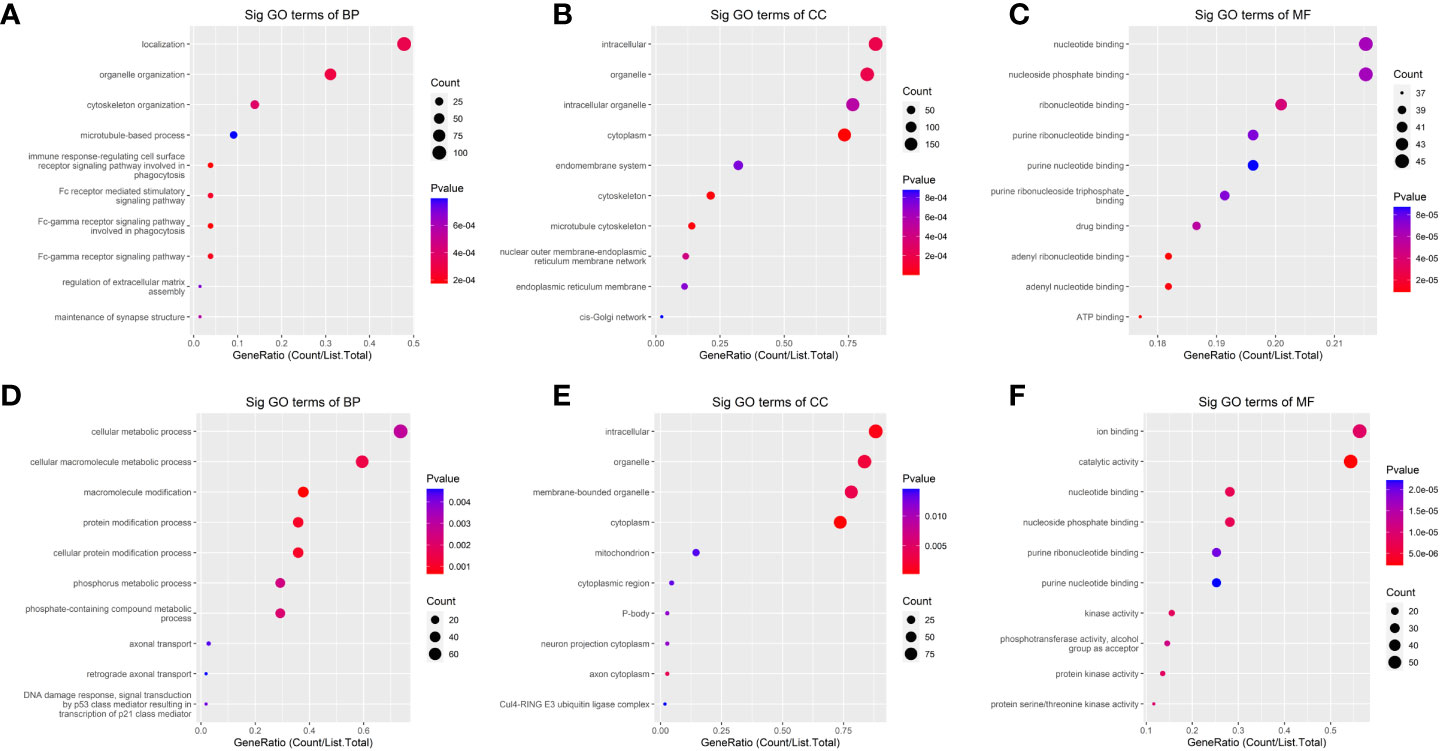
Figure 4 The Gene ontology (GO)-term enrichment analysis of significantly altered m6A-modified circRNAs between the in situ and the peritoneal metastasis lesions of the HGSCO patients. (A–C) upregulation of m6A-modified circRNAs. (D–F) downregulation of m6A-modified circRNAs.
Meanwhile, the HIF-1 and MAPK signaling pathways have been reported to be closely related with the cancer metastasis (25, 26), therefore, these two pathways were further analyzed. The results showed that the HIF-1 signaling pathway was associated with the upregulated m6A-modified circRNAs, while MAPK signaling pathway was associated with the downregulated m6A-modified circRNAs (Figure 5).
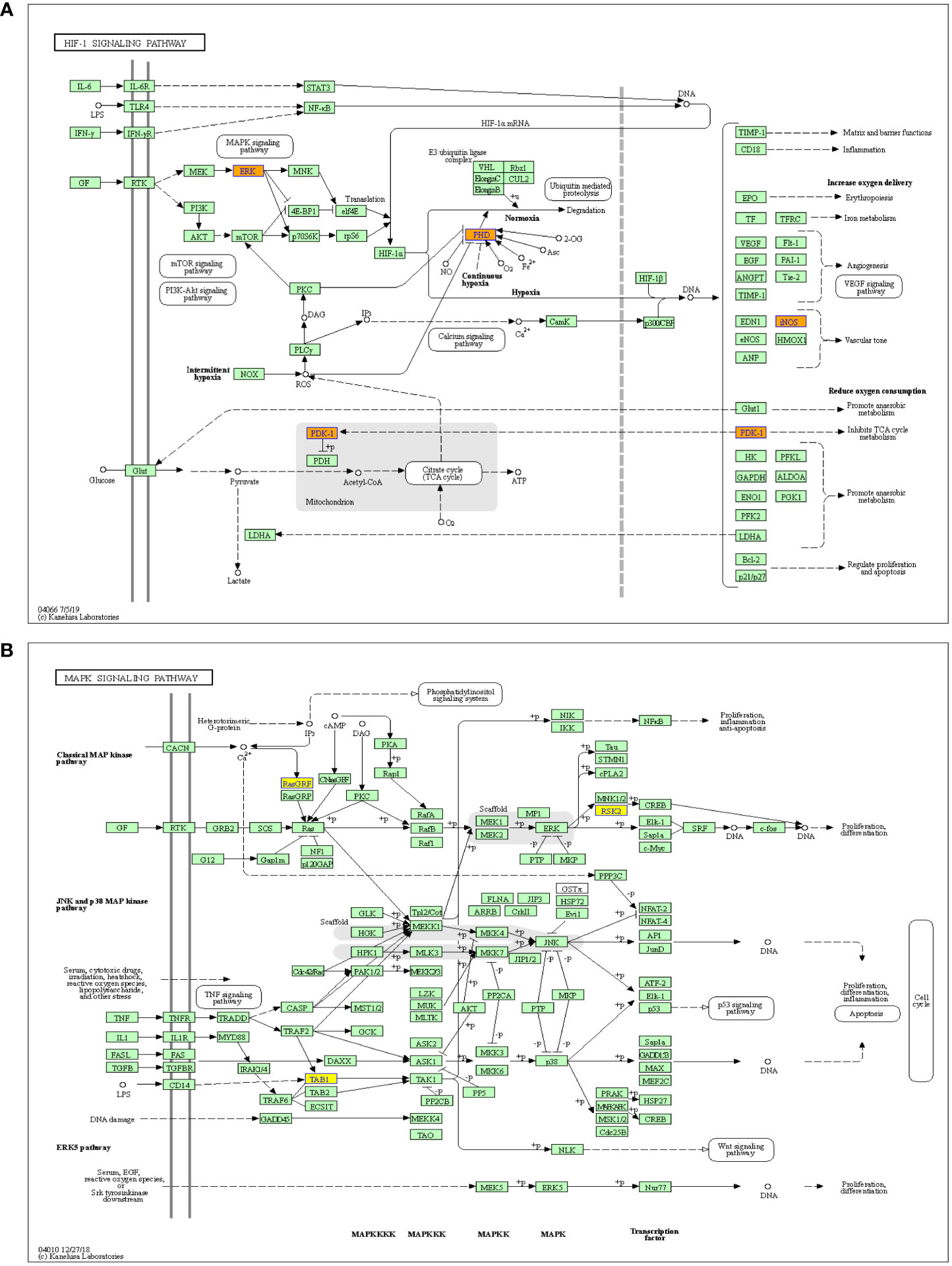
Figure 5 Pathway analysis of significantly altered m6A-modified circRNAs between the in situ and the peritoneal metastasis lesions of the HGSCO patients. (A) The up-regulated m6A-modified circRNAs involved in HIF-1 pathway. (B) The down-regulated m6A-modified circRNAs play important roles in MAPK signaling pathway.
Discussion
The overall survival rate of HGSOC has not been improved in the last three decades, and most patients develop recurrent disease within 3 years and died from the disease within 5 years. HGSOC is extremely regulated by epigenetic modifications (27). M6A plays an important role in numerous biological processes, which contributes to cancer development, cancer cell proliferation and cancer stem cell self-renewal (28, 29). The expression profile and potential function of circRNAs in different cancers have been identified, including HGSOC (13, 30, 31). As a new epitranscriptomic marker, m6A is recognized as a reversible and dynamic RNA modification in eukaryotes. Recently, m6A epigenetic modification in circRNA has attracted attention as a novel epigenetic event (22, 23, 32). However, the role of m6A epigenetic modification in circRNAs during HGSOC metastasis has not been characterized.
As far as we know, our current work was the first to explore the m6A-circRNA profile during peritoneal metastasis of HGSOC by using MeRIP-Seq. Our results support the concept of a dynamic characteristic of m6A epigenetic modification during peritoneal metastasis of HGSOC, which is associated with HGSOC development.
A genome-wide profile of m6A-modified circRNAs between the in situ and the peritoneal metastasis lesions were obtained by using MeRIP-seq. A total of 2089 m6A-modified circRNAs were shared in the peritoneal metastasis and the in situ lesions from HGSOC patients, whereas1293 of m6A-modified circRNAs were identified in the in situ lesions only but lacking in the peritoneal metastasis lesions, and 1541 m6A-modified circRNAs were identified in the peritoneal metastasis lesions only but lacking in the in situ lesions of the HGSOC patients.
Our results demonstrated that the expression of differentially regulated circRNAs was decreased in the peritoneal metastasis lesions than in the situ lesions by RNA sequencing, indicating that m6A may downregulate the circRNA expressions during the peritoneal metastasis of HGSCO.
Many questions have been raised with the findings of m6A-modified circRNA profile that need to be addressed in the future work, including the significance of m6A-modification on exons where the circRNAs originate. A recent work reported that m6A-modified circRNAs are more commonly encoded by single exons, which is likely to be longer than the multiexon circRNAs in human embryonic stem cells (18). Interestingly, this phenomenon has also been validated in our current work, which showed that single m6A-modified circRNA peaks per gene was predominant for both the peritoneal metastasis and the in situ lesions from the HGSCO patients, indicating that m6A methylation was abundant in circRNAs originated from single exons in both the peritoneal metastasis and the in situ lesions with a slight downregulation in the peritoneal metastasis lesion.
Studies have reported different rules which may control the biogenesis of m6A-modification in circRNAs compared with the mRNAs (18). Therefore, further studies are necessary to prove whether m6A-modified circRNAs present distinct modification patterns compared with the mRNAs between the peritoneal metastasis and the in situ lesions of the HGSOC patients.
A key function of circRNAs is to affect the biological roles of cells by regulating the target gene expressions. Thus, regulating the m6A-modification status in circRNAs may control the target gene expression of circRNAs and the cellular biological functions (33). CircRNAs mainly affect miRNAs and reduce their expression in regulating cytokine levels (34). Cancer is one of the leading causes of death worldwide, and the factors responsible for its progression need to be elucidated (35). CircRNAs could affect gene expression via targeting miRNAs to treat cancer (36). Therefore, in the present study, the GO and KEGG pathway enrichment analyses were carried out to explore the possible functions of changed m6A-modified circRNAs by combining analysis of differentially m6A-modified circRNAs and differentially expressed circRNAs associated with peritoneal metastasis of the HGSOC patients. We found that HIF-1 signaling was associated with the up-regulated m6A-modified circRNAs, while MAPK signaling was associated with the downregulated m6A-modified circRNAs, which will need to be further studied in the future.
In conclusion, the level of m6A abundance in total circRNAs was slightly increased in the peritoneal metastasis lesion from HGSOC than the in situ lesion, and m6A-modfication may downregulate the circRNA expressions during peritoneal metastasis of HGSCO. The bioinformatics analysis predicted the potential functions of m6A-modified circRNAs and the involved relevant signaling pathways, with HIF-1 pathway associated with the upregulated, while MAPK pathway associated with the downregulated m6A-modified circRNAs during peritoneal metastasis of HGSOC. Our findings provided evidences regarding the involvement of m6A-modifications in circRNAs of peritoneal metastasis of HGSOC, which might provide a new insight in the molecular target of HGSCO metastasis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
All animal procedures were approved by the Research Ethics Committee of the Second Hospital of Hebei Medical University.
Author contributions
Conceptualization, methodology, design, and writing: LG, NX, DQ, XY, and SZ. Supervision: HZ. All authors contributed to the article and approved the submitted version.
Funding
This research has been supported by Health Research Funding of Shaanxi Province Grant (Number: 2022D063); Innovation Project of Tangdu Hospital (Grant Number: 2018QYTS009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liu P, Fu R, Chen K, Zhang L, Wang S, Liang W, et al. ETV5-mediated upregulation of lncRNA CTBP1-DT as a ceRNA facilitates HGSOC progression by regulating miR-188-5p/MAP3K3 axis. Cell Death Dis (2021) 12(12):1146. doi: 10.1038/s41419-021-04256-9
2. Sun C, Cao W, Qiu C, Li C, Dongol S, Zhang Z, et al. MiR-509-3 augments the synthetic lethality of PARPi by regulating HR repair in PDX model of HGSOC. J Hematol Oncol (2020) 13(1):9. doi: 10.1186/s13045-020-0844-0
3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
4. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
5. Arend RC, Scalise CB, Gordon ER, Davis AM, Foxall ME, Johnston BE, et al. Metabolic alterations and WNT signaling impact immune response in HGSOC. Clin Cancer Res (2022) 28(7):1433–45. doi: 10.1158/1078-0432.CCR-21-2984
6. The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature (2011) 474(7353):609–15. doi: 10.1038/nature10166
7. Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res (2008) 14(16):5198–208. doi: 10.1158/1078-0432.CCR-08-0196
8. Wang C, Armasu SM, Kalli KR, Maurer MJ, Heinzen EP, Keeney GL, et al. Pooled clustering of high-grade serous ovarian cancer gene expression leads to novel consensus subtypes associated with survival and surgical outcomes. Clin Cancer Res (2017) 23(15):4077–85. doi: 10.1158/1078-0432.CCR-17-0246
9. Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform (2017) 18(4):547–57. doi: 10.1093/bib/bbw045
10. Han Y, Zhang H, Bian C, Chen C, Tu S, Guo J, et al. Circular RNA expression: Its potential regulation and function in abdominal aortic aneurysms. Oxid Med Cell Longev (2021) 2021:9934951. doi: 10.1155/2021/9934951
11. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal (2021) 35(7):e23866. doi: 10.1002/jcla.23866
12. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
13. Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends (2019) 13(2):204–11. doi: 10.5582/bst.2019.01021
14. Su H, Wang G, Wu L, Ma X, Ying K, Zhang R. Transcriptome-wide map of m(6)A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics (2020) 21(1):39. doi: 10.1186/s12864-020-6462-y
15. Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, et al. m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res (2020) 30(3):211–28 . doi: 10.1038/s41422-020-0279-8
16. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res (2018) 46(D1):D303–7. doi: 10.1093/nar/gkx1030
17. Wang S, Chai P, Jia R, Jia R. Novel insights on m(6)A RNA methylation in tumorigenesis: A double-edged sword. Mol Cancer (2018) 17(1):101. doi: 10.1186/s12943-018-0847-4
18. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-Type-Specific methylation patterns that are distinct from mRNAs. Cell Rep (2017) 20(9):2262–76. doi: 10.1016/j.celrep.2017.08.027
19. Shi H, Wei J, He C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell (2019) 74(4):640–50. doi: 10.1016/j.molcel.2019.04.025
20. Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res (2019) 79(7):1285–92. doi: 10.1158/0008-5472.CAN-18-2965
21. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
22. Li P, Yu H, Zhang G, Kang L, Qin B, Cao Y, et al. Identification and characterization of N6-methyladenosine CircRNAs and methyltransferases in the lens epithelium cells from age-related cataract. Invest Ophthalmol Vis Sci (2020) 61(10):13. doi: 10.1167/iovs.61.10.13
23. Zhang Y, Geng X, Xu J, Li Q, Hao L, Zeng Z, et al. Identification and characterization of N6-methyladenosine modification of circRNAs in glioblastoma. J Cell Mol Med (2021) 25(15):7204–17. doi: 10.1111/jcmm.16750
24. Hu S, Chang J, Ruan H, Zhi W, Wang X, Zhao F, et al. Cantharidin inhibits osteosarcoma proliferation and metastasis by directly targeting miR-214-3p/DKK3 axis to inactivate β-catenin nuclear translocation and LEF1 translation. Int J Biol Sci (2021) 17(10):2504–22. doi: 10.7150/ijbs.51638
25. Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: A new perspective. Cancer (2014) 120(22):3446–56. doi: 10.1002/cncr.28864
26. Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science (2016) 352(6282):175–80. doi: 10.1126/science.aaf4405
27. Matthews BG, Bowden NA, Wong-Brown MW. Epigenetic mechanisms and therapeutic targets in chemoresistant high-grade serous ovarian cancer. Cancers (Basel) (2021) 13(23):5993. doi: 10.3390/cancers13235993
28. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis (2018) 9(2):124. doi: 10.1038/s41419-017-0129-x
29. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res (2018) 28(5):507–17. doi: 10.1038/s41422-018-0034-6
30. Zhang N, Yang Z, Jin Y, Cheng S, Yang J, Wang Y. Low expression of circular RNA hsa_circ_0078607 predicts poor prognosis in high-grade serous ovarian cancer. Cancer Manag Res (2021) 13:2877–83. doi: 10.2147/CMAR.S300738
31. Bachmayr-Heyda A, Auer K, Sukhbaatar N, Aust S, Deycmar S, Reiner AT, et al. Small RNAs and the competing endogenous RNA network in high grade serous ovarian cancer tumor spread. Oncotarget (2016) 7(26):39640–53. doi: 10.18632/oncotarget.9243
32. Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS, et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer (2022) 21(1):51. doi: 10.1186/s12943-022-01521-z
33. Fu XD. Non-coding RNA: A new frontier in regulatory biology. Natl Sci Rev (2014) 1(2):190–204. doi: 10.1093/nsr/nwu008
34. Ashrafizadeh M, Zarrabi A, Mostafavi E, Aref AR, Sethi G, Wang L, et al. Non-coding RNA-based regulation of inflammation. Semin Immunol (2022) 10:101606. doi: 10.1016/j.smim.2022.101606
35. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol (2022) 15(1):83. doi: 10.1186/s13045-022-01305-4
Keywords: high grade serious carcinoma of ovary peritoneal metastasis, circular RNA, N6-methyladenosine, m6A-modified circRNAs, HGSOC
Citation: Guo L, Xu N, Qiu D, Yang X, Zhao S and Zhao H (2022) Comprehensive analysis of m6A-modified circRNAs in peritoneal metastasis of high grade serious carcinoma of ovary. Front. Oncol. 12:988578. doi: 10.3389/fonc.2022.988578
Received: 07 July 2022; Accepted: 03 August 2022;
Published: 20 September 2022.
Edited by:
Gautam Sethi, National University of Singapore, SingaporeReviewed by:
Ajaikumar B. Kunnumakkara, Indian Institute of Technology Guwahati, IndiaMilad Ashrafizadeh, Sabancı University, Turkey
Manoj Garg, Amity University, India
Copyright © 2022 Guo, Xu, Qiu, Yang, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxi Zhao, zhaohx@fmmu.edu.cn
 Lin Guo
Lin Guo