
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 02 November 2022
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.986867
A correction has been applied to this article in:
Corrigendum: Machine learning prediction model for post- hepatectomy liver failure in hepatocellular carcinoma: a multicenter study
 Jitao Wang1,2†
Jitao Wang1,2† Tianlei Zheng3,4†
Tianlei Zheng3,4† Yong Liao1†
Yong Liao1† Shi Geng3†
Shi Geng3† Jinlong Li1†
Jinlong Li1† Zhanguo Zhang5
Zhanguo Zhang5 Dong Shang6
Dong Shang6 Chengyu Liu1
Chengyu Liu1 Peng Yu7
Peng Yu7 Yifei Huang8
Yifei Huang8 Chuan Liu2
Chuan Liu2 Yanna Liu9
Yanna Liu9 Shanghao Liu2
Shanghao Liu2 Mingguang Wang2
Mingguang Wang2 Dengxiang Liu2
Dengxiang Liu2 Hongrui Miao5
Hongrui Miao5 Shuang Li6
Shuang Li6 Biao Zhang6
Biao Zhang6 Anliang Huang6
Anliang Huang6 Yewei Zhang10*
Yewei Zhang10* Xiaolong Qi2*
Xiaolong Qi2* Shubo Chen1*
Shubo Chen1*Introduction: Post-hepatectomy liver failure (PHLF) is one of the most serious complications and causes of death in patients with hepatocellular carcinoma (HCC) after hepatectomy. This study aimed to develop a novel machine learning (ML) model based on the light gradient boosting machines (LightGBM) algorithm for predicting PHLF.
Methods: A total of 875 patients with HCC who underwent hepatectomy were randomized into a training cohort (n=612), a validation cohort (n=88), and a testing cohort (n=175). Shapley additive explanation (SHAP) was performed to determine the importance of individual variables. By combining these independent risk factors, an ML model for predicting PHLF was established. The area under the receiver operating characteristic curve (AUC), sensitivity, specificity, positive predictive value, negative predictive value, and decision curve analyses (DCA) were used to evaluate the accuracy of the ML model and compare it to that of other noninvasive models.
Results: The AUCs of the ML model for predicting PHLF in the training cohort, validation cohort, and testing cohort were 0.944, 0.870, and 0.822, respectively. The ML model had a higher AUC for predicting PHLF than did other non-invasive models. The ML model for predicting PHLF was found to be more valuable than other noninvasive models.
Conclusion: A novel ML model for the prediction of PHLF using common clinical parameters was constructed and validated. The novel ML model performed better than did existing noninvasive models for the prediction of PHLF.
In 2020, primary liver cancer was the sixth most commonly diagnosed cancer and the third leading cause of cancer-related deaths worldwide, as approximately 906,000 new cases and 830,000 deaths occurred in 2020 (1). More than 50% of the world’s total new cases of liver cancer each year are attributed to hepatitis B, which has a high incidence in China (2). Radical liver resection remains the first choice of treatment for hepatocellular carcinoma (HCC) (3). Post-hepatectomy liver failure (PHLF) is the most common cause of postoperative death among patients who undergo hepatectomy for HCC (4). The incidence of PHLF has been reported to be 1.2%-32% and is attributed to different etiologies and surgical procedures (5, 6) as the most common cause of early death after liver surgery (7).
A variety of comprehensive scoring systems and nomogram prediction models can be used to help predict PHLF in patients with HCC (8–10). However, no universally recognized method for the prediction of PHLF has been established. Machine learning (ML), one of the most important branches of artificial intelligence (AI), has undergone rapid development and is being widely used in the field of disease prediction, where it has achieved remarkable results in clinical practice (11). ML is widely used in cancer research, where it is applied to clinical data, radiomics, and genomics to develop predictive models for efficient and accurate decision making (12–14). ML uses computational algorithms to learn from and analyze large amounts of data in a short period of time. Therefore, ML may outperform traditional risk stratification tools via the integration of different algorithms such as decision trees, artificial neural networks, random forests, support vector machines, extreme gradient boosting, and light gradient boosting machines (LightGBM) (15). LightGBM uses a histogram-based decision tree algorithm. Compared with other ML models, the LightGBM model is characterized by fast training speed and low memory usage. ML based on LightGBM has only recently been introduced in research involving liver disease (16–18), and the LightGBM model has not yet been used to predict PHLF.
In this study, a novel ML model based on the LightGBM algorithm, namely ML PHLF, was constructed. This novel model may replace traditional scoring systems and facilitate the assessment of liver function and reduction of the incidence of PHLF and postoperative mortality after radical hepatectomy.
This retrospective study was performed using a multicenter database of patients who underwent radical hepatectomy for HCC at the following hospitals: The Xingtai People’s Hospital, The Second Affiliated Hospital of Nanjing Medical University, Fifth Medical Center of People's Liberation Army (PLA) General Hospital, The First Affiliated Hospital of Dalian Medical University, and Tongji Hospital Affiliated to Huazhong University of Science and Technology. Two independent investigators (JW and JL) reviewed the baseline data, laboratory parameters, treatment records, and pathological findings. All patients were randomly divided into the training, validation, and testing cohorts at a ratio of 7:2:1. This study was performed in accordance with the ethical guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (2022–006).
Patients aged > 18 years with confirmed HCC based on histopathological examination of the tumor specimen and no history of anticancer therapy, including transarterial chemoembolization, ablation, or targeted drugs, were included in this study. Patients who underwent other surgical procedures at the time of hepatectomy and those with insufficient data on important indicators, such as total bilirubin (TBIL) and international normalized ratio (INR) on or after the fifth postoperative day, were excluded from the study.
Patient demographic data, including age, weight, body mass index (BMI), sex, presence of hypertension, etiology of liver disease, and cirrhosis, were retrieved from the medical records. Data on the surgical method (open or minimally invasive), extent of liver resection (major resection: ≥ 3 segments; minor resection:< 3 segments), requirement of intraoperative blood transfusion, number of tumors, maximum tumor diameter, and intraoperative blood loss were extracted from the preoperative and surgical records. Laboratory indicators included red blood cell (RBC) count, white blood cell (WBC) count, platelet (PLT) count, TBIL, direct bilirubin (DBIL), albumin (ALB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), serum alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), creatinine (Cr), prothrombin time (PT), and INR. Portal hypertension was defined as the presence of varicose veins or a PLT count<100 x 109/L and a spleen diameter > 12 cm. According to previous literature, the model for end-stage liver disease (MELD) score (19) was calculated as:
The fibrosis-4 (FIB-4) index (20) was calculated as:
The albumin-bilirubin (ALBI) score (21) was calculated as:
The aminotransferase-to-platelet ratio index (APRI) score (22) was calculated as:
Finally, the Child-Turcotte-Pugh (CTP) score (23) was calculated and obtained. Based on the Chinese Society of Hepatology guidelines for the diagnosis and treatment of liver cirrhosis, the diagnosis of liver cirrhosis was made using preoperative clinical variables such as the etiology, history, clinical manifestations, complications, laboratory results, imaging examinations, or liver biopsy histology (24).
The International Study Group of Liver Surgery (ISGLS) diagnostic criteria for PHLF were used in this study (5). PHLF was defined as an increase in TBIL and INR on or after the fifth postoperative day when compared to preoperative levels, after the exclusion of biliary obstruction as a cause for increased TBIL or INR.
A total of 835 patients, including 192 with PHLF and 683 without PHLF, were included in this study. Twenty-five clinical variables, including sex, age, weight, liver disease etiology, cirrhosis, portal hypertension, PLT count, RBC count, WBC count, TBIL, ALB, AST, ALT, DBIL, Cr, PT, INR, AFP, CEA, tumor size, tumor number, surgical approach, extent of resection, intraoperative blood loss, and intraoperative blood transfusion, were used in this study. To verify the performance of the model, 70% of the dataset was used as the training set, 10% was used as the validation set, and 20% was used as the testing set. Data from the training and validation sets were applied to LightGBM, which computed the value of each variable using a decision tree to generate a prediction model for PHLF (Figure 1).
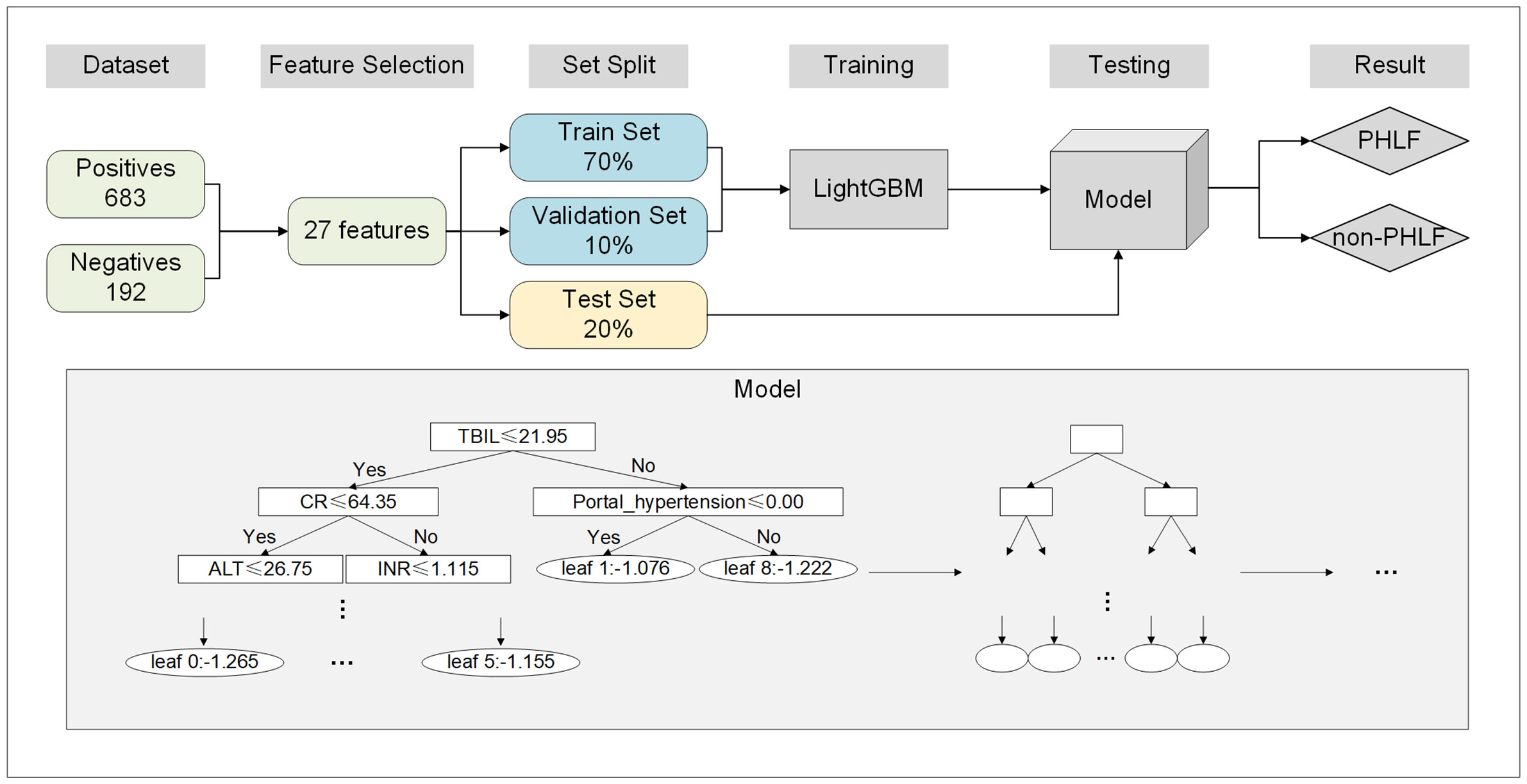
Figure 1 Flowchart of the development of the ML model. ML, machine earning; PHLF, post-hepatectomy liver failure.
The Shapley additive explanation (SHAP), a game-theoretic approach to interpreting the output of the ML PHLF (25, 26), was used to quantitatively measure the importance of each variable and describe the overall relationship between PHLF and all variables. To obtain the best ML model for PHLF, the LightGBM algorithm was optimized by adjusting the number of iterations, number of leaves, and maximum depth of the tree. The optimal number of trees, maximum tree depth, and number of leaves obtained were combined with the hyperparameters adjusted by the validation set to construct an optimal LightGBM model. In addition, the LightGBM algorithm can speed up the training process without affecting the performance of the model. This overall increase in speed is the result of a combination of gradient-based one-sided sampling and exclusive feature bundling. Subsequently, the LightGBM model is used to establish an accurate PHLF diagnosis model with a favorable area under the receiver operating characteristic curve (AUROC). The AUROCs of the training, verification, and testing cohorts were determined.
Continuous variables with normal distribution are presented as median and interquartile range or mean and standard deviation. These variables were compared using Student’s t-test. Non-normal variables were analyzed using the Mann–Whitney rank sum test. Categorical variables are presented as numbers and frequencies (%). The chi-squared test or Fisher’s exact test were used to analyze categorical variables. The predictive performance of the ML PHLF model was assessed using AUROC, sensitivity, specificity, positive predictive value, and negative predictive value (NPV). Decision curve analyses (DCA) were used to measure the clinical utility of each model by calculating the net benefit at various threshold probabilities. R software (version 4.1.2) or Python software (version 3.7.9) was used for data analysis and model building. Statistical significance was set at P< 0.05.
A total of 875 patients were enrolled in this study and randomly assigned to the training (n=612), validation (n=88), and testing (n=175) cohorts at a ratio of 7:1:2. The baseline characteristics of the three groups were not significantly different (Table 1).
The top five factors associated with PHLF were PLT count, age, Cr, INR, and AFP (Figure 2A). The top 20 variables and the correlation between high or low SHAP values and the predicted PHLF are presented in Figure 2B.
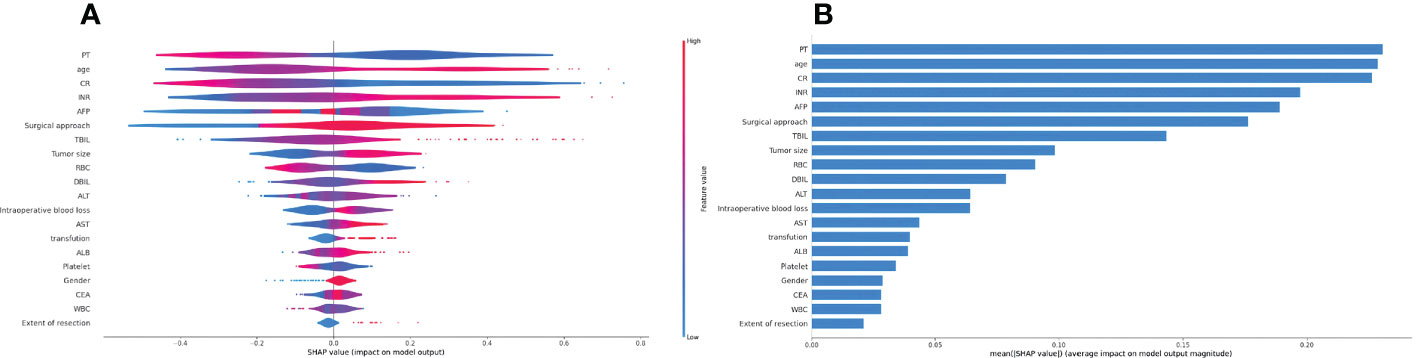
Figure 2 Summary Shapley additive explanations plot revealing the impact of individual clinical variables. (A) The importance matrix plot of clinical variables is derived using the LightGBM model. The matrix plot ranks the importance of the variables selected for the final analysis, revealing the contribution of each variable to PHLF versus non-PHLF. (B) The SHAP summary plot of the LightGBM model is shown. The higher the SHAP value for each clinical variable, the higher risk of PHLF. LightGBM, light gradient boosting machines; PHLF, post-hepatectomy liver failure.
In the training, validation, and testing cohorts, the number of patients with PHLF were 137 (22.4%), 15 (17.1%), and 40 (22.9%), respectively. The areas under the curve (AUCs) of the ML model for detecting PHLF in the training, validation, and testing cohorts were 0.944 (95% confidence interval [CI], 0.924-0.964), 0.870 (95% CI, 0.791-0.950), and 0.822 (95% CI, 0.755-0.888), respectively (Figure 3). The ML PHLF model identified PHLF in the training cohort with a sensitivity, specificity, and NPV of 87.6%, 85.9%, and 96.0%, respectively. The sensitivity, specificity, and NPV of the ML PHLF model were 100%, 64.4%, and 100%, respectively, in the validation cohort and 87.5%, 64.4%, and 94.6%, respectively, in the testing cohort (Tables 2–4).
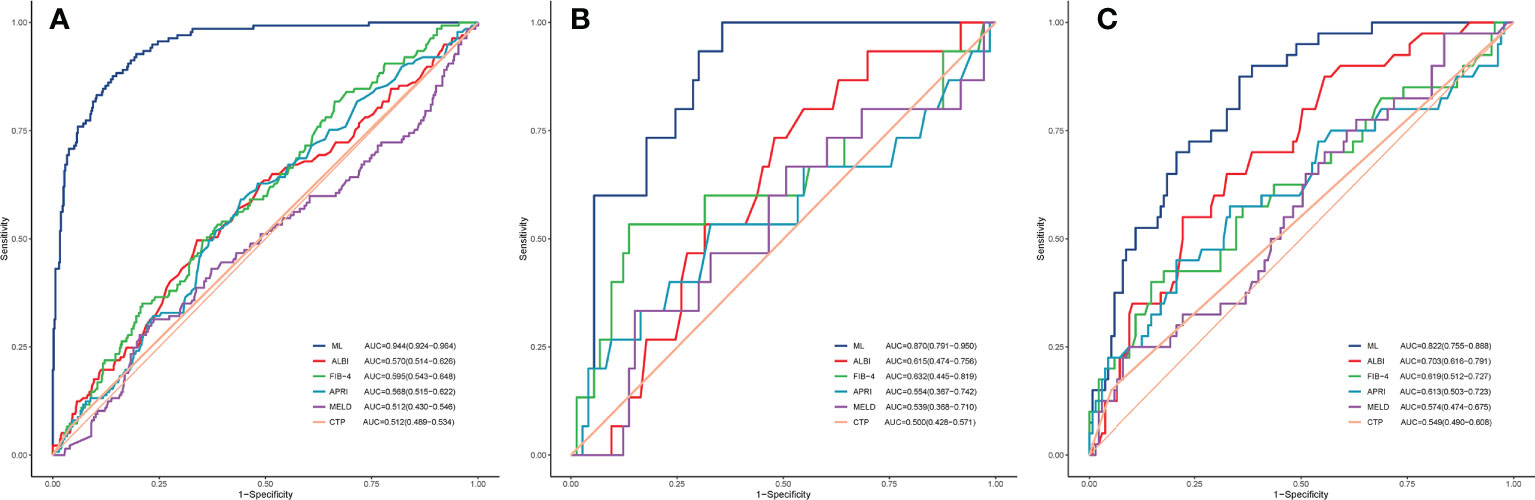
Figure 3 ROC curves. The ROC curves of the FIB-4 score, APRI score, CTP score, MELD score, and ALBI score are compared with that of the ML model in the training (A), validation (B), and testing (C) cohorts. ROC, receiver operating characteristic curves; ML, machine learning; FIB-4, fibrosis-4; APRI, aminotransferase to platelet ratio index; CTP, Child-Turcotte-Pugh; MELD, model for end-stage liver disease; ALBI, albumin–bilirubin.
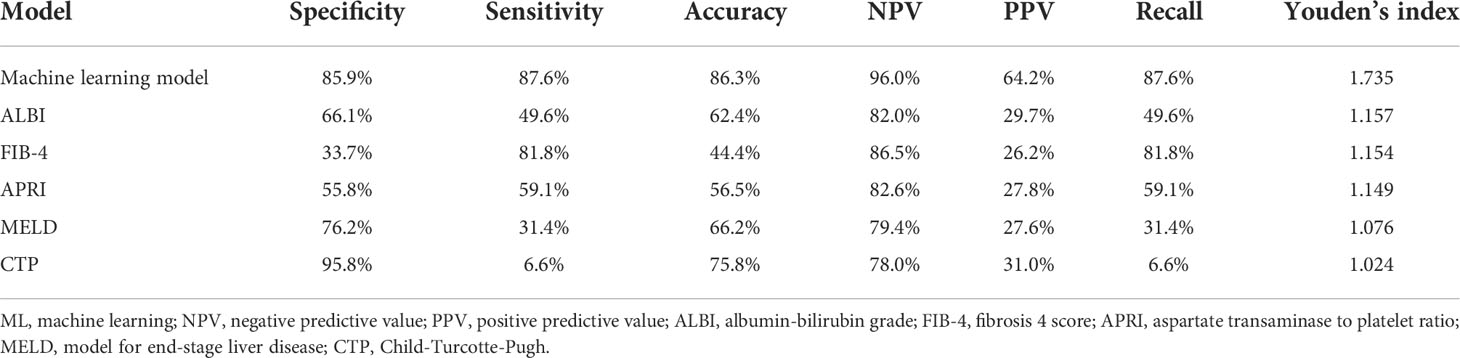
Table 2 Predictive power of the ML model and routine clinical models using data of the training cohort.
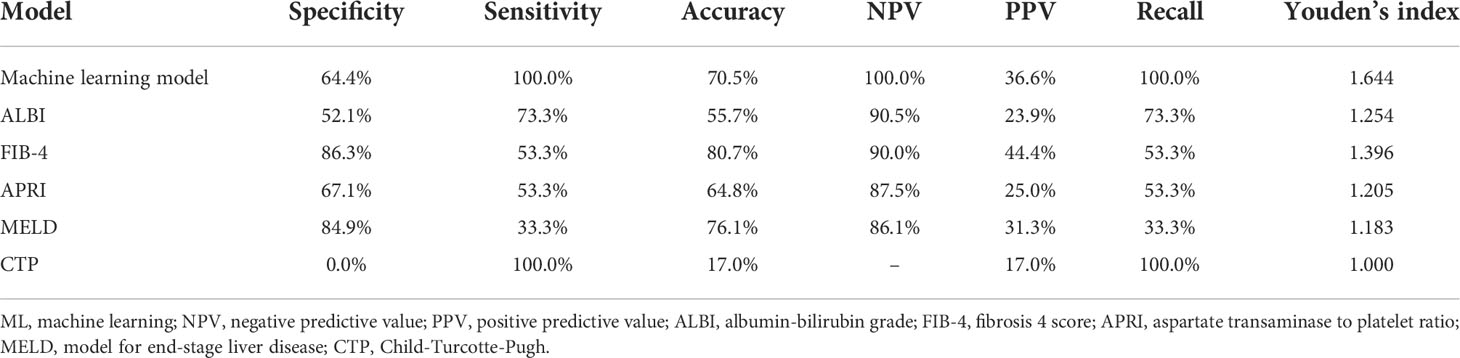
Table 3 Predictive power of the ML model and routine clinical models using data of the validation cohort.
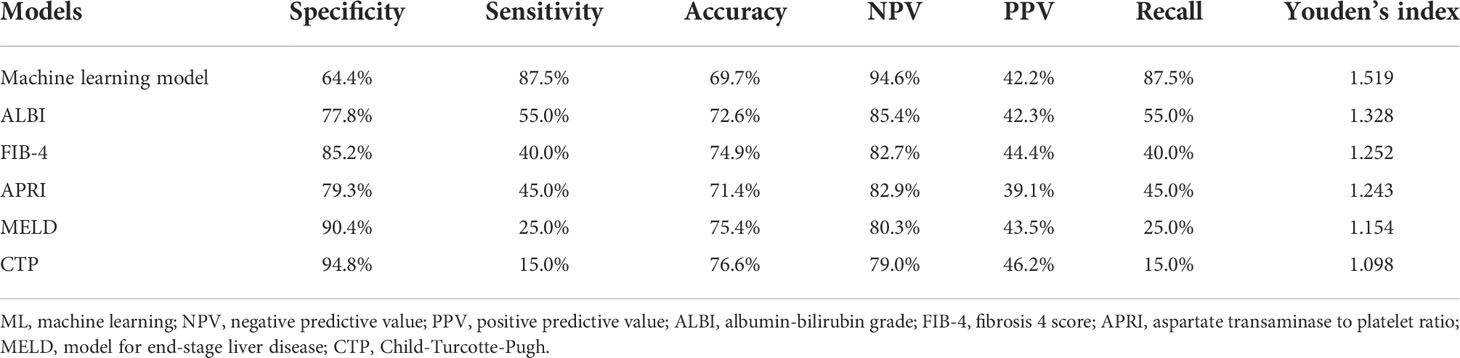
Table 4 Predictive power of the ML model and routine clinical models using data of the testing cohort.
We further compared the diagnostic performance of the ML PHLF model with that of routine clinical models, such as ALBI, FIB-4, APRI, MELD, and CTP. The ML PHLF model had the highest AUC for the prediction of PHLF among the noninvasive models (Figure 3). In addition, the AUCs for the ALBI, FIB-4, APRI, MELD, and CTP score were 0.570, 0.595, 0.568, 0.512, and 0.512, respectively, in the training cohort (Figure 3A); 0.615, 0.632, 0.554, 0.539, and 0.500, respectively, in the validation cohort (Figure 3B); and 0.703, 0.619, 0.613, 0.574, and 0.549, respectively, in the testing cohort (Figure 3C). The diagnostic performances of routine clinical models in the training, validation, and testing cohorts are summarized in Tables 2, 3, and 4, respectively.
The ML PHLF model added more value than did the FIB-4, APRI, ALBI, MELD, or CTP score for predicting PHLF in the training cohort (Figure 4A). The results were similar in the validation and testing cohorts. The novel ML PHLF model was more reliable than the traditional models (Figure 4B and Figure 4C).
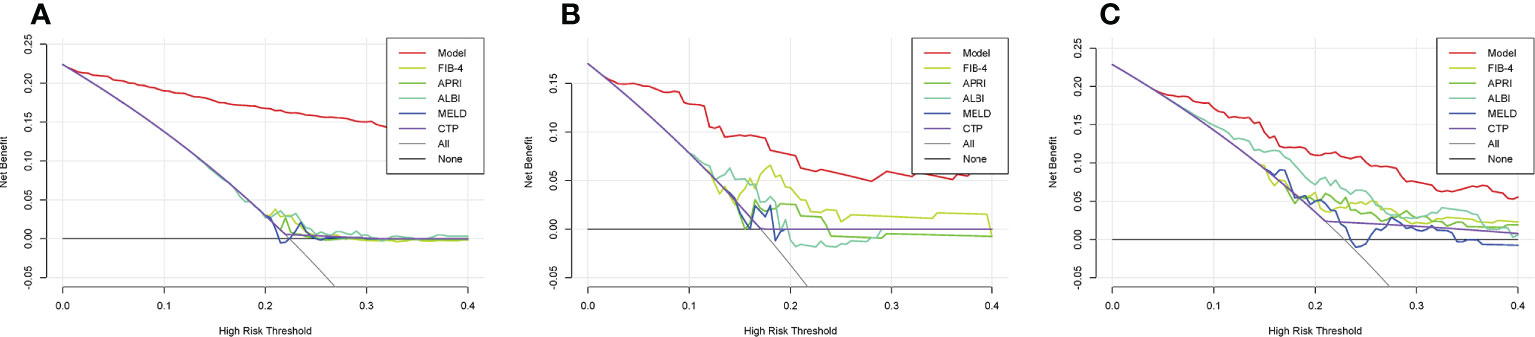
Figure 4 DCA curves. The DCA curves of the FIB-4 score, APRI score, CTP score, MELD score, and ALBI score are compared with that of the ML model in the training (A), validation (B), and testing (C) cohorts. sDCA, decision curve analysis; ML, machine learning; FIB-4, fibrosis-4; APRI, aminotransferase-to-platelet ratio index; CTP, Child-Turcotte-Pugh; MELD, model for end-stage liver disease; ALBI, albumin–bilirubin.
The ML model is composed of 29 decision trees based on the LightGBM algorithm. Owing to the large number of trees and the complex structure of each tree, only the first three and last two decision trees are shown in Supplementary Figure S1. To increase the clinical utility, a web calculator application based on the novel ML model has been developed (http://www.pan-chess.cn/calculator/PHLF_score) (Supplementary Figure S2).
In this study, a novel ML PHLF model for predicting the risk of PHLF was developed based on the LightGBM algorithm using the data of 875 patients with HCC who underwent liver resection. This novel model exhibited the best AUC when compared to existing noninvasive prediction models and good decision making in the training, validation, and testing cohorts in this study. This valuable and reliable predictive model for PHLF may be effective in optimizing personalized treatment options for patients with HCC, allowing for early identification of patients with HCC at high risk for PHLF.
Radical liver resection remains the first-choice treatment for HCC. With the update of new surgical techniques, optimization of innovative surgical instruments, and advancement of surgical intensive care medicine, the safety of liver resection has significantly improved. Therefore, perioperative mortality after hepatectomy has decreased (27). However, PHLF remains a serious complication for patients with HCC after hepatectomy. Accurate identification of patients with a high risk of PHLF is critical. Therefore, development of a predictive model for PHLF is crucial for clinical decision making.
Previously reported, noninvasive models for the prediction of PHLF are mainly based on laboratory indicators. The FIB-4, APRI, CTP score, MELD, and ALBI are widely used scoring systems for the evaluation of liver function and have been confirmed to predict the occurrence of PHLF (28–32). However, the predictive efficacy of these traditional noninvasive models that are based on simple laboratory indicators is relatively poor, whereas AI-based combined models using multiple clinical parameters have greater predictive potential.
ML is a field of AI that uses data-driven mining of complex datasets to predict future outcomes (33–35). The use of various ML algorithms to perform disease risk prediction has become a research hotspot in the field of medical big data. Various complex algorithms can be used to deeply mine the relationships between disease variables. The ML model has two advantages over other models, including the use of nonlinear functions and the consideration of the possible effects between all variables. ML algorithms have been increasingly applied to pertinent issues in the field of liver surgery (16). Mai et al. (36) developed an artificial neural network-based model to predict the risk of PHLF in patients with HCC undergoing partial hepatectomy. The predictive performance of the model exceeded that of traditional logistic regression models and commonly-used scoring systems. However, no research regarding ML models developed based on LightGBM that predict PHLF has been reported.
Twenty-five clinically meaningful variables were used to develop the ML PHLF model according to the SHAP analysis in this study. Specifically, both the importance matrix plot and the SHAP results indicate that PLT count, age, Cr, INR, and AFP are the five most important contributors to the final model. The preoperative PLT count was identified as the most important factor. A meta-analysis of 13 studies (37) assessed the effects of perioperative PLT count on PHLF and mortality using two PLT count cutoffs (100 and 150 platelets/nL). Patients with a perioperative PLT count< 150/nL (four studies, 817 patients; odds ratio [OR]: 4.79; 95% CI, 2.89-7.94) and those with a PLT count< 100/nL (four studies, 949 patients; OR: 4.65; 95% CI: 2.608.31) had a high risk of developing PHLF (37). As shown in previous studies, PLT count (38), age (39), Cr (40), INR (41), and AFP (42) are all predictors of PHLF.
The ML PHLF model is more accurate for the prediction of PHLF than are existing models and is convenient to use. The AUCs of the ML PHLF model for detecting PHLF in the training, validation, and testing cohorts were 0.944, 0.870, and 0.822, respectively, confirming that the ML PHLF model has good predictive value for PHLF in different groups. The ML PHLF model had the highest predictive value for AUC among traditional scoring systems in all three cohorts. The ML model identified PHLF in the training cohort with a sensitivity, specificity, and NPV of 87.6%, 85.9%, and 96.0%, respectively. The sensitivity, specificity, and NPV of the ML PHLF model were 100%, 64.4%, and 100%, respectively, in the validation cohort and 87.5%, 64.4%, and 94.6%, respectively, in the testing cohort. When compared to the sensitivity, specificity, and NPV of traditional scoring systems, those of the ML PHLF model were the highest. The ML PHLF model outperformed traditional noninvasive models according to the DCA curves. To facilitate the use of this model in the clinic, a free web calculator to predict the risk of PHLF has been developed (http://www.pan-chess.cn/calculator/PHLF_score).
This is the first multicenter study to explore the development and validation of a LightGBM-based model for the prediction of PHLF in patients with HCC. The ML PHLF model is based on routine clinical parameters obtained in patients with HCC. With the advantages of convenient data collection, availability, and objectiveness, the novel model is suitable for the prediction of PHLF in most clinical situations, showing good interpretability and consistency with clinical experience and demonstrating good reliability.
However, this study has some limitations. First, a selection bias was unavoidable; however, this offset has been minimized via the multicenter design. Second, the ML PHLF model is poorly interpretable, a black box, and prone to overfitting. Therefore, interpretable ML algorithms will be assessed in follow-up studies. Last, the novel ML model predicts the overall risk of PHLF as defined by the ISGLS criteria. Prospective multicenter studies are required to determine the predictive value of ML PHLF models in CTP class B and C subgroups and other PHLF diagnostic criteria, such as the 50-50 criteria.
In conclusion, an ML PHLF model using common clinical parameters was constructed and validated based on the LightGBM algorithm. Compared to other noninvasive models, this novel model has the best PHLF-predictive ability. This model can be used to help accurately predict the risk of PHLF, screen high-risk PHLF subgroups, and help surgeons determine personalized treatment options.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Ethics Committee of Xingtai People’s Hospital of Hebei Province. The patients/participants provided their written informed consent to participate in this study.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by TZ, YoL, SG, JL, ZZ, DS, CheL, PY, YH, ChuL, YaL, ShaL, MW, DL, HM, ShuL, BZ, and AH. The first draft of the manuscript was written by JW and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.986867/full#supplementary-material
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis b and c viruses worldwide. Int J Cancer (2018) 142:2471–7. doi: 10.1002/ijc.31280
3. Farges O, Goutte N, Bendersky N, Falissard B. Incidence and risks of liver resection: An all-inclusive French nationwide study. Ann Surg (2012) 256:697–704. doi: 10.1097/SLA.0b013e31827241d5
4. van Mierlo KM, Schaap FG, Dejong CH, Olde Damink SW. Liver resection for cancer: New developments in prediction, prevention and management of postresectional liver failure. J Hepatol (2016) 65:1217–31. doi: 10.1016/j.jhep.2016.06.006
5. Rahbari NN, Garden OJ, Padbury R, Brooke-Smith M, Crawford M, Adam R, et al. Posthepatectomy liver failure: a definition and grading by the international study group of liver surgery (ISGLS). Surgery (2011) 149:713–24. doi: 10.1016/j.surg.2010.10.001
6. Paugam-Burtz C, Janny S, Delefosse D, Dahmani S, Dondero F, Mantz J, et al. Prospective validation of the “fifty-fifty” criteria as an early and accurate predictor of death after liver resection in intensive care unit patients. Ann Surg (2009) 249:124–8. doi: 10.1097/SLA.0b013e31819279cd
7. Rahbari NN, Reissfelder C, Koch M, Elbers H, Striebel F, Büchler MW, et al. The predictive value of postoperative clinical risk scores for outcome after hepatic resection: A validation analysis in 807 patients. Ann Surg Oncol (2011) 18:3640–9. doi: 10.1245/s10434-011-1829-6
8. Xiang F, Liang X, Yang L, Liu X, Yan S. CT radiomics nomogram for the preoperative prediction of severe post-hepatectomy liver failure in patients with huge (≥ 10 cm) hepatocellular carcinoma. World J Surg Oncol (2021) 19:344. doi: 10.1186/s12957-021-02459-0
9. Cai S, Lin X, Sun Y, Lin Z, Wang X, Lin N, et al. Quantitative parameters obtained from gadobenate dimeglumine-enhanced MRI at the hepatobiliary phase can predict post-hepatectomy liver failure and overall survival in patients with hepatocellular carcinoma. Eur J Radiol (2022) 154:110449. doi: 10.1016/j.ejrad.2022.110449
10. Meng D, Liang C, Zheng Y, Wang X, Liu K, Lin Z, et al. The value of gadobenate dimeglumine-enhanced biliary imaging from the hepatobiliary phase for predicting post-hepatectomy liver failure in HCC patients. Eur Radiol (2022) 31. doi: 10.1007/s00330-022-08874-5
11. Kawaguchi T, Tokushige K, Hyogo H, Aikata H, Nakajima T, Ono M, et al. A data mining-based prognostic algorithm for NAFLD-related hepatoma patients: A nationwide study by the Japan study group of NAFLD. Sci Rep (2018) 8:10434. doi: 10.1038/s41598-018-28650-0
12. Kawaguchi T, Yoshio S, Sakamoto Y, Hashida R, Koya S, Hirota K, et al. Impact of decorin on the physical function and prognosis of patients with hepatocellular carcinoma. J Clin Med (2020) 9:936. doi: 10.3390/jcm9040936
13. Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics (2017) 37:505–15. doi: 10.1148/rg.2017160130
14. Mattonen SA, Davidzon GA, Benson J, Leung A, Vasanawala M, Horng G, et al. Bone marrow and tumor radiomics at (18)F-FDG PET/CT: Impact on outcome prediction in non-small cell lung cancer. Radiology (2019) 293:451–9. doi: 10.1148/radiol.2019190357
15. Rufo DD, Debelee TG, Ibenthal A, Negera WG. Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM). Diagn (Basel) (2021) 11:1714. doi: 10.3390/diagnostics11091714
16. Veerankutty FH, Jayan G, Yadav MK, Manoj KS, Yadav A, Nair S, et al. Artificial intelligence in hepatology, liver surgery and transplantation: Emerging applications and frontiers of research. World J Hepatol (2021) 13:1977–90. doi: 10.4254/wjh.v13.i12.1977
17. Kurosaki K, Uesawa Y. Development of in silico prediction models for drug-induced liver malignant tumors based on the activity of molecular initiating events: Biologically interpretable features. J Toxicol Sci (2022) 47:89–98. doi: 10.2131/jts.47.89
18. Grissa D, Nytoft Rasmussen D, Krag A, Brunak S, Juhl Jensen L. Alcoholic liver disease: A registry view on comorbidities and disease prediction. PloS Comput Biol (2020) 16:e1008244. doi: 10.1371/journal.pcbi.1008244
19. Freeman RB Jr. Model for end-stage liver disease (MELD) for liver allocation: A 5-year score card. Hepatology (2008) 47:1052–7. doi: 10.1002/hep.22135
20. Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology (2006) 43:1317–25. doi: 10.1002/hep.21178
21. Johnson PJ, Berhane S, Kagebayashi C, Satomura S, Teng M, Reeves HL, et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the ALBI grade. J Clin Oncol (2015) 33:550–8. doi: 10.1200/JCO.2014.57.9151
22. Shiha G, Ibrahim A, Helmy A, Sarin SK, Omata M, Kumar A, et al. Asian-Pacific association for the study of the liver (APASL) consensus guidelines on invasive and non-invasive assessment of hepatic fibrosis: a 2016 update. Hepatol Int (2017) 11:1–30. doi: 10.1007/s12072-016-9760-3
23. Pinato DJ, Stebbing J, Ishizuka M, Khan SA, Wasan HS, North BV, et al. A novel and validated prognostic index in hepatocellular carcinoma: The inflammation based index (IBI). J Hepatol (2012) 57:1013–20. doi: 10.1016/j.jhep.2012.06.022
24. Chinese Society of Hepatology, Chinese Medical Association. Guideline for diagnosis and treatment of liver cirrhosis. J Clin Hepatol (2019) 35(11):2048–425. doi: 10.3748/wjg.v25.i36.5403
25. Hathaway QA, Roth SM, Pinti MV, Sprando DC, Kunovac A, Durr AJ, et al. Machine-learning to stratify diabetic patients using novel cardiac biomarkergs and integrative genomics. Cardiovasc Diabetol (2019) 18:78. doi: 10.1186/s12933-019-0879-0
26. Liu W, Zhang L, Xin Z, Zhang H, You L, Bai L, et al. A promising preoperative prediction model for microvascular invasion in hepatocellular carcinoma based on an extreme gradient boosting algorithm. Front Oncol (2022) 12:852736. doi: 10.3389/fonc.2022.852736
27. Russell MC. Complications following hepatectomy. Surg Oncol Clin N Am (2015) 24:73–96. doi: 10.1016/j.soc.2014.09.008
28. Wang YY, Zhong JH, Su ZY, Huang JF, Lu SD, Xiang BD, et al. Albumin-bilirubin versus child-pugh score as a predictor of outcome after liver resection for hepatocellular carcinoma. Br J Surg (2016) 103:725–34. doi: 10.1002/bjs.10095
29. Fagenson AM, Gleeson EM, Pitt HA, Lau KN. Albumin-bilirubin score vs model for end-stage liver disease in predicting post-hepatectomy outcomes. J Am Coll Surg (2020) 230:637–45. doi: 10.1016/j.jamcollsurg.2019.12.007
30. Andreatos N, Amini N, Gani F, Margonis GA, Sasaki K, Thompson VM, et al. Albumin-bilirubin score: Predicting short-term outcomes including bile leak and post-hepatectomy liver failure following hepatic resection. J Gastrointest Surg (2017) 21:238–48. doi: 10.1007/s11605-016-3246-4
31. Malinchoc M, Kamath PS, Gordon FD, Peine CJ, Rank J, Borg tPC. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology (2000) 31:864–71. doi: 10.1053/he.2000.5852
32. Zhou P, Chen B, Miao XY, Zhou JJ, Xiong L, Wen Y, et al. Comparison of FIB-4 index and child-pugh score in predicting the outcome of hepatic resection for hepatocellular carcinoma. J Gastrointest Surg (2020) 24:823–31. doi: 10.1007/s11605-019-04123-1
33. Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol (2019) 20:e262–262e273. doi: 10.1016/S1470-2045(19)30149-4
34. Waljee AK, Higgins PD. Machine learning in medicine: a primer for physicians. Am J Gastroenterol (2010) 105:1224–6. doi: 10.1038/ajg.2010.173
35. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med (2019) 380:1347–58. doi: 10.1056/NEJMra1814259
36. Mai RY, Lu HZ, Bai T, Liang R, Lin Y, Ma L, et al. Artificial neural network model for preoperative prediction of severe liver failure after hemihepatectomy in patients with hepatocellular carcinoma. Surgery (2020) 168:643–52. doi: 10.1016/j.surg.2020.06.031
37. Mehrabi A, Golriz M, Khajeh E, Ghamarnejad O, Probst P, Fonouni H, et al. Meta-analysis of the prognostic role of perioperative platelet count in posthepatectomy liver failure and mortality. Br J Surg (2018) 105:1254–61. doi: 10.1002/bjs.10906
38. Xu B, Li XL, Ye F, Zhu XD, Shen YH, Huang C, et al. Development and validation of a nomogram based on perioperative factors to predict post-hepatectomy liver failure. J Clin Transl Hepatol (2021) 9:291–300. doi: 10.14218/JCTH.2021.00013
39. Chin KM, Allen JC, Teo JY, Kam JH, Tan EK, Koh Y, et al. Predictors of post-hepatectomy liver failure in patients undergoing extensive liver resections for hepatocellular carcinoma. Ann Hepatobiliary Pancreat Surg (2018) 22:185–96. doi: 10.14701/ahbps.2018.22.3.185
40. Dasari B, Hodson J, Roberts KJ, Sutcliffe RP, Marudanayagam R, Mirza DF, et al. Developing and validating a pre-operative risk score to predict post-hepatectomy liver failure. HPB (Oxf) (2019) 21:539–46. doi: 10.1016/j.hpb.2018.09.011
41. Silva A, Greensmith M, Praseedom RK, Jah A, Huguet EL, Harper S, et al. Early derangement of INR predicts liver failure after liver resection for hepatocellular carcinoma. Surgeon (2022) 20:e288–95. doi: 10.1016/j.surge.2022.01.002
Keywords: hepatocellular carcinoma, liver resection, post-hepatectomy liver failure, artificial intelligence, machine learning
Citation: Wang J, Zheng T, Liao Y, Geng S, Li J, Zhang Z, Shang D, Liu C, Yu P, Huang Y, Liu C, Liu Y, Liu S, Wang M, Liu D, Miao H, Li S, Zhang B, Huang A, Zhang Y, Qi X and Chen S (2022) Machine learning prediction model for post- hepatectomy liver failure in hepatocellular carcinoma: A multicenter study. Front. Oncol. 12:986867. doi: 10.3389/fonc.2022.986867
Received: 05 July 2022; Accepted: 14 October 2022;
Published: 02 November 2022.
Edited by:
Wentao Wang, Sichuan University, ChinaReviewed by:
Zenichi Morise, Fujita Health University, JapanCopyright © 2022 Wang, Zheng, Liao, Geng, Li, Zhang, Shang, Liu, Yu, Huang, Liu, Liu, Liu, Wang, Liu, Miao, Li, Zhang, Huang, Zhang, Qi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shubo Chen, Y3NiODE2MEAxMjYuY29t; Xiaolong Qi, cWl4aWFvbG9uZ0B2aXAuMTYzLmNvbQ==; Yewei Zhang, emhhbmd5ZXdlaUBuam11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.